Fusion of ER membranes by atlastin and interaction of ER with growing microtubule ends and dynein cooperate to generate distinct ER morphologies during the cell cycle.
Abstract
In metazoans the endoplasmic reticulum (ER) changes during the cell cycle, with the nuclear envelope (NE) disassembling and reassembling during mitosis and the peripheral ER undergoing extensive remodeling. Here we address how ER morphology is generated during the cell cycle using crude and fractionated Xenopus laevis egg extracts. We show that in interphase the ER is concentrated at the microtubule (MT)-organizing center by dynein and is spread by outward extension of ER tubules through their association with plus ends of growing MTs. Fusion of membranes into an ER network is dependent on the guanosine triphosphatase atlastin (ATL). NE assembly requires fusion by both ATL and ER-soluble N-ethyl-maleimide–sensitive factor adaptor protein receptors. In mitotic extracts, the ER converts into a network of sheets connected by ER tubules and loses most of its interactions with MTs. Together, these results indicate that fusion of ER membranes by ATL and interaction of ER with growing MT ends and dynein cooperate to generate distinct ER morphologies during the cell cycle.
Introduction
The mechanisms by which organelles are shaped and remodeled during the cell cycle are largely unknown. We have started to address this problem for the ER. In interphase, the ER is a continuous membrane system consisting of the nuclear envelope (NE) and a peripheral network of ER tubules and interdispersed sheets (Shibata et al., 2009; Chen et al., 2013; English and Voeltz, 2013a; Goyal and Blackstone, 2013). ER tubules exhibit high membrane curvature in cross section and are shaped by members of two evolutionarily conserved protein families, the reticulons and DP1/Yop1p (Voeltz et al., 2006). These abundant membrane proteins stabilize the high curvature of tubules (Shibata et al., 2008, 2009) and may also be involved in generating peripheral ER sheets, as they also localize to sheet edges (Shibata et al., 2010).
The generation of a continuous tubular network requires that the tubules be connected by membrane fusion to form three-way junctions. Fusion is mediated by a class of membrane-anchored dynamin-like GTPases called atlastins (ATLs) in metazoans (Hu et al., 2009; Orso et al., 2009). ATL-mediated ER fusion is supported by the observation that depletion of ATLs leads to long, nonbranched ER tubules in tissue culture cells (Hu et al., 2009) and ER fragmentation in Drosophila melanogaster (Orso et al., 2009). Long, nonbranched tubules are also observed when dominant-negative fragments of ATL are overexpressed in tissue culture cells (Hu et al., 2009). In addition, antibodies against ATL inhibit ER network formation in Xenopus laevis egg extracts (Hu et al., 2009), and proteoliposomes containing purified D. melanogaster ATL undergo GTP-dependent fusion in vitro (Orso et al., 2009; Bian et al., 2011; Liu et al., 2012). However, it is unclear whether additional ER fusion mechanisms exist and whether Rab proteins have a role in the formation of a tubular ER network, as suggested by experiments in Caenorhabditis elegans and X. laevis egg extracts (Audhya et al., 2007; English and Voeltz, 2013b).
The ER network is very dynamic, with tubules continuously forming and retracting. In metazoans, ER tubules can be “pulled out” of a membrane reservoir by molecular motors moving along microtubules (MTs) or by the tips of growing MTs (Du et al., 2004; Grigoriev et al., 2008; Friedman et al., 2010). However, because ER tubules can be formed in the absence of MTs (Dreier and Rapoport, 2000; Voeltz et al., 2006), and the ER network does not immediately collapse upon MT depolymerization (Terasaki et al., 1986), the cytoskeleton does not seem to be necessary for the generation of the tubular ER network, per se. Rather, the cytoskeleton may be involved in the spatial distribution of the ER network in cells, as the density of the ER is generally higher toward the center of the cell, where the MT-organizing center (MTOC) is located.
Although the morphology of the peripheral ER in interphase cells is well characterized, the changes occurring during mitosis have been controversial. Some studies suggest that peripheral ER sheets convert into fenestrated sheets and tubules (Anderson and Hetzer, 2007; Puhka et al., 2007, 2012), whereas others propose that tubules transform into sheets (Poteryaev et al., 2005; Lu et al., 2009, 2011). These experiments were all done in intact cells, which round up during mitosis, making the analysis of the peripheral ER morphology difficult. How the NE is reformed after mitosis is also unclear. It has been proposed that NE reformation is exclusively driven by ER fusion, with no other membrane fusion reaction required (Anderson and Hetzer, 2007). However, other data suggest that NE formation may require additional fusion by unidentified SNARE proteins that are normally involved in vesicular transport. This hypothesis would explain why dominant-negative forms of the SNARE complex disassembly factor NSF or of its cofactor αSNAP inhibit NE formation in X. laevis egg extracts (Baur et al., 2007).
X. laevis egg extracts are a powerful system to study the formation of an ER network and address how ER morphology changes during the cell cycle (Allan, 1995; Waterman-Storer et al., 1995; Dreier and Rapoport, 2000; Voeltz et al., 2006). Here, we have used this system to show that the fusion of ER membranes by ATL and the interaction of the ER with dynein and the plus ends of growing MTs cooperate to generate distinct ER morphologies during interphase and mitosis. We also found that NE formation requires not only ATL-mediated membrane fusion but also a fusion reaction mediated by ER SNAREs. The two distinct fusion systems cooperate at different stages to reform the NE at the end of mitosis.
Results
A role for dynein and MT plus tips in ER network distribution during interphase
We used crude X. laevis egg extracts to study ER morphology at different stages of the cell cycle and to avoid limitations associated with imaging the ER in intact cells during mitosis. The crude extract is essentially undiluted cytoplasm that maintains a vigorous energy metabolism and contains large amounts of ER membranes (Niethammer et al., 2008). The extract can easily be driven into either interphase or mitosis. A mitotic extract was obtained by preparing it in the presence of the Ca2+ chelator EGTA (cytostatic factor [CSF] extract). An interphase extract was then generated by addition of Ca2+, which releases the cell cycle arrest (Murray et al., 1989). At the same time, lysolecitin-permeabilized sperm was added, providing both centrosomes as nucleation sites for MTs and allowing the eventual formation of a NE around the decondensing chromatin.
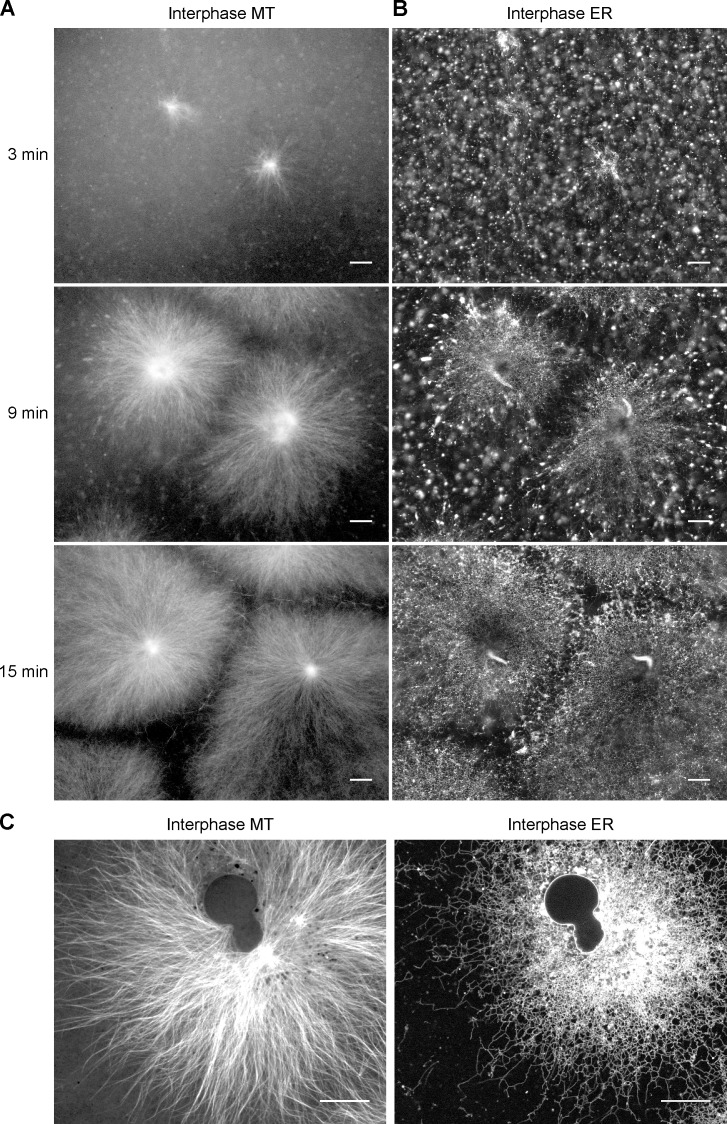
To better mimic normal cytoplasm, the extracts were prepared in the absence of the actin-depolymerizing agent cytochalasin. Fluorescently labeled tubulin was added to monitor the formation of MTs. We optimized the experimental conditions to allow the growth of large interphase asters, i.e., star-like MT assemblies that nucleate at the centrosomes of the MTOCs, similar to those that normally form in interphase of X. laevis eggs and in early embryos (Mitchison et al., 2013). The MTs are polarized with their minus ends close to the MTOC and the growing plus ends pointing outwards. To stain ER membranes, a lipophilic dye (DiIC18 or DiOC18) was preincubated with a small aliquot of extract and then diluted into fresh extract. Imaging in real time showed that MT asters indeed emanate from the sperm centrosomes (Fig. 1 A and Video 1). Simultaneously, an ER network was generated (Fig. 1 B and Video 2). Initially, there was a predominant movement of membranes toward the MTOCs, but after a short time, the ER network started to spread outwards. Even after the ER network stopped spreading, it remained dynamic, with tubules continuously forming and retracting (Video 2). After ∼30 min, the chromatin of the sperm had significantly decondensed and nuclei with a visible NE were generated (Fig. 1 C). At later time points, the ER network was denser at the MTOC than at the periphery (Fig. 1 C; and see Fig. 2 B and Fig. 5, C and D), a spatial distribution that is strikingly similar to that in tissue culture cells. As reported previously (Dreier and Rapoport, 2000), addition of the MT-depolymerizing drug nocodazole did not prevent formation of an ER network (Fig. S1), indicating that MTs are not required for tubule formation or fusion. However, in the absence of MTs the network was evenly distributed throughout the imaged area (Fig. S1), indicating that MTs are necessary for the inhomogeneous distribution of the ER network.
