Significance
Growing evidence suggests that successful intervention in many human cancers will require combinations of therapeutic agents. Critical to this effort will be a detailed understanding of the crosstalk between signaling networks that modulate proliferation, cell death, drug sensitivity, and acquired resistance. Here we investigated DNA-damage signaling elicited by small-molecule inhibitors against MAP/ERK kinase (MEK) and PI3K in melanoma cells. This work, performed using cutting-edge mass spectrometry proteomics, uncovered a burst of signaling among proteins in the DNA-damage pathway upon initiation of the cell-death program by agents targeting the RAS–RAF–MEK and PI3K–AKT–mTOR pathways. These signals may prove important to the short- and long-term sensitivity of tumor cells to MEK- and PI3K-targeted therapies.
Keywords: phosphoproteomics, PI3 kinase, PRKDC
Abstract
Targeted therapeutics that block signal transduction through the RAS–RAF–MEK and PI3K–AKT–mTOR pathways offer significant promise for the treatment of human malignancies. Dual inhibition of MAP/ERK kinase (MEK) and phosphatidylinositol 3-kinase (PI3K) with the potent and selective small-molecule inhibitors GDC-0973 and GDC-0941 has been shown to trigger tumor cell death in preclinical models. Here we have used phosphomotif antibodies and mass spectrometry (MS) to investigate the effects of MEK/PI3K dual inhibition during the period immediately preceding cell death. Upon treatment, melanoma cell lines responded by dramatically increasing phosphorylation on proteins containing a canonical DNA damage-response (DDR) motif, as defined by a phosphorylated serine or threonine residue adjacent to glutamine, [s/t]Q. In total, >2,000 [s/t]Q phosphorylation sites on >850 proteins were identified by LC-MS/MS, including an extensive network of DDR proteins. Linear mixed-effects modeling revealed 101 proteins in which [s/t]Q phosphorylation was altered significantly in response to GDC-0973/GDC-0941. Among the most dramatic changes, we observed rapid and sustained phosphorylation of sites within the ABCDE cluster of DNA-dependent protein kinase. Preincubation of cells with the inhibitors of the DDR kinases DNA-dependent protein kinase or ataxia-telangiectasia mutated enhanced GDC-0973/GDC-0941–mediated cell death. Network analysis revealed specific enrichment of proteins involved in RNA metabolism along with canonical DDR proteins and suggested a prominent role for this pathway in the response to MEK/PI3K dual inhibition.
Dysregulation of the RAS–RAF–MAP/MEK and PI3K–AKT–mTOR pathways represents a common theme in human cancer. The importance of these interconnected pathways is highlighted by the frequency of mutational activation of pathway members including RAS, RAF, and PI3K, as well as inactivation of the inhibitory phosphatase and tensin homolog (PTEN) (1, 2). Targeted therapies that block signaling through the RAS–RAF–MEK pathway, including specific inhibitors of oncogenic forms of BRAF (e.g., BRAF-V600E, which is observed in ∼50% of melanomas) and of the downstream effector MEK have shown clinical efficacy in melanoma and other tumor types (3, 4). Likewise, suppression of cell-survival signaling through inhibition of PI3K has been shown to kill cancer cells (5, 6). Although inhibition of either pathway individually can elicit measureable responses, feedback through signal-transduction networks often limits the effectiveness of single-agent therapies. Mounting evidence suggests that dual inhibition of the RAS–RAF–MEK and PI3K–AKT–mTOR pathways will demonstrate improved efficacy over single-agent therapies.
At the heart of the RAS–RAF–MEK and PI3K–AKT–mTOR pathways is a web of phosphorelay networks in which individual phosphorylation sites serve as nodes. Many key nodes reside on protein kinases, where phosphorylation individually and in aggregate modulates the amplitude and specificity of downstream signaling. Our understanding of these networks has been shaped by studies using phosphospecific antibodies against these individual, site-specific phosphorylation events including ERK1/2 at Thr202/Tyr204 (7, 8) and AKT at Thr308 (9, 10). Although this strategy has proven successful, the generation of sensitive phosphospecific reagents capable of reading out signal unambiguously remains a challenge. Likewise, multiply phosphorylated sequences or those occurring adjacent to other posttranslational modifications can confound data interpretation. A key limitation is that phosphospecific antibodies are intended to interrogate only a single node in a signal-transduction network, so that even when multiplexed they provide only a narrow portal through which to view the dynamic system.
Mass spectrometry (MS) proteomics provides a platform to dissect signaling networks in breadth and depth. Although theoretically the phosphorylated peptides can be profiled directly from digested cell lysates, interrogating signal-transduction networks requires enrichment of modified peptides from the cellular milieu. One approach involves immunoaffinity enrichment (IAE) with antibodies recognizing classes of phosphopeptides, such as phosphotyrosine (11). IAE methods also have been reported for assaying phosphorylation in the AKT (12) and DNA damage-response (DDR) signaling networks (13, 14), using antibodies that recognize phosphorylation within a local sequence context. For substrates of AKT family kinases, phosphorylation occurs within an RXRXX[s/t] sequence, where X represents any amino acid and [s/t] refers to the phosphorylated serine or threonine (12). For substrates of ataxia-telangiectasia mutated (ATM) and other DDR kinases, phosphorylation occurs on serine or threonine adjacent to glutamine, [s/t]Q (15). In contrast to phosphospecific antibodies directed against a single sequence, motif-specific antibodies recognize a degenerate motif and permit enrichment of an array of peptides. Likewise, these reagents provide a readout for screening conditions by immunoblot before phosphopeptide enrichment and LC-MS/MS.
In the current study, we noticed that MEK/PI3K dual inhibition in melanoma lines resulted in a marked DDR. Using motif-directed IAE and MS proteomics, we investigated the signaling elicited by small-molecule inhibitors of MEK and PI3K currently in clinical development to establish a molecular understanding of this response.
