Abstract
This review traces the evolution of the cytochrome bc complexes from their early spread among prokaryotic lineages and up to the mitochondrial cytochrome bc1 complex (complex III) and its role in apoptosis. The results of phylogenomic analysis suggest that the bacterial cytochrome b6f-type complexes with short cytochromes b were the ancient form that preceded in evolution the cytochrome bc1-type complexes with long cytochromes b. The common ancestor of the b6f-type and the bc1-type complexes probably resembled the b6f-type complexes found in Heliobacteriaceae and in some Planctomycetes. Lateral transfers of cytochrome bc operons could account for the several instances of acquisition of different types of bacterial cytochrome bc complexes by archaea. The gradual oxygenation of the atmosphere could be the key evolutionary factor that has driven further divergence and spread of the cytochrome bc complexes. On one hand, oxygen could be used as a very efficient terminal electron acceptor. On the other hand, auto-oxidation of the components of the bc complex results in the generation of reactive oxygen species (ROS), which necessitated diverse adaptations of the b6f-type and bc1-type complexes, as well as other, functionally coupled proteins. A detailed scenario of the gradual involvement of the cardiolipin-containing mitochondrial cytochrome bc1 complex into the intrinsic apoptotic pathway is proposed, where the functioning of the complex as an apoptotic trigger is viewed as a way to accelerate the elimination of the cells with irreparably damaged, ROS-producing mitochondria.
Keywords: bioenergetics, molecular evolution, ubiquinol:cytochrome c oxidoreductase, ubiquinone, plastoquinone, cytochrome c, cardiolipin, cell death, photosynthesis, apoptosome
The most amazing combinations can result if you shuffle the pack enough.
Mikhail Bulgakov, The Master and Margarita1
1. Introduction
Cytochrome bc complexes (electrogenic quinol:cytochrome c/plastocyanin oxidoreductases) play key roles in respiration (cytochrome bc1 complex, alias mitochondrial complex III, hereafter bc1) and photosynthesis (cytochrome b6f complex, hereafter b6f). These enzymes catalyze electron transfer (ET) from diverse membrane quinols to high-potential redox carriers (routinely c-type cytochromes) and use the energy released to translocate protons across energy-converting membranes, see [1–5] for reviews. Thereby one of the membrane-adjoining water phases becomes positively charged (p-side), whereas the other one charges negatively (n-side). The resulting transmembrane difference in electrochemical potentials of proton consists of chemical (ΔpH) and electrical (Δψ) components and reaches approx. 200–250 mV under physiological conditions [6].
As revealed by X-ray crystallography [4, 7–13], the mitochondrial cytochrome bc1 complex is a intertwined dimer (see further reviews in this BBA issue). The catalytic core of each bc1 monomer is formed by three subunits: the membrane-embedded cytochrome b, the [Fe2S2] cluster-carrying iron-sulfur Rieske protein, and cytochrome c1. The catalytic, hydrophilic domains of the two latter subunits are anchored in the membrane by single hydrophobic α-helices. Each cytochrome b is a bundle of 8 α-helices that accommodates two (proto)hemes, one close to the p-side of the membrane, bp, and other close to the n-side of the membrane, bn. Because heme bp usually has a lower midpoint redox potential than heme bn, the two hemes are also denoted as the low- and high-potential hemes (bl and bh, respectively). The number of subunits in the bc1 varies between 3 catalytic subunits in some bacteria and 11 subunits in the mitochondrial bc1. It is noteworthy that the X-ray structure of the simplest bc1 of Rhodobacter capsulatus, which contains only 3 subunits, matches the structure of the three catalytic subunits of the mitochondrial bc1 [5, 11].
The cytochrome b6f complexes of green plants and cyanobacteria, although evolutionarily related to the cytochrome bc1 complexes [14], differ structurally from them [2, 5, 15–17]. Specifically, the cytochrome b of the bc1 (formed by 8 transmembrane helices) corresponds to the two subunits of the b6f: the N-terminal part of the cytochrome b is homologous to the cytochrome b6 (4 transmembrane helices), whereas the next 3 transmembrane helices (the C-terminal part) are homologous to the subunit IV (PetD) of the cytochrome b6f complex [1, 5, 14, 18]. The cytochrome b6 subunit, besides accommodating two b-type hemes, as in the bc1, carries an additional c-type heme (denoted cn or ci) that does not have a counterpart in the bc1 [5, 15, 16, 19–22]. The iron atom of this heme is connected to the propionate of heme bn by a water bridge. The subunit IV also binds single molecules of chlorophyll a and β-carotene [5, 23–26]. In addition, the cytochrome f, which accepts electrons from the mobile FeS domain of the Rieske protein, although it carries a c-type heme, is structurally unrelated to the cytochrome c1 of the bc1 [5, 27].
Hence, structurally, the bc1 and the b6f have in common only the cytochrome b and the iron-sulfur Rieske protein. Accordingly, upon superposition of the structures of the bc1 and the b6f complexes, the cytochrome b and Rieske protein of the cytochrome bc1 complex overlap with the cytochrome b6, subunit IV and Rieske protein of the cytochrome b6f complex. Thereby the two hemes of cytochromes b/b6 and the FeS clusters overlap almost exactly [5, 15]. Because of that, Kramer and co-workers have suggested referring to bc1 and b6f complexes together as to Rieske/cyt b complexes [28]. Still, we would use here the more traditional term “cytochrome bc complex” to address both the bc1 and the b6f.
The operation of the cytochrome bc complexes can be described by the Mitchell’s Q-cycle mechanism [29, 30]. The quinol-oxidizing center, so-called catalytic center P, is formed by a loop that connects the third and the forth helices of cytochrome b/b6, a highly conserved P[DE]W[FY] motif of the cytochrome b loop that connects its fifth and sixth helices (or is provided by the subunit IV in case of the b6f-type complexes) and by residues of the [Fe2S2] cluster-carrying domain of the Rieske protein (hereafter the FeS domain) [4, 5]. Upon quinol oxidation, one electron is accepted by the FeS domain to be transferred to cytochrome c1 (or cytochrome f) whereas the other electron, via heme bp and heme bn, crosses the bilayer and reduces a quinone molecule in the catalytic center N close to the opposite side of the membrane. The reduction of cytochrome c1 (or cytochrome f) by the FeS domain requires a 60°–70° rotation of the domain [4, 5, 8, 20, 31]. As a result of Q-cycle, one quinol molecule QNH2 is produced in center(s) N per each two molecules of substrate quinol QPH2 that are oxidized in center(s) P. Since the nascent QNH2 molecules can be also oxidized in center(s) P, two protons are ultimately translocated across the membrane per each electron that passes through the cytochrome bc complex
The cytochrome bc1 complexes are functional dimers, capable of electron exchange between the monomers via closely placed heme bp [32–34]. In the bc1-type complexes, under physiological conditions of a half-reduced ubiquinone pool, a total of two electrons seem to be continuously present in the dimeric cytochrome b moiety [33, 35], owing to a possibility of electron equilibration with the membrane quinol pool via centers N, see [1] and references therein. If center N is “preloaded” with an electron, oxidation of a quinol molecule in the corresponding center P would result in immediate quinol formation in the center N [33, 35].
Both the complexes of the bc1 type and the b6f type seem to be involved in regulation of cellular homeostasis, albeit differently. It is well established that the cytochrome b6f complex plays a key role in regulating the flow of excitation energy between the two photosystems of green plants, see [5, 36, 37] for reviews. By tuning the distribution of light between the two photosystems, plants prevent their overreduction and production of reactive oxygen species (ROS), see [5, 37].
Recently the mitochondrial cytochrome bc1 complex (complex III) has drawn attention because of its possible involvement in triggering apoptosis in animal cells. It has been shown that the intrinsic apoptosis pathway is triggered by the increase in the production of ROS by the components of the mitochondrial ET chain [38–41], specifically by the cytochrome bc1 complex. The ROS appear to trigger the apoptosis in several different ways, which are not fully understood, see [42, 43] for recent reviews. First, they can increase the permeability of the inner mitochondrial membrane (IMM), either by directly damaging it or by inducing the formation of mitochondrial permeability transition pores, ultimately leading to the swelling of mitochondria and to the rupturing of the outer mitochondrial membrane (OMM). Second, ROS can trigger the formation of pores in the OMM, which is mediated by BAX and BAK proteins [44–46]. These pores can be large enough to permit escape of proteins from the intramembrane space into the cell cytosol [40]. Thus, both rupturing of the OMM and formation of pores in it lead to the escape into the cell cytoplasm of the proteins residing in the intramembrane space. In vertebrates, one of such proteins is the small cytochrome c, which, within mitochondria, serves as an electron acceptor from the cytochrome bc1 complex. When in the cytoplasm, cytochrome c interacts with the apoptotic protease activating factor (Apaf-1). This interaction induces oligomerization of the Apaf-1 proteins into an apoptosome, followed by the activation of the pro-caspase-9, which triggers the apoptosis [39, 40, 47].
In this review, the evolution of cytochrome bc complexes is considered in a general context of the gradual oxygenation of the atmosphere. It is suggested that cytochrome bc complexes could have evolved from a simpler membrane-anchored oxidoreductase that was similar to the modern membrane dehydrogenases which catalyze electron exchange between diverse water-soluble substrates and the membrane quinone pool. The emergence of the cytochrome bc1 complexes may have been prompted by the emergence of chlorophyll-based photosynthesis, whereby the b6f-type enzymes with a split cytochrome b preceded in evolution the bc1-type complexes. The intrinsic apoptotic pathway may have emerged as a means to accelerate the elimination of the cells with irreparably damaged, ROS-generating cytochrome bc1 complexes. By considering concurrently the evolution of cytochrome bc complexes and their role in apoptosis we attempt to bring the apoptotic function of these enzymes into a broader evolutionarily context.
2. Evolution of the cytochrome bc complexes: From membrane-anchored electron translocases to electrogenic ubiquinol:cytochrome c oxidoreductases
2.1. Evolutionary relations between the cytochrome bc complexes of the b6f type and the bc1 type
The very presence of two distinct types of cytochrome bc complexes, namely of the bc1-type complexes and the b6f-type complexes, has prompted questions on the evolutionary relations between them. In the first comprehensive review on the evolution of cytochrome bc complexes, Cramer and coworkers have noted that “the larger cytochrome b could be a fusion product of, or has undergone fission to yield the cytochrome b6 and subunit IV polypeptide” [18]. Accordingly, these authors discussed two possibilities on the presence of the ancestral cytochrome bc complex in the common ancestor of archaea, bacteria and eukarya, previously postulated by Woese and co-workers [48]. Since cytochrome bc1 complex had been described both in archaea (Sulfolobus acidocaldarius [49]) and in many bacteria, one possibility was that it could emerge before the separation of the three domains of life. Alternatively, since no eukarya-specific form of this enzyme had ever been described, archaea could obtain the cytochrome bc complex via lateral gene transfer (LGT) from bacteria. In both cases, eukaryotes were assumed to obtain their bc1-type and b6f-type complexes via endosymbiotic events [18]. Later, however, the phylogeny of cytochrome bc complexes, with few exceptions, was correlated with the 16S rRNA-based phylogeny, which suggested that the respective genes had been vertically inherited without a significant LGT [50–53]. Based on this suggestion, it has been concluded that the Last Universal Common Ancestor of the cellular life forms (LUCA), the common ancestor of bacteria and archaea [54, 55], already contained a bc1-type complex; the b6f-type enzymes were assumed to emerge within bacteria after their separation from archaea [50, 51, 53, 56]. However, the suggestion that the primordial cytochrome bc complex was a bc1-type enzyme and was present already in the LUCA [50, 51, 53, 56] appears to be inconsistent with current knowledge about the LUCA and its environment. Specifically, cytochrome bc complexes, for proper functioning, require high-potential electron acceptors. Before the emergence of oxygenic photosynthesis in the ancestors of cyanobacteria, all environments on Earth had been highly reduced [57, 58], so that there should have been no abiotic electron acceptors for the cytochrome bc complexes.
In an attempt to clarify the evolutionary relations among various types of cytochrome bc complexes, a comprehensive phylogenomic analysis was performed here as described in Supplementary Materials. As noted above, the only subunits that are shared by the bc1-type and the b6f-type complexes are the Rieske iron-sulfur protein and the cytochrome b. The redox carriers that accept electrons from the FeS domain are not homologous in different lineages and therefore were not included into the analysis; the evolutionary histories of some of them are addressed separately below. The Rieske protein, although ubiquitously present, carries a weak phylogenetic signal [59]; its inclusion into analysis did not significantly affect the tree topology (unpublished observation). Therefore, the phylogenetic tree has been built using only the sequences of “long” cytochromes b of the bc1-type complexes and the corresponding sequences of “short” cytochromes b plus subunit IV of the b6f-type complexes, see Fig. S1 of File 1 of Supplementary Materials for further details.
An unrooted phylogenetic tree of cytochromes b is shown in Fig. 1 and, in an alternative projection, in Fig. S2 of Supplementary Materials (File 1). In addition, File 2 of Supplementary Materials contains a detailed, zoomable (PDF) version of the unrooted tree, where the bootstrap values and the results of the Approximate likelihood-ratio test (aLRT), calculated as in [60, 61], respectively, are indicated (separated by slashes) for the each branch. The multiple alignment that was used for construction of the tree is provided as File 3 of Supplementary Materials.
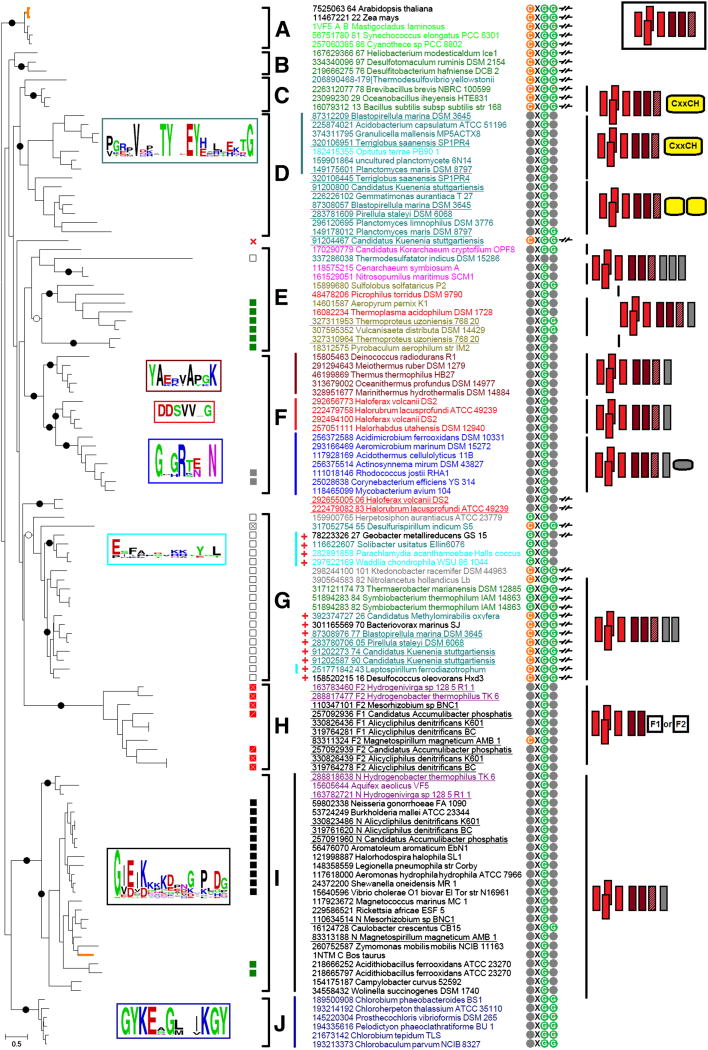
Figure 1.
Phylogenetic tree of cytochromes b.
Each protein is indicated by its NCBI’s GenInfo identifier (gi number), followed by the name of the source organism; in two instances, the proteins are labeled by their PDB codes (1VF5 and 1NTM). The colors of protein names indicate the taxonomical positions of the respective species. The detailed correspondence between colors and taxons is provided in Table S1 of Supplementary Materials (File 1). Black dots mark branches with strong bootstrap support > 70%, white dots show branches with good bootstrap support > 50%. Alternative schematic representations of the same tree are given as Fig. S2 in File 1 of Supplementary Materials (with indicated chemical nature of the predominant pool quinone) and also provided as a separate File 2 of Supplementary Materials (with bootstrap values and aLRT results indicated).
Clades marked on the trees are as follows: A, cyanobacterial and plant clade; eukaryotic sequences are marked by thick orange branches; B, clade with cytochrome b of Heliobacterium modesticaldum and related proteins; C, clade of Bacilli members and Thermodesulfovibrio yellowstonii; D and G, unusual clades, see the main text; E, mostly archaeal clade; F, clade with sequences from Deinococcus-Thermus bacteria, actinobacteria and haloarchaea; H, fusions between cytochrome b and different sets of redox domains; I, proteobacterial clade with mitochondrial cytochromes b and proteins from Aquificae (the mitochondrial cytochrome is indicated by an orange-colored branch), J, Chlorobi clade.
Specific marks are as follows:
1) Complexes with subunit IV as a separate protein are marked with symbol -/ /-. The gi’s of the pairs “cytochrome b - subunit IV” are separated with a space (for instance, “7525063 64” means that cytochrome b6-like and subunit IV-like proteins have gi’s 7525063 and 7525064, respectively).
2) In the heme cn binding motif, Cys residue is shown in orange, Gly residues are shown in green and other residues are marked grey.
3) Squares with different filling show the following deviations from the typical quinone-binding motif P[DE]W[FY] in the subunit IV (or in the C-terminal part of the “long” cytochrome b): black square, PVW[FY], green square, PPW[FY], gray square, PDIY, white square, P W[FY], white square with a cross, G WF, red square with a cross, [LV]DW[FY], red square with a slash, FDW[FY].
4) The red cross symbol indicates the absence of subunit IV.
5) If the genome contains sequences coding for unusual cytochromes ñ (see the main text), the proteins from this genome are marked with the “red plus” sign.
6) Lines of different colors before the names of the proteins indicate different conservative linkers between the cytochrome b6-like parts and subunit IV-like parts of the full-length cytochromes b. Frames of the same color to the left show sequence logo diagrams for these linkers. The sequences 118575215 (Cenarchaeum symbiosum) and 161529051 (Nitrosopumilus maritimus) lack linker parts.
7) Figures after the species names depict divergence from the typical structure shown in a rectangle in the top right corner. Four red rectangles correspond to a 4-helical bundle (cytochrome b6-like part), two dark red rectangles depict two well-aligned helices of the subunit IV, hatched dark rectangles indicate an unaligned third helix of subunit IV. Vertical grey rectangles correspond to additional helices after subunit IV, yellow rectangles with round edges signify domains with heme-binding sites (cytochrome c-like domains), the small grey rectangle with round edges indicates a small domain conserved in actinobacteria. Finally, rectangles with marks “F1” and “F2” in clade H indicate two types of fusions with different sets of domains (see File 1 of Supplementary materials).
As could be seen from the data presented, cytochrome b sequences separate into several subfamilies (clades), members of which not only show sequence similarity, but also, in many cases, share specific, functionally relevant traits. Before discussing these traits, we would like to note that, in a number of genomes, several homologs of the cytochrome b could be identified. Analysis of the respective operons showed the genes of Rieske proteins upstream of the cytochrome b gene(s) in most of them, so that these multiple operons most likely code for cytochrome bc complexes, as already noted earlier [52, 62, 63]. The organisms with several operons are found both within bacteria (e.g. among Planctomycetes, with up to 4 operons in Candidatus Kuenenia stuttgartiensis, see Table S2 in File 1 of the Supplementary Materials and [62, 63]) and archaea (e.g. among Halobacteria, with up to 3 operons in Haloferax volcanii DS2, see Table S3 in File 1 of the Supplementary Materials and [52]). The phylogenetic tree in Fig. 1 shows that the multiple copies of the bc-encoding operons do not result from duplication within these phyla. For example, in the case of Planctomycetes, the respective cytochrome b sequences do not cluster together but belong to separate clades (D and G). Moreover, sequences from clade D are full-length cytochromes b while sequences from clade G are split cytochromes b of the b6f type. In the case of archaeal cytochrome bc complexes, the split cytochrome b branches next to the clade G, while the “fused” archaeal cytochromes b belong to the clades E and F.
Most cytochrome bc1 complexes contain a quinol-binding motif P[DE]W[FY] either in the cytochrome b (“long” cytochromes b) or in the subunit IV (in case of split cytochromes b). Still, some clades show consistent modifications of this motif. For example, the members of the clade G share the PW[FY] motif without the acidic (D or E) residue (see Fig. 1).
The main feature of the phylogenetic tree in Fig. 1 is that the split cytochromes b of the b6f-type complexes form several separate groups. First, split cytochromes b are found in Cyanobacteria (and chloroplasts), Firmicutes (Clostridia and Bacilli), and in Thermodesulfobacteria, these are clades A, B, and C in the upper part of the tree. Second, split cytochromes b make up the clade G. It contains sequences from organisms belonging to a number of diverse bacterial phyla: Chloroflexi, Chrysiogenetes, Proteobacteria, Acidobacteria, Chlamydiae, Firmicutes, Nitrospirae, Planctomycetes and even from a member of the candidate division NC10, Candidatus Methylomirabilis oxyfera [64].
The short cytochromes b, which form clades A, B, and C have a conserved CxGG motif close to the N-terminus, just before the first transmembrane helix. The cysteine residue was shown to bind the third heme (cn) in the b6f-type complexes of plants and cyanobacteria [15, 16], as well as in the cytochrome bc complexes of Firmicutes [65, 66]. The abundance of this CxGG motif within clade G (see Fig. 1) might indicate the presence of the third heme also in the cytochromes b belonging to this clade, which, unfortunately, have not yet been purified and characterized.
