Abstract
During meiosis, the stable pairing of the homologous chromosomes is mediated by the assembly of the synaptonemal complex (SC). Its tripartite structure is well conserved in Metazoa and consists of two lateral elements (LEs) and a central region (CR) that in turn is formed by several transverse filaments (TFs) and a central element (CE). In a previous article, we have shown that not only the structure, but also the major structural proteins SYCP1 (TFs) and SYCP3 (LEs) of the mammalian SC are conserved in metazoan evolution. In continuation of this work, we now investigated the evolution of the mammalian CE-specific proteins using phylogenetic and biochemical/cytological approaches. In analogy to the observations made for SYCP1 and SYCP3, we did not detect homologs of the mammalian CE proteins in insects or nematodes, but in several other metazoan clades. We were able to identify homologs of three mammalian CE proteins in several vertebrate and invertebrate species, for two of these proteins down to the basal-branching phylum of Cnidaria. Our approaches indicate that the SC arose only once, but evolved dynamically during diversification of Metazoa. Certain proteins appear to be ancient in animals, but successive addition of further components as well as protein loss and/or replacements have also taken place in some lineages.
Keywords: meiosis, synaptonemal complex, Metazoa, mammals, Hydra, phylogeny
SEXUAL reproduction was established as the beneficial mode of propagation during evolution of animals. Most of the metazoan species reproduce sexually meaning via formation of a new organism by syngamy, that is the fusion of two gametes from different genders. The formation of the gametes, in turn, is dependent on meiosis, a specialized type of cell division that is responsible for the reduction of the chromosome set from the original diploid state of the gonia to the haploid state of differentiated sperms or eggs. During the meiotic cell cycle, a germ cell progenitor passes through two successive rounds of chromosome segregation (called meiosis I and II) after a single round of DNA replication. In the most crucial process of meiosis I, the homologous chromosomes have to pair and exchange genetic material (homologous recombination) as a requirement for their accurate segregation into two daughter cells. An important feature of this evolutionarily well-conserved pairing process is the assembly of the synaptonemal complex (SC), a proteinaceous structure that connects the two chromosomes of a homologous pair like a zipper during prophase of meiosis I (synapsis). The successful synapsis of the chromosomes is essential for proper homologous recombination in mammals, preventing missegregation of the chromosomes that results in aneuploid germ cells or even cell death (for reviews, see Hassold and Hunt 2001; Page and Hawley 2004; Costa and Cooke 2007; Handel and Schimenti 2010; Bolcun-Filas and Schimenti 2012). Electron microscopic data of animals from several different phyla illustrated the nearly ubiquitous existence of the SC in meiosis and the evolutionary conservation of its tripartite structure (for reviews, see Gillies 1975; von Wettstein et al. 1984; Page and Hawley 2004). With the shape similar to that of a ladder, the SC consists of two parallel rod-like lateral elements (LEs) that are linked during synapsis by transverse filaments (TFs) that are arranged in a crosswise fashion. The TFs from opposing LEs overlap in the center of the SC, thereby forming the central element (CE). Together, the TFs and the CE constitute the central region (CR) of the SC (for review, see Page and Hawley 2004).
In mammals, seven different SC protein components have been identified so far (for review, see Fraune et al. 2012a) that are essential for the correct assembly of the SC. The LEs are composed of the proteins SYCP2 and SYCP3 [1500 and 254 amino acids (aa) in mouse, respectively] (Lammers et al. 1994; Offenberg et al. 1998). Dimers of the large coiled coil protein SYCP1 (993 aa in the mouse) form the TFs (Meuwissen et al. 1992). In addition, four rather small proteins locate specifically to the CE: SYCE1, SYCE2, SYCE3, and Tex12 (329, 171, 88, and 123 aa in mouse, respectively) (Costa et al. 2005; Hamer et al. 2006; Schramm et al. 2011). These CE proteins are essential for initiation and elongation of the synapsis. A complex made of SYCE1 and SYCE3 is postulated to initiate synapsis by allowing the initial interaction of opposing TFs (Bolcun-Filas et al. 2007; Schramm et al. 2011). Both proteins localize in a continuous pattern along the SC, similar to SYCP1 (Costa et al. 2005; Schramm et al. 2011). Disruption of either SYCE1 or SYCE3 leads to a complete disruption of synapsis (Bolcun-Filas et al. 2009; Schramm et al. 2011). In contrast, SYCE2 and Tex12 present a rather punctate localization pattern and the corresponding knockout spermatocytes still exhibit short stretches of CE-like structures during the meiotic prophase I substages of zygotene and pachytene (Costa et al. 2005; Hamer et al. 2006, 2008; Bolcun-Filas et al. 2007). SYCE2 and Tex12 are therefore proposed to be essential for elongation of synapsis (Bolcun-Filas et al. 2007; Hamer et al. 2008).
Recently, we have shown that the main structural SC components SYCP1 (TFs) and SYCP3 (LEs) of the mouse are ancient in Metazoa and present in a variety of different organisms, even in the early-branching Hydra lineage (Cnidaria) (Fraune et al. 2012b). This opened the possibility that the entire SC could be of ancient origin, meaning that not only the main structural components of the LEs and TFs, but also the CE components of the mammalian SC might have been present in the last ancestor of metazoans. To test this hypothesis, we analyzed the evolutionary history of the mouse CE through a phylogenetic approach. We identified homologs of three of the four CE proteins in various species that belong to metazoan lineages, which are distantly related to mammals. This points to a very ancient origin of the corresponding components in Metazoa. More precisely, we show that SYCE2 and Tex12 were present in the ancestor of Eumetazoa. In contrast, the phylogeny of SYCE1 indicates that this protein is slightly more recent and emerged in the ancestor of Bilateria, whereas SYCE3 emerged much later in the vertebrate lineage. The two candidate components SYCE2 and Tex12 found in the cnidarian Hydra were analyzed experimentally to confirm their potential role in the assembly of the SC in this basal early-diverging animal lineage.
Materials and Methods
Dataset assembly
We used the four characterized mouse CE proteins SYCE1 (RefSeq: NP_001137237), SYCE2 (RefSeq: NP_082230), SYCE3 (RefSeq: NP_001156352), and Tex12 (RefSeq: NP_079963); CONA (RefSeq: NP_650719), a CR protein described in Drosophila melanogaster; and the Caenorhabditis elegans CR proteins SYP-2, SYP-3, and SYP-4 (RefSeq: NP_504462, NP_492345, and NP_491960) as seeds to query public sequence databases. Homologous sequences available in the nr database at the National Center for Biotechnology Information (NCBI) (http://blast.ncbi.nlm.nih.gov) were identified using the BLASTp program (Matrix: BLOSUM45; default values for all other parameters; Altschul et al. 1997). To ensure that all homologs were correctly sampled, we used the PSI-BLAST program (Matrix: BLOSUM45; default values for all other parameters; Altschul et al. 1997). Convergence was reached after three iterations. Additional or more divergent homologs were retrieved from the nr/nt, est, wgs, and ongoing genome projects data available at the NCBI, Ensembl database, release 71 (http://www.ensembl.org/index.html), the Dana-Farber Cancer Institute (DFCI) (http://compbio.dfci.harvard.edu), and the InParanoid 7 project (http://inparanoid.sbc.su.se/cgi-bin/index.cgi; Ostlund et al. 2010) using tBLASTn and BLASTp. All BLASTp and tBLASTn searches were repeated several times by using each newly detected homolog as seed for a new search. The absence of any homologous sequence in a given species/lineage, for which the complete genome is available, was checked by screening the corresponding genome with the tBLASTn program. The sequences retrieved were used for reciprocal BLAST analyses to ensure that they represented putative homologs of CE proteins (and not false positives).
