Abstract
The ratio of genetic diversity on X chromosomes relative to autosomes in organisms with XX/XY sex chromosomes could provide fundamental insight into the process of genome evolution. Here we report this ratio for 24 cynomolgus monkeys (Macaca fascicularis) originating in Indonesia, Malaysia, and the Philippines. The average X/A diversity ratios in these samples was 0.34 and 0.20 in the Indonesian–Malaysian and Philippine populations, respectively, considerably lower than the null expectation of 0.75. A Philippine population supposed to derive from an ancestral population by founding events showed a significantly lower ratio than the parental population, suggesting a demographic effect for the reduction. Taking sex-specific mutation rate bias and demographic effect into account, expected X/A diversity ratios generated by computer simulations roughly agreed with the observed data in the intergenic regions. In contrast, silent sites in genic regions on X chromosomes showed strong reduction in genetic diversity and the observed X/A diversity ratio in the genic regions cannot be explained by mutation rate bias and demography, indicating that natural selection also reduces the level of polymorphism near genes. Whole-genome analysis of a female cynomolgus monkey also supported the notion of stronger reduction of genetic diversity near genes on the X chromosome.
Keywords: sex chromosome; genetic diversity; primate; natural selection; coalescent theory, kernel-ABC
CONTRASTING the pattern of molecular evolution and genetic diversity on autosomes and sex chromosomes provides evolutionary insights into the organization of genomes (reviewed in Vicoso and Charlesworth 2006). The different level of genetic diversity between autosomes and X chromosomes in organisms with mammalian-like sex determination systems (XX/XY system) is a debated issue (Andolfatto 2001 Vicoso and Charlesworth 2006; Hammer et al. 2008, 2010; Keinan et al. 2009). Although the ratio of X to autosomal genetic diversity (X/A diversity ratio) is expected to be 0.75 under “idealized” population genetic processes in the XX/XY system, many evolutionary factors influence the ratio. These factors include natural selection, demography, sex-biased reproductive success, sex-biased migration rate, and male/female mutation rate differences (reviewed in Caballero 1994; Vicoso and Charlesworth 2006). For example, natural selection (both positive and negative selection) could reduce genetic diversity on functional sites and linked neutral sites; the magnitude of the effect depends on many factors, such as effective population size, strength of selection (selection and dominance coefficients), and recombination rate (Charlesworth 2012). Those effects could be different between autosomes and the X chromosome and potentially increase or decrease the X/A diversity ratio (Begun and Whitley 2000; Vicoso and Charlesworth 2009). Selection-free processes such as demography could also drastically change the ratio. Previous theoretical studies showed that the ratio would increase after a population expansion and decrease after a population reduction or bottleneck (Pool and Nielsen 2007; Pool and Nielsen 2008). Similarly, the ratio would also decrease when the mutation rate per generation is higher in males than in females, presumably owing to more germ-line cell divisions in males than in females, an effect known as male-driven molecular evolution (Miyata et al. 1987). Because so many factors could bias the X/A diversity ratio, it is often challenging to find the actual cause of bias.
The estimates on the X/A genetic diversity ratios in humans have been conflicting among studies, crossing the boundary of the neutral expectation of 0.75, after controlling mutation rate bias (Hammer et al. 2008; Bustamante and Ramachandran 2009; Keinan et al. 2009). In nonhuman primates, however, some studies have reported much lower ratios. For example, a recent genome-wide resequencing study in central chimpanzees (Pan troglodytes) found a ratio of near 0.5 (Hvilsom et al. 2012). A similar ratio was observed in Chinese and Indian rhesus macaques (Macaca mulatta), using shotgun sequencing data (Gibbs et al. 2007). Hvilsom et al. (2012) proposed that a larger effective population size in chimpanzees than that in humans may have led the low X/A diversity ratio because of strong natural selection in chimpanzees. In other primate studies, the results were mixed; an analysis by Pool and Nielsen (2007) showed that the ratios were >0.75 in presumably parental populations of humans, chimpanzees, and orangutans, whereas derived populations, which are supposed to have emerged from parental populations, showed X/A diversity ratios <0.75. On the other hand, a study of bonobo (Pan paniscus) population genomics showed that the ratio was significantly >0.75, which might be due to high reproductive variance in bonobo males (Prufer et al. 2012).
Because the previous estimate in macaques was based on shotgun sequencing, for which the error rate is relatively higher than that for individual resequencing, data estimated by resequencing methods would be helpful for a comparison of evolutionary processes between humans and nonhuman primates. Moreover, multiple factors such as demography and natural selection have not been evaluated in a single study. In particular, the estimation of genetic diversity is strongly affected by random genetic drift, which could increase the variance of the diversity ratios without any biological mechanisms and makes it difficult to evaluate the statistical significance of findings. To overcome the problem, we employed coalescent simulation, which is an effective way to control such a stochastic effect.
In a previous study we have sequenced autosomal regions of 24 cynomolgus monkeys (Macaca fascicularis) originating in Indonesia, Malaysia, and the Philippines, and quantified the genetic diversity in these populations at the single-nucleotide polymorphism (SNP) level for autosomal 27 coding sequence (CDS) and 27 intergenic sequence (IGS) regions (Osada et al. 2010). The study showed strong genetic differentiation between the Indonesian–Malaysian populations and Philippine populations, but almost no genetic differentiation between the Indonesian and Malaysian populations. Whereas the Indonesian–Malaysian population showed negatively skewed Tajima D statistics, the Tajima D statistics in the Philippine population was highly skewed toward positive values, suggesting population expansion in the Indonesian–Malaysian population and population contraction in the Philippine population (Osada et al. 2010). The average autosomal genetic diversity in the IGS regions among those macaques was ∼0.35%, which was much higher than that among humans (Table 1). We sequenced 10 additional X-chromosomal loci to investigate the level of genetic diversity at sex-linked loci in nonhuman primates, and found that the X/A diversity in macaques is lower than that reported previously for primates. We evaluate and discuss the potential causes of the strong reduction of X chromosome diversity in macaques.