The clade G is the only clade that contains both short and long cytochromes b. The respective group of sequences (see Fig. 1) includes a short cytochrome b of Geobacter metallireducens that lacks the cysteine residue (with a GxGx motif instead of a CxGG motif) and three long cytochromes b of Solibacter usitatus, Parachlamydia acanthamoebae and Waddlia chondrophila, all with the same GxGx motif.
The cytochromes b, which belong to the clade G, co-occur with large proteins possessing a NAD(P)-binding domain and many conservative cysteine residues capable of binding FeS-clusters (Fig. S3 in File 1 of the Supplementary Materials). In prokaryotic genomes, these proteins, which apparently have not been yet experimentally characterized, share operons with ferredoxin-NAD(P)H oxidoreductases and are mostly (and, perhaps, erroneously) annotated as subunits of glutamate synthase. In some cases, the genes encoding proteins of this family are found within the operons of cytochrome bc complexes (see Table S2 in File 1 of the Supplementary Materials and [52, 62, 63]. As noted by Kartal and co-workers in the case of Candidatus Kuenenia stuttgartiensis, the absence of a signal sequence or predicted transmembrane segments indicates that this large protein subunit should be located in the cytoplasm [62, 63]. In addition, presence of cytochromes b that belong to the clade G often correlates with the presence of a protein that shows similarity (e-value ~10−10) to a typical cytochrome c Pfam domain (PF00034, Cytochrome_C) and contains several heme-binding motifs, see Fig. S4 in File 1 of the Supplementary Materials and [62, 63]. This occurrence of specific genes within operons suggests that the b6f-type complexes forming the clade G may be viewed as a separate subfamily of cytochrome bc complexes of the same rank as the bc1-type complexes of aerobic organisms and the b6f-type complexes of phototrophs. In Candidatus Kuenenia stuttgartiensis, deep sequencing showed that the cytochrome bc complexes of this bacterium were expressed at the transcriptional and protein levels, albeit in different amounts; the complex that corresponds to #5 in Table S2 (File 1 of the Supplementary Materials) was the major species [62, 63].
A number of bacteria have cytochrome b-containing, “fused” genes (Table S4, File 1 of the Supplementary Materials) along with the “classical” bc-complex operons (Table S5, File 1 of the Supplementary Materials) in their genomes. The two identified types of fusions are labeled F1 (strings 3b, 4b and 5b in Table S4) and F2. F1- and F2-type cytochromes b form a separate clade on the phylogenetic tree (clade H on Fig. 1), while all cytochromes b from “classical” operons of the same organisms fall within the clade I. Operons with fused cytochromes b do not contain Rieske proteins and have only very short linkers between the b6-like and the subunit IV-like parts of cytochrome b. The functions of these cytochromes b remain to be established.
On the phylogenetic tree (Fig. 1), archaeal sequences belong mostly to the clades E and F. The clade F, besides archaeal sequences, contains also the sequences of cytochromes b from Actinobacteria and the Deinococcus-Thermus group. Outside these clades, only haloarchaeal sequences with rather uncommon operon structure were found (see #1 and #3 in Table S3 in File 1 of the Supplementary Materials and [52]); these sequences do not group with other archaeal proteins and appear to be a result of a separate LGT from bacteria.
In sum, the phylogenetic tree in Fig. 1 contains several clades of bc1-type complexes with “long” cytochromes b and four separate clades of b6f-type complexes. Hence, following the rationale of Cramer and co-workers [18], one should choose between two possibilities, namely: (1) the fusion scenario according to which the ancestral version of the cytochrome bc complex was a b6f-type complex with a split cytochrome b, so that the fusion of a small cytochrome b with subunit IV took place several times in different lineages and (2) the fission scenario, where the ancestral version contained a long cytochrome b that split several times in the course of evolution. Although the fission scenario is the one that dominates the current literature on the evolution of cytochrome bc complexes [50–53], the fusion scenario looks more appealing because of following reasons:
Since the split cytochromes b make several clades (see Fig. 1), the fission scenario implies independent splits of the cytochrome b gene in the same point in several lineages, followed by independent appearances of the conserved, heme cn-binding CxGG motif in the same positions within these lineages. Such coincidences seem extremely unlikely;
The fission scenario implies that the split should have affected the quinol-oxidizing site P of cytochrome bc complexes. While there are many catalytic quinol-oxidizing and quinone-reducing sites in different enzymes, the center P of cytochrome bc complexes is unique in its ability to catalyze bifurcated quinol oxidation, where one electron is accepted by the FeS cluster, whereas the other electron is taken up by the heme bn. In the bc1-type complex, this catalytic site is formed by the CD loop of cytochrome b and a highly conserved P[DE]W[FY] motif, which is provided either by the EF loop of cytochrome b in the bc1-type enzymes or by subunit IV in the b6f-type complexes [1, 4, 5]. The consequence of this arrangement is that the fission scenario implies several independent splits through the key catalytic site of the enzyme.
Superposition of the structures of the bc1-type and b6f-type complexes shows that the QN quinone in the cytochrome bc1 complex is bound in the position that is occupied by heme cn in the b6f-type complex [15]. Proteins, generally, have a characteristic tight packing of amino acids in their cores, which prevents their fast unfolding and degradation in response to thermal fluctuations [67]. Because of this tight packing, insertion of a porphyrin ring, e.g. of the cn heme, into a pre-existing, functional protein is hard to imagine [68]. Alternatively, a loss of the cn heme could prompt its functional replacement by a smaller quinone ring, which, by filling the gap, would stabilize the protein. The protein cavity at the QN site of the bc1-type complexes seems not to provide a tight encasement for the quinone ring; it has been argued that water molecules are involved as bridges to fix the respective semiquinone in its binding site [69]. Certain “looseness” of the QN site might reflect the presence of a large heme cn in the ancestral, b6f-type form of the enzyme.
The clade-specific conservation of the linkers between the cytochrome b6-like and subunit IV-like parts of full-length cytochromes b and the absence of any conservation in this region between different clades (Fig. 1) are consistent with the hypothesis of several independent fusions. If the long version of cytochrome b were present in the common ancestor of the cytochrome bc complexes, one would expect a similar conservation pattern in this region in all branches of the tree (or the absence of any sequence conservation). Remarkably, archaeal sequences do not show conservation in this region, and two members of Thaumarchaeota (Caldiarchaeaum subterraneum and Nitrosopumilis maritimus) even have a truncation in this region.
Components of cytochrome bc complexes are prone to fuse together; the cytochrome b gene is often fused to the gene of cytochrome c1 or its functional analogs. These fusions are particularly widespread among Gram-positive bacteria, see e.g. [70]; the organisms in clade H provide further examples of fused cytochromes b, see Table S4 in the File 1 of the Supplementary Materials. Protein fusions are generally more common in evolution than fissions [71, 72], as they typically facilitate the interaction between functionally coupled proteins. Such fusions could occur through mutations in the stop-codons, in which case no frame shift would happen [73]. Fusion of two genes into a single gene of a long cytochrome b should be advantageous, because it could fix, within one gene/protein sequence, the catalytically important parts of the protein, namely the heme-binding sites provided by the four-helical bundle of the small cytochrome b and the quinol-binding motif P[DE]W[FY], provided by the subunit IV.
Last, but not least, the process of fusion can be traced within the clade G (see Fig. 1). As already mentioned, this clade contains a group that includes a short cytochrome b and three long cytochromes b. This group has a very high bootstrap value of 92; since it falls within the clade G that otherwise contains only short cytochromes b, the long cytochromes in Solibacter usitatus, Parachlamydia acanthamoebae and Waddlia chondrophila must have resulted from a (relatively recent?) fusion event.
The current model of evolution of cytochrome bc complexes via fission [50–53] assumes that all split cytochromes b belong to a single clade (the “green clade” [51]), which, in turn, implies that the split of a long cytochrome b happened only once [51, 59]. Therefore, the presence of four separate clades of split cytochromes b (Fig. 1) has been checked (1) by building the tree using the neighbor-joining method [74] instead of otherwise used maximum likelihood method, and (2) by constructing a tree using full-length alignments of cytochromes b except for the obviously unalignable N- and C-terminal regions. Both of these trees yielded several separate clades of b6f-type complexes (DVD, unpublished observations). While presenting the first results of our phylogenomic analysis [75], we have emphasized our inability to reproduce the single clade of split cytochromes b as presented in [51, 59]. Most recently, in this special issue of BBA:Bioenergetics, Baymann and co-workers have published an updated phylogenetic tree [53]. In this latest tree, split cytochromes b are unified not in a “green clade” but in a “Cys-group”, named for the cysteine ligand of heme cn in the b6f-type complexes. Unlike the “green clade”, the “Cys-group”, which is encased by a thick red contour line in Fig 1 of [53], actually encompasses three separate clades with split cytochromes b. The arrows in Figure S5 of Supplementary File 1 demonstrate how the single large “Cys-group” (Fig. S5A) can be transformed into three small “Cys-groups” (Fig. S5B) just by swapping the branches depicted by bold lines. In the resulting projection, the tree from [53] is generally consistent with phylogenetic trees presented here, specifically with the radial tree that is depicted in Fig. S2 of the Supplementary File 1.
Thus, there are only minor differences in the topology between our tree shown in Fig. 1 (and also in Fig. S2 and in File 2 of the Supplementary Materials) and the tree from [53], these differences are discussed in the caption to Fig. S5 of Supplementary File 1. In summary, the two most recent and complete phylogenetic trees of cytochrome b indicate the presence of several clades of split cytochromes b, all containing cysteine ligands for the heme cn in their sequences. Since several independent fissions followed by insertions of similar hemes and the emergence of same heme-binding motif in several lineages are hardly probable, the data in Fig. 1 and Fig. S2, as well as in Fig. 1 from [53], can be viewed as supporting the evolutionary primacy of the b6f-type complexes and the emergence of the bc1-type complexes from several independent fusion events.
2.2. Cytochrome bc complexes and the LUCA
As noted above, contemporary evolutionary scenarios imply the presence of a bc1-type complex already in the LUCA, because the initially discovered archaeal cytochrome bc complexes were of the bc1 type [50–53, 56]. This view, however, has been weakened by the identification of the b6f-type complexes in some archaeal genomes, see [52] and Table S3 (File 1 of Supplementary Materials). As a general principle, presence of a certain enzyme in bacteria and archaea cannot be alone considered an ultimate evidence for the presence of its ancestor in the LUCA because of the possibility of an LGT between the domains [76]. Moreover, multiple operons of cytochrome bc complexes in many bacterial and archaeal genomes, as well as the affiliation of these operons with different phylogenetic clades (see Fig. 1), indicate that cytochrome bc complexes are prone to the LGT. It is well established that a significant fraction of archaeal metabolic enzymes have been obtained via the LGT from bacteria [76, 77]. There are no reasons to assume that cytochrome bc complexes were an exception; they could also be obtained by some archaeal lineages via the LGT from bacteria.
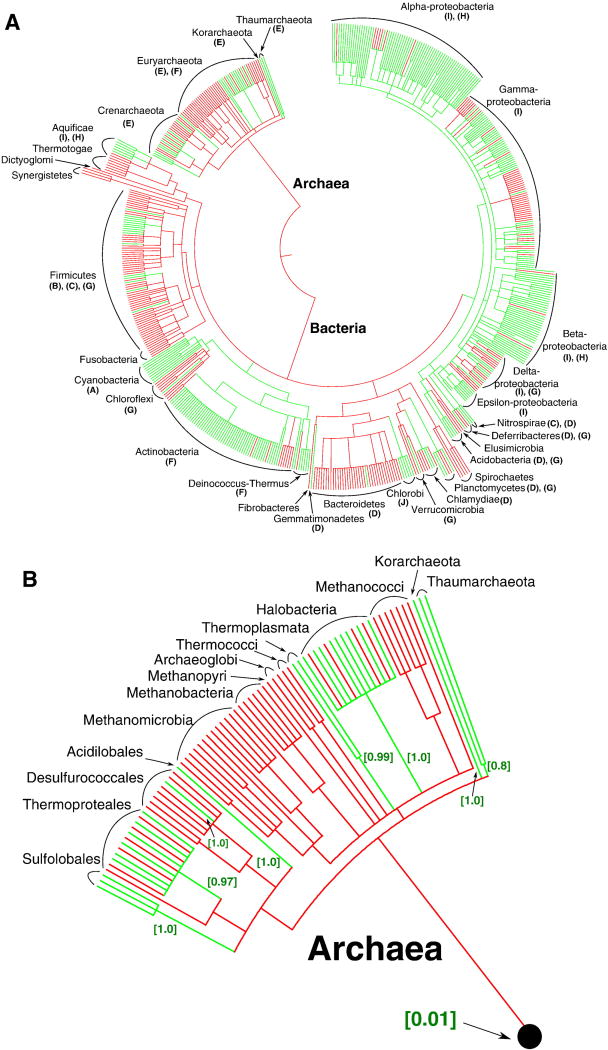
As mentioned above, the scenario on the presence of a bc1-type complex in the LUCA was based on an apparent similarity in the phylogenetic trees of 16S rRNA and cytochrome bc complexes, which implied predominantly vertical inheritance of this enzyme complex [50–53, 56]. Fig. 2A shows the data on the presence and absence of the cytochrome b of the bc1 and b6f-type complexes (COG1290), mapped upon the phylogenetic tree of prokaryotes. Presence of at least one member of the COG1290 in the genome is shown with green color, whereas the absence is shown with red color. The letters in brackets refer to the cytochrome bc complex clades (see Fig. 1), representatives of which are found in respective phyla. It is evident that cytochrome bc complexes are absent from many prokaryotic phyla. When particular phyla contain cytochrome bc complexes, these complexes can belong to different clades. The presence of up to 4 diverse cytochrome bc complexes in bacterial genomes and up to 3 cytochrome bc complexes in archaeal genomes indicates that these enzyme complexes are subject to extensive LGT. One of the bc1 operons in Haloferax volcanii DS2 (#5 in Table S3) is even located on a plasmid, which points to the possible mechanism of the LGT of the cytochrome bc complexes. Altogether, this pattern indicates that the cytochrome bc complexes are prone to the LGT and may have been acquired by those organisms which either have access to high-potential electron acceptors or contain photosynthetic reaction centers whose functioning is accompanied by generation of high-potential electron vacancies.
Figure 2.
Presence and absence of cytochrome b (COG1290), used as a marker of the cytochrome bc1 complexes, mapped on the ribosomal protein-based phylogenetic tree of prokaryotes [233, 234]. Branch lengths do not exactly reflect the evolutionary distance between the nodes. The assignment of proteins from the RefSeq release 45 (Jan 07, 2011) to the Clusters of Orthologous Groups (COGs) [236] was taken from the NCBI FTP site (ftp://ftp.ncbi.nih.gov/pub/wolf/COGs/Prok1202/). The redundancy in the list of complete genomes from the RefSeq release 45 was reduced by manually removing species of the same genera, which resulted in a list of 582 prokaryotic species. Taxonomy data from the NCBI (http://www.ncbi.nlm.nih.gov/taxonomy) [235] to the level of family were used to map the taxonomy for these genomes on the aforementioned large-scale tree. For calculations, the set of 582 bacterial genomes was further reduced to a compact set of 102 bacterial genomes. Within bacteria, we selected genomes which contained cytochromes b (68 genomes), as well as genomes of closely related species which did not contain cytochrome b. In calculations with the COUNT software, a full sample of 115 archaeal genomes and a compact set of 102 bacterial genomes from all major phyla were used, which resulted in a sample of 217 genomes. 29 COGs which occur in at least half of major bacterial and archaeal phyla and do not contain more than 10 members in each genome were selected randomly. These COGs were used as a reference for the estimation of typical rates of gene losses, gene gains and other parameters in COUNT by the “Gain-Loss-Duplication” model with default parameters. For the reference COGs and the cytochrome b COG1290, respectively, the occurrences in each of 217 sampled genomes were calculated as described above. The rates of gene losses and gains were optimized on a subset of 217 genomes chosen to satisfy the computational requirements of the program.
(A) Phylogenetic tree of prokaryotes. For the phyla that contain cytochromes b, the letters in brackets indicate the clades in Figure 1 that include cytochrome b sequences found in these phyla.
(B) Enlarged archaeal clade. The estimated probabilities of independent acquisition of cytochrome(s) b in each group are given in square brackets.
We have performed an estimation of the probabilities of the alternative evolutionary scenarios for cytochrome b using the COUNT software [78]. This software requires two types of the input data, namely: (1) a phylogenetic tree of species (“the tree of life”) and (2) a table of occurrences of different protein families including the family of interest in the complete genomes. The evolutionary history of the cytochrome b was reconstructed with the posterior probability analysis implemented in the COUNT software (see the caption to Fig. 2 for details). The overall results of this analysis are included in Fig. 2A. The cytochrome b (both “long” and “split”) is represented with the COG1290. The presence of at least one member of the COG1290 in a particular genome is shown by the green color of terminal branches, the absence of the COG1290 members is shown by the red color. The green basal branches indicate that the ancestor of the respective phyla likely contained a cytochrome bc complex with a probability of more than 50%, as estimated by the COUNT software. As seen in Fig. 2, the cytochrome bc complex was likely present in the ancestors of two bacterial clades, namely the large clade that includes cyanobacteria, Chloroflexi and Actinobacteria, and the clade that includes proteobacteria and several other bacterial groups. The probability of the presence of a cytochrome bc complex in the common ancestor of the former, “cyanobacterial” clade was estimated by COUNT as 93% whereas its presence in the common ancestor of the latter, “proteobacterial” group was estimated as 85%. COUNT estimated the probability of the presence of a cytochrome bc complex in the common ancestor of bacteria as 7%, which suggests that cytochrome bc complexes were acquired by the majority of bacterial phyla after their radiation from the bacterial common ancestor. Detailed results for archaea are depicted in Fig. 2B. Independent gains of the cytochrome bc complex in archaeal phyla show probabilities mostly exceeding 90%, as depicted in the figure. In sum, COUNT assessed the probability of the presence of a cytochrome bc complex in the LUCA as 1%.
The likely absence of a bc1-type complex in the LUCA is also supported by the following arguments:
As discussed in the previous section, phylogenomic data, as well as structural considerations, suggest that the b6f-type complexes should have preceded in evolution the bc1-type complexes. The b6f-type complexes, however, are almost exclusively affiliated with the bacterial domain. The few archaeal b6f-type complexes represent clear-cut cases of the LGT from specific bacterial phyla (see Fig. 1 and [52]). Hence, emergence of the b6f-type complexes within bacteria, to the best of our knowledge, has not been challenged. Accordingly, the bc1-type complexes, which seem to be evolutionary derivatives of the b6f-type complexes, should have also emerged within bacteria and, hence, were unlikely to be present in the LUCA.
In Fig. 1, the archaeal clades fall within the bacterial clades, which is compatible with the emergence of cytochrome bc complex within bacteria and its LGT to archaea. It is noteworthy, however, that the branches in the archaeal clade E in Fig. 1 are longer than most other individual terminal branches, while the branch that separates the clade E from other clades is relatively short. This pattern might reflect the consequences of an LGT from bacteria to archaea: a bacterial bc complex, after being transferred to archaea, finds itself in a quite different physico-chemical environment (as archaeal membranes differ fundamentally from the bacterial ones [79]), not to mention a new genomic environment. Such abrupt changes could prompt fast adaptation of the laterally transferred cytochrome bc complexes to the new environments, accounting for the long branches within the clade.
The structure of clade F support the suggestion that haloarcheal bc1 genes were obtained by LGT from bacteria [52]. The opposite hypothesis would imply two independent LGT events from a halobacterial ancestor to the ancestors of the Actinobacteria and Deinococcus-Thermus phyla.
The presence of cytochrome bc complexes in archaea correlates in most cases (except for Candidatus Korarchaeum cryptofilum and Thermoplasma acidophilum) with the presence of the cytochrome oxidase genes, another likely subject of LGT into Archaea [80].
One of the earlier arguments in favor of the long cytochrome b as the ancestral, LUCA-encoded form was the fact that archaeal and mitochondial (proteobacterial) sequences both have 13 residues between the histidine heme ligands in 4th helix of cytochrome b; the b6f complexes of chloroplasts and cyanobacteria with only 14 residues between the corresponding histidine residues were considered an exception [14, 18]. However, with more genomes sequenced, it now appears that the vast majority of the cytochrome b sequences actually have 14 residues between these histidine residues. This fact has been already noted in [52] but no evolutionary consequences were drawn from it. This trait argues against evolutionary oldness of archaeal bc1 complexes and might indicate their acquisition via the LGT from a particular bacterial lineage that had 13 amino acids between the respective histidine residues.
In addition to the presented phylogenomic evidence, the presence of cytochrome bc complexes in the LUCA is unlikely from (bio)geochemical considerations. For proper functioning, cytochrome bc complexes require high-potential electron acceptors with redox potentials of at least 100 mV. Before the emergence of oxygenic photosynthesis in the ancestors of cyanobacteria, all environments on Earth were highly reduced, as inferred from geological evidence (reviewed in [58]) and also from biochemical considerations. Indeed, the redox state of the cytoplasm in modern organisms, except perhaps cyanobacteria, is kept highly reduced and an essential part of organismal energy, especially in aerobic organisms, is used to keep it that way. Wald has explained this phenomenon by the emergence of cellular metabolism under highly reducing conditions and by difficulties of its re-tuning for operation at high oxygen levels [57]. From the redox state of cysteine residues within modern cells, the redox potential of primordial environments can be inferred as low as −200 to −400 mV, see [81] and references therein. Under such reducing conditions, potential abiotic electron acceptors for cytochrome bc complex, if even occasionally formed, would be promptly reduced by inorganic reactants, e.g. by sulfur compounds (see also the next section).