For each CE protein, the retrieved sequences were aligned using ClustalO (Sievers et al. 2011) implemented in the Seaview program, version 4.4.0 (Gouy et al. 2010). A preliminary neighbor-joining (NJ) tree was inferred with the same program (default parameters). Based on this tree, the closest homologs of the sequences that were experimentally demonstrated as part of the SC (e.g., the mouse sequences) were selected, realigned, and used to build a specific hidden Markov model (HMM) profile using the HMMer 3.0 webserver (http://hmmer.janelia.org). The resulting profile was used to query the nr database (hmmsearch option) as well as to individually verify all the other sequences present in the initial alignment by starting from the closest and progressing to the more distantly related sequences (according to the NJ tree). New and verified sequences were added to the new alignment step by step and used to update the HMM profile. This procedure was repeated iteratively until no further homologs could be identified (Supporting Information, Table S1, Table S2, Table S3, and Table S4).
Phylogenetic analysis
To reduce potential tree reconstruction artifacts linked to the overrepresentation of a few lineages (such as placental mammals), a taxonomically balanced subset of homologs was selected for final phylogenetic analyses. The sequences were aligned using MAFFT version 7 with the linsi option, which allows accurate alignment reconstructions (Katoh et al. 2002). Alignments were inspected using Seaview (Gouy et al. 2010) and ambiguously aligned regions were removed with the Block Mapping and Gathering with Entropy (BMGE, default parameters; Criscuolo and Gribaldo 2010). Maximum likelihood (ML) trees were inferred with PhyML version 3.0.1 (Guindon et al. 2010) with the LG model (Le and Gascuel 2008), option NNI+SPR and a gamma distribution to take into account heterogeneous evolutionary rates (four categories of sites and an estimated alpha parameter) as suggested by the proposed model tool implemented in TreeFinder, version 2011 (aicc criterion; Jobb et al. 2004). The robustness of the resulting tree was assessed with the nonparametric bootstrap procedure implemented in PhyML (100 replicates of the original alignment). Bayesian phylogenetic trees (Bayesian inference, BI) were constructed using MrBayes 3.2.1 (Ronquist et al. 2012) with a mixed amino-acid-substitution model and gamma distribution (four categories of sites and an estimated alpha parameter). The search was run with four independent chains for 1 million generations. Trees were sampled every 100 generations. The first 2000 trees were discarded as “burn-in.” The branch robustness was estimated by calculation of posterior probabilities.
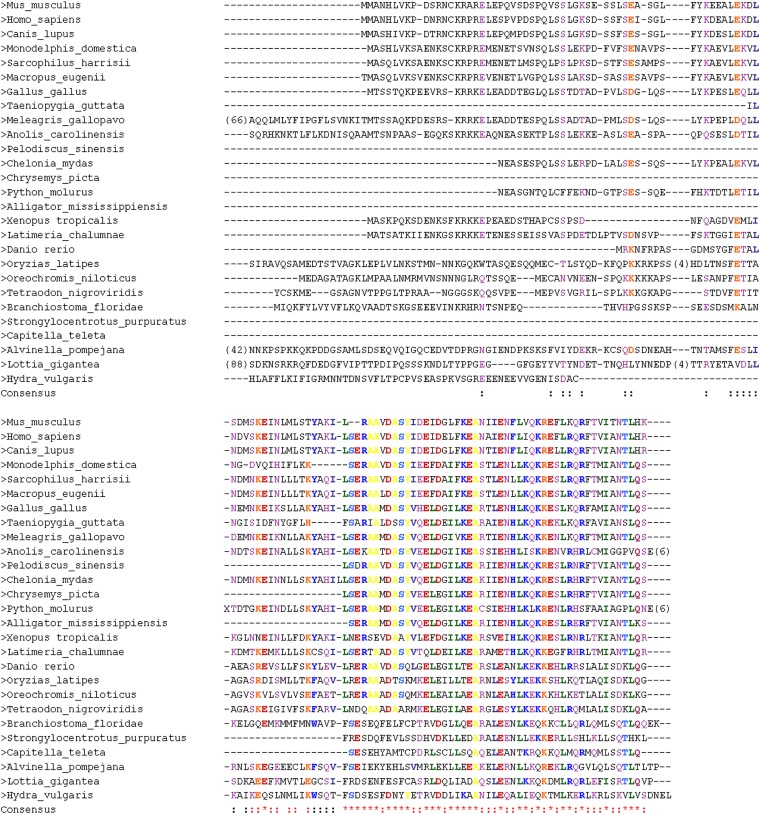
Annotated sequence alignments were designed using CHROMA version 1.0 (Goodstadt and Ponting 2001). The identity threshold for grouping of the residues was set to 60%. Seven groups were created, depending on different features of the amino acids: identical, charged, Ser/Thr, aliphatic, aromatic, polar, and hydrophobic. Sparse regions longer than four residues were removed if at least 80% of the sequences were blank gaps at these positions. The number of removed residues is indicated by numbers in parentheses in the corresponding sequences at those positions (Figure 1, Figure 2, Figure S1, and Figure S2).
Figure 1.
Multiple alignment of SYCE2 homologs. Positions kept for phylogenetic inferences are indicated in red. Positions removed by BMGE are indicated in black.
Figure 2.
Multiple alignment of Tex12 homologs. Positions kept for phylogenetic inferences are indicated in red. Positions removed by BMGE are indicated in black.
Animal strains and culture conditions
For expression analysis, animals from the strain AEP (Martin et al. 1997), belonging to the Hydra vulgaris group, were cultured at 18° following standard procedure. Induction of testes formation was achieved by feeding the animals daily for at least 1 week and starving them afterward (Wittlieb et al. 2006).
Sequence information of H. vulgaris AEP was obtained from transcriptome data on the Compagen_NG server (Hemmrich et al. 2012).
Isolation of RNA, reverse transcription, PCR, and cloning of cDNA
RNA extraction from whole animals or tissues of Hydra AEP was accomplished using the peqGOLD TriFast RNA Extraction kit (peqLab, Erlangen). cDNA was obtained by reverse transcription of 1 µg RNA with an oligo(dT) primer and M-MLV reverse transcriptase (Promega, Mannheim, Germany) and used as template for cloning of full-length cDNA sequences or expression analysis in different tissue fractions (head, midpiece, foot, and testis). Full-length cDNA of HySyce2 and HyTex12 was amplified with Phusion DNA polymerase (Thermo Scientific, St. Leon Roth, Germany) and the following sequence-specific primer pairs: Hy_Syce2_ATG 5′ (ATGACTAACAAACGCAAGTTTGTGAG), Hy_Syce2_TGA 3′ (CTACTGCAATGATGGATAGGTAGCTTG) at 66° and HyTex12 5′ (CTGAACATGTGTAAAAATGTCTCAG), HyTex12 3′ (CAGTTTTAATATTTAACTGTTAAAA-GTGTTAATAG) at 61°. Subsequently, the cDNA was cloned into the pSC-B-amp/kan PCR cloning Vector (Agilent Technologies, Böblingen, Germany) and sequenced. Comparison of different independent cloning attempts with sequence data on the public databases was performed to verify the obtained cDNA sequences from single-read sequencing.
Tissue-specific expression profiles of HySyce2 and HyTex12 were likewise analyzed by Phusion PCR using the same primer pairs. Actin expression was traced as an internal loading control with actin-specific primers (Hym_actin 5′: 5′-AAGCTCTTCCCTTGAGAAATC-3′; Hym_actin 3′: 5′-CCAAAATAGATCCTCCGATCC-3′, at 60°).
