Summary
Glutamate transporters are integral membrane proteins that catalyze neurotransmitter uptake from the synaptic cleft into the cytoplasm of glial cells and neurons1. Their mechanism involves transitions between extracellular- (outward-) and intracellular- (inward-) facing conformations, whereby substrate binding sites become accessible to the opposite sides of the membrane2. This process has been proposed to entail trans-membrane movements of three discrete transport domains within a trimeric scaffold3. Using single-molecule fluorescence resonance energy transfer (smFRET) imaging4, we have directly observed large-scale transport domain movements in a bacterial homologue of glutamate transporters for the first time. We find that individual transport domains alternate between periods of quiescence and periods of rapid transitions, reminiscent of bursting patterns first recorded in single ion channels using patch-clamp methods5,6. We suggest that the switch to the dynamic mode in glutamate transporters is due to separation of the transport domain from the trimeric scaffold, which precedes domain movements across the bilayer. This spontaneous dislodging of the substrate-loaded transport domain is approximately 100-fold slower than subsequent trans-membrane movements and may be rate determining in the transport cycle.
Main Text
In the brain, glutamate mediates excitatory synaptic transmission, responsible for learning, memory formation and cognition1,7. Glutamate transporters are electrochemically driven pumps that maintain a low neurotransmitter background at glutamatergic synapses, allowing for repeated rounds of signaling and preventing excitotoxicity8. The sodium/aspartate symporter from Pyrococcus horikoshii, GltPh, is the only glutamate transporter homologue with known three-dimensional structures of both outward- and inward-facing states3,9. Correspondingly, this system has served as a valuable model for establishing the structural and dynamic underpinnings of the transport cycle10-12. Because GltPh originates from a hyper-thermophilic archaeon, it has a slow turnover time of ∼100 seconds at room temperature12, suggesting that the dynamic processes required for transport may fall within the time regime accessible through smFRET imaging4,13.
Crystal structures have shown that GltPh is a homo-trimer where each protomer comprises two domains: a rigid trimerization domain14, which serves as a scaffold and mediates the inter-protomer interactions, and a transport domain, which can move ∼15 Å across the bilayer3. High extracellular and low intracellular chemical potentials of sodium ions (Na+) drive substrate binding and unbinding to the transporter, respectively15. By contrast, large-scale transport domain movements, hypothesized to mediate the translocation of substrate aspartate (Asp) and coupled Na+ ions across the cellular membranes3,16, are expected to occur spontaneously3.
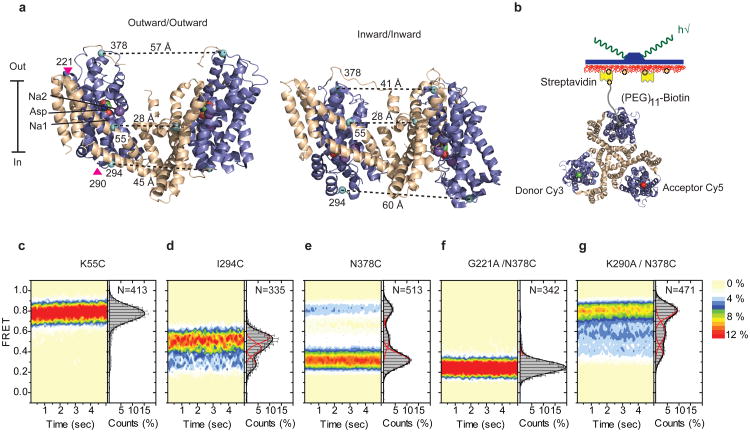
We probed the dynamics of the transport domain movements using wide-field, total internal reflection smFRET imaging17. To detect the relative motions of two transport domains, we introduced single cysteine mutations at positions of low sequence conservation and high solvent accessibility that exhibited inter-protomer distance changes >20 Å in the crystal structures of the outward- and inward-facing GltPh (Fig. 1a). The three cysteines present in each GltPh trimer were derivatized with a mixture of maleimide-activated Cy3, Cy5 and biotin-(PEG)11 (Methods). This labeling strategy ensured that only trimers containing one of each of these moieties were immobilized on streptavidin-decorated surfaces and yielded detectable FRET signals (Fig. 1b). The constructs pursued showed selective labeling reactivity, >80 % functional activity, and biotin-specific surface attachment (Supplementary Figs. 1 and 2). To parallel previous crystallographic studies, GltPh mutants were first characterized in the presence of saturating concentrations of Na+ and Asp (200 mM and 100 μM, respectively).
