Abstract
Significance: Regulation of the activity of the chloroplast ATP synthase is largely accomplished by the chloroplast thioredoxin system, the main redox regulation system in chloroplasts, which is directly coupled to the photosynthetic reaction. We review the current understanding of the redox regulation system of the chloroplast ATP synthase. Recent Advances: The thioredoxin-targeted portion of the ATP synthase consists of two cysteines located on the central axis subunit γ. The redox state of these two cysteines is under the influence of chloroplast thioredoxin, which directly controls rotation during catalysis by inducing a conformational change in this subunit. The molecular mechanism of redox regulation of the chloroplast ATP synthase has recently been determined. Critical Issues: Regulation of the activity of the chloroplast ATP synthase is critical in driving efficiency into the ATP synthesis reaction in chloroplasts. Future Directions: The molecular architecture of the chloroplast ATP synthase, which confers redox regulatory properties requires further investigation, in light of the molecular structure of the enzyme complex as well as the physiological significance of the regulation system. Antioxid. Redox Signal. 19, 1846–1854.
Introduction
The FoF1-ATP synthase (FoF1) is ubiquitously found in energy transducing membranes such as chloroplast and cyanobacterial thylakoid membranes, mitochondrial inner membranes, and bacterial plasma membranes. This enzyme catalyzes the synthesis of ATP from ADP and inorganic phosphate using the electrochemical proton gradient formed across these membranes by photosynthetic or respiratory electron transfer reaction (7, 79). FoF1, a critical enzyme for energy conversion in the cell, consists of a membrane-peripheral component F1, which in turn is formed of five different subunits with a stoichiometry of α3β3γ1δ1ɛ1 (80), and a membrane-embedded part Fo, which consists of three different subunits with stoichiometry of a1b2c10–15 (46, 51, 62). The soluble F1 possesses ATP hydrolysis activity and is thus called F1-ATPase. The minimum catalytic core of F1 maintaining ATP hydrolysis activity is the α3β3γ complex (40), in which the F1 catalytic sites are predominantly located on three β subunits. Based on the characterization of the catalytic cooperativity observed within these three catalytic sites, Boyer and his coworker proposed the rotational catalysis model (20), in which relative rotational movement of α3β3 core against the γ subunit occurs during catalytic reaction, which was initially viewed with some skepticism. In 1994, however, determination of the crystal structure of the mitochondrial α3β3γ complex at 2.8 Å resolution (1) revealed an alternating hexagonal arrangement of three α and three β subunits around an α-helical domain containing the N- and C-terminal regions of the γ subunit, which occurs as a central axis in the complex. Since three β subunits in the structure showed different conformations due to different nucleotide binding situations at three catalytic sites, the reported structure strongly supported the idea of rotation of the γ subunit against the α3β3 ring during catalysis. Since then, further experiments have allowed confirmation of the rotation of the γ subunit in the α3β3 core (13, 55). Finally, direct visualization of the counter-clockwise continuous rotation of the γ subunit coupled with ATP hydrolysis reaction was observed under an optical microscope by attaching a fluorescent-labeled actin filament to the γ subunit of the α3β3γ complex immobilized on a glass surface (48) (Fig. 1). Rotation analyses of this enzyme were then extensively studied, and the discrete 120° step rotation of γ per single molecule of ATP consumption, and 80° and 40° substeps within this 120° step, determined (78). In addition, further analyses revealed that ADP release occurs at the 240° position of γ rotated from the ATP binding position (2, 4). ATP synthase has since been acknowledged as a molecular motor enzyme in the cell.
FIG. 1.
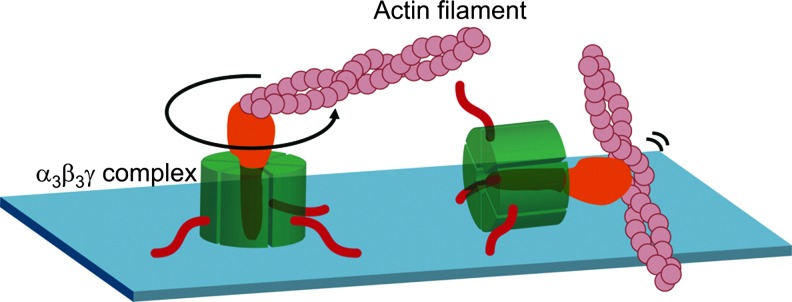
Direct visualization of rotation of the γ subunit of F1. A fluorescein-labeled actin filament was attached on the γ subunit of F1 molecule by using biotin and streptavidin interaction. For this purpose, two cysteines were introduced into the γ subunit and biotinylated. Red lines are His-tags used to immobilize the protein on the glass surface. Counter-clockwise rotation of the filament in the presence of MgATP was observed under the fluorescent microscope. F1, membrane-peripheral part of ATP synthase.
Regulation of ATP Synthase Activity
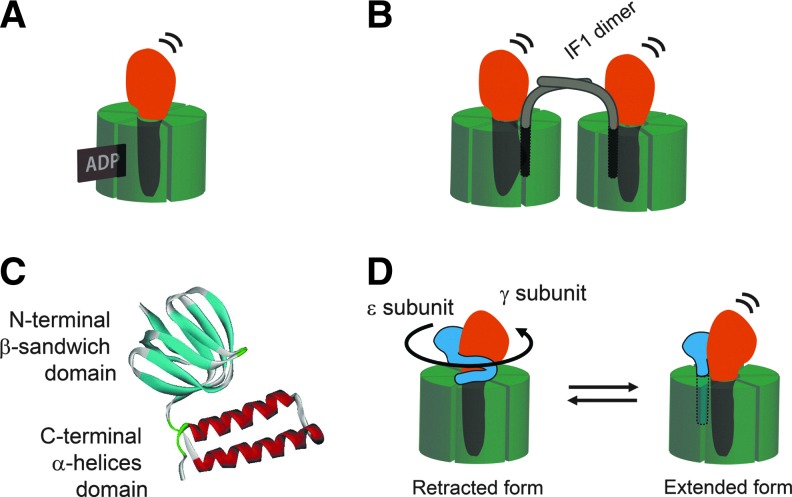
Since ATP synthase can hydrolyze ATP when the electrochemical proton gradient across the membrane is not available for ATP synthesis, prevention of this wasteful reverse reaction seems to be critical for this enzyme complex, and multiple regulatory mechanisms exist for this purpose, irrespective of the origin of the enzyme. The most common regulatory mechanism for ATP synthase is known as ADP inhibition. Once MgADP has been produced by the hydrolysis reaction, it becomes tightly bound to the catalytic site, strongly preventing subsequent ATP hydrolysis, but not ATP synthesis (14, 45, 68) (Fig. 2A). To recover from this ADP inhibition, ATP binding to the noncatalytic sites on the α subunits is required (39). Some detergents, such as lauryldimethylamine oxide (15) and octylglucoside (50), and anions (11, 69), efficiently recover the enzyme from this inhibition. Based on the single-molecule analysis of the catalytic turnover of thermophilic F1-ATPase using rotation assay, Hirono-Hara et al. clearly indicated that ADP inhibition can be observed as frequent long pauses at 80° within the 120° catalytic step during rotation (21).
