Abstract
Despite their importance as a biofuel production platform, only a very limited number of butanol-tolerant bacteria have been identified thus far. Here, we extensively explored butanol- and isobutanol-tolerant bacteria from various environmental samples. A total of 16 aerobic and anaerobic bacteria that could tolerate greater than 2.0% (vol/vol) butanol and isobutanol were isolated. A 16S rRNA gene sequencing analysis revealed that the isolates were phylogenetically distributed over at least nine genera: Bacillus, Lysinibacillus, Rummeliibacillus, Brevibacillus, Coprothermobacter, Caloribacterium, Enterococcus, Hydrogenoanaerobacterium, and Cellulosimicrobium, within the phyla Firmicutes and Actinobacteria. Ten of the isolates were phylogenetically distinct from previously identified butanol-tolerant bacteria. Two relatively highly butanol-tolerant strains CM4A (aerobe) and GK12 (obligate anaerobe) were characterized further. Both strains changed their membrane fatty acid composition in response to butanol exposure, i.e., CM4A and GK12 exhibited increased saturated and cyclopropane fatty acids (CFAs) and long-chain fatty acids, respectively, which may serve to maintain membrane fluidity. The gene (cfa) encoding CFA synthase was cloned from strain CM4A and expressed in Escherichia coli. The recombinant E. coli showed relatively higher butanol and isobutanol tolerance than E. coli without the cfa gene, suggesting that cfa can confer solvent tolerance. The exposure of strain GK12 to butanol by consecutive passages even enhanced the growth rate, indicating that yet-unknown mechanisms may also contribute to solvent tolerance. Taken together, the results demonstrate that a wide variety of butanol- and isobutanol-tolerant bacteria that can grow in 2.0% butanol exist in the environment and have various strategies to maintain structural integrity against detrimental solvents.
INTRODUCTION
Due to fluctuations in the price of crude oil and an increase in environmental concerns, microbial fuel production from renewable feedstock has become a global priority. n-Butanol (hereafter referred to as butanol) is considered to be a good alternative to the traditional biofuel ethanol due to the advantages of butanol's high energy content, high octane rating, low volatility, and miscibility with gasoline and diesel oil (1, 2).
Butanol is produced biologically through an acetone-butanol-ethanol (ABE) fermentation process by obligate anaerobic clostridia (3). Recently, vigorous efforts have been made to develop alternative processes to generate C3- to C5-chain alcohols, including butanol and 2-methylpropan-1-ol (hereafter referred to as isobutanol), through metabolic engineering using Escherichia coli as the platform microorganism (4). However, production is very limited due to the high toxicity of butanol and isobutanol to microorganisms (5, 6). These solvents are known to cause an increase in plasma membrane fluidity by intercalating into the membrane and breaking the hydrogen bonds between lipid tails, resulting in a loss of membrane potential and a decline in cell growth (7, 8, 9). In fact, the known butanol-producing microorganisms, including clostridia, genetically engineered E. coli, and Saccharomyces cerevisiae, are highly sensitive to butanol (10, 11, 12), and this toxicity has long been a critical issue in practical biobutanol production.
To date, only a few butanol-tolerant bacterial species with the ability to grow in greater than 2.0% (vol/vol) butanol have been found (5, 10, 13, 14, 15). The previously identified butanol-tolerant microorganisms were predominantly screened from culture collections that included well-known strains that tolerate other organic solvents (e.g., toluene and benzene) (5, 10, 14). This strategy was effective but resulted in the collection of taxonomically limited butanol-tolerant bacteria, such as the genera Clostridium, Pseudomonas, Zymomonas, Bacillus, Lactobacillus, and Enterococcus (5, 7, 10, 13, 14, 15). Furthermore, as there have been very few attempts to isolate butanol-tolerant bacteria from natural environments, very little is known about the butanol-tolerant bacteria that are present in nature.
In this study, we sought to extensively screen butanol- and isobutanol-tolerant microorganisms that can grow in greater than 2.0% butanol from various environmental samples and investigated the phylogenetic positions and tolerance to butanol and isobutanol of the microorganisms. Among those isolates, two strains that showed a relatively high tolerance were further characterized by examining cell surface structures, fatty acid compositions, and genes associated with butanol tolerance.
MATERIALS AND METHODS
Sampling, enrichment, and isolation.
Butanol- and isobutanol-tolerant microorganisms were enriched in the presence of these solvents. Cultivation was performed under both aerobic and strictly anaerobic conditions. To prevent evaporation of the solvents, a 50-ml, tightly capped conical tube (for aerobic cultivation) and 50-ml serum vial sealed with a butyl rubber stopper and aluminum crimp (for strictly anaerobic cultivation) were used. Evaporation was negligible during the cultivation period under both the aerobic and anaerobic conditions and thus did not affect the evaluation of solvent tolerance.
To enrich and isolate aerobic bacteria, samples were collected from freshwater sediments, grease-contaminated soils, cabbage-field soils, vegetable wastes, and composts in Ibaraki Prefecture, Japan. After the samples were sonicated in water and centrifuged at 400 × g for 5 min, the supernatant was inoculated into fresh medium containing butanol (0.1 to 3.0% [vol/vol]); the butanol-containing medium consisted of (per liter) 5 g glucose, 1 g tryptone, 0.5 g yeast extract, 1 g (NH4)2SO4, 0.75 g KH2PO4, and 0.78 g K2HPO4 supplemented with 7 ml basal salt solution and 1 ml vitamin solution, as described previously (16). Two different aerobic enrichments, short- and long-term cultivations, were performed. For short-term cultivation, 50 μl of inoculum was added to 2 ml of fresh medium containing 0.5 to 3.0% butanol. After incubation at 30°C with shaking for 48 h, the cultures were spread onto agar plates with the same medium containing 2.0% butanol, and colonies were isolated and purified. For long-term cultivation (3 to 9 months in total), 1 ml of each inoculum was added to 50 ml of fresh medium containing 0.1% butanol. After incubation at room temperature with shaking for 48 to 72 h, 2 to 3% of the culture fluid was transferred into fresh medium containing 1.0% butanol and incubated for 5 days. The culture was then repeatedly transferred into fresh medium with increasing concentrations of butanol of up to 9.0% in a stepwise manner. After consecutive transfers, the cultures were spread onto agar plates as described above.
