Abstract
Isoprenoid compounds constitute an immensely diverse group of acyclic, monocyclic and polycyclic compounds that play important roles in all living organisms. Despite the diversity of their structures, this plethora of natural products arises from only two 5-carbon precursors, isopentenyl diphosphate (IPP) and dimethylallyl diphosphate (DMAPP). This review will discuss the enzymes in the mevalonate (MVA) and methylerythritol phosphate (MEP) biosynthetic pathways leading to IPP and DMAPP with a particular focus on MEP synthase (DXR) and IPP isomerase (IDI), which are potential targets for the development of antibiotic compounds. DXR is the second enzyme in the MEP pathway and the only one for which inhibitors with antimicrobial activity at pharmaceutically relevant concentrations are known. All of the published DXR inhibitors are fosmidomycin analogues, except for a few bisphosphonates with moderate inhibitory activity. These far, there are no other candidates that target DXR. IDI was first identified and characterised over 40 years ago (IDI-1) and a second convergently evolved isoform (IDI-2) was discovered in 2001. IDI-1 is a metalloprotein found in Eukarya and many species of Bacteria. Its mechanism has been extensively studied. In contrast, IDI-2 requires reduced flavin mononucleotide as a cofactor. The mechanism of action for IDI-2 is less well defined. This review will describe how lead inhibitors are being improved by structure-based drug design and enzymatic assays against DXR to lead to new drug families and how mechanistic probes are being used to address questions about the mechanisms of the isomerases.
Keywords: DXR, IDI, isomerase, isopentenyl, isoprenoid, MEP, mevalonate, MVA, reductoisomerase
1. INTRODUCTION
Isoprenoid compounds constitute an immensely diverse group of acyclic, monocyclic and polycyclic compounds that play important roles in all living organisms [1]. They function as regulators of gene expression, constituents of membranes, vitamins, antimicrobial agents, mating pheromones, reproductive hormones, components of signal transduction pathways, and constituents of electron transport and photo-synthetic machinery [2].
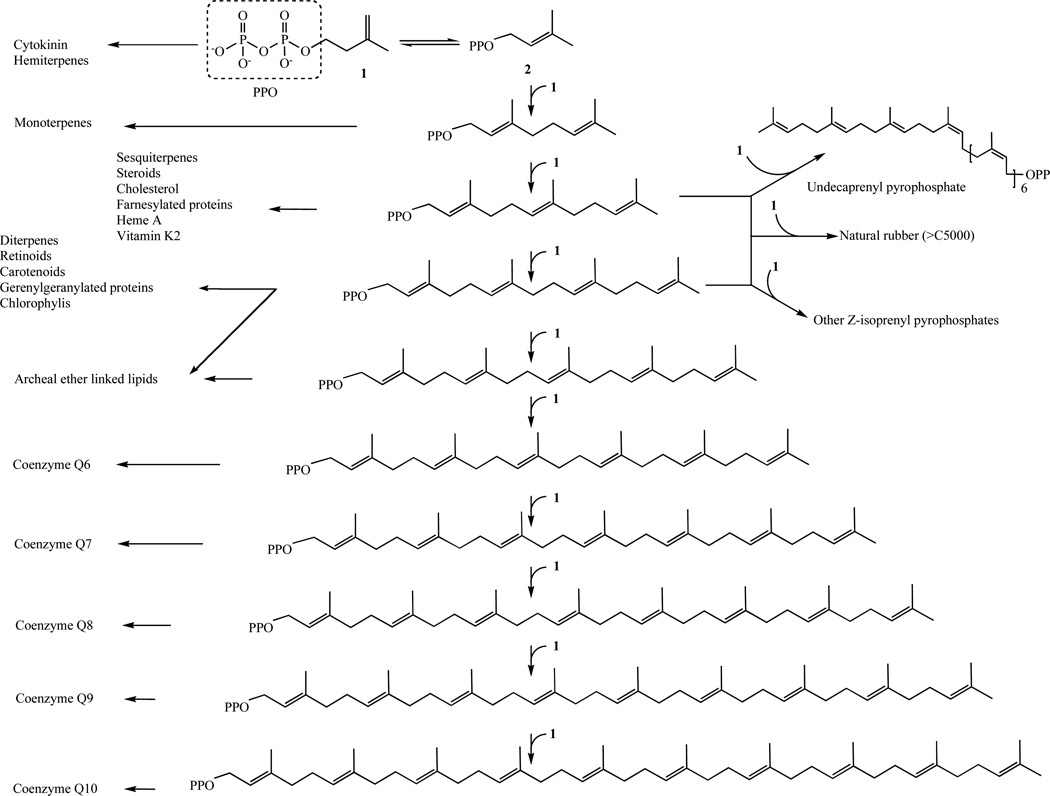
Despite the diversity of their structures, this plethora of natural products arises from only two 5-carbon precursors, isopentenyl diphosphate (IPP, (1)) and dimethylallyl diphosphate (DMAPP, (2)) (Fig. 1) [3].
Fig. 1.
Biosynthetic pathways catalyzed by isoprenyl diphosphate synthases and the final reaction products. Adapted from Wang and Ohnuma [3].
2. THE MEVALONATE PATHWAY
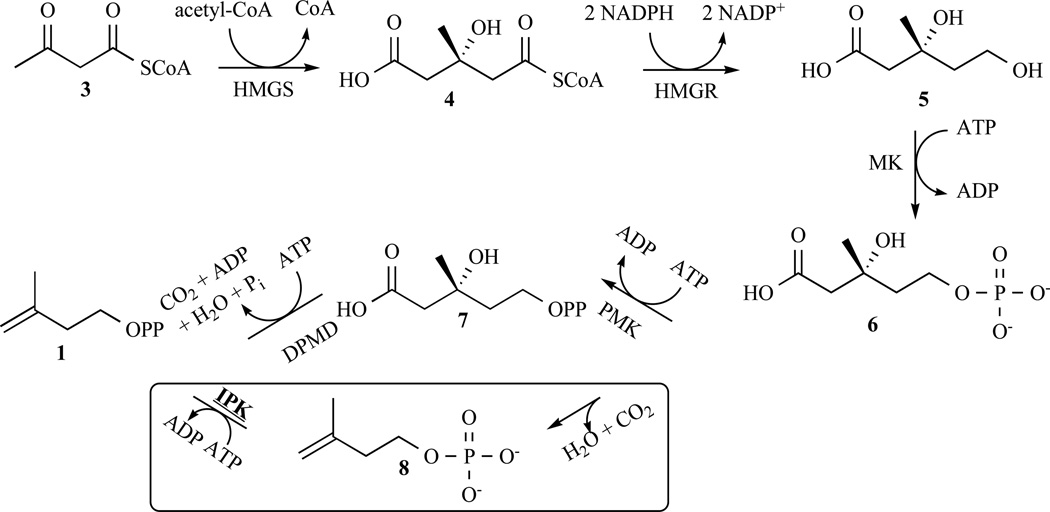
Following the discovery of the mevalonate pathway (MEV pathway) in the 1960s [4], it was widely accepted that it was the sole route to IPP and DMAPP in all living organisms. The first step is the formation of the acetoacetyl-CoA by the condensation of two molecules of acetyl-CoA catalyzed by a thiolase. In the following reaction, a third molecule of acetyl-CoA condenses with the aceto-acetyl-CoA to form 3-hydroxy-3-methylglutaryl-CoA (HMG-CoA (4)); the reaction is catalyzed by the HMG-CoA synthase (HMGS). Then, HMG-CoA is reduced to 3R-mevalonate (5) during a reaction, which consumes two molecules of NADPH. This third stage of the pathway is catalysed by HMG-CoA reductase (HMGR). A series of three reactions then converts mevalonate into IPP with the consumption of one molecule of ATP. The first two steps are catalyzed by mevalonate kinase (MK) and phosphomevalonate kinase (PMK), which converts the primary hydroxyl group in 3R-mevalonate to a diphosphate ester. The third reaction, catalysed by the diphosphomevalonate decarboxylase (DPMD), phosphorylates the tertiary hydroxyl group in (7) and is followed by a fragmentation to give IPP (Fig. 2). The mevalonate pathway and its inhibition were studied [5, 6], especially HMGR as a target for the statins class of antihypercholesterolemia drugs but also targeting Alzheimer’s disease and Parkinson’s disease [7].
Fig. 2.
Overview of the well-described mevalonate pathway. The alternative route described by Grochowski et al. in Archae is represented in the box.
In 2006, Grochowski et al. reported that the translated protein from open reading frame MJ0044 in Methanocal-dococcus jannaschii (MJ) catalyzes the ATP-dependent phosphorylation of isopentenyl phosphate (IP) rather than mevalonate or mevalonate phosphate, and the protein was assigned as an IP kinase (IPK) [8]. The M. jannaschii protein and homologous proteins from Thermoplasma acidophilum and Methanothermobacter thermautotrophicus have high catalytic efficiencies for phosphorylation of IP (∼106 M−1s−1) [9] similar to those of established enzymes in the isoprenoid pathway. Grochowski et al. suggested an alternative mevalonate route in Archaea where decarboxylation of mevalonate 5-phosphate (6) produces isopentenyl 5-phosphate (IP (8)) the substrate for the isopentenyl 5-phosphate kinase (IPK), to give IPP (Fig. 2). In 2010, Mabanglo and coworkers described the first crystal structure [10] of this enzyme, which is now under investigation [9, 11].
3. THE MEP PATHWAY
3.1. General Description of the Pathway
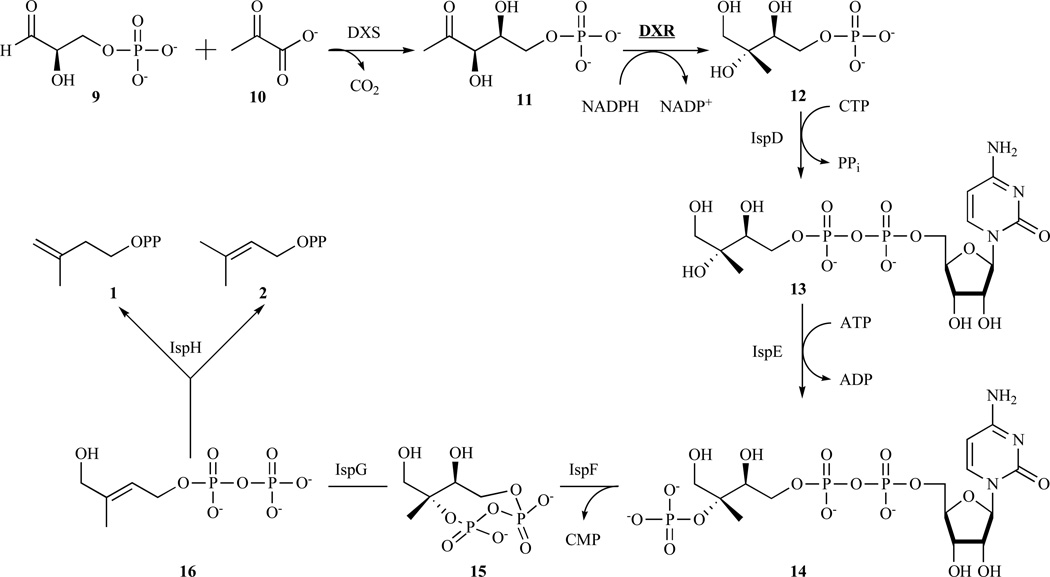
The MEP pathway [12, 13] starts with the condensation of glyceraldehyde phosphate (9) and pyruvate (10) to afford 1-deoxyxylulose 5-phosphate (DXP) (11) in a reaction catalysed by 1-deoxyxylulose 5-phosphate synthase (DXS) [14, 15]. A sigmatropic rearrangement of 1-deoxyxylulose 5-phosphate followed by reduction of the intermediate aldehyde, catalysed by 1-deoxy-D-xylulose 5-phosphate reductoisomerase (DXR), gives the branched polyol, 2-C-methyl-D-erythritol 4-phosphate (MEP) (12) [16, 17]. This enzyme and its inhibition will be further developed in section 3.2. The 2-C-methyl-D-erythritol 4-phosphate is converted into cyclic diphosphate (15) by a series of three enzyme-catalysed reaction steps. Specifically, IspD converts the branched polyol and CTP into diphosphocytidyl derivative (CDP-ME) (13) [18]. Then, the IspE phosphorylates the tertiary hydroxyl group in CDP-ME to produce 4-diphosphocytidyl-2-C-methyl-D-erythritol 2- phosphate (CDP-ME2P) (14) [19], which is then converted into 2-C-methyl-D-erythritol 2,4-cyclodiphosphate (MECDP) (15) with release of CMP by the catalytic action of the IspF protein [20, 21]. Cyclic diphosphate (15) contains the 5-carbon branched motif required for IPP and DMAPP but is in a more highly oxidised state. This problem is addressed by the consecutive action of two structurally and mechanistically unique iron–sulphur enzymes specified by the ispG and ispH genes. More specifically, the IspG protein catalyses the reductive opening (two-electron reduction) of the eight-membered ring of 15, which yields 1-hydroxy-2-methyl-2-(E)-butenyl 4-diphosphate (HMBDP) (16) [22–25]. The residual primary hydroxyl group of 16 is then reductively removed by IspH protein [26] to give IPP and DMAPP (Fig. 3).
Fig. 3.
Overview of the mevalonate-independent pathway. Interestingly, IPP (1) and DMAPP (2) are obtained in a kinetically controlled reaction.
It is now firmly established that the non-mevalonate pathway is the single source of terpenoids in a large number of eubacteria [27]. Notably, Gram-positive cocci, Coxiella burnetii and Borrelia burgdorferi are the only major bacterial pathogens using the mevalonate pathway.
