Abstract
Specialized Candida albicans cell surface proteins called adhesins mediate binding of the fungus to host cells. The mammalian transglutaminase (TG) substrate and adhesin, Hyphal wall protein 1 (Hwp1), is expressed on the hyphal form of C. albicans where it mediates fungal adhesion to epithelial cells. Hwp1 is also required for biofilm formation and mating thus the protein functions in both fungal-host and self-interactions. Hwp1 is required for full virulence of C. albicans in murine models of disseminated candidiasis and of esophageal candidiasis. Previous studies correlated TG activity on the surface of oral epithelial cells, produced by epithelial TG (TG1), with tight binding of C. albicans via Hwp1 to the host cell surfaces. However, the contribution of other Tgs, specifically tissue TG (TG2), to disseminated candidiasis mediated by Hwp1 was not known. A newly created hwp1 null strain in the wild type SC5314 background was as virulent as the parental strain in C57BL/6 mice, and virulence was retained in C57BL/6 mice deleted for Tgm2 (TG2). Further, the hwp1 null strains displayed modestly reduced virulence in BALB/c mice as did strain DD27-U1, an independently created hwp1Δ/Δ in CAI4 corrected for its ura3Δ defect at the URA3 locus. Hwp1 was still needed to produce wild type biofilms, and persist on murine tongues in an oral model of oropharyngeal candidiasis consistent with previous studies by us and others. Finally, lack of Hwp1 affected the translocation of C. albicans from the mouse intestine into the bloodstream of mice. Together, Hwp1 appears to have a minor role in disseminated candidiasis, independent of tissue TG, but a key function in host- and self-association to the surface of oral mucosa.
Introduction
Candida albicans is often found in the oral cavity and gastrointestinal (GI) tract of healthy individuals as a member of the normal flora. Impairment of the immune system allows for the fungus to overgrow, germinate and produce filamentous cells capable of invading the local mucosal surface. Germination and production of filamentous hyphal cells is accompanied by the expression of adhesion proteins that facilitate host cell interactions. Hyphal wall protein 1 (Hwp1; Accession number AAC96368) [1], Als1p (Agglutinin-like sequence 1 protein) and Als3p are involved in the binding of C. albicans to oral epithelial cells [2–6].
Hwp1 participates in the formation of covalent bonds to primary amines and to a buccal epithelial cell (BEC) surface protein catalyzed by human transglutaminases (TGs) [1,7,8]. TGs are multi-functional enzymes widely distributed in the body that modify glutamyl residues in proteins [9–11]. These enzymes catalyze the formation of isodipeptide bonds between an acceptor glutamine residue and a donor lysine residue in other proteins, a reaction known as transamidation. TG1 (keratinocyte transglutaminase) is active on the surface of BECs [12] where it functions in assembly of the cornified cell envelope, a scaffold of cross-linked proteins that gives mucosal cells their barrier properties. We found the participation of Hwp1 in the formation of a stabilized (heat and detergent-resistant) TG-dependent adhesion to BECs [1], and speculated that TG1 on the surface of BECs catalyzed this reaction. In support of this conclusion, an hwp1Δ/Δ strain is unable to cause oropharyngeal candidiasis in mice [13]. However, we were unable to verify the role of TG1 directly as Tgm1-/- mice die of dehydration soon after birth [14].
Although a mechanism of adhesion based on transglutaminase activity on BECs could explain the requirement for Hwp1 in oral candidiasis, the functional requirement for Hwp1 in disseminated candidiasis in mice [1,15] where active TG1 is not expressed [16] is not clear. TG2 (tissue transglutaminase) is widely distributed in the body (e.g. liver, kidneys, extracellular matrix, muscle) [9,10,16] therefore, we hypothesized that TG2 may have a role in virulence in a disseminated candidiasis animal model. In vitro, both native and recombinant Hwp1 are substrates for guinea pig liver TG2 [1], therefore, it is plausible that tissue TG2 catalyzes transamidation reactions involving Hwp1. To test this hypothesis, we created a new hwp1 null strain (hwp1Δ/Δ) in the wild type C. albicans strain SC5314 using the flp recombinase method [17] to avoid URA3-associated confounding effects [18–20]. We tested the virulence of the new hwp1Δ/Δ strain in wild type C57BL/6 and in Tgm2-/- mice, and did not observe a TG2-dependent requirement for Hwp1. We re-tested the virulence of the newly created null strain in BALB/c mice as before [15] and observed that virulence was attenuated, and reintroducing HWP1 at its native locus did not restore full virulence of the reconstituted strain (hwp1Δ/+HWP1). In light of these results, other Hwp1-phenotypes were tested with the newly created deletion strain. Biofilm formation was deficient in the hwp1Δ/Δ strain as reported by others [21,22] and restored upon reintroducing a functional copy of HWP1 at its native locus. The knock-out strain was also deficient in causing disease in an oropharyngeal model of candidiasis [23]. Lastly, we examined the role of Hwp1 in gut translocation of C. albicans to the bloodstream of C57BL/6 mice using the experimental model previously developed [24]. This infection model mimics a common infection route in at-risk candidiasis patients [24–26], and the role of Hwp1 in this model in unknown. The hwp1Δ/Δ strain was capable of colonizing the mouse gut to equal levels as HWP1+ strains. However, the hwp1 null strain was less virulent relative to the wild type or the reconstituted strain in this model, thus defining a new phenotype for Hwp1 in murine candidiasis.
Our results suggested that Hwp1 has adapted C. albicans for localized infections of the oral mucosa and is less important for disseminated disease. The fungus is still capable of invading kidneys and livers during invasive disease in the absence of Hwp1 or TG2. The wild type genetic background of SC5314 has allowed us to discern subtle in vivo phenotypes that were previously masked, perhaps due to unknown in vivo fitness defects of the previous parental strain (CAI4 [27]). Our results suggest that Hwp1 engenders C. albicans with niche-specific capabilities to inhabit the oral cavity where fungal-host and fungal-fungal interactions are needed for successful colonization and localized superficial tissue invasion that typifies oropharyngeal candidiasis.
Materials and Methods
Ethics statement
All animal studies were approved by the Johns Hopkins University Animal Care and Use Committee (ACUC). The approved protocol was in compliance with the Animal Welfare Act regulations and Public Health Service (PHS) Policy. For virulence/survival studies, C. albicans-infected mice were closely monitored for signs of distress. Moribund mice exhibiting signs of illness (ruffled fur, lethargy) were humanely sacrificed by CO2 inhalation according to the recommendations of the Johns Hopkins University ACUC. The studies were performed under the ACUC protocol number MO11M261.
Candida albicans strains
Clinical isolate, SC5314 [28], was used as source for genomic DNA and for the construction of HWP1 deletion mutants. The hwp1Δ/hwp1Δ strain constructed in CAI4 [27], DD27-U1[29], was kindly provided by William Fonzi (Georgetown University, Washington, DC). The organisms were stored at -80OC and cultured in liquid or agar-containing plates of yeast peptone dextrose (YPD) medium at 30°C according to standard methods [30].
Candida albicans deletion strains
SC5314 was used as parent to delete the coding region for HWP1 using the SAT1 flipper system for gene disruption in C. albicans [17,31]. Plasmid pSFS1 (provided by Joachim Morschhäuser, University of Würzburg, Würzburg, Germany), a derivative of pSFS2 [17], was used to construct the disruption cassette for HWP1 (orf19.1321; www.candidagenome.org) as follows: the region between bp -491 to +35 (relative to ATG; 5’ amplicon) was amplified using the PCR from SC5314 genomic DNA with oligonucleotides H1Ap and H2Xh (Table S1), and the region between bp +1860 to +2249 (3’ amplicon) of HWP1 was amplified with oligonucleotides H3SII and H4 using Platinum Taq High Fidelity (Invitrogen/Life Technologies, Grand Island, NY) according to the manufacturer’s recommendations. The 3’ amplicon contained sequences downstream of the polyadenylation site of the HWP1 mRNA [32], across a SacI site. The HWP1 rescue fragment was also generated by the PCR (bp -362 to bp 517 downstream of the stop codon) using oligonucleotides H1Ap and H5Xh to add an ApaI site and an XhoI site at the 5’ and 3’ ends of the amplicon, respectively. The disruption plasmid was constructed by first cloning the 3’ HWP1 amplicon between the SacII and SacI sites of pSFS2 to generate pSFSH3. This plasmid was then used to clone both the 5’ HWP1 amplicon between the ApaI and XhoI sites, and the rescue fragment between the ApaI and SalI sites. The cloned sequences were verified by sequencing for authenticity and PCR-induced errors. The disruption plasmid, pSFSH35, contained 5’ and 3’ HWP1 sequences interrupted by the SAT1 flipper cassette, while the rescue plasmid, pSFSH329, contained HWP1 sequences up- and downstream of the coding region followed by the SAT1 flipper cassette and 3’ HWP1 sequences. Gel-purified linear disruption fragment generated by digesting pSFSH35 with KpnI and SacI was used to transform SC5314 by electroporation [17]. Transformants were selected on YPD plates containing 250 µg/mL of nourseothricin (clonNAT, Werner BioAgents, Jena, Germany) at 30°C for 2 days. Two independent transformants integrated the disruption cassette at a single allele of HWP1, and were genotypically HWP1/hwp1Δ::SAT1-FLP determined by Southern blot analysis (see Figure S2). Both transformants were cultured in YPD for 24 h at 30°C in the presence of 0.4% BSA (Sigma-Aldrich, St. Louis, MO; product number B4287) to induce loss of the SAT1 cassette via intramolecular recombination at the FRT sites by the site-specific recombinase FLP, and to re-establish nourseothricin sensitivity. Loss of the SAT1-FLP cassette was verified by Southern blotting, and both single disruption derivatives (HWP1/hwp1Δ::FRT) were subjected to a second round of transformation with the linearized deletion construct. Integration of the disruption cassette at the intact HWP1 allele was verified by Southern blotting, and two derivatives were grown in YPD 0.4% BSA to induce loss of the SAT1-FLP cassette at the second HWP1 allele. Nourseothricin sensitive strains were subsequently verified by Southern blotting. The double disruption strain SCH1211 (hwp1Δ::FRT/hwp1Δ::FRT; “hwp1Δ/Δ”) was used as recipients for the rescue DNA fragment harbored in pSFSH329 (digested with BssHII release the rescue HWP1 gene fragment). Integration of the rescue DNA fragment at a single (hwp1Δ::FRT allele was verified by Southern blotting. Reintegrant derivatives were grown in YPD 0.4% BSA to generate strains sensitive to nourseothricin and with a single functionally corrected HWP1 verified by northern blotting or reverse transcription-PCR (RT-PCR) (Figure 1 and data not shown).
