Abstract
Microalgal biofuels offer great promise in contributing to the growing global demand for alternative sources of renewable energy. However, to make algae-based fuels cost competitive with petroleum, lipid production capabilities of microalgae need to improve substantially. Recent progress in algal genomics, in conjunction with other “omic” approaches, has accelerated the ability to identify metabolic pathways and genes that are potential targets in the development of genetically engineered microalgal strains with optimum lipid content. In this review, we summarize the current bioeconomic status of global biofuel feedstocks with particular reference to the role of “omics” in optimizing sustainable biofuel production. We also provide an overview of the various databases and bioinformatics resources available to gain a more complete understanding of lipid metabolism across algal species, along with the recent contributions of “omic” approaches in the metabolic pathway studies for microalgal biofuel production.
Introduction
In recent years, research initiatives on renewable bioenergy and biofuels have been gaining momentum, not only due to fast depletion of finite reserves of fossil fuels but also because of the associated concerns for the environment and future energy security. Use of biofuels derived from biomass feedstocks is considered to be promising transport fuels to substitute petroleum products (Ravindranath et al., 2011). According to a recent estimate (IEA, 2012) the world production of biofuels grew from 16 billion liters in 2000 to more than 100 billion liters in 2011, which accounted for 3% of the global transport fuels. In the United States between the period 2009 and 2011, the combined production of two principal liquid biofuels viz., ethanol and biodiesel from non-renewable sources, registered an increase of over 30% from 11454 to 14915 million gallons (EIA, 2012). Presently, biofuels contribute about 10 Mtoe (Million Tonnes of Oil Equivalent) to the European Union road transportation of which 80% is biodiesel, mostly derived from rapeseed, and the remaining is bioethanol obtained from wheat, maize, sugarbeet, and sugarcane (EASAC, 2012). Global bioethanol and biodiesel production are projected to the tune of 180 and 42 billion liters in 2021, respectively. While United States, Brazil, and the European Union are expected to remain as top producers of ethanol, the European Union is expected to retain its lead position as the largest producer as well as consumer of biodiesel (OECD-FAO, 2012). Domestic policy incentives aiming at energy security and reduction of CO2 emission, along with mandatory blending targets (varying from 5%–20% of the petroleum fuel) stipulated by many countries are considered to be the driving forces for future higher demand of biofuels. Besides, promotion of biofuel programs by several developing countries to support rural livelihood enhancement programmes, reclaimation of waste land, and local energy security are expected to augment the growth of global biofuel industry. Presently, while much of the first-generation biofuel feedstock come from agricultural crops such as maize, sugarcane, sugarbeet, rapeseed, and oil palm, their large-scale adoption for biofuel production is perceived to have adverse impacts on security and agri-biodiversity (Chisti, 2008; Schenk et al., 2008). On the other hand, second-generation biofuel, including lignocellulosic biomass derived from agricultural residues, and non-food dedicated energy crops grown on marginal lands overcome some of the aforementioned limitations of the first-generation crops (Immerzeel et al., 2013). However, the technologies for second-generation biofuels have not fully matured, and despite several successful pilot-scale operations, they are yet to be available commercially (EASAC, 2012).
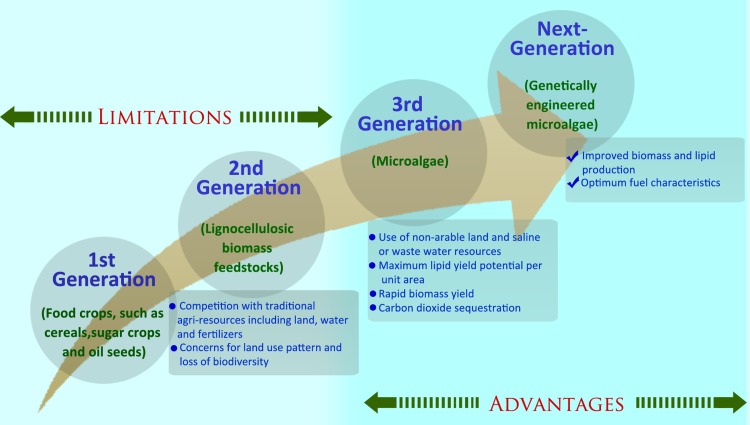
In this context, many recent studies have conclusively advocated the oleaginous fast growing microalgae having multiple advantages over land plants, as the most promising alternative feedstocks for the production of third-, and next-generation biofuels (Chisti, 2007; Lam and Lee, 2012, Mata et al., 2010) (Fig. 1). Algae can be grown on waste land that is unsuitable for cultivation and utilize a wide variety of water sources including waste and saline water, thus mitigating the problem of competition with agricultural resources. Algae efficiently recycle carbon-rich flue emissions and are responsible for more than 40% of global carbon fixation. Algal strains have the potential to accumulate significant amount of neutral lipids, particularly triacylglycerol (TAG), with a magnitude higher than that of potential biofuel crops such as soybean, jatropha, oil palm, and sunflower. In addition to oil accumulation, the rate of microalgal biomass production is quite high as compared to land plants. Furthermore, the low complexity of the cell structure allows easier engineering of algal strains for efficient production of biofuel precursors and other valuable bioactive co-products (Chisti, 2007; Lam and Lee, 2012, Malcata, 2011; Mata et al., 2010; Schenk et al., 2008).
FIG. 1.
Schematic representation of the advantages and limitations of different biofuel feedstock derived from renewable resources.
Notwithstanding these many advantages, the relatively high cost of algal biofuels (US $300–2600 per barrel) compared to petroleum (US $40–80 per barrel) indicates further improvements in technology are required to make it cost-competitive and commercially scalable (Hannon et al., 2010; Pienkos and Darzins, 2009; Singh et al., 2011; Wijffels and Barbosa, 2010). One of the potential solutions to the above problem is to enhance the lipid production capabilities of microalgae by inducing nutrition-deficient conditions in order to channel metabolic fluxes towards lipid biosynthesis. However, several studies have reported that cultivating algal strains under such controlled stress regimes have resulted in impeded cell growth with low lipid productivity (Courchesne et al., 2009). The recent advancements in molecular biology techniques have led to speculation that lipid accumulation in microalgae can alternatively be augmented without applying the aforementioned stress by appropriately modifying their genomes through genetic engineering (Radakovits et al., 2010). Significant progress has been made towards the successful overexpression or knock-out of genes from oleaginous microalgal species with high potential for biofuel production (Courchesne et al., 2009; Hu et al., 2008; Radakovits et al., 2010). While such efforts demonstrate the feasibility of genetic engineering in improving algal biofuels, its effectiveness will be dependent on a deep understanding of the target genes and metabolic pathways responsible for lipid accumulation in microalgae (Khozin-Goldberg and Cohen, 2011).
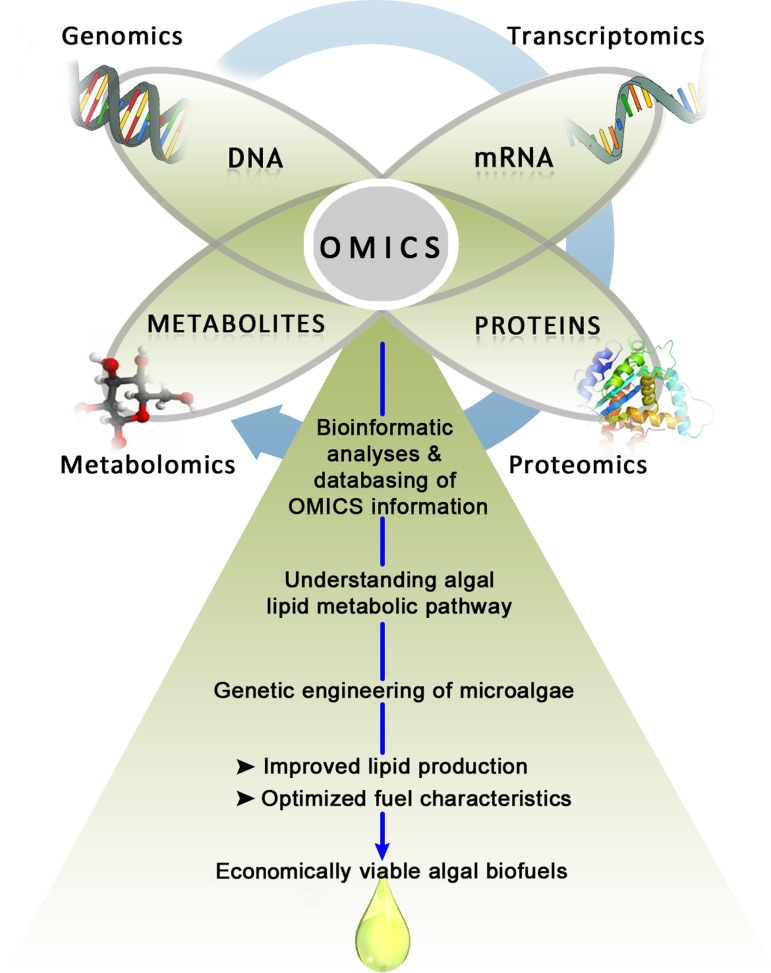
However, compared with to higher plants and other eukaryotes, knowledge of lipid biosynthetic pathways in microalgae remains far from complete (Hu et al., 2008). Access to multiple microalgal genome sequences now provides a wealth of opportunities for application of “omic” approaches to unravel algal lipid metabolism and identify gene targets for the development of potentially engineered strains with optimized lipid content (Fig. 2) (Beer et al., 2009; Georgianna and Mayfield, 2012; Mukhopadhyay et al., 2008; Rodriguez-Moya and Gonzalez, 2010; Yu et al., 2011). The term “omics” in biological sciences widely encompasses genomics, transcriptomics, proteomics, and metabolomics which refers to the study of DNA, mRNA, proteins and metabolites, respectively. While “omics” is now indispensable in the search of novel biomarkers for drug and vaccine development (Hu et al., 2011; Ozdemir et al., 2011), its application to microalgae is just beginning to emerge in the last few years. Furthermore, to date, no review attempt has been undertaken to evaluate the successful integration of “omic” approaches in algal biofuel research. This review is mainly focused on the role of “omics” in improving the development of sustainable biofuel production from microalgae. First, a comprehensive overview of the recent advances in algal genomics is presented, followed by a series of bioinformatics resources available for exploring lipid metabolic pathways in microalgae. Some recent studies employing various “omic” approaches for identification and characterization of putative genes responsible for microalgal biofuel production are also discussed.