Moreover, the enzymes of the non-mevalonate isoprenoid pathway are essential in malaria parasites and in numerous pathogenic bacteria, which cause a wide variety of infectious diseases including tuberculosis that is estimated to cause around a million fatalities per year (Table 1) [28]. Due to the absence of the non-mevalonate pathway in humans, any anti-infective drugs designed to interrupt isoprenoid biosynthesis in the respective pathogens should be exempt from target-related toxicity, and understanding the structures and mechanisms of the pathway enzymes is important for inhibitor design.
Table 1.
Distribution of Isoprenoid Biosynthetic Enzymes in Major Human Bacterial Pathogens. As for Reference Human Utilizes the MEV Pathway and Type I IPP Isomerase to Produce Isoprenoid Compounds
| Microorganism | MEP pathway | MEV pathway | IPP isomerase | Examples of diseases |
|---|---|---|---|---|
| Gram-positive cocci | ||||
| Staphylococcus aureus | − | + | Type II | Impetigo follicularis, furunculosis |
| Staphylococcus epidermidis | − | + | Type II | Wound infections |
| Streptococcus pyogenes | − | + | Type II | Scarler fever, toxic-shock syndrome |
| Streptococcus agalactiae | − | + | Type II | Neonatal septicaemia |
| Streptococcus viridans | − | + | Type II | Endocarditis lenta |
| Streptococcus pneumoniae | − | + | Type II | Pneumonia |
| Enterococcus faecalis | − | + | Type II | Endocarditis, Bacteraemia |
| Gram-negative cocci | ||||
| Neisseria meningitis | + | − | − | Meningitis |
| Neisseria gonorrhoea | + | − | − | Gonorrhoea |
| Gram-positive rods | ||||
| Listeria monocytogenes | + | + | Type II | Listeriosis |
| Bacillus anthracis | + | − | Type II | Anthrax |
| Clostridium botulinum | + | − | − | Botulism |
| Clostridium tetani | + | − | − | Tetanus |
| Gram-negative rods | ||||
| E. coli | + | − | Type I | Enterocolitis |
| Salmonella typhi | + | − | Type I | Typhus |
| Salmonella paratyphi | + | − | Type I | Bacteraemia |
| Shigella sonnei | + | − | Type I | Typhus |
| Gram-negative/spiral-shaped bacteria | ||||
| Vibrio cholerae | + | − | Type I | Cholera |
| Helicobacter pylori | + | − | - | Gastritis Type B |
| Camphylobacter jejuni | + | − | - | Enterocolitis |
| Spirochaetal bacteria | ||||
| Borrelia burgdorferi | − | + | Type II | Lyme disease |
| Acid-fast rods | ||||
| Mycobacterium tuberculosis | + | − | Type I | Tuberculosis |
| Mycobacterium leprae | + | − | Type I | Leprosy |
| Obligate intracellular bacteria | ||||
| Rickettsia prowazecki | − | − | Type II | Typhus |
| Rickettsia rickettsii | − | − | Type II | Rocky mountain spotted fever |
| Coxiella burnetii | − | + | Type I | Q fever |
At this time, there is no report of inhibitors of enzymes of the MEP pathway, other than DXR, with proven antimicrobial activity at pharmaceutically relevant concentrations.
3.2. Enzyme Inhibition in Drug Design: The Case of DXR
Identification of the Escherichia coli dxr gene and successful expression of the recombinant 1-deoxy-D-xylulose 5-phosphate reductoisomerase (DXR, IspC, EC 1.1.1.86) were first reported in 1998 [29]. DXR is a well-studied 42–45 kDa enzyme with NADPH and a divalent cation (Mg2+, Mn2+ or Co2+) as cofactors. The enzyme typically has a pH optimum in the range 7–8 (for more details [30]). Table 2 summarizes the kinetic data available for DXR through mid-2011.
Table 2.
Enzymatic Characteristics of DXR form Several Microorganisms. Temperature is the Temperature at which Enzymatic Assays were Conducted
| Km (µM) | kcat (s−1) | kcat/Km (µM−1s−1) | Temperature (°C) | Reference | |
|---|---|---|---|---|---|
| Z. mobilis | 300 | 14 | 0.05 | 40 | [31] |
| S. coelicolor | 190 | 19.2 | 0.10 | 25 | [32] |
| E. coli | 720 | 21.7 | 0.03 | 37 | |
| 115 | 116 | 0.99 | 37 | [33] | |
| Synechocystis sp.PCC6803 | 134 | 5 | 0.04 | 37 | [34] |
| M. tuberculosis | 42 | 2.1 | 0.05 | 25 | [35] |
| F. tularensis | 104 | 2 | 0.02 | 22 | [36] |
| T. maritima | 110 | 5.5 | 0.05 | 85 | [37] |
| 40 | 0.29 | 0.01 | 50 |
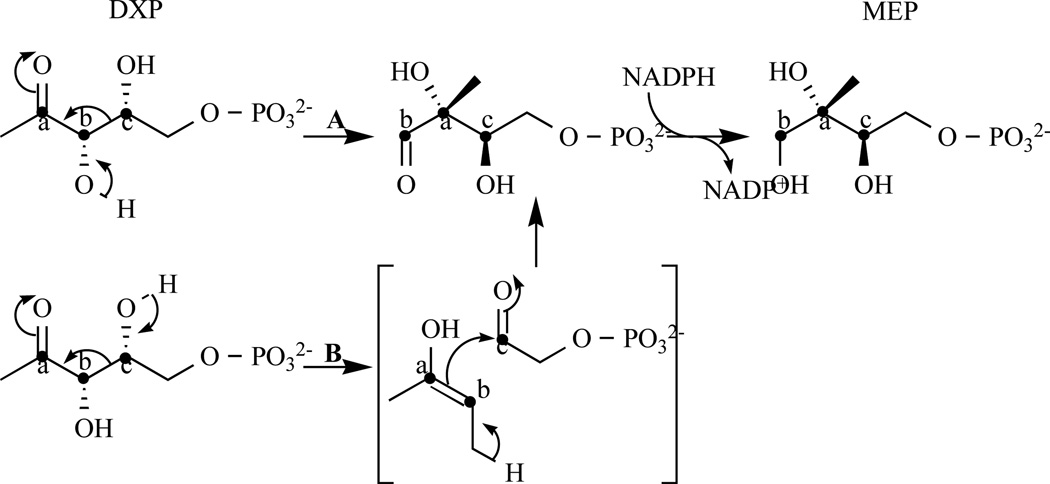
Two mechanisms have been proposed for the DXR-catalysed reaction, an α-ketol rearrangement (Fig. 4 path A) or a retroaldol/aldol rearrangement (Fig. 4 path B). Despite much effort, the catalytic mechanism of DXR remains elusive [37–44].
Fig. 4.
Representation of path A: α-ketol or path B retroaldol/aldol rearrangement mechanisms. This latter is preferred by Munos et al. [44].
3.2.1. First Inhibition Studies to Design New Antimalarial Drugs
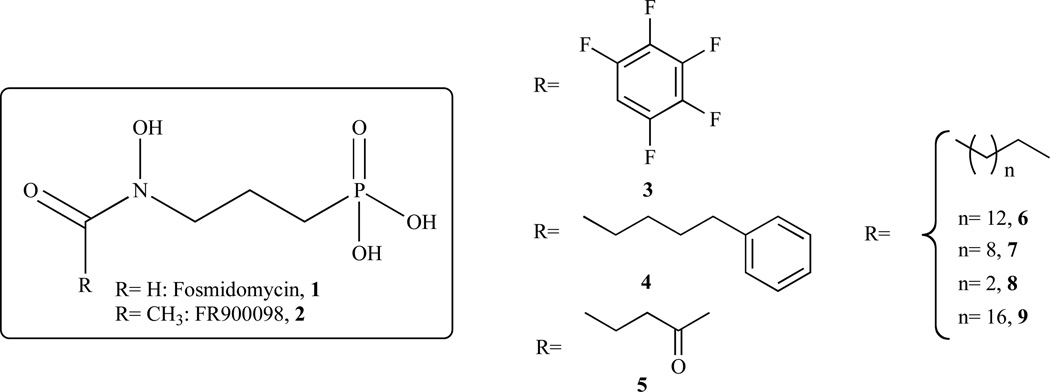
Fosmidomycin (FR-31564; 3-(N-formyl-N-hydroxy-amino)propyl phosphonic acid (Fig. 5, 1)) was identified as a natural antibiotic from Streptomyces lavendulae [45]. Shortly after the discovery of DXR, it was established that this antibacterial compound is a mixed (competitive and non-competitive) inhibitor of E. coli DXR (ec-DXR) with a Ki of 38 nM [46]. When fosmidomycin was tested as an inhibitor of the Z. Mobilis DXR (zm-DXR), it was determined to be a competitive inhibitor with a Ki of 600 nM [31]. A related natural product, FR900098 (Fig. 5, 2), is also an effective inhibitor of DXR and has shown greater antimalarial activity than fosmidomycin in a mouse model [47]. Subsequent work has shown that fosmidomycin is a slow, tight-binding inhibitor of DXR, with an initial phase of inhibition that is competitive with the substrate (DXP) and a second phase of inhibition that is non-competitive [33]. A conformational change in the enzyme has been proposed for this second phase and, in 2005, Mac Sweeney, et al. solved the X-ray crystal structure of the ec-DXR in a ternary complex with fosmidomycin and NADPH representing this closed conformation of the enzyme [48].
Fig. 5.
Chemical structures of selected compounds synthesized and evaluated by Ortman et al. Chemical structures of the reference inhibitors are represented in the box.
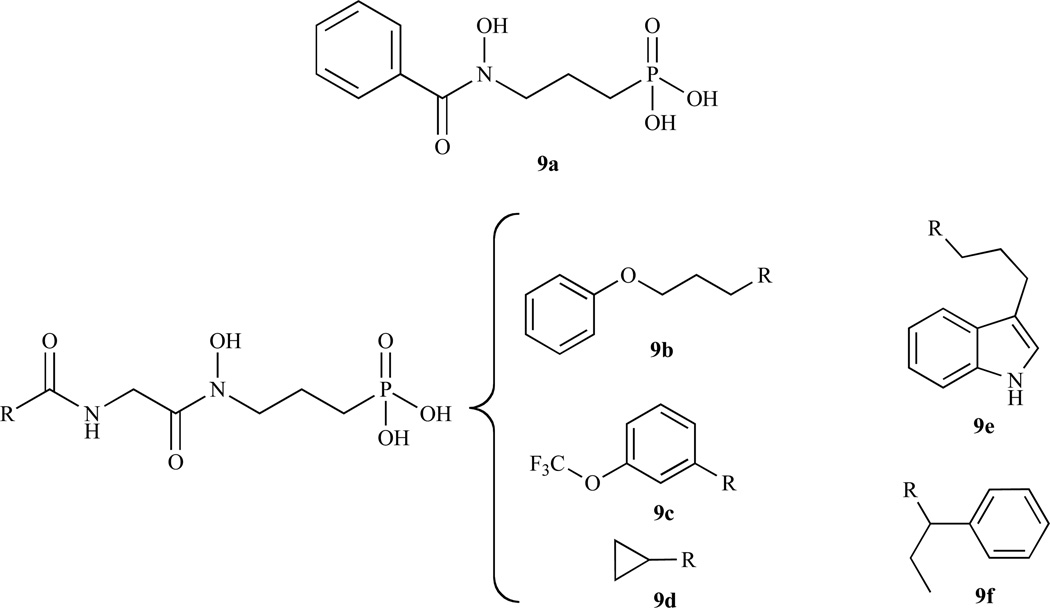
In 2006, Proteau and co-workers synthesised and evaluated fosmidomycin analogues as inhibitors of Synechocystis sp. PCC6803 DXR (spcc-DXR). The broad range of inhibition potencies of these analogues (go through and replace w/analogues) provided information about structural requirements for the design of more potent inhibitors [49]. Singh and coworkers published a review that summarizes the developments in DXR inhibition to 2007 [50]. Although fosmidomycin was shown to be effective in several clinical trials, high doses of the compound are required to achieve the desired results. This seems in part to be due to the highly polar nature of this molecule [51]. Therefore, Ortmann et al. addressed the question by synthesising more lipophilic and larger acyl residues and evaluating them against ec-DXR [52] (Fig. 5, Table 3). Unfortunately, these derivatives/analogues invariably have considerably reduced DXR inhibitory activity and exhibit no antibacterial activity. In addition, high-throughput screening of 32.000 compounds only yielded 30 hits with IC50< 20 µM. However, the structures of these hits were not disclosed and the active compounds therefore cannot be confirmed [53].
Table 3.
