Abstract
The human intestine, colonized by a dense community of resident microbes, is a frequent target of bacterial pathogens. Undisturbed, this intestinal microbiota provides protection from bacterial infections. Conversely, disruption of the microbiota with oral antibiotics often precedes the emergence of several enteric pathogens1–4. How pathogens capitalize upon the failure of microbiota-afforded protection is largely unknown. Here we show that two antibiotic-associated pathogens, Salmonella typhimurium and Clostridium difficile, employ a common strategy of catabolizing microbiota-liberated mucosal carbohydrates during their expansion within the gut. S. typhimurium accesses fucose and sialic acid within the lumen of the gut in a microbiota-dependent manner, and genetic ablation of the respective catabolic pathways reduces its competitiveness in vivo. Similarly, C. difficile expansion is aided by microbiota-induced elevation of sialic acid levels in vivo. Colonization of gnotobiotic mice with a sialidase-deficient mutant of the model gut symbiont Bacteroides thetaiotaomicron (Bt) reduces free sialic acid levels resulting in a downregulation of C. difficile’s sialic acid catabolic pathway and impaired expansion. These effects are reversed by exogenous dietary administration of free sialic acid. Furthermore, antibiotic treatment of conventional mice induces a spike in free sialic acid and mutants of both Salmonella and C. difficile that are unable to catabolize sialic acid exhibit impaired expansion. These data show that antibiotic-induced disruption of the resident microbiota and subsequent alteration in mucosal carbohydrate availability are exploited by these two distantly related enteric pathogens in a similar manner. This insight suggests new possibilities for therapeutic approaches for preventing diseases caused by antibiotic-associated pathogens.
The intestinal microbiota is composed of trillions of microbial cells that together form a complex, dynamic, and highly competitive ecosystem5,6. Limited nutrients and high microbial densities likely play a key role in protecting the host against invading microbes7. Carbohydrates derived from diet or host play a well-established role in sustaining the resident members of the microbiota8–10, and more recently have been shown to play important roles in gut microbiota-pathogen dynamics11–14. Oral antibiotic use is one of the leading risk factors for disease associated with Salmonella spp. and Clostridium difficile, consistent with increased enteric vulnerability upon disruption of the resident microbiota1–4,15. In addition, mouse models of S. typhimurium or C. difficile infection commonly require disruption of the intestinal microbiota with antibiotics to promote pathogen expansion within the lumen of the gut and to initiate disease16–19. Deciphering the numerous mechanisms by which the microbiota prevents bacterial pathogen expansion and how microbiota disruption enables pathogens to circumvent these mechanisms remains an important task.
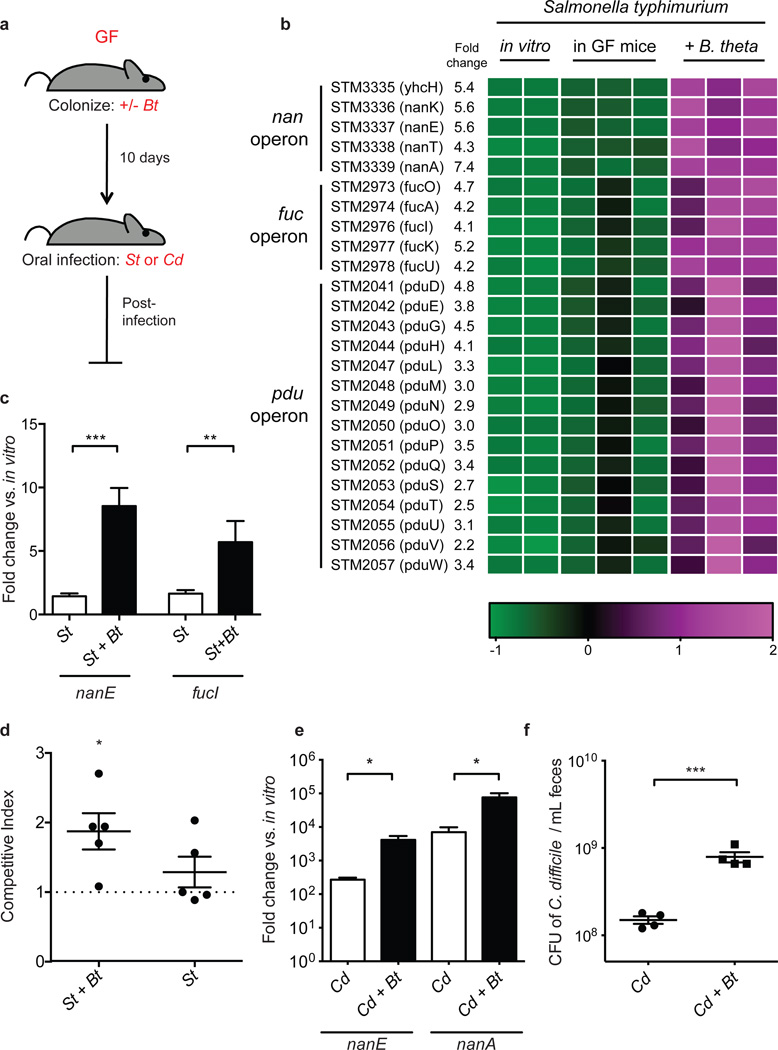
We used transcriptional profiling of Salmonella typhimurium from orally infected gnotobiotic mice to gain insight into the pathogen’s biology while inhabiting the gastrointestinal tract. Our goal was to reveal adaptations of the pathogen within a ‘low-complexity’ gnotobiotic microbiota that might be relevant to antibiotic-induced microbiota disruption. Mice that were monoassociated with the model gut symbiont Bacteroides thetatiotaomicron (Bt) were used as a simplified model of a microbiota that is susceptible to pathogen emergence within the gut. Five days after S. typhimurium infection of the Bt-monoassociated or germ-free (GF) mice (Fig. 1a), cecal contents were collected and subjected to transcriptional profiling using a custom S. typhimurium GeneChip. In the presence of Bt, all 59 S. typhimurium genes that displayed significantly altered expression relative to infection of GF mice were upregulated (Supplementary Table 1). Functional classification of these genes revealed enriched COG categories: “carbohydrate metabolism and transport” and “secondary metabolites biosynthesis, transport, and catabolism” (Supplementary Fig. 2). Genes encoding host mucin carbohydrate metabolism pathways are prominently represented in this gene set, including three operons encoding catabolic pathways for sialic acid, fucose, and the fucose catabolite propanediol, (nan, fuc and pdu, respectively) (Fig. 1b). We surveyed expression of genes within the nan and fuc operons 1 day after S. typhimurium infection in GF or Bt-monoassociated mice, to determine if these operons identified by expression profiling on day 5 post-infection also display high expression earlier in the infection. S. typhimurium nanE and fucI are significantly upregulated 1 day after infection of Bt-monoassociated mice relative to infection of GF mice (nanE, 6.0-fold, p=1.47 × 10−5; fucI, 3.5-fold, p=0.0028) (Fig. 1c) when S. typhimurium densities and host pathology are similar between colonization states (Supplementary Fig. 3, 4). These data are consistent with S. typhimurium catabolizing sialic acid and fucose in the lumen of the gut in a Bt-dependent manner soon after infection.
