Abstract
Deep mitochondrial divergence within species may result from cryptic speciation, from phylogeographic isolation or from endosymbiotic bacteria like Wolbachia that manipulate host reproduction. Phengaris butterflies are social parasites that spend most of their life in close relationship with ants. Previously, cryptic speciation has been hypothesised for two Phengaris species based on divergent mtDNA sequences. Since Phengaris species are highly endangered, the existence of cryptic species would have drastic consequences for conservation and management. We tested for cryptic speciation and alternative scenarios in P. teleius and P. nausithous based on a comprehensive sample across their Palaearctic ranges using COI gene sequences, nuclear microsatellites and tests for Wolbachia. In both species a deep mitochondrial split occurring 0.65–1.97 myrs ago was observed that did not correspond with microsatellite data but was concordant with Wolbachia infection. Haplotypes previously attributed to cryptic species were part of the Wolbachia-infected clades. In both species remaining phylogeographic structure was largely consistent between mitochondrial and nuclear genomes. In P. teleius several mitochondrial and nuclear groups were observed in East Asia while a single haplogroup and nuclear cluster prevailed across continental Eurasia. Neutrality tests suggested rapid demographic expansion into that area. In contrast, P. nausithous had several mitochondrial and nuclear groups in Europe, suggesting a complex phylogeographic history in the western part of the species range. We conclude that deep intraspecific divergences found in DNA barcode studies do not necessarily need to represent cryptic speciation but instead can be due to both infection by Wolbachia and phylogeographic structure.
Introduction
Cryptic species, i.e. the presence of phylogenetically distinct units within a morphologically defined taxon [1], are a common phenomenon among all animal taxa and biogeographical regions. They can seriously confuse taxonomy based solely on morphological characters [2]. More importantly, cryptic speciation affects our understanding of biodiversity and its conservation [1]. Arthropods are expected to contain many cryptic species and in particular parasitic species seem to have a higher evolutionary potential than free-living species and are potential candidates for cryptic speciation [3]. DNA barcoding using the mitochondrial gene Cytochrome c Oxidase I (COI) has become a standard method to assign unknown individuals to species, to assess biodiversity, and to discover new species including cryptic units within well-defined morphospecies [4], [5]. For some of the cryptic units it has additionally been shown that they correspond well with a divergent ecological niche [6].
However, the sole use of mtDNA sequences as a tool for species detection and delimitation can be problematic [7], [8]. Patterns of deep divergence of mitochondrial DNA sequences within species may be due to historical processes like introgression between species [9], or phylogeographic isolation [10]. Furthermore, in invertebrates the assumption of neutral evolution of mtDNA may not be met due to the presence of endosymbiotic bacteria [11], [12], [13]. The common microbial endoparasite Wolbachia often manipulates the reproductive system of its host thus enhancing its own transmission to the next generation [11], [14]. When a Wolbachia infection has no detrimental fitness effects on the host [15], [16] and the host has no Wolbachia suppressing elements [17], the infection can spread through whole populations and species to fixation [17], [18]. As a consequence of the maternal inheritance of the infection this may lead to a selective sweep and fixation of the mitochondrial haplotype of infected individuals [15]. However, a number of empirical studies have found that selfish genetic elements like Wolbachia are maintained within populations at relatively low frequencies [17], [19]. Under which conditions Wolbachia persists at low frequency, thus maintaining mitochondrial polymorphism, is less clear as fixation frequency depends on various factors like reproductive fitness effects, population size and structure, infection and transmission frequency, bacterial density and/or phage presence [15], [20], [21], [22]. Furthermore, fitness effects of Wolbachia on host individuals can be conditional on environmental factors [23], [24]. An important consequence of Wolbachia infection is its influence on mtDNA patterns which may seriously undermine the power of DNA barcoding for species detection. It can either mask species diversity due to mtDNA introgression between species [18], [25]. Or, in contrast, it can promote high mtDNA divergence due to long lasting reproductive isolation between infected and uninfected lineages and may even lead to the formation of new species [26]. Wolbachia may also become lost because of inefficient transmission [27] which may further complicate the interpretation. Hence, to assess whether an observed mtDNA haplotype pattern was the result of Wolbachia infection, additional analyses are needed including tests for the presence of Wolbachia and the use of additional nuclear markers [9], [28], [29].
Species with a parasitic lifestyle are both potential candidates for cryptic speciation [3] and particularly prone to be horizontally infected by endoparasites like Wolbachia [30]. Butterflies of the genus Phengaris Doherty, 1891 (formerly Maculinea van Eecke, 1915; Lepidoptera: Lycaenidae) exhibit a parasitic phase within their life cycle [31]. In their last larval stage caterpillars show numerous evolutionary adaptations to an intricate nest parasitism of Myrmica ant species [32]. Phengaris are rare and threatened species listed in the European Habitats Directive and of high importance for nature conservation [33]. Thus, cryptic biodiversity could potentially have impacts on the evaluation of their vulnerability and conservational status and on management strategies. In Europe, populations have suffered from local extinctions for decades, mainly because of changes in local farming practices of grasslands, the main habitat of the species [34]. Evidence for a high infection rate with endoparasites exists for two Phengaris species. In populations of P. alcon from Poland and Lithuania as well as P. arion from Poland and Italy all screened samples turned out to be infected with Wolbachia strains of supergroup B and A, respectively [35], [36]. Out of the total of 11 Wolbachia supergroups these two are currently the only ones known to occur in butterflies, where supergroup B is prevalent [37]. In the genus Phengaris 12 species are currently recognized [38], [39]. The two co-occurring, closely related species studied in detail here, Phengaris teleius (Bergsträsser, [1779]) and Phengaris nausithous (Bergsträsser, [1779]), share Sanguisorba officinalis (Rosaceae) as their only foodplant. P. teleius is morphologically variable and a number of subspecies have been described from Asia [40]. In contrast in P. nausithous most authors only recognize the nominate form (but see Rákosy et al. [41] who recognize subspecies kijevensis Sheljuzhko, 1928 in Eastern Europe). Both species have wide and overlapping distribution areas in temperate regions of the Palaearctic [42]. Phylogenetic hypotheses of the genus based on morphological, ecological [43] as well as molecular evidence [38], [44], [45] have been formulated. Cryptic speciation has explicitly been hypothesized for P. nausithous [44], P. teleius [45] or both [38] based on a limited number of divergent sequences. Alternatively, however, high sequence variation and the large morphological variability of P. teleius could be a result of its phylogeographic history, e.g. by isolation in different pleistocene refugia, or may involve endosymbiotic bacteria. However, a comprehensive phylogenetic and phylogeographic analysis that also considers the potential contribution of Wolbachia infection is still lacking.
Here we present a phylogenetic analysis of Phengaris teleius and P. nausithous based on a comprehensive sample across their Palaearctic ranges. We use COI sequences of mitochondrial DNA as well as nuclear microsatellite markers to test for cryptic speciation. Furthermore, we test whether distinct lineages can be explained by an association with Wolbachia infections. Finally, we use the data to analyse phylogeographic patterns of P. teleius and P. nausithous. Our study thus provides further insights into postglacial movement patterns of Palaearctic insect species.
Materials and Methods
Sampling
Phengaris teleius (Bergsträsser, [1779]) including the subspecies sinalcon Murayama, 1992 [46], obscurata Staudinger, 1892 [47], euphemia Staudinger, 1887 [48], hosonoi Takahasi, 1973 [49], kazamoto Druce, 1875 [50], ogumae Matsumura, 1910 [51] and daisensis Matsumura, 1926 [52] and Phengaris nausithous (Bergsträsser, [1779]) were sampled throughout their distribution ranges from 44 and 36 populations, respectively (Fig. 1, Table S1). Note that P. teleius and P. nausithous co-occurred in 19 populations. Up to 10 individuals were sampled per species and location. Hand netted adults (N = 110) were killed with potassium cyanide and kept either in glassine envelopes or in 99.8% ethanol. Caterpillars (N = 149) taken from the food plant Sanguisorba officinalis were conserved in ethanol. We could not assess sex ratio as sample sizes per population were too low and more than half of the specimens were larvae, for which sex could not be assessed. Collection permits were obtained from Struktur- und Genehmigungsdirektion Nord (Koblenz, Germany), Regierungspräsidium Leipzig (Germany), Regierung von Unterfranken (Würzburg, Germany), Thüringer Landesverwaltungsamt (Weimar, Germany), and specimen collectors’ own collection permits, if required in respective countries.