FIG. 2.
Regulatory mechanism for ATP synthase. (A) Tightly bound ADP inhibits ATP hydrolysis activity. (B) IF1 is the intrinsic ATPase inhibitor for MF1. (C) The ɛ subunit is composed of two domains: the N-terminal β-sandwich domain and C-terminal α-helices domain. The latter domain is structurally variable. The NMR structure of the ɛ subunit from Thermosynechococcus elongatus BP-1 (PDB:2RQ6) is shown. For more detail, see Ref. (67). (D) The extended conformation of the ɛ subunit inhibits the ATPase. IF1, intrinsic ATPase inhibitor protein; MF1, F1 part of mitochondrial ATP synthase.
ATP synthase is a multi-subunit complex and several accessory subunits participate in the regulatory function. Recently, dimerization of FoF1 molecule on the mitochondrial membranes has been reported to function as an inhibitory regulation mechanism. In the case of the mitochondrial FoF1, acidic pH-dependent dimerization of IF1 (9) and its binding to the F1-β subunit is known to achieve dimerization of FoF1 on the membrane, which inhibits ATP hydrolysis activity (17) (Fig. 2B). In contrast, the ɛ subunit works as an intrinsic inhibitor of ATP hydrolysis activity in bacterial and chloroplast ATP synthase. Based on the structural analysis of the ɛ subunit, the C-terminal helix-turn-helix domain undergoes a large conformational change from a retracted to an extended form (54, 77) (Fig. 2C, D). The conformational change of this ɛ subunit must be induced by a change in certain physiological conditions, such as ATP concentrations (28, 77) for bacterial ATP synthase, and membrane potential for chloroplast ATP synthase (24). The conformational change of the ɛ subunit in the C-terminal helices is linked to the inhibitory function of this subunit (27, 66). The ɛ subunit of the chloroplast ATP synthase is regarded as a stronger inhibitor for ATP hydrolysis activity than the bacterial one (53, 70). Single-molecule analysis of the function of the ɛ subunit of cyanobacterial F1during rotation revealed that the ɛ subunit completely stops the rotation of the γ subunit of cyanobacterial F1 (32), whereas the bacterial ɛ subunit decreases the average rotation speed in F1-ATPase by way of increasing pause duration (59). In addition, based on rigorous rotation analysis on the difference between ADP inhibition and ɛ inhibition, we found that inhibition of the latter is mechanically more stable (31). Hence, inhibition of the ATP hydrolysis activity by the ɛ subunit is thought to be the regulatory mechanism, which also prevents futile ATP hydrolysis. These various shut-down mechanisms, which prevent ATP hydrolysis activity must work in a complementary fashion.
Chloroplast ATP Synthase Is a Thiol Enzyme, Which Is Subject To Redox Regulation by Thioredoxin
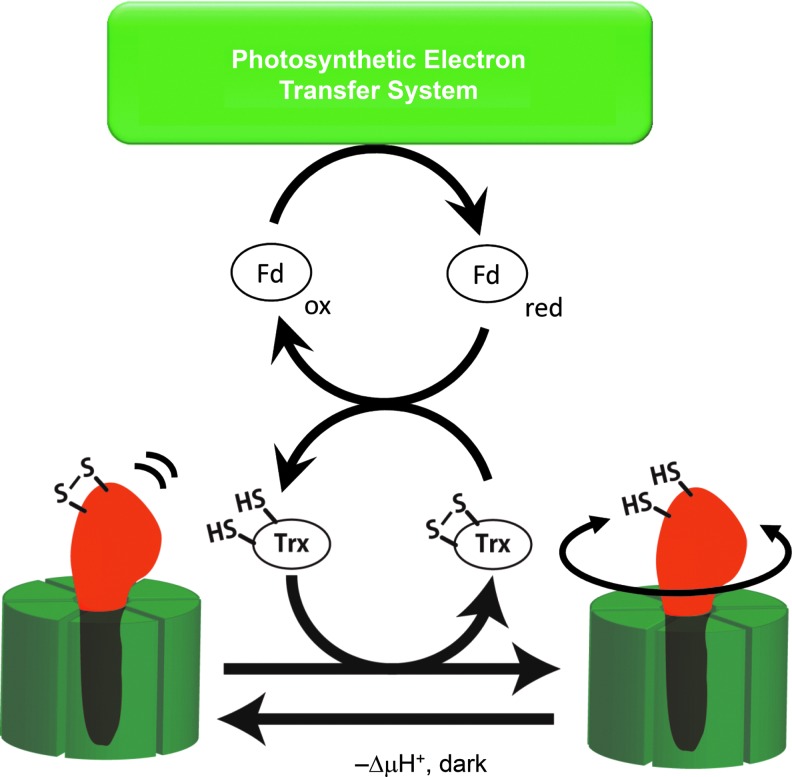
Chloroplast ATP synthase (CFoCF1) is classed as a thiol enzyme; a disulfide bond located on the γ subunit is reduced by reducing equivalents supplied by thioredoxin (Trx), which is, in turn, reduced by the photosynthetic electron transfer reaction (44) (Fig. 3). Trx was first identified in Escherichia coli as a ribonucleotide reductase cofactor and supplier of reducing equivalents (37). The significance of Trx in chloroplasts as a mediator of reducing equivalents was first characterized by the Buchanan's group (8, 73), leading to the establishment of the idea of thiol modulation of chloroplast enzymes (57, 73). In the case of the catalytic moiety of the chloroplast ATP synthase, CF1, activation of the ATP hydrolysis activity of the isolated enzyme was first demonstrated by chemical reduction (41). McKinney et al. then reported that the reduced form Trx can activate the isolated CF1 ATPase (42). Hereafter, critical cysteines (Cys199 and Cys205) for this thiol modulation were determined as part of four cysteines on the γ subunit of spinach CF1 (43, 47). Interestingly, the region containing the two regulatory cysteines, the so-called inserted sequence, is comprised of a total of about 40 amino acid residues, and is only present in the γ subunit of chloroplast ATP synthases obtained from higher plants and green algae (23). In contrast, the γ subunit of cyanobacteria includes the insertion sequence consisting of 30 amino acids, although the region lacks 9 amino acids, including two regulatory cysteines (Fig. 4) (71).
FIG. 3.
Trx-dependent activation of CF1-ATPase. The reduced Trx reduces CF1- γ subunit. Under suboptimal conditions for ATP synthesis, the γ subunit is immediately oxidized to prevent ATP hydrolysis activity. CF1, F1 part of CFoCF1; Trx, thioredoxin.
FIG. 4.