To enrich and isolate strictly anaerobic bacteria, samples for enrichment were collected from oil-contaminated soils, thermophilic and mesophilic anaerobic digesters, bovine rumens, hot springs, and bovine manure composts in Hokkaido Prefecture, Japan. The oil-contaminated soil or bovine manure compost samples were mixed well with 10 mM phosphate-buffered 150 mM NaCl. A portion of a 5-ml sample or slurry was inoculated into fresh medium containing 2.0% or 5.0% butanol or isobutanol. The medium was prepared based on a modified Widdel medium (17) with the following composition (per liter), as described previously (18): 5 g glucose, 1 g yeast extract, 0.53 g NH4Cl, 0.14 g KH2PO4, 0.2 g MgCl2 · 6H2O, 0.15 g CaCl2 · 2H2O, and 2.52 g NaHCO3 supplemented with 1 ml selenium and tungsten solution, 1 ml trace element solution, and 2 ml vitamin solution. Cultivation was performed in 50-ml serum vials containing 20 ml of medium under an atmosphere of N2/CO2 (80:20) at 37°C or 55°C. The cultures were transferred to fresh media five times at intervals of approximately 1 month. The Hungate roll tube technique (19) was used to isolate single colonies using the above-mentioned medium solidified with 2% Noble agar. Each colony was inoculated into a liquid medium containing 2.0% butanol to verify tolerance to butanol.
Phylogenetic analysis based on 16S rRNA gene sequences.
The 16S rRNA gene sequences (E. coli positions 28 to 1491) of each isolate were determined as previously described (20). All sequences were aligned with their relatives using ARB software (21) and the SILVA database (22). A phylogenetic tree was constructed by the neighbor-joining method (23) using the MEGA 4.0 program (24). The robustness of the tree's topology was assessed by a bootstrap analysis (25) based on 1,000 replications.
Butanol and isobutanol tolerance assay.
To obtain better growth for determining solvent tolerance and physiological traits, the media for the aerobic and anaerobic solvent-tolerant isolates were altered as follows. For the aerobic strains, 5 g/liter glucose, 20 g/liter tryptone, 5 g/liter yeast extract, 1.5 g/liter KH2PO4, and 1.56 g/liter K2HPO4 were supplemented with 7 ml basal salt solution and 1 ml vitamin solution (16). For the anaerobic strains, 10 g/liter glucose, 5 g/liter yeast extract, 0.53 g/liter NH4Cl, 0.14 g/liter KH2PO4, 0.2 g/liter MgCl2 · 6H2O, 0.15 g/liter CaCl2 · 2H2O, and 2.52 g/liter NaHCO3 were supplemented with 1 ml selenium and tungsten solution, 1 ml trace element solution, and 2 ml vitamin solution (18). The cultivation temperature for each isolate was the same temperature used in the enrichment.
Tolerance to butanol and isobutanol of the isolates was assessed based on cellular growth in the presence of the solvents as previously described (5, 10, 13, 15). In particular, butanol-tolerant bacteria were defined in this study as those that could grow in greater than 2.0% butanol, as all of the previously identified butanol-tolerant microorganisms can grow in at least 2.0% butanol (5, 10, 13, 14, 15). The maximum concentrations of butanol and isobutanol that allowed the isolates to grow were determined. Cells grown without solvent were inoculated into fresh liquid media amended with butanol or isobutanol at a concentration of 1.0 to 5.5% in increments of 0.5%. The optical density at 600 nm (OD600) was monitored using a spectrophotometer, and the specific growth rates were calculated from the linear range of exponential growth. The concentration of butanol or isobutanol in culture was measured by high-performance liquid chromatography equipped with a cation-exchange column and a refractive index (RI) detector. To examine cellular adaptation to butanol, all the isolates were grown with 2.0% butanol and inoculated into the subsequent medium. After 2 to 15 successive transfers, the butanol tolerance of the isolates was evaluated by the same methods as described above. Ultimately, two isolates (strains CM4A and GK12) with a relatively high butanol tolerance were further characterized as described below.
TEM.
Changes in the cell surface morphology of strains CM4A and GK12 in the late exponential growth phase and grown with or without butanol were observed by transmission electron microscopy (TEM) (Hitachi H-7600) (26). The thickness of the extracellular capsule of strain CM4A was measured from the transmission electron micrographs of 10 randomly selected cells (two spots per single cell; n = 20).
Cell surface hydrophobicity measurement.
The cell surface hydrophobicity of strain CM4A grown with or without butanol was assessed by the bacterial adhesion to hydrocarbon (BATH) test (27). In brief, 0.2 ml of organic solvent was added to 5 ml of a late-exponential-phase cell suspension, which was adjusted to an OD600 of 1.0. The solution was then vortex agitated for 30 s and left to stand for 15 min to allow separation. Butanol, n-hexane, n-tetradecane, toluene, and xylene were used as the solvents in the assay. The percentage of cells that adhered to the solvent was calculated by the following formula: [1 − (OD600 of aqueous phase after mixing/OD600 of initial suspension)] × 100 (28).
Fatty acid analysis.
An analysis of whole-cell lipid extracts from strains CM4A and GK12 grown with or without butanol was performed using previously described methods (29). Briefly, cells in the late exponential phase of growth were directly methanolyzed. The products containing fatty acid methyl esters were then extracted with n-hexane and analyzed by gas chromatography-mass spectrometry (M7200A GC/3DQMS system; Hitachi, Japan).
Cloning and expression of the cyclopropane fatty acid (CFA) synthase gene (cfa) in E. coli.
The cfa gene of strain CM4A was amplified and cloned into the pET-28b expression vector (Novagen) to produce an N-terminal His6-tagged fusion protein. The primers used for the PCR amplification, direct sequencing, and cloning of the cfa gene are listed in Table S1 in the supplemental material. E. coli DH5α cells were transformed and subsequently plated onto LB medium supplemented with 30 μg/ml kanamycin. The expression and activity of the recombinant protein in the positive clone, designated E. coli/pCFA, were confirmed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and by the above-described fatty acid analysis, respectively. E. coli/pCFA and the negative-control clone E. coli/pET28, which was transformed with an empty pET-28b vector, were subjected to the solvent tolerance assay as described above. Briefly, the cells were grown on LB medium containing 0.7 to 1.0% butanol or isobutanol, 0.01 mM isopropyl-1-thio-β-d-galactopyranoside (IPTG), and 30 μg/ml kanamycin.
Nucleotide sequence accession numbers.
The GenBank/EMBL/DDBJ accession numbers of the 16S rRNA gene sequences of the isolates are AB669587 to AB669596 and AB537978 to AB537983, and the accession number of the cfa gene of strain CM4A is AB669597.
RESULTS AND DISCUSSION
Isolation of butanol- and isobutanol-tolerant bacteria.
After enrichment in the presence of butanol under aerobic and anaerobic conditions, a total of 16 microorganisms that could grow in the presence of greater than 2.0% butanol and isobutanol were isolated from various environmental samples (Table 1). All isolates comprised heterotrophic mesophilic bacteria, except for a thermophilic bacterium, strain GK5. To date, no thermophilic microorganisms tolerating butanol have been found. The obligate anaerobic isolates (GK3, GK5, GK12, YN1, YN3, and YN5) were obtained from either thermophilic anaerobic digesters or oil-contaminated soil. In contrast, aerobic strains (CM4A, CM9A, SK4A, SK4D, SK5A, SK7A, SK9A, FW5A, CP3A, and CY2C) were isolated from all the samples used in this study, even freshwater sediments and cabbage-field soils in which organic solvents were likely not present.