Figure 1. Deletion and reconstitution of HWP1 expression in SC5314.
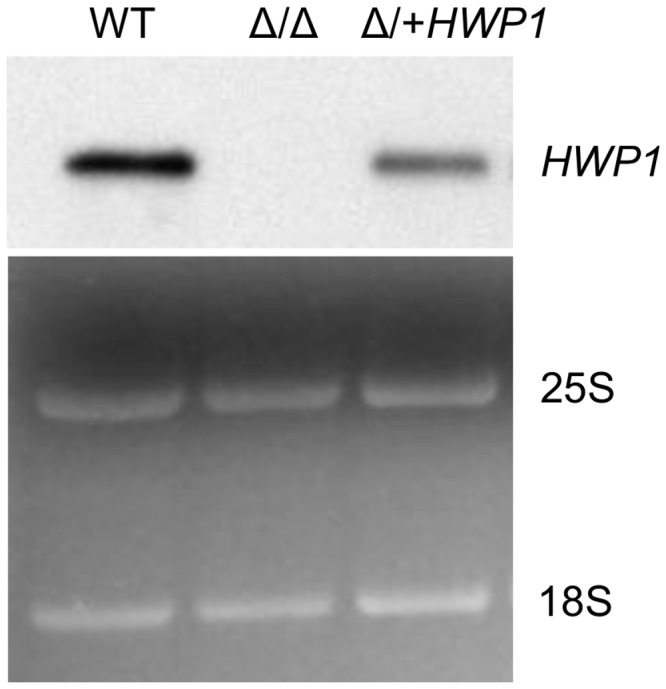
Northern blot analysis of the parental SC5314 (WT), hwp1 null SCH1211 (Δ/Δ) and HWP1 put-back HR615 (Δ/+HWP1) strains. Five µg of total RNA was separated in a standard formaldehyde gel and transferred to a nylon membrane prior to hybridization with a biotinylated riboprobe to 5’ HWP1 sequences (top panel). Lower panel shows the rRNA bands stained with ethidium bromide as loading control.
Southern blot analysis
Genomic DNA was prepared from overnight cultures using a MasterPure Yeast DNA Purification Kit (Epicentre Biotechnologies, Madison, WI). Three to five µg of DNA was digested to completion with NdeI (Invitrogen/Life Technologies, Grand Island, NY) and the fragments separated in an agarose gel as per standard methods. The DNA fragments were blotted onto a BrightStar-Plus (Ambion/Life Technologies) nylon membrane and probed with a biotinylated (BrightStar Psoralen-Biotin labeling kit, Ambion/Life Technologies) HWP1 amplicon (nt -491 to +35 relative to the ATG) overnight at 42°C in ULTRAHyb solution (Ambion/Life Technologies). DNA bands were visualized by incubating the membrane with Strepavidin conjugated to alkaline phosphatase and chemiluminescence reagents (BrightStar BioDetect Kit, Ambion/Life Technologies), followed by exposure of the membrane to X-ray film (Kodak BioMax XAR).
Northern blotting
Total RNA was prepared using the hot acid phenol method [33] from C. albicans germ tubes grown in YPD with 10% bovine serum for two hr at 37°C. Five µg of total RNA was separated in a standard formaldehyde gel and transferred to a charged nylon membrane (BrightStar-Plus, Amibon/Life Technologies). The membrane was probed with a biotinylated (BrightStar Psoralen-Biotin Kit, Ambion/Life Technologies) antisense transcript (riboprobe) containing HWP1 sequences from +11 to +623 relative to the ATG start site generated according to the manufacturer’s recommendations (MAXIscript Kit, Ambion/Life Technologies). The membrane was hybridized overnight at 68°C in ULTRAHyb solution (Ambion/Life Technologies). RNA bands were visualized by incubating the membrane with Strepavidin conjugated to alkaline phosphatase and chemiluminescence reagents (BrightStar BioDetect Kit, Ambion/Life Technologies), followed by exposure of the membrane to X-ray film (Kodak BioMax XAR).
Germ tube TG assays
Expression of Hwp1 on the surface of germ tubes was visualized by cross-linking 5-(biotinamido)pentylamine (5-BPA) (Pierce/Thermo Scientific, Rockford, IL) as before [1] with slight modifications. Human recombinant TG2 (hTG2; Zedira, Darmstadt, Germany) (12.5 U/mL) was used in 400 µL reactions containing 2 x 107 germ tubes in reaction buffer (100 mM Tris-HCl pH 7.5, 10 mM CaCl2, 1 mM DTT and 1 mM EDTA, 25 µM 5-BPA) for 15 min at 37°C. The reactions were stopped by adding 1.0 mL of 10 mM EDTA, pH 8.0, and the germ tubes washed twice with dH2O. Germ tubes (1 x 105) were spotted onto poly-L-lysine coated slides, allowed to air dry and incubated with Streptavidin conjugated to FITC (Invitrogen/Life Technologies) at 1:50 dilution in phosphate buffered saline (PBS, GIBCO/Life Technologies). After 30 min incubation at 37°C, the slides were washed in PBS and rinsed with water. A drop of VECTASHIELD mounting medium with DAPI (VECTOR Laboratories Inc, Burlingame, CA) was added to the germ tubes before observing under epifluorescence using the FITC or DAPI excitation cubes (Zeiss LSM 410 confocal microscope; Carl Zeiss Microscopy, Jena, Germany). Cell images were acquired with an Olympus camera running cellSense 1.6 software (Olympus America). Captured images were minimally manipulated with ImageJ64 (v.1.46r; open-source software; http://imagej.nih.gov.ij).
Virulence studies
Female BALB/c mice (6-8 weeks) were purchased from Charles River Laboratories (Frederick, MD) one week prior to experiments to allow the mice to acclimatize to the housing at JHU. C57BL/6 Tgm2-/- [34] breeder mice were purchased from the European Mouse Mutant Archive (Munich, Germany) and bred in-house to generate animals for our studies. Wild type female (6-8 weeks) C57BL/6 controls were purchased from The Jackson Laboratory (Bar Harbor, ME) at least one week prior to experiments.
Disseminated candidiasis studies [35] were performed as follows: C. albicans strains were grown in YPD with vigorous shaking for approximately 24 h at 30°C, washed twice with PBS and adjusted to an OD600 of 0.3-0.32 (3.5-4.0 x 106 cfu/mL) in sterile PBS. We found that adjusting the yeast suspensions to OD600 of 0.3-0.32 (vs. counting by hemocytometer) produced consistent results across experiments. One hundred microliters of the yeast suspensions was injected into the tail veins of mice, and these observed for signs of illness over a 10-14 day period. Moribund mice were humanely euthanized by CO2 inhalation. For histological observations, C57BL/6 and Tgm2-/- mice injected with SCH1211 (hwp1Δ::FRT/hwp1Δ::FRT) were sacrificed at day 3 post inoculation. Kidneys were fixed in buffered formalin, embedded in Paraffin, and sections stained with Periodic-acid Schiff (PAS) to observe the fungal elements at the site of infection.
The role of Hwp1 in gut-translocation and dissemination of C. albicans was tested in mice as previously described [24] (University of Texas Southwestern Medical Center, Dallas, TX) with one modification. Briefly, 6-8 week old female C57BL/6 mice were gut-colonized with C. albicans after first reducing the GI bacterial and fungal flora with penicillinG (1500 U/mL) and streptomycin (2 mg/mL), and fluconazole (250 µg/mL) in their drinking water. After verifying the depletion of the gut normal flora, the drinking water was replaced with water containing a suspension of C. albicans at 107 cfu/mL plus penicillinG and streptomycin as above for 5 days. After verifying gut-colonization of C. albicans as CFU/g of stools, the mice were immunosuppressed by four (vs. three in the original protocol) intraperitoneal injections of cyclophosphamide (150 mg/kg/dose) every other day. Groups of mice (n=7) were observed for signs of illness prior to sacrifice to determine the fungal burden in the livers. Serial dilutions of homogenized livers were spread onto YPD agar with vancomycin at 10 µg/mL and gentamycin at 100 µg/mL, trypicase soy agar (TSA) and MacConkey’s agar plates (microbiological media from Becton-Dickinson). Creamy white colonies were enumerated to determine CFU/g of liver.
Candida biofilms
C. albicans biofilms were produced on silicone squares as before [22] with slight modifications. Silicone squares (1.5 x 1.5 cm) (Bentec Medical, Inc., Woodland, CA) were conditioned overnight at 37°C in bovine calf serum (Hyclone), rinsed with PBS and placed in non-tissue culture 6-well plates containing 2 mL of Spider medium (1% nutrient broth; Remel/Thermo Scientific; 1% mannitol and 2% potassium monophosphate, the latter two reagents from Sigma-Aldrich). C. albicans cultures were grown for approximately 18 h in YPD at 30°C, washed with PBS and adjusted to an OD600 of 1.0 in Spider medium. Two mL of each yeast culture was added to the silicone squares to produce a final cell suspension at OD600 equal to 0.5. After 90 min incubation at 37°C with shaking at 60 rpm, the silicone squares were briefly rinsed in PBS and transferred to new 6-well plates containing 4 mL of fresh Spider medium. The plates were further incubated at 37°C for 48 h with shaking. The silicone squares were subsequently rinsed in PBS and set to dry for 2-3 days at room temperature inside a fume hood prior to weighing. The biofilms were generated twice in triplicate for each strain. The biofilms produced by the hwp1Δ/Δ strains had a tendency to partially or completely slide off the silicone square just prior to drying, and these squares were excluded from measurements. The data represent measurements of 3-5 biofilms.