Figure 1.
ER network formation in crude interphase X. laevis egg extracts. Demembranated sperm was added to a crude interphase X. laevis egg extract containing Alexa fluor 488–labeled tubulin and the hydrophobic dye DiIC18. The formation of MT asters (A) and ER network (B) was followed over time by confocal fluorescence microscopy (for full time course, see Videos 1 and 2). Sperm was added at time zero. Bars, 30 µm. (C) As in A and B, but after 60 min, a time point at which the NE has formed. Bars, 20 µm.
Figure 2.
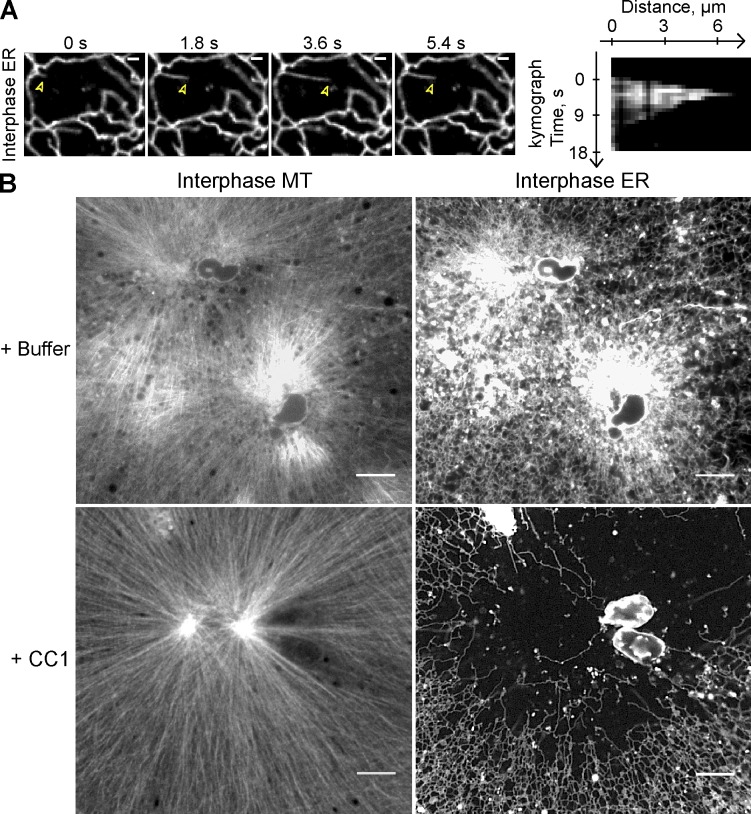
Dynein is required for movement of ER membranes toward the minus end of MTs. (A) Demembranated sperm was added after 10 min to a crude interphase X. laevis egg extract containing Alexa fluor 488–labeled tubulin and the hydrophobic dye DiIC18. The extension of membrane tubules was followed over time. Arrowheads indicate the tip of extending ER membrane tubule. The kymograph shows the movement of the same tubule. Bars, 2 µm. (B) As in A, but the extract was incubated for 30 min with and without 6 µM of the CC1 of p150 glued to inhibit dynein function. Network formation was visualized by confocal fluorescence microscopy. Bars, 10 µm.
Figure 5.
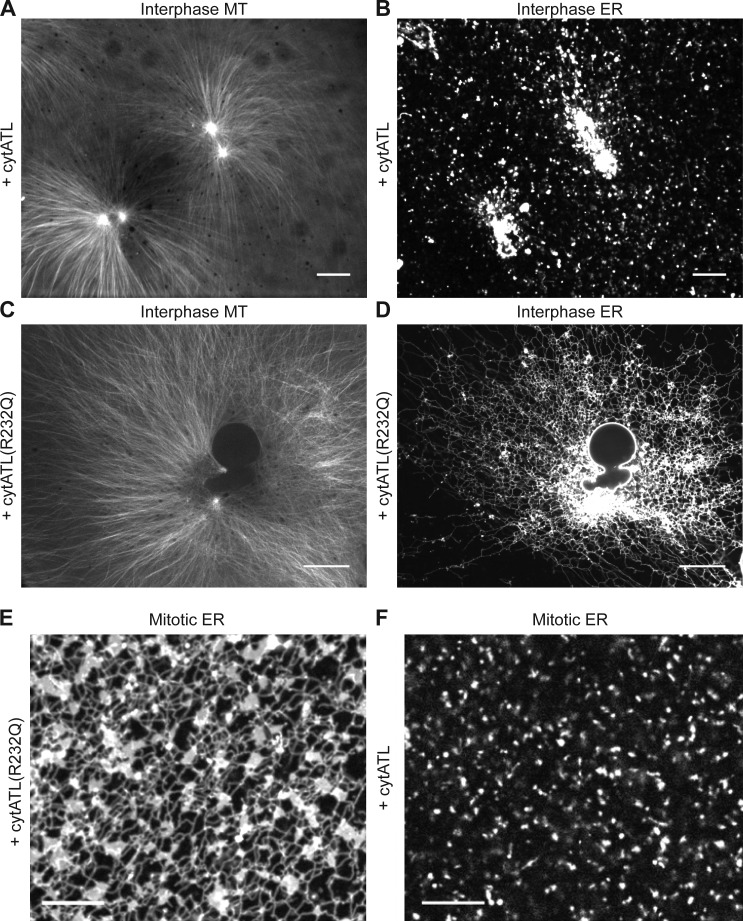
ATL is required for ER network formation in crude interphase and mitotic extracts. (A and B) Demembranated sperm was added to a crude interphase X. laevis egg extract containing Alexa fluor 488–labeled tubulin and the hydrophobic dye DiIC18. The extract also contained 2 µM of the cytoplasmic fragment of X. laevis ATL2 (cytATL) to inhibit ATL-mediated membrane fusion. Labeled MT asters (A) and ER membranes (B) were visualized by confocal fluorescence microscopy. Bars, 20 µm. (C and D) As in A and B, but with 2 µM of a point mutant of cytATL (cytATL(R232Q)) that does not inhibit fusion. Bars, 20 µm. (E and F) Inhibition of ATL-mediated fusion in mitotic extracts. 2 µM cytATL(R232Q) (E) or cytATL (F) were added to CSF extracts and the membranes were stained with DiIC18. Bars, 10 µm.
Rapid movement of ER membranes toward the center of the growing MT aster early in network formation suggested an involvement of the minus end–directed molecular motor cytoplasmic dynein. Indeed, the speed at which many tubules moved along MTs in the minus direction was 1.27 ± 0.27 µm/s (Fig. 2 A), consistent with them being pulled by dynein (Video 3; Allan and Vale, 1991; Lane and Allan, 1999). Occasionally, the membrane tubules rapidly retracted and then started to regrow along the same track. Kymographs showed that dynein extended membrane tubules for several micrometers (Fig. 2 A), much longer than the run length of a single dynein motor on MTs (Reck-Peterson et al., 2006; Ross et al., 2006).