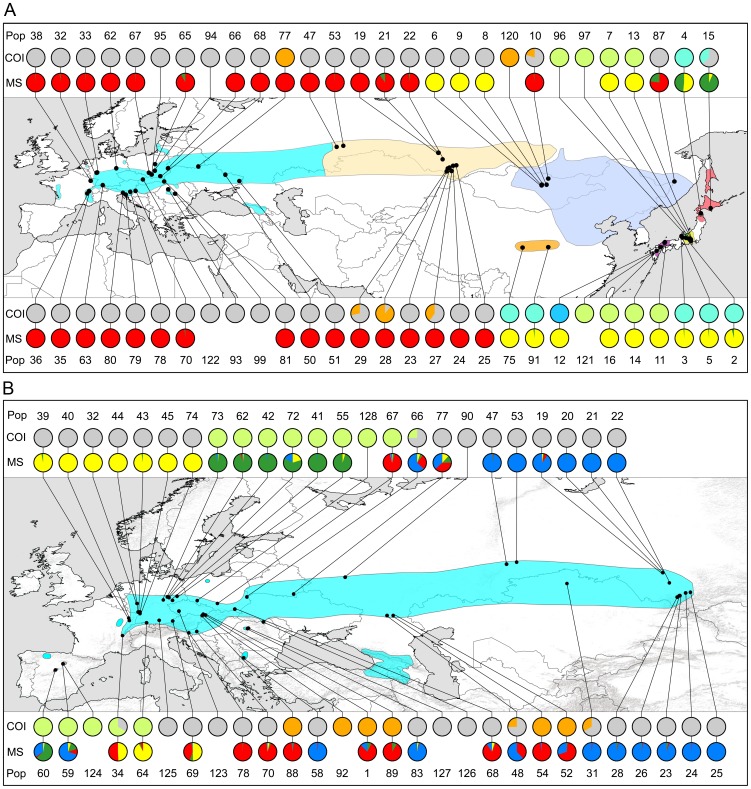
Figure 1. Distribution range, sampling sites and genetic structure at mitochondrial and nuclear genomes.
COI = cytochrome oxidase I (for details see Figure 2) and MS = microsatellites (for details see Figure 4) for Phengaris teleius (A) and P. nausithous (B). For details on populations, see Table S1. Distribution ranges are based on records (Europe: [42], [105]; Asia: [106], [107], [108], [109] and expert knowledge (Asia: personal communication Kosterin, Wang). Shading color corresponds to subspecific affiliation of P. teleius according to Table S1 (cyan: nominate species, peach: P. t. obscurata, blue: P. t. euphemia, orange: P. t. sinalcon, pink: P. t. ogumae, yellow: P. t. kazamoto, green: P. t. hosonoi, violet: P. t. daisensis).
DNA Barcoding and Tests for Wolbachia Infection
Total genomic DNA was extracted using the QIAGEN DNeasy Blood & Tissue Kit (QIAGEN, Hilden, Germany), following the manufacturer’s instructions. Two fragments of the COI gene were amplified using the primer combinations: LCO – Nancy and Tonya – Hobbes [45]. PCR was performed in 20 µl reactions, containing 0.5 pmol of each primer, 200 µM dNTPs, PCR buffer, 1.875 mM MgCl2, and 0.8 units Fermentas Taq DNA polymerase (Fermentas, Leon-Rot, Germany). The thermocycler protocol was: denaturation at 95°C (2 min) followed by 37 cycles of 95°C (1 min), 47°C (1 min) and 72°C (1.5 min), and a subsequent final extension step at 72°C (10 min). PCR-products were directly cycle-sequenced using the ABI BigDye Terminater v3.1 cycle sequencing Kit using the same primers. Products were sequenced on an Applied Biosystems 3130×l Genetic Analyzer (Applied Biosystems, Foster City, USA). About 10% of the fragments were treated with a multiple tube approach and 20% of the fragments were sequenced in both directions which did not show any mismatches. Sequences were obtained for 147 samples of P. teleius and 112 samples of P. nausithous. GenBank accession numbers for all concatenated sequences are provided in Table S1.
We tested all individuals for infection with Wolbachia performing two independent PCR screens for the Wolbachia surface protein (wsp) following Zhou et al. [53]. PCR products were visualized on 1.5% agarose gels and scored for the presence of Wolbachia infections (Table S1). The wsp-genes of Wolbachia endosymbionts of 8 P. teleius and 3 P. nausithous were sequenced to determine allele and supergroup correspondence, using the Wolbachia wsp Database [54] and BLAST.
mtDNA Sequence Analysis
COI fragments of 147 and 112 individuals of P. teleius and P. nausithous, respectively, were manually concatenated and aligned with BioEdit [55]. To avoid the inclusion of mitochondrial pseudogenes [56], translated amino acid sequences were tested for substitutions and stop-codons using the program MEGA 5 [57]. In both P. teleius and P. nausithous ten non-synonymous substitutions were found leading to a change in the amino acid sequence. However, we did not regard these substitutions as indicative for a pseudogene because the mutations occurred in parts of the protein known for their high amino acid variability [58], [59] or because substitutions led to amino acids of similar characteristics. Additionally, we added all published COI/COII sequences of P. teleius and P. nausithous available from GenBank as of 1 June 2013 (Table S1). These also included all publicly available barcode sequences from the barcode of life database (BOLD; [60]). Note that the test for Wolbachia infection could not be performed for sequences retrieved from GenBank. As outgroup taxa sequences of further Phengaris species (P. arion (Linnaeus, 1758), P. alcon ([Denis & Schiffermüller], 1775), P. albida Leech, 1893, P. atroguttata (Oberthür, 1876), P. daitozana Wileman, 1908) were taken from GenBank (Table S1).
A haplotype analysis was carried out using TCS 1.21 [61]. Prior to this analysis, parts of our alignment which were only available for a minority of sequences (alignment positions 1–60, 649–766 and 1193–2210) were removed, and sequences were sorted according to the number of non-ambiguous sites in decreasing order. All short sequences (below 680 bp, i.e. all short barcode sequences) which were included in the first haplotype analysis were removed from further analysis due to the low level of overlap resulting in 157 sequences for P. teleius and 120 for P. nausithous. Gene evolution was visualized with a haplotype network using statistical parsimony as implemented in TCS 1.21 using default options.
Phylogenetic trees were inferred applying two criteria, i.e. unweighted Maximum Parsimony (MP), and Maximum Likelihood (ML), using the consensus haplotype sequences of the complete alignment. MP analysis was conducted in MEGA 5 [57] doing a heuristic search (Close-Neighbor-Interchange algorithm). Initial trees were obtained by random addition of sequences (10 replicates). All codon positions were included and alignment gaps were treated as missing data. For ML inference, the Tamura-Nei model [62] with a gamma distribution for rate variation among sites (G = 0.084) was selected using jModelTest 0.1.1 [63] as the best fitting evolutionary model. Tree searches were performed with PhyML version 3.0 [64] using the SPR search option and a BIONJ starting tree. Branch support for MP- and ML-trees was estimated by bootstrapping the dataset 500 times.
Average sequence divergence for COI was calculated as uncorrected pairwise p-distances of all haplotype sequence pairs within and between clades using MEGA 5 [57]. Because fossil data of Phengaris are not available, and geological events cannot be linked with branching events in our trees, we calculated age estimates of splitting events by using three COI substitution rates reported for arthropods, i.e. 1.3% [65], 2.3% [66], and 3.5% per million years [67].
We estimated nucleotide diversity π [68]. To test whether sequence diversity was concordant with expectations of neutral evolution we computed Tajima’s D [68] and Fu’s F [69] as implemented in Arlequin 3.5.1.2 [70]. Deviations from neutral evolution may suggest recent demographic expansions or bottlenecks. For these analyses only nucleotide positions represented in every sample were included (P. teleius N = 845; P. nausithous N = 768). Furthermore, we excluded samples from divergent “Wolbachia” clades.
Nuclear Microsatellite Analysis
Samples were genotyped at eight nuclear microsatellite loci (Macu1, Macu3, Macu7, Macu8, Macu9, Macu11, Macu15, Macu16; [71]). Loci were amplified in three reactions with a multiplex PCR kit (QIAGEN) using fluorescent labelled primers and separated on an Applied Biosystems 3130×l Genetic Analyzer (Applied Biosystems, Foster City, USA). Individuals for which fewer than four loci yielded interpretable results were excluded from the analysis resulting in a data set of 143 P. teleius and 109 P. nausithous genotypes.
We used a Bayesian clustering method to assess population structure of individual multilocus genotypes separately for each species using STRUCTURE 2.3 [72]. For each K ranging from 1 to 10, we performed 10 replicate runs with 100.000 steps after a burn-in period of 50.000 steps. We used the admixture model without prior population information and with correlated allele frequencies. Most likely K values were estimated following Evanno et al. [73]; see Fig. S1. The program CLUMPP 1.1.1 [74] was used to estimate the mean cluster assignment across replicate runs. For the resulting main clusters we calculated gene diversity (H e), allelic richness (A r), private allelic richness (pA r), and shared allelic richness (sA r) using ADZE 1.0 [75]. Differentiation among clusters was quantified as θ, an estimator of Wright’s F ST [76] and as standardized G’ST ([77], eq. 4b) calculated in Fstat 2.9.3.2. [78]. Individuals with highly ambiguous cluster membership (inferred ancestry <0.8; P. teleius: N = 4; P. nausithous: N = 24) were excluded from this analysis.
We assessed the relationship between nuclear and mitochondrial genomes by correlating inter-individual genetic distances and testing the significance by a Mantel test with 1000 randomizations in R version 2.12.2 [79]. For microsatellites, genetic distances were quantified as proportion of shared alleles calculated with MSA v. 3.0 [80]. For COI sequences we used Maximum Composite Likelihood estimates with pairwise deletion of missing data and gamma distributed substitution rates among sites, calculated in MEGA 5.
Results
Wolbachia Infection
In P. teleius we found 19 out of 147 (13%) individuals investigated to be infected with Wolbachia, while in P. nausithous we found 6 out of 112 (5.4%) (Table S1). The Wolbachia wsp genes had one allele each in P. teleius and P. nausitous (GenBank-accession no. JX470438, JX470439). The sequence from P. teleius is identical with allele 431 found in Heteroptera from Japan [81]. The sequence from P. nausithous differs only slightly from three known alleles (264, 266, 436) detected in Lepidoptera and Hemiptera also originating from Japan [81], [82] and was submitted as new allele 639 to the Wolbachia wsp database. The two wsp alleles are very distinct (nucleotide p-distance: 9.4%; protein p-distance: 14%), however, both are affiliated with Wolbachia supergroup B.