Utilization of such dedicated proton translocators as cytochrome bc complexes for ATP synthesis requires proton-tight membranes that can maintain proton potential of 200–250 mV [6]. In modern organisms, this function is performed by sophisticated lipid bilayers that are mostly formed by complex, two-tail phospholipids. Since both the phospholipids and the enzymes of their biosynthesis are completely different in archaea and bacteria [79], it has been suggested that modern-type lipids have emerged separately in Archaea and Bacteria [82]. Accordingly, LUCA may have used simpler, most likely, single-tailed lipids [83]. Simple membranes of single-tailed fatty acids are by factor of approx. 106 more conductive to monovalent cations than membranes that are formed of two-tail phospholipids [84]. In turn, even modern phospholipid membranes are 106–109 times more conductive for protons than for other monovalent cations [85]. Therefore the ability of primitive membranes of the LUCA to maintain high proton potential is questionable. The phylogenomic reconstructions performed for such ubiquitous membrane energy-converting enzymes as rotary ATPases [86–88] and pyrophosphatases [89, 90] have revealed that the ancestral forms of these enzymes must have translocated sodium. It has been speculated that the membranes of the LUCA could be tight enough to maintain sodium potential, so that sodium-dependent bioenergetics may have operated already at the stage of the LUCA [91]. However, the cytochrome bc complex is built in such a way that precludes it from translocating sodium, which apparently leaves cytochrome bc complex without energy-conserving function at the stage of the LUCA.
2.3. Scenario for the emergence of cytochrome bc complexes
As discussed in the previous section, the current scenario of the emergence of a bc1 complex at the stage of LUCA [50–52, 56, 92] leads to certain inconsistencies when compared with the available phylogenomic data and (bio)geochemical considerations. These difficulties, however, could be overcome by an alternative evolutionary scenario for the cytochrome bc complexes where the ancestral, b6f-type complexes with split cytochromes b emerged within bacteria, underwent transitions into bc1-type complexes with a long cytochrome b in some lineages, and were then transferred to archaea via several independent LGT events.
In the current scenario of the evolution of the cytochrome bc complexes, the ancestral bc1-type form of the complex has been suggested to emerge at the stage of the LUCA from an interaction between a Rieske-type iron sulfur protein and a large cytochrome b [50–52, 92]. The origin of a large, 8-helix cytochrome b and its possible function in the LUCA before being recruited into the bc1 has remained enigmatic in this scenario. It has been suggested that the primordial “construction kit” of protein “modules” has contained two different cytochrome b modules, namely a four-helical module to be used in diverse dehydrogenases/oxidoreductases and a large, 8-helical module to be used only in the ancestral cytochrome bc complex [92].
The evolutionary primacy of the b6f-type complexes suggests that emergence of the ancestral form of the cytochrome bc complex could start from a four-helical cytochrome b (see Fig. 3). A bundle formed of 4 alpha-helices represents one of the widespread protein folds; this is one of few folds which are found both in water-soluble and in membrane proteins [93]. Binding of two hemes has been shown to stabilize the fold [94]; it has been shown that of the half of the de novo four-helical proteins from designed combinatorial libraries could bind the heme [95]. In the SCOP database [96], the fold “heme-binding four-helical bundle” comprises three superfamilies; the four-helix cytochrome b of cytochrome bc complex, together with the membrane cytochrome of the formate dehydrogenase makes the superfamily of “transmembrane di-heme cytochromes”. Membrane cytochromes with such fold usually serve as membrane anchors for large, protruding subunits where a distal substrate-binding site is connected by an electron-transferring “wire” of iron sulfur clusters with the membrane, as e.g. in formate dehydrogenase [97] or Ni-Fe hydrogenase [98].
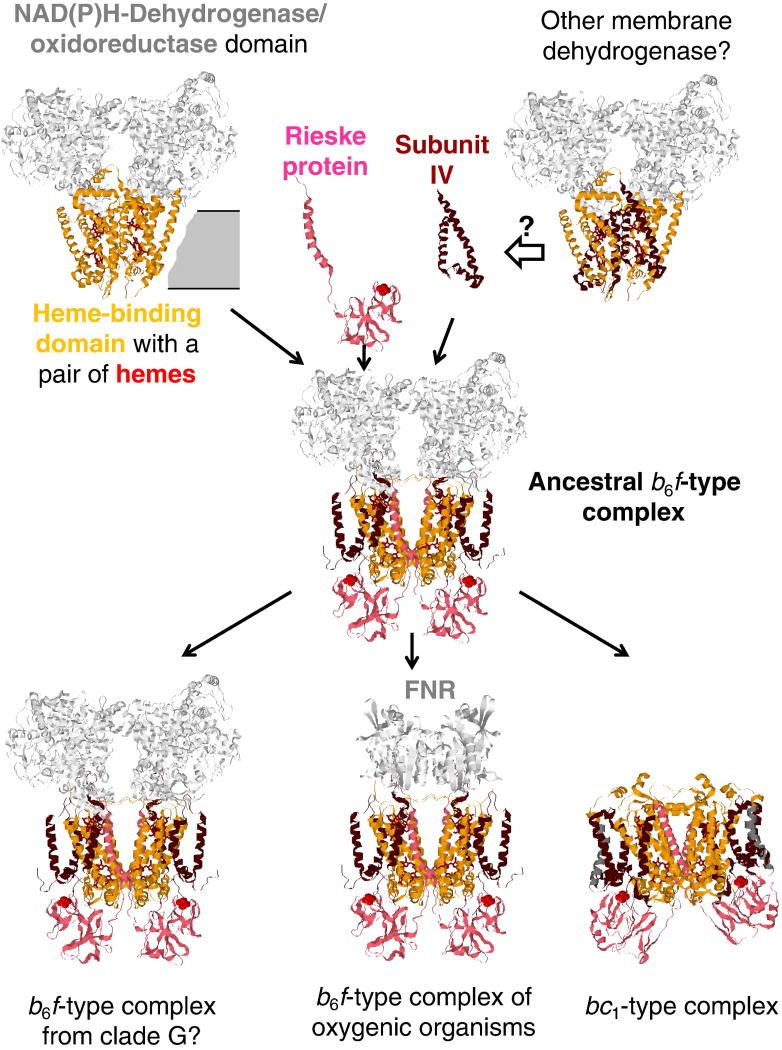
Figure 3.
Evolutionary scenario for the cytochrome bc complexes. Cytochrome b6-like parts (the 4-helical bundle) are colored orange, subunit IV-like parts are colored dark red, the Rieske proteins are colored pink. The three-helix subunit IV is arbitrarily suggested to be recruited from a membrane dehydrogenase, see text for further details.
In some of such enzymes the protruding parts are facing the exterior of the prokaryotic cell (formate dehydrogenase, Ni-Fe hydrogenase), while in others they look into the cytoplasm (e.g. fumarate reductase). Functionally, the enzymes that protrude out of the cell interact with simple electron donors, such as formate and hydrogen, and reduce quinones within the membrane, while the enzymes which protrude into the cytoplasm connect the membrane quinone pool with cellular metabolites such as e.g. succinate or fumarate. Acting together, such enzymes accomplish a quinone-mediated translocation of reducing equivalents across the cellular membrane. Depending on the metabolic situation, the cell could benefit from either a sink for excess electrons or electrons for biosynthesis, so that a system of reversible, differently oriented oxidoreductases could catalyze ET in both directions, perhaps, already in the LUCA [75, 99]. Primitive membranes, while leaky for protons [84, 86, 91, 100], could already represent a significant hydrophobic barrier for reducing equivalents (electrons). By invoking large porphyrin rings as electron carriers, the desolvation penalty for electrons could be decreased and the transfer of electrons across the membrane could be accelerated, see e.g. [101]. Further acceleration could be achieved by translocating a proton together with an electron; such phenomenon, the mechanism of which is not quite clear, has been described for menaquinol:fumarate reductases of Wolinella succinogenes [102, 103] and of Bacillus subtilis [104]. The recruitment of two hemes, which seems to happen independently in several protein families [105], could facilitate, by providing two electron vacancies, the electronic coupling with quinols, which are two-electron carriers. A joint action of several differently oriented, membrane-anchored dehydrogenases, which released protons into periplasm upon oxidation of external electron donors – and trapped cytosolic protons upon reducing intracellular substrates – would lead to the generation of membrane proton potential, which, could pave the way to proton-dependent bioenergetics [97] - but only after the cell membranes became proton-tight [91].
As already noted, potential inorganic electron acceptors for cytochrome bc complexes should have been absent before the oxygenation of earth [58] that took place some 2.5 Gyr ago [106]. However, the transition from a membrane electron translocase to a primordial cytochrome bc complex could be driven by the appearance of biogenic high-potential electron acceptors, produced e.g. upon photosynthesis (A. Bogachev, personal communication). Indeed, the essence of (bacterio)chlorophyll-based photosynthesis is using the energy of light quanta for separating electric charges at the so called “special” pair of (bacterio)chlorophyll molecules within photochemical reaction center (PRC), see [107, 108] for reviews. As a result of such separation, an electron is removed from the special pair to reduce low-potential acceptors, such as NAD(P)+ or quinones, while a high-potential electron vacancy (hole) remains at the (bacterio)chlorophyll moiety. In most modern phototrophic organisms, the bc1-type and b6f-type complexes are involved in re-reducing these oxidized (bacterio)chlorophyll molecules. It is tempting to speculate that this function could have been the initial function of the first (mena)quinol-oxidizing cytochrome bc complexes of a b6f type. This suggestion is consistent with the evolutionary primacy of the b6f-type complexes, as inferred from phylogenomic analysis (see above) and the affiliation of many b6f-type complexes with photosynthetic reaction centers (see [51]). The emergence of first such complexes within phototrophic membranes can also explain the involvement of a chlorophyll molecule and a carotenoid molecule as structural elements of the b6f-type complexes of green plants and cyanobacteria [15, 16]. While, as it has been already noted, an insertion of large chlorophyll and carotenoid molecules in a pre-formed, tightly folded membrane protein is unlikely from the viewpoint of protein physics, the recruitment of such bulky cofactors upon the very formation of the protein seems quite plausible. It is noteworthy that (bacterio)chlorophyll-based photosynthesis, according to current views, emerged within the bacterial lineage, after its separation from archaea [109, 110].
Fig. 3 suggests that the ancestor of the first b6f-type complex could have been a membrane oxidoreductase that, possibly, interacted with cytoplasmic NAD(P)H pool, with its membrane-anchoring subunit belonging to the “transmembrane di-heme cytochrome” fold. It is noteworthy that unlike the bc1-type complexes, the b6f-type complexes seem to be functionally coupled to oxidoreductases. Specifically, the ferredoxin:NADP+ oxidoreductase (FNR) is a functional counterpart of the plant b6f-type complex [111–114], whereas the b6f-type complexes which belong to the clade G, as already discussed, contain in their operons a gene coding for a large, NAD(P)H-binding oxidoreductase subunit (see Table S2 and Figure S3 in File 1 of Supplementary Materials).
The transition from a membrane oxidoreductase to the cytochrome bc complex should have included recruitment of a three-helix protein (the ancestor of the subunit IV) and a Rieske protein. The three-helical subunit IV, the evolutionary origin of which remains unclear, provided the quinol-binding P[DE]W[FY] motif that forms the catalytic site [115, 116], where the bifurcated oxidation of a quinol molecule takes place. Since the subunit IV is also involved in binding of the cn heme [15, 16], the recruitment of subunit IV and heme cn may have occurred simultaneously. The recruitment of a Rieske protein with its mobile FeS domain should have secured the bifurcation of electron flows and facilitated the delivery of electrons to the high-potential electron vacancies at primordial photosynthetic reaction centers. This module is present also in other enzymes, e.g. arsenite oxidase [59]. It is noteworthy that the evolutionary scenario in Fig. 3 shows similarity with the assembly order of the modern cytochrome b6f complexes, see [5] in this volume of BBA:Bioenergetics.
Hence, as shown in Fig. 3, the ancestral form of the cytochrome bc complex could structurally and functionally resemble the b6f-type complexes of anaerobic, menaquinone-containing organisms, such as still unexplored enzymes from clade G organisms or the b6f-type complex from the heliobacterial clade B in Fig. 1. Specifically, the photosynthetic machineries of Heliobacillus mobilis and Heliobacterium modesticaldum are harbored on large operons [117, 118], potentially capable of LGT. Since Heliobacteriaceae are the only phototrophs among Firmicutes (Gram-positive bacteria), it has been argued that heliobacteria most likely obtained their photosynthetic genes via the LGT from now extinct phototrophic, anoxygenic ancestors of cyanobacteria, supposedly the first organisms that used bacteriochlorophyll-based photosynthesis [109]. It is conceivable that these procyanobacteria also harbored the first b6f-type cytochrome bc complexes. However, while the cyanobacteria proper should have underwent dramatic changes in response to the oxygenation [109, 119, 120], which they could not evade, the strictly anaerobic heliobacteria retained not only the ancestral version of the homodimeric PRC, but apparently, an ancestral version of the b6f-type complex, which is coded by the same operon as the ancient PRC [117] and, perhaps, also stems from the anoxygenic ancestors of cyanobacteria. It is noteworthy that a separate operon in the genome of Heliobacterium modesticaldum codes for a tandem of a large NAD(P)-binding oxidoreductase, which is found within the operons of the b6f-type complexes of the clade G (see Table S2 and Figure S3 in File 1 of the Supplementary Materials), and a FNR.
Based on the available data on the properties of the b6f-type complex of heliobacteria [66, 121–123] and on the data for the b6f-type complexes of clade G [62, 63], it is possible to infer that the ancestral cytochrome bc complex should have possessed, in addition to a split, two-subunit cytochrome b, a low-potential heme cn with a Em value of ~ −100 mV, a low-potential version of the Rieske FeS cluster with a Em value of ~150 mV, a multiheme cytochrome c as an acceptor of electrons from the Rieske protein, and, most likely, a further FeS-cluster(s)-containing subunit (NAD(P)H oxidoreductase?) localized on the cytoplasmic, n-side of the membrane. Kartal and co-workers have speculated that modern b6f-type complexes of Candidatus Kuenenia stuttgartiensis (clade G in Fig. 1) might even reduce NAD(P)+ via the NAD(P)H oxidoreductase subunit [62, 63]. Under primordial highly reducing conditions, coupling of a Q-cycle to the reduction of NAD(P)+ should be considered a possibility.
Most likely, the ancestral b6f-type complex had a conserved P[DE]W[FY] motif in its subunit IV. This motif is found in clades A, B, and C, as well as in the majority of long cytochromes b, see Fig. 1. As it also follows from Fig. 1, the cytochrome b of the ancestral enzyme, most likely contained seven transmembrane helices, four of cytochrome b and three of subunit IV. It is noteworthy that the absence of the heme cn-binding CxGG motif correlates with the presence of a long cytochrome b (see Fig. 1). There are only few split cytochromes b without the conserved CxGG motif, all of them within clade G (see Fig. 1). In the b6f-type complexes, heme cn serves as a linker between the two subunits of cytochrome b, so that the loss of heme cn may have prompted the fusion of four-helical cytochromes b and the respective subunits IV, which, apparently, happened independently in several lineages (see Fig. 1). In many long cytochromes b, a part of the CxGG motif is still present either as a single cysteine residue (e.g. in Magnetospirillum magneticum AMB-1) or as a pair of glycine residues (in all the members of group J). The eighth and ninth additional helices in the b6f-type complexes from clade G do not show significant sequence similarity with the additional, eighth helix of long cytochromes b of some bc1-type complexes of groups F and I; these additional helices seem to be later acquisitions.
It is worthwhile to discuss here the possible peculiarities of the Q-cycle mechanism in the primordial cytochrome bc complex. As reviewed by Lancaster [124, 125], modern dehydrogenases, which are membrane-anchored by four-helix cytochromes b, are very flexible with regard to the interactions between the hemes and membrane quinone molecules, so that many topological variants of quinone/quinol processing were realized by nature. The transition to the Q-cycle mechanism should have required oxidation of a (mena)quinol molecule from the p-side of the membrane, electron transfer across the membrane and the reduction of a quinone [or possibly NAD(P)+] molecule from the n-side of the membrane. However, since one of the two electrons released upon quinol oxidation would leave the complex via the FeS domain, only one electron would be injected into the two-heme cytochrome b moiety. This electron should have crossed the membrane against the backpressure of the membrane potential and then reduce a two-electron carrier from the other side of the membrane in a reaction which, generally, should be thermodynamically unfavorable. Known membrane dehydrogenases are dimeric enzymes [124, 125]. If, in the ancient cytochrome bc complex, the cytochrome b hemes of two monomers were close enough for an electron exchange between them, then they could cooperate in performing a two-electron reduction. It is tempting to speculate that the need for cooperation may have driven the establishment of a short edge-to-edge distance between the two bp hemes of the bc1-type and the b6f-type complexes, which enables the apparent electron exchange between the monomers in the modern complexes [20, 32, 34, 126]. The ET against membrane potential backpressure could be also kinetically facilitated by a concomitant proton transfer in the same direction, as described for menaquinol:fumarate reductases [102–104]. And indeed the transmembrane ET seems to be electrostatically compensated both in the bc1–type [127, 128] and in the b6f-type [129, 130] complexes. Kinetically helpful could also be the injection of an electron into an enzyme with pre-reduced cn/bn heme moieties; then a thermodynamically favorable two-electron reduction of a menaquinone molecule in center N (or, perhaps, of a NAD(P)+ molecule) would take place after each quinol oxidation in center P. Since semiquinone species have not been reported for the centers N of the b6f-type complexes, it is believed that the cn/bn heme system performs a two-electron reduction of a quinone molecules in center N, surpassing thus the stage of a stabilized semiquinone [2, 20, 21]. In the primordial b6f-type complex, a cn/bn heme moiety should have exchanged electrons with both the membrane menaquinol pool and the NAD(P)+/NAD(P)H pool (via the cytoplasmic dehydrogenase module), as shown in Fig. 4A.
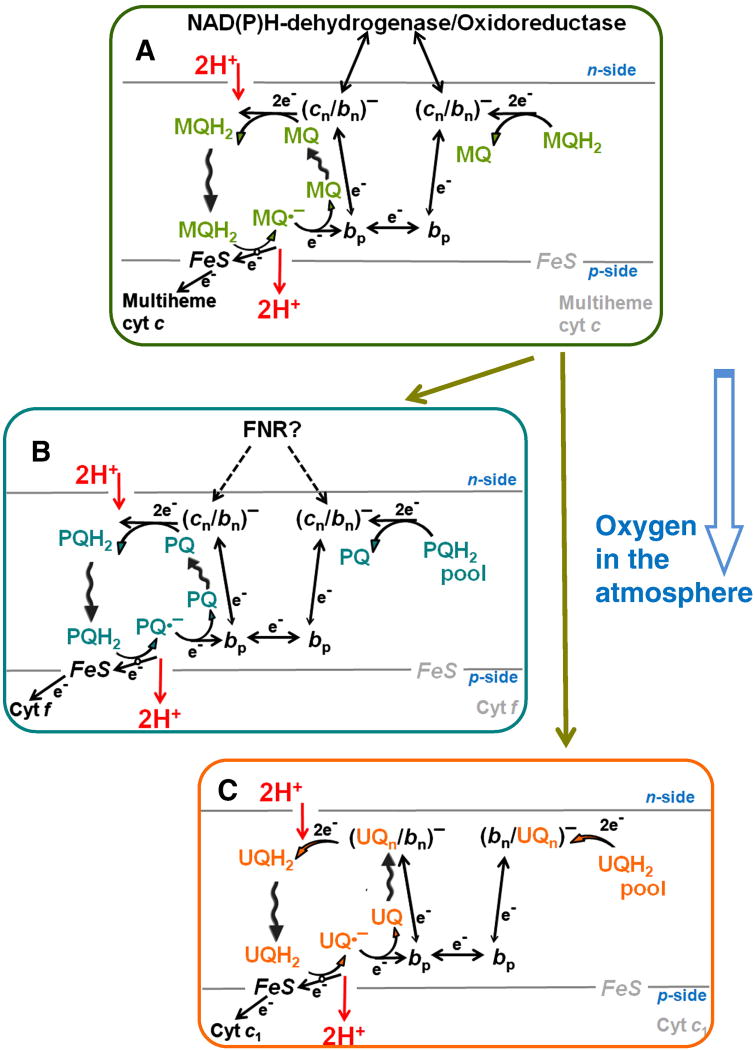
Figure 4.
Q-cycle mechanisms in different cytochrome bc complexes. A, a menaquinone (MQ)-dependent b6f-type complex of anoxic organisms; B, a plastoquinone (PQ)-dependent b6f-type of oxygenic organisms; C, an ubiquinone (UQ)-dependent bc1–type complex of aerobic organisms; see text for further details and references.
2.4. Oxygenation of the atmosphere and diversification of cytochrome bc complexes
The menaquinol-oxidizing b6f-type complexes of modern anaerobes differ substantially both from the bc1-type complexes of aerobic organisms and from the b6f-type complexes of oxygenic plants and cyanobacteria. It has been argued that the appearance of oxygen in the atmosphere some 2.5 Gyr ago, led to the replacement of low-potential menaquinone by high-potential quinones, namely ubiquinone in some bacteria, plastoquinone in cyanobacteria, and caldariellaquinone in certain archaea [18, 131–133]. The oxygenation should have also prompted major modifications in the energy-converting enzymes [18, 52, 109, 120, 132]. Specifically for the cytochrome bc complexes, the Em values of the redox components involved should have been adjusted to the ~150 mV increase in the redox potential of the pool quinone [18, 52, 132]. In addition, the electron escape from the redox components to oxygen (leading to the formation of the potentially deleterious ROS) should have been prevented, which could be achieved by minimizing the number of auto-oxidizable components in the electron transport chain. As discussed in the following sections, the bc1-type complexes of aerobic organisms and the b6f-type complexes of oxygenic phototrophs found different solutions while adapting to the oxygenated atmosphere.
