Abstract
In this work, we describe a periplasmic protein that is essential for flagellar rotation in Rhodobacter sphaeroides. This protein is encoded upstream of flgA, and its expression is dependent on the flagellar master regulator FleQ and on the class III flagellar activator FleT. Sequence comparisons suggest that this protein is a distant homologue of FlgT. We show evidence that in R. sphaeroides, FlgT interacts with the periplasmic regions of MotB and FliL and with the flagellar protein MotF, which was recently characterized as a membrane component of the flagellum in this bacterium. In addition, the localization of green fluorescent protein (GFP)-MotF is completely dependent on FlgT. The Mot− phenotype of flgT cells was weakly suppressed by point mutants of MotB that presumably keep the proton channel open and efficiently suppress the Mot− phenotype of motF and fliL cells, indicating that FlgT could play an additional role beyond the opening of the proton channel. The presence of FlgT in purified filament-hook-basal bodies of the wild-type strain was confirmed by Western blotting, and the observation of these structures under an electron microscope showed that the basal bodies from flgT cells had lost the ring that covers the LP ring in the wild-type structure. Moreover, MotF was detected by immunoblotting in the basal bodies obtained from the wild-type strain but not from flgT cells. From these results, we suggest that FlgT forms a ring around the LP ring, which anchors MotF and stabilizes the stator complex of the flagellar motor.
INTRODUCTION
The bacterial flagellum is a complex rotary motor driven by the electrochemical potential. The rotating part of the motor includes the export apparatus, the C ring, the rod, the hook, and the filament (for a review, see references 1 and 2). The stator is a proton channel and couples proton flow with torque generation (3–5). Many alkalophilic and marine species use a Na+-dependent motor to generate torque (6–9).
In Escherichia coli and Salmonella, the FliG, FliM, and FliN flagellar proteins are part of the rotor and form a cytoplasmic structure named the C ring (10–13). The C-terminal domain of FliG forms the interface between the rotor and the stator. Several charged residues located in this domain are involved in rotation and also in recruiting the stator complexes around the flagellum (12, 14–18).
Two proteins, MotA and MotB, form the flagellar stator. A functional proton channel has a MotA4/MotB2 stoichiometry, and approximately 11 complexes are arranged around the flagellar rotor (19–25). MotA has four transmembrane helices (TM1 to TM4), with short periplasmic loops between TM1 and TM2 and between TM3 and TM4 and a large cytoplasmic loop between TM2 and TM3 (25–28). MotB has a single TM helix with a short N-terminal cytoplasmic region and a large periplasmic domain (29). The periplasmic C-terminal region of MotB contains a peptidoglycan binding domain that anchors the stator to the cell wall (30, 31).
It has been shown that electrostatic interactions between charged amino acids of FliG and MotA are important for motor rotation. These residues are localized at the interface of the rotor and the stator (14, 15, 32). In particular, it has been proposed that the interaction between glutamic acid 98 (E98) of MotA and arginine 281 (R281) of FliG is important for torque generation, whereas the interaction between arginine 90 (R90) of MotA and aspartic acid 289 (D289) of FliG seems to be critical for recruiting the MotA/MotB complexes around the rotor (16).
Disulfide cross-linking studies of the MotA4/MotB2 complexes indicate that TM helices 2, 3, and 4 of MotA together with the TM helix of MotB form the proton channel (24, 25, 33). In particular, a conserved aspartic acid residue in the TM helix of MotB (D32) is crucial for motor functioning (34, 35). It has been proposed that protons are translocated through the MotA/MotB channel using this residue as the proton-binding site. It has been reported that charge-neutralizing mutations of this residue provoke a conformational change in the cytoplasmic loop of MotA where the charged residues E98 and R90 are localized (36). From this, it has been proposed that flagellar rotation involves rounds of protonation and deprotonation of the conserved D32 residue of MotB that modulate the electrostatic interactions at the MotA-FliG interface (4, 37, 38).
When MotA and MotB were gradually expressed, stepwise increases in the rotation speed were detected, indicating that the MotA/MotB complexes were incorporated in a stepwise fashion into the previously formed flagellar rotor (39). Recently, using total internal reflection fluorescence microscopy, it was observed that green fluorescent protein (GFP)-MotB associated with the motor is rapidly exchanged with GFP-MotB molecules in a mobile pool (40).
Free membrane-bound stator complexes show a very low ability to conduct ions (20, 41), indicating that activation of the proton channel occurs when stator complexes are recruited by the basal body. The amphipathic alpha helix segment immediately after the TM helix of the MotB periplasmic domain has been proposed to act as a plug that keeps the proton channel closed. Activation occurs when the stator complex interacts with the basal body, promoting a conformational change of the periplasmic domain of MotB that alters the position of this region, thus releasing the plug (42–46).
R. sphaeroides has two different full sets of flagellar genes. One of these sets, called Fla1, is constitutively expressed under laboratory conditions. It was previously shown that Fla1 was acquired by horizontal gene transfer (47). With the exception of motAB, the genes encoding the proteins of flagellar system 1 are found in a single cluster in chromosome I (positions 1132643 to 1189294). The flagellar genes encoding the Fla2 flagella seem to be the native flagellar genes and are also found mainly in a cluster located in a different position of chromosome I (positions 2492976 to 2525148). The genes encoding the filament protein FlaA and the regulatory proteins FlaF and FlbT are located in a plasmid (plasmid A). The Fla2 genes include all the previously characterized genes present in E. coli and Salmonella and also two potential regulatory genes (47).
The Fla2 genes are expressed only in mutants that have been selected to swim in the absence of Fla1 (47); these mutants assemble multiple polar flagella that are powered by H+-dependent motors (48). The detailed structure of the Fla2 flagellum has not yet been described.
In contrast, several studies of the flagellum encoded by the Fla1 genes have been reported. These studies have made evident the differences of this flagellum from its counterparts found in enteric bacteria. In the case of flagellar system 1, it is known that the products of the Fla1 genes assemble a single subpolar flagellum, which rotates in a clockwise (CW) direction to produce smooth swimming. Reorientation of the cell occurs when flagellar rotation stops briefly (49, 50). Most of the genes encoding the structural components of this flagellum show good similarity to their counterparts reported in other bacterial species. Accordingly, the function of these genes is also conserved (51–55). fliL is one of the flagellar genes that have a low similarity to their counterparts and the function of which is not well conserved. In R. sphaeroides, FliL is essential for flagellar rotation of the Fla1 flagellum (56). This Mot− phenotype has also been observed for fliL mutants of Caulobacter crescentus and Silicibacter sp. (57, 58) but not for fliL mutants of Salmonella enterica, Proteus mirabilis, and Pseudomonas putida. For the latter species, the lack of FliL was associated with a Fla− phenotype (59); however, in S. enterica, FliL is required for swarming (60, 61), and in Proteus mirabilis, FliL is involved in swarming differentiation (62).
In R. sphaeroides, we observed that the Mot− phenotype of a fliL mutant strain was suppressed by secondary mutations in motB. Given that these changes affected three residues within the plug segment, it was proposed that FliL could be relevant for the activation of the proton channel (56).
In addition, we have recently characterized a protein that is a new essential component of the flagellar motor (Fla1) of R. sphaeroides. This transmembrane protein, named MotF, is localized at the base of the flagellum in wild-type cells but not in a mutant strain lacking the master regulator of the flagellar hierarchy. Noteworthy, the Mot− phenotype elicited by the absence of motF was also suppressed by point mutations in motB that affect the plug region of the MotB protein and by all the motB alleles isolated as second-site suppressors of ΔfliL3::aadA. Therefore, it was proposed that FliL and MotF together could participate in the activation of the proton channel. However, experimental evidence showed that these proteins do not interact with each other or with MotB (56, 63).
In this work, we describe a distant homologue of FlgT present in the Fla1 flagellum of R. sphaeroides. This protein forms a ring that is similar to the H ring previously observed in basal bodies of Vibrio alginolyticus (64). In addition, we show evidence of a direct interaction of FlgT with MotB, FliL, and probably MotF, explaining the Mot− phenotype of flgT cells. The role of FlgT in the recruitment of the stator complexes around the flagellar motor is discussed.
MATERIALS AND METHODS
Plasmids, bacterial strains, and growth conditions.
Plasmids and bacterial strains used in this work are listed in Table 1. R. sphaeroides WS8 (65) was grown in Sistrom's minimal medium (66) at 30°C in the dark with shaking at 200 rpm. Escherichia coli was grown in LB medium at 37°C. Swimming assays were carried out with bacteria grown in liquid medium or on swimming plates with Sistrom's medium and 0.2% agar. When required, the following antibiotics were added: kanamycin (25 μg/ml), tetracycline (1 μg/ml), and spectinomycin (50 μg/ml) for R. sphaeroides, and kanamycin (50 μg/ml), spectinomycin (50 μg/ml), and ampicillin (100 μg/ml) for E. coli. Saccharomyces cerevisiae was grown at 30°C in YPDA culture medium (1% yeast extract, 2% peptone, 2% dextrose, and 0.003% adenine) or in synthetic defined (SD) minimal medium (Clontech) complemented with the appropriated supplements.