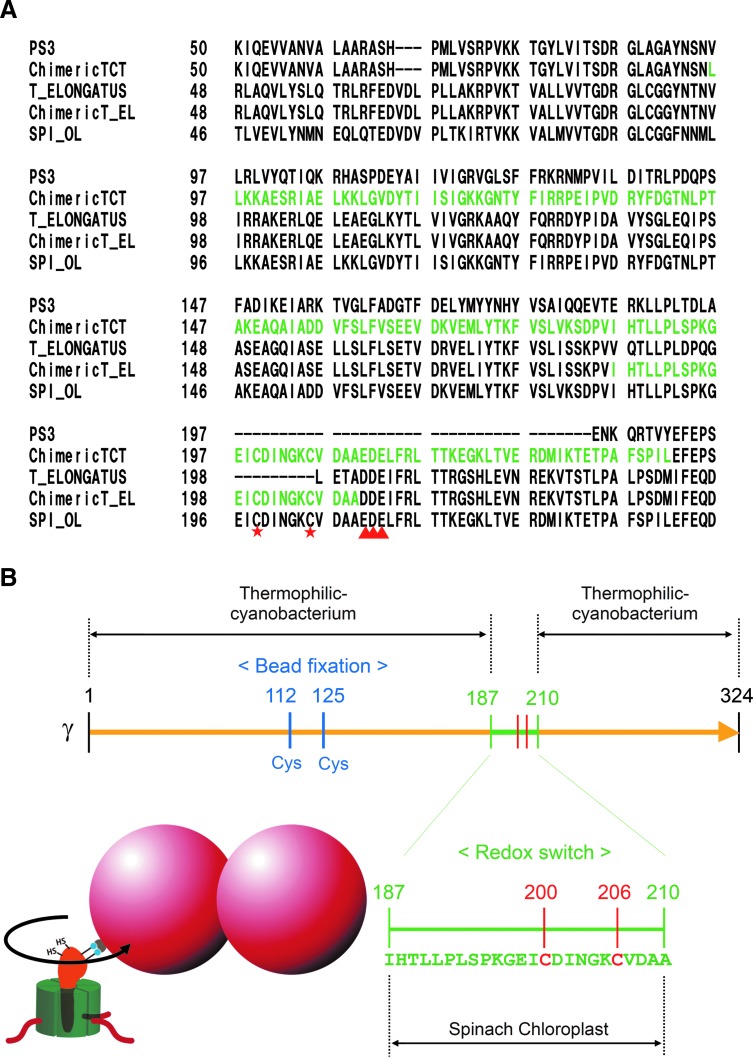
Alignment of the γ subunit of ATP synthase from various organisms. The N-terminal α-helix is shaded in yellow, and the C-terminal α-helix in orange. The insertion region observed only in photosynthetic organisms is shaded in green. The colorless region is mainly the globular domain. Two cysteines for redox regulation are labeled with red stars. E_COLI, Eschericia coli; PS3, thermophilic Bacillus PS3; BOVINE_MT, mitochondria from Bos taurus; SYN6803, cyanobacteria Synechocystis sp. PCC6803; ANA7120, cyanobacteria Anabaena sp. PCC7120; T_ELONGATUS, cyanobacteria T. elongatus; ARA_ATPC1, Arabidopsis thaliana chloroplast type 1; ARA_ATPC2, A. thaliana chloroplast type 2; SPI_OL, chloroplast form Spinacia oleracea.
In higher plant chloroplasts, four Trx isoforms, Trx-f, Trx-m, Trx-x, and Trx-y, have been identified to date [see reviews such as (38, 57)]. Among these, Trx-f and Trx-m were found before the genome project of Arabidopsis thaliana (3), and have been well studied (61, 74). For activation/reduction of CF1, Trx-f rather than Trx-m is regarded to act as the physiological reductant (58), based on kinetic analysis of thiol modulation of CFoCF1 in the thylakoid membranes. The efficiency and rate of the dithiol–disulfide exchange reaction within the reduced form Trx and an oxidized form target protein must be defined by the protein–protein interaction between Trx isoforms and target proteins (18). Several critical amino acid residues have been identified on the Trx molecule, which are important for interaction with the target proteins. However, no crystal structures of cocrystals of chloroplast Trx and target protein have been obtained to date; thus, the critical interacting amino acid residues remain to be identified. Interestingly, the mutant Trx, which lacks two critical cysteines can assist the reduction of the disulfide bond on the γ subunit in the presence of high concentrations of DTT. We therefore concluded that Trx displays an affinity to this region and induces the conformational change of a certain region on the γ subunit, which may assist the reduction of the disulfide bond, although the interaction between Trx and the regulatory region of the γ subunit has not been fully elucidated (63).
Molecular Switch on the γ Subunit Governed by Trx
The reduction of the disulfide bond located on the γ subunit has been a subject of extensive biochemical studies predominantly with the isolated CF1 (10, 60), leading to the elucidation of the link between the enzyme activity and the redox state of the regulatory cysteines. Following the rotation experiments reported by Noji et al. (48), we proceeded to visualize the rotation of the γ subunit in the authentic CF1 complex and successfully observed the unidirectional counterclockwise rotation (22). The redox regulation of rotation of the γ subunit was then studied using the chimeric thermophilic bacterial α3β3γ complex designated as α3β3γTCT, the γ subunit being constructed by fusion of the N- and C-terminal regions of γ from thermophilic bacteria, separated by a central region composed of the regulatory region of the spinach γ (5) (Fig. 5A). By using this α3β3γTCT complex, the relation between the suppressed enzyme activity under oxidizing conditions and the suppressed rotation movement was characterized, although further experiments were difficult due to the poor stability of this chimeric complex (6). More recently, the regulatory region of the spinach γ subunit was introduced into the α3β3γ complex of thermophilic cyanobacterial F1, and the redox regulation of rotation was thoroughly investigated (29) (Fig. 5A, B). Thanks to the optimal properties of this stable redox switch harboring chimeric complex, the pause position during rotation under oxidizing conditions could be assigned to 80° within the 120° catalytic step, which is the same as the position of ADP inhibition. In addition, the lower activity and rotation frequency of the oxidized form of the enzyme was attributed to a longer pause duration and a relatively shorter rotation duration [see Fig. 6 of Ref. (29)]. Thus, suppression of ATP hydrolysis activity under oxidizing conditions, which will occur in the dark in the natural enzyme, must be accomplished using the ADP inhibition properties.
FIG. 5.
Analysis of the redox regulation of rotation using the cyanobacterial α3β3γ complex containing the regulatory region of the γ subunit of spinach CF1. (A) Sequence alignment of the chimeric γ subunit. The chimeric construct part alone is shown. Chimeric TCT is the γ subunit constructed with thermophilic Bacillus PS3 and spinach γ subunit (5). Chimeric T_EL is the γ subunit constructed with T. elongatus and spinach γ subunits (29). In both sequences, those from spinach γ subunit are shown in green letters. Other labels are same as those shown in Fig. 4. Two cysteines for redox regulation are labeled with red star and EDE sequence with red triangle. (B) Experimental setup for analysis of the redox regulation of rotation. Four cysteines were introduced on the γ subunit; two cysteines at positions of 112 and 125 were for the fixation of the probe beads and those at 200 and 206 (originally 199 and 205 on the spinach γ) were for redox regulation. Amino acid numbering is based on the sequence of T. elongatus BP-1. Due to the limit of the restriction sites on DNA, 24 amino acids were introduced from spinach γ. Rotation of the γ subunit was observed using the attached polystyrene beads on the γ subunit. For more detail, see Ref. (29).
FIG. 6.