Figure 1. Bt facilitates S. typhimurium and C. difficile carbohydrate utilization during emergence.
a, Schematic of mouse infection experiments. Germ-free, GF; B. thetaiotaomicron, Bt; S. typhimurium, St; C. difficile, Cd.
b, S. typhimurium operons displaying significant differences in gene expression levels in vivo in the presence and absence of Bt, 5 days post-infection. Colors indicate the deviation of each gene's signal above (purple) and below (green) its mean expression value across all six in vivo samples and duplicate in vitro growths conducted in minimal medium.
c, Induction of S. typhimurium nanE and fucI in cecal contents 1 day post-infection relative to growth in LB broth [n = 9 and 4 for St and St+Bt, respectively].
d, Competitive index of wild-type St/St-ΔnanAΔfucI in Bt-monoassociated (St+Bt) and ex-germ-free (St) mice 1 day post-infection. Horizontal bars indicate the geometric means of CI values, and individual CI values are represented with dots [n = 5/group].
e, Induction of C. difficile nan genes in cecal contents 3 days post-infection relative to growth in minimal medium containing 0.5% glucose [n = 4/group].
f, C. difficile density in feces 1 day post-infection [n = 4/group].
Error bars indicate SEM.
We next constructed mutant strains of S. typhimurium to quantitatively assess the requirement of sialic acid and fucose during expansion in vivo. Deletion of nanA and fucI, the first committed steps in the sialic acid and fucose utilization pathways, abolished growth of the strains on the respective sugars (Supplementary Fig. 5). In competition experiments, Bt-monoassociated mice coinfected with wildtype S. typhimurium and a nanA/fucI double mutant strain (St-ΔnanAΔfucI) revealed a significant disadvantage of the mutant on days 1 and 2 after infection (day 1, CI = 1.87, p=0.028; day 2, CI = 1.45, p=0.016; Fig. 1d). This mutant, however, displayed no competitive disadvantage when competing with wild-type S. typhimurium within GF mice, consistent with S. typhimurium’s sialic acid and fucose use being microbiota-dependent (day 1, p=0.26). The competitive index was not significantly different between the two colonization conditions (Supplementary Fig. 5), however this is likely due to the small amount of free sialic acid present in the GF mouse gut (see Fig. 2a).
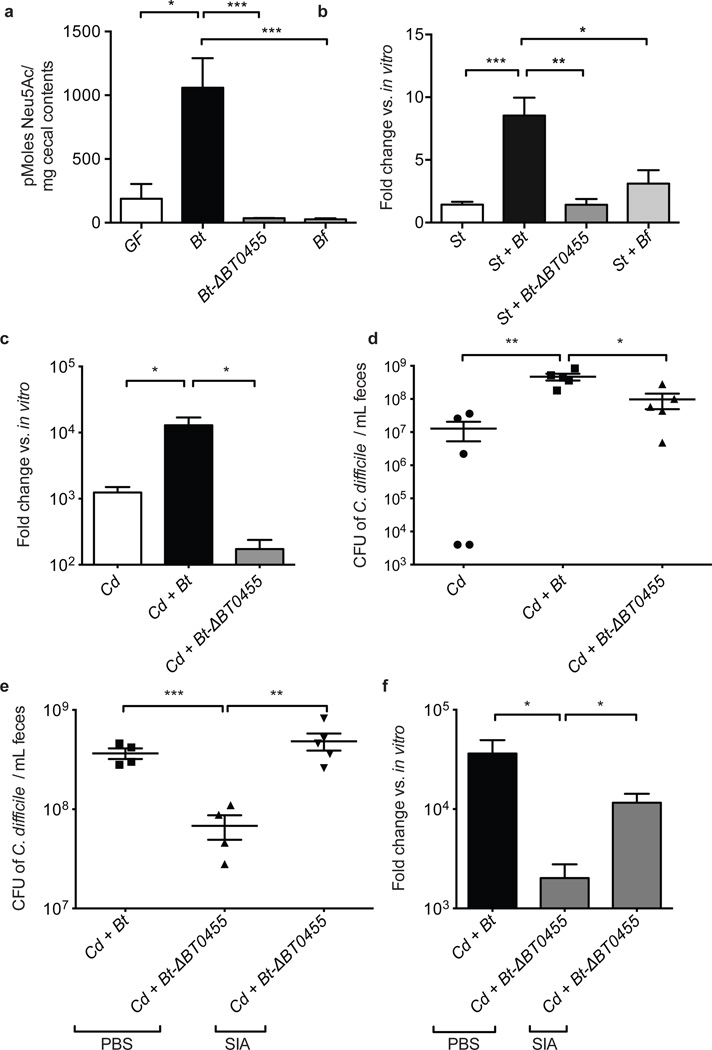
Figure 2. Bt-liberated sialic acid promotes emergence of S. typhimurium and C. difficile.