Results
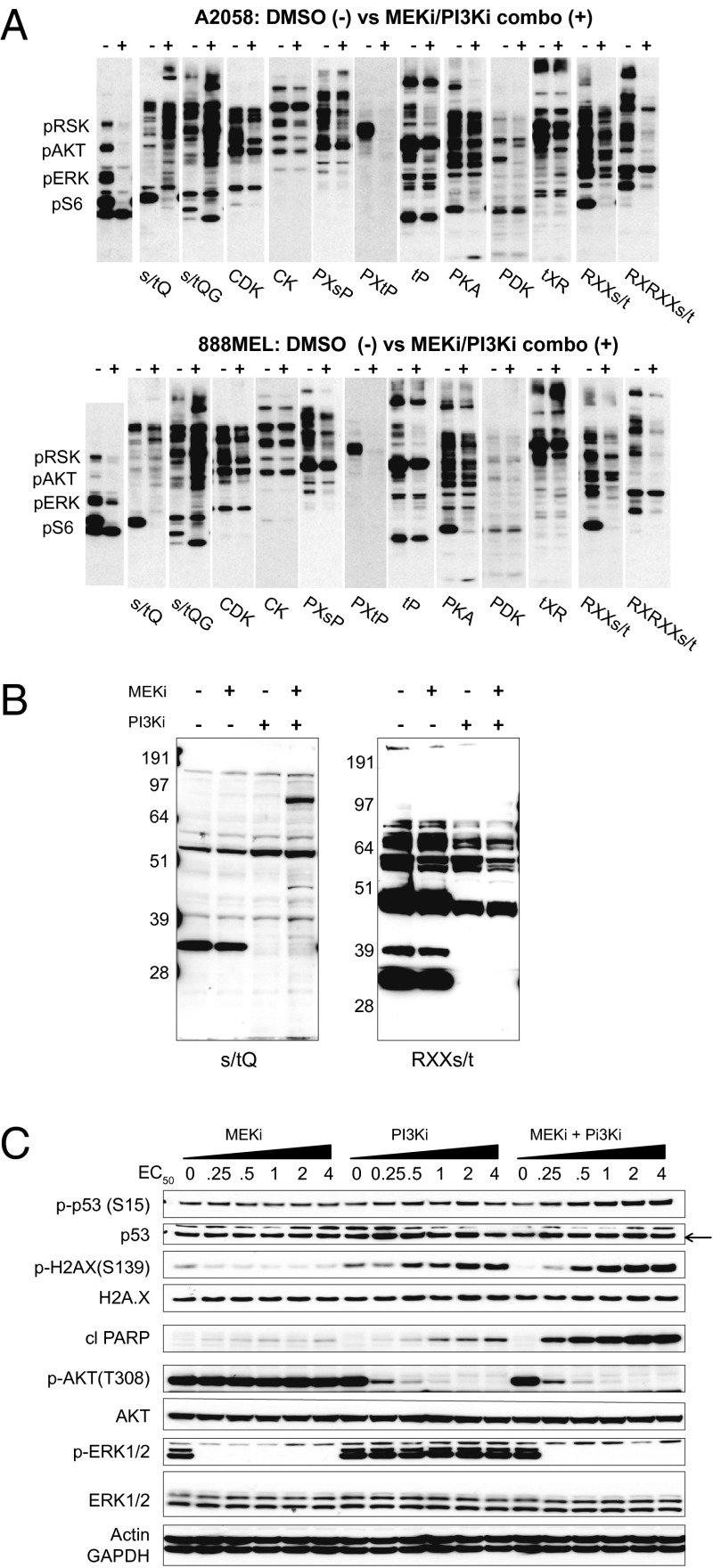
Published studies have shown that the compounds GDC-0973 (MEKi) and GDC-0941 (PI3Ki) potently and selectively inhibit kinase activities of MEK and PI3K, respectively (16, 17). Recent work indicates that concurrent inhibition of the RAS–RAF–MEK and PI3K–AKT–mTOR pathways is more potent at killing cells than individual agents (17, 18). In 4-d viability assays, A2058 (BRAF-V600E, PTEN-deficient; EC50: 2.5 µM GDC-0973, 2.5 µM GDC-0941) and 888MEL (BRAF-V600E; EC50: 0.05 µM GDC-0973, 2.5 µM GDC-0941) cells displayed synergistic decreases in cell number after combined pathway inhibition (17). To investigate the mechanism of cell death, A2058 and 888MEL cells were treated with 4× EC50 concentrations of MEKi and PI3Ki (hereafter referred to as “combo” treatment). After 6-h treatment, cell lysates were subjected to multiplexed Western blot profiling. Consistent with PI3K inhibition, permissive (RXX[s/t]) and rigid (RXRXX[s/t]) AKT-substrate motif antibodies showed robust decreases for the majority of observable bands (Fig. 1A). Diminished signal also was observed for the PXtP, PXsP, and MAPK substrate motifs, consistent with MEK inhibition blocking canonical MAPK signaling. Also striking were elevated levels of DDR phosphorylation reported by two ATM-substrate motif antibodies, [s/t]Q and [s/t]QG (Fig. 1A). To understand the individual contributions of MEK and PI3K inhibition, A2058 cells were left untreated or were treated with MEKi, PI3Ki, or the combination (4× EC50). AKT-substrate motif phosphosignals were decreased by PI3Ki alone or in combination with MEKi. ATM-substrate phosphomotif blots showed a single, low molecular weight feature which similarly decreased in cells when PI3K was inhibited (i.e., in cells treated with PI3Ki alone or combo). With this exception, immunoblots showed increased DDR signaling in cells treated with combo but not in cells treated with either single agent alone (Fig. 1B).
Fig. 1.
Dual inhibition of MEK and PI3K induces phosphorylation of DDR substrates. (A) A2058 or 888MEL melanoma cells were treated for 6 h with DMSO or GDC-0973+GDC-0941 (MEKi/PI3Ki combo; 4× EC50) and were subjected to KinomeView Profiling. A2058 and 888MEL cells were treated with 10 μM GDC-0973 + 10 μM GDC-0941 or 0.2 µM GDC-0973 + 10 µM GDC-0941, respectively. Blots were probed using an antibody mixture recognizing pRSK, pAKT, pERK, and pS6 or phosphomotif antibodies (e.g., DDR substrates with the [s/t]Q motif and AKT substrates with the RXX[s/t] and RXRXX[s/t] motifs). (B) A2058 lysates probed with the [s/t]Q and RXX[s/t] antibodies after treatment with DMSO, 10 μM GDC-0973 (MEKi), 10 μM GDC-0941 (PI3Ki), or the combination. (C) Dose response of A2058 cells to increasing concentrations of MEKi and PI3Ki alone or in combination. Blots were performed against DDR (p53 pSer15, histone 2AX pSer139), cell survival/cell death (AKT pThr308, cleaved PARP), and cell signaling (ERK1/2 pThr202/Tyr204) markers and controls. Actin and GAPDH served as loading controls.