Figure 1. FRET efficiency changes reflect relative orientations of the transport domains.
a, GltPh protomer pairs in symmetrical outward- and inward-facing states viewed within the membrane plane. Trimerization and transport domains are colored wheat and blue, respectively. Bound Asp and Na+ ions are emphasized as spheres and colored by atom type. Introduced cysteines are highlighted in cyan with inter-protomer distances above the dotted lines. Magenta arrows mark sites of mutations altering state distributions. b, Labeling and surface-immobilization strategies. c-g, FRET efficiency population histograms for Asp/Na+-bound transporters. Introduced mutations are indicated above the panels. The number of molecules analyzed (N) is shown. Population contour plots (left) are color-coded from tan (lowest) to red (highest population) with the color scale shown next to the graphs. In the cumulative population histogram (right), the solid black lines are fits to the sums of individual Gaussian functions (red lines).
For a control K55C construct labeled within the rigid trimerization domain, individual smFRET trajectories exhibited stable, high FRET prior to photobleaching (τFRET ∼40 seconds) (Supplementary Fig. 3). FRET efficiency distributions, calculated from hundreds of trajectories, yielded a single, narrow peak centered at ∼0.8 (Fig. 1c; Supplementary Table 1). These data indicated robust fluorophore performance18 and confirmed that the trimerization domain lacks significant dynamics3,14. By contrast, GltPh labeled on I294C or N378C, located on the cytoplasmic and extracellular surfaces of the transport domain, respectively, exhibited clear evidence of discrete FRET states. In both constructs, low- and high-FRET states were observed (I294C: ∼0.35 and ∼0.55; N378C: ∼0.35 and ∼0.8) (Fig. 1d and e), consistent with the inter-protomer distances in outward- and inward-facing GltPh structures (Fig. 1a and Supplementary Table 1). Here, the intracellular GltPh-I294C construct preferentially populated a high-FRET state, while the extracellular GltPh-N378C construct favored a low-FRET state (Fig. 1d and e). These data suggest that the individual GltPh protomers preferentially reside in outward-facing conformations, but also adopt less favored inward-facing orientations. In both systems, intermediate-FRET states were also observed (Fig. 1d and e). This state was better resolved in GltPh-N378C (∼0.55), where it could be attributed to a configuration in which neighboring subunits adopt inward- and outward-facing orientations (Supplementary Methods; Supplementary Fig. 4; Supplementary Table 1). Such a configuration is consistent with a recent crystal structure of an asymmetric GltPh trimer19. Importantly, similar FRET states and FRET state occupancies were obtained when GltPh-N378C was reconstituted into liposomes (Supplementary Methods, Supplementary Fig. 5).
To validate our FRET state assignments, we introduced mutations distal from the site of fluorophore attachment in GltPh-N378C to either stabilize or destabilize the outward-facing state (Fig. 1a; Supplementary Fig. 6). A G221A mutation was introduced within the hinge region between the transport and trimerization domains, to hinder local rearrangements accompanying inward transport domain motions3. A K290A mutation was introduced to disrupt a network of polar interactions at the interface between the transport and trimerization domains in the outward-facing structure11. Consistent with expectations, we observed dramatic increases and decreases in the low-FRET, outward-facing state population for the G221A and K290A mutants, respectively (Fig. 1f and g). We conclude that transitions between FRET states in GltPh-N378C reflect motions of the transport domains between outward- and inward-facing orientations. This construct was pursued to examine the impact of ligand binding on conformational dynamics.