2.4.1. Emergence of the b6f-type complexes of oxygenic organisms
One of the potentially oxidizable points in the menaquinol-dependent b6f-type complexes is the center N, due to the low Em value of the cn heme of ca. −150 mV [66, 123]. In the plant b6f-type complex, after the transition to plastoquinol (Em ~100 mV), the Em value of cytochrome cn was apparently elevated by 200 mV to ~50 mV due to the replacement of a negatively charged Glu, supposedly serving as an iron ligand in the cn heme, by a neutral phenylalanine residue [66, 123]. As a result, electron escape from heme cn to oxygen should have diminished. Accordingly, the Em value of the Rieske protein also increased by ~150 mV in the plastoquinol-processing complex [22, 66, 123]. The low-potential multiheme cytochrome, which is present in anaerobes [52, 62, 66, 122, 123] and was supposedly present in the ancesteral b6f-type complex, was replaced by cytochrome f with a high-potential, not auto-oxidizable heme. The sequence of cytochrome f does not show significant similarity to any other protein, so its evolutionary origin remains enigmatic [2, 20].
As argued elsewhere [35], the kinetic data on plant b6f-type complexes indicate that under physiological conditions one electron is “shared” by hemes bn and cn of each monomer. In this case, the injection of only one electron into the cytochrome b moiety after quinol oxidation in center P is sufficient for a quinol formation in center N. The origin of electrons for the bn/cn heme system in the b6f-type complexes of oxygenic organisms is ambiguous. Apparently, they can come from the plastoquinol pool (see Fig. 4B), but only when this pool is reduced. It has been argued that the ferredoxin-NADP+ reductase (FNR), which seems to be an integral part of the b6f-type complex of plants and cyanobacteria, might also participate in reducing the heme bn/cn system, particularly under conditions of an oxidized plastoquinone pool, see [2, 20, 35] for reviews. This reaction, however, would be accompanied by a loss of about 400 meV of free energy (because of a large energy gap between the redox potentials of NADPH or ferredoxin and the bn/cn heme system of oxygenic organisms), so that the use of this mechanism must be tightly regulated. The available data on the variable coupling of the FNR to the membranes of plants and cyanobacteria, reviewed in [134], might reflect their ability to regulate the coupling between the cytochrome b6f-complexes and FNR depending on the redox poise of the plastoquinol pool and energy status of the cell.
2.4.2. Emergence of the bc1-type complexes of aerobic organisms
Within the suggested evolutionary framework, the transition to the bc1-type complexes of aerobic bacteria, as shown in Fig. 4C, was driven by replacement of menaquinone by ubiquinone [18, 131–133]. Owing to the much higher intrinsic kinetic stability of ubisemiquinone as compared to plasto- and menasemiquinone [135], the redox potential of the stabilized semiquinone in centers N of modern bc1-type complexes (~100–150 mV at pH 7.0 [136–138]) is high enough to prevent an electron escape to oxygen. Under the physiological conditions of a half-reduced ubiquinone pool, a bc1 dimer seems to contain one reduced heme bn (heme b150 with an apparent Em of ~ 150 mV) [1, 139] and one EPR-silent ubisemiquinone [140, 141], owing to an electron exchange with the membrane ubiquinol pool via centers N [33]. It is not clear yet whether an electron is shared by heme bn and semiquinone in each monomer or, asymmetrically, one semiquinone is stabilized in one monomer and one heme bn is fully reduced in the other monomer. Anyhow, the apparent Em of such a pre-reduced state is about 100–150 mV [33], so that oxygen cannot “quench” it. Due to the high apparent Em, this “activated” state should be fully populated under physiological conditions. Cleavage of the link between the two parts of the bc1 of Rb. sphaeroides, aimed at “engineering” of a b6f-type complex, led to the destabilization of the QN semiquinone and to the disappearance of the heme b150 state [142]. This observation indicates that the conservation of the link within phylogenetic groups (see Fig. 1) might be related to its involvement in the stabilization of the QN site in some of these groups and that the emergence of a long, “linked” cytochrome b could be a precondition of semiquinone stabilization in center N. Hence, in the case of well-studied bc1 of proteobacteria and mitochondria, there is no obvious need for delivery of electrons into centers N by external oxidoreductases, which could compromise the efficiency of energy conversion. Accordingly, there are no reports on such electron donations in the cytochrome bc1 complexes of aerobic organisms. Furthermore, in eukaryotic bc1 complexes, the cytoplasmic side of cytochromes b is covered by two bulky “core” subunits which function as mitochondrial processing peptidases in some species [143]. These subunits prevent the access of external, cytoplasmic electron donors to the site N.
Similarly to the b6f-type complexes of oxygenic organisms, the Em values of the Rieske FeS cluster should increase by ~150 mV to fit the Em value of the ubiquinol/ubiquinone redox pair [18, 52, 66, 132, 133]. Accordingly, the low-potential, multiheme cytochrome(s) of anaerobic cytochrome bc complexes was/were replaced by a single-heme cytochrome c1 in the bc1-type complexes of certain proteobacteria. The origin of cytochrome c1 could be traced to a two-heme c4-type proteobacterial cytochrome [144]. This large protein, apparently, has “collapsed”, as a result of numerous deletions, into a smaller protein with a single, high-potential heme not prone to oxidation by oxygen.
2.4.3. Emergence of the mitochondrial-type cytochrome c (cytochrome c2)
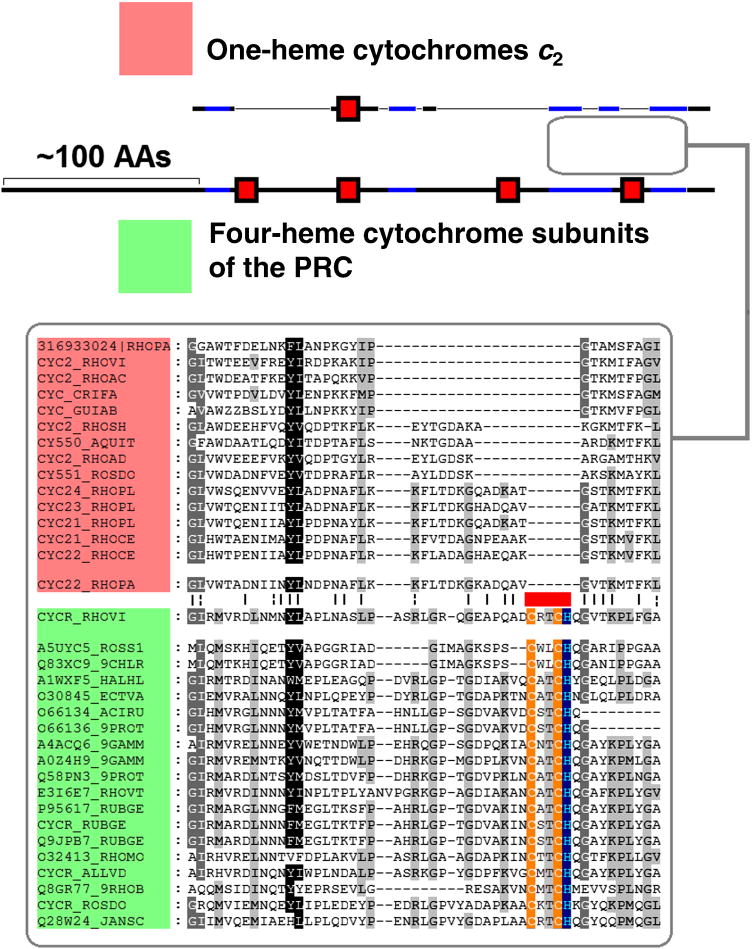
The need to replace low-potential, autooxidizable redox carriers by high-potential non-oxidizable counterparts in aerobic organisms was not limited to the constituents of the bc1 complex, but should have affected all members of ET chains. One of the proteins that may have emerged upon the transition to aerated environments was the small cytochrome c with Em ≥ 300 mV that transfers electrons from the bc1 complex to multiple electron acceptors including the cytochrome oxidases and the PRCs of phototrophic proteobacteria (cytochrome c2, according to recent classification [145]). This small protein has been extensively studied during the early years of molecular biology and served as a model system for numerous phylogenetic studies, see e.g. [146–148]. Dickerson separated all cytochromes c into three groups, namely short, medium and long; he suggested that that the “long” cytochromes, found within α-proteobacteria, were derived from the “medium” cytochromes by insertions [147]. More recent analysis indicated the emergence of cytochromes c2 within α-proteobacteria [145]. Since the “long” cytochromes c2 are found only among α-proteobacteria, they could be then closer to the ancient form, while the widespread shorter cytochromes could result from deletions. This tentative scenario contradicts the classical scenario of Dickerson [147], but corresponds to the “collapse” mechanism, as suggested for the evolution of cytochrome c1 [144]. It is noteworthy that in some phototrophic α-proteobacteria, cytochromes c2 serve as direct electron donors to the PRC, whereas in other α-proteobacteria this function is performed by a tetraheme cytochrome c subunit of the PRC. Comparison of the “long” cytochromes c2 with the tetraheme PRC subunits reveals a certain degree of similarity between two stretches; the similarity is most pronounced upon comparison of the longest cytochromes c2 of Rhodopseudomonas palustris with the tetraheme cytochrome of the closely related Blastochloris viridis, formerly Rhodopseudomonas viridis (Fig. 5). Although the extent of the similarity is low and statistically insignificant, it still can reflect the collapse of a tetraheme cytochrome c into a single-heme cytochrome c2. Indeed, the similarity is found between two proteins which perform the same function in photosynthesis, serving as immediate electron donors to the PRC. The PRC of Blastochloris viridis contains a menaquinone as a primary electron acceptor QA [149] and therefore might represent an evolutionarily older form as compared to the PRC of Rps. palustris that contains an ubiquinone molecule as QA [150]. The collapse of a larger cytochrome should have been accompanied by the loss of the three hemes. The alignment in Fig. 5 indicates a deletion of one of the heme-binding sites. Generally, the heme loss could proceed gradually; a loss of one of the four hemes in course of evolution has been reported for the PRC cytochrome subunits in members of genus Rhodovulvum [151, 152]. The complete genome of Rp. palustris contains several “longest” known isoforms of cytochrome c2 [153]. It is tempting to speculate that this genome has retained the intermediate forms reflecting the (multistep?) transition from a large tetraheme cytochrome c to a smaller, versatile single-heme cytochrome c2.
Figure 5.
Comparison of the one-heme cytochome c2 from Rhodopseudomonas palustris and its closest homologs (colored red) and the four-heme PRC cytochome subunit from Blastochloris viridis and its homologs (colored green). The heme-binding sites are marked by red rectangles. The part of the multiple alignment between C-terminal parts of the proteins is shown below in the box. The black stretches are the regions which are not aligned on multiple alignment, while the blue stretches show regions with detectable similarity that are included in the multiple alignment.
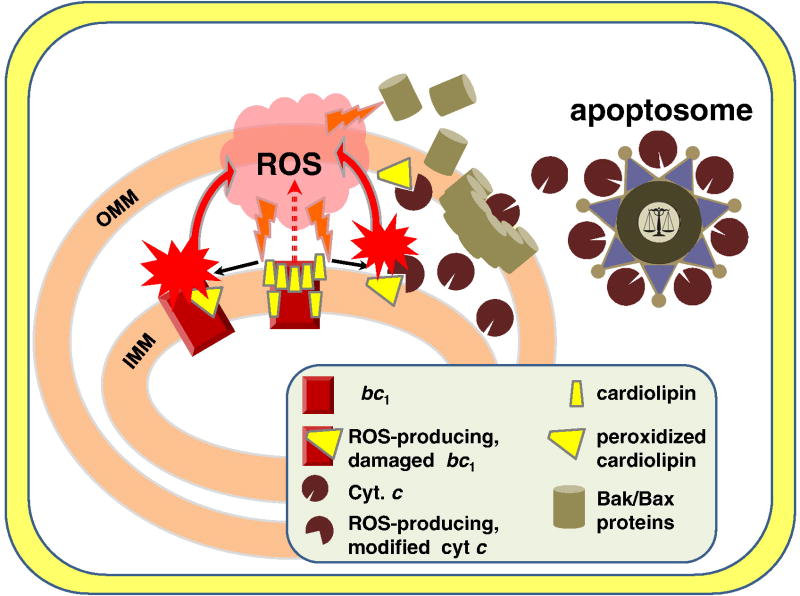
3. Evolution of apoptosis as a strategy to diminish the oxygen-caused damage to consortia of cells
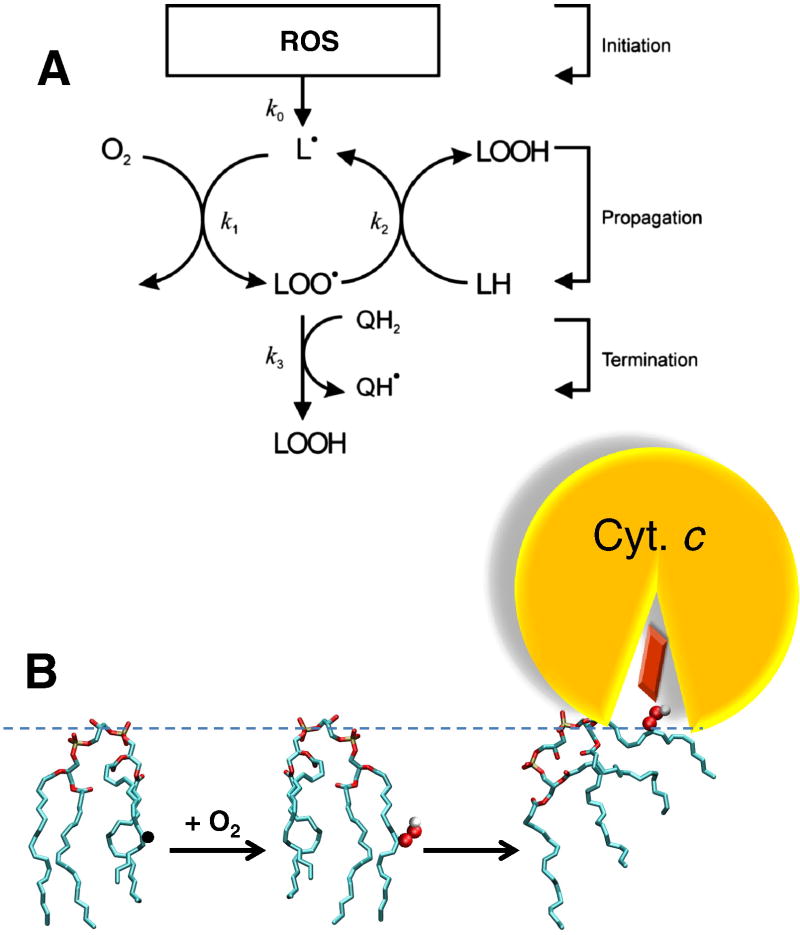
As discussed in the previous section, evolution of the cytochrome bc complexes both in aerobic prokaryotes and in oxygenic phototrophs was accompanied by “defusing” the potential sources of ROS within these enzymes. Still, one source of ROS could not be deactivated completely. The oxidation of a quinol molecule in the center P of all cytochrome bc complexes is accompanied by a transient formation of a low-potential unstable ubisemiqunone UQp•− that promptly reduces the low-potential heme bp, see Fig. 4C, Fig. S6 in Supplementary Materials, and refs. [120, 154–156] for reviews. The redox potential of this ubisemiquinone cannot be increased (e.g. via its stabilization by the surrounding amino acid side chains), without losses in the thermodynamic efficiency of the Q-cycle. Instead, the cytochrome bc complexes are particularly fine-tuned to minimize the electron escape to oxygen in center P [120, 155]. Specifically, the lifetime of the semiquinone in center P is kept very short, so that that this semiquinone could be measured only under very special, steady state conditions [3]. However, when the oxidation of cytochrome b via center N is blocked, which can happen in the presence of inhibitors, or under the backpressure of membrane potential, or as a result of a conformational distortion of the bc1, or in response to an abrupt change in the redox balance of the ET chain, the probability of ubisemiquinone in center P could transiently increase, so that electrons can occasionally escape to oxygen yielding superoxide and other ROS [120, 155–159]. Specifically, the ROS yield increases in response to the oxidation of the membrane ubiquinol pool [156, 158]. Under such oxidized conditions, the bc1 can get out from the kinetically optimized activated mode (see Section 2.3. and [33, 35]), which could lead to an increased probability of UQp•− accumulation. Transient oxidation of ubiquinol pool can hardly be avoided after traumas and during reperfusion (the restoration of blood flow to an organ or to tissue, e.g. after a heart attack, ischemia or a stroke). ROS can damage membrane components and, specifically, the bc1 itself, which would deregulate the fine tuning in this enzyme. Yin and co-workers have shown recently that gradual destruction of the bc1 structural integrity by different means – e.g. heat inactivation or proteinase K digestion – all led to a concurrent increase in superoxide production [159]. Hence, the bc1 can get into a vicious cycle – occasional generation of ROS could eventually damage the bc1 itself or, by affecting its neighbors in the membrane, change its conformation, which would lead to a further increase in the ROS production and further functional damage to the bc1. One possible strategy to save other cells from the ROS-generating vicious cycle in damaged mitochondria is by dismantling the initially affected cell – i.e. apoptosis [38].
3.1. Role of the bc1–bound cardiolipin molecules in triggering apoptosis in vertebrates
While some apoptotic mechanisms may already be present in bacteria [160–162], they are much better studied in the cells of multicellular organisms, where mitochondria, the descendants of endosymbiotically obtained α-proteobacteria, were found to be involved in the so-called intrinsic apoptotic pathway [38, 40, 163, 164]. Under physiological conditions, ROS, apparently, can serve as triggers of apoptosis [38, 157, 165]. While ROS generation could occur both in the NADH dehydrogenase (complex I) and in the cytochrome bc1 complex (complex III) [154], the latter is far more dangerous as a source of ROS. ROS are generated in complex I under the conditions of reverse electron flow, which implies high membrane potential, high succinate/fumarate ratio and a low NADH/NAD+ ratio [157, 166, 167]. These conditions are anything but physiological. In addition, any damage to the membrane, caused by ROS, would decrease the membrane potential and thereby diminish the ROS production in complex I. The situation with the ROS production in the bc1 (mitochondrial complex III) is different from that in complex I. The ROS are produced by the forward electron flow [156]. Generally, a drop in membrane potential should stop the production of ROS also in an intact bc1 [166]. If it is not the case, the ROS-producing cell must be eliminated [165, 168].
Comparison of the apoptotic pathways in different multicellular organisms shows that the evolution of apoptotic cascades in vertebrates has led to development of mechanisms which diminish the ROS-induced damage to other cells by triggering the apoptosis ASAP, i.e. even before the disruption of the affected mitochondria. Specifically, a signal amplification cascade seems to operate within mitochondria of vertebrates, see Fig. 6. That cascade involves molecules of cardiolipin (CL), a four-tail lipid that is particularly susceptible to the ROS-induced peroxidation. CL molecules are the main targets of peroxidation in mitochondrial membranes [169, 170]. Their susceptibility to peroxidation is due to the fact that one molecule of CL usually carries four linoleate chains, (i.e. contains eight unsaturated bonds; a scheme of radical propagation upon peroxidation of unsaturated lipid bonds is shown in Fig. 6A). The high number of double bonds in CL is important because it enables packing of the CL molecules in the bilayer. To form a stable membrane bilayer, amphipilic molecules must have cylindrical shape with approximately similar widths of the polar head and the hydrophobic tails [171]. This condition is fulfilled for a CL molecule with unsaturated fatty acids but not for CL molecules with peroxidized, partially polar chains. Therefore, a peroxidized CL molecule would tend to stick out from the bilayer and can interact with cytochrome c molecules at the membrane surface, see Fig. 6B [172–177]. By inserting into the cytochrome c, a fatty acid chain of CL opens the heme-binding cleft, breaks the methionine-iron bond and makes the heme accessible to external ligands, such as oxygen and peroxy groups [177, 178], so that cytochrome c starts to generate ROS, including even singlet oxygen [179]. The ROS apparently accelerate formation of a pore in the outer membrane and the release of cytochrome c into the cytoplasm, see [41, 177].
Figure 6.
Oxidation of cardiolipin in the mitochondrial membrane.
A, general scheme of lipid peroxidation, adapted from [237]; L is a lipid molecule, QH2and QH• are membrane ubiquinol and its semiquinone form, respectively. B, transformations of a cardiolipin molecule upon peroxidation according to [177, 178]. The black dot indicates the position of an unpaired electron, see the main text for further details.
The breakage of the methionine-iron bond should also decrease the Em value of cytochrome c [180, 181], so that the ability of superoxide generation by such modified cytochrome molecules cannot be excluded. Ironically, this conversion is exactly the reverse of what has happened in the course of evolution, when the autooxidizable cytochromes were replaced by non-oxidizable, high-potential cytochromes, the hemes of which were tightly wrapped in polypeptide chains to prevent the access of dioxygen molecules to the heme iron, as discussed in previous sections.
