Abstract
SIRT1 is an NAD+-dependent deacetylase that is implicated in prevention of many age-related diseases including metabolic disorders. Since SIRT1 deacetylase activity is dependent on NAD+ levels and the development of compounds that directly activate SIRT1 has been controversial, indirectly activating SIRT1 through enhancing NAD+ bioavailability has received increasing attention. NAD+ levels are reduced in obesity and the aged, but the underlying mechanisms remain unclear. We recently showed that hepatic microRNA-34a (miR-34a), which is elevated in obesity, directly targets and decreases SIRT1 expression. Here we further show that miR-34a reduces NAD+ levels and SIRT1 activity by targeting NAMPT, the rate-limiting enzyme for NAD+ biosynthesis. A functional binding site for miR-34a is present in the 3′ UTR of NAMPT mRNA. Hepatic overexpression of miR-34a reduced NAMPT/NAD+ levels, increased acetylation of the SIRT1 target transcriptional regulators, PGC-1α, SREBP-1c, FXR, and NF-κB, and resulted in obesity-mimetic outcomes. The decreased NAMPT/NAD+ levels were independent of miR-34a effects on SIRT1 levels since they were also observed in SIRT1 liver-specific knockout mice. Further, the miR-34a-mediated decreases were reversed by treatment with the NAD+ intermediate, nicotinamide mononucleotide. Conversely, antagonism of miR-34a in diet-induced obese mice restored NAMPT/NAD+ levels and alleviated steatosis, inflammation, and glucose intolerance. Anti-miR-34a-mediated increases in NAD+ levels were attenuated when NAMPT was downregulated. Our findings reveal a novel function of miR-34a in reducing both SIRT1 expression and activity in obesity. The miR-34a/NAMPT axis presents a potential target for treating obesity- and aging-related diseases involving SIRT1 dysfunction like steatosis and type 2 diabetes.
Keywords: miR-34a, steatosis, diabetes, resveratrol, sirtuins, deacetylation
Introduction
Sirtuin1 (SIRT1) is an NAD+-dependent protein deacetylase (Imai et al. 2000) that extends lifespan in lower organisms and protects against many age-related diseases in mammals (Guarente 2006; Houtkooper et al. 2012; Finkel et al. 2009; Haigis & Sinclair 2010; Guarente 2011). By modulating the acetylation status and activity of transcriptional regulatory proteins, histones, and metabolic enzymes, SIRT1 enhances mitochondrial function and oxidative metabolism and counteracts obesity. In this regard, activating SIRT1 pharmacologically should be beneficial in treating obesity and age-related diseases. However, there has been debate and continuing controversy about the mechanisms of direct activation of SIRT1 by natural and synthetic compounds including resveratrol (Pacholec et al. 2010; Canto & Auwerx 2012; Kaeberlein et al. 2005; Hubbard et al. 2013). Since SIRT1 activity is dependent on NAD+ levels (Imai et al. 2000; Imai 2011), the therapeutic potential of modulating bioavailability of NAD+ to enhance SIRT1 activity has received increasing attention.
Recent studies have shown that the pathophysiology of age-related metabolic disorders like obesity and type 2 diabetes were ameliorated by administration of a key NAD+ intermediate, nicotinamide mononucleotide (NMN), or a NAD+ precursor, nicotinamide riboside (NR), or by downregulation of a major NAD+ consumer, poly ADP ribose polymerase-1 (Yoshino et al. 2011; Canto et al. 2012; Bai et al. 2011). Importantly, cellular levels of NAD+ and nicotinamide phosphoribosyl transferase (NAMPT), the key enzyme in the salvage pathway for NAD+ biosynthesis, were decreased in metabolic tissues from high fat (HF) diet-induced obese mice and aged mice (Yoshino et al. 2011), but the underlying mechanisms of reduced NAMPT/NAD+ levels in obesity and the aged remain unclear.
MicroRNAs (miRs) are negative gene regulators that have been implicated in many biological processes including development, differentiation, cell proliferation, and metabolism (Neilson & Sharp 2008). Emerging evidence indicates that miRs have crucial functions in metabolic regulation and that miRs are aberrantly expressed in metabolic disease (Rottiers & Naar 2012; Lee & Kemper 2010). Our group and others recently reported that miR-34a directly targets SIRT1 and reduces its expression (Lee et al. 2010; Yamakuchi et al. 2008). We further showed that hepatic miR-34a levels are highly elevated in both dietary and leptin-deficient ob/ob obese mice (Lee et al. 2010). Consistent with these initial findings, recent microRNA microarray analyses identified miR-34a as the most highly elevated hepatic miR in dietary and ob/ob obese mice (Trajkovski et al. 2011). Although it is known that miR-34a reduces the protein levels of SIRT1 (Lee et al. 2010; Yamakuchi et al. 2008), whether SIRT1 deacetylase activity is also affected by miR-34a remains unknown.
Here we report a surprising finding that elevated miR-34a in obesity reduces hepatic NAD+ levels and SIRT1 activity by directly targeting NAMPT. Further, in vivo silencing of miR-34a in diet-induced obese mice demonstrates the therapeutic feasibility of targeting miR-34a for treatment of diseases of obesity and aging including steatosis and type 2 diabetes.
Results
Inverse correlation between miR-34a and NAMPT/NAD+ levels
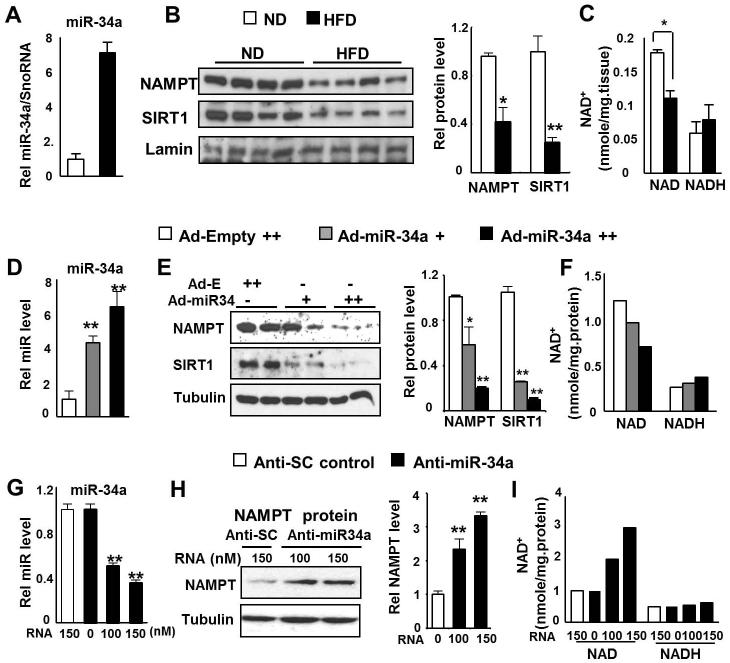
Our group and others recently showed that miR-34a binds to the 3′UTR of the SIRT1 mRNA and downregulates its expression (Lee & Kemper 2010; Lee et al. 2010; Yamakuchi et al. 2008). Surprisingly, a nucleotide sequence imperfectly complementary to the miR-34a seed sequence was also detected in the 3′ UTR of mouse Nampt mRNA (Fig. S1). The miR-34a seed sequence in the NAMPT gene is well conserved between mouse and human (not shown). We, therefore, examined whether elevated miR-34a levels in obesity correlate inversely with NAMPT levels. In dietary obese mice, hepatic miR-34a levels were elevated (Fig. 1A), NAMPT and SIRT1 expression levels were decreased and NAD+ levels were also significantly reduced (Fig. 1B, 1C, Fig. S2). In hepatocytes, overexpression of miR-34a decreased expression of NAMPT and SIRT1 and reduced NAD+ levels (Fig. 1D-F, Fig. S3A). Conversely, antisense inhibition of miR-34a increased NAMPT protein and NAD+ levels (Fig. 1G-I, Fig. S3B). These results indicate an inverse correlation between hepatic miR-34a and NAMPT/NAD+ levels, suggesting that miR-34a may target NAMPT.