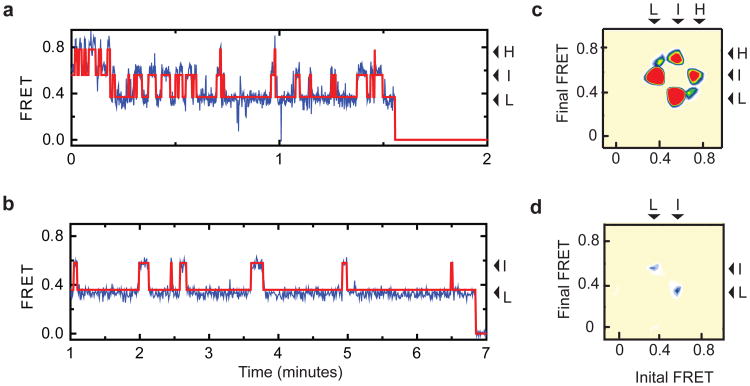
In the absence and in the presence of Na+ and Asp, the FRET state values were largely similar whereas ligands shifted the state occupancies in favor of the low-FRET state (Supplementary Fig. 5). Strikingly, individual smFRET traces exhibited much more frequent transitions in the apo compared to the bound transporter (Fig. 2a and b). To quantify the dynamics, we idealized the smFRET trajectories using a model containing three kinetically linked, non-zero FRET states (Methods, Supplementary Fig. 7). The quality of idealizations and the average rates of the conformational transitions were assessed by inspection of individual trajectories as well as transition density plots, which report on the frequencies of transitions between distinct FRET states20. In apo GltPh, a comparable number of transitions occurred between low-, intermediate- and high-FRET states with an average frequency of ∼0.5 s−1 (Fig. 2c), suggesting that the apo transporter samples outward- and inward-facing states relatively rapidly. For the substrate-loaded transporter, the frequency of transitions was reduced by more than an order of magnitude (∼0.02 s−1). Under these conditions, we primarily observed transitions between low- and intermediate-FRET states (Fig. 2d), consistent with transport domains moving inward one at a time. The paucity of direct transitions from low- to high-FRET states in the presence and in the absence of Na+ and Asp is in line with previous functional studies14,21, suggesting that individual protomers undergo conformational transitions, and thus function, independently of each other.
Figure 2. Dynamics in the apo and substrate bound transporter.
a-b, Shown are smFRET trajectories (blue), acquired for apo (a) and Na+/Asp-bound (b) GltPh-N378C. Overlaid are idealizations generated in QuB (red). Arrows mark population averages for the low- (L), intermediate- (I) and high- (H) FRET efficiencies. Data in panel b was collected using 400 ms integration time. c-d, Transition density plots for the apo (c) and Na+/Asp-bound (d) transporters show that transitions occur between three distinct FRET states (L, I and H) with an average frequency of ∼0.5 s−1 and ∼0.02 s−1, respectively. Initial and final FRET values for each transition are accumulated into two-dimensional histograms. Color scale is from tan (lowest frequency) to red (highest frequency).
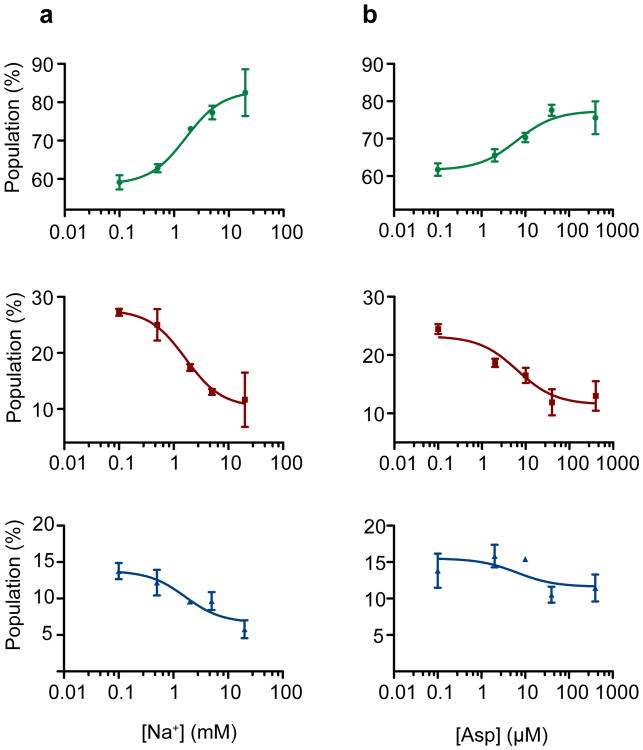
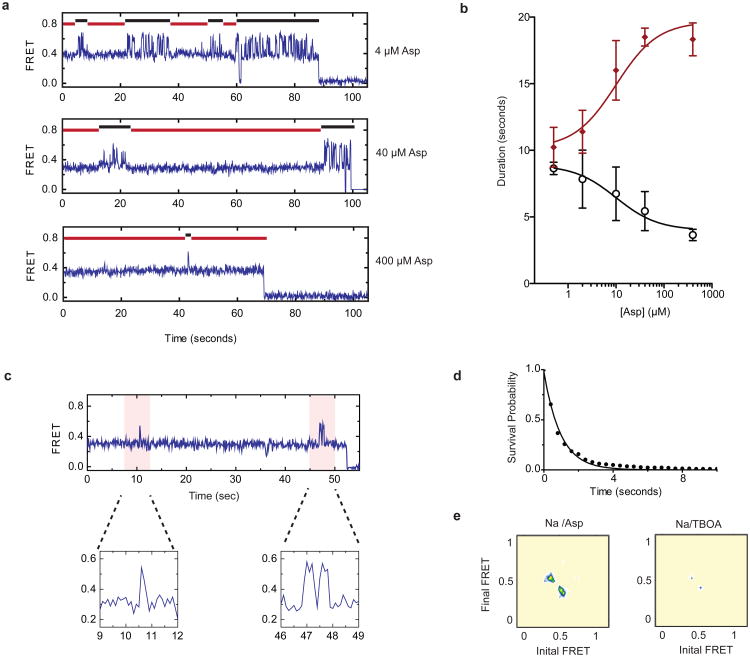
To assess the concentration dependence of the ligand-induced effects, we performed two titrations on GltPh-N378C: a Na+ titration in the presence of 10 μM Asp and an Asp titration in the presence of 2 mM Na+. At the population level, we observed a gradual stabilization of the low-FRET, outward-facing GltPh configuration as concentrations of both ligands increased (Fig. 3). Consistent with previous bulk experiments10, these binding isotherms yielded apparent dissociation constants (KD-s) of approximately 2 mM for Na+ and 7 μM for Asp (Supplementary Methods). The titration data also showed a gradual decrease in the average transition frequency. At intermediate Asp concentrations, smFRET trajectories showed molecules reversibly switching between periods of quiescence and periods of rapid transitions, likely due to apo protomers (Fig. 4a, top panel). Increased substrate concentrations shortened the durations of the dynamic periods and increased the lifetimes of the stable states (Fig. 4a, lower panels; Fig. 4b). The midpoints of these changes (∼10 μM) were consistent with the KD of Asp binding obtained from the population data. From the apparent lifetimes of the dynamic and non-dynamic modes, we estimate the ligand binding and dissociation rates of kon ∼104 M−1 s−1 and koff ∼0.1 s−1, respectively, at 2 mM Na+ (Supplementary Methods). A substrate residence time of ∼10 seconds is consistent with previous estimates12.