Table 1.
Characteristics of the isolates
| Bacterial type and strain | Origin | Enrichment conditionsa | Phylum | Closest relative species (accession no.) | Similarity (%) | Tolerance (%)b |
|
|---|---|---|---|---|---|---|---|
| Butanol | Isobutanol | ||||||
| Butanol-tolerant bacteria isolated under aerobic conditions | |||||||
| CM4A | Grease-contaminated soil | 30°C/4 mo/4.0% | Firmicutes | Enterococcus faecalis (AB012212) | 99.6 | 3.5 | 4.0 |
| CM9A | Grease-contaminated soil | 30°C/9 mo/9.0% | Firmicutes | Rummeliibacillus pycnus (AB271739) | 98.4 | 2.0 | 2.5 |
| SK4A | Freshwater sediment | 30°C/3 mo/4.0% | Firmicutes | Bacillus amyloliquefaciens (AB255669) | 99.6 | 2.0 | 2.5 |
| SK4D | Freshwater sediment | 30°C/4 mo/4.0% | Firmicutes | Enterococcus faecalis (AB012212) | 99.6 | 2.5 | 3.0 |
| SK5A | Freshwater sediment | 30°C/7 mo/5.0% | Firmicutes | Enterococcus faecalis (AB012212) | 99.5 | 3.0 | 3.5 |
| SK7A | Freshwater sediment | 30°C/9 mo/7.0% | Firmicutes | Lysinibacillus xylanilyticus (FJ477040) | 98.5 | 2.5 | 3.0 |
| SK9A | Freshwater sediment | 30°C/9 mo/9.0% | Firmicutes | Brevibacillus reuszeri (AB112715) | 99.8 | 2.0 | 2.5 |
| FW5A | Vegetable waste | 30°C/2 days/0.5% | Firmicutes | Bacillus amyloliquefaciens (AB255669) | 97.9 | 2.0 | 2.5 |
| CP3A | Cabbage-field soil | 30°C/2 days/3.0% | Firmicutes | Bacillus mycoides (AB021192) | 98.3 | 2.5 | 3.0 |
| CY2C | Compost | 30°C/2 days/2.0% | Actinobacteria | Cellulosimicrobium cellulans (X83809) | 98.1 | 2.0 | 2.5 |
| Butanol-tolerant bacteria isolated under anaerobic conditions | |||||||
| GK3 | Thermophilic anaerobic digester | 37°C/5 mo/2.0% | Firmicutes | Garciella nitratireducens (AY176772) | 92.8 | 2.0 | 2.5 |
| GK5 | Thermophilic anaerobic digester | 55°C/4 mo/2.0% | Firmicutes | Coprothermobacter proteolyticus (X69335) | 98.8 | 2.0 | 2.0 |
| GK12 | Thermophilic anaerobic digester | 37°C/5 mo/2.0% | Firmicutes | Eubacterium cylindroides (L34617) | 91.2 | 3.0 | 3.5 |
| YN1 | Oil-contaminated soil | 37°C/5 mo/2.0% | Firmicutes | Caloribacterium cisternae (JF262044) | 96.7 | 2.0 | 2.0 |
| YN3 | Oil-contaminated soil | 37°C/5 mo/2.0% | Firmicutes | Hydrogenoanaerobacterium saccharovorans (EU158190) | 98.0 | 2.0 | 2.0 |
| YN5 | Oil-contaminated soil | 37°C/4 mo/2.0% | Firmicutes | Clostridium pasteurianum (FR870440) | 93.6 | 2.5 | 3.0 |
The conditions of the enrichment culturesare given as follows: temperature/culture period/final butanol concentration.
The values represent the maximum butanol or isobutanol concentration allowing growth and were based on three independent replicates. The temperature at which tolerance was tested was the same temperature used for enrichment.
Phylogenetic identification of the butanol-tolerant microorganisms obtained.
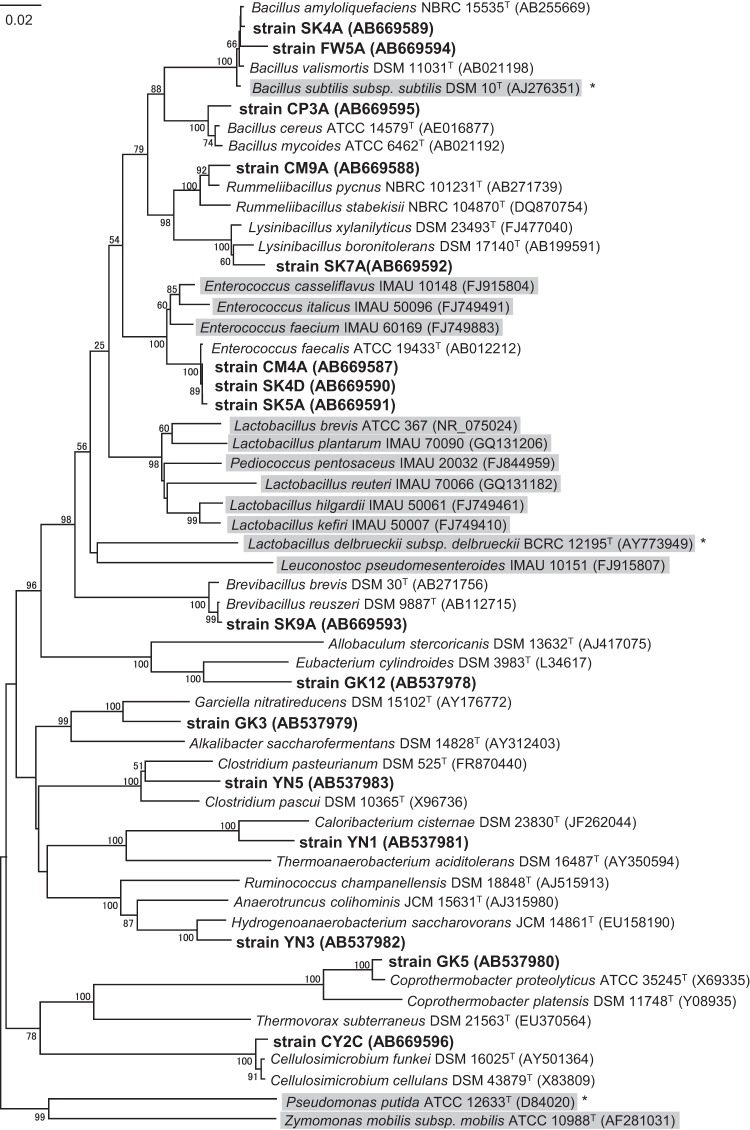
The phylogenetic analysis based on 16S rRNA gene sequencing indicated that the isolates obtained were affiliated with the genera Bacillus, Lysinibacillus, Brevibacillus, Rummeliibacillus, Coprothermobacter, Caloribacterium, Enterococcus, and Hydrogenoanaerobacterium within the phylum Firmicutes and the genus Cellulosimicrobium within the phylum Actinobacteria (Table 1). Strains CM4A, SK4D, and SK5A, belonging to the genus Enterococcus, exhibited a relatively higher tolerance than did the other isolates. This finding is consistent with the results of a previous study showing that many lactic acid bacteria, including Enterococcus species, can tolerate butanol (5, 15). Similarly, strains SK4A, FW5A, and CP3A were closely related to the known butanol-tolerant bacterium Bacillus subtilis (13). In contrast, the other 10 isolates were phylogenetically distinct from the previously identified butanol-tolerant bacteria (Fig. 1). In particular, strains GK3, GK12, and YN5 were phylogenetically novel, showing less than 94% sequence similarity to the most closely related species. This result indicates that there is a wider variety of butanol- and isobutanol-tolerant bacteria that can grow in the presence of greater than 2.0% solvent than previously recognized.
