Background: Unusual fatty acids occur in higher plants through the action of divergent Δ12 fatty acid desaturases (FADs).
Results: Two new and one conserved group of Δ12 FADs were identified in Santalaceae plants.
Conclusion: Santalaceae FADs modify common and acetylenic fatty acids and show plasticity of activities.
Significance: This is the first report of FADs contributing to ximenynic acid formation.
Keywords: Enzyme Catalysis, Gene Expression, Lipids, Membrane Enzymes, Phylogenetics, Polyunsaturated Fatty Acids
Abstract
Plants in the Santalaceae family, including the native cherry Exocarpos cupressiformis and sweet quandong Santalum acuminatum, accumulate ximenynic acid (trans-11-octadecen-9-ynoic acid) in their seed oil and conjugated polyacetylenic fatty acids in root tissue. Twelve full-length genes coding for microsomal Δ12 fatty acid desaturases (FADs) from the two Santalaceae species were identified by degenerate PCR. Phylogenetic analysis of the predicted amino acid sequences placed five Santalaceae FADs with Δ12 FADs, which include Arabidopsis thaliana FAD2. When expressed in yeast, the major activity of these genes was Δ12 desaturation of oleic acid, but unusual activities were also observed: i.e. Δ15 desaturation of linoleic acid as well as trans-Δ12 and trans-Δ11 desaturations of stearolic acid (9-octadecynoic acid). The trans-12-octadecen-9-ynoic acid product was also detected in quandong seed oil. The two other FAD groups (FADX and FADY) were present in both species; in a phylogenetic tree of microsomal FAD enzymes, FADX and FADY formed a unique clade, suggesting that are highly divergent. The FADX group enzymes had no detectable Δ12 FAD activity but instead catalyzed cis-Δ13 desaturation of stearolic acid when expressed in yeast. No products were detected for the FADY group when expressed recombinantly. Quantitative PCR analysis showed that the FADY genes were expressed in leaf rather than developing seed of the native cherry. FADs with promiscuous and unique activities have been identified in Santalaceae and explain the origin of some of the unusual lipids found in this plant family.
Introduction
Higher plants produce a wide range of lipids de novo for incorporation into essential components such as phospholipid membranes and cuticular wax, as well as seed lipids that provide energy during germination. Some plants produce unusual fatty acids in their seed oils including those containing acetylenic moieties, where there are one or more carbon-carbon triple bonds within the fatty acid chain. The production of unusual fatty acids is considered to be a defense mechanism against predators or pathogens (1). Many higher plant species that produce acetylenic fatty acids have long been used for medicinal purposes, and recent studies have shown that acetylenic lipids have potent antimicrobial (2), antifungal (3), insecticidal (4), and anticancer (5) properties. The native cherry Exocarpos cupressiformis and sweet quandong Santalum acuminatum belong to the Santalaceae family, which share the presence of ximenynic or santalbic acid (trans-11-octadecen-9-ynoic acid, see Fig. 1A) at significant levels in the triacylglycerol fraction of seed kernels and conjugated polyacetylenic fatty acids in root tissue (6–8). The remaining fatty acids in seed oil include oleic acid, and small quantities of linoleic, linolenic, and stearolic (9-octadecynoic acid, Fig. 1B) acids are reportedly present in the oil of several members within the family (9).
FIGURE 1.
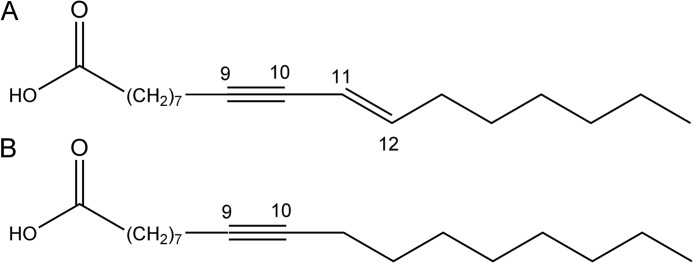
Molecular structure of unusual fatty acids identified in Santalaceae seed oils ximenynic acid (A) and stearolic acid (B).
In higher plants, much of the production of unusual fatty acids results from the action of divergent orthologs of the Arabidopsis thaliana FAD2-type Δ12 fatty acid desaturases (FADs2; 10) although most of the enzymes in this family modify the Δ12 position of an 18-carbon fatty acid. With regard to acetylenic bond formation in plant fatty acids, there have been two distinct types of FADs reported: the bifunctional acyl-lipid Δ6 acetylenase/desaturase identified in moss (11) and divergent forms of FAD2-type Δ12 acetylenases that are found in several higher plant families (12–14). In an attempt to identify the FADs involved in the biosynthesis of ximenynic acid and related polyacetylenic fatty acids in the Santalaceae, we investigated the Δ12 FADs of E. cupressiformis and S. acuminatum and tested their functions by recombinant expression in yeast and leaves of Nicotiana benthamiana. The range of FAD2-like desaturases from Santalaceae were found to exhibit unique activities and substrate plasticity toward olefinic and acetylenic substrates, illustrating the diversity in function that has evolved from this family of FADs in higher plants.
EXPERIMENTAL PROCEDURES
Plant Collection and Chemicals
Developing seeds and mature leaf tissue of E. cupressiformis were collected at Black Mountain (Australian Capital Territory, Australia), and developing seeds and mature leaf tissue of S. acuminatum were obtained from suburban Adelaide (South Australia). Quandong oil was extracted from mature S. acuminatum seed (Blackwood Seeds, Inman Valley, South Australia).
Molecular Characterization of Santalaceae FADs
Isolated total RNA from developing seeds and mature leaf and genomic DNA from mature leaf were used as templates for PCR with the following degenerate primer pair: PCdegFW, 5′-CAYGARTGYGGNCAYCAYGC and PCdegRV, 5′CCNCKNARCCARRCCCAYTC (12). Full-length nucleic acid sequences coding for putative Δ12 FADs were obtained using 3′- and 5′-RACE or genome walking using primers given in supplemental Table S1.
Functional Analysis of Santalaceae FADs in Saccharomyces cerevisiae and Nicotiana benthamiana
E. cupressiformis fatty acid desaturases (EcFADs) were amplified by RT-PCR from total RNA isolated from developing seed or by PCR from quandong genomic DNA with primer pairs containing restriction sites (supplemental Table S1) for directional cloning into pYES2 (Invitrogen) or pYES-DEST52 (Invitrogen) for EcFADX-1A and EcFADX-1B. SaFAD2 and SaFADY with a 6×His tag fused to the N terminus of the protein for yeast expression were amplified by PCR using the primers indicated in supplemental Table S1. The resulting PCR products were ligated into pENTRTM-d-TOPO® (Invitrogen), sequence verified, and then transferred to pYES-DEST52 using the Gateway®LR clonaseTM kit.
Clones containing S. acuminatum fatty acid desaturases SaFADs and EcFADs were transformed into S. cerevisiae strains ole1 L8-14C oleate desaturase-deficient (MATα, ole1:LEU2, leu2-3, leu2-112, trp1-1, ura3-52, his4) (15) and INVSc1 (MATα his3Δ1 leu2 trp1-289 ura3-52; Invitrogen) using the Sigma Yeast Transformation Kit and inoculated into cultures as described previously (16). Individual fatty acid substrates were added to yeast cultures in 20% tergitol, Nonidet P-40 at a final concentration of 200 μm during induction with galactose.