Figure 3. Na+ ions and Asp favor the outward facing state.
Populations of low- (top, green), intermediate- (center, red) and high- (bottom, blue) FRET states as a function of a, Na+ ions added in the presence of 10 μM Asp and b, Asp added in the presence of 2 mM Na+. The titrations yielded dissociation constants (KD s) of 1.6± 0.3 mM and 6.5± 2.5 μM and Hill coefficients of 1.3± 0.3 and 0.9 ± 0.3, respectively. Shown are averages and standard deviations from at least three independent datasets (each containing at least 250 molecules). Solid lines through the data points are the results of global fitting of the data to Hill equation.
Figure 4. Modulation of dynamics by substrate and inhibitor binding.
a, SmFRET traces obtained in the presence of 2 mM Na+ and increasing Asp concentrations. b, The apparent durations of the dynamic (black) and quiescent (red) periods as a function of Asp concentration. Shown are averages and standard deviations for 3 independent datasets containing at least 250 molecules each. c, A smFRET trace obtained in the presence of 200 mM Na+ and 100 μM Asp. Expanded views of the flicker events (shaded in pink) are shown below the trace. d, Survival plot of the observed flickers. Solid line is a fit to a single exponential decay. e, Transition density plots for flicker events in saturating Asp (left) or TBOA (right). Average transition frequencies are ∼ 0.02 s−1 and < 0.005 s−1, respectively. Data in panels d and e were collected with 400 ms integration time.
Remarkably, dynamic and non-dynamic periods were also observed in the absence of both Na+ and Asp. Visual inspection of smFRET trajectories revealed molecules where 1] rapid dynamics persisted throughout the entire observation periods; 2] stable FRET states predominated; and 3] dynamic and non-dynamic periods were reversibly visited (Supplementary Fig. 8). Consequently, biphasic FRET state lifetimes were observed (Supplementary Fig. 9), wherein the longer dwell times reflected the durations of non-dynamic periods and the shorter dwell times reflected the lifetimes of the FRET states during rapid dynamics. On average, dynamic and non-dynamic periods were approximately equally represented (Fig. 4b). The addition of Na+ and Asp shortened the dynamic periods. However, periods of dynamics persisted, even as the substrate binding sites became saturated (Fig. 4b). In the presence of 200 mM Na+ (a Na+ concentration at which the KD for Asp is ∼1 nM10) and 100 μM Asp, when we expect the fraction of the unbound transporters to be ∼1 in 105, we still observed brief dynamic periods. Such bursts were typified by one or two transient excursions to an inward-facing state with an average dwell time of ∼ 1 s (Fig. 4c and Supplementary Figure 8), before returning to outward-facing conformations (Fig. 4d). “Flickers” of this kind occurred every ∼200 s and constituted about half of all transitions observed out of the outward-facing, low-FRET state. The remainder of the transitions out of the low-FRET state lead to long-lasting intermediate- or high-FRET states (Supplementary Fig. 8). To confirm the that such flickers were due to the rapid transitions of the substrate-loaded transport domains, we performed identical experiments in the presence of saturating concentrations of D,L-threo-β-benzyloxyaspartate (TBOA)22, which binds to the substrate-binding site in a manner that blocks transport10. Compared to Asp, TBOA reduced flickers by > 80 % (Fig. 4e), suggesting that these excursions reflect translocation of Asp across the bilayer. Notably, similar dynamic signatures were also observed for GltPh within the context of proteoliposomes (Supplementary Fig. 8).