Fig 1.
Neighbor-joining tree based on 16S rRNA gene sequences, showing the relationship between the isolates (boldface type), their relatives, and other butanol-tolerant bacteria (shaded clusters). Due to the lack of sequences for butanol-tolerant Bacillus subtilis (13), Lactobacillus delbrueckii (10), and Pseudomonas putida (14) strains, as indicated by asterisks, their type strains are shown in the tree. The bootstrap values that were above 50% are shown at the nodes. Bar, 0.02 substitution per nucleotide position.
Solvent tolerance of the isolates.
To further evaluate the ability of the isolates to tolerate butanol and isobutanol, the maximum concentrations of solvents that allowed the isolates to grow were determined. Most of the isolates (all of the aerobic strains and three anaerobic strains [GK3, GK12, and YN5]) grew slightly better with isobutanol than with butanol (Table 1). None of the isolates tested assimilated or degraded butanol and isobutanol (data not shown).
Of the isolates, the aerobic strain CM4A exhibited the highest tolerance, growing in the presence of 3.5% butanol and 4.0% isobutanol. The anaerobic isolate, strain GK12, also showed a relatively higher tolerance, with the ability to grow in 3.0% butanol and 3.5% isobutanol. The tolerance levels of the two strains were comparable or even greater than the tolerance of previously identified butanol-tolerant microorganisms (5). More importantly, both strains showed clear exponential growth and reached high cell densities, even in the presence of 2.5% butanol (Fig. 2), which was not observed for the previously reported butanol-tolerant bacteria (5, 15). Therefore, we selected strains CM4A and GK12 for further analyses.
Fig 2.
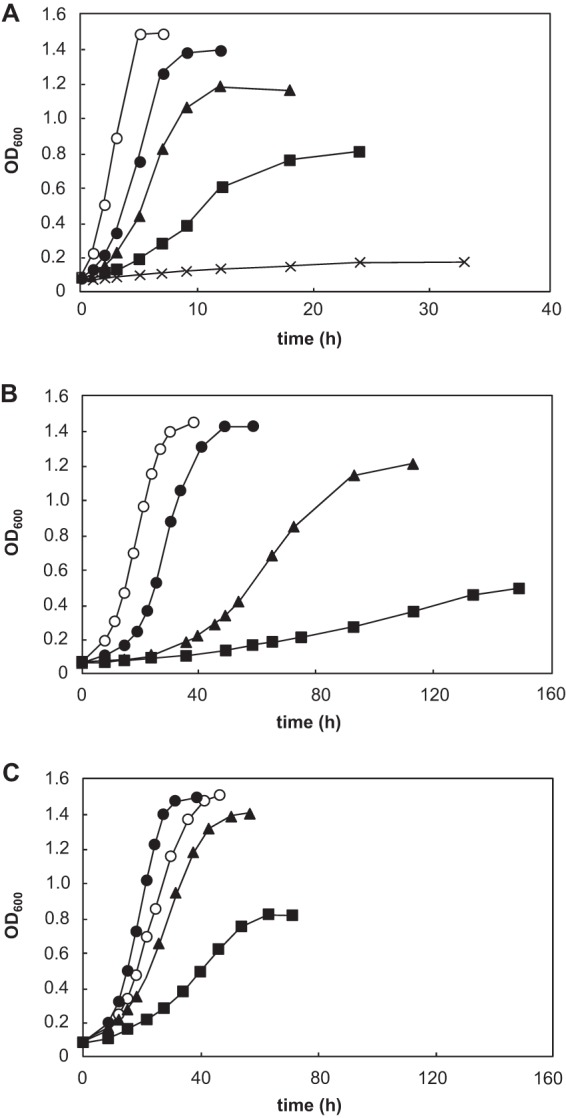
Effect of butanol on the growth of strain CM4A (A), strain GK12 (B), and the butanol-adapted cells of strain GK12 (C). Butanol concentrations of 0% (○), 2% (●), 2.5% (▲), 3.0% (■), and 3.5% (×) were tested. The values represent the means of triplicate experiments.
Morphological characterization of strains CM4A and GK12.
The cell morphologies were investigated by TEM. As shown in Fig. 3, changes in the cell surface properties in the presence of a high butanol concentration (2.0%) were evident in strain CM4A. The cells exhibited a diplococcus shape with a capsule structure (Fig. 3A and C), and the thickness of the extracellular capsule increased 2-fold when the cells were grown with butanol (P < 0.01 by t test) (Fig. 3B and D). To date, these changes in cell surface properties in response to butanol exposure have not been reported. The BATH test (27) was performed to clarify whether the cell surface hydrophobicity of strain CM4A was altered by the increased extracellular capsule thickness. Cells grown with 2% butanol exhibited a significantly lower affinity for butanol and other organic solvents than did the cells grown without butanol (P < 0.01 by t test) (see Table S2 in the supplemental material). This result suggests that the changes in the capsule structure of strain CM4A decrease the cell surface hydrophobicity and thus may function as a physical defense against butanol. Previous studies have reported that the solvent-tolerant microorganisms Rhodococcus rhodochrous and Staphylococcus sp. produce extracellular hydrophilic compounds that may prevent hydrophilic solvents, such as n-hexadecane and toluene, from intercalating into the membrane (30, 31). In contrast to strain CM4A, such a change in morphology was not observed in strain GK12 (data not shown).
Fig 3.
