Main Text
Introductory paragraph
Prostate cancers remain indolent in the majority of patients but behave aggressively in a minority1,2. However, the molecular basis for this clinical heterogeneity remains incompletely understood3-5. Here, we characterize a novel lncRNA termed SChLAP1 (Second Chromosome Locus Associated with Prostate-1, HGNC #48603) overexpressed in a subset of prostate cancers. SChLAP1 levels independently predicted for poor patient outcomes, including metastasis and prostate cancer specific mortality. In vitro and in vivo gain-of-function and loss-of-function experiments indicated that SChLAP1 is critical for cancer cell invasiveness and metastasis. Mechanistically, SChLAP1 antagonized the genome-wide localization and regulatory functions of the SWI/SNF chromatin-modifying complex. These results suggest that SChLAP1 contributes to the development of lethal cancer at least in part by antagonizing tumor-suppressive functions of the SWI/SNF complex.
Manuscript text
With over 200,000 diagnoses per year, 1 in 6 U.S. men are diagnosed with prostate cancer during their lifetime. Yet, only 20% of prostate cancer patients have a high-risk cancer that represents possibly lethal disease1,2,4. While mutational events in key genes characterizes a subset of lethal prostate cancers3,5,6, the molecular basis for aggressive disease remains poorly understood.
Long non-coding RNAs (lncRNAs) are RNA species >200bp in length that are frequently polyadenylated and associated with transcription by RNA polymerase II7. lncRNA-mediated biology has been implicated in a wide variety of cellular processes and in cancer, lncRNAs are emerging as a prominent layer of transcriptional regulation, often by collaborating with epigenetic complexes7-10.
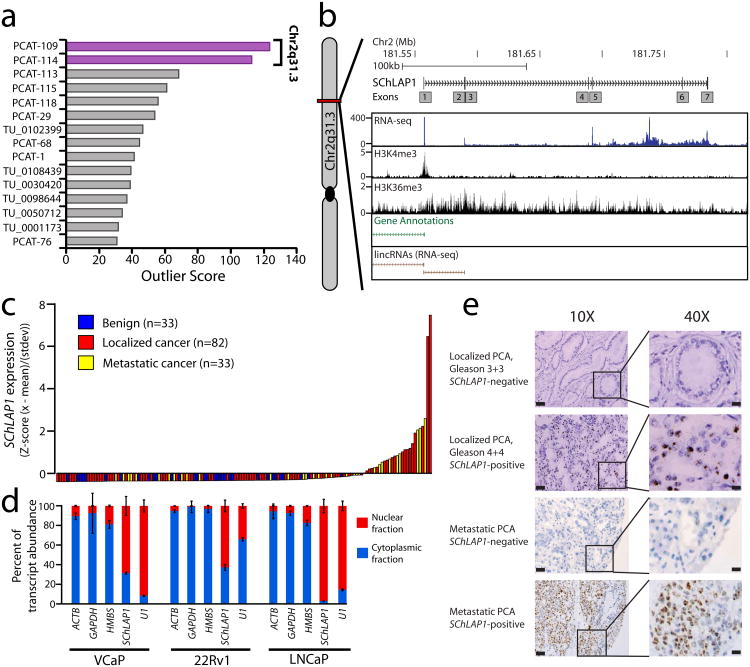
Here, we hypothesized that prostate cancer aggressiveness was governed by uncharacterized lncRNAs and sought to discover lncRNAs associated with aggressive disease. We previously used RNA-Seq to describe 121 novel lncRNA loci (out of >1,800) that were aberrantly expressed in prostate cancer tissues11. Because only a fraction of prostate cancers present with aggressive clinical features2, we performed cancer outlier profile analysis11 (COPA) to nominate intergenic lncRNAs selectively upregulated in a subset of cancers (Supplementary Table 1). We observed that only two, PCAT-109 and PCAT-114, which are both located in a “gene desert” on Chromosome 2q31.3 (Supplementary Fig. 1), showed striking outlier profiles and ranked among the best outliers in prostate cancer11 (Fig. 1a).
Figure 1. Discovery of SChLAP1 as a prostate cancer lncRNA.
(a) Cancer outlier profile analysis (COPA) for intergenic lncRNAs. (b) A representation of the SChLAP1 gene and its annotations in current databases. An aggregated representation of current gene annotations for Ensembl, ENCODE, UCSC, Ref-Seq, and Vega shows no annotation for SChLAP1. ChIP-Seq data for H3K4me3 and H3K36me3 show enrichment at the SChLAP1 gene. Also, RNA-Seq data showing an outlier sample for SChLAP1 illustrates its expression. (c) qPCR for SChLAP1 on a panel of benign prostate (n=33), localized prostate cancer (n=82), and metastatic prostate cancer (n=33) samples. qPCR data is normalized to the average of (GAPDH + HMBS) and represented as standardized expression values. (d) Fractionation of prostate cell lysates demonstrates nuclear expression of SChLAP1. U1 is a positive control for nuclear gene expression. (e)In situ hybridization of SChLAP1 in human prostate cancer. SChLAP1 staining is shown for both localized and metastatic tissues.
Of the two, PCAT-114 was expressed at higher levels in prostate cell lines, and in the PCAT-114 region we defined a 1.4 kb, polyadenylated gene composed of up to seven exons and spanning nearly 200kb on Ch2q31.3 (Fig. 1b and Supplementary Fig. 2a). We named this gene Second Chromosome Locus Associated with Prostate-1 (SChLAP1) after its genomic location. Published prostate cancer ChIP-Seq data12 confirmed that the transcriptional start site (TSS) of SChLAP1 was marked by H3K4 trimethylation (H3K4me3) and its gene body harbored H3K36 trimethylation (H3K36me3) (Fig. 1b), an epigenetic signature consistent with lncRNAs13. We observed numerous SChLAP1 splicing isoforms of which three (termed isoforms #1, #2, and #3, respectively) constituted the vast majority (>90%) of transcripts in the cell (Supplementary Fig. 2b,c).
Using quantitative PCR (qPCR), we validated that SChLAP1 was highly expressed in ∼25% of prostate cancers (Fig. 1c). SChLAP1 prevalence was more frequent in metastatic compared to localized prostate cancers and was associated with ETS gene fusions in this cohort but not other molecular events (Supplementary Fig. 2d,e). A computational analysis of the SChLAP1 sequence suggested no coding potential, which was confirmed experimentally by in vitro translation assays of three SChLAP1 isoforms (Supplementary Fig. 3). Additionally, we found that SChLAP1 transcripts were located in the nucleus (Fig. 1d). We confirmed the nuclear localization of SChLAP1 in human samples (Fig. 1e) using an in situ hybridization (ISH) assay in formalin-fixed paraffin-embedded (FFPE) prostate cancers (Supplementary Fig. 4a,b and Supplementary Note).
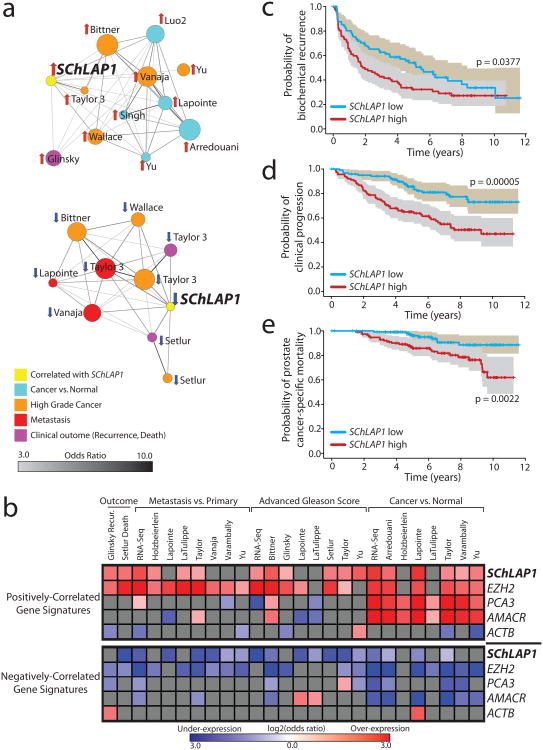
An analysis of SChLAP1 expression in localized tumors demonstrated a striking correlation with higher Gleason scores, a histopathological measure of aggressiveness (Supplementary Fig. 4c,d and Supplementary Table 2). Next, we performed a network analysis of prostate cancer microarray data in the Oncomine14 database using signatures of SChLAP1-correlated or -anti-correlated genes, given that SChLAP1 is not measured by expression microarrays (Supplementary Table 3a and Online Methods). We found a remarkable association with enriched concepts related to prostate cancer progression (Fig. 2a and Supplementary Table 3b). For comparison, we next incorporated disease signatures using prostate RNA-seq data as well as additional known prostate cancer genes: EZH2, a metastasis gene15, PCA3, a lncRNA biomarker4, AMACR, a tissue biomarker4, and β-actin (ACTB) as a control (Supplementary Fig. 5, Supplementary Tables 3c-i, and Supplementary Note). A heat-map visualization of significant comparisons confirmed a strong association of SChLAP1-correlated genes, but not PCA3- and AMACR-correlated genes, with high-grade and metastatic cancers (Fig. 2b). Kaplan-Meier analysis similarly showed significant associations between the SChLAP1 signature and biochemical recurrence16 and overall survival17 (Supplementary Fig. 6a,b).
Figure 2. SChLAP1 expression characterizes aggressive prostate cancer.
