Abstract
Cardiovascular risk factors and atherosclerosis precursors were examined in 365 Turkish children and adolescents. Study participants were recruited at five different state schools. We tested single and multi-locus effects of six polymorphisms from five candidate genes, chosen based on prior known association with lipid levels in adults, for association with low (≤10th percentile) high density lipoprotein cholesterol (HDL-C) and high (≥90th percentile) triglycerides (TG), and the related continuous outcomes. We observed an association between CETP variant rs708272 and low HDL-C (allelic p=0.020, genotypic p=0.046), which was supported by an independent analysis, PRAT (PRAT control p=0.027). Sex-stratified logistic regression analysis showed that the B2 allele of rs708272 decreased odds of being in the lower tenth percentile of HDL-C measurements (OR=0.36, p=0.02) in girls; this direction of effect was also seen in boys but was not significant (OR=0.64, p=0.21). Logistic regression analysis also revealed that the T allele of rs6257 (SHBG) decreased odds of being in the top tenth percentile of TG measurements in boys (OR=0.43, p=0.03). Analysis of lipid levels as a continuous trait revealed a significant association between rs708272 (CETP) and LDL-C levels in males (p=0.02) with the B2B2 genotype group having the lowest mean LDL-C; the same direction of effect was also seen in females (p=0.05). An effect was also seen between rs708272 and HDL-C levels in girls (p=0.01), with the B2B2 genotype having the highest mean HDL-C levels. Multi-locus analysis, using quantitative multifactor dimensionality reduction (qMDR) identified the previously mentioned CETP variant as the best single locus model, and overall model, for predicting HDL-C levels in children. This study provides evidence for association between CETP and low HDL-C phenotype in children, but the results appear to be weaker in children than previous results in adults and may also be subject to gender effects.
Introduction
Prospective epidemiological studies have shown that unfavorable serum lipid levels, such as raised levels of total cholesterol (TC), triglyceride (TG), low-density lipoprotein cholesterol (LDL-C), together with decreased levels of high-density lipoprotein cholesterol (HDL-C), are important risk factors for cardiovascular disease (CVD) and form a target for therapeutic intervention (Musunuru et al., 2010). In addition, cardio-metabolic risk factors in childhood confer substantial risk for future CVD in adulthood (Magnussen et al., 2010). In recognition of the potential implications for public health, screening for cardiovascular risk factors in children is now recommended by the American Academy of Pediatrics (AAP), American Heart Association (AHA), and National Heart, Lung, and Blood Institute (Expert Panel on Integrated Guidelines for Cardiovascular Health and Risk Reduction in Children and Adolescents, 2011; Daniels et al., 2008; Weintraub et al., 2011). However, lipid levels change with age, puberty, race, and gender among children and adolescents (Hickman et al., 1998). For example, HDL-C levels decrease and TG levels increase after puberty (Agirbasli et al., 2010; Berenson et al., 1981) and the trajectory of these changes and how they affect the risk of CVD are not well understood.
Adding to the complexity of using dyslipidemia (low HDL-C, elevated non-HDL-C and TG) as a predictor of CVD risk is the fact that dyslipidemia itself is a complex trait caused by multiple environmental and genetic factors and their interactions (Evans et al., 2011; Teslovich et al., 2010). Recently, several genetic loci that influence lipid levels in adulthood have been reported (Oliveira et al., 2010); alleles in ATP binding cassette transporter (ABCA1), hepatic lipase C514T (LIPC), lipoprotein lipase S447X (LPL), and cholesterol ester transfer protein (CETP) Taq1B affect HDL-C and triglyceride levels in adults (Hodoglugil et al., 1999; Pan et al., 2012; Rip et al., 2006; Weissglas-Volkov et al., 2010) Similarly, sex hormone levels and sex hormone binding globulin (SHBG) have important effects on lipid levels (particularly HDL-C) in children and adolescents (Agirbasli et al., 2010; Garcés et al., 2010). Additionally, genetic variations in SHBG (rs1799941 and rs6257) associate with circulating sex hormone levels in adults (Dunning et al., 2004; Eriksson et al., 2006; Haiman et al., 2005).
In this study, we assessed the relationship of six variants in five genes that previously associated with lipid levels in adults. We tested the single and multi-locus effects of the gene polymorphisms (rs328 in LPL, rs708272 in CETP, rs1800588 in LIPC, rs1800977 in ABCA1, and rs1799941, rs6257 in SHBG) on the concentrations of the serum lipids and body mass index (BMI) among Turkish children and adolescents.
Materials and Methods
Cardiovascular risk factors and atherosclerosis precursors were examined in Turkish children and adolescents in 2007. The subject demographics are displayed in Table 1.
Table 1.
Demographics and Biochemical Characteristics for High Triglyceride (High TG) and Low High Density Lipoprotein Cholesterol (Low HDL-C) Cases and Controls by Gender
| |
Males |
Females |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Risk determinant | High TG (n=36) | Non-High TG (n=134) | P3 | Low HDL-C (n=27) | Non-Low HDL-C (n=143) | P3 | High TG (n=11) | Non-High TG (n=179) | P3 | Low HDL-C (n=18) | Non-Low HDL-C (n=172) | P3 |
| Age1 | 12.5 (0.58) | 12.8 (0.29) | 0.462 | 15.3 (0.40) | 12.3 (0.28) | <0.0001 | 13.6 (0.97) | 13.2 (0.25) | 0.95 | 13.1 (0.74) | 13.2 (0.25) | 0.877 |
| BMI1 | 19.8 (0.94) | 18.5 (0.29) | 0.472 | 22.3 (0.73) | 18.1 (0.31) | <0.0001 | 22.6 (1.38) | 18.8 (0.27) | 0.01 | 20.8 (1.06) | 18.8 (0.27) | 0.066 |
| Fasting glucose1 | 78.0 (2.10) | 79.3 (0.77) | 0.464 | 73.1 (2.51) | 80.2 (0.72) | 0.0005 | 71.7 (2.14) | 75.5 (0.63) | 0.153 | 70.1 (2.09) | 75.8 (0.63) | 0.006 |
| Insulin resistance (IR)2 | [6 (30)] | [19 (115)] | 0.708* | [8 (19)] | [17 (126)] | 0.024* | [1 (10)] | [15 (164)] | 0.63* | [0 (18)] | [16 (156)] | 0.190* |
Mean (SE) of risk determinant are presented, unless otherwise indicated.
Insulin Resistance: [IR Cases (IR Controls)], IR Case definition is HOMA-IR ≥3.16.
P value presented is Student's t test p value or Wilcoxon Rank Sum test p value where appropriate, unless otherwise indicated.
P value presented is from Chi-square test or Fisher's Exact test of association where appropriate.
Study participants
Five different state elementary and secondary schools were selected. Elementary and secondary schools (Cengelkoy, Mehmet Rauf, Yavuzturk, Dorduncu Murat, and Remzi Bayraktar) were all located in the Uskudar region of Istanbul, Turkey. Subjects were instructed to fast for at least 12 hours prior to screening. Their compliance was ascertained by an interview on the day of examination. The fasting status was based on self-report. Screening took place at schools during normal school hours and blood samples were drawn at 9 AM. Students who did not want to participate in the survey were excluded (approximately 10% for elementary school children and 20% for secondary school students). The Institutional Review Board of Marmara University, Istanbul, and the Educational Board approved the study protocol. Informed consent was obtained from parents or guardians. Subjects were given case numbers and identities were kept confidential.