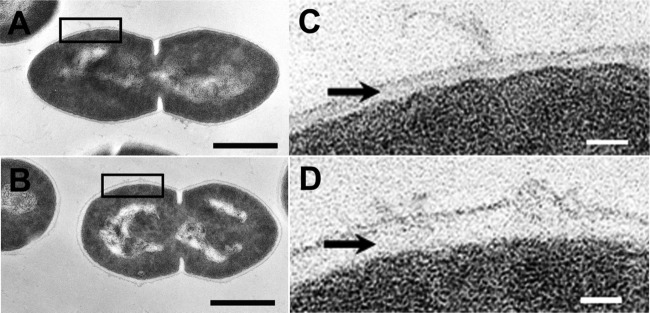
(A to D) Transmission electron micrographs of strain CM4A in the absence (A and C) and presence (B and D) of 2.0% butanol. The transmission electron micrographs in panels C and D are magnified views of the black boxes in panels A and B, respectively. The black arrows point to the positions of the capsule. The average capsule thicknesses were 17 ± 3.7 nm and 32 ± 3.7 nm in the absence and presence, respectively, of 2.0% butanol (P < 0.01 by t test). Bars, 0.5 μm (A and B) and 50 nm (C and D).
Changes in the fatty acid compositions of strains CM4A and GK12 in the presence of butanol.
To elucidate the underlying mechanism of solvent tolerance, we further investigated the physiological response to butanol of strains CM4A and GK12. Exposure to solvents impairs the integrity and stability of the cytoplasmic membrane, and several solvent-tolerant microorganisms are known to reestablish membrane fluidity and decrease solvent permeability by altering their membrane lipid compositions (7, 32, 33). Thus, to evaluate the responses of the cell membrane to high concentrations of butanol, changes in the fatty acid compositions of the highly solvent-tolerant strains CM4A and GK12 were examined at a concentration of up to 2.5% butanol.
In the absence of butanol, strain CM4A contained saturated fatty acids (44% of total), together with unsaturated fatty acids (56%), such as palmitic acid (C16:0) and cis-vaccenic acid (C18:1ω7c). When the cells were grown with butanol, the proportion of C16:0 and CFAs (cis-11,12-methylene octadecanoic acid [cyclo-C19:0]) increased, whereas the proportion of C18:1ω7c decreased, resulting in an increase in the proportion of total saturated fatty acids and CFAs by up to 56% (Table 2). Although the cis-hexadecenoic acid (C16:1ω7c) level relatively increased with 2.0% butanol but decreased with 2.5%, the total unsaturated fatty acid proportion tended to decrease in a butanol dose-dependent manner (Table 2). In contrast, strain GK12 did not contain unsaturated fatty acids but rather saturated fatty acids and saturated 1,1-dimethoxy alkanes (DMAs) (34). In the presence of butanol, the proportions of longer-chain fatty acids (stearic acid [C18:0] and arachidic acid [C20:0]) significantly increased (Table 3). Although the proportion of C18:0 DMAs relatively increased with 2.0% butanol but decreased with 2.5%, the total proportion of alkyl groups of a length of 18 increased in a butanol dose-dependent manner (Table 3). Because increases in the acyl chain length and proportion of saturated fatty acids and the cyclization of unsaturated fatty acids promote more-rigid membrane structures (35, 36), these changes in response to butanol exposure in both strains may compensate for the increased membrane fluidity imposed by butanol. Other microorganisms, such as Clostridium acetobutylicum and Pseudomonas putida, have also been reported to maintain membrane rigidity by increasing both the mean length of the acyl chain and the proportion of saturated fatty acids and by synthesizing CFAs in the presence of solvents (37, 38, 39, 40), which is in accordance with our findings.
Table 2.
Changes in the fatty acid composition of strain CM4A in the presence of 2.0% and 2.5% butanol
| Fatty acida | Fatty acid compositionb (%) of strain CM4A grown with: |
||
|---|---|---|---|
| No solvents | 2.0% butanol | 2.5% butanol | |
| C9:1c | ND | ND | 1.1 ± 0.3 |
| C14:0 | 5.7 ± 0.2 | 6.9 ± 0.6 | 6.6 ± 0.2 |
| C14:1ω7c | 0.6 ± 0.1 | 0.8 ± 0.1 | ND |
| C15:0 | 0.1 ± 0.0 | 0.1 ± 0.0 | ND |
| C16:0 | 35.7 ± 1.3 | 41.0 ± 0.8 | 45.8 ± 0.7 |
| C16:1c | ND | ND | 0.9 ± 0.0 |
| C16:1ω7c | 8.9 ± 0.2 | 9.7 ± 0.4 | 7.9 ± 0.3 |
| C18:0 | 1.9 ± 0.1 | 1.2 ± 0.0 | 1.7 ± 0.2 |
| C18:1ω7c | 46.4 ± 1.5 | 38.5 ± 1.6 | 33.6 ± 0.4 |
| Cyclo-C19:0 | 0.6 ± 0.1 | 1.8 ± 0.2 | 2.4 ± 0.1 |
| Total saturated fatty acidsd | 43.4 ± 1.4 | 49.2 ± 1.3 | 54.1 ± 0.5 |
Abbreviations: CX:YωZc, a fatty acid containing X carbon atoms with Y double bonds at position Z, counted from the methyl terminus in the cis configuration; cyclo-C19:0, cis-11,12-methylene octadecanoic acid.
Each fatty acid composition value is given as a percentage of the whole-cell lipids. Values are means ± standard deviations of three independent measurements. ND, not detected.
The double-bond positions of C9:1 and C16:1 were not identified.
Myristic acid (C14:0), pentadecanoic acid (C15:0), palmitic acid (C16:0), and stearic acid (C18:0) were added together to calculate total saturated fatty acids.
Table 3.