To test for a role of dynein, we inhibited it by blocking its interaction with the essential cofactor dynactin. This was achieved by adding to the extract the purified coiled-coil region 1 (CC1) of the dynactin subunit p150 glued (King et al., 2003; the purity of CC1 is shown in Fig. S2). CC1 addition did not affect growth of MT asters, but no minus end–directed movements of ER membranes were observed and the ER network no longer concentrated around the MTOCs (Fig. 2 B). Rather, tubules tended to accumulate at the aster periphery, although sometimes the network was evenly distributed (unpublished data), as in extracts treated with nocodazole. NE formation still occurred with the same rate, as determined by time-lapse imaging (unpublished data), but only few ER tubules were seen in the vicinity of nuclei, in striking contrast to the controls. We therefore conclude that cytoplasmic dynein is not required for ER tubule or NE formation, but is largely responsible for concentrating the ER network at the MTOC.
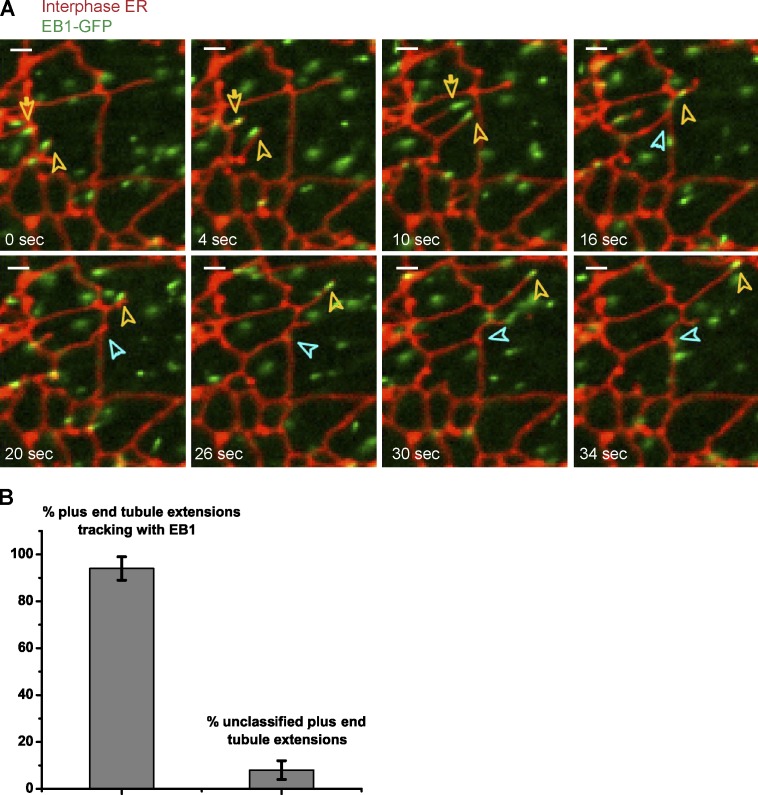
Next we analyzed in more detail the plus end–directed movement of ER tubules. To label the growing plus ends of MTs, we added to the extract a purified GFP fusion of the MT plus end tracking protein EB-1 (end-binding protein 1; EB-1–GFP; Morrison et al., 1998; Juwana et al., 1999; Tirnauer et al., 1999; the purity of EB-1–GFP is shown in Fig. S2). Simultaneous imaging of the plus ends of MTs and membranes showed that the leading ends of essentially all outward moving ER tubules tracked together with EB-1–GFP comets (Fig. 3 and Video 4). Thus, plus tip–dependent movement is likely the main mechanism by which the ER network expands in X. laevis egg extracts. This observation is consistent with an earlier study in which tip tracking of membrane tubules was observed in egg extract by differential interference contrast microscopy (Waterman-Storer et al., 1995).
Figure 3.
The leading ends of ER tubules track with the plus ends of MTs. (A) Demembranated sperm was added for 30 min to a crude interphase X. laevis egg extract containing the hydrophobic dye DiIC18 and a GFP fusion of the plus end tracking protein EB-1. The movements of ER tubules and of the plus ends of MTs were followed over time by confocal fluorescence microscopy (for full time course, see Video 4). Yellow arrows indicate the leading ends of ER tubules (red) that track with the plus ends of growing microtubules (green). The blue arrow indicates the site of attachment between one of the extending ER tubules and another tubule to form a three-way junction. Bars, 2 µm. (B) Quantification of experiments such as shown in A. The percentage of outward moving ER tubules that tracked with EB-1 is shown. Error bars indicate the range within three independent experiments, with 50 tubule-extension events analyzed in each experiment.
ER network morphology in mitosis
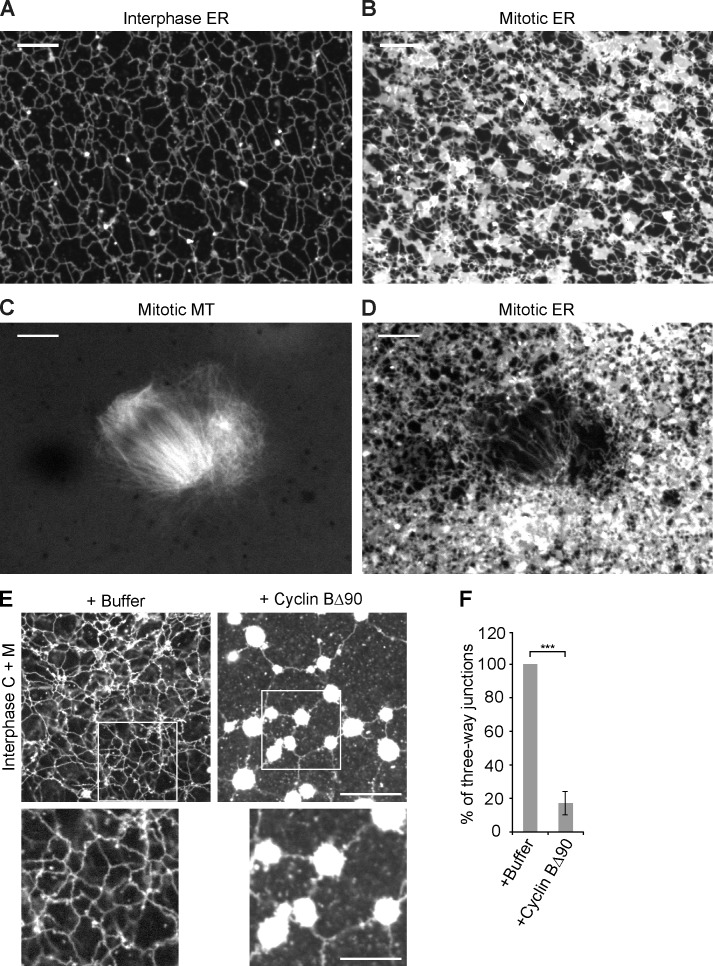
Next we analyzed ER morphology in X. laevis extracts that were kept in metaphase of the cell cycle (CSF extracts) by omitting Ca2+ addition. In contrast to previous findings (Allan and Vale, 1991), an ER network formed, but it was drastically different from that in interphase (Fig. 4, compare A with B). It contained numerous membrane sheets, which largely replaced three-way junctions between tubules. The addition of lysolecitin-permeabilized sperm led to formation of meiosis II spindles, which were visualized with fluorescently labeled tubulin (Fig. 4 C). No membranes were seen inside the spindle (Fig. 4 D), indicating that most interactions with MTs are lost during mitosis. Only few tubules were observed along the side of the spindle (Fig. 4 D), although some membranes accumulated at the spindle poles. These results are consistent with recent observations in tissue culture cells (Smyth et al., 2012) and are in agreement with the observed detachment of mitotic X. laevis membranes from stabilized MTs (Niclas et al., 1996).
Figure 4.
The ER network undergoes tubule-to-sheet conversion during the transition from interphase to meiosis and mitosis. (A) A crude CSF extract was driven into interphase by addition of Ca2+ ions. The ER network was stained with the hydrophobic dye DiIC18 and visualized by confocal fluorescence microscopy. Bar, 10 µm. (B) As in A, but the meiotic (CSF) state of the extract was maintained by omitting Ca2+. Bar, 20 µm. (C and D) As in B, but with addition of demembranated sperm. C shows the MTs stained with Alexa fluor 488–labeled tubulin, and D shows the membranes stained with DiIC18. Bars, 10 µm. (E) Interphase cytosol, light membranes, and an energy regenerating system were mixed with cyclin BΔ90 to generate a mitotic state. A control was performed with buffer. The membranes were stained with octadecyl rhodamine and visualized by confocal microscopy. The bottom panels show magnified views of the three-way junctions highlighted in the boxed area. Bars: (top) 20 µm; (bottom) 10 µm. (F) The number of three-way junctions in E was counted and expressed as a percentage of the control. The data plotted are the mean ± SD of three independent experiments. ***, P < 0.001, Student’s t test.