a, Levels of free sialic acid in cecal contents in GF and gnotobiotic mice monoassociated for 10 days [n=3, 3, 5, 5, respectively].
b, Fold change of expression of S. typhimurium nanE in cecal contents 1 day post-infection relative to growth in vitro [n = 9, 4, 5, 5, respectively].
c, Induction of C. difficile nanE expression in cecal contents 3 days post-infection relative to growth in minimal medium containing 0.5% glucose [n = 4/group].
d, C. difficile density in feces 1 day post-infection [n = 5/group].
e, C. difficile density 1 day post-infection in feces of PBS or exogenous free sialic acid (SIA) treated mice. [n = 4–5/group].
f, Induction of C. difficile nanE gene expression 1 day post-infection in feces of PBS or exogenous free sialic acid (SIA) treated mice relative to growth in minimal medium containing 0.5% glucose [n = 5/group].
Error bars indicate SEM.
C. difficile possesses a sialic acid catabolic operon, like S. typhimurium, but encodes no apparent genes for fucose consumption (Supplementary Fig. 6). To identify whether C. difficile also expresses sialic acid catabolism genes during its expansion within the gut, we quantified the expression of two genes within the nan operon, nanE and nanA, by qRT-PCR of RNA extracted from gnotobiotic mouse cecal contents. C. difficile nanE and nanA displayed elevated expression in Bt-monoassociated mice relative to expression levels observed when C. difficile colonized GF mice alone (nanE, 15-fold higher expression, p= 0.02; nanA 11-fold higher expression, p=0.039; Fig. 1e). The presence of Bt in the gut of gnotobiotic mice resulted in an increased density of C. difficile one day post-infection compared to infection of GF mice (1.5 × 108 vs. 7.9 × 108 CFU/ml; p= 0.0009; Fig. 1f).
Many commensal and pathogenic bacteria can utilize sialic acids from their hosts as a source of energy, carbon, and nitrogen20. However, some bacteria, such as Bt, encode the sialidase required to cleave and release this terminal sugar from the mucosal glycoconjugates, but lack the catabolic pathway (i.e., nan operon) required to consume the liberated monosaccharide. Presumably, the release of sialic acids allows Bt to access highly coveted underlying carbohydrates in the mucus21,22. Conversely, S. typhimurium and C. difficile encode the nan operon but each lacks the sialidase required for sialic acid liberation23,24.
We quantified levels of free sialic acids in the ceca of Bt-monoassociated and GF mice. Bt-monoassociated mice exhibited a significantly higher concentration of the common sialic acid N-acetylneuraminic acid (Neu5Ac) versus GF mice, consistent with Bt’s ability to liberate but not consume the monosaccharide (1059 pmoles/mg, Bt-associated; 188 pmoles/mg, GF; p=0.029; Fig. 2a). Colonization of mice with Bt-ΔBT0455 (a mutant strain of Bt lacking a predicted cell surface sialidase that achieves the same density as wt in vivo; Supplementary Fig. 7) did not result in increased free sialic acid, nor did colonization with Bacteroides fragilis (Bf), which encodes both a sialidase and the nan operon and is therefore able to catabolize Neu5Ac (Fig. 2a). Expression of S. typhimurium’s nan operon was reduced upon infection of gnotobiotic mice colonized with Bt-ΔBT0455 or B. fragilis, consistent with S. typhimurium’s dependence upon elevated levels of microbiota liberated sialic acid (Fig. 2b).
Loss of Bt-liberated sialic acid impacts C. difficile in a manner similar to that observed with S. typhimurium. Nan gene expression in C. difficile was lower in mice colonized with the sialidase-deficient mutant Bt-ΔBT0455 relative to expression in the presence of Bt-colonized mice (nanE, 75-fold higher expression, p= 0.0187; Fig. 2c). Furthermore, C. difficile density decreased in infected mice colonized with Bt-ΔBT0455 mutant relative to densities in mice colonized with wild-type Bt, (9.7 × 107 vs. 4.6 × 108 CFU/ml; p= 0.0143) illustrating the importance of Bt-liberated sialic acid in C. difficile expansion in vivo (Fig. 2d; Supplementary Fig. 8). Free sialic acid was orally administered to Bt-ΔBT0455 and C. difficile co-colonized mice to determine if exogenous administration of the monosaccharide could reverse the decrease in C. difficile density by complementing the sialidase deficiency in this model. C. difficile densities increased 1 day post-infection in Bt-ΔBT0455 monoassociated mice fed free sialic acid compared to unsupplemented controls (4.8 × 108 vs. 6.8 × 107 CFU/ml; p=0.0066) reaching densities similar to those observed in the presence of wild-type Bt (Fig. 2e). Furthermore, expression of C. difficile nanE increases in the sialic acid-fed Bt-ΔBT0455-associated mice, further demonstrating that sialic acid use by C. difficile occurs concomitant with its increased densities in vivo (nanE, 58-fold higher expression over PBS-treated controls, p= 0.019; Fig. 2f). Notably, free sialic acid administration to GF mice infected with C. difficile resulted in higher densities of the pathogen in the lumen of the gut, confirming the important role of this monosaccharide in vivo (Supplementary Fig. 9). These data demonstrate that sialic acid catabolism by C. difficile promotes higher densities of the pathogen and depends upon the availability of the liberated monosaccharide within the lumen of the gut.