Biochemical analysis
Lipid and glucose levels were measured by enzymatic colorimetric-assay method using Cobas Integra 800 kits (Roche® Diagnostic). Analyses were performed in an accredited laboratory (Centro Laboratories, Istanbul, Turkey). The laboratory is accredited by DAR (Deutscher Akkreditierungs Rat) according to ISO 15189 of the clinical laboratory accreditation committee. Measurement uncertainties of each method at the time of analysis were 1.5% for total cholesterol, 1.8% for triglyceride, 2.7% for HDL-C, and 1.6% for glucose. Homeostasis model assessment-estimated insulin resistance (HOMA-IR) is calculated using the formula (glucose (mmol/L)×fasting insulin (μU/mL)/22.5).
Lipid levels
We used the National Heart Lung and Blood Institute Growth and Health Study (NGHS) to calculate the age and sex-specific lipid percentiles (NGHS Coordinating Center, 1998). TG level ≥90th percentile or HDL-C level ≤10th percentile from NGHS data were defined as ‘high TG’ and ‘low HDL-C’, respectively (Table 1). In addition, we used alternative cutoffs to assess the robustness of our results: TG level ≥75th percentile and HDL-C level ≤25th percentile (Supplementary Table S1; supplementary data are available online at www.liebertpub.com/omi).
Determination of the genotypes
Two mL of venous blood from study subjects were collected in tubes containing EDTA and then stored at +4°C. Genomic DNA was isolated from peripheral blood leukocytes using High Pure PCR Template Preparation Kit (Roche Diagnostics GmbH, Mannheim, Germany) within 1 week of sample collection. LPL stop codon polymorphism rs328 (S447Ter (terminal codon), ABCA1 promoter polymorphism rs1800977 (C14T), LIPC promoter polymorphism rs1800588 (C514T), and CETP promoter polymorphism rs708272 were analyzed using polymerase chain reaction (PCR)-restriction fragment length polymorphism (RFLP). A base substitution from G (B1) to A (B2) in intron 1 of the CETP gene leads to 3 genotypes, B1B1, B1B2, or B2B2 at Taq1B site (5454G>A). SHBG polymorphisms, rs1799941 and rs6257 genotyping was performed by real-time PCR amplification and fluorescent probe melting point analysis on the Light Cycler™ 1.5 instrument (Roche® Diagnostics GmbH) (Table 2).
Table 2.
Genotyped Single Nucleotide Polymorphisms (SNPs) in Study Cohort
| Chromosome | Gene | SNP | Alleles |
|---|---|---|---|
| 8 | LPL | rs328 | S/X1 |
| 9 | ABCA1 | rs1800977 | C/T |
| 15 | LIPC | rs1800588 | C/T |
| 16 | CETP | rs708272 | B1/B22 |
| 17 | SHBG | rs1799941 | A/G |
| 17 | SHBG | rs6257 | C/T |
rs328 is an LPL gene stop codon SNP, S (serine amino acid) 447Ter (termination codon). LPL S447X (rs328) involves a C→G change at nucleotide 1595 of the LPL gene, which leads to a change in amino acid 447 from a serine (S) to a stop codon (X). Heterozygotes for the X447 allele are displayed as SX and S447 homozygotes are listed as SS.
rs708272 is a silent base change SNP; a base substitution from G (B1) to A (B2) in intron 1 of the CETP gene leads to 3 variants, B1B1, B1B2 or B2B2 at Taq1B site (5454G>A).
Primers and hybridization probes were designed using Gen Bank cDNA sequence. For the PCR reaction, 50 ng genomic DNA was amplified with the Light Cycler® Fast Start DNA Master Hyb Probe Kit (Roche® Diagnostics GmbH). Each 20 μL reaction contained 1X Fast Start DNA Master Hyb Probe, 1.5 mM MgCl2, 0.3 μM each primer, and 0.15 μM hybridization probe. After initial denaturation step at 95°C for 10 min, amplification was performed using 45 cycles of denaturation at 95°C for 5 sec, annealing at 55°C for 20 sec, and extension at 72°C for 20 sec. This was followed by a melting curve analysis of 95°C for 0 sec, 40°C for 30 sec, and a slow ramp (0.1°C/sec) to 85°C with continuous fluorescent acquisition.
Statistical analyses
Prior to association testing, all SNPs were tested for deviations from Hardy Weinberg Equilibrium (HWE), as this test can serve as a means to assess genotyping error. This was done using the software package PLINK (Purcell et al., 2007). A HWE p<0.01 was considered to indicate a significant deviation from equilibrium. For continuous data analyses, the Shapiro Wilkes test was used to determine whether data were normally distributed. In cases where data were found to be non-normal, (p<0.05), the data were log transformed in an attempt to make the distribution normal. When successful, log transformed data were analyzed using appropriate methods; when transformation did not bring data to normality, non-parametric analytical techniques were used. Specifically, for continuous normally distributed data, the analysis of variance (ANOVA) test was used to determine if there was a significant difference in variable mean by SNP genotype; in the case of non-normal data, the nonparametric Kruskal-Wallis (K-W) test was employed. ANOVA and K-W analyses were performed using the statistical package STATA 10 (StataCorp. 2007).
For case/control analyses (Low HDL and High TG), allelic and genotypic Chi-square tests were used to investigate associations between SNP genotype–phenotype in the full dataset. The Fisher's Exact test was implemented when the Chi-square test was inappropriate due to small sample size (cell n<5). Chi-square and Fisher's Exact tests were carried out in PLINK.
The Prevalence-based Association Test (PRAT) is a single locus test that is methodologically different from the chi-square test of association and is based on Hardy-Weinberg principles (Ryckman et al., 2008). PRAT tests the expected case and control genotype frequency distributions against expected values estimated from prevalence of the phenotypes and the allele frequencies within case and control groups. Depending on the genetic model of the phenotype, PRAT has been shown to be as or more powerful than standard tests for association, and serves to validate associations detected using chi-square analyses. PRAT analyses were corrected for multiple tests using permutation (n=1000). PRAT analysis was carried out in the software package PLATO (Grady et al., 2010).
Logistic regression analysis was used to reveal the direction of single SNP effects on low HDL-C and high TG phenotypes separately among males and females, in order to reveal possible gender-specific effects. Logistic regression results were adjusted for age, body mass index (BMI), and insulin resistance (HOMA-IR). Logistic regression analysis was performed using the STATA 10 software package. Corrections for multiple testing were done using the false discovery rate (FDR) for tests that did not generate an empirical/permutation p value. FDR q was set at 0.15.