(a) Network representation of genes positively or negatively correlated with SChLAP1 in localized prostate cancers using Oncomine concepts analysis and visualized with the Force Directed Layout algorithm in the Cytoscape36 tool. Node sizes reflect the number of genes that comprise each molecular concept and node names are labeled according to the author of the primary study as detailed in Supplementary Table 3b. The nodes are colored according to the concept categories indicated in the figure legend. Edges are drawn between nodes with statistically significant enrichment (p-value < 1e-6, odds ratio > 3.0) and darker edge shading implies higher odds ratio. (b) Heatmap representation of comparisons between co-expression gene signatures and molecular concepts. Comparisons to positively (top portion) and negatively correlated (bottom portion) gene signatures are shown separately. Comparisons that do not reach statistical significance (q > 0.01 or odds ratio < 2) are shown in grey. Associations with over-expression concepts are colored red, and under-expression concepts blue. (c-e) Kaplan-Meier analyses of prostate cancer outcomes in the Mayo Clinic cohort. SChLAP1 expression was measured using Affymetrix exon arrays and patients were stratified according to their SChLAP1 expression. Patient outcomes were analyzed for biochemical recurrence (c), clinical progression to systemic disease (d), and prostate cancer-specific mortality (e). The shaded regions represent the 95% confidence interval.
To evaluate SChLAP1 levels with clinical outcomes directly, we next used SChLAP1 expression to stratify 235 radical prostatectomy localized prostate cancer patients from the Mayo Clinic18 (Supplementary Fig. 6c and Online Methods). Samples were evaluated for three clinical endpoints: biochemical recurrence (BCR), clinical progression to systemic disease (CP), and prostate cancer-specific mortality (PCSM) (Supplementary Table 4). At the time of this analysis, patients had a median follow-up of 8.1 years.
SChLAP1 was a powerful single-gene predictor of aggressive prostate cancer (Fig. 2c-e). SChLAP1 expression was highly significant when distinguishing CP and PCSM (p = 0.00005 and p = 0.002, respectively) (Fig. 2d,e). For the BCR endpoint, high SChLAP1 expression was associated with a rapid median time-to-progression (1.9 vs 5.5 years for SChLAP1 high and low patients, respectively) (Fig. 2c). We further confirmed that this association with rapid BCR using an independent cohort (Supplementary Fig. 6d). Multivariable and univariable regression analyses of the Mayo Clinic data demonstrated that SChLAP1 expression is an independent predictor of prostate cancer aggressiveness with highly significant hazard ratios for predicting BCR, CP, and PCSM (HR or 3.045, 3.563, and 4.339, respectively, p < 0.01) which were comparable to other clinical factors such as advanced clinical stage and the Gleason histopathological score (Supplementary Fig. 7 and Supplementary Note).
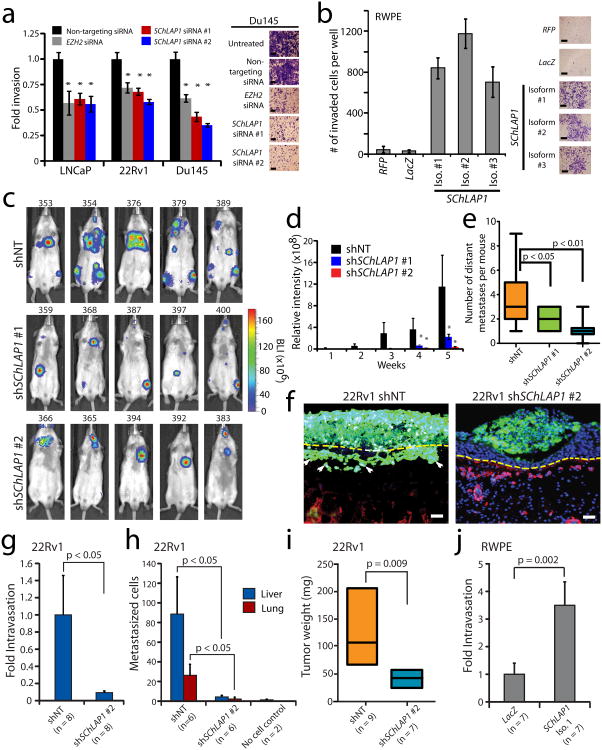
To explore the functional role for SChLAP1, we performed siRNA knockdowns to compare the impact of SChLAP1 depletion to that of EZH2, which is essential for cancer cell aggressiveness15. Remarkably, knockdown of SChLAP1 dramatically impaired cell invasion and proliferation in vitro at a level comparable to EZH2 (Fig. 3a and Supplementary Fig. 8a,b). Overexpression of a siRNA-resistant SChLAP1 isoform rescued the in vitro invasive phenotype of 22Rv1 cells treated with siRNA-2 (Supplementary Fig. 8c,d). Next, overexpression of three SChLAP1 isoforms in RWPE benign immortalized prostate cells dramatically increased the ability of these cells to invade in vitro but did not impact cell proliferation (Fig. 3b and Supplementary Fig. 8e,f).
Figure 3. SChLAP1 coordinates cancer cell invasion in vitro and metastatic seeding in vivo.
(a) siRNA knockdown of SChLAP1 in vitro in three prostate cell lines (LNCaP, 22Rv1, Du145) impairs cellular invasion through Matrigel in a Boyden chamber assay. EZH2 siRNA serves as a positive control. (b) Overexpression of SChLAP1 in RWPE cells results in increased cellular invasion through Matrigel in Boyden chamber assays. (c) Intracardiac injection of 22Rv1 cells with stable SChLAP1 knockdown in severe combined immunodeficient (SCID) mice. Example luciferase bioluminescence images from 22Rv1 shNT, shSChLAP1 #1, and shSChLAP1 #2 mice five weeks following intracardiac injection. Mouse IDs are given above each image. (d) The relative intensity of whole-mouse luciferase signal is plotted for 22Rv1 shNT (n=9), shSChLAP1 #1 (n=14) and shSChLAP1 #2 (n=14) intracardiac injection experiments. (e) The number of gross metastatic sites observed by luciferase signal in 22Rv1 shSChLAP1 cells or shNT controls. Independent foci of luciferase signal were averaged for shNT (n=9), shSChLAP1 #1 (n=14) and shSChLAP1 #2 (n=14) mice. (f) Invasion of 22Rv1-shNT and 22Rv1 shSChLAP1 cells in the chick chorioallantoic membrane (CAM) assay. 22Rv1 cells are labeled with GFP. The image is counterstained with chicken collagen IV for vasculature (RFP) and DAPI for nuclei. (g-i) Using the CAM assay, 22Rv1 shSChLAP1 cells demonstrate decreased intravasation (g), metastatic spread to the liver and lungs (h), and reduced tumor weight (i). (j) Quantification of intravasation of RWPE-LacZ and RWPE-SChLAP1 cells in the CAM assay. All data in this figure are represented as mean +/- S.E.M. Statistical significance was determined by a two-tailed Student's t-test. An asterisk (*) indicates a p-value < 0.05.
To test SChLAP1 in vivo, we performed intracardiac injection of 22Rv1 cells stably knocking down SChLAP1 (Supplementary Fig. 9a) and observed that SChLAP1 depletion impaired metastatic seeding and growth by luciferase signaling at both proximal (lungs) and distal sites (Fig. 3c,d). Indeed, 22Rv1 shSChLAP1 cells displayed both fewer gross metastatic sites overall as well as smaller metastatic tumors when they did form (Fig. 3d,e). Histopathological analysis of the metastatic 22Rv1 tumors, regardless of SChLAP1 knockdown, showed uniformly high-grade epithelial cancer (Supplementary Fig. 9b). Interestingly, shSChLAP1 subcutaneous xenografts displayed slower tumor progression; however this was due to delayed tumor engraftment rather than decreased tumor growth kinetics with no change in Ki67 staining observed between shSChLAP1 and shNT cells (Supplementary Fig. 9c-i).
Next, using the chick chorioallantoic membrane (CAM) assay19, we found that 22Rv1 shSChLAP1 #1 cells, which have depleted expression of both isoforms 1 and 2, demonstrated a greatly reduced ability to invade, intravasate and metastasize distant organs (Fig. 3f-h). Additionally, shSChLAP1 cells also showed decreased tumor growth (Fig. 3i). Importantly, overexpression of RWPE-SChLAP1 isoform #1 cells partially recapitulated these results, displaying a markedly increased ability to intravasate (Fig. 3j). RWPE-SChLAP1 cells did not generate distant metastases or cause altered tumor growth in this model (data not shown). Together, the murine metastasis and CAM data strongly implicate SChLAP1 in tumor invasion and metastasis through cancer cell intravasation, extravasation, and subsequent tumor cell seeding.
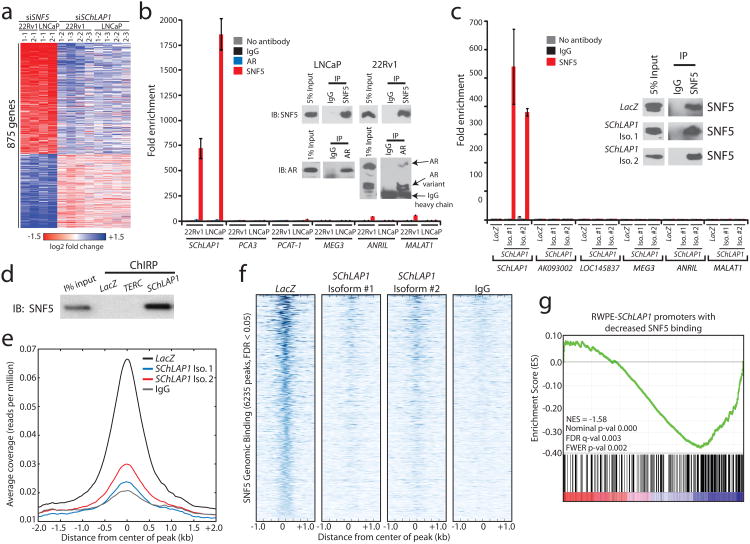
To elucidate mechanisms of SChLAP1 function, we profiled 22Rv1 and LNCaP SChLAP1-knockdown cells, which revealed 165 upregulated and 264 downregulated genes (q-value < 0.001) (Supplementary Fig. 10a and Supplementary Table 5a). After ranking genes according to differential expression20, we employed Gene Set Enrichment Analysis (GSEA)21 to search for enrichment across the Molecular Signatures Database (MSigDB)22. Among the highest ranked concepts we noticed genes positively or negatively correlated with the SWI/SNF complex23, which was independently confirmed using gene signatures generated from our RNA-Seq data (Supplementary Fig. 10b-e, and Supplementary Table 5b,c). Importantly, SChLAP1-regulated genes were inversely correlated with these datasets, suggesting that SChLAP1 and SWI/SNF function in opposing manners.