Table 1. Genetic diversity of X chromosomal and autosomal loci.
| Locus ID: Gene symbol | Rhesus macaque genomea | NIMb | NPb | Length | πSAc | πSIMc | πSPc |
|---|---|---|---|---|---|---|---|
| CDS28: AMELX | Chr X: 8,970,276–8,970,952 | 29 | 18 | 677 | 0.00059 | 0.00101 | 0.00000 |
| CDS29: UTX | Chr X: 42,828,710–42,829,485 | 29 | 18 | 746 | 0.00122 | 0.00181 | 0.00000 |
| CDS30: UBE1X | Chr X: 45,033,665–45,034,444 | 29 | 18 | 780 | 0.00113 | 0.00123 | 0.00036 |
| CDS31: RPS4X | Chr X: 71,201,776–71,202,514 | 29 | 18 | 740 | 0.00026 | 0.00026 | 0.00043 |
| CDS32: ACSL4 | Chr X: 108,393,547–108,394,254 | 29 | 18 | 708 | 0.00128 | 0.00104 | 0.00120 |
| CDS33: RBMX | Chr X: 134,951,606–134,952,319 | 29 | 18 | 714 | 0.00018 | 0.00000 | 0.00000 |
| X chr. CDS average | 727.5 | 0.00078 | 0.00089 | 0.00033 | |||
| IGS28 | Chr X: 2,035,502–2,036,174 | 27 | 18 | 681 | 0.00193 | 0.00223 | 0.00031 |
| IGS29 | Chr X: 34,279,437–34,280,302 | 29 | 18 | 866 | 0.00183 | 0.00203 | 0.00066 |
| IGS30 | Chr X: 40,194,693–40,195,387 | 29 | 18 | 695 | 0.00194 | 0.00221 | 0.00151 |
| IGS31 | Chr X: 74,880,899–74,881,659 | 29 | 18 | 749 | 0.00061 | 0.00072 | 0.00039 |
| X chr. IGS average | 747.8 | 0.00158 | 0.00180 | 0.00072 | |||
| Autosomal CDS averaged | 0.00388 | 760.9 | 0.00388 | 0.00414 | 0.00231 | ||
| Autosomal IGS averaged | 0.00352 | 735.4 | 0.00352 | 0.00359 | 0.00244 |
Coordinates of the rhesus macaque draft genome sequence (rheMac2).
Number of sampled chromosomes; subscripts represent Indonesian–Malaysian (IM), and Philippine (P) origins.
Nucleotide diversity at silent sites (πS); subscripts represent all samples (A), Indonesian–Malaysian (IM), and Philippine (P) origins.
Autosomal data from Osada et al. (2010): average of 54 autosomal loci.
Materials and Methods
DNA samples
Blood sample was obtained from adult M. fascicularis individuals who were bred and raised at Tsukuba Primate Research Center, National Institute of Biomedical Innovation (NIBIO). All individuals are F1 crosses of macaques captured. Briefly, three different breeding subpopulations were organized according to the country of origin: Malaysian, Indonesian, and the Philippine subpopulations. Each subpopulation consists of ∼18 male and 180 female feral M. fascicularis that were imported from 1978 to 1980. Mating pairs were selected from the same breeding subpopulation to maintain the gene pool of each founder population. To avoid inbreeding, mating pairs were set between males and females belonging to different import lots (Honjo et al. 1984). Precise sampling location of the Indonesian and Philippine individuals are unknown except for Malaysian individuals, which were captured at the south of Kuala Lumpur. They were housed in indoor individual cages under constant conditions at temperature of 25 ± 2° and humidity of 50 ± 5%. Monkeys were cared for and treated humanely in accordance with the Guiding Principles for Animal Experiments using nonhuman primates formulated by the Primate Society of Japan and with the guidelines provided by the Animal Care and Use Committee of NIBIO. A regular health check was done once in a year and 5 ml of blood was drawn from saphenous vein under ketamine anesthesia. We reused these blood samples for this research to avoid an additional invasive sampling.
DNA sequencing and data analysis
We sequenced DNA samples of eight, seven, and eight M. fascicularis individuals from Indonesia, peninsular Malaysia, and the Philippines, respectively (Osada et al. 2010). Except for one individual from Malaysia, all individuals were females. Primer pairs were designed to amplify six CDS and four IGS regions (Supporting Information, Table S1). All loci were distributed across nonpseudoautosomal regions of X chromosomes. DNA fragments were amplified using PCR and directly sequenced using ABI 3730 sequencers. Potential SNPs were visually inspected using the ABI sequence analysis software. All analyzed sequences were deposited in the public database (DDBJ/EMBL/Genbank: AB739064–AB739302). The DNA sequences were aligned using the ClustalW software implemented in MEGA5.0 (Thompson et al. 1994; Tamura et al. 2011), and population genetics summary statistics for each locus were estimated using DnaSP 5.0 (Librado and Rozas 2009). Haplotypes were estimated using the PHASE software (Stephens et al. 2001). Permutation tests were performed as follows: each sample was randomly assigned to each population, statistics (X/A diversity ratio) were calculated for 10,000 iterations, and observed statistics were compared with the null distribution. Male to female mutation rate ratio (α) was estimated using
where R is the substitution ratio on the X chromosome to autosomes (Miyata et al. 1987).