Given that neither GDC-0941 nor GDC-0973 causes genotoxicity or directly damages DNA in mutagenicity, clastogenicity, or micronucleus formation assays, we speculated that increased [s/t]Q motif phosphorylation was a consequence of cell death. In a dose–response experiment, we first confirmed that pThr202/pTyr204 ERK1/2 and pThr308 AKT were reduced by MEKi or PI3Ki as single agents, respectively (Fig. 1C). Immunoblots against pSer15 p53 and pSer139 of the histone variant H2AX (γH2AX) revealed dose-dependent activation of DDR signaling, most notably in combo-treated cells. H2AX and p53 phosphorylation correlated with increased poly-ADP ribose polymerase (PARP) cleavage in both dose–response (Fig. 1C) and time-course (Fig. S1A) experiments. Increased signals for cleaved PARP, pSer15 p53, pSer139 H2AX, and [s/t]Q phosphomotif blots were also notable across five of six additional melanoma cell lines (C32, 888MEL, Colo829, Hs695T, and G361, but not LOX-IMVI), all carrying the BRAF-V600E mutation (Fig. S1B and Table S1). To confirm that induction of the DDR was not unique to these particular inhibitors, experiments were carried out using combinations of either GDC-0973 and the PI3K/mTOR inhibitor GDC-0980 (19) or GDC-0941 and the BRAF inhibitor PLX-4720 (20). As expected, inhibition of various signaling nodes resulted in a similar extent of PARP cleavage, DDR signaling, and p53/H2AX phosphorylation (Fig. S1C). Additionally, caspase inhibition by z-Val-Ala-Asp-fluoromethylketone partially protected cells from the effects of dual inhibition, decreasing PARP cleavage and H2AX phosphorylation (Fig. S1D). Taken together, activation of DDR signaling was dependent on efficient and sustained inhibition of the MEK/PI3K pathway and was coincident with the induction of apoptosis across multiple treatments, time points, and cell lines.
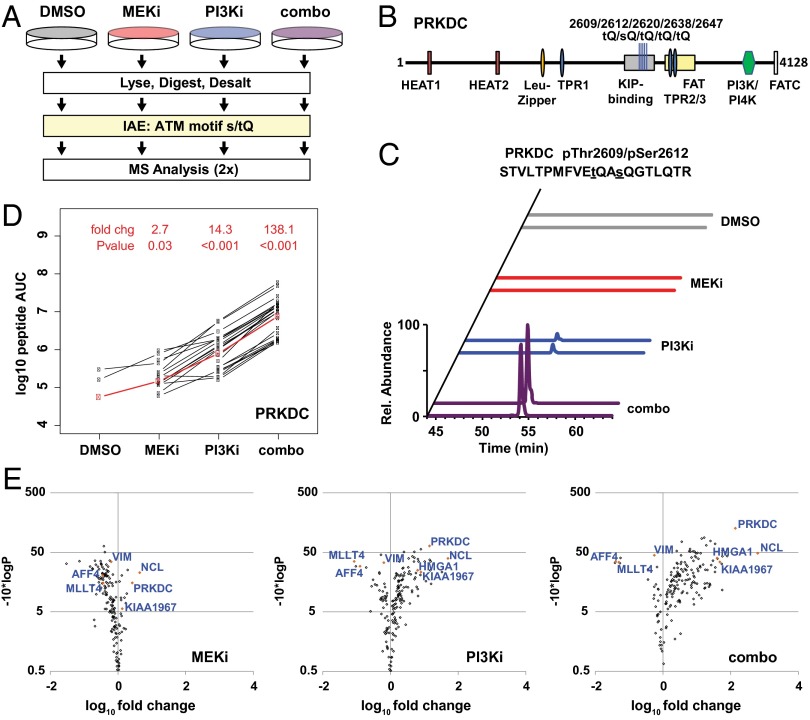
We next used IAE and MS proteomics to profile DDR signaling at the level of individual substrates (Fig. 2A). In total, 1,066 unique phosphopeptides were identified via 4,314 phosphopeptide spectral matches (phosphoPSMs) with false discovery rates (FDRs) of 0.74% and 2.07% at the peptide (pepFDR) and protein (protFDR) levels, respectively (Fig. S2A). Using the Ascore algorithm (AScore) to optimize phosphorylation-site localization (21), we identified 690 unique phosphopeptides (3,072 PSMs) in which the phosphorylation was assigned to a serine or threonine residing immediately adjacent to a glutamine [Ascore algorithm values (AScore) >13; 95% confidence score for localization]. Filtering the data to contain only the 1,004 phosphopeptides (4,219 PSMs) bearing either a [s/t]Q motif phosphorylation event or an available [S/T]Q motif (unphosphorylated) was sufficient to eliminate the remaining decoy peptides from the dataset. Conversely, identified phosphopeptides lacking an [S/T]Q motif (62 unique, 95 PSMs) displayed an FDR of >33%. This result suggests that the majority of 314 unique phosphopeptides which carry a [S/T]Q motif but in which the modification is not explicitly localized within the motif represent high-confidence hits in which reassignment of the phosphorylation event may be appropriate even when localization scores remain ambiguous.
Fig. 2.
Characterization of DDR signaling by LC-MS/MS and LiME analysis. (A) Diagram of IAE and MS analysis of [s/t]Q motif phosphopeptides in the combined-effects experiment. (B) PRKDC protein (4,128 residues), including domains and the five [s/t]Q phosphorylation sites from the combined-effects experiment. (C) Normalized extracted ion chromatograms (± 10 ppm) for duplicate injections of 819.0374 m/z ion from PRKDC pThr2609/pSer2612. (D) LiME plot showing label-free AUC for peaks from each phosphopeptide (black lines). The LiME model is shown in red. Fold changes and P values are reported for each treatment (MEKi, PI3Ki, or combo) relative to DMSO treatment. (E) LiME data displayed as volcano plots. The log10 fold change versus T−10*log10 (P value) is shown for each protein in a treatment group (MEKi, PI3Ki, or combo) relative to DMSO treatment. Selected proteins are identified, including several top hits denoted by orange symbols.
For a subset of identified proteins, the number of phosphoPSMs increased dramatically after combo treatment relative to no treatment (control) or treatment with either single agent. Among these proteins were known DDR substrates including the DNA-dependent protein kinase (PRKDC), Nucleolin (NUCL), high-mobility group AT-hook 1 (HMGA1), and K1967 (Dataset S1). For each, no phosphoPSMs were observed in untreated cells. A total of five phosphorylation sites were assigned to the ABCDE cluster of PRKDC (22–24); many were based on both singly and multiply modified forms (Fig. 2B). In line with spectral-count data, extracted ion chromatograms for the doubly phosphorylated pThr2609/pSer2612 peptide from PRKDC increased >10-fold in combo-treated cells compared with either single agent (Fig. 2C). Enhanced phosphorylation of pThr2609 and pSer2612 on PRKDC following MEKi + PI3Ki was observed in six additional melanoma lines (Fig. S2B). In contrast, many proteins (e.g., G3P/GAPDH; >90phosphoPSMs per sample) displayed little or no difference in the frequency of [s/t]Q phosphoPSMs between untreated and treated groups.