FIG. 2.
Schematic overview of the “omic” approaches for improved algal biofuel production.
Update on Sequenced Microalgal Genomes
Significant advances in next-generation sequencing technology have facilitated rapid accumulation of microalgal genomic sequences along with EST (expressed sequence tag) and transcriptome data sets (Grossman, 2005; Radakovits et al., 2010; Tirichine and Bowler, 2011). To date, the whole genome sequences of more than ten microalgae have been generated (Guarnieri et al., 2011; Liu and Benning, 2012;) (Table 1). These includes the Cyanidioschyzon merolae 10D (Matsuzaki et al., 2004), Phaeodactylum tricornutum CCP1055/1 (Bowler et al., 2008), Thalassiosira pseudonana CCMP1335 (Armbrust et al., 2004), Guillardia theta CCMP2712 (Curtis et al., 2012), Chlamydomonas reinhardtii CC-503 (Merchant et al., 2007), Ostreococcus tauri OTH95 (Derelle et al., 2006), Ostreococcus lucimarinus CCE9901 (Palenik et al., 2007), two strains of Micromonas pusilla, RCC299 and CCMP1545 (Worden et al., 2009), Bathycoccus prasinos RCC1105 (Moreau et al., 2012), Volvox carteri UTEX2908 (Prochnik et al., 2010), Chlorella vulgaris NC64A (Blanc et al., 2010), Coccomyxa subellipsoidea C-169 (Blanc et al., 2012), Ectocarpus siliculosus EC32 (Cock et al., 2010), Aureococcus anophagefferens CCMP1984 (Gobler et al., 2011), Nannochloropsis gaditana (Radakovits et al., 2012), and Bigelowiella natans CCMP2755(Curtis et al., 2012). Other algal genomes in the sequencing pipeline are Ostreococcus sp RCC809, Botryococcus braunii Berkeley strain, Dunaliella salina CCAP19/18, Galdieria sulphuraria, Chondrus crispus, Fragilariopsis cylindrus CCMP1102, Pseudo-nitzchia multiseries CLN-47, Emiliana huxleyi CCMP1516 (Radakovits et al., 2010; Tirichine and Bowler, 2011). Several organelle (mitochondria or/and plastid) genomes in microalgae have also been sequenced, including those for D. salina CCAP19/18 (Smith et al., 2010), Botryococcus braunii (Weiss et al., 2010; 2011), Nephroselmis olivaceae (Turmel et al., 1999), Chaetosphaeridium globosum (Turmel et al., 2002), Mesostigma viride (Lemieux et al., 2000), Cyanophora paradoxa (Stirewalt et al., 1995), Cyanidium caldarium (Glockner et al., 2000), Gracilaria tenuistipitata (Hagopian et al., 2004), Porphyra purpurea (Reith and Munholland, 1995), and Odontella sinensis (Kowallik et al., 1995).
Table 1.
An Overview on Genomic Characteristics of Complete and Ongoing Microalgal Sequencing Projects
| Class | Organism | Strain | Organelle | Status | Genome size | No of chromosomes | GC (%) | Protein coding genes | Genes with introns (%) | Mean intron length (bp) | Institution/Reference | Web link |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Chlorophytes | Chlamydomonas reinhardtii | CC-503 | Nucleus | complete | 121Mb | 17 | 64 | 15,143 | 92 | 373 | Merchant et al., 2007 | http://genome.jgi-psf.org/Chlre3/Chlre3.home.html |
| Volvox carteri | UTEX2908 | Nucleus | complete | 138 Mb | 14 | 56 | 14,520 | 92 | 358 | Prochnik et al., 2010 | http://genome.jgi-psf.org/Volca1/Volca1.home.html | |
| Chlorella vulgaris | NC64A | Nucleus | complete | 46.2 Mb | 12 | 67 | 9,791 | 209 | Blanc et al., 2010 | http://genome.jgi-psf.org/ChlNC64A_1/ChlNC64A_1.home.html | ||
| Coccomyxa subellipsoidea | C-169 | Nucleus | complete | 48.8 Mb | 20 | 53 | 9,851 | 5.0 | Blanc et al., 2012 | http://genome.jgi-psf.org/Chlvu1/Chlvu1.home.html | ||
| Dunaliella salina | CCAP19/18 | Plastid | complete | 269Kb | 32.1 | 34 | Smith et al., 2010 | |||||
| Mitochondria | complete | 28.3Kb | 34.4 | 42 | ||||||||
| Nucleus | ongoing | 300Mb | Joint Genome Institute | |||||||||
| Botryococcus braunii | Berkeley | Nucleus | ongoing | 166.2 (±2.2)Mb | 54.4 | Weiss et al., 2010 | ||||||
| Yamanaka | Nucleus | ongoing | 166.0 (±.4)Mb | Weiss et al., 2011 | ||||||||
| Songkla | Nucleus | ongoing | 211.3 (±1.7)Mb | |||||||||
| Nephroselmis olivaceae | Plastid | complete | 200.7Kb | 42.1 | 127 | Turmel et al., 1999 | ||||||
| Mesostigma viride | Plastid | complete | 118.3Kb | 30.1 | 135 | Lemieux et al., 2000 | ||||||
| Prasinophytes | Ostreococcus lucimarinus | CCE9901 | Nucleus | complete | 13.2Mb | 21 | 43 | 7,651 | 187 | Palenik et al., 2007 | http://genome.jgi-psf.org/Ost9901_3/Ost9901_3.home.html | |
| Ostreococcus tauri | OTH95 | Nucleus | complete | 12.6Mb | 20 | 58 | 7,892 | 39 | 126 | Derelle et al., 2006 | http://genome.jgi-psf.org/Ostta4/Ostta4.home.html | |
| Ostreococcus sp | RCC809 | Nucleus | ongoing | 13.6Mb | 7,773 | Joint Genome Institute | http://genomeportal.jgi-psf.org/OstRCC809_2/OstRCC809_2.home.html | |||||
| Micromonas pusilla | RCC299 | Nucleus | complete | 20.9Mb | 17 | 64 | 10,056 | 2.2 | 163 | Worden et al., 2009 | http://genome.jgi.doe.gov/MicpuN3/MicpuN3.home.html | |
| CCMP1545 | Nucleus | complete | 21.9Mb | 19 | 65 | 10,575 | 2.1 | 187 | http://genome.jgi-psf.org/MicpuC2/MicpuC2.home.html | |||
| Bathycoccus prasinos | RCC1105 | Nucleus | complete | 15Mb | 19 | 48 | 7,847 | Moreau et al., 2012 | ||||
| Rhodophytes | Cyanidioschyzon merolae | 10D | Nucleus | complete | 16.5Mb | 20 | 55 | 5,331 | 0.5 | 248 | Matsuzaki et al., 2004 | http://merolae.biol.s.u-tokyo.ac.jp/ |
| Galdieria sulphuraria | Nucleus | ongoing | 15Mb | Michigan State University | http://genomics.msu.edu/galdieria/ | |||||||
| Chondrus crispus | Nucleus | ongoing | Genoscope | http://www.genoscope.cns.fr/spip/ | ||||||||
| Cyanidium caldarium | Plastid | complete | 164.9Kb | 32.7 | 232 | Glockner et al., 2000 | ||||||
| Gracilaria tenuistipitata | Plastid | complete | 183.8Kb | 29.1 | 231 | Hagopian et al., 2004 | ||||||
| Porphyra purpurea | Plastid | complete | 191.0Kb | 33 | 251 | Reith and Munholland, 1995 | ||||||
| Stramenopiles | Phaeodactylum tricornutum | CCP1055/1 | Nucleus | complete | 27.4Mb | 49 | 10,402 | 47 | 123 | Bowler et al., 2008 | http://genome.jgi-psf.org/Phatr2/Phatr2.home.html | |
| Thalassiosira pseudonana | CCMP1335 | Nucleus | complete | 34.5 | 24 | 47 | 11,242 | 992 | Armbrust et al., 2004 | http://genome.jgi-psf.org/thaps1/thaps1.home.html | ||
| Fragilariopsis cylindrus | CCMP1102 | Nucleus | ongoing | 81Mb | Joint Genome Institute | http://genome.jgi-psf.org/Fracy1/Fracy1.home.html | ||||||
| Pseudo-nitzchia multiseries | CLN-47 | Nucleus | ongoing | 218Mb | Joint Genome Institute | http://genome.jgi.doe.gov/Psemu1/Psemu1.home.html | ||||||
| Ectocarpus siliculosus | EC32 | Nucleus | complete | 195.8Mb | 53.6 | 16,256 | 703.8 | Cock et al., 2010 | ||||
| Aureococcus anophagefferens | CCMP1984 | Nucleus | complete | 57Mb | 11,501 | Gobler et al., 2011 | http://genome.jgi.doe.gov/Auran1/Auran1.home.html | |||||
| Nannochloropsis gaditana | Nucleus | complete | 29Mb | 54.2 | 9,052 | 1.62 | Radakovits et al., 2012 | http://nannochloropsis.genomeprojectsolutions-databases.com | ||||
| Odontella sinensis | Plastid | complete | 119.7Kb | 31.8 | 178 | Kowallik et al., 1995 | ||||||
| Cryptophytes | Guillardia theta | CCMP2712 | Nucleus | complete | 87.2Mb | 53 | 24,840 | 80 | 110 | Curtis et al., 2012 | ||
| Chlorarachniophytes | Bigelowiella natans | CCMP2755 | Nucleus | complete | 94.7Mb | 45 | 21,708 | 86 | 184 | Curtis et al., 2012 | ||
| Haptophytes | Emiliana huxleyi | CCMP1516 | Nucleus | ongoing | 167.7Mb | Joint Genome Institute | http://genome.jgi-psf.org/Emihu1/Emihu1.home.html | |||||
| Glaucophytes | Cyanophora paradoxa | Plastid | complete | 135.5Kb | 30.4 | 192 | Stirewalt et al., 1995 |
In addition to the assembled genomic sequences, a cornucopia of ESTs is currently available in public databases to explore the functional characterization of microalgal genomes (Table 2). ESTs represent tags of the expressed region of a genome and are extensively used for identification of protein coding genes, prediction of gene structure, alternate splicing analysis, and characterization of putative SNPs (Nagaraj et al., 2006). There are a number of public resources that store the sequenced ESTs from various algal species. A recently developed TBestDB provides complete information on ESTs obtained from the genomes of Chlamydomonas incerta, Mesostigma viride, Nephroselmis olivaceae, Cyanophora paradoxa, Glaucocystis nostochinearum, Pavlova lutheri, Isochrysis galbana, and Scenedesmus obliquus (O'Brien et al., 2007). Another EST database portal dedicated only for diatom species presently contains 130,000 and 77,000 ESTs (as of November 2008) for P. tricornutum and T. pseudonana, respectively (Maheswari et al., 2005; 2008). In addition to these databases, ESTs for several other microalgae are freely available in the NCBI (National Center for Biotechnology Information) (http://www.ncbi.nlm.nih.gov/dbEST) (Archibald et al., 2003; Bachvaroff et al., 2004; Crepineau et al., 2000; Hackett et al., 2005; Henry et al., 2004; Lluisma and Ragan, 1997; Nikaido et al., 2000; Scala et al., 2002; Wahlund et al., 2004; Weber et al., 2004).