Fig. 1. Inverse correlation between miR-34a and NAD+ levels.
(A-C) HF diet-induced obese mouse experiments. (A) Hepatic miR-34a levels in lean and dietary obese mice. (B) Hepatic NAMPT/SIRT1 protein levels in lean and diet-induced obese mice were detected and the ratio to laminin is plotted in the right panel. (C) Hepatic NAD+ and NADH levels are shown. (D-F) Overexpression of miR-34a. Primary mouse hepatocytes were infected with Ad-viral vectors as indicated and miR-34a levels (D), NAMPT/SIRT1 protein levels (E) and NAD+ and NADH levels (F) were measured. (G-I) Downregulation of miR-34a. Conversely, hepatocytes were transfected with anti-miR-34a and miR-34a levels (G), NAMPT protein levels (H), and the NAD+ and NADH levels (I) were measured. The mean and SEM are plotted (n=4), *, p<0.05, **, p<0.01.
MiR-34a directly targets the 3′UTR of NAMPT mRNA
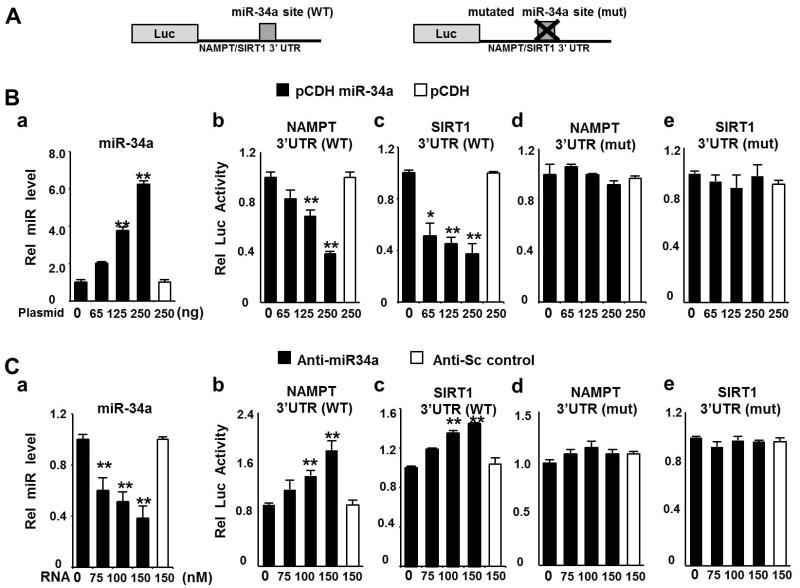
To determine whether NAMPT is a direct target of miR-34a, the wild type (WT) or a mutated sequence of the predicted miR-34a site in the NAMPT 3′UTR was inserted into a luciferase reporter (Fig. 2A) for comparison with the 3′UTR of the known miR-34a target, SIRT1 (Lee et al. 2010; Yamakuchi et al. 2008). Overexpression of miR-34a (Fig. 2B-a) inhibited the activities of luciferase reporters containing either the NAMPT or SIRT1 WT sequence (Fig. 2B-b, c), but not those of reporters containing mutated 3′ UTRs (Fig. 2B-d, e). Conversely, downregulation of miR-34a with antisense-miR-34a (Fig. 2C-a) increased reporter activity for the WT sequences (Fig. 2C-b, c), but not the mutated sequences (Fig. 2C-d, e) while the control RNA did not affect reporter activity. These results indicate that miR-34a negatively regulates NAMPT and that the inhibition is likely mediated by direct binding of miR-34a to the 3′ UTR of NAMPT mRNA.
Fig. 2. MiR-34a directly binds to the 3′ UTR of NAMPT mRNA.
(A) Schematic of luciferase (Luc) reporter plasmids with the wild type (WT) or mutated (mut) miR-34a site in the 3′UTR of NAMPT or SIRT1 inserted. (B) Hepa1c1c7 cells were transfected with an expression plasmid for miR-34a, along with the indicated luciferase reporter plasmids. (C) Cos-1 cells were transfected with control or anti-miR-34a and the indicated luciferase reporter plasmids. The miR-34a levels (a) or reporter activities (b-e) were measured. Values for luciferase activities were normalized to β-galactosidase activities. The mean and SEM are plotted (n=3), *, p<0.05, **, p<0.01.
Hepatic overexpression of miR-34a decreases NAMPT/NAD+ levels and SIRT1 activity
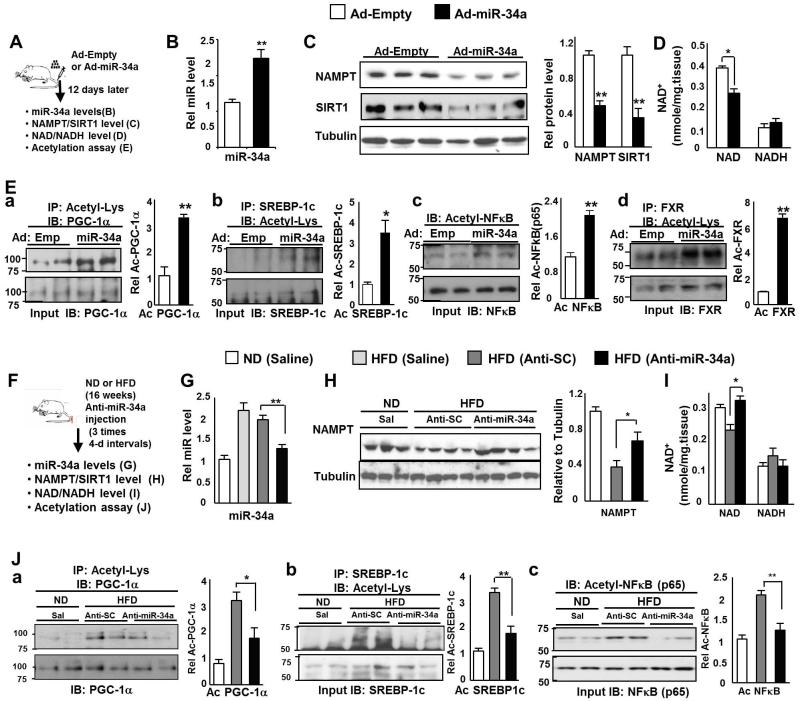
To explore the effects of miR-34a on NAMPT in vivo, miR-34a was adenovirally overexpressed in lean mice (Fig. 3A). Hepatic overexpression of miR-34a (Fig. 3B) decreased expression of NAMPT (Fig. 3C) and also significantly reduced NAD+ levels about 30% (Fig. 3D). Since NAD+ is required for SIRT1-mediated deacetylation (Imai et al. 2000; Imai 2011) and NAD+ levels are decreased by miR-34a, we next asked if SIRT1 deacetylase activity is also affected by measuring acetylation levels of the known SIRT1 target transcriptional regulators, PGC-1α, SREBP-1c, and NF-κB, which are involved in fatty acid β-oxidation, lipogenesis, and inflammation, respectively (Rodgers et al. 2005; Ponugoti et al. 2010; Walker et al. 2010; Yoshizaki et al. 2009). Overexpression of miR-34a led to elevated acetylation of each of these transcriptional regulators (Fig. 3E). Elevated acetylation was also detected for FXR (Fig. 3E), which is also a target of SIRT1 and drives beneficial transcriptional programs of hepatic lipid metabolism (Kemper et al. 2009). These results indicate that overexpression of miR-34a in lean mice reduces NAD+ levels and SIRT1 deacetylase activity.