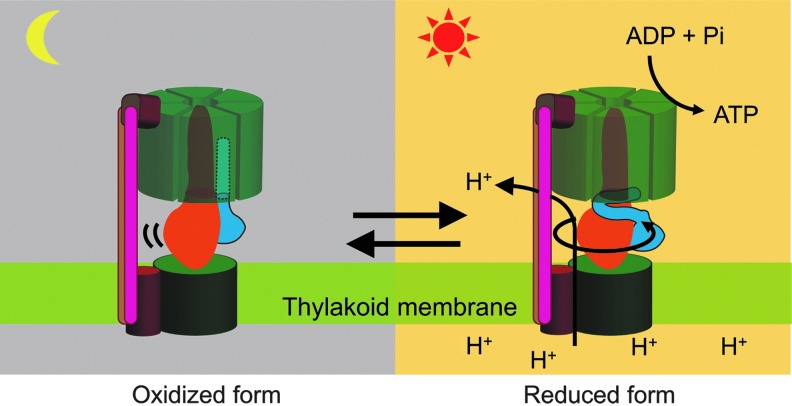
Oxidized enzyme in the dark is the inactive enzyme for ATP hydrolysis. In the light, CFoCF1 synthesize ATP using ΔμH+ across thylakoid membranes. In contrast, inactivation of the enzyme in the dark is critical to maintain ATP levels in the cell. CFoCF1, chloroplast ATP synthase.
Although, as described above, the majority of the molecular mechanism of redox regulation of rotation of this motor enzyme has already been characterized, there remain a number of interesting features relating to this regulatory system. For example, deletion of three negatively charged residues on the insertion region (210Glu-211Asp-212Glu) (Fig. 5A, red triangles) from the whole γ subunit sequence of spinach CF1 results in an apparent reversal of redox regulation of the enzyme activity; apparently, inactivated by reduction and activated by oxidation (34, 67). Correlation between the redox state of the redox switch and the inhibitory effect by the ɛ subunit has also been studied (12, 60). In the case of the isolated CF1, reduction of the γ subunit enhances the dissociation of the ɛ subunit from γ. Although structural aspects of the regulatory region of the γ subunit are extrapolated based on the calculation of the molecular model of the globular domain of the γ subunit (56), there remains limited information pertaining to the conformational change of the γ subunit by reduction and oxidation of two regulatory cysteines. Therefore, it is still difficult to explain the change in affinity of the ɛ subunit to the γ subunit in a redox-dependent manner. Hence, further structural insights into this motor enzyme are required to fully understand the molecular machinery that underpins regulation.
Molecular Evolution of the Thiol Switch of ATP Synthase; The Cyanobacterum Is the Ancestor of the Chloroplast
As described above, the γ subunit of cyanobacterial FoF1 also bears the inserted sequence found in spinach CF1-γ, although the sequence lacks 9 amino acids including two regulatory Cys residues involved in redox regulation (72) (Fig. 4). This insertion region is not present in the bacterial and mitochondrial F1 (MF1)-γ counterparts. Phylogenetic tree analysis also shows that the γ subunits of photosynthetic organisms have a shared ancestor (23), even though the cyanobacterial γ subunit lacks part of the regulatory region (72). The γ subunits of proteobacterial F1 are phylogenetically close to photosynthetic organisms, and the γ subunits of the mitochondrial ATPase form the original subfamily (23). This independence is in good agreement with the observed difference in the ɛ subunit as well. In the MF1, the counterpart of the ɛ subunit of the bacterial and chloroplast F1 is the δ subunit, and MF1 possesses the additional unique ɛ subunit, which binds to the γ subunit just on the upper region of the δ subunit binding domain (19). The ɛ subunit of MF1bound to the γ subunit is thought to disturb the conformational change of the δ subunit in the F1-ATPase complex. The apparent difference in the mitochondrial γ subunit on the phylogenetic tree from those of bacteria and chloroplast γ subunit [see Fig. 3 of Ref. (23)] might be reflected in this characteristic structural difference.
To understand the structural evolution of the γ subunit and the evolution of the regulatory mechanism, we have focused on the insertion region of the cyanobacterial F1γ subunit, and have prepared a mutant complex with the subunit γ lacking this insertion (32, 64). We have then studied the special characteristics of the mutant complex at both the single-molecule and whole-cell levels. Deletion of this insertion region from the γ subunit results in acceleration of the ATP hydrolysis activity (32). By detailed analysis of rotation behavior of the mutant complex, we concluded that this region confers the ability to frequently shift into an ADP-inhibition state; this is a highly conserved regulatory mechanism, which prevents ATP synthase from carrying out the reverse reaction (64). We then prepared the mutant strain of Synechocystis sp. PCC6803, whose γ subunit of ATP synthase lacks the insertion region. The intracellular ATP levels in cyanobacterial cells under light and dark conditions were then measured to determine the physiological significance of ADP inhibition in preventing the intrinsic ATP hydrolysis activity of CF1. The intracellular ATP level decreased sharply in both the wild-type and the mutant cells in darkness. Amounts of ATP in the wild-type cells decreased to 50% of the original level measured in the light when transferred into dark, whereas those in the insertion deletion mutant cells decreased to 20% or less. When the cells were subjected to light again, the ATP levels of both cells recovered to the original levels immediately [see Fig. 6 of Ref. (64)]. We therefore concluded that the physiological significance of this insertion must be in conferring the special capability to shift into the ADP inhibition state, which allows intracellular ATP levels to be maintained due to the shut-down of ATP hydrolysis activity, a property that must be especially critical for photosynthetic organisms.
Why can the insertion region located on the bottom of the γ subunit affect the ADP inhibition, which occurs at the catalytic site located on the β subunit? To examine the possible signal transfer pathway from the γ subunit to the catalytic sites, we recently introduced multiple Cys residues into the applicable positions on two central α-helices of the γ subunit, which enable us to restrict the relative movement of the α-helices and conformational change of γ (65). Consequently, we found that the conformational changes of both the central α -helices and the lower globular domain of the γ subunit are important for regulation of the activity.
Physiological Significance of Redox Regulation of ATP Synthase
The relation between ATP synthesis activity in chloroplasts and the redox state of the chloroplast ATP synthase was established first in class II chloroplasts, that is, the chloroplasts lacking outer membranes (26). In the study, four forms of the enzyme were suggested, namely, reduced active, reduced inactive, oxidized active, and oxidized inactive. By illumination in the presence of reductants, the oxidized inactive form enzyme, whose activity is supposed to be the basal level after the preparation of the chloroplasts, turns into the reduced active form enzyme. This form shows significant activities both in ATP hydrolysis and ATP synthesis. In contrast, the inactive species show neither ATP hydrolysis nor synthesis activities. Although the activation process was clearly described in their study, the physiological impact of the redox regulation of ATP synthase was not. Further progress in the study of regulation of chloroplast ATP synthase is well summarized in Ref. (49) especially in light of the electrochemical potential, the oxidation state of the γ subunit, and the bound nucleotide(s) at the catalytic sites.