Table 2.
Number of ESTs Available for Various Microalgal Species
| Class | Organism | No of ESTs | Reference |
|---|---|---|---|
| Chlorophytes | Chlamydomonas incerta | 5,124 | O'Brien et al., 2007 |
| Mesostigma viride | 5,615 | O'Brien et al., 2007 | |
| Nephroselmis olivaceae | 126 | O'Brien et al., 2007 | |
| Scenedesmus obliquus | 6,615 | O'Brien et al., 2007 | |
| Acetabularia acetabulum | 941 | Henry et al., 2004 | |
| Stramenopiles | Laminaria digitata | 500 | Crepineau et al., 2000 |
| Phaeodactylum tricornutum | 130,000 | Maheswari et al., 2005; 2008 | |
| Thalassiosira pseudonana | 77,000 | Maheswari et al., 2005; 2008 | |
| Rhodophytes | Porphyra yezoensis | 10,154 | Nikaido et al., 2000 |
| Galdieria sulphuraria | 5,270 | Weber et al., 2004 | |
| Gracilaria gracilis | 200 | Lluisma and Ragan, 1997 | |
| Glaucophytes | Cyanophora paradoxa | 9,867 | O'Brien et al., 2007 |
| Glaucocystis nostochinearum | 8,475 | O'Brien et al., 2007 | |
| Haptophytes | Emiliania huxleyi | 3,000 | Wahlund et al., 2004 |
| Pavlova lutheri | 7,590 | O'Brien et al., 2007 | |
| Isochrysis galbana | 12,205 | O'Brien et al., 2007 | |
| Chlorachniophytes | Bigelowiella natans | 3,472 | Archibald et al., 2003 |
| Dinoflagellates | Amphidinium carterae | 2,143 | Bachvaroff et al., 2004 |
| Lingulodinium polyedrum | 1,012 | Hackett et al., 2005 | |
| Alexandrium tamarense | 6,723 | Hackett et al., 2005 |
It is pertinent to note here that, despite the aforementioned progress in algal genomics, the rationale for sequencing these algal strains was not in the context of their lipid synthesis and hence, there still remains a paucity of biofuel-relevant genomic information (Blankenship, 2010; Rismani-Yazdi et al., 2011). The recently reported endeavours in algal transcriptome profiling have successfully demonstrated the feasibility of bypassing the bottleneck of genome sequencing and have improved our understanding of lipid metabolism in some unsequenced oleaginous microalgal species (Guarnieri et al., 2011; Rismani-Yazdi et al., 2011). Transcriptomics is an excellent tool to quantify the relative abundance of mRNA levels in a single cell or a population of cells. Unlike a genomic approach, it offers a more exhaustive view of the genes that are being actively expressed in response to particular environmental conditions and thus can be employed to identify putative gene targets that can be engineered to augment lipid content in microalgae (Liu and Benning, 2012; Zhang et al., 2010). In this regard, the first de novo transcriptomics study on a potential biofuel algal feedstock, D. tertiolecta, revealed a repertoire of enzymes involved in the biosynthesis and catabolism of fatty acids, TAG, and starch (Rismani-Yazdi et al., 2011). Furthermore, the analyses revealed that the reconstructed metabolic pathways of this particular organism were similar to C. reinhardtii (Hu et al., 2008; Riekhof et al., 2005) and higher plants (Durrett et al., 2008), and demonstrated its inherent genetic ability of linking starch metabolism with ethanol fermentation through the glycolysis pathway.
Subsequently, the same approach was employed to unravel the TAG biosynthetic pathway in one more unsequenced oleaginous microalga, C. variabilis UTEX395 under Nitrogen-replete and -deplete conditions (Guarnieri et al., 2011). In addition to these studies, a plethora of transcriptome information is readily available for the model alga, C. reinhardtii under various genetic and physiological stress (Boyle et al., 2012; Castruita et al., 2011; Fang et al., 2012; Gonzalez-Ballester et al., 2010; Miller et al., 2010). For example, Boyle et al. (2012) conducted genome-wide expression analysis to explore the molecular mechanisms underlying the induction of TAG accumulation in C. reinhardtii. They identified three vital acyltransferase genes including DGAT1, DGATT1 and PDAT1 to be likely responsible for TAG accumulation. Interestingly, they also observed that following N-deprivation, Chlamydomonas cells switch their metabolism from converting acetate to glucose into a more direct incorporation of acetate into fatty acids by downregulating glyoxylate cycle activity and gluconeogenesis. Likewise, Miller et al. (2010) confirmed the direct involvement of NRR1 and PDAT genes in nitrogen assimilation and TAG accumulation in C. reinhardtii. Taken together, these analyses corroborate the significance of transcriptome sequencing as an emerging powerful approach for identification of genes responsible for algal biofuel production.
Databases and Other Bioinformatics Resources for Studying Lipid Metabolic Pathways in Microalgae
The growing compendium of genome sequences for an increasing number of organisms have resulted in a concomitant need for the development of bioinformatics resources that will not only serve as a knowledge base of various metabolic pathways but also facilitate comparative genomic analysis for functional annotation and interpretation of newly identified genes at proteomic and metabolic levels. Metabolic pathways represent a programmed sequence of molecular events underlying a specific biological activity. As discussed earlier, elucidation of biological pathways, in particular lipid metabolic pathways is useful for genetic engineering efforts directed towards augmenting lipid accumulation in microalgae (Georgianna and Mayfield, 2012; Khozin-Goldberg and Cohen, 2011; Yu et al., 2011). While several such databases specific for lipid pathway analyses have been constructed for plants (Beisson et al., 2003; Caspi et al., 2008; Child et al., 2012; Duvick et al., 2007; Mao et al., 2009; Mekhedov et al., 2000; Sucaet and Deva, 2011), very few to date are available for the study of microalgae (Lopez et al., 2011; May et al., 2009). It is likely that the dearth of algal genomic data as compared to higher plants has contributed to the present status of limited pathway databases and web resources. However, integration of plant lipid pathway databases in comparative genomic analyses might reveal the homologous enzymes responsible for lipid biosynthesis and regulation in microalgae (Beisson et al., 2003; Misra et al., 2012). In this section, we provide a brief overview of the various databases and bioinformatics tools with particular reference to explore lipid metabolic pathways in microalgae, along with few selected plant-specific databases relevant for microalgal biofuel research.
KEGG (Kyoto Encyclopedia of Genes and Genomes) (http://www.genome.jp/kegg/) is one of the most widely used comprehensive resource of metabolic pathways including for several organisms (Kanehisa et al., 2010). The database also provides reference pathways that serve as a framework for construction of organism-independent biochemical pathways from the user input whole genome. To further enrich the KEGG pathway information, KEGG ortholog prediction tool is offered to categorize orthologous and paralogous gene groups from evolutionarily related organisms. Recent genome-wide studies have employed KEGG pathway database to identify genes and reconstruct major lipid biosynthetic pathways in various oleaginous microalgal species (Hashimoto et al., 2008; Misra et al., 2012; Rismani-Yazdi et al., 2011; Smith et al., 2012). MetaCyc (http://metacyc.org) is also a popular database that contains experimentally verified metabolic pathway information for more than 600 organisms including species ranging from lower microorganisms, to plants and human (Caspi et al., 2008). MetaCyc is also suitable as a reference database to enrich functional characterization of newly sequenced algal putative genes involved in lipid biosynthesis.
ChlamyCyc is the only available algae-specific web-based database (http://chlamycyc.mpimp-golm.mpg.de), specialized for the comprehensive analyses of metabolic pathways and fundamental cellular processes in C. reinhardtii (May et al., 2009). It was developed as a part of the GoFORSYS (German Systems Biology research initiative) (http://www.goforsys.de) project and currently provides curated experimental information on a total of 253 pathways, 2851 enzymes, 1419 enzymatic reactions, 1416 compounds, and 928 literature citations. The web-portal also houses the known and predicted biochemical pathways from other widely recognized pathway databases such as PlantGDB (Duvick et al., 2007), PlnTFDB (Riano-Pachon et al., 2007), Quantprime (Arvidsson et al., 2008), ProMEX (Hummel et al., 2007), and MapMan (Usadel et al., 2005). To facilitate phylogenomic analyses, Inparanoid (Remm et al., 2001) and OrthoMCL-DB (Chen et al., 2006) databases are also integrated into the tool. Further, to contribute towards the functional annotation of uncharacterized genes and gene products, a web version of the standard BLAST software (Altschul et al., 1990) is provided within the ChlamyCyc.