To directly analyze the morphological changes of the ER during the transition from interphase to mitosis, we induced a mitotic state by adding purified nondegradable cyclin B protein (cyclin BΔ90) to an interphase extract (the purity of cyclin BΔ90 is shown in Fig. S2 and the mitotic state is confirmed by immunoblotting with MPM-2 antibodies, which recognize mitotically phosphorylated proteins; Fig. S3 A) and by the generation of condensed chromosomes (Fig. S3 B; Murray et al., 1989). The ER morphology was similar to that observed in a CSF extract (Fig. S3 C). Real-time imaging showed examples of three-way junctions converting into sheets upon addition of cyclin BΔ90 (Video 5). The existence of an ER network and the tubule-to-sheet conversion in mitosis were probably missed in a previous study (Allan and Vale, 1991) because only MT-attached membranes were analyzed, which are scarce in mitotic extracts.
Similar results were obtained with a more defined system, in which isolated X. laevis egg membranes were mixed with membrane-depleted cytosol. With interphase cytosol added, a tubular network with numerous three-way junctions was formed (Fig. 4 E). When cyclin BΔ90 was included in the reaction, large bright membrane structures, probably sheets, were formed, which were interconnected by tubules (Fig. 4 E). This coincided with a drastic reduction of the number of three-way junctions between tubules (Fig. 4 F). Again, real-time imaging showed that three-way junctions converted into sheets (Fig. S3, D and E; and Videos 6 and 7). It seems that initially small sheets form along the tubules, which then merge at three-way junctions. The resulting structures fuse further, so that the number of three-way junctions is ultimately drastically reduced. Collectively, our results show that an ER network is maintained in mitosis, but the tubules are mainly connected by small sheets instead of three-way junctions.
ATL-mediated fusion of ER membranes
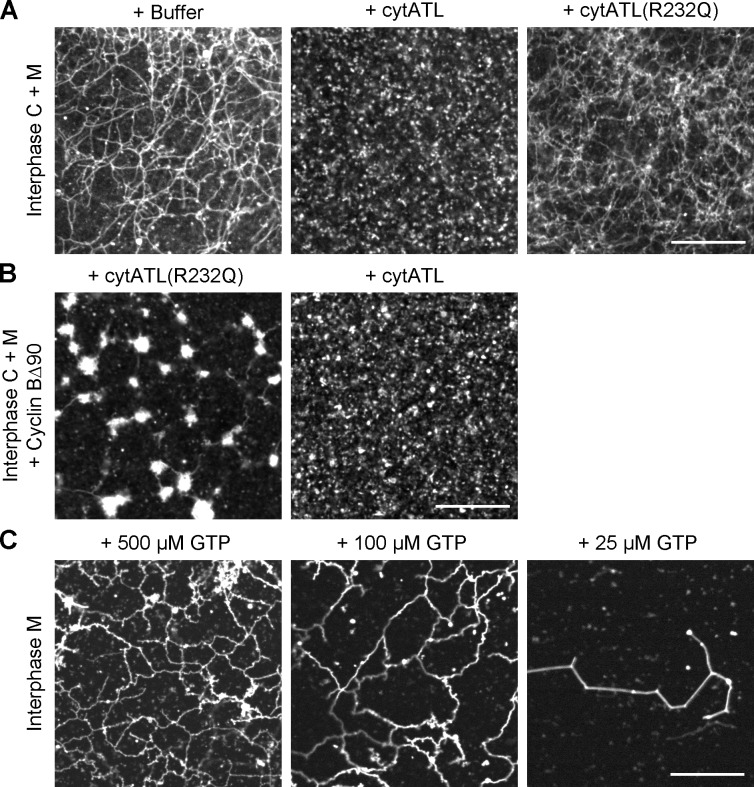
To test the role of ATL in ER network formation, we used a purified cytoplasmic fragment of X. laevis ATL (cytATL) as a dominant-negative reagent. This fragment contains the GTPase domain and the three-helix bundle, but lacks the trans-membrane segments and cytosolic tail of ATL (Bian et al., 2011; Byrnes and Sondermann, 2011). The equivalent fragment of Drosophila ATL inhibits the fusion of proteoliposomes (Orso et al., 2009; Bian et al., 2011; Liu et al., 2012), and the expression of the cytoplasmic domain of human ATL1 in tissue culture cells generates long, nonbranched tubules (Hu et al., 2009). Addition of 2 µM of purified X. laevis cytATL to interphase extracts completely blocked ER network formation (Fig. 5, A and B; the purity of cytATL is shown in Fig. S2). Membranes stained with a fluorescent dye accumulated at the MTOC, indicating that the nonfused membranes can still interact with dynein, but no tubules were seen and no plus end–directed membrane movement was observed. In addition, no NE was formed. As a control, we added cytATL containing a single point mutation (R232Q); the equivalent mutation abolishes the dimerization of human ATL1 (Byrnes and Sondermann, 2011). Addition of X. laevis cytATL(R232Q) had no effect on ER network formation (Fig. 5, C and D; the purity of cytATL(R232Q) is shown in Fig. S2). Inhibition of ATL function by cytATL also blocked ER network formation in mitotic extracts (Fig. 5 F). Again, the mutant fragment had no effect (Fig. 5 E). Thus, the fusion of precursor membranes into a network appears to completely rely on ATL.
Next we tested the effect of ATL inhibition in a network formation reaction with isolated membranes, which consist of vesicles of ∼100 nm diameter (Lourim and Krohne, 1993; Wiese et al., 1997; Dreier and Rapoport, 2000). Wild-type cytATL inhibited ER network formation, regardless of whether the reaction was performed in the presence or absence of membrane-depleted interphase cytosol or in the presence of mitotic cytosol (Fig. 6, A and B; and Fig. S4 A). Again, the mutant fragment had no effect. These results further demonstrate that ATL is required for ER network formation both in interphase and mitosis.
Figure 6.
ATL is required for ER network formation in a fractionated system. (A) Interphase cytosol, light membranes, and an energy regenerating system were mixed and incubated for 30 min with buffer, 2 µM of the cytoplasmic fragment of X. laevis ATL2 (cytATL), or 2 µM of mutant fragment (cytATL(R232Q)). The membranes were stained with octadecyl rhodamine and visualized by confocal microscopy. (B) As in A, but the mitotic state was generated by addition of cyclin BΔ90. (C) Interphase ER network formation with DiOC18 prelabeled light membranes was performed at different GTP concentrations for 15 min at room temperature. Bars, 20 µm.
Further support for this conclusion is provided by experiments in which we varied the GTP concentration in a network formation reaction with isolated membranes (Fig. 6 C). At 500 µM GTP, the network was completely normal, at 100 µM it was slightly reduced, and at 25 µM it almost did not form at all. The half-maximum GTP concentration required for ER network formation was thus about the same as the Km value determined for the purified cytoplasmic domain of human ATL1 (145 µM; Bian et al., 2011).
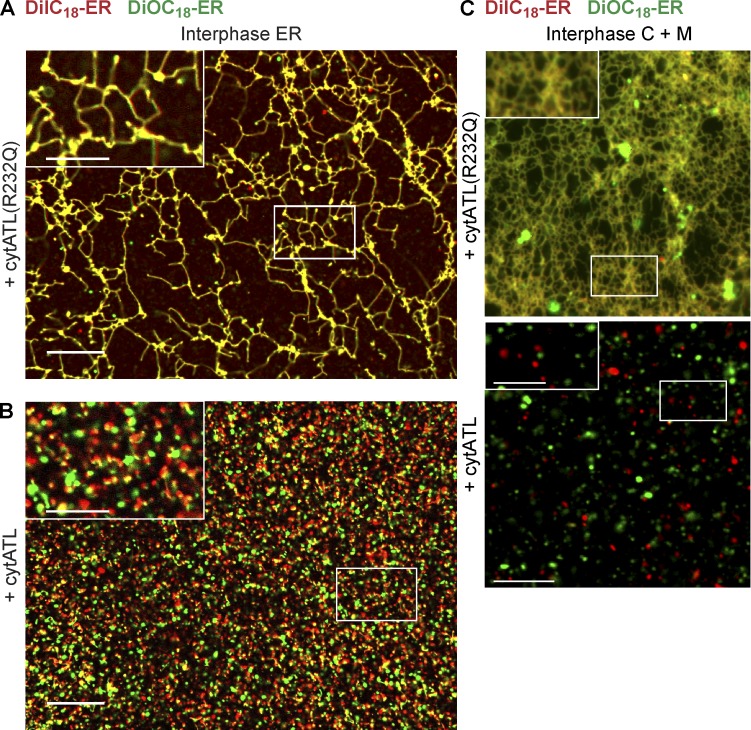
To confirm that cytATL blocks membrane fusion, we assembled an interphase network in a crude extract with starting membranes that were prestained with different fluorescent dyes (red and green). Whereas the membranes mixed when the reaction was performed with the inactive cytATL(R232Q) mutant, resulting in a yellow-colored ER network (Fig. 7 A), the red and green colored membrane structures remained separate in the presence of cytATL (Fig. 7 B). Similar results were obtained when prelabeled red and green membranes were incubated with cytosol in the presence of cytATL or cytATL(R232Q) (Fig. 7 C). Thus, cytATL blocks the mixing of the lipid bilayers, providing strong evidence that ER network formation depends on ATL-mediated membrane fusion.