The SWI/SNF complex regulates gene transcription as a multi-protein system that physically move nucleosomes at gene promoters24. Loss of SWI/SNF functionality promotes cancer progression and multiple SWI/SNF components are somatically inactivated in cancer24,25. SWI/SNF mutations do occur in prostate cancer albeit not commonly3, and down-regulation of SWI/SNF complex members characterizes subsets of prostate cancer23,26. Thus, antagonism of SWI/SNF activity by SChLAP1 is consistent with the oncogenic behavior of SChLAP1 and the tumor suppressive behavior of the SWI/SNF complex.
To directly test whether SChLAP1 antagonizes SWI/SNF-mediated regulation, we performed siRNA knockdown of SNF5 (also known as SMARCB1) (Supplementary Fig. 10f), an essential subunit that facilitates SWI/SNF binding to histone proteins24,25,27, and confirmed predicted expression changes for several SChLAP1 or SNF5-regulated genes (Supplementary Fig. 10g,h). A comparison of genes regulated by knockdown of SNF5 to genes regulated by SChLAP1 demonstrated an antagonistic relationship where SChLAP1 knockdown affected the same genes as SNF5 but in the opposing direction (Fig. 4a and Supplementary Tables 5d-h). We used GSEA to quantify and verify the significance of these findings (FDR < 0.05) (Supplementary Fig. 10i-k). Furthermore, a shared SNF5-SChLAP1 signature of co-regulated genes was highly enriched for prostate cancer clinical signatures for disease aggressiveness (Supplementary Fig. 11 and Supplementary Table 5i).
Figure 4. SChLAP1 antagonizes SNF5 function and attenuates SNF5 genome-wide localization.
(a) Heatmap results for SChLAP1 or SNF5 knockdown in LNCaP and 22Rv1 cells. (b) RNA immunoprecipitation (RIP) of SNF5 or AR in SNF5 in 22Rv1 and LNCaP cells. Inset Western blots demonstrate pulldown efficiency. (c) RIP analysis of SNF5 in RWPE cells overexpressing LacZ, SChLAP1 isoform #1, or SChLAP1 isoform #2. Inset Western blots demonstrate pulldown efficiency. (d) Pulldown of SChLAP1 RNA using Chromatin Isolation by RNA Purification (ChIRP) recovers SNF5 protein in RWPE-SChLAP1 isoform 1 cells. LacZ and TERC serve as controls. (e) A global representation of SNF5 genomic binding over ±2kb window surrounding each SNF5 ChIP-Seq peak in RWPE-LacZ, RWPE-SChLAP1 isoform 1, and RWPE-SChLAP1 isoform 2 cells. (f) A heatmap representation of SNF genomic binding at target sites in RWPE-LacZ, RWPE-SChLAP1 isoform 1, and RWPE-SChLAP1 isoform 2 cells. A ±1kb interval surrounding the called SNF5 peak is shown. (g) Gene set enrichment analysis results showing significant enrichment of ChIP-Seq promoter peaks with >2-fold loss of SNF5 binding for underexpressed genes in RWPE-SChLAP1 cells.
Mechanistically, although SChLAP1 and SNF5 mRNA levels were comparable (Supplementary Fig. 12a), SChLAP1 knockdown or overexpression did not alter SNF5 protein abundance (Supplementary Fig. 12b), suggesting that SChLAP1 regulates SWI/SNF activity post-translationally. To explore this possibility, we performed RNA immunoprecipitation assays (RIP) for SNF5. We found that endogenous SChLAP1, but not other cytoplasmic or nuclear lncRNAs7,28, robustly co-immunoprecipitated with SNF5 in both native (Fig. 4b) and UV-crosslinked conditions (Supplementary Fig. 12c) as well as with a second SNF5 antibody (Supplementary Fig. 12d). In contrast, SChLAP1 did not co-immunoprecipitate with androgen receptor (Fig. 4b). Furthermore, both SChLAP1 isoform #1 and isoform #2 co-immunoprecipitated with SNF5 in RWPE overexpression models (Fig. 4c and Supplementary Fig. 12e). SNRNP70 binding to the U1 RNA was used as a technical control in all cell lines (Supplementary Fig. 12f,g). Finally, pulldown of the SChLAP1 RNA in RWPE-SChLAP1 isoform #1 cells robustly recovered SNF5 protein, confirming this interaction (Fig. 4d and Supplementary Fig. 12h).
To address whether SChLAP1 modulated SWI/SNF genomic binding, we performed ChIP-Seq of SNF5 in RWPE-LacZ and RWPE-SChLAP1 cells and called significantly enriched peaks with respect to an IgG control (Supplementary Table 6a and Online Methods). Western blot validations confirmed SNF5 pull-down by ChIP (Supplementary Fig. 13a), After aggregating called peaks from all samples, we found 6,235 genome-wide binding sites for SNF5 (FDR < 0.05, Supplementary Table 6b), which were highly enriched for sites near gene promoters (Supplementary Fig. 13b), supporting previous studies of SWI/SNF binding29-31.
A comparison of SNF5 binding across these 6,235 genomic sites demonstrated a dramatic decrease in SNF5 genomic binding as a result of SChLAP1 overexpression (Fig. 4e,f and Supplementary Fig. 13c). Of the 1,299 SNF5 peaks occurring within 1kb of a gene promoter, 390 decreased ≥2-fold in relative SNF5 binding (Supplementary Fig. 13d and Supplementary Table 6c). To verify these findings independently, we performed ChIP for SNF5 in 22Rv1 sh-SChLAP1 cells, with the hypothesis that knockdown of SChLAP1 should increase SNF5 genomic binding compared to controls. We found that 9 of 12 target genes showed a substantial increase in SNF5 binding (Supplementary Fig. 14a), confirming our predictions.
Finally, we used expression profiling of RWPE-LacZ and RWPE-SChLAP1 cells to characterize the relationship between SNF5 binding and SChLAP1-mediated gene expression changes. After identifying a gene signature with highly significant expression changes (Supplementary Table 6d), we intersected this signature with the ChIP-Seq data. We observed that a significant subset of genes with ≥2-fold relative decrease in SNF5 genomic binding were dysregulated when SChLAP1 was overexpressed (Supplementary Fig. 14b). Decreased SNF5 binding was primarily associated with downregulation of target gene expression (Supplementary Table 6e), although the SWI/SNF complex is known to regulate expression in either direction24,25. An integrative GSEA analysis of the microarray and SNF5 ChIP-Seq data demonstrated a significant enrichment for genes that were repressed when SChLAP1 was overexpressed (q-value = 0.003, Fig. 4g). Overall, these data argue that SChLAP1 overexpression antagonizes SWI/SNF complex function by attenuating the genomic binding of this complex, thereby impairing its ability to regulate gene expression properly.
Here, we have discovered SChLAP1, a highly prognostic lncRNA that is abundantly expressed in ∼25% of prostate cancers and aided the discrimination of aggressive from indolent forms of this disease. Mechanistically, we find that SChLAP1 coordinates cancer cell invasion in vitro and metastatic spread in vivo. Moreover, we characterize an antagonistic SChLAP1-SWI/SNF axis in which SChLAP1 impairs SNF5-mediated gene expression regulation and genomic binding (Supplementary Fig. 14c). Thus, while other lncRNAs such as HOTAIR and HOTTIP are known to assist epigenetic complexes such as PRC2 and MLL by facilitating their genomic binding and enhancing their functions8,9,32, SChLAP1 is the first lncRNA, to our knowledge, that impairs a major epigenetic complex with well-documented tumor suppressor function23-25,33-35. Taken together, our discovery of SChLAP1 has broad implications for cancer biology and provides supporting evidence for the role of lncRNAs in the progression of aggressive cancers.
Data Deposition
Sequences for SChLAP1 isoforms #1-7 have been deposited to GenBank as accession numbers JX117418 – JX117424. Microarray data have been deposited to GEO as accession number GSE40386.
Online Methods
Experimental studies
Cell lines
All cell lines were obtained from the American Type Culture Collection (Manassas, VA). Cell lines were maintained using standard media and conditions. Specifically, VCaP and Du145 cells were maintained in DMEM (Invitrogen) plus 10% fetal bovine serum (FBS) plus 1% penicillin-streptomycin. LNCaP and 22Rv1 were maintained in RPMI 1640 (Invitrogen) plus 10% FBS and 1% penicillin-streptomycin. RWPE cells were maintained in KSF media (Invitrogen) plus 10ng/mL EGF (Sigma) and bovine pituitary extract (BPE) and 1% penicillin-streptomycin. All cell lines were grown at 37°C in a 5% CO2 cell culture incubator. All cell lines were genotyped for identity at the University of Michigan Sequencing Core and tested routinely for Mycoplasma contamination.