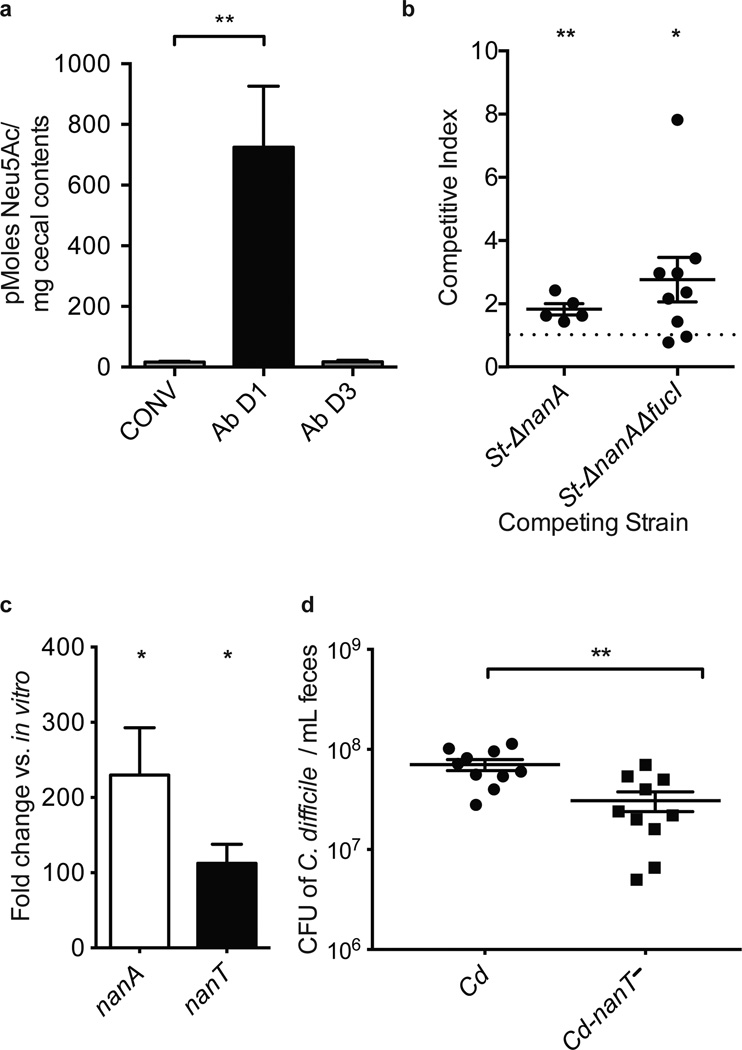
To determine if sialic acid use is relevant to pathogen proliferation in an antibiotic-treated complex microbiota, we quantified free sialic acids in the ceca of conventional mice before and after antibiotic treatment. Levels of free Neu5Ac were very low within untreated conventional mice, consistent with efficient partitioning of Neu5Ac between members of an undisturbed complex microbiota (Fig. 3a). However, antibiotic-treated mice exhibited elevated levels of free sialic acid 1 day after treatment (725 pmoles/mg 1 day post-streptomycin compared to 17 pmoles/mg in untreated mice; p=0.0019), a time frame that coincides with pathogen expansion and acute microbiota disturbance (Supplementary Fig. 10)25. The pool of free sialic acids decreased by day 3 post-treatment, consistent with recovery of the microbiota after antibiotic treatment25 (Fig. 3a). St-ΔnanA and St-ΔnanAΔfucI mutants both showed a competitive defect relative to wild-type St 1 day after infection in antibiotic-treated conventional mice (St-ΔnanA, CI=1.83 p=0.0095; St-Δ nanAΔfucI, CI=2.77, p=0.036), consistent with sialic acid and fucose utilization providing an advantage to S. typhimurium during emergence (Fig. 3b). The lack of significance of the phenotype in the fucI single mutant suggests possible redundancy for the functional significance of the fucose catabolic pathway in this experimental model (Supplemental Fig. 11). To test whether C. difficile relies upon sialic acid catabolism in post-antibiotic expansion, we quantified the expression of the nan operon in antibiotic-treated conventional mice 1 day post-infection. Coincident with expansion of C. difficile, the nan operon was highly induced compared to basal expression in vitro (nanA, 230-fold, p= 0.0358; nanT, 112-fold, p= 0.0217) confirming that C. difficile expresses this operon at high levels during its post-antibiotic-expansion within a complex microbiota (Fig. 3c). As a test of sialic acid catabolism importance in C. difficile proliferation, we constructed a nanT-mutant strain of C. difficile (Cd-nanT−) that is deficient in sialic acid consumption (Supplementary Fig. 4). Cd-nanT− was significantly compromised in post-antibiotic expansion of conventional mice relative to wt Cd (3.1 × 107 vs. 7.0 × 107 CFU/ml; p= 0.0023) demonstrating the importance of sialic acid catabolism to C. difficile in attaining high densities in the context of an antibiotic-disrupted complex microbiota (Fig. 3d).