In 1994, Ort and coworkers carried out a random mutagenesis study of A. thaliana in which mutant plants were selected that grew poorly under low irradiance, but performed satisfactorily at high irradiance (16). From this study, they found that the cfq mutant, impaired in effective reduction of the γ subunit by Trx, showed a lower efficiency of ATP formation (75, 76). The cfq mutant showed a phenotype in which the γ subunit was poorly reduced. Proper redox regulation must therefore be important for ATP synthesis as well, although there are several reports that ATP synthase can become active and support high rates of ATP synthesis even in the oxidized state (26, 52). Similar to the cfq mutant, a Trx-insensitive γ subunit isoform, ATPC2, in A. thaliana was recently identified (30) (see Fig. 4). The mutant plant expressing only ATPC2 showed high ATP synthesis activity in the light and dark although the change in the internal ATP level was not measured in vivo. The physiological significance of this γ subunit isoform should be further investigated as the authors showed that expression of ATPC2 occurs in root and that it is involved in root development.
Irrespective of the significance of the redox regulation switch for CFoCF1, there had been no in vivo reports pertaining to the reduction of the disulfide bond on the γ subunit of the CFoCF1 complex by reducing equivalents generated in vivo. For example, photosynthetic activation of CFoCF1 was indirectly measured using the electrochromic shift of carotenoids in chloroplasts (35). By using this technique, activation of the sunflower CFoCF1 in the light, which was assessed from the measurement of electrochromic shifts in leaf thylakoids, was reported. The authors then concluded that the catalytic activation of CFoCF1 is not rate limiting for the induction of carbon assimilation under field conditions (36). Although the actual redox states of CF1 could not be determined in their measurements, this study is certainly the first effort to determine the redox state of CFoCF1 in vivo. In contrast, the relevance of the electrochemical proton gradient in chloroplasts to activation of ATP synthase was studied using a combined method of spectrophotometric and fluorescence measurements (25). In this study, the authors suggested that the Calvin-Benson cycle is predominantly controlled by the ATP concentration in chloroplasts, and thiol enzymes in chloroplasts are in the reduced active state even in the dark-adapted leaves, based on their kinetic analysis.
We then directly assessed the reduction level of the γ subunit in the leaf and in intact chloroplasts using the thiol modifier 4-acetoamido-4′-maleimidylstilbene-2,2′-disulfonate and a specific antibody against the γ subunit (33). In this study, we clearly showed that the γ subunits in the dark-adapted leaves and in the intact chloroplasts exist in an oxidized state. In addition, we reported that the reduction of the γ subunit in chloroplasts is not a prerequisite for efficient ATP synthesis in the light, as previously suggested (26). Furthermore, we found that the disulfide bond formation on the γ subunit occurs spontaneously in chloroplasts, irrespective of the light/dark conditions and the reduced form Trx conclusively forces to shift the redox state of the γ subunit to the reduced form. The observed tendency of the equilibrium shift to the oxidation state of CF1 in chloroplasts must provide an advantage in conferring the ability to shut down prejudicial ATP hydrolysis activity, thus avoiding futile ATP hydrolysis under inappropriate conditions for photophosphorylation (Fig. 6).
Conclusions
The chloroplast ATP synthase is a unique molecular motor possessing a redox switch to control its motion. The role of this switch is important in carrying out efficient ATP synthesis and avoiding futile ATP hydrolysis under the transient light/dark conditions in nature. However, the physiological significance of this switch is not understood very well due to the unclear phenotype of the redox switch mutants analyzed to date. In contrast, recent research progress in the control of the molecular motion of F1 motor by this redox switch has been remarkable using mainly the F1 subcomplex α3β3γ. Various properties of cyanobacterial ATPase come down to those of the chloroplast enzyme except the redox regulation system and by mere introduction of the redox switch into the γ subunit, the cyanobacterial enzyme is able to adopt major characteristics of the chloroplast ATPase. Hence, we successfully analyzed the significance of the redox regulation system in vitro. For a complete understanding of this regulatory system, insights into the conformational change of the redox region as well as the structural information of the whole regulated enzyme complex are required.
Abbreviations Used
- CF1
F1 part of CFoCF1
- CFoCF1
chloroplast ATP synthase
- F1
membrane-peripheral part of ATP synthase
- Fo
membrane-embedded part of ATP synthase
- IF1
intrinsic ATPase inhibitor protein
- MF1
F1 part of mitochondrial ATP synthase
- Trx
thioredoxin
Acknowledgments
We thank T. Yonejima for his artistic work to prepare the figures. This work was financially supported by the Core Research of Evolutional Science & Technology program (CREST) from the Japan Science and Technology Agency (JST) and partly supported by Grant-in-aid for Scientific Research (No. 22651048 to T.H.) from the Japan Society for the Promotion of Science. E.S. is a research fellow of the Japan Society for the Promotion of Science.
References
- 1.Abrahams J. Leslie A. Lutter R. Walker J. Structure at 2.8 Å resolution of F1-ATPase from bovine heart mitochondria. Nature. 1994;370:621–628. doi: 10.1038/370621a0. [DOI] [PubMed] [Google Scholar]
- 2.Adachi K. Oiwa K. Nishizaka T. Furuike S. Noji H. Itoh H. Yoshida M. Kinosita K., Jr. Coupling of rotation and catalysis in F1-ATPase revealed by single-molecule imaging and manipulation. Cell. 2007;130:309–321. doi: 10.1016/j.cell.2007.05.020. [DOI] [PubMed] [Google Scholar]