Inhibition Power of Selected Among the Huge Amount of Synthesized Compounds. N.d. Stands for Not Determined
| Studied microor ganism |
IC50 (jiM) |
Reference | ||||
|---|---|---|---|---|---|---|
| 1 | E.coli | P.falciparum | 0.22 | 0.14 | [57] | |
| 2 | 0.13 | 0.015 | ||||
| 2007 | 3 | E.coli | 1.3 | [52] | ||
| 4 | 5.1 | |||||
| 5 | 5.4 | |||||
| 6 | 5.6 | |||||
| 7 | 9.0 | |||||
| 8 | 10.0 | |||||
| 9 | < 30 | |||||
| 2008 | 9a | E.coli | P.falciparum | 0.13 | 0.061 | [54] |
| 9b | 1.0 | 0.40 | ||||
| 9c | 5.1 | 2.9 | ||||
| 9d | 7.1 | 3.3 | ||||
| 9e | 17 | 6.6 | ||||
| 9f | 20 | 9.3 | ||||
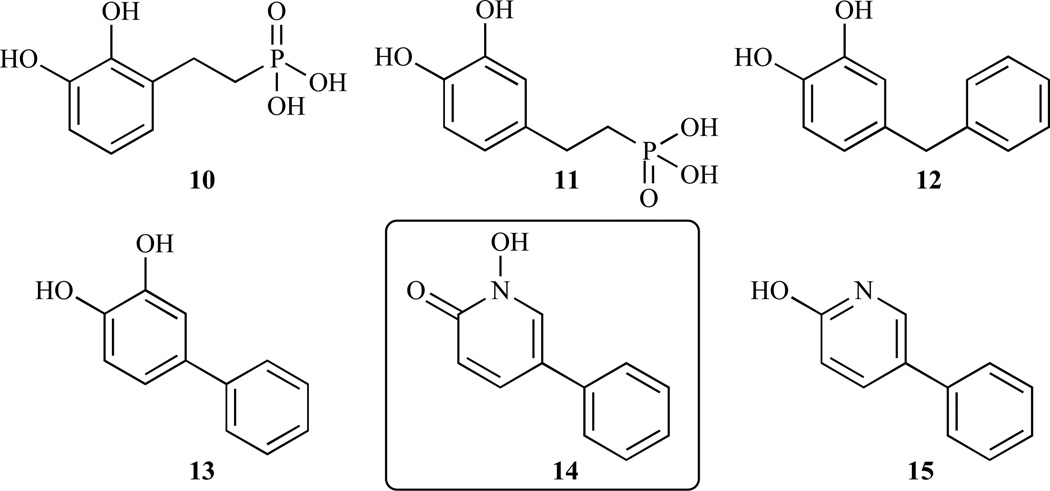
| 2009 | 10 | E.coli | 24.8 | [55] | ||
| 11 | 4.5 | |||||
| 12 | 44.7 | |||||
| 13 | 22.4 | |||||
| 14 | 1.4 | |||||
| 15 | 75 | |||||
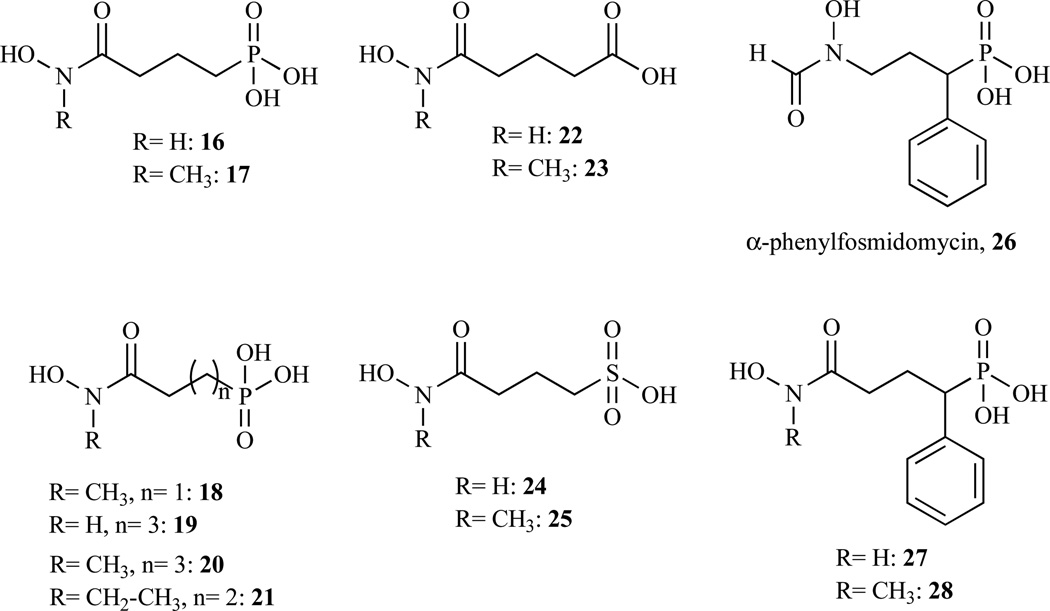
| 2010 | 16 | E. coli | 0.17 | [56] | ||
| 17 | 0.049 | |||||
| 18 | 19 | |||||
| 19 | 0.27 | |||||
| 20 | 0.11 | |||||
| 21 | 6.5 | |||||
| 22 | 720 | |||||
| 23 | 25 | |||||
| 24 | 1000 | |||||
| 25 | 48 | |||||
| 26 | E. coli | n.d. | [57] | |||
| 27 | E.coli | P.falciparum | 0.59 | 0.012 | ||
| 28 | 0.24 | 0.003 | ||||
| 2011 | 29 | E.coli | 472 | [58] | ||
| 30 | 730 | |||||
| 31 | 408 | |||||
| 32 | 7.1 | [59] | ||||
| 33 | 10.0 | |||||
| 34 | 4.6 | |||||
| 35 | 0.84 | |||||
| 36 | 15.9 | |||||
In order to expand the structure-activity relationships of non-hydrolysable phosphate mimics, linear modifications of the fosmidomycin structure was carried out by Link and coworkers in 2008 [54]. A set of phosphonic acids with inhibitory activity in the range from 0.1 to 20 µM against ec-DXR is presented in Fig. 6 and Table 3.
Fig. 6.
Chemical structures of selected compounds synthesized and evaluated by Link et al.
Later, by using coordination chemistry combined with a structure based approach, Deng et al. obtained a large series of compounds that were tested against ec-DXR (Fig. 7, Table 3) [55]. Amongst them, they identified for the first time, a strong, lipophilic DXR inhibitor (compound 14), which has a structure that is distinctly different than fosmidomycin.
Fig. 7.
Chemical structures of selected compounds synthesized and evaluated by Deng et al. Compound 14 is strong, lipophilic inhibitor and has a distinct structure from fosmidomycin.
Nevertheless, fosmidomycin remained the preferred scaffold for the design of new inhibitors of DXR. Additional structural variations around the phosphonate anchor and the spacer of this compound gave new whose structures are given in Fig. 8 and IC50 are presented in Table 3 [56, 57]. Indeed, a new highly active and selective pf-DXR inhibitor with a unique chemical structure (compound 28) was developed by Behrendt et al. [57]. Molecule 28 is an α-phenyl phosphonic acid derivative of 17. Data are not available for the corresponding α-phenyl derivative of fosmidomycin (26).
Fig. 8.
Chemical structures of selected compounds synthesized and evaluated by Zinglé et al. (16–25) and by Behrendt et al. (26–28).
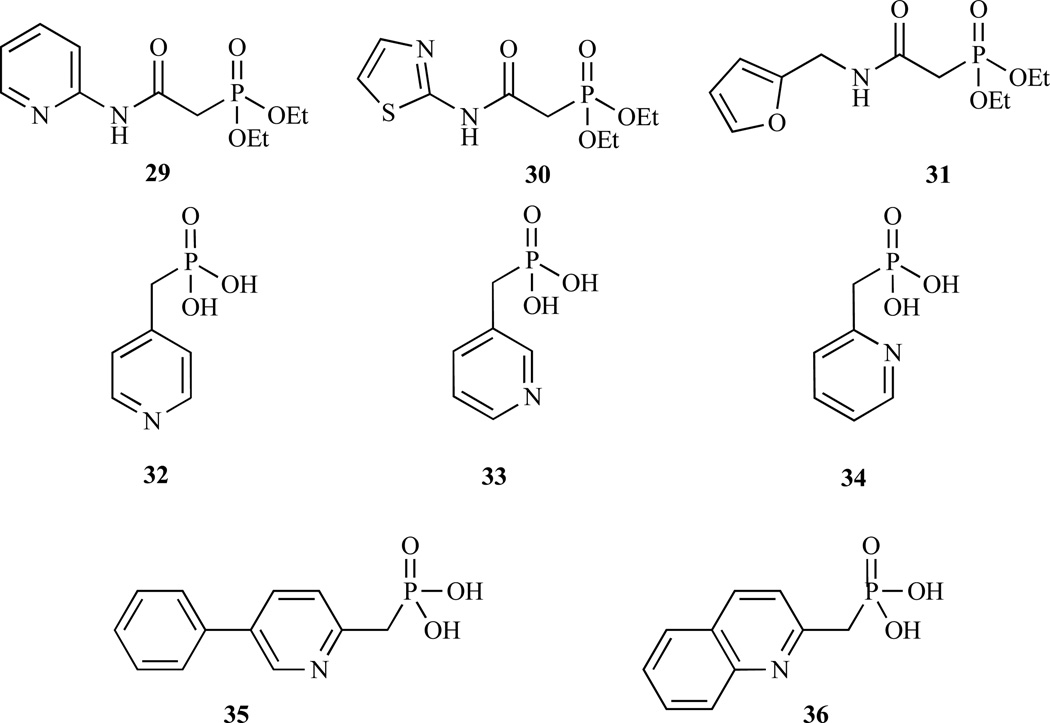
A series of phosphonated N-heteroarylcarboxamides have also been synthesised and evaluated against ec-DXR but all are poor inhibitors. The most active compounds are presented in Fig. 9 and Table 3 (29–31) [58].
Fig. 9.
Most recent chemical structures of selected compounds synthesized and evaluated against DXR.
More recently a new class of pyridine/quinolone containing phosphonates that inhibits DXR (Fig. 9, 32–36 Table 3) with IC50 as low as 0.84 µM were identified. These compounds provide a new scaffold for further inhibitor development. Moreover, they can be used to identify potential hydrophobic pockets in DXR [59, 60].
3.2.2. Structural Basis for Future Design and Development of Clinically Effective Inhibitors
Reuter and co-workers published the first structure of DXR [61]. This structure provided an initial look at the general fold of the protein. The enzyme is a homodimer with three distinct domains: a N-terminal domain presenting a typical Rossmann folded NADPH binding site, a central connecting domain, and a C-terminal helical domain. The V-shaped central domain contains most of the active site residues, as well as a flexible loop region that appears to function as a lid over the active site. Due to this intrinsic flexibility, it has been proposed that binding of cation, substrate/inhibitor could induce a conformational change [62]. From 2002 till 2005, five other articles have been published reporting crystal structures of the Z. mobilis DXR [63] and E. coli DXR [48, 62, 64, 65].
In 2005, a fragment-based approach was performed by Mercklé et al. [66] to understand the inhibition of DXR. None of the inhibitors they tested showed time-dependent inhibition of DXR suggesting that these compounds are not able to take up a conformation that is capable of inducing full-loop closure [48, 67]. Moreover, a close structural analogue of fosmidomycin ((S)-N-hydroxy-4-(phosphoryloxy) methyloxazolidin-2-one) showed neither cooperative nor time dependent inhibition of DXR, despite the fact that modelling and docking studies suggested it should be able to bind to the enzyme. These results highlighted two significant problems with simulated-docking strategies with this enzyme. The first is that standard force-field implementations are inadequate for simulating metal binding, and secondly the large changes in flexible loop conformation that accompany inhibitor binding can be difficult to simulate.
Nevertheless, in the absence of crystallographic structures from pathogenic microorganisms such as P. falciparum or M. tuberculosis during this period, molecular modelling continued to be used. This allowed several research groups to gain insight into the structure and function of the enzyme and also facilitated structure-based inhibitor design. In 2007, the first model of mt-DXR was built and evaluated on the basis of its ability to explain several site-directed mutagenesis data [68]. Furthermore, a comparison of this homology model with the X-ray structure published later [69, 70] shows excellent agreement and validates the theoretical approach. Similarly, a 3-D model of pf-DXR was published in 2010. This model was validated using structure-checking programs and protein docking studies and subjected to structure-function analysis of its active site and to ligand docking studies. It was also used to develop an efficient screening method to identify potential lead compounds for use in the rational design of novel DXR inhibitors, where IC50s could be estimated [71]. A further validation of this model will be possible when a full account of a preliminary X-ray crystallographic study of pf-DXR is published [72]. The recent crystal structure of the hyperthermophile T. maritima DXR (tm-DXR) reveals a new extra space available for potential drug design. Moreover, the structure adopted the closed form by rigid domain rotation [48, 69] but without the flexible loop over the active site, to give a novel conformation for this enzyme [37].
Very recently, with further improvements to the NMR experimental procedure and sample conditions, it has been possible to solve a part of the flexible loop of ec-DXR and further experiments might provide information about the conformational changes in the protein during substrate and cofactor binding. The results published by Englebert et al. represent one of the relatively few NMR reports of a large ternary homodimeric complex. In particular, the experimental procedures reported will provide helpful clues for NMR-based drug-design for large flexible molecular targets [73].
In conclusion, biochemical approaches (e.g. [74]), together with computational approaches and high-throughput screening assays (e.g. [75]), should provide additional inhibitors of DXR. While much remains to be learned, considerable progress has been made in recent years and the prospects of obtaining clinically effective DXR inhibitors seems attainable.
4. ISOMERISATION AND ELONGATION OF THE POLYPRENYL CHAIN
DMAPP and IPP are both required as building blocks for the biosynthesis of terpenes. Since the primary product of the mevalonate pathway is IPP, an IPP isomerase is absolutely required for the formation of DMAPP as a prerequisite for elaboration of the downstream terpene pathways. On the other hand, the non-mevalonate pathway can function even in the absence of an isomerase since DMAPP and IPP can both be produced by IspH enzyme [26].