Figure 3. S. typhimurium and C. difficile utilize mucin-derived monosaccharides resulting from antibiotic treatment of conventional mice.
a, Levels of free sialic acid in cecal contents of conventional mice (CONV), antibiotic-treated mice 1 day (Ab D1) and 3 days (Ab D3) post-treatment [n= 8, 9, 3, respectively].
b, Competitive index of wt S. typhimurium versus S. typhimurium mutants in cecal contents (St-ΔnanA) or feces (St-ΔnanAΔfucI) of antibiotic-treated conventional mice. Horizontal bars indicate the geometric means of CI values, and individual CI values are represented with dots [n=5 and 9, respectively].
c, Induction of C. difficile nanA and nanT expression in fecal samples 1 day post-infection of antibiotic-treated conventional mice relative to growth in minimal medium containing 0.5% glucose [n = 4/group].
d, Density of wt C. difficile or a mutant deficient in sialic acid consumption (Cd-nanT−) 3 days post-infection in feces of antibiotic-treated conventional mice. [n = 10/group].
Error bars indicate SEM.
Recent studies have illustrated that enteric bacterial pathogens can subvert aspects of host inflammation to hold potential competitors within the microbiota at bay and enable pathogen proliferation7,26–28. Our results indicate that the antibiotic-associated pathogens S. typhimurium and C. difficile exploit increases in mucosal carbohydrate availability that occur upon disruption of the competitive ecosystem in which nutrients are typically efficiently consumed by endogenous community members. The transient post-antibiotic increase in monosaccharides liberated by the resident microbiota from host mucus provides a window of opportunity for these pathogens to expand to densities sufficient to induce self-promoting host inflammation (Supplementary Fig. 1). Implicit in these findings are new potential therapeutic strategies to combat post-antibiotic pathogen expansion.
Online-Only Methods
Bacterial Strains and Culture Conditions
B. thetaiotaomicron (ATCC 29148, also known as VPI-5482), was grown anaerobically (6% H2, 20% CO2, 74% N2) overnight in TYG medium (1% tryptone, 0.5% yeast extract, 0.2% glucose, w/v) supplemented with 100 mM potassium phosphate buffer, (pH 7.2), 4.1 mM cysteine, 200 µM histidine, 6.8 µM CaCl2, 140 nM FeSO4, 81 µM MgSO4, 4.8 mM NaHCO3, 1.4 mM NaCl, 1.9 µM hematin, plus 5.8 µM Vitamin K3.
All strains of S. typhimurium were derived from wild-type strain SL1344, which is naturally streptomycin-resistant. Using the methods of Datsenko and Wanner29, mutant strains were first constructed in strain LT2, verified by PCR and then transduced into SL1344 using P22 phage transduction. Mutant strains and primers utilized in their generation are listed in Supplementary Table 2. Growth defects were not observed on glucose for either mutant, and the presence of sialic acid did not pose a toxicity issue with the nanA mutant as has been previously reported for E. coli30 consistent with its polarity that compromises nanT expression (Supplementary Fig 5e). For colonization experiments, S. typhimurium strains were grown in Luria-Bertani (LB) broth at 37°C with aeration or on LB agar plates, with the appropriate antibiotics (200 µg/ml streptomycin, 30 µg/ml kanamycin). Minimal medium used for transcriptional profiling consisted of 100 mM KH2PO4 (pH 7.2), 15 mM NaCl, 8.5 mM (NH4)2SO4, 4 mM L-cysteine, 1.9 mM hematin+200 mM L-histidine, 100 mM MgCl2, 1.4 mM FeSO4, 50 mM CaCl2, 1 mg ml−1 vitamin K3, and 5 ng ml−1 vitamin B12, and 0.5% glucose (w/v). For evaluation of growth on various monosaccharides, strains were grown in M9 minimal media supplemented with 0.02% w/v histidine. Fecal densities (CFU) of S. typhimurium were quantified by duplicate sampling with 1 µl loops, and subsequent dilution and spot plating on plain LB agar for gnotobiotic experiments and LB agar with streptomycin for conventional experiments.
C. difficile strain 630 was utilized in all C. difficile experiments and was cultured in Reinforced Clostridial Medium (RCM) + cysteine (Becton Dickinson, MD) anaerobically (6% H2, 20% CO2, 74% N2). C. difficile growth curves were generated using minimal medium (MM) composed of ammonium sulfate, sodium carbonate, calcium chloride, magnesium chloride, manganese chloride, cobalt chloride, histidine hematin, vitamin B12, vitamin K1, FeSO4, and 1% Bacto Tryptone diluted 1:1 with 1% or 0.5% carbon source. OD600 was monitored using a BioTek PowerWave 340 plate reader (BioTek, Winooski, VT) every 30 minutes, at 37°C anaerobically (6% H2, 20% CO2, 74% N2). Fecal densities (CFU) of C. difficile were quantified by duplicate sampling with 1 µl loops and subsequent dilution and spot plating on blood-BHI supplemented with erythromycin. For quantification of C. difficile CFU in conventional mice, 1ul of feces was serially diluted in PBS and plated onto CDMN plates, composed of Clostridium difficile Agar Base (Oxoid) with 7% v/v of Defibrinated Horse Blood (Lampire Biological Laboratories), supplemented with 32 mg/L Moxalactam (Santa Cruz Biotechnology) and 12 mg/L Norfloxacin (Sigma-Aldrich). Plates were incubated overnight at 37°C in an anaerobic chamber (Coy). Colonies identified as C. difficile were validated by colony PCR.
To construct the nanT null mutant (Cd-nanT−), the ClosTron method for targeted gene disruption in C. difficile and detailed protocol were used31, 32. SOEing PCRs with primers IBS, EBS1d, EBS2 and EBS (see Supplementary Table 2) were used to assemble and amplify the product for intron targeting, as outlined in the TargeTron users’ manual (Sigma Aldrich). The retargeting sequence was digested with BsrGI/HindIII and cloned into pMTL007C-E2. The resulting plasmid was transformed into HB101/pRK24 for conjugation into JIR809433 (a generous gift from Aimee Shen) to generate Cd-nanT−.