Table 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s) | Source or reference |
|---|---|---|
| Strains | ||
| E. coli | ||
| LMG194 | Protein expression strain | Invitrogen |
| Rosetta | Protein expression strain | Novagen |
| S17-1 | recA endA thi hsdR RP4-2 Tc::Mu::Tn7 Smr | 70 |
| TOP10 | Cloning strain | Invitrogen |
| R. sphaeroides | ||
| FS3 | ΔfliL3::aadA | 56 |
| SF3 | flgT::aadA | This work |
| SF4 | flgT::aadA ΔmotB1::Kan | This work |
| SF5 | flgT::aadA motBsup10 [MotB(S62P)] | This work |
| SF6 | flgT::aadA motBsup11 [MotB(F63S)] | This work |
| SP12 | ΔfleT1::aadA | 85 |
| SP13 | ΔfleQ1::Kan | 85 |
| WS8 | Wild-type strain | 65 |
| S. cerevisiae AH109 | Yeast reporter strain for HIS3, ADE2, and lacZ | Clontech |
| Plasmids | ||
| pBAD/Myc-HisA | Expression vector of His-tagged proteins; Apr | Invitrogen |
| pBAD/Myc-HisA-flgT | Vector expressing FlgT-His6 | This work |
| pGAD-flgT | pGAD derivative expressing GAL4 AD-FlgT | This work |
| pGAD-fliLp | pGADT7 derivative expressing GAL4 AD-FliLp | 56 |
| pGAD-motBp | pGADT7 derivative expressing GAL4 AD-MotBp | This work |
| pGAD-motFp | pGADT7 derivative expressing GAL4 AD-MotFp | This work |
| pGAD-motFpΔ77–98 | pGADT derivative expressing GAL4 AD MotFpΔ77–98 | This work |
| pGAD-T7 | GAL4 activation domain; LEU2 | Clontech |
| pGBD-flgT | pGBD derivative expressing GAL4 BD-FlgT | This work |
| pGBKT7 | GAL4 DNA binding domain; TRP1 | Clontech |
| pJQ200mp18 | Mobilizable suicide vector; Gmr | 68 |
| pPIRL | Plasmid that encodes tRNAs for rare codons; Cmr | 75 |
| pRK415 | Vector used for expression in R. sphaeroides; Tcr | 74 |
| pRK415_flgT | pRK415 expressing FlgT | This work |
| pRK_gfp-fliL | pRK415 expressing the fusion GFP-FliL | 56 |
| pRK_gfp-motF | pRK415 expressing the GFP-MotF fusion | 63 |
| pRKmotBsup2 | pRK415 expressing motBsup2 [MotB(A67E)] | 56 |
| pRKmotBsup4 | pRK415 expressing motBsup4 [MotB(F63L)] | 56 |
| pRKmotBsup5 | pRK415 expressing motBsup5 [MotB(A67D)] | 56 |
| pRKmotBsup6 | pRK415 expressing motBsup6 [MotB(A67T)] | 56 |
| pRKmotBsup8 | pRK415 expressing motBsup8 [MotB(A67G)] | 56 |
| pTZ19R | Cloning vector; Apr | Pharmacia |
| pTZ19R Bam− | pTZ19R derivative without BamHI site | Laboratory collection |
| pTZ_FlgT | flgT cloned into pTZ19R without BamHI site | This work |
| pWM5 | pUC derivative carrying the omega-Spcr cassette | 67 |
Oligonucleotides.
The oligonucleotides used in this work are listed in Table S1 in the supplemental material.
Isolation of mutant strains.
Strain SF3 was obtained by a double recombination event replacing the wild-type allele in WS8 cells by the flgT::aadA allele. For this, flgT was amplified by PCR using the oligonucleotides RSP0035B and ORF12, and the product of this reaction was cloned into pTZ19R Bam−. The resultant plasmid was named pTZ_flgT. The aadA gene was obtained by PCR as an internal portion of the omega-Spcr cassette that removed the known transcriptional termination signals using plasmid pWM5 (67) as the template. The PCR product containing the aadA gene was cloned between the BamHI sites of pTZ_flgT that are located 9 bp apart within the coding region of flgT. Finally, the fragment carrying the flgT::aadA allele was subcloned into pJQ200 mp18 (68) and introduced into WS8 by conjugation (69) with strain S17-1 (70). The double recombination events were selected as described previously. Strain SF4 was isolated by conjugation between SF3 and S17-1 carrying a pJQ200 derivative with the ΔmotB1::Kan allele (56).
Motility assays.
A 4-μl sample of a culture in stationary phase was placed onto the surface of plates containing Sistrom's minimal medium with 0.2% agar. Swimming was evaluated after 48 h of incubation at 30°C in a humidity chamber. Swimming cells were observed by dark-field microscopy using a 3-μl sample of a cell culture in exponential phase (optical density at 600 nm [OD600] = 0.8) grown aerobically in Sistrom's medium.
Detection of FliC in the supernatant and cellular fraction.
R. sphaeroides was grown heterotrophically until exponential phase. When the culture reached an OD600 of 0.8, a sample of 1 ml was transferred into an Eppendorf tube; after strong vortexing, the sample was centrifuged at 13,000 rpm. The supernatant was subjected to an additional step of centrifugation before the soluble protein was precipitated by the addition of trichloroacetic acid (TCA) to a final concentration of 25%. Soluble proteins were precipitated for 15 min at 4°C, followed by centrifugation at 13,000 rpm for 15 min at 4°C. The cellular pellet and the TCA-precipitated protein were resuspended in 100 μl and 20 μl of sample buffer, respectively. Ten microliters of these samples was analyzed by Western blotting using anti-FliC antibodies.
Detection of FlgE in the supernatant and cellular fraction.
R. sphaeroides cells were grown heterotrophically until exponential phase. When the culture reached an OD600 of 0.8, a sample of 1.5 ml was transferred into an Eppendorf tube. The sample was centrifuged at 3,000 rpm for 5 min twice. The supernatant was subject to an additional step of centrifugation at 13,000 rpm. Soluble proteins were precipitated as described above. The cellular pellet and the TCA-precipitated protein were resuspended in 150 μl and 15 μl of sample buffer, respectively. Ten microliters of these samples was analyzed by Western blotting using anti-FlgE antibodies.
Tertiary structure prediction of FlgT.
The I-Tasser Web server was used to generate potential structural models of FlgT (http://zhanglab.ccmb.med.umich.edu/I-TASSER/) (71–73).
Plasmid constructs used in this work.
pRK_flgT was obtained by cloning flgT into pRK415 (74). For this, flgT was amplified by PCR using the oligonucleotides pRK6086F and pRK6086R. The 1,360-bp product was then cloned into pRK415 as an XbaI-EcoRI fragment. In this construct, flgT is transcribed from the lac promoter (lacp) present in pRK415. To obtain pBAD/Myc-HisA-flgT, a PCR product that encompasses nucleotide 61 to 1083 was obtained by using oligonucleotides pBADmyc6086SPS-fw and pBADmyc6086SPS-rv. The product of 1,023 bp was purified and cloned into pBAD/Myc-HisA. The resultant plasmid expresses the mature protein FlgT (amino acids 21 to 361) fused to a C-terminal His6 tag. The plasmids used for the double-hybrid experiments are described in “Yeast two-hybrid assay” below.
Protease sensitivity assay.
The protease sensitivity assay was performed as described previously (63).
Protein overexpression and purification.
E. coli strain LMG194 carrying plasmid pPIRL (75) was transformed with pBAD/Myc-His-flgT. A culture of this strain grown overnight was diluted 1:100 in fresh medium and incubated at 37°C until it reached an OD600 of 0.5. At this moment, arabinose was added to a final concentration of 0.2%. After a further 3 h of incubation, cells were collected and resuspended in TGED buffer (0.01 M Tris-HCl [pH 7.9], 15% glycerol, 0.1 mM EDTA, 1 mM dithiothreitol [DTT]). Lysozyme was added (0.2 mg/ml final concentration), and the mixture was incubated for 30 min on ice. The cell suspension was sonicated on ice using three cycles of 10 s. Cell debris were removed by at least three steps of centrifugation (14,000 rpm for 5 min). The supernatant was mixed with nickel-nitrilotriacetic acid (Ni2+-NTA) agarose beads and incubated for 1 h on ice, with the tube being inverted sporadically. The mixture was used to load a propylene column and washed with 50 volumes of TGED buffer with 10 mM imidazole. The protein was eluted by using TGED buffer with 100 mM imidazole. Glutathione S-transferase fused to the periplasmic domain of MotB (GST-MotBp) and GST-FliLp were purified by using plasmids and protocols previously reported (56).
FlgT antibodies.
Polyclonal antibodies were raised in female BALB/c mice against FlgT-His6, as described previously (76).
Immunoblotting.
Proteins were separated by SDS–12% PAGE (77), blotted onto nitrocellulose, and tested with the appropriate antibody after blocking in 5% nonfat milk–phosphate-buffered saline (PBS) (76). The membrane was incubated for 2 h with anti-FliC (1:30,000), anti-FliH (1:5,000), anti-GST (1:20,000) (Pierce/GE), anti-CheY3 (1:10,000), anti-GFP (1:5,000), anti-FlgE (1:10,000), anti-MotF (1:5,000), or anti-FlgT (1:5,000) gamma globulins. Detection was carried out with a secondary antibody conjugated to alkaline phosphatase and CDP-Star (Applied Biosystems). The amount of protein loaded for each blot is indicated for each experiment.
Pulldown assays.
According to previously reported protocols (78), 13 μg of GST-FliLp, 16 μg of GST-MotBp, or 8 μg of GST bound to glutathione agarose beads in PBS (pH 7.4) was mixed with FlgT-His6 to yield a 1:1 molar ratio of GST-FliLp, GST-MotBp, or GST to FlgT-His6. The total volume was adjusted to 250 μl with PBS (pH 7.4), and glycerol was added to a final concentration of 10%. The mixture was incubated for 2 h at 4°C with constant agitation. After incubation, the beads were collected by centrifugation (1 min at 3,000 rpm), and the supernatant was carefully removed. The beads were washed with 1 volume of PBS (pH 7.4). Finally, 100 μl of elution buffer (10 mM reduced l-glutathione, 100 mM Tris-HCl [pH 8]) was added, and after 10 min, the sample was centrifuged. An aliquot of 15 μl of the supernatant was loaded onto a 12% SDS-PAGE gel to be analyzed by Western blotting using anti-FlgT antibodies. A sample of 10 μl was analyzed with anti-GST antibodies.
Yeast two-hybrid assay.