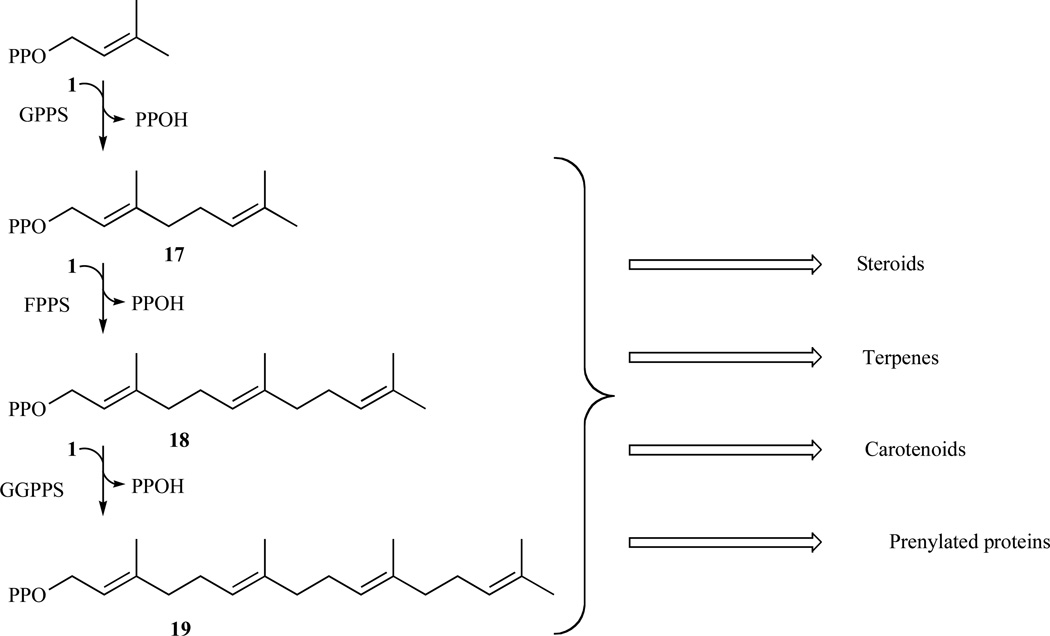
The five-carbon compounds IPP and DMAPP are condensed to form the 10-carbon geranyl diphosphate (GPP) (17). GPP serves as the precursor for the synthesis of all monoterpenes. The addition of another IPP unit to GPP yields the 15-carbon farnesyl diphosphate (FPP) (18). The enzyme FPP synthase catalyzes the synthesis of both GPP and FPP in mammals [76]. FPP sits at the branch-point between sterol and longer-chain nonsterol synthesis. The enzyme squalene synthase catalyzes the head-to-head condensation of two FPP molecules to form the sterol precursor squalene [77]. Subsequent cyclization steps lead to sterol synthesis. Geranylgeranyl diphosphate synthase catalyzes the addition of IPP to FPP to form GGPP (19) [78] (Fig. 10).
Fig. 10.
Elongation of isoprenoids in order to obtain complex and diverse essential molecules.
Both farnesyl and geranylgeranyl residues appended posttranslationally to large family of proteins in Eukaryotes. The hydrophobic groups enhance membrane association essential for protein function. These postransational modifications are extensively studied especially in the case of cancer treatment [79] and bone resorption [80].
In this section of the review, we will focus on isopentenyl diphosphate isomerase (IDI) and inhibition studies to help understand the mechanism of the isomerisation of IPP to DMAPP.
4.1. Isopentenyl Diphosphate Isomerase
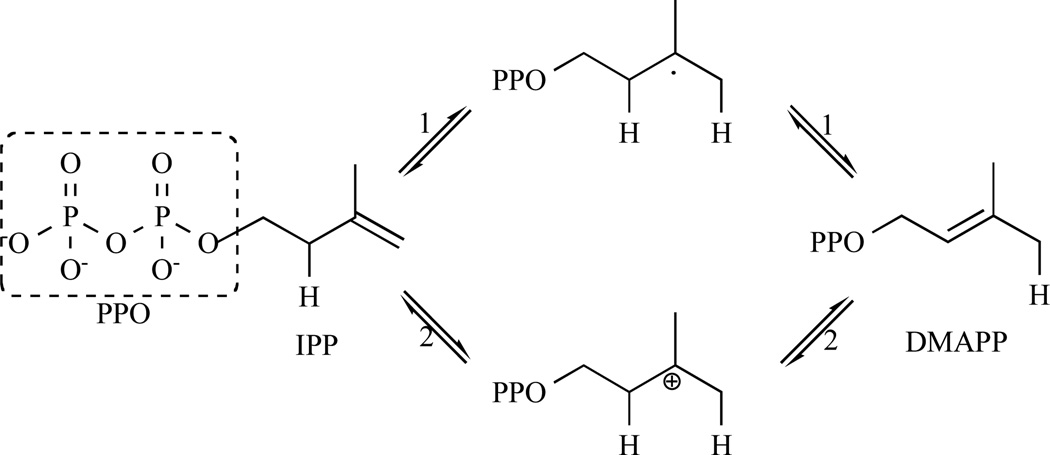
Isopentenyl diphosphate:dimethylallyl diphosphate (IPP: DMAPP) isomerase (IDI; EC 5.3.3.2) is a key enzyme involved in the biosynthesis of isoprenoids and catalyses the isomerisation of IPP into DMAPP (Fig. 11).
Fig. 11.
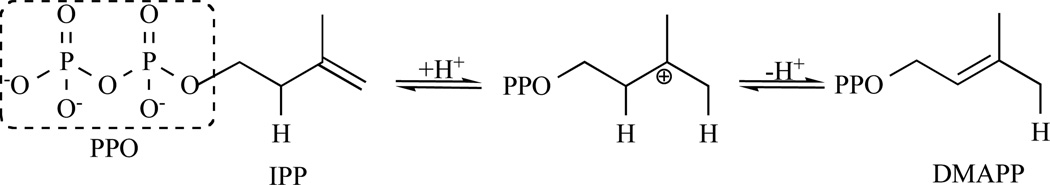
Representation of the catalytic mechanism of IDI-1 via protonation of the IPP and deprotonation of the carbocationic transition state.
Two types of IDIs are reported. They show no sequence similarity but catalyse the same reaction. Type I IDI (IDI-1) is a metalloprotein containing divalent cations, Zn2+ and Mg2+, as cofactors [81–83]. Crystal structures of free and metal-bound Escherichia coli IDI-1 show that Zn2+ is involved in the active conformation folding, with the metal occupying a coordination site composed of three histidines and two glutamates [84]. Several lines of evidence have established the mechanism for isomerisation catalysed by IDI-1. In particular, studies with IPP analogues have provided extensive support for substrate protonation to generate a transient carbocationic intermediate. Epoxide and diene analogues (3,4-epoxy-3-methylbutyl diphosphate (eIPP) and 3-methylene-4-penten-1-yl diphosphate (vIPP), respectively) were shown to irreversibly inhibit the enzyme through formation of covalent adducts with active site cysteine and glutamate residues [85, 86]. In both cases, protonation serves to activate the analogue for attack by an active site nucleophile. N, N-dimethyl-2-amino-1-ethyl diphosphate (nIPP), a reactive intermediate analogue with a positively charged ammonium group, binds to the type I enzyme with subnanomolar affinity [87]. In addition, IPP and DMAPP analogues substituted with strong electron-withdrawing fluorine groups are poor substrates for isomerisation [87, 88]. These studies provide evidence for a mechanism that involves protonation at the double bond of IPP, followed by deprotonation of the carbocationic intermediate to generate DMAPP as the product [89–93].
It is important to note that type 2 IPP isomerases are essential enzymes in several classes of microorganisms using exclusively the mevalonate pathway, including Streptococci, Staphylococci, and Enterococci (Table 1). The occurrence of methicillin-resistant Staphylococci (MRSA) and vancomycin-resistant Enterococci (VRE) is increasing rapidly [94–96], and novel chemotherapeutic strategies are urgently needed. IPP isomerase, and more precisely IDI-2, inhibitors could thus possibly serve as selective drugs for infections by drug-resistant strains. In contrast to IDI-1, IDI-2 was discovered recently and its mechanism is still under investigation. This section will highlight the recent advances made on this enzyme.
4.1.1. Identification and Characterisation of IDI-2
In 2001, Kaneda et al. isolated an open reading frame (ORF), orfD, encoding an unknown protein. The recombinant product of orfD was purified as a soluble 37 kDa protein. The enzyme catalysed the isomerisation of IPP into DMAPP in the presence of both FMN and NADPH in bacteria and was classified as Type 2 IDI for the FMN- and NAD(P)H-dependent enzyme [97]. Since then different research groups focused their attention on this new interesting enzyme. They all concluded that IDI-2 requires reduced flavin mononucleotide and MgCl2 as cofactors. Table 4 summarises the kinetic and crystal data available in the literature until 2011. A crystallographic state of the art of the enzymes involved in isoprenoid biosynthesis was published in 2008 [98].
Table 4.
Available Kinetic and Crystal Data for IDI-2 Until 2011. N.d. Stands for Not Determined.
| Km (µM) | kcat (s−1) | kcat/Km (M−1.s−1) | Optimal temperature (°C) | Crystal Structure(s) | Reference | |
|---|---|---|---|---|---|---|
| B. subtilis | 670 | 25 | 3.7 × 104 | 37 | 1P0K, 1P0N [99] | [100] |
| n.d. | 0.4 | n.d. | n.d. | [27] | ||
| S. shibatae | 63 | 0.2 | 3.2 × 103 | 60 | 2ZR(U–Z) [101] | [102] |
| Synechocystis sp.6803 | 52 | 0.2 | 4.4 × 103 | 37 | n.d. | [103] |
| M. thermoautotrophicus | 64 | 1.6 | 2.5 × 104 | 70 | n.d. | [104] |
| S. aureus | 16.8 | 0.7 | 4.1 × 104 | n.d. | n.d. | [105] |
| 19 | 1.3 | 6.8 × 104 | n.d. | n.d. | [106] | |
| Streptomyces sp. CL190 | 450 | 0.7 | 1.6 × 103 | 35 | n.d. | |
| T. thermophilus | 5.6 | 0.2 | 3.2 × 104 | n.d. | 3DH7 [107] | [108] |
| M. jannashii | 15.3 × 103 | 191 | 1.3 × 104 | 85–95 | n.d. | [109] |
| T. kodakaraensis | 84 | 76 | 9.0 × 105 | 80 | n.d. | [110] |
Steinbacher and co-workers reported the first structure of IDI-2 from B. subtilis (bs-IDI2) at 1.9 Å resolution (PDB ID: 1P0K) in 2003 [99]. The monomer of IDI-2 consists of a regular TIM barrel (α8β8 barrel) domain. The N-terminal region forms an irregular structure and an anti-parallel two-stranded β-sheet that function as a support at one side of the barrel, a structural feature commonly observed for TIM barrels. The FMN cofactor is located in the standard phosphate binding (SPB) region of the TIM barrel as described for several FMN-dependent proteins [111]. However, due to the conformational flexibility of the enzyme, electron density was not detected for several amino acid residues at the active site of bs-IDI2. In 2008, we solved the first complete structure of IDI-2 from T. thermophilus (tt-IDI2) in complex with inorganic pyrophosphate (PDB ID: 3DH7) [107] confirming the theoretical model obtained in 2005 [112].
4.1.2. Inhibition Studies to Further Understand the Enzymatic Mechanism
Most flavoproteins carry out reactions with net redox changes: including oxidations, reductions, monooxygenations, dioxygenations and electron transfers. However, there are a number of unusual flavoproteins that catalyze reactions with no net redox change. These fall in four groups: those that utilize two-electron flavin chemistry, others that involve free radical flavin chemistry, a number where the role of flavin is not yet clear and the remainder that intriguingly do not appear to involve the flavin directly in catalysis (for review [113]).
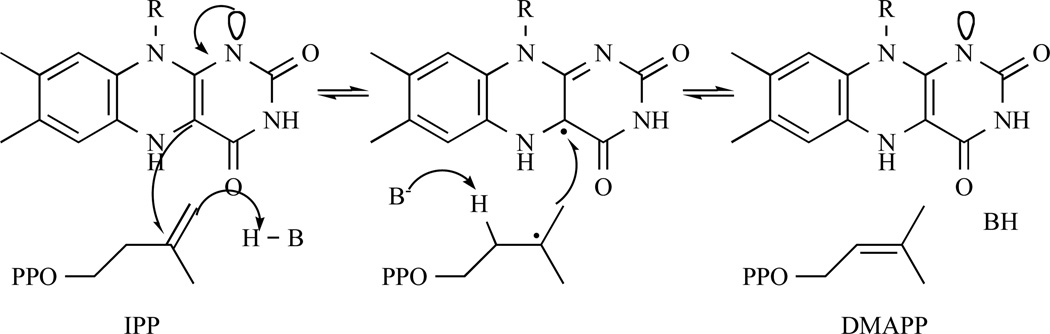
The first mechanism proposed for IDI-2 by Kaneda et al. in 2001 was based on this of IDI-1, which involved a carbocationic intermediate [106] (Fig. 12 (2)). In 2004, Hemmi and co-workers performed spectrometric analyses and enzyme assays under anaerobic conditions, proving that the reduced form of the flavin co-enzyme (FMNH2) is sufficient for the enzyme to catalyse the isomerase reaction. Moreover, using 5-deaza FMN as cofactor, S. shibatae IDI-2 (ss-IDI2) was nearly devoid of IPP isomerase suggesting the possibility of a redox role of FMN. Based on these results, they proposed a reaction mechanism, similar to the one reported for UDP-galactopyranose mutase [114], in which a radical intermediate is generated [115].
Fig. 12.
Representation of the two putative IDI-2 mechanisms through a radical rearrangement (1) or through a protonation/deprotonation mechanism similar to IDI-1 (2).
Indeed, detection of trace amounts of isolated neutral flavin semiquinone in reaction mixtures by electron paramagnetic resonance (EPR) provided initial evidence for a radical mechanism (Fig. 13). However, no coupled substrate radical was observed. Subsequent attempts to detect intermediates in stopped-flow studies under single turnover conditions failed and several radical clock mechanistic probes also failed to reveal the existence of radical substrates intermediates [108, 116].
Fig. 13.
Representation of the radical mechanism suggested by Hemmi et al. and similar to the UDP-galactopyranose mutase mechanism.