We conclude that dynamic heterogeneity, which manifests kinetically as periods of quiescence punctuated by periods of rapid transitions, is an intrinsic property of GltPh arising from spectroscopically hidden isomerization of the protein23. We posit that quiescent periods of substrate-bound GltPh reflect stable conformations that closely resemble crystal structures of the outward- and inward-facing states3,9. In these states, the transport and trimerization domains are closely packed. Dynamic periods reflect trans-membrane movements of individual transport domains. These movements are enabled by a critical protein isomerization, which involves dislodging of the transport domain from the trimeric scaffold (Supplementary Fig. 10a). Such an “unlocked” configuration has been captured crystallographically19 showing disrupted packing and polar interactions at the domain interface. Consistent with this model, the interfacial K290A mutation (Supplementary Fig. 6) dramatically increases the population of molecules in the dynamic mode (Supplementary Fig. 10b-e). The existence of unlocked intermediates with potentially increased hydration at the domain interface24 may help explain the well-known capacity of glutamate transporters and GltPh to catalyze bidirectional anion fluxes25,26. Our data also show that binding of Na+ and Asp significantly increases the lifetimes of stable outward- and inward-facing states, suggesting that there is an allosteric coupling between the substrate-binding site and the domain interface.
Mammalian glutamate transporters are architecturally similar to GltPh, and their transport cycle is anticipated to proceed via similar intermediates. Current kinetic models for the mammalian transporters suggest that substrate translocation is relatively fast27,28. However, in GltPh we find that rapid translocation is preceded by a slow isomerization step that occurs on a time scale comparable to the transporter turnover rate (∼0.01 s−1). Therefore, under our experimental conditions, isomerization of the substrate-loaded GltPh may constitute a key, rate-limiting step in the transport cycle, while return of the unloaded transport domain to an outward-facing state is relatively fast (∼0.5 s−1). The apparent differences between GltPh and the mammalian transporters may, at least in part, be due to the fact that only ensemble properties have thus far been obtained in the latter system. Here, the application of the glutamate substrate to cell membranes expressing the mammalian transporters induced transient electric currents decaying to lower steady state levels. These currents are thought to reflect rapid substrate translocation followed by slower transport cycle events, including relocation of the unloaded transporter. We speculate that the transient currents observed in these experiments reflect translocation of the already unlocked transporter, while the slower process of dislodging of the transport domain is masked by steady state, asynchronous events.
Future efforts must be aimed at delineating the temporal relationship between transport domain translocation and substrate release and the correspondence between transport domain dynamics and anion conductance. Such efforts will be greatly aided by the establishment of imaging strategies wherein stable trans-membrane gradients can be maintained over extended periods, the advent of fluorophores that are highly stable in the lipid environment, and technologies enabling the detection of single transport events.