Another web-based analysis suite known as Algal Functional Annotation Tool (http://pathways.mcdb.ucla.edu) has been recently developed for functional annotation of multiple algal genome sequences (Lopez et al., 2011). Currently the tool provides annotation for C. reinhardtii, the model green microalgae, with a provision to include additional algal genomes. Annotation includes assigning pathways, ontology, and protein family terms to the predicted proteins by integrating several bioinformatics resources including KEGG (Kanehisa et al., 2010), MetaCyc (Caspi et al., 2008), Pfam (Finn et al., 2010), Reactome (Matthews et al., 2009), Panther (Thomas et al., 2003), Gene Ontology (Ashburner et al., 2000), InterPro (Hunter et al., 2009), MapMan Ontology (Thimm et al., 2004), and KOG (Tatusov et al., 2003). High-throughput gene expression data for various environmental conditions is also provided along with an integrated search tool to identify the functionally related genes. Additionally, the tool enables dynamic visualization of genes on KEGG pathway maps.
Although research over the past few years has led to significant advancement in our understanding of lipid metabolism in higher plants, much remains to be learned about these processes in microalgae. Nevertheless, as the core metabolic pathways are presumed to be conserved in microalgae and higher plant species, particularly in the model plant Arabidopsis thaliana (Hu et al., 2008), the homologous sequences and the corresponding pathways responsible for lipid accumulation in microalgae can be accurately inferred to the greatest extent using the metabolic pathway information of plants as reference. Towards this end, the Arabidopsis Lipid Gene (ALG) database (http://www.plantbiology.msu.edu/lipids/genesurvey/index.html) that provides a comprehensive catalog of the Arabidopsis lipid biosynthetic genes will be highly beneficial (Beisson et al, 2003). The ALG database is an updated version of the previously determined repository of plant lipid biosynthetic genes (http://www.canr.msu.edu/lgc) (Mekhedov et al., 2000), and currently includes 600 encoded proteins that have been classified according to biological function, subcellular localization, and alternate splicing. The database uses TargetP (Emanuelsson et al., 2000) for subcellular localization prediction. Additionally, information of on 3700 ESTs stored in the database will certainly assist in designing gene silencing or disruption experiments. The potential of ALG database has been successfully demonstrated in several comparative genomic studies where the putative genes involved in lipid biosynthesis in Arabidopsis were used to identify corresponding homologous genes involved in TAG biosynthesis in several oleaginous microalgal and plant species for biofuel implications (Li et al., 2010; Misra et al., 2012; Sharma and Chauhan, 2012). Other databases developed to facilitate genome-wide investigations in plant biofuel feedstock species are the BFGR (Biofuel Feedstock Genomic Resource) and pDAWG. BFGR database (http://bfgr.plantbiology.msu.edu) provides high-quality uniform and integrated functional annotation of genes as well as transcripts sequences from 54 lignocellulosic biofuel feedstock species (Childs et al., 2012). pDAWG (http://csbl1.bmb.uga.edu/pDAWG/) is a prototype of a one-stop shop database that offers complete information on plant cell wall genes/proteins, notably information related to phylogenomic, sequence and structure-function (Mao et al., 2009). Presently the database comprises complete genomes of 7 higher plants and 12 microalgal species, including 6 green algae (O. lucimarinus, O. tauri, C. reinhardtii, V. carteri, M. pusilla strain CCMP1545, and M. pusilla strain RCC299), 2 diatoms (T. pseudonana and P. tricornutum), 2 Stramenopiles (Phytophthora ramorum and P. sojae), brown tide algae (A. anophagefferens), and red algae (C. merolae). The above databases, although designed to expedite the process of functional annotation of genes putatively involved in plant lipid biosynthesis, additionally provide a platform for performing comparative genomic analyses in order to identify corresponding homologous genes of algal lipid metabolism. Further, they also serve as ideal templates to develop and enrich the relatively less explored algae lipid pathway databases and web resources.
Recent Application of “Omics” to Identify Genes and Metabolic Pathways Involved in Microalgal Biofuel Production
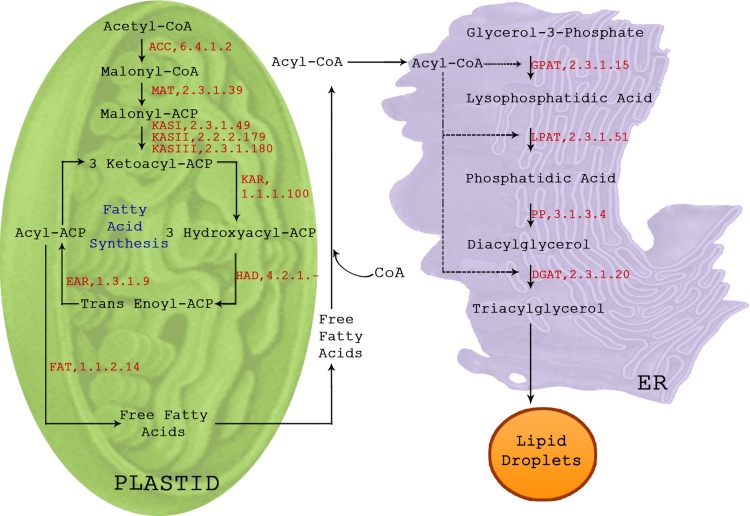
The global synthesis pathway of TAG begins with the basic fatty acid precursor, acetyl-CoA, and continues through fatty acid biosynthesis, complex lipid assembly, and saturated fatty acid modification processes until TAG bodies are finally formed. A simplified overview of TAG biosynthetic pathway is shown as Figure 3. Acetyl-CoA carboxylase (ACC) is the initial rate-limiting enzyme that catalyzes carboxylation of acetyl-CoA to produce malonyl-CoA in both eukaryotes and prokaryotes (Sasaki and Nagano, 2004). Two physically distinct types of ACC enzymes have been reported to occur in nature. Plastidic heteromeric ACC composed of four subunits, including biotin carboxyl carrier protein (BCCP), biotin carboxylase (BC), and α, β monomers of carboxyltransferase (CT), is common among prokaryotes. In contrast, the eukaryotic cytosolic homomeric ACC is composed of a single polypeptide with each of these subunits functioning as distinct domains (Cronan and Waldrop, 2002; Huerlimann and Heimann, 2012). Once malonyl-CoA is synthesized, the malonyl group is transferred by malonyl-CoA:ACP transacylase to the acyl carrier protein (ACP) of the fatty acid synthase (FAS) multienzyme complex in order to form malonyl-ACP. Fatty acid biosynthesis is catalyzed by two types of FAS. The type I FAS as found in vertebrates, yeast, and some bacteria contain all the active sites on one or two multifunctional polypeptide, and in type II FAS of many bacteria and plants, the active site resides in discrete gene products. The β-ketoacyl-ACP synthase (KAS) components of the type II FAS enzyme complex carry out the condensation steps of the process. FabH (KAS III) catalyzes the first elongation step of fatty acid biosynthetic process using acetyl-CoA and malonyl-ACP as substrates to produce acetoacetyl-ACP. The cycle is completed by the sequential action of β-ketoacyl-ACP reductase (KAR), β-hydroxyacyl-ACP dehydratases (HAD), and enoyl-ACP reductase (ENR) to form acyl-ACP. The subsequent two carbon condensation steps are further carried over by FabB (KAS I) and FabF (KAS II), producing palmitic (C16:0) or a stearic acid (C18:0) as the final products (Gonzalez-Mellado et al., 2010; Li et al., 2009).
FIG. 3.
Schematic overview of triacylglycerol biosynthetic pathway in microalgae.
Completion of the de novo fatty acid synthesis is accomplished in one of three ways. Either the newly synthesized fatty acid is hydrolyzed by fatty acyl-ACP thioesterase (FAT) enzymes, further modified by desaturases, or directly transferred to complex lipid formation using plastid acyltransferases (prokaryotic lipid pathway). Free fatty acids thus produced from plastid eventually enters smooth endoplasmic reticulum (ER) for further elongation, modification, or participation in the synthesis of membrane lipids or storage TAGs (eukaryotic lipid pathway). TAG biosynthetic pathway involves sequential esterification of glycerol chain by a class of acyltransferases. The initial esterification at the sn-1 position of glycerol-3-phosphate is catalyzed by glycerol-3-phosphate acyltransferases (GPAT), followed by esterification at the sn-2 position by lysophosphatidic acid acyltransferase (LPAT) to form phosphatidic acid, subsequently dephosphorylated by phosphatidic acid phosphatase (PAP), and finally esterification at the sn-3 position by diacylglycerol acyltransferase (DGAT) to form TAG (for an extensive review of plant and microalgal lipid biosynthesis, refer to Courchesne et al., 2009; Hu et al., 2008; Ohlrogge and Browse, 1995).
The use of high-throughput methods to evaluate gene expression and availability of whole genome sequences in public domain has immensely facilitated characterization of genes and enzymes underpinning various crucial metabolic pathways of microalgae. However, the current understanding of the underlying molecular mechanisms responsible for lipid accumulation in microalgae is yet inadequate as compared to higher plants (Hu et al., 2008).
Systems biology represents the ultimate approach for a thorough understanding of metabolic pathways by harmoniously integrating the various “omic” platforms. Instead of focusing on individual genes, proteins, or metabolites one at a time, it takes into account the interconnections between these elements during regulation of a biological activity (Rodriguez-Moya and Gonzalez, 2010). Recently, several studies have successfully implemented “omic” approaches in parallel to identify the differently expressed genes and enzymes underpinning the metabolic pathways that are likely to be involved in algal lipid accumulation (Table 3). These genes and gene products represent the most potential targets for metabolic pathway reconstruction in order to develop engineered microorganisms with desired fuel-grade properties (Guarnieri et al., 2011; Lei et al., 2012; Misra et al., 2012; Nguyen et al., 2011; Radakovits et al., 2012; Riekhof et al., 2005; Rismani-Yazdi et al., 2011; Smith et al., 2012; Valenzuela et al., 2012).
Table 3.