To interrogate phosphopeptide results systematically, we assembled a filtered list of phosphoPSMs from confidently identified spectra containing an [S/T]Q motif, regardless of whether phosphorylation was formally assigned to the motif. Areas under the curve (AUCs) were determined using the VistaQuant algorithm (25) in either direct or cross-quantitation (XQuant) mode. To identify proteins most significantly affected by the stimulus and to address the challenges posed by missing values, we used linear mixed-effects modeling (LiME). LiME, an improvement over ad hoc cutoffs or simple feature averaging, takes advantage of inherent replicate structure of the data and leverages information from a series of biological conditions to identify the significantly affected proteins (26, 27). For PRKDC, LiME analysis revealed a systematic increase in peak area for the [s/t]Q containing phosphoPSMs, with >100-fold increase (P < 0.001) between combo and control treatments (Fig. 2D). This change and other protein-level fold changes were calculated for each treatment group relative to the control using a mixed-effect model, with P values determined based on the fit between individual phosphoPSMs and the model. Volcano plots show that PRKDC, NUCL, HMGA1, and K1967 each displayed >30-fold change and a P value < 0.001 between combo and control treatments (Fig. 2E). Although less pronounced, similar trends were observed for PRKDC and NUCL in lysates from cells treated with GDC-0973 (MEKi) or GDC-0941 (PI3Ki) as single agents (Fig. 2E). These LiME plots, their associated fold changes, and P values are provided in Dataset S2.
Examining the original [s/t]Q immunoblot results (Fig. 1 A and B), we noted an abundant band which decreased following PI3K inhibition and combo treatment. In the spectral-counting data (Dataset S1) and the LiME results (Dataset S2), we noted a similar pattern among a subset of [s/t]Q phosphopeptides. Two notable examples were pSer1799 on AFAD/MLLT4 (Fig. S3 A and C) and pSer836 on AFF4 (Fig. S3 B and D). The [s/t]Q phosphomotif of these phosphopeptides is nested within an adjacent AKT-substrate motif (RXRXX[s/t]). Given the effects of PI3Ki on AKT-substrate phosphorylation, these data indicate that features that decrease in ATM-substrate motif blots likely represent inhibited phosphorylation within these nested motifs. In total, 192 proteins were found to carry phosphoPSMs nested within an RXX[s/t]Q motif, including several others that similarly decreased upon PI3K or dual inhibition (e.g., UCKL1 and IF4B/EIF4B; Dataset S2).
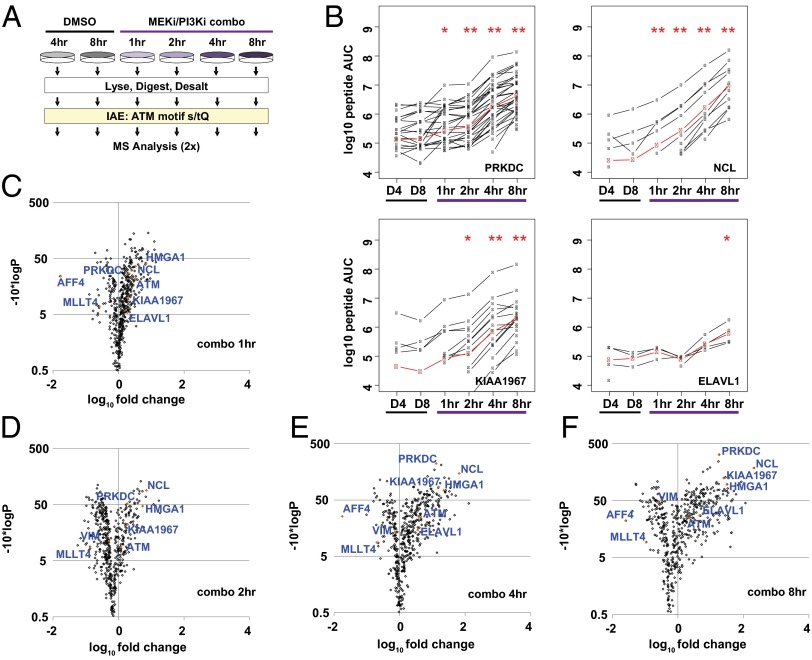
To confirm our initial findings and build a temporal understanding of this unique DDR, we initiated a time-course experiment using A2058 cells (Fig. 3A). Two groups of control cells were left untreated, and four groups were dosed with the combo treatment (4 × EC50, 10 μM each) concurrent with the addition of fresh medium. For treated cells, lysates were prepared after 1, 2, 4, and 8 h. To ascertain the effects of cell proliferation in this context, untreated cultures were lysed 4 and 8 h after the addition of fresh medium. Immunoblots against the [s/t]Q motif, pSer15 p53, and pSer139 H2AX confirmed the effectiveness of the treatment (Fig. S4A). Phosphopeptides bearing the [s/t]Q motif were captured and analyzed, yielding 3,100 unique phosphopeptides and 932 proteins with a pepFDR and a protFDR of 0.26% and 2.15%, respectively (Fig. S4B). Using the presence of an [S/T]Q motif as an orthogonal filter decreased the pepFDR to 0.05% (protFDR 0.47%; 2,898 unique phosphopeptides). Among these, the AScore localized phosphorylation to the [s/t]Q motif within 1,916 unique phosphopeptides.
Fig. 3.
Temporal profiling of DDR signaling upon MEK/PI3K dual inhibition. (A) Diagram depicting IAE of [s/t]Q motif phosphopeptides and MS analysis in the time course. (B) LiME plots for PRKDC, NUCL, K1967, and ELAV1 in each sample (black lines). The LiME model (red) was used to determine protein-level fold changes and P values relative to 4-h DMSO treatment (D4). *P < 0.01, **P < 0.001 relative to 4-h DMSO. (C–F) LiME data for 1-, 2-, 4-, and 8-h combo samples displayed as volcano plots vs. 4-h DMSO treatment. Selected proteins are identified, including several top hits denoted by orange symbols.