Inference of demography
We estimated the demography of macaques using kernel approximate Bayesian computation (kernel-ABC) (Fukumizu et al. 2011; Nakagome et al. 2012). Kernel-ABC is a reproducing kernel Hilbert space (RKHS)-based method, used to obtain an approximation of the posterior estimate by using summary statistics where we can handle higher-dimensional summary statistics than those of conventional ABC methods. The sequence data from Indonesia–Malaysian M. fascicularis (2N = 30) and Philippine M. fascicularis (2N = 18) were summarized into site frequency spectrum (SFS) and haplotype frequency spectrum (HFS). SFS and HFS were calculated for the Indonesia–Malaysian population (SFSIM and HFSIM) and for the Philippine population (SFSP and HFSP). We further transformed the histograms of SFS and HFS into a matrix of SFS (2D-SFS) and HFS (2D-HFS), respectively. These summary statistics were measured for each locus and merged into a set of 2D-SFS and 2D-HFS across loci (Figure S1). The matrices contain the variant sites or haplotypes that are polymorphic in either or both population. The allelic state (ancestral or derived allele) at each segregating site was determined by the alignment with an orthologous sequence from the human genome (hg19). Based on the observed summary statistics, we estimated posterior means of the parameters by kernel-ABC.
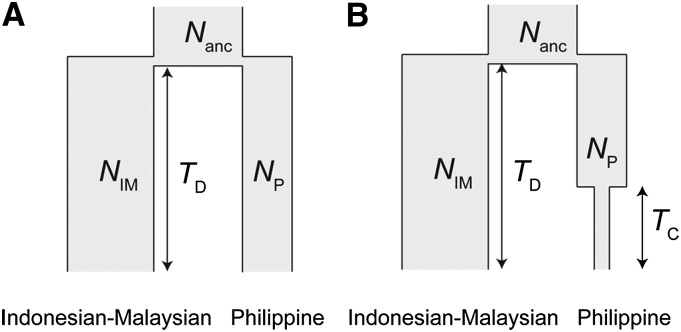
We modeled demography for the population divergence between the Indonesia–Malaysian and the Philippine populations as models 1 and 2 (Figure 1). Model 1 consisted of four parameters (NIM, the effective population size in the Indonesia–Malaysian population; NP, the effective population size in the Philippine population; Nanc, the effective population size in the ancestral population; TD, the divergence time). In model 2, one parameter, TC (contraction time in the Philippine population), was added into model 1. The prior density for each parameter was given by a log-normal distribution (LN) whose variance was a square of the mean (μ): Nanc ∼ LN(μ = 100000, μ2 = 1000002), NIM ∼ LN(μ = 1000000, μ2 = 10000002), NP ∼ LN(μ = 50000, μ2 = 500002), TD ∼ LN(μ = 50000, μ2 = 500002), and TC ∼ LN(μ = 10000, μ2 = 100002). To attain these models in all simulations, the parameter of NIM, NP, or TC was conditional on NIM > Nanc, NP < Nanc, or TC < TD, respectively (see File S1). The algorithm of kernel-ABC is as follows:
Sample a set of parameters () from the priors.
Simulate data () using .
Compute the summary statistics () for , and go to step 1.
Repeat the steps 1–3 20,000 times.
Compute posterior means of the parameters as a predictor by the kernel ridge regression of onto based on .
We repeated this algorithm 100 times and calculated the mean and the standard deviation (SD) of the posterior estimate for each parameter. We used the Gaussian radial base function kernel,. The bandwidth () in the kernel function and the regularization parameter (, where n is the number of simulations) were chosen by 10-fold cross-validation (Fukumizu et al. 2011; Nakagome et al. 2012).
Figure 1.
Demographic models of the Indonesian–Malaysian and Philippine populations. (A) Model 1: the size of the two populations changes from the common ancestors at the same time TD. (B) Model 2: the Philippine population experiences one more population size reduction.
All coalescent simulations were performed using the program package ms that generates samples from the coalescent model under neutrality (Hudson 2002). We simulated data for each locus under a set of parameters (), and merged summary statistics into a set of 2D-SFS and 2D-HFS across 26 IGS loci (). The recombination rate in each locus was assumed to follow exponential distribution whose mean was equal to 7.34 × 10−9, which was estimated from a rhesus macaque family data (Rogers et al. 2006; Dumont and Payseur 2008), while the mutation rate was calculated from the average number of substitutions in each locus between the human (hg18) and rhesus macaque (rheMac2) genomes, assuming the divergence between human and M. fascicularis of 25 million years ago (MYA). Both of the recombination and mutation rates were scaled four times of NIM with a sequenced genomic size of each locus and a generation time of 6 years. (See Figure S2.)
Model selection by kernel-ABC
We selected a model under the kernel-ABC framework. Let be an observed summary statistics. To compare two alternative models of and , a Bayes factor (BF) is given as
where is the observed data. Here we introduce an approximated Bayes factor (aBF), in which data are summarized by a set of summary statistics, . It is
where is an approximated marginal likelihood of a given model. An estimator of the approximated marginal likelihood is given by
| (1) |
where is a set of summary statistics for the ith simulated data, is an indicator function, and n is the total number of simulations. Assuming the equal prior probabilities for the two models, the ratio of the acceptance rate under to that under is an estimator of BF between and . However, if a set of summary statistics is high dimensional or continuous, it should be hard to get samples that satisfy , and hence is expected to get close to 0. Therefore we utilized a kernel density estimate of the marginal likelihood by replacing in (1) with the normalized Gaussian kernel function, , where p is the dimensionality of summary statistics.
The choice of is crucial in kernel density estimation. Large and small values lead oversmoothing and overfitting to the simulated samples, respectively. This bias-variance trade-off is compromised by choosing an optimal value of by the 10-fold cross-validation with 20,000 data as follows. Let be a set of indices in the ath subsample (e.g., ) and be all subsamples except for the ath subsample (e.g., ). The kernel density estimate of the marginal likelihood by using the learning data of is
As a criterion of the cross-validation, we utilized the cross-entropy, which measures divergence between two densities. Here, the cross-entropy of the true density and the estimated density is
However, the true density is not available. We therefore replace the expectation over the true density with the empirical distribution constructed by the test data of . Namely,
We chose , which minimizes the average of the 10 estimates for the cross-entropy given by this scheme.