Statistical methods
Survival studies in BALB/c or C57BL/6 mice were analyzed for significance using the Log-rank (Mantel-Cox) test derived from Kaplan-Meier survival curves. Mean biofilm dry weights were analyzed by one-way ANOVA using Tukey’s multiple comparisons test assuming an alpha value of 0.05. Gut colonization levels (CFU/g of stool) were analyzed by one-way ANOVA of log-transformed data and applying Tukey’s multiple comparisons test. For all statistical analyses, a P value <0.05 was considered significant. Statistical analyses were performed using GraphPad Prism (v. 6.01, GraphPad Software, Inc).
Results
Disruption of HWP1 in wild type C. albicans
An open question regarding the contribution of Hwp1 to virulence is whether the TG substrate reactivity of the protein has a role in disseminated candidiasis. To address this question, we generated new hwp1 null strains in wild type C. albicans SC5314 to avoid unknown in vivo auxotrophic requirements that could complicate virulence studies [18,20]. Also, the use of SC5314 vs. another parental strain for gene knock-outs, BWP17 [36], obviated the manipulation of a strain generated post multiple rounds of transformations and with reported chromosome abnormalities [37–39]. Because chromosomal copy number affects virulence or fitness in vivo in confounding ways [40], HWP1 was disrupted in SC5314 at both alleles using the flp recombinase system [17] (Figure S1). Two independent hwp1Δ/Δ strains were created (SCH1211 and SCH2822), and one disruptant, SCH1211, was corrected for HWP1 expression at its native locus using the same flp recombinase molecular system (Figure 1, and Information S1 and S2). Re-introducing HWP1 also restored germ tube surface expression of Hwp1 (Information S3) and the amount of protein paralleled the expression of a single allele (compare HWP1 RNA in lanes 1 vs. 3 in Figure 1; compare SC5134 and HR615 fluorescence brightness in S3).
The role of Hwp1 in disseminated candidiasis
Virulence of the hwp1Δ/Δ strain in Tgm2-/- mice
The newly constructed hwp1Δ/Δ strain SCH1211 and the parental SC5314 were tested for virulence [35] in wild type C57BL/6 and Tgm2-/- mice to determine whether TG activity participated in C. albicans tissue infection such as in the kidneys where TG2 is active [16]. Disseminated candidiasis was established by intravenous injection of yeasts; 3.5 x 105 yeasts per animal as per published methods [35]. Wild type or Tgm2-/- mice infected with SCH1211 succumbed to candidiasis at equal rates and at the same rate as wild type C57BL/6 mice infected with SC5315 (Figure 2A). Median survival rates for all groups of mice were 6 days. Histological analysis of kidneys from wild type and Tgm2-/- mice infected with SCH1211 revealed the invasion of tissue to similar degrees (Figure 2B). We observed identical survival kinetics when we tested a second, independently generated hwp1Δ/Δ strain SCH2283 in wild type and Tgm2-/- mice (Supporting Information, Figure S4). The use of an independent hwp1Δ/Δ strain discounted the possibility of a SCH1211-specific phenotype independent of the HWP1 deletion. The similar rate of survival among all the mice suggested that Hwp1 was dispensable for disseminated candidiasis in the C57BL/6 genetic background.
Figure 2. The hwp1 SCH1211 null strain retains virulence in wild type and Tgm2-/- mice.
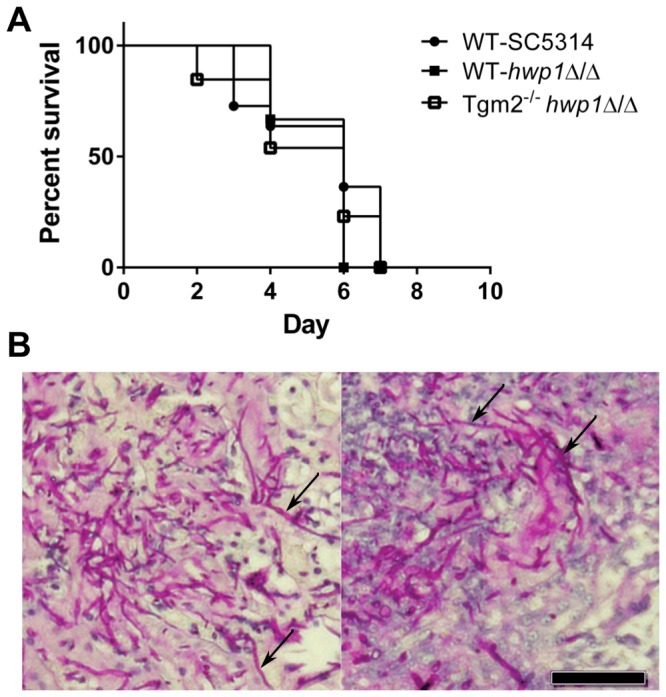
(A) Wild type C57BL/6 (n=6) and Tgm2 knock-out (n=13) mice were infected with 3-3.5 x 105 SCH1211 yeasts and observed for signs of illness over an eight day period. C57BL/6 mice (n=11) infected with the wild type parental strain, SC5314, served as comparator. The survival kinetics among the groups of mice was indistinguishable (P>0.05). The median survival day for all groups was 6 days. (B) Histological analysis of kidneys from C57BL/6 (WT, left panel) or Tgm2 knock-out (Tgm2-/-, right panel) mice sacrificed 3-days post infection with SCH1211. Kidney sections were stained with period acid Schiff to visualize fungal elements. Arrows point to hyphae penetrating the tissue. Size bar=50 µm.
Virulence of the hwp1Δ/Δ strain in BALB/c mice
As a consequence of the virulence results using C57BL/6 mice, we wanted to re-test the designation of Hwp1 as a virulence factor in disseminated candidiasis. Previous HWP1 deletion virulence studies were performed in BALB/c mice utilizing strains generated in CAI4 [27] with URA3 at the ENO locus [15] or at its native locus [29]. We repeated the virulence studies in BALB/c mice with the newly created strains, and found that the absence of HWP1 expression attenuated the virulence of C. albicans (Figure 3) that was statistically different from that of SC5314 (Log-rank test, P=0.01). The median survival of mice infected with SCH1211 was 5 days vs. 3 days for mice infected with SC5314. Mice that received the reconstituted strain HR615 had a 6 day survival median and the rate of death was not different from that of mice infected with SCH1211 (P=0.87). We also tested the hwp1Δ/Δ strain DD27-U1 created in a CAI4 background with URA3 at its native locus and corrected for the partial IRO1 deletion [29]. Mice infected with DD27-U1 had a median survival of 5.5 days and a similar rate of death relative to SCH1211 (P=0.73) and HR615 (P=0.73). Our results here contrasted with our previous survival data that show a median survival of >10 days in BALB/c mice infected with hwp1Δ/Δ strains even though the inoculum was higher [15]. Our current results are in line with data reported by Skarkey, et al. [29] who show an attenuated virulence for DD27-U1 in BALB/c mice. Although survival rates of mice infected with hwp1Δ/Δ strains were statistically different from that of SC5314, the relatively small boost in the survival rate of mice infected with the former strains appeared to diminish the biological significance of Hwp1’s role in disseminated candidiasis.
Figure 3. hwp1 null strains display attenuated virulence in BALB/c mice.
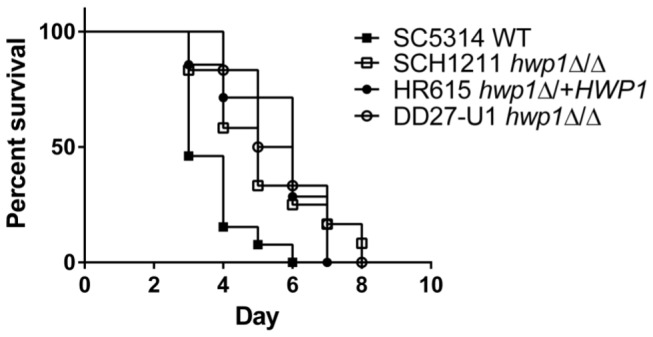
BALB/c mice infected with hwp1Δ/Δ strains SCH1211 (n=11) or DD27-U1 (n=6) were attenuated in virulence in intravenously infected BALB/c mice. Reintroducing HWP1 at its native site in SCH1211 (HR615, n=7) did not restore full virulence of the strain to wild type levels (SC5314, n=13). The rate of survival among the strains deleted for HWP1 (SCH1211 and DD27-U1) or expressing a single allele (HR615) was statistically different (P<0.05) relative to the wild type SC5314.
Hwp1 and in vitro/in vivo biofilms
Formation of biofilms on a silicone surface
In light of the virulence studies results, we tested other phenotypes associated with Hwp1. Hwp1 may have a significant role in the establishment of C. albicans at specific anatomical sites, e.g. the oral cavity, but have a minor role in invasive disease. Re-validating other Hwp1-associated phenotypes using our newly created hwp1Δ/Δ strains would bolster this hypothesis. A well-established function for Hwp1 is the mediation of C. albicans self-association in a three-dimensional colonial structure typical of biofims [3,21,22,41–43]. We used the silicone-square method [22,42] to quantify biofilm formation among the strains. Both hwp1 null strains, SCH1211 and SCH2283, were defective in biofilm formation, and this defect was corrected by reintroducing a single copy of HWP1 into SCH1211 (Figure 4). Our strains retained the in vitro biofilm defect phenotype observed by others [21], confirming the important role of Hwp1 in biofilm formation.
Figure 4. Biofilm formation is defective in newly created hwp1 null strains.