Increases in [s/t]Q motif-containing phosphoPSMs were observed again after MEK/PI3K dual inhibition (Dataset S1). In addition to the initial five sites on PRKDC, three additional [s/t]Q sites (pThr398, pThr1865, and pThr4102) residing outside the KIP-binding domain were identified (Fig. S4C). Notable was the triply phosphorylated pThr2638/pThr2645/pThr2647 PRKDC, which was not detectable in untreated cells but increased in a time-dependent manner after treatment (Fig. S4 D and E). In contrast to the initial experiment, [s/t]Q phosphoPSMs were detected on PRKDC and other DDR substrates in untreated cells (Dataset S1). This finding is consistent with data showing that pSer15 p53 and pSer139 H2AX were detectable in both 4-h and 8-h DMSO samples and increased in a time-dependent manner following treatment. The [s/t]Q immunoblots displayed a single dominant feature in control cells to such an extent that it obscured most other [s/t]Q phosphorylated species (Fig. S4A). Although the additional phosphoPSMs in this experiment may be explained by the enhanced sensitivity of the Orbitrap Velos, the increased levels of phosphorylation on certain proteins at baseline was attributed to cell proliferation and the DDR-associated replicative stress (28).
The XQuant algorithm was used again to generate label-free areas for LiME analysis (Fig. S5 and Dataset S2). For proteins with significant increases in the combined effects experiment, time-dependent increases in label-free AUC were observed beginning 1 h after MEK/PI3K dual inhibition (Fig. 3B). For each protein, the mixed-effect model increased by >10-fold at 4 h and >30-fold at 8 h relative the respective control values. Elevated [s/t]Q phosphorylation of other proteins, such as embryonic lethal, abnormal vision (Drosophila)-like 1 (ELAV1), was delayed until later timepoints. Volcano plots from individual timepoints (relative to 4-h DMSO) show PRKDC, NUCL, HMGA1, and K1967 emerging after 2 h and becoming among the most significantly altered proteins 4 h after MEK/PI3K dual inhibition (Fig. 3 C–F). Elevated phosphorylation was not observed when comparing DMSO at 8 h vs. 4 h (Fig. S4F). Ingenuity Pathway Analysis was used to generate a network-level view of proteins with [s/t]Q motif phosphopeptides and P values less than 1E−5 (−10*log, P >5; 8-h combo vs. 4-h DMSO treatment). Although the proteins observed to change upon MEK/PI3K dual inhibition were highly interconnected, a series of central nodes including PRKDC, ATM, H2AX, and ELAV1 emerged (Fig. S6). In addition, several proteins were highly interconnected with the phosphorylation dataset despite not having [s/t]Q phosphopeptides detected within this analysis. Among these were p53/tumor protein 53 (TP53), PARP1, sirtuin 7 (SIRT7), and lamin A (Fig. S6). Although immunoblotting demonstrated time-dependent elevation of pSer15 p53/TP53 and PARP cleavage (Fig. S1A), additional work is required to understand the potential roles of SIRT7 and lamin A.
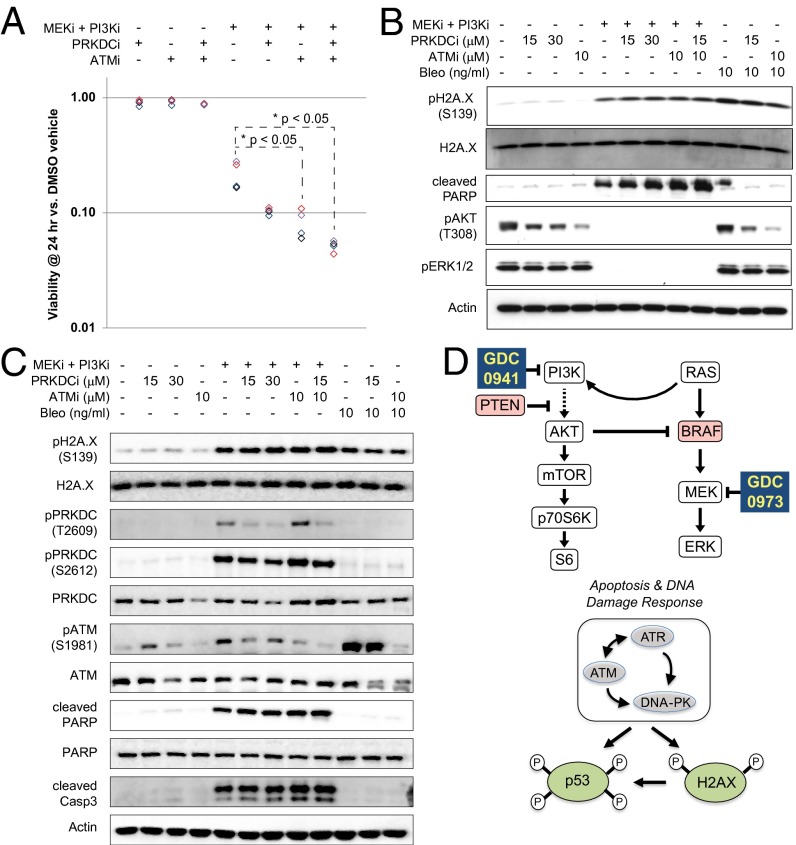
Because PRKDC and ATM are important after DNA damage, we predicted that their inhibition might enhance the cytotoxic effects of MEKi and PI3Ki. A2058 cells were pretreated for 1.5 h with NU7026 (PRKDCi; 15 μM) and/or KU55933 (ATMi; 10 μM) and then were treated with either DMSO or GDC-0973 and GDC-0941 combo (1× EC50) for 24 h. In viability assays, cells pretreated with PRKDCi and/or ATMi showed little change unless treated with MEKi and PI3Ki. In contrast, the viability of cells treated with MEKi and PI3Ki decreased by ∼80% even without pretreatment (Fig. 4A). Viability in combo-treated cells was decreased further by ATMi alone or in combination with PRKDCi.
Fig. 4.