Figure 7.
ATL mediates the fusion of membranes into an ER network. (A) A crude interphase extract was preincubated for 5 min with 2 µM of an inactive mutant of the cytosolic fragment of ATL (ATL(R232Q)). Then, membranes were added that were separately prestained with DiIC18 (red) and DiOC18 (green). The sample was imaged after 30-min incubation by confocal microscopy with a short exposure time (40 ms). (B) As in A, but with wild-type cytosolic fragment of ATL (cytATL). Insets in A and B show a magnified view of the boxed areas. (C) As in A and B, but with membrane-depleted interphase cytosol and a mixture of light membranes that were prestained with either DiIC18 or DiOC18. Insets show a magnified view of the boxed areas. Bars: (A and B) 20 µm; (A and B, insets) 10 µm; (C) 10 µm; (C, insets) 5 µm.
NE formation requires additional membrane fusion by ER SNAREs
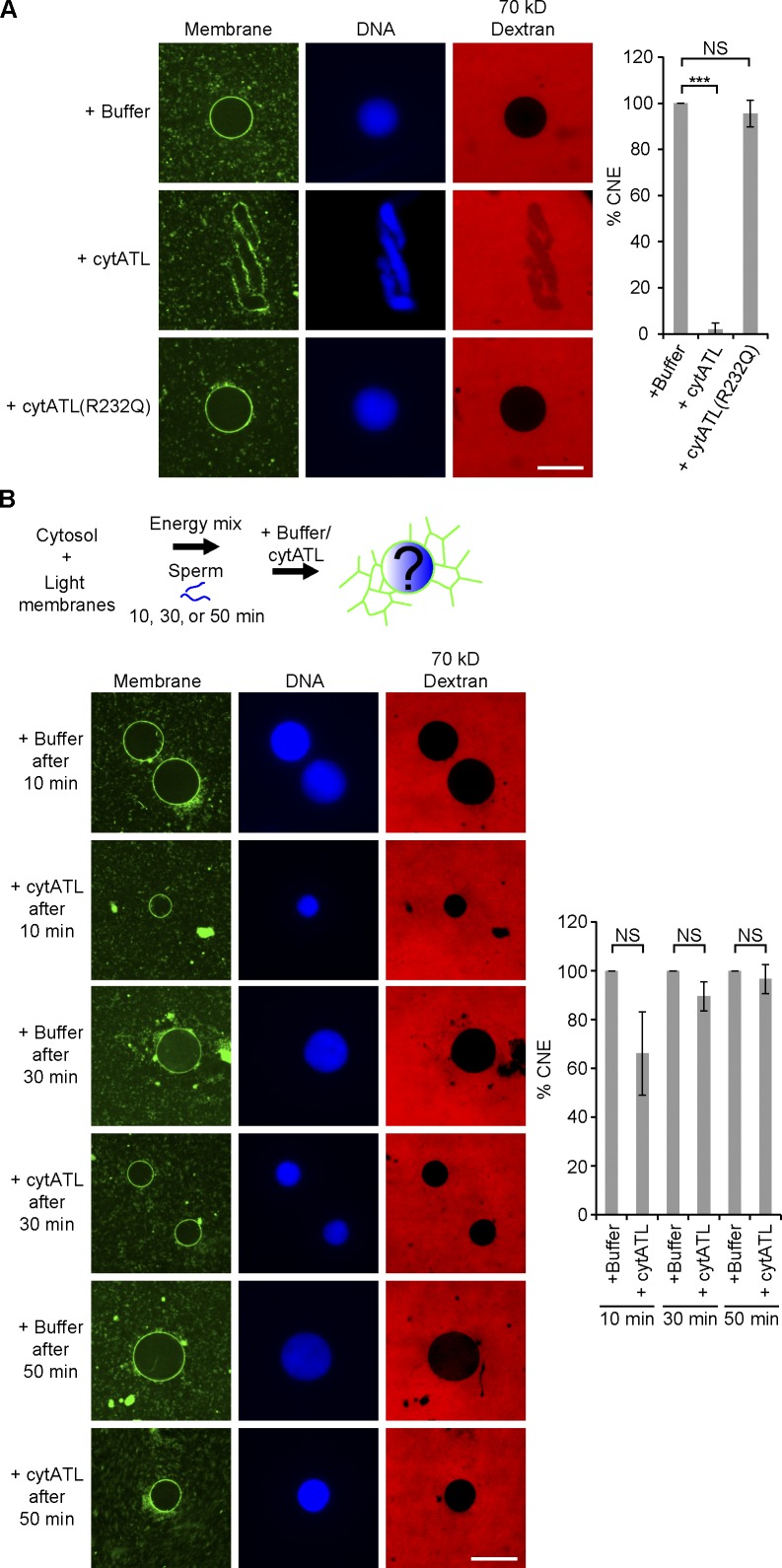
Experiments with crude extracts suggested that ATL was required for NE formation (Fig. 5). To test this more rigorously, we used a partially fractionated system. Lysolecitin-permeabilized sperm was added to reactions containing isolated membranes and cytosol, and NE formation was monitored by the generation of a smooth stained membrane surface on chromatin and by exclusion of fluorescently labeled 70-kD dextran. Control reactions and reactions containing the inactive ATL mutant formed NEs (Fig. 8 A). In contrast, addition of cytATL at the beginning of the reaction blocked formation of intact NEs (Fig. 8 A). Vesicles still accumulated on the chromatin, but did not fuse to form a smooth surface, and 70-kD dextran was not excluded. In addition, sperm chromatin was only partially decondensed, presumably because proteins required for decondensation could not be actively concentrated in the nucleus. Thus, ATL is required for NE formation, likely by promoting fusion of ER vesicles or tubules.
Figure 8.
ATL is required for NE fusion. (A) Interphase cytosol and light membranes were mixed with buffer, 2 µM of the cytoplasmic fragment of X. laevis ATL2 (cytATL), or 2 µM of mutant fragment (cytATL(R232Q)). Demembranated sperm and an energy regenerating system were added. The samples were incubated at room temperature for 1.5 h and mixed with fluorescently labeled 70-kD dextran for 5 min on ice. After fixation with a solution containing Hoechst to stain chromatin and DHCC to stain membranes, the samples were analyzed by confocal microscopy. Bar, 20 µm. The number of closed nuclei (CNE) was counted based on membrane continuity and exclusion of dextran. Fusion is expressed as the percentage of the control. At least 100 nuclei were counted for each sample. The data plotted are the mean ± SD of three independent experiments. ***, P < 0.001; NS (not significant), P > 0.05; Student’s t test. (B) Interphase cytosol, light membranes, sperm, and an energy regenerating system were mixed. After 10, 30, or 50 min of incubation at room temperature, buffer or 2 µM cytATL was added. Samples were fixed after 1.5 h and analyzed by confocal microscopy. Bar, 20 µm. NS, not significant (P > 0.05, Student’s t test).
To test if ATL was required for NE formation at later stages of the reaction, we added cytATL after 10 min, a time point at which the ER network has formed (unpublished data) but the NE membrane has not completely fused around chromatin. In this case, cytATL only had a minor effect on NE formation (Fig. 8 B). Addition of cytATL at even later time points had essentially no effect on NE formation (Fig. 8 B). It therefore appears that another fusion reaction is required to complete envelope formation. However, the growth of the completed NE does seem to require ATL function, as the nuclei remained relatively small (Fig. 8 B).