Antibodies
HySYCE2 and HyTex12 antibodies were generated against the full-length HySYCE2 and HyTex12. The proteins were expressed as His-tagged fusion proteins from the pET21a vector (Novagen, Darmstadt, Germany) and purified through a Nickel-nitrilotriacetic acid (Ni-NTA) agarose matrix (Qiagen, Hilden, Germany). Immunization of a rabbit and a guinea pig was performed by Seqlab (Göttingen, Germany). The obtained antisera of the final bleedings were affinity purified through the HiTrap system (GE Healthcare, Munich), following the manufacturer’s protocol. An α-actin antibody (A4700) was purchased from Sigma (Steinheim, Germany).
SDS–PAGE and immunoblot analysis
Protein fractions from different tissues of Hydra (head, midpiece, foot, and testis) were separated in a 15% (vol/vol) acrylamide SDS–PAGE and subsequently transferred to nitrocellulose membranes by the semi-dry Western blotting system (Matsudaira 1987). Detection of the proteins HySYCE2 and actin was performed as previously described (Fraune et al. 2012b). We used a rabbit α-HySYCE2 (1:2000) that recognized a protein in the testis tissue that matches the expected molecular weight of HySYCE2 (17.6 kDa) and a mouse α-actin antibody (1:10,000) that, according to the manufacturer, has a broad species reactivity. A peroxidase-conjugated secondary antibody (1:10,000) of Dianova (Hamburg) was applied for final detection of the protein with the Western Lightning Plus-ECL (PerkinElmer, Waltham, MA). The α-HyTex12 antibody was not effective in Western blot analysis but it recognized its target protein on chromosome spreads.
In situ hybridization
Whole-mount in situ hybridization on Hydra was conducted following the standard protocol (Grens et al. 1996) with minor modification as reported recently (Fraune et al. 2012b). Digoxigenin (DIG)-labeled RNA probes were synthesized from the full-length cDNA of HySyce2 (459 bp) and HyTex12 (336 bp).
Immunocytochemistry
Meiotic chromosome spreads were prepared following the dry-down procedure (De Boer et al. 2009), but with slight adaptions to the Hydra tissue (Fraune et al. 2012b). Immunostaining of the chromosome spreads was carried out as indicated in De Boer et al. (2009) with the help of following affinity-purified antibodies: rabbit α-HySYCE2 (1:200), rabbit α-HyTex12 (1:300), guinea pig α-HyTex12 (1:300), guinea pig α-HySYCP1 (1:300), rabbit α-HySYCP1 (1:900), guinea pig α-HySYCP3 (1:75), and rabbit α-HySYCP3 (1:600). Secondary antibodies were purchased from Dianova and applied as recommended by the manufacturer.
Microscopy and imaging
A Leica TCS-SP2 confocal laser scanning microscope (Leica, Wetzlar, Germany) equipped with a ×63/1.40 HCX PL APO lbd.BL oil immersion objective was used for confocal imaging. The images are 2D projections from a series of ∼20 optical sections per cell, generated by the maximum projection algorithm (Leica), and they were pseudocolored using the Leica TCS-SP2 software. Processing of the digital images was done with Adobe Photoshop CS5 (Adobe Systems).
Results
Mouse CE proteins SYCE1, SYCE2, and TEX12 are ancient in Metazoa
Our in-depth survey of sequence databases revealed many homologs of the mouse SYCE1, SYCE2, and Tex12 components in representatives of distantly related animal lineages. More precisely, homologs could be identified in most Bilateria lineages, namely in Chordata (vertebrates and Cephalochordata) and the invertebrate lineage of Echinodermata, all belonging to Deuterostomia as well as in Lophotrochozoa (Mollusca and Annelida) (Table S1, Table S2, and Table S3). Conversely, we could not identify SYCE1 homologs in any bird species, although complete genome sequences are available for this lineage. This reflects either the loss or the nonhomologous replacement or a faster evolutionary rate (beyond recognition) of the SYCE1 components in this lineage. Interestingly, we also detected SYCE2 and Tex12, but not SYCE1 homologs in representatives of the basal-branching phylum of Cnidaria, namely Hydra and/or Nematostella, and a SYCE2 divergent homolog in the crustacean species Daphnia pulex (Ecdysozoa). The situation was different for the last CE component, because we detected SYCE3 homologs only in vertebrate species (Table S4) despite an intense search in nonvertebrate sequence data.
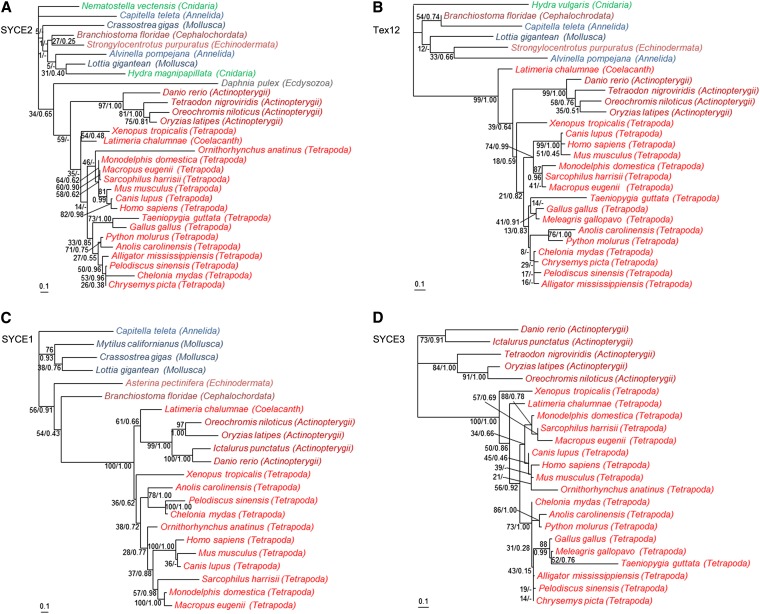
The multiple alignments show that the central regions of SYCE2 (aa 59–153 of the mouse; aa 67–161 of the human) and Tex12 (aa 61–121 of the mouse; aa 61–123 of the human) are relatively well conserved across metazoans, whereas their N- and C termini are highly variable (Figure 1 and Figure 2). In contrast, SYCE1 harbors a larger conserved region that corresponds to a large part of the central coiled coil domain of the protein (aa 58–268 of the mouse; aa 56–266 of the human; Figure S1). The ML and BI phylogenies of SYCE2 and Tex12 are not fully resolved, especially for the deepest nodes [weak bootstrap values (BVs) and Bayesian posterior probabilities (PPs] (Figure 3, A and B). This is not surprising, given the relatively restricted number of positions that have been kept for phylogenetic analyses (Figure 1 and Figure 2) and the relatively long branches associated to some invertebrate species (i.e., D. pulex, Figure 3A or Alvinella pompejana and Capitella teleta, Figure 3B), indicating fast evolutionary rates. The recovered relationships among vertebrate lineages, however, are globally consistent with recently published phylogenies (Philippe et al. 2009; Simakov et al. 2013). In the case of SYCE1, more positions could be kept for the phylogenetic analysis (Figure S1) and, unsurprisingly, the resulting ML and BI trees are more resolved (Figure 3C) and consistent with the global phylogeny of Metazoa. Finally, the ML and BI trees of SYCE3 (based on the conserved region of aa 1–86 of the mouse and the human; Figure S2) are globally consistent with the phylogeny of vertebrates (Figure 3D), suggesting that this component emerged in the ancestor of this lineage.