Microsomal fractions were prepared from induced yeast expressing tagged proteins via differential centrifugation of homogenized pellets. Microsomal protein (40 μg) was separated on a 4–12% Tris-Bis gel and transferred to Hybond-C Extra nitrocellulose membranes. The first and second antibodies were monoclonal anti-polyhistidine antibody produced in mouse (Sigma Aldrich) and anti-mouse IgG (Fab-specific) peroxidase antibody produced in goat (Sigma Aldrich). Chemiluminescent peroxidase substrate-1 (Sigma Aldrich) was used to detect horseradish peroxidase activity on the second antibody.
Recombinant Agrobacterium tumefaciens strain AGL1 containing SaFAD2, FADX, FADY, viral suppressor 35S:V2, 35S:NbhpFAD2.1, and 35S:LEC2 (17) using primers for directional cloning given in supplemental Table S1 were infiltrated into 5–6-week-old N. benthamiana leaves according to Wood et al. (18). After 5 days at 24 °C, protein expression in the infiltrated regions were confirmed by presence of a GFP signal, and leaf sections were harvested for lipid analysis.
Lipid Analysis
Fatty acid methyl esters (FAMEs) of yeast and N. benthamiana leaf lipids were prepared and analyzed according to Horne et al. (16) except by omission of the first washing step. Varian 1200 GC-MS conditions for analysis were as follows: injections were made in the split mode using helium as the carrier gas, and the oven temperature was raised from 60 to 180 °C at 20 °C per min, then 190 °C at 10 °C per min, and then to 260 °C at 75 °C per min and held for 5 min (Method 1). Alternatively, the initial oven temperature of 90 °C was raised to 190 °C at 20 °C per min, then to 190 °C at 2.5 °C per min, then to 200 °C at 20 °C per min, and finally to 260 °C at 40 °C per min (Method 2). Injector and transfer line were held at 250 °C. Picolinyl ester derivatives were prepared as described (19).
Synthesis of Fatty Acid Standards
cis and trans-12-octadecene-9-ynoic and cis-13, cis-15 and 17-octadecene-9-ynoic acids were synthesized based on established methods as detailed in the supplemental “Materials and Methods.” The products were confirmed by GC-MS of methyl esters and picolinyl ester derivatives of the corresponding deuterated fatty acids.
Quantitative Real-time PCR of E. cupressiformis Membrane FADs
Quantitative PCR analysis of E. cupressiformis FAD expression was compared with reference genes actin and ubiquitin using the Bio-Rad CFX96 real-time detection system with primer sequences given in supplemental Table S1 at a final concentration of 500 nm.
RESULTS
Twelve FAD2-type Fatty Acid Desaturases Identified from Santalaceae
We identified three distinct partial DNA sequences from S. acuminatum and five from E. cupressiformis using degenerate primers corresponding to conserved regions of FAD2-type FADs. Ultimately, three full-length genes were obtained for S. acuminatum, whereas nine full-length genes were obtained for E. cupressiformis, which included four pairs of genes sharing over 98% nucleic acid sequence identity within pairs, suggesting the occurrence of a recent genome duplication event.
The ORFs for all 12 DNA sequences coded for polypeptides of between 379 and 409 amino acid residues in length and the Kyte and Doolittle hydrophobicity profiles indicated each had six putative transmembrane or membrane-associated regions. The gene names were given a two-letter abbreviation for the species, Sa and Ec, followed by the type of fatty acid desaturase, i.e. FAD2, FADX, or FADY. Accession numbers for the Santalaceae desaturases are as follows: SaFAD2, KF556636; SaFADX, KF556637; SaFADY, KF556638; EcFAD2-1A, KF556639; EcFAD2-1B, KF556640; EcFAD2-2A, KF556641; EcFAD2-2B, KF556642; EcFADX-1A, KF556643; EcFADX-1B, KF556644; EcFADX-2, KF556645; EcFADY-A, KF556646; and EcFADY-B, KF556647.
The closest BLASTP result for SaFAD2 protein sequence against the non-redundant protein sequences in GenbankTM was 75% identity to the characterized Δ12 FAD from Davidia involucrata (20), whereas for SaFADY, the closest sequences had 63% identity to the Sesamum indicum FAD (21). SaFADX was also divergent from most of the FAD2 sequences in GenbankTM with 65% identity to a Δ12 FAD from Sapium sebiferum (Triadica sebifera) (22) as the closest match. Furthermore, FADX and FADY sequence identities were as distant from the Santalaceae FAD2 as they were from other plant FAD2 sequences (data not shown).
Two Divergent FAD2 Groups within Santalaceae
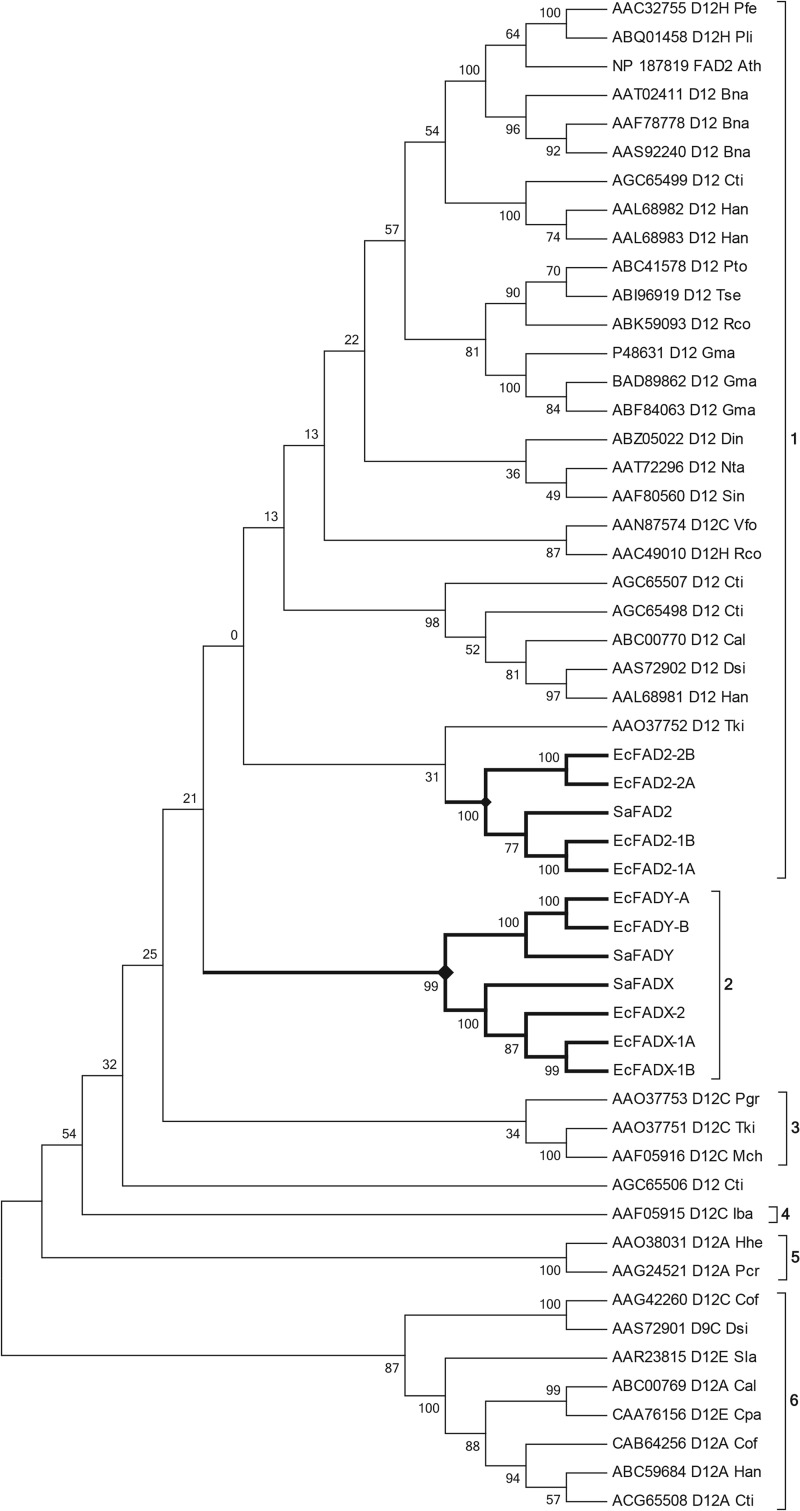
Phylogenetic analysis of the predicted amino acid sequences of the 12 Santalaceae genes was conducted against other reported FAD2-type FADs from higher plants using MEGA (version 5.2) (23) and inferred using the maximum likelihood method, which showed the Santalaceae FADs clustering into three groups: Sa- and EcFAD2s were associated with the well characterized microsomal Δ12 FAD lineage, and the other two groups (FADX, FADY) formed a separate clade, albeit with low bootstrap support, from the Δ12 FAD lineage and the divergent FAD clades (Fig. 2). There was strong support for two clusters of Santalaceae proteins (FADX, FADY) within the divergent group.