Methods
DNA manipulations, protein expression, purification and labeling
Single cysteine mutations were introduced within a cysteineless GltPh variant, in which seven unconserved residues have been replaced with histidines resulting in improved expression levels (termed GltPh from hereon for brevity)9, using QuikChange kit (Strategene). Constructs were verified by DNA sequencing and transformed into E. coli DH10-B cells (Invitrogen). Proteins were expressed as C-terminal (His)8 fusions as described previously3,9. Briefly, isolated cell membranes were re-suspended in Buffer A, containing 20 mM Hepes/NaOH, pH 7.4, 200 mM NaCl, 0.1 mM L-aspartate, 0.1 mM Tris(2-carboxyethyl)phosphine (TCEP). Membranes were solubilized in the presence of 40 mM n-dodecyl β-D-maltopyranoside (DDM) for 1 hour at 4 °C. Solubilized transporters were purified by metal-affinity chromatography in buffer A supplemented with 1 mM DDM and eluted in 250 mM imidazole. The (His)8-tag was cleaved by thrombin and proteins were further purified by size exclusion chromatography (SEC) in Buffer A supplemented with 1 mM DDM. GltPh was labeled at a concentration of 40 μM in Buffer A with a mixture of maleimide-activated Cy3, Cy5 and biotin-PEG11 at 50, 100 and 25 μM final concentrations, respectively (molar ratio of ∼ 1:2:0.5). Labeled proteins were quenched with 10 mM 2-mercaptoethanol and subsequently purified away from excess reagents by size exclusion chromotography. The extent of labeling was determined by measuring absorbance at 552 nm and 650 nm, for Cy3 and Cy5, respectively.
Protein reconstitution into liposomes and transport assays
Labeled and unlabeled GltPh variants were reconstituted into liposomes and assayed as previously described10,12. Briefly, liposomes, prepared from 3:1 (w/w) mixture of E. coli total lipid extract and egg yolk phosphotidylcholine (Avanti Polar Lipids) in a buffer containing 20 mM Tris/HEPES, pH 7.4 and 200 mM KCl, were destabilized by addition of Triton X-100 at a detergent to lipid ratio of 0.5:1 (w/w). For transport assays, proteins were added at final protein to lipid ratio of 1:400 (w/w) and incubation for 30 minutes at room temperature. Detergents were removed by repeated incubations with Biobeads as described12. The proteoliposomes were extruded through 400 nm filters prior to analysis12. To measure the uptake of radioactive substrate, proteoliposomes were diluted into reaction buffer containing 20 mM Tris/HEPES, pH 7.4, 200 mM NaCl and 0.3 μM [3H]Asp at room temperature. In control experiments, NaCl was replaced with KCl. For smFRET experiments, protein to lipid ratio of 1:1000 (w/w) was used and proteoliposomes were extruded through 100 nm filters.
smFRET experiments
All experiments were performed using a home-built prism-based total internal reflection fluorescence microscope constructed around a Nikon TE2000 Eclipse inverted microscope body. The samples were illuminated with a 532 nm laser (Laser Quantum), Cy3 and Cy5 fluorescence were separated using a 650DCXR dichroic filter (Chroma) mounted in a DualView apparatus (Photometrics). Imaging data were acquired using MetaMorph acquisition software (Molecular Devices) and an Evolve 512 EMCCD (Photometrics). Before the experiments, the passivated microfluidic imaging chambers prepared and coated with streptavidin as previously described31. Briefly, the microfluidic channel was prepared by incubating with a buffer solution containing 10 mM Tris/acetate, pH 7.5 and 50 mM KCl, 10 μM BSA, 10 μM 25 nt DNA, 8 μM streptavidin (Invitrogen), 1% (v/v) glycerol for 5 minutes. The channel was then rinsed thoroughly in Buffer A. All imaging experiments were performed in Buffer A (unless otherwise stated), supplemented with 2 mM cyclooctatetraene (Sigma), 5 mM BME, an enzymatic oxygen scavenger system comprising 1 unit/ml glucose oxidase (Sigma), 8 units/ml catalase (Sigma) and 0.1% glucose18. If not otherwise specified, all data were collected at an imaging rate of 10 s−1 (100 ms integration time).
Analysis of smFRET data
Fluorescence trajectories were selected for analysis using custom-made MATLAB- (Mathworks) encoded software30,31 according to the following criteria: a single catastrophic photobleaching event; >8:1 signal-to-background noise ratio; a FRET lifetime of at least 15 frames. FRET trajectories were calculated from the acquired intensities, ICy3 and ICy5, using the formula FRET = ICy5/(ICy3 + ICy5). Population contour plots were constructed by superimposing the FRET data from individual traces. Histograms of these population data were fitted to the sum of three Gaussian functions for all mutants, except K55C, for which only a single Gaussian function was used. The Gaussian means, widths and amplitudes were optimized in Origin (OriginLab). The relative populations, the dwell times of each FRET state and the transition frequencies between states were obtained by idealizing the smFRET traces using QuB32,33 (Supplementary Methods). Transition density plots were generated as previously described20. The global fits of the titration curves to Hill equations were performed in Prism34 (Supplementary Methods). The dwell time survival plots were fitted to exponential decay functions and the logarithmic histograms of the dwell times were fitted to transformed probability density functions35, respectively, using Origin (Supplementary Methods).