Figure 3.
Unrooted maximum likelihood phylogenies of CE proteins. (A) SYCE2 (30 sequences, 75 aa positions kept). (B) Tex12 (27 sequences, 59 aa positions kept). (C) SYCE1 (22 sequences, 150 aa positions kept). (D) SYCE3 (23 sequences, 81 aa positions kept). Numbers at branches correspond to bootstrap values (given in percentages) and posterior probabilities (given in fractions) inferred with PhyML and MrBayes, respectively (e.g. 100/1.00). A dash indicates that the corresponding branch is not recovered in the consensus Bayesian tree (e.g. 46/-). Bars, average number of substitutions per site. Color code: Cnidaria, green; Lophotrochozoa, blue; Ecdysozoa, gray; and Deuterostomia, red. Sublineages are indicated by different shades of the same color.
The presence of homologs of SYCE2, Tex12, and SYCE1 in most bilaterian lineages was suspected, as we had previously proved the ancient and monophyletic origin of the two bona fide structural SC components SYCP1 and SYCP3 (Fraune et al. 2012b). Indeed, our data suggest that these three CE components were likewise present in the last common ancestor of Bilateria. The origin of SYCE2 and Tex12 can even be pushed back to the ancestor of Eumetazoa, given that homologs of these two components have been detected in Cnidaria representatives. At this step, a more ancient origin, however, cannot be proposed, because (despite many attempts) we neither detected homologs of these two components in Porifera or in Placozoa (two basal-branching metazoan lineages) nor in protist lineages closely related to Metazoa (i.e., Choanoflagellida, Ichthyosporea, etc.) for which complete genome sequences are available. In contrast, SYCE3 seems to be of much more recent origin, likely being an innovation of vertebrates.
Despite our extensive survey of sequence databases, we did not detect any homolog of the mammalian CE components in ecdysozoan species, including D. melanogaster and C. elegans. Instead, alternative CR proteins that do not share any sequence homology with other proteins were characterized in these model organisms for meiosis (MacQueen et al. 2002; Colaiacovo et al. 2003; Smolikov et al. 2007, 2009; Page et al. 2008; Schild-Prüfert et al. 2011). As the single exception to the remarkable situation of the ecdysozoan lineage, we identified a potential homolog of SYCE2 in the Crustacea D. pulex. Regarding its very long branch in the SYCE2 phylogenetic tree (Figure 3A), the evolutionary distance between this protein and the other SYCE2 sequences, however, seems to be peculiarly large and suggests a fast evolutionary rate.
In D. melanogaster, the Corona (CONA) protein seems to stabilize the interaction of the TFs in the center of the SC, which is comparable to the function of the mammalian SYCE2/Tex12 complex (Page et al. 2008). Besides C(3)G, it is the only identified CR protein in D. melanogaster so far. According to the authors’ own statement, CONA is only conserved within the genus of Drosophila (Page et al. 2008). Consistently, we did not find homologous sequences beyond the border of this group using CONA as an alternative seed for our phylogenetic analysis.
A similar situation was reported in C. elegans. In total, four different SYP proteins (SYP-1, SYP-2, SYP-3, and SYP-4) have been identified to localize in the CR of the SC (MacQueen et al. 2002; Colaiacovo et al. 2003; Smolikov et al. 2007, 2009; Schild-Prüfert et al. 2011). However, it is yet unclear which proteins fulfill CE-like functions (Smolikov et al. 2009). According to the model proposed by Schild-Prüfert et al. (2011) SYP-1, which forms homodimers (or higher ordered structures), is most likely to be an essential module of the TFs (Schild-Prüfert et al. 2011) and was not included in the analysis. Using SYP-2, SYP-3, and SYP-4 as seeds for a homology search, we did not detect any homolog outside the genus Caenorhabiditis.
Expression of SYCE2 and TEX12 in Hydra meiosis
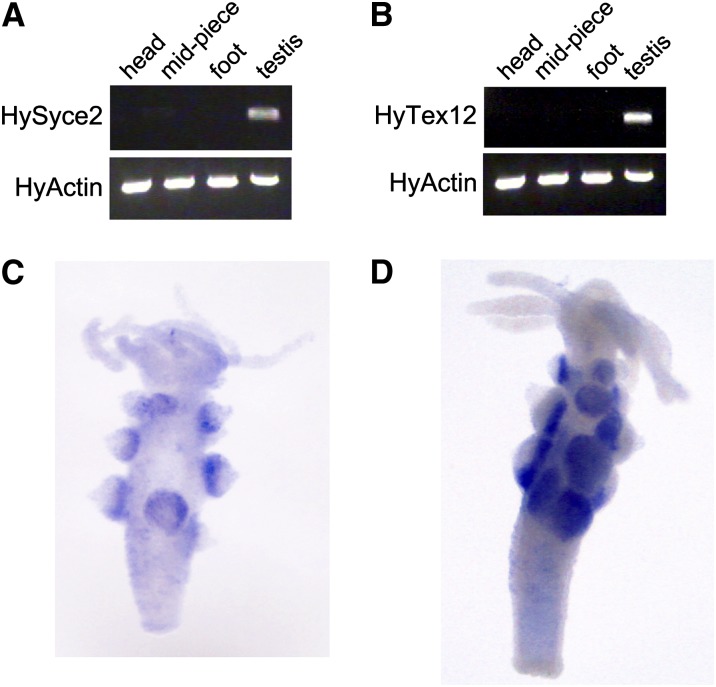
The presence of SYCE2 and Tex12 in Cnidaria and Bilateria indicates that they were present in their last common ancestor, i.e., the Eumetazoa ancestor. Under the assumption that these proteins have not undergone functional changes, the cnidarian SYCE2 and Tex12 homologs are expected to be part of a SC in these animals. The competing hypothesis is that the SC is of more recent origin, but could have been built by the recruitment of ancient preexisting proteins. In this case, the cnidarian SYCE2 and Tex12 homologs should be involved in other functions. To discriminate between these two hypotheses, we tested the potential meiotic role of the putative SYCE2 and Tex12 homologs in H. vulgaris (strain AEP). We first cloned and sequenced the putative full-length cDNA of Hydra Syce2 (GenBank: KC580661) and Tex12 (GenBank: KC580662) and subsequently performed an expression analysis on the level of mRNA as well as on the protein level. We applied RT–PCR on isolated mRNA from four different tissue fractions of Hydra—head, midpiece, foot, and testis—and could selectively amplify Hydra Syce2 and Tex12 cDNA in the testis fraction by using sequence-specific primers. A weak signal was also observed in the midpiece lane for HySyce2. This is not surprising, as Hydra testes grow as conical swellings along the body column and testis leftovers might have remained at the midpiece tissue during preparation. Amplification of Hydra actin served as an internal control for RT–PCR (Figure 4, A and B). By whole-mount in situ hybridization with DIG-labeled RNA probes that were complementary to the mRNAs of Hydra Syce2 and Tex12, we could localize the transcripts to the basal cell layer of the conical testes of Hydra (Figure 4, C and D). In accordance with our findings, previous histological analysis of Hydra testis identified this region to be the location of spermatocytes (Kuznetsov et al. 2001).
Figure 4.
Expression analysis of HySyce2 and HyTex12. Testis-specific synthesis of HySyce2 (A) and HyTex12 (B) mRNA is shown by RT–PCR as well as by in situ hybridization (C and D).