Phylogenetic Inference in Phengaris Teleius and P. nausithous
The final COI+COII alignment contained 282 sequences (157 P. teleius, 120 P. nausithous, 5 outgroup) with a total length of 2253 bases of which 333 (14.8%) sites were variable and 210 were parsimony informative (9.3%). No indels were detected. In total 124 unique haplotypes were observed (Table S1), 72 in P. teleius and 52 in P. nausithous. Of these, 3 haplotypes (N50, N51, N52) were observed exclusively in barcode sequences, which were excluded from further analysis. However, these haplotypes only differed in single nucleotide positions from other haplotypes (N06, N42, and N49, respectively).
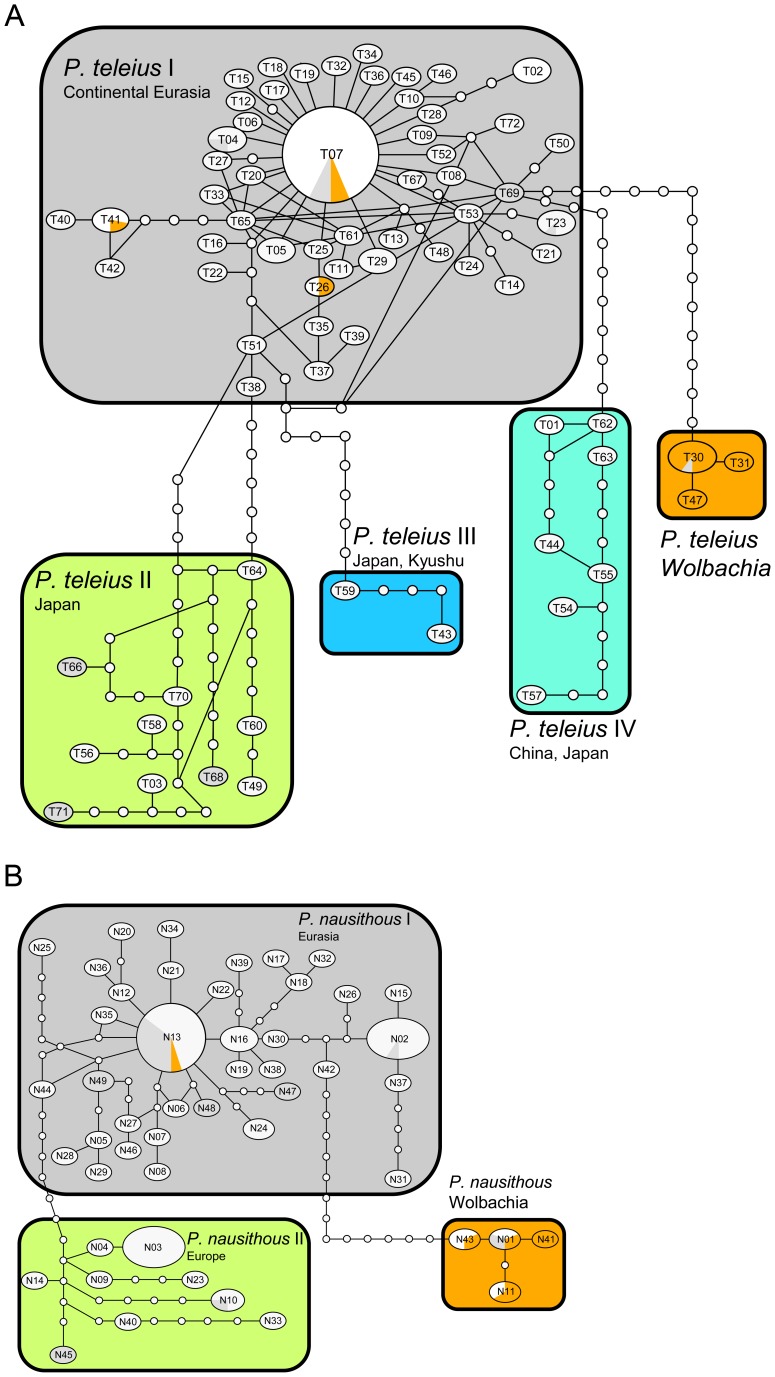
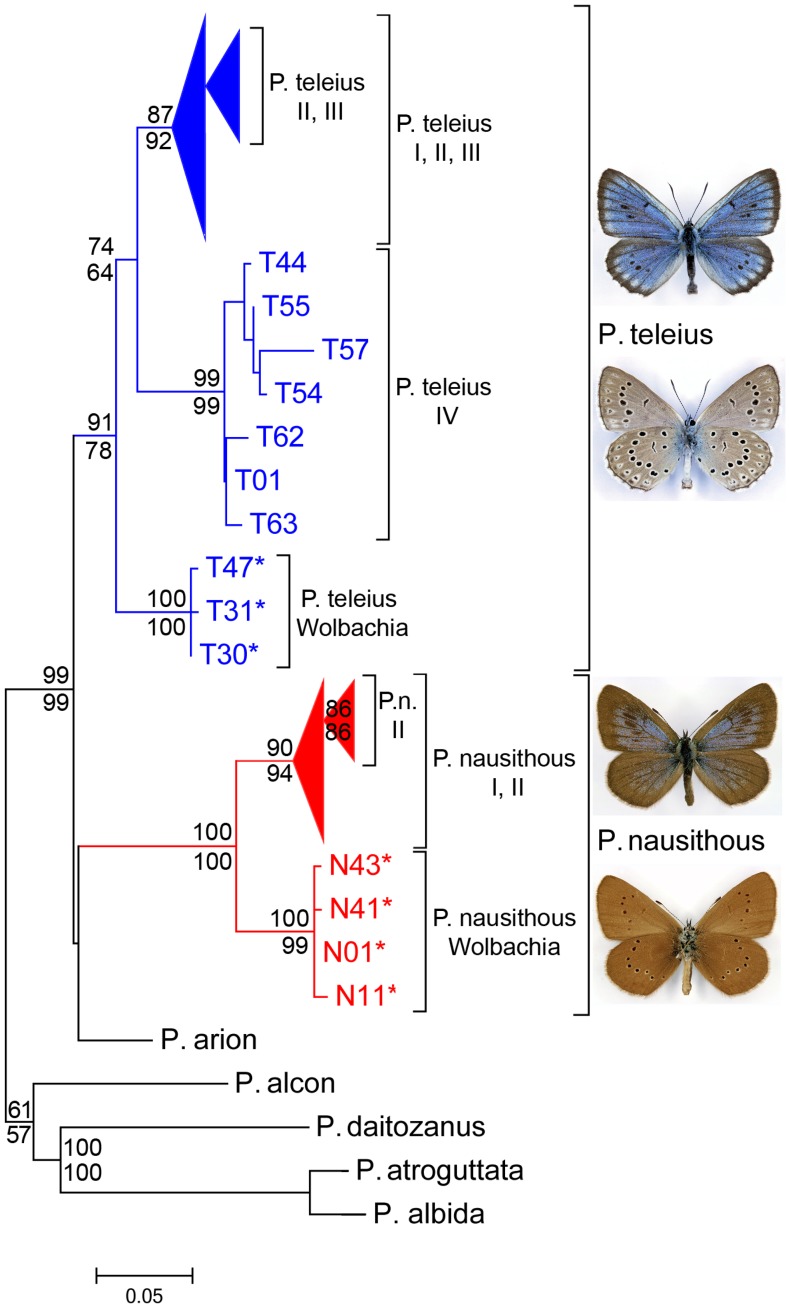
The COI haplotype network calculation resulted in independent networks for each outgroup species, P. teleius, P. nausithous, and within both study species a clade dominated by Wolbachia-infected individuals under the 95% parsimony limit (0.956 = 13 steps). A slightly relaxed parsimony limit (0.949 = 14 steps in P. nausithous; 0.942 = 15 steps in P. teleius) led to a connection between the “Wolbachia” and the respective remaining haplotypes (Fig. 2). Thus, in both species there is a majority phylogroup plus several long branching groups, one of which is characterised by Wolbachia infection. Phylogenetic inference using both ML and MP yielded essentially the same results, with both Phengaris teleius and P. nausithous being monophyletic (Fig. 3, Fig. S2, Fig. S3). Together with P. arion the two species formed a clade clearly separated from other members of the genus. However, only in the parsimony analysis were P. teleius and P. nausithous supported as sister species (Fig. S3). In both species there is a basal “Wolbachia” clade sister to four (P. teleius) or two (P. nausithous) further haplogroups.
Figure 2. COI haplotype networks for Phengaris teleius (A) and P. nausithous (B).
Circle size is proportional to haplotypes frequency (Table S1). The proportion of individuals infected with Wolbachia is indicated by a colored pie chart. Note that in several haplotypes samples could not be tested for Wolbachia since the corresponding sequence was extracted from Genbank (grey shading).
Figure 3. Phylogram for haplotypes of Phengaris teleius and P. nausithous based on ML analysis for mitochondrial COI.
Bootstrap values in percent (>50%) are given above branches (based on ML analysis) and below branches (MP analysis). Bootstrap values within subclades are not shown (see Figure S2, S3). *haplotypes associated with Wolbachia.