Matchmaker GAL4 two-hybrid system 3 (Clontech) was used to test the interactions between FlgT and FlgT, FlgT and MotBp, FlgT and FliL, FlgT and MotFp, and FlgT and MotFpΔ77–98. The region encoding the mature FlgT polypeptide was amplified by PCR using the oligonucleotides 6086DHFW and 6086DHRv. The product of this reaction was cloned into pGBKT7, which encodes the DNA binding domain (BD) of GAL4. The resultant plasmid, pGBD-flgT, expresses FlgT fused to the BD of GAL4. The oligonucleotides 6086DHFW and GAD6086Rv amplified the same region of flgT, but this PCR product was cloned into pGADT7, which encodes the activation domain (AD) of GAL4. The resultant plasmid, pGAD-flgT, expresses FlgT fused to the AD of GAL4. Two versions of MotFp were fused to the AD of GAL4. The first one carries the complete MotFp (residues 75 to 239). To obtain this plasmid, a region of motF was amplified by using oligonucleotides DH67CoilF and RSP_0067-GST Rvs STOP, and the amplification product was then cloned into pGADT7 to obtain plasmid pGAD-MotFp. motF was also amplified by using oligonucleotides pG67FCOOH and RSP_0067-GST Rvs STOP; in this case, the region encoding residues 77 to 98 was excluded. The PCR product was then cloned into pGADT7. The resultant plasmid was called pGAD-MotFpΔ77–98. Plasmid pGAD-motBp was constructed by amplifying the DNA region encoding the periplasmic segment of MotB using oligonucleotides ADBDmotB1 and ADmotB2. The product of this reaction was cloned into pGADT7. Interactions were examined by introducing the plasmids expressing the proteins to be tested into the reporter strain AH109. The double transformants were selected as tryptophan (Trp) and leucine (Leu) prototrophs. Transformants were grown overnight in SD minimal medium without Leu and Trp but supplemented with histidine (His) and adenine (Ade). Aliquots of the cultures were washed once with SD minimal medium without supplements and then normalized to an OD600 of 0.5. Immediately, 10-fold serial dilutions were made in the same medium. From these dilutions, 10-μl aliquots were seeded onto selection plates lacking Trp, Leu, and His or lacking Trp, Leu, His, and Ade.
Microscopy.
A sample of an exponentially growing culture was placed onto a slide with an agarose pad containing Sistrom's medium. Epifluorescence images were taken by using a Nikon Eclipse 600 microscope equipped with a Hamamatsu Orca-ER cooled charge-coupled-device (CCD) camera, and images were acquired for 3 s.
Transmission electron microscopy (EM).
Cells were removed from an exponentially growing culture and collected by centrifugation at 3,000 rpm. The cell pellet was gently resuspended in one-third of the original volume of Tris-EDTA (TE) buffer. An aliquot was placed onto the surface on a carbon-coated grid and stained with 1% phosphotungstic acid (pH 7). Filament-hook-basal body samples were prepared as previously described (79), negatively stained with 2% phosphotungstic acid (pH 7.0), and observed with a JEM-1200EXII electron microscope (JEOL, Tokyo). Micrographs were taken with an 11-Mp Gatan digital camera at an accelerating voltage of 80 kV.
RESULTS
Deletion of RSP_6086 impairs flagellar motility.
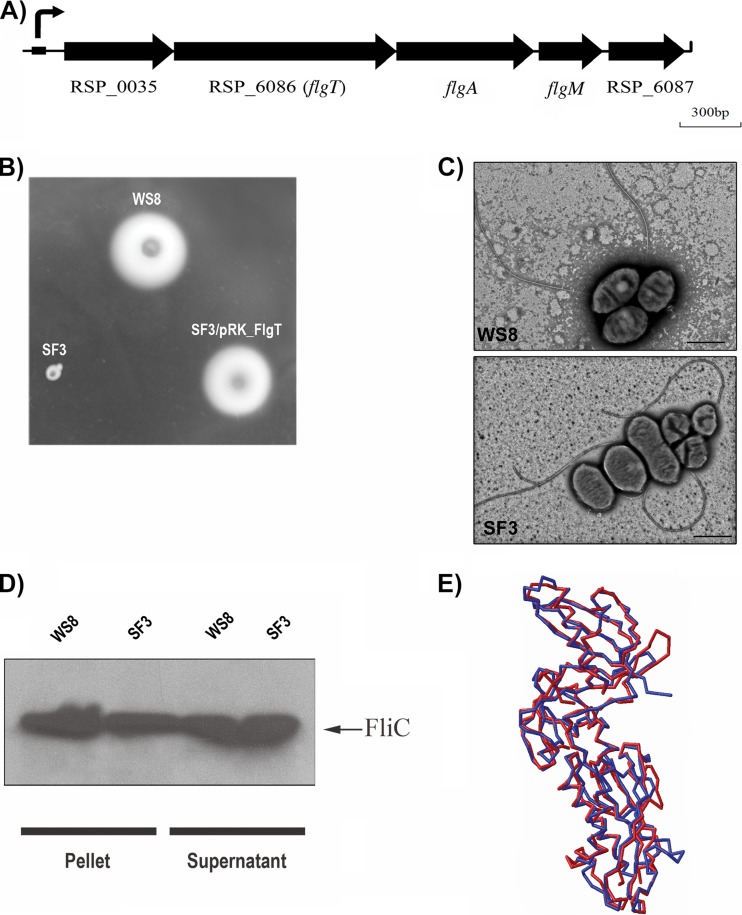
In the genome of R. sphaeroides, RSP_6086 is located within a cluster of flagellar genes and is flanked by flgA and RSP_0035. As shown in Fig. 1A, RSP_6086 is part of a putative operon that also includes flgM and RSP_6087. The product encoded by RSP_0035 is similar to the flagellar protein FlgP that has been characterized in Campylobacter jejuni and Vibrio cholerae (80, 81). In a BLASTP search, RSP_6086 shows high values of similarity only with hypothetical proteins present in other strains of R. sphaeroides and a few species of the Rhodobacteraceae family. However, PSI-BLAST of the protein encoded by RSP_6086 against the nr protein database retrieved FlgT from Pseudoalteromonas sp. and other hypothetical proteins. A second iteration retrieved the FlgT protein from V. alginolyticus. These results suggest that this protein could be a distant homologue of FlgT so that a simple alignment does not reveal the relationship between them (see Fig. S1 in the supplemental material). Nevertheless, a prediction of the tertiary structure of RSP_6086 or R. sphaeroides FlgT (FlgTRs), as we have named it in this work, shows a structure very similar to that of FlgT from V. alginolyticus recently elucidated at a 2-Å resolution (82) (Fig. 1E).
Fig 1.
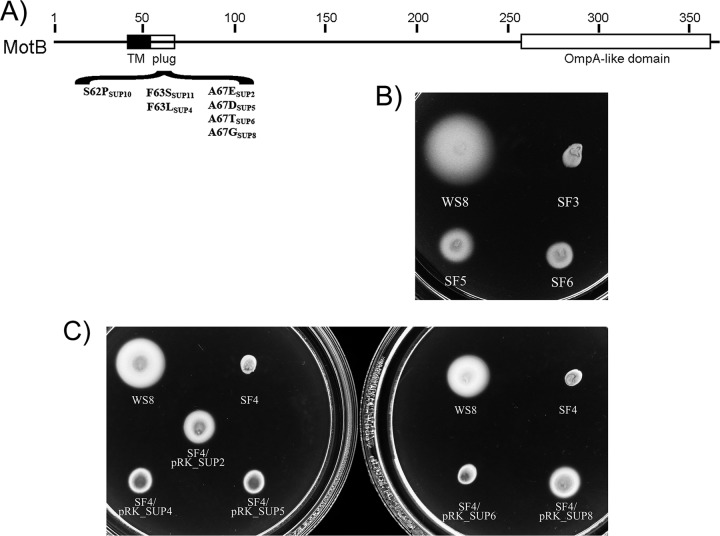
Gene context, phenotype of the SF3 (flgT::aadA) mutant strain, and tertiary structure comparison with FlgT from V. alginolyticus. (A) Gene arrangement of the flagellar operon containing RSP_6086 (flgT). The arrows indicate the direction of gene transcription. The black box indicates the regulatory region that contains the sigma 54-dependent flgAp promoter characterized previously (85). The arrow above the black box symbolizes transcription from this promoter. (B) Swimming plate inoculated with the indicated strains. (C) Electron micrograph of WS8 and SF3 cells showing the presence of flagellar filaments. Bar = 1 μm. (D) Supernatant and pellet fractions obtained after strong vortexing of WS8 and SF3. Samples were analyzed by immunoblotting using anti-FliC antibodies. (E) Tertiary structure prediction of FlgTRs (red line) superimposed with the structure of FlgT from V. alginolyticus (82) (blue line).
Given that FlgT has so far been found only in bacterial species carrying other flagellar components that are absent in R. sphaeroides, i.e., MotX and MotY, and that this flagellar gene cluster was acquired by horizontal gene transfer, we tested if the gene encoding FlgT plays a role in the flagellar motility of R. sphaeroides. For this, the chromosomal copy of flgT was inactivated by the insertion of a nonpolar cassette that confers spectinomycin resistance. The resultant strain, named SF3, was unable to swim when observed under a microscope or when it was inoculated onto soft-agar plates (Fig. 1B). Motility was fully recovered when a plasmid expressing flgT was introduced into SF3 cells (Fig. 1B).
Visualization of SF3 cells by transmission electron microscopy showed the presence of a single subpolar flagellum that is similar in length and appearance to that formed by WS8 cells (Fig. 1C). Furthermore, Western blot analyses showed that the amount of extracellular flagellin produced by SF3 is comparable to the amount detected for the wild-type strain (Fig. 1D). To determine if there is a defect in flagellar assembly or if the flagellum is being released into the culture medium, as occurs in V. alginolyticus and V. cholerae, respectively (64, 83), we counted the number of cells with attached flagellum after 4′,6-diamidino-2-phenylindole (DAPI) staining. Also, we detected the amount of FlgE present in the supernatant of cell cultures from wild-type or flgT cells. We did not notice substantial differences in the amounts of FlgE in the culture medium (see Fig. S2 in the supplemental material) or in the amounts of cells with attached flagellum between the wild-type strain and SF3 (data not shown), indicating that the phenotype of the flgT mutant in R. sphaeroides is different from the phenotypes previously reported for V. cholerae and V. alginolyticus.
Together, these results indicate that FlgT is required for flagellar rotation in R. sphaeroides.
FlgT is a periplasmic protein.