Changes in the fatty acid and DMA compositions of nonadapted and butanol-adapted cells of strain GK12 in the presence of butanol
| Fatty acid or DMAa | Fatty acid or DMA compositionb (%) of strain GK12 |
||||||
|---|---|---|---|---|---|---|---|
| Nonadapted cells grown with: |
Butanol-adapted cellsc grown with: |
||||||
| No solvents | 2.0% butanol | 2.5% butanol | No solvents | 2.0% butanol | 2.5% butanol | 3.0% butanol | |
| C14:0 | 24.5 ± 1.1 | 6.6 ± 0.5 | 1.5 ± 0.8 | 27.0 ± 1.6 | 26.1 ± 0.4 | 5.2 ± 0.7 | 1.1 ± 0.4 |
| C16:0 | 23.5 ± 1.4 | 23.7 ± 2.2 | 21.6 ± 1.1 | 25.1 ± 0.3 | 27.8 ± 0.6 | 31.2 ± 0.9 | 19.2 ± 0.4 |
| C18:0 | 10.8 ± 0.6 | 30.4 ± 0.7 | 42.1 ± 1.3 | 3.8 ± 0.5 | 5.2 ± 0.5 | 35.2 ± 0.2 | 48.1 ± 0.8 |
| C20:0 | 2.8 ± 0.6 | 10.1 ± 1.3 | 14.1 ± 1.3 | 2.6 ± 0.5 | 4.7 ± 0.5 | 6.6 ± 0.2 | 19.4 ± 0.6 |
| C14:0 DMA | 4.9 ± 1.3 | 0.6 ± 0.2 | ND | 5.5 ± 0.5 | 2.7 ± 1.2 | ND | ND |
| C16:0 DMA | 18.0 ± 1.7 | 1.8 ± 0.5 | 1.6 ± 0.4 | 10.3 ± 1.3 | 4.2 ± 0.3 | 2.1 ± 0.3 | 1.1 ± 0.4 |
| C18:0 DMA | 15.5 ± 0.8 | 26.8 ± 2.2 | 19.1 ± 1.9 | 25.7 ± 0.6 | 29.3 ± 0.5 | 19.6 ± 0.4 | 11.2 ± 0.9 |
| Total C14 | 29.4 ± 1.4 | 7.2 ± 0.4 | 1.5 ± 0.7 | 32.5 ± 1.4 | 28.8 ± 1.1 | 5.2 ± 0.7 | 1.1 ± 0.4 |
| Total C16 | 41.5 ± 1.9 | 25.5 ± 2.0 | 23.3 ± 1.0 | 35.5 ± 1.2 | 32.0 ± 0.6 | 33.3 ± 0.8 | 20.3 ± 0.5 |
| Total C18 | 26.3 ± 0.9 | 57.3 ± 2.0 | 61.2 ± 2.0 | 29.4 ± 0.7 | 34.5 ± 0.6 | 54.8 ± 0.4 | 59.3 ± 1.0 |
Abbreviations: CX:Y, fatty acid containing X carbon atoms with Y double bonds; total X, the sum of fatty acids and DMAs with an acyl chain length of X.
Each fatty acid composition value is shown as a percentage of whole-cell lipids. Values are means ± standard deviations of three independent measurements. ND, not detected.
Cell adaptation was achieved by 15 consecutive passages with 2.0% butanol.
Overall, butanol dose-dependent changes in the saturated and CFA levels and acyl chain length in strains CM4A and GK12, respectively, were clearly observed (Tables 2 and 3). In particular, strain GK12 exhibited a more marked alteration in membrane composition than strain CM4A. In general, an increase in the length of the acyl chain has a smaller effect on the fluidity of the lipid bilayer than does the saturation of fatty acids (36). Because strain GK12 lacked unsaturated fatty acids, this strain would have to markedly alter the length of the acyl chain of its saturated fatty acids to maintain membrane fluidity.
Heterologous expression of the cfa gene derived from strain CM4A improved solvent tolerance in E. coli.
The results of fatty acid analysis indicated that CFAs may contribute to the butanol tolerance of strain CM4A. To verify this phenomenon, we examined whether the cyclization of unsaturated fatty acids among the membrane lipids could enhance the tolerance of E. coli to butanol and isobutanol. Recombinant strain E. coli/pCFA was generated by the introduction of the cfa gene derived from strain CM4A. The deduced 388-amino-acid sequence resulting from the cfa gene (AB669597) of strain CM4A is 36% identical to the E. coli cfa gene (AM946981) product. The expression of the recombinant protein and the increase in the proportion of CFAs (cyclo-C19:0) were confirmed by SDS-PAGE and fatty acid analysis, respectively (see Fig. S2 and Table S3 in the supplemental material). A solvent tolerance assay showed that the relative growth rate of recombinant E. coli/pCFA in the presence of butanol or isobutanol was significantly higher than the growth rate of the negative-control strain E. coli/pET28 (P < 0.01 by t test) (Fig. 4; see Fig. S1 in the supplemental material). These findings strongly demonstrate that CFA synthesis due to cfa gene transformation enhanced the tolerance of E. coli to butanol and isobutanol. CFA synthase is known to directly modify unsaturated fatty acids in the membrane and therefore does not require energy or carbon for de novo fatty acid synthesis (35). The enhancement of butanol tolerance by reinforcing CFA synthesis thus did not compete with butanol production, at least in terms of energy and carbon consumption. Introduction of the cfa gene into a candidate microbial platform may serve as a new strategy to improve butanol production efficiency.
Fig 4.
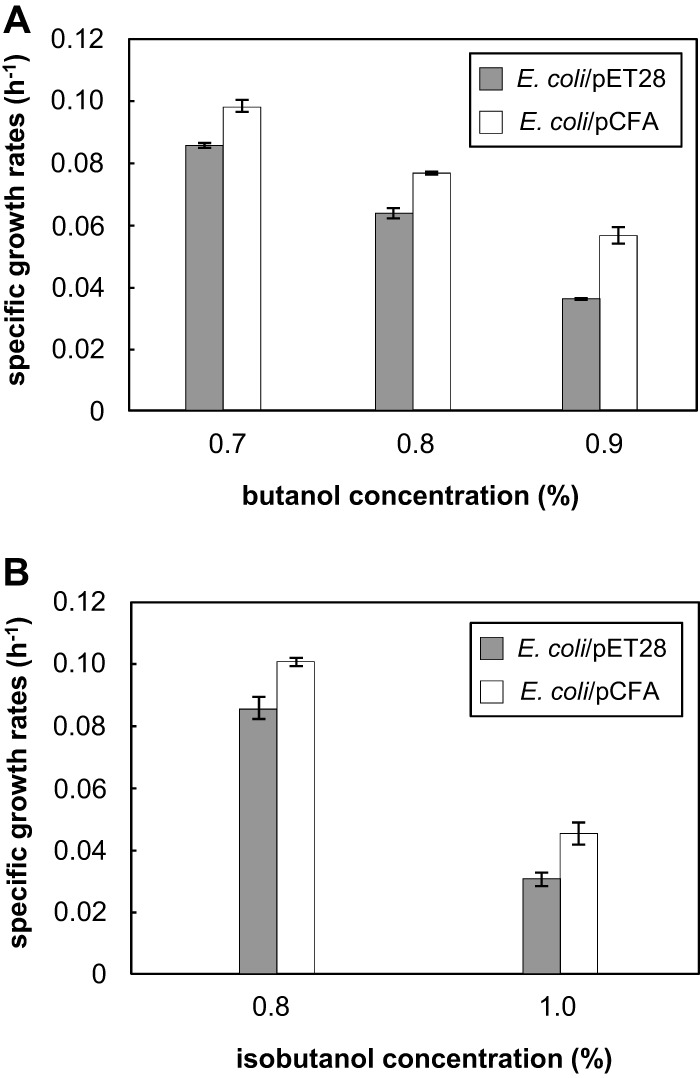
Butanol (A) and isobutanol (B) tolerance of E. coli/pCFA and the control strain E. coli/pET28 showing the differences in growth rates in the presence of butanol and isobutanol. To induce heterologous gene expression, 0.01 mM IPTG was added to each strain. Values are means ± standard deviations (SD) (error bars) of triplicate experiments. The growth rates of the E. coli/pCFA and E. coli/pET28 strains without solvent were equal.