In P. teleius the majority of haplotypes formed a star-like network with many single steps (haplogroup P. teleius I) (Fig. 2). This phylogroup was distributed throughout continental Eurasia except for one haplotype which occurred in the most northern Japanese population (Hokkaido; ssp. ogumae) (Fig. 1). Three additional long-branched clades were geographically confined to Eastern Asia: P. teleius II to Honshū (ssp. kazamoto and daisensis), P. teleius III to Kyushu (ssp. daisensis) and P. teleius IV to China (ssp. sinalcon) and Japan (Hokkaido and Northern Honshū; ssp. ogumae, kazamoto and hosonoi). In the long branched P. teleius “Wolbachia” clade most individuals (94%; N = 15/16) were infected, significantly more than within the rest of P. teleius (2.8%; 4/141; X2-test: p<0.0001, Fig. 3). This clade was geographically restricted to Belarus, the Russian Altai, and Mongolia (Fig. 1). The subspecies within P. teleius showed no clear correspondence to haplogroups since subspecies either consisted of several haplotypes (ssp. kazamoto, daisensis and ogumae), or haplogroups harboured several subspecies (P. teleius I, II and IV).
In P. nausithous the majority haplogroup I was distributed through most of the species range. One additional clade was formed (P. nausithous II) by European haplotypes from Poland, Eastern Germany, Southern Germany, the Western Alps and Spain (Fig. 1, Fig. 2). In P. nausithous the “Wolbachia” clade harboured 56% infected individuals (N = 5/9) in contrast to the rest of P. nausithous (<1%; 1/110; X2-test: p<0.0001, Fig. 3). Infected individuals originated from Eastern Europe and Western Asia (Fig. 1). Subspecies kijevensis [41] had haplotypes of two clades, P. nausithous “Wolbachia” and P. nausithous I.
Sequence Divergence and Nucleotide Diversity
Average sequence divergence between Phengaris teleius and P. nausithous was 4.19% ±0.54%, placing the split between the species at the end of the Pliocene or beginning of the Pleistocene (Table 1). In both species the sequence divergence between haplotypes of the “Wolbachia” clades and all other haplotypes was similar and translated into estimated ages between 0.65 and 1.97 myrs.
Table 1. Sequence divergence values and estimated node dates for prominent splits of recovered phylogenetic trees (Figure 3).
| Split | Sequence divergence(%) between clades | MYA (Evolutionary rateof COI 1.3% per 1Million year) | MYA (Evolutionary rateof COI 2.3% per 1Million year) | MYA (Evolutionary rateof COI 3.5% per 1Million year) |
| P. teleius versus P. nausithous | 4.19±0.54 | 3.22 | 1.82 | 1.19 |
| P. teleius I-IV versus P. teleius“Wolbachia” | 2.56±0.47 | 1.97 | 1.11 | 0.73 |
| P. teleius IV versus P. teleius I-III | 2.16±0.37 | 1.66 | 0.94 | 0.62 |
| P. teleius II versus P. teleius I+III+IV | 1.39±0.22 | 1.07 | 0.60 | 0.40 |
| P. teleius III versus P. teleius I+II+IV | 1.52±0.26 | 1.17 | 0.66 | 0.43 |
| P. nausithous I-II versus P. nausithous“Wolbachia” | 2.28±0.43 | 1.75 | 0.99 | 0.65 |
| P. nausithous I versus P. nausithous II | 1.42±0.30 | 1.09 | 0.62 | 0.41 |
Nucleotide diversity was π = 5.52 (including Wolbachia infected individuals: 7.73) for P. teleius and π = 5.41 (6.82) for P. nausithous (Table 2). Neutrality tests for different geographic areas revealed contrasting results for the two species. P. teleius showed low π combined with significantly negative Tajima’s D or Fu’s F for continental Asia and Europe suggestive of rapid demographic expansion in that area, whereas samples from Japan showed high π and no deviation from neutrality. In P. nausithous, clade P. nausithous I showed significant deviation from neutrality suggesting demographic expansion in the eastern part of the range, whereas in the western part P. nausithous II conformed to a neutral model.
Table 2. Nucleotide diversity π, Tajima’s D, and Fu’s F estimates at mtDNA COI of different phylogenetic clusters and geographic zones.
| Sample pool | N | S | π ± s.d. | Tajima’s D | P value | Fu’s F | P value |
| P. teleius I+II+III+IV | 141 | 88 | 5.52±2.96 | −2.07 | 0.002 | −25.18 | 0.000 |
| P. teleius I+partly IV (Continental Eurasia) | 117 | 57 | 2.56±1.53 | −2.40 | 0.000 | −26.83 | 0.000 |
| P. teleius II+III+partly IV (Japan) | 24 | 50 | 14.58±7.54 | 0.35 | 0.694 | −00.71 | 0.414 |
| P. nausithous I+II | 110 | 40 | 5.41±2.91 | −0.89 | 0.177 | −20.01 | 0.000 |
| P. nausithous I | 76 | 33 | 2.21±1.37 | −2.13 | 0.002 | −27.07 | 0.000 |
| P. nausithous II | 34 | 12 | 2.52±1.55 | −0.44 | 0.325 | −01.03 | 0.330 |
N = number of sequences.
S = number of polymorphic sites.
Nuclear Microsatellite Analysis
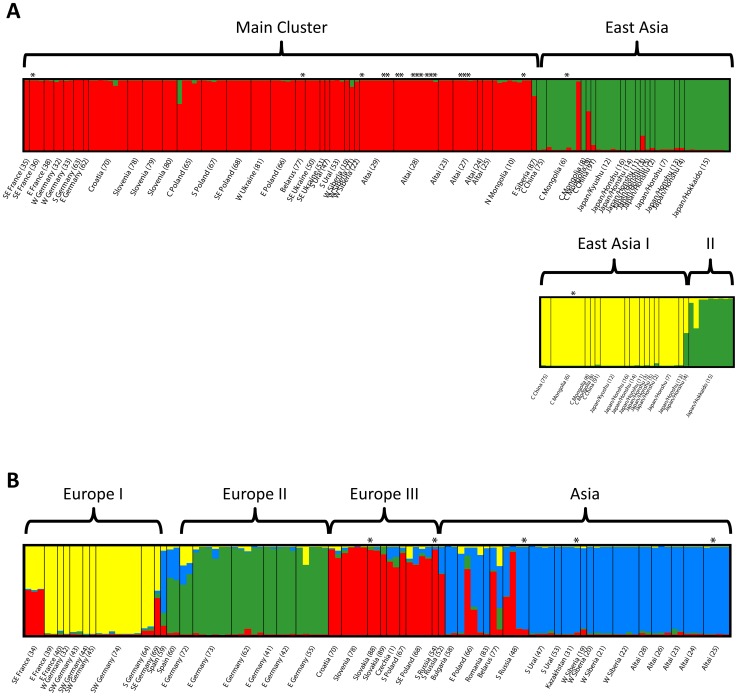
In P. teleius, the STRUCTURE analyses revealed consistent outcomes with K = 2, separating two geographically coherent clusters (Fig. 4, Fig. S1). The “Main Cluster” was formed by all samples from Europe and extended to continental Asia. The second cluster “East Asia” was formed by all samples from Japan, China and Central Mongolia. A few individuals showed admixture in the border region of the two clusters (Fig. 1). In additional separate STRUCTURE analyses of the two clusters the East Asian cluster was again split into two groups separating Hokkaido from the rest. Within the “Main Cluster” no further substructure was found as a peak of ΔK = 12 at K = 4 was very low compared to the other analyses (Fig. S1) and the resulting groups showed a high degree of admixture and no clear geographic pattern (data not shown). The “Main Cluster” in P. teleius corresponded largely to haplogroup P. teleius I obtained in the COI analysis, while the different Japanese clades and the “Wolbachia” clade were not retrieved in the microsatellite analysis.
Figure 4. Results of STRUCTURE analysis of microsatellite genotypes for Phengaris teleius (A) and P. nausithous (B).
Small bars represent individuals and their cluster membership coefficients. For details see text, Figure 1, and Figure S1; for population details see Table S1. *individuals infected with Wolbachia.
In P. nausithous the STRUCTURE analysis revealed four clusters (Fig. 4, Fig. S1). Three clusters corresponded to areas in Europe (I, II, III) comprising western, central and eastern European populations, respectively. The fourth and largest cluster extended from Eastern Europe into Asia. Admixture was observed in contact zones of the clusters in specimens from France, Czech Republic, Belarus, Poland, SW Germany, and E Germany. Specimens from peripheral sites in Spain, Germany and Southern Russia also appeared admixed (Fig. 1). In STRUCTURE analyses at lower values of K, a strong East-West split was found. At K = 2, a western cluster comprising Europe I+II and an eastern cluster comprising Europe III+Asia was formed and at K = 3, Europe III was separated from Asia (data not shown).
Genetic differentiation between clusters was strong in both species (P. teleius: θ = 0.265 (SE 0.063), G’ST = 0.671; P. nausithous: θ = 0.143 (SE 0.02), G’ST = 0.497) although lower in P. nausithous as expected from the larger number of clusters. Genetic variation within clusters is shown in Table 3. In P. teleius, cluster East Asia I was the most genetically diverse as indicated by higher values of H e, Ar and private Ar. In P. nausithous, the four clusters showed similar levels of genetic variation.