Analysis of the primary sequence of FlgT with SignalP (84) revealed a sequence resembling a signal peptide (SP) at the N terminus. To test if the localization of FlgT is indeed periplasmic, we carried out a protease sensitivity assay. Polyclonal antibodies were raised against the purified FlgT-His6 protein. As shown in Fig. 2A, FlgT was clearly identified in spheroplasts obtained from wild-type cells. In the presence of proteinase K (PK), the band corresponding to FlgT disappeared, indicating that the protein was hydrolyzed by the action of the enzyme. As a control, using anti-FliH antibodies, we verified that in the same samples, the amount of the cytoplasmic protein FliH did not change regardless of the presence of proteinase K (Fig. 2A).
Fig 2.
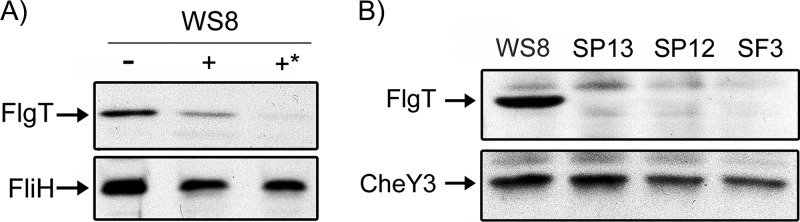
Subcellular localization and expression of FlgT in different strains. (A) The presence of FlgT in the periplasm of wild-type cells was analyzed by using a protease sensitivity assay. For this, spheroplasts from 15 ml of WS8 cells growing exponentially (OD600 = 0.8) were obtained by treatment with lysozyme and EDTA, and spheroplasts were incubated in the presence of proteinase K (100 μg/ml) for 20 min (+) and 40 min (+*). A control without proteinase K was also included and incubated with Tris buffer for 40 min (−). The resulting samples containing 5 μg of protein were analyzed by Western blotting using specific antibodies. As a control, the same samples were tested for the presence of the cytoplasmic protein FliH. (B) Five micrograms of total cell extracts of the indicated strains was analyzed by Western blotting using anti-FlgT or anti-CheY3 antibodies.
FlgT is expressed within the flagellar hierarchy.
To establish if flgT belongs to the flagellar hierarchy, we carried out a Western blot assay to detect FlgT in strains SP12 (ΔfleT1::aadA) and SP13 (ΔfleQ1::Kan). In R. sphaeroides, FleQ is the master regulator of the flagellar hierarchy and activates the expression of the class II operon fleT-fliFGHIJ. FleT, together with FleQ, activates the class III genes, which include most of the genes encoding the basal body, the export apparatus, the hook, and the flagellar regulators FliA (sigma 28 factor) and FlgM (anti-sigma 28 factor) (85).
As shown in Fig. 2B, FlgT was detected in the cell extract from the wild-type strain but not in that from strain SP13 or SP12, suggesting that flgT belongs to the flagellar hierarchy and that its expression is dependent on the transcriptional activators FleQ and FleT. The same extracts were tested by using an anti-CheY3 antibody as a control. It is known that cheY3 is importantly expressed from a σ70 promoter (86); therefore, the amount of this protein should be similar between these strains, as shown in Fig. 2B. In addition, to rule out that FlgT could be unstable in the SP13 and SP12 backgrounds, we carried out a Western blot assay of total cell extracts from strains SP13 and SP12 expressing flgT from the lac promoter present in pRK415. No differences in the amount of FlgT were detected between these strains and WS8 (see Fig. S3 in the supplemental material).
FlgT interacts with the periplasmic regions of MotB and FliL.
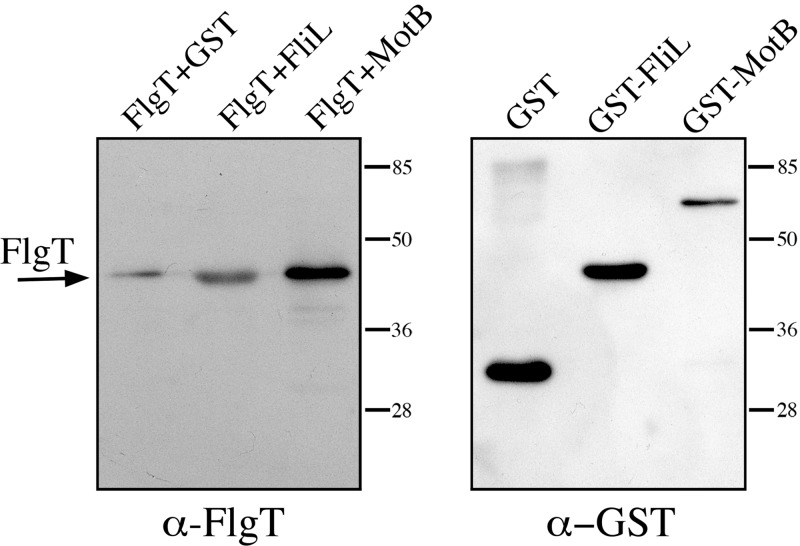
Since the phenotype of the SF3 strain is Mot−, we hypothesized that other flagellar proteins also involved in flagellar rotation, such as MotB, FliL, and MotF, could interact with FlgT. To test this idea, we carried out pulldown assays using the periplasmic regions of the transmembrane proteins MotB and FliL, both fused to GST (glutathione S-transferase), as bait and FlgT-His6 as prey. As a negative control, we used GST. The assay revealed a positive interaction between FliL and FlgT and also between MotB and FlgT (Fig. 3).
Fig 3.
FlgT interactions tested by pulldown. Shown are pulldowns of FlgT-His6 with GST alone (29 kDa), GST-FliL (48 kDa), and GST-MotB (63 kDa). After coprecipitation, the sample was divided in two and probed with anti-FlgT and anti-GST antibodies by immunoblotting.
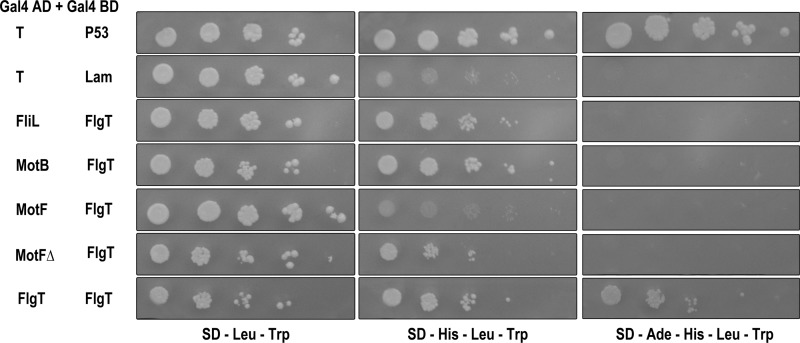
The interactions detected by pulldown experiments were confirmed by using a yeast two-hybrid assay. For this experiment, the mature form of FlgT was fused to the DNA binding domain of the yeast transcriptional activator GAL4 (BD-FlgT), whereas the periplasmic domains of FliL and MotB (FliLp and MotBp, respectively) were fused to the activation domain of GAL4 (AD-FliLp and AD-MotBp, respectively).
For this assay, the plasmids expressing the proteins to be tested were cotransformed into the reporter strain AH109. In this strain, HIS3 and ADE2 genes are under the positive control of GAL4. A positive interaction brings together the AD (activation) and BD (DNA binding) domains of GAL4 and, as a consequence, the expression of HIS3 and ADE2, which, depending on the strength of the interaction, allows growth of AH109 cells in the absence of histidine and/or adenine.
As shown in Fig. 4, the AH109 strains coexpressing AD-MotBp and BD-FlgT or coexpressing AD-FliLp and BD-FlgT were able to grow in the absence of histidine but not adenine. This result confirms that FlgT interacts with MotBp and FliLp but suggests that these interactions are relatively weak.
Fig 4.
FlgT interactions tested by double-hybrid assays. AH109 yeast cells were transformed with the plasmids indicated on the left. The pair AD-T and BD-P53 and the pair AD-T and BD-Lam are the positive and negative controls, respectively. Serial dilutions of cultures of the transformant cells were inoculated onto plates containing the growth medium indicated at the bottom, which is synthetic medium (SD) lacking leucine (−Leu), tryptophan (−Trp), histidine (−His), or adenine (−Ade). Pictures were taken after 10 days of incubation at 30°C.
Using the same assay, we tested if FlgT could interact with itself. For this, we constructed the plasmid encoding the AD-FlgT fusion, and it was cotransformed with the plasmid encoding BD-FlgT into AH109 stain. The resulting strain was able to grow in the absence of histidine and adenine, indicating a strong interaction (Fig. 4).
FlgT interacts with a fragment of MotF.
We have recently reported that in R. sphaeroides, besides FliL and MotA/B, the presence of the flagellar protein MotF is required for flagellar rotation. MotF is an inner membrane protein that has a long periplasmic region (63). To test the interaction of FlgT with MotF, we cloned the periplasmic domain of MotF (MotFp) fused to the activation domain of GAL4 (AD-MotFp). The plasmid expressing this fusion protein was cotransformed with the plasmid expressing BD-FlgT into AH109. The resultant yeast strain was unable to grow in the absence of histidine and adenine, indicating that these proteins do not interact (Fig. 4). A version of MotF carrying a deletion of a region adjacent to the transmembrane domain (MotFpΔ77–98) was also tested in this assay. It was previously observed that this region of MotF is essential for flagellar rotation and also for the proper localization of the fusion protein GFP-MotF (63). As shown in Fig. 4, the AH109 strain coexpressing AD-MotFpΔ77–98 and BD-FlgT was able to grow in the absence of histidine (Fig. 4), suggesting that MotF interacts with FlgT and that residues 77 to 98 hinder this interaction.
FlgT stabilizes the localization of MotF.