FIGURE 2.
Maximum likelihood phylogenetic analysis (bootstrap 1000 replicates) of desaturase protein sequences isolated from Exocarpos cupressiformis (Ec) and Santalum acuminatum (Sa) with those for reported phosphatidylcholine-type fatty acid desaturases. Ath, Arabidopsis thaliana; Bna, Brassica napus; Cal, Crepis alpina; Cof, Calendula officinalis; Cpa, Crepis palestina; Cti, Carthamus tinctorius; Dca, Daucus carota; Din, Davidia involucrate; Dsi, Dimorphotheca sinuate; Gma, Glycine max; Han, Helianthus annus; Hhe, Hedera helix; Iba, Impatiens balsamina; Mch, Momordica charantia; Nta, Nicotiana tabacum; Pcr, Petroselinum crispum; Pfe, Physaria fendleri; Pli, Physaria lindheimeri; Pto, Populus tomentosa; Ptr, Populus trichocarpa; Rco, Ricinus communis; Rhi, Rudbeckia hirta; Sin, Sesamum indicum; Sla, Stokesia lavis; Tki, Trichosanthes kirilowii; Tse, Triadica sebifera; Vfo, Vernicia fordii; Vvi, Vitis vinifera; Xbr, Xerochrysum bracteatum. Santalaceae fatty acid desaturase branch lines are in boldface type and indicated by filled diamonds. Enzyme activities are indicated to the right hand side of the tree. 1, Δ12 fatty acid desaturases; 2, Santalaceae divergent fatty acid desaturases; 3, Cucurbitaceae and Lythraceae fatty acid conjugases; 4, Balsaminaceae fatty acid conjugase; 5, Apiaceae/Araliaceae fatty acid acetylenases; 6, Asteraceae divergent fatty acid desaturases.
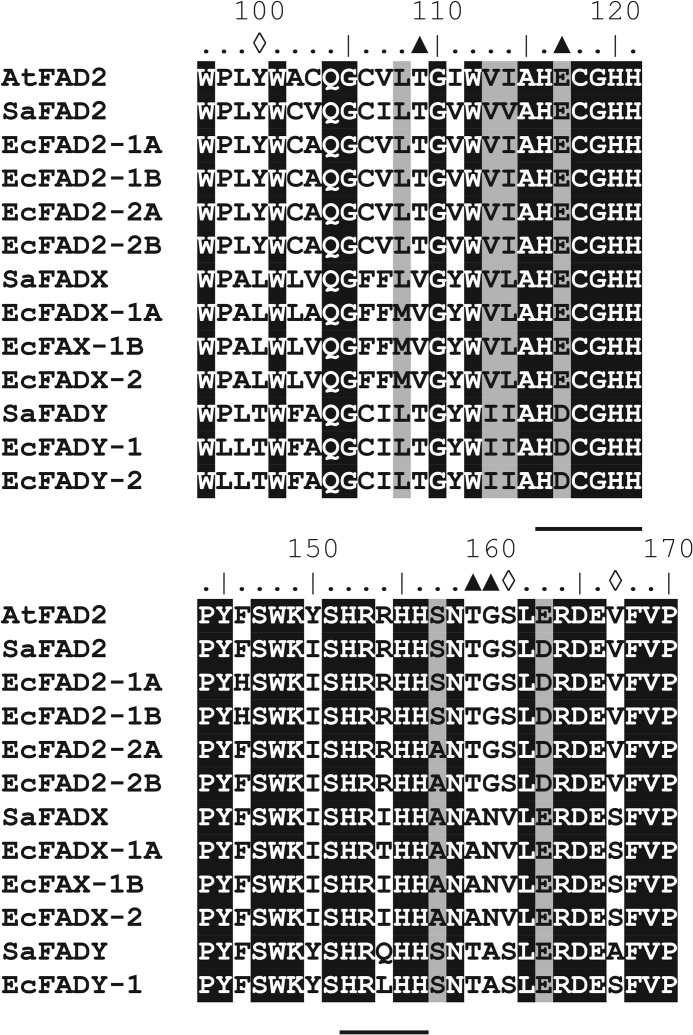
The amino acid residues that comprised the three conserved histidine box regions of the Santalaceae FAD2 sequences were identical to those of Δ12 FADs such as the archetypal A. thaliana FAD2 (Fig. 3 for the first two box motifs). However, the Santalaceae FADYs were distinctively different in having an unusual motif in the first histidine box: HDCGH containing aspartate in the second position, whereas glutamate in the equivalent position is conserved among all microsomal Δ12 FADs and their divergent family members with one other exception (see “Discussion”). Intriguingly, the microsomal Δ15 FAD of higher plants also have the HDCGH motif in the second histidine box; however, other features of the Δ15 FADs were not shared with the FADY sequences. The FADX and FADY sequences displayed further divergence from conserved residues in the proximity of the histidine boxes of Δ12 FADs (Fig. 3), suggesting they may catalyze unusual reactions.
FIGURE 3.
Selected regions of amino acid sequence alignment between Santalaceae fatty acid desaturases and A. thaliana FAD2. A, residues 97–121 incorporating the first “histidine box.” B, residues 144–170 incorporating the second histidine box. Dark shaded sequence represents complete conservation of residue and light shading indicates conservation of character. Locations of the key amino acid residues in the Santalaceae sequences which differ from highly conserved residues in all microsomal Δ12 fatty acid desaturases (including divergent forms) are indicated by open diamonds and where they differ from Δ12 fatty acid desaturases but not other divergent enzymes, such as fatty acid acetylenases and conjugases, are indicated by filled triangles.
Two FAD Groups Exhibit Activity on Both Olefinic and Acetylenic Fatty Acids
All Santalaceae FADs were functionally assessed by expression in S. cerevisiae, and their activities were determined in the presence and absence of exogenously supplied fatty acids. The putative Δ12 FADs coded by SaFAD2, and the four EcFAD2s all exhibited considerable Δ12 FAD activity on oleic acid-linked substrates naturally occurring within the yeast to form linoleic acid, as expected based on the phylogenetic analysis and sequence examination (Table 1 and Fig. 4A). In particular, SaFAD2 exhibited high desaturation activity in yeast. The Santalaceae FAD2s also converted palmitoleate to palmitolinoleic acid in yeast (Table 1 and Fig. 4A), whereas none of the products were detected in yeast expressing the empty vector (Fig. 4B). Δ12 FAD activity of SaFAD2 was also observed by transient expression in N. benthamiana leaf with silencing of the endogenous FAD2.1 using a hairpin construct and resulted in 160% increase in linoleic acid content (n = 3; p < 0.05) compared with leaf expressing the FAD2.1 silencing hairpin only.