Coinhibition of ATM and PRKDC enhances cell killing by GDC-0973 and GDC-0941. (A) Luminescent-based viability assays performed on A2058 cells pretreated for 90 min with DMSO, 15 μM NU7026 (PRKDCi), 10 μM KU55933 (ATMi), or 15 μM NU7026 + 10 μM KU55933. Cells subsequently were treated for 24 h with either DMSO or GDC-0973+GDC-0941 (MEKi + PI3Ki; 2.5 μM each). Average values (n = 6 technical replicates) are reported relative to DMSO for n = 4 replicate experiments. *P < 0.05 determined by paired student's t test followed by Bonferroni correction. (B) Blots of A2058 lysates treated for 3 h with DMSO, MEKi + PI3Ki (2.5 μM each), 15–30 μM PRKDCi, and/or 10 μM ATMi, as indicated. As a control for DDR, cells also were treated with bleomycin (10 ng/mL). Blots were probed against H2AX pSer139, cleaved PARP, AKT pThr308, and ERK1/2 pThr202/Tyr204, with total H2AX and actin as controls. (C) Blots of A2058 lysates treated as in B and probed for PRKDC pThr2609, PRKDC pSer2609, ATM pSer1981, and cleaved caspase 3, with total H2AX, PRKDC, ATM, PARP, and actin serving as controls. (D) Diagram depicting the mechanism of action for GDC-0973 and GDC-0941in melanoma cells. MAPK and PI3K pathways are drivers of cell proliferation and survival, here through the loss of PTEN tumor-suppressor activity and/or oncogenic activation of BRAF (red). Mutational activation of RAS, PI3K, and/or AKT also can signal through these pathways. Dual inhibition of the MEK/PI3K pathways decreases cell-survival signaling leading to apoptosis, activation of DNA damage-response kinases (DNA-PK, ATM, and ATR), and phosphorylation of downstream biomarkers H2AX and p53.
To validate these findings, immunoblots were performed to assess markers of DDR signaling and cell death. Pretreatment with PRKDCi and ATMi decreased the levels of pThr308 AKT but did not alter the levels of pSer139 H2AX, pThr202/pTyr204 ERK1/2, or cleaved PARP (Fig. 4B). Dual inhibition of MEK/PI3K eliminated both pThr202/pTyr204 ERK1/2 and pThr308 AKT signals while increasing the levels of pSer139 H2AX and cleaved PARP. Bleomycin, a classical DNA-damaging agent, affected only pSer139 H2AX. As predicted by viability assays, coadministration of MEKi and PI3Ki with PRKDCi and/or ATMi enhanced cleavage of PARP (Fig. 4 B and C) and caspase 3 (Fig. 4C). Phosphorylation of H2AX Ser139 increased after MEKi + PI3Ki to a extent similar to the increase seen in cells treated with bleomycin (Fig. 4 B and C) and, interestingly, was unaffected by preinhibition of PRKDC or ATM. Western blots against pThr2609/pSer2612 PRKDC and pSer1981 ATM confirmed inhibited phosphorylation in the DDR network, particularly with coadministration of ATMi/PRKDCi (Fig. 4C).
Discussion
In this work we have used motif-specific IAE and MS to assess known DDR phosphorylation events (13, 14, 29, 30) while simultaneously interrogating signals unique to this model in which coadministration of GDC-0973 with GDC-0941 has proven efficacious (Fig. 4D and Dataset S3) (17, 18). Initially we were intrigued by the observation of elevated [s/t]Q motif phosphorylation, given that the DDR is driven by kinases with known homologies to PI3K including ATM and PRKDC. Early strategies for targeting PI3K coinhibited the activities of ATM and PRKDC (31, 32). PI3K-mTOR dual inhibitors (e.g., NVP-BEZ235, PI-103) also reportedly inhibit ATM and PRKDC in biochemical and cell-culture models (33, 34). Used here, GDC-0941 is a pan-class I PI3K inhibitor with enzyme IC50 values of 0.003 μM (p110α), 0.033 μM (p110β), 0.003 μM (p110δ), or 0.075 μM (p110γ), making it >400 times more selective for p110α/δ than PRKDC (16). Instead of inhibition, immunoblotting and MS suggested activation of PRKDC and ATM as a consequence of MEK/PI3K dual inhibition. This activation also was seen with GDC-0941 alone, albeit to a lesser extent. Recent work from breast cancer models similarly showed that PI3Kα siRNA or the pan-class I–selective NVP-BKM120 increased polyADP ribosylation, pSer139 H2AX, pSer2056 PRKDC, and pSer1981 ATM (35, 36). The authors noted the absence of Rad51 foci after PI3Kα inhibition or knockdown and speculated that PRKDC activation may represent a feedback response (36). In line with a previous report that PRKDC controls prosurvival signaling by modulating pSer473 AKT (37), our data show that PRKDC/ATM inhibition decreases pT308 AKT (Fig. 4B). When these data are taken together, one prediction is that loss of pAKT triggers a feedback loop leading to activation of PRKDC. Alternatively, DNA cleaved during the initial phases of apoptosis may activate PRKDC to initiate nonhomologous end joining as a late-stage effort to avoid apoptosis (38). The nature of such a signal remains to be elucidated, although the consequences of such signaling might be hypothesized to create an environment in which acquired resistance may emerge.
Beyond the known roles of p53, PARP, ATM, and H2AX in the DDR, the [s/t]Q substrates significantly altered by MEK/PI3K dual inhibition include modules controlling protein acetylation, chromatin status, and RNA metabolism. Notable was the number of [s/t]Q substrates previously found to associate with SIRT7 (39) and ELAV1 (40). Several sirtuin family members play roles in the DDR, including SIRT1 which is controlled by ELAV1 (i.e., HuR) and affects cell survival after DNA damage (41). ELAV1 and a cluster of differentially regulated [s/t]Q substrates (e.g., NUCL, NFIP2, DHX9, HNRPU, K1967) comprise the messenger ribonucleoprotein particle complex (Fig. S6, Table S1, and Datasets S1 and S2), which packages and exports mRNA from the nucleus and itself is regulated by the mTOR and ribosomal S6 kinase (42). SIRT7 is associated with the TPR of the nuclear pore complex, THOC2 (which controls RNA export by binding spliced mRNAs), the ribosome biogenesis factor BMS1, and the ribonuclease POP1. Other [s/t]Q substrates from our study affect transcription by RNA polymerase II (RTF1, CCNC, and MED1), RNA transport (FXR1), splicing (LSM2), and translation (4EBP1). Considering these in aggregate, our data suggest that the cell-death program elicited by GDC-0973 and GDC-0941 may impinge upon multiple aspects of RNA metabolism.
Despite these advances, several key questions remain. It is unclear whether the DDR downstream of MEKi + PI3Ki derives from bona fide double-strand breaks as a consequence of cell death or instead is a noncanonical response to chromatin remodeling or oxidative stress (43, 44). Additional work also will be required to elucidate the sensors of [s/t]Q phosphorylation events and to determine whether common themes exist between subsets of DDR substrates. Understanding the full effects of the inhibition of the RAS–RAF–MEK and PI3K–AKT–mTOR pathways on cell death and acquired resistance remains critical to development of targeted therapeutic agents.