Recent Application of Integrated “Omics” Approaches to Identify Genes Involved in Lipid Accumulation for Microalgal Biofuel Production
| Organism | Objective | Key results | Reference |
|---|---|---|---|
| Nannochloropsis gaditana | To present a draft genome sequence and a genetic transformation method for the marine microalga N. gaditana | Genes required for glycerolipid biogenesis were identified in addition to genes specific to N. gaditana genome and those conserved across stramenopile lineage. | Radakovits et al., 2012 |
| Chlamydomonas reinhardtii | To annotate glycerolipid biosynthesis genes based on similarity to homologs from Arabidopsis thaliana | Reconstruction of the major glycerolipid biosynthetic pathways. A novel BTA1Cr gene encoding a Chlamydomonas Betaine lipid synthase was identified and isolated. | Riekhof et al., 2005 |
| C. reinhardtii | To analyze purified oil bodies by using proteomics approach | LC-MS/MS analysis identified 248 proteins of which 33 were putatively involved in lipid metabolism, particularly acyl- lipids and sterol. Besides the gene already known to regulate TAG biosynthesis in C. reinhardtii, 19 novel genes were predicted including acyltransferases, hydrolase/lipases, lipid trafficking proteins and proteins of sterol/ergosterol metabolism. | Nguyen et al., 2011 |
| Cyanidioschyzon merolae | To study lipid biosynthetic pathway by genomic and biochemical analyses | Genes involved in the synthesis of fatty acids and lipids were identified. Genomic analysis indicated that C. merolae possess a standard pathway of biosynthesis of glycerolipids except for the absence of plant-type galactosyltransferase enzymes, which underlines the marked difference in lipid biosynthesis pathway between red algae and the green lineage. | Sato and Moriyama, 2007 |
| Haematococcus pluvialis | To study the correlation between fatty acid synthesis and gene expression patterns under different stress conditions | Good correlation between ACP, KAS, FATA gene expression and fatty acid synthesis was observed. In response to stress treatment, the fatty acid synthesis was upregulated while there was no change in the quality of the synthesized fatty acids. | Lei et al., 2012 |
| Diatom species including Thalassiosira pseudonana, Phaeodactylum tricornutum and Fragilariopsis cylindrus | To perform comprehensive in silico evaluation of the genes underlying the carbon metabolism pathways of diatoms | The study identified a total of 164 genes including 51 genes from T. pseudonana, 55 from P. tricornutum and 58 from F. cylindrus. Among them, the enzyme PFK was indicated to play a pivotal role in carbon metabolism and thus a potential target for genetic engineering to enhance biofuel production in microalgae. Further, subcellular localization of the putative enzymes demonstrated the lower half and upper half of glycolysis to be conserved in mitochondria and diatoms, respectively. The results highlight the significant role of intracellular trafficking in the regulation of carbon flux. | Smith et al., 2012 |
| P. tricornutum | To study gene expression profiles during nutrient-deprivation and lipid-accumulation conditions | Genes/proteins up- and downregulated were identified. The potential role of carbon fixation pathways in lipid accumulation is also described. | Valenzuela et al., 2012 |
| Chlorella | To study the distribution, domain structure, evolution and expressivity of autophagy genes (ATG) using genome-wide comparative analysis | The Chlorella genome was observed to encode all crucial ATG genes, which was further corroborated by expression profiles of the putative genes as determined by RT-PCR analysis. However, the receptor protein in cytoplasm-to-vacuole targeting and mitophagy were found to be absent in microalgae. | Jiang et al., 2012 |
| Eukaryotic genomes (fifty-six species) | To examine desaturases and elongases by phylogenetics and motif structure prediction approaches | The predicted 275 desaturases and 265 elongases homologs were characterized into 4 and 2 distinct subfamilies, respectively. The resulting Hidden Markov Model profile can be applied to other comparative genomic studies for functional annotation of novel genes. | Hashimoto et al., 2008 |
| Seven microalgal species including C. reinhardtii, T. pseudonana, P. tricornutum, C. merolae, Ostreococcus lucimarinus and O. tauri | To characterize the fatty acid desaturases on the basis of conserved histidine-rich motifs and phylogenetic profiles | 67 desaturase genes were predicted and annotated along with reconstruction of the pathways involved in unsaturated fatty acid biosynthesis. A novel microsomal Δ12 desaturase was identified. | Chi et al., 2008 |
| Dunaliella tertiolecta | To compare transcriptome and proteome of D. tertiolecta grown under N-replete and N-deplete conditions and high salt concentrations. | Several overexpressed genes and enzymes possibly involved in biosynthesis and catabolism of fatty acids, TAG and starch were discovered and the relevant pathways were reconstructed. Additionally, the pathways associated with biofuel precursor production were found to be interlinked to each other. | Rismani-Yazdi et al., 2011 |
| Chlorella vulgaris UTEX 395 | To demonstrate the utility of a transcriptome as a search model for proteomic analysis in an unsequenced microalga | Various genes and gene products involved in TAG biosynthetic pathway were identified. | Guarnieri et al., 2011 |
| Five microalgal species including C. reinhardtii, Volvox carteri, O.lucimarinus, O. tauri and C. merolae | To perform comparative genomic analysis including subcellular localization, physicochemical characterization, exon-intron pattern, motif/ domain predictions of 398 putative lipid genes involved in lipid biosynthesis | The results indicated that although each of the algal species maintains the basic genomic repertoire required for lipid biosynthesis, they possess additional lineage-specific gene groups and the core lipid metabolic pathways in all the studied species are carried out by a comparable number of orthologous proteins. | Misra et al., 2012 |
| Seven microalgal species including C. reinhardtii, V. carteri, O. lucimarinus, O. tauri, C. merolae, P. tricornutum, T. pseudonana | To perform sequence-structure analyses of plastid-located glycerol-3-phosphate acyltransferase (GPAT), a vital enzyme of TAG biosynthetic pathway | The phylogenetic tree along with motif and gene structure analyses indicated a close evolutionary relationship between red algae/diatom and green algae/plant lineages. Further subtle variation in amino acids of fatty acyl binding site were identified that might influence the substrate selectivity of GPAT. | Misra et al., 2013 |
Despite these commendable efforts, it is important to note that many of these studies were focused on the model organism C. reinhardtii or on single algal strains that are not potential biofuel feedstock (Fan et al., 2011). Thus, species representing different clades will probably have to be further explored in order to gain a comprehensive understanding of microalgal lipid metabolism. In addition, comparative genome-wide analyses of algal species showing variations in their fatty acid composition and accumulation will unveil the target genes and underpinning evolutionary processes responsible for the different phenotypes. Furthermore, a comparative analysis of lipid biosynthetic pathways in microalgae and higher plants is another promising area that is relatively underexploited. Although the core fundamental pathways are found to be largely conserved, recent molecular and biochemical studies have reported striking dissimilarities regarding the biosynthetic origin of TAG between microalgae and plants (Fan et al., 2011; Liu and Benning, 2012). In view of these observations, genome data of microalgae is beginning to be mined using phylogenomics approach to determine the variation in gene contents and differential subcellular localization of TAG synthesis between algal species and higher plants such as Arabidopsis, thereby resolving several fundamental questions on algal evolution (Misra et al., 2012; 2013; Sato and Moriyama, 2007).
The ever increasing accumulation of genomic data has changed our perspective of the complexity of pathways involved in microalgal biofuel precursor production. Although the TAG biosynthetic pathway still appears to be the principal lipid metabolic process, it is now clear that other carbon fixation pathways such as autophagy comes into play and eventually signifies alternative pathways for lipid accumulation. Whereas considerable information is available on processes involving metabolic fluxes towards lipid accumulation, less is known about the autophagy genes and corresponding enzymes responsible for carbon fixation in microalgae. Only one such analysis has been recently performed for a green microalga, Chlorella (Jiang et al., 2012). Hence, a thorough knowledge of these additional routes and their precise role in biofuel precursor production will be useful.
The majority of the omic-based studies undertaken so far have primarily addressed identification of gene targets for improving lipid production in microalgae Now it is apparent that modification of the fatty acid profile to include more stearic acid (C18:0) and oleic acid (C18:1) is also indispensable for improving the algal-derived biofuel properties (Knothe, 2009). As a fundamental step in their direction, in silico studies aimed at prediction of candidate genes whose combination determines the inherent fatty ester composition of microalgae has been reported (Chi et al., 2008; Hashimoto et al., 2008). These studies conclusively highlight the potential utility of the emerging “omic” approaches in providing greater insights into genome structure and lipid metabolism of various microalgal species.
Conclusion
A comprehensive understanding of metabolic pathways that direct carbon flux towards lipid accumulation is central in establishing genetic engineering strategies for optimizing biofuel production in microalgae. Several recent studies have employed omic-based approaches to unravel the fundamental genetic and cellular processes involved in the synthesis of biofuel precursors from diverse species of algae. Despite these efforts, several challenges still remain to be addressed to make algal-derived fuels cost-competitive with petroleum. For instance, a majority of the genomic studies has been reported for microalgal species of low oil content, particularly for the model microalga C. reinhardtii. This is perhaps due to the paucity of biofuel-relevant genomic data for other oleaginous microalgae. Therefore, access to more algal genomes is required to facilitate identification of novel genes and gene products in the metabolic pathways that could be appropriately harnessed for optimum biofuel production. Undoubtedly, with the progress in genome sequencing and “omics” technologies, appropriate bioinformatics resources including databases must also be made available to organize, visualize, and logically interpret the large datasets. Integration of complementary “omic” approaches is envisaged to provide the much needed insights into the algal lipid metabolism for making biofuel production commercially scalable.
Acknowledgments
The authors gratefully acknowledge the valuable suggestions of the reviewers for improvement of the manuscript. This work was partially funded by the Department of Biotechnology, Government of India. N.M. acknowledges the support of Council for Scientific and Industrial Research, India for granting Senior Research Fellowship. The authors also thank Director, CSIR-IMMT for his support and providing laboratory facilities.
Author Disclosure Statement
The authors declare that no conflicting financial interests exist.