TABLE 1.
Summary of the identified fatty acid products and mean percent conversion of substrates (and S.D. where number of replicates was three or greater) by Santalaceae fatty acid desaturases expressed in S. cerevisiae
Mean percentage conversion is calculated based on weight percent of product as a proportion of the total weight of product and substrate, averaged from three independent experiments.
| Gene/Substrate | Product | Conversion (%, S.D.) |
|---|---|---|
| SaFAD2 | ||
| Palmitoleic | Palmitolinoleic | 26.7 (2.9) |
| Oleic | Linoleic | 63.3 (5.8) |
| Linoleic | Linolenic | 0.2 |
| Stearolic | Trans-12-octadecen-9-ynoic | 35.0 (12.3) |
| Stearolic | Ximenynic | 1.3 (0.9) |
| EcFAD2-1A | ||
| Palmitoleic | Palmitolinoleic | 4.1 |
| Oleic | Linoleic | 23.0 |
| Stearolic | Trans-12-octadecen-9-ynoic | 4.8 |
| EcFAD2-2A | ||
| Palmitoleic | Palmitolinoleic | 10.0 |
| Oleic | Linoleic | 34.0 |
| Linoleic | Linolenic | trace |
| Stearolic | Trans-12-octadecen-9-ynoic | 18.0 |
| SaFADX | ||
| Stearolic | Cis-13-octadecen-9-ynoic | 18.7 (13.1) |
| EcFADX-1B | ||
| Stearolic | Cis-13-octadecen-9-ynoic | 0.7 |
| EcFADX-2 | ||
| Stearolic | Cis-13-octadecen-9-ynoic | 0.8 |
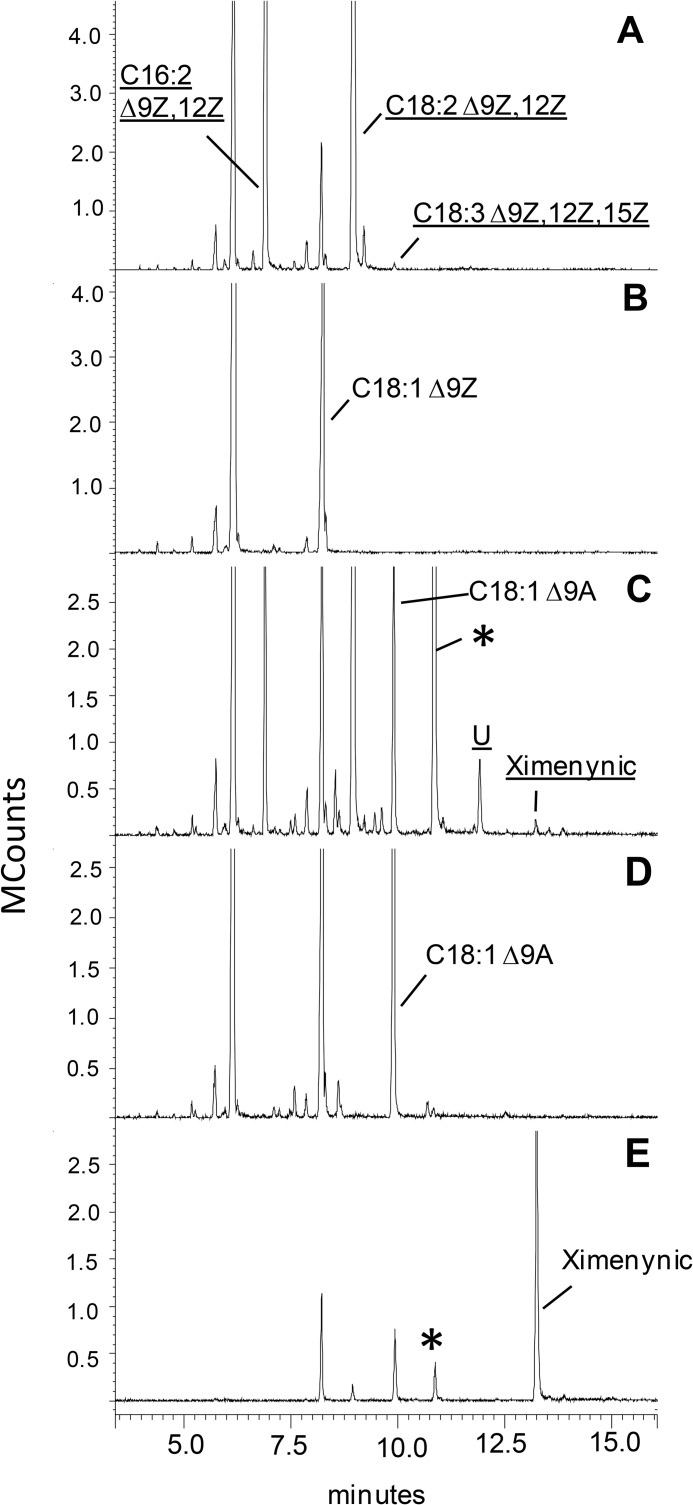
FIGURE 4.
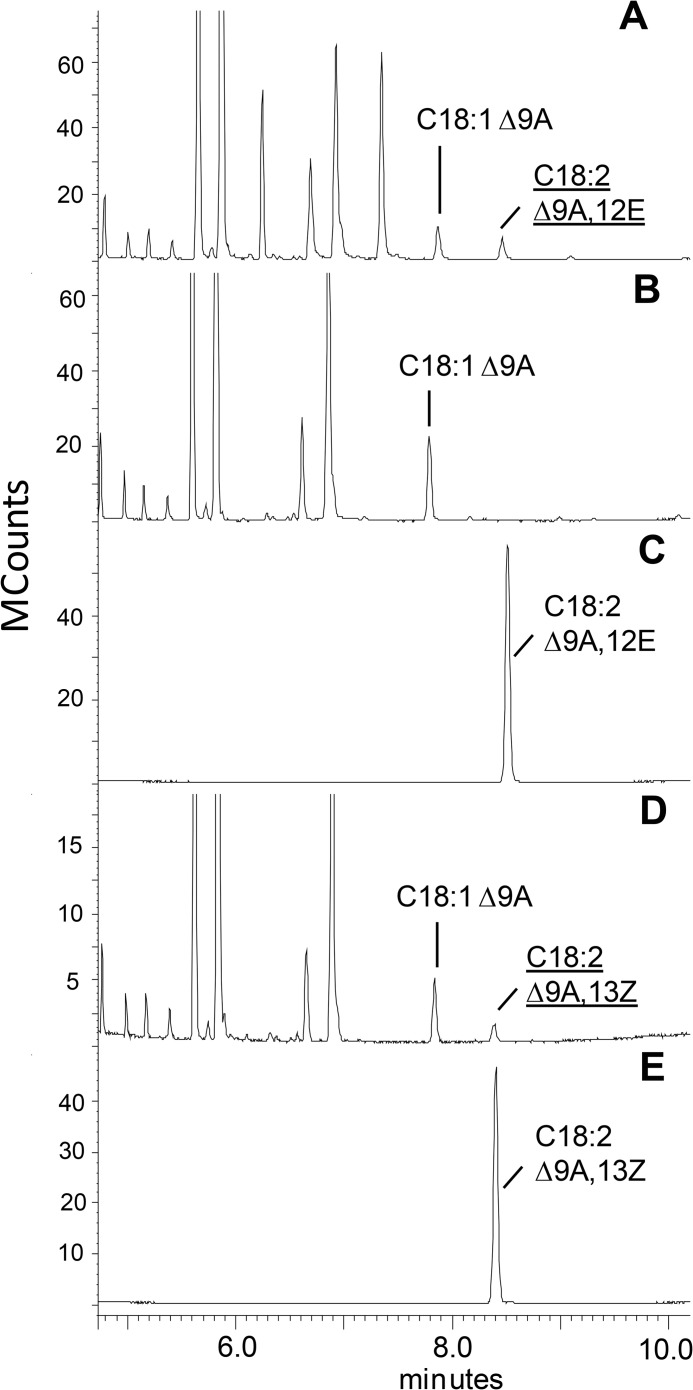
A gas chromatography/mass spectrometry analysis of FAMEs is shown using analysis method 1 of yeast expressing SaFAD2 (A) or yeast expressing the empty vector pYES2 with no added substrates (B). Shown is yeast expressing SaFAD2 (C) and empty vector pYES2 (D) where both samples were supplemented with stearolic acid (C18:1 9A) during gene expression and an extract of S. acuminatum seed oil (E). In C, the product designated with an asterisk was identified as trans-12-octadecen-9-ynoic acid, but the product denoted as U was not conclusively identified. Fatty acids are represented by XX:YΔZ where XX is the carbon chain length, Y is the total number of carbon-carbon double or triple bonds in the molecule, and Z is the position of insertion of the unsaturation. A indicates an acetylenic bond. New products formed in the sample are indicated by underlined letters. Results are shown for the INVSc1 yeast strain.