In which part of mitochondrial membrane the CL molecules could be oxidized? The oxidation is unlikely to take place within the membrane bilayer where lipids are well protected from peroxidation by the pool ubiquinol molecules, potent antioxidants. Indeed, the bottleneck in the radical propagation is the step of the hydrogen atom transfer (reaction 1 in Fig. 6A). This reaction is very slow. For a polyunsaturated linoleate acid, its rate constant is as small as 60 M−1s−1. Ubiquinol molecules interact with peroxides (reaction 3 in Fig. 6A) much faster, with a rate constant as high as 3×105 M−1s−1, see [182] and references therein. Because the concentration of ubiquinol in the mitochondrial membrane is comparable with the concentration of polyunsaturated lipid chains, ubiquinol should fully protect the bilayer lipids from peroxidation [183]. Therefore the only lipid molecules that are susceptible to peroxidation are those molecules that are not accessible for the pool ubiquinol molecules, i.e. lipid molecules that are trapped between membrane protein complexes, primarily, molecules of CL. Most recent data show that a large part, if not the majority, of CL molecules in mitochondrial membranes are associated with protein complexes [184, 185]. Cardiolipin molecules were identified in the crystal structures of many energy-converting enzymes, such as ADP/ATP carrier [186], succinate dehydrogenase [187], cytochrome bc1 complex [17, 188–190], cytochrome c oxidase [191], and formate dehydrogenase [192], see also [184, 185, 193, 194] for reviews. The mitochondrial respiratory supercomplex as a whole appears to include dozens of CL molecules [184].
It should be stressed that CL molecules are indispensable since they perform several specific functions in energy converting membranes [195, 196]. First, owing to their four hydrophobic tails, they can “glue” membrane proteins with each other [197–199], which is why they are so abundant within supercomplexes of membrane enzymes [184, 185]. Second, CL molecules, due to their high second pK of about 8.0, can serve as proton traps and guide protons to the active sites of energy converting enzymes [195, 200, 201].
Obviously, the fastest way to accelerate the apoptosis is to use a ROS-generating cytochrome bc1 complex proper as a trigger of a signal amplification cascade that involves oxidized CL molecules. It seems that exactly this strategy is realized in vertebrates. It is noteworthy that in all the aforementioned CL-containing structures, with the exception of the cytochrome bc1 complex, CL is bound at the protein/membrane interface. In the bovine complex III, however, one of the CL binding sites is deeply buried within the protein (Fig. 7A). Specifically, in the 1PP9 structure [202], the site contains two CL molecules and one molecule of phosphatidylcholine. Structural and sequence analysis of the residues that bind these CL molecules within the cytochrome bc1 complex (see Figs S6–S10 in File 1 of Supplementary Materials) showed that the number of charged residues, which bind the phosphate groups of the CL molecules, has increased upon the evolution from the b6f-type complexes, via the bc1-type complexes of α-proteobacteria, the predecessors of mitochondria, to mitochondrial bc1-type complexes.
Figure 7.
Clusters of occluded cardiolipin molecules in the cytochrome bc1 complexes. Cardiolipin molecules are colored by element: carbon – cyan, oxygen – red and phosphorus – yellow. The 9.5 kD subunit (subunit G in the bovine bc1 and subunit H in the yeast bc1) is colored orange, its positively charged residues are colored blue. The positively charged residues, provided by cytochromes b and c1, are colored violet, see also Figs S7–S11 in Supplementary Materials. A, bovine cytochrome bc1 complex, PDB record 1PP9 [202]; B, yeast cytochrome bc1 complex, PDB record 3CX5 [189]. The figure was produced with the help of the VMD software package [238].
While in prokaryotes the ligands for these CL molecules are provided by subunits of cytochromes b and c1, the eukaryotic organisms use one more 9.5 kD subunit subunit to fix these CL molecules. With help of this subunit, protein completely encases the lipid molecules by a kind of a “belt”. A comparison of two structures in Fig. 7 shows that the extent of trapping of these CL molecules increased upon the evolutionary transition from yeasts to vertebrates. In mammals (Fig. 7A), the unusually bent, laterally running α-helical “belt” fully encases the CL patch and additionally stabilizes it by providing three positively charged residues. On the contrary, in the structure of yeast enzyme (Fig. 7B), the belt contains less positively charged residues and seems to be not so tight which should permit a faster exchange of CL molecules with those in the lipid phase.
Thus, upon the evolution from bacteria to vertebrates, a very special CL-binding site has evolved within the cytochrome bc1 complex where the CL molecules are tightly bound close to the major source of ROS. In response to ROS generation by a mammalian cytochrome bc1 complex, this patch of CL molecules, where eight linoleate chains are tightly packed in the site so that the local concentration of double bonds is an order of magnitude higher than that in the lipid phase on the average, would be oxidized pretty soon. The conformation of the trapped CL molecules would change, affecting the conformation of the cytochrome bc1 complex, which, according to already cited data of Yin et al. [159] could lead to the further increase in the ROS production. The ROS should also prompt the oxidation of other CL molecules abundant in the vicinity of the cytochrome bc1 complex. If not tightly bound, these molecules would “slip out” of the bilayer, so that their fatty acid tails could convert molecules of cytochrome c into peroxidases. The resulting increase in generation of ROS would then trigger the formation of the inner membrane pore, swelling of mitochondrial matrix, disruption of outer mitochondrial membrane, the release of cytochrome c molecules to cytosol, their interaction with the Apaf-1 protein, and activation of apoptotic caspases [38, 47, 165].
4. Some medical implications and outlook
The scheme in Fig. 8 shows that mitochondria of vertebrates use several amplification cascades to accelerate the transmission of the suicide message from the interior of mitochondria to the apoptotic machinery in the cytoplasm. As it follows from the scheme, the apoptotic cascade could be most efficiently stopped by specifically inhibiting the peroxidation of CL molecules, which might explain why apoptotic reactions can be prevented or diminished by some antioxidants. It has been shown that mitochondrially targeted peptide antioxidants could simultaneously prevent lipid peroxidation and the cell death [203]. Shao et al. have studied the impact of propofol, a low-molecular phenolic anesthetic on diverse mitochondrial activities as well as on lipid peroxidation and CL content. Propofol treatment had strong dose-dependent protection attenuating alteration of these parameters [204]. An ubiquinone analog idebenone (6-(10-hydroxydecyl)-2,3-dimethoxy-5-methyl-1,4-benzoquinone) has been shown to protect the mitochondrial membrane against lipid peroxidation and improve the overall brain function after vascular disorders due to strokes or experimental cerebral ischemia [205–208]. Another ubiquinol analog, decylubiquinol, also blocked ROS production and prevented activation of the mitochondrial permeability transition and the cell death [209]. Yet another mitochondrially targeted ubiquinol analog 10-(6′-ubiquinolyl)decyltriphenylphosphonium (MitoQ) has been shown to prevent the lipid peroxidation and the apoptotic reactions [210, 211]. Even more pronounced effects were observed with mitochondrially targeted plastoquinone derivatives (SkQs) [169, 212–221]. In vitro, the antioxidant effect of SkQ1 correlates with prevention of the peroxidation of CL [214, 219]. The experiments in vivo have shown a multiplicity of effects. SkQs prolonged lifespan of various organisms, from fungi to mammals. These compounds helped animals to survive after kidney ischemia, stroke, and heart attack and decelerated development of many age-related diseases and traits, including cataract and some other eye diseases, balding, achromotrichia, lordokyphosis, and myeloid shift of the blood [212–214, 216, 217, 221–227].
Figure 8.
Triggering of the intrinsic apoptotic pathway by a mitochondrion in a mammalian cell. The ROS occasionally generated in the bc1 (red dashed arrow), by oxidizing cardiolipin and eventually damaging the bc1 itself, produce two potent sources of ROS, thus accelerating the triggering of the apoptosis (see the main text for further details). Potential positive feedback loops, namely the increase in the yield of ROS upon damaging of the bc1 [159] and the transformation of cytochrome c into a peroxidase by oxidized cardiolipin (CL) molecules [177, 178] are emphasized. See the text for further details.
Recently mitochondria-targeted antioxidant, a cationic rhodamine derivative linked to a plastoquinone molecule (SkQR1), was studied in the model of open focal trauma of rat brain sensorimotor cortex. It was found that injections of SkQR1 after the trauma improved performance in a test characterizing neurological deficit and decreased the volume of the damaged cortical area [225]. Most recently, it has been shown that under the conditions of experimental traumatic brain injury the activation of ROS production was followed by selective oxidation of cardiolipin, whose peroxidized molecules have been identified and quantified by electrospray ionization mass spectrometry analysis [228]. By applying a brain-permeable mitochondria-targeted electron scavenger XJB-5–131 (a conjugate of 4-amino TEMPO and a chemically modified segment of a membrane-targeting peptide antibiotic Gramicidin S), it was possible to prevent the CL oxidation in the brain, achieve a substantial reduction in neuronal death both in vitro and in vivo, and markedly reduce behavioral deficits and cortical lesion volume [228].
Supposedly, all the aforementioned antioxidants can quench the radical states of protein-bound CL molecules, unlike bulky, hydrophobic molecules of natural ubiquinol that should protect from oxidation the lipids of in the membrane bilayer, but are apparently unable to insert into membrane proteins or intercalate between such proteins. Indeed, small and amphiphilic molecules of propofol, peptide antioxidants and XJB-5–131 should be able to reach even the occluded CL molecules, particularly, the CL molecules that are trapped within the bc1. Idebenone, decylubiquinone, MitoQ and SkQs contain 10-carbon atom chains and show clear structural similarity with 10-nonyl acridine orange which preferably binds to CL [229, 230].
The therapeutic effect of such small antioxidants (see above) might be due to the fact that the ROS generation in the cytochrome bc1 complex could be triggered by two different causes. On the one hand, ROS could result from a breakdown of the cytochrome bc1 complex itself or other potentially ROS-generating enzymes. Such cases seem to be irreparable and prompt apoptosis or mitophagy [43] should be the best solution. On the other hand, transient bursts of ROS could be caused by abrupt changes in the redox balance of the mitochondrial electron transfer system [120, 156, 159, 165, 231], e.g. after a trauma, ischemia, or a stroke. If during this transient period the CL molecules, particularly those interacting with the cytochrome bc1 complex, are protected from peroxidation, the system could restore its redox balance and return to normality. It is conceivable that the therapeutic impact of penetrating antioxidants is due to their ability to postpone the activation of the cardiolipin - ROS - cytochrome c - Apaf1 cascade.
Summing up, the overall evolution of cytochrome bc complex seems to represent a chain of exaptation events, when the preexisting systems are recruited to perform a new function, which is followed by their adaptive changes [232]. The cytochrome bc complex could develop from a pre-existing membrane-anchored dehydrogenase in response to a “redox challenge” from biogenic photosynthesis. The emergence of the oxygenic photosynthesis and the subsequent increase in the atmospheric oxygen content could be interpreted as events that shaped the bc1-type complexes of mitochondria and b6f-type complexes of chloroplasts as we know them. The respective selection pressure might explain the differences between the cytochrome bc complexes of anaerobic, oxygenic and aerobic organisms, respectively. Apparently, those organisms that dwelled in oxygenated habitats should have adapted to minimize the production of ROS by their enzymes.
The evolutionary history of the bc1-type complexes and their functionally coupled proteins, such as cytochrome c2, made, however, a remarkable twist after the emergence of multicellular organisms. In such organisms, the increased level of ROS in a particular cell – e.g. under conditions of oxidative stress – would not cause the death of whole organism, provided that the affected cell is promptly eliminated. To accelerate apoptosis of a damaged cell, its cytochrome bc1 complexes and molecules of cytochrome c2, apparently, undergo transformations which are reverse to what has happened with these proteins in the course of evolution. As a result, these enzymes start to produce more ROS under certain conditions, so that the apoptosis of the damaged cell takes place earlier, and the neighboring cells avoid being damaged by ROS.
Supplementary Material
Highlights.
Cytochrome bc complex could have evolved from a membrane-anchored dehydrogenase
The first cytochrome bc complex - a b6f-type enzyme with a split cytochrome b?
Was the divergence of cytochrome bc complexes driven by oxygenation of the atmosphere?
Oxidation of cardiolipin molecules within the mitochondrial bc1 may impair its function
Impaired cytochrome bc1 produces reactive oxygen species, which trigger apoptosis
Acknowledgments
Very useful discussions with Drs. D. Frishman, E.V. Koonin, K.S. Makarova and Y.I. Wolf are greatly appreciated. Of a particularly value for us was the suggestion of Dr. A.V. Bogachev on the emergence of first high-potential electron acceptors in the framework of biogenic photosynthesis. We are grateful to an anonymous Reviewer for pointing out the binding of the cn heme in the b6f- type complexes by the CXGG motif. This study was supported by the Mitoengineering Institute, Moscow, Russia and grants from the Deutsche Forschungsgemeinschaft (DFG-Mu-1285/1-10, DFG-436-RUS 113/963/0-1), the COST Action CM0902 of the EU and the Russian Government (No 02.740.11.5228) to A.Y.M., from the Deutscher Akademischer Austausch Dienst to D.V.D. and from the Russian Foundation for Basic Research (RFBR 0-04-91331) to V.P.S. M.Y.G is supported by the Intramural Research Program of the NIH at the National Library of Medicine.
Footnotes
Translated from Russian by Michael Glenny, 1967, Collins and Harvill Press, London
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Berry EA, Guergova-Kuras M, Huang LS, Crofts AR. Structure and function of cytochrome bc complexes. Ann Rev Biochem. 2000;69:1005–1075. doi: 10.1146/annurev.biochem.69.1.1005. [DOI] [PubMed] [Google Scholar]
- 2.Cramer WA, Hasan SS, Yamashita E. The Q cycle of cytochrome bc complexes: a structure perspective. Biochim Biophys Acta. 2011;1807:788–802. doi: 10.1016/j.bbabio.2011.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Al-Attar S, de Vries S. Energy transduction by respiratory metallo-enzymes: From molecular mechanism to cell physiology. Coord Chem Rev. 2012;257:64–80. [Google Scholar]
- 4.Berry EA, De Bari H, Huang LS. Unanswered questions about the structure of cytochrome bc complexes. Biochim Biophys Acta. 2013 doi: 10.1016/j.bbabio.2013.04.006. [DOI] [PubMed] [Google Scholar]
- 5.Hasan SS, Yamashita E, Cramer WA. Transmembrane signaling and assembly of the cytochrome bf-lipidic charge transfer complex. Biochim Biophys Acta. 2013 doi: 10.1016/j.bbabio.2013.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cramer WA, Knaff DB. Energy Transduction in Biological Membranes: A Textbook of Bioenergetics. Springer-Verlag; 1990. [Google Scholar]
- 7.Xia D, Yu CA, Kim H, Xia JZ, Kachurin AM, Zhang L, Yu L, Deisenhofer J. Crystal structure of the cytochrome bc1 complex from bovine heart mitochondria. Science. 1997;277:60–66. doi: 10.1126/science.277.5322.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang Z, Huang L, Shulmeister VM, Chi YI, Kim KK, Hung LW, Crofts AR, Berry EA, Kim SH. Electron transfer by domain movement in cytochrome bc1. Nature. 1998;392:677–684. doi: 10.1038/33612. [DOI] [PubMed] [Google Scholar]
- 9.Iwata S, Lee JW, Okada K, Lee JK, Iwata M, Rasmussen B, Link TA, Ramaswamy S, Jap BK. Complete structure of the 11-subunit bovine mitochondrial cytochrome bc1 complex. Science. 1998;281:64–71. doi: 10.1126/science.281.5373.64. [DOI] [PubMed] [Google Scholar]
- 10.Lange C, Hunte C. Crystal structure of the yeast cytochrome bc1 complex with its bound substrate cytochrome c. Proc Natl Acad Sci U S A. 2002;99:2800–2805. doi: 10.1073/pnas.052704699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Berry EA, Huang LS. Observations concerning the quinol oxidation site of the cytochrome bc1 complex. FEBS Letters. 2003;555:13–20. doi: 10.1016/s0014-5793(03)01099-8. [DOI] [PubMed] [Google Scholar]
- 12.Esser L, Elberry M, Zhou F, Yu CA, Yu L, Xia D. Inhibitor-complexed structures of the cytochrome bc1 from the photosynthetic bacterium Rhodobacter sphaeroides. J Biol Chem. 2008;283:2846–2857. doi: 10.1074/jbc.M708608200. [DOI] [PubMed] [Google Scholar]
- 13.Xia D, Esser L, Tang WK, Zhou F, Zhou Y, Yu L, Yu CA. Structural analysis of cytochrome bc1 complexes: Implications to the mechanism of function. Biochim Biophys Acta. 2012 doi: 10.1016/j.bbabio.2012.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Widger WR, Cramer WA, Herrmann RG, Trebst A. Sequence homology and structural similarity between cytochrome b of mitochondrial complex III and the chloroplast b6-f complex: position of the cytochrome b hemes in the membrane. Proc Natl Acad Sci U S A. 1984;81:674–678. doi: 10.1073/pnas.81.3.674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stroebel D, Choquet Y, Popot JL, Picot D. An atypical haem in the cytochrome b6f complex. Nature. 2003;426:413–418. doi: 10.1038/nature02155. [DOI] [PubMed] [Google Scholar]
- 16.Kurisu G, Zhang H, Smith JL, Cramer WA. Structure of the cytochrome b6f complex of oxygenic photosynthesis: tuning the cavity. Science. 2003;302:1009–1014. doi: 10.1126/science.1090165. [DOI] [PubMed] [Google Scholar]
- 17.Berry EA, Huang LS, Saechao LK, Pon NG, Valkova-Valchanova M, Daldal F. X-Ray structure of Rhodobacter capsulatus cytochrome bc1: comparison with its mitochondrial and chloroplast counterparts. Photosynth Res. 2004;81:251–275. doi: 10.1023/B:PRES.0000036888.18223.0e. [DOI] [PubMed] [Google Scholar]
- 18.Furbacher PN, Tae GS, Cramer WA. Evolution and origins of the cytochrome bc1 and b6f complexes. In: Baltscheffsky H, editor. Origin and evolution of biological energy conversion. Wiley-VCH; New York: 1996. pp. 221–253. [Google Scholar]