In an approach similar to RT–PCR, we could also detect the corresponding protein product of HySyce2 in the testis tissue by applying Western blot analysis. A specific and strong protein band of the predicted molecular mass of Hydra SYCE2 (17.6 kDa) appeared in the testis lane. Here as well, with the α-SYCE2 antibody, a faint protein band was also detected in the midpiece fraction (Figure 5). The raised α-Tex12 antibody was not effective in Western blot analysis.
Figure 5.
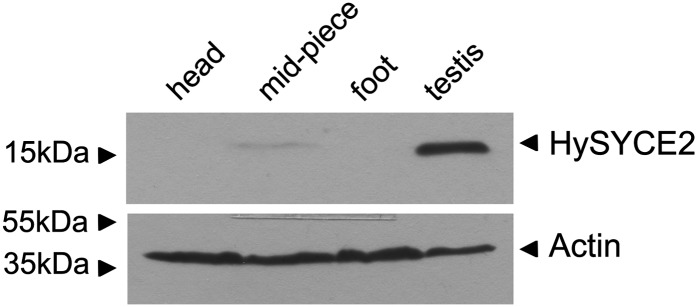
Protein expression of HySYCE2. HySYCE2 protein expression in the testis tissue is verified by immunoblotting.
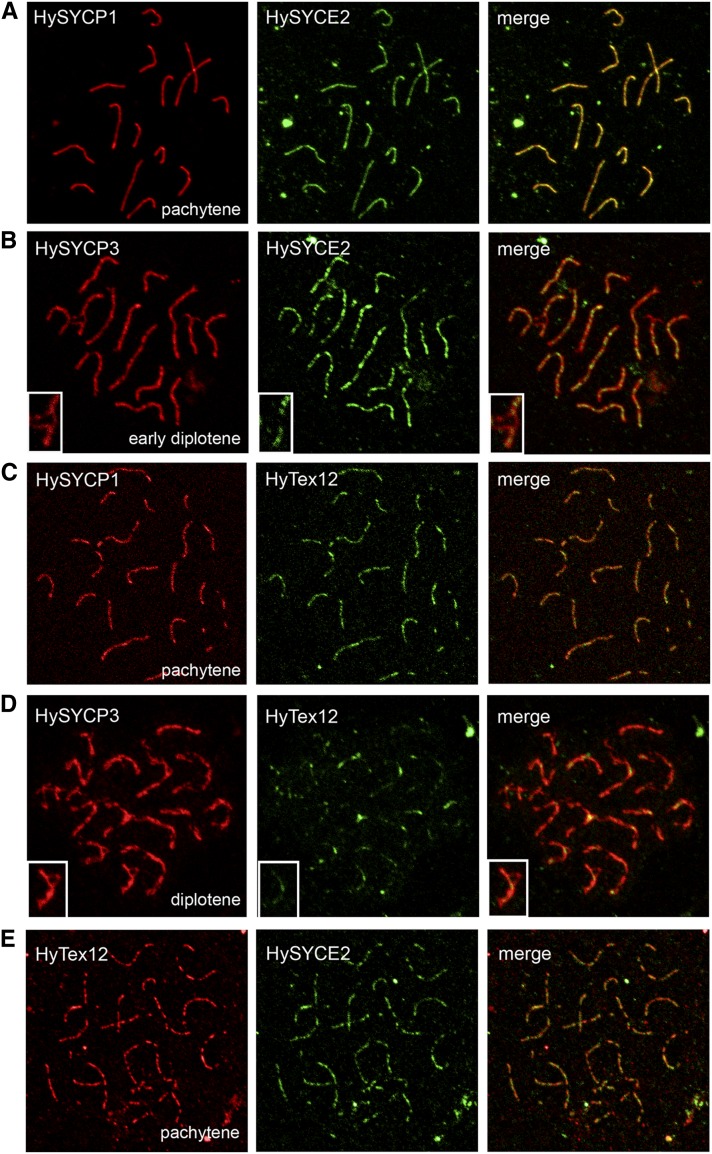
Although the HyTex12 protein product could not be detected by Western blot, the results indicated that Hydra Syce2 and Tex12 indeed are selectively expressed in meiotic cells, consistent with a role in Hydra meiosis. More detailed information about the localization of Hydra SYCE2 and Tex12 proteins was finally obtained by immunofluorescence analysis. The antibodies raised against the putative CE proteins were tested on chromosome spread preparations of Hydra testis tissue in at least two independent immunofluorescence experiments with a minimum of three slides. The resulting images of Figure 6 provide a representative picture of the observations in the confocal laser scanning microscope. As expected, the antibodies stained 15 SCs of Hydra pachytene spermatocytes that correlate in number to the quantity of homologous chromosome pairs in H. vulgaris (Zacharias et al. 2004). Colocalization of HySYCE2 and HySYCP1—a marker protein for the CR of the SC (Fraune et al. 2012b)—showed that the localization of the proteins fully overlap, but HySYCE2 exhibited a more punctate pattern than HySYCP1 along the synapsed SC in pachytene spermatocytes (Figure 6A). In contrast, HySYCE2 and HySYCP3—a marker protein for the chromosome axes (Fraune et al. 2012b)—only colocalize in regions where the chromosomes are synapsed via the CR. On unsynapsed axes in early diplotene, only HySYCP3 can be detected (Figure 6B, insets). Similar results were obtained for HyTex12, as its antibody worked effectively in immunofluorescence analysis. HyTex12 colocalized with HySYCP1 in pachytene spermatocytes (Figure 6C), but disappeared from the chromosome axes—marked by HySYCP3—in diplotene at sites where the SC disassembles and the chromosome axes become unsynapsed across long sections (Figure 6D, insets). At sites where the SC-mediated connection of the chromosome axes starts loosening, remains of HyTex12 can also be detected to localize between the two parallel running axes (marked by HySYCP3) of the bivalents (Figure 6D). Finally, double-label immunofluorescence microscopy of HySYCE2 and HyTex12 in a pachytene spermatocyte confirmed the punctate colocalization pattern of the two proteins within the cnidarian SC (Figure 6E).
Figure 6.
Immunolocalization of HySYCE2 and HyTex12 on chromosome spread preparations. HySYCE2 (A) and HyTex12 (C) colocalize with HySYCP1 to the fully synapsed chromosomes during pachytene, but exhibit a more punctate localization pattern. In early diplotene and diplotene, colocalization of HySYCE2 (B) and HyTex12 (D) with HySYCP3 is restricted to synapsed regions of the chromosomes. Double labeling of HySYCE2 and HyTex12 confirms their punctate colocalization pattern (E).
Comparing our findings on the localization pattern of Hydra SYCE2 and Tex12 with the organization of the mammalian SC, both proteins obviously are cnidarian SC components specific to the CR. These results, therefore, strongly support the hypothesis of an ancestral SC in the ancestor of Eumetazoa formed by at least HySYCP1, HySYCP3 (Fraune et al. 2012b), as well as HySYCE2 and HyTex12.
Discussion
The evolutionary origin of mammalian CE-specific proteins of the SC
Our phylogenetic analyses indicate that three of the four mouse CE proteins appear to be conserved in the majority of metazoan clades. Especially homologs of SYCE2 and Tex12 could be traced back to the ancestor of Eumetazoa. Although we could not detect any homologous sequences in the oldest phyla of Porifera and Placozoa, we assume that these two proteins evolved at the time of metazoan origin. Our previously reported results, which described such an early origin of the main structural SC components SYCP1 and SYCP3 (Fraune et al. 2012b), support this assumption.