- 3.Arabidopsis Genome Initiative. Analysis of the genome sequence of the flowering plant. Arabidopsis thaliana. Nature. 2000;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- 4.Ariga T. Muneyuki E. Yoshida M. F1-ATPase rotates by an asymmetric, sequential mechanism using all three catalytic subunits. Nat Struct Mol Biol. 2007;14:841–846. doi: 10.1038/nsmb1296. [DOI] [PubMed] [Google Scholar]
- 5.Bald D. Noji H. Stumpp MT. Yoshida M. Hisabori T. ATPase activity of a highly stable α3β3γ subcomplex of thermophilic F1 can be regulated by the introduced regulatory region of γ subunit of chloroplast F1. J Biol Chem. 2000;275:12757–12762. doi: 10.1074/jbc.275.17.12757. [DOI] [PubMed] [Google Scholar]
- 6.Bald D. Noji H. Yoshida M. Hirono-Hara Y. Hisabori T. Redox regulation of the rotation of F1-ATP synthase. J Biol Chem. 2001;276:39505–39507. doi: 10.1074/jbc.C100436200. [DOI] [PubMed] [Google Scholar]
- 7.Boyer PD. The ATP synthase—a splendid molecular machine. Annu Rev Biochem. 1997;66:717–749. doi: 10.1146/annurev.biochem.66.1.717. [DOI] [PubMed] [Google Scholar]
- 8.Buchanan BB. Wolosiuk RA. Photosynthetic regulatory protein found in animal and bacterial cells. Nature. 1976;264:669–670. doi: 10.1038/264669a0. [DOI] [PubMed] [Google Scholar]
- 9.Cabezon E. Arechaga I. Jonathan P. Butler G. Walker JE. Dimerization of bovine F1-ATPase by binding the inhibitor protein, IF1. J Biol Chem. 2000;275:28353–28355. doi: 10.1074/jbc.C000427200. [DOI] [PubMed] [Google Scholar]
- 10.Dann MS. McCarty RE. Characterization of the activation of membrane-bound and soluble CF1 by thioredoxin. Plant Physiol. 1992;99:153–160. doi: 10.1104/pp.99.1.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Du ZY. Boyer PD. On the mechanism of sulfite activation of chloroplast thylakoid ATPase and the relation of ADP tightly bound at a catalytic site to the binding change mechanism. Biochemistry. 1990;29:402–407. doi: 10.1021/bi00454a014. [DOI] [PubMed] [Google Scholar]
- 12.Duhe RJ. Selman BR. The dithiothreitol-stimulated dissociation of the chloroplast coupling factor 1 ɛ-subunit is reversible. Biochim Biophys Acta. 1990;1017:70–78. doi: 10.1016/0005-2728(90)90180-c. [DOI] [PubMed] [Google Scholar]
- 13.Duncan TM. Bulygin VV. Zhou Y. Hutcheon ML. Cross RL. Rotation of subunits during catalysis by Escherichia coli F1-ATPase. Proc Natl Acad Sci U S A. 1995;92:10964–10968. doi: 10.1073/pnas.92.24.10964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dunham KR. Selman BR. Regulation of spinach chloroplast coupling factor 1 ATPase activity. J Biol Chem. 1981;256:212–218. [PubMed] [Google Scholar]
- 15.Dunn SD. Tozer RG. Zadorozny VD. Activation of Escherichia coli F1-ATPase by lauryldimethylamine oxide and ethylene glycol: relationship of ATPase activity to the interaction of the ɛ and β subunits. Biochemistry. 1990;29:4335–4340. doi: 10.1021/bi00470a011. [DOI] [PubMed] [Google Scholar]
- 16.Gabrys H. Kramer DM. Crofts AR. Ort DR. Mutants of chloroplast coupling factor reduction in Arabidopsis. Plant Physiol. 1994;104:769–776. doi: 10.1104/pp.104.2.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garcia JJ. Morales-Rios E. Cortes-Hernandez P. Rodriguez-Zavala JS. The inhibitor protein (IF1) promotes dimerization of the mitochondrial F1F0-ATP synthase. Biochemistry. 2006;45:12695–12703. doi: 10.1021/bi060339j. [DOI] [PubMed] [Google Scholar]
- 18.Geck MK. Larimer FW. Hartman FC. Identification of residues of spinach thioredoxin f that influence interactions with target enzymes. J Biol Chem. 1996;271:24736–24740. doi: 10.1074/jbc.271.40.24736. [DOI] [PubMed] [Google Scholar]
- 19.Gibbons C. Montgomery M. Leslie A. Walker J. The structure of the central stalk in bovine F1-ATPase at 2.4 Å resolution. Nat Struct Biol. 2000;7:1055–1061. doi: 10.1038/80981. [DOI] [PubMed] [Google Scholar]
- 20.Gresser MJ. Myers JA. Boyer PD. Catalytic site cooperativity of beef heart mitochondrial F1 adenosine triphosphatase. Correlations of initial velocity, bound intermediate, and oxygen exchange measurements with an alternating three-site model. J Biol Chem. 1982;257:12030–12038. [PubMed] [Google Scholar]
- 21.Hirono-Hara Y. Noji H. Nishiura M. Muneyuki E. Hara KY. Yasuda R. Kinosita K., Jr. Yoshida M. Pause and rotation of F1-ATPase during catalysis. Proc Natl Acad Sci U S A. 2001;98:13649–13654. doi: 10.1073/pnas.241365698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hisabori T. Kondoh A. Yoshida M. The γ subunit in chloroplast F1-ATPase can rotate in a unidirectional and counter-clockwise manner. FEBS Lett. 1999;463:35–38. doi: 10.1016/s0014-5793(99)01602-6. [DOI] [PubMed] [Google Scholar]
- 23.Hisabori T. Ueoka-Nakanishi H. Konno H. Koyama F. Molecular evolution of the modulator of chloroplast ATP synthase: origin of the conformational change dependent regulation. FEBS Lett. 2003;545:71–75. doi: 10.1016/s0014-5793(03)00395-8. [DOI] [PubMed] [Google Scholar]
- 24.Johnson EA. McCarty RE. The carboxyl terminus of the ɛ subunit of the chloroplast ATP synthase is exposed during illumination. Biochemistry. 2002;41:2446–2451. doi: 10.1021/bi011939f. [DOI] [PubMed] [Google Scholar]
- 25.Joliot P. Joliot A. Quantification of the electrochemical proton gradient and activation of ATP synthase in leaves. Biochim Biophys Acta. 2008;1777:676–683. doi: 10.1016/j.bbabio.2008.04.010. [DOI] [PubMed] [Google Scholar]
- 26.Junesch U. Gräber P. Influence of the redox state and the activation of the chloroplast ATP synthase on proton-transport-coupled ATP synthesis/hydrolysis. Biochim Biophys Acta. 1987;893:275–288. [Google Scholar]
- 27.Kato-Yamada Y. Yoshida M. Hisabori T. Movement of the helical domain of the ɛ subunit is required for the activation of thermophilic F1-ATPase. J Biol Chem. 2000;275:35746–35750. doi: 10.1074/jbc.M006575200. [DOI] [PubMed] [Google Scholar]
- 28.Kato S. Yoshida M. Kato-Yamada Y. Role of the ɛ subunit of thermophilic F1-ATPase as a sensor for ATP. J Biol Chem. 2007;282:37618–37623. doi: 10.1074/jbc.M707509200. [DOI] [PubMed] [Google Scholar]