- 19.Zhang H, Primak A, Cape J, Bowman MK, Kramer DM, Cramer WA. Characterization of the high-spin heme x in the cytochrome b6f complex of oxygenic photosynthesis. Biochemistry. 2004;43:16329–16336. doi: 10.1021/bi048363p. [DOI] [PubMed] [Google Scholar]
- 20.Cramer WA, Zhang H, Yan J, Kurisu G, Smith JL. Transmembrane traffic in the cytochrome b6f complex. Annu Rev Biochem. 2006;75:769–790. doi: 10.1146/annurev.biochem.75.103004.142756. [DOI] [PubMed] [Google Scholar]
- 21.Baymann F, Giusti F, Picot D, Nitschke W. The ci/bH moiety in the b6f complex studied by EPR: a pair of strongly interacting hemes. Proc Natl Acad Sci U S A. 2007;104:519–524. doi: 10.1073/pnas.0606369104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.de Lacroix de Lavalette A, Barucq L, Alric J, Rappaport F, Zito F. Is the redox state of the ci heme of the cytochrome b6f complex dependent on the occupation and structure of the Qi site and vice versa? J Biol Chem. 2009;284:20822–20829. doi: 10.1074/jbc.M109.016709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pierre Y, Breyton C, Lemoine Y, Robert B, Vernotte C, Popot JL. On the presence and role of a molecule of chlorophyll a in the cytochrome b6f complex. J Biol Chem. 1997;272:21901–21908. doi: 10.1074/jbc.272.35.21901. [DOI] [PubMed] [Google Scholar]
- 24.Zhang H, Huang D, Cramer WA. Stoichiometrically bound beta-carotene in the cytochrome b6f complex of oxygenic photosynthesis protects against oxygen damage. J Biol Chem. 1999;274:1581–1587. doi: 10.1074/jbc.274.3.1581. [DOI] [PubMed] [Google Scholar]
- 25.Dashdorj N, Zhang H, Kim H, Yan J, Cramer WA, Savikhin S. The single chlorophyll a molecule in the cytochrome b6f complex: unusual optical properties protect the complex against singlet oxygen. Biophys J. 2005;88:4178–4187. doi: 10.1529/biophysj.104.058693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Baniulis D, Yamashita E, Zhang H, Hasan SS, Cramer WA. Structure-function of the cytochrome b6f complex. Photochem Photobiol. 2008;84:1349–1358. doi: 10.1111/j.1751-1097.2008.00444.x. [DOI] [PubMed] [Google Scholar]
- 27.Martinez SE, Huang D, Szczepaniak A, Cramer WA, Smith JL. Crystal structure of chloroplast cytochrome f reveals a novel cytochrome fold and unexpected heme ligation. Structure. 1994;2:95–105. doi: 10.1016/s0969-2126(00)00012-5. [DOI] [PubMed] [Google Scholar]
- 28.Kramer DM, Nitschke W, Cooley JW. The cytochrome bc1 and related bc complexes: The Rieske/cytochrome b complex as the functional core of a central electron/proton transfer complex. In: Hunter CN, Daldal F, Thurnauer MC, Beatty JT, editors. The Purple Phototrophic Bacteria. Springer; Dordrecht: 2009. pp. 451–473. [Google Scholar]
- 29.Mitchell P. Protonmotive redox mechanism of the cytochrome b-c1 complex in the respiratory chain: protonmotive ubiquinone cycle. FEBS Lett. 1975;56:1–6. doi: 10.1016/0014-5793(75)80098-6. [DOI] [PubMed] [Google Scholar]
- 30.Mitchell P. Possible molecular mechanisms of the protonmotive function of cytochrome systems. J Theor Biol. 1976;62:327–367. doi: 10.1016/0022-5193(76)90124-7. [DOI] [PubMed] [Google Scholar]
- 31.Kim H, Xia D, Yu CA, Xia JZ, Kachurin AM, Zhang L, Yu L, Deisenhofer J. Inhibitor binding changes domain mobility in the iron-sulfur protein of the mitochondrial bc1 complex from bovine heart. Proc Natl Acad Sci U S A. 1998;95:8026–8033. doi: 10.1073/pnas.95.14.8026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gopta OA, Feniouk BA, Junge W, Mulkidjanian AY. The cytochrome bc1 complex of Rhodobacter capsulatus: ubiquinol oxidation in a dimeric Q-cycle? FEBS Lett. 1998;431:291–296. doi: 10.1016/s0014-5793(98)00768-6. [DOI] [PubMed] [Google Scholar]
- 33.Mulkidjanian AY. Proton translocation by the cytochrome bc1 complexes of phototrophic bacteria: introducing the activated Q-cycle. Photochem Photobiol Sci. 2007;6:19–34. doi: 10.1039/b517522d. [DOI] [PubMed] [Google Scholar]
- 34.Swierczek M, Cieluch E, Sarewicz M, Borek A, Moser CC, Dutton PL, Osyczka A. An electronic bus bar lies in the core of cytochrome bc1. Science. 2010;329:451–454. doi: 10.1126/science.1190899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mulkidjanian AY. Activated Q-cycle as a common mechanism for cytochrome bc1 and cytochrome b6f complexes. Biochim Biophys Acta. 2010;1797:1858–1868. doi: 10.1016/j.bbabio.2010.07.008. [DOI] [PubMed] [Google Scholar]
- 36.Vener AV, van Kan PJ, Rich PR, Ohad I, Andersson B. Plastoquinol at the quinol oxidation site of reduced cytochrome bf mediates signal transduction between light and protein phosphorylation: thylakoid protein kinase deactivation by a single-turnover flash. Proc Natl Acad Sci U S A. 1997;94:1585–1590. doi: 10.1073/pnas.94.4.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kallas T. Cytochrome b6f complex at the heart of energy transduction and redox signaling. In: Eaton-Rye JJ, Tripathy BC, Sharkey TD, editors. Photosynthesis: Plastid Biology, Energy Conversion and Carbon Assimilation. Springer; Dordrecht: 2012. pp. 501–560. [Google Scholar]
- 38.Skulachev VP. Why are mitochondria involved in apoptosis? Permeability transition pores and apoptosis as selective mechanisms to eliminate superoxide-producing mitochondria and cell. FEBS Lett. 1996;397:7–10. doi: 10.1016/0014-5793(96)00989-1. [DOI] [PubMed] [Google Scholar]
- 39.Green DR, Reed JC. Mitochondria and apoptosis. Science. 1998;281:1309–1312. doi: 10.1126/science.281.5381.1309. [DOI] [PubMed] [Google Scholar]
- 40.Wang C, Youle RJ. The role of mitochondria in apoptosis. Annu Rev Genet. 2009;43:95–118. doi: 10.1146/annurev-genet-102108-134850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huttemann M, Pecina P, Rainbolt M, Sanderson TH, Kagan VE, Samavati L, Doan JW, Lee I. The multiple functions of cytochrome c and their regulation in life and death decisions of the mammalian cell: From respiration to apoptosis. Mitochondrion. 2011;11:369–381. doi: 10.1016/j.mito.2011.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cloonan SM, Choi AM. Mitochondria: sensors and mediators of innate immune receptor signaling. Curr Opin Microbiol. 2013 doi: 10.1016/j.mib.2013.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Osiewacz HD, Bernhardt D. Mitochondrial quality control: Impact on aging and life span - a mini-review. Gerontology. 2013 doi: 10.1159/000348662. [DOI] [PubMed] [Google Scholar]
- 44.Shimizu S, Narita M, Tsujimoto Y. Bcl-2 family proteins regulate the release of apoptogenic cytochrome c by the mitochondrial channel VDAC. Nature. 1999;399:483–487. doi: 10.1038/20959. [DOI] [PubMed] [Google Scholar]
- 45.Wei MC, Zong WX, Cheng EH, Lindsten T, Panoutsakopoulou V, Ross AJ, Roth KA, MacGregor GR, Thompson CB, Korsmeyer SJ. Proapoptotic BAX and BAK: a requisite gateway to mitochondrial dysfunction and death. Science. 2001;292:727–730. doi: 10.1126/science.1059108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kushnareva Y, Andreyev AY, Kuwana T, Newmeyer DD. Bax Activation Initiates the Assembly of a multimeric catalyst that facilitates Bax pore formation in mitochondrial outer membranes. PLoS Biol. 2012;10:e1001394. doi: 10.1371/journal.pbio.1001394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Reubold TF, Wohlgemuth S, Eschenburg S. Crystal structure of full-length Apaf-1: how the death signal is relayed in the mitochondrial pathway of apoptosis. Structure. 2011;19:1074–1083. doi: 10.1016/j.str.2011.05.013. [DOI] [PubMed] [Google Scholar]
- 48.Woese CR, Kandler O, Wheelis ML. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc Natl Acad Sci U S A. 1990;87:4576–4579. doi: 10.1073/pnas.87.12.4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lubben M, Kolmerer B, Saraste M. An archaebacterial terminal oxidase combines core structures of two mitochondrial respiratory complexes. EMBO J. 1992;11:805–812. doi: 10.1002/j.1460-2075.1992.tb05117.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schutz M, Brugna M, Lebrun E, Baymann F, Huber R, Stetter KO, Hauska G, Toci R, Lemesle-Meunier D, Tron P, Schmidt C, Nitschke W. Early evolution of cytochrome bc complexes. J Mol Biol. 2000;300:663–675. doi: 10.1006/jmbi.2000.3915. [DOI] [PubMed] [Google Scholar]
- 51.Nitschke W, van Lis R, Schoepp-Cothenet B, Baymann F. The “green” phylogenetic clade of Rieske/cytb complexes. Photosynth Res. 2010;104:347–355. doi: 10.1007/s11120-010-9532-1. [DOI] [PubMed] [Google Scholar]
- 52.Baymann F, Schoepp-Cothenet B, Lebrun E, van Lis R, Nitschke W. Phylogeny of Rieske/cytb complexes with a special focus on the Haloarchaeal enzymes. Genome Biol Evol. 2012;4:720–729. doi: 10.1093/gbe/evs056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ten Brink F, Schoepp-Cothenet B, van Lis R, Nitschke W, Baymann F. Multiple Rieske/cytb complexes in a single organism. Biochim Biophys Acta. 2013 doi: 10.1016/j.bbabio.2013.03.003. [DOI] [PubMed] [Google Scholar]
- 54.Lazcano A, Forterre P. The molecular search for the last common ancestor. J Mol Evol. 1999;49:411–412. [PubMed] [Google Scholar]
- 55.Koonin EV. Comparative genomics, minimal gene-sets and the last universal common ancestor. Nat Rev Microbiol. 2003;1:127–136. doi: 10.1038/nrmicro751. [DOI] [PubMed] [Google Scholar]
- 56.Castresana J, Saraste M. Evolution of energetic metabolism: the respiration-early hypothesis. Trends Biochem Sci. 1995;20:443–448. doi: 10.1016/s0968-0004(00)89098-2. [DOI] [PubMed] [Google Scholar]
- 57.Wald G. The Origins of Life. Proc Natl Acad Sci U S A. 1964;52:595–611. doi: 10.1073/pnas.52.2.595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Williams RJP, Frausto da Silva JJR. The Chemistry of Evolution: The Development of our Ecosystem. Elsevier; Amsterdam: 2006. [Google Scholar]
- 59.Lebrun E, Santini JM, Brugna M, Ducluzeau AL, Ouchane S, Schoepp-Cothenet B, Baymann F, Nitschke W. The Rieske protein: a case study on the pitfalls of multiple sequence alignments and phylogenetic reconstruction. Mol Biol Evol. 2006;23:1180–1191. doi: 10.1093/molbev/msk010. [DOI] [PubMed] [Google Scholar]
- 60.Anisimova M, Gascuel O. Approximate likelihood-ratio test for branches: A fast, accurate, and powerful alternative. Syst Biol. 2006;55:539–552. doi: 10.1080/10635150600755453. [DOI] [PubMed] [Google Scholar]
- 61.Anisimova M, Gil M, Dufayard JF, Dessimoz C, Gascuel O. Survey of branch support methods demonstrates accuracy, power, and robustness of fast likelihood-based approximation schemes. Syst Biol. 2011;60:685–699. doi: 10.1093/sysbio/syr041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kartal B, de Almeida NM, Maalcke WJ, Op den Camp HJ, Jetten MS, Keltjens JT. How to make a living from anaerobic ammonium oxidation. FEMS Microbiol Rev. 2013;37:428–461. doi: 10.1111/1574-6976.12014. [DOI] [PubMed] [Google Scholar]
- 63.Kartal B, Maalcke WJ, de Almeida NM, Cirpus I, Gloerich J, Geerts W, Op den Camp HJ, Harhangi HR, Janssen-Megens EM, Francoijs KJ, Stunnenberg HG, Keltjens JT, Jetten MS, Strous M. Molecular mechanism of anaerobic ammonium oxidation. Nature. 2011;479:127–130. doi: 10.1038/nature10453. [DOI] [PubMed] [Google Scholar]
- 64.Ettwig KF, Butler MK, Le Paslier D, Pelletier E, Mangenot S, Kuypers MM, Schreiber F, Dutilh BE, Zedelius J, de Beer D, Gloerich J, Wessels HJ, van Alen T, Luesken F, Wu ML, van de Pas-Schoonen KT, Op den Camp HJ, Janssen-Megens EM, Francoijs KJ, Stunnenberg H, Weissenbach J, Jetten MS, Strous M. Nitrite-driven anaerobic methane oxidation by oxygenic bacteria. Nature. 2010;464:543–548. doi: 10.1038/nature08883. [DOI] [PubMed] [Google Scholar]
- 65.Yu J, Le Brun NE. Studies of the cytochrome subunits of menaquinone:cytochrome c reductase (bc complex) of Bacillus subtilis. Evidence for the covalent attachment of heme to the cytochrome b subunit. J Biol Chem. 1998;273:8860–8866. doi: 10.1074/jbc.273.15.8860. [DOI] [PubMed] [Google Scholar]
- 66.Baymann F, Nitschke W. Heliobacterial Rieske/cytb complex. Photosynth Res. 2010;104:177–187. doi: 10.1007/s11120-009-9524-1. [DOI] [PubMed] [Google Scholar]
- 67.Finkelstein A, Ptitsyn O. Protein Physics: A Course of Lectures. Academic Press; 2002. [Google Scholar]
- 68.Mulkidjanian AY, Junge W. On the origin of photosynthesis as inferred from sequence analysis - A primordial UV-protector as common ancestor of reaction centers and antenna proteins. Photosynthesis Research. 1997;51:27–42. [Google Scholar]
- 69.Dikanov SA, Samoilova RI, Kolling DR, Holland JT, Crofts AR. Hydrogen bonds involved in binding the Qi-site semiquinone in the bc1 complex, identified through deuterium exchange using pulsed EPR. J Biol Chem. 2004;279:15814–15823. doi: 10.1074/jbc.M313417200. [DOI] [PubMed] [Google Scholar]
- 70.Yu J, Hederstedt L, Piggot PJ. The cytochrome bc complex (menaquinone:cytochrome c reductase) in Bacillus subtilis has a nontraditional subunit organization. J Bacteriol. 1995;177:6751–6760. doi: 10.1128/jb.177.23.6751-6760.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Marcotte EM, Pellegrini M, Ng HL, Rice DW, Yeates TO, Eisenberg D. Detecting protein function and protein-protein interactions from genome sequences. Science. 1999;285:751–753. doi: 10.1126/science.285.5428.751. [DOI] [PubMed] [Google Scholar]
- 72.Enright AJ, Iliopoulos I, Kyrpides NC, Ouzounis CA. Protein interaction maps for complete genomes based on gene fusion events. Nature. 1999;402:86–90. doi: 10.1038/47056. [DOI] [PubMed] [Google Scholar]
- 73.Hawkins AR, Lamb HK. The molecular biology of multidomain proteins. Selected examples. Eur J Biochem. 1995;232:7–18. doi: 10.1111/j.1432-1033.1995.tb20775.x. [DOI] [PubMed] [Google Scholar]
- 74.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 75.Dibrova DV, Galperin MY, Mulkidjanian AY. The cytochrome bc1 complex and the evolution of membrane bioenergetics. Biochim Biophys Acta. 2012;1817:S91. [Google Scholar]
- 76.Koonin EV, Makarova KS, Aravind L. Horizontal gene transfer in prokaryotes: quantification and classification. Annu Rev Microbiol. 2001;55:709–742. doi: 10.1146/annurev.micro.55.1.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Makarova KS, Aravind L, Galperin MY, Grishin NV, Tatusov RL, Wolf YI, Koonin EV. Comparative genomics of the Archaea (Euryarchaeota): evolution of conserved protein families, the stable core, and the variable shell. Genome Res. 1999;9:608–628. [PubMed] [Google Scholar]
- 78.Csuros M. Count: evolutionary analysis of phylogenetic profiles with parsimony and likelihood. Bioinformatics. 2010;26:1910–1912. doi: 10.1093/bioinformatics/btq315. [DOI] [PubMed] [Google Scholar]
- 79.Pereto J, Lopez-Garcia P, Moreira D. Ancestral lipid biosynthesis and early membrane evolution. Trends Biochem Sci. 2004;29:469–477. doi: 10.1016/j.tibs.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 80.Hemp J, Gennis RB. Diversity of the heme-copper superfamily in archaea: insights from genomics and structural modeling. Results Probl Cell Differ. 2008;45:1–31. doi: 10.1007/400_2007_046. [DOI] [PubMed] [Google Scholar]
- 81.Raina S, Missiakas D. Making and breaking disulfide bonds. Annu Rev Microbiol. 1997;51:179–202. doi: 10.1146/annurev.micro.51.1.179. [DOI] [PubMed] [Google Scholar]
- 82.Koonin EV, Martin W. On the origin of genomes and cells within inorganic compartments. Trends Genet. 2005;21:647–654. doi: 10.1016/j.tig.2005.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mulkidjanian AY, Galperin MY. Evolutionary origins of membrane proteins. In: Frishman D, editor. Structural Bioinformatics of Membrane Proteins. Springer; Heidelberg: 2010. pp. 1–28. [Google Scholar]
- 84.Mansy SS. Membrane transport in primitive cells. Cold Spring Harb Perspect Biol. 2010;2:a002188. doi: 10.1101/cshperspect.a002188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Deamer DW. The first living systems: a bioenergetic perspective. Microbiol Mol Biol Rev. 1997;61:239–261. doi: 10.1128/mmbr.61.2.239-261.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Mulkidjanian AY, Galperin MY, Makarova KS, Wolf YI, Koonin EV. Evolutionary primacy of sodium bioenergetics. Biol Direct. 2008;3:13. doi: 10.1186/1745-6150-3-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Mulkidjanian AY, Dibrov P, Galperin MY. The past and present of sodium energetics: may the sodium-motive force be with you. Biochim Biophys Acta. 2008;1777:985–992. doi: 10.1016/j.bbabio.2008.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Dibrova DV, Galperin MY, Mulkidjanian AY. Characterization of the N-ATPase, a distinct, laterally transferred Na+-translocating form of the bacterial F-type membrane ATPase. Bioinformatics. 2010;26:1473–1476. doi: 10.1093/bioinformatics/btq234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Luoto HH, Baykov AA, Lahti R, Malinen AM. Membrane-integral pyrophosphatase subfamily capable of translocating both Na+ and H+ Proc Natl Acad Sci U S A. 2013;110:1255–1260. doi: 10.1073/pnas.1217816110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Luoto HH, Belogurov GA, Baykov AA, Lahti R, Malinen AM. Na+-translocating membrane pyrophosphatases are widespread in the microbial world and evolutionarily precede H+-translocating pyrophosphatases. J Biol Chem. 2011;286:21633–21642. doi: 10.1074/jbc.M111.244483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Mulkidjanian AY, Galperin MY, Koonin EV. Co-evolution of primordial membranes and membrane proteins. Trends Biochem Sci. 2009;34:206–215. doi: 10.1016/j.tibs.2009.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Baymann F, Lebrun E, Brugna M, Schoepp-Cothenet B, Giudici-Orticoni MT, Nitschke W. The redox protein construction kit: pre-last universal common ancestor evolution of energy-conserving enzymes. Philos Trans R Soc Lond B Biol Sci. 2003;358:267–274. doi: 10.1098/rstb.2002.1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Neumann S, Fuchs A, Mulkidjanian A, Frishman D. Current status of membrane protein structure classification. Proteins. 2010;78:1760–1773. doi: 10.1002/prot.22692. [DOI] [PubMed] [Google Scholar]
- 94.Choma CT, Lear JD, Nelson MJ, Dutton PL, Robertson DE, Degrado WF. Design of a heme-binding four-helix bundle. J Am Chem Soc. 1994;116:856–865. [Google Scholar]
- 95.Rojas NR, Kamtekar S, Simons CT, McLean JE, Vogel KM, Spiro TG, Farid RS, Hecht MH. De novo heme proteins from designed combinatorial libraries. Protein Sci. 1997;6:2512–2524. doi: 10.1002/pro.5560061204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Andreeva A, Howorth D, Chandonia JM, Brenner SE, Hubbard TJ, Chothia C, Murzin AG. Data growth and its impact on the SCOP database: new developments. Nucleic Acids Res. 2008;36:D419–425. doi: 10.1093/nar/gkm993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Jormakka M, Byrne B, Iwata S. Protonmotive force generation by a redox loop mechanism. FEBS Lett. 2003;545:25–30. doi: 10.1016/s0014-5793(03)00389-2. [DOI] [PubMed] [Google Scholar]
- 98.Pandelia ME, Lubitz W, Nitschke W. Evolution and diversification of Group 1 [NiFe] hydrogenases. Is there a phylogenetic marker for O2-tolerance? Biochim Biophys Acta. 2012;1817:1565–1575. doi: 10.1016/j.bbabio.2012.04.012. [DOI] [PubMed] [Google Scholar]
- 99.Hagerhall C. Succinate: quinone oxidoreductases. Variations on a conserved theme. Biochim Biophys Acta. 1997;1320:107–141. doi: 10.1016/s0005-2728(97)00019-4. [DOI] [PubMed] [Google Scholar]
- 100.Dibrova DV, Chudetsky MY, Galperin MY, Koonin EV, Mulkidjanian AY. The role of energy in the emergence of biology from chemistry. Orig Life Evol Biosph. 2012;42:459–468. doi: 10.1007/s11084-012-9308-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Krishtalik LI. Intramembrane electron transfer: Processes in the photosynthetic reaction center. Biochim Biophys Acta. 1996;1273:139–149. [Google Scholar]