Reagents and Mice
Germ-free Swiss-Webster mice were maintained in gnotobiotic isolators and fed an autoclaved standard diet (Purina LabDiet 5K67) or a polysaccharide-deficient diet34, in accordance with A-PLAC, the Stanford IACUC. All animals were 6–12 weeks of age and both genders were used. For all experiments involving C. difficile colonization of germ-free mice, the diet was switched to polysaccharide-deficient chow one day before inoculation with C. difficile. Conventional Swiss-Webster mice (RFSW, Taconic) were used for S. typhimurium and C. difficile antibiotic-treated experiments. Number of animals per group was chosen as the minimum likely required for conclusions of biological significance, established from prior experience. Randomization was not possible in the gnotobiotic setting and blinding was not applicable.
Conventional mice were orally gavaged with 20 mg streptomycin dissolved in water 24 hours before infection, and starved 18 hours before infection. Mice were infected via oral gavage of 14 hour overnight cultures of S. typhimurium resuspended in PBS. For single infections of gnotobiotic mice, 108 cfu of S. typhimurium were gavaged. For S. typhimurium competitive index experiments, pure cultures of wild-type and mutant bacteria were diluted to equal densities, mixed in a 1:1 ratio and serial diluted in PBS to a total of 103 cfu/200 µl. Each mouse was orally gavaged with 200 µl of this dilution. Throughout the experiment, fecal samples were taken and dilutions were plated on LB agar plates containing streptomycin, which allows for growth of both the wild-type and mutant strains. Colonies from these plates were then patched onto LB agar + kanamycin plates to determine the proportion of kanamycin-resistant (mutant) cells. With each sample, the ratio of kanamycin-sensitive (wild-type) bacteria to kanamycin-resistant (mutant) bacteria was divided by the kans/kanr ratio determined from the original inoculum to produce the competitive index. Significance was evaluated using one-sample t-tests with a theoretical mean of 1. All competitive indices were determined for fecal samples with the exception of St-ΔnanA, which was surveyed in cecal contents.
For C. difficile experiments involving conventional mice, antibiotics were administered in the water for 3 days, starting 6 days before inoculation including: kanamycin (0.4 mg/mL), gentamycin (0.035 mg/mL), colistin (850 U/mL), metronidazole (0.215 mg/mL) and vancomycin (0.045 mg/mL)19. Mice were then switched to regular water for 2 days, and administered 1mg of clindamycin by oral gavage 1 day before inoculation with C. difficile. Inoculations were by oral gavage at a density of 108 CFU from overnight cultures.
For sialic acid administration experiments, N-acetylneuraminic acid (Calbiochem or Santa Cruz Biotechnology) was administered in the water at a 1% concentration. Additionally, mice were orally gavaged 1mg of sialic acid twice a day. The amount of sialic acid in the cecal contents were calculated to equal approximately 700 pmoles/mg of cecal contents, which mirrors the average concentration of free sialic acids we quantified post-antibiotic treatment (725 pmoles/mg).
Expression analysis
Genome-wide transcriptional profiling of S. typhimurium was conducted using custom-made GeneChips, which contain probes for all annotated coding sequences for S. typhimurium LT2. RNA was purified from cecal contents and in vitro culture and cDNA was prepared, fragmented and labeled as described36.
GeneChip data were RMA-MS normalized as described37 and log2 transformed. Statistical significance for differential gene expression was determined using Significance Analysis of Microarrays (SAM)38. The delta parameter was adjusted to achieve a FDR nearest to 10%, and this delta value was used to select significantly-regulated genes.
qRT-PCR analysis was performed on RNA extracted from cecal or fecal contents by phenol-chloroform extraction and bead beating. Superscript II (Invitrogen) was utilized to convert RNA to cDNA, and SYBR Green (ABgene) in a MX3000P thermocycler (Strategene) was utilized. Fold changes were normalized to in vitro growths in Minimal Medium containing 0.5% glucose (MM-G) for C. difficile and LB for S. typhimurium.
Quantification of sialic acids
All steps were carried out at 4°C to minimize enzymatic hydrolysis. Approximately 200 mg of flash-frozen cecal contents were weighed out and resuspended in 400 µl dH2O. Samples were vortexed for 30 minutes at max speed and centrifuged for 15 minutes at 14,000 × g in the tabletop centrifuge. The supernatant was stored, and the pellet resuspended in an additional 400 µl dH2O. The tubes were vortexed individually until the pellet was dispersed, and then all samples were vortexed for 30 minutes, centrifuged, and supernatants were pooled. This process was repeated once more for a total volume of approximately 1 ml. 700µl of each sample was filtered through a Pall 1K MWCO filter for 9 hours at 7,000 × g. Samples were derivatized with DMB (1,2-diamino-4,5-methylene-dioxybenzene) as described previously39. The resulting product was analyzed by reverse-phase HPLC using a C18 column (Dionex) at a flow rate of 0.9 ml/min, using a gradient of 5% to 11% acetonitrile in 7% methanol. The excitation and emission were 373 and 448 nm, respectively. The DMB-derivatized sialic acids were identified and quantified by comparing elution times and peak areas to known standards.
16S rRNA microbial community composition analysis
Fecal DNA was isolated and amplicons generated of the 16S rRNA V4 region (515F, 806R). Samples were sequenced at Medical Genome Facility, Mayo Clinic, Rochester, MN using the MiSeq (Illumina) platform40. Data analysis was done using QIIME41. Single end reads were analyzed to determine OTUs (Operational Taxonomic Units) at 97% sequence similarity using uclust. Taxonomy was assigned using RDP classifier against the GreenGenes database and a phylogenetic tree was built using FastTree. The OTU table was rarified to a sequencing depth of 900 for each set of samples. Beta diversity was determined using unweighted and weighted UniFrac42.