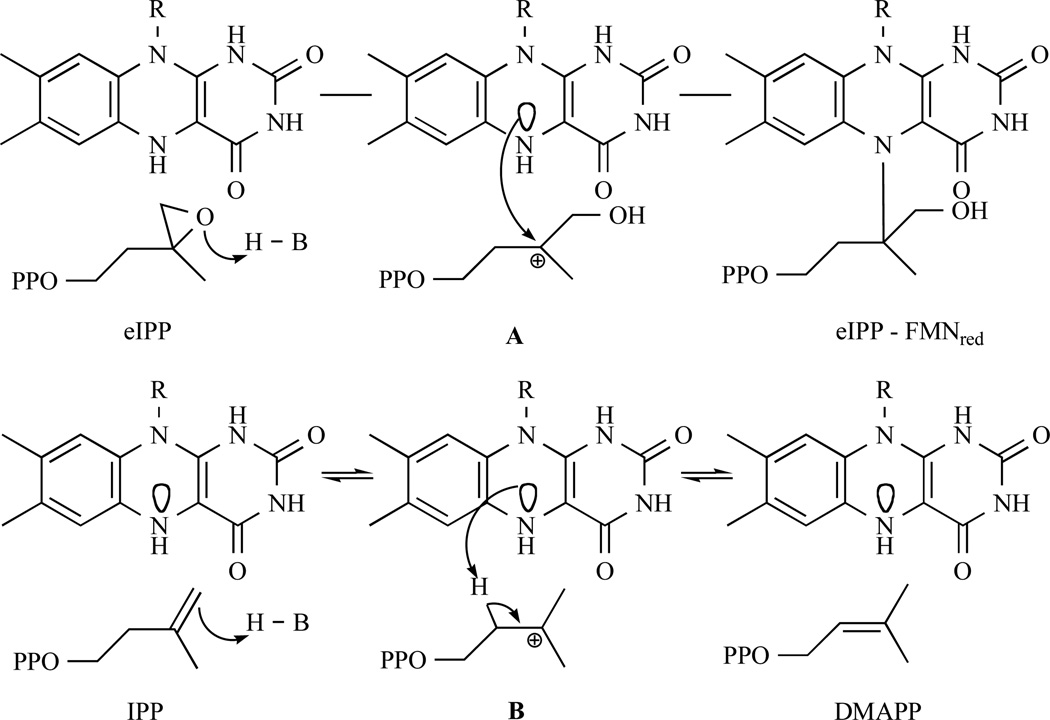
In 2006, Eguchi and co-workers studied the inhibition of M. jannaschii IDI-2 (mj-IDI2) by eIPP, a mechanism-based inhibitor of IDI-1. The mechanism for inhibition of wild type E. coli IDI-1 by eIPP involves the protonation of the epoxide ring in eIPP, which alkylates the nucleophilic thiolate moiety of a cysteine (Cys-67) in the active site [89] Eguchi and co-workers assumed that if protonation of the double bond in IPP was the first step in the reaction catalysed by IDI-2, as in the case of IDI-1, eIPP would inhibit the enzyme by alkylation of a near-by nucleophilic group of the IDI-2 active site (Fig. 14a). Upon incubation of racemic eIPP with mj-IDI2, they observed a time- and concentration-dependent decrease of the enzymatic activity (KI = 56.6 mM, kinact = 0.10 s−1, kinact/KI = 1.76 s−1 M−1) suggesting that the IDI-2 reaction proceeded via a carbocation-type intermediate as in the case of IDI-1. During the isomerisation, the aromatic ring of the reduced flavin should stabilise the carbocation intermediate [109] (Fig. 14b).
Fig. 14.
a. Representation of the inhibition mechanism of eIPP against IDI-2. First epoxide is activated by protonation and then an attack of a near-by nucleophilic group of the IDI-2 active centre form covalent bond. b. IDI-2 reaction is similar to IDI-1, which proceeds via carbocation-type intermediate. The FMNH2 is acting as a nucleophilic group.
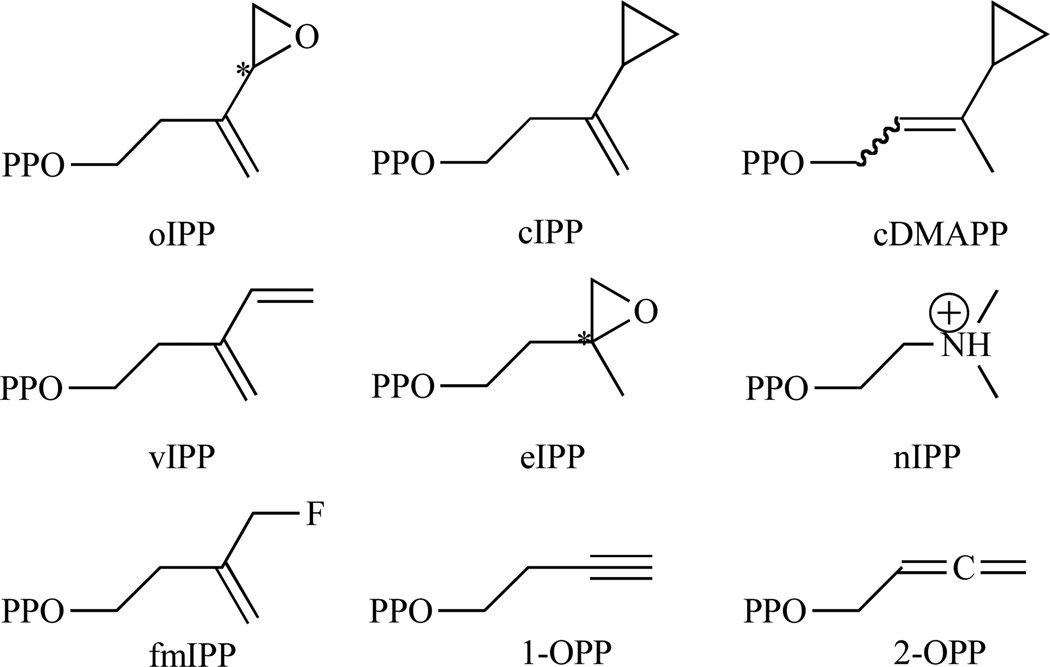
To further probe the mechanism of the IDI-2 reaction, additional epoxy and new cyclopropyl substrate analogues (oIPP, cIPP and cDMAPP), designed as mechanism-based irreversible inhibitors, were synthesised and evaluated against IDI-2 from T. thermophilus (tt-IDI2) by Walker and coworkers [117, 118]. They observed by 1H NMR that cIPP isomerized to cDMAPP and that a similar experiment starting with cDMAPP resulted in the transient production of cIPP, with the unstable cDMAPP analogue undergoing non-enzymatic solvolysis. They concluded that these diphosphates were alternative substrates that do not irreversibly inhibit IDI-2. In contrast, incubation of reduced IDI-2 with oIPP resulted in a rapid first-order irreversible loss of activity of the enzyme (KI = 1.4 µM, kinact = 0.37 min−1). Combined with mass spectrometry and UV spectroscopy, those results showed that the epoxide ring is activated by protonation at oxygen rather than by hydrogen atom addition to the double bond of oIPP, followed by trapping of a radical intermediate by flavin semiquinone [117, 118]. This mechanism is consistent with the previous one suggested by Hoshino [109] and similar to the one adopted for IDI-1. So additional IDI-1 irreversible inhibitors (Fig. 15), epoxy, diene and fluorinated substrate analogues, were analysed as mechanistic probes for IDI-2 by Poulter and coworkers [119].
Fig. 15.
Chemical structures of the studied IDI-2 inhibitors (substrate or product analogues).
eIPP (KI = 48.6 µM, kinact = 0.041 min−1), vIPP (KI = 8.0 µM, kinact = 1.2 min−1) and 3-(fluoromethyl)-3-buten-1-yl diphosphate (fmIPP) (KI = 7.4 µM, kinact = 0.044 min−1) all inactivate tt-IDI2 through formation of covalent adducts with the reduced flavin. At this point, UV-visible spectra of the inactivated complexes were consistent with modification of the isoalloxazine ring at N5. Two other alkyne (3-butyn-1-yl diphosphate (1-OPP), KI = 48 µM) and allene (2,3-butadien-1-yl diphosphate (2-OPP), KI = 36 µM) substrate analogues for the enzyme were synthesised to distinguish between the mechanisms initiated by proton transfer and hydrogen atom transfer to the double bond in IPP and suggested a protonation-deprotonation mechanism for the enzyme-catalysed isomerisation of IPP and DMAPP [120]. Consistent with a carbocationic mechanism, the alkyne and allene analogues were not substrates. Inhibition constants are summarised in Table 4.
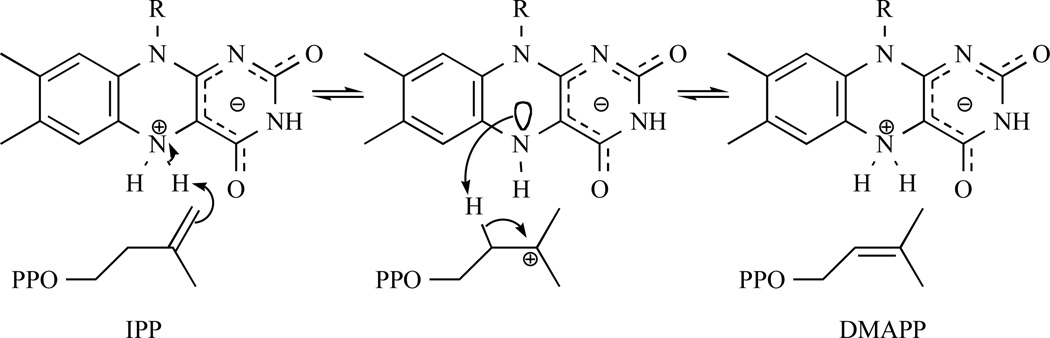
Based on the latest results, Poulter and Hemmi’s groups performed UV, structural and mutagenic studies and demonstrated that reduced FMN, not amino acid residues, acts as a general acid-base catalyst in the protonation-deprotonation mechanism [101, 119]. They proposed a mechanism where the N5 nitrogen of FMN seems the most plausible candidate for the catalyst and that this mechanism should be distinct from the established one for human type 1 IDI because of their antarafacial and assumed suprafacial nature, IDI-1 and IDI-2 respectively (Fig. 16).
Fig. 16.
Latest suggested IDI-2 mechanism where the N5 nitrogen of FMN seems the most plausible candidate for the catalyst.
Additional studies performed on IDI-2 from Staphilococcus aureus IDI-2 (sa-IDI2) by the Liu research group are consistent with the growing body of experimental evidence suggesting that the flavin coenzyme of IDI-2 serves a novel function as an acid-base catalyst [105, 116, 121].
5. CONCLUSION
The structural diversity of isoprenoids is immense, including acyclic, monocyclic and polycyclic compounds. Terpenoids play important roles in all living organisms. They function as regulators of gene expression, constituents of membranes, vitamins, antimicrobial agents, mating pheromones, reproductive hormones, components of signal transduction pathways, and constituents of electron transport and photosynthetic machinery. In light of the many diverse functions of terpenes, it is not surprising that their biosynthesis has been subject of intense investigation for more than four decades.
The first pathway starting from mevalonate and leading to the building blocks of large isoprenoids was discovered in 1950s. Only recently, an alternative route was discovered requiring decarboxylation of mevalonate 5-phosphate to produce the substrate for the isopentenyl 5-phosphate kinase (IPK), which would catalyze formation of IPP.
The second pathway starting from 1-deoxy-xylulose 5-phosphate is present in some human pathogens and plants and totally absent from human species. This is why the enzymes of this pathway were extensively characterised in order to design new anti-infectious agents or herbicides. The most representative example is the alternative treatment of malaria and tuberculosis. Indeed, multi-drug resistant strains of pathogens have recently forced many countries to change national treatment protocols.
This review summarises the latest improvements made on a better comprehension of two key enzymes (DXR and IDI) involved in the biosynthesis of isoprenoids by pathogenic microorganisms.
Table 4.
Inhibition Parameters of the Studied Inhibitors. N.d. Stands for Not Determined
| Protein studied | KI (µM) | kinact (min−1) | kinact/KI (s−1.mM−1) | References | ||
|---|---|---|---|---|---|---|
| Inhibitors | eIPP | mj-IDI2 | 56.5 (mM) | 0.10 (s−1) | 0.002 | [109] |
| tt-IDI2 | 48.6 ± 8.2 | 0.041 ± 0.01 | 0.014 | [117] | ||
| oIPP | tt-IDI2 | 1.4 ± 0.3 | 0.37 ± 0.07 | 4.4 | [119] | |
| vIPP | tt-IDI2 | 8.0 ± 2.0 | 1.2 ± 0.1 | 2.5 | ||
| fmIPP | tt-IDI2 | 7.4 ± 0.9 | 0.044 ± 0.002 | 0.1 | ||
| nIPP | tt-IDI2 | 5.1 ± 0.5 | n.d. | n.d. | [108] | |
| 1-OPP | tt-IDI2 | 48 ± 6 | n.d. | n.d. | [120] | |
| 2-OPP | tt-IDI2 | 36 ± 5 | n.d. | n.d. | ||
ACKNOWLEDGEMENTS
JDR and CDP acknowledge support from NIH grant GM 25521. Authors wish to thank M.W. Janczek, F. Riviere and H. Vanobbergen for scientific and fruitful discussions.