SChLAP1 or control-expressing cell lines were generated by cloning SChLAP1 or control into the pLenti6 vector (Invitrogen) using pcr8 non-directional Gateway cloning (Invitrogen) as an initial cloning vector and shuttling to pLenti6 using LR clonase II (Invitrogen) according to the manufacturer's instructions. Stably-transfected RWPE and 22Rv1 cells were selected using blasticidin (Invitrogen) for one week. For LNCAP and 22Rv1 cells with stable knockdown of SChLAP1, cells were transfected with SChLAP1 or non-targeting shRNA lentiviral constructs for 48 hours. GFP+ cells were selected with 1ug/mL puromycin for 72 hours. All lentiviruses were generated by the University of Michigan Vector Core.
Tissue Samples
Prostate tissues were obtained from the radical prostatectomy series and Rapid Autopsy Program at the University of Michigan tissue core37. These programs are part of the University of Michigan Prostate Cancer Specialized Program Of Research Excellence (S.P.O.R.E.). All tissue samples were collected with informed consent under an Institutional Review Board (IRB) approved protocol at the University of Michigan. (SPORE in Prostate Cancer (Tissue/Serum/Urine) Bank Institutional Review Board # 1994-0481).
RNA isolation and cDNA synthesis
Total RNA was isolated using Trizol and an RNeasy Kit (Invitrogen) with DNase I digestion according to the manufacturer's instructions. RNA integrity was verified on an Agilent Bioanalyzer 2100 (Agilent Technologies, Palo Alto, CA). cDNA was synthesized from total RNA using Superscript III (Invitrogen) and random primers (Invitrogen).
Quantitative Real-time PCR
Quantitative Real-time PCR (qPCR) was performed using Power SYBR Green Mastermix (Applied Biosystems, Foster City, CA) on an Applied Biosystems 7900HT Real-Time PCR System. All oligonucleotide primers were obtained from Integrated DNA Technologies (Coralville, IA) and are listed in Supplementary Table 7. The housekeeping genes, GAPDH, HMBS, and ACTB, were used as loading controls. Fold changes were calculated relative to housekeeping genes and normalized to the median value of the benign samples.
Reverse-transcription PCR
Reverse-transcription PCR (RT-PCR) was performed for primer pairs using Platinum Taq High Fidelity polymerase (Invitrogen). PCR products were resolved on a 1.0% agarose gel. PCR products were either sequenced directly (if only a single product was observed) or appropriate gel products were extracted using a Gel Extraction kit (Qiagen) and cloned into pcr4-TOPO vectors (Invitrogen). PCR products were bidirectionally sequenced at the University of Michigan Sequencing Core using either gene-specific primers or M13 forward and reverse primers for cloned PCR products. All oligonucleotide primers were obtained from Integrated DNA Technologies (Coralville, IA) and are listed in Supplementary Table 7.
RNA-ligase-mediated rapid amplification of cDNA ends (RACE)
5′ and 3′ RACE was performed using the GeneRacer RLM-RACE kit (Invitrogen) according to the manufacturer's instructions. RACE PCR products were obtained using Platinum Taq High Fidelity polymerase (Invitrogen), the supplied GeneRacer primers, and appropriate gene-specific primers indicated in Supplementary Table 7. RACE-PCR products were separated on a 1.5% agarose gels. Gel products were extracted with a Gel Extraction kit (Qiagen), cloned into pcr4-TOPO vectors (Invitrogen), and sequenced bidirectionally using M13 forward and reverse primers at the University of Michigan Sequencing Core. At least three colonies were sequenced for every gel product that was purified.
siRNA knockdown studies
Cells were plated in 100mM plates at a desired concentration and transfected with 20uM experimental siRNA oligos or non-targeting controls twice, at 8 hours and 24 hours post-plating. Knockdowns were performed with Oligofectamine in OptiMEM media. Knockdown efficiency was determined by qPCR. siRNA sequences (in sense format) for knockdowns were as follows:
SChLAP1 siRNA 1: CCAAUGAUGAGGAGCGGGA
SChLAP1 siRNA 2: CUGGAGAUGGUGAACCCAA
SNF5 siRNA 5: GUGACGAUCUGGAUUUGAA
SNF5 siRNA 7: GAUGACGCCUGAGAUGUUU
72 hours post-transfection, cells were trypsinized, counted with a Coulter counter, and diluted to 1 million cells/mL.
Overexpression studies
SChLAP1 full length transcript was amplified from LNCaP cells and cloned into the pLenti6 vector (Invitrogen) along with LacZ controls. Insert sequences were confirmed by Sanger sequencing at the University of Michigan Sequencing Core. Lentiviruses were generated at the University of Michigan Vector Core. The benign immortalized prostate cell line RWPE was infected with lentiviruses expressing SChLAP1 or LacZ and stable pools and clones were generated by selection with blasticidin (Invitrogen). Similarly, the immortalized cancer cell line 22Rv1 was infected with lentiviruses expressing SChLAP1 or LacZ and stable pools were generated by selection with blasticidin (Invitrogen).
Cell proliferation assays
72 hours post-transfection with siRNA, cells were trypsinized, counted with a Coulter counter, and diluted to 1 million cells/mL. For proliferation assays, 10,000 cells were plated in 24-well plates and grown in regular media. 48 and 96 hours post-plating, cells were harvested by trypsinizing and counted using a Coulter counter. All assays were performed in quadruplicate.
Basement Membrane Matrix Invasion Assays
For invasion assays, cells were treated with the indicated siRNAs and 72 hours post-transfection, cells were trypsinized, counted with a Coulter counter, and diluted to 1 million cells/mL. Cells were seeded onto the basement membrane matrix (EC matrix, Chemicon, Temecula, CA) present in the insert of a 24 well culture plate. Fetal bovine serum was added to the lower chamber as a chemo-attractant. After 48 hours, the non-invading cells and EC matrix were gently removed with a cotton swab. Invasive cells located on the lower side of the chamber were stained with crystal violet, air-dried and photographed. For colorimetric assays, the inserts were treated with 150 μl of 10% acetic acid and the absorbance measured at 560nm using a spectrophotometer (GE Healthcare).
shRNA knockdown
The prostate cancer cell lines LNCaP and 22Rv1 were seeded at 50-60% confluency and allowed to attach over night. Cells were transfected with SChLAP1 or non-targeting shRNA lentiviral constructs as described previously for 48 hours. GFP+ cells were drug-selected using 1 ug/mL puromycin for 72 hours. 48 hours post-selection cells were harvested for protein and RNA using RIPA buffer or trizol, respectively. RNA was processed as described above.
Gene expression profiling
Expression profiling was performed using the Agilent Whole Human Genome Oligo Microarray (Santa Clara, CA), according to previously published protocols38. All samples were run in technical triplicates comparing knockdown samples treated with SChLAP1 siRNA compared to treatments with non-targeting control siRNA. Expression data was analyzed using the SAM method as described previously20.
Murine intracardiac and subcutaneous in vivo models
All experimental procedures were approved by the University of Michigan Committee for the Use and Care of Animals (UCUCA). Intracardiac injection model: 5 × 105 cells from one of three experimental cell lines (22Rv1 shNT, 22Rv1 shSChLAP1 #1, shSChLAP1 #2, all with luciferase constructs incorporated) were introduced to CB-17 severe combine immunodefiecient mice (CB-17 SCID) at 6 weeks of age. Female mice were used to minimize endogenous androgen production that may stimulate xenografted prostate cells. 15 mice were used per cell line in order to ensure adequate statistical power to distinguish phenotypes between groups. Mice used in these studies were randomized by double-blind injection of cell line samples into mice and were monitored for tumor growth by researchers blinded to the study design. Beginning one week post injection, bioluminescent imaging of mice was performed weekly using a CCD IVIS system with a 50-mm lens (Xenogen Corp.) and the results were analyzed using LivingImage software (Xenogen). When the mice reached determined endpoint, whole body region of interest (ROI) of 1 × 1010 photons, or became fatally ill, the animal was euthanized and the lung and liver resected. Half of the resected specimen was put in an immunohistochemistry cassette and placed in 10% buffered formalin phosphate (Fisher Scientific) for 24 hours, and then transferred to 70% ethanol until further analysis. The other half of each specimen was snap frozen in liquid nitrogen and stored in -80°C. A specimen was disregarded if the tumor was localized to the heart only. After accounting for these considerations, there were 9 mice analyzed for 22Rv1 shNT cells, 14 mice each analyzed for 22Rv1 shSChLAP1 #1 and #2 cells. Subcutaneous injection model: 1 × 106 cells from one of the three previously described experimental cell lines were introduced to mice (CB-17 SCID), ages 5-7 weeks, with a Matrigel scaffold (BD Matrigel Matrix, BD Biosciences) in the posterior dorsal flank region (n = 10 per cell line). Tumors were measured weekly using a digital caliper, and endpoint was determined as a tumor volume of 1000 mm3. When endpoint was reached, or the animal became fatally ill, the mouse was euthanized and the primary tumor resected. The resected specimen was divided in half: one half in 10% buffer formalin and the other half snap frozen. For histological analyses, FFPE-fixed mouse livers and lungs were sectioned on a microtome into 5uM sections onto glass slides. Slides were stained with hematoxalyn and eosin using standard methods and analyzed by a board-certified pathologist (LPK).
Immunoblot Analysis
Cells were lysed in RIPA lysis buffer (Sigma, St. Louis, MO) supplemented with HALT protease inhibitor (Fisher). Western blotting analysis was performed with standard protocols using Polyvinylidene Difluoride membrane (GE Healthcare, Piscataway, NJ) and the signals visualized by enhanced chemiluminescence system as described by the manufacturer (GE Healthcare).
Protein lysates were boiled in sample buffer, and 10 ug protein was loaded onto a SDS-PAGE gel and run for separation of proteins. Proteins were transferred onto Polyvinylidene Difluoride membrane (GE Healthcare) and blocked for 90 minutes in blocking buffer (5% milk, 0.1% Tween, Tri-buffered saline (TBS-T)). Membranes were incubated overnight at 4C with primary antibody. Following 3 washes with TBS-T, and one wash with TBS, the blot was incubated with horseradish peroxidase-conjugated secondary antibody and the signals visualized by enhanced chemiluminescence system as described by the manufacturer (GE Healthcare).