Since logarithm of the Bayes factor gives a rough approximation to Bayesian information criterion (BIC; Kass and Raftery 1995), aBF can be regarded as a criterion for model selection. We computed the kernel density estimate of the marginal likelihood. To estimate the marginal likelihoods of the models, we generated 3 million simulated samples based on the kernel-ABC algorithm from step 1 to step 4. Among these samples, 100,000 were used to standardize summary statistics and the remaining samples were used to compute kernel density estimate of the approximate marginal likelihoods.
Analysis of a resequenced individual genome
Short-read sequences of a female Vietnamese cynomolgus monkey were retrieved from the public database (SRA023856). The reads were mapped to the reference genome sequence of rhesus macaque (rheMac2) using Bowtie2 software (Langmead and Salzberg 2012) and SNPs were called using SAMtools (Li et al. 2009) with a mismatch tuning parameter (-C) of 50. To measure the density of SNPs, we considered only biallelic sites that have ≥10-fold and <150-fold coverage and genotyping quality scores ≥30. The number of variant sites (heterozygous sites in a sample) and invariant sites (homozygous sites in a sample) were summed across nonoverlapping bins of 1 kb according to the distance from the nearest exons. For the first bin within 1 kb from exons, the first 100 bp were filtered out to reduce the effect of potential functional sequences such as splicing signals. Genome alignment data of the human and rhesus macaque were downloaded from the UCSC genome browser website (rheMac2.hg19.net.axt) and the divergent sites were binned in the same way.
Results
We sequenced 10 X-chromosomal loci in nonpseudoautosomal regions of 24 M. fascicularis individuals originating in Indonesia, Malaysia, and the Philippines. Each locus contained ∼600–800 bp of DNA sequence. Six loci harbored at least one exon (CDS regions) and four loci were at least 100 kb away from any annotated exons (IGS regions). The level of polymorphism was measured at silent (synonymous and noncoding) sites.
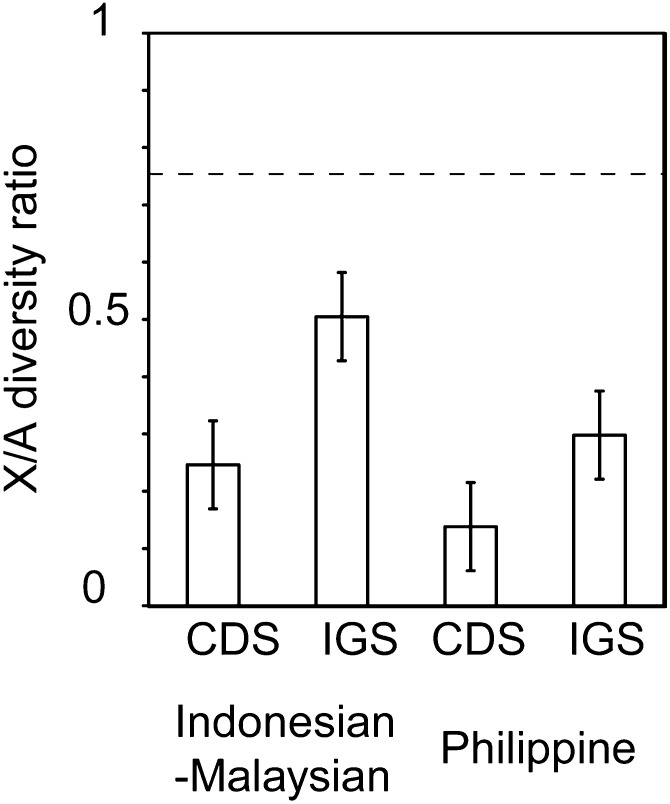
Autosomal data were obtained from the previous study (Osada et al. 2010), which analyzed the same individuals using the same sequencing method. Overall, X-chromosomal loci showed lower levels of polymorphism (Table 1); the X/A ratio of average genetic diversity was 0.30 when all regional population data were combined. When two groups were separately analyzed, the ratio became 0.34 and 0.20 in the Indonesian–Malaysian and Philippine populations, respectively (Figure 2). In all populations, the X/A diversity ratios were much lower than those reported for other primates including humans (e.g., Pool and Nielsen 2007; Hammer et al. 2008; Keinan et al. 2009; Hvilsom et al. 2012). In addition, Philippine macaques, which are thought to have originated by migration from continental populations (e.g., Smith et al. 2007), showed lower X/A diversity ratio than Indonesian–Malaysian macaques. The difference between the Indonesian–Malaysian and Philippine populations was statistically significant (P = 0.002; permutation test).
Figure 2.
X/A diversity ratio in the Indonesian–Malaysian and Philippine populations. CDS and IGS data are shown separately. The vertical bars represent the standard errors that were estimated using 1000 times bootstrap resampling. The horizontal dashed line denotes the null expectation of 0.75.
We further investigated the effect of natural selection on macaque X chromosomes and autosomes. When natural selection is affecting the level of polymorphisms, the level of reduction would be different between the CDS regions and IGS regions. In the Indonesian–Malaysian population, the X/A diversity ratios in the CDS (silent sites) and IGS regions were 0.24 (0.07) and 0.50 (0.10), respectively, while in the Philippine population, the ratios were 0.14 (0.07) and 0.29 (0.10), respectively (standard errors estimated using the bootstrap resampling are shown in parentheses; Figure 1). The results show that the reduction of X chromosome diversity relative to autosomal diversity is stronger in the CDS regions than in the IGS regions.