Multi-locus analysis for low HDL-C and high TG
Recognizing the complexity of both HDL-C and TG, we performed quantitative multifactor dimensionality reduction (qMDR) analysis in order to reveal the presence of multi-locus interactions, or epistasis in our phenotypes (Gui et al., 2013). Multifactor dimensionality reduction (MDR) is a well-known nonparametric method for detecting epistatic interactions with or without the presence of a significant single locus effect in dichotomous data (Ritchie et al., 2001), and the quantitative version (qMDR) is optimized to use the same principles for continuous outcomes. In qMDR, for each single/multi locus genotype combination, the mean trait value is calculated and then compared with the overall mean trait value in the total sample. If the mean value from the genotype group is larger than the overall mean, then the genotype is considered “high-risk;” otherwise it is labeled “low-risk.” The trait outcome is then compared between high and low risk groups using a T-test, the corresponding t-statistic is used to define the “training t-statistic” using only 1/10 of the data and the “testing t-statistic” using the remaining 9/10 of the data. The “testing t-statistic” is the metric that is used to identify the best overall model. The null distribution of the testing t-statistic follows the normal distribution with mean 0. The normal distribution was used to estimate empirical model p values based on the testing t-statistic with n-1 (359) degrees of freedom. In order to increase our power, we analyzed HDL-C and TG as continuous variables using qMDR in the full dataset only.
Results
A total of 365 children and adolescents were enrolled in the study (175 boys, mean age 12.8±3.4 and 190 girls, mean age 13.2±3.3). Genetic analysis was performed in 360 subjects as DNA was not available for 5 male subjects. Demographics are displayed in Table 1 for all subjects with available genetic data (n=360).
Distribution of gene polymorphisms
The chromosomal location, allele definitions, and frequencies, and HWE p values for genotyped SNPs are presented in Tables 2 and 3. All HWE SNP p values were p≥0.01).
Table 3.
Genotype Distrisbution and Hardy Weinberg Equilibrium (HWE) Analysis for all SNPs
| Gene | SNP | Genotype categories | Genotype counts (%) | HWE P1 |
|---|---|---|---|---|
| ABCA1 | rs1800977 | CC | 163 (45.3) | 0.27 |
| CT | 166 (46.1) | |||
| TT | 31 (8.6) | |||
| LPL | rs328 | SX | 69 (19.2) | 0.06 |
| SS | 291 (80.8) | |||
| CETP | rs708272 | B1B1 | 108 (30.0) | 0.60 |
| B1B2 | 174 (48.3) | |||
| B2B2 | 78 (21.7) | |||
| LIPC | rs1800588 | CC | 244 (67.8) | 1.00 |
| CT | 105 (29.2) | |||
| TT | 11 (3.1) | |||
| SHBG | rs1799941 | AA | 8 (2.2) | 1.00 |
| AG | 93 (25.8) | |||
| GG | 259 (71.9) | |||
| rs6257 | CC | 4 (1.1) | 0.01 | |
| CT | 119 (33.1) | |||
| TT | 237 (65.8) |
Analyses were performed in all subjects (male and female combined).
Hardy Weinberg Equilibrium test p value is presented.
Dichotomous analysis of low HDL-c and high TG by genotype groups
Overall 47 (36 males, 11 females) and 45 (27 males, 18 females) subjects had high TG and low HDL-C respectively (Table 1). Chi-square analysis in the full dataset revealed a significant association between rs708272, located in CETP, and low HDL-C (Table 4). The allelic association (p=0.020) remained significant after correction for multiple testing. PRAT analysis validated the association between rs708272 and low HDL-C (PRAT control p=0.027, Table 5), and also revealed a significant association between rs6257, located in SHBG, and low HDL-C (PRAT case p=0.030, Table 5). Evidence for this latter association was not revealed by the chi-square analysis. None of the SNPs were shown to be associated with high TG in the full dataset.
Table 4.
Single Variant Assocation Analysis Genotyped SNPs with Low HDL-C and High TG
| |
|
|
Low HDL-C |
High TG |
||||
|---|---|---|---|---|---|---|---|---|
| Gene | SNP | Genotype categories | Genotype counts cases (%) | Genotype counts controls (%) | P1(allelic2, genotypic3) | Genotype counts cases (%) | Genotype counts controls (%) | P1 (allelic2, genotypic3) |
| ABCA1 | rs1800977 | CC | 22 (48.9) | 141 (44.8) | (0.278, 0.283) | 23 (48.9) | 140 (44.7) | (0.674, 0.867) |
| CT | 22 (48.9) | 144 (45.7) | 20 (42.6) | 146 (46.6) | ||||
| TT | 1 (2.2) | 30 (9.5) | 4 (8.5) | 27 (8.7) | ||||
| LPL | rs328 | SX | 6 (13.3) | 63 (20.0) | (0.315, 0.197) | 6 (12.8) | 63 (20.1) | (0.258, 0.232) |
| SS | 39 (86.7) | 252 (80.0) | 41 (87.2) | 250 (79.9) | ||||
| CETP | rs708272 | B1B1 | 18 (40.0) | 90 (28.6) | (0.020, 0.046) | 14 (29.8) | 94 (30.0) | (0.985, 0.996) |
| B1B2 | 23 (51.1) | 151 (47.9) | 23 (48.9) | 151 (48.2) | ||||
| B2B2 | 4 (8.9) | 74 (23.5) | 10 (21.3) | 68 (21.8) | ||||
| LIPC | rs1800588 | CC | 26 (57.8) | 218 (69.2) | (0.356, 0.096) | 32 (68.1) | 212 (67.7) | (0.680, 0.265) |
| CT | 19 (42.2) | 86 (27.3) | 12 (25.5) | 93 (29.7) | ||||
| TT | 0 (0) | 11 (3.5) | 3 (6.4) | 8 (2.6) | ||||
| SHBG | rs1799941 | AA | 0 (0) | 8 (2.5) | (0.254, 0.648) | 1 (2.1) | 7 (2.2) | (0.393, 0.549) |
| AG | 10 (22.2) | 83 (26.3) | 15 (31.9) | 78 (24.9) | ||||
| GG | 35 (77.8) | 224 (71.2) | 31 (66.0) | 228 (72.8) | ||||
| rs6257 | CC | 0 (0) | 4 (1.3) | (0.130, 0.143) | 1 (2.1) | 3 (1.0) | (0.116, 0.169) | |
| CT | 21 (46.7) | 98 (31.1) | 20 (42.6) | 99 (31.6) | ||||
| TT | 24 (53.3) | 213 (67.6) | 26 (55.3) | 211 (67.4) | ||||
Chi-Square or Fisher's Exact test p value is presented where appropriate for indicated test, either allelic or genotypic.
Allelic test: Compares the frequencies of alleles at the specified marker in cases versus controls using a Chi-Square test with 1 degree of freedom; M versus m (M=major allele, m=minor allele).
Genotypic test: Compares the frequencies of genotypes at a specifed marker in cases versus controls using a Chi-Square test with 2 degrees of freedom; MM versus Mm versus mm (M=major allele, m=minor allele).
Table 5.