The Santalaceae FAD2 proteins also showed plasticity of FAD activity; SaFAD2 and EcFAD2–2 enzymes displayed minor Δ15 desaturation activity producing linolenic from linoleic acid; the products were identified by GC-MS comparison with an authentic standard, and picolinyl ester derivatives showed diagnostic ions for the gaps of 40 mass units in α-linolenic acid, each representing a double bond and methylene group at m/z = 220 and 260, 260 and 300, and 300 and 340. When supplied with exogenous stearolic acid, SaFAD2 produced ximenynic acid (Fig. 4C), which was unequivocally identified as the product by comparison of retention time (Fig. 4E) and mass spectra with an authentic standard (Fig. 5, A and B), and was generated in four independent experiments with Santalaceae FAD2 expressed in two yeast strains: ole1 and INVSc1 (Table 1). The product was absent in the vector control experiments (Fig. 4D).
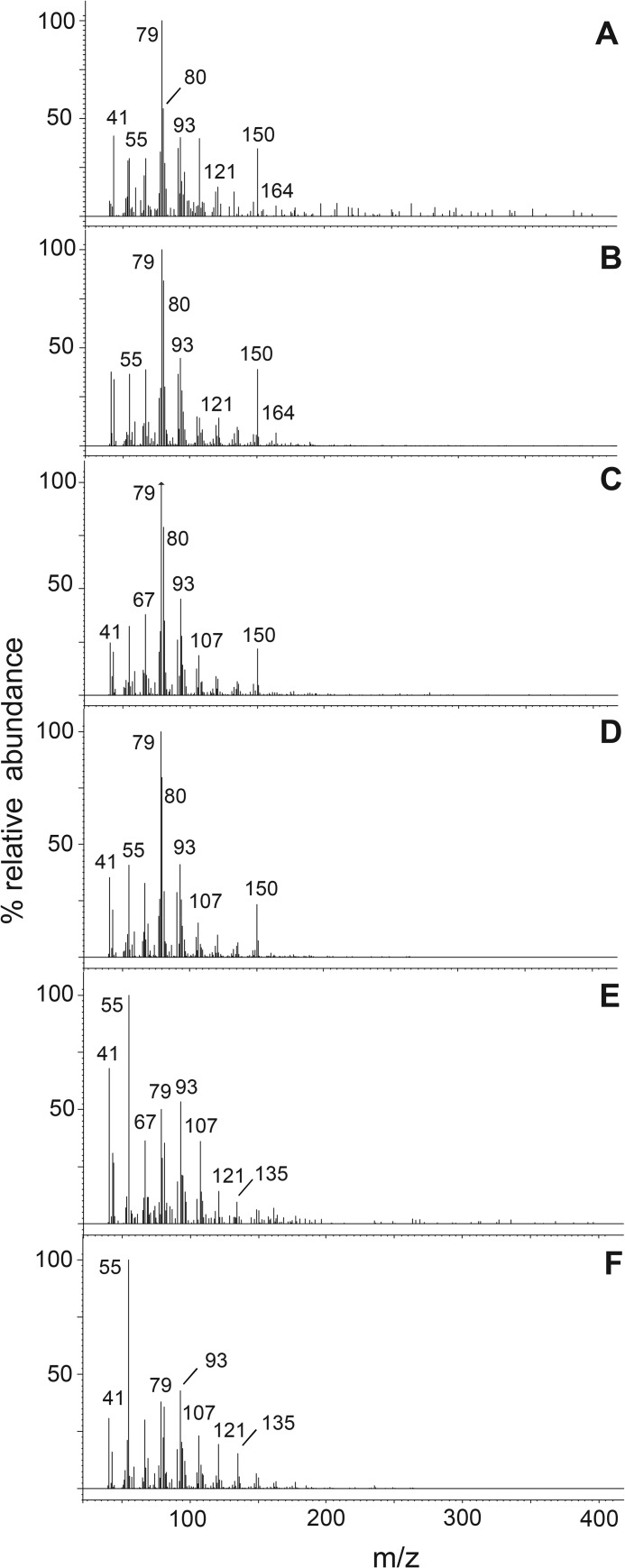
FIGURE 5.
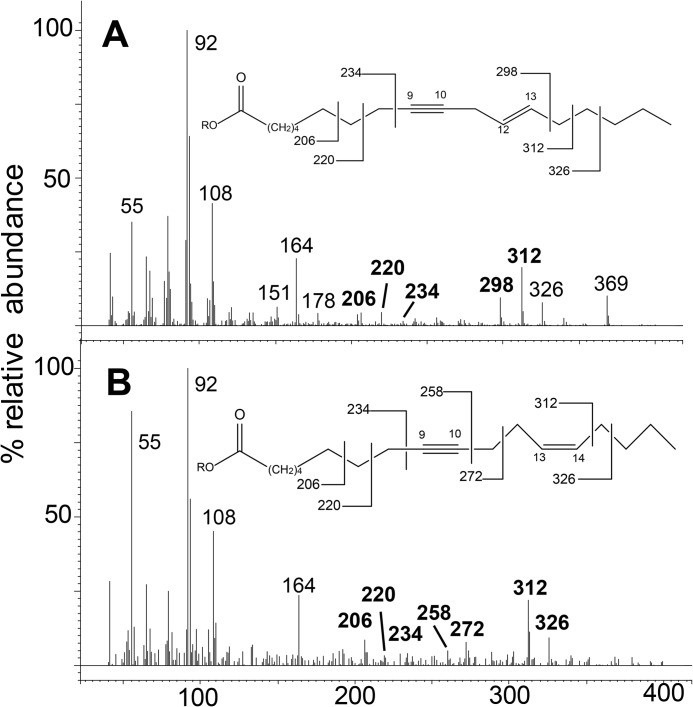
Mass spectra obtained over 40–400 atomic mass units (amu) of FAME derivatives of metabolites and standards using analysis method 2. A, yeast lipid product of SaFAD2 acting on stearolic acid eluting at retention time 13. 228 min. B, authentic standard of ximenynic acid eluting at 13.252 min (analysis method 1). C, yeast lipid product of SaFAD2 acting on stearolic acid eluting at 8.478 min. D, synthesized standard of trans-12-octadecen-9-ynoic acid eluting at 8.500 min. E, yeast lipid product of SaFADX acting on stearolic acid eluting at 8.384 min. F, synthesized standard of cis-13-octadecen-9-ynoic acid eluting at 8.405 min.