We further investigated whether the observed ratios can be explained by two particular evolutionary factors: demography and male-driven evolution. We first estimate the demography of macaques using 26 autosomal IGS data obtained in the previous study, using the kernel-ABC method. One IGS that lacked an outgroup sequence was not used for the demography estimation. The kernel-ABC method would efficiently infer past demographic parameters using high-dimensional summary statistics (Nakagome et al. 2012). The previous study and our preliminary kernel-ABC analysis (see File S1) suggested the population expansion in the Indonesian–Malaysian population and population contraction in the Philippine population after their split. In addition, the Philippine population showed strongly biased Tajima’s D statistics toward positive values (Osada et al. 2010), which motivated us to consider two specific demographic models. The first model (model 1) has four parameters, where both Indonesian–Malaysian and Philippine populations change their size when they split from the ancestral population at time TD (see Figure 1A and Materials and Methods). Because we did not have any prior information whether the split of the Philippine population is sufficiently recent to cause the positively biased Tajima’s D statistics, the second model (model 2) with five parameters, where the Philippine population additionally experienced population contraction at time TC after the split (Figure 1B), was also evaluated. For model 2, four values of population contraction intensity (75, 50, 25, and 10% of original population size, NP) were evaluated. To select the best model, we computed the kernel density estimate of the marginal likelihood in each model and evaluated aBF among these models. As shown in Table 2, the model 2 with 25% population size showed the highest marginal likelihood; the aBF was 1.66 against model 1, 1.17 against the model 2 with 75% size, 1.49 against that with 50% size, and 11.0 against that with 10% size. We therefore adopted model 2 with 25% population size and used those estimated parameters for further coalescent simulations.
Table 2. Estimated demographic parameter and marginal likelihood values using the kernel-ABC method.
| Parameters | Model 1 | Model 2 | Model 2 | Model 2 | Model 2 |
|---|---|---|---|---|---|
| Population size change (% of original size) | − | 75% | 50% | 25% | 10% |
| Nanc | 99,000 (1,312) | 99,136 (1,238) | 98,770 (1,197) | 98,913 (1,477) | 103,225 (1,733) |
| NIM | 813,107 (71,977) | 753,119 (67,751) | 770,335 (73,752) | 881,195 (57,651) | 906,049 (62,448) |
| NP | 39,330 (996) | 41,633 (907) | 50,490 (1,044) | 70,999 (1,658) | 88,790 (2,262) |
| TD (generation) | 24,543 (717) | 25,290 (638) | 25,340 (595) | 25,019 (689) | 25,359 (1,085) |
| TC (generation) | − | 8,345 (600) | 7,876 (553) | 5,682 (359) | 3,143 (296) |
| Approximated marginal likelihood (loge-scaled) | −70.636 | −70.281 | −70.527 | −70.128 | −72.527 |
Standard deviation is shown in parentheses.
To assess the reliability of demographic inference by the kernel-ABC method, we generated samples that have parameter values similar to our estimated parameter values for both model 1 and model 2 (25% population size) as well as a simpler constant population size model. Detailed procedure is described in File S1. As shown in Table S2, the kernel-ABC method reconstructed these demographic models reasonably well.
We also estimated mutation rate bias between X chromosomes and autosomes using divergence data between humans and macaques. In the IGS regions, the X to autosomal divergence ratio was 0.81, which corresponds to the male mutation rate bias (α) of 3.8 (see Materials and Methods). Although this value is slightly greater than the value estimated using the whole-genome sequence of rhesus macaque (α = 2.7) (Gibbs et al. 2007), we applied our estimated value because our test becomes more conservative.
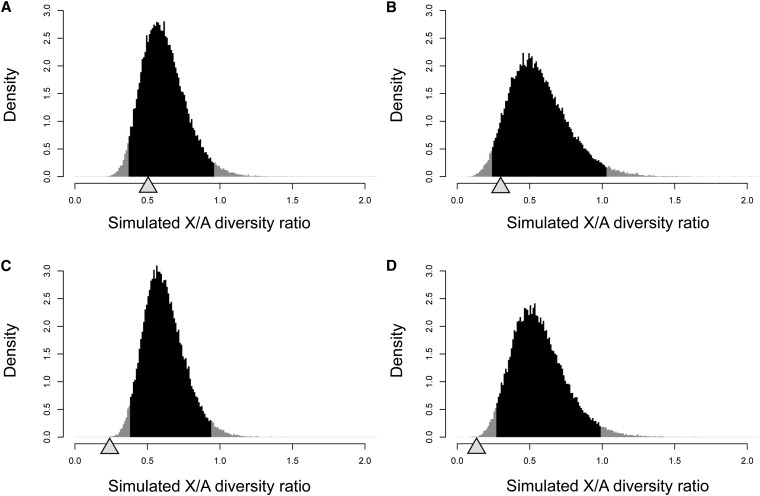
Using the estimated demographic parameters and mutation rate difference, we simulated 50,000 data sets for IGS and CDS that have the same number of loci, number of individuals, and number of sites as the real data, assuming that the effective population size on the X chromosome is 75% of that on autosomes. The X/A diversity ratios in the pseudo populations of Indonesian–Malaysian and Philippine M. fascicularis were estimated from the simulated data set. Because the recombination rate on macaque X chromosomes has been unknown, we performed simulations under two different recombination rates on the X chromosome. In the first case, the recombination rate between autosomes and the X chromosomes were set to be equal. In the second case, the recombination rate of the X chromosome was one-half that of autosomes. The observed values and simulated distributions with equal recombination rate are shown in Figure 3. In the simulated data set, the Philippine population showed slightly lower ratios than the Indonesian–Malaysian population both in IGS and CDS regions on average. While observed ratios in the IGS regions fell in the expected distributions, CDS regions showed significant reduction from the expected distribution in the both populations (P = 0.00028 and P = 0.00076 in Indonesian–Malaysian and Philippine population, respectively; two-tailed test), indicating that natural selection is further reducing the X/A diversity ratio in the CDS regions. Simulation results with a low recombination rate on the X chromosomes were qualitatively the same as the result of the first case, and the results are presented in Figure S3.
Figure 3.