Prevalence Based Association Test (PRAT) Results for Single Variant Assocation Between Genotyped SNPs and Selected Phenotypes (Low HDL-C/High TG)
| |
|
Low HDL-C |
High TG |
||
|---|---|---|---|---|---|
| Gene | SNP | PRAT P cases1 | PRAT P controls2 | PRAT P cases1 | PRAT P controls2 |
| ABCA1 | rs1800977 | 0.167 | 0.701 | 0.930 | 0.497 |
| LPL | rs328 | 0.547 | 0.190 | 0.424 | 0.175 |
| CETP | rs708272 | 0.097 | 0.027 | 1.000 | 0.789 |
| LIPC | rs1800588 | 0.048 | 0.224 | 0.336 | 0.507 |
| SHBG | rs1799941 | 0.403 | 0.317 | 0.693 | 0.516 |
| rs6257 | 0.030 | 0.962 | 0.136 | 0.638 | |
PRAT p value from case only analysis in selected phenotype, p value presented is permutation based p value (n=1000).
PRAT p value from control only anlaysis in selected phenotype, p value presented is permutaiton based p value (n=1000).
In order to determine the direction of effect of associated SNPs, and also to elucidate SNPs that had differing directional effects by gender that may have caused them to be overlooked in pooled analyses, we performed logistic regression analysis, adjusted for age, BMI, and HOMA-IR, for both dichotomous phenotypes in males and females, separately (Table 6). We constructed our logistic regression model by categorizing SNP genotypes into three groups, with the homozygous major genotype serving as the referent genotype. The model assumed an additive effect for each additional minor allele. The models describe the effects of each additional minor allele on high TG and low HDL-C. The minor allele of rs708272 (B2 allele) associated with decreased odds of being in the lower tenth percentile of HDL-C measurements (OR=0.36, p=0.02) in females; this direction of effect was also evident in males but the OR was not significant (OR=0.64, p=0.21). The rs6257 minor allele (T allele) associated with decreased odds of being in the top tenth percentile of TG measurements in males only (OR=0.43, p=0.03); this effect was not seen in females. Additional analyses using the alternative cutoffs (TG ≥75th percentile and HDL-C ≤25th percentile) produced similar results for all statistical tests (Supplementary Tables S1–S4). In the case of rs708272 and HDL-C, the results were more statistically significant as might be expected with increased numbers in the low HDL-C category.
Table 6.
Logistic Regression Results of Low HDL-C and High TG among Girls and Boys
| |
|
Low HDL-C |
High TG |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| |
|
Males |
Females |
Males |
Females |
||||||||
| GENE | SNP | OR1 | 95% Confidence interval | P2 | OR1 | 95% Confidence interval | P2 | OR1 | 95% Confidence interval | P2 | OR1 | 95% Confidence interval | P2 |
| ABCA1 | rs1800977 | 0.58 | 0.27–1.25 | 0.16 | 0.83 | 0.34–1.99 | 0.67 | 0.79 | 0.44–1.42 | 0.43 | 0.52 | 0.16–1.65 | 0.27 |
| LPL | rs328 | 0.73 | 0.21–2.55 | 0.62 | 0.44 | 0.09–2.12 | 0.31 | 0.54 | 0.17–1.71 | 0.30 | 0.66 | 0.12–3.57 | 0.63 |
| CETP | rs708272 | 0.64 | 0.32–1.28 | 0.21 | 0.36 | 0.16–0.82 | 0.02 | 1.16 | 0.67–2.00 | 0.59 | 0.56 | 0.22–1.48 | 0.24 |
| LIPC | rs1800588 | 1.05 | 0.46–2.43 | 0.90 | 1.77 | 0.71–4.46 | 0.22 | 0.92 | 0.45–1.86 | 0.81 | 1.60 | 0.50–5.12 | 0.43 |
| SHBG | rs1799941 | 2.52 | 0.78–8.10 | 0.12 | 1.27 | 0.45–3.62 | 0.65 | 0.62 | 0.29–1.30 | 0.20 | 0.71 | 0.21–2.38 | 0.58 |
| rs6257 | 0.52 | 0.21–1.30 | 0.16 | 0.60 | 0.23–1.56 | 0.30 | 0.43 | 0.21–0.90 | 0.03 | 1.00 | 0.28–3.55 | 1.00 | |
All analyses were adjusted for age, BMI, and HOMA-IR measurements.
Logistic regression models tested were coded additively using three independant genotype categories for each marker, with the homozygous major genotype group as the reference. Genotype group categorizates were as follows; homozygous major=0, heterozygote=1, homozygous minor=2. OR presented above represent the change in odds per each 1 minor allele change in genotype group.
OR=Odds Ratio; 2P value presented is from adjusted logistic regression analysis for association with the specified phenotype.
Quantitative analysis of lipids and BMI by genotype groups
In order to further explore the impact of gender in the context of SNP genotype and biometric measurements in our cohort, we performed exploratory analysis using BMI and lipid concentrations as continuous outcomes in males and females separately (Tables 7–9).
Table 7.
Association of Gene Polymorphisms with BMI for Males and Females
| |
|
|
Males |
Females |
||
|---|---|---|---|---|---|---|
| Gene | SNP | Genotype | Average BMI (SE1) | P2 | Average BMI (SE) | P2 |
| ABCA1 | rs1800977 | CC | 18.8 (0.48) | 0.42 | 18.9 (0.38) | 0.34 |
| CT | 19.0 (0.45) | 18.9 (0.39) | ||||
| TT | 18.0 (0.87) | 21.0 (1.60) | ||||
| LPL | rs328 | SX | 18.8 (0.33) | 0.82 | 18.8 (0.30) | 0.15 |
| SS | 18.8 (0.77) | 19.8 (0.57) | ||||
| CETP | rs708272 | B1B1 | 18.4 (0.48) | 0.56 | 20.0 (0.48) | 0.01 |
| B1B2 | 19.1 (0.46) | 18.3 (0.40) | ||||
| B2B2 | 18.6 (0.72) | 19.1 (0.51) | ||||
| LIPC | rs1800588 | CC | 18.6 (0.36) | 0.64 | 19.1 (0.34) | 0.52 |
| CT | 19.1 (0.63) | 18.9 (0.48) | ||||
| TT | 19.7 (1.74) | 17.2 (1.21) | ||||
| SHBG | rs1799941 | AA | 16.9 (2.27) | 0.32 | 16.6 (0.81) | 0.15 |
| AG | 19.7 (0.81) | 18.7 (0.52) | ||||
| GG | 18.6 (0.32) | 19.2 (0.32) | ||||
| rs6257 | CC | 14.8 (0.03) | 0.18 | 18.9 (2.84) | 0.52 | |
| CT | 19.1 (0.50) | 19.3 (0.43) | ||||
| TT | 18.7 (0.39) | 18.9 (0.34) | ||||
Standard Error (SE) of the Mean; 2p value is from Kruskal Wallis test.
Table 9.