Fig. 3. Effects of miR-34a on hepatic NAMPT/NAD+ levels and SIRT1 activity.
(A-E) MiR-34a overexpression experiments in lean mice. (A) Experimental outline. (B) Hepatic miR-34a levels are shown. (C) NAMPT and SIRT1 protein levels are shown and the ratio of NAMPT or SIRT1 to tubulin is plotted (right panel). (D) Hepatic NAD+ and NADH levels are shown. (E) Acetylation of transcriptional regulators were detected by IP/IB and the ratio of acetylated to total protein is plotted in panels to the right. The mean and SEM are plotted (n=4), *, p<0.05, **, p<0.01, NS, statistically not significant. (F-J) Anti-miR-34a experiments in dietary obese mice. (F) Experimental outline. (G) Hepatic miR-34a levels are shown. (H) NAMPT protein levels are shown and the ratio of NAMPT to tubulin is plotted (right panel). (I) Hepatic NAD+ and NADH levels are shown. (J) Acetylated protein levels were detected by IP/IB and the ratio of acetylated to total protein is plotted in panels to the right. The mean and SEM are plotted (n=3-5), *, p<0.05; **, p<0.01, NS, statistically not significant.
In vivo silencing of miR-34a in obesity restores NAMPT/NAD+ levels and SIRT1 activity
Since elevated miR-34a reduced NAD+ levels and SIRT1 activity, we tested the idea that antagonism of abnormally elevated miR-34a in obesity might restore NAD+ levels and SIRT1 activity. In this study, we used 2′-O-methoxyethyl phosphorothioate-modified RNA antisense oligonucleotides in order to downregulate miR-34a in vivo. Mice were fed normal chow or HF chow for 20 weeks and given three injections of antisense-miR-34a oligonucleotides, control scrambled RNA, or saline, at 4-day intervals (Fig. 3F). Anti-miR-34a treatment of obese mice significantly reduced hepatic miR-34a levels toward the levels observed in lean mice (Fig. 3G) and increased hepatic levels of NAMPT (Fig. 3H, Fig. S4) and NAD+ (Fig. 3I). Further, the acetylation levels of SIRT1 target transcriptional regulators, PGC-1α, SREBP-1c, and NF-κB, were significantly decreased (Fig. 3J). These results indicate that downregulation of the elevated miR-34a in obese mice increases NAD+ levels and SIRT1 activity toward those observed in lean mice.
NAMPT is important for miR-34a effects on reduced hepatic NAD+ levels
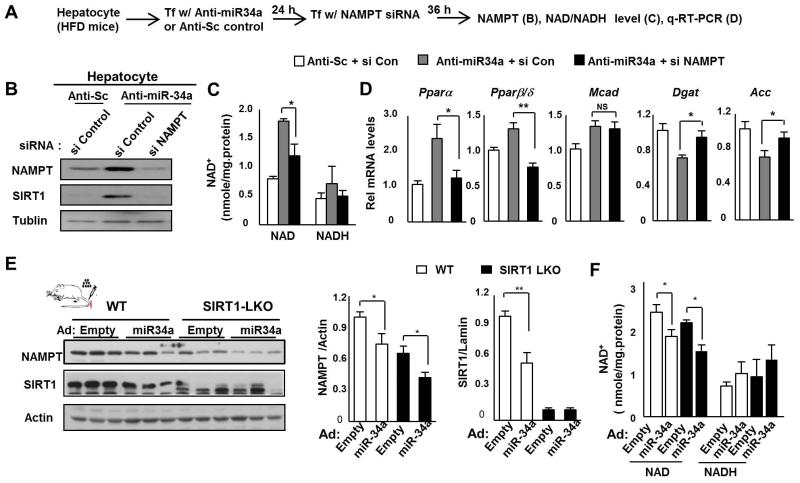
The results from in vivo gain- and loss-functional experiments above indicate that miR-34a reduces hepatic NAMPT/NAD+ levels and consequently SIRT1 activity. MiR-34a targets multiple genes, including Sirt1 and Nampt, which could contribute to the overall downstream metabolic effects, including NAD+ levels (Lee et al. 2010; Yamakuchi et al. 2008; Fu et al. 2012). We therefore assessed the relative contribution of NAMPT to anti-miR-34a-mediated effects on NAD+ levels by downregulating NAMPT in primary hepatocytes from obese mice (Fig. 4A). Anti-miR-34a treatment increased NAMPT protein levels and treatment with NAMPT siRNA effectively reduced NAMPT and also SIRT1 levels (Fig. 4B). Anti-miR-34a-mediated increases in NAD+ levels were significantly reversed (Fig. 4C) and acetylation levels of a known SIRT1 target, NF-κB, were increased by NAMPT siRNA (Fig. S5). Anti-miR-34a treatment increased levels of mRNA for the β-oxidation genes, PPARα and PPARβ/δ and Mcad, and decreased levels for the lipogenic genes, Dgat and Acc (Fig. 4D). Downregulation of the NAMPT completely reversed the increased PPARα and PPARβ/δ mRNA levels and partially reversed the decreased Dgat and Acc mRNA levels, but had little effect on Mcad mRNA (Fig 4D). These results indicate that NAMPT is important for anti-miR-34a-mediated increases in NAD+ levels and the beneficial effects on a subset of lipid metabolic genes. This was particularly evident for PPARα and PPARβ/δ, nuclear receptors that drive lipid-lowering and insulin-sensitizing transcriptional programs.
Fig. 4. NAMPT is important for anti-miR-34a-mediated increases in NAD+ levels.
(A-D) Hepatocyte experiments. (A) Experimental outline. Effects of NAMPT siRNA on NAMPT/SIRT1 protein levels (B), NAD+ and NADH levels (C), and mRNA levels of β-oxidation and lipogenic genes (D) are shown. The mean and SEM are plotted (n=4), *, p<0.05; **, p<0.01, NS, statistically not significant. (E, F) SIRT1-LKO experiments. WT littermates and SIRT-LKO mice were tail vein injected with Ad-empty or Ad-miR-34a and 1 week later, livers were collected for measuring NAMPT/SIRT1 protein levels (E) and hepatic NAD+ and NADH levels (F). The mean and SEM are plotted (n=3), *, p<0.05 and **, p<0.01.
To further assess the direct contribution of NAMPT to miR-34a effects on decreased NAD+ levels, we determined the effects of miR-34a on NAMPT/NAD+ levels in SIRT1 liver-specific knockout (LKO) mice. Hepatic NAMPT protein and NAD+ levels are significantly decreased in SIRT1-LKO mice compared to those in wild type (WT) littermates and importantly, miR-34a-mediated decreases in NAMPT and NAD+ levels were still observed in SIRT1-LKO mice (Fig. 4E, F). These results suggest that miR-34a reduces hepatic NAD+ levels by directly targeting NAMPT in a SIRT1-independent manner although miR-34a effects on SIRT1 expression also likely contribute to reduced NAMPT/NAD+ levels.