Primary antibodies used were:
SNF5 (1:1000, Millipore, ABD22, rabbit)
SNF5 (1:1000, Abcam, ab58209, mouse)
ACTB (1:5000, Sigma, rabbit)
AR (1:1000. Millipore, 06-680, rabbit)
RNA immunoprecipitation
RIP assays were performed using a Millipore EZ-Magna RIP RNA-Binding Protein Immunoprecipitation kit (Millipore, #17-701) according to the manufacturer's instructions. RIP-PCR was performed as qPCR, as described above, using total RNA as input controls. 1:150th of RIP RNA product was used per PCR reaction. Antibodies used for RIP were Rabbit polyclonal IgG (Millipore, PP64), SNRNP70 (Millipore, CS203216), SNF5 (Millipore, ABD22), SNF5 (Abcam, ab58209), and AR (Millipore, 06-680, rabbit), using 5 – 7 ug of antibody per RIP reaction. All RIP assays were performed in biological duplicate. For UV-crosslinked RIP experiments, cells were subjected to 400J of 254nM UV light twice and then harvested for RIP experiments as above.
Chromatin immunoprecipitation
ChIP assays were performed as described previously11,12, using antibodies for SNF5 (Millipore ABD22) and Rabbit IgG (Millipore PP64B). Briefly, approximately 10^6 cells were crosslinked per antibody for 10-15 minutes with 1% formaldehyde and the crosslinking was inactivated by 0.125M glycine for 5 minutes at room temperature. Cells were rinsed with cold PBS three times and cell pellets were resuspended in lysis buffer plus protease inhibitors. Chromatin was sonicated to an average length of 500bp, centrifuged to remove debris, and supernatants containing chromatin fragments were incubated with protein A/G beads to reduce non-specific binding. Then, beads were removed and supernatants were incubated with 6ug of antibody overnight at 4C. Beads were added and incubated with protein-chromatin-antibody complexes for 2 hours at 4C, washed twice with 1× dialysis buffer and four times with IP wash buffer, and eluted in 150 ul IP elution buffer. 1:10th of the ChIP reaction was taken for protein evaluation for validation of ChIP pull-down. Reverse crosslinking was performed by inclubating the eluted product with 0.3 M NaCl at 65C overnight. ChIP product was cleaned up with the USB PrepEase kit (USB). ChIP experiments were validated for specificity by Western blotting.
ChIP-Seq experiments
Paired-end ChIP-Seq libraries were generated following the Illimuna ChIP-Seq protocol with minor modifications. The ChIP DNA was subjected to end-repair and A base addition before ligating with Illumina adaptors. Samples were purified using Ampure beads (Beckman Coulter Inc., Brea CA) and PCR-enriched with a combination of specific index primers and PE2.0 primer under the following conditions: 98C (30 sec), 65C (30 sec), and 72C (40 sec with a 4 sec increment per cycle). After 14 cycles of amplification a final extension at 72C for 5 minutes was carried out. The barcoded libraries were size-selected using a 3% NuSieve Agarose gele (Lonza, Allendale, NJ) and subjected to an additional PCR enrichment step. The libraries were analyzed and quantitated using Bio-Analyzer (Agilent Technologies, Santa Clara, CA) before subjecting it to paired-end sequencing using the Illumina Hi-Seq platform.
CAM assays
CAM assays were performed as previously described39. Briefly, fertilized eggs were incubated in a rotary humidified incubator at 38°C for 10 days. CAM was released by applying mild amount of low pressure to the hole over the air sac and cutting a 1 cm2 window encompassing a second hole near the allantoic vein. Approximately 2 million cells in 50μl of media were implanted in each egg, windows were sealed and the eggs were returned to a stationary incubator.
For local invasion and intravasation experiments, the upper and lower CAM were isolated after 72hr. The upper CAM were processed and stained for chicken collagen IV (immunofluorescence) or human cytokeratin (immunohistochemistry) as previously described39.
For metastasis assay, the embryonic livers were harvested on day 18 of embryonic growth and analyzed for the presence of tumor cells by quantitative human Alu-specific PCR. Genomic DNA from lower CAM and livers were prepared using Puregene DNA purification system (Qiagen) and quantification of human-Alu was performed as described39. Fluorogenic TaqMan qPCR probes were generated as described above and used to determine DNA copy number.
For xenograft growth assay with RWPE cells, the embryos were sacrificed on day 18 and the extra-embryonic xenograft were excised and weighed.
In situ hybridization
ISH assays were performed as a commercial service from Advanced Cell Diagnostics, Inc. Briefly, cells in the clinical specimens are fixed and permeablized using xylenes, ethanol, and protease to allow for probe access. Slides are boiled in pretreatment buffer for 15 min and rinsed in water. Next, two independent target probes are hybridized to the SChLAP1 RNA at 40C for 2 hours, with this pair of probes creating a binding site of a preamplifier. After this, the preamplifier is hybridized to the target probes at 30C and amplified with 6 cycles of hybridization followed by 2 washes. Cells are counter-stained to visualize signal. Finally, slides are H&E stained, dehydrated with 100% ethanol and xylene, and mounted in a xylene-based mounting media.
In vitro translation
Full length SChLAP1, PCAT-1, or GUS positive control were cloned into the PCR2.1 entry vector (Invitrogen). Insert sequences were confirmed by Sanger sequencing at the University of Michigan Sequencing Core. In vitro translation assays were performed with the TnT Quick Coupled Transcription/Translation System (Promega) with 1mM methionine and Transcend Biotin-Lysyl-tRNA (Promega) according to the manufacturer's instructions.
ChIRP Assay
ChIRP assays were performed as previously described40. Briefly, antisense DNA probes targeting the SChLAP1 full-length sequence were designed using the online designer at http://www.singlemoleculefish.com. Fifteen probes spanning the entire transcript and unique to the SChLAP1 sequence were chosen. Additionally, ten probes were designed against TERC RNA as a positive control and twenty-four probes were designed against LacZ RNA as a negative control. All probes were synthesized with 3′ biotinylation (IDT). Sequences of all probes are listed in Supplementary Table 8. RWPE cells overexpressing SChLAP1 isoform 1 were grown to 80% confluency in 100mm cell culture dishes. Two dishes were used for each probe set. Prior to harvesting, the cells were rinsed with 17times;PBS and crosslinked with 1% glutaraldehyde (Sigma) for 10 min at room temperature. Crosslinking was quenched with 0.125M glycine for 5 min at room temperature. The cells were rinsed twice with 1×PBS, collected and pelleted at 1500×g for 5 min. Nuclei were isolated using the Pierce NE-PER Nuclear Protein Extraction Kit. The nuclear pellet was resuspended in 100mg/ml cell lysis buffer (50 mM Tris, pH 7.0, 10 mM EDTA, 1% SDS, and added before use: 1 mM dithithreitol (DTT), phenylmethylsulphonyl fluoride (PMSF), protease inhibitor and Superase-In (Invitrogen)). The lysate was placed on ice for 10 min and sonicated using a Bioruptor (Diagenode) at the highest setting with 30 sec on and 45 sec off cycles until lysates were completely solubilized. Cell lysates were diluted in twice the volume of hybridization buffer (500 mM NaCl, 1% SDS, 100 mM Tris, pH 7.0, 10 mM EDTA, 15% formamide, and added before use: DTT, PMSF, protease inhibitor, and Superase-In) and 100pmol/ml probes were added to the diluted lysate. Hybridization was carried out by end-over-end rotation at 37 °C for 4 hours. Magnetic streptavidin C1 beads were prepared by washing three times in cell lysis buffer and then added to each hybridization reaction at 100ul per 100pmol of probes. The reaction was incubated at 37°C for 30 min with end-over-end rotation. Bead–probe–RNA complexes were captured with magnetic racks (Millipore) and washed five times with 1mL wash buffer (2×SSC, 0.5% SDS, fresh PMSF added). After the last wash, 20% of the sample was used for RNA isolation and 80% of the sample was used for protein isolation. For RNA elution, beads were resuspended in 200μl of RNA proteinase K buffer (100 mM NaCl, 10 mM Tris, pH 7.0, 1 mM EDTA, 0.5% SDS) and 1mg/ml proteinase K (Ambion). The sample was incubated at 50°C for 45 min and then boiled for 10 min. RNA was isolated using 500ul of Trizol reagent using the miRNeasy kit (Qiagen) with on-column DNase digestion (Qiagen). RNA was eluted with 10ul H2O and then analyzed by qRT–PCR for the detection of enriched transcripts. For protein elution, beads were resuspended in 3× the original volume of DNase buffer (100 mM NaCl and 0.1% NP-40), and protein was eluted with a cocktail of 100 ug/ml RNase A (Sigma-Aldrich), 0.1 Units/microliter RNase H (Epicenter), and 100 U/ml DNase I (Invitrogen) at 37°C for 30 min. The eluted protein sample was supplemented with NuPAGE® LDS Sample Buffer (Novex) and NuPAGE® Sample Reducing Agent (Novex) to a final concentration of 1× each and then boiled for 10 min before SDS-PAGE Western blot analysis using a SNF5 antibody (Millipore).
RNA-Seq Library Preparation
Total RNA was extracted from healthy and cancer cell lines and patient tissues, and the quality of the RNA were assessed with the Agilent Bioanalyzer. Transcriptome libraries from the mRNA fractions were generated following the RNA-Seq protocol (Illumina). Each sample was sequenced in a single lane with the Illumina Genome Analyzer II (with a 40- to 80-nt read length) or with the Illumina HiSeq 2000 (with a 100-nt read length) according to published protocols11,41. For strand-specific library construction, we employed the dUTP method of second-strand marking as described previously42.