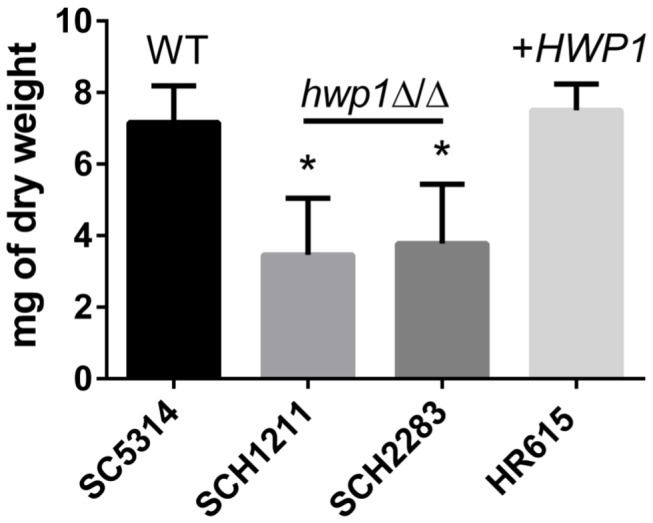
C. albicans strains were induced to form biofilms on the surface of silicone squares for 60 hr in Spider medium at 37°C. Two independently created hwp1 null strains, SCH1211 and SCH2283, generated less biomass that were less adherent to the silicone squares. Reintroduction of a single HWP1 allele into SCH1211 restored wild type biofilm biomass levels (HR615). The dry weights of the hwp1 null mutants were statistically different relative to that produced by wild type SC5314 (asterisks, P<0.05; One-way ANOVA, multiple comparisons). The mean dry weight of HR615 was not statistically different relative to that produced by SC5314. The data represent means of 3-5 measurements from two independent experiments performed in triplicate.
Formation of oral biofilms
Pseudomembranous oropharyngeal candidiasis lesions are considered a form of in vivo biofilm [3], and several animal models mimic this localized infection of the oral cavity [23,44–46]. These models are based on the superficial infection of murine tongues post oral inoculation of C. albicans in immunosuppressed or immunocompromised mice. The need for Hwp1 to form in vivo oral biofilms further supports the role of Hwp1 as a niche-specific virulence attribute required for colonization and local invasion of tissue. To test the capability of the hwp1Δ/Δ strain to form in vivo biofilms, BALB/c mice were orally inoculated [23] with SCH1211 and HR615, and sacrificed 3 days later to determine fungal burdens of the tongues. Two of 6 mice inoculated with SCH1211 were lost post-procedure; however the remaining 4 mice did not develop oral candidiasis (no CFU). In contrast, 6 of 7 mice infected with HR615 had infected tongues (mean CFU/g tongue, 4.2 x 105 +/- 2 x 105) at comparable levels reported for SC5314 [23]. Previous OPC studies in mice found that lack of HWP1 expression impaired the strain’s ability to establish an infection of the tongue [3], and our results confirmed the in vivo biofilm defective phenotype of our hwp1Δ/Δ strain. Published data [13] together with our results here also demonstrated that URA3 function did not influence the establishment and local invasion of oral tissue in mice further supporting the independent and key role of Hwp1 in OPC. Indeed, CAI4 (ura3Δ/Δ) is capable of producing OPC in immunocompromised gnotobiotic mice [47] showing that URA3 function is nonessential in this infection model.
Gut translocation studies
Intestinal colonization of C57BL/6 mice with C. albicans prior to translocation studies
A common route of infection in hospitalized patients is the translocation of endogenous C. albicans from the intestinal tract into the blood stream associated with antibiotic use and hematological immunosuppression or chemotherapy that result in the loss of intestinal barrier function [48]. The requirement for Hwp1 to promote dissemination of C. albicans from the GI tract is unknown, and may define a new role for the fungal adhesin. We used the murine model that mimics this route of infection developed by Koh and colleagues [24]. Mice are treated with antibiotics and fluconazole to allow the colonization of C. albicans in their GI tracts prior to immunosuppression. We found that all three C. albicans strains, SC5314, SCH1211 and HR615, colonized the intestinal track of C57BL/6 mice to similar levels (Figure 5A). The median SC5314 CFU/g of stool, 2.03 x 107, was comparable to that achieved in C3H/HeN mice (2.24 x 107 CFU/g of stool) [24]. SCH1211 and HR615 colonized the mice at slightly higher numbers (3.06 x 107 and 3.94 x 107 CFU/g of stool, respectively; the levels of gut colonization were not statistically different among the groups by one-way ANOVA, Tukey’s multiple comparisons test, alpha=0.5) suggesting that Hwp1 was not influential in gut colonization of mice. Hwp1 is also not a factor in colonizing the GI tract of gnotobiotic mice [13].
Figure 5. Hwp1 is required for full virulence in an animal model of gut translocation candidiasis.
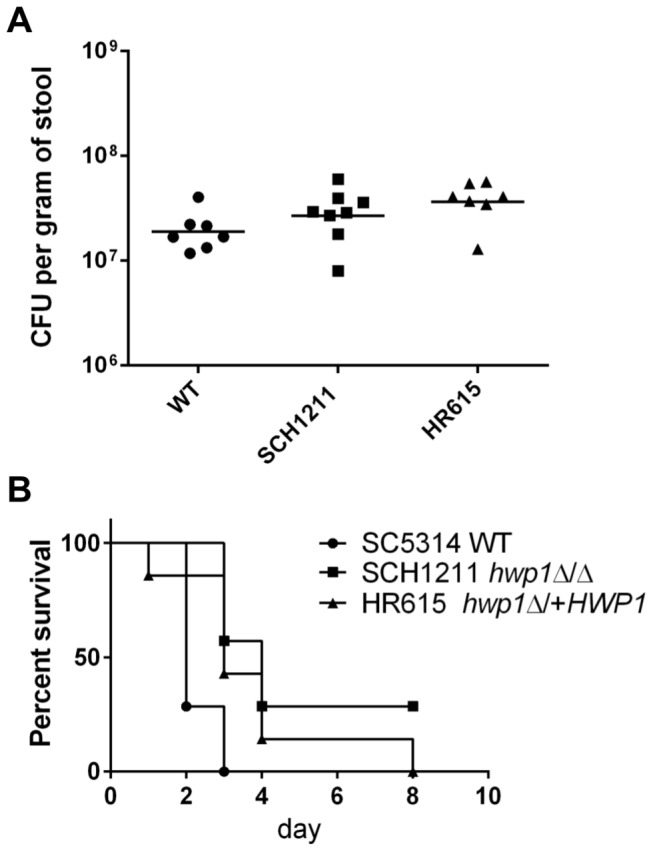
C57BL/6 mice were fed C. albicans in their drinking water to establish GI colonization followed by immunosuppression to induce neutropenia and intestinal mucosa damage. (A) Gut colonization levels are independent upon HWP1 expression. Colonization levels in mice were assessed by measuring the fungal burden in the stools (CFU/g) of individual mice. Mean CFU/g of stool of mice fed SCH1211 (n=8) or HR615 (n=7) were not statistically different relative to SC5314 (WT, n=7). (B) Survival of mice post immunosuppression with cyclophosphamide. Mice colonized with the hwp1 null strain SCH1211 were less virulent relative to SC5314 (P=0.003). Single expression of HWP1 (HR615) did not restore wild type survival kinetics in mice (P=0.035), although none of the mice colonized with HR615 survived to the end of the observation period. C. albicans was recovered from the livers of all the mice at the time of sacrifice (data not shown), indicating translocation from the GI tract.
Gut translocation and dissemination of C. albicans to the liver
The gut-colonized mice were treated with intraperitoneal injections of cyclophosphamide to induce immunosuppression/mucosal damage, and allow translocation of C. albicans into the blood stream and dissemination to the liver. We modified the method of Koh et al. [24] to add an additional cyclophosphamide injection after pilot experiments revealed inconsistent translocation of C. albicans to the liver of mice treated with the published number of cyclophosphamide injections. All of the mice colonized with strains expressing at least one allele of HWP1 (n=14) died by day 8 post-immunosuppression while 2 of the 7 mice colonized with the hwp1Δ/Δ strain survived to the end of the study (Figure 5B). The survival rate between SC5314 and SCH1211-colonized mice was statistically different (P=0.003); however the survival rates of mice harboring SCH1211 or HR615 were not different from each other (P=0.21). C. albicans was recovered from the livers of all the mice at the time of death demonstrating translocation from the GI tract to the organ independent of HWP1 expression. The mice colonized with SCH1211 did not have statistically different median CFU/g of liver compared to mice colonized with SC5314 or HR615 (one-way ANOVA with Tukey’s multiple comparisons test, alpha=0.5; data not shown). These results suggested that wild type hyphal surface levels of Hwp1 were needed for rapid translocation of C. albicans from the mouse gut into the blood stream. However, once translocation had occurred, the lack of Hwp1 did not prevent dissemination to and establishment of C. albicans in the liver.
Discussion
The studies here were initiated to address the role of TG-mediated reactions as a possible mechanism for the requirement of Hwp1 in disseminated candidiasis. We expected to protect mice infected with the hwp1Δ/Δ strain from death in a Tgm2-/- background relative to wild type mice. However, our results discounted a TG-mediated mechanism in the virulence of C. albicans driven by Hwp1. The equal rates of deaths among the wild type C57BL/6 and Tgm2-/- mice, regardless of the infecting strain, supported the null hypothesis. Further, histological analysis of infected kidneys revealed invasion of tissue by the hwp1Δ/Δ strain independent upon Tgm2 expression status. However, the possibility existed that transamidation-unrelated factors confounded the survival data. Tgm2-/- mice are less sensitive to bacterial endotoxin [49], and TG2 is elevated during inflammatory processes [50–53]. A dysregulated immune response may have obscured any benefit in survival in Tgm2-/- mice. The similar rates of death observed in C57BL/6 mice infected with either SC5314 or SCH1211 lessened the association between Hwp1 and virulence (whether the protein participated in transamidation reactions in tissue or not). And although the use of Tgm2-/- mice had limitations, the lack of TG2 activity was not sufficient to improve survival to a significant level even in the absence of Hwp1. Thus, we concluded that host tissue TG2 activity in organs such as the liver and kidneys [16] was dispensable or played a small role in the successful dissemination and tissue invasion by C. albicans. Host transamidation activity did not appear to promote dissemination of C. albicans via reactions involving Hwp1.