Both NAMPT and SIRT1 form a positive regulatory loop that controls hepatic NAD+ levels
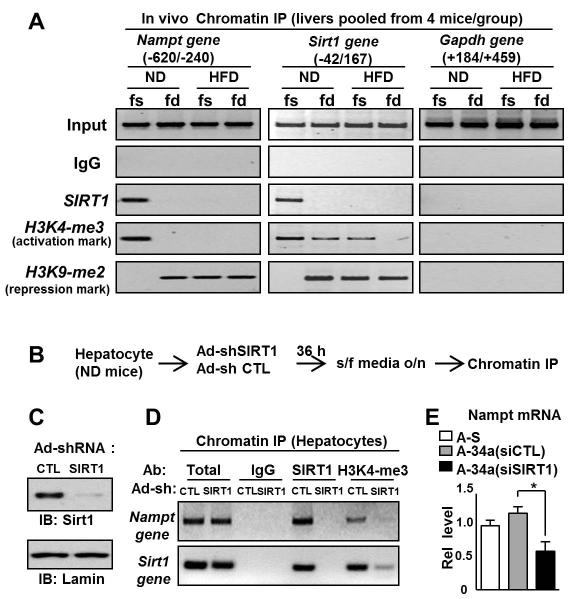
To better understand the SIRT1/NAMPT regulatory loop that controls hepatic NAD+ levels, we examined the effects of fasting on SIRT1 occupancy and histone modifications at the Nampt and Sirt1 genes by chromatin immunoprecipitation (ChIP) assays. In lean mice, overnight fasting resulted in dramatically increased occupancy of SIRT1 at the Nampt gene (Fig. 5A). In addition, in fasted animals, increased histone H3K4 tri-methylation, a gene activation histone mark, and decreased histone H3K9 di-methylation, a gene repression histone mark, were observed (Fig. 5A). Strikingly however, these fasting-mediated dramatic effects on SIRT1 occupancy and histone modifications leading to Nampt gene activation were not detected in dietary obese mice (Fig. 5A). Interestingly, similar effects on SIRT1 occupancy and histone modifications were detected at the Sirt1 gene in lean and obese mice (Fig. 5A).
Fig. 5. Both NAMPT and SIRT1 form a positive regulatory loop that controls NAD+ levels.
(A) ChIP assays in vivo. Lean or diet-induced obese mice were fed or fasted for 16 h and livers from four mice per experimental group were pooled and ChIP assays were performed. SIRT1 occupancy and gene activation or repression histone modifications at the Nampt and Sirt1 genes and the Gapdh gene as a control were detected by semi-qPCR analysis. (B-E) ChIP assays in hepatocytes. (B) Experimental outline. (C) Effects of Ad-shSIRT1 on expression of SIRT1 are shown. Primary hepatocytes isolated from lean mice were infected with Ad-shSIRT1 or Ad-sh control (CTL) and incubated in serum-free media overnight, and ChIP assays (D) and q-RTPCR (E) were performed. The mean and SEM are plotted (n=3), *, p<0.05.
To assess the functional role of SIRT1 in the regulation of the Nampt genes, endogenous SIRT1 in primary mouse hepatocytes was downregulated by siRNA (Fig. 5B, C) and ChIP assays were performed. Levels of the gene activation histone mark, histone H3K4-me3, at the Nampt and Sirt1 promoters were markedly reduced by downregulation of SIRT1 (Fig. 5D). Consistent with these results, expression of the Nampt gene was decreased in primary hepatocytes by downregulation of SIRT1 (Fig. 5E). These results indicate that SIRT1 under conditions mimicking fasting in hepatocyte is recruited to the both Nampt and Sirt1 genes and leads to gene induction.
These findings collectively indicate that during fasting SIRT1 induces Nampt and Sirt1 in lean mice, but not in obese mice. This results in a positive regulatory loop in which SIRT1 induces its own expression and that of Nampt which leads to increased NAD+ levels and therefore increased SIRT1 activity. In obesity, at least in part due to elevated miR34a, this positive regulatory loop is disrupted by downregulation of both Sirt1 and Nampt.
Overexpression of miR34a results in obesity-mimetic outcomes
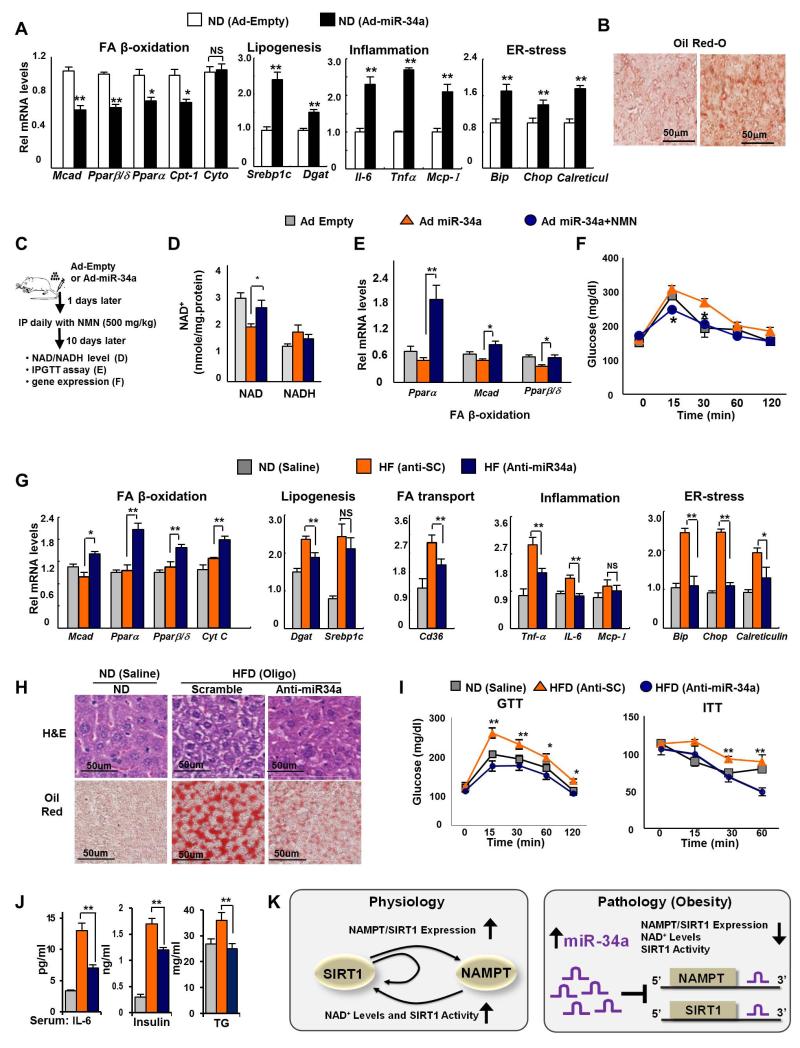
Adenoviral-mediated hepatic overexpression of miR-34a increased acetylation of PGC-1α, FXR, SREBP-1c, and NF-κB (Fig. 3A-E) which should result in decreased transcriptional activities of PGC-1α and FXR and increased activities of SREBP-1c and NF-κB (Rodgers et al. 2005; Ponugoti et al. 2010; Walker et al. 2010; Yoshizaki et al. 2009; Kemper et al. 2009). As expected from these changes in SIRT1 target transcriptional regulators, the β-oxidation genes, Pparα and Pparα, in addition to Mcad and Cpt, were downregulated and lipogenic, inflammation, and ER-stress genes were upregulated in mice overexpressing miR-34a (Fig. 6A, Fig. S6). Consistent with these transcriptional changes, fat deposition in the liver detected by Oil Red-O (Fig. 6B) and H&E staining was increased and glucose tolerance was significantly compromised (Fig. S7. S8).
Fig. 6. In vivo gain- or loss-of-function miR-34a experiments.