Statistical analyses
The Student’s t-test was used for statistical calculations, and * indicates p < 0.05, ** indicates p < 0.01 and *** indicates p < 0.001. Error bars indicate SEM. n indicates the number of mice used per condition. Normal distribution was assumed for all data, and no deviations were noted. Grubbs’ test was used to identify and eliminate statistical outliers.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Erica Sonnenburg for comments on the manuscript, Michelle St. Onge for technical assistance, and Aimee Shen, Nigel Minton, and Rob Knight for valuable help and reagents. This research was supported by R01-DK085025 (to J.L.S.), NSF graduate fellowships (to K.M.N. and J.A.F) and a Stanford Graduate Fellowship (to K.M.N.). Justin L. Sonnenburg, Ph.D. holds an Investigators in the Pathogenesis of Infectious Disease Award from the Burroughs Wellcome Fund (J.L.S.).
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
AUTHOR CONTRIBUTIONS
K.M.N., J.A.F., D.M.M. and J.L.S. designed experiments. K.M.N., J.A.F., S.K.H., J.B.L., P.C.K., N.N., B.C. and J.L.S. performed experiments. K.M.N., J.A.F., P.C.K., S.G., N.N., D.M.M. and J.L.S. analyzed data. D.M.M. and B.C.W. contributed reagents. K.M.N., J.A.F. and J.L.S. wrote the paper.
DATA DEPOSITION
Microbiota enumeration (16S rRNA) datasets have been deposited in the EMBL European Nucleotide Archive (ENA) under accession # ERP003629 and can also be found in the QIIME Database under the study ID 1958 (http://www.microbio.me/qiime/). GeneChip datasets are available in the GEO database under accession number GSE49076.
The authors declare no competing financial interests.
REFERENCES
- 1.Doorduyn Y, Van Den Brandhof WE, Van Duynhoven YT, Wannet WJ, Van Pelt W. Risk factors for Salmonella Enteritidis and Typhimurium (DT104 and non-DT104) infections in The Netherlands: predominant roles for raw eggs in Enteritidis and sandboxes in Typhimurium infections. Epidemiology and infection. 2006;134:617–626. doi: 10.1017/S0950268805005406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pavia AT, et al. Epidemiologic evidence that prior antimicrobial exposure decreases resistance to infection by antimicrobial-sensitive Salmonella. The Journal of infectious diseases. 1990;161:255–260. doi: 10.1093/infdis/161.2.255. [DOI] [PubMed] [Google Scholar]
- 3.Pepin J, et al. Emergence of fluoroquinolones as the predominant risk factor for Clostridium difficile-associated diarrhea: a cohort study during an epidemic in Quebec. Clin Infect Dis. 2005;41:1254–1260. doi: 10.1086/496986. [DOI] [PubMed] [Google Scholar]
- 4.Kelly CP, Pothoulakis C, LaMont JT. Clostridium difficile colitis. N Engl J Med. 1994;330:257–262. doi: 10.1056/NEJM199401273300406. [DOI] [PubMed] [Google Scholar]
- 5.Backhed F, Ley RE, Sonnenburg JL, Peterson DA, Gordon JI. Host-bacterial mutualism in the human intestine. Science (New York, N.Y. 2005;307:1915–1920. doi: 10.1126/science.1104816. [DOI] [PubMed] [Google Scholar]
- 6.Qin J, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stecher B, et al. Like will to like: abundances of closely related species can predict susceptibility to intestinal colonization by pathogenic and commensal bacteria. PLoS pathogens. 2010;6:e1000711. doi: 10.1371/journal.ppat.1000711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chang DE, et al. Carbon nutrition of Escherichia coli in the mouse intestine. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:7427–7432. doi: 10.1073/pnas.0307888101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sonnenburg JL, et al. Glycan foraging in vivo by an intestine-adapted bacterial symbiont. Science (New York, N.Y. 2005;307:1955–1959. doi: 10.1126/science.1109051. [DOI] [PubMed] [Google Scholar]
- 10.Martens EC, Chiang HC, Gordon JI. Mucosal glycan foraging enhances fitness and transmission of a saccharolytic human gut bacterial symbiont. Cell host & microbe. 2008;4:447–457. doi: 10.1016/j.chom.2008.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fabich AJ, et al. Comparison of carbon nutrition for pathogenic and commensal Escherichia coli strains in the mouse intestine. Infection and immunity. 2008;76:1143–1152. doi: 10.1128/IAI.01386-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kamada N, et al. Regulated virulence controls the ability of a pathogen to compete with the gut microbiota. Science (New York, N.Y. 2012;336:1325–1329. doi: 10.1126/science.1222195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pacheco AR, et al. Fucose sensing regulates bacterial intestinal colonization. Nature. 2012;492:113–117. doi: 10.1038/nature11623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maltby R, Leatham-Jensen MP, Gibson T, Cohen PS, Conway T. Nutritional Basis for Colonization Resistance by Human Commensal Escherichia coli Strains HS and Nissle 1917 against E. coli O157:H7 in the Mouse Intestine. PLoS ONE. 2013;8:e53957. doi: 10.1371/journal.pone.0053957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dethlefsen L, Huse S, Sogin ML, Relman DA. The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLoS Biol. 2008;6:e280. doi: 10.1371/journal.pbio.0060280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hapfelmeier S, Hardt WD. A mouse model for S. typhimurium-induced enterocolitis. Trends in microbiology. 2005;13:497–503. doi: 10.1016/j.tim.2005.08.008. [DOI] [PubMed] [Google Scholar]