Statistical analyses for experimental studies
All data are presented as means ± S.E.M. All experimental assays were performed in duplicate or triplicate. Statistical analyses shown in figures represent Fisher's exact tests or two-tailed t-tests, as indicated. For details regarding the statistical methods employed during microarray, RNA-Seq and ChIP-Seq data analysis, see Bioinformatic analyses.
Bioinformatics Analysis
Nomination of SChLAP1 as an outlier using RNA-Seq data
We nominated SChLAP1 as a prostate cancer outlier using the methodology detailed in Prensner JR et al., Nature Biotechnology 2011. Briefly, a modified COPA analysis was performed on the 81 tissue samples in the cohort. RPKM expression values were used and shifted by 1.0 in order to avoid division by zero. The COPA analysis had the following steps: 1) gene expression values were median centered, using the median expression value for the gene across the all samples in the cohort. This sets the gene's median to zero. 2) The median absolute deviation (MAD) was calculated for each gene, and then each gene expression value was scaled by its MAD. 3) The 80, 85, 90, 98 percentiles of the transformed expression values were calculated for each gene and the average of those four values was taken. Then, genes were rank ordered according to this “average percentile”, which generated a list of outliers genes arranged by importance. 4) Finally, genes showing an outlier profile in the benign samples were discarded.
LNCaP ChIP-Seq data
Sequencing data from GSE14097 were downloaded from GEO. Reads from the LNCAP H3K4me3 and H3K36me3 ChIP-Seq samples were mapped to human genome version hg19 using BWA 0.5.943. Peak calling was performed using MACS44 according to the published protocols45. Data was visualized using the UCSC Genome Browser46.
RWPE ChIP-Seq data
Sequencing data from RWPE SNF5 ChIP-Seq samples were mapped to human genome version hg19 using BWA 0.5.943. Although we performed paired-end sequencing, the ChIP-Seq reads were processed as single-end to adhere to our preexisting analysis protocol. Basic read alignment statistics are listed in Supplementary Table 6A. Peak calling was performed respect to an IgG control using the MACS algorithm44. We bypassed the model-building step of MACS (using the ‘--nomodel’ flag) and specified a shift size equal to half the library fragment size determined by the Agilent Bioanalyzer (using the ‘--shiftsize’ option). For each sample we ran the CEAS program and generated genome-wide reports47. We retained peaks with an false discovery rate (FDR) less than 5% (peak calling statistics across multiple FDR thresholds are shown in Supplementary Table 6B). We then aggregated SNF5 peaks from the RWPE-LacZ, RWPE-SChLAP1 Isoform #1, and RWPE-SChLAP1 Isoform #2 samples using the “union” of the genomic peak intervals. We intersected peaks with RefSeq protein-coding genes and found that 1,299 peaks occurred within one kilobase of transcription start sites (TSSs). We counted the number of reads overlapping each of these promoter peaks across each sample using a custom python script and used the DESeq R package version 1.6.148 to compute the normalized fold change between RWPE-LacZ and RWPE-SChLAP1 (both isoforms). We observed that 389 of the 1,299 promoter peaks had at least a 2-fold average decrease in SNF5 binding. This set of 389 genes was subsequently used as a gene set for Gene Set Enrichment Analysis (GSEA) (Supplementary Table 6C).
Microarray Gene Expression Analysis
Microarray experiments
We performed two-color microarray gene expression profiling of 22Rv1 and LNCaP cells treated with two independent siRNAs targeting SChLAP1 as well as control non-targeting siRNAs. These profiling experiments were run in technical triplicate for a total of 12 arrays (6 from 22Rv1 and 6 from LNCaP). Additionally, we profiled 22Rv1 and LNCaP cells treated with independent siRNAs targeting SWI/SNF protein SNF5 (SMARCB1) as well as control non-targeting siRNAs. These profiling experiments were run as biological duplicates for a total of 4 arrays (2 cell lines × 2 independent siRNAs × 1 protein). Finally, we profiled of RWPE cells expressing two different SChLAP1 isoforms as well as the control LacZ gene. These profiling experiments were run in technical duplicate for a total of 4 arrays (2 from RWPE-SChLAP1 isoform #1 and 2 from RWPE-SChLAP1 isoform #2).
Processing to determine ranked gene expression lists
All of the microarray data were represented as base-2 log fold-change between targeting versus control siRNAs. We used the CollapseDataset tool provided by the GSEA package to convert from Agilent Probe IDs to gene symbols. Genes measured by multiple probes were consolidated using the median of probes. We then ran one-class SAM analysis from the Multi-Experiment Viewer application and ranked all genes by the difference between observed versus expected statistics. These ranked gene lists was imported to GSEA version 2.07.
SChLAP1 siRNA knockdown microarrays
For the 22Rv1 and LNCaP SChLAP1 knockdown experiments we ran the GseaPreRanked tool to discover enriched gene sets in the Molecular Signatures Database (MSigDB) version 3.022. Lists of positively and negatively enriched concepts were interpreted manually.
SNF5 siRNA knockdown microarrays
For each SNF5 protein knockdown we nominated genes that were altered by an average of at least 2-fold. These signatures of putative SNF5 target genes were then used to assess enrichment of SChLAP1-regulated genes using the GseaPreRanked tool. Additionally, we nominated genes that changed by an average 2-fold or greater across SNF5 knockdown experiments and quantified the enrichment for SChLAP1 target genes using GSEA.
RWPE SChLAP1 expression microarrays
The RWPE-SChLAP1 versus RWPE-LacZ expression profiles were ranked using SAM analysis as described above. A total of 1,245 genes were significantly over- or under-expressed and are shown in Supplementary Table 6D. A q-value of 0.0 in this SAM analysis signifies that no permutation generated a more significant difference between observed and expected gene expression ratios. The ranked gene expression list was used as input to the GseaPreRanked tool and compared against SNF5 ChIP-Seq promoter peaks that decreased by >2-fold in RWPE-SChLAP1 cells. Of the 389 genes in the ChIP-Seq gene set, 250 were profiled by the Agilent HumanGenome microarray chip and present in the GSEA gene symbol database. An expression profile across these 250 genes is in Supplementary Table 6E.
RNA-Seq data
We assembled an RNA-Seq cohort from prostate cancer tissues sequenced at multiple institutions. We included data 12 primary tumors and 5 benign tissues published in GEO as GSE2226049, 16 primary tumors and 3 benign tissues released in dbGAP as study phs000310.v1.p150, and 17 benign, 57 primary, 14 metastatic tumors sequenced by our own institution and released as dbGAP study phs000443.v1.p1. Supplementary Table 1A shows sample information, and Supplementary Table 1B shows sequencing library information.
RNA-Seq alignment and gene expression quantification
Sequencing data were aligned using Tophat version 1.3.151 against the Ensembl GRCh37 human genome build. Known introns (Ensembl release 63) were provided to Tophat. Gene expression across the Ensembl version 63 genes and the SChLAP1 transcript was quantified by HT-Seq version 0.5.3p3 using the script htseq-count (www-huber.embl.de/users/anders/HTSeq/). Reads were counted without respect to strand to avoid bias between unstranded and strand-specific library preparation methods. This bias results from the inability to resolve reads in regions where two genes on opposite strands overlap in the genome.
RNA-Seq differential expression analysis
Differential expression analysis was performed using R package DESeq version 1.6.148. Read counts were normalized using the estimateSizeFactors function and variance was modeled by the estimateDispersions function. Differentially expression statistics were computed by the nbinomTest function. We called differentially expressed genes by imposing adjusted p-value cutoffs for cancer versus benign (padj < 0.05), metastasis versus primary (padj < 0.05), and gleason 8+ versus 6 (padj < 0.10). Heatmap visualizations for these analyses are presented as Supplementary Fig. 5.
RNA-Seq correlation analysis
Read count data were normalized using functions from the R package DESeq version 1.6.1. Adjustments for library size were made using the estimateSizeFactors function and variance was modeled using the estimateDispersions function using the parameters “method=blind” and “sharingMode=fit-only”. Next, the raw read count data was converted to pseudo-counts using the getVarianceStabilizedData function. Gene expression levels were then mean-centered and standardized using the scale function in R. Pearson correlation coefficients were computed between each gene of interest and all other genes. Statistical significance of Pearson correlations was determined by comparison to correlation coefficients achieved by 1,000 random permutations of the expression data. We controlled for multiple hypothesis testing using the qvalue package in R. The 253-gene SChLAP1 correlation signature was determined by imposing a cutoff of q < 0.05.
Oncomine Concepts Analysis of SChLAP1 Signature
We separated the 253 genes with expression levels significantly correlated to SChLAP1 into positively and negatively correlated gene lists. We imported these gene lists into Oncomine as custom concepts. We then nominated significantly associated Prostate Cancer concepts with Odds Ratio > 3.0 and p-value < 10-6. We exported these results as nodes and edges of a concept association network, and visualized the network using Cytoscape version 2.8.2. The node positions were computed using the Force Directed Layout algorithm in Cytoscape using the odds ratio as the edge weight. Node positions were subtly altered manually to enable better visualization of node labels.