Mechanisms involved in the butanol adaptation of strain GK12.
Microorganisms are known to adapt to organic solvents during exposure to nonlethal levels of organic solvents (14, 41). To verify whether the isolates could adapt to butanol, the microorganisms were subjected to successive subculturing with nonlethal concentrations of butanol. Only strain GK12 exhibited significant adaptation after 15 consecutive passages in the presence of 2.0% butanol (see Fig. S3 in the supplemental material). Interestingly, the butanol-treated GK12 cells grew even faster in the presence of 2.0% butanol than in the absence of butanol (Fig. 2C and Fig. S3 in the supplemental material). In addition, when the butanol-treated cells were grown in 2.5% or 3.0% butanol, the cell population density in the stationary phase was higher than that of the untreated cells, clearly demonstrating the ability of the cells to adapt to butanol. Conversely, the enhanced tolerance was completely abolished (data not shown) by repeatedly culturing the butanol-treated cells in the absence of butanol, suggesting that the butanol adaption of strain GK12 was due to physiological responses rather than genetic mutations. To our knowledge, no other microorganism can grow faster in the presence of such toxic and nonmetabolizable organic solvents than in the absence of those solvents.
To elucidate the adaptation mechanisms, we analyzed the fatty acid and DMA compositions of the adapted cells in comparison to the nonadapted cells. Similar to the nonadapted cells, the adapted cells increased their acyl chain lengths in a butanol dose-dependent manner (Table 3). However, considerable differences in the compositions were observed between the nonadapted and adapted cells grown with 2.0% butanol. In particular, the proportion of short chains (myristic acid [C14:0]) in the butanol-adapted cells was much higher than in the nonadapted cells, whereas the proportion of long chains (C18:0 and C20:0) in the butanol-adapted cells was lower than in the nonadapted cells. It should also be noted that, when comparing the adapted cells grown with or without 2.0% butanol, there were no significant differences in the proportions of short and long chains; rather, such proportions were similar to those of the nonadapted cells grown without butanol (Table 3). These results strongly imply that 2.0% butanol-adapted cells may use alternative strategies to acquire butanol tolerance instead of altering the chain lengths of fatty acids and DMAs, at least at the concentration (2.0%) to which the cells had adapted. Interestingly, the proportions of long chains and short chains markedly increased and decreased, respectively, when the adapted cells were grown with 2.5% butanol (Table 3), showing that a 0.5% increase was critical for the collapse of the homeostatic state of the fatty acid composition.
A similar finding was reported for the adaptation of Pseudomonas putida to toluene (42). Changes in the membrane fatty acid compositions via the isomerization of cis- to trans-unsaturated fatty acids in response to solvent exposure were observed in nonadapted P. putida cells but not in adapted cells, suggesting that alternative mechanisms in the adapted cells, such as a solvent efflux system, allowed for improved solvent tolerance (43).
Conclusions.
We successfully isolated a wide variety of butanol- and isobutanol-tolerant bacteria from environmental samples. Analyses of tolerance to these detrimental solvents using two representative strains suggested that the organisms perhaps maintain their structural integrity by increasing the extracellular capsule thickness, altering the membrane fatty acid components, and adaptation via unknown mechanisms. These findings provide further strategies for developing potential solvent-tolerant platforms for microbial fuel production.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the New Energy and Industrial Technology Development Organization (NEDO) program in Japan.
We thank Mizuho Muramatsu at National Institute of Advanced Industrial Science and Technology (AIST) for the fatty acid analysis.
Footnotes
Published ahead of print 6 September 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.02900-13.
REFERENCES
- 1.Lee SY, Park JH, Jang SH, Nielsen LK, Kim J, Jung KS. 2008. Fermentative butanol production by clostridia. Biotechnol. Bioeng. 101:209–228 [DOI] [PubMed] [Google Scholar]
- 2.Schwarz WH, Gapes JR. 2006. Butanol-rediscovering a renewable fuel. BioWorld Europe 1:16–19 [Google Scholar]
- 3.Jones DT, Woods DR. 1986. Acetone-butanol fermentation revisited. Microbiol. Mol. Biol. Rev. 50:484–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Atsumi S, Hanai T, Liao JC. 2008. Non-fermentative pathways for synthesis of branched-chain higher alcohols as biofuels. Nature 451:86–89 [DOI] [PubMed] [Google Scholar]
- 5.Li J, Zhao JB, Zhao M, Yang YL, Jiang WH, Yang S. 2010. Screening and characterization of butanol-tolerant micro-organisms. Lett. Appl. Microbiol. 50:373–379 [DOI] [PubMed] [Google Scholar]
- 6.Liu S, Qureshi N. 2009. How microbes tolerate ethanol and butanol. New Biotechnol. 26:117–121 [DOI] [PubMed] [Google Scholar]
- 7.Huffer S, Clark ME, Ning JC, Blanch HW, Clark DS. 2011. The role of alcohols in growth, lipid composition, and membrane fluidity of yeast, bacteria, and archaea. Appl. Environ. Microbiol. 77:6400–6408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ingram LO, Buttke TM. 1984. Effects of alcohols on micro-organisms. Adv. Microb. Physiol. 25:253–300 [DOI] [PubMed] [Google Scholar]
- 9.Vollherbst-Schneck K, Sands JA, Montenecourt BS. 1984. Effect of butanol on lipid composition and fluidity of Clostridium acetobutylicum ATCC 824. Appl. Environ. Microbiol. 47:193–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Knoshaug EP, Zhang M. 2009. Butanol tolerance in a selection of microorganisms. Appl. Biochem. Biotechnol. 153:13–20 [DOI] [PubMed] [Google Scholar]
- 11.Qureshi N, Saha BC, Cotta MA. 2007. Butanol production from wheat straw hydrolysate using Clostridium beijerinckii. Bioproc. Biosyst. Eng. 30:419–427 [DOI] [PubMed] [Google Scholar]
- 12.Woods DR. 1995. The genetic engineering of microbial solvent production. Trends Biotechnol. 13:259–264 [DOI] [PubMed] [Google Scholar]