- 102.Madej MG, Nasiri HR, Hilgendorff NS, Schwalbe H, Lancaster CR. Evidence for transmembrane proton transfer in a dihaem-containing membrane protein complex. EMBO J. 2006;25:4963–4970. doi: 10.1038/sj.emboj.7601361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Madej MG, Muller FG, Ploch J, Lancaster CR. Limited reversibility of transmembrane proton transfer assisting transmembrane electron transfer in a dihaem-containing succinate:quinone oxidoreductase. Biochim Biophys Acta. 2009;1787:593–600. doi: 10.1016/j.bbabio.2009.02.011. [DOI] [PubMed] [Google Scholar]
- 104.Azarkina N, Konstantinov AA. Electrogenicity of succinate: Menaquinone oxidoreductase from Bacillus subtilis depends on the direction of electron transfer. Biochim Biophys Acta. 2010;1797:112. [Google Scholar]
- 105.Berry EA, Walker FA. Bis-histidine-coordinated hemes in four-helix bundles: how the geometry of the bundle controls the axial imidazole plane orientations in transmembrane cytochromes of mitochondrial complexes II and III and related proteins. J Biol Inorg Chem. 2008;13:481–498. doi: 10.1007/s00775-008-0372-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Hazen RM, Bekker A, Bish DL, Bleeker W, Downs RT, Farquhar J, Ferry JM, Grew ES, Knoll AH, Papineau D, Ralph JP, Sverjensky DA, Valley JW. Needs and opportunities in mineral evolution research. Am Mineral. 2011;96:953–963. [Google Scholar]
- 107.Govindjee J, Whitmarsh Govindjee. Photosynthesis. Academic Press; New York: 1982. Introduction to photosynthesis: Energy conversion by plants and bacteria; pp. 1–16. [Google Scholar]
- 108.Crofts AR, Wraight CA. The electrochemical domain of photosynthesis. Biochim Biophys Acta. 1983;726:149–185. [Google Scholar]
- 109.Mulkidjanian AY, Koonin EV, Makarova KS, Mekhedov SL, Sorokin A, Wolf YI, Dufresne A, Partensky F, Burd H, Kaznadzey D, Haselkorn R, Galperin MY. The cyanobacterial genome core and the origin of photosynthesis. Proc Natl Acad Sci U S A. 2006;103:13126–13131. doi: 10.1073/pnas.0605709103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Hohmann-Marriott MF, Blankenship RE. Evolution of photosynthesis. Annu Rev Plant Biol. 2011;62:515–548. doi: 10.1146/annurev-arplant-042110-103811. [DOI] [PubMed] [Google Scholar]
- 111.Baniulis D, Zhang H, Zakharova T, Hasan SS, Cramer WA. Purification and crystallization of the cyanobacterial cytochrome b6f complex. Methods Mol Biol. 2011;684:65–77. doi: 10.1007/978-1-60761-925-3_7. [DOI] [PubMed] [Google Scholar]
- 112.Zhang H, Whitelegge JP, Cramer WA. Ferredoxin:NADP+ oxidoreductase is a subunit of the chloroplast cytochrome b6f complex. J Biol Chem. 2001;276:38159–38165. doi: 10.1074/jbc.M105454200. [DOI] [PubMed] [Google Scholar]
- 113.Szymanska R, Dluzewska J, Slesak I, Kruk J. Ferredoxin:NADP+ oxidoreductase bound to cytochrome b6f complex is active in plastoquinone reduction: implications for cyclic electron transport. Physiol Plant. 2011;141:289–298. doi: 10.1111/j.1399-3054.2010.01434.x. [DOI] [PubMed] [Google Scholar]
- 114.Iwai M, Takizawa K, Tokutsu R, Okamuro A, Takahashi Y, Minagawa J. Isolation of the elusive supercomplex that drives cyclic electron flow in photosynthesis. Nature. 2010;464:1210–1213. doi: 10.1038/nature08885. [DOI] [PubMed] [Google Scholar]
- 115.Zito F, Finazzi G, Delosme R, Nitschke W, Picot D, Wollman FA. The Qo site of cytochrome b6f complexes controls the activation of the LHCII kinase. EMBO J. 1999;18:2961–2969. doi: 10.1093/emboj/18.11.2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Hunte C, Koepke J, Lange C, Rossmanith T, Michel H. Structure at 2.3 A resolution of the cytochrome bc1 complex from the yeast Saccharomyces cerevisiae co-crystallized with an antibody Fv fragment. Structure Fold Des. 2000;8:669–684. doi: 10.1016/s0969-2126(00)00152-0. [DOI] [PubMed] [Google Scholar]
- 117.Xiong J, Inoue K, Bauer CE. Tracking molecular evolution of photosynthesis by characterization of a major photosynthesis gene cluster from Heliobacillus mobilis. Proc Natl Acad Sci U S A. 1998;95:14851–14856. doi: 10.1073/pnas.95.25.14851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Swingley WD, Chen M, Cheung PC, Conrad AL, Dejesa LC, Hao J, Honchak BM, Karbach LE, Kurdoglu A, Lahiri S, Mastrian SD, Miyashita H, Page L, Ramakrishna P, Satoh S, Sattley WM, Shimada Y, Taylor HL, Tomo T, Tsuchiya T, Wang ZT, Raymond J, Mimuro M, Blankenship RE, Touchman JW. Niche adaptation and genome expansion in the chlorophyll d-producing cyanobacterium Acaryochloris marina. Proc Natl Acad Sci U S A. 2008;105:2005–2010. doi: 10.1073/pnas.0709772105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Zhaxybayeva O, Hamel L, Raymond J, Gogarten JP. Visualization of the phylogenetic content of five genomes using dekapentagonal maps. Genome Biol. 2004;5:R20. doi: 10.1186/gb-2004-5-3-r20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Rutherford AW, Osyczka A, Rappaport F. Back-reactions, short-circuits, leaks and other energy wasteful reactions in biological electron transfer: redox tuning to survive life in O2. FEBS Lett. 2012;586:603–616. doi: 10.1016/j.febslet.2011.12.039. [DOI] [PubMed] [Google Scholar]
- 121.Liebl U, Rutherford AW, Nitschke W. Evidence for a unique Rieske iron-sulfur center in Heliobacterium chlorum. FEBS Letters. 1990;261:427–430. [Google Scholar]
- 122.Yue H, Kang Y, Zhang H, Gao X, Blankenship RE. Expression and characterization of the diheme cytochrome c subunit of the cytochrome bc complex in Heliobacterium modesticaldum. Arch Biochem Biophys. 2012;517:131–137. doi: 10.1016/j.abb.2011.11.012. [DOI] [PubMed] [Google Scholar]
- 123.Ducluzeau AL, Chenu E, Capowiez L, Baymann F. The Rieske/cytochrome b complex of Heliobacteria. Biochim Biophys Acta. 2008;1777:1140–1146. doi: 10.1016/j.bbabio.2008.04.026. [DOI] [PubMed] [Google Scholar]
- 124.Lancaster CR. Wolinella succinogenes quinol:fumarate reductase-2.2-A resolution crystal structure and the E-pathway hypothesis of coupled transmembrane proton and electron transfer. Biochim Biophys Acta. 2002;1565:215–231. doi: 10.1016/s0005-2736(02)00571-0. [DOI] [PubMed] [Google Scholar]
- 125.Lancaster CR. Succinate:quinone oxidoreductases: an overview. Biochimica et Biophysica Acta. 2002;1553:1–6. doi: 10.1016/s0005-2728(01)00240-7. [DOI] [PubMed] [Google Scholar]
- 126.Osyczka A, Moser CC, Daldal F, Dutton PL. Reversible redox energy coupling in electron transfer chains. Nature. 2004;427:607–612. doi: 10.1038/nature02242. [DOI] [PubMed] [Google Scholar]
- 127.Mulkidjanian A, Mamedov MD, Drachev LA. Slow electrogenic events in the cytochrome bc1-complex of Rhodobacter sphaeroides. The electron transfer between cytochrome b hemes can be non-electrogenic. FEBS Lett. 1991;284:227–231. doi: 10.1016/0014-5793(91)80691-u. [DOI] [PubMed] [Google Scholar]
- 128.Klishin SS, Junge W, Mulkidjanian AY. Flash-induced turnover of the cytochrome bc1 complex in chromatophores of Rhodobacter capsulatus: binding of Zn2+ decelerates likewise the oxidation of cytochrome b, the reduction of cytochrome c1 and the voltage generation. Biochim Biophys Acta. 2002;1553:177–182. doi: 10.1016/s0005-2728(01)00250-x. [DOI] [PubMed] [Google Scholar]
- 129.Joliot P, Joliot A. Electrogenic events associated with electron and proton transfers within the cytochrome b6/f complex. Biochim Biophys Acta-Bioenergetics. 2001;1503:369–376. doi: 10.1016/s0005-2728(00)00232-2. [DOI] [PubMed] [Google Scholar]
- 130.Finazzi G. Redox-coupled proton pumping activity in cytochrome b6f, as evidenced by the pH dependence of electron transfer in whole cells of Chlamydomonas reinhardtii. Biochemistry. 2002;41:7475–7482. doi: 10.1021/bi025714w. [DOI] [PubMed] [Google Scholar]
- 131.Nitschke W, Kramer DM, Riedel A, Liebl U. From naphtho- to benzoquinones — (r)evolutionary reorganisations of electron transfer chains. In: Mathis P, editor. Photosynthesis: From Light to Biosphere. I. Kluwer Acad. Publ; Dordrecht: 1995. pp. 945–950. [Google Scholar]
- 132.Schoepp-Cothenet B, Lieutaud C, Baymann F, Vermeglio A, Friedrich T, Kramer DM, Nitschke W. Menaquinone as pool quinone in a purple bacterium. Proc Natl Acad Sci U S A. 2009;106:8549–8554. doi: 10.1073/pnas.0813173106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Schoepp-Cothenet B, van Lis R, Atteia A, Baymann F, Capowiez L, Ducluzeau AL, Duval S, Ten Brink F, Russell MJ, Nitschke W. On the universal core of bioenergetics. Biochim Biophys Acta. 2013;1827:79–93. doi: 10.1016/j.bbabio.2012.09.005. [DOI] [PubMed] [Google Scholar]
- 134.Busch KB, Deckers-Hebestreit G, Hanke GT, Mulkidjanian AY. Dynamics of bioenergetic microcompartments. Biol Chem. 2013;394:163–88. doi: 10.1515/hsz-2012-0254. [DOI] [PubMed] [Google Scholar]
- 135.Pethig R, Gascoyne PR, McLaughlin JA, Szent-Gyorgyi A. Ascorbate-quinone interactions: electrochemical, free radical, and cytotoxic properties. Proc Natl Acad Sci U S A. 1983;80:129–132. doi: 10.1073/pnas.80.1.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.de Vries S, Berden JA, Slater EC. Properties of a semiquinone anion located in the QH2:cytochrome c oxidoreductase segment of the mitochondrial respiratory chain. FEBS Lett. 1980;122:143–148. doi: 10.1016/0014-5793(80)80422-4. [DOI] [PubMed] [Google Scholar]
- 137.Robertson DE, Prince RC, Bowyer JR, Matsuura K, Dutton PL, Ohnishi T. Thermodynamic properties of the semiquinone and its binding site in the ubiquinol- cytochrome c (c2) oxidoreductase of respiratory and photosynthetic systems. J Biol Chem. 1984;259:1758–1763. [PubMed] [Google Scholar]
- 138.Meinhardt SW, Yang XH, Trumpower BL, Ohnishi T. Identification of a stable ubisemiquinone and characterization of the effects of ubiquinone oxidation-reduction status on the Rieske iron-sulfur protein in the three-subunit ubiquinol-cytochrome c oxidoreductase complex of Paracoccus denitrificans. J Biol Chem. 1987;262:8702–8706. [PubMed] [Google Scholar]
- 139.Rich PR, Jeal AE, Madgwick SA, Moody AJ. Inhibitor effects on redox-linked protonations of the b haems of the mitochondrial bc1 complex. Biochim Biophys Acta. 1990;1018:29–40. doi: 10.1016/0005-2728(90)90106-e. [DOI] [PubMed] [Google Scholar]
- 140.Siedow JN, Power S, de la Rosa FF, Palmer G. The preparation and characterization of highly purified, enzymically active complex III from baker’s yeast. J Biol Chem. 1978;253:2392–2399. [PubMed] [Google Scholar]
- 141.de la Rosa FF, Palmer G. Reductive titration of CoQ-depleted Complex III from Baker’s yeast. Evidence for an exchange-coupled complex between QH. and low-spin ferricytochrome b. FEBS Letters. 1983;163:140–143. doi: 10.1016/0014-5793(83)81181-8. [DOI] [PubMed] [Google Scholar]
- 142.Kuras R, Guergova-Kuras M, Crofts AR. Steps toward constructing a cytochrome b6 f complex in the purple bacterium Rhodobacter sphaeroides: an example of the structural plasticity of a membrane cytochrome. Biochemistry. 1998;37:16280–16288. doi: 10.1021/bi9813476. [DOI] [PubMed] [Google Scholar]
- 143.Teixeira PF, Glaser E. Processing peptidases in mitochondria and chloroplasts. Biochim Biophys Acta. 2013;1833:360–370. doi: 10.1016/j.bbamcr.2012.03.012. [DOI] [PubMed] [Google Scholar]
- 144.Baymann F, Lebrun E, Nitschke W. Mitochondrial cytochrome c1 is a collapsed di-heme cytochrome. Proc Natl Acad Sci U S A. 2004;101:17737–17740. doi: 10.1073/pnas.0407442101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Bertini I, Cavallaro G, Rosato A. Evolution of mitochondrial-type cytochrome c domains and of the protein machinery for their assembly. J Inorg Biochem. 2007;101:1798–1811. doi: 10.1016/j.jinorgbio.2007.02.001. [DOI] [PubMed] [Google Scholar]
- 146.Margoliash E. Primary structure and evolution of cytochrome c. Proc Natl Acad Sci U S A. 1963;50:672–679. doi: 10.1073/pnas.50.4.672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Dickerson RE. Evolution and gene transfer in purple photosynthetic bacteria. Nature. 1980;283:210–212. doi: 10.1038/283210a0. [DOI] [PubMed] [Google Scholar]
- 148.Pettigrew GW, Moore GR. Cytochrome c - Biological aspects. Springer-Verlag; Berlin: 1987. [Google Scholar]
- 149.Deisenhofer J, Epp O, Miki K, Huber R, Michel H. X-Ray structure analysis of a membrane protein complex: Electron density map at 3 A resolution and the model of the chromophores of the photosynthetic reaction center from Rhodopseudomonas viridis. J Mol Biol. 1984;180:385–398. doi: 10.1016/s0022-2836(84)80011-x. [DOI] [PubMed] [Google Scholar]
- 150.Collins MD, Jones D. Distribution of isoprenoid quinone structural types in bacteria and their taxonomic implication. Microbiol Rev. 1981;45:316–354. doi: 10.1128/mr.45.2.316-354.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Tsukatani Y, Matsuura K, Masuda S, Shimada K, Hiraishi A, Nagashima KV. Phylogenetic distribution of unusual triheme to tetraheme cytochrome subunit in the reaction center complex of purple photosynthetic bacteria. Photosynth Res. 2004;79:83–91. doi: 10.1023/B:PRES.0000011922.56394.92. [DOI] [PubMed] [Google Scholar]
- 152.Alric J, Tsukatani Y, Yoshida M, Matsuura K, Shimada K, Hienerwadel R, Schoepp-Cothenet B, Nitschke W, Nagashima KV, Vermeglio A. Structural and functional characterization of the unusual triheme cytochrome bound to the reaction center of Rhodovulum sulfidophilum. J Biol Chem. 2004;279:26090–26097. doi: 10.1074/jbc.M400361200. [DOI] [PubMed] [Google Scholar]
- 153.Larimer FW, Chain P, Hauser L, Lamerdin J, Malfatti S, Do L, Land ML, Pelletier DA, Beatty JT, Lang AS, Tabita FR, Gibson JL, Hanson TE, Bobst C, Torres JL, Peres C, Harrison FH, Gibson J, Harwood CS. Complete genome sequence of the metabolically versatile photosynthetic bacterium Rhodopseudomonas palustris. Nat Biotechnol. 2004;22:55–61. doi: 10.1038/nbt923. [DOI] [PubMed] [Google Scholar]
- 154.Turrens JF. Mitochondrial formation of reactive oxygen species. J Physiol. 2003;552:335–344. doi: 10.1113/jphysiol.2003.049478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Mulkidjanian AY. Ubiquinol oxidation in the cytochrome bc1 complex: reaction mechanism and prevention of short-circuiting. Biochim Biophys Acta. 2005;1709:5–34. doi: 10.1016/j.bbabio.2005.03.009. [DOI] [PubMed] [Google Scholar]
- 156.Bleier L, Drose S. Superoxide generation by complex III: From mechanistic rationales to functional consequences. Biochim Biophys Acta. 2012 doi: 10.1016/j.bbabio.2012.12.002. [DOI] [PubMed] [Google Scholar]
- 157.Andreyev AY, Kushnareva YE, Starkov AA. Mitochondrial metabolism of reactive oxygen species. Biochemistry (Mosc) 2005;70:200–214. doi: 10.1007/s10541-005-0102-7. [DOI] [PubMed] [Google Scholar]
- 158.Drose S, Brandt U. The mechanism of mitochondrial superoxide production by the cytochrome bc1 complex. J Biol Chem. 2008;283:21649–21654. doi: 10.1074/jbc.M803236200. [DOI] [PubMed] [Google Scholar]
- 159.Yin Y, Yang S, Yu L, Yu CA. Reaction mechanism of superoxide generation during ubiquinol oxidation by the cytochrome bc1 complex. J Biol Chem. 2010;285:17038–17045. doi: 10.1074/jbc.M110.104364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Koonin EV, Aravind L. Origin and evolution of eukaryotic apoptosis: the bacterial connection. Cell Death Differ. 2002;9:394–404. doi: 10.1038/sj.cdd.4400991. [DOI] [PubMed] [Google Scholar]
- 161.Koonin EV, Galperin MY. Sequence - Evolution - Function: Computational Approaches in Comparative Genomics. Kluwer Academic Publisher; Boston: 2003. [PubMed] [Google Scholar]
- 162.Lewis K. Programmed death in bacteria. Microbiol Mol Biol Rev. 2000;64:503–514. doi: 10.1128/mmbr.64.3.503-514.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Zamzami N, Susin SA, Marchetti P, Hirsch T, Gomez-Monterrey I, Castedo M, Kroemer G. Mitochondrial control of nuclear apoptosis. J Exp Med. 1996;183:1533–1544. doi: 10.1084/jem.183.4.1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Petit PX, Susin SA, Zamzami N, Mignotte B, Kroemer G. Mitochondria and programmed cell death: back to the future. FEBS Lett. 1996;396:7–13. doi: 10.1016/0014-5793(96)00988-x. [DOI] [PubMed] [Google Scholar]
- 165.Skulachev VP. Cytochrome c in the apoptotic and antioxidant cascades. FEBS Lett. 1998;423:275–280. doi: 10.1016/s0014-5793(98)00061-1. [DOI] [PubMed] [Google Scholar]
- 166.Korshunov SS, Skulachev VP, Starkov AA. High protonic potential actuates a mechanism of production of reactive oxygen species in mitochondria. FEBS Lett. 1997;416:15–18. doi: 10.1016/s0014-5793(97)01159-9. [DOI] [PubMed] [Google Scholar]
- 167.Kushnareva Y, Murphy AN, Andreyev A. Complex I-mediated reactive oxygen species generation: modulation by cytochrome c and NAD(P)+ oxidation-reduction state. Biochem J. 2002;368:545–553. doi: 10.1042/BJ20021121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Skulachev VP. Bioenergetic aspects of apoptosis, necrosis and mitoptosis. Apoptosis. 2006;11:473–485. doi: 10.1007/s10495-006-5881-9. [DOI] [PubMed] [Google Scholar]
- 169.Skulachev VP, Anisimov VN, Antonenko YN, Bakeeva LE, Chernyak BV, Erichev VP, Filenko OF, Kalinina NI, Kapelko VI, Kolosova NG, Kopnin BP, Korshunova GA, Lichinitser MR, Obukhova LA, Pasyukova EG, Pisarenko OI, Roginsky VA, Ruuge EK, Senin, Severina, Skulachev MV, Spivak IM, Tashlitsky VN, Tkachuk VA, Vyssokikh MY, Yaguzhinsky LS, Zorov DB. An attempt to prevent senescence: a mitochondrial approach. Biochim Biophys Acta. 2009;1787:437–461. doi: 10.1016/j.bbabio.2008.12.008. [DOI] [PubMed] [Google Scholar]
- 170.Ji J, Tyurina YY, Tang M, Feng W, Stolz DB, Clark RS, Meaney DF, Kochanek PM, Kagan VE, Bayir H. Mitochondrial injury after mechanical stretch of cortical neurons in vitro: biomarkers of apoptosis and selective peroxidation of anionic phospholipids. J Neurotrauma. 2012;29:776–788. doi: 10.1089/neu.2010.1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Israelachvili JN, Mitchell DJ, Ninham BW. Theory of self-assembly of lipid bilayers and vesicles. Biochim Biophys Acta. 1977;470:185–201. doi: 10.1016/0005-2736(77)90099-2. [DOI] [PubMed] [Google Scholar]
- 172.Tuominen EK, Wallace CJ, Kinnunen PK. Phospholipid-cytochrome c interaction: evidence for the extended lipid anchorage. J Biol Chem. 2002;277:8822–8826. doi: 10.1074/jbc.M200056200. [DOI] [PubMed] [Google Scholar]
- 173.Petrosillo G, Casanova G, Matera M, Ruggiero FM, Paradies G. Interaction of peroxidized cardiolipin with rat-heart mitochondrial membranes: induction of permeability transition and cytochrome c release. FEBS Lett. 2006;580:6311–6316. doi: 10.1016/j.febslet.2006.10.036. [DOI] [PubMed] [Google Scholar]
- 174.Kagan VE, Borisenko GG, Tyurina YY, Tyurin VA, Jiang J, Potapovich AI, Kini V, Amoscato AA, Fujii Y. Oxidative lipidomics of apoptosis: redox catalytic interactions of cytochrome c with cardiolipin and phosphatidylserine. Free Radic Biol Med. 2004;37:1963–1985. doi: 10.1016/j.freeradbiomed.2004.08.016. [DOI] [PubMed] [Google Scholar]
- 175.Bayir H, Fadeel B, Palladino MJ, Witasp E, Kurnikov IV, Tyurina YY, Tyurin VA, Amoscato AA, Jiang J, Kochanek PM, DeKosky ST, Greenberger JS, Shvedova AA, Kagan VE. Apoptotic interactions of cytochrome c: redox flirting with anionic phospholipids within and outside of mitochondria. Biochim Biophys Acta. 2006;1757:648–659. doi: 10.1016/j.bbabio.2006.03.002. [DOI] [PubMed] [Google Scholar]