Table 3. Mean estimates of genetic diversity across 8 microsatellite loci in clusters identified in the STRUCTURE analysis.
| Species/cluster | N indivi-duals | H e (SD) | A | Ar (SE) | private Ar (SE) | % private alleles |
| P. teleius | ||||||
| Main Cluster | 103 | 0.55 (0.26) | 11.8 | 4.7 (1.0) | 2.8 (1.0) | 60% |
| East Asia I | 26 | 0.67 (0.21) | 9.0 | 5.4 (1.0) | 3.3 (1.0) | 61% |
| East Asia II | 9 | 0.41 (0.31) | 3.3 | 3.2 (0.7) | 1.5 (0.7) | 47% |
| P. nausithous | ||||||
| Europe I | 16 | 0.70 (0.23) | 8.3 | 6.7 (1.5) | 3.4 (1.4) | 51% |
| Europe II | 21 | 0.70 (0.24) | 9.6 | 7.1 (1.0) | 2.4 (0.6) | 34% |
| Europe III | 11 | 0.70 (0.23) | 6.4 | 6.2 (1.3) | 2.6 (1.0) | 42% |
| Asia | 37 | 0.71 (0.30) | 13.9 | 7.8 (1.5) | 3.5 (0.9) | 45% |
H e expected heterozygosity, A mean number of alleles, Ar allelic richness based on 7 and 10 individuals, for P. teleius and P. nausithous, respectively.
Genetic divergence was largely consistent between nuclear and mitochondrial genomes, but influenced by the inclusion of Wolbachia haplogroups which did not form similarly divergent microsatellite clusters. In P. teleius genetic distances of microsatellites and COI sequences were not correlated when all haplotypes were considered (r = 0.083, Mantel-p = 0.12), but became significantly positively correlated when Wolbachia haplotypes were removed (r = 0.405, Mantel-p = 0.001). For P. nausithous genetic distances were correlated both overall (r = 0.269, Mantel-p = 0.001) and without Wolbachia haplotypes (r = 0.177, Mantel-p = 0.001).
Discussion
Phylogenetic Inference and Wolbachia Infection
Our phylogenetic analysis based on mtDNA COI sequences revealed that P. teleius and P. nausithous were clearly separated and formed well supported monophyletic clades. However, within both species we found highly distinct evolutionary lineages. These clades were not concordant with known subspecies nor did they represent spatially contiguous groups. Such an intraspecific phylogenetic pattern could be the result of either recent, secondary contact of formerly geographically separated populations of the species or it could be evidence for intrinsic reproductive barriers among sympatric cryptic species [83]. Indeed, the observed average sequence divergence between haplotypes of the “Wolbachia” clades and the rest of the species (2.28–2.56%) for P. teleius and P. nausithous resembled the divergence that has been reported between species [6]. Similar levels of divergence have already been found in Phengaris teleius and P. nausithous and have led to the hypothesis of cryptic species [44], [45]. However, the divergent haplogroups were strongly associated with Wolbachia infections in contrast to the remaining haplotypes. A similar pattern has been already described within other butterfly species [84], [85]. In fact, the COI sequences which led to the hypotheses of cryptic speciation within P. teleius ([45]: specimen Uk-08-J627) and within P. nausithous ([44]: specimen ZD-99-S301) corresponded perfectly to haplotypes that were associated with Wolbachia in our new samples originating from the same regions. This suggests that these specimens were also likely to be infected with Wolbachia. Our interpretation is corroborated by the fact that divergent haplotypes of infected and uninfected individuals co-occurred at several localities and that in both species the mtDNA Wolbachia clades were not reflected in the nuclear genome. Cryptic speciation should result in similar patterns across different genomes [28]. Thus, these inconsistencies between mitochondrial and nuclear data sets are evidence against cryptic species.
Our results suggest that the Wolbachia infection took place between 0.7–2.0 mya and 0.6–1.7 mya in P. teleius and P. nausithous, respectively, and well after species diversification, which we estimated between 1.2 and 3.2 mya, a time span consistent with previous estimates using external calibration points for chronology estimation [44]. However, the infection persists only in a minority of individuals from few populations. Hence, mitochondrial sequences of infected and uninfected parts of the populations accumulated substantial divergence, resulting in well separated clades in the inferred phylogeny. A similar phylogenetic pattern has been shown for other butterfly species [18], [85], [86], [87]. In 44% of the specimens of the P. nausithous “Wolbachia” clade no infection was detected. These individuals might indeed lack an infection, which can happen when Wolbachia is not efficiently transmitted to the next generation [88]. Thus, a negative Wolbachia test in particular samples does not disprove Wolbachia infection as causal for lineage divergence. However, we cannot exclude that the PCR-screening for Wolbachia might have produced false negatives, e.g. due to mutations in the primer binding sites.
Within 19 populations examined, P. teleius and P. nausithous co-occurred in the same locality. Three of these populations harbour a Wolbachia infection, either hosted by P. teleius (populations 28 and 77) or hosted by P. nausithous (population 25), but never hosted by both species at any locality. Our observation suggests low rates, or lack of horizontal transmission between the two sister species although there is general evidence from non-LTR retrotransposons for recent horizontal transmission between Phengaris species [89].
It has been shown that different Wolbachia strains can have different effects on the fitness of their hosts, ranging from positive to detrimental [90], [91]. In both species infected individuals were found across large parts of the distribution ranges from Belarus to Mongolia and from Slovakia to the South Ural Mountains, for P. teleius and P. nausithous, respectively. Because the Wolbachia infections were present within populations at low frequency in wide distributional areas infected individuals might experience a positive fitness effect due to the presence of Wolbachia [22]. Transmission rates into the next host generation seem to be imperfect, since the infection did not sweep through whole populations [22]. This effect could also depend on certain environmental conditions (e.g. Wolbachia density was highest in Leptopilina wasps at high temperatures, [23]). Indeed, in P. teleius only populations inhabiting steppe habitats with relatively hot and dry conditions in summer harboured Wolbachia infected individuals. Furthermore, in P. teleius two adults were Wolbachia infected both of which were male which might be an indication for a CI strain in P. teleius [11]. In P. nausithous, however, all four Wolbachia infected adults were females which might be an indication for a male-killing or feminization strain in that species [11]. For a better characteriziation of Wolbachia strains infecting P. teleius and P. nausithous and for clarification of its reproductive mechanisms and its fitness effects on hosts and populations further studies are needed, such as MLST genotyping [37], VNTR molecular screening [92], analyses of sex ratios and egg hatch-rates [19], or demographic models [87].
Phylogeography of Phengaris Teleius and P. nausithous
In both species the samples that were not affected by Wolbachia showed considerable divergence in both the mitochondrial and the nuclear genome. However, the two species showed contrasting geographical patterns of differentiation and likely evolutionary scenarios.
In P. teleius there was little mtDNA variation across the western part of its range (P. teleius I). However, in East Asia three divergent haplogroups were found (P. teleius II, III, and IV). Although not fully concordant with the mtDNA pattern, the nuclear microsatellite data also revealed a stronger sub-structuring in Eastern Asia (East Asia I+II). This pattern might be well explained by the following scenario. After speciation of P. teleius between 1.2 and 3.2 mya, which likely took place in Central or Eastern Asia [40], [93], lineages may have spread and diversified throughout Eurasia. However, climatic conditions in one of the last glacial phases [94] could have eliminated the species from Europe and from most parts of continental Asia. In the Far East of continental Asia and Japan the species may have found larger or more suitable refugial areas [95], concordant with the high genetic diversity in that area. This phylogeographic hypothesis is also corroborated by the fact that all described subspecies in P. teleius are restricted to the Eastern Palaearctic and mainly to Japan [40]. Recolonization of continental Asia and Europe may then have started by founder individuals surviving in East Asia or Japan. The presence of isolated refugia in this area is likely given the complex topography and may be mirrored e.g. in the haplotypes T40, T41 and T42, or T02, forming distinct groups within P. teleius I (Fig. 2) and geographically confined to the Hustai Mountains in Central Mongolia (Pops. 6, 8, 9), or Hokkaido (Pop. 15). Low Tajima’s D and Fu’s F values coupled with the low nucleotide diversity (π) values of East Asia corroborate such a recent range expansion. Similar east-to-west colonization routes of butterflies have been suggested for an Eastern clade of Melitaea cinxia coming from Far Eastern populations and migrating into Scandinavia [96] as well as for Coenonympha hero which seems to have had a glacial centre of survival in the Southern Ural Mountains and expanded from there westwards to Europe [97]. However, although Eastern Asia clearly emerges as a centre of diversification within P. teleius in our analyses, the mtDNA phylogenetic clades and the microsatellite clusters were not congruent with the morphologically defined subspecies. Similar patterns were found e.g. in Aglais urticae, a nymphalid butterfly, and could be due to ecological differentiation occurring more rapidly than evolution at the mtDNA level [98].
In contrast to P. teleius, in P. nausithous the mitochondrial as well as the nuclear data set showed a stronger structuring of genetic diversity in the western part of its distribution. However, the divergence of the geographically separated haplogroups P. nausithous I and II was not concordant with the microsatellite clusters Europe I, II and III and their geographic patterns.