References
- Altschul SF. Gish W. Miller W. Myers EW. Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Archibald JM. Rogers MB. Toop M. Ishida K. Keeling PJ. Lateral gene transfer and the evolution of plastid-targeted proteins in the secondary plastid containing alga Bigelowiella natans. Proc Natl Acad Sci USA. 2003;100:7678–7683. doi: 10.1073/pnas.1230951100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armbrust EV. Berges JA. Bowler C, et al. The genome of the diatom Thalassiosira pseudonana: Ecology, evolution and metabolism. Science. 2004;306:79–86. doi: 10.1126/science.1101156. [DOI] [PubMed] [Google Scholar]
- Arvidsson S. Kwasniewski M. Riano-Pachon DM. Mueller-Roeber B. QuantPrime—A flexible tool for reliable high-throughput primer design for quantitative PCR. BMC Bioinformatics. 2008;9:465–480. doi: 10.1186/1471-2105-9-465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M. Ball CA. Blake JA, et al. Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachvaroff TR. Concepcion GT. Rogers CR. Herman EM. Delwiche CF. Dinoflagellate expressed sequence tag data indicate massive transfer of chloroplast genes to the nuclear genome. Protist. 2004;155:65–78. doi: 10.1078/1434461000165. [DOI] [PubMed] [Google Scholar]
- Beer LL. Boyd ES. Peters JW. Posewitz MC. Engineering algae for bio hydrogen and biofuel production. Curr Opin Biotechnol. 2009;20:264–271. doi: 10.1016/j.copbio.2009.06.002. [DOI] [PubMed] [Google Scholar]
- Beisson F. Koo AJK. Ruuska S, et al. Arabidopsis genes involved in acyl lipid metabolism. A 2003 census of the candidates, a study of the distribution of expressed sequence tags in organs, and a web-based database. Plant Physiol. 2003;132:682–697. doi: 10.1104/pp.103.022988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc G. Agarkova I. Grimwood J, et al. The genome of the polar eukaryotic microalga Coccomyxa subellipsoidea reveals traits of cold adaptation. Genome Biol. 2012;13:R39–R51. doi: 10.1186/gb-2012-13-5-r39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc G. Duncan G. Agarkova I, et al. The Chlorella variabilis NC64A reveals adaptation to photosymbiosis, coevolution with viruses, and cryptic sex. Plant Cell. 2010;22:2943–2955. doi: 10.1105/tpc.110.076406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blankenship RE. Early evolution of photosynthesis. Plant Physiol. 2010;154:434–438. doi: 10.1104/pp.110.161687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowler C. Allen AE. Badger JH, et al. The Phaeodactylum genome reveals the evolutionary history of diatom genomes. Nature. 2008;465:239–244. doi: 10.1038/nature07410. [DOI] [PubMed] [Google Scholar]
- Boyle NR. Page MD. Liu B, et al. Three acyltransferases and a nitrogen responsive regulator are implicated in nitrogen starvation-induced triacylglycerol accumulation in Chlamydomonas. J Biol Chem. 2012;287:15811–15825. doi: 10.1074/jbc.M111.334052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspi R. Foerster H. Fulcher CA, et al. The MetaCyc Database of metabolic pathways and enzymes and the BioCyc collection of Pathway/Genome Databases. Nucleic Acids Res. 2008;36:D623–D631. doi: 10.1093/nar/gkm900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castruita M. Casero D. Karpowicz SJ, et al. Systems biology approach in Chlamydomonas reveals connections between copper nutrition and multiple metabolic steps. Plant Cell. 2011;23:1273–1292. doi: 10.1105/tpc.111.084400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F. Mackey AJ. Stoeckert CJ., Jr Roos DS. OrthoMCL-DB: Querying a comprehensive multi-species collection of ortholog groups. Nucleic Acids Res. 2006;34:D363–368. doi: 10.1093/nar/gkj123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi X. Zhang X. Guan X. Ding L. Li Y. Wang M, et al. Fatty acid biosynthesis in eukaryotic photosynthetic microalgae: Identification of a microsomal delta 12 desaturase in Chlamydomonas reinhardtii. J Microbiol. 2008;46:189–201. doi: 10.1007/s12275-007-0223-3. [DOI] [PubMed] [Google Scholar]
- Childs KL. Konganti K. Buell CR. The Biofuel Feedstock Genomics Resources: A web-based portal and database to enable functional genomics of plant biofuel feedstock species. Database Article ID bar061. 2012:1–9. doi: 10.1093/database/bar061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chisti Y. Biodiesel from microalgae. Biotechnol Adv. 2007;25:294–306. doi: 10.1016/j.biotechadv.2007.02.001. [DOI] [PubMed] [Google Scholar]
- Chisti Y. Biodiesel from microalgae beats bioethanol. Trends Biotechnol. 2008;26:126–131. doi: 10.1016/j.tibtech.2007.12.002. [DOI] [PubMed] [Google Scholar]
- Cock JM. Sterck L. Rouze P, et al. The Ectocarpus genome and the independent evolution of multicellularity in brown algae. Nature. 2010;465:617–621. doi: 10.1038/nature09016. [DOI] [PubMed] [Google Scholar]
- Courchesne NM. Parisien A. Wang B. Lan CQ. Enhancement of lipid production using biochemical, genetic and transcription factor engineering approaches. J Biotechnol. 2009;141:31–41. doi: 10.1016/j.jbiotec.2009.02.018. [DOI] [PubMed] [Google Scholar]
- Crepineau F. Roscoe T. Kaas R. Kloareg B. Boyen C. Characterisation of complementary DNAs from the expressed sequence tag analysis of life cycle stages of Laminaria digitata (Phaeophyceae) Plant Mol Biol. 2000;43:503–513. doi: 10.1023/a:1006489920808. [DOI] [PubMed] [Google Scholar]
- Cronan JE. Waldrop GL. Multi-subunit acetyl-CoA carboxylase. Prog Lipid Res. 2002;41:407–435. doi: 10.1016/s0163-7827(02)00007-3. [DOI] [PubMed] [Google Scholar]
- Curtis BA. Tanifuji G. Burki F, et al. Algal genomes reveal evolutionary mosaicism and the fate of nucleomorphs. Nature. 2012;492:59–65. doi: 10.1038/nature11681. [DOI] [PubMed] [Google Scholar]
- Derelle E. Ferraz C. Rombaut S, et al. Genome analysis of the smallest free-living eukaryote Ostreococcus tauri unveils many unique features. Proc Natl Acad Sci USA. 2006;103:11647–11652. doi: 10.1073/pnas.0604795103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durrett TP. Benning C. Ohlrogge J. Plant triacylglycerols as feedstocks for the production of biofuels. Plant J. 2008;54:593–607. doi: 10.1111/j.1365-313X.2008.03442.x. [DOI] [PubMed] [Google Scholar]
- Duvick J. Fu A. Muppirala U, et al. PlantGDB: A resource for comparative plant genomics. Nucleic Acids Res. 2007;36:D959–D965. doi: 10.1093/nar/gkm1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- EASAC. The current status of biofuels in the European Union, their environmental impacts and future prospects. EASAC policy report. 2012:19. [Google Scholar]
- EIA. Washington: 2012. Biofuels Issues and Trends. [Google Scholar]
- Emanuelsson O. Nielsen H. Brunak S. von Heijne G. Predicting subcellular localization of proteins based on their N-terminal amino acid sequences. J Mol Biol. 2000;300:1005–1016. doi: 10.1006/jmbi.2000.3903. [DOI] [PubMed] [Google Scholar]
- Fan J. Andre C. Xu C. A chloroplast pathway for the de novo biosynthesis of triacylglycerol in Chlamydomonas reinhardtii. FEBS Lett. 2011;585:1985–1991. doi: 10.1016/j.febslet.2011.05.018. [DOI] [PubMed] [Google Scholar]
- Fang W. Si Y. Douglass S, et al. Transcriptome-wide changes in Chlamydomonas reinhardtii gene expression regulated by carbon dioxide and the CO2-concentrating mechanism regulator CIA5/CCM1. Plant Cell. 2012;24:1876–1893. doi: 10.1105/tpc.112.097949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RD. Mistry J. Tate J, et al. The Pfam protein families database. Nucleic Acids Res. 2010;38:D211–D222. doi: 10.1093/nar/gkp985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgianna DR. Mayfield SP. Exploiting diversity and synthetic biology for the production of algal biofuels. Nature. 2012;488:329–335. doi: 10.1038/nature11479. [DOI] [PubMed] [Google Scholar]
- Glockner G. Rosenthal A. Valentine K. The structure and gene repertoire of an ancient red algal plastid genome. J Mol Evol. 2000;51:382–390. doi: 10.1007/s002390010101. [DOI] [PubMed] [Google Scholar]
- Gobler CJ. Berry DL. Dyhrman ST, et al. Niche of harmful alga Aureococcus anophagefferens revealed through ecogenomics. Proc Natl Acad Sci USA. 2011;108:4352–4357. doi: 10.1073/pnas.1016106108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Ballester D. Casero D. Cokus S. Pellegrini M. Merchant SS. Grossmann AR. RNA-seq analysis of sulfur-deprived Chlamydomonas cells reveals aspects of acclimation critical for cell survival. Plant Cell. 2010;22:2058–2084. doi: 10.1105/tpc.109.071167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Mellado D. von Wettstein-Knowles P. Garces R. Martinez-Force E. The role of β-ketoacyl-Acyl carrier protein synthase III in the condensation steps of fatty acid biosynthesis in sunflower. Planta. 2010;231:1277–1289. doi: 10.1007/s00425-010-1131-z. [DOI] [PubMed] [Google Scholar]
- Grossman AR. Paths towards agal genomics. Plant Physiol. 2005;137:410–427. doi: 10.1104/pp.104.053447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarnieri MT. Nag A. Smolinski SL. Darzins A. Seibert M. Pienkos PT. Examination of triacylglycerol biosynthetic pathways via de novo transcriptomic and proteomic analyses in an unsequenced microalgae. PLoS ONE. 2011;6:1–13. doi: 10.1371/journal.pone.0025851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackett JD. Scheetz TE. Yoon HS, et al. Insights into a dinoflagellate genome through expressed sequence tag analysis. BMC Genomics. 2005;6:80–93. doi: 10.1186/1471-2164-6-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagopian JC. Reis M. Kitajima JP. Bhattacharya D. de Oliveira MC. Comparative analysis of the complete plastid genome sequence of the red alga Gracilaria tenuistipitata var. liui provides insights into the rvolution of rhodoplasts and their relationship to other plastids. J Mol Evol. 2004;59:464–477. doi: 10.1007/s00239-004-2638-3. [DOI] [PubMed] [Google Scholar]