Quantitative Multifactor Dimensionality Reduction (qMDR) Analysis of HDL-C in Full Dataset
| Model1 | Training T-statistic2 | Testing T-statistic3 | CVC4 | P5 |
|---|---|---|---|---|
| rs708272 | 2.644 | 2.133 | 8/10 | 0.034 |
| rs1800977 – rs6257 | 3.338 | 1.586 | 4/10 | 0.114 |
| rs1800977 – rs708272 – rs6257 | 4.445 | 0.363 | 6/10 | 0.717 |
| rs1800977 – rs708272 – rs328 – rs6257 | 5.581 | 0.251 | 3/10 | 0.802 |
Defines the genetic single or multi-locus model being analyzed; 2Students t-statistic based on comparison of HDL-c measurments between high and low risk groups defined by selected SNP(s) genotype in the training dataset; 3Students t-statistic cased on comparison of HDL-c measurments between high and low risk groups defined by selected SNP(s) genotype in the testing dataset; 4Cross Validation Consistency of the selected model;
Empirical p value calculated from the Student's t probablity distribution using the Testing t-statistic and 359 degrees of freedom.
SNP rs708272 associated with BMI in females only (p=0.01), with the B1B1 genotype group having the highest average BMI measurements (Table 7). Analysis of lipid levels in males and females, separately, revealed a significant association between rs708272 and LDL-C levels in males (p=0.02) with the B2B2 genotype group having the lowest mean LDL-C; the same direction of effect was also seen in females with marginal significance (p=0.05). An effect was also seen between rs708272 and HDL-C levels in females (p=0.01), with the B2B2 genotype group having the highest mean HDL-C levels, this direction of effect was also seen in males but was not significant (p=0.68) (Table 8).
Table 8A.
Distribution of Lipid Levels at Genotype Groups for Males
| |
|
|
Males |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Gene | SNP | Genotype | Total cholesterol1 | P2 | LDL-C1 | P2 | HDL-C1 | P2 | TG1 | P2 |
| ABCA1 | rs1800977 | CC | 142.4 (3.3) | 0.16 | 72.9 (2.4) | 0.24 | 52.7 (1.8) | 0.14 | 71.4 (4.3) | 0.30 |
| CT | 146.4 (2.9) | 74.8 (2.4) | 55.6 (1.8) | 72.6 (4.1) | ||||||
| TT | 156.2 (7.0) | 82.1 (5.1) | 60.0 (3.2) | 62.2 (7.2) | ||||||
| LPL | rs328 | SX | 147.1 (2.3) | 0.23 | 75.4 (1.8) | 0.72 | 55.0 (1.4) | 0.97 | 72.1 (3.2) | 0.28 |
| SS | 141.0 (5.0) | 73.1 (3.9) | 54.5 (2.5) | 65.0 (5.0) | ||||||
| CETP | rs708272 | B1B1 | 145.4 (3.7) | 0.35 | 76.8 (3.0) | 0.02 | 53.9 (2.1) | 0.68 | 69.2 (5.3) | 0.64 |
| B1B2 | 148.6 (3.1) | 77.7 (2.4) | 53.9 (1.8) | 73.6 (4.3) | ||||||
| B2B2 | 140.9 (4.5) | 66.1 (2.9) | 58.8 (2.6) | 66.6 (4.3) | ||||||
| LIPC | rs1800588 | CC | 144.2 (2.5) | 0.38 | 72.7 (1.9) | 0.05 | 55.3 (1.4) | 0.83 | 70.4 (3.5) | 0.63 |
| CT | 150.7 (4.0) | 80.8 (3.2) | 54.4 (2.4) | 70.5 (4.8) | ||||||
| TT | 143.8 (14.1) | 73.8 (10.9) | 51.3 (4.4) | 80.8 (12.9) | ||||||
| SHBG | rs1799941 | AA | 164.7 (13.2) | 0.25 | 84.3 (19.1) | 0.77 | 65.3 (6.2) | 0.45 | 72.7 (18.2) | 0.69 |
| AG | 149.2 (3.7) | 75.9 (3.1) | 55.1 (2.6) | 78.3 (7.9) | ||||||
| GG | 144.6 (2.5) | 74.4 (1.9) | 54.7 (1.4) | 68.6 (2.8) | ||||||
| rs6257 | CC | 155.5 (38.5) | 0.86 | 80.0 (23.0) | 0.64 | 58.0 (12.0) | 0.04 | 88.0 (17.0) | 0.35 | |
| CT | 144.2 (3.1) | 76.4 (2.7) | 50.9 (1.9) | 76.5 (5.8) | ||||||
| TT | 146.8 (2.8) | 74.1 (2.1) | 57.1 (1.5) | 67.4 (2.9) | ||||||
Mean (Standard Error of the mean) of selected lipid is presented; 2p value for normally distributed variables is from ANOVA test, Kruskal Wallis p value is presented for variables that are not normally distributed.
Table 8B.
Distribution of Lipid Levels at Genotype Groups for Females
| |
|
|
Females |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Gene | SNP | Genotype | Total cholesterol1 | P2 | LDL-C1 | P2 | HDL-C1 | P2 | TG1 | P2 |
| ABCA1 | rs1800977 | CC | 153.2 (2.5) | 0.77 | 80.8 (2.2) | 0.25 | 57.0 (1.3) | 0.41 | 77.1 (4.2) | 0.62 |
| CT | 151.3 (2.6) | 77.1 (2.2) | 59.6 (1.4) | 72.8 (3.2) | ||||||
| TT | 154.8 (5.3) | 81.4 (4.1) | 57.1 (2.8) | 81.3 (9.4) | ||||||
| LPL | rs328 | SX | 151.4 (1.9) | 0.25 | 78.7 (1.6) | 0.50 | 57.6 (1.0) | 0.17 | 75.1 (3.0) | 0.64 |
| SS | 156.5 (4.1) | 80.8 (3.7) | 60.4 (1.9) | 76.4 (4.9) | ||||||
| CETP | rs708272 | B1B1 | 152.3 (3.2) | 0.65 | 81.4 (2.8) | 0.05 | 54.7 (1.5) | 0.01 | 81.0 (5.0) | 0.29 |
| B1B2 | 153.7 (2.4) | 80.9 (2.2) | 58.5 (1.2) | 71.8 (2.8) | ||||||
| B2B2 | 149.7 (3.5) | 72.3 (2.8) | 62.3 (2.2) | 75.1 (7.0) | ||||||
| LIPC | rs1800588 | CC | 153.0 (2.1) | 0.58 | 79.5 (1.9) | 0.97 | 58.7 (1.1) | 0.49 | 74.0 (2.9) | 0.54 |
| CT | 150.5 (3.1) | 78.7 (2.6) | 56.8 (1.7) | 74.6 (3.5) | ||||||
| TT | 159.6 (4.5) | 75.8 (11.2) | 60.4 (2.4) | 117.6 (48.4) | ||||||
| SHBG | rs1799941 | AA | 155.4 (8.3) | 0.32 | 78.6 (8.1) | 0.33 | 62.8 (5.1) | 0.56 | 70.0 (11.6) | 0.38 |
| AG | 156.7 (3.6) | 83.3 (3.3) | 59.0 (1.8) | 72.6 (4.9) | ||||||
| GG | 150.4 (1.9) | 77.4 (1.7) | 57.7 (1.1) | 76.7 (3.1) | ||||||
| rs6257 | CC | 196.5 (10.5) | 0.05 | 112.0 (2.0) | 0.10 | 64.0 (17.0) | 0.67 | 104.5 (21.5) | 0.10 | |
| CT | 151.4 (3.1) | 79.3 (2.8) | 57.2 (1.6) | 73.8 (5.8) | ||||||
| TT | 152.2 (2.0) | 78.6 (1.8) | 58.6 (1.1) | 75.6 (2.6) | ||||||
Mean (Standard Error of the mean) of selected lipid is presented; 2p value for normally distributed variables is from ANOVA test, Kruskal Wallis p value is presented for variables that are not normally distributed.