Materials and Methods
Melanoma cell lines were cultured and protein lysates were prepared under standard conditions following treatment with GDC-0973 and/or GDC-0941, as indicated. For MS studies, phosphopeptides were captured using [s/t]Q phosphomotif antibodies using PTMscan protocols (Cell Signaling Technology) (14) and were analyzed on an Orbitrap XL or Orbitrap-Velos. Database searches were performed using Mascot, peak areas determined using VistaQuant (25), and protein-level effects were assessed using LiME analysis (27). A detailed overview of the methods used here is presented in SI Materials and Methods and in Datasets S1–S3.
Supplementary Material
Acknowledgments
We thank T. O'Brien, R. Yauch, J. Baughman, and the Microchemistry and Proteomics Lab for providing feedback on the manuscript. Phosphopeptide IAE studies were carried out under limited license of PTMscan from Cell Signaling Technology.
Footnotes
Conflict of interest statement: All authors are employees and shareholders of Genentech Inc., a member of the Roche group.
This article is a PNAS Direct Submission.
Data deposition: The large data tables provided as Supporting Information also are available at http://research-pub.gene.com/proteomics_data.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1309473110/-/DCSupplemental.
References
- 1.Downward J. Targeting RAS signalling pathways in cancer therapy. Nat Rev Cancer. 2003;3(1):11–22. doi: 10.1038/nrc969. [DOI] [PubMed] [Google Scholar]
- 2.Engelman JA, Luo J, Cantley LC. The evolution of phosphatidylinositol 3-kinases as regulators of growth and metabolism. Nat Rev Genet. 2006;7(8):606–619. doi: 10.1038/nrg1879. [DOI] [PubMed] [Google Scholar]
- 3.Roberts PJ, Der CJ. Targeting the Raf-MEK-ERK mitogen-activated protein kinase cascade for the treatment of cancer. Oncogene. 2007;26(22):3291–3310. doi: 10.1038/sj.onc.1210422. [DOI] [PubMed] [Google Scholar]
- 4.Sebolt-Leopold JS, Herrera R. Targeting the mitogen-activated protein kinase cascade to treat cancer. Nat Rev Cancer. 2004;4(12):937–947. doi: 10.1038/nrc1503. [DOI] [PubMed] [Google Scholar]
- 5.Garcia-Echeverria C, Sellers WR. Drug discovery approaches targeting the PI3K/Akt pathway in cancer. Oncogene. 2008;27(41):5511–5526. doi: 10.1038/onc.2008.246. [DOI] [PubMed] [Google Scholar]
- 6.Liu P, Cheng H, Roberts TM, Zhao JJ. Targeting the phosphoinositide 3-kinase pathway in cancer. Nat Rev Drug Discov. 2009;8(8):627–644. doi: 10.1038/nrd2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zimmermann S, Moelling K. Phosphorylation and regulation of Raf by Akt (protein kinase B) Science. 1999;286(5445):1741–1744. doi: 10.1126/science.286.5445.1741. [DOI] [PubMed] [Google Scholar]
- 8.Kyriakis JM, Avruch J. Mammalian mitogen-activated protein kinase signal transduction pathways activated by stress and inflammation. Physiol Rev. 2001;81(2):807–869. doi: 10.1152/physrev.2001.81.2.807. [DOI] [PubMed] [Google Scholar]
- 9.Alessi DR, et al. Mechanism of activation of protein kinase B by insulin and IGF-1. EMBO J. 1996;15(23):6541–6551. [PMC free article] [PubMed] [Google Scholar]
- 10.Sarbassov DD, Guertin DA, Ali SM, Sabatini DM. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science. 2005;307(5712):1098–1101. doi: 10.1126/science.1106148. [DOI] [PubMed] [Google Scholar]
- 11.Rush J, et al. Immunoaffinity profiling of tyrosine phosphorylation in cancer cells. Nat Biotechnol. 2005;23(1):94–101. doi: 10.1038/nbt1046. [DOI] [PubMed] [Google Scholar]
- 12.Moritz A, et al. Akt-RSK-S6 kinase signaling networks activated by oncogenic receptor tyrosine kinases. Sci Signal. 2010;3(136):ra64. doi: 10.1126/scisignal.2000998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Matsuoka S, et al. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007;316(5828):1160–1166. doi: 10.1126/science.1140321. [DOI] [PubMed] [Google Scholar]
- 14.Stokes MP, et al. Profiling of UV-induced ATM/ATR signaling pathways. Proc Natl Acad Sci USA. 2007;104(50):19855–19860. doi: 10.1073/pnas.0707579104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim ST, Lim DS, Canman CE, Kastan MB. Substrate specificities and identification of putative substrates of ATM kinase family members. J Biol Chem. 1999;274(53):37538–37543. doi: 10.1074/jbc.274.53.37538. [DOI] [PubMed] [Google Scholar]
- 16.Folkes AJ, et al. The identification of 2-(1H-indazol-4-yl)-6-(4-methanesulfonyl-piperazin-1-ylmethyl)-4-morpholin-4-yl-thieno[3,2-d]pyrimidine (GDC-0941) as a potent, selective, orally bioavailable inhibitor of class I PI3 kinase for the treatment of cancer. J Med Chem. 2008;51(18):5522–5532. doi: 10.1021/jm800295d. [DOI] [PubMed] [Google Scholar]
- 17.Hoeflich KP, et al. Intermittent administration of MEK inhibitor GDC-0973 plus PI3K inhibitor GDC-0941 triggers robust apoptosis and tumor growth inhibition. Cancer Res. 2012;72(1):210–219. doi: 10.1158/0008-5472.CAN-11-1515. [DOI] [PubMed] [Google Scholar]
- 18.Hoeflich KP, et al. In vivo antitumor activity of MEK and phosphatidylinositol 3-kinase inhibitors in basal-like breast cancer models. Clin Cancer Res. 2009;15(14):4649–4664. doi: 10.1158/1078-0432.CCR-09-0317. [DOI] [PubMed] [Google Scholar]
- 19.Wallin JJ, et al. GDC-0980 is a novel class I PI3K/mTOR kinase inhibitor with robust activity in cancer models driven by the PI3K pathway. Mol Cancer Ther. 2011;10(12):2426–2436. doi: 10.1158/1535-7163.MCT-11-0446. [DOI] [PubMed] [Google Scholar]