(A,B) In vivo m-34a overexpression experiments. Lean mice were injected via the tail vein with Ad-miR-34a or empty adenoviral vector. (A) The mRNA levels of metabolic genes, measured by q-RTPCR, are shown. (B) Staining of liver tissue from mice with Oil Red-O is shown. The mean and SEM are plotted (n=4), *, p<0.05, **, p<0.01, NS, statistically not significant. (C-F) In vivo Ad-miR-34a/NMN experiments. (C) Experimental outline. Effects of treatment with NMN on hepatic NAD+ and NADH levels (D), expression of FA β-oxidation genes (E), and glucose tolerance (F). (G-J) Anti-miR-34a in vivo silencing. Mice were fed normal diets (ND) of HF diets (HFD) for 20 weeks and injected with anti-miR34a oligonucleotides or control scrambled RNA (SC), and control mice fed normal diets were injected with saline. (G) The mRNA levels of metabolic genes, measured by q-RTPCR, are shown. (H) Staining of liver tissue by H&E and Oil Red-O is shown. (I) GTT and ITT: Serum glucose levels, after injection of glucose (GTT) or insulin (ITT), are shown. (J) Serum cytokine, insulin, and TG levels are shown. The mean and SEM are plotted (n=3-5), *, p<0.05; **, p<0.01, NS, statistically not significant. (K) The SIRT1 and NAMPT loop negatively regulated by miR-34a. During fasting under physiological conditions, SIRT1 is recruited to the Nampt and Sirt1 genes, leading to epigenetic activation of these genes. Induced NAMPT, in turn, increases NAD+ levels and SIRT1 mediated deacetylation of its target transcriptional regulators, including PGC-1α, SREBP-1c, and NF-κB, resulting in increased lipid oxidation and reduced lipogenesis and inflammation (left panel). In fatty liver in obesity, however, elevated hepatic miR-34a inhibits both NAMPT and SIRT1 expression by binding to the 3′UTRs of mRNA, which effectively disrupts the feed-forward positive regulatory loop controlling NAD+ levels and SIRT1 deacetylase activity, resulting in detrimental transcriptional and metabolic outcomes (right panel).
Recent studies have shown that treatment with NAD+ intermediates or precursors that increase cellular NAD+ levels, including NMN or NR, counteracts obesity and improves insulin sensitivity in diabetic obese mice (Yoshino et al. 2011; Canto et al. 2012; Bai et al. 2011). Therefore, we asked whether detrimental effects due to elevated miR-34a levels can be reversed by treatment with NMN (Fig. 6C). The decreases in NAMPT/NAD+ levels and Pparα, Pparβ/δ and Mcad mRNA levels observed as a result of hepatic overexpression of miR-34a in mice were significantly reversed and glucose tolerance was significantly improved by NMN treatment (Fig. 6D-F). Similar results were observed in primary mouse hepatocytes (Fig. S9). Conversely, treatment with an inhibitor of NAMPT, FK866, also resulted in decreased NAD+ levels comparable to overexpression of miR-34a in hepatocytes (Fig. S10). Treatment with the SIRT1 activator, resveratrol, also reversed the decreased NAMPT/NAD+ levels resulting from miR-34a expression consistent with the induction of the Nampt gene by SIRT1 (Fig. S11). These results collectively indicate that in lean mice, overexpression of miR34a results in obesity-mimetic gene expression patterns that are associated with liver steatosis and glucose intolerance and that these detrimental effects of miR-34a are likely due to decreased NAD+ levels, at least in part, by direct targeting of NAMPT.
In vivo silencing of miR-34a results in beneficial functional outcomes
In vivo antagonism of miR-34a in obese mice resulted in decreased acetylation of PGC-1α, FXR, SREBP-1c, and NF-κB in mouse liver (Fig. 3F-J) which should result in increased transcriptional activities of PGC-1α and FXR and decreased activities of SREBP-1c and NF-κB (Rodgers et al. 2005; Ponugoti et al. 2010; Walker et al. 2010; Yoshizaki et al. 2009; Kemper et al. 2009). Consistent with these expectations, in vivo anti-miR-34a treatment significantly increased expression of the β-oxidation genes and decreased expression of lipogenesis, inflammation, and ER-stress genes (Fig. 6G). Notably, transcriptional responses of most of these genes to anti-miR-34a in obese mice were the opposite of the responses to overexpression of miR-34a in lean mice (Fig. 6A). Consistent with improved transcriptional outcomes, fat deposits in the liver detected by Oil Red-O staining (Fig. 6H), as previously shown (Fu et al. 2012), and by H&E staining (Fig. 6H) were substantially decreased by anti-miR-34a treatment. Notably, infiltration of macrophages into the liver was also decreased (Fig. S12), suggesting inflammation is ameliorated. Consistent with anti-miR-34-mediated beneficial transcriptional and metabolic outcomes, glucose tolerance and insulin sensitivity were improved by anti-miR-34a treatment (Fig. 6I) to levels similar to those in lean mice.
In order to further assess the therapeutic value of antisense-miR-34a oligonucleotides, we also measured the serum levels of IL-6, insulin, and triglycerides. Highly elevated serum levels of IL-6, insulin, and triglycerides in obese mice were significantly decreased by anti-miR-34a treatment (Fig. 6J). In contrast, serum levels of transaminases, AST and ALT, were not increased, indicating that anti-miR-34a treatment does not cause liver toxicity (Fig. S13). These in vivo antagonism studies show that anti-miR-34a treatment of diet-induced obese mice restores hepatic NAD+ levels and SIRT1 activity and these changes are associated with protection from diet-induced liver steatosis, inflammation, and improved glucose homeostasis. Further, these findings suggest the therapeutic potential of targeting the novel miR-34a/NAMPT axis to treat SIRT1-related diseases of obesity and aging like liver steatosis and type 2 diabetes.
Discussion
In this manuscript, we demonstrate that miR-34a directly targets NAMPT and reduces hepatic NAD+ levels and SIRT1 deacetylase activity. In diet-induced obese animals, which have elevated miR-34a levels and reduced NAMPT/NAD+ levels, downregulation of miR-34a by treatment with antisense oligonucleotides restored the NAMPT/NAD+ levels toward to those in lean mice. Downregulation of miR-34a also decreased acetylation of the SIRT1 target transcriptional regulators, PGC-1α, FXR, SREBP-1c, and NF-κB, resulting in increased expression of β-oxidation genes and decreased expression of lipogenesis, ER-stress, and inflammation genes. Antisense-miR-34a and NAMPT siRNA experiments in primary hepatocytes from dietary obese mice, together with functional studies in SIRT1-LKO mice, indicate that the direct miR-34a effects on NAMPT expression and consequently NAD+ levels are likely major contributors to the overall effects of miR-34a on decreased SIRT1 activity.
Recent studies have shown that SIRT1 and NAMPT work cooperatively in the regulation of metabolism including glucose-regulated insulin secretion at pancreatic β-cells and circadian regulation of NAD+ biosynthesis (Nakahata et al. 2009; Ramsey et al. 2009; Imai & Kiess 2009). Our findings suggest that NAMPT and SIRT1 constitute a positive regulatory loop under nutrient-deprived conditions (Fig. 6K). During fasting, SIRT1 binds to the Nampt and Sirt1 genes leading to gene activation. Induced NAMPT, in turn, raises NAD+ levels, which results in increased SIRT1 deacetylase activities on its target transcriptional regulators like PGC-1α and SREBP-1c and consequently, results in increased lipid oxidation and inhibited lipid synthesis (Rodgers et al. 2005; Ponugoti et al. 2010; Walker et al. 2010; Yoshizaki et al. 2009; Kemper et al. 2009). In contrast, elevated miR-34a in obesity inhibits both NAMPT and SIRT1 expression by directly binding to the 3′UTRs of their mRNAs and thus, effectively disrupts the NAMPT and SIRT1 loop. These events in obesity lead to reduced NAD+ levels and SIRT1 activity, resulting in transcriptional responses of decreased lipid oxidation and increased lipogenesis and inflammation. Since SIRT1 function at the target genes is normally associated with gene silencing by deacetylating histones, our findings that SIRT1 acts as a positive regulator of both Nampt and Sirt1 genes are intriguing. A recent study has shown that SIRT1 directly upregulates breast cancer-associated aromatase expression by modulating acetylation status of ERRα (Holloway et al. 2013). It will be important to elucidate how SIRT1 is recruited to the Nampt and Sirt1 genes and how SIRT1 functions as a positive gene regulator.