Multi-locus analysis
We performed exploratory multi locus analysis, using qMDR, on HDL-C and TG as continuous variables in the full dataset in order to explore possible higher order interaction effects between SNPs with and without the presence of main effects (Table 9). qMDR identified the single locus model containing rs708272 as the best model for prediction of HDL-C in the full dataset (CVC=8/10, empirical p=0.034) (Fig. 1). None of the identified multi-locus models for HDL-C were statistically significant. qMDR analysis of TG did not identify any significant single or multi-locus models (data not shown).
FIG. 1.
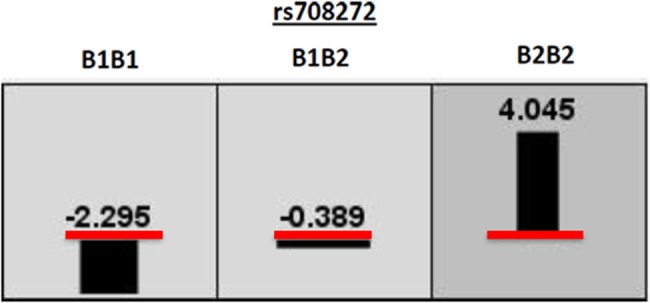
Quantitative Multifactor Dimensionality Reduction graphical representaion of Best Model for HDL-C. Red line indicates the global average of HDL-C measurement. *Numbers and bar height show the difference between the global average of HDL-C and the genotype specific average of HDL-C for a given genotype. Bar width indicates proportion of data that falls within the specified genotype group.
Discussion
Dyslipidemic lipoprotein levels in adolescence are associated with an increased risk of atherosclerosis in young adulthood and adolescent lipid levels are more strongly associated with high carotid intima media thickness in adulthood than a potential change in lipid levels later in life (Magnussen et al., 2009). Thus, dyslipidemia in children and adolescents increases risk of atherosclerosis even if lipid levels improve by adulthood (Magnussen et al., 2009). Therefore, the challenge of whether the SNPs and genes already identified affect lipid variation in childhood or adolescence is a crucial one.
Plasma lipids are intensely studied risk factors for cardiovascular disease in adults. Using a candidate gene approach, we examined six SNPs, previously found to associate with HDL-C or TG levels (Dunning et al., 2004; Eriksson et al., 2006; Haiman et al., 2005; Hodoglugil et al., 1999; Pan et al., 2012; Rip et al., 2006; Weissglas-Volkov et al., 2010) in adult populations, for association with dyslipidemia in children. An association is observed at CETP (rs708272) with HDL-C and BMI (in girls) levels. The T allele of rs6257 (SHBG) decreased odds of being in the top tenth percentile of TG measurements in boys. However, none of the identified multi-locus models for HDL-C or TG were statistically significant. Instead, qMDR identified the single locus model containing CETP rs708272 as the best model for prediction of HDL-C.
CETP is responsible for the exchange of cholesteryl esters (CE) and TG between circulating lipoproteins; specifically CETP mediates the exchange of CE and TG between HDL-C and apolipoprotein B containing molecules such as LDL-C. Inhibition of CETP activity increases plasma HDL-C levels (Hewing et al., 2012). Alternatively, greater CETP activity increases the CE content of LDL-C, and decreases CE content of HDL-C (Esteves-Gonzales et al., 2010).
Similar to our observations in children and adolescents, a previous study among Turkish adults showed that CETP Taq1B polymorphism associated with HDL-C levels. In a previous study of an adult Turkish cohort (n=1585), elevated HDL-C levels associated with B2B2 genotype of Taq1B polymorphism (p<0.0001). After adjusting for age and sex, B2B2 individuals had 15.9% higher HDL-C levels than B1B1 individuals (Ozsait et al., 2008).
This cross-sectional study obviously comes with limitations, given the smaller number of subjects available. Environmental risk factors (i.e., diet or physical activity) can alter epigenetic patterns, thereby partially ablating or enhancing genetic effects. Lipid level variation in children is due to multiple genes and environmental factors. Hence we performed exploratory multi-locus analysis in addition to searching for single markers with main effects. However, our data did not provide evidence for significant interactive effects.
Comparatively little is known about genetic loci affecting lipid levels in children, and despite a higher heritability in adolescents than in adults, most published studies have focused on adults. Dyslipidemia (low HDL-C, elevated non-HDL-C and TG) begins in childhood (Expert Panel on Integrated Guidelines for Cardiovascular Health and Risk Reduction in Children and Adolescents, 2011; Berenson et al., 1981; Hickman et al., 1998), but genetic studies of these factors in children are rare. Lipid levels are known to be substantially influenced by genetic variation, with high heritability (70%–80%) in adolescents (Middelberg et al., 2012). Lipid levels normally change with age among children and adolescents. Association studies suggest that some of the previously identified SNPs associate differently with lipid traits in adolescents compared to adults (Middelberg et al., 2012). The reason(s) for the inconsistency may include developmental changes in lipid levels, or greater interactions of lipids with environmental differences in adults (Middelberg et al., 2012). Results of studies such as ours help to illuminate the relationship(s) between gene polymorphisms and their phenotypic effects; and in an era of endemic childhood obesity, this and similar studies are critical to preventing CVD throughout life.
We examined a representative sample of healthy students from Istanbul, Turkey. Turkish population sets a unique sample, since low HDL-C is reported to be more common among Turkish adults than any other population (Mahley et al, 1995; 2001; Onat et al., 1999). Turkish children paradoxically have lower BMI, higher triglyceride, and lower HDL-C levels compared to the age and gender matched Bogalusa Heart Study population (Agirbasli et al., 2005). The underlying reason(s) for low HDL-C and high triglycerides in Turks remain unclear. Mahley et al. (1995) reported that low HDL-C levels in the Turkish population remain even after adjustment for environmental factors such as diet and smoking. These observations suggest that genetic factors are important in determining HDL-C levels in the Turkish population starting early in life. The allelic associations found in our data are consistent with previous reports. The B2B2 genotype in CETP was associated with decreased LDL-C, reduced BMI (in girls), and increased HDL-C (in girls).
CETP is an important molecule which is on the focus of HDL-C lowering clinical trials. The B2 allele has previously been associated with higher HDL-C levels in Chinese adults (Pan et al., 2012). In a study on children, CETP variation has contributed to the variation in HDL-C levels in Spanish prepubescent girls (López-Simón et al., 2009). Allele frequencies are similar to those in our study. The associations obtained in this study have the same directional effect as previous studies; theB2B2 genotype of CETP was associated with favorable effects on LDL-C and HDL-C levels among children and adolescents. The allelic direction of the association was the same in different populations (Chinese or Spanish) and age groups (adults versus children) (López-Simón et al., 2009; Pan et al., 2012). In a cohort of Greek children, Taq1B B2 allele carriers also had higher HDL-C levels (p<0.001) (Smart et al., 2010).