Our experimental data proved that Hydra SYCE2 and Tex12, in fact, are components of the cnidarian SC. The expression analysis demonstrated clearly the testis-specific synthesis of Hydra SYCE2 and Tex12. Antibodies raised against the proteins recognized SCs in Hydra spermatocytes. The punctate colocalization pattern of Hydra SYCE2 and Tex12 and their restriction to synapsed regions of the chromosome axes are therefore consistent with what is known from their mammalian homologs. In mouse, SYCE2 and Tex12 specifically localize to CE in a discontinuous pattern forming the so-called elongation complex that is responsible for the extension of synapsis along the entire chromosome length (Costa et al. 2005; Hamer et al. 2006, 2008; Bolcun-Filas et al. 2007). Recently, it was also reported that human SYCE2 and Tex12 form very stable and constitutive complexes under different experimental conditions. The regions located from aa 57 to 165 of human SYCE2 and 49 to 123 of human Tex12 were defined as essential for the capability to polymerize (Davies et al. 2012) and nicely correspond to the most conserved parts of the proteins in our analysis (Figure 1 and Figure 2). It is therefore justified to assume that SYCE2 and Tex12 fulfill a similar function in the SC of different metazoans.
In mammals, SYCE1 and SYCE3 form the so-called initiation complex that, together with SYCP1, is involved in early synapsis steps between the axes of the homologous chromosomes. However, neither SYCE1 nor SYCE3 could be detected in Hydra. Homologs of SYCE1 were identified in different invertebrate clades including Mollusca and Annelida, suggesting an ancient origin, but an origin at the time of the rise of SYCE2 and Tex12 could not be demonstrated. This could be due to either too little sequence data or more likely to a true absence of SYCE1 homologs in nonbilaterian lineages. In contrast, SYCE3 is missing in all analyzed invertebrate genomes and therefore is most likely specific to the vertebrate lineage.
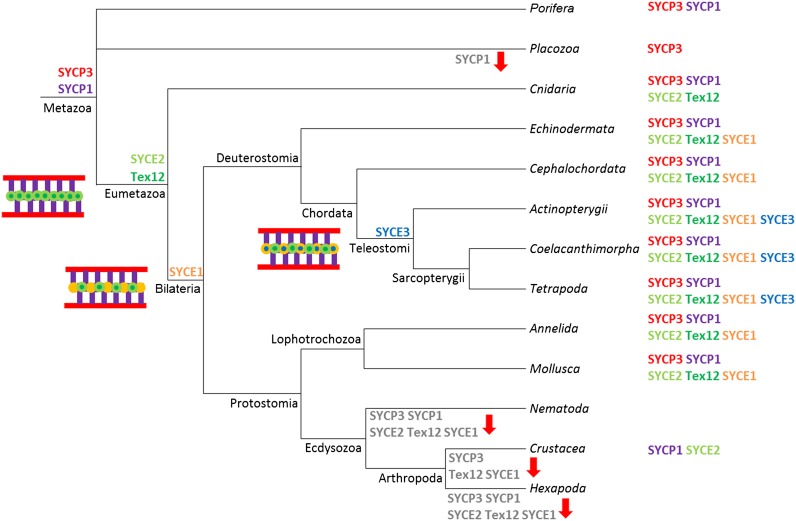
The picture that emerges from our analysis suggests that the mammalian SC is formed by very ancient (SYCE2, Tex12, and SYCE1) and much more recent elements (SYCE3). Although the structure of the SC is ancient in Metazoa, its composition has undergone a dynamic evolutionary history during the diversification of animals, which is summarized in the form of a model in Figure 7. The model illustrates the early origin of the bona fide structural SC proteins SYCP1 and SYCP3 (Fraune et al. 2012b) and of the elongation complex proteins SYCE2 and Tex12 in the ancestor of Eumetazoa. Further components were added step by step to the CE in the ancestor of Bilateria (SYCE1) and vertebrates (SYCE3). Finally, the absence of any SC protein homolog in Nematoda and Hexapoda also reflects the dynamic evolution of the SC and could be interpreted as either nonhomologous replacement of the ancestral components or their fast evolution beyond recognition (for further discussion, see section below).
Figure 7.
Model of the dynamic evolution of the SC in Metazoa. Given bioinformatic and experimental results of the preceding Fraune et al. (2012b) and the present study, the model shows that a basic SC of SYCP1, SYCP3, SYCE2, and Tex12 emerged in the last common ancestor of eumetazoan species. SYCE1 and SYCE3 evolved subsequently at the time of bilaterian and vertebrate emergence. Corresponding schematic illustrations of potential SC structures at the different evolutionary stages are shown. Homologs that were identified in the different taxonomic lineages of present-day organisms are listed next to the taxon name. Potential losses of homologs in certain lineages are indicated by the gray font of the respective protein names and a red arrow at the corresponding branch.
The significance of the initiation complex and the elongation complex
Regarding the phylogenetic data, the elongation complex of the SC seems to be evolutionarily older than the initiation complex. However, our knowledge about the function of SYCE2/Tex12 and SYCE1/SYCE3 was obtained from investigations of only the mammalian proteins and their corresponding knockout mice. As the other available metazoan model systems for meiosis and the SC, e.g., D. melanogaster and C. elegans, do not possess homologous CR proteins (see sections above and below), our present results provide a starting point for discussing the potential significance of the different CE components during evolution.
In previous polymerization studies using a heterologous system, it was shown that the mammalian TF protein SYCP1 is capable of self-assembling to higher ordered structures that resemble SCs, including the electron-dense region of the CE without support of any other SC protein (Öllinger et al. 2005). Beyond this, SYCP1 is recruited to the chromosome axes in mice even in the absence of single CE proteins (Hamer et al. 2008; Bolcun-Filas et al. 2009; Schramm et al. 2011). However, homologous chromosomes fail to start synapsis in the Syce1 and Syce3 knockout mice (Bolcun-Filas et al. 2009; Schramm et al. 2011). The absence of SYCE1 and SYCE3 homologs in Cnidaria could imply that different mechanisms of synapsis initiation are at work in this early-diverging animal lineage. Whether SYCP1 alone would be sufficient to initiate synapsis in vivo has not been investigated. However, our results also suggest that synapsis takes place in the absence of a mammalian-like initiation complex in these species. In mammals, it has been shown that the protein SYCE2 of the elongation complex is able to bind to SYCP1 (Costa et al. 2005). Whether SYCE2 of the cnidarians can bind to SYCP1 is not known. A direct binding of the protein to the TFs in this clade, however, could provide an explanation for the occurrence of an elongated synapsis even in the absence of a mammalian-like initiation complex made of SYCE1 and SYCE3.
The fact that the elongation complex is evolutionarily more conserved than the initiation complex could further be related to the process of homologous recombination. Knockout mice of the characterized CR proteins revealed a tight interdependency between the recombination machinery that is highly conserved in evolution (Cole et al. 2010) and the assembly of the CR (for review, see Fraune et al. 2012a). It is hypothesized that the CR might function as an essential platform for the recruitment of the recombination machinery to the chromosomes to generate crossovers, as close physical contacts between the CR and recombination nodules could be observed in the electron microscope (Schmekel and Daneholt 1998). In mammals, SYCP1 and SYCE2 were described to interact directly with RAD51, a RecA homolog responsible for the catalysis of the DNA strand exchange (Tarsounas et al. 1999; Moens et al. 2002; Bolcun-Filas et al. 2009), which reveals ∼82% sequence identity between mouse (RefSeq: NP_035364.1) and Hydra (RefSeq: XP_002169171.1) (our unpublished data). If, together with SYCP1, SYCE2 is a linker protein between the CR and the homologous recombination machinery, it seems sensible that this protein and its tight interaction partner are conserved during metazoan evolution.