- 13.Kataoka N, Tajima T, Kato J, Rachadech W, Vangnai A. 2011. Development of butanol-tolerant Bacillus subtilis strain GRSW2-B1 as a potential bioproduction host. AMB Express 1:10. 10.1186/2191-0855-1-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ruhl J, Schmid A, Blank LM. 2009. Selected Pseudomonas putida strains can grow in the presence of high butanol concentrations. Appl. Environ. Microbiol. 75:4653–4656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ting CNW, Wu J, Takahashi K, Endo A, Zhao H. 2012. Screened butanol-tolerant Enterococcus faecium capable of butanol production. Appl. Biochem. Biotechol. 168:1672–1680 [DOI] [PubMed] [Google Scholar]
- 16.Hanada S, Kawase Y, Hiraishi A, Takaichi S, Matsuura K, Shimada K, Nagashima KVP. 1997. Porphyrobacter tepidarius sp. nov., a moderately thermophilic aerobic photosynthetic bacterium isolated from a hot spring. Int. J. Syst. Evol. Microbiol. 47:408–413 [DOI] [PubMed] [Google Scholar]
- 17.Pfennig N, Widdel F, Trüper HG. 1981. The dissimilatory sulfate-reducing bacteria, p 926–940 In Starr MP, Stolp H, Tr̈uper HG, Balows A, Schlegel HG. (ed), The prokaryotes, vol 1 Springer, New York, NY [Google Scholar]
- 18.Sekiguchi Y, Kamagata Y, Nakamura K, Ohashi A, Harada H. 2000. Syntrophothermus lipocalidus gen. nov., sp. nov., a novel thermophilic, syntrophic, fatty-acid-oxidizing anaerobe which utilizes isobutyrate. Int. J. Syst. Evol. Microbiol. 50:771–779 [DOI] [PubMed] [Google Scholar]
- 19.Hungate R. 1969. A roll tube method for cultivation of strict anaerobes. Methods Microbiol. 3:117–132 [Google Scholar]
- 20.Tamaki H, Hanada S, Sekiguchi Y, Tanaka Y, Kamagata Y. 2009. Effect of gelling agent on colony formation in solid cultivation of microbial community in lake sediment. Environ. Microbiol. 11:1827–1834 [DOI] [PubMed] [Google Scholar]
- 21.Ludwig W, Strunk O, Westram R, Richter L, Meier H. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363–1371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig W, Peplies J, Glöckner FO. 2007. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 35:7188–7196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saitou N, Nei M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406–425 [DOI] [PubMed] [Google Scholar]
- 24.Tamura K, Dudley J, Nei M, Kumar S. 2007. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596–1599 [DOI] [PubMed] [Google Scholar]
- 25.Felsenstein J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791 [DOI] [PubMed] [Google Scholar]
- 26.Toju H, Hosokawa T, Koga R, Nikoh N, Meng XY, Kimura N, Fukatsu T. 2010. “Candidatus Curculioniphilus buchneri,” a novel clade of bacterial endocellular symbionts from weevils of the genus Curculio. Appl. Environ. Microbiol. 76:275–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yamashita S, Satoi M, Iwasa Y, Honda K, Sameshima Y, Omasa T, Kato J, Ohtake H. 2007. Utilization of hydrophobic bacterium Rhodococcus opacus B-4 as whole-cell catalyst in anhydrous organic solvents. Appl. Microbiol. Biotechnol. 74:761–767 [DOI] [PubMed] [Google Scholar]
- 28.Reid G, Cuperus PL, Bruce AW, van der Mei HC, Tomeczek L, Khoury AH, Busscher HJ. 1992. Comparison of contact angles and adhesion to hexadecane of urogenital, dairy, and poultry lactobacilli: effect of serial culture passages. Appl. Environ. Microbiol. 58:1549–1553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hanada S, Takaichi S, Matsuura K, Nakamura K. 2002. Roseiflexus castenholzii gen. nov., sp. nov., a thermophilic, filamentous, photosynthetic bacterium that lacks chlorosomes. Int. J. Syst. Evol. Microbiol. 52:187–193 [DOI] [PubMed] [Google Scholar]
- 30.Iwabuchi N, Sunairi M, Anzai H, Nakajima M, Harayama S. 2000. Relationships between colony morphotypes and oil tolerance in Rhodococcus rhodochrous. Appl. Environ. Microbiol. 66:5073–5077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zahir Z, Seed KD, Dennis JJ. 2006. Isolation and characterization of novel organic solvent-tolerant bacteria. Extremophiles 10:129–138 [DOI] [PubMed] [Google Scholar]
- 32.Torres S, Pandey A, Castro GR. 2011. Organic solvent adaptation of Gram positive bacteria: applications and biotechnological potentials. Biotechnol. Adv. 29:442–452 [DOI] [PubMed] [Google Scholar]
- 33.Weber FJ, De Bont JA. 1996. Adaptation mechanisms of microorganisms to the toxic effects of organic solvents on membranes. Biochim. Biophys. Acta 1286:225–245 [DOI] [PubMed] [Google Scholar]
- 34.Rainey F, Andersson M, Lassila ELN, Ulrych U, Busse H, Weiss N, Mikkola R, Salkinoja-Salonen M. 2000. Frigoribacterium faeni gen. nov., sp. nov., a novel psychrophilic genus of the family Microbacteriaceae. Int. J. Syst. Evol. Microbiol. 50:355–363 [DOI] [PubMed] [Google Scholar]
- 35.Grogan DW, Cronan JE., Jr 1997. Cyclopropane ring formation in membrane lipids of bacteria. Microbiol. Mol. Biol. Rev. 61:429–441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mykytczuk NCS, Trevors JT, Leduc LG, Ferroni GD. 2007. Fluorescence polarization in studies of bacterial cytoplasmic membrane fluidity under environmental stress. Prog. Biophys. Mol. Biol. 95:60–82 [DOI] [PubMed] [Google Scholar]
- 37.Heipieper HJ, De Bont JA. 1994. Adaptation of Pseudomonas putida S12 to ethanol and toluene at the level of fatty acid composition of membranes. Appl. Environ. Microbiol. 60:4440–4444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Heipieper HJ, Neumann G, Cornelissen S, Meinhardt F. 2007. Solvent-tolerant bacteria for biotransformations in two-phase fermentation systems. Appl. Microbiol. Biotechnol. 74:961–973 [DOI] [PubMed] [Google Scholar]
- 39.Lepage C, Fayolle F, Hermann M, Vandecasteele JP. 1987. Changes in membrane lipid composition of Clostridium acetobutylicum during acetone-butanol fermentation: effects of solvents, growth temperature and pH. Microbiology 133:103–110 [Google Scholar]
- 40.Pini CV, Bernal P, Godoy P, Ramos JL, Segura A. 2009. Cyclopropane fatty acids are involved in organic solvent tolerance but not in acid stress resistance in Pseudomonas putida DOT-T1E. Microb. Biotechnol. 2:253–261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Weber FJ, Ooijkaas LP, Schemen RM, Hartmans S, De Bont JA. 1993. Adaptation of Pseudomonas putida S12 to high concentrations of styrene and other organic solvents. Appl. Environ. Microbiol. 59:3502–3504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Neumann G, Kabelitz N, Zehnsdorf A, Miltner A, Lippold H, Meyer D, Schmid A, Heipieper HJ. 2005. Prediction of the adaptability of Pseudomonas putida DOT-T1E to a second phase of a solvent for economically sound two-phase biotransformations. Appl. Environ. Microbiol. 71:6606–6612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Isken S, de Bont JAM. 1996. Active efflux of toluene in a solvent-resistant bacterium. J. Bacteriol. 178:6056–6058 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.