Association of Correlation Signatures with Oncomine Concepts
We applied our RNA-Seq correlation analysis procedure on the genes SChLAP1, EZH2, PCA3, AMACR, ACTB. For each gene we created signatures from the top 5 percent of positively and negatively correlated genes (Supplementary Table 3). We performed a large meta-analysis of these correlation signatures across Oncomine datasets corresponding to disease outcome (Glinsky Prostate, Setlur Prostate), metastatic disease (Holzbeierlein Prostate, Lapointe Prostate, LaTulippe Prostate, Taylor Prostate 3, Vanaja Prostate, Varambally Prostate, and Yu Prostate), advanced gleason score (Bittner Prostate, Glinsky Prostate, Lapointe Prostate, LaTulippe Prostate, Setlur Prostate, Taylor Prostate 3, and Yu Prostate), and localized cancer (Arredouani Prostate, Holzbeierlein Prostate, Lapointe Prostate, LaTulippe Prostate, Taylor Prostate 3, Varambally Prostate, and Yu Prostate). We also incorporated our own concept signatures for metastasis, advanced Gleason score, and localized cancer determined from our RNA-Seq data. For each concept we downloaded the gene signatures corresponding to the Top 5 Percent of genes up- and down-regulated. Pairwise signature comparisons were performed using a one-sided Fisher's Exact Test. We controlled for multiple hypothesis testing using the qvalue package in R. We considered concept pairs with q < 0.01 and odds ratio > 2.0 as significant. In cases where a gene signature associates with both the over- and under-expression gene sets from a single concept, only the most significant result (as determined by odds ratio) is shown.
Analysis of SChLAP1-SNF5 expression signatures
The siSCHLAP1 and siSNF5 gene signatures were generated from Agilent gene expression microarray datasets. For each cell line we obtained a single vector of per-gene fold changes by averaging technical replicates and then taking the median across biological replicates. We merged the individual cell line results using the median of the changes in 22Rv1 and LNCaP. Venn diagram plots were produced using the BioVenn website (http://www.cmbi.ru.nl/cdd/biovenn/)52. We then compared the top 10% up-regulated and down-regulated genes for siSChLAP1 and siSNF5 to gene signatures downloaded from the Taylor Prostate 3 dataset in the Oncomine database. We performed signature comparison using one-sided Fisher's Exact Tests and controlled for multiple testing using the R package “qvalue”. Signature comparisons with q < 0.05 were considered significantly enriched. We plotted the odds ratios from significant comparison using the “heatmap.2” function in the “gplots” R package.
Kaplan-Meier Survival Analysis Based on SChLAP1 Gene Signature
We downloaded prostate cancer expression profiling data and clinical annotations from GSE8402 published by Setlur et. al.17. We intersected the 253-gene SChLAP1 signature with the genes in this dataset and 80 genes in common. We then assigned SChLAP1 expression scores to each patient sample in the cohort using the un-weighted sum of standardized expression levels across the 80 genes. Given that we observed SChLAP1 expression in approximately 20% of prostate cancer samples, we used the 80th percentile of SChLAP1 expression scores as the threshold for “high” versus “low” scores. We then performed 10-year survival analysis using the survival package in R and computed statistical significance using the log-rank test.
Additionally, we imported the 253-gene SChLAP1 signature into Oncomine in order to download the expression data for 167 of the 253 genes profiled by the Glinsky prostate dataset16. We assigned SChLAP1 expression scores in a similar fashion and designated the top 20% of patients as “high” for SChLAP1. We performed survival analysis using the time to biochemical PSA recurrence and computed statistical significance as above.
PhyloCSF Analysis
46-way multi-alignment FASTA files for SChLAP1, HOTAIR, GAPDH, and ACTB were obtained using the “Stitch Gene blocks” tool within the Galaxy bioinformatics framework (usegalaxy.org). We evaluated each gene for its likelihood to represent a protein-coding region using the PhyloCSF software (version released 2012-10-28). Each gene was evaluated using the phylogeny from 29 mammals (available by default within PhyloCSF) in any of the 3 reading frames. Scores are measured in decibans and reflect the likelihood that a predicted protein coding sequence is preferred over its non-coding counterpart.
Mayo Clinic Cohort Analyses
Study Design
Patients were selected from a cohort of high-risk radical prostatectomy (RP) patients from the Mayo Clinic. The cohort was defined as 1010 high-risk men that underwent RP between 2000 -2006, of which 73 patients developed clinical progression (defined as patients with systemic disease as evidenced by positive bone or CT scan)53. High-risk of recurrence was defined as pre-operative PSA >20 ng/ml, pathological Gleason score 8-10, seminal vesicle invasion (SVI), or GPSM score >=1054. The sub-cohort incorporated all 73 CP progression patients and a 20% random sampling of the entire cohort (202 men including 19 with CP). The total case-cohort study was 256 patients, of which tissue specimens were available for 235 patients. The sub-cohort was previously used to validate a genomic classifier (GC) for predicting Clinical Progression53.
Tissue Preparation
Formalin-fixed paraffin embedded (FFPE) samples of human prostate adenocarcinoma prostatectomies were collected from patients with informed consent at the Mayo Clinic according to an institutional review board-approved protocol. Pathological review of H&E tissue sections was used to guide macrodissection of tumour from surrounding stromal tissue from three to four 10 μm sections. The index lesion was considered the dominant lesion by size.
RNA Extraction and Microarray Hybridization
For validation cohort, total RNA was extracted and purified using a modified protocol for the commercially available RNeasy FFPE nucleic acid extraction kit (Qiagen Inc., Valencia, CA). RNA concentrations were calculated using a Nanodrop ND-1000 spectrophotometer (Nanodrop Technologies, Rockland, DE). Purified total RNA was subjected to whole-transcriptome amplification using the WT-Ovation FFPE system according to the manufacturer's recommendation with minor modifications (NuGen, San Carlos, CA). For the validation only the Ovation® FFPE WTA System was used. Amplified products were fragmented and labelled using the Encore™ Biotin Module (NuGen, San Carlos, CA) and hybridized to Affymetrix Human Exon (HuEx) 1.0 ST GeneChips following manufacturer's recommendations (Affymetrix, Santa Clara, CA).
Microarray Expression Analysis
The normalization and summarization of the microarray samples was done with the frozen Robust Multiarray Average (fRMA) algorithm using custom frozen vectors. These custom vectors were created using the vector creation methods as described previously55. Quantile normalization and robust weighted average methods were used for normalization and summarization, respectively, as implemented in fRMA.
Statistical Analysis
Given the exon/intron structure of isoform 1 of SChLAP1, all probe selection regions (or PSRs) that fall within the genomic span of SChLAP1 were inspected for overlapping with any of the exons of this gene. One PSR, 2518129, was found fully nested within the third exon of SChLAP1 and was used for further analysis as a representative PSR for this gene. The PAM (Partition Around Medoids) unsupervised clustering method was used on the expression values of all clinical samples to define two groups of high and low expression of SChLAP1.
Statistical analysis on the association of SChLAP1 with clinical outcomes was done using three endpoints (i) Biochemical Recurrence, defined as two consecutive increases of >=0.2ng/ml after RP, (ii) Clinical Progression, defined as a positive CT or bone scan and (iii) Prostate Cancer Specific Mortality (or PCSM).
For CP end point, all patients with CP were included in the survival analysis, whereas the controls in the sub-cohort were weighted in a 5-fold manner in order to be representative of patients from the original cohort. For PCSM end point, patients from the cases who did not die by PCa were omitted, and weighting was applied in a similar manner. For BCR, since the case-cohort was designed based on CP endpoint, resampling of BCR patients and sub-cohort was done in order to have a representative of the selected BCR patients from the original cohort.
Supplementary Material
Acknowledgments
We thank Oscar Alejandro Balbin, Scott A. Tomlins, Chad Brenner, Scott Deroo, and Sameek Roychowdhury for helpful discussions. This work was supported in part by the NIH Prostate Specialized Program of Research Excellence grant P50CA69568, the Early Detection Research Network grant UO1 CA111275, the US National Institutes of Health R01CA132874-01A1, and the Department of Defense grant PC100171 (A.M.C.). A.M.C. is supported by the Doris Duke Charitable Foundation Clinical Scientist Award, the Prostate Cancer Foundation, and the Howard Hughes Medical Institute. A.M.C. is an American Cancer Society Research Professor. A.M.C. is Taubman Scholar of the University of Michigan. F.Y.F. was supported by the Prostate Cancer Foundation and the D.O.D. grant PC094231. Q.C. was supported by a D.O.D. Postdoctoral Fellowship grant PC094725. J.R.P. was supported by the D.O.D. Predoctoral Fellowship PC094290. M.K.I was supported by the D.O.D. Predoctoral Fellowship BC100238. J.R.P., M.K.I., and A.S. are Fellows of the University of Michigan Medical Scientist Training Program.
Footnotes
Disclosures and Competing Financial Interests: The University of Michigan has filed a patent on lncRNAs in prostate cancer, including SChLAP1, in which A.M.C., J.R.P. and M.K.I. are named as co-inventors. Wafergen, Inc. has a non-exclusive license for creating commercial research assays for the detection of lncRNAs including SChLAP1. GenomeDx Biosciences Inc. has licensed lncRNAs including SChLAP1 for the molecular analysis of clinical prostate cancer samples. A.M.C. is a co-founder and advisor to Compendia Biosceiences, which supports the Oncomine database. He also serves on the Scientific Advisory Board of Wafergen; Life Technologies or Wafergen had no role in the design or experimentation of this study, nor have they participated in the writing of the manuscript. I.A.V., E.D., N.E., M.G., and T.J.T. are employees of GenomeDx Biosciences Inc.
Author Contributions: J.R.P., M.K.I., A.S. and A.M.C. designed the project and directed experimental studies. J.R.P, Q.C., W.C., S.M.D., B.C., S.H., R.M., L.P., T.M. and A.S. performed in vitro studies. X.W. performed in vitro translation assays. I.A.A. and A.S. performed CAM assays. R.B., N.M. and K.P. performed in vivo studies. L.P.K. and W.Y. performed histopathological analyses. M.K.I. performed bioinformatics analysis. X.J. and X.C. performed gene expression microarrays. J.S. and F.Y.F. facilitated biological sample procurement. F.Y.F. performed clinical analyses. For the Mayo Clinic Cohort, R.B.J. provided clinical samples and outcomes data. T.J.T. and E.D. generated and analyzed expression profiles for the Mayo Clinic cohort. E.D., N.E., M.G., and I.A.V. performed statistical analyses of SChLAP1 expression in the Mayo Clinic cohort. J.R.P., M.K.I., A.S. and A.M.C. interpreted data and wrote the manuscript.