The wild type virulence of the hwp1Δ/Δ strains in C57BL/6 mice was an unexpected result of our studies. C57BL/6 and BALB/c are considered to be less susceptible to intravenous C. albicans challenge among mice of different genetic backgrounds tested in this model of candidiasis [54–58], and we observed similar survival kinetics between the mouse strains injected with the wild type SC5314. However, BALB/c and C57BL/6 mice can produce different immunological responses to C. albicans infections and complicate the interpretation of virulence data [59–63]. The susceptibility to C. albicans infection can be difficult to qualify when survival kinetics reveal an attenuated virulence in a given mouse strain. The inability to observe the same attenuated virulence of SCH1211 in C57BL/6 mice may reflect differences of genetic effects between C57BL/6 and BALB/c mice that govern susceptibility to candidiasis. At least two and perhaps more genetic loci are linked to susceptibility to infection with C. albicans [58], and how these factors affect survival kinetics is unknown. This can be a caveat when working with C57BL/6 transgenic knock-out mice which are not historically used in C. albicans virulence studies. If the role of Hwp1 in disseminated candidiasis was weak at best, this effect was masked in the C57BL/6 genetic background.
Consideration of the virulence data independently of mouse strain suggested that Hwp1 was not a major factor in dissemination of C. albicans regardless of Tgm2 expression. The attenuated virulence of the hwp1 null strain observed in BALB/c mice, not corrected by re-introducing a single allele of HWP1, has been noted by others [29], and further supports a diminished role for Hwp1 in disseminated candidiasis. Previous virulence studies utilizing hwp1Δ/Δ strains [1,15] attributed a greater effect to Hwp1 in systemic candidiasis due to the large shift in survival rates and to the survival of a subset of mice to the end of the study. These latter hwp1Δ/Δ strains were created in CAI4 and not corrected for the large deletion that encompasses URA3, a region of IRO1, a putative transcriptional factor, and perhaps the 3’ untranslated region of ORF19.1717 ([20] and www.candidagenome.org). Loss of a functional IRO1 is known to affect the virulence of C. albicans [64]. Full restoration of ORF19.1717-URA3-IRO1 in CAI4 generates an hwp1Δ/Δ strain (DD27-U1) with attenuated virulence BALB/c mice [29], a phenotype that we recapitulated here and observed with our newly created hwp1Δ/Δ strains in SC5314. Together, the data suggested that strains not fully restored at the URA3 locus confused previous results of virulence studies using hwp1Δ/Δ strains and attributed a greater virulence function to Hwp1. URA3/CAI4-related effects on virulence of C. albicans has been addressed in the literature [15,18,20,29,65,66]; however the studies here highlighted once again the importance of selecting the appropriate tools to address virulence questions. Small or nuanced contributions to phenotypes can be missed in vivo by confounding host and fungal factors. The data also highlighted the complex relationship between C. albicans and the mammalian host, likely defined by the host’s niche and genetic factors that modulate infection.
HWP1 is part of a core of 8 genes induced during filamentation of C. albicans that is independent of media [67]. HWP1 together with 3 other filamentation core genes, ALS3, ECE1, and RBT1, code for hyphal wall proteins that function in biofilm formation [21,22,68]. Expression of ALS3, ECE1, HWP1 and RBT1 is induced in fungal cells within biofilms, when they are in contact with plastic and with oral epithelial cells [67]. ALS3 and RBT1 deletion mutants have no role or a reduced effect, respectively, on the virulence of C. albicans in BALB/c mice in a systemic model of candidiasis [69,70]; presently there are no reports describing an ECE1 murine virulence study. Hwp1, Rbt1, Als3p are required for C. albicans mating at wild type levels [21,42], another type of self-interaction in addition that which occurs in “pathogenic” biofilms [71]. The phenotypic parallels between hwp1Δ/Δ als3Δ/Δ and rbt1Δ/Δ strains suggest that these gene products function primarily for self (Hwp1, Als3p and Rbt1p) and in host-fungal interactions on the surface of epithelial or endothelial cells (Hwp1 and/or Als3p, respectively) at host niches (oral and esophageal mucosa) where a biofilm phenotype is the dominant “infectious” morphology. However, the role of these proteins to individually promote candidiasis that involves deep tissue invasion in mice appears to be minimal. Finally, C. albicans clinical isolates expressing an allele of HWP1 (HWP1-2) form poor biofilms in vitro but are nevertheless recovered from the blood stream of patients [72], supporting our conclusions here that Hwp1 is important for biofilms but is not important in promoting systemic candidiasis.
Wild type levels of Hwp1 were needed for the rapid translocation of C. albicans from the gut into the blood stream and infection of murine livers. However, the hwp1 null strain was able to colonize murine guts at wild type levels similarly to that observed for rbt1 deletion mutants [73]. Both of these genes are expressed in C. albicans cells found in the cecum of C57BL/6 mice [73,74] although the cells are predominantly in the yeast morphology. HWP1 expression has also been detected in fungal cells recovered from the oral cavities of asymptomatic individuals [75] presumably growing as a commensal organism (yeasts) of the mouth. It is not clear whether HWP1 expression originates in yeast cells at these anatomical niches or in low numbers of hyphal cells that are part of the commensal population. C. albicans cells unable to form filaments and express HWP1 (cph1Δ/Δ efg1Δ/Δ) [76,77] do not translocate from the mouse gut to the liver as well as SC514; in contrast, cells locked in the filamentous morphology (tup1Δ/Δ) appear more virulent relative to wild type cells in this animal model of candidiasis [24]. These results did not distinguish between the ability of C. albicans to undergo morphological transitions or the expression of adhesins (e.g. Hwp1, Als3p) associated with the hyphal morphology as factors affecting gut translocation. Because hwp1 null strains are not deficient in filamentation ([1] Figure 2 and Figure S3), we were able to consider hyphae formation and HWP1 expression as separate variables in our studies. The results here implied that expression of adhesins and perhaps other proteins associated with the hyphal form are the key variables essential for C. albicans translocation. The mechanism by which Hwp1 aids translocation is not yet understood but we propose two models that are not mutually exclusive: (A) a critical amount of C. albicans self-aggregation is necessary to achieve a threshold fungal burden traversing the damaged intestinal mucosa from the lumen into the blood stream. Dissemination to the liver of the hwp1 null strain was delayed as a consequence of decreased cell numbers trafficking from the GI tract to the blood stream. In support of this hypothesis, the flocculent tup1Δ/Δ cells are more virulent in this gut translocation candidiasis model relative to wild type even when gut colonization levels are two logs below wild type [24]. (B) Alternatively, Hwp1 contributed to interactions with epithelial cells lining the intestinal mucosa and the lack of Hwp1 hampered the initial binding and subsequent translocation of the fungus into the blood stream. Hwp1also participates in adhesion of C. albicans to oral epithelial cells in a non-TG dependent manner [1], therefore it is plausible that the lack of such fungal-host interactions may affect the kinetics of GI translocation.
Expression of a single allele of HWP1 in HR615 restored biofilm formation both in vitro and in vivo but did not fully correct the hwp1 null phenotype in two out of three animal models that involved dissemination via the blood stream. The factors that control wild type dissemination of C. albicans are likely different from those that influence the establishment of superficial biofilms on mucosal surfaces. The results observed with HR615 suggested that Hwp1 may participate in yet undefined functions that aid dissemination via the blood stream that require wild type Hwp1 expression levels.
Taken together, our results here support a new functional model for Hwp1 and similar fungal adhesins: Hwp1 engenders C. albicans with niche-specific properties to form biofilms at oral/mucosal surfaces with localized penetration of the underlying tissue. Hwp1 and other hyphal proteins organize and maintain the three-dimensional structure of the biofilm which is held in place by cross-linking Hwp1 to the surface of oral epithelial cells by the action of host TG. Thus Hwp1 performs the dual function of self-aggregating C. albicans into pathogenic biofilms and adhesion of the biofilm/hyphal cells to the surface of host tissue via TG activity. HWP1 gene expression differs from other niche-specific genes in that HWP1 is normally expressed in hyphal cells regardless of host niche. Many of the currently known differentially-expressed niche-specific genes code for enzymes (e.g. SAPs, phospholipases, and carbon metabolic proteins) [78–87] that enhance the growth fitness of C. albicans as infection takes hold and progresses at given anatomical sites. Hwp1 plays an important niche-defined structural role in C. albicans in vivo colonial morphology that is not specifically associated with fungal fitness or invasive growth advantage. Ultimately, the ability to combine fitness-enhancing niche-specific gene expression with the appropriate infectious colonial morphology adapts C. albicans for survival and growth at ecologically diverse anatomical sites found within its mammalian host.
Supporting Information
DNA primers used for generating the HWP1 disruption and reconstitution cassettes. Oligonucleotides used to generate the hwp1Δ:SAT1 and HWP1:SAT1 gene cassettes for the construction of deletion and reconstitution (put-back) strains in pSFS1, respectively. Underlined nucleotides introduce an ApaI site at the 5’ end of the amplicon. Double underlined nucleotides introduce XhoI sites; nucleotides in bold introduce a SacII site. The nucleotides in small letters indicate the SacI site in the 3’ region downstream of HWP1.
(DOCX)
Construction of the hwp1 deletion and reconstituted strains. (A) Schematic of the HWP1 genomic locus (middle construct) with integration of the HWP1-disruption cassette. NdeI restriction sites are shown in bold above the DNA constructs. The PCR amplicon used to detect HWP1 sequences in Southern blots is labeled “HWP1 probe”. The size marker at top indicates 300 nt. Gene designations: CaFLP, sequences coding for the C. albicans flp recombinase; CaSAT1, sequences coding for C. albicans nourseothricin resistance. The arrow above the inducible SAP2 promoter (SAP2p) indicates the direction of transcription of the CaFLP gene. Dashed green lines indicate the double crossover event leading to integration of the disruption cassette (bottom schematic). (B) Reconstitution of HWP1 expression at its native locus. hwp1Δ::FRT at the HWP1 genomic locus (bottom) with integration of the reconstitution cassette (top). Dashed green lines indicate the sites of the double crossovers restoring expression of HWP1. Size marker, 300 nt. The schematics were generated using Gene Construction Kit (v. 3.5, Texco BioSoftware, Inc.).