- 17.Lawley TD, et al. Host transmission of Salmonella enterica serovar Typhimurium is controlled by virulence factors and indigenous intestinal microbiota. Infection and immunity. 2008;76:403–416. doi: 10.1128/IAI.01189-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lawley TD, et al. Antibiotic treatment of clostridium difficile carrier mice triggers a supershedder state, spore-mediated transmission, and severe disease in immunocompromised hosts. Infection and immunity. 2009;77:3661–3669. doi: 10.1128/IAI.00558-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen X, et al. A mouse model of Clostridium difficile-associated disease. Gastroenterology. 2008;135:1984–1992. doi: 10.1053/j.gastro.2008.09.002. [DOI] [PubMed] [Google Scholar]
- 20.Vimr ER, Kalivoda KA, Deszo EL, Steenbergen SM. Diversity of microbial sialic acid metabolism. Microbiol Mol Biol Rev. 2004;68:132–153. doi: 10.1128/MMBR.68.1.132-153.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martens EC, Chiang HC, Gordon JI. Mucosal glycan foraging enhances fitness and transmission of a saccharolytic human gut bacterial symbiont. Cell host & microbe. 2008;4:447–457. doi: 10.1016/j.chom.2008.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marcobal A, et al. Consumption of human milk oligosaccharides by gut-related microbes. J. Agric. Food Chem. 2010;58:5334–5340. doi: 10.1021/jf9044205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hoyer LL, Hamilton AC, Steenbergen SM, Vimr ER. Cloning, sequencing and distribution of the Salmonella typhimurium LT2 sialidase gene, nanH, provides evidence for interspecies gene transfer. Molecular microbiology. 1992;6:873–884. doi: 10.1111/j.1365-2958.1992.tb01538.x. [DOI] [PubMed] [Google Scholar]
- 24.Sebaihia M, et al. The multidrug-resistant human pathogen Clostridium difficile has a highly mobile, mosaic genome. Nature genetics. 2006;38:779–786. doi: 10.1038/ng1830. [DOI] [PubMed] [Google Scholar]
- 25.Stecher B, et al. Salmonella enterica serovar typhimurium exploits inflammation to compete with the intestinal microbiota. PLoS Biol. 2007;5:2177–2189. doi: 10.1371/journal.pbio.0050244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Winter SE, et al. Gut inflammation provides a respiratory electron acceptor for Salmonella. Nature. 2010;467:426–429. doi: 10.1038/nature09415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lupp C, et al. Host-mediated inflammation disrupts the intestinal microbiota and promotes the overgrowth of Enterobacteriaceae. Cell host & microbe. 2007;2:204. doi: 10.1016/j.chom.2007.08.002. [DOI] [PubMed] [Google Scholar]
- 28.Barman M, et al. Enteric salmonellosis disrupts the microbial ecology of the murine gastrointestinal tract. Infection and immunity. 2008;76:907–915. doi: 10.1128/IAI.01432-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A. 2000;97(12):6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vimr ER, Troy FA. Identification of an Inducible Catabolic System for Sialic Acids (Nan) in Escherichia-Coli. Journal of Bacteriology. 1985;164(2):845–853. doi: 10.1128/jb.164.2.845-853.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Heap JT, et al. The ClosTron: Mutagenesis in Clostridium refined and streamlined. J Microbiol Methods. 2010;80(1):49–55. doi: 10.1016/j.mimet.2009.10.018. [DOI] [PubMed] [Google Scholar]
- 32.Adams CM, et al. Structural and functional studies of the CspB protease required for Clostridium spore germination. PLoS Pathogens. doi: 10.1371/journal.ppat.1003165. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.O'Connor JR, et al. Construction and analysis of chromosomal Clostridium difficile mutants. Mol Microbiol. 2006;61(5):1335–1351. doi: 10.1111/j.1365-2958.2006.05315.x. [DOI] [PubMed] [Google Scholar]
- 34.Sonnenburg ED, et al. Specificity of polysaccharide use in intestinal bacteroides species determines diet-induced microbiota alterations. Cell. 2010;141(7):1241–1252. doi: 10.1016/j.cell.2010.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen X, et al. A mouse model of Clostridium difficile-associated disease. Gastroenterology. 2008;135(6):1984–1992. doi: 10.1053/j.gastro.2008.09.002. [DOI] [PubMed] [Google Scholar]
- 36.Sonnenburg JL, et al. Glycan foraging in vivo by an intestine-adapted bacterial symbiont. Science. 2005;307(5717):1955–1959. doi: 10.1126/science.1109051. [DOI] [PubMed] [Google Scholar]
- 37.Stevens JR, et al. Statistical issues in the normalization of multi-species microarray data; Proceedings of Conference on Applied Statistics in Agriculture; 2008. pp. 47–62. [Google Scholar]
- 38.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001;98(9):5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Manzi AE, Diaz S, Varki A. High-pressure liquid chromatography of sialic acids on a pellicular resin anion-exchange column with pulsed amperometric detection: a comparison with six other systems. Anal Biochem. 1990;188(1):20–32. doi: 10.1016/0003-2697(90)90523-c. [DOI] [PubMed] [Google Scholar]
- 40.Caporaso JG, et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 2012;6(8):1621–1624. doi: 10.1038/ismej.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Caporaso JG, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7(5):335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lozupone C, Hamady M, Knight R. UniFrac--an online tool for comparing microbial community diversity in a phylogenetic context. BMC Bioinformatics. 2006;7:371. doi: 10.1186/1471-2105-7-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.