References and Notes
- 1.Etzioni R, Cha R, Feuer EJ, Davidov O. Asymptomatic incidence and duration of prostate cancer. Am J Epidemiol. 1998;148:775–85. doi: 10.1093/oxfordjournals.aje.a009698. [DOI] [PubMed] [Google Scholar]
- 2.Cooperberg MR, Moul JW, Carroll PR. The changing face of prostate cancer. J Clin Oncol. 2005;23:8146–51. doi: 10.1200/JCO.2005.02.9751. [DOI] [PubMed] [Google Scholar]
- 3.Grasso CS, et al. The mutational landscape of lethal castration-resistant prostate cancer. Nature. 2012 doi: 10.1038/nature11125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Prensner JR, Rubin MA, Wei JT, Chinnaiyan AM. Beyond PSA: the next generation of prostate cancer biomarkers. Sci Transl Med. 2012;4:127rv3. doi: 10.1126/scitranslmed.3003180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Taylor BS, et al. Integrative genomic profiling of human prostate cancer. Cancer Cell. 2010;18:11–22. doi: 10.1016/j.ccr.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berger MF, et al. The genomic complexity of primary human prostate cancer. Nature. 2011;470:214–20. doi: 10.1038/nature09744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Prensner JR, Chinnaiyan AM. The emergence of lncRNAs in cancer biology. Cancer Discov. 2011;1:391–407. doi: 10.1158/2159-8290.CD-11-0209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rinn JL, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–23. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tsai MC, et al. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329:689–93. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kotake Y, et al. Long non-coding RNA ANRIL is required for the PRC2 recruitment to and silencing of p15(INK4B) tumor suppressor gene. Oncogene. 2011;30:1956–62. doi: 10.1038/onc.2010.568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Prensner JR, et al. Transcriptome sequencing across a prostate cancer cohort identifies PCAT-1, an unannotated lincRNA implicated in disease progression. Nat Biotechnol. 2011;29:742–9. doi: 10.1038/nbt.1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yu J, et al. An integrated network of androgen receptor, polycomb, and TMPRSS2-ERG gene fusions in prostate cancer progression. Cancer Cell. 2010;17:443–54. doi: 10.1016/j.ccr.2010.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guttman M, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–7. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rhodes DR, et al. Oncomine 3.0: genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia. 2007;9:166–80. doi: 10.1593/neo.07112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Varambally S, et al. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature. 2002;419:624–9. doi: 10.1038/nature01075. [DOI] [PubMed] [Google Scholar]
- 16.Glinsky GV, Glinskii AB, Stephenson AJ, Hoffman RM, Gerald WL. Gene expression profiling predicts clinical outcome of prostate cancer. J Clin Invest. 2004;113:913–23. doi: 10.1172/JCI20032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Setlur SR, et al. Estrogen-dependent signaling in a molecularly distinct subclass of aggressive prostate cancer. J Natl Cancer Inst. 2008;100:815–25. doi: 10.1093/jnci/djn150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nakagawa T, et al. A tissue biomarker panel predicting systemic progression after PSA recurrence post-definitive prostate cancer therapy. PLoS One. 2008;3:e2318. doi: 10.1371/journal.pone.0002318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Asangani IA, et al. Characterization of the EZH2-MMSET histone methyltransferase regulatory axis in cancer. Mol Cell. 2013;49:80–93. doi: 10.1016/j.molcel.2012.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001;98:5116–21. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–50. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liberzon A, et al. Molecular signatures database (MSigDB) 3.0. Bioinformatics. 2011;27:1739–40. doi: 10.1093/bioinformatics/btr260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shen H, et al. The SWI/SNF ATPase Brm is a gatekeeper of proliferative control in prostate cancer. Cancer Res. 2008;68:10154–62. doi: 10.1158/0008-5472.CAN-08-1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roberts CW, Orkin SH. The SWI/SNF complex--chromatin and cancer. Nat Rev Cancer. 2004;4:133–42. doi: 10.1038/nrc1273. [DOI] [PubMed] [Google Scholar]
- 25.Reisman D, Glaros S, Thompson EA. The SWI/SNF complex and cancer. Oncogene. 2009;28:1653–68. doi: 10.1038/onc.2009.4. [DOI] [PubMed] [Google Scholar]
- 26.Sun A, et al. Aberrant expression of SWI/SNF catalytic subunits BRG1/BRM is associated with tumor development and increased invasiveness in prostate cancers. Prostate. 2007;67:203–13. doi: 10.1002/pros.20521. [DOI] [PubMed] [Google Scholar]
- 27.Dechassa ML, et al. Architecture of the SWI/SNF-nucleosome complex. Mol Cell Biol. 2008;28:6010–21. doi: 10.1128/MCB.00693-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Derrien T, et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012;22:1775–89. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.De S, et al. Dynamic BRG1 recruitment during T helper differentiation and activation reveals distal regulatory elements. Mol Cell Biol. 2011;31:1512–27. doi: 10.1128/MCB.00920-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Euskirchen GM, et al. Diverse roles and interactions of the SWI/SNF chromatin remodeling complex revealed using global approaches. PLoS Genet. 2011;7:e1002008. doi: 10.1371/journal.pgen.1002008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yen K, Vinayachandran V, Batta K, Koerber RT, Pugh BF. Genome-wide Nucleosome Specificity and Directionality of Chromatin Remodelers. Cell. 2012;149:1461–73. doi: 10.1016/j.cell.2012.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gupta RA, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–6. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jones S, et al. Frequent mutations of chromatin remodeling gene ARID1A in ovarian clear cell carcinoma. Science. 2010;330:228–31. doi: 10.1126/science.1196333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Varela I, et al. Exome sequencing identifies frequent mutation of the SWI/SNF complex gene PBRM1 in renal carcinoma. Nature. 2011;469:539–42. doi: 10.1038/nature09639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Versteege I, et al. Truncating mutations of hSNF5/INI1 in aggressive paediatric cancer. Nature. 1998;394:203–6. doi: 10.1038/28212. [DOI] [PubMed] [Google Scholar]
- 36.Cline MS, et al. Integration of biological networks and gene expression data using Cytoscape. Nat Protoc. 2007;2:2366–82. doi: 10.1038/nprot.2007.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rubin MA, et al. Rapid (“warm”) autopsy study for procurement of metastatic prostate cancer. Clin Cancer Res. 2000;6:1038–45. [PubMed] [Google Scholar]
- 38.Tomlins SA, et al. Role of the TMPRSS2-ERG gene fusion in prostate cancer. Neoplasia. 2008;10:177–88. doi: 10.1593/neo.07822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Asangani IA, et al. Characterization of the EZH2-MMSET Histone Methyltransferase Regulatory Axis in Cancer. Mol Cell. 2012 doi: 10.1016/j.molcel.2012.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chu C, Qu K, Zhong FL, Artandi SE, Chang HY. Genomic maps of long noncoding RNA occupancy reveal principles of RNA-chromatin interactions. Mol Cell. 2011;44:667–78. doi: 10.1016/j.molcel.2011.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Maher CA, et al. Chimeric transcript discovery by paired-end transcriptome sequencing. Proc Natl Acad Sci U S A. 2009;106:12353–8. doi: 10.1073/pnas.0904720106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Levin JZ, et al. Comprehensive comparative analysis of strand-specific RNA sequencing methods. Nat Methods. 2010;7:709–15. doi: 10.1038/nmeth.1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–60. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang Y, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Feng J, Liu T, Zhang Y. Using MACS to identify peaks from ChIP-Seq data. Curr Protoc Bioinformatics. 2011;Chapter 2:Unit 2 14. doi: 10.1002/0471250953.bi0214s34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kent WJ, et al. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shin H, Liu T, Manrai AK, Liu XS. CEAS: cis-regulatory element annotation system. Bioinformatics. 2009;25:2605–6. doi: 10.1093/bioinformatics/btp479. [DOI] [PubMed] [Google Scholar]
- 48.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kannan K, et al. Recurrent chimeric RNAs enriched in human prostate cancer identified by deep sequencing. Proc Natl Acad Sci U S A. 2011;108:9172–7. doi: 10.1073/pnas.1100489108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pflueger D, et al. Discovery of non-ETS gene fusions in human prostate cancer using next-generation RNA sequencing. Genome Res. 2011;21:56–67. doi: 10.1101/gr.110684.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–11. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hulsen T, de Vlieg J, Alkema W. BioVenn - a web application for the comparison and visualization of biological lists using area-proportional Venn diagrams. BMC Genomics. 2008;9:488. doi: 10.1186/1471-2164-9-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Buerki C, et al. Validation of a genomic-clinical classifier for predicting clinical progression in high-risk prostate cancer; ASCO Annual Meeting; 2012. Abstract #4565. [Google Scholar]
- 54.Blute ML, Bergstralh EJ, Iocca A, Scherer B, Zincke H. Use of Gleason score, prostate specific antigen, seminal vesicle and margin status to predict biochemical failure after radical prostatectomy. J Urol. 2001;165:119–25. doi: 10.1097/00005392-200101000-00030. [DOI] [PubMed] [Google Scholar]
- 55.Vergara IA, et al. Genomic “Dark Matter” in Prostate Cancer: Exploring the Clinical Utility of ncRNA as Biomarkers. Front Genet. 2012;3:23. doi: 10.3389/fgene.2012.00023. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.