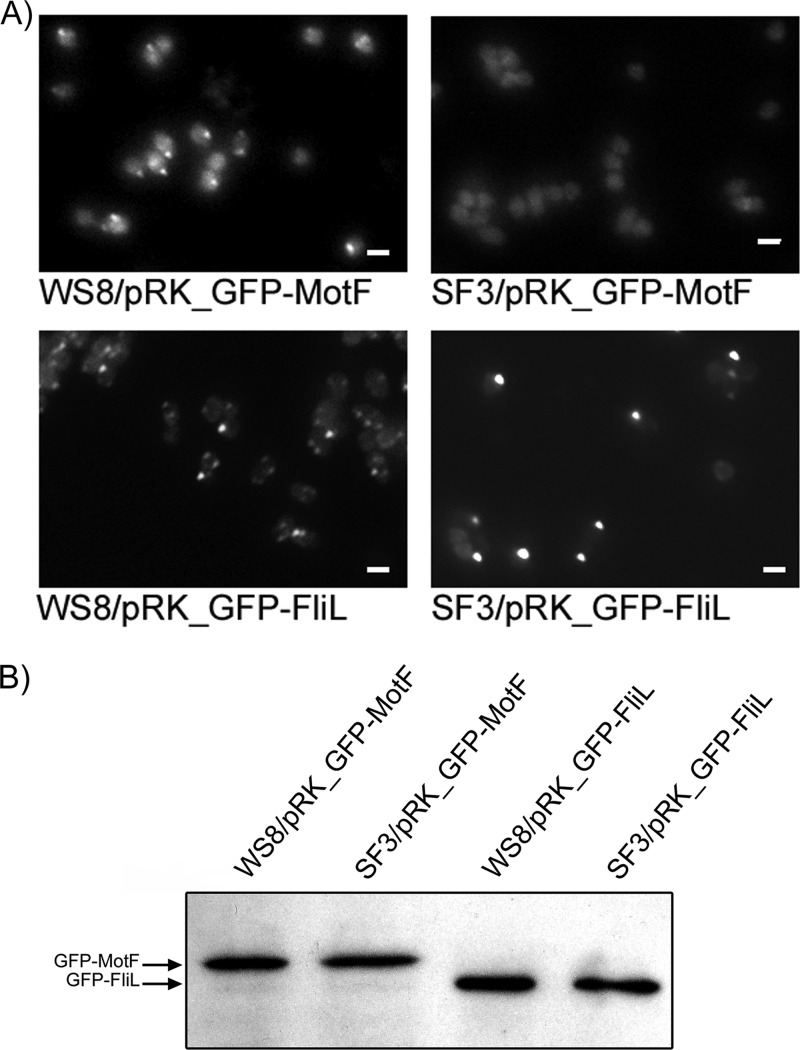
The positive interaction between a fragment of the periplasmic regions of MotF and FlgT, as well as the interaction detected between FlgT and FliLp, prompted us to evaluate the localization of these proteins in vivo. Our efforts to obtain a fluorescent version of FlgT were not successful given that FlgT-mCherry was indistinguishable from the intrinsic fluorescence of the cells, and the FlgT-cyan fluorescent protein (CFP) fusion in which the SP of FlgT was replaced by a sequence predicted to be recognized by the twin-arginine translocation pathway did not show a periplasmic fluorescence pattern. Nevertheless, we proceeded to determine the localization of GFP-MotF and GFP-FliL in the absence of FlgT (strain SF3). As shown in Fig. 5A, GFP-FliL mainly formed a single fluorescent focus in wild-type and SF3 cells. This pattern is similar to what has been previously reported (56), indicating that FliL localizes regardless of the presence of FlgT. In contrast, GFP-MotF formed a single fluorescent focus in the wild-type strain, in accordance with previous observations (63), but no fluorescent foci were detected in SF3 cells, suggesting that FlgT is required to recruit MotF to the base of the flagellum. An image with the outlines of the cell bodies was included to help visualize the cell contours (see Fig. S4 in the supplemental material). To confirm the proper expression of GFP-MotF in strain SF3, we tested its presence by Western blotting. As shown in Fig. 5B, in SF3 cells, both GFP-FliL and GFP-MotF are present in amounts similar to those detected in WS8 cells.
Fig 5.
GFP-MotF and GFP-FliL localization in strain SF3. (A) Representative images of GFP-MotF and GFP-FliL in WS8 and SF3 cells. Bar = 1 μm. (B) Western blot analysis of GFP-FliL and GFP-MotF expressed in WS8 and SF3 strains. A total cell extract containing 5 μg of protein was subjected to SDS-PAGE and tested by Western blotting using anti-GFP antibodies.
Pseudorevertants of flgT map in motB.
Suppressor mutants provide indirect but important information about the function of a protein. To obtain a better notion about the role of FlgT in flagellar rotation, we isolated two pseudorevertant strains that reestablish the swimming ability of SF3 cells. Soft-agar plates were inoculated with independent cultures of this strain and incubated in a humidity chamber for 10 to 14 days. After this time, a small halo appeared around the inoculation point. The cells from these halos were purified, and their swimming ability was tested; we noticed that both suppressor strains SF5 and SF6 were able to swim, although the expansion halo was only approximately 50% of that of the wild type (Fig. 6A). Microscopic observations of these mutants showed that, in comparison with the wild type, only 15 to 20% of the cells were motile and that cells swam at a reduced speed. SF5 showed a swimming speed of 8.6 ± 3.3 μm/s, and SF6 showed a swimming speed of 9.9 ± 3.8 μm/s, whereas the wild-type strain showed a swimming velocity of 29 ± 4.1 μm/s. To identify the mutation that enabled these strains to recover a Mot+ phenotype, we sequenced motA, motB, fliM, fliN, and fliG. By comparing these sequences with those of the wild type, we found a single change in the coding sequence of motB in both suppressor strains. The point mutations replaced serine and phenylalanine residues at positions 62 and 63 with proline and serine, respectively.
Fig 6.
Mutations in MotB promote the swimming of SF3. (A) Map of the coding region of MotB indicating relevant features. Black box, transmembrane domain (TM); open box with a middle black line, plug region; open box, OmpA-like domain. The changes in each suppressor are indicated. (B) Swimming plate inoculated with strains WS8, SF3, SF5, and SF6. (C) Swimming plates inoculated with WS8, SF4, and SF4 expressing the indicated mutant allele of motB.
It should be noted that both mutations affect the region of MotB known as the plug. For R. sphaeroides, it was proposed previously that the plug region comprises amino acids 56 to 67 of MotB (56). Several mutations isolated previously as suppressors of the Mot− phenotype of the ΔfliL3::aadA and motF::aadA alleles also mapped in this region. It was proposed previously that these changes modify the plug and, as a consequence, keep the proton channel open, making the presence of any other flagellar protein required to promote a conformational change in MotB unnecessary (56, 63).
Hence, if FlgT also participates in the activation of the proton channel, the motB versions isolated as suppressor mutants of the ΔfliL3::aadA allele should also rescue the swimming phenotype of strain SF4 (flgT::aadA ΔmotB1::Kan). To test this idea, of the eight mutant versions of motB isolated as suppressor alleles of fliL, five were expressed in SF4, from the lacp promoter of pRK415. It should be noticed that we did not test SUP1 and SUP7 because they show a change in the same residues as SUP2 and SUP4, respectively; SUP3 was not included since this allele barely supports swimming of strain FS3 (ΔfliL::aadA) (56). We believe that the function of MotB is compromised by the change present in SUP3 [MotB(A56E)], given that it maps in the boundary of the predicted TM helix. As shown in Fig. 6B, the mutant versions of motB restore swimming of SF4 cells, but the expansion halo was severely reduced compared to that of wild-type strain WS8. Inspection of these strains under a microscope showed that, as occurs with the original SF3 suppressor strains, only a fraction of the population of the cells swam and at a reduced speed. Therefore, we conclude that although the motB suppressor alleles promote swimming of SF4 cells, suppression is not as efficient as it is for the ΔfliL3::aadA and motF::aadA alleles (56, 63), suggesting that FlgT should have an additional role beyond the process of MotB activation.
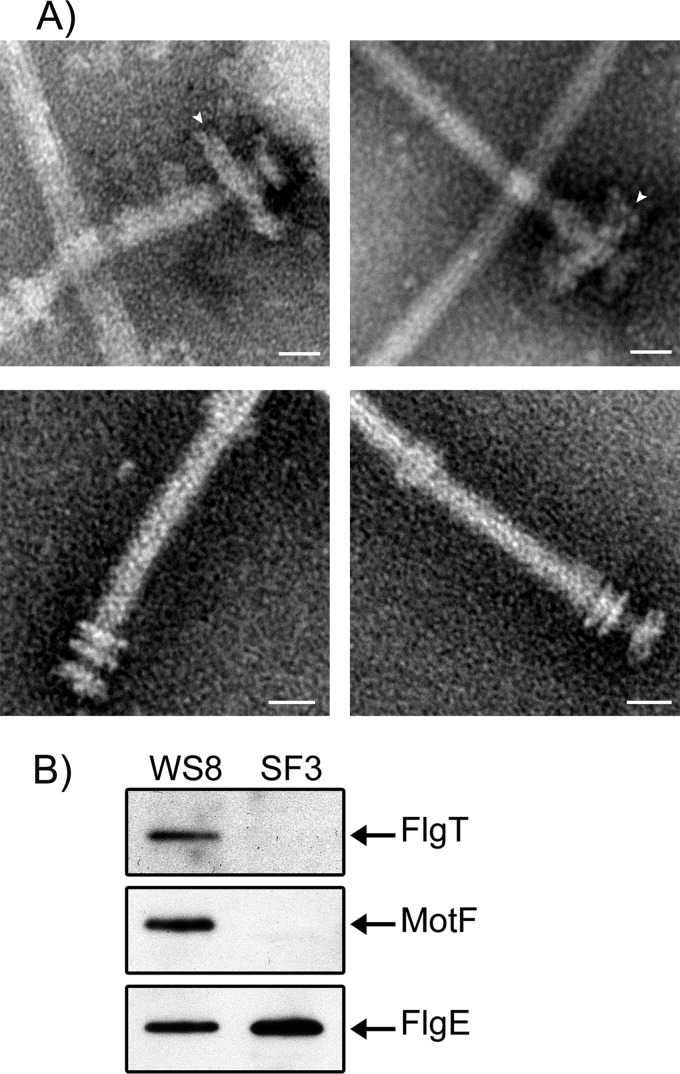
FlgT is required to form a ring that covers the LP rings.