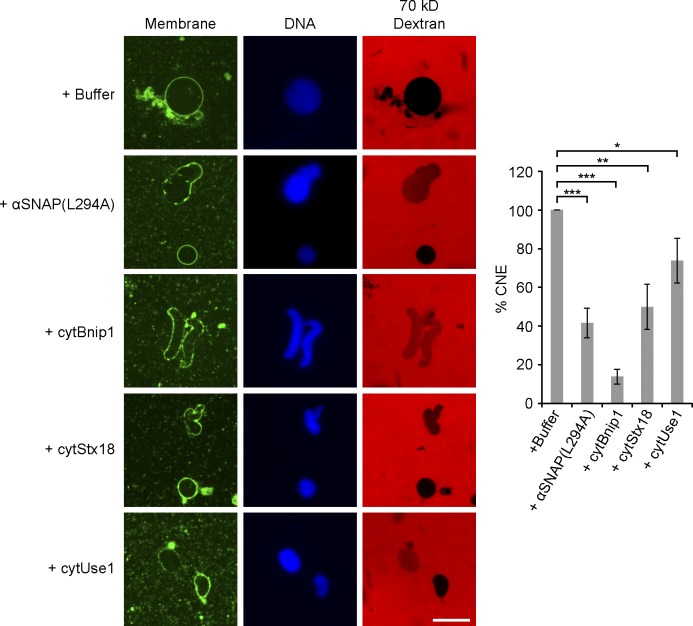
Previous experiments suggested that NSF and its cofactor αSNAP are required for NE assembly (Baur et al., 2007). We confirmed that a purified mutant of αSNAP (αSNAP(L294A)) partially inhibited NE (Fig. 9), but not ER network (Fig. S4 B), formation (the purity of αSNAP(L294A) is shown in Fig. S2). Because NSF and αSNAP generally cooperate with SNARE proteins, we tested whether ER SNARE–mediated membrane fusion is involved in NE formation. We used cytoplasmic domains of SNARE proteins as dominant-negative reagents, as these should interfere with the assembly of endogenous SNARE proteins during fusion. Purified cytosolic fragments of Bnip1 (homologue of yeast Sec20p) and syntaxin 18 (homologue of yeast Ufe1p) partially inhibited NE formation, particularly at early time points (Fig. 9; the purity of the fragments is shown in Fig. S2). A cytoplasmic fragment of the ER SNARE Use1 had a more modest effect (Fig. 9). Even at later time points of the reaction, the nuclei remained smaller than in the control (unpublished data). None of the SNARE fragments had an effect on ER network formation (Fig. S4 B). The addition of αSNAP(L294A) or the SNARE fragments to a preformed ER network blocked NE formation around subsequently added sperm, although they left the ER network intact (Fig. S5). Collectively, these results suggest that NE formation requires both ATL- and ER SNARE–mediated membrane fusion.
Figure 9.
ER SNARE function is required for NE fusion. NE assembly was examined by mixing interphase cytosol, light membranes, sperm, and an energy regenerating system with buffer, 50 µM dominant-negative αSNAP mutant (αSNAP(L294A)), 20 µM of the cytoplasmic fragment of Bnip1 (cytBnip1), 30 µM of the cytoplasmic fragment of syntaxin 18 (cytStx18), or 70 µM of the cytoplasmic fragment of Use1 (cytUse1). Fluorescently labeled dextran was added to detect nuclei with closed NE. The samples were also stained for membranes and DNA. Bar, 20 µm. The percentage of NE fusion was determined as described in Fig. 8 A and the data plotted are the mean ± SD of three independent experiments. *, P < 0.05; **, P < 0.005; and ***, P < 0.001; Student’s t test.
Discussion
Our results suggest how the ER network achieves its inhomogeneous spatial distribution in interphase cells. Using X. laevis egg extracts, we show that the concentration of the ER network at the MTOC depends on the presence of dynamic MTs and is caused by two opposing processes: minus end–directed movement of membranes mediated by cytoplasmic dynein and plus end–directed movement of membrane tubules by association with the growing tips of MTs. Dynein-mediated movement occurs with small vesicles and larger membrane structures, as well as with tubules. Plus end–directed movement was observed only with tubules, not with vesicles, so it is possible that only tubules associate with the tips of growing MTs. In extracts, we observed that membrane fragments move initially toward the minus end of MTs, followed by massive movement of tubules and the entire network in the MT plus end direction. However, at later time points when nuclei were formed, the network was denser around the MTOC, suggesting that dynein-mediated movements ultimately predominate. We also show that in mitosis the association of ER membranes with MTs is strongly reduced, consistent with other data (Niclas et al., 1996; Smyth et al., 2012). As a result, the ER network is more evenly distributed in mitosis than in interphase.
These results extend previous observations, which showed that dynein is involved in ER movement in X. laevis interphase extracts (Allan and Vale, 1991; Allan, 1995; Niclas et al., 1996; Steffen et al., 1997; Lane and Allan, 1999). It was also reported that in the presence of taxol-stabilized MTs, all tubules were moved toward minus ends of MTs (Allan and Vale, 1991). Because the MTs used were not dynamic, the authors could not detect MT plus tip–associated movements. The previous study also found that in mitotic X. laevis extracts no ER membranes associated with taxol-stabilized MTs (Allan and Vale, 1991), consistent with our observations. In tissue culture cells, motor-driven sliding of ER tubules along MTs as well as MT tip–associated movements of tubules have been observed (Waterman-Storer and Salmon, 1998; Grigoriev et al., 2008; Friedman et al., 2010). Dynein-driven movements of ER membranes have been observed in mammalian cells and in Ustilago maydis (Wedlich-Söldner et al., 2002; Woźniak et al., 2009). Interestingly, in mammalian cells, conventional kinesin also plays a role, as its inhibition reduced the outwards movement of ER tubules by ∼50% (Woźniak et al., 2009). In contrast, our results suggest that kinesin does not play a role in X. laevis eggs, as there are no or only few sliding movements toward the plus end of MTs. This is consistent with the observation that kinesin is present on membranes but is activated only at later stages of development (Lane and Allan, 1999).
Our results contribute to the discussion on how ER morphology changes during mitosis (Poteryaev et al., 2005; Anderson and Hetzer, 2007; Puhka et al., 2007, 2012; Lu et al., 2009, 2011). They support the idea that ER tubules are partially converted into sheets during the transition from interphase to mitosis (Poteryaev et al., 2005; Lu et al., 2009, 2011). Many of the three-way tubular junctions are converted into small sheets. These changes occur without contribution from MTs. Whether they are caused by partial inactivation of the reticulons and DP1 remains to be tested.
We found that ER network formation in X. laevis extracts is completely blocked by the addition of a cytoplasmic fragment of ATL, but not by a mutant fragment that cannot form ATL dimers. This is true for both interphase and mitotic extracts. The wild-type ATL fragment blocks membrane fusion, as confirmed by the observation that it prevents the mixing of two populations of differently colored membranes. Importantly, the ATL fragment also completely inhibited ER network formation when the reaction was performed with membranes alone, in the absence of cytosol; the vesicles remained small and did not fuse with other small vesicles. Given that the same cytoplasmic ATL fragment inhibits the fusion of proteoliposomes that contain only ATL (Bian et al., 2011; Liu et al., 2012), it appears that ER network formation relies on ATL-mediated membrane fusion. Previous experiments with ATL antibodies already hinted at a role of ATL in ER network formation (Hu et al., 2009), but its specific function remained unclear and the lengthy preincubation time of membranes and antibodies made the conclusions less convincing. In addition, these experiments did not address the role of ATL during mitosis. The conclusion that ATL-mediated membrane fusion is required for ER network formation is consistent with the fact that depletion of ATLs leads to long, nonbranched ER tubules in tissue culture cells and ER fragmentation in Drosophila melanogaster (Hu et al., 2009; Orso et al., 2009). In crude X. laevis extracts and in the fractionated system, ATL inhibition results instead in membrane structures that seem to be small sheets and small vesicles, respectively, perhaps because these are the precursors to ER network formation in these systems or because ATL inhibition is more complete. ATL may not only be a fusogen but may also regulate the function of the reticulons and DP1. This could explain why inactivation of ATL in crude extracts prevents the formation of tubules from sheet-like structures.
The role of Rab proteins in ER fusion remains uncertain. Rab proteins were previously implicated on the basis of the inhibitory effect of GDP dissociation inhibitor on network formation (Audhya et al., 2007; English and Voeltz, 2013b). However, no rescue experiments with Rab-GTP were reported, so it is possible that GDP dissociation inhibitor inhibited network formation indirectly. In addition, removal of Rab10 from the membranes did not entirely correlate with inhibition of ER network formation. Support for ATL being the exclusive ER fusogen comes from the observation that ATL inhibition blocks all network formation and that the half-maximum concentration of GTP required for network formation is about the same as the Km value of purified ATL1220 (145 µM), whereas Rab proteins generally have much lower GTP dissociation constants (Simon et al., 1996).