(TIF)
Southern blot analysis of the HWP1 deletion and reconstitution strains used in this study. SC5314 and subsequent HWP1 deletion/reconstitution derivatives are shown in order from left to right. The expected HWP1-hybridizing NdeI DNA fragments (arrows at left) are as follows: HWP1, 6.3 Kb, hwp1Δ::SAT1, 3.9 Kb, hwp1Δ::FRT, 4.5 Kb, HWP1::SAT1, 6.3 Kb, and HWP1::FRT, 6.7 Kb . The strains in bold lettering were used in the studies described here.
(TIF)
Expression of Hwp1 on the surface of C. albicans germ tubes. The primary amine, 5-(biotinamido)pentylamine and human recombinant TG2 were used to visualize Hwp1 on the fungal surfaces (see Materials and Methods). The expression level of Hwp1 on the surface of HR615 appeared decreased relative to SC5314 consistent with the introduction of a single copy of HWP1 at its native locus in SCH1211 (see Figure 1, northern analysis of SCH1211 and HR615). Left column, light images; right column, FITC and DAPI (nuclear staining) images combined. Size bar, 20 µm.
(TIF)
Virulence of SCH2283 (hwp1Δ/Δ) in C57BL/6 wild type and Tgm2 knock-out mice. Survival curves of wild type (n=5) or Tgm2-/- (n=7) mice injected with a second HWP1 deletion strain, SCH2283. Survival rates between the animal groups were indistinguishable, P=0.86. The median survival for both groups of mice was 3 days.
(TIF)
Acknowledgments
The authors wish to thank Joachim Morschhäuser and William Fonzi for generously providing plasmids and strains, respectively. We also thank Theodore White for critical reading of the manuscript.
Funding Statement
The study was funded by a grant from the National Institute of Dental & Craniofacial Research, DE021972, to JFS. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Staab JF, Bradway SD, Fidel PL, Sundstrom P (1999) Adhesive and mammalian transglutaminase substrate properties of Candida albicans Hwp1. Science 283: 1535-1538. doi: 10.1126/science.283.5407.1535. PubMed: 10066176. [DOI] [PubMed] [Google Scholar]
- 2. Coleman DA, Oh SH, Zhao X, Zhao H, Hutchins JT et al. (2009) Monoclonal antibodies specific for Candida albicans Als3 that immunolabel fungal cells in vitro and in vivo and block adhesion to host surfaces. J Microbiol Methods 78: 71-78. doi: 10.1016/j.mimet.2009.05.002. PubMed: 19427882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Fanning S, Xu W, Solis N, Woolford CA, Filler SG et al. (2012) Divergent targets of Candida albicans biofilm regulator Bcr1 In vitro and In vivo. Eukaryot Cell 11: 896-904. doi: 10.1128/EC.00103-12. PubMed: 22544909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Fu Y, Rieg G, Fonzi WA, Belanger PH, Edwards JE Jr. et al. (1998) Expression of the Candida albicans gene ALS1 in Saccharomyces cerevisiae induces adherence to endothelial and epithelial cells. Infect Immun 66: 1783-1786. PubMed: 9529114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Oh SH, Cheng G, Nuessen JA, Jajko R, Yeater KM et al. (2005) Functional specificity of Candida albicans Als3p proteins and clade specificity of ALS3 alleles discriminated by the number of copies of the tandem repeat sequence in the central domain. Microbiology 151: 673-681. doi: 10.1099/mic.0.27680-0. PubMed: 15758214. [DOI] [PubMed] [Google Scholar]
- 6. Zhao X, Oh SH, Cheng G, Green CB, Nuessen JA et al. (2004) ALS3 and ALS8 represent a single locus that encodes a Candida albicans adhesin; functional comparisons between Als3p and Als1p. Microbiology 150: 2415-2428. doi: 10.1099/mic.0.26943-0. PubMed: 15256583. [DOI] [PubMed] [Google Scholar]
- 7. Ponniah G, Rollenhagen C, Bahn YS, Staab JF, Sundstrom P (2007) State of differentiation defines buccal epithelial cell affinity for cross-linking to Candida albicans Hwp1. J Oral Pathol Med 36: 456-467. doi: 10.1111/j.1600-0714.2007.00565.x. PubMed: 17686003. [DOI] [PubMed] [Google Scholar]
- 8. Staab JF, Bahn YS, Tai CH, Cook PF, Sundstrom P (2004) Expression of transglutaminase substrate activity on Candida albicans germ tubes through a coiled, disulfide-bonded N-terminal domain of Hwp1 requires C-terminal glycosylphosphatidylinositol modification. J Biol Chem 279: 40737-40747. doi: 10.1074/jbc.M406005200. PubMed: 15262971. [DOI] [PubMed] [Google Scholar]
- 9. Lorand L, Conrad SM (1984) Transglutaminases. Mol Cell Biochem 58: 9-35. doi: 10.1007/BF00240602. PubMed: 6143256. [DOI] [PubMed] [Google Scholar]
- 10. Lorand L, Graham RM (2003) Transglutaminases: crosslinking enzymes with pleiotropic functions. Nat Rev Mol Cell Biol 4: 140-156. doi: 10.1038/nrm1014. PubMed: 12563291. [DOI] [PubMed] [Google Scholar]
- 11. Nemes Z, Steinert PM (1999) Bricks and mortar of the epidermal barrier. Exp Mol Med 31: 5-19. doi: 10.1038/emm.1999.2. PubMed: 10231017. [DOI] [PubMed] [Google Scholar]
- 12. Steinert PM, Chung SI, Kim SY (1996) Inactive zymogen and highly active proteolytically processed membrane-bound forms of the transglutaminase 1 enzyme in human epidermal keratinocytes. Biochem Biophys Res Commun 221: 101-106. doi: 10.1006/bbrc.1996.0552. PubMed: 8660317. [DOI] [PubMed] [Google Scholar]
- 13. Sundstrom P, Balish E, Allen CM (2002) Essential role of the Candida albicans transglutaminase substrate, hyphal wall protein 1, in lethal oroesophageal candidiasis in immunodeficient mice. J Infect Dis 185: 521-530. doi: 10.1086/338836. PubMed: 11865405. [DOI] [PubMed] [Google Scholar]
- 14. Matsuki M, Yamashita F, Ishida-Yamamoto A, Yamada K, Kinoshita C et al. (1998) Defective stratum corneum and early neonatal death in mice lacking the gene for transglutaminase 1 (keratinocyte transglutaminase). Proc Natl Acad Sci U S A 95: 1044-1049. doi: 10.1073/pnas.95.3.1044. PubMed: 9448282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sundstrom P, Cutler JE, Staab JF (2002) Reevaluation of the role of HWP1 in systemic candidiasis by use of Candida albicans strains with selectable marker URA3 targeted to the ENO1 locus. Infect Immun 70: 3281-3283. doi: 10.1128/IAI.70.6.3281-3283.2002. PubMed: 12011025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Itoh M, Kawamoto T, Tatsukawa H, Kojima S, Yamanishi K et al. (2011) In situ detection of active transglutaminases for keratinocyte type (TGase 1) and tissue type (TGase 2) using fluorescence-labeled highly reactive substrate peptides. J Histochem Cytochem Off J Histochem Soc 59: 180-187. doi: 10.1369/jhc.2010.957225. PubMed: 20876521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Reuss O, Vik A, Kolter R, Morschhäuser J (2004) The SAT1 flipper, an optimized tool for gene disruption in Candida albicans . Gene 341: 119-127. doi: 10.1016/j.gene.2004.06.021. PubMed: 15474295. [DOI] [PubMed] [Google Scholar]
- 18. Brand A, MacCallum DM, Brown AJ, Gow NA, Odds FC (2004) Ectopic expression of URA3 can influence the virulence phenotypes and proteome of Candida albicans but can be overcome by targeted reintegration of URA3 at the RPS10 locus. Eukaryot Cell 3: 900-909. doi: 10.1128/EC.3.4.900-909.2004. PubMed: 15302823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Cheng S, Nguyen MH, Zhang Z, Jia H, Handfield M et al. (2003) Evaluation of the roles of four Candida albicans genes in virulence by using gene disruption strains that express URA3 from the native locus. Infect Immun 71: 6101-6103. doi: 10.1128/IAI.71.10.6101-6103.2003. PubMed: 14500538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Staab JF, Sundstrom P (2003) URA3 as a selectable marker for disruption and virulence assessment of Candida albicans genes. Trends Microbiol 11: 69-73. doi: 10.1016/S0966-842X(02)00029-X. PubMed: 12598128. [DOI] [PubMed] [Google Scholar]
- 21. Ene IV, Bennett RJ (2009) Hwp1 and related adhesins contribute to both mating and biofilm formation in Candida albicans . Eukaryot Cell 8: 1909-1913. doi: 10.1128/EC.00245-09. PubMed: 19837954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Nobile CJ, Nett JE, Andes DR, Mitchell AP (2006) Function of Candida albicans adhesin Hwp1 in biofilm formation. Eukaryot Cell 5: 1604-1610. doi: 10.1128/EC.00194-06. PubMed: 17030992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Solis NV, Filler SG (2012) Mouse model of oropharyngeal candidiasis. Nat Protoc 7: 637-642. doi: 10.1038/nprot.2012.011. PubMed: 22402633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Koh AY, Köhler JR, Coggshall KT, Van Rooijen N, Pier GB (2008) Mucosal damage and neutropenia are required for Candida albicans dissemination. PLOS Pathog 4: e35. doi: 10.1371/journal.ppat.0040035. PubMed: 18282097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Maccallum DM (2012) Hosting infection: experimental models to assay Candida virulence. International journal of microbiology 2012: 363764. PubMed: 22235206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Spellberg B (2008) Novel insights into disseminated candidiasis: pathogenesis research and clinical experience converge. PLOS Pathog 4: e38. doi: 10.1371/journal.ppat.0040038. PubMed: 18282100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Fonzi WA, Irwin MY (1993) Isogenic strain construction and gene mapping in Candida albicans . Genetics 134: 717-728. PubMed: 8349105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Gillum AM, Tsay EY, Kirsch DR (1984) Isolation of the Candida albicans gene for orotidine-5'-phosphate decarboxylase by complementation of S. cerevisiae ura3 and E. coli pyrF mutations. Molecular General Genet MGG 198: 179-182. doi: 10.1007/BF00328721. [DOI] [PubMed] [Google Scholar]