We isolated flagella from wild-type WS8 and SF3 cells, and the purified flagella were analyzed by electron microscopy (EM). Figure 7A (top) shows WS8 basal bodies with a large, seemingly two-layered ring. This large ring is missing in SF3 basal bodies, which are shown in Fig. 7A (bottom). It should be noted that the LP ring is now visible. The diameter of the large ring in the basal bodies of wild-type cells was approximately 64 ± 10.7 nm, whereas the diameter of the LP rings detected in the basal bodies of SF3 cells was 24 ± 1 nm. Western blot analyses carried out with these samples showed the presence of FlgT in basal bodies of WS8 but not in samples from SF3 (Fig. 7B). The same samples were tested with anti-MotF and with anti-FlgE antibodies as a loading control. MotF was detected only in samples obtained from WS8 (Fig. 7B), suggesting that this protein is an integral part of the basal body of R. sphaeroides and that it is stabilized by FlgT.
Fig 7.
Electron micrographs and immunoblot analysis of purified filament-hook-basal bodies from strains WS8 and SF3. (A) Images of purified HBBs from strains WS8 (top) and SF3 (bottom). A white arrow denotes the H ring. Bar = 20 nm. (B) Western blot analysis of the purified basal bodies from WS8 and SF3 samples. One-tenth of the total sample (approximately 10 μg of total protein) of purified HBBs was analyzed by immunoblotting using specific antibodies.
The same ring covering the LP ring was also present in basal bodies of strain VR1 (motF::aadA); the presence of FlgT in this sample was confirmed by Western blotting (data not shown). This result suggests that the absence of MotF does not affect the recruitment of FlgT in the basal body or the formation of this structure.
DISCUSSION
R. sphaeroides has several ORFs of unknown function embedded in the Fla1 cluster. In this work, we show that one of these ORFs, RSP_6086, encodes a protein essential for flagellar rotation. This protein is conserved in all species of R. sphaeroides sequenced so far and also in some other species from the Rhodobacteraceae family. A more sensitive search using PSI-BLAST showed that RSP_6086 encodes a protein that seems to be a distant homologue of the FlgT protein previously reported for V. cholerae and V. alginolyticus (64, 87). FlgTRs is only 17% identical to FlgT from V. alginolyticus (see Fig. S1 in the supplemental material). Nevertheless, a comparison of their tertiary structures shows a good match between them (TM score, 0.91) (Fig. 1E). The few differences between them may represent specific adaptations of FlgTRs. In addition, the ring observed in the basal bodies of wild-type R. sphaeroides strain WS8 is highly similar in size and appearance to the H ring of V. alginolyticus (64). These facts support the idea that RSP_6086 is indeed a homologue of FlgT.
FlgT has been found in Vibrio and several species of Shewanella; these bacteria belong to the Gammaproteobacteria class. Although this is the first report of a structure of this type in a bacterium that belongs to a different class, it should be noted that FlgT is a component of the Fla1 system, which was acquired by horizontal gene transfer from a gammaproteobacterium (47). Nonetheless, this is the first time in which an exclusively proton-energized motor includes the H ring. It was reported previously that Shewanella oneidensis MR-1, which encodes FlgT in its genome (GI:24374769), simultaneously uses MotAB and PomAB stators, but only the latter are essential for swimming (88).
So far, FlgT has been identified in bacteria having motX and motY genes; in fact, it has been shown that the H ring stabilizes the presence of MotX and MotY in the basal body of V. alginolyticus (64, 82). R. sphaeroides does not have genes similar to motX and motY; therefore, in R. sphaeroides, FlgT could be important for recruiting other proteins such as MotF, MotB, and possibly other proteins with unknown function. In addition, it is possible that in the absence of MotX and MotY, FlgT could have evolved at a higher rate, leading to its low degree of conservancy.
The isolation of purified hook-basal bodies of R. sphaeroides was previously reported; however, the structure of the H ring was overlooked (89, 90).
Our results suggest that flgT expression is dependent on the FleT and FleQ flagellar activator proteins, placing this gene within the flagellar hierarchy either as a class III gene or within the FliA-dependent class IV. However, it seems to be the second gene of an operon that starts with the putative flgP gene (previously reported as the flgA operon). Since the promoter of this operon has been shown to be part of class III, flgT should also belong to this flagellar class (85).
We show that in the absence of FlgT, R. sphaeroides cells are able to assemble flagella but are paralyzed, indicating that this protein should be related to motor function. In contrast, in V. alginolyticus and V. cholerae, the absence of flgT reduces both flagellation and motility (64, 82, 87), indicating that the function of these proteins has diverged.
In line with the idea that FlgTRs is required for motor function, we found that FlgT is able to interact with the periplasmic domains of MotB and FliL, indicating a direct role in the activation and/or recruitment of the stator complexes. In addition, we obtained evidence suggesting that FlgT also interacts with MotF. This result was supported by the requirement of FlgT to recruit MotF to the flagellar basal body. Previously, it was proposed that FliL and MotF could participate in the opening of the proton channel; however, no interaction could be detected between the periplasmic regions of FliL and MotF and the periplasmic region of MotB (56, 63). Our results allow us to propose that FliL and MotF promote the opening of the proton channel through FlgT.
The interaction between FlgT and the periplasmic region of MotF in the double-hybrid assay could be detected only when a version of MotF lacking amino acids 77 to 98 was used. Given that other results support the interaction between these proteins, the negative effect that this part of MotF exerts on the FlgT-MotF interaction may reflect an artifact caused by the conformation that MotF acquires in the absence of the flagellar-basal body environment or some form of regulation of the interaction between these two proteins.
The location of the MotF protein within the flagellar structure is unknown; however, our results suggest that MotF is part of the basal body and presumably, due to its interaction with FlgT, must be part of the nonrotating components of the flagellum.
We also show that single mutations in motB rescue the swimming ability of SF3 cells to some degree. These changes in MotB mapped to the region immediately after the TM region. Several mutations that we isolated previously as suppressors of the Mot− phenotype of the ΔfliL3::aadA and motF::aadA alleles also mapped to this region. It has been proposed that these changes modify the plug, and as a consequence, the proton channel remains open, hence making the presence of any other protein that promotes a conformational change in MotB dispensable (56, 63). The possibility that FlgT could be required only to open the proton channel seems unlikely, since the motB mutant alleles do not suppress the Mot− phenotype of flgT cells efficiently. We previously determined that the motB mutant alleles that act as second-site suppressors of ΔfliL3::aadA can efficiently restore swimming of the motF::aadA strain (63), suggesting that these mutations compensate for the swimming defect caused by the absence of fliL and motF through an unspecific mechanism. In this work, we show that the motB suppressors of the ΔfliL3::aadA allele were able to promote swimming of SF4 cells, but these strains showed a swimming halo that was 70 to 30% smaller than that produced by WS8 cells. This result is in strong contrast with the effect that these mutant versions of motB exerted on the swimming behavior of strains FS5 (ΔfliL3::aadA ΔmotB1::Kan) and VR2 (motF::aadA ΔmotB1::Kan); in these cases, the expression of these alleles promoted a swimming halo that was only 28 to 1% smaller than that of the wild type (56, 63). These results suggest that FlgT could play a role beyond the opening of the proton channel. This function could be related to the recruitment or stabilization of the MotA/B complexes, as suggested by the interaction between FlgT and MotB.
Undoubtedly, the interaction of the stator complex with the flagellar structure should be robust, and other flagellar components might participate in stabilizing the stator complexes. In Salmonella, it has been demonstrated that interactions between MotA and FliG are important for the assembly of the stator complexes (16). In addition, in E. coli, using a disulfide cross-linking approach, it was suggested that FlgI, which forms the P ring, interacts with MotB (91). In V. alginolyticus, colocalization experiments suggested that MotX and MotY could interact with the stator complexes (92, 93). These examples suggest that MotA and MotB interact with the flagellar motor in more than a single way, probably to allow a better performance of the motor or to allow additional levels of control. In R. sphaeroides, MotB could be recruited or stabilized to the flagellar motor through its interaction with FlgT, but other interactions have to exist in order to explain the weak swimming of the flgT strain complemented with the mutant alleles of motB.
In addition, it should be noted that even though FlgT interacts with FliL, the absence of FlgT does not affect the localization pattern of GFP-FliL, indicating that FliL should interact with at least an additional flagellar component that keeps its normal pattern of localization even in the absence of FlgT. This suggests that FliL and MotF may have different roles in the opening of the proton channel.
How FlgT, FliL, and MotF interact with MotB to activate the opening of the proton channel is still largely an open question, and it is possible that other proteins may intervene in this process that different bacteria have adapted to suit their needs.
Supplementary Material
ACKNOWLEDGMENTS
We thank Teresa Ballado, Yael González, and Georgina Díaz Herrera for technical assistance. We also thank Diego González Halphen for very useful suggestions to improve this work. We also thank the IFC Molecular Biology Unit for sequencing facilities as well as Fernando García, who is responsible for the IFC Microscopy Unit.
This work was supported in part by grants from the Consejo Nacional de Ciencia y Tecnología (SEP-CONACYT 106081) and DGAPA/UNAM (IN206811). This work is part of the requisites to obtain a doctoral degree by S.F. (Doctorado en Ciencias Biomédicas, Universidad Nacional Autónoma de México), who was supported during his studies by a fellowship from CONACyT.
Footnotes
Published ahead of print 20 September 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.00760-13.