- Hannon M. Gimpel J. Tran M. Rasala B. Mayfield S. Biofuels from algae: Challenges and potential. Biofuels. 2010;1:763–784. doi: 10.4155/bfs.10.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto K. Yoshizawa AC. Okuda S. Kuma K. Goto S. Kanehisa M. The repertoire of desaturases and elongases reveals fatty acid variations in 56 eukaryotic genomes. J Lipid Res. 2008;49:183–191. doi: 10.1194/jlr.M700377-JLR200. [DOI] [PubMed] [Google Scholar]
- Henry IM. Wilkinson MD. Hernandez JM. Schwarz-Sommer Z. Grotewold E. Mandoli DF. Comparison of ESTs from juvenile and adult phases of the giant unicellular green alga Acetabularia acetabulum. BMC Plant Biol. 2004;4:3–18. doi: 10.1186/1471-2229-4-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Q. Sommerfeld M. Jarvis E, et al. Microalgal triacylglycerols as feedstocks for biofuel production: Perspectives and advances. Plant J. 2008;54:621–639. doi: 10.1111/j.1365-313X.2008.03492.x. [DOI] [PubMed] [Google Scholar]
- Hu ZZ. Huang H. Wu CH, et al. Omics-based molecular target and biomarker identification. Methods Mol Biol. 2011;719:547–571. doi: 10.1007/978-1-61779-027-0_26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huerlimann R. Heimann K. Comprehensive guide to acetyl-carboxylases in algae. Crit Rev Biotechnol. 2012:1–17. doi: 10.3109/07388551.2012.668671. [DOI] [PubMed] [Google Scholar]
- Hummel J. Niemann M. Wienkoop S, et al. ProMEX: A mass spectral reference database for proteins and protein phosphorylation sites. BMC Bioinformatics. 2007;8:216–224. doi: 10.1186/1471-2105-8-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter S. Apweiler R. Attwood TK, et al. InterPro: The integrative protein signature database. Nucleic Acids Res. 2009;37:D211–D215. doi: 10.1093/nar/gkn785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- IEA. Paris, France: 2012. Tracking Clean Energy Progress: Energy Technology Perspectives 2012 excerpt as IEA input to the Clean Energy Ministerial. [Google Scholar]
- Jiang Q. Zhao L. Dai J. Wu Q. Analysis of autophagy genes in microalgae: Chlorella as a potential model to study mechanism of autophagy. PLoS ONE. 2012;7:1–16. doi: 10.1371/journal.pone.0041826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M. Goto S. Furumichi M. Tanabe M. Hirakawa M. KEGG for representation and analysis of molecular networks involving diseases and drugs. Nucleic Acids Res. 2010;38:D355–D360. doi: 10.1093/nar/gkp896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khozin-Goldberg I. Cohen Z. Unraveling algal lipid metabolism: Recent advances in gene identification. Biochimie. 2011;93:91–100. doi: 10.1016/j.biochi.2010.07.020. [DOI] [PubMed] [Google Scholar]
- Knothe G. Improving biodiesel fuel properties by modifying fatty ester composition. Energ Environ Sci. 2009;2:759–766. [Google Scholar]
- Kowallik KV. Stoebe B. Schaffran I. Kroth-Pancic P. Freier U. The chloroplast genome of a chlorophyll a+c containing alga, Odontella sinensis. Plant Mol Biol Rep. 1995;13:336–342. [Google Scholar]
- Lam MK. Lee KT. Microalgae biofuels: A critical review of issues, problems and the way forward. Biotechnol Adv. 2012;30:673–690. doi: 10.1016/j.biotechadv.2011.11.008. [DOI] [PubMed] [Google Scholar]
- Lei A. Chen H. Shen G. Hu Z. Chen L. Wang J. Expression of fatty acid synthesis genes and fatty acid accumulation in Haematococcus pluvialis under different stressors. Biotechnol Biofuels. 2012;5:18–29. doi: 10.1186/1754-6834-5-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemieux C. Otis C. Turmel M. Ancestral chloroplast genome in Mesostigma viride reveals an early branch of green plant evolution. Nature. 2000;403:649–652. doi: 10.1038/35001059. [DOI] [PubMed] [Google Scholar]
- Li L. Li H. Li J, et al. A genome-wide survey of maize lipid-related genes: Candidate genes mining, digital gene expression profiling and co-location with QTL for maize kernel oil. Sci China Life Sci. 2010;53:690–700. doi: 10.1007/s11427-010-4007-3. [DOI] [PubMed] [Google Scholar]
- Li MJ. Li AQ. Xia H, et al. Cloning and sequence analysis of putative type II fatty acid synthase genes from Arachis hypogaea. J Biosci. 2009;34:227–238. doi: 10.1007/s12038-009-0027-1. [DOI] [PubMed] [Google Scholar]
- Liu B. Benning C. Lipid metabolism in microalgae distinguishes itself. Curr Opin Biotechnol. 2012;24:1–10. doi: 10.1016/j.copbio.2012.08.008. [DOI] [PubMed] [Google Scholar]
- Lluisma AO. Ragan MA. Expressed sequence tags (ESTs) from the marine red alga Gracilaria gracilis. J Appl Phycol. 1997;9:287–293. [Google Scholar]
- Lopez D. Casero D. Cokus SJ. Merchant SS. Pellegrini M. Algal Functional Annotation Tool: A web-based analysis suite to functionally interpret large gene lists using integrated annotation and expression data. BMC Bioinformatics. 2011;12:282–292. doi: 10.1186/1471-2105-12-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maheswari U. Mock T. Armbrust EV. Bowler C. Update of the Diatom EST Database: A new tool for digital transcriptomics. Nucleic Acids Res. 2008;37:D1001–D1005. doi: 10.1093/nar/gkn905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maheswari U. Montsant A. Goll J, et al. Nucleic Acids Res. 2005;33:D344–D347. doi: 10.1093/nar/gki121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malcata FX. Microalgae and biofuels: A promising partnership? Trends Biotechnol. 2011;29:542–549. doi: 10.1016/j.tibtech.2011.05.005. [DOI] [PubMed] [Google Scholar]
- Mao F. Yin Y. Zhou F, et al. pDAWG: An integrated database for plant cell wall genes. Bioenerg Res. 2009;2:209–216. [Google Scholar]
- Mata TM. Martins AA. Caetano NS. Microalgae for biodiesel production and other applications: A review. Renew Sust Energ Rev. 2010;14:217–232. [Google Scholar]
- Matsuzaki M. Misumi O. Shin-i T, et al. Genome sequence of the ultrasmall unicellular red alga Cyanidioschyzon merolae 10D. Nature. 2004;428:653–657. doi: 10.1038/nature02398. [DOI] [PubMed] [Google Scholar]
- Matthews L. Gopinath G. Gillespie M, et al. Reactome knowledgebase of human biological pathways and processes. Nucleic Acids Res. 2009;7:D619–D622. doi: 10.1093/nar/gkn863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May P. Christian JO. Kempa S. Walther D. ChlamyCyc: An integrative systems biology database and web-portal for Chlamydomonas reinhardtii. BMC Genomics. 2009;10:209–220. doi: 10.1186/1471-2164-10-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mekhedov S. de Ilarduya OM. Ohlrogge J. Toward a functional catalog of the plant genome. A survey of genes for lipid biosynthesis. Plant Physiol. 2000;122:389–402. doi: 10.1104/pp.122.2.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merchant SS. Prochnik SE. Vallon O, et al. The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science. 2007;318:245–250. doi: 10.1126/science.1143609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller R. Wu G. Deshpande RR, et al. Changes in transcript abundance in Chlamydomonas reinhardtii following nitrogen deprivation predict diversion of metabolism. Plant Physiol. 2010;154:1737–1752. doi: 10.1104/pp.110.165159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misra N. Panda PK. In search of actionable targets for agrigenomics and microalgal biofuel production: Sequence-structural diversity studies on algal and higher plants with a focus on GPAT protein. OMICS J Integr Biol. 2013;17:173–186. doi: 10.1089/omi.2012.0094. [DOI] [PubMed] [Google Scholar]
- Misra N. Panda PK. Parida BK. Mishra BK. Phylogenomic study of lipid genes involved in microalgal biofuel production—Candidate gene mining and metabolic pathway analyses. Evol Bioinform. 2012;8:545–564. doi: 10.4137/EBO.S10159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreau H. Verhelst B. Couloux A, et al. Gene functionalities and genome structure in Bathycoccus prasinos reflect cellular specializations at the base of the green lineage. Genome Biol. 2012;13:R74–R90. doi: 10.1186/gb-2012-13-8-r74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay A. Redding AM. Rutherford BJ. Keasling JD. Importance of systems biology in engineering microbes for biofuel production. Curr Opin Biotechnol. 2008;19:228–234. doi: 10.1016/j.copbio.2008.05.003. [DOI] [PubMed] [Google Scholar]
- Nagaraj SH. Gasser RB. Ranganathan S. A hitchhiker's guide to expressed sequence tag (EST) analysis. Brief Bioinform. 2006;8:6–21. doi: 10.1093/bib/bbl015. [DOI] [PubMed] [Google Scholar]
- Nguyen HM. Baudet M. Cuine S, et al. Proteomic profiling of oil bodies isolated from the unicellular green microalga Chlamydomonas reinhardtii: With focus on proteins involved in lipid metabolism. Proteomics. 2011;11:4266–4273. doi: 10.1002/pmic.201100114. [DOI] [PubMed] [Google Scholar]
- Nikaido I. Asamizu E. Nakajima M. Nakamura Y. Saga N. Tabata S. Generation of 10,154 expressed sequence tags from a leafy gametophyte of a marine red alga, Porphyra yezoensis. DNA Res. 2000;7:223–227. doi: 10.1093/dnares/7.3.223. [DOI] [PubMed] [Google Scholar]