A further new product of SaFAD2 incubation with stearolic acid was putatively identified as trans-12-octadecen-9-ynoic acid by picolinyl derivatization (Fig. 6A) and then confirmed by GC retention time and FAME MS comparison with synthetic references (Fig. 5, C and D; Fig. 7, A and C). Interestingly, this compound was also present in quandong seed oil (Fig. 4E). All EcFAD2s also displayed the v+3 desaturation activity toward stearolic acid at a lower conversion rate (Table 1).
FIGURE 6.
Mass spectra of picolinyl derivatives of desaturation products generated by yeast supplemented with stearolic acid and expressing SaFAD2 (A) or SaFADX (B). Diagnostic mass spectral peaks are shown in boldface type, and an explanation for the fragmentation pattern is shown with each mass spectrum where R = 3-picolinyl.
FIGURE 7.
Shown is a gas chromatography/mass spectrometry analysis of FAME using analysis method 2 of yeast expressing SaFAD2 (A) yeast expressing the empty vector pYES2 (B) supplemented with stearolic acid. C, synthesized standard of trans-12-octadecen-9-ynoic acid (C18:2 Δ9A 12E). D, yeast expressing SaFADX supplemented with stearolic acid. E, synthesized standard of cis-13-octadecen-9-ynoic acid (C18:2 Δ9A 13Z). Fatty acids are represented by XX:YΔZ where XX is the carbon chain length, Y is the total number of carbon-carbon double or triple bonds in the molecule, and Z is the position of insertion of the unsaturation. A indicates an acetylenic bond. New products formed in the sample are indicated by underlined names. Results shown for the INVSc1 yeast strain.
Santalaceae FADX expressed in yeast had no detectable Δ12 desaturase activity either acting on native fatty acids or when supplied with exogenous substrates but instead showed cis-Δ13 desaturation activity on stearolic acid (Fig. 7D). Identification of the cis-13-octadecen-9-ynoic acid product was made first via the picolinyl derivative (Fig. 6B) and then confirmed by the retention time of the methyl ester of a synthesized standard (Fig. 7E) and comparison of FAME mass spectra (Fig. 5, E and F). Standards of cis-12-, cis-15-, and 17-octadecen-9-ynoic acids were also prepared, but none of these products matched the retention times/mass spectra of unknown metabolites of Santalaceae FAD2 and FADX incubated with stearolic acid (data not shown). No other activity was detected for this enzyme group despite testing them against a wide range of fatty acid substrates.
By contrast and despite many attempts, no activity could be detected for the Santalaceae FADY genes expressed in both yeast and N. benthamiana leaf. A mutated version of the SaFADY gene which altered the encoded amino acid residue from D106E (numbering based on A. thaliana FAD2) and returned a glutamate residue in place of the unusual aspartate to the first histidine box, was assessed in yeast; however, no activity was observed from this mutant gene. Expression of N-terminal polyhistidine-tagged versions of SaFADY and SaFAD2 in yeast was assessed by Western blot. Both protein products were detected in yeast microsomal fractions prepared from cells harvested at 18 h post induction (data not shown). Therefore, lack of activity of SaFADY in yeast was not due to lack of expression or localization to endoplasmic reticulum.
Santalaceae FAD Expression Profile Differs between Developing Seed and Leaf Tissue
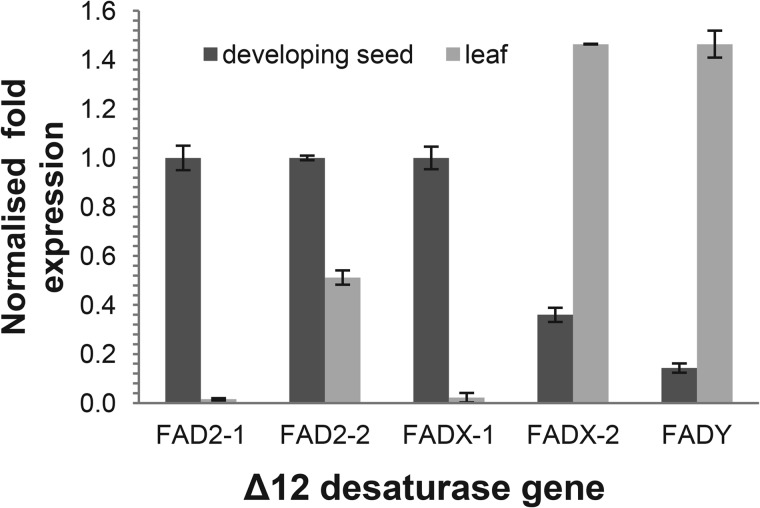
Relative quantitative PCR was conducted in E. cupressiformis leaf and developing seed RNA on the four EcFAD pairs: EcFAD2-1A/B; EcFAD2-2A/B; EcFADY-A/B; EcFADX-1A/B and the single gene EcFADX-2, which were normalized against the expression of ubiquitin and actin. Due to the high DNA sequence identity between all four pairs of EcFADs extending through to the 3′-UTR region, it was not possible to design specific primers to distinguish between them; therefore, the expression levels of the eight EcFADs were determined as four pairs. The expression patterns in developing seed and mature leaf tissue showed a consistent trend with the Δ12 FADs EcFAD2-1 and -2 and FADX-1 all significantly more highly expressed in developing seed than leaf, and FADY and FADX-2 with a much higher expression in leaf compared with developing seed (Fig. 8).
FIGURE 8.
Relative expression levels of E. cupressiformis fatty acid desaturases genes determined by quantitative PCR analysis of developing seed and mature leaf RNA. Amplified PCR products were sequence-verified. All Ct values were obtained from triplicate PCR reactions, and PCR efficiencies for all genes based on standard curves were calculated to be between 95 and 105%. ΔΔCT scores were normalized against all three reference genes.
DISCUSSION
Herein we have described the isolation and characterization of twelve full-length genes coding for microsomal Δ12 fatty acid FADs from two Santalaceae species that accumulate high levels of acetylenic fatty acids in their seed oils. Based on phylogenetic and conserved residue analysis of amino acid sequences, two new divergent clusters of FADs of the Δ12 type were identified from the Santalaceae, and the remaining sequences were grouped within the well characterized FAD2 family from plants. Santalaceae Δ12 FADs are likely to have key roles in the modification of acetylenic fatty acids and contribute to the production of seed oil and root lipids.
The Santalaceae FAD2 enzymes showed significant plasticity of activity: the enzymes demonstrated efficient cis-Δ12 and cis-Δ15 desaturation of olefinic fatty acids and, notably, also exhibited trans-Δ12 and trans-Δ11 (production of ximenynic acid) FAD activities when incubated with stearolic acid. Flax FAD2s have also been shown to produce small amounts of linolenic acid (24), and together with the present study, strengthens the suggestion that the evolutionary origin of the Δ15 FAD family may be the Δ12 FAD. Plant microsomal Δ12 FADs do exhibit functional plasticity. A small number of amino acid changes around the catalytic site results in changed enzymatic activities (25, 26). Within the major Δ12 FAD cluster (group 1) given in Fig. 2, there are enzymes with divergent activities that include conjugase and hydroxylase; however, it is unusual to observe such promiscuity in catalytic activity in a plant microsomal Δ12 FAD as observed with the Santalaceae FAD2 enzymes.