- 29.Kim Y. Konno H. Sugano Y. Hisabori T. Redox regulation of rotation of the cyanobacterial F1-ATPase containing thiol regulation switch. J Biol Chem. 2011;286:9071–9078. doi: 10.1074/jbc.M110.200584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kohzuma K. Dal Bosco C. Kanazawa A. Dhingra A. Nitschke W. Meurer J. Kramer DM. Thioredoxin-insensitive plastid ATP synthase that performs moonlighting functions. Proc Natl Acad Sci U S A. 2012;109:3293–3298. doi: 10.1073/pnas.1115728109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Konno H. Isu A. Kim Y. Murakami-Fuse T. Sugano Y. Hisabori T. Characterization of the relationship between ADP- and ɛ-induced inhibition in cyanobacterial F1-ATPase. J Biol Chem. 2011;286:13423–13429. doi: 10.1074/jbc.M110.155986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Konno H. Murakami-Fuse T. Fujii F. Koyama F. Ueoka-Nakanishi H. Pack CG. Kinjo M. Hisabori T. The regulator of the F1 motor: inhibition of rotation of cyanobacterial F1-ATPase by the ɛ subunit. EMBO J. 2006;25:4596–4604. doi: 10.1038/sj.emboj.7601348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Konno H. Nakane T. Yoshida M. Ueoka-Nakanishi H. Hara S. Hisabori T. Thiol modulation of the chloroplast ATP synthase is dependent on the energization of thylakoid membranes. Plant Cell Physiol. 2012;53:626–634. doi: 10.1093/pcp/pcs018. [DOI] [PubMed] [Google Scholar]
- 34.Konno H. Yodogawa M. Stumpp MT. Kroth P. Strotmann H. Motohashi K. Amano T. Hisabori T. Inverse regulation of F1-ATPase activity by a mutation at the regulatory region on the γ subunit of chloroplast ATP synthase. Biochem J 352 Pt. 2000;3:783–788. [PMC free article] [PubMed] [Google Scholar]
- 35.Kramer DM. Crofts AR. Activation of the chloroplast ATPase measured by the electrochromic change in leaves of intact plants. Biochim Biophys Acta. 1989;976:28–41. [Google Scholar]
- 36.Kramer DM. Wise RR. Frederick JR. Alm DM. Hesketh JD. Ort DR. Crofts AR. Regulation of coupling factor in field-grown sunflower: a Redox model relating coupling factor activity to the activities of other thioredoxin-dependent chloroplast enzymes. Photosynth Res. 1990;26:213–222. doi: 10.1007/BF00033134. [DOI] [PubMed] [Google Scholar]
- 37.Laurent TC. Moore EC. Reichard P. Enzymatic synthesis of deoxyribonucleotides. IV. Isolation and characterization of thioredoxin, the hydrogen donor from Escherichia coli B. J Biol Chem. 1964;239:3436–3444. [PubMed] [Google Scholar]
- 38.Lemaire SD. Michelet L. Zaffagnini M. Massot V. Issakidis-Bourguet E. Thioredoxins in chloroplasts. Curr Genet. 2007;51:343–365. doi: 10.1007/s00294-007-0128-z. [DOI] [PubMed] [Google Scholar]
- 39.Matsui T. Muneyuki E. Honda M. Allison WS. Dou C. Yoshida M. Catalytic activity of the α3β3γ complex of F1-ATPase without noncatalytic nucleotide binding site. J Biol Chem. 1997;272:8215–8221. doi: 10.1074/jbc.272.13.8215. [DOI] [PubMed] [Google Scholar]
- 40.Matsui T. Yoshida M. Expression of the wild-type and the Cys-/Trp-less α3β3γ complex of thermophilic F1-ATPase in Escherichia coli. Biochim Biophys Acta. 1995;1231:139–146. doi: 10.1016/0005-2728(95)00070-y. [DOI] [PubMed] [Google Scholar]
- 41.McCarty RE. Racker E. Partial resolution of the enzymes catalyzing photophosphorylation. 3. Activation of adenosine triphosphatase and 32P-labeled orthophosphate-adenosine triphosphate exchange in chloroplasts. J Biol Chem. 1968;243:129–137. [PubMed] [Google Scholar]
- 42.McKinney DW. Buchanan BB. Wolosiuk RA. Activation of chloroplast ATPase by reduced thioredoxin. Phytochemistry. 1978;17:794–795. [Google Scholar]
- 43.Miki J. Maeda M. Mukohata Y. Futai M. The γ-subunit of ATP synthase from spinach chloroplasts. Primary structure deduced from the cloned cDNA sequence. FEBS Lett. 1988;232:221–226. doi: 10.1016/0014-5793(88)80421-6. [DOI] [PubMed] [Google Scholar]
- 44.Mills JD. Mitchell P. Schurmann P. Modulation of coupling Factor ATPase Activity in intact chloroplasts, the role of the thioredoxin system. FEBS Lett. 1980;112:173–177. [Google Scholar]
- 45.Minkov IB. Fitin AF. Vasilyeva EA. Vinogradov AD. Mg2+-induced ADP-dependent inhibition of the ATPase activity of beef heart mitochondrial coupling factor F1. Biochem Biophys Res Commun. 1979;89:1300–1306. doi: 10.1016/0006-291x(79)92150-8. [DOI] [PubMed] [Google Scholar]
- 46.Mitome N. Suzuki T. Hayashi S. Yoshida M. Thermophilic ATP synthase has a decamer c-ring: indication of noninteger 10:3 H+/ATP ratio and permissive elastic coupling. Proc Natl Acad Sci U S A. 2004;101:12159–12164. doi: 10.1073/pnas.0403545101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nalin CM. McCarty RE. Role of a disulfide bond in the γ subunit in activation of the ATPase of chloroplast coupling factor 1. J Biol Chem. 1984;259:7275–7280. [PubMed] [Google Scholar]
- 48.Noji H. Yasuda R. Yoshida M. Kinosita K., Jr. Direct observation of the rotation of F1-ATPase. Nature. 1997;386:299–302. doi: 10.1038/386299a0. [DOI] [PubMed] [Google Scholar]
- 49.Ort DR. Oxborough K. In situ regulation of chlloroplast coupling factor activity. Annu Rev Plant Physiol Plant Mol Biol. 1992;43:269–291. [Google Scholar]
- 50.Pick U. Bassilian S. Activation of magnesium ion specific adenosinetriphosphatase in chloroplast coupling factor 1 by octyl glucoside. Biochemistry. 1982;21:6144–6152. doi: 10.1021/bi00267a019. [DOI] [PubMed] [Google Scholar]
- 51.Pogoryelov D. Yu J. Meier T. Vonck J. Dimroth P. Muller DJ. The c15 ring of the Spirulina platensis F-ATP synthase: F1/F0 symmetry mismatch is not obligatory. EMBO Rep. 2005;6:1040–1044. doi: 10.1038/sj.embor.7400517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ponomarenko S. Volfson I. Strotmann H. Proton gradient-induced changes of the interaction between CF0 and CF1 related to activation of the chloroplast ATP synthase. FEBS Lett. 1999;443:136–138. doi: 10.1016/s0014-5793(98)01681-0. [DOI] [PubMed] [Google Scholar]
- 53.Richter ML. Patrie WJ. McCarty RE. Preparation of the ɛ subunit and ɛ subunit-deficient chloroplast coupling factor 1 in reconstitutively active forms. J Biol Chem. 1984;259:7371–7373. [PubMed] [Google Scholar]
- 54.Rodgers AJ. Wilce MC. Structure of the γ−ɛ complex of ATP synthase. Nat Struct Biol. 2000;7:1051–1054. doi: 10.1038/80975. [DOI] [PubMed] [Google Scholar]
- 55.Sabbert D. Engelbrecht S. Junge W. Intersubunit rotation in active F-ATPase. Nature. 1996;381:623–625. doi: 10.1038/381623a0. [DOI] [PubMed] [Google Scholar]
- 56.Samra HS. Gao F. He F. Hoang E. Chen Z. Gegenheimer PA. Berrie CL. Richter ML. Structural analysis of the regulatory dithiol-containing domain of the chloroplast ATP synthase γ subunit. J Biol Chem. 2006;281:31041–31049. doi: 10.1074/jbc.M603315200. [DOI] [PubMed] [Google Scholar]