REFERENCES
- 1.Sacchettini JC, Poulter CD. Creating isoprenoid diversity. Science. 1997;277:1788–1789. doi: 10.1126/science.277.5333.1788. [DOI] [PubMed] [Google Scholar]
- 2.Cane DE. In: Comprehensive Natural Products Chemistry. Barton D, Nakanishi K, editors. Oxford: Elsevier; 1999. [Google Scholar]
- 3.Wang KC, Ohnuma S. Isoprenyl diphosphate synthases. Biochim. Biophys. Acta. 2000;1529:33–48. doi: 10.1016/s1388-1981(00)00136-0. [DOI] [PubMed] [Google Scholar]
- 4.Bloch K. The biological synthesis of cholesterol. Science. 1965;150:19–28. doi: 10.1126/science.150.3692.19. [DOI] [PubMed] [Google Scholar]
- 5.Matsumi R, Atomi H, Driessen AJM, van der Oost J. Isoprenoid biosynthesis in Archaea--biochemical and evolutionary implications. Res. Microbiol. 2011;162:39–52. doi: 10.1016/j.resmic.2010.10.003. [DOI] [PubMed] [Google Scholar]
- 6.Miziorko HM. Enzymes of the mevalonate pathway of isoprenoid biosynthesis. Arch. Biochem. Biophys. 2011;505:131–143. doi: 10.1016/j.abb.2010.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Willey JZ, Elkind MS. 3-Hydroxy-3-methylglutaryl-coenzyme A reductase inhibitors in the treatment of central nervous system diseases. Arch. Neurol. 2010;67:1062–1067. doi: 10.1001/archneurol.2010.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grochowski LL, Huimin X, White RH. Methanocaldococcus jannaschii uses a modified mevalonate pathway for biosynthesis of isopentenyl diphosphate. J. Bacteriol. 2006;188:3192–3198. doi: 10.1128/JB.188.9.3192-3198.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen M, Poulter CD. Characterization of thermophilic archaeal isopentenyl phosphate kinases. Biochemistry. 2010;49:207–217. doi: 10.1021/bi9017957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mabanglo MF, Schubert HL, Chen M, Hill CP, Poulter CD. X-ray structures of isopentenyl phosphate kinase. ACS Chem. Biol. 2010;5:517–527. doi: 10.1021/cb100032g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dellas N, Noel JP. Mutation of archaeal isopentenyl phosphate kinase highlights mechanism and guides phosphorylation of additional isoprenoid monophosphates. ACS Chem. Biol. 2010;5:589–601. doi: 10.1021/cb1000313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rohmer M, Knani M, Simonin P, Sutter B, Sahm H. Isoprenoid biosynthesis in bacteria: a novel pathway for the early steps leading to isopentenyl diphosphate. Biochem. J. 1993;295(Pt 2):517–524. doi: 10.1042/bj2950517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eisenreich W, Bacher A, Arigoni D, Rohdich F. Biosynthesis of isoprenoids via the non-mevalonate pathway. Cell Mol. Life Sci. 2004;61:1401–26. doi: 10.1007/s00018-004-3381-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sprenger GA, Schorken U, Wiegert T, Grolle S, de Graaf AA, Taylor SV, Begley TP, Bringer-Meyer S, Sahm H. Identification of a thiamin-dependent synthase in Escherichia coli required for the formation of the 1-deoxy-D-xylulose 5-phosphate precursor to isoprenoids, thiamin, and pyridoxol. Proc. Natl. Acad. Sci. USA. 1997;94:12857–12862. doi: 10.1073/pnas.94.24.12857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lois LM, Campos N, Putra SR, Danielsen K, Rohmer M, Boronat A. Cloning and characterization of a gene from Escherichia coli encoding a transketolase-like enzyme that catalyzes the synthesis of D-1-deoxyxylulose 5-phosphate, a common precursor for isoprenoid, thiamin, and pyridoxol biosynthesis. Proc. Natl. Acad. Sci. USA. 1998;95:2105–2110. doi: 10.1073/pnas.95.5.2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Takahashi S, Kuzuyama T, Watanabe H, Seto H. A 1-deoxy-D-xylulose 5-phosphate reductoisomerase catalyzing the formation of 2-C-methyl-D-erythritol 4-phosphate in an alternative non-mevalonate pathway for terpenoid biosynthesis. Proc. Natl. Acad. Sci. USA. 1998;95:9879–9884. doi: 10.1073/pnas.95.17.9879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fox DT, Poulter CD. Synthesis and evaluation of 1-deoxy-D-xylulose 5-phosphoric acid analogues as alternate substrates for methylerythritol phosphate synthase. J. Org. Chem. 2005;70:1978–1985. doi: 10.1021/jo048022h. [DOI] [PubMed] [Google Scholar]
- 18.Rohdich F, Wungsintaweekul J, Fellermeier M, Sagner S, Herz S, Kis K, Eisenreich W, Bacher A, Zenk MH. Cytidine 5’-triphosphate-dependent biosynthesis of isoprenoids: YgbP protein of Escherichia coli catalyzes the formation of 4-diphosphocytidyl-2-C-methylerythritol. Proc. Natl. Acad. Sci. USA. 1999;96:11758–11763. doi: 10.1073/pnas.96.21.11758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Luttgen H, Rohdich F, Herz S, Wungsintaweekul J, Hecht S, Schuhr CA, Fellermeier M, Sagner S, Zenk MH, Bacher A, Eisenreich W. Biosynthesis of terpenoids: YchB protein of Escherichia coli phosphorylates the 2-hydroxy group of 4-diphosphocytidyl-2C-methyl-D-erythritol. Proc. Natl. Acad. Sci. USA. 2000;97:1062–1067. doi: 10.1073/pnas.97.3.1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Herz S, Wungsintaweekul J, Schuhr CA, Hecht S, Luttgen H, Sagner S, Fellermeier M, Eisenreich W, Zenk MH, Bacher A, Rohdich F. Biosynthesis of terpenoids: YgbB protein converts 4-diphosphocytidyl-2C-methyl-D-erythritol 2-phosphate to 2C-methyl-D-erythritol 2,4-cyclodiphosphate. Proc. Natl. Acad. Sci. USA. 2000;97:2486–2490. doi: 10.1073/pnas.040554697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Takagi M, Kuzuyama T, Kaneda K, Dairi T, Seto H. Studies on the non-mevalonate pathway: formation of 2-C-methyl-D-erythritol 2,4-cyclodiphosphate from 2-phospho-4-(cytidine 5’-diphospho)-2-C-methyl-D-erythritol. Tetrahedron Lett. 2000;41:3395–3398. [Google Scholar]
- 22.Hecht S, Eisenreich W, Adam P, Amslinger S, Kis K, Bacher A, Arigoni D, Rohdich F. Studies on the non-mevalonate pathway to terpenes: the role of the GcpE (IspG) protein. Proc. Natl. Acad. Sci. USA. 2001;98:14837–14842. doi: 10.1073/pnas.201399298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kollas AK, Duin EC, Eberl M, Altincicek B, Hintz M, Reichenberg A, Henschker D, Henne A, Steinbrecher I, Ostrovsky DN, Hedderich R, Beck E, Jomaa H, Wiesner J. Functional characterization of GcpE, an essential enzyme of the non-mevalonate pathway of isoprenoid biosynthesis. FEBS Lett. 2002;532:432–436. doi: 10.1016/s0014-5793(02)03725-0. [DOI] [PubMed] [Google Scholar]
- 24.Seemann M, Bui BT, Wolff M, Tritsch D, Campos N, Boronat A, Marquet A, Rohmer M. Isoprenoid biosynthesis through the methylerythritol phosphate pathway: the (E)-4-hydroxy-3-methylbut-2-enyl diphosphate synthase (GcpE) is a [4Fe–4S] protein. Angew. Chem. Int. Ed. Engl. 2002;41:4337–4339. doi: 10.1002/1521-3773(20021115)41:22<4337::AID-ANIE4337>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 25.Lee M, Gräwert T, Quitterer F, Rohdich F, Eppinger J, Eisenreich W, Bacher A, Groll M. Biosynthesis of isoprenoids: crystal structure of the [4Fe–4S] cluster protein IspG. J. Mol. Biol. 2010;404:600–10. doi: 10.1016/j.jmb.2010.09.050. [DOI] [PubMed] [Google Scholar]
- 26.Gräwert T, Span I, Bacher A, Groll M. Reductive dehydroxylation of allyl alcohols by IspH protein. Angew. Chem. Int. Ed. Engl. 2010;49:8802–8809. doi: 10.1002/anie.201000833. [DOI] [PubMed] [Google Scholar]
- 27.Laupitz R, Hecht S, Amslinger S, Zepeck F, Kaiser J, Richter G, Schramek N, Steinbacher S, Huber R, Arigoni D, Bacher A, Eisenreich W, Rohdich F. Biochemical characterization of Bacillus subtilis type II isopentenyl diphosphate isomerase, and phylogenetic distribution of isoprenoid biosynthesis pathways. Eur. J. Biochem. 2004;271:2658–2669. doi: 10.1111/j.1432-1033.2004.04194.x. [DOI] [PubMed] [Google Scholar]
- 28.Rohdich F, Bacher A, Eisenreich W. Isoprenoid biosynthetic pathways as anti-infective drug targets. Biochem. Soc. Trans. 2005;33:785–791. doi: 10.1042/BST0330785. [DOI] [PubMed] [Google Scholar]
- 29.Takahashi S, Kuzuyama T, Watanabe H, Seto H. A 1-deoxy-D-xylulose 5-phosphate reductoisomerase catalyzing the formation of 2-C-methyl-D-erythritol 4-phosphate in an alternative non-mevalonate pathway for terpenoid biosynthesis. Proc. Natl. Acad. Sci. USA. 1998;18:9879–9884. doi: 10.1073/pnas.95.17.9879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Proteau Ph.J. 1-Deoxy-D-xylulose 5-phosphate reductoisomerase: an overview. Bioorg. Chem. 2004;32:483–493. doi: 10.1016/j.bioorg.2004.08.004. [DOI] [PubMed] [Google Scholar]
- 31.Grolle S, Bringer-Meyer S, Sahm H. Isolation of the dxr gene of Zymomonas mobilis and characterization of the 1-deoxy-D-xylulose 5-phosphate reductoisomerase. FEMS Microbiol. lett. 2000;191:131–137. doi: 10.1111/j.1574-6968.2000.tb09329.x. [DOI] [PubMed] [Google Scholar]
- 32.Cane DE, Chow C, Lillo A, Kang I. Molecular cloning, expression and characterization of the first three genes in the mevalonate-independent isoprenoid pathway in Streptomyces coelicolor . Bioorg. Med. Chem. 2001;9:1467–1477. doi: 10.1016/s0968-0896(01)00050-5. [DOI] [PubMed] [Google Scholar]
- 33.Koppish AT, Fox DT, Blagg BSJ, Poulter CD. E. coli MEP synthase: steady-state kinetic analysis and substrate binding. Biochemistry. 2002;41:236–243. doi: 10.1021/bi0118207. [DOI] [PubMed] [Google Scholar]
- 34.Yin X, Proteau Ph.J. Characterization of native and histidinetagged deoxyxylulose 5-phosphate reductoisomerase from the cyanobacterium Synechocystis sp. PCC6803 . Biochim. Biophys. Acta. 2003;1652:75–81. doi: 10.1016/j.bbapap.2003.08.005. [DOI] [PubMed] [Google Scholar]
- 35.Argyrou A, Blanchard JS. Kinetic and chemical mechanism of Mycobacterium tuberculosis 1-deoxy-D-xylulose-5-phosphate isomeroreductase. Biochemistry. 2004;43:4375–4384. doi: 10.1021/bi049974k. [DOI] [PubMed] [Google Scholar]
- 36.Jawaid S, Seidle H, Zhou W, Abdirahman H, Abadeer M, Hix JH, van Hoek ML, Couch RD. Kinetic characterization and phosphoregulation of the Francisella tularensis 1-deoxy-D-xylulose 5-phosphate reductoisomerase (MEP synthase) PLoS ONE. 2009;4(e8288):1–9. doi: 10.1371/journal.pone.0008288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Takenoya M, Ohtaki A, Noguchi K, Endo K, Sasaki Y, Ohsawa K, Yajima S, Yohda M. Crystal structure of 1-deoxy-d-xylulose 5-phosphate reductoisomerase from the hyperthermophile Thermotoga maritima for insights into the coordination of conformational changes and an inhibitor binding. J. Struct. Biol. 2010;2:165–170. doi: 10.1016/j.jsb.2010.03.015. [DOI] [PubMed] [Google Scholar]
- 38.Wong A, Munos JW, Devasthali V, Johnson KA, Liu H-w. Study of 1-deoxy-D-xylulose-5-phosphate reductoisomerase: synthesis and evaluation of fluorinated substrate analogues. Org. Lett. 2004;6:3625–3628. doi: 10.1021/ol048459b. [DOI] [PubMed] [Google Scholar]
- 39.Phaosiri C, Proteau Ph.J. Substrate analogs for the investigation of deoxyxylulose 5-phosphate reductoisomerase inhibition: synthesis and evaluation. Bioorg. Med. Chem. Lett. 2004;14:5309–5312. doi: 10.1016/j.bmcl.2004.08.023. [DOI] [PubMed] [Google Scholar]
- 40.Fox DT, Poulter CD. Synthesis and evaluation of 1-deoxy-D-xylulose 5-phosphoric acid analogues as alternate substrates for methylerythritol phosphate synthase. J. Org. Chem. 2005;70:1978–1985. doi: 10.1021/jo048022h. [DOI] [PubMed] [Google Scholar]
- 41.Fox DT, Poulter CD. Mechanistic studies with 2-C-methyl-D-erythritol 4-phosphate synthase from Escherichia coli . Biochemistry. 2005;44:8360–8368. doi: 10.1021/bi047312p. [DOI] [PubMed] [Google Scholar]
- 42.Hoeffler JF, Tritsh D, Grosdemange-Billiard C, Rohmer M. Isoprenoid biosynthesis via the methylerythritol phosphate pathway Mechanistic investigations of the 1-deoxy-D-xylulose 5-phosphate reductoisomerase. Eur. J. Biochem. 2002;269:4446–4457. doi: 10.1046/j.1432-1033.2002.03150.x. [DOI] [PubMed] [Google Scholar]