For P. nausithous a likely scenario is that after speciation in Central Asia it subsequently spread towards Western Asia and Europe where it diversified. Haplogroup P. nausithous II located in central and western Europe has slightly increased nucleotide diversity and non-significant Tajima’s D and Fu’s F values which suggest that the species survived during Pleistocene ice ages within European glacial refugia. For P. nausithous haplogroup I recent range expansion into the Eastern parts of its distribution range is likely to have started from a limited set of individuals indicated by significantly low Tajima’s D and Fu’s F values of Eurasian samples. The microsatellite clusters are also in line with the survival of P. nausithous in several European refugia. Three major European glacial refugia for animal species have been identified on the Iberian, Italian and Balkan peninsulas [94], [99], [100]. Cluster Asia and Europe III represent genetic groups that likely survived in the Balkans. Europe II represents a refugium located on the Iberian Peninsula and Europe I possibly represents a refugium on the Italian peninsular which today is not populated by the species anymore [42]. However, admixed populations in the contact zones of clusters Europe I and III, admixed populations in Spain and the distribution of cluster Europe III both east and west of the Alps indicate a complex phylogeographic history in western and central Europe. The East European (Europe III) and Asian clusters are overlapping as evidenced by several admixed populations located in East Poland, Belarus, and South Russia. Overall, P. nausithous shows complex phylogeographic patterns especially in contact zones and peripheral areas which deserve further analysis based on denser sampling.
Conclusions
Based on mtDNA barcoding, nuclear microsatellite analyses and Wolbachia screening we reject the hypothesis of cryptic speciation within Phengaris teleius and P. nausithous. The major splits in the mtDNA phylogeny in both species can be explained by Wolbachia infections. Furthermore, geographic isolation during Pleistocene glaciations contributed to differentiation of mitochondrial and nuclear genomes.
Our study has shown that DNA barcoding studies can deliver robust information on cryptic species only in combination with tests for Wolbachia infections and additional analysis on nuclear markers, especially in groups with high prevalence of Wolbachia-infection [101].
Our study has some important implications for nature conservation. Wolbachia may constitute a risk for the stability of Phengaris populations, as an introduction of Wolbachia infected individuals into small populations might lead to a selective sweep and to full fixation of the associated introduced genotype within the population [22] which also means the elimination of the locally adapted genetic composition and diversity. Therefore, future reintroduction programs for insect species, like Phengaris butterflies, should include screening for the presence of Wolbachia in order not to introduce possibly detrimental elements into small and already threatened populations [87].
P. teleius showed increased genetic structuring in Eastern Asia whereas P. nausithous was more structured in Western Eurasia likely indicating opposing refugial areas during past glacial maxima. The phylogeographic clusters identified may represent locally adapted gene pools. Thus, reintroduction of the species into extinct European populations should include individuals from the same genetic group in order not to introduce possibly maladapted individuals. In P. nausithous four geographically separated microsatellite groups were observed in a small area in Europe. More detailed studies are necessary on the extent and delineation of these European clusters and the affiliation of peripheral populations.
Supporting Information
Determination of the most likely K of the STRUCTURE analyses.
(DOC)
Maximum Likelihood cladogram.
(DOC)
Maximum Parsimony cladogram.
(DOC)
Phengaris material used for analysis.
(DOC)
Microsatellite genotypes of Phengaris teleius and P. nausithous .
(DOC)
Acknowledgments
We are very grateful to the following people for help in collecting samples: Alban Pfeifer, Alexander Blinov, Anatoli Kulak, Christian Anton, Dirk Immisch, Dusan Zitnan, Franc Rebeušek, Holger Loritz, Julien Dabry, Karsten Wesche, Katrin Ronnenberg, László Rákosy, Marko Eigner, Martin Musche, Masaya Yago, Michal Zapletal, Miguel L. Munguira, Oleg Kosterin, Olga Novikova, Priska Koelman, Rainer Wendt, Sergey Popov, Viktoria Wagner, Wang Min, Xiushan Li, and Zdravko Kolev. Antti Roine kindly permitted the reproduction of Phengaris images from Lepibase [102]. We thank David Nash, Niklas Wahlberg and two anonymous reviewers for their comments which helped to improve the manuscript. This publication made use of the Wolbachia MLST website [54], [103], [104]. We also thank the graduate school HIGRADE for support.
Data accessibility:
- DNA-sequences: GenBank accessions: JX311049–JX311307, JX470438, JX470439.
- Microsatellite genotypes: Supplement material, Table S2.
Funding Statement
The study was financially supported by the VolkswagenStiftung (http://www.volkswagenstiftung.de/) (Sylvia Ritter, Az 1/82 751). Furthermore, research has been conducted within the project CLIMIT (Climit Change Impacts on Insects and their Mitigation; http://www.climit-project.net/) funded by Deutsches Zentrum für Luft- und Raumfahrt-Bundesministerium für Bildung und Forschung (www.pt-dlr.de/?) (Germany), the Natural Environment Research Council (http://www.nerc.ac.uk/) and the Department of Environment, Food & Rural Affairs (http://www.defra.gov.uk/) (UK), the French National Research Agency (http://www.agence-nationale-recherche.fr/) (France), Formas (http://www.formas.se/) (Sweden), and Swedish Environmental Protection Agency (http://www.naturvardsverket.se/) (Sweden) through the FP6 BiodivERsA Eranet (http://www.biodiversa.org/) and was also funded within the EU funded project SCALES (http://www.scales-project.net/) (FP7 grant agreement no. 226852). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Bickford D, Lohman DJ, Sodhi NS, Ng PK, Meier R, et al. (2007) Cryptic species as a window on diversity and conservation. Trends Ecol Evol 22: 148–155. [DOI] [PubMed] [Google Scholar]
- 2. Pfenninger M, Schwenk K (2007) Cryptic animal species are homogeneously distributed among taxa and biogeographical regions. BMC Evol Biol 7: 121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Huyse T, Poulin R, Theron A (2005) Speciation in parasites: a population genetics approach. Trends Parasitol 21: 469–475. [DOI] [PubMed] [Google Scholar]
- 4. Hebert PD, Penton EH, Burns JM, Janzen DH, Hallwachs W (2004) Ten species in one: DNA barcoding reveals cryptic species in the neotropical skipper butterfly Astraptes fulgerator . Proc Natl Acad Sci U S A 101: 14812–14817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hebert PD, Cywinska A, Ball SL, deWaard JR (2003) Biological identifications through DNA barcodes. Proc Biol Sci 270: 313–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. McBride CS, Van Velzen R, Larsen TB (2009) Allopatric origin of cryptic butterfly species that were discovered feeding on distinct host plants in sympatry. Mol Ecol 18: 3639–3651. [DOI] [PubMed] [Google Scholar]
- 7. Galtier N, Nabholz B, Glemin S, Hurst GDD (2009) Mitochondrial DNA as a marker of molecular diversity: a reappraisal. Mol Ecol 18: 4541–4550. [DOI] [PubMed] [Google Scholar]
- 8. Duplouy A, Hurst GDD, O’Neill SL, Charlat S (2010) Rapid spread of male-killing Wolbachia in the butterfly Hypolimnas bolina . J Evol Biol 23: 231–235. [DOI] [PubMed] [Google Scholar]
- 9. Munoz AG, Baxter SW, Linares M, Jiggins CD (2011) Deep mitochondrial divergence within a Heliconius butterfly species is not explained by cryptic speciation or endosymbiotic bacteria. BMC Evol Biol 11: 358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wiemers M, Fiedler K (2007) Does the DNA barcoding gap exist? - a case study in blue butterflies (Lepidoptera: Lycaenidae). Front Zool 4: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Werren JH, Baldo L, Clark ME (2008) Wolbachia: master manipulators of invertebrate biology. Nat Rev Microbiol 6: 741–751. [DOI] [PubMed] [Google Scholar]
- 12. Russell JA, Funaro CF, Giraldo YM, Goldman-Huertas B, Suh D, et al. (2012) A veritable menagerie of heritable bacteria from ants, butterflies, and beyond: broad molecular surveys and a systematic review. Plos One 7: e51027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hurst GD, Jiggins FM (2005) Problems with mitochondrial DNA as a marker in population, phylogeographic and phylogenetic studies: the effects of inherited symbionts. Proc R Soc Biol Sci Ser B 272: 1525–1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Werren JH, Zhang W, Guo LR (1995) Evolution and phylogeny of Wolbachia: reproductive parasites of arthropods. Proc R Soc Biol Sci Ser B 261: 55–63. [DOI] [PubMed] [Google Scholar]
- 15. Caspari E, Watson GS (1959) On the evolutionary importance of cytoplasmic sterility in mosquitos. Evolution 13: 568–570. [Google Scholar]
- 16. Hoffmann AA, Turelli M (1988) Unidirectional incompatibility in Drosophila simulans - Inheritance, geographic variation and fitness effects. Genetics 119: 435–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hatcher MJ (2000) Persistence of selfish genetic elements: population structure and conflict. Trends Ecol Evol 15: 271–277. [DOI] [PubMed] [Google Scholar]
- 18. Narita S, Nomura M, Kato Y, Fukatsu T (2006) Genetic structure of sibling butterfly species affected by Wolbachia infection sweep: evolutionary and biogeographical implications. Mol Ecol 15: 1095–1108. [DOI] [PubMed] [Google Scholar]
- 19. Hurst GDD, Jiggins FM, von der Schulenburg JHG, Bertrand D, West SA, et al. (1999) Male-killing Wolbachia in two species of insect. P R Soc B 266: 735–740. [Google Scholar]
- 20. Bordenstein SR, Marshall ML, Fry AJ, Kim U, Wernegreen JJ (2006) The tripartite associations between bacteriophage, Wolbachia, and arthropods. Plos Pathog 2: 384–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Egas M, Vala F, Breeuwer JAJ (2002) On the evolution of cytoplasmic incompatibility in haplodiploid species. Evolution 56: 1101–1109. [DOI] [PubMed] [Google Scholar]