The evolutionary origin of alternative metazoan CE proteins
Considering the evolutionary trees of the metazoan CE proteins, the broad absence of ecdysozoan species is striking. As only SC components of D. melanogaster (CONA) and C. elegans (SYP-2, SYP-3, and SYP-4) are characterized from this clade (and invertebrates in general), these were the only starting options to explore the origin of these apparently nonhomologous CR proteins. Surprisingly, our search for homologs did not retrieve any sequences beyond the genus of either Drosophila or Caenorhabditis, pointing to the very recent and genus-specific origin of these proteins and asking the question about SC components being present in closely related lineages. As we could find neither any homologs that would bridge the sequence divergence between the mammalian CE proteins (or other mammalian proteins) nor any ecdysozoan CR protein, we are currently not in the position to make statements about the origin of these alternative CR proteins or their evolutionary relationship to the mammalian CE. However, two hypotheses may explain the available data: First, CONA of D. melanogaster and the SYP proteins of C. elegans are indeed nonhomologous proteins, but functional analogs to SYCE1, SYCE2, SYCE3, and/or Tex12, arisen independently by convergent evolution. The second hypothesis is that the CONA and SYP proteins derive from the CR proteins (SYCP1, SYCE1, SYCE2, and Tex12) that were present in the last common ancestor of Bilateria, but have diverged in the fast evolving Ecdysozoa to such a high degree that the homology is no longer recognizable. In fact, we favor the second hypothesis. The few crustacean sequences found in the case of SYCE2 (in this study) and SYCP1 (Fraune et al. 2012b) might be interpreted as an indicator for the existence of homologs in the ecdysozoan clade which, however, reveal a high evolutionary distance/divergence. A definitive answer, however, would require additional genomic and experimental data for these lineages.
Conclusion
We had previously demonstrated that the main structural components SYCP1 and SYCP3 of the mammalian SC are ancient in metazoans (Fraune et al. 2012b). Here, we show that this is also the case for three of the four proteins comprising the CE. Furthermore, we could clearly verify the testis-specific expression of the Hydra SYCE2 and Tex12 and showed that their localization pattern and dynamics in spermatocytes corresponds to that in mammals, pointing to a homology in sequence and conservation in function. Therefore, we conclude that not only the ladder-like structure of the SC, but also protein components of the three SC domains, namely, the LE (SYCP3), the TFs (SYCP1), and the CE (SYCE2 and Tex12) are conserved in Metazoa and presumably are indispensable for the basic functionality of the SC. The diversification of the main metazoan lineages was finally accompanied by a dynamic evolution of the SC, indicated by the nonhomologous replacement or the very fast divergence of some components in Ecdysozoa and the addition of further components in Bilateria (SYCE1) and vertebrates (SYCE3) (Figure 7).
Supplementary Material
Acknowledgments
We thank Jörg Schulz (Biocenter, University of Würzburg) for generous help at the beginning of this project, Thomas Bosch (University of Kiel) and his group for their support in issues regarding the culturing and treatment of Hydra, and Nicola Jones (Biocenter, University of Würzburg) for critical reading of the manuscript. J.F. and R.B. are supported by a grant of the Priority Program SPP1384: Mechanisms of genome haploidization (Deutsche Forschungsgemeinschaft). C.B-A. is supported by the Investissement d’Avenir grant (ANR-10-BINF-01-01) and is a member of the Institut Universitaire de France. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Communicating editor: N. Hollingsworth
Literature Cited
- Altschul S. F., Madden T. L., Schaffer A. A., Zhang J., Zhang Z., et al. , 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25: 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolcun-Filas E., Schimenti J. C., 2012. Genetics of meiosis and recombination in mice. Int Rev Cell Mol Biol 298: 179–227 [DOI] [PubMed] [Google Scholar]
- Bolcun-Filas E., Costa Y., Speed R., Taggart M., Benavente R., et al. , 2007. SYCE2 is required for synaptonemal complex assembly, double strand break repair, and homologous recombination. J. Cell Biol. 176: 741–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolcun-Filas E., Hall E., Speed R., Taggart M., Grey C., et al. , 2009. Mutation of the mouse Syce1 gene disrupts synapsis and suggests a link between synaptonemal complex structural components and DNA repair. PLoS Genet. 5: e1000393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colaiacovo M. P., MacQueen A. J., Martinez-Perez E., McDonald K., Adamo A., et al. , 2003. Synaptonemal complex assembly in C. elegans is dispensable for loading strand-exchange proteins but critical for proper completion of recombination. Dev. Cell 5: 463–474 [DOI] [PubMed] [Google Scholar]
- Cole F., Keeney S., Jasin M., 2010. Evolutionary conservation of meiotic DSB proteins: more than just Spo11. Genes Dev. 24: 1201–1207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa Y., Speed R., Öllinger R., Alsheimer M., Semple C. A., et al. , 2005. Two novel proteins recruited by synaptonemal complex protein 1 (SYCP1) are at the centre of meiosis. J. Cell Sci. 118: 2755–2762 [DOI] [PubMed] [Google Scholar]
- Costa Y., Cooke H. J., 2007. Dissecting the mammalian synaptonemal complex using targeted mutations. Chromosome Res. 15: 579–589 [DOI] [PubMed] [Google Scholar]
- Criscuolo A., Gribaldo S., 2010. BMGE (Block Mapping and Gathering with Entropy): a new software for selection of phylogenetic informative regions from multiple sequence alignments. BMC Evol. Biol. 10: 210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies O. R., Maman J. D., Pellegrini L., 2012. Structural analysis of the human SYCE2–TEX12 complex provides molecular insights into synaptonemal complex assembly. Open Biol 2: 120099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Boer E., Lhuissier F. G., Heyting C., 2009. Cytological analysis of interference in mouse meiosis. Methods Mol. Biol. 558: 355–382 [DOI] [PubMed] [Google Scholar]
- Fraune J., Schramm S., Alsheimer M., Benavente R., 2012a The mammalian synaptonemal complex: protein components, assembly and role in meiotic recombination. Exp. Cell Res. 318: 1340–1346 [DOI] [PubMed] [Google Scholar]
- Fraune J., Alsheimer M., Volff J. N., Busch K., Fraune S., et al. , 2012b Hydra meiosis reveals unexpected conservation of structural synaptonemal complex proteins across metazoans. Proc. Natl. Acad. Sci. USA 109: 16588–16593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillies C. B., 1975. Synaptonemal complex and chromosome structure. Annu. Rev. Genet. 9: 91–109 [DOI] [PubMed] [Google Scholar]
- Goodstadt L., Ponting C. P., 2001. CHROMA: consensus-based colouring of multiple alignments for publication. Bioinformatics 17: 845–846 [DOI] [PubMed] [Google Scholar]
- Gouy M., Guindon S., Gascuel O., 2010. SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol. Biol. Evol. 27: 221–224 [DOI] [PubMed] [Google Scholar]
- Grens A., Gee L., Fisher D. A., Bode H. R., 1996. CnNK-2, an NK-2 homeobox gene, has a role in patterning the basal end of the axis in Hydra. Dev. Biol. 180: 473–488 [DOI] [PubMed] [Google Scholar]
- Guindon S., Dufayard J., Lefort V., Anisimova M., Hordijk W., et al. , 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 59: 307–321 [DOI] [PubMed] [Google Scholar]