REFERENCES
- 1.Berg HC. 2003. The rotary motor of bacterial flagella. Annu. Rev. Biochem. 72:19–54 [DOI] [PubMed] [Google Scholar]
- 2.Macnab RM. 2003. How bacteria assemble flagella. Annu. Rev. Microbiol. 57:77–100 [DOI] [PubMed] [Google Scholar]
- 3.Minamino T, Imada K, Namba K. 2008. Molecular motors of the bacterial flagella. Curr. Opin. Struct. Biol. 18:693–701 [DOI] [PubMed] [Google Scholar]
- 4.Terashima H, Kojima S, Homma M. 2008. Flagellar motility in bacteria: structure and function of flagellar motor. Int. Rev. Cell Mol. Biol. 270:39–85 [DOI] [PubMed] [Google Scholar]
- 5.Kojima S, Blair DF. 2004. The bacterial flagellar motor: structure and function of a complex molecular machine. Int. Rev. Cytol. 233:93–134 [DOI] [PubMed] [Google Scholar]
- 6.Yorimitsu T, Homma M. 2001. Na(+)-driven flagellar motor of Vibrio. Biochim. Biophys. Acta 1505:82–93 [DOI] [PubMed] [Google Scholar]
- 7.Li N, Kojima S, Homma M. 2011. Sodium-driven motor of the polar flagellum in marine bacteria Vibrio. Genes Cells 16:985–999 [DOI] [PubMed] [Google Scholar]
- 8.Dibrov PA, Kostryko VA, Lazarova RL, Skulachev VP, Smirnova IA. 1986. The sodium cycle. I. Na+-dependent motility and modes of membrane energization in the marine alkalotolerant Vibrio alginolyticus. Biochim. Biophys. Acta 850:449–457 [DOI] [PubMed] [Google Scholar]
- 9.Gonzalez Y, Venegas D, Mendoza-Hernandez G, Camarena L, Dreyfus G. 2010. Na(+)- and H(+)-dependent motility in the coral pathogen Vibrio shilonii. FEMS Microbiol. Lett. 312:142–150 [DOI] [PubMed] [Google Scholar]
- 10.Francis NR, Sosinsky GE, Thomas D, DeRosier DJ. 1994. Isolation, characterization and structure of bacterial flagellar motors containing the switch complex. J. Mol. Biol. 235:1261–1270 [DOI] [PubMed] [Google Scholar]
- 11.Yamaguchi S, Aizawa S, Kihara M, Isomura M, Jones CJ, Macnab RM. 1986. Genetic evidence for a switching and energy-transducing complex in the flagellar motor of Salmonella typhimurium. J. Bacteriol. 168:1172–1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Paul K, Gonzalez-Bonet G, Bilwes AM, Crane BR, Blair D. 2011. Architecture of the flagellar rotor. EMBO J. 30:2962–2971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhao R, Amsler CD, Matsumura P, Khan S. 1996. FliG and FliM distribution in the Salmonella typhimurium cell and flagellar basal bodies. J. Bacteriol. 178:258–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lloyd SA, Blair DF. 1997. Charged residues of the rotor protein FliG essential for torque generation in the flagellar motor of Escherichia coli. J. Mol. Biol. 266:733–744 [DOI] [PubMed] [Google Scholar]
- 15.Zhou J, Lloyd SA, Blair DF. 1998. Electrostatic interactions between rotor and stator in the bacterial flagellar motor. Proc. Natl. Acad. Sci. U. S. A. 95:6436–6441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Morimoto YV, Nakamura S, Hiraoka KD, Namba K, Minamino T. 2013. Distinct roles of highly conserved charged residues at the MotA-FliG interface in bacterial flagellar motor rotation. J. Bacteriol. 195:474–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee LK, Ginsburg MA, Crovace C, Donohoe M, Stock D. 2010. Structure of the torque ring of the flagellar motor and the molecular basis for rotational switching. Nature 466:996–1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kojima S, Nonoyama N, Takekawa N, Fukuoka H, Homma M. 2011. Mutations targeting the C-terminal domain of FliG can disrupt motor assembly in the Na(+)-driven flagella of Vibrio alginolyticus. J. Mol. Biol. 414:62–74 [DOI] [PubMed] [Google Scholar]
- 19.Blair DF, Berg HC. 1990. The MotA protein of E. coli is a proton-conducting component of the flagellar motor. Cell 60:439–449 [DOI] [PubMed] [Google Scholar]
- 20.Stolz B, Berg HC. 1991. Evidence for interactions between MotA and MotB, torque-generating elements of the flagellar motor of Escherichia coli. J. Bacteriol. 173:7033–7037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tang H, Braun TF, Blair DF. 1996. Motility protein complexes in the bacterial flagellar motor. J. Mol. Biol. 261:209–221 [DOI] [PubMed] [Google Scholar]
- 22.Reid SW, Leake MC, Chandler JH, Lo CJ, Armitage JP, Berry RM. 2006. The maximum number of torque-generating units in the flagellar motor of Escherichia coli is at least 11. Proc. Natl. Acad. Sci. U. S. A. 103:8066–8071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Khan S, Dapice M, Reese TS. 1988. Effects of mot gene expression on the structure of the flagellar motor. J. Mol. Biol. 202:575–584 [DOI] [PubMed] [Google Scholar]
- 24.Braun TF, Blair DF. 2001. Targeted disulfide cross-linking of the MotB protein of Escherichia coli: evidence for two H(+) channels in the stator complex. Biochemistry 40:13051–13059 [DOI] [PubMed] [Google Scholar]
- 25.Braun TF, Al-Mawsawi LQ, Kojima S, Blair DF. 2004. Arrangement of core membrane segments in the MotA/MotB proton-channel complex of Escherichia coli. Biochemistry 43:35–45 [DOI] [PubMed] [Google Scholar]
- 26.Blair DF, Berg HC. 1991. Mutations in the MotA protein of Escherichia coli reveal domains critical for proton conduction. J. Mol. Biol. 221:1433–1442 [DOI] [PubMed] [Google Scholar]
- 27.Dean GE, Macnab RM, Stader J, Matsumura P, Burks C. 1984. Gene sequence and predicted amino acid sequence of the MotA protein, a membrane-associated protein required for flagellar rotation in Escherichia coli. J. Bacteriol. 159:991–999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhou J, Fazzio RT, Blair DF. 1995. Membrane topology of the MotA protein of Escherichia coli. J. Mol. Biol. 251:237–242 [DOI] [PubMed] [Google Scholar]
- 29.Chun SY, Parkinson JS. 1988. Bacterial motility: membrane topology of the Escherichia coli MotB protein. Science 239:276–278 [DOI] [PubMed] [Google Scholar]
- 30.Hizukuri Y, Morton JF, Yakushi T, Kojima S, Homma M. 2009. The peptidoglycan-binding (PGB) domain of the Escherichia coli Pal protein can also function as the PGB domain in E. coli flagellar motor protein MotB. J. Biochem. 146:219–229 [DOI] [PubMed] [Google Scholar]
- 31.Roujeinikova A. 2008. Crystal structure of the cell wall anchor domain of MotB, a stator component of the bacterial flagellar motor: implications for peptidoglycan recognition. Proc. Natl. Acad. Sci. U. S. A. 105:10348–10353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou J, Blair DF. 1997. Residues of the cytoplasmic domain of MotA essential for torque generation in the bacterial flagellar motor. J. Mol. Biol. 273:428–439 [DOI] [PubMed] [Google Scholar]
- 33.Kim EA, Price-Carter M, Carlquist WC, Blair DF. 2008. Membrane segment organization in the stator complex of the flagellar motor: implications for proton flow and proton-induced conformational change. Biochemistry 47:11332–11339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Che YS, Nakamura S, Kojima S, Kami-ike N, Namba K, Minamino T. 2008. Suppressor analysis of the MotB(D33E) mutation to probe bacterial flagellar motor dynamics coupled with proton translocation. J. Bacteriol. 190:6660–6667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhou J, Sharp LL, Tang HL, Lloyd SA, Billings S, Braun TF, Blair DF. 1998. Function of protonatable residues in the flagellar motor of Escherichia coli: a critical role for Asp 32 of MotB. J. Bacteriol. 180:2729–2735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kojima S, Blair DF. 2001. Conformational change in the stator of the bacterial flagellar motor. Biochemistry 40:13041–13050 [DOI] [PubMed] [Google Scholar]
- 37.Sowa Y, Berry RM. 2008. Bacterial flagellar motor. Q. Rev. Biophys. 41:103–132 [DOI] [PubMed] [Google Scholar]
- 38.Blair DF. 2003. Flagellar movement driven by proton translocation. FEBS Lett. 545:86–95 [DOI] [PubMed] [Google Scholar]
- 39.Blair DF, Berg HC. 1988. Restoration of torque in defective flagellar motors. Science 242:1678–1681 [DOI] [PubMed] [Google Scholar]
- 40.Leake MC, Chandler JH, Wadhams GH, Bai F, Berry RM, Armitage JP. 2006. Stoichiometry and turnover in single, functioning membrane protein complexes. Nature 443:355–358 [DOI] [PubMed] [Google Scholar]
- 41.Wilson ML, Macnab RM. 1990. Co-overproduction and localization of the Escherichia coli motility proteins MotA and MotB. J. Bacteriol. 172:3932–3939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hosking ER, Vogt C, Bakker EP, Manson MD. 2006. The Escherichia coli MotAB proton channel unplugged. J. Mol. Biol. 364:921–937 [DOI] [PubMed] [Google Scholar]
- 43.Van Way SM, Hosking ER, Braun TF, Manson MD. 2000. Mot protein assembly into the bacterial flagellum: a model based on mutational analysis of the motB gene. J. Mol. Biol. 297:7–24 [DOI] [PubMed] [Google Scholar]
- 44.Morimoto YV, Che YS, Minamino T, Namba K. 2010. Proton-conductivity assay of plugged and unplugged MotA/B proton channel by cytoplasmic pHluorin expressed in Salmonella. FEBS Lett. 584:1268–1272 [DOI] [PubMed] [Google Scholar]
- 45.O'Neill J, Xie M, Hijnen M, Roujeinikova A. 2011. Role of the MotB linker in the assembly and activation of the bacterial flagellar motor. Acta Crystallogr. D Biol. Crystallogr. 67:1009–1016 [DOI] [PubMed] [Google Scholar]
- 46.Kojima S, Imada K, Sakuma M, Sudo Y, Kojima C, Minamino T, Homma M, Namba K. 2009. Stator assembly and activation mechanism of the flagellar motor by the periplasmic region of MotB. Mol. Microbiol. 73:710–718 [DOI] [PubMed] [Google Scholar]