We show that NE formation requires ATL-mediated fusion of ER membranes, consistent with previous conclusions that generation of an ER network is a prerequisite (Anderson and Hetzer, 2007). However, the previous study proposed that no additional fusion reaction is required for NE formation, whereas we found that ER SNARE function is likely needed in addition. Our results are consistent with results that showed an inhibitory effect of dominant-negative mutants of NSF or αSNAP on NE formation (Baur et al., 2007). We now extend these findings by demonstrating that dominant-negative constructs of ER SNAREs also inhibit NE formation. The results lead to a model in which NE formation would begin with rapid ATL-mediated fusion of ER membranes into a network. ER tubules would then use ER SNAREs to fuse with a different vesicle population, resulting in the formation of flat membrane sheets on the chromatin surface and ultimately in a continuous spherical envelope surface (Dreier and Rapoport, 2000). The nature of this vesicle population remains unclear, but the requirement of distinct membrane populations for NE formation is consistent with previous results (Collas and Poccia, 1996). Finally, the ER network would provide additional membrane material, allowing the NE to grow in diameter. Whether this growth phase also requires ER SNAREs remains to be clarified.
Materials and methods
Plasmids used in this study
Maltose-binding protein–cyclin BΔ90 (MBP-cyclin BΔ90) lacking the N-terminal destruction box of cyclin B was a gift from R. King (Harvard Medical School, Boston, MA). Bovine αSNAP cloned into pET28b (EMD Millipore) was provided by R. Jahn (Max Planck Institute, Munich, Germany). Codon-optimized cytATL (residues 1–462) of X. laevis ATL, a protein most closely related to human ATL2, was synthesized (GenScript) and cloned into pGEX-4T-3 (GE Healthcare). X. laevis Bnip1 (GenBank accession no. BC054986), Syntaxin 18 (Stx18; GenBank accession no. BC092152), and Use1 (GenBank accession no. BC110785) cDNA clones were purchased from Thermo Fisher Scientific. The cytoplasmic regions of these proteins were predicted by TMHMM (version 2.0; Krogh et al., 2001). CytBnip1 (residues 1–202), cytStx18 (residues 1–303), and cytUse1 (1–231) were amplified and cloned into pET28b. Point mutations in the respective constructs were generated using the QuickChange Site-Directed Mutagenesis kit (Agilent Technologies). All plasmid inserts were confirmed by DNA sequence analysis.
Protein purification and probes
All proteins were expressed in BL21 CodonPlus (DE3) RIPL (Agilent Technologies) or BL21 Escherichia coli strains (New England Biolabs, Inc.). The CC1 fragment of dynactin was recombinantly expressed and purified as previously described (King et al., 2003). In brief, CC1 was expressed in BL21 E. coli. Proteins in an E. coli lysate were precipitated with 25% (wt/vol) NH4SO4. The pellet was dissolved in phosphate buffer, pH 7.2, containing 2 mM DTT, 1 mM EDTA, 0.1 mM PMSF, and 0.75 M KCl, heated at 100°C for 3 min, and clarified by centrifugation. The supernatant was subjected to anion exchange chromatography (monoQ-HR; GE Healthcare) and peak fractions were collected. A fusion of human EB1 and His-tagged GFP (EB1-GFP) was purified as described previously (Petry et al., 2011). EB-1-GFP was expressed in BL21 E. coli and purified using Ni-NTA (QIAGEN) affinity chromatography. The protein was further purified by size-exclusion chromatography (Superdex 200; GE Healthcare). CytATL and cytATL(R232Q) were purified as described previously with minor changes (Bian et al., 2011). In brief, the protein was isolated using glutathione Sepharose 4B resin (GE Healthcare) and the GST tag was removed by incubation with thrombin (MP Biomedicals) at 4°C overnight. The protein was further purified using anion-exchange chromatograph (HiTrap Q HP; GE Healthcare) and gel filtration (Superdex 200). MBP-cyclin BΔ90 was purified using amylose resin (New England Biolabs, Inc.). αSNAP(L294A), cytBnip1, cytStx18, and cytUse1 were purified using Ni-NTA chromatography (Thermo Fisher Scientific). The proteins were further purified by ion-exchange chromatography on either HiTrap Q HP for αSNAP(L294A) and cytStx18 or on HiTrap SP HP for cytBnip1 and cytUse1, followed by gel filtration on Superdex 200. The proteins were concentrated and subjected to centrifugation at 100,000 g for 10 min at 4°C before snap freezing in liquid nitrogen. The final storage buffer for all proteins was 20 mM Hepes, pH 7.5, 150 mM KCl, 250 mM sucrose, and 1 mM DTT. Calf brain tubulin was purified as described previously (Williams and Lee, 1982). In brief, a calf brain homogenate supplemented with ATP, GTP, and 30% glycerol was subjected to two cycles of MT polymerization at 37°C, followed by MT pelleting and subsequent depolymerization of MTs in ice-cold buffer. Tubulin was further purified by cation-exchange chromatography using a phosphocellulose column (P11 Cellulose Phosphate; Whatman).
Tubulin was fluorescently labeled as previously described (Hyman et al., 1991). In brief, tubulin was polymerized at 37°C in the presence of 10% dimethyl sulfoxide. MTs were isolated by pelleting through a 60% glycerol cushion. After resuspension, they were incubated with Alexa Fluor 488 NHS-ester (Invitrogen) at a 10:1 dye/tubulin ratio for 1 h at 37°C. Labeled MTs were isolated by pelleting though a 60% glycerol cushion, depolymerized in ice-cold buffer, and clarified by centrifugation. Labeled tubulin was supplemented with 1 mM GTP and polymerized at 37°C for 30 min in the presence of 30% glycerol. MTs were isolated again by pelleting though a 60% glycerol cushion, and then depolymerized in ice-cold buffer, aliquoted, snap frozen in liquid nitrogen, and stored at −80°C.
Preparation of crude X. laevis egg extract
Metaphase-arrested crude extracts from X. laevis eggs (CSF extract) were prepared as described previously (Desai et al., 1999), with the following modifications. Eggs were dejellyed at 18°C. All subsequent steps were performed at 4°C. The eggs were washed several times with 10 mM Hepes/KOH, pH 7.7, 0.1 M KCl, 1 mM MgCl2, 0.1 mM CaCl2, and 50 mM sucrose. They were then washed repeatedly in the same buffer containing 5 mM EGTA and 2 mM MgCl2. After addition of protease inhibitors (LPC: 10 µg/ml leupeptin, 10 µg/ml pepstatin, and 10 µg/ml chymostatin [all from Sigma-Aldrich]), the eggs were subjected to a crush spin. The extract was used within 5 h. Note that no cytochalasin B or energy regenerating system was added.
For the experiment in Fig. S3 C, the interphase extract was prepared in the absence of EGTA and presence of cycloheximide (Sigma-Aldrich). In brief, the interphase crude extract was obtained by crushing eggs in lysis buffer (50 mM Hepes/KOH, pH 7.7, 50 mM KCl, 2.5 mM MgCl2, and 250 mM sucrose) containing 50 µg/ml cycloheximide and protease inhibitors (Newmeyer and Wilson, 1991; Dreier and Rapoport, 2000; Chan and Forbes, 2006; Voeltz et al., 2006).
Interphase extracts were converted to mitotic extracts by addition of 0.1 mg/ml MBP-cyclin BΔ90, followed by a 40-min incubation at room temperature (Murray et al., 1989). The mitotic status was verified by immunoblotting with anti-MPM2 antibodies (EMD Millipore). Alternatively, chromosome condensation was followed, as described previously (Maresca and Heald, 2006). The interphase cytosol was incubated with buffer or MBP-cyclin BΔ90 and 20× energy regenerating system (2.5 mg/ml creatine phosphokinase, 250 mM phosphocreatine, 50 mM ATP, and 10 mM GTP) for 40 min at room temperature. Then, demembranated sperm (105 sperms/µl) was added and the reactions were further incubated at room temperature for 60 min. Aliquots were withdrawn at specific time points and fixed with a solution containing10 µg/ml Hoechst 33258 (Invitrogen). Chromosomes were visualized using a fluorescence microscope.
Fractionation of X. laevis egg extracts
To prepare cytosol and membranes, a crude extract prepared in the absence of EGTA, but presence of 50 µg/ml cycloheximide and 10 µg/ml cytochalasin B (Sigma-Aldrich), was supplemented with 1 mM DTT and centrifuged at 200,000 g at 4°C for 1 h. The lipid layer was carefully aspirated and the cytosol was further centrifuged at 200,000 g at 4°C for 30 min. A light membrane layer was separated from a heavier membrane layer and washed with ELB200 (50 mM Hepes/KOH, pH 7.5, 200 mM KCl, 2.5 mM MgCl2, and 250 mM sucrose) containing 1 mM DTT and LPC. The membranes were sedimented at 40,000 g for 20 min at 4°C and resuspended in the same buffer in a volume corresponding to 1/10 or 1/20 of that of the cytosol. Both cytosol and membranes were snap frozen in liquid nitrogen and stored in aliquots at −80°C.