- 22. Jansen VAA, Turelli M, Godfray HCJ (2008) Stochastic spread of Wolbachia . P R Soc B 275: 2769–2776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mouton L, Henri H, Bouletreau M, Vavre F (2006) Effect of temperature on Wolbachia density and impact on cytoplasmic incompatibility. Parasitology 132: 49–56. [DOI] [PubMed] [Google Scholar]
- 24. Reynolds KT, Thomson LJ, Hoffmann AA (2003) The effects of host age, host nuclear background and temperature on phenotypic effects of the virulent Wolbachia strain popcorn in Drosophila melanogaster . Genetics 164: 1027–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Whitworth TL, Dawson RD, Magalon H, Baudry E (2007) DNA barcoding cannot reliably identify species of the blowfly genus Protocalliphora (Diptera: Calliphoridae). P R Soc B 274: 1731–1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bordenstein SR, O’Hara FP, Werren JH (2001) Wolbachia-induced incompatibility precedes other hybrid incompatibilities in Nasonia . Nature 409: 707–710. [DOI] [PubMed] [Google Scholar]
- 27. Hurst GDD, Werren JH (2001) The role of selfish genetic elements in eukaryotic evolution. Nat Rev Genet 2: 597–606. [DOI] [PubMed] [Google Scholar]
- 28. Dasmahapatra KK, Elias M, Hill RI, Hoffman JI, Mallet J (2010) Mitochondrial DNA barcoding detects some species that are real, and some that are not. Mol Ecol Resources 10: 264–273. [DOI] [PubMed] [Google Scholar]
- 29. Smith MA, Bertrand C, Crosby K, Eveleigh ES, Fernandez-Triana J, et al. (2012) Wolbachia and DNA barcoding insects: patterns, potential, and problems. Plos One 7: e36514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Heath BD, Butcher RDJ, Whitfield WGF, Hubbard SF (1999) Horizontal transfer of Wolbachia between phylogenetically distant insect species by a naturally occurring mechanism. Curr Biol 9: 313–316. [DOI] [PubMed] [Google Scholar]
- 31. Pierce NE, Braby MF, Heath A, Lohman DJ, Mathew J, et al. (2002) The ecology and evolution of ant association in the Lycaenidae (Lepidoptera). Annu Rev Entomol 47: 733–771. [DOI] [PubMed] [Google Scholar]
- 32. Thomas JA, Settele J (2004) Evolutionary biology - Butterfly mimics of ants. Nature 432: 283–284. [DOI] [PubMed] [Google Scholar]
- 33. van Swaay C, Collins S, Dušej G, Maes D, Munguira ML, et al. (2012) Dos and Don’ts for butterflies of the Habitats Directive of the European Union. Nature Conservation 1: 73–153. [Google Scholar]
- 34.Munguira ML, Martín J, editors (1999) Action plan for Maculinea butterflies in Europe. Nature and environment. Strasbourg: Council of Europe.
- 35. Sielezniew M, Rutkowski R, Ponikwicka-Tyszko D, Ratkiewicz M, Dziekanska I, et al. (2012) Differences in genetic variability between two ecotypes of the endangered myrmecophilous butterfly Phengaris ( = Maculinea) alcon- the setting of conservation priorities. Insect Conserv Divers 5: 223–236. [Google Scholar]
- 36. Patricelli D, Sielezniew M, Ponikwicka-Tyszko D, Ratkiewicz M, Bonelli S, et al. (2013) Contrasting genetic structure of rear edge and continuous range populations of a parasitic butterfly infected by Wolbachia . BMC Evol Biol 13: 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Salunke BK, Salunkhe RC, Dhotre DP, Walujkar SA, Khandagale AB, et al. (2012) Determination of Wolbachia diversity in butterflies from Western Ghats, India, by a multigene approach. Appl Environ Microbiol 78: 4458–4467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Fric Z, Wahlberg N, Pech P, Zrzavy J (2007) Phylogeny and classification of the Phengaris-Maculinea clade (Lepidoptera: Lycaenidae): total evidence and phylogenetic species concepts. Syst Entomol 32: 558–567. [Google Scholar]
- 39. Wang M, Settele J (2010) Notes on and key to the genus Phengaris (s. str.) (Lepidoptera, Lycaenidae) from mainland China with description of a new species. Zookeys 48: 21–28. [Google Scholar]
- 40. Sibatani A, Saigusa T, Hirowatari T (1994) The genus Maculinea van Eecke, 1915 (Lepidoptera: Lycaenidae) from the East Palaearctic Region. Tyô to Ga 44: 157–220. [Google Scholar]
- 41. Rákosy L, Tartally A, Goia M, Mihali C, Varga Z (2010) The Dusky Large Blue Maculinea nausithous kijevensis (Sheljuzhko, 1928) in the Transylvanian basin: New data on taxonomy and ecology. Nota lepid 33: 31–37. [Google Scholar]
- 42. Wynhoff I (1998) The recent distribution of the European Maculinea species. J Insect Conserv 2: 15–27. [Google Scholar]
- 43. Pech P, Fric Z, Konvicka M, Zrzavy J (2004) Phylogeny of Maculinea blues (Lepidoptera: Lycaenidae) based on morphological and ecological characters: evolution of parasitic myrmecophily. Cladistics-the International Journal of the Willi Hennig Society 20: 362–375. [DOI] [PubMed] [Google Scholar]
- 44. Als TD, Vila R, Kandul NP, Nash DR, Yen SH, et al. (2004) The evolution of alternative parasitic life histories in large blue butterflies. Nature 432: 386–390. [DOI] [PubMed] [Google Scholar]
- 45. Ugelvig LV, Vila R, Pierce NE, Nash DR (2011) A phylogenetic revision of the Glaucopsyche section (Lepidoptera: Lycaenidae), with special focus on the Phengaris-Maculinea clade. Mol Phylogenet Evol 61: 237–243. [DOI] [PubMed] [Google Scholar]
- 46. Murayama S (1992) Some new Lycaenid species of Chinese Rhopalocera. Nature Insects 27: 37–41. [Google Scholar]
- 47. Staudinger O (1892) Lepidopteren des Kentei-Gebirges. Deutsche Entomologische Zeitschrift Iris 5: 300–393. [Google Scholar]
- 48.Staudinger O (1887) Neue Arten und Varietäten von Lepidopteren aus dem Amurgebiet. In: Romanoff NM, editor. Mémoires sur les lépidoptères. St. Pétersbourg: Imprimerie de M.M. Stassuléwitch. 126–232.
- 49. Takahashi A (1973) Maculinea teleius Bergsträsser at high elevation in the Central Honshû, Japan, with description of a new subspecies. Tyô to Ga 23: 75–85. [Google Scholar]
- 50. Druce HH (1875) Descriptions of new species of diurnal lepidoptera. Cistula Entomologica 1: 357–363. [Google Scholar]
- 51. Matsumura S (1910) Lycaeniden Japans. Entomol Z 23: 221–222. [Google Scholar]
- 52. Matsumura S (1926) Some new and unrecorded Lycaenids-species from Japan, Corea and Formosa. Insecta Matsumurana 1: 23–31. [Google Scholar]
- 53. Zhou W, Rousset F, O’Neil S (1998) Phylogeny and PCR-based classification of Wolbachia strains using wsp gene sequences. Proc R Soc Biol Sci Ser B 265: 509–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Jolley KA, Chan MS, Maiden MC (2004) mlstdbNet - distributed multi-locus sequence typing (MLST) databases. BMC Bioinformatics 5: 86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp Ser 41: 95–98. [Google Scholar]
- 56. Williams ST, Knowlton N (2001) Mitochondrial pseudogenes are pervasive and often insidious in the snapping shrimp genus Alpheus . Mol Biol Evol 18: 1484–1493. [DOI] [PubMed] [Google Scholar]
- 57. Tamura K, Peterson D, Peterson N, Stecher G, Nei M, et al. (2011) MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28: 2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Lunt DH, Zhang DX, Szymura JM, Hewitt GM (1996) The insect cytochrome oxidase I gene: evolutionary patterns and conserved primers for phylogenetic studies. Insect Mol Biol 5: 153–165. [DOI] [PubMed] [Google Scholar]
- 59. Kim I, Lee EM, Seol KY, Yun EY, Lee YB, et al. (2006) The mitochondrial genome of the Korean hairstreak, Coreana raphaelis (Lepidoptera: Lycaenidae). Insect Mol Biol 15: 217–225. [DOI] [PubMed] [Google Scholar]
- 60. Ratnasingham S, Hebert PD (2007) BOLD: The Barcode of Life Data System (http://www.barcodinglife.org). Mol Ecol Notes. 7: 355–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Clement M, Posada D, Crandall KA (2000) TCS: a computer program to estimate gene genealogies. Mol Ecol 9: 1657–1660. [DOI] [PubMed] [Google Scholar]
- 62. Tamura K, Nei M (1993) Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol 10: 512–526. [DOI] [PubMed] [Google Scholar]
- 63. Posada D (2008) jModelTest: Phylogenetic Model Averaging. Mol Biol Evol 25: 1253–1256. [DOI] [PubMed] [Google Scholar]
- 64. Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, et al. (2010) New Algorithms and Methods to Estimate Maximum-Likelihood Phylogenies: Assessing the Performance of PhyML 3.0. Syst Biol 59: 307–321. [DOI] [PubMed] [Google Scholar]
- 65. Quek S-P, Davies SJ, Itino T, Pierce NE (2004) Codiversification in ant-plant mutualism: stem texture and the evolution of host use in Crematogaster (Formicidae: Myrmicinae) inhabitants of Macaranga (Euphorbiaceae). Evolution 58: 554–570. [PubMed] [Google Scholar]
- 66. Brower AVZ (1994) Rapid morphological radiation and convergence among races of the butterfly Heliconius erato inferred from patterns of mitochondrial DNA evolution. Proc Natl Acad Sci U S A 91: 6491–6495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Papadopoulou A, Anastasiou I, Vogler AP (2010) Revisiting the insect mitochondrial molecular clock: The mid-Aegean trench calibration. Mol Biol Evol 27: 1659–1672. [DOI] [PubMed] [Google Scholar]
- 68. Tajima F (1989) Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123: 585–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Fu YX (1996) New statistical tests of neutrality for DNA samples from a population. Genetics 143: 557–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Excoffier L, Lischer HEL (2010) Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resources 10: 564–567. [DOI] [PubMed] [Google Scholar]
- 71. Zeisset I, Als TD, Settele J, Boomsma JJ (2005) Microsatellite markers for the large blue butterflies Maculinea nausithous and Maculinea alcon (Lepidoptera: Lycaenidae) and their amplification in other Maculinea species. Mol Ecol Notes 5: 165–168. [Google Scholar]
- 72. Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155: 945–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14: 2611–2620. [DOI] [PubMed] [Google Scholar]
- 74. Jakobsson M, Rosenberg NA (2007) CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics 23: 1801–1806. [DOI] [PubMed] [Google Scholar]
- 75. Szpiech ZA, Jakobsson M, Rosenberg NA (2008) ADZE: a rarefaction approach for counting alleles private to combinations of populations. Bioinformatics 24: 2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Weir BS, Cockerham CC (1984) Estimating F-statistics for the analysis of population structure. Evolution 38: 1358–1370. [DOI] [PubMed] [Google Scholar]
- 77. Hedrick PW (2005) A standardized genetic differentiation measure. Evolution 59: 1633–1638. [PubMed] [Google Scholar]
- 78.Goudet J (2001) FSTAT, a program to estimate and test gene diversities and fixation indices (version 2.9.3). Available from http://www.unil.ch/izea/softwares/fstat.html. Updated from Goudet (1995).