Preparation of structural figures
All structural renderings were generated using PyMol36 and coordinates deposited in protein data base: accession number 2NWX10 for the outward-facing state and 3KBC3 for the inward-facing state.
Supplementary Material
Acknowledgments
We would like to thank Daniel S. Terry for his help with the design of smFRET experiments and discussions; Zhou Zhou for the synthesis of cyanine fluorophores; Elka Georgieva for initial biochemical characterization of the single-cysteine mutants; Grégory Verdon, Alessio Accardi and Nicolas Reyes for helpful discussions and comments on the manuscript. The work was supported in part by National Institute of Health grant 5U54GM087519.
Footnotes
Supplementary Information is linked to the online version of the paper.
Author contributions: N.A. purified GltPh mutants, carried out the experiments and analyzed the data. R.B.A. prepared reagents for smFRET experiments. N.A., O.B. and S.C.B. together designed, analyzed and interpreted the experiments and wrote the manuscript.
The authors declare no competing financial interests
References
- 1.Danbolt NC. Glutamate uptake. Prog Neurobiol. 2001;65(1):1. doi: 10.1016/s0301-0082(00)00067-8. [DOI] [PubMed] [Google Scholar]
- 2.Krishnamurthy H, Piscitelli CL, Gouaux E. Unlocking the molecular secrets of sodium-coupled transporters. Nature. 2009;459(7245):347. doi: 10.1038/nature08143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Reyes N, Ginter C, Boudker O. Transport mechanism of a bacterial homologue of glutamate transporters. Nature. 2009;462(7275):880. doi: 10.1038/nature08616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weiss S. Fluorescence spectroscopy of single biomolecules. Science. 1999;283(5408):1676. doi: 10.1126/science.283.5408.1676. [DOI] [PubMed] [Google Scholar]
- 5.Sakmann B, Patlak J, Neher E. Single acetylcholine-activated channels show burst-kinetics in presence of desensitizing concentrations of agonist. Nature. 1980;286(5768):71. doi: 10.1038/286071a0. [DOI] [PubMed] [Google Scholar]
- 6.Cull-Candy SG, Parker I. Rapid kinetics of single glutamate-receptor channels. Nature. 1982;295(5848):410. doi: 10.1038/295410a0. [DOI] [PubMed] [Google Scholar]
- 7.Tzingounis AV, Wadiche JI. Glutamate transporters: confining runaway excitation by shaping synaptic transmission. Nat Rev Neurosci. 2007;8(12):935. doi: 10.1038/nrn2274. [DOI] [PubMed] [Google Scholar]
- 8.Zerangue N, Kavanaugh MP. Flux coupling in a neuronal glutamate transporter. Nature. 1996;383(6601):634. doi: 10.1038/383634a0. [DOI] [PubMed] [Google Scholar]
- 9.Yernool D, Boudker O, Jin Y, Gouaux E. Structure of a glutamate transporter homologue from Pyrococcus horikoshii. Nature. 2004;431(7010):811. doi: 10.1038/nature03018. [DOI] [PubMed] [Google Scholar]
- 10.Boudker O, et al. Coupling substrate and ion binding to extracellular gate of a sodium-dependent aspartate transporter. Nature. 2007;445(7126):387. doi: 10.1038/nature05455. [DOI] [PubMed] [Google Scholar]
- 11.Boudker O, Verdon G. Structural perspectives on secondary active transporters. Trends Pharmacol Sci. 2010;31(9):418. doi: 10.1016/j.tips.2010.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ryan RM, Compton EL, Mindell JA. Functional characterization of a Na+− dependent aspartate transporter from Pyrococcus horikoshii. J Biol Chem. 2009;284(26):17540. doi: 10.1074/jbc.M109.005926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Blanchard SC. Single-molecule observations of ribosome function. Curr Opin Struct Biol. 2009;19(1):103. doi: 10.1016/j.sbi.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Groeneveld M, Slotboom DJ. Rigidity of the subunit interfaces of the trimeric glutamate transporter GltT during translocation. J Mol Biol. 2007;372(3):565. doi: 10.1016/j.jmb.2007.06.067. [DOI] [PubMed] [Google Scholar]
- 15.Reyes N, Oh S, Boudker O. Binding thermodynamics of a glutamate transporter homolog. Nat Struct Mol Biol. 2013 doi: 10.1038/nsmb.2548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Groeneveld M, Slotboom DJ. Na(+):aspartate coupling stoichiometry in the glutamate transporter homologue Glt(Ph) Biochemistry. 2010;49(17):3511. doi: 10.1021/bi100430s. [DOI] [PubMed] [Google Scholar]