- 43.Munos JW, Pu X, Liu H-w. Synthesis and analysis of a fluorinated product analogue as an inhibitor for 1-deoxy-D-xylulose 5-phosphate reductoisomerase. Bioorg. Med. Chem. Lett. 2008;18:3090–3094. doi: 10.1016/j.bmcl.2007.11.062. [DOI] [PubMed] [Google Scholar]
- 44.Munos JW, Pu X, Mansoorabadi SO, Kim HJ, Liu H-w. A secondary kinetic isotope effect study of the 1-deoxy-D-xylulose-5-phosphate reductoisomerase-catalyzed reaction: evidence for a retroaldol-aldol rearrangement. J. Am. Chem. Soc. 2009;131:2048–2049. doi: 10.1021/ja807987h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Okuhara M, Kuroda Y, Goto T, Okamoto M, Terano H, Kohsaka M, Aoki H, Imanaka H. Studies on new phosphonic acid antibiotics. III. Isolation and characterization of FR-31564, FR-32863 and FR-33289. J. Antibiot. (Tokyo) 1980;33:24–28. doi: 10.7164/antibiotics.33.24. [DOI] [PubMed] [Google Scholar]
- 46.Kuzuyama T, Shimizu T, Takahashi S, Seto H. Fosmidomycin, a specific inhibitor of 1-deoxy-D-xylulose 5-phosphate reductoisomerase in the non-mevalonate pathway for terpenoid biosynthesis. Tetrahedron Lett. 1998;39:7913–7916. [Google Scholar]
- 47.Jomaa H, Wiesner J, Sanderbrand S, Altincicek B, Weide-meyer C, Hintz M, Türbachova I, Eberl M, Zeidler J, Lichtenhaler HK, Soldati D, Beck E. Inhibitors of the non-mevalonate pathway of isoprenoid biosynthesis as antimalarial drugs. Science. 1999;285:1573–1576. doi: 10.1126/science.285.5433.1573. [DOI] [PubMed] [Google Scholar]
- 48.Mac Sweeney A, Lange R, Fernandes RPM, Shulz H, Dale GE, Douangamath A, Proteau Ph.J, Oefner C. The crystal structure of E.coli 1-deoxy-D-xylulose-5-phosphate reductoisomerase in a ternary complex with the antimalarial compound fosmidomycin and NADPH reveals a tight-binding closed enzyme conformation. J. Mol. Biol. 2005;345:115–127. doi: 10.1016/j.jmb.2004.10.030. [DOI] [PubMed] [Google Scholar]
- 49.Woo Y-h, Fernandes RPM, Proteau Ph.J. Evaluation of fosmidomycin analogs as inhibitors of the Synechocystis sp. PCC6803 1-deoxy-D-xylulose 5-phosphate reductoisomerase. Bioorg. Med. Chem. 2006;14:2375–2385. doi: 10.1016/j.bmc.2005.11.012. [DOI] [PubMed] [Google Scholar]
- 50.Singh N, Chevé G, Avery MA, McCurdy CR. Targeting the methyl erythritol phosphate (MEP) pathway for novel antimalarial, antibacterial and herbicidal drug discovery: inhibition of 1-deoxy-D-xylulose-5-phosphate reductoisomerase (DXR) enzyme. Curr. Pharm. Des. 2007;11:1161–1177. doi: 10.2174/138161207780618939. [DOI] [PubMed] [Google Scholar]
- 51.Missinou MA, Borrmann S, Schindler A, Issifou S, Adeg-nika AA, Matsiegui PB, Binder R, Lell B, Wiesner J, Baranek T, Jomaa H, Kremsner PG. Fosmidomycin for malaria. Lancet. 2002;360:1941–1942. doi: 10.1016/S0140-6736(02)11860-5. [DOI] [PubMed] [Google Scholar]
- 52.Ortmann R, Wiesner J, Silber K, Klebe G, Jomaa H, Schlitzer M. Novel deoxyxylulosephosphate-reductoisomerase inhibitors: fosmidomycin derivatives with spacious acyl residues. Arch. Pharm. Chem. Life Sci. 2007;340:483–490. doi: 10.1002/ardp.200700149. [DOI] [PubMed] [Google Scholar]
- 53.Gottlin EB, Benson RE, Conary S, Antonio B, Duke K, Payne ES, Ashraf SS, Christensen DJ. High-throughput screen for inhibitors of 1-deoxy-d-xylulose 5-phosphate reductoisomerase by surrogate ligand competition. J. Biomol. Sreen. 2003;8:332–339. doi: 10.1177/1087057103008003011. [DOI] [PubMed] [Google Scholar]
- 54.Gieµmann D, Heidler Ph, Haemers T, Van Calenbergh S, Reichenberg A, Jomaa H, Weidemeyer C, Sanderbrand S, Wiesner J, Link A. Towards new antimalarial drugs: synthesis of non-hydrolyzable phosphate mimics as feed for a predictive QSAR study on 1-deoxy-D-xylulose-5-phosphate reductoisomerase inhibitors. Chem. Biodiv. 2008;5:643–656. doi: 10.1002/cbdv.200890060. [DOI] [PubMed] [Google Scholar]
- 55.Deng L, Sundriyal S, Rubio V, Shi Z-z, Song Y. Coordination chemistry based approach to lipophilic inhibitors of 1-deoxy-D-xylulose-5-phosphate reductoisomerase. J. Med. Chem. 2009;52:6539–6542. doi: 10.1021/jm9012592. [DOI] [PubMed] [Google Scholar]
- 56.Zinglé C, Kuntz L, Tritsh D, Grosdemange-Billiard C, Rohmer M. Isoprenoid biosynthesis via the methylerythritol phosphate pathway: structural variations around phosphonate anchor and spacer of fosmidomycin, a potent inhibitor of deoxyxylulose phosphate reductoisomerase. J. Org. Chem. 2010;75:3203–3207. doi: 10.1021/jo9024732. [DOI] [PubMed] [Google Scholar]
- 57.Behrendt CT, Kunfermann A, Illarionova V, Matheeussen A, Gräwert T, Groll M, Rohdich F, Bacher A, Eisenreich W, Fischer M, Maes L, Kurz T. Synthesis and antiplasmodial activity of highly active reverse analogues of the antimalarial drug candidate fosmidomycin. ChemMedChem. 2010;5:1673–1676. doi: 10.1002/cmdc.201000276. [DOI] [PubMed] [Google Scholar]
- 58.Bodill T, Conibear AC, Blatch GL, Lobb KA, Kaye PT. Synthesis and evaluation of phosphonated N-heteroarylcarboxamides as DOXP-reductoisomerase (DXR) inhibitors. Bioorg. Med. Chem. 2011;19:1321–1327. doi: 10.1016/j.bmc.2010.11.062. [DOI] [PubMed] [Google Scholar]
- 59.Deng L, Endo K, Kato M, Cheng G, Yajima S, Song Y. Structures of 1-Deoxy-D-Xylulose-5-Phosphate Reductoisomerase/Lipophilic Phosphonate Complexes. ACS Med. Chem. Lett. 2011;2:165–170. doi: 10.1021/ml100243r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Deng L, Diao J, Chen P, Pujari V, Yao Y, Cheng G, Crick DC, Prasad BV, Song Y. Inhibition of 1-deoxy-D-xylulose-5-phosphate reductoisomerase by lipophilic phosphonates: SAR, QSAR, and crystallographic studies. J. Med. Chem. 2011 doi: 10.1021/jm200363d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Reuter K, Sanderbrand S, Jomaa H, Wiesner J, Steinbrecher I, Beck E, Hintz M, Klebe G, Stubbs MT. Crystal structure of 1-deoxy-D-xylulose-5-phosphate reductoisomerase, a crucial enzyme in the non-mevalonate pathway of isoprenoid biosynthesis. J. Biol. Chem. 2002;277:5378–5384. doi: 10.1074/jbc.M109500200. [DOI] [PubMed] [Google Scholar]
- 62.Yajima S, Nonaka T, Kuzuyama T, Seto H, Ohsawa K. Crystal structure of 1-deoxy-D-xylulose 5-phosphate reductoisomerase complexed with cofactors: implications of a flexible loop movement upon substrate binding. J. Biochem. (Tokyo) 2002;131:313–317. doi: 10.1093/oxfordjournals.jbchem.a003105. [DOI] [PubMed] [Google Scholar]
- 63.Ricagno S, Grolle S, Bringer-Meyer S, Sahm H, Lindqvist Y, Schneider G. Crystal structure of 1-deoxy-d-xylulose-5-phosphate reductoisomerase from Zymomonas mobilis at 1.9-A resolution. Biochim. Biophys. Acta. 2004;1698:37–44. doi: 10.1016/j.bbapap.2003.10.006. [DOI] [PubMed] [Google Scholar]
- 64.Steinbacher S, Kaiser J, Eisenreich W, Huber R, Bacher A, Rohdich F. Structural basis of fosmidomycin action revealed by the complex with 2-C-methyl-D-erythritol 4-phosphate synthase (IspC). Implications for the catalytic mechanism and anti-malaria drug development. J. Biol. Chem. 2003;278:18401–18407. doi: 10.1074/jbc.M300993200. [DOI] [PubMed] [Google Scholar]
- 65.Yajima S, Hara K, Sanders JM, Yin F, Ohsawa K, Wiesner J, Jomaa H, Oldfield E. Crystallographic structures of two bisphosphonate: 1-deoxyxylulose-5-phosphate reductoisomerase complexes. J. Am. Chem. Soc. 2004;126:10824–10825. doi: 10.1021/ja040126m. [DOI] [PubMed] [Google Scholar]
- 66.Mercklé L, de Andrés-Gómez A, Dick B, Cox RJ, Godfrey CRA. A fragment-based approach to understanding inhibition of 1-deoxy-D-xylulose-5-phosphate reductoisomerase. ChemBioChem. 2005;6:1866–1874. doi: 10.1002/cbic.200500061. [DOI] [PubMed] [Google Scholar]
- 67.Fernandes RPM, Phaosiri C, Proteau Ph.J. Mutation in the flexible loop of 1-deoxy-D-xylulose 5-phosphate reductoisomerase broadens substrate utilization. Arch. Biochem. Biophys. 2005;444:159–164. doi: 10.1016/j.abb.2005.10.004. [DOI] [PubMed] [Google Scholar]
- 68.Singh N, Avery MA, McCurdy CR. Towards Mycobacterium tuberculosis DXR inhibitor design: homology modeling and molecular dynamics simulations. J. Comput. Aided Mol. Des. 2007;21:511–522. doi: 10.1007/s10822-007-9132-0. [DOI] [PubMed] [Google Scholar]
- 69.Henriksson LM, Unge T, Carlsson J, Åqvist J, Mowbray SL, Jones TA. Structures of Mycobacterium tuberculosis 1-deoxy-D-xylulose-5-phosphate reductoisomerase provide new insights into catalysis. J. Biol. Chem. 2007;282:19905–19916. doi: 10.1074/jbc.M701935200. [DOI] [PubMed] [Google Scholar]
- 70.Andaloussi M, Henriksson LM, Więckowska A, Lindh M, Björkelid C, Larsson AM, Suresh S, Iyer H, Srinivasa BR, Bergfors T, Unge T, Mowbray SL, Larhed ML, Jones AT, Karlen AB. Design, synthesis, and X-ray crystallographic studies of α-aryl substituted fosmidomycin analogues as inhibitors of Mycobacterium tuberculosis 1-deoxy-d-xylulose 5-phosphate reductoisomerase. J. Med. Chem. 2011 doi: 10.1021/jm2000085. [DOI] [PubMed] [Google Scholar]
- 71.Goble J, Adendorff MR, de Beer TAP, Stephens LL, Blatch GL. The malarial drug target Plasmodium falciparum 1-deoxy-D-xylulose-5-phosphate reductoisomerase (PfDXR): development of a 3-D model for identification of novel, structural and functional features and for inhibitor screening. Protein Pept. Lett. 2010;17:109–120. doi: 10.2174/092986610789909548. [DOI] [PubMed] [Google Scholar]
- 72.Umeda T, Tanaka N, Kusakabe Y, Nakanishi M, Kitade Y, Nakamura KT. Crystallization and preliminary X-ray crystallographic study of 1-deoxy-D-xylulose 5-phosphate reductoisomerase from Plasmodium falciparum . Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2010;66:330–332. doi: 10.1107/S1744309110001739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Englebert NE, Richter C, Wiesner J, Hintz M, Jomaa H, Schwalbe H. NMR studies of DOXP reductoisomerase and its inhibitor complex. ChemBioChem. 2011;12:468–476. doi: 10.1002/cbic.201000465. [DOI] [PubMed] [Google Scholar]
- 74.Odom AR, van Voorhis WC. Functional genetic analysis of the Plasmodium falciparum deoxyxylulose 5-phosphate reductoisomerase gene. Mol. Biochem. Parasitol. 2010;170:108–111. doi: 10.1016/j.molbiopara.2009.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Humnabadkar V, Jha R, Ghatnekar N, de Sousa SM. A high-throughput screening assay for simultaneous selection of inhibitors of Mycobacterium tuberculosis 1-deoxy-D-xylulose-5-phosphate synthase (Dxs) or 1-deoxy-D-xylulose 5-phosphate reductoisomerase (Dxr) J. Biomol. Screen. 2011;16:303–312. doi: 10.1177/1087057110394845. [DOI] [PubMed] [Google Scholar]
- 76.Poulter CD, Rilling HC. The prenyl transfer-reaction. Enzymatic and mechanistic studies of 1’-4 coupling reaction in the terpene biosynthetic-pathway. Acc. Chem. Res. 1978;11:307–313. [Google Scholar]
- 77.Beytia E, Qureshi AA, Porter JW. Squalene synthetase. 3. Mechanism of the reaction. J. Biol. Chem. 1973;248:1856–1867. [PubMed] [Google Scholar]
- 78.Kandutsch AA, Paulus H, Levin E, Bloch K. Purification Of Geranylgeranyl Pyrophosphate Synthetase From Micrococcus lysodeikticus . J. Biol. Chem. 1964;239:2507–2515. [PubMed] [Google Scholar]
- 79.Wiemer AJ, Hohl RJ, Wiemer DF. The intermediate enzymes of isoprenoid metabolism as anticancer targets. Anticancer Agents Med. Chem. 2009;9:526–542. doi: 10.2174/187152009788451860. [DOI] [PubMed] [Google Scholar]
- 80.Dunford JE. Molecular targets of the nitrogen containing bisphosphonates: the molecular pharmacology of prenyl synthase inhibition. Curr. Pharm. Des. 2010;16:2961–2969. doi: 10.2174/138161210793563617. [DOI] [PubMed] [Google Scholar]