Numerous studies have shown beneficial effects of a natural SIRT1 activator, resveratrol, in prevention of many aging-related diseases (Baur et al. 2006; Lagouge et al. 2006). Resveratrol was recently shown to inhibit cAMP-specific phosphodiesterases and a cAMP-effector protein, Epac1, was identified as a key mediator of the effects of resveratrol, which results in the activation of AMPK and SIRT1 (Park et al. 2012). In addition to resveratrol, SIRT1 is also activated by synthetic compounds, including SRT1720 (Milne et al. 2007; Feige et al. 2008). In contrast to these indirect mechanisms for activation of SIRT1, Hubbard et al. recently demonstrated that all previously known sirtuin-activating compounds, including resveratrol, directly activate SIRT1 through a common allosteric mechanism and that remarkably, a single amino acid in the N-terminal domain in SIRT1 is critical for SIRT1 activation by all these compounds (Hubbard et al. 2013). Thus, there continues to be significant debate and controversy around the mechanism and specificity of direct SIRT1 activation by these natural and synthetic compounds (Pacholec et al. 2010; Canto & Auwerx 2012; Kaeberlein et al. 2005). Therefore, the therapeutic potential of indirectly activating SIRT1 through increasing NAD+ bioavailability is an attractive alternative to direct activation of SIRT1. In this regard, the miR-34a/NAMPT regulatory axis identified in the present study reveals novel potential targets to indirectly increase SIRT1 activity through increasing NAD+ levels.
MicroRNAs often directly downregulate multiple targets critically involved in common physiological and pathological processes (Rottiers & Naar 2012; Lee & Kemper 2010). In addition to targeting SIRT1 and NAMPT, miR-34a also targets β-Klotho which is a membrane coreceptor for an intestinal peptide hormone FGF19 (Fu et al. 2012). We have shown that downregulation of miR-34a in obese mice restores the impaired FGF19 signaling observed in dietary obese mice and improved metabolic responses, such as increased glycogen synthesis and inhibited bile acid and glucose synthesis and decreased lipid synthesis (Fu et al. 2012). Further, βKL is also the membrane coreceptor for FGF21 in liver and adipose tissue (Kuro-o 2008). FGF21 plays an important role in mediating hepatic responses during prolonged fasting (Fisher et al. 2011) and regulates energy metabolism in part by activating AMPK (Chau et al. 2010). Importantly, AMPK enhances oxidative metabolism by increasing NAMPT/NAD+ levels and SIRT1 activity (Canto et al. 2009). Therefore, targeting the elevated miR-34a in obesity thus restores both SIRT1 levels and activity, via targeting SIRT1 and NAMPT, and also improves the responsiveness to FGF19 and possibly FGF21. These multiple beneficial effects may provide a substantial advantage for anti-miR-34a therapy compared to targeting the individual signaling pathways.
Mammalian sirtuins (SIRT1-7) have been intensively studied because of their great potential to benefit human health (Guarente 2006; Houtkooper et al. 2012; Finkel et al. 2009; Haigis & Sinclair 2010; Guarente 2011). Mounting evidence indicates that SIRT1 improves many age-related diseases including metabolic, cardiovascular, and neurodegenerative cognitive diseases, and prevents the onset of these diseases (Guarente 2006; Houtkooper et al. 2012; Finkel et al. 2009; Haigis & Sinclair 2010). Recent studies have provided evidence for beneficial functions of SIRT1 in protection from diet-induced liver steatosis and inflammation (Purushotham et al. 2009; Pfluger et al. 2008). NAD+ is an essential cofactor for not only SIRT1 but also for other sirtuins, PARPs, CD38, and mono-ADP-robosylransferases, and many other proteins (Imai et al. 2000; Imai 2011; Bai et al. 2011; Aksoy et al. 2006) and thus, regulation of cellular NAD+ levels by miR-34a may be important for controlling the activities of these enzymes in addition to SIRT1. Since NAMPT and NAD+ levels are reduced in aged mice as well as obese mice (Yoshino et al. 2011), it will be interesting to see whether there is a functional link between elevated miR-34s and NAMPT/NAD+ levels in aged animals as well. The present study identifying miR-34a as a novel molecular target for restoring NAD+ levels and consequently the activity of SIRT1, and possibly other proteins that require NAD+ as an essential cofactor, potentially provides a new therapeutic option for treating SIRT1-related diseases of aging, as well as obesity. The beneficial effects of downregulating miR-34a in diet-induced obese mice demonstrate the feasibility of such an exciting therapeutic approach.
Experimental Procedures
Reagents
Anti-miR-34a (2′-O-methoxyethyl phosphorothioate-modified RNA oligonucleotide), control RNA, and primers for detecting miR-34a were purchased from Applied Biosystems. The siRNAs were purchased from Ambion. Antibodies for NAMPT (OMNI379) and SIRT1 (07-131) were from Enzo Life Science and Millipore, Inc. respectively, and acetylated NF-κB (3045) and acetyl-Lys (9441) antibodies were purchased from Cell Signaling Technology. Antibodies for lamin (sc-20680), tubulin (sc-5274), PGC-1α (sc-13067), SREBP-1c (sc-8984), NF-κB (sc-109R), and FXR (sc-13063) were from Santa Cruz Biotechnology. Antibodies for H3K4-me3 (#07-521) and H3K9-me2 (#07-030) were from Millipore, Inc.
Animal Experiments
Eight to 12 week old male BALB/c mice were fed normal chow or HF chow (60% fat, D12451, Research Diet, Inc.) for 16-20 weeks. SIRT1-LKO mice with deletion of the exon 4 were described previously (Purushotham et al. 2009). Anti-miR-34a experiments were performed as described previously (Fu et al. 2012). Ad-miR-34a (0.5-1.0×109 active viral particles in 200 μl PBS) was injected via the tail vein of mice as previously described (Ponugoti et al. 2010; Kemper et al. 2009). Ad-miR-34a has been described previously (Lee et al. 2010). For NMN experiments, mice were injected via the tail vein with Ad-miR-34a and i.p. treated with 500 mg/kg/day. For GTT and ITT, mice were fasted for 6 h and injected i.p. with glucose solution (Sigma, Inc, 2 g/kg) or insulin (Sigma, Inc, 2 units/kg) and blood glucose levels were measured using an Accu-chek Aviva glucometer (Roche, Inc). All animal protocols were approved by the Institutional Animal Care and Biosafety Committees at University of Illinois at Urbana-Champaign.
Histological Microscopy
Frozen liver sections were stained with Oil Red O and paraffin-embedded liver sections were stained with hematoxylin and eosin.
Blood Chemistry
Serum IL-6, insulin, and AST/ALT levels were measured by ELISA.
MiR-34a Measurement and q-RTPCR
For measuring miR-34a, RNA was isolated using Trizol (Invitrogen) and the amount of miR-34a was normalized to that of control SnoRNA. The amount of mRNA for each gene was determined by q-RTPCR and was normalized to that of 36B4 mRNA. The list of q-RTPCR primer sequences is shown in Fig. S14.
NAD+ and NADH Measurement
NAD+ and NADH levels were measured by enzymatic reactions according to the manufacturer’s instructions (Biovision, Inc., K808-200).
Acetylation Levels of SIRT1 Target Proteins
Acetylation levels of PGC-1α, FXR, and SREBP-1c were detected by the IP/IB method as previously described (Ponugoti et al. 2010; Kemper et al. 2009). IP buffer contains 1 μM TSA and 10 mM nicotinamide to inhibit deacetylation. NF-κB acetylation was detected by IB using acetyl NF-κB (Lys-310) antibody.
Cell-Based Luciferase Reporter Assays
Luciferase reporter assays were performed as described previously (Lee et al. 2010; Fu et al. 2012). An Spe I/Hind III fragment containing the 3′UTR of NAMPT was inserted into the pMIR plasmid (Invitrogen) and mutations in the miR-34a site were generated using site-directed mutagenesis (Stratagene). Positive clones were confirmed by sequencing. Hepa1c1c7 cells, which express low levels of endogenous miR-34a, were used for miR-34a overexpression experiments, whereas Cos-1 cells, which express high levels of miR-34a, were used for anti-miR-34a studies.