Distribution of X/A diversity ratio in the simulated data sets. The grey and black areas represent the density of X/A diversity ratio in the simulated data sets. The black areas indicate 95% confidence intervals. The shaded triangles point out the observed values. Same recombination rate distribution was assumed between autosomes and the X chromosomes (see also Figure S3). (A) IGS in the Indonesian–Malaysian population, (B) IGS in the Philippine population, (C) CDS in the Indonesian–Malaysian population, and (D) CDS in the Philippine population.
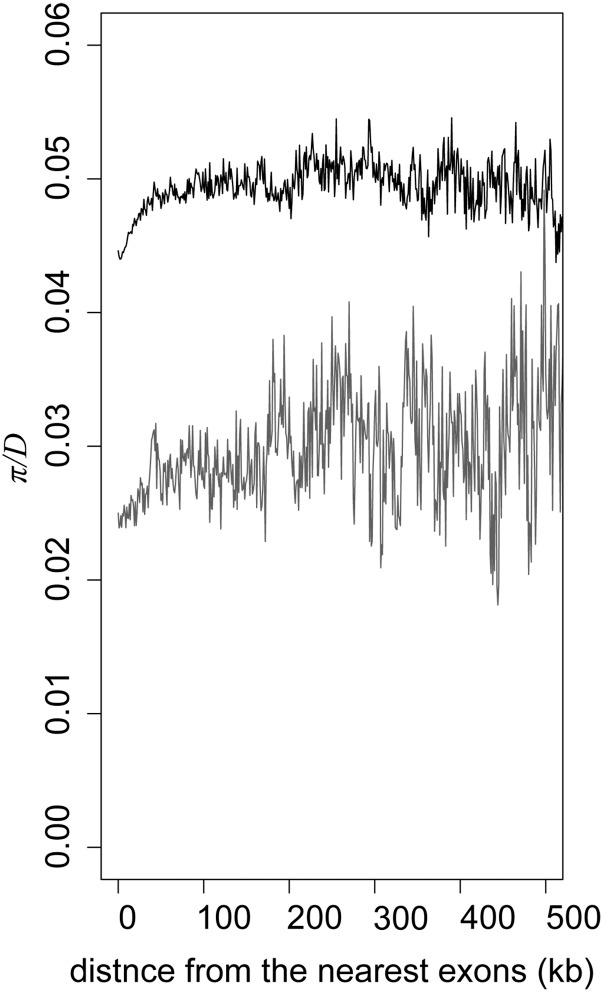
To further support the evidence of stronger reduction of polymorphisms near CDS on the X chromosomes than on autosomes, we utilized a recently determined female Vietnamese M. fascicularis genome sequence using massively parallel sequencing (Yan et al. 2011). We mapped the short-read sequences to the rhesus macaque genome sequence and estimated the rate of heterozygous SNPs on autosomes (0.29%) and the X chromosome (0.15%), which are fairly close to the values in the previous report using a different read mapping and SNP calling pipeline (Yan et al. 2011). The number of SNPs between two sequenced chromosomes was summed into bins according to the distance from the nearest exons. The levels of polymorphisms were normalized using the divergence to the human genome. As shown in Figure 4, the reduction of polymorphisms near exons was observed both on autosomes and the X chromosome. In the bins 400–500 kb away from exons, the genome-wide X/A diversity ratio in the Vietnamese macaque was 0.61, which was similar to the level of Indonesian–Malaysian population data. Notably, the magnitude of reduction was stronger on the X chromosomes than on autosomes. The X/A diversity ratio (without normalized by divergence) in the bins within 1 kb from exons (excluding the first 100 bp from exons) to the bins 400–500 kb away from exons was 0.56 and 0.47 on autosomes and the X chromosome, respectively.
Figure 4.
Reduction of polymorphisms near exons on autosomes and the X chromosome. The number of differences per site between two chromosomes (π) of Vietnamese M. fascicularis, normalized by the divergence between humans and macaques (D), were plotted. The data were summed across 1-kb-size bins with no overlap.
Discussion
The X/A diversity ratio in macaques was considerably lower than the values reported in humans and chimpanzees and also lower than the previously reported ratio for rhesus macaques determined using shotgun sequencing. Among many factors that could contribute to the reduced X/A diversity ratio in macaques, we have discussed three particular factors in this article.
First, mutation rate bias certainly contributes the reduction of X/A diversity ratio. In particular, male-driven evolution, which posits a higher mutation rate in males, potentially decreases the X/A diversity ratio (Miyata et al. 1987). Many molecular evolution studies have shown that the estimated mutation rate on X chromosomes is lower than that on autosomes in primates, indicating that the mutation rate in males relative to females (α) is high and certainly contributes to the observed reduction of the ratio in macaques. Previous studies estimated that the value of α ranges from 2 to 6 in primate species (Makova and Li 2002; Tcsaa 2005; Gibbs et al. 2007; Presgraves and Yi 2009). A recent whole-genome sequencing study of human families also showed the similar level of mutation rate increase in males (Kong et al. 2012). We estimated the mutation rate bias using the divergence data between humans and macaques and controlled the mutation rate bias using the value in the following computer simulations.
As suggested by the previous studies, population size contraction also leads the reduction of X/A diversity ratio. To estimate the effect of demography, we applied the kernel-ABC method under a relatively simple model using the data in the autosomal IGS regions. In particular, five different levels of population contraction in the Philippine populations were examined. We found that the strongest levels of population contraction (10% of original size) were unlikely compared with milder ones. Although four models with mild or no contraction showed statistically indistinguishable marginal likelihood values, the model with 25% of the original size showed the highest marginal likelihood and we used the model to evaluate the effect of demography. As expected, the X/A diversity ratios in the Philippine population were reduced compared with the ratios in the Indonesian–Malaysian population in the simulated data set. Although the observed ratios were smaller than the mean of simulated distribution, they were not significantly deviated from the expected distribution in the both populations. The result indicates that the observed reduction in the IGS regions can be explained by those two factors: mutation rate bias and demography. However, we should note that the observed values were smaller than the mode of expected distribution, which implies that we might have underestimated the mutation rate bias α, that the actual demography was more complex than our model, or that other unknown factors are contributing to the ratio. For example, male-biased founding of the Philippine populations potentially reduces the relative genetic diversity on the X chromosome and such a scenario is likely, considering the matrilocal nature of macaque monkeys (Tosi et al. 2003).