- 29. Sharkey LL, Liao WL, Ghosh AK, Fonzi WA (2005) Flanking direct repeats of hisG alter URA3 marker expression at the HWP1 locus of Candida albicans . Microbiology 151: 1061-1071. doi: 10.1099/mic.0.27487-0. PubMed: 15817775. [DOI] [PubMed] [Google Scholar]
- 30. Rose MD, Winston F, Hieter P (1990) Method in yeast genetics: a laboratory course manual. Cold Spring Harbor: Cold Spring Harbor Laboratory Publishing. [Google Scholar]
- 31. Morschhäuser J, Michel S, Staib P (1999) Sequential gene disruption in Candida albicans by FLP-mediated site-specific recombination. Mol Microbiol 32: 547-556. doi: 10.1046/j.1365-2958.1999.01393.x. PubMed: 10320577. [DOI] [PubMed] [Google Scholar]
- 32. Staab JF, Sundstrom P (1998) Genetic organization and sequence analysis of the hypha-specific cell wall protein gene HWP1 of Candida albicans . Yeast 14: 681-686. doi: 10.1002/(SICI)1097-0061(199805)14:7. PubMed: 9639315. [DOI] [PubMed] [Google Scholar]
- 33. Schmitt ME, Brown TA, Trumpower BL (1990) A rapid and simple method for preparation of RNA from Saccharomyces cerevisiae . Nucleic Acids Res 18: 3091-3092. doi: 10.1093/nar/18.10.3091. PubMed: 2190191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. De Laurenzi V, Melino G (2001) Gene disruption of tissue transglutaminase. Mol Cell Biol 21: 148-155. doi: 10.1128/MCB.21.1.148-155.2001. PubMed: 11113189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. MacCallum DM, Odds FC (2005) Temporal events in the intravenous challenge model for experimental Candida albicans infections in female mice. Mycoses 48: 151-161. doi: 10.1111/j.1439-0507.2005.01121.x. PubMed: 15842329. [DOI] [PubMed] [Google Scholar]
- 36. Wilson RB, Davis D, Mitchell AP (1999) Rapid hypothesis testing with Candida albicans through gene disruption with short homology regions. J Bacteriol 181: 1868-1874. PubMed: 10074081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ahmad A, Kabir MA, Kravets A, Andaluz E, Larriba G et al. (2008) Chromosome instability and unusual features of some widely used strains of Candida albicans . Yeast 25: 433-448. doi: 10.1002/yea.1597. PubMed: 18509849. [DOI] [PubMed] [Google Scholar]
- 38. Arbour M, Epp E, Hogues H, Sellam A, Lacroix C et al. (2009) Widespread occurrence of chromosomal aneuploidy following the routine production of Candida albicans mutants. FEMS Yeast Res 9: 1070-1077. doi: 10.1111/j.1567-1364.2009.00563.x. PubMed: 19732157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Selmecki A, Bergmann S, Berman J (2005) Comparative genome hybridization reveals widespread aneuploidy in Candida albicans laboratory strains. Mol Microbiol 55: 1553-1565. doi: 10.1111/j.1365-2958.2005.04492.x. PubMed: 15720560. [DOI] [PubMed] [Google Scholar]
- 40. Chen X, Magee BB, Dawson D, Magee PT, Kumamoto CA (2004) Chromosome 1 trisomy compromises the virulence of Candida albicans . Mol Microbiol 51: 551-565. doi: 10.1046/j.1365-2958.2003.03852.x. PubMed: 14756793. [DOI] [PubMed] [Google Scholar]
- 41. Ganguly S, Mitchell AP (2011) Mucosal biofilms of Candida albicans . Curr Opin Microbiol 14: 380-385. doi: 10.1016/j.mib.2011.06.001. PubMed: 21741878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Nobile CJ, Schneider HA, Nett JE, Sheppard DC, Filler SG et al. (2008) Complementary adhesin function in C. albicans biofilm formation. Curr Biol CB 18: 1017-1024. doi: 10.1016/j.cub.2008.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ramage G, Saville SP, Wickes BL, López-Ribot JL (2002) Inhibition of Candida albicans biofilm formation by farnesol, a quorum-sensing molecule. Appl Environ Microbiol 68: 5459-5463. doi: 10.1128/AEM.68.11.5459-5463.2002. PubMed: 12406738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kamai Y, Kubota M, Hosokawa T, Fukuoka T, Filler SG (2001) New model of oropharyngeal candidiasis in mice. Antimicrob Agents Chemother 45: 3195-3197. doi: 10.1128/AAC.45.11.3195-3197.2001. PubMed: 11600377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Naglik JR, Fidel PL Jr., Odds FC (2008) Animal models of mucosal Candida infection. FEMS Microbiol Lett 283: 129-139. doi: 10.1111/j.1574-6968.2008.01160.x. PubMed: 18422625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Costa AC, Pereira CA, Junqueira JC, Jorge AO (2013) Recent mouse and rat methods for the study of experimental oral candidiasis. Virulence 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Balish E (2009) A URA3 null mutant of Candida albicans (CAI-4) causes oro-oesophageal and gastric candidiasis and is lethal for gnotobiotic, transgenic mice (Tgepsilon26) that are deficient in both natural killer and T cells. J Med Microbiol 58: 290-295. doi: 10.1099/jmm.0.004846-0. PubMed: 19208876. [DOI] [PubMed] [Google Scholar]
- 48. Mattiuzzi G, Ostrosky-Zeichner L (2010) Fungal and Parasitic Infections. In: Kleinberg M. Comtemporay Hematology: Managing infections in patients with hematological malignancies. New York: Humana Press. [Google Scholar]
- 49. Falasca L, Farrace MG, Rinaldi A, Tuosto L, Melino G et al. (2008) Transglutaminase type II is involved in the pathogenesis of endotoxic shock. J Immunol 180: 2616-2624. PubMed: 18250473. [DOI] [PubMed] [Google Scholar]
- 50. Falasca L, Iadevaia V, Ciccosanti F, Melino G, Serafino A et al. (2005) Transglutaminase type II is a key element in the regulation of the anti-inflammatory response elicited by apoptotic cell engulfment. J Immunol 174: 7330-7340. PubMed: 15905580. [DOI] [PubMed] [Google Scholar]
- 51. Kim SY (2006) Transglutaminase 2 in inflammation. Front Biosci J Virtual Libr 11: 3026-3035. doi: 10.2741/2030. [DOI] [PubMed] [Google Scholar]
- 52. Mohan K, Pinto D, Issekutz TB (2003) Identification of tissue transglutaminase as a novel molecule involved in human CD8+ T cell transendothelial migration. J Immunol 171: 3179-3186. PubMed: 12960346. [DOI] [PubMed] [Google Scholar]
- 53. Park KS, Kim DS, Ko C, Lee SJ, Oh SH et al. (2011) TNF-alpha mediated NF-kappaB activation is constantly extended by transglutaminase 2. Front Biosci 3: 341-354. doi: 10.2741/s155. PubMed: 21196314. [DOI] [PubMed] [Google Scholar]
- 54. Romani L (2001) Animal models for candidiasis. Current Protocols in Immunology. John Wiley & Sons, Inc. p. 19.16.11-19.16.16. [DOI] [PubMed] [Google Scholar]
- 55. Hector RF, Domer JE, Carrow EW (1982) Immune responses to Candida albicans in genetically distinct mice. Infect Immun 38: 1020-1028. PubMed: 6759403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Ashman RB, Fulurija A, Papadimitriou JM (1996) Strain-dependent differences in host response to Candida albicans infection in mice are related to organ susceptibility and infectious load. Infect Immun 64: 1866-1869. PubMed: 8613406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Ashman RB, Papadimitriou JM (1987) Murine candidiasis. Pathogenesis and host responses in genetically distinct inbred mice. Immunol Cell Biol 65 ( 2): 163-171. doi: 10.1038/icb.1987.18. [DOI] [PubMed] [Google Scholar]
- 58. Radovanovic I, Mullick A, Gros P (2011) Genetic control of susceptibility to infection with Candida albicans in mice. PLOS ONE 6: e18957. doi: 10.1371/journal.pone.0018957. PubMed: 21533108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Carvalho A, Giovannini G, De Luca A, D'Angelo C, Casagrande A et al. (2012) Dectin-1 isoforms contribute to distinct Th1/Th17 cell activation in mucosal candidiasis. Cell Mol Immunol 9: 276-286. doi: 10.1038/cmi.2012.1. PubMed: 22543832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Mosci P, Pietrella D, Ricci G, Pandey N, Monari C et al. (2013) Mouse strain-dependent differences in estrogen sensitivity during vaginal candidiasis. Mycopathologia 175: 1-11. doi: 10.1007/s11046-012-9589-9. PubMed: 23054331. [DOI] [PubMed] [Google Scholar]
- 61. Schofield DA, Westwater C, Balish E (2005) Divergent chemokine, cytokine and beta-defensin responses to gastric candidiasis in immunocompetent C57BL/6 and BALB/c mice. J Med Microbiol 54: 87-92. doi: 10.1099/jmm.0.45755-0. PubMed: 15591261. [DOI] [PubMed] [Google Scholar]
- 62. Wellington M, Dolan K, Krysan DJ (2009) Live Candida albicans suppresses production of reactive oxygen species in phagocytes. Infect Immun 77: 405-413. doi: 10.1128/IAI.00860-08. PubMed: 18981256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Zou Y, Zhang H, Li H, Chen H, Song W et al. (2012) Strain-dependent production of interleukin-17/interferon-gamma and matrix remodeling-associated genes in experimental Candida albicans keratitis. Mol Vis 18: 1215-1225. PubMed: 22665968. [PMC free article] [PubMed] [Google Scholar]
- 64. Chibana H, Uno J, Cho T, Mikami Y (2005) Mutation in IRO1 tightly linked with URA3 gene reduces virulence of Candida albicans . Microbiol Immunol 49: 937-939. PubMed: 16237272. [DOI] [PubMed] [Google Scholar]