- 81.Ramos-Valdivia AC, van der Heijden R, Verpoorte R, Camara B. Purification and characterization of two isoforms of isopentenyl-diphosphate isomerase from elicitor-treated Cinchona robusta cells. Eur. J. Biochem. 1997;249:161–170. doi: 10.1111/j.1432-1033.1997.t01-1-00161.x. [DOI] [PubMed] [Google Scholar]
- 82.Carrigan CN, Poulter CD. Zinc is an essential cofactor for type I isopentenyl diphosphate: dimethylallyl diphosphate isomerase. J. Am. Chem. Soc. 2003;125:9008–9009. doi: 10.1021/ja0350381. [DOI] [PubMed] [Google Scholar]
- 83.Lee S, Poulter CD. Escherichia coli type I isopentenyl diphosphate isomerase: structural and catalytic roles for divalent metals. J. Am. Chem. Soc. 2006;128:11545–11550. doi: 10.1021/ja063073c. [DOI] [PubMed] [Google Scholar]
- 84.Durbecq V, Sainz G, Oudjama Y, Clantin B, Bompard-Gilles C, Tricot C, Caillet J, Stalon V, Droogmans L, Villeret V. Crystal structure of isopentenyl diphosphate:dimethylallyl diphosphate isomerase. EMBO J. 2001;20:1530–1537. doi: 10.1093/emboj/20.7.1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lu XJ, Christensen DJ, Poulter CD. Isopentenyl-diphosphate isomerase: irreversible inhibition by 3-methyl-3,4-epoxybutyl diphosphate. Biochemistry. 1992;31:9955–9960. doi: 10.1021/bi00156a014. [DOI] [PubMed] [Google Scholar]
- 86.Wu Z, Wouters J, Poulter CD. Isopentenyl diphosphate isomerase. Mechanism-based inhibition by diene analogues of isopentenyl diphosphate and dimethylallyl diphosphate. J. Am. Chem. Soc. 2005;127:17433–17438. doi: 10.1021/ja056187h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Muehlbacher M, Poulter CD. Isopentenyl-diphosphate isomerase: inactivation of the enzyme with active-site-directed irreversible inhibitors and transition-state analogues. Biochemistry. 1988;27:7315–7328. doi: 10.1021/bi00419a021. [DOI] [PubMed] [Google Scholar]
- 88.Reardon JE, Abeles RH. Mechanism of action of isopentenyl pyrophosphate isomerase: evidence for a carbonium ion intermediate. Biochemistry. 1986;25:5609–5616. doi: 10.1021/bi00367a040. [DOI] [PubMed] [Google Scholar]
- 89.Wouters J, Oudjama Y, Barkley SJ, Tricot C, Stalon V, Droogmans L, Poulter CD. Catalytic mechanism of Escherichia coli isopentenyl diphosphate isomerase involves Cys-67, Glu-116, and Tyr-104 as suggested by crystal structures of complexes with transition state analogues and irreversible inhibitors. J. Biol. Chem. 2003;278:11903–11908. doi: 10.1074/jbc.M212823200. [DOI] [PubMed] [Google Scholar]
- 90.Wouters J, Oudjama Y, Ghosh S, Stalon V, Droogmans L, Oldfield E. Structure and mechanism of action of isopentenylpyrophosphate-dimethylallylpyrophosphate isomerase. J. Am. Chem. Soc. 2003;125:3198–3199. doi: 10.1021/ja029171p. [DOI] [PubMed] [Google Scholar]
- 91.Wouters J, Oudjama Y, Stalon V, Droogmans L, Poulter CD. Crystal structure of the C67A mutant of isopentenyl diphosphate isomerase complexed with a mechanism-based irreversible inhibitor. Proteins. 2004;54:216–221. doi: 10.1002/prot.10573. [DOI] [PubMed] [Google Scholar]
- 92.Wouters J, Yin F, Song Y, Zhang Y, Oudjama Y, Stalon V, Droogmans L, Morita CT, Oldfield E. A crystallographic investigation of phosphoantigen binding to isopentenyl pyrophosphate/dimethylallyl pyrophosphate isomerase. J. Am. Chem. Soc. 2005;127:536–537. doi: 10.1021/ja040207i. [DOI] [PubMed] [Google Scholar]
- 93.de Ruyck J, Durisotti V, Oudjama Y, Wouters J. Structural role for Tyr-104 in Escherichia coli isopentenyl-diphosphate isomerase: site-directed mutagenesis, enzymology, and protein crystallography. J. Biol. Chem. 2006;281:17864–17869. doi: 10.1074/jbc.M601851200. [DOI] [PubMed] [Google Scholar]
- 94.Alou L, Cafini F, Sevillano D, Unzueta I, Prieto J. In vitro activity of mupirocin and amoxicillin-clavulanate alone and in combination against staphylococci including those resistant to methicillin. Int. J. Antimicrob. Agents. 2004;23:513–516. doi: 10.1016/j.ijantimicag.2003.09.030. [DOI] [PubMed] [Google Scholar]
- 95.Weigel LM, Clewell DB, Gill SR, Clark NC, McDougal LK, Flannagan SE, Kolonay JF, Shett J, Killgore GE, Tenover FC. Genetic analysis of a high-level vancomycin-resistant isolate of Staphylococcus aureus . Science. 2003;302:1569–1571. doi: 10.1126/science.1090956. [DOI] [PubMed] [Google Scholar]
- 96.Moaddab SR, Rafi A. Prevalence of vancomycin and high level aminoglycoside resistant enterococci among high-risk patients. Southeast Asian J. Trop. Med. Public Health. 2003;34:849–854. [PubMed] [Google Scholar]
- 97.Kaneda K, Kuzuyama T, Takagi M, Hayakawa Y, Seto H. An unusual isopentenyl diphosphate isomerase found in the mevalonate pathway gene cluster from Streptomyces sp. strain CL190. Proc. Natl. Acad. Sci. U S A. 2001;98:932–937. doi: 10.1073/pnas.020472198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.de Ruyck J, Wouters J. Structure-based drug design targeting biosynthesis of isoprenoids: a crystallographic state of the art of the involved enzymes. Curr. Protein Pept. Sci. 2008;9:117–137. doi: 10.2174/138920308783955261. [DOI] [PubMed] [Google Scholar]
- 99.Steinbacher S, Kaiser J, Gerhardt S, Eisenreich W, Huber R, Bacher A, Rohdich F. Crystal structure of the type II isopentenyl diphosphate:dimethylallyl diphosphate isomerase from Bacillus subtilis . J. Mol. Biol. 2003;329:973–982. doi: 10.1016/s0022-2836(03)00527-8. [DOI] [PubMed] [Google Scholar]
- 100.Takagi M, Kaneda K, Shimizu T, Hayakawa Y, Seto H, Kuzuyama T. Bacillus subtilis ypgA gene is fni, a non-essential gene encoding type 2 isopentenyl diphosphate isomerase. Biosci. Biotechnol. Biochem. 2004;68:132–137. doi: 10.1271/bbb.68.132. [DOI] [PubMed] [Google Scholar]
- 101.Unno H, Yamashita S, Ikeda Y, Sekiguchi S-y, Yoshida N, Yoshimura T, Kusunoki M, Nakayama T, Nishino T, Hemmi H. New role of flavin as a general acid-base catalyst with no redox function in type 2 isopentenyl-diphosphate isomerase. J. Biol. Chem. 2009;284:9160–9167. doi: 10.1074/jbc.M808438200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Yamashita S, Hemmi H, Ikeda Y, Nakayama T, Nishino T. Type 2 isopentenyl diphosphate isomerase from a thermoacidophilic archaeon Sulfolobus shibatae . Eur. J. Biochem. 2004;271:1087–1093. doi: 10.1111/j.1432-1033.2004.04010.x. [DOI] [PubMed] [Google Scholar]
- 103.Barkley SJ, Desai SB, Poulter CD. Type II isopentenyl diphosphate isomerase from Synechocystis sp. strain PCC 6803. J. Bacteriol. 2004;186:8156–8158. doi: 10.1128/JB.186.23.8156-8158.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Barkley SJ, Cornish RM, Poulter CD. Identification of an Archaeal type II isopentenyl diphosphate isomerase in Methanothermobacter thermautotrophicus . J. Bacteriol. 2004;186:1811–1817. doi: 10.1128/JB.186.6.1811-1817.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kittleman W, Thibodeaux CJ, Liu Y-N, Zhang H, Liu H-W. Characterization and mechanistic studies of type II isopentenyl diphosphate:dimethylallyl diphosphate isomerase from Staphylococcus aureus . Biochemistry. 2007;46:8401–8413. doi: 10.1021/bi700286a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kaneda K, Kuzuyama T, Takagi M, Hayakawa Y, Seto H. An unusual isopentenyl diphosphate isomerase found in the mevalonate pathway gene cluster from Streptomyces sp. strain CL190. Proc. Natl. Acad. Sci. U S A. 2001;98:932–937. doi: 10.1073/pnas.020472198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.de Ruyck J, Pouyez J, Rothman SC, Poulter CD, Wouters J. Crystal structure of type 2 isopentenyl diphosphate isomerase from Thermus thermophilus in complex with inorganic pyrophosphate. Biochemistry. 2008;47:9051–9053. doi: 10.1021/bi801159x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Rothman SC, Helm TR, Poulter CD. Kinetic and spectroscopic characterization of type II isopentenyl diphosphate isomerase from Thermus thermophilus: evidence for formation of substrate-induced flavin species. Biochemistry. 2007;46:5437–5445. doi: 10.1021/bi0616347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Hoshino T, Tamegai H, Kakinuma K, Eguchi T. Inhibition of type 2 isopentenyl diphosphate isomerase from Methanocaldococcus jannaschii by a mechanism-based inhibitor of type 1 isopentenyl diphosphate isomerase. Bioorg. Med. Chem. 2006;14:6555–6559. doi: 10.1016/j.bmc.2006.06.011. [DOI] [PubMed] [Google Scholar]
- 110.Siddiqui MA, Yamanaka A, Hirooka K, Bamaba T, Kobayashi A, Imanaka T, Fukusaki E, Fujiwara S. Enzymatic and structural characterization of type II isopentenyl diphosphate isomerase from hyperthermophilic archaeon Thermococcus kodakaraensis . Biochem. Biophys. Res. Commun. 2005;331:1127–1136. doi: 10.1016/j.bbrc.2005.04.029. [DOI] [PubMed] [Google Scholar]
- 111.Nagano N, Orengo CA, Thornton JM. One fold with many functions: the evolutionary relationships between TIM barrel families based on their sequences, structures and functions. J. Mol. Biol. 2002;321:741–765. doi: 10.1016/s0022-2836(02)00649-6. [DOI] [PubMed] [Google Scholar]
- 112.de Ruyck J, Rothman SC, Poulter CD, Wouters J. Structure of Thermus thermophilus type 2 isopentenyl diphosphate isomerase inferred from crystallography and molecular dynamics. Biochem. Biophys. Res. Commun. 2005;338:1515–1518. doi: 10.1016/j.bbrc.2005.10.114. [DOI] [PubMed] [Google Scholar]
- 113.Bornemann S. Flavoenzymes that catalyse reactions with no net redox change. Nat. Prod. Rep. 2002;19:761–772. doi: 10.1039/b108916c. [DOI] [PubMed] [Google Scholar]
- 114.Huang Z, Zhang Q, Liu H-w. Reconstitution of UDP-galactopyranose mutase with 1-deaza-FAD and 5-deaza-FAD: analysis and mechanistic implications. Bioorg. Chem. 2003;31:494–502. doi: 10.1016/j.bioorg.2003.08.002. [DOI] [PubMed] [Google Scholar]
- 115.Hemmi H, Ikeda Y, Yamashita Y, Nakayama T, Nishino T. Catalytic mechanism of type 2 isopentenyl diphosphate:dimethylallyl diphosphate isomerase: verification of a redox role of the flavin cofactor in a reaction with no net redox change. Biochem. Biophys. Res. Commun. 2004;322:905–910. doi: 10.1016/j.bbrc.2004.08.013. [DOI] [PubMed] [Google Scholar]
- 116.Thibodeaux CJ, Mansoorabadi SO, Kittleman W, Chang W-c, Liu H-w. Evidence for the involvement of acid/base chemistry in the reaction catalyzed by the type II isopentenyl diphosphate/dimethylallyl diphosphate isomerase from Staphylococcus aureus . Biochemistry. 2008;47:2547–2558. doi: 10.1021/bi701467g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Johnston JB, Walker JR, Rothman SC, Poulter CD. Type-2 isopentenyl diphosphate isomerase. Mechanistic studies with cyclopropyl and epoxy analogues. J. Am. Chem. Soc. 2007;129:7740–7741. doi: 10.1021/ja072501r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Walker JR, Rothman SC, Poulter CD. Synthesis and evaluation of substrate analogues as mechanism-based inhibitors of type II isopentenyl diphosphate isomerase. J. Org. Chem. 2008;73:726–729. doi: 10.1021/jo702061d. [DOI] [PubMed] [Google Scholar]
- 119.Rothman SC, Johnston JB, Lee S, Walker JR, Poulter CD. Type II isopentenyl diphosphate isomerase: irreversible inactivation by covalent modification of flavin. J. Am. Chem. Soc. 2008;130:4906–4913. doi: 10.1021/ja7108954. [DOI] [PubMed] [Google Scholar]
- 120.Sharma NK, Pan J-j, Poulter CD. Type II isopentenyl diphosphate isomerase: probing the mechanism with alkyne/allene diphosphate substrate analogues. Biochemistry. 2010;49:6228–6233. doi: 10.1021/bi100844e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Thibodeaux CJ, Chang W-c, Liu H-w. Linear free energy relationships demonstrate a catalytic role for the flavin mononucleotide coenzyme of the type II isopentenyl diphosphate:dimethylallyl diphosphate isomerase. J. Am. Chem. Soc. 2010;132:9994–9996. doi: 10.1021/ja104090m. [DOI] [PMC free article] [PubMed] [Google Scholar]