- 57.Schurmann P. Buchanan BB. The ferredoxin/thioredoxin system of oxygenic photosynthesis. Antioxid Redox Signal. 2008;10:1235–1274. doi: 10.1089/ars.2007.1931. [DOI] [PubMed] [Google Scholar]
- 58.Schwarz O. Schurmann P. Strotmann H. Kinetics and thioredoxin specificity of thiol modulation of the chloroplast H+-ATPase. J Biol Chem. 1997;272:16924–16927. doi: 10.1074/jbc.272.27.16924. [DOI] [PubMed] [Google Scholar]
- 59.Sekiya M. Hosokawa H. Nakanishi-Matsui M. Al-Shawi MK. Nakamoto RK. Futai M. Single molecule behavior of inhibited and active states of Escherichia coli ATP synthase F1 rotation. J Biol Chem. 2010;285:42058–42067. doi: 10.1074/jbc.M110.176701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Soteropoulos P. Suss KH. McCarty RE. Modifications of the γ subunit of chloroplast coupling factor 1 alter interactions with the inhibitory ɛ subunit. J Biol Chem. 1992;267:10348–10354. [PubMed] [Google Scholar]
- 61.Soulie JM. Buc J. Meunier JC. Pradel J. Ricard J. Molecular properties of chloroplastic thioredoxin f and the photoregulation of the activity of fructose 1,6-bisphosphatase. Eur J Biochem. 1981;119:497–502. doi: 10.1111/j.1432-1033.1981.tb05635.x. [DOI] [PubMed] [Google Scholar]
- 62.Stock D. Leslie AG. Walker JE. Molecular architecture of the rotary motor in ATP synthase. Science. 1999;286:1700–1705. doi: 10.1126/science.286.5445.1700. [DOI] [PubMed] [Google Scholar]
- 63.Stumpp MT. Motohashi K. Hisabori T. Chloroplast thioredoxin mutants without active-site cysteines facilitate the reduction of the regulatory disulphide bridge on the γ-subunit of chloroplast ATP synthase. Biochem J. 1999;341:157–163. [PMC free article] [PubMed] [Google Scholar]
- 64.Sunamura E. Konno H. Imashimizu-Kobayashi M. Sugano Y. Hisabori T. Physiological impact of intrinsic ADP inhibition of cyanobacterial FoF1 conferred by the inherent sequence inserted into the γ subunit. Plant Cell Physiol. 2010;51:855–865. doi: 10.1093/pcp/pcq061. [DOI] [PubMed] [Google Scholar]
- 65.Sunamura E. Konno H. Imashimizu M. Mochimaru M. Hisabori T. A conformational change of the γ subunit indirectly regulates the activity of cyanobacterial F1-ATPase. J Biol Chem. 2012;287:38695–38704. doi: 10.1074/jbc.M112.395053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tsunoda SP. Rodgers AJ. Aggeler R. Wilce MC. Yoshida M. Capaldi RA. Large conformational changes of the ɛ subunit in the bacterial F1F0 ATP synthase provide a ratchet action to regulate this rotary motor enzyme. Proc Natl Acad Sci U S A. 2001;98:6560–6564. doi: 10.1073/pnas.111128098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ueoka-Nakanishi H. Nakanishi Y. Konno H. Motohashi K. Bald D. Hisabori T. Inverse regulation of rotation of F1-ATPase by the mutation at the regulatory region on the γ subunit of chloroplast ATP synthase. J Biol Chem. 2004;279:16272–16277. doi: 10.1074/jbc.M400607200. [DOI] [PubMed] [Google Scholar]
- 68.Vasilyeva EA. Minkov IB. Fitin AF. Vinogradov AD. Kinetic mechanism of mitochondrial adenosine triphosphatase. ADP-specific inhibition as revealed by the steady-state kinetics. Biochem J. 1982;202:9–14. doi: 10.1042/bj2020009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wakagi T. Ohta T. Implications of the existence of two states of beef liver mitochondrial adenosine triphosphatase as revealed by kinetic studies. J Biochem. 1981;89:1205–1213. [PubMed] [Google Scholar]
- 70.Weber J. Dunn SD. Senior AE. Effect of the ɛ-subunit on nucleotide binding to Escherichia coli F1-ATPase catalytic sites. J Biol Chem. 1999;274:19124–19128. doi: 10.1074/jbc.274.27.19124. [DOI] [PubMed] [Google Scholar]
- 71.Werner-Grune S. Gunkel D. Schumann J. Strotmann H. Insertion of a ‘chloroplast-like’ regulatory segment responsible for thiol modulation into γ-subunit of F0F1-ATPase of the cyanobacterium Synechocystis 6803 by mutagenesis of atpC. Mol Gen Genet. 1994;244:144–150. doi: 10.1007/BF00283515. [DOI] [PubMed] [Google Scholar]
- 72.Werner S. Schumann J. Strotmann H. The primary structure of the γ-subunit of the ATPase from Synechocystis 6803. FEBS Lett. 1990;261:204–208. doi: 10.1016/0014-5793(90)80671-5. [DOI] [PubMed] [Google Scholar]
- 73.Wolosiuk RA. Buchanan BB. Thioredoxin and glutathione regulate photosynthesis in chloroplasts. Nature. 1977;266:565–567. [Google Scholar]
- 74.Wolosiuk RA. Crawford NA. Yee BC. Buchanan BB. Isolation of three thioredoxins from spinach leaves. J Biol Chem. 1979;254:1627–1632. [PubMed] [Google Scholar]
- 75.Wu G. Ort DR. Mutation in the cysteine bridge domain of the γ-subunit affects light regulation of the ATP synthase but not photosynthesis or growth in Arabidopsis. Photosynth Res. 2008;97:185–193. doi: 10.1007/s11120-008-9315-0. [DOI] [PubMed] [Google Scholar]
- 76.Wu G. Ortiz-Flores G. Ortiz-Lopez A. Ort DR. A point mutation in atpC1 raises the redox potential of the Arabidopsis chloroplast ATP synthase γ-subunit regulatory disulfide above the range of thioredoxin modulation. J Biol Chem. 2007;282:36782–36789. doi: 10.1074/jbc.M707007200. [DOI] [PubMed] [Google Scholar]
- 77.Yagi H. Kajiwara N. Tanaka H. Tsukihara T. Kato-Yamada Y. Yoshida M. Akutsu H. Structures of the thermophilic F1-ATPase ɛ subunit suggesting ATP-regulated arm motion of its C-terminal domain in F1. Proc Natl Acad Sci U S A. 2007;104:11233–11238. doi: 10.1073/pnas.0701045104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yasuda R. Noji H. Yoshida M. Kinosita K., Jr. Itoh H. Resolution of distinct rotational substeps by submillisecond kinetic analysis of F1-ATPase. Nature. 2001;410:898–904. doi: 10.1038/35073513. [DOI] [PubMed] [Google Scholar]
- 79.Yoshida M. Muneyuki E. Hisabori T. ATP synthase—a marvellous rotary engine of the cell. Nat Rev Mol Cell Biol. 2001;2:669–677. doi: 10.1038/35089509. [DOI] [PubMed] [Google Scholar]
- 80.Yoshida M. Sone N. Hirata H. Kagawa Y. Ui N. Subunit structure of adenosine triphosphatase. Comparison of the structure in thermophilic bacterium PS3 with those in mitochondria, chloroplasts, and Escherichia coli. J Biol Chem. 1979;254:9525–9533. [PubMed] [Google Scholar]