- 20.Tsai J, et al. Discovery of a selective inhibitor of oncogenic B-Raf kinase with potent antimelanoma activity. Proc Natl Acad Sci USA. 2008;105(8):3041–3046. doi: 10.1073/pnas.0711741105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Beausoleil SA, Villén J, Gerber SA, Rush J, Gygi SP. A probability-based approach for high-throughput protein phosphorylation analysis and site localization. Nat Biotechnol. 2006;24(10):1285–1292. doi: 10.1038/nbt1240. [DOI] [PubMed] [Google Scholar]
- 22.Meek K, Dang V, Lees-Miller SP. DNA-PK: The means to justify the ends? Adv Immunol. 2008;99:33–58. doi: 10.1016/S0065-2776(08)00602-0. [DOI] [PubMed] [Google Scholar]
- 23.Ding Q, et al. Autophosphorylation of the catalytic subunit of the DNA-dependent protein kinase is required for efficient end processing during DNA double-strand break repair. Mol Cell Biol. 2003;23(16):5836–5848. doi: 10.1128/MCB.23.16.5836-5848.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meek K, Douglas P, Cui X, Ding Q, Lees-Miller SP. trans Autophosphorylation at DNA-dependent protein kinase’s two major autophosphorylation site clusters facilitates end processing but not end joining. Mol Cell Biol. 2007;27(10):3881–3890. doi: 10.1128/MCB.02366-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bakalarski CE, et al. The impact of peptide abundance and dynamic range on stable-isotope-based quantitative proteomic analyses. J Proteome Res. 2008;7(11):4756–4765. doi: 10.1021/pr800333e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Clough T, et al. Protein quantification in label-free LC-MS experiments. J Proteome Res. 2009;8(11):5275–5284. doi: 10.1021/pr900610q. [DOI] [PubMed] [Google Scholar]
- 27.Clough T, Thaminy S, Ragg S, Aebersold R, Vitek O. Statistical protein quantification and significance analysis in label-free LC-MS experiments with complex designs. BMC Bioinformatics. 2012;13(Suppl 16):S6. doi: 10.1186/1471-2105-13-S16-S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu S, et al. Distinct roles for DNA-PK, ATM and ATR in RPA phosphorylation and checkpoint activation in response to replication stress. Nucleic Acids Res. 2012;40(21):10780–10794. doi: 10.1093/nar/gks849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bennetzen MV, et al. Site-specific phosphorylation dynamics of the nuclear proteome during the DNA damage response. Mol Cell Proteomics. 2010;9(6):1314–1323. doi: 10.1074/mcp.M900616-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bensimon A, et al. ATM-dependent and -independent dynamics of the nuclear phosphoproteome after DNA damage. Sci Signal. 2010;3(151):rs3. doi: 10.1126/scisignal.2001034. [DOI] [PubMed] [Google Scholar]
- 31.Powis G, et al. Wortmannin, a potent and selective inhibitor of phosphatidylinositol-3-kinase. Cancer Res. 1994;54(9):2419–2423. [PubMed] [Google Scholar]
- 32.Sarkaria JN, et al. Inhibition of phosphoinositide 3-kinase related kinases by the radiosensitizing agent wortmannin. Cancer Res. 1998;58(19):4375–4382. [PubMed] [Google Scholar]
- 33.Mukherjee B, et al. The dual PI3K/mTOR inhibitor NVP-BEZ235 is a potent inhibitor of ATM- and DNA-PKCs-mediated DNA damage responses. Neoplasia. 2012;14(1):34–43. doi: 10.1593/neo.111512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kong D, Yaguchi S, Yamori T. Effect of ZSTK474, a novel phosphatidylinositol 3-kinase inhibitor, on DNA-dependent protein kinase. Biol Pharm Bull. 2009;32(2):297–300. doi: 10.1248/bpb.32.297. [DOI] [PubMed] [Google Scholar]
- 35.Ibrahim YH, et al. PI3K inhibition impairs BRCA1/2 expression and sensitizes BRCA-proficient triple-negative breast cancer to PARP inhibition. Cancer Discov. 2012;2(11):1036–1047. doi: 10.1158/2159-8290.CD-11-0348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Juvekar A, et al. Combining a PI3K inhibitor with a PARP inhibitor provides an effective therapy for BRCA1-related breast cancer. Cancer Discov. 2012;2(11):1048–1063. doi: 10.1158/2159-8290.CD-11-0336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bozulic L, Surucu B, Hynx D, Hemmings BA. PKBalpha/Akt1 acts downstream of DNA-PK in the DNA double-strand break response and promotes survival. Mol Cell. 2008;30(2):203–213. doi: 10.1016/j.molcel.2008.02.024. [DOI] [PubMed] [Google Scholar]
- 38.Mukherjee B, et al. DNA-PK phosphorylates histone H2AX during apoptotic DNA fragmentation in mammalian cells. DNA Repair (Amst) 2006;5(5):575–590. doi: 10.1016/j.dnarep.2006.01.011. [DOI] [PubMed] [Google Scholar]
- 39.Tsai YC, Greco TM, Boonmee A, Miteva Y, Cristea IM. Functional proteomics establishes the interaction of SIRT7 with chromatin remodeling complexes and expands its role in regulation of RNA polymerase I transcription. Mol Cell Proteomics. 2012;11(5):60–76. doi: 10.1074/mcp.A111.015156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Butter F, Scheibe M, Mörl M, Mann M. Unbiased RNA-protein interaction screen by quantitative proteomics. Proc Natl Acad Sci USA. 2009;106(26):10626–10631. doi: 10.1073/pnas.0812099106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gorospe M, de Cabo R. AsSIRTing the DNA damage response. Trends Cell Biol. 2008;18(2):77–83. doi: 10.1016/j.tcb.2007.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ma XM, Yoon SO, Richardson CJ, Jülich K, Blenis J. SKAR links pre-mRNA splicing to mTOR/S6K1-mediated enhanced translation efficiency of spliced mRNAs. Cell. 2008;133(2):303–313. doi: 10.1016/j.cell.2008.02.031. [DOI] [PubMed] [Google Scholar]
- 43.Guo Z, Kozlov S, Lavin MF, Person MD, Paull TT. ATM activation by oxidative stress. Science. 2010;330(6003):517–521. doi: 10.1126/science.1192912. [DOI] [PubMed] [Google Scholar]
- 44.Soutoglou E, Misteli T. Activation of the cellular DNA damage response in the absence of DNA lesions. Science. 2008;320(5882):1507–1510. doi: 10.1126/science.1159051. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.