- O'Brien E. Koski LB. Zhang Y, et al. TBestDB: A taxonomically broad database of expressed sequence tags (ESTs) Nucleic Acids Res. 2007;35:D445–D451. doi: 10.1093/nar/gkl770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- OECD–FAO. OECD-FAO Agricultural Outlook 2012–2021. 2012. http://dx.doi.org/10.1787/agr_outlook-2012-en. [May 21;13 ]. http://dx.doi.org/10.1787/agr_outlook-2012-en
- Ohlrogge J. Browse J. Lipid biosynthesis. Plant Cell. 1995;7:957–970. doi: 10.1105/tpc.7.7.957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozdemir V. Pang T. Knoppers BM, et al. Vaccines of the 21st century and vaccinomics: Data-enabled science meets global health to spark collective action for vaccine innovation. OMICS J Integr Biol. 2011;15:523–527. doi: 10.1089/omi.2011.03ed. [DOI] [PubMed] [Google Scholar]
- Palenik B. Grimwood J. Aerts A, et al. The tiny eukaryote Ostreococcus provides genomic insights into the paradox of plankton speciation. Proc Natl Acad Sci USA. 2007;104:7705–7710. doi: 10.1073/pnas.0611046104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pienkos PT. Darzins A. The promise and challenges of microalgal-derived biofuels. Biofuel Biprod Bior. 2009;3:431–440. [Google Scholar]
- Prochnik SE. Umen J. Nedelcu AM, et al. Genomic analysis of complexity in the multicellular green alga Volvox carteri. Science. 2010;329:223–226. doi: 10.1126/science.1188800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radakovits R. Jinkerson RE. Darzins A. Posewitz MC. Genetic engineering of algae for enhanced biofuel production. Eukaryot Cell. 2010;9:486–501. doi: 10.1128/EC.00364-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radakovits R. Jinkerson RE. Fuerstenberg SI, et al. Draft genome sequence and genetic transformation of the oleaginous alga: Nannochloropsis gaditana. Nat Commun. 2012;3:686–711. doi: 10.1038/ncomms1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravindranath NH. Lakshmi CS. Manuvie R. Blachandra P. Biofuel production and implications for land use, food production and environment in India. Energ Policy. 2011;39:5737–5745. [Google Scholar]
- Reith M. Munholland J. Complete nucleotide sequence of the Porphyra purpurea chloroplast genome. Plant Mol Biol Rep. 1995;13:333–335. [Google Scholar]
- Remm M. Storm CE. Sonnhammer EL. Automatic clustering of orthologs and in-paralogs from pairwise species comparisons. J Mol Biol. 2001;314:1041–1052. doi: 10.1006/jmbi.2000.5197. [DOI] [PubMed] [Google Scholar]
- Riano-Pachon DM. Ruzicic S. Dreyer I. Mueller-Roeber B. PlnTFDB: An integrative plant transcription factor database. BMC Bioinformatics. 2007;8:42–52. doi: 10.1186/1471-2105-8-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riekhof WR. Sears BB. Benning C. Annotation of genes involved in glycerolipid biosynthesis in Chlamydomonas reinhardtii: Discovery of the betaine lipid synthase BTA1Cr. Eukaryot Cell. 2005;4:242–252. doi: 10.1128/EC.4.2.242-252.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rismani-Yazdi H. Haznedaroglu BZ. Bibby K. Peccia J. Transcriptome sequencing and annotation of the microalgae Dunaliella tertiolecta: Pathway description and gene discovery for production of next-generation biofuels. BMC Genomics. 2011;12:148–165. doi: 10.1186/1471-2164-12-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Moya M. Gonzalez R. Systems biology approaches for the microbial production of biofuels. Biofuels. 2010;1:291–310. [Google Scholar]
- Sasaki Y. Nagano Y. Plant acetyl-CoA carboxylase: Structure, biosynthesis, regulation and gene manipulation for plant breeding. Biosci Biotechnol Biochem. 2004;68:1175–1184. doi: 10.1271/bbb.68.1175. [DOI] [PubMed] [Google Scholar]
- Sato N. Moriyama T. Genomic and biochemical analysis of lipid biosynthesis in the unicellular rhodophyte Cyanidioschyzon merolae: Lack of a plastidic desaturation pathway results in the coupled pathway of galactolipid synthesis. Eukaryot Cell. 2007;6:1006–1017. doi: 10.1128/EC.00393-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scala S. Carels N. Falciatore A. Chiusano ML. Bowler C. Genome properties of the diatom Phaeodactylum tricornutum. Plant Physiol. 2002;129:993–1002. doi: 10.1104/pp.010713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenk PM. Thomas-Hall SR. Stephens E, et al. Second generation biofuels: High-efficiency microalgae for biodiesel production. Bioenergy Res. 2008;1:20–43. [Google Scholar]
- Sharma A. Chauhan R. In Silico identification and comparative genomics of candidate genes involved in biosynthesis and accumulation of seed oil in plants. Comp Funct Genom Article ID. 2012;914843:1–14. doi: 10.1155/2012/914843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A. Nigam PS. Murphy JD. Mechanism and challenges in commercialisation of algal biofuels. Bioresour Technol. 2011;102:26–34. doi: 10.1016/j.biortech.2010.06.057. [DOI] [PubMed] [Google Scholar]
- Smith DR. Lee RW. Cushman JC. Magnuson JK. Tran D. Polle JEW. The Dunaliella salina organelle genomes: Large sequences, inflated with intronic and intergenic DNA. BMC Plant Biol. 2010;10:83–97. doi: 10.1186/1471-2229-10-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SR. Abbriano RM. Hildebrand M. Comparative analysis of diatom genomes reveals substantial differences in the organization of carbon portioning pathways. Algal Res. 2012;1:2–16. [Google Scholar]
- Stirewalt VL. Michalowski CB. Loffelhardt W. Bohnert HJ. Bryant DA. Nucleotide sequence of the cyanelle genome from Cyanophora paradoxa. Plant Mol Biol Rep. 1995;13:327–332. [Google Scholar]
- Sucaet Y. Deva T. Evolution and application of pathway resources and databases. Brief Bioinform. 2011;12:530–544. doi: 10.1093/bib/bbq083. [DOI] [PubMed] [Google Scholar]
- Tatusov RL. Fedorova ND. Jackson JD, et al. The COG database: An updated version includes eukaryotes. BMC Bioinformatics. 2003;4:41–55. doi: 10.1186/1471-2105-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thimm O. Blasing O. Gibon Y, et al. MAPMAN: A user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J. 2004;37:914–939. doi: 10.1111/j.1365-313x.2004.02016.x. [DOI] [PubMed] [Google Scholar]
- Thomas PD. Campbell MJ. Kejariwal A, et al. PANTHER: A library of protein families and sub families indexed by function. Genome Res. 2003;13:2129–2141. doi: 10.1101/gr.772403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirichine L. Bowler C. Decoding algal genomes: Tracing back the history of photosynthetic life on earth. Plant J. 2011;66:45–57. doi: 10.1111/j.1365-313X.2011.04540.x. [DOI] [PubMed] [Google Scholar]
- Turmel M. Otis C. Lemieux C. The complete chloroplast DNA sequence of the green alga Nephroselmis olivaceae: Insights into the architecture of ancestral chloroplast genomes. Proc Natl Acad Sci USA. 1999;96:10248–10253. doi: 10.1073/pnas.96.18.10248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turmel M. Otis C. Lemieux C. The chloroplast and mitochondrial genome sequences of the chlorophyte Chaetosphaeridium globosum: Insights into the timing of the events that restructured organelle DNAs within the green algal lineage that led to land plants. Proc Natl Acad Sci USA. 2002;99:11275–11280. doi: 10.1073/pnas.162203299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usadel B. Nagel A. Thimm O, et al. Extension of the visualization tool MapMan to allow statistical analysis of arrays, display of corresponding genes, and comparison with known responses. Plant Physiol. 2005;138:1195–1204. doi: 10.1104/pp.105.060459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valenzuela J. Mazurie A. Carlson RP, et al. Potential role of multiple carbon fixation pathways during lipid accumulation in Phaeodactylum tricornutum. Biotechnol Biofuels. 2012;5:40–57. doi: 10.1186/1754-6834-5-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahlund TM. Hadaegh AR. Clark R. Nguyen B. Fanelli M. Read BA. Analysis of expressed sequence tags from calcifying cells of marine coccolithophorid (Emiliania huxleyi) Mar Biotechnol. 2004;6:278–290. doi: 10.1007/s10126-003-0035-3. [DOI] [PubMed] [Google Scholar]
- Weber A. Oesterhelt C. Gross W, et al. EST-analysis of the thermo-acidophilic red microalga Galdieria sulphuraria reveals potential for lipid A biosynthesis and unveils the pathway of carbon export from rhodoplasts. Plant Mol Biol. 2004;55:17–32. doi: 10.1007/s11103-004-0376-y. [DOI] [PubMed] [Google Scholar]
- Weiss TL. Johnston JS. Fujisawa K. Okada S. Devarenne TP. Genome size and phylogenetic analysis of the A and L races of Botryococcus braunii. J Appl Phycol. 2011;23:833–839. [Google Scholar]
- Weiss TL. Johnston JS. Fujisawa K, et al. Phylogenetic placement, genome size, and GC content of the liquid-hydrocarbon-producing green microalga Botryococcus braunii strain Berkeley (SHOWA) Chlorophyta. J Phycol. 2010;46:534–540. [Google Scholar]
- Wijffels RH. Barbosa M. An outlook on microalgal biofuels. Science. 2010;329:796–799. doi: 10.1126/science.1189003. [DOI] [PubMed] [Google Scholar]
- Worden AZ. Lee JH. Mock T, et al. Evolution and dynamic adaptations revealed by genomes of the marine picoeukaryotes micromonas. Science. 2009;324:268–272. doi: 10.1126/science.1167222. [DOI] [PubMed] [Google Scholar]
- Yu WL. Ansari W. Schoepp NG. Hannon MJ. Mayfield SP. Burkart MD. Modifications of the metabolic pathways of lipid and triacylglycerol production in microalgae. Microb Cell Fact. 2011;10:91–102. doi: 10.1186/1475-2859-10-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W. Li F. Nie L. Integrating multiple ‘omics’ analysis for microbial biology: Application and methodologies. Microbiology. 2010;156:287–301. doi: 10.1099/mic.0.034793-0. [DOI] [PubMed] [Google Scholar]