Combined Anti-miR-34a and siRNA Experiments
Primary hepatocytes from dietary obese mice were prepared as described (Fu et al. 2012) and transfected with anti-miR-34a or control RNA (100 nM) and 24 h later, cells were further transfected with NAMPT siRNA (5 nM) and 36 h later, harvested for further analyses.
Chromatin Immunoprecipitation (ChIP) Assays
Lean mice or diet-induced obese mice were fasted or fed overnight (16 h), livers were collected, and ChIP assays were performed. For ChIP assays in hepatocytes, primary hepatocytes were isolated and SIRT1 was downregulated by Ad-shSIRT1 infection and ChIP assays were performed. Primer sequences and detailed mouse liver ChIP procedures are described in Fig. S15.
Supplementary Material
Acknowledgments
This study was supported by a grant from the Korean Health 21 R&D Project, Ministry of Health and Welfare (A102065) to K.W. Lee and a grant from MRC 2012-051426 to Y. Kang, and grants from National Institutes of Health (DK62777 and DK95842) and a Basic Science Award from the American Diabetes Association to J. K. Kemper.
Footnotes
Author contributions/Conflict of Interest: SC, KL, YK, BK, and JK designed research; SC, TF, SS, DK, and EY performed experiments; SC, TF, KL, YK, XL, BK, and JK analyzed data; and BK and JK wrote the paper. All of the authors declare no conflict of interest.
References
- Aksoy P, Escande C, White TA, Thompson M, Soares S, Benech JC, Chini EN. Regulation of SIRT 1 mediated NAD dependent deacetylation: a novel role for the multifunctional enzyme CD38. Biochem.Biophys.Res.Commun. 2006;349:353–359. doi: 10.1016/j.bbrc.2006.08.066. [DOI] [PubMed] [Google Scholar]
- Bai P, Canto C, Oudart H, Brunyanszki A, Cen Y, Thomas C, Yamamoto H, Huber A, Kiss B, Houtkooper RH, Schoonjans K, Schreiber V, Sauve AA, Menissier-de Murcia J, Auwerx J. PARP-1 inhibition increases mitochondrial metabolism through SIRT1 activation. Cell.Metab. 2011;13:461–468. doi: 10.1016/j.cmet.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baur JA, Pearson KJ, Price NL, Jamieson HA, Lerin C, Kalra A, Prabhu VV, Allard JS, Lopez-Lluch G, Lewis K, Pistell PJ, Poosala S, Becker KG, Boss O, Gwinn D, Wang M, Ramaswamy S, Fishbein KW, Spencer RG, Lakatta EG, Le Couteur D, Shaw RJ, Navas P, Puigserver P, Ingram DK, de Cabo R, Sinclair DA. Resveratrol improves health and survival of mice on a high-calorie diet. Nature. 2006;444:337–342. doi: 10.1038/nature05354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canto C, Auwerx J. Targeting sirtuin 1 to improve metabolism: all you need is NAD(+)? Pharmacol.Rev. 2012;64:166–187. doi: 10.1124/pr.110.003905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canto C, Gerhart-Hines Z, Feige JN, Lagouge M, Noriega L, Milne JC, Elliott PJ, Puigserver P, Auwerx J. AMPK regulates energy expenditure by modulating NAD+ metabolism and SIRT1 activity. Nature. 2009;458:1056–1060. doi: 10.1038/nature07813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canto C, Houtkooper RH, Pirinen E, Youn DY, Oosterveer MH, Cen Y, Fernandez-Marcos PJ, Yamamoto H, Andreux PA, Cettour-Rose P, Gademann K, Rinsch C, Schoonjans K, Sauve AA, Auwerx J. The NAD(+) precursor nicotinamide riboside enhances oxidative metabolism and protects against high-fat diet-induced obesity. Cell.Metab. 2012;15:838–847. doi: 10.1016/j.cmet.2012.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chau MD, Gao J, Yang Q, Wu Z, Gromada J. Fibroblast growth factor 21 regulates energy metabolism by activating the AMPK-SIRT1-PGC-1alpha pathway. Proc.Natl.Acad.Sci.U.S.A. 2010;107:12553–12558. doi: 10.1073/pnas.1006962107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feige JN, Lagouge M, Canto C, Strehle A, Houten SM, Milne JC, Lambert PD, Mataki C, Elliott PJ, Auwerx J. Specific SIRT1 activation mimics low energy levels and protects against diet-induced metabolic disorders by enhancing fat oxidation. Cell Metab. 2008;8:347–358. doi: 10.1016/j.cmet.2008.08.017. [DOI] [PubMed] [Google Scholar]
- Finkel T, Deng CX, Mostoslavsky R. Recent progress in the biology and physiology of sirtuins. Nature. 2009;460:587–591. doi: 10.1038/nature08197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher FM, Estall JL, Adams AC, Antonellis PJ, Bina HA, Flier JS, Kharitonenkov A, Spiegelman BM, Maratos-Flier E. Integrated regulation of hepatic metabolism by fibroblast growth factor 21 (FGF21) in vivo. Endocrinology. 2011;152:2996–3004. doi: 10.1210/en.2011-0281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu T, Choi SE, Kim DH, Seok S, Suino-Powell KM, Xu HE, Kemper JK. Aberrantly elevated microRNA-34a in obesity attenuates hepatic responses to FGF19 by targeting a membrane coreceptor beta-Klotho. Proc.Natl.Acad.Sci.U.S.A. 2012 doi: 10.1073/pnas.1205951109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarente L. Sirtuins as potential targets for metabolic syndrome. Nature. 2006;444:868–874. doi: 10.1038/nature05486. [DOI] [PubMed] [Google Scholar]
- Guarente L. Sirtuins, aging, and metabolism. Cold Spring Harb.Symp.Quant.Biol. 2011;76:81–90. doi: 10.1101/sqb.2011.76.010629. [DOI] [PubMed] [Google Scholar]
- Haigis MC, Sinclair DA. Mammalian sirtuins: biological insights and disease relevance. Annu.Rev.Pathol. 2010;5:253–295. doi: 10.1146/annurev.pathol.4.110807.092250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holloway KR, Barbieri A, Malyarchuk S, Saxena M, Nedeljkovic-Kurepa A, Cameron Mehl M, Wang A, Gu X, Pruitt K. SIRT1 positively regulates breast cancer associated human aromatase (CYP19A1) expression. Mol.Endocrinol. 2013;27:480–490. doi: 10.1210/me.2012-1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houtkooper RH, Pirinen E, Auwerx J. Sirtuins as regulators of metabolism and healthspan. Nat.Rev.Mol.Cell Biol. 2012;13:225–238. doi: 10.1038/nrm3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard BP, Gomes AP, Dai H, Li J, Case AW, Considine T, Riera TV, Lee JE, E SY, Lamming DW, Pentelute BL, Schuman ER, Stevens LA, Ling AJ, Armour SM, Michan S, Zhao H, Jiang Y, Sweitzer SM, Blum CA, Disch JS, Ng PY, Howitz KT, Rolo AP, Hamuro Y, Moss J, Perni RB, Ellis JL, Vlasuk GP, Sinclair DA. Evidence for a common mechanism of SIRT1 regulation by allosteric activators. Science. 2013;339:1216–1219. doi: 10.1126/science.1231097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai S. Dissecting systemic control of metabolism and aging in the NAD World: the importance of SIRT1 and NAMPT-mediated NAD biosynthesis. FEBS Lett. 2011;585:1657–1662. doi: 10.1016/j.febslet.2011.04.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai S, Armstrong CM, Kaeberlein M, Guarente L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature. 2000;403:795–800. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- Imai S, Kiess W. Therapeutic potential of SIRT1 and NAMPT-mediated NAD biosynthesis in type 2 diabetes. Front.Biosci. 2009;14:2983–2995. doi: 10.2741/3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaeberlein M, McDonagh T, Heltweg B, Hixon J, Westman EA, Caldwell SD, Napper A, Curtis R, DiStefano PS, Fields S, Bedalov A, Kennedy BK. Substrate-specific activation of sirtuins by resveratrol. J.Biol.Chem. 2005;280:17038–17045. doi: 10.1074/jbc.M500655200. [DOI] [PubMed] [Google Scholar]