- 47.Poggio S, Abreu-Goodger C, Fabela S, Osorio A, Dreyfus G, Vinuesa P, Camarena L. 2007. A complete set of flagellar genes acquired by horizontal transfer coexists with the endogenous flagellar system in Rhodobacter sphaeroides. J. Bacteriol. 189:3208–3216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Martinez-del Campo A, Ballado T, Camarena L, Dreyfus G. 2011. In Rhodobacter sphaeroides, chemotactic operon 1 regulates rotation of the flagellar system 2. J. Bacteriol. 193:6781–6786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Armitage JP, Macnab RM. 1987. Unidirectional, intermittent rotation of the flagellum of Rhodobacter sphaeroides. J. Bacteriol. 169:514–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pilizota T, Brown MT, Leake MC, Branch RW, Berry RM, Armitage JP. 2009. A molecular brake, not a clutch, stops the Rhodobacter sphaeroides flagellar motor. Proc. Natl. Acad. Sci. U. S. A. 106:11582–11587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Garcia N, Campos A, Osorio A, Poggio S, Gonzalez-Pedrajo B, Camarena L, Dreyfus G. 1998. The flagellar switch genes fliM and fliN of Rhodobacter sphaeroides are contained in a large flagellar gene cluster. J. Bacteriol. 180:3978–3982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ballado T, Camarena L, Gonzalez-Pedrajo B, Silva-Herzog E, Dreyfus G. 2001. The hook gene (flgE) is expressed from the flgBCDEF operon in Rhodobacter sphaeroides: study of an flgE mutant. J. Bacteriol. 183:1680–1687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gonzalez-Pedrajo B, de la Mora J, Ballado T, Camarena L, Dreyfus G. 2002. Characterization of the flgG operon of Rhodobacter sphaeroides WS8 and its role in flagellum biosynthesis. Biochim. Biophys. Acta 1579:55–63 [DOI] [PubMed] [Google Scholar]
- 54.Shah DS, Sockett RE. 1995. Analysis of the motA flagellar motor gene from Rhodobacter sphaeroides, a bacterium with a unidirectional, stop-start flagellum. Mol. Microbiol. 17:961–969 [DOI] [PubMed] [Google Scholar]
- 55.Shah DS, Armitage JP, Sockett RE. 1995. Rhodobacter sphaeroides WS8 expresses a polypeptide that is similar to MotB of Escherichia coli. J. Bacteriol. 177:2929–2932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Suaste-Olmos F, Domenzain C, Mireles-Rodriguez JC, Poggio S, Osorio A, Dreyfus G, Camarena L. 2010. The flagellar protein FliL is essential for swimming in Rhodobacter sphaeroides. J. Bacteriol. 192:6230–6239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jenal U, White J, Shapiro L. 1994. Caulobacter flagellar function, but not assembly, requires FliL, a non-polarly localized membrane protein present in all cell types. J. Mol. Biol. 243:227–244 [DOI] [PubMed] [Google Scholar]
- 58.Belas R, Horikawa E, Aizawa S, Suvanasuthi R. 2009. Genetic determinants of Silicibacter sp. TM1040 motility. J. Bacteriol. 191:4502–4512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Segura A, Duque E, Hurtado A, Ramos JL. 2001. Mutations in genes involved in the flagellar export apparatus of the solvent-tolerant Pseudomonas putida DOT-T1E strain impair motility and lead to hypersensitivity to toluene shocks. J. Bacteriol. 183:4127–4133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Attmannspacher U, Scharf BE, Harshey RM. 2008. FliL is essential for swarming: motor rotation in absence of FliL fractures the flagellar rod in swarmer cells of Salmonella enterica. Mol. Microbiol. 68:328–341 [DOI] [PubMed] [Google Scholar]
- 61.Partridge JD, Harshey RM. 2013. More than motility: Salmonella flagella contribute to overriding friction and facilitating colony hydration during swarming. J. Bacteriol. 195:919–929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lee YY, Patellis J, Belas R. 2013. Activity of Proteus mirabilis FliL is viscosity dependent and requires extragenic DNA. J. Bacteriol. 195:823–832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ramirez-Cabrera V, Poggio S, Domenzain C, Osorio A, Dreyfus G, Camarena L. 2012. A novel component of the Rhodobacter sphaeroides Fla1 flagellum is essential for motor rotation. J. Bacteriol. 194:6174–6183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Terashima H, Koike M, Kojima S, Homma M. 2010. The flagellar basal body-associated protein FlgT is essential for a novel ring structure in the sodium-driven Vibrio motor. J. Bacteriol. 192:5609–5615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sockett RE, Foster JCA, Armitage JP. 1990. Molecular biology of the Rhodobacter sphaeroides flagellum. FEMS Symp. 53:473–479 [Google Scholar]
- 66.Sistrom WR. 1962. The kinetics of the synthesis of photopigments in Rhodopseudomonas sphaeroides. J. Gen. Microbiol. 28:607–616 [DOI] [PubMed] [Google Scholar]
- 67.Metcalf WW, Wanner BL. 1993. Construction of new beta-glucuronidase cassettes for making transcriptional fusions and their use with new methods for allele replacement. Gene 129:17–25 [DOI] [PubMed] [Google Scholar]
- 68.Quandt J, Hynes MF. 1993. Versatile suicide vectors which allow direct selection for gene replacement in gram-negative bacteria. Gene 127:15–21 [DOI] [PubMed] [Google Scholar]
- 69.Davis J, Donohue TJ, Kaplan S. 1988. Construction, characterization, and complementation of a Puf− mutant of Rhodobacter sphaeroides. J. Bacteriol. 170:320–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Simon R, Priefer U, Phüler A. 1983. A broad host range mobilization system for in vivo genetics engineering: transposon mutagenesis in Gram-negative bacteria. Biotechnology 1:784–791 [Google Scholar]
- 71.Zhang Y. 2008. I-TASSER server for protein 3D structure prediction. BMC Bioinformatics 9:40. 10.1186/1471-2105-9-40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Roy A, Kucukural A, Zhang Y. 2010. I-TASSER: a unified platform for automated protein structure and function prediction. Nat. Protoc. 5:725–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Roy A, Yang J, Zhang Y. 2012. COFACTOR: an accurate comparative algorithm for structure-based protein function annotation. Nucleic Acids Res. 40:W471–W477. 10.1093/nar/gks372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Keen NT, Tamaki S, Kobayashi D, Trollinger D. 1988. Improved broad-host-range plasmids for DNA cloning in Gram-negative bacteria. Gene 70:191–197 [DOI] [PubMed] [Google Scholar]
- 75.Bao K, Cohen SN. 2001. Terminal proteins essential for the replication of linear plasmids and chromosomes in Streptomyces. Genes Dev. 15:1518–1527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Harlow E, Lane D. 1988. Antibodies. A laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 77.Laemmli UK. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685 [DOI] [PubMed] [Google Scholar]
- 78.Lane MC, O'Toole PW, Moore SA. 2006. Molecular basis of the interaction between the flagellar export proteins FliI and FliH from Helicobacter pylori. J. Biol. Chem. 281:508–517 [DOI] [PubMed] [Google Scholar]
- 79.Aizawa SI, Dean GE, Jones CJ, Macnab RM, Yamaguchi S. 1985. Purification and characterization of the flagellar hook-basal body complex of Salmonella typhimurium. J. Bacteriol. 161:836–849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sommerlad SM, Hendrixson DR. 2007. Analysis of the roles of FlgP and FlgQ in flagellar motility of Campylobacter jejuni. J. Bacteriol. 189:179–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Martinez RM, Dharmasena MN, Kirn TJ, Taylor RK. 2009. Characterization of two outer membrane proteins, FlgO and FlgP, that influence Vibrio cholerae motility. J. Bacteriol. 191:5669–5679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Terashima H, Li N, Sakuma M, Koike M, Kojima S, Homma M, Imada K. 2013. Insight into the assembly mechanism in the supramolecular rings of the sodium-driven Vibrio flagellar motor from the structure of FlgT. Proc. Natl. Acad. Sci. U. S. A. 110:6133–6138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Martinez RM, Jude BA, Kirn TJ, Skorupski K, Taylor RK. 2010. Role of FlgT in anchoring the flagellum of Vibrio cholerae. J. Bacteriol. 192:2085–2092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Petersen TN, Brunak S, von Heijne G, Nielsen H. 2011. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods 8:785–786 [DOI] [PubMed] [Google Scholar]
- 85.Poggio S, Osorio A, Dreyfus G, Camarena L. 2005. The flagellar hierarchy of Rhodobacter sphaeroides is controlled by the concerted action of two enhancer-binding proteins. Mol. Microbiol. 58:969–983 [DOI] [PubMed] [Google Scholar]
- 86.Martin AC, Gould M, Byles E, Roberts MA, Armitage JP. 2006. Two chemosensory operons of Rhodobacter sphaeroides are regulated independently by sigma 28 and sigma 54. J. Bacteriol. 188:7932–7940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Cameron DE, Urbach JM, Mekalanos JJ. 2008. A defined transposon mutant library and its use in identifying motility genes in Vibrio cholerae. Proc. Natl. Acad. Sci. U. S. A. 105:8736–8741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Paulick A, Koerdt A, Lassak J, Huntley S, Wilms I, Narberhaus F, Thormann KM. 2009. Two different stator systems drive a single polar flagellum in Shewanella oneidensis MR-1. Mol. Microbiol. 71:836–850 [DOI] [PubMed] [Google Scholar]
- 89.West MA, Dreyfus G. 1997. Isolation and ultrastructural study of the flagellar basal body complex from Rhodobacter sphaeroides WS8 (wild type) and a polyhook mutant PG. Biochem. Biophys. Res. Commun. 238:733–737 [DOI] [PubMed] [Google Scholar]
- 90.Kobayashi K, Saitoh T, Shah DS, Ohnishi K, Goodfellow IG, Sockett RE, Aizawa SI. 2003. Purification and characterization of the flagellar basal body of Rhodobacter sphaeroides. J. Bacteriol. 185:5295–5300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hizukuri Y, Kojima S, Homma M. 2010. Disulphide cross-linking between the stator and the bearing components in the bacterial flagellar motor. J. Biochem. 148:309–318 [DOI] [PubMed] [Google Scholar]
- 92.Okabe M, Yakushi T, Homma M. 2005. Interactions of MotX with MotY and with the PomA/PomB sodium ion channel complex of the Vibrio alginolyticus polar flagellum. J. Biol. Chem. 280:25659–25664 [DOI] [PubMed] [Google Scholar]
- 93.Terashima H, Fukuoka H, Yakushi T, Kojima S, Homma M. 2006. The Vibrio motor proteins, MotX and MotY, are associated with the basal body of Na-driven flagella and required for stator formation. Mol. Microbiol. 62:1170–1180 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.