- 65. Lay J, Henry LK, Clifford J, Koltin Y, Bulawa CE et al. (1998) Altered expression of selectable marker URA3 in gene-disrupted Candida albicans strains complicates interpretation of virulence studies. Infect Immun 66: 5301-5306. PubMed: 9784536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Correia A, Lermann U, Teixeira L, Cerca F, Botelho S et al. (2010) Limited role of secreted aspartyl proteinases Sap1 to Sap6 in Candida albicans virulence and host immune response in murine hematogenously disseminated candidiasis. Infect Immun 78: 4839-4849. doi: 10.1128/IAI.00248-10. PubMed: 20679440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Martin R, Albrecht-Eckardt D, Brunke S, Hube B, Hünniger K et al. (2013) A core filamentation response network in Candida albicans is restricted to eight genes. PLOS ONE 8: e58613. doi: 10.1371/journal.pone.0058613. PubMed: 23516516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Nobile CJ, Andes DR, Nett JE, Smith FJ, Yue F et al. (2006) Critical role of Bcr1-dependent adhesins in C. albicans biofilm formation in vitro and in vivo. PLOS Pathog 2: e63. doi: 10.1371/journal.ppat.0020063. PubMed: 16839200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Braun BR, Head WS, Wang MX, Johnson AD (2000) Identification and characterization of TUP1-regulated genes in Candida albicans . Genetics 156: 31-44. PubMed: 10978273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Cleary IA, Reinhard SM, Miller CL, Murdoch C, Thornhill MH et al. (2011) Candida albicans adhesin Als3p is dispensable for virulence in the mouse model of disseminated candidiasis. Microbiology 157: 1806-1815. doi: 10.1099/mic.0.046326-0. PubMed: 21436220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Park YN, Daniels KJ, Pujol C, Srikantha T, Soll DR (2013) Candida albicans forms a specialized "sexual" as well As "pathogenic" biofilm. Eukaryot Cell, 12: 1120–31. PubMed: 23771904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Padovan AC, Chaves GM, Colombo AL, Briones MR (2009) A novel allele of HWP1, isolated from a clinical strain of Candida albicans with defective hyphal growth and biofilm formation, has deletions of Gln/Pro and Ser/Thr repeats involved in cellular adhesion. Med Mycol Off Publ International Society For Human Animal Mycology 47: 824-835. doi: 10.3109/13693780802669574. PubMed: 19184714. [DOI] [PubMed] [Google Scholar]
- 73. White SJ, Rosenbach A, Lephart P, Nguyen D, Benjamin A et al. (2007) Self-regulation of Candida albicans population size during GI colonization. PLOS Pathog 3: e184. doi: 10.1371/journal.ppat.0030184. PubMed: 18069889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Rosenbach A, Dignard D, Pierce JV, Whiteway M, Kumamoto CA (2010) Adaptations of Candida albicans for growth in the mammalian intestinal tract. Eukaryot Cell 9: 1075-1086. doi: 10.1128/EC.00034-10. PubMed: 20435697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Naglik JR, Fostira F, Ruprai J, Staab JF, Challacombe SJ et al. (2006) Candida albicans HWP1 gene expression and host antibody responses in colonization and disease. J Med Microbiol 55: 1323-1327. doi: 10.1099/jmm.0.46737-0. PubMed: 17005778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Braun BR, Johnson AD (2000) TUP1, CPH1 and EFG1 make independent contributions to filamentation in Candida albicans . Genetics 155: 57-67. PubMed: 10790384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Lane S, Birse C, Zhou S, Matson R, Liu H (2001) DNA array studies demonstrate convergent regulation of virulence factors by Cph1, Cph2, and Efg1 in Candida albicans . J Biol Chem 276: 48988-48996. doi: 10.1074/jbc.M104484200. PubMed: 11595734. [DOI] [PubMed] [Google Scholar]
- 78. Hube B, Hess D, Baker CA, Schaller M, Schäfer W et al. (2001) The role and relevance of phospholipase D1 during growth and dimorphism of Candida albicans. Microbiology 147: 879-889. PubMed: 11283284. [DOI] [PubMed] [Google Scholar]
- 79. Jackson BE, Wilhelmus KR, Hube B (2007) The role of secreted aspartyl proteinases in Candida albicans keratitis. Invest Ophthalmol Vis Sci 48: 3559-3565. doi: 10.1167/iovs.07-0114. PubMed: 17652724. [DOI] [PubMed] [Google Scholar]
- 80. Lorenz MC, Fink GR (2001) The glyoxylate cycle is required for fungal virulence. Nature 412: 83-86. doi: 10.1038/35083594. PubMed: 11452311. [DOI] [PubMed] [Google Scholar]
- 81. Naglik JR, Moyes D, Makwana J, Kanzaria P, Tsichlaki E et al. (2008) Quantitative expression of the Candida albicans secreted aspartyl proteinase gene family in human oral and vaginal candidiasis. Microbiology 154: 3266-3280. doi: 10.1099/mic.0.2008/022293-0. PubMed: 18957581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Naglik JR, Rodgers CA, Shirlaw PJ, Dobbie JL, Fernandes-Naglik LL et al. (2003) Differential expression of Candida albicans secreted aspartyl proteinase and phospholipase B genes in humans correlates with active oral and vaginal infections. J Infect Dis 188: 469-479. doi: 10.1086/376536. PubMed: 12870130. [DOI] [PubMed] [Google Scholar]
- 83. Samaranayake YH, Dassanayake RS, Cheung BP, Jayatilake JA, Yeung KW et al. (2006) Differential phospholipase gene expression by Candida albicans in artificial media and cultured human oral epithelium. APMIS Acta Pathol Microbiol Immunol Scand 114: 857-866. PubMed: 17207086. [DOI] [PubMed] [Google Scholar]
- 84. Schaller M, Schackert C, Korting HC, Januschke E, Hube B (2000) Invasion of Candida albicans correlates with expression of secreted aspartic proteinases during experimental infection of human epidermis. J Invest Dermatol 114: 712-717. doi: 10.1046/j.1523-1747.2000.00935.x. PubMed: 10733678. [DOI] [PubMed] [Google Scholar]
- 85. Barelle CJ, Priest CL, Maccallum DM, Gow NA, Odds FC et al. (2006) Niche-specific regulation of central metabolic pathways in a fungal pathogen. Cell Microbiol 8: 961-971. doi: 10.1111/j.1462-5822.2005.00676.x. PubMed: 16681837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Staib P, Kretschmar M, Nichterlein T, Hof H, Morschhäuser J (2000) Differential activation of a Candida albicans virulence gene family during infection. Proc Natl Acad Sci U S A 97: 6102-6107. doi: 10.1073/pnas.110031497. PubMed: 10811913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Schofield DA, Westwater C, Warner T, Nicholas PJ, Paulling EE et al. (2003) Hydrolytic gene expression during oroesophageal and gastric candidiasis in immunocompetent and immunodeficient gnotobiotic mice. J Infect Dis 188: 591-599. doi: 10.1086/377182. PubMed: 12898449. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
DNA primers used for generating the HWP1 disruption and reconstitution cassettes. Oligonucleotides used to generate the hwp1Δ:SAT1 and HWP1:SAT1 gene cassettes for the construction of deletion and reconstitution (put-back) strains in pSFS1, respectively. Underlined nucleotides introduce an ApaI site at the 5’ end of the amplicon. Double underlined nucleotides introduce XhoI sites; nucleotides in bold introduce a SacII site. The nucleotides in small letters indicate the SacI site in the 3’ region downstream of HWP1.
(DOCX)
Construction of the hwp1 deletion and reconstituted strains. (A) Schematic of the HWP1 genomic locus (middle construct) with integration of the HWP1-disruption cassette. NdeI restriction sites are shown in bold above the DNA constructs. The PCR amplicon used to detect HWP1 sequences in Southern blots is labeled “HWP1 probe”. The size marker at top indicates 300 nt. Gene designations: CaFLP, sequences coding for the C. albicans flp recombinase; CaSAT1, sequences coding for C. albicans nourseothricin resistance. The arrow above the inducible SAP2 promoter (SAP2p) indicates the direction of transcription of the CaFLP gene. Dashed green lines indicate the double crossover event leading to integration of the disruption cassette (bottom schematic). (B) Reconstitution of HWP1 expression at its native locus. hwp1Δ::FRT at the HWP1 genomic locus (bottom) with integration of the reconstitution cassette (top). Dashed green lines indicate the sites of the double crossovers restoring expression of HWP1. Size marker, 300 nt. The schematics were generated using Gene Construction Kit (v. 3.5, Texco BioSoftware, Inc.).
(TIF)
Southern blot analysis of the HWP1 deletion and reconstitution strains used in this study. SC5314 and subsequent HWP1 deletion/reconstitution derivatives are shown in order from left to right. The expected HWP1-hybridizing NdeI DNA fragments (arrows at left) are as follows: HWP1, 6.3 Kb, hwp1Δ::SAT1, 3.9 Kb, hwp1Δ::FRT, 4.5 Kb, HWP1::SAT1, 6.3 Kb, and HWP1::FRT, 6.7 Kb . The strains in bold lettering were used in the studies described here.
(TIF)
Expression of Hwp1 on the surface of C. albicans germ tubes. The primary amine, 5-(biotinamido)pentylamine and human recombinant TG2 were used to visualize Hwp1 on the fungal surfaces (see Materials and Methods). The expression level of Hwp1 on the surface of HR615 appeared decreased relative to SC5314 consistent with the introduction of a single copy of HWP1 at its native locus in SCH1211 (see Figure 1, northern analysis of SCH1211 and HR615). Left column, light images; right column, FITC and DAPI (nuclear staining) images combined. Size bar, 20 µm.
(TIF)
Virulence of SCH2283 (hwp1Δ/Δ) in C57BL/6 wild type and Tgm2 knock-out mice. Survival curves of wild type (n=5) or Tgm2-/- (n=7) mice injected with a second HWP1 deletion strain, SCH2283. Survival rates between the animal groups were indistinguishable, P=0.86. The median survival for both groups of mice was 3 days.
(TIF)