- 17.Roy R, Hohng S, Ha T. A practical guide to single-molecule FRET. Nat Methods. 2008;5(6):507. doi: 10.1038/nmeth.1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dave R, Terry DS, Munro JB, Blanchard SC. Mitigating unwanted photophysical processes for improved single-molecule fluorescence imaging. Biophys J. 2009;96(6):2371. doi: 10.1016/j.bpj.2008.11.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Verdon G, Boudker O. Crystal structure of an asymmetric trimer of a bacterial glutamate transporter homolog. Nat Struct Mol Biol. 2012;19(3):355. doi: 10.1038/nsmb.2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McKinney SA, Joo C, Ha T. Analysis of single-molecule FRET trajectories using hidden Markov modeling. Biophys J. 2006;91(5):1941. doi: 10.1529/biophysj.106.082487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grewer C, et al. Individual subunits of the glutamate transporter EAAC1 homotrimer function independently of each other. Biochemistry. 2005;44(35):11913. doi: 10.1021/bi050987n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shimamoto K, et al. DL-threo-beta-benzyloxyaspartate, a potent blocker of excitatory amino acid transporters. Mol Pharmacol. 1998;53(2):195. doi: 10.1124/mol.53.2.195. [DOI] [PubMed] [Google Scholar]
- 23.Liu S, Bokinsky G, Walter NG, Zhuang X. Dissecting the multistep reaction pathway of an RNA enzyme by single-molecule kinetic “fingerprinting”. Proc Natl Acad Sci U S A. 2007;104(31):12634. doi: 10.1073/pnas.0610597104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stolzenberg S, Khelashvili G, Weinstein H. Structural Intermediates in a Model of the Substrate Translocation Path of the Bacterial Glutamate Transporter Homologue GltPh. J Phys Chem B. 2013;116(18):5372. doi: 10.1021/jp301726s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fairman WA, et al. An excitatory amino-acid transporter with properties of a ligandgated chloride channel. Nature. 1995;375(6532):599. doi: 10.1038/375599a0. [DOI] [PubMed] [Google Scholar]
- 26.Wadiche JI, Amara SG, Kavanaugh MP. Ion fluxes associated with excitatory amino acid transport. Neuron. 1995;15(3):721. doi: 10.1016/0896-6273(95)90159-0. [DOI] [PubMed] [Google Scholar]
- 27.Grewer C, Watzke N, Wiessner M, Rauen T. Glutamate translocation of the neuronal glutamate transporter EAAC1 occurs within milliseconds. Proc Natl Acad Sci U S A. 2000;97(17):9706. doi: 10.1073/pnas.160170397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Otis TS, Kavanaugh MP. Isolation of current components and partial reaction cycles in the glial glutamate transporter EAAT2. J Neurosci. 2000;20(8):2749. doi: 10.1523/JNEUROSCI.20-08-02749.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Blanchard SC, et al. tRNA dynamics on the ribosome during translation. Proc Natl Acad Sci U S A. 2004;101(35):12893. doi: 10.1073/pnas.0403884101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhao Y, et al. Single-molecule dynamics of gating in a neurotransmitter transporter homologue. Nature. 2010;465(7295):188. doi: 10.1038/nature09057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Munro JB, Altman RB, O'Connor N, Blanchard SC. Identification of two distinct hybrid state intermediates on the ribosome. Mol Cell. 2007;25(4):505. doi: 10.1016/j.molcel.2007.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Qin F, Auerbach A, Sachs F. A direct optimization approach to hidden Markov modeling for single channel kinetics. Biophys J. 2000;79(4):1915. doi: 10.1016/S0006-3495(00)76441-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Qin F. Restoration of single-channel currents using the segmental k-means method based on hidden Markov modeling. Biophys J. 2004;86(3):1488. doi: 10.1016/S0006-3495(04)74217-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Motulsky Harvey, Christopoulos Arthur. Fitting Models to Biological Data using Linear and Nonlinear Regression A Practical Guide to Curve Fitting. Oxford University Press; New York: 2004. [Google Scholar]
- 35.Sigworth FJ, Sine SM. Data transformations for improved display and fitting of single-channel dwell time histograms. Biophys J. 1987;52(6):1047. doi: 10.1016/S0006-3495(87)83298-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.The PyMOL Molecular Graphics System, Version 1.5.0.4. Schrödinger, LLC; [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.