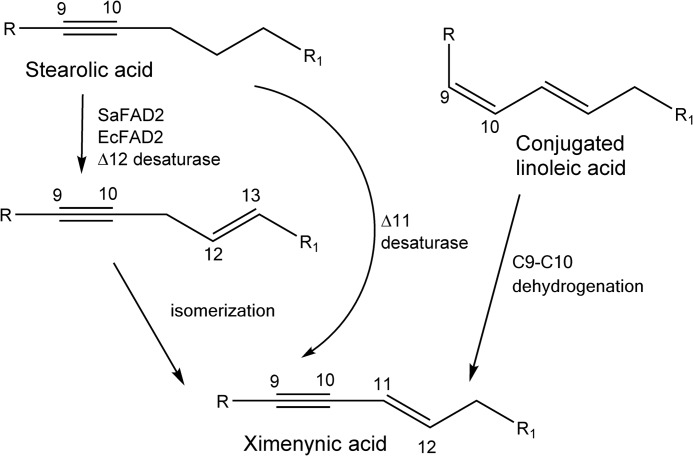
Quantitative PCR analysis on the E. cupressiformis FAD genes showed much higher relative expression of EcFAD2 genes in developing seed compared with leaf tissue, which is consistent with their having a role in seed lipid modification. Mature seed oils of Santalum spp. and E. cupressiformis contain mainly ximenynic and oleic acids (7, 8) with <2% linoleic acid and <5% stearolic acid (7, 27). In considering the biosynthetic origin of ximenynic acid in the Santalaceae, Morris and Marshall (27) have proposed that the fatty acid arises by Δ12 desaturation of stearolic acid followed by isomerization of the Δ12 double bond into the trans-Δ11 position (Fig. 9). Additionally, Liu et al. (9) have suggested a route proceeding via a conjugated linoleic acid intermediate, which undergoes dehydrogenation at the Δ9 position. To test these possibilities, we have incubated the Santalaceae FADs expressed in yeast with trans-12- and cis-12-octadecen-9-ynoic acids as well as various conjugated linoleic acid isoforms and found no conversion into ximenynic acid (data not shown). Therefore, the available data indicate formation of ximenynic acid by direct trans-Δ11-desaturation of stearolic acid (Fig. 9).
FIGURE 9.
Potential pathways for the formation of ximenynic acid from stearolic or conjugated linoleic acid and those identified as catalyzed by Santalaceae Δ12 fatty acid desaturases. R = HOOC(CH2)7− and R1 = −C4H9.
As a further example of the divergent nature of the Santalaceae FADs, the FADX enzymes were found to lack Δ12 desaturase activity when expressed in yeast but instead displayed an unusual desaturation activity toward stearolic acid by inserting a cis-double bond in the Δ13 position. Although the FADX-1 genes of E. cupressiformis are preferentially expressed in developing seeds, the Δ9yne Δ13ene product of this gene on stearolic acid was not detected in seed oil and may undergo further modification.
The second group of divergent Santalaceae FADs, the FADY type, were strongly expressed in leaf but no activity could be detected for the enzyme under heterologous expression. Although the identification of FAD isoforms that are expressed predominantly in vegetative tissues versus seed-specific forms is not unusual, for example, soybean and olive each possess two FADs showing this expression pattern (28, 29), the FADYs all possessed an aspartate in place of the conserved glutamate in the first histidine box motif, a corresponding enzymatic function to which has, to date, only been identified in one type of Δ12 FAD, the Momordica charantia (MomoFADX) fatty acid conjugase (30) and closely related sequences. MomoFADX appeared to be inactive when expressed in yeast, but produced α-eleostearic acid (cis-9, trans-11, trans-13-octadecatrienoic acid) when expressed in somatic soybean embryo. Furthermore, when the first histidine box aspartate of MomoFADX was converted to the more common glutamate, production of α-eleostearic acid increased (31). However, in our case, expression of Santalaceae FADY genes in a plant system, i.e. transient N. benthamiana leaf or mutation of the SaFADY first histidine box asparate to glutamate did not produce any new fatty acid products.
Ximenynic acid contains an acetylenic bond at the Δ9 position, which provokes the question: what enzyme catalyzes formation of the triple bond? Arguing against the possible involvement of Santalaceae Δ12 FADs in this transformation is the knowledge that the vast majority of these enzymes insert a double bond between C12-C13 position or above, however, there have been two exceptions reported: a conjugase from Calendula officinalis (32) and a fatty acid desaturase/hydroxylase from Dimorphotheca spp. (33). Nevertheless, Δ9 acetylenase activity was not detected in the Santalaceae FAD enzymes.
New catalytic activities among the plant microsomal Δ12 FADs are reported intermittently; many of these new activities catalyze the modifications of fatty acids that have resulted in the remarkable diversity of plant seed oils. The Δ12 FAD group has had multiple gene duplication events, which then evolved to acquire novel functions other than Δ12 desaturation, and contrasts strongly with the lack of functional diversity observed among the microsomal Δ15 FADs in higher plants. The present study has revealed new activities from the plant Δ12 FAD enzymes, i.e. trans-Δ11 (ximenynic acid formation), trans-Δ12, and cis-Δ13 desaturations when acting on stearolic acid and Δ15 desaturation when acting on olefinic fatty acids.
Acknowledgments
We thank Surinder Singh and Thomas Vanhercke for helpful comments on the manuscript.
This work was supported by the CSIRO Grains Research Development Corporation Crop Biofactories Initiative.

This article contains supplemental “Materials and Methods,” Table S1, and additional references.
- FAD
- fatty acid desaturase
- FAME
- fatty acid methyl ester
- Sa
- S. acuminatum
- Ec
- E. cupressiformis
- amu
- atomic mass unit
- SaFAD
- S. acuminatum fatty acid desaturase
- EcFAD
- E. cupressiformis fatty acid desaturase.