Prelabeled light membranes were prepared as described previously (Hetzer et al., 2000). In brief, the light membrane fraction was incubated with 200 µg/ml DiOC18 (3,3′-dioctadecyloxacarbocyanine perchlorate; Invitrogen) for 20 min on ice. Subsequently, it was washed with 50 vol of ELB200 plus 1 mM DTT and LPC and centrifuged at 40,000 g for 20 min at 4°C. Aliquots of the membranes were snap frozen in liquid nitrogen and stored at −80°C.
Demembranated sperm preparation
Sperm chromatin was prepared as described previously (Murray, 1991). Testes isolated from male frogs were rinsed first in cold MMR buffer (5 mM Hepes/NaOH, pH 7.8, 100 mM NaCl, 2 mM KCl, 1 mM MgCl2, 2 mM CaCl2, and 0.1 mM EDTA) and then in NPB buffer (15 mM Hepes/KOH, pH 7.4, 1 mM EDTA, pH 8.0, 0.5 mM spermidine trihydrochloride, 0.2 mM spermine tetrahydrochloride, 1 mM DTT, 10 µg/ml leupeptin, 0.3 mM PMSF, and 250 mM sucrose). A razor blade was used to macerate the testes. Fragments of testes were resuspended in NPB and filtered through cheesecloth. Sperm was obtained by spinning the filtered solution at 3,000 rpm for 10 min. After further washing steps with NPB, the sperm was resuspended in 1 ml NPB. For demembranation, lysolecithin was added to 0.5 mg/ml for 5 min at room temperature. Demembranated sperm chromatin was rinsed with NPB containing 3% BSA followed by NPB containing 0.3% BSA. The sample was finally resuspended in NPB in buffer containing 0.3% BSA and 30% (wt/vol) glycerol, but lacking PMSF. Sperm was counted using a hemocytometer and adjusted to a density of 108/µl. Aliquots were snap frozen in liquid nitrogen and stored at −80°C.
Assembly of interphase ER and MT asters in crude extracts
To visualize ER membranes, the CSF extract was prelabeled with 100 µg/ml DiIC18 (1,1’-dioctadecyl-3,3,3′,3′-tetramethylindocarbocyanine perchlorate) or DiOC18 and incubated for 45 min at 18°C. Reactions were prepared by adding 1:10 vol of prelabeled CSF extract to a fresh extract. Alexa fluor 488– or Rhodamine-labeled calf brain tubulin was included at 50 µg/ml concentration to follow MT assembly. Recombinant EB1-GFP at 100 nM concentration was added in experiments where MT plus ends were visualized. The reactions were supplemented with 0.4 mM CaCl2 and incubated at 18°C for 7 min to release the metaphase arrest, and then placed on ice for 2 min, followed by addition of demembrenated sperm (105/µl) to nucleate MT asters. An aliquot of the reaction (8 µl) was placed between two 22 × 22-mm PEG-pretreated glass coverslips. The samples were sealed and mounted over a metal slide with a 20-mm-diameter hole using VALAP (1:1:1 mix of vaseline, paraffin, and lanolin). They were imaged immediately using a spinning disk confocal fluorescence microscope.
Mitotic ER was visualized in the same way, except that CaCl2 was omitted to preserve the cell cycle arrest. Alternatively, the CSF extract was driven into interphase by addition of 0.4 mM CaCl2, and then supplemented with 0.1 mg/ml of cyclin BΔ90 to induce mitosis (Murray et al., 1989).
In vitro ER network formation using isolated cytosol and membranes
ER network formation from isolated cytosol and membranes was performed as described previously with minor modifications (Dreier and Rapoport, 2000). In general, cytosol and membranes were mixed at a 20:1 volume ratio. Protein or buffer were added in ∼0.5 µl to the desired concentration to prevent excess dilution of the egg extract. After incubation on ice for 5 min, a 20× energy regenerating system was added and the reaction was further incubated for 30 min at room temperature. An aliquot of the reaction was mixed with octadecyl rhodamine (Invitrogen) at 10 µg/ml and applied to a passivated no. 1.5 coverslip sandwich sealed with VALAP. For ER networks formed from interphase membranes, 0.5–1 µl of DiOC18-prelabled membranes were resuspended in 10 µl ELB200 and 1 mM DTT. After addition of protein or buffer, a 20× regenerating system or GTP was added to concentrations indicated in Fig. 6 C and the sample was incubated at room temperature for 15 min. An aliquot of the reaction was directly applied to a passivated no. 1.5 coverslip sandwich sealed with VALAP.
Coverslip passivation
Glass coverslips were sonicated in isopropanol for 5 min, rinsed with water, sonicated in water for 5 min, air dried, and etched using an oxygen plasma etcher (500-II Plasma System; Technics) for 4 min at 200-W power. One side of the glass coverslip was then layered with a 100-mg/ml ethanol solution of 5,000 MW methoxy-polyethylene glycol-silane (Jemkem Technology USA) and incubated at 60°C for at least 5 h. Coverslips were rinsed with ethanol, rinsed with water, air dried in a clean room, and stored in a sealed container until use.
Fluorescence microscopy
All samples were visualized using a spinning disk confocal head (CSU-X1; Yokogawa Corporation of America) with Borealis modification (Spectral Applied Research) and a quad bandpass 405/491/561/642 dichroic mirror (Semrock). The confocal was mounted on a Ti inverted microscope (Nikon) equipped with a 60× Plan Apo NA 1.4 oil immersion objective or 60× CFI Plan Apochromat NA 1.2 water immersion objective and the Perfect Focus System for continuous maintenance of focus (Nikon). Green fluorescence images were collected using a 491-nm solid-state laser controlled with an AOTF (Spectral Applied Research) and ET525/50 emission filter (Chroma Technology Corp.). Red fluorescence images were collected using a 561-nm solid-state laser controlled with an AOTF (Spectral Applied Research) and ET620/60 emission filter (Chroma Technology Corp.). Blue fluorescence was excited with a 395/25 filter and a wide-field Hg halide light source and collected with a 460/25 filter through the confocal head. All images were acquired with a cooled CCD camera (ORCA AG; Hamamatsu Photonics) controlled with MetaMorph software (version 7.0; Molecular Devices) and archived using ImageJ (National Institutes of Health) and Photoshop CS5 (Adobe). In some cases, linear adjustments were applied to enhance the contrast of images using levels in the image adjustments function of ImageJ and Photoshop.
Nuclear assembly
Nuclear assembly reactions were performed as described previously (Newmeyer and Wilson, 1991). Typically, 19 µl of the interphase cytosol, 1 µl of membranes, and ∼0.5–1 µl of protein or buffer were mixed on ice for 5 min. A 20× energy regenerating system and 0.4 µl of sperm (105 demembranated sperms/µl) were added and the reaction was incubated at room temperature for 1.5 h. Alternatively, an ER network was first formed for 20 min at room temperature using interphase cytosol, membranes, and a 20× energy regenerating system. About 0.5–1 µl of protein or buffer were added and an aliquot was saved for visualization of the ER network. Sperm chromatin was then added and the reaction was incubated at room temperature. After 1.5 h, 2 µl of the mixture was mixed with a 70-kD rhodamine B–labeled dextran (Invitrogen), which was purified using gel filtration (Superdex 200). After a 5-min incubation on ice, the nuclei were fixed with a solution containing 10 mM Hepes, pH 7.5, 200 mM sucrose, 10 µg/ml Hoechst 33258 (Invitrogen), 12% (wt/vol) paraformaldehyde, and 10 µg/ml DHCC (3,3-dihexyloxacarbocyanine iodide; Sigma-Aldrich). The number of closed nuclei was counted based on a continuous DHCC staining and exclusion of a 70-kD dextran. At least 100 nuclei were counted for NE fusion, and the percentage of closed nuclei was determined relative to a control in an independent experiment. The data plotted are the mean ± SD of three independent experiments.
Online supplemental material
Fig. S1 shows that the ER network formed in the presence of nocodazole is evenly distributed. Fig. S2 shows the purity of the proteins used in this study. Fig. S3 shows a mitotic ER network assembled using the fractionated cytosol and membranes. Fig. S4 demonstrates that cytATL, but not ER SNARE mutants, inhibit interphase ER network formation. Fig. S5 shows that ER SNARE mutants prevent NE assembly from a preformed ER network. Video 1 shows the formation of an interphase MT aster and Video 2 shows the simultaneous interphase ER network formation. Video 3 shows the minus-directed movement of ER tubules along MTs. Video 4 demonstrates that extension of ER tubules is associated with the plus ends of growing MTs. Videos 5–7 show the tubule-to-sheet conversion of the ER during the transition from interphase to mitosis. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201308001/DC1. Additional data are available in the JCB DataViewer at http://dx.doi.