- Kemper JK, Xiao Z, Ponugoti B, Miao J, Fang S, Kanamaluru D, Tsang S, Wu S, Chiang CM, Veenstra TD. FXR acetylation is normally dynamically regulated by p300 and SIRT1 but constitutively elevated in metabolic disease states. Cell Metabolism. 2009;10:392–404. doi: 10.1016/j.cmet.2009.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuro-o M. Endocrine FGFs and Klothos: emerging concepts. Trends Endocrinol.Metab. 2008;19:239–245. doi: 10.1016/j.tem.2008.06.002. [DOI] [PubMed] [Google Scholar]
- Lagouge M, Argmann C, Gerhart-Hines Z, Meziane H, Lerin C, Daussin F, Messadeq N, Milne J, Lambert P, Elliott P, Geny B, Laakso M, Puigserver P, Auwerx J. Resveratrol improves mitochondrial function and protects against metabolic disease by activating SIRT1 and PGC-1alpha. Cell. 2006;127:1109–1122. doi: 10.1016/j.cell.2006.11.013. [DOI] [PubMed] [Google Scholar]
- Lee J, Kemper JK. Controlling SIRT1 expression by microRNAs in health and metabolic disease. Aging (Albany NY) 2010;2:527–534. doi: 10.18632/aging.100184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Padhye A, Sharma A, Song G, Miao J, Mo YY, Wang L, Kemper JK. A pathway involving farnesoid X receptor and small heterodimer partner positively regulates hepatic sirtuin 1 levels via microRNA-34a inhibition. J.Biol.Chem. 2010;285:12604–12611. doi: 10.1074/jbc.M109.094524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milne JC, Lambert PD, Schenk S, Carney DP, Smith JJ, Gagne DJ, Jin L, Boss O, Perni RB, Vu CB, Bemis JE, Xie R, Disch JS, Ng PY, Nunes JJ, Lynch AV, Yang H, Galonek H, Israelian K, Choy W, Iffland A, Lavu S, Medvedik O, Sinclair DA, Olefsky JM, Jirousek MR, Elliott PJ, Westphal CH. Small molecule activators of SIRT1 as therapeutics for the treatment of type 2 diabetes. Nature. 2007;450:712–716. doi: 10.1038/nature06261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakahata Y, Sahar S, Astarita G, Kaluzova M, Sassone-Corsi P. Circadian control of the NAD+ salvage pathway by CLOCK-SIRT1. Science. 2009;324:654–657. doi: 10.1126/science.1170803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neilson JR, Sharp PA. Small RNA regulators of gene expression. Cell. 2008;134:899–902. doi: 10.1016/j.cell.2008.09.006. [DOI] [PubMed] [Google Scholar]
- Pacholec M, Bleasdale JE, Chrunyk B, Cunningham D, Flynn D, Garofalo RS, Griffith D, Griffor M, Loulakis P, Pabst B, Qiu X, Stockman B, Thanabal V, Varghese A, Ward J, Withka J, Ahn K. SRT1720, SRT2183, SRT1460, and resveratrol are not direct activators of SIRT1. J.Biol.Chem. 2010;285:8340–8351. doi: 10.1074/jbc.M109.088682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SJ, Ahmad F, Philp A, Baar K, Williams T, Luo H, Ke H, Rehmann H, Taussig R, Brown AL, Kim MK, Beaven MA, Burgin AB, Manganiello V, Chung JH. Resveratrol ameliorates aging-related metabolic phenotypes by inhibiting cAMP phosphodiesterases. Cell. 2012;148:421–433. doi: 10.1016/j.cell.2012.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfluger PT, Herranz D, Velasco-Miguel S, Serrano M, Tschop MH. Sirt1 protects against high-fat diet-induced metabolic damage. Proc.Natl.Acad.Sci.U.S.A. 2008;105:9793–9798. doi: 10.1073/pnas.0802917105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponugoti B, Kim DH, Xiao Z, Smith Z, Miao J, Zang M, Wu SY, Chiang CM, Veenstra TD, Kemper JK. SIRT1 deacetylates and inhibits SREBP-1C activity in regulation of hepatic lipid metabolism. J.Biol.Chem. 2010;285:33959–33970. doi: 10.1074/jbc.M110.122978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purushotham A, Schug TT, Xu Q, Surapureddi S, Guo X, Li X. Hepatocyte-specific deletion of SIRT1 alters fatty acid metabolism and results in hepatic steatosis and inflammation. Cell Metab. 2009;9:327–338. doi: 10.1016/j.cmet.2009.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsey KM, Yoshino J, Brace CS, Abrassart D, Kobayashi Y, Marcheva B, Hong HK, Chong JL, Buhr ED, Lee C, Takahashi JS, Imai S, Bass J. Circadian clock feedback cycle through NAMPT-mediated NAD+ biosynthesis. Science. 2009;324:651–654. doi: 10.1126/science.1171641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodgers JT, Lerin C, Haas W, Gygi SP, Spiegelman BM, Puigserver P. Nutrient control of glucose homeostasis through a complex of PGC-1alpha and SIRT1. Nature. 2005;434:113–118. doi: 10.1038/nature03354. [DOI] [PubMed] [Google Scholar]
- Rottiers V, Naar AM. MicroRNAs in metabolism and metabolic disorders. Nat.Rev.Mol.Cell Biol. 2012;13:239–250. doi: 10.1038/nrm3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trajkovski M, Hausser J, Soutschek J, Bhat B, Akin A, Zavolan M, Heim MH, Stoffel M. MicroRNAs 103 and 107 regulate insulin sensitivity. Nature. 2011;474:649–653. doi: 10.1038/nature10112. [DOI] [PubMed] [Google Scholar]
- Walker AK, Yang F, Jiang K, Ji JY, Watts JL, Purushotham A, Boss O, Hirsch ML, Ribich S, Smith JJ, Israelian K, Westphal CH, Rodgers JT, Shioda T, Elson SL, Mulligan P, Najafi-Shoushtari H, Black JC, Thakur JK, Kadyk LC, Whetstine JR, Mostoslavsky R, Puigserver P, Li X, Dyson NJ, Hart AC, Naar AM. Conserved role of SIRT1 orthologs in fasting-dependent inhibition of the lipid/cholesterol regulator SREBP. Genes Dev. 2010;24:1403–1417. doi: 10.1101/gad.1901210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamakuchi M, Ferlito M, Lowenstein CJ. miR-34a repression of SIRT1 regulates apoptosis. Proc.Natl.Acad.Sci.U.S.A. 2008;105:13421–13426. doi: 10.1073/pnas.0801613105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshino J, Mills KF, Yoon MJ, Imai S. Nicotinamide mononucleotide, a key NAD(+) intermediate, treats the pathophysiology of diet-and age-induced diabetes in mice. Cell.Metab. 2011;14:528–536. doi: 10.1016/j.cmet.2011.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshizaki T, Milne JC, Imamura T, Schenk S, Sonoda N, Babendure JL, Lu JC, Smith JJ, Jirousek MR, Olefsky JM. SIRT1 exerts anti-inflammatory effects and improves insulin sensitivity in adipocytes. Mol.Cell.Biol. 2009;29:1363–1374. doi: 10.1128/MCB.00705-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.