- 176.Vladimirov YA, Proskurnina EV, Izmailov DY, Novikov AA, Brusnichkin AV, Osipov AN, Kagan VE. Mechanism of activation of cytochrome c peroxidase activity by cardiolipin. Biochemistry (Mosc) 2006;71:989–997. doi: 10.1134/s0006297906090070. [DOI] [PubMed] [Google Scholar]
- 177.Kagan VE, Bayir HA, Belikova NA, Kapralov O, Tyurina YY, Tyurin VA, Jiang J, Stoyanovsky DA, Wipf P, Kochanek PM, Greenberger JS, Pitt B, Shvedova AA, Borisenko G. Cytochrome c/cardiolipin relations in mitochondria: a kiss of death. Free Radic Biol Med. 2009;46:1439–1453. doi: 10.1016/j.freeradbiomed.2009.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Kagan VE, Tyurin VA, Jiang J, Tyurina YY, Ritov VB, Amoscato AA, Osipov AN, Belikova NA, Kapralov AA, Kini V, Vlasova, Zhao Q, Zou M, Di P, Svistunenko DA, Kurnikov IV, Borisenko GG. Cytochrome c acts as a cardiolipin oxygenase required for release of proapoptotic factors. Nat Chem Biol. 2005;1:223–232. doi: 10.1038/nchembio727. [DOI] [PubMed] [Google Scholar]
- 179.Miyamoto S, Nantes IL, Faria PA, Cunha D, Ronsein GE, Medeiros MH, Di Mascio P. Cytochrome c-promoted cardiolipin oxidation generates singlet molecular oxygen. Photochem Photobiol Sci. 2012;11:1536–1546. doi: 10.1039/c2pp25119a. [DOI] [PubMed] [Google Scholar]
- 180.Winkler JR, Wittung-Stafshede P, Leckner J, Malmstrom BG, Gray HB. Effects of folding on metalloprotein active sites. Proc Natl Acad Sci U S A. 1997;94:4246–4249. doi: 10.1073/pnas.94.9.4246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Tezcan FA, Winkler JR, Gray HB. Effects of ligation and folding on reduction potentials of heme proteins. J Am Chem Soc. 1998;120:13383–13388. [Google Scholar]
- 182.Pratt DA, Tallman KA, Porter NA. Free radical oxidation of polyunsaturated lipids: New mechanistic insights and the development of peroxyl radical clocks. Acc Chem Res. 2011;44:458–467. doi: 10.1021/ar200024c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Lokhmatikov AV, Voskoboynikova NE, Cherepanov DA, Steinhoff HJ, Skulachev VP, Mulkidjanian AY. Oxidation of cardiolipin in liposomes: A new insight into the primary steps of mitochondria-triggered apoptosis. Biochim Biophys Acta. 2012;1817:S100. [Google Scholar]
- 184.Althoff T, Mills DJ, Popot JL, Kuhlbrandt W. Arrangement of electron transport chain components in bovine mitochondrial supercomplex I1III2IV1. EMBO J. 2011;30:4652–4664. doi: 10.1038/emboj.2011.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Mileykovskaya E, Penczek PA, Fang J, Mallampalli VK, Sparagna GC, Dowhan W. Arrangement of the respiratory chain complexes in Saccharomyces cerevisiae supercomplex III2IV2 revealed by single particle cryo-electron microscopy. J Biol Chem. 2012;287:23095–23103. doi: 10.1074/jbc.M112.367888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Pebay-Peyroula E, Dahout-Gonzalez C, Kahn R, Trezeguet V, Lauquin GJ, Brandolin G. Structure of mitochondrial ADP/ATP carrier in complex with carboxyatractyloside. Nature. 2003;426:39–44. doi: 10.1038/nature02056. [DOI] [PubMed] [Google Scholar]
- 187.Yankovskaya V, Horsefield R, Tornroth S, Luna-Chavez C, Miyoshi H, Leger C, Byrne B, Cecchini G, Iwata S. Architecture of succinate dehydrogenase and reactive oxygen species generation. Science. 2003;299:700–704. doi: 10.1126/science.1079605. [DOI] [PubMed] [Google Scholar]
- 188.Lange C, Nett JH, Trumpower BL, Hunte C. Specific roles of protein-phospholipid interactions in the yeast cytochrome bc1 complex structure. EMBO J. 2001;20:6591–6600. doi: 10.1093/emboj/20.23.6591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Solmaz SR, Hunte C. Structure of complex III with bound cytochrome c in reduced state and definition of a minimal core interface for electron transfer. J Biol Chem. 2008;283:17542–17549. doi: 10.1074/jbc.M710126200. [DOI] [PubMed] [Google Scholar]
- 190.Poyry S, Cramariuc O, Postila PA, Kaszuba K, Sarewicz M, Osyczka A, Vattulainen I, Rog T. Atomistic simulations indicate cardiolipin to have an integral role in the structure of the cytochrome bc1 complex. Biochim Biophys Acta. 2013;1827:769–778. doi: 10.1016/j.bbabio.2013.03.005. [DOI] [PubMed] [Google Scholar]
- 191.Tsukihara T, Shimokata K, Katayama Y, Shimada H, Muramoto K, Aoyama H, Mochizuki M, Shinzawa-Itoh K, Yamashita E, Yao M, Ishimura Y, Yoshikawa S. The low-spin heme of cytochrome c oxidase as the driving element of the proton-pumping process. Proc Natl Acad Sci U S A. 2003;100:15304–15309. doi: 10.1073/pnas.2635097100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Jormakka M, Tornroth S, Byrne B, Iwata S. Molecular basis of proton motive force generation: structure of formate dehydrogenase-N. Science. 2002;295:1863–1868. doi: 10.1126/science.1068186. [DOI] [PubMed] [Google Scholar]
- 193.Palsdottir H, Hunte C. Lipids in membrane protein structures. Biochim Biophys Acta. 2004;1666:2–18. doi: 10.1016/j.bbamem.2004.06.012. [DOI] [PubMed] [Google Scholar]
- 194.Arias-Cartin R, Grimaldi S, Arnoux P, Guigliarelli B, Magalon A. Cardiolipin binding in bacterial respiratory complexes: Structural and functional implications. Biochim Biophys Acta. 2012;1817:1937–1949. doi: 10.1016/j.bbabio.2012.04.005. [DOI] [PubMed] [Google Scholar]
- 195.Haines TH. A new look at cardiolipin. Biochim Biophys Acta. 2009;1788:1997–2002. doi: 10.1016/j.bbamem.2009.09.008. [DOI] [PubMed] [Google Scholar]
- 196.Mileykovskaya E, Dowhan W. Cardiolipin membrane domains in prokaryotes and eukaryotes. Biochim Biophys Acta. 2009;1788:2084–2091. doi: 10.1016/j.bbamem.2009.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Mikel’saar K, Severina, Skulachev VP. Phospholipids and oxidative phosphorylation. Usp Sovrem Biol. 1974;78:348–370. [PubMed] [Google Scholar]
- 198.Zhang M, Mileykovskaya E, Dowhan W. Gluing the respiratory chain together. Cardiolipin is required for supercomplex formation in the inner mitochondrial membrane. J Biol Chem. 2002;277:43553–43556. doi: 10.1074/jbc.C200551200. [DOI] [PubMed] [Google Scholar]
- 199.Mileykovskaya E, Zhang M, Dowhan W. Cardiolipin in energy transducing membranes. Biochemistry (Mosc) 2005;70:154–158. doi: 10.1007/s10541-005-0095-2. [DOI] [PubMed] [Google Scholar]
- 200.Haines TH. Anionic lipid headgroups as a proton-conducting pathway along the surface of membranes: a hypothesis. Proc Natl Acad Sci U S A. 1983;80:160–164. doi: 10.1073/pnas.80.1.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Mulkidjanian AY, Heberle J, Cherepanov DA. Protons @ interfaces: implications for biological energy conversion. Biochim Biophys Acta. 2006;1757:913–930. doi: 10.1016/j.bbabio.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 202.Huang LS, Cobessi D, Tung EY, Berry EA. Binding of the respiratory chain inhibitor antimycin to the mitochondrial bc1 complex: a new crystal structure reveals an altered intramolecular hydrogen-bonding pattern. J Mol Biol. 2005;351:573–597. doi: 10.1016/j.jmb.2005.05.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Zhao K, Zhao GM, Wu D, Soong Y, Birk AV, Schiller PW, Szeto HH. Cell-permeable peptide antioxidants targeted to inner mitochondrial membrane inhibit mitochondrial swelling, oxidative cell death, and reperfusion injury. J Biol Chem. 2004;279:34682–34690. doi: 10.1074/jbc.M402999200. [DOI] [PubMed] [Google Scholar]
- 204.Shao H, Li J, Zhou Y, Ge Z, Fan J, Shao Z, Zeng Y. Dose-dependent protective effect of propofol against mitochondrial dysfunction in ischaemic/reperfused rat heart: role of cardiolipin. Br J Pharmacol. 2008;153:1641–1649. doi: 10.1038/bjp.2008.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Koyama T, Zhu MY, Kinjo M, Araiso T. Protective effects of idebenone against alterations in dynamic microstructure induced by lipid peroxidation in rat cardiac mitochondria. Jpn Heart J. 1991;32:91–100. doi: 10.1536/ihj.32.91. [DOI] [PubMed] [Google Scholar]
- 206.Senin U, Parnetti L, Barbagallo-Sangiorgi G, Bartorelli L, Bocola V, Capurso A, Cuzzupoli M, Denaro M, Marigliano V, Tammaro AE, Fioravanti M. Idebenone in senile dementia of Alzheimer type: a multicentre study. Arch Gerontol Geriatr. 1992;15:249–260. doi: 10.1016/0167-4943(92)90060-h. [DOI] [PubMed] [Google Scholar]
- 207.Suno M, Nagaoka A. Inhibition of lipid peroxidation by a novel compound (CV-2619) in brain mitochondria and mode of action of the inhibition. Biochem Biophys Res Commun. 1984;125:1046–1052. doi: 10.1016/0006-291x(84)91389-5. [DOI] [PubMed] [Google Scholar]
- 208.Zs -Nagy I. Chemistry, toxicology, pharmacology and pharmacokinetics of idebenone: a review. Arch Gerontol Geriatr. 1990;11:177–186. doi: 10.1016/0167-4943(90)90063-c. [DOI] [PubMed] [Google Scholar]
- 209.Armstrong JS, Whiteman M, Rose P, Jones DP. The Coenzyme Q10 analog decylubiquinone inhibits the redox-activated mitochondrial permeability transition: role of mitochondrial complex III. J Biol Chem. 2003;278:49079–49084. doi: 10.1074/jbc.M307841200. [DOI] [PubMed] [Google Scholar]
- 210.Jauslin ML, Meier T, Smith RA, Murphy MP. Mitochondria-targeted antioxidants protect Friedreich Ataxia fibroblasts from endogenous oxidative stress more effectively than untargeted antioxidants. FASEB J. 2003;17:1972–1974. doi: 10.1096/fj.03-0240fje. [DOI] [PubMed] [Google Scholar]
- 211.Kelso GF, Porteous CM, Coulter CV, Hughes G, Porteous WK, Ledgerwood EC, Smith RA, Murphy MP. Selective targeting of a redox-active ubiquinone to mitochondria within cells: antioxidant and antiapoptotic properties. J Biol Chem. 2001;276:4588–4596. doi: 10.1074/jbc.M009093200. [DOI] [PubMed] [Google Scholar]
- 212.Agapova LS, Chernyak BV, Domnina LV, Dugina VB, Efimenko AY, Fetisova EK, Ivanova OY, Kalinina NI, Khromova NV, Kopnin BP, Kopnin PB, Korotetskaya MV, Lichinitser MR, Lukashev AL, Pletjushkina OY, Popova EN, Skulachev MV, Shagieva GS, Stepanova EV, Titova EV, Tkachuk VA, Vasiliev JM, Skulachev VP. Mitochondria-targeted plastoquinone derivatives as tools to interrupt execution of the aging program. 3. Inhibitory effect of SkQ1 on tumor development from p53-deficient cells. Biochemistry (Mosc) 2008;73:1300–1316. doi: 10.1134/s0006297908120031. [DOI] [PubMed] [Google Scholar]
- 213.Anisimov VN, Bakeeva LE, Egormin PA, Filenko OF, Isakova EF, Manskikh VN, Mikhelson VM, Panteleeva AA, Pasyukova EG, Pilipenko DI, Piskunova TS, Popovich IG, Roshchina NV, Rybina OY, Saprunova VB, Samoylova TA, Semenchenko AV, Skulachev MV, Spivak IM, Tsybul’ko EA, Tyndyk ML, Vyssokikh MY, Yurova MN, Zabezhinsky MA, Skulachev VP. Mitochondria-targeted plastoquinone derivatives as tools to interrupt execution of the aging program. 5. SkQ1 prolongs lifespan and prevents development of traits of senescence. Biochemistry (Mosc) 2008;73:1329–1342. doi: 10.1134/s0006297908120055. [DOI] [PubMed] [Google Scholar]
- 214.Antonenko YN, Avetisyan AV, Bakeeva LE, Chernyak BV, Chertkov VA, Domnina LV, Ivanova OY, Izyumov DS, Khailova LS, Klishin SS, Korshunova GA, Lyamzaev KG, Muntyan MS, Nepryakhina OK, Pashkovskaya AA, Pletjushkina OY, Pustovidko AV, Roginsky VA, Rokitskaya TI, Ruuge EK, Saprunova VB, Severina, Simonyan RA, Skulachev IV, Skulachev MV, Sumbatyan NV, Sviryaeva IV, Tashlitsky VN, Vassiliev JM, Vyssokikh MY, Yaguzhinsky LS, Zamyatnin AA, Jr, Skulachev VP. Mitochondria-targeted plastoquinone derivatives as tools to interrupt execution of the aging program. 1. Cationic plastoquinone derivatives: synthesis and in vitro studies. Biochemistry (Mosc) 2008;73:1273–1287. doi: 10.1134/s0006297908120018. [DOI] [PubMed] [Google Scholar]
- 215.Antonenko YN, Roginsky VA, Pashkovskaya AA, Rokitskaya TI, Kotova EA, Zaspa AA, Chernyak BV, Skulachev VP. Protective effects of mitochondria-targeted antioxidant SkQ in aqueous and lipid membrane environments. J Membr Biol. 2008;222:141–149. doi: 10.1007/s00232-008-9108-6. [DOI] [PubMed] [Google Scholar]
- 216.Bakeeva LE, Barskov IV, Egorov MV, Isaev NK, Kapelko VI, Kazachenko AV, Kirpatovsky VI, Kozlovsky SV, Lakomkin VL, Levina SB, Pisarenko OI, Plotnikov EY, Saprunova VB, Serebryakova LI, Skulachev MV, Stelmashook EV, Studneva IM, Tskitishvili OV, Vasilyeva AK, Victorov IV, Zorov DB, Skulachev VP. Mitochondria-targeted plastoquinone derivatives as tools to interrupt execution of the aging program. 2. Treatment of some ROS- and age-related diseases (heart arrhythmia, heart infarctions, kidney ischemia, and stroke) Biochemistry (Mosc) 2008;73:1288–1299. doi: 10.1134/s000629790812002x. [DOI] [PubMed] [Google Scholar]
- 217.Neroev VV, Archipova MM, Bakeeva LE, Fursova A, Grigorian EN, Grishanova AY, Iomdina EN, Ivashchenko Zh N, Katargina LA, Khoroshilova-Maslova IP, Kilina OV, Kolosova NG, Kopenkin EP, Korshunov SS, Kovaleva NA, Novikova YP, Philippov PP, Pilipenko DI, Robustova OV, Saprunova VB, Senin, Skulachev MV, Sotnikova LF, Stefanova NA, Tikhomirova NK, Tsapenko IV, Shchipanova AI, Zinovkin RA, Skulachev VP. Mitochondria-targeted plastoquinone derivatives as tools to interrupt execution of the aging program. 4. Age-related eye disease. SkQ1 returns vision to blind animals. Biochemistry (Mosc) 2008;73:1317–1328. doi: 10.1134/s0006297908120043. [DOI] [PubMed] [Google Scholar]
- 218.Severin FF, Severina, Antonenko YN, Rokitskaya TI, Cherepanov DA, Mokhova EN, Vyssokikh MY, Pustovidko AV, Markova OV, Yaguzhinsky LS, Korshunova GA, Sumbatyan NV, Skulachev MV, Skulachev VP. Penetrating cation/fatty acid anion pair as a mitochondria-targeted protonophore. Proc Natl Acad Sci U S A. 2010;107:663–668. doi: 10.1073/pnas.0910216107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Skulachev VP, Antonenko YN, Cherepanov DA, Chernyak BV, Izyumov DS, Khailova LS, Klishin SS, Korshunova GA, Lyamzaev KG, Pletjushkina OY, Roginsky VA, Rokitskaya TI, Severin FF, Severina, Simonyan RA, Skulachev MV, Sumbatyan NV, Sukhanova EI, Tashlitsky VN, Trendeleva TA, Vyssokikh MY, Zvyagilskaya RA. Prevention of cardiolipin oxidation and fatty acid cycling as two antioxidant mechanisms of cationic derivatives of plastoquinone (SkQs) Biochim Biophys Acta. 2010 doi: 10.1016/j.bbabio.2010.03.015. [DOI] [PubMed] [Google Scholar]
- 220.Severina II, Severin FF, Korshunova GA, Sumbatyan NV, Ilyasova TM, Simonyan RA, Rogov AG, Trendeleva TA, Zvyagilskaya RA, Dugina VB, Domnina LV, Fetisova EK, Lyamzaev KG, Vyssokikh MY, Chernyak BV, Skulachev MV, Skulachev VP, Sadovnichii VA. In search of novel highly active mitochondria-targeted antioxidants: Thymoquinone and its cationic derivatives. FEBS Lett. 2013;587:2018–2024. doi: 10.1016/j.febslet.2013.04.043. [DOI] [PubMed] [Google Scholar]
- 221.Skulachev MV, Antonenko YN, Anisimov VN, Chernyak BV, Cherepanov DA, Chistyakov VA, Egorov MV, Kolosova NG, Korshunova GA, Lyamzaev KG, Plotnikov EY, Roginsky VA, Savchenko AY, Severina, Severin FF, Shkurat TP, Tashlitsky VN, Shidlovsky KM, Vyssokikh MY, Zamyatnin AA, Jr, Zorov DB, Skulachev VP. Mitochondrial-targeted plastoquinone derivatives. Effect on senescence and acute age-related pathologies. Curr Drug Targets. 2011;12:800–826. doi: 10.2174/138945011795528859. [DOI] [PubMed] [Google Scholar]
- 222.Obukhova LA, Skulachev VP, Kolosova NG. Mitochondria-targeted antioxidant SkQ1 inhibits age-dependent involution of the thymus in normal and senescence-prone rats. Aging (Albany NY) 2009;1:389–401. doi: 10.18632/aging.100043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Shipounova IN, Svinareva DA, Petrova TV, Lyamzaev KG, Chernyak BV, Drize NI, Skulachev VP. Reactive oxygen species produced in mitochondria are involved in age-dependent changes of hematopoietic and mesenchymal progenitor cells in mice. A study with the novel mitochondria-targeted antioxidant SkQ1. Mech Ageing Dev. 2010;131:415–421. doi: 10.1016/j.mad.2010.06.003. [DOI] [PubMed] [Google Scholar]
- 224.Plotnikov EY, Chupyrkina AA, Jankauskas SS, Pevzner IB, Silachev DN, Skulachev VP, Zorov DB. Mechanisms of nephroprotective effect of mitochondria-targeted antioxidants under rhabdomyolysis and ischemia/reperfusion. Biochim Biophys Acta. 2011;1812:77–86. doi: 10.1016/j.bbadis.2010.09.008. [DOI] [PubMed] [Google Scholar]
- 225.Isaev NK, Novikova SV, Stelmashook EV, Barskov IV, Silachev DN, Khaspekov LG, Skulachev VP, Zorov DB. Mitochondria-targeted plastoquinone antioxidant SkQR1 decreases trauma-induced neurological deficit in rat. Biochemistry (Mosc) 2012;77:996–999. doi: 10.1134/S0006297912090052. [DOI] [PubMed] [Google Scholar]
- 226.Skulachev VP. Mitochondria-targeted antioxidants as promising drugs for treatment of age-related brain diseases. J Alzheimers Dis. 2012;28:283–289. doi: 10.3233/JAD-2011-111391. [DOI] [PubMed] [Google Scholar]
- 227.Kapay NA, Popova OV, Isaev NK, Stelmashook EV, Kondratenko RV, Zorov DB, Skrebitsky VG, Skulachev VP. Mitochondria-targeted plastoquinone antioxidant SkQ1 prevents amyloid-beta-induced impairment of long-term potentiation in rat hippocampal slices. J Alzheimers Dis. 2013;36:377–383. doi: 10.3233/JAD-122428. [DOI] [PubMed] [Google Scholar]
- 228.Ji J, Kline AE, Amoscato A, Samhan-Arias AK, Sparvero LJ, Tyurin VA, Tyurina YY, Fink B, Manole MD, Puccio AM, Okonkwo DO, Cheng JP, Alexander H, Clark RS, Kochanek PM, Wipf P, Kagan VE, Bayir H. Lipidomics identifies cardiolipin oxidation as a mitochondrial target for redox therapy of brain injury. Nat Neurosci. 2012;15:1407–1413. doi: 10.1038/nn.3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Mileykovskaya E, Dowhan W. Visualization of phospholipid domains in Escherichia coli by using the cardiolipin-specific fluorescent dye 10-N-nonyl acridine orange. J Bacteriol. 2000;182:1172–1175. doi: 10.1128/jb.182.4.1172-1175.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Mileykovskaya E, Dowhan W, Birke RL, Zheng D, Lutterodt L, Haines TH. Cardiolipin binds nonyl acridine orange by aggregating the dye at exposed hydrophobic domains on bilayer surfaces. FEBS Lett. 2001;507:187–190. doi: 10.1016/s0014-5793(01)02948-9. [DOI] [PubMed] [Google Scholar]
- 231.Skulachev VP. Role of uncoupled and non-coupled oxidations in maintenance of safely low levels of oxygen and its one-electron reductants. Q Rev Biophys. 1996;29:169–202. doi: 10.1017/s0033583500005795. [DOI] [PubMed] [Google Scholar]
- 232.Darwin C. On the Origin of Species by Means of Natural Selection or, The Preservation of Races in the Struggle for Life. John Murray; London: 1859. [Google Scholar]
- 233.Ciccarelli FD, Doerks T, von Mering C, Creevey CJ, Snel B, Bork P. Toward automatic reconstruction of a highly resolved tree of life. Science. 2006;311:1283–1287. doi: 10.1126/science.1123061. [DOI] [PubMed] [Google Scholar]
- 234.Yutin N, Puigbo P, Koonin EV, Wolf YI. Phylogenomics of prokaryotic ribosomal proteins. PLoS One. 2012;7:e36972. doi: 10.1371/journal.pone.0036972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Federhen S. The NCBI Taxonomy database. Nucleic Acids Res. 2012;40:D136–143. doi: 10.1093/nar/gkr1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Tatusov RL, Koonin EV, Lipman DJ. A genomic perspective on protein families. Science. 1997;278:631–637. doi: 10.1126/science.278.5338.631. [DOI] [PubMed] [Google Scholar]
- 237.Kagan VE, Tsuchiya M, Serbinova E, Packer L, Sies H. Interaction of the pyridoindole stobadine with peroxyl, superoxide and chromanoxyl radicals. Biochem Pharmacol. 1993;45:393–400. doi: 10.1016/0006-2952(93)90075-8. [DOI] [PubMed] [Google Scholar]
- 238.Humphrey W, Dalke A, Schulten K. VMD: Visual molecular dynamics. Journal of Molecular Graphics. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.