REFERENCES
- 1. Minto R. E., Blacklock B. J. (2008) Biosynthesis and function of polyacetylenes and allied natural products. Prog. Lipid Res. 47, 233–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Panthama N., Kanokmedhakul S., Kanokmedhakul K. (2010) Polyacetylenes from the roots of Polyalthia debilis. J. Nat. Prod. 73, 1366–1369 [DOI] [PubMed] [Google Scholar]
- 3. Li X. C., Jacob M. R., Khan S. I., Ashfaq M. K., Babu K. S., Agarwal A. K., Elsohly H. N., Manly S. P., Clark A. M. (2008) Potent in vitro antifungal activities of naturally occurring acetylenic acids. Antimicrob. Agents Chemother. 52, 2442–2448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cantrell C. L., Pridgeon J. W., Fonczek F. R., Becnel J. J. (2010) Structure-activity relationship studies on derivatives of Eudesmanolides from Inula helenium as toxicants against Aedes aegypti larvae and adults. Chem. Biodivers. 7, 1681–1697 [DOI] [PubMed] [Google Scholar]
- 5. Dembitsky V. M. (2006) Anticancer activity of natural and synthetic acetylenic lipids. Lipids 41, 883–924 [DOI] [PubMed] [Google Scholar]
- 6. Hatt H. H., Triffett A. C., Wailes P. C. (1960) Acetylenic acids from fats of the Olacaceae and Santalaceae. 4. The occurrence of octadeca-trans-11, trans-13-dien-9-ynoic acid in plant lipids. Aust. J. Chem. 13, 488–497 [Google Scholar]
- 7. Hatt H. H., Triffett A. C., Wailes P. C. (1959) Acetylenic acids from fats of Santalaceae and Olacaceae – Seed and root oils of Exocarpus-cupressiformis Labill. Aust. J. Chem. 12, 190–195 [Google Scholar]
- 8. Bu'Lock J. D., Smith G. N. (1963) Acetylenic fatty acids in seeds and seedlings of sweet quandong. Phytochemistry 2, 289–296 [Google Scholar]
- 9. Liu Y., Longmore R. B., Kailis S. G. (1997) Proximate and fatty acid composition changes in developing sandalwood (Santalum spicatum) seeds. J. Sci. Food Agric. 75, 27–30 [Google Scholar]
- 10. Dwyer J. M., Stymne S., Green A. G., Carlsson A. S. (2008) High value oils from plants. Plant J. 54, 640–655 [DOI] [PubMed] [Google Scholar]
- 11. Sperling P., Lee M., Girke T., Zahringer U., Stymne S., Heinz E. (2000) A bifunctional δ-fatty acyl acetylenase/desaturase from the moss Ceratodon purpureus. A new member of the cytochrome b5 superfamily. Eur. J. Biochem. 267, 3801–3811 [DOI] [PubMed] [Google Scholar]
- 12. Lee M., Lenman M., Banaś A., Bafor M., Singh S., Schweizer M., Nilsson R., Liljenberg C., Dahlqvist A., Gummeson P. O., Sjödahl S., Green A., Stymne S. (1998) Identification of non-heme diiron proteins that catalyze triple bond and epoxy group formation. Science 280, 915–918 [DOI] [PubMed] [Google Scholar]
- 13. Cahoon E. B., Schnurr J. A., Huffman E. A., Minto R. E. (2003) Fungal responsive fatty acid acetylenases occur widely in evolutionarily distant plant families. Plant J. 34, 671–683 [DOI] [PubMed] [Google Scholar]
- 14. Cao S., Zhou X. R., Wood C. C., Green A. G., Singh S. P., Liu L., Liu Q. (2013) A large and functionally diverse family of Fad2 genes in safflower (Carthamus tinctorius L.). BMC Plant Biol. 13, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Stukey J. E., McDonough V. M., Martin C. E. (1990) The OLE1 gene of Saccharomyces cerevisiae encodes the δ 9 fatty acid desaturase and can be functionally replaced by the rat stearoyl-CoA desaturase gene. J. Biol. Chem. 265, 20144–20149 [PubMed] [Google Scholar]
- 16. Horne I., Gibb N., Damcevski K., Glover K., Haritos V. S. (2010) Two conserved Z9-octadecanoic acid desaturases in the red flour beetle, Tribolium castaneum. Gene 468, 41–47 [DOI] [PubMed] [Google Scholar]
- 17. Naim F., Nakasugi K., Crowhurst R. N., Hilario E., Zwart A. B., Hellens R. P., Taylor J. M., Waterhouse P. M., Wood C. C. (2012) Advanced engineering of lipid metabolism in Nicotiana benthamiana using a draft genome and the V2 viral silencing suppressor protein. PLoS One 7, e52717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wood C. C., Petrie J. R., Shrestha P., Mansour M. P., Nichols P. D., Green A. G., Singh S. P. (2009) A leaf-based assay using interchangeable design principles to rapidly assemble multistep recombinant pathways. Plant Biotechnol. J. 7, 914–924 [DOI] [PubMed] [Google Scholar]
- 19. Harvey D. J. (1982) Picolinyl esters as derivatives for the structural determination of long chain branched and unsaturated fatty acids. Biol. Mass Spectrom. 9, 33–38 [Google Scholar]
- 20. Lei N., Peng S., Niu B., Chen J., Zhou J., Tang L., Xu Y., Wang S., Chen F. (2010) Molecular cloning and characterization of a novel microsomal oleate desaturase gene DiFAD2 from Davidia involucrata Baill. Biol. Plant. 54, 41–46 [Google Scholar]
- 21. Jin U. H., Lee J. W., Chung Y. S., Lee J. H., Yi Y. B., Kim Y. K., Hyung N. I., Pyee J. H., Chung C. H. (2001) Characterization and temporal expression of a [omega]-6 fatty acid desaturase cDNA from sesame (Sesamum indicum L.) seeds. Plant Sci. 161, 935–941 [Google Scholar]
- 22. Niu B., Ye H., Xu Y., Wang S., Chen P., Peng S., Ou Y., Tang L., Chen F. (2007) Cloning and characterization of a novel δ12-fatty acid desaturase gene from the tree Sapium sebiferum. Biotechnol. Lett. 29, 959–964 [DOI] [PubMed] [Google Scholar]
- 23. Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. (2011) MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Khadake R. M., Ranjekar P. K., Harsulkar A. M. (2009) Cloning of a novel omega-6 desaturase from flax (Linum usitatissimum L.) and its functional analysis in Saccharomyces cerevisiae. Mol. Biotechnol. 42, 168–174 [DOI] [PubMed] [Google Scholar]
- 25. Broadwater J. A., Whittle E., Shanklin J. (2002) Desaturation and hydroxylation. Residues 148 and 324 of Arabidopsis FAD2, in addition to substrate chain length, exert a major influence in partitioning of catalytic specificity. J. Biol. Chem. 277, 15613–15620 [DOI] [PubMed] [Google Scholar]
- 26. Blacklock B. J., Scheffler B. E., Shepard M. R., Jayasuriya N., Minto R. E. (2010) Functional diversity in fungal fatty acid synthesis: the first acetylenase from the Pacific golden chanterelle, Cantharellus formosus. J. Biol. Chem. 285, 28442–28449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Morris L. J., Marshall M. O. (1966) Occurrence of stearolic acid in Santalaceae seed oils. Chemistry Industry 12, 460–461 [PubMed] [Google Scholar]
- 28. Heppard E. P., Kinney A. J., Stecca K. L., Miao G. H. (1996) Developmental and growth temperature regulation of two different microsomal w-6 desaturase genes in soybeans. Plant Physiol. 110, 311–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hernández M. L., Mancha M., Martínez-Rivas J. M. (2005) Molecular cloning and characterization of genes encoding two microsomal oleate desaturases (FAD2) from olive. Phytochemistry 66, 1417–1426 [DOI] [PubMed] [Google Scholar]
- 30. Cahoon E. B., Carlson T. J., Ripp K. G., Schweiger B. J., Cook G. A., Hall S. E., Kinney A. J. (1999) Biosynthetic origin of conjugated double bonds: production of fatty acid components of high-value drying oils in transgenic soybean embryos. Proc. Natl. Acad. Sci. U.S.A. 96, 12935–12940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Rawat R., Yu X. H., Sweet M., Shanklin J. (2012) Conjugated fatty acid synthesis. Residues 111 and 115 influence product partitioning of Momordica charantia conjugase. J. Biol. Chem. 287, 16230–16237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Qiu X., Reed D. W., Hong H., MacKenzie S. L., Covello P. S. (2001) Identification and analysis of a gene from Calendula officinalis encoding a fatty acid conjugase. Plant Physiol. 125, 847–855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Cahoon E. B., Kinney A. J. (2004) Dimorphecolic acid is synthesized by the coordinate activities of two divergent Δ12-oleic acid desaturases. J. Biol. Chem. 279, 12495–12502 [DOI] [PubMed] [Google Scholar]