- Hamer G., Gell K., Kouznetsova A., Novak I., Benavente R., et al. , 2006. Characterization of a novel meiosis-specific protein within the central element of the synaptonemal complex. J. Cell Sci. 119: 4025–4032 [DOI] [PubMed] [Google Scholar]
- Hamer G., Wang H., Bolcun-Filas E., Cooke H. J., Benavente R., et al. , 2008. Progression of meiotic recombination requires structural maturation of the central element of the synaptonemal complex. J. Cell Sci. 121: 2445–2451 [DOI] [PubMed] [Google Scholar]
- Handel M. A., Schimenti J. C., 2010. Genetics of mammalian meiosis: regulation, dynamics and impact on fertility. Nat. Rev. Genet. 11: 124–136 [DOI] [PubMed] [Google Scholar]
- Hassold T., Hunt P., 2001. To err (meiotically) is human: the genesis of human aneuploidy. Nat. Rev. Genet. 2: 280–291 [DOI] [PubMed] [Google Scholar]
- Hemmrich G., Khalturin K., Boehm A. M., Puchert M., Anton-Erxleben F., et al. , 2012. Molecular signatures of the three stem cell lineages in Hydra and the emergence of stem cell function at the base of multicellularity. Mol. Biol. Evol. 29: 3267–3280 [DOI] [PubMed] [Google Scholar]
- Jobb G., von Haeseler A., Strimmer K., 2004. TREEFINDER: a powerful graphical analysis environment for molecular phylogenetics. BMC Evol. Biol. 4: 18. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Katoh K., Misawa K., Kuma K.-I., Miyata T., 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30: 3059–3066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuznetsov S., Lyanguzowa M., Bosch T. C., 2001. Role of epithelial cells and programmed cell death in Hydra spermatogenesis. Zoology 104: 25–31 [DOI] [PubMed] [Google Scholar]
- Lammers J. H., Offenberg H. H., van Aalderen M., Vink A. C., Dietrich A. J., et al. , 1994. The gene encoding a major component of the lateral elements of synaptonemal complexes of the rat is related to X-linked lymphocyte-regulated genes. Mol. Cell. Biol. 14: 1137–1146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le S. Q., Gascuel O., 2008. An improved general amino acid replacement matrix. Mol. Biol. Evol. 7: 1307–1320 [DOI] [PubMed] [Google Scholar]
- MacQueen A. J., Colaiacovo M. P., McDonald K., Villeneuve A. M., 2002. Synapsis-dependent and -independent mechanisms stabilize homolog pairing during meiotic prophase in C. elegans. Genes Dev. 16: 2428–2442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin V. J., Littlefield C. L., Archer W. E., Bode H. R., 1997. Embryogenesis in Hydra. Biol. Bull. 192: 345–363 [DOI] [PubMed] [Google Scholar]
- Matsudaira P., 1987. Sequence from picomole quantities of proteins electroblotted onto polyvinylidene difluoride membranes. J. Biol. Chem. 262: 10035–10038 [PubMed] [Google Scholar]
- Meuwissen R. L., Offenberg H. H., Dietrich A. J., Riesewijk A., van Iersel M., et al. , 1992. A coiled-coil related protein specific for synapsed regions of meiotic prophase chromosomes. EMBO J. 11: 5091–5100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moens P. B., Kolas N. K., Tarsounas M., Marcon E., Cohen P. E., et al. , 2002. The time course and chromosomal localization of recombination-related proteins at meiosis in the mouse are compatible with models that can resolve the early DNA-DNA interactions without reciprocal recombination. J. Cell Sci. 115: 1611–1622 [DOI] [PubMed] [Google Scholar]
- Offenberg H. H., Schalk J. A., Meuwissen R. L., van Aalderen M., Kester H. A., et al. , 1998. SCP2: a major protein component of the axial elements of synaptonemal complexes of the rat. Nucleic Acids Res. 26: 2572–2579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Öllinger R., Alsheimer M., Benavente R., 2005. Mammalian protein SCP1 forms synaptonemal complex-like structures in the absence of meiotic chromosomes. Mol. Biol. Cell 16: 212–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostlund G., Schmitt T., Forslund K., Kostler T., Messina D. N., et al. , 2010. InParanoid 7: New algorithms and tools for eukaryotic orthology analysis. Nucleic Acids Res. 38: D196–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page S. L., Hawley R. S., 2004. The genetics and molecular biology of the synaptonemal complex. Annu. Rev. Cell Dev. Biol. 20: 525–558 [DOI] [PubMed] [Google Scholar]
- Page S. L., Khetani R. S., Lake C. M., Nielsen R. J., Jeffress J. K., et al. , 2008. Corona is required for higher-order assembly of transverse filaments into full-length synaptonemal complex in Drosophila oocytes. PLoS Genet. 4: e1000194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippe H., Derelle R., Lopez P., Pick K., Borchiellini C., et al. , 2009. Phylogenomics revives traditional views on deep animal relationships. Curr. Biol. 19: 706–712 [DOI] [PubMed] [Google Scholar]
- Ronquist F., Teslenko M., van der Mark P., Ayres D. L., Darling A., et al. , 2012. MrBayes 3.2: efficient bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61: 539–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schild-Prüfert K., Saito T. T., Smolikov S., Gu Y., Hincapie M., et al. , 2011. Organization of the synaptonemal complex during meiosis in Caenorhabditis elegans. Genetics 189: 411–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmekel K., Daneholt B., 1998. Evidence for close contact between recombination nodules and the central element of the synaptonemal complex. Chromosome Res. 6: 155–159 [DOI] [PubMed] [Google Scholar]
- Schramm S., Fraune J., Naumann R., Hernandez-Hernandez A., Hoog C., et al. , 2011. A novel mouse synaptonemal complex protein is essential for loading of central element proteins, recombination, and fertility. PLoS Genet. 7: e1002088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievers F., Wilm A., Dineen D., Gibson T. J., Karplus K., et al. , 2011. Fast, scalable generation of high-quality protein multiple sequence alignments using clustal omega. Mol. Syst. Biol. 7: 539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simakov O., Marletaz F., Cho S. J., Edsinger-Gonzales E., Havlak P., et al. , 2013. Insights into bilaterian evolution from three spiralian genomes. Nature 493: 526–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smolikov S., Eizinger A., Schild-Prüfert K., Hurlburt A., McDonald K., et al. , 2007. SYP-3 restricts synaptonemal complex assembly to bridge paired chromosome axes during meiosis in Caenorhabditis elegans. Genetics 176: 2015–2025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smolikov S., Schild-Prüfert K., Colaiacovo M. P., 2009. A yeast two-hybrid screen for SYP-3 interactors identifies SYP-4, a component required for synaptonemal complex assembly and chiasma formation in Caenorhabditis elegans meiosis. PLoS Genet. 5: e1000669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarsounas M., Morita T., Pearlman R. E., Moens P. B., 1999. RAD51 and DMC1 form mixed complexes associated with mouse meiotic chromosome cores and synaptonemal complexes. J. Cell Biol. 147: 207–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Wettstein D., Rasmussen S. W., Holm P. B., 1984. The synaptonemal complex in genetic segregation. Annu. Rev. Genet. 18: 331–411 [DOI] [PubMed] [Google Scholar]
- Wittlieb J., Khalturin K., Lohmann J. U., Anton-Erxleben F., Bosch T. C., 2006. Transgenic Hydra allow in vivo tracking of individual stem cells during morphogenesis. Proc. Natl. Acad. Sci. USA 103: 6208–6211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zacharias H., Anokhin B., Khalturin K., Bosch T. C., 2004. Genome sizes and chromosomes in the basal metazoan Hydra. Zoology 107: 219–227 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.