In this study, we used humans as an outgroup species and polarized SFS using the information. However, misinference of the ancestral state may have a significant impact on our analysis, because the divergence between humans and macaques is relatively large (∼6%). We repeated our analyses using the draft genome sequence of more closely related taxa, baboon (Papio anubis), as an outgroup. Although we observed several cases of ancestral state misinference, we confirmed that the same model (model 2 with 25% size) was selected as the best model and observed only a slight difference in the estimated parameters. Overall conclusions of our study were not affected by the choice of the outgroup species (Figure S4).
Finally, natural selection could reduce the diversity on X chromosomes more than on autosomes. Such a process would be more effective in the genomic regions proximal to functional elements. Our observation that the X/A diversity ratio was significantly lower in CDS regions than in IGS regions agrees well with the previous observations in humans (Hammer et al. 2010; Hernandez et al. 2011). Despite the small sample size, the whole-genome sequence analysis of M. fascicularis individuals also displayed more reduction of genetic diversity on the X chromosome near CDS.
The prediction of genetic diversity level under natural selection depends on many parameters. Previous studies suggested that background selection affects the genetic diversity more on autosomes than on the X chromosomes when the strength of deleterious mutations are not very weak and recessive (Begun and Whitley 2000), or the sex-averaged recombination rate is higher on the X chromosome than autosomes (Vicoso and Charlesworth 2009). However, sex-averaged recombination rates are generally lower on the X chromosomes than on autosomes in mammals (Jensen-Seaman et al. 2004), and it could reduce the X/A diversity ratio in mammals under natural selection (Hammer et al. 2010). In addition, theoretical studies showed that, within some range of selection coefficient, genetic diversity of linked sites could be reduced by weak purifying selection as strength of selection increases (e.g., Williamson and Orive 2002, Figure 3). Therefore, distinguishing the model of hitchhiking by advantageous mutations and the model of background selection is difficult at this point.
We also note that the level of reduction of X/A genetic diversity near CDS in the whole-genome sequence data is much weaker than that in our 10-locus data. Because silent sites of our CDS data contain many synonymous sites, which could be direct targets of purifying selection (Lu and Wu 2005), the natural selection on the genes in our CDS data may be confounding the effect of direct and indirect forces. Further studies using genome-wide pattern of polymorphisms of many individuals of nonhuman primates would be necessary to conclude the importance of natural selection for explaining the reduction of X chromosome diversity near CDS.
In summary, we found a significant reduction of X/A diversity ratio by resequencing 24 M. fascicularis individuals. The stronger reduction in the derived population suggested that a demographic effect is one of the causes of the reduction. We also found evidence that natural selection is reducing the diversity on X chromosomes more than that on autosomes around CDS regions. A higher mutation rate in males than in females is another factor contributing to the decreased X/A diversity ratio in macaques. These above factors are not mutually exclusive and we conclude that they jointly produced the low X/A diversity ratio. The amount of data presented in this study is rather limited. However, genome-wide polymorphism data are still hard to obtain in nonmodel organisms. The framework of this study will help future studies of natural variations of nonmodel organisms on autosomes and sex chromosomes and reveal important biological mechanisms behind them.
Supplementary Material
Acknowledgments
We are grateful to Hideki Innan and two anonymous reviewers for many helpful comments on the manuscript. S.N. was supported as a Grant-in-Aid for the Japan Society for the Promotion of Science (JSPS) Research Fellow (24-3234).
Footnotes
Communicating editor: B. A. Payseur
Literature Cited
- Andolfatto P., 2001. Contrasting patterns of X-linked and autosomal nucleotide variation in Drosophila melanogaster and Drosophila simulans. Mol. Biol. Evol. 18: 279–290 [DOI] [PubMed] [Google Scholar]
- Begun D. J., Whitley P., 2000. Reduced X-linked nucleotide polymorphism in Drosophila simulans. Proc. Natl. Acad. Sci. USA 97: 5960–5965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustamante C. D., Ramachandran S., 2009. Evaluating signatures of sex-specific processes in the human genome. Nat. Genet. 41: 8–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caballero A., 1994. Developments in the prediction of effective population size. Heredity 73: 657–679 [DOI] [PubMed] [Google Scholar]
- Charlesworth B., 2012. The effects of deleterious mutations on evolution at linked sites. Genetics 190: 5–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumont B. L., Payseur B. A., 2008. Evolution of the genomic rate of recombination in mammals. Evolution 62: 276–294 [DOI] [PubMed] [Google Scholar]
- Fukumizu K., Song L., Gretton A., 2011. Kernel Bayes’ Rule, Advances in Neural Information Processing Systems, Vol. 24, pp. 1549–1557 Curran Associates, Inc., Red Hook, NY. [Google Scholar]
- Gibbs R. A., Rogers J., Katze M. G., Bumgarner R., Weinstock G. M., et al. , 2007. Evolutionary and biomedical insights from the rhesus macaque genome. Science 316: 222–234 [DOI] [PubMed] [Google Scholar]
- Hammer M. F., Mendez F. L., Cox M. P., Woerner A. E., Wall J. D., 2008. Sex-biased evolutionary forces shape genomic patterns of human diversity. PLoS Genet. 4: e1000202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammer M. F., Woerner A. E., Mendez F. L., Watkins J. C., Cox M. P., et al. , 2010. The ratio of human X chromosome to autosome diversity is positively correlated with genetic distance from genes. Nat. Genet. 42: 830–831 [DOI] [PubMed] [Google Scholar]