Lipid levels are mainly stable during childhood, but vary between sexes during puberty. Studies reported that HDL-C levels decrease in boys at puberty, while remaining unchanged in girls (Garces et al., 2010; Lopez et al., 1996). This change may be attributable to an increase in testosterone levels in boys at puberty. SHBG modulates the availability of biologically active free testosterone and estradiol and their metabolic clearance rates (Rosner et al., 1991); and Hergenc et al. (1999) reported that Turkish men and women had significantly lower levels of SHBG (p<0.001 in men and <0.05 in women) compared to Germans.
We observed that the T allele of rs6257 (SHBG) decreased the odds of being in the top tenth percentile of TG measurements in boys. We previously reported that SHBG levels is an important determinant of HDL-C levels and insulin resistance in children (Agirbasli et al., 2010) and SHBG gene polymorphisms associate with circulating SHBG levels in adults (Dunning et al., 2004; Haiman et al., 2005). SHBG gene polymorphism may modulate lipid levels in children through its effects on circulating SHBG levels.
Lessons learned
This study provides evidence for associations between CETP and SHBG SNPs with lipid levels in children. The study supports previous results from adults in the same population, and also provided evidence of gender differences (Hergenc et al.,1999; Ozsait B et al., 2008). The information gained from this study may provide further insight into the genetic architecture of lipid profiles in Turkish children and adolescents. Although individual SNPs showed only modest effects on lipid variation, in combination, they may provide valuable information about lipid profiles (Kathiresan et al., 2008).
Conclusions
This study suggests that genetic factors are important in determining HDL-C levels starting early in life, and our results highlight the persistence of these genetic effects even after accounting for environmental factors, such as age, HOMA-IR, and BMI, which are known to have a significant impact on lipid profiles. Our observations underscore the importance of individualized prevention programs to improve pediatric dyslipidemia-related causes of early atherosclerosis, and highlight the need for further investigation of candidate genes, CETP and SHBG, in pediatric populations.
Future studies may identify high-order gene–gene and gene–environment interactions, related to lipid levels in children.
Supplementary Material
Acknowledgments
The authors would like to thank the schools, students, and parents for their participation. The study was funded by the Scientific Research Council of Turkey (TUBITAK) Project Number: 109S282. SMW was partially supported by US NIH grant P20 GM103534.
Author Disclosure Statement
The authors have no conflicts to disclose.
References
- Agirbasli M. Ciliv G. Cakir S. Srinivasan S. Berenson GS. Ozme S. Bodymass index and lipid levels in children from Ankara, Turkey versus Bogalusa, Louisiana. Prev Med. 2005;41:843–845. doi: 10.1016/j.ypmed.2005.08.001. [DOI] [PubMed] [Google Scholar]
- Agirbasli M. Agaoglu NB. Orak N, et al. Sex hormones, insulin resistance and high-density lipoprotein cholesterol levels in children. Horm Res Paediatr. 2010;73:166–174. doi: 10.1159/000284357. [DOI] [PubMed] [Google Scholar]
- Berenson GS. Srinivasan SR. Cresanta JL. Foster TA. Webber LS. Dynamic changes of serum lipoproteins in children during adolescence and sexual maturation. Am J Epidemiol. 1981;113:157–170. doi: 10.1093/oxfordjournals.aje.a113080. [DOI] [PubMed] [Google Scholar]
- Daniels SR. Greer FR Committee on Nutrition. Lipid screening and cardiovascular health in childhood. Pediatrics. 2008;122:198–208. doi: 10.1542/peds.2008-1349. [DOI] [PubMed] [Google Scholar]
- Dunning AM. Dowsett M. Healey CS, et al. Polymorphisms associated with circulating sex hormone levels in postmenopausal women. J Natl Cancer Inst. 2004;96:936–945. doi: 10.1093/jnci/djh167. [DOI] [PubMed] [Google Scholar]
- Eriksson AL. Lorentzon M. Mellström D, et al. SHBG gene promoter polymorphisms in men are associated with serum sex hormone-binding globulin, androgen and androgen metabolite levels, and hip bone mineral density. J Clin Endocrinol Metab. 2006;91:5029–5037. doi: 10.1210/jc.2006-0679. [DOI] [PubMed] [Google Scholar]
- Estévez-González MD. Saavedra-Santana P. López-Ríos L. Chirino R. Cebrero-García E. Peña-Quintana L. Betancor-León P. HDL cholesterol levels in children with mild hypercholesterolemia: Effect of consuming skim milk enriched with olive oil and modulation by the TAQ 1B polymorphism in the CETP gene. Ann Nutr Metab. 2010;56:288–293. doi: 10.1159/000290405. [DOI] [PubMed] [Google Scholar]
- Evans D. Aberle J. Beil FU. The relative importance of common and rare genetic variants in the development of hypertriglyceridemia. Expert Rev Cardiovasc Ther. 2011;9:637–644. doi: 10.1586/erc.11.53. [DOI] [PubMed] [Google Scholar]
- Expert Panel on Integrated Guidelines for Cardiovascular Health and Risk Reduction in Children and Adolescents; National Heart, Lung, and Blood Institute. Expert panel on integrated guidelines for cardiovascular health and risk reduction in children and adolescents: Summary report. Pediatrics. 2011;128:S213–256. doi: 10.1542/peds.2009-2107C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcés C. Oya I. Lasunción MA. López-Simón L. Cano B. de Oya M. Sex hormone-binding globulin and lipid profile in pubertal children. Metabolism. 2010;59:166–171. doi: 10.1016/j.metabol.2009.06.033. [DOI] [PubMed] [Google Scholar]
- Grady BJ. Torstenson ES. Dudek SM. Giles J. Sexton D. Ritchie MD. Finding unique filter sets in PLATO: A precursor to efficient interaction analysis in GWAS data. Pacific Symposium on Biocomputing. 2010:315–326. PM 19908384, PMC2903053. [PMC free article] [PubMed] [Google Scholar]
- Gui J. Moore JH. Williams SM, et al. A simple and computationally efficient approach to multifactor dimensionality reduction analysis of gene-gene interactions for quantitative traits. PLoS One. 2013;8:e66545. doi: 10.1371/journal.pone.0066545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haiman CA. Riley SE. Freedman ML. Setiawan VW. Conti DV. Le Marchand L. Common genetic variation in the sex steroid hormone-binding globulin (SHBG) gene and circulating levels among postmenopausal women: The Multiethnic Cohort. J Clin Endocrinol Metab. 2005;90:2198–2204. doi: 10.1210/jc.2004-1417. [DOI] [PubMed] [Google Scholar]
- Hergenç G. Schulte H. Assmann G. von Eckardstein A. Associations of obesity markers, insulin, and sex hormones with HDL-cholesterol levels in Turkish and German individuals. Atherosclerosis. 1999;145:147–156. doi: 10.1016/s0021-9150(99)00027-1. [DOI] [PubMed] [Google Scholar]