- 79.R Development Core Team (2011) R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. ISBN 3–900051–07–0, URL http://www.R-project.org/.
- 80. Dieringer D, Schlötterer C (2003) Microsatellite Analyser (MSA): A platform independent analysis tool for large microsatellite data sets. Mol Ecol Notes 3: 167–169. [Google Scholar]
- 81. Kikuchi Y, Fukatsu T (2003) Diversity of Wolbachia endosymbionts in heteropteran bugs. Appl Environ Microbiol 69: 6082–6090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Tagami Y, Miura K (2004) Distribution and prevalence of Wolbachia in Japanese populations of Lepidoptera. Insect Mol Biol 13: 359–364. [DOI] [PubMed] [Google Scholar]
- 83. Avise JC, Arnold J, Ball RM, Bermingham E, Lamb T, et al. (1987) Intraspecific phylogeography: the mitochondrial DNA bridge between population genetics and systematics. Annu Rev Ecol Syst 18: 489–522. [Google Scholar]
- 84. Lohman DJ, Peggie D, Pierce NE, Meier R (2008) Phylogeography and genetic diversity of a widespread Old World butterfly, Lampides boeticus (Lepidoptera: Lycaenidae). BMC Evol Biol 8: 301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Charlat S, Duplouy A, Hornett EA, Dyson EA, Davies N, et al. (2009) The joint evolutionary histories of Wolbachia and mitochondria in Hypolimnas bolina . BMC Evol Biol 9: 64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Gompert Z, Nice CC, Fordyce JA, Forister ML, Shapiro AM (2006) Identifying units for conservation using molecular systematics: the cautionary tale of the Karner blue butterfly. Mol Ecol 15: 1759–1768. [DOI] [PubMed] [Google Scholar]
- 87. Nice CC, Gompert Z, Forister ML, Fordyce JA (2009) An unseen foe in arthropod conservation efforts: The case of Wolbachia infections in the Karner blue butterfly. Biol Conserv 142: 3137–3146. [Google Scholar]
- 88. Hurst GDD, Jiggins FM, Robinson SJW (2001) What causes inefficient transmission of male-killing Wolbachia in Drosophila? Heredity 87: 220–226. [DOI] [PubMed] [Google Scholar]
- 89. Novikova O, Sliwinska E, Fet V, Settele J, Blinov A, et al. (2007) CRI clade of non-LTR retrotransposons from Maculinea butterflies (Lepidoptera: Lycaenidae): evidence for recent horizontal transmission. BMC Evol Biol 7: 93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Bordenstein SR, Werren JH (2000) Do Wolbachia influence fecundity in Nasonia vitripennis? Heredity 84: 54–62. [DOI] [PubMed] [Google Scholar]
- 91. Sarakatsanou A, Diamantidis AD, Papanastasiou SA, Bourtzis K, Papadopoulos NT (2011) Effects of Wolbachia on fitness of the Mediterranean fruit fly (Diptera: Tephritidae). J Appl Entomol 135: 554–563. [Google Scholar]
- 92. Schneider DI, Garschall KI, Parker AG, Abd-Alla AMM, Miller WJ (2013) Global Wolbachia prevalence, titer fluctuations and their potential of causing cytoplasmic incompatibilities in tsetse flies and hybrids of Glossina morsitans subgroup species. J Invertebr Pathol 112: S104–S115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Fiedler K (1998) Lycaenid-ant interactions of the Maculinea type: tracing their historical roots in a comparative framework. J Insect Conserv 2: 3–14. [Google Scholar]
- 94. Schmitt T (2007) Molecular biogeography of Europe: Pleistocene cycles and postglacial trends. Front Zool 4: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Tsukada M (1982) Cryptomeria japonica - Glacial refugia and late-glacial and postglacial migration. Ecology 63: 1091–1105. [Google Scholar]
- 96. Wahlberg N, Saccheri I (2007) The effects of Pleistocene glaciations on the phylogeography of Melitaea cinxia (Lepidoptera: Nymphalidae). Eur J Entomol 104: 675–684. [Google Scholar]
- 97. Cassel A, Tammaru T (2003) Allozyme variability in central, peripheral and isolated populations of the scarce heath (Coenonympha hero: Lepidoptera, Nymphalidae): Implications for conservation. Conserv Genet 4: 83–93. [Google Scholar]
- 98. Vandewoestijne S, Baguette M, Brakefield PM, Saccheri IJ (2004) Phylogeography of Aglais urticae (Lepidoptera) based on DNA sequences of the mitochondrial COI gene and control region. Mol Phylogenet Evol 31: 630–646. [DOI] [PubMed] [Google Scholar]
- 99. Hewitt GM (2000) The genetic legacy of the Quaternary ice ages. Nature 405: 907–913. [DOI] [PubMed] [Google Scholar]
- 100. Taberlet P, Fumagalli L, Wust-Saucy AG, Cosson JF (1998) Comparative phylogeography and postglacial colonization routes in Europe. Mol Ecol 7: 453–464. [DOI] [PubMed] [Google Scholar]
- 101. Gerth M, Geissler A, Bleidorn C (2011) Wolbachia infections in bees (Anthophila) and possible implications for DNA barcoding. Syst Biodiv 9: 319–327. [Google Scholar]
- 102.Roine A (2000) LepiBase 2.0. Butterflies of Europe - Species and Habitat. Vanha-Ulvila: Antti Roine.
- 103. Jolley KA, Maiden MC (2010) BIGSdb: Scalable analysis of bacterial genome variation at the population level. BMC Bioinformatics 11: 595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Wolbachia MLST databases. Available: http://pubmlst.org/wolbachia/. Last accessed: 19 Sep 2013.
- 105.Kudrna O, Harpke A, Lux K, Pennerstorfer J, Schweiger O, et al.. (2011) Distribution Atlas of Butterflies in Europe. Halle (Saale): Gesellschaft für Schmetterlingsschutz. 576 p. [Google Scholar]
- 106.Lukhtanov V, Lukhtanov A (1994) Die Tagfalter Nordwestasiens. Herbipoliana 3.
- 107.Tshikolovets VV, Bidzilya A, Golovushkin M (2002) The butterflies of Transbaikal Siberia. Kyiv-Brno: Author’s edition. 320 p. [Google Scholar]
- 108.Tshikolovets VV, Yakovlev RV, Bálint Z (2009) The butterflies of Mongolia. Kiev: Tshikolovets Publ.
- 109.Tshikolovets VV, Yakovlev RV, Kosterin OE (2009) The butterflies of Altai, Sayan and Tuva. Kiev: Tshikolovets Publ.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Determination of the most likely K of the STRUCTURE analyses.
(DOC)
Maximum Likelihood cladogram.
(DOC)
Maximum Parsimony cladogram.
(DOC)
Phengaris material used for analysis.
(DOC)
Microsatellite genotypes of Phengaris teleius and P. nausithous .
(DOC)