- Hernandez R. D., Kelley J. L., Elyashiv E., Melton S. C., Auton A., et al. , 2011. Classic selective sweeps were rare in recent human evolution. Science 331: 920–924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honjo S., Cho F., Terao K., 1984. Establishing the cynomolgus monkey as a laboratory animal. Adv. Vet. Sci. Comp. Med. 28: 51–80 [DOI] [PubMed] [Google Scholar]
- Hudson R. R., 2002. Generating samples under a Wright–Fisher neutral model of genetic variation. Bioinformatics 18: 337–338 [DOI] [PubMed] [Google Scholar]
- Hvilsom C., Qian Y., Bataillon T., Li Y., Mailund T., et al. , 2012. Extensive X-linked adaptive evolution in central chimpanzees. Proc. Natl. Acad. Sci. USA 109: 2054–2059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen-Seaman M. I., Furey T. S., Payseur B. A., Lu Y., Roskin K. M., et al. , 2004. Comparative recombination rates in the rat, mouse, and human genomes. Genome Res. 14: 528–538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kass R. E., Raftery A. E., 1995. Bayes factors. J. Am. Stat. Assoc. 90: 773–795 [Google Scholar]
- Keinan A., Mullikin J. C., Patterson N., Reich D., 2009. Accelerated genetic drift on chromosome X during the human dispersal out of Africa. Nat. Genet. 41: 66–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong A., Frigge M. L., Masson G., Besenbacher S., Sulem P., et al. , 2012. Rate of de novo mutations and the importance of father’s age to disease risk. Nature 488: 471–475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Salzberg S. L., 2012. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9: 357–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., et al. , 2009. The sequence alignment/map format and samtools. Bioinformatics 25: 2078–2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Librado P., Rozas J., 2009. DNAsp v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25: 1451–1452 [DOI] [PubMed] [Google Scholar]
- Lu J., Wu C. I., 2005. Weak selection revealed by the whole-genome comparison of the X chromosome and autosomes of human and chimpanzee. Proc. Natl. Acad. Sci. USA 102: 4063–4067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makova K. D., Li W. H., 2002. Strong male-driven evolution of DNA sequences in humans and apes. Nature 416: 624–626 [DOI] [PubMed] [Google Scholar]
- Miyata T., Hayashida H., Kuma K., Mitsuyasu K., Yasunaga T., 1987. Male-driven molecular evolution: a model and nucleotide sequence analysis. Cold Spring Harb. Symp. Quant. Biol. 52: 863–867 [DOI] [PubMed] [Google Scholar]
- Nakagome, S., K. Fukumizu, and S. Mano, 2012 Kernel approximate Bayesian computation for population genetic inferences. ArXiv e-prints: arXiv:1205.3246, http://arxiv.org/abs/1205.3246 [DOI] [PubMed]
- Osada N., Uno Y., Mineta K., Kameoka Y., Takahashi I., et al. , 2010. Ancient genome-wide admixture extends beyond the current hybrid zone between Macaca fascicularis and M. mulatta. Mol. Ecol. 19: 2884–2895 [DOI] [PubMed] [Google Scholar]
- Pool J. E., Nielsen R., 2007. Population size changes reshape genomic patterns of diversity. Evolution 61: 3001–3006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pool J. E., Nielsen R., 2008. The impact of founder events on chromosomal variability in multiply mating species. Mol. Biol. Evol. 25: 1728–1736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presgraves D. C., Yi S. V., 2009. Doubts about complex speciation between humans and chimpanzees. Trends Ecol. Evol. 24: 533–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prufer K., Munch K., Hellmann I., Akagi K., Miller J. R., et al. , 2012. The bonobo genome compared with the chimpanzee and human genomes. Nature 486: 527–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers J., Garcia R., Shelledy W., Kaplan J., Arya A., et al. , 2006. An initial genetic linkage map of the rhesus macaque (Macaca mulatta) genome using human microsatellite loci. Genomics 87: 30–38 [DOI] [PubMed] [Google Scholar]
- Smith D. G., Mcdonough J. W., George D. A., 2007. Mitochondrial DNA variation within and among regional populations of longtail macaques (Macaca fascicularis) in relation to other species of the fascicularis group of macaques. Am. J. Primatol. 69: 182–198 [DOI] [PubMed] [Google Scholar]
- Stephens M., Smith N. J., Donnelly P., 2001. A new statistical method for haplotype reconstruction from population data. Am. J. Hum. Genet. 68: 978–989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Peterson D., Peterson N., Stecher G., Nei M., et al. , 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28: 2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tcsaa C., 2005. Initial sequence of the chimpanzee genome and comparison with the human genome. Nature 437: 69–87 [DOI] [PubMed] [Google Scholar]
- Thompson J. D., Higgins D. G., Gibson T. J., 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22: 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tosi A. J., Morales J. C., Melnick D. J., 2003. Paternal, maternal, and biparental molecular markers provide unique windows onto the evolutionary history of macaque monkeys. Evolution 57: 1419–1435 [DOI] [PubMed] [Google Scholar]
- Vicoso B., Charlesworth B., 2006. Evolution on the X chromosome: unusual patterns and processes. Nat. Rev. Genet. 7: 645–653 [DOI] [PubMed] [Google Scholar]
- Vicoso B., Charlesworth B., 2009. Recombination rates may affect the ratio of X to autosomal noncoding polymorphism in african populations of Drosophila melanogaster. Genetics 181: 1699–1701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson S., Orive M. E., 2002. The genealogy of a sequence subject to purifying selection at multiple sites. Mol. Biol. Evol. 19: 1376–1384 [DOI] [PubMed] [Google Scholar]
- Yan G., Zhang G., Fang X., Zhang Y., Li C., et al. , 2011. Genome sequencing and comparison of two nonhuman primate animal models, the cynomolgus and chinese rhesus macaques. Nat. Biotechnol. 29: 1019–1023 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.