- Hewing B. Fisher A. Rationale for cholesteryl ester transfer protein inhibition. Curr Opin Lipidol. 2012;23:372–376. doi: 10.1097/MOL.0b013e328353ef1d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickman TB. Briefel RR. Carroll MD, et al. Distributions and trends of serum lipid levels among United States children and adolescents ages 4–19 years: Data from the Third National Health and Nutrition Examination Survey. Prev Med. 1998;27:879–890. doi: 10.1006/pmed.1998.0376. [DOI] [PubMed] [Google Scholar]
- Hodoglugil U. Williamson DW. Mahley RW. Polymorphisms in the hepatic lipase gene affect plasma HDL-cholesterol levels in a Turkish population. J Lipid Res. 2010;51:422–430. doi: 10.1194/jlr.P001578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kathiresan S. Melander O. Anevski D, et al. Polymorphisms associated with cholesterol and risk of cardiovascular events. N Engl J Med. 2008;358:1240–1249. doi: 10.1056/NEJMoa0706728. [DOI] [PubMed] [Google Scholar]
- Lopez Martinez D. Gil A. Porres A, et al. Lipoprotein profile in children and adolescents of the Autonomous Community of Madrid. Med Clin (Barc) 1996;107:366–370. [PubMed] [Google Scholar]
- López-Simón L. de Oya M. Lasunción MA, et al. Genetic determinants of plasma HDL-cholesterol levels in prepubertal children. Clin Chim Acta. 2009;403:203–206. doi: 10.1016/j.cca.2009.03.002. [DOI] [PubMed] [Google Scholar]
- Magnussen CG. Venn A. Thomson R, et al. The association of pediatric low- and high-density lipoprotein cholesterol dyslipidemia classifications and change in dyslipidemia status with carotid intima-media thickness in adulthood evidence from the cardiovascular risk in Young Finns study, the Bogalusa Heart study, and the CDAH (Childhood Determinants of Adult Health) study. J Am Coll Cardiol. 2009;53:860–869. doi: 10.1016/j.jacc.2008.09.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnussen CG. Koskinen J. Chen W, et al. Pediatric metabolic syndrome predicts adulthood metabolic syndrome, subclinical atherosclerosis, and type 2 diabetes mellitus but is no better than body mass index alone: The Bogalusa Heart Study and the Cardiovascular Risk in Young Finns Study. Circulation. 2010;122:1604–1611. doi: 10.1161/CIRCULATIONAHA.110.940809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahley RW. Palaoglu KE. Atak Z, et al. Turkish Heart Study: Lipids, lipoproteins, and apolipoproteins. J Lipid Res. 1995;36:839–859. [PubMed] [Google Scholar]
- Mahley RW. Arslan P. Pekcan G, et al. Plasma lipids in Turkish children: Impact of puberty, socioeconomic status, and nutrition on plasma cholesterol and HDL. J Lipid Res. 2001;42:1996–2006. [PubMed] [Google Scholar]
- Middelberg RP. Heath AC. Madden PA. Montgomery GW. Martin NG. Whitfield JB. Evidence of differential allelic effects between adolescents and adults for plasma high-density lipoprotein. Plos One. 2012;7:e35605. doi: 10.1371/journal.pone.0035605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musunuru K. Kathiresan S. Genetics of coronary artery disease. Annu Rev Genomics Hum Genet. 2010;11:91–108. doi: 10.1146/annurev-genom-082509-141637. [DOI] [PubMed] [Google Scholar]
- NGHS Coordinating Center. Baltimore Maryland Medical Research; 1998. NHLBI Growth and Health Study (NGHS) data monitoring report. [Google Scholar]
- Oliveira FL. Patin RV. Escrivão MA. Atherosclerosis prevention and treatment in children and adolescents. Expert Rev Cardiovasc Ther. 2010;8:513–528. doi: 10.1586/erc.09.170. [DOI] [PubMed] [Google Scholar]
- Onat A. Yıldırım B. Uslu N. Plasma lipoproteins and apolipoproteins in Turkish adults: Overall levels, associations with other risk parameters and HDL's role as a marker of coronary risk in women. Arch Turk Soc Cardiol. 1999;27:72–79. [Google Scholar]
- Ozsait B. Kömürcü Bayrak E. Poda M, et al. CETP TaqIB polymorphism in Turkish adults: Association with dyslipidemia and metabolic syndrome. Anadolu Kardiyol Derg. 2008;8:324–330. [PubMed] [Google Scholar]
- Pan SL. Wang F. Lu ZP, et al. Cholesteryl ester transfer protein TaqIB polymorphism and its association with serum lipid levels and longevity in Chinese Bama Zhuang population. Lipids Health Dis. 2012;11:26. doi: 10.1186/1476-511X-11-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S. Neale B. Todd-Brown K, et al. PLINK: A toolset for whole-genome association and population-based linkage analysis. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rip J. Nierman MC. Ross CJ, et al. Lipoprotein lipase S447X; A naturally occurring gain of function mutation. Arterioscler Thromb Vasc Biol. 2006;26:1236–1245. doi: 10.1161/01.ATV.0000219283.10832.43. [DOI] [PubMed] [Google Scholar]
- Ritchie MD. Hahn LW. Roodi N, et al. Multifactor-dimensionality reduction reveals high-order interactions among estrogen-metabolism genes in sporadic breast cancer. Am J Hum Genet. 2001;69:1138–1147. doi: 10.1086/321276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosner W. Plasma steroid-binding proteins. Endocrinol Metab Clin North Am. 1991;20:697–720. [PubMed] [Google Scholar]
- Ryckman KK. Jiang L. Li C. Bartlett J. Haines JL. Williams SM. A prevalence-based association test for case-control studies. Genet Epidemiol. 2008;32:600–605. doi: 10.1002/gepi.20342. [DOI] [PubMed] [Google Scholar]
- Smart MC. Dedoussis G. Louizou E, et al. APOE, CETP and LPL genes show strong association with lipid levels in Greek children. Nutr Metab Cardiovasc Dis. 2010;20:26–33. doi: 10.1016/j.numecd.2009.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- StataCorp. College Station, TX: Stata Corp LP; 2007. Stata Statistical Software: Release 10. [Google Scholar]
- Teslovich TM. Musunuru K. Smith AV, et al. Biological, clinical and population relevance of 95 loci for blood lipids. Nature. 2010;466:707–713. doi: 10.1038/nature09270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weintraub WS. Daniels SR. Burke LE, et al. American Heart Association Advocacy Coordinating Committee; Council on Cardiovascular Disease in the Young; Council on the Kidney in Cardiovascular Disease; Council on Epidemiology and Prevention; Council on Cardiovascular Nursing; Council on Arteriosclerosis; Thrombosis and Vascular Biology; Council on Clinical Cardiology, and Stroke Council. Value of primordial and primary prevention for cardiovascular disease: A policy statement from the American Heart Association. Circulation. 2011;124:967–990. doi: 10.1161/CIR.0b013e3182285a81. [DOI] [PubMed] [Google Scholar]
- Weissglas-Volkov D. Pajukanta P. Genetic causes of high and low serum cholesterol. J Lipid Res. 2010;51:2032–2057. doi: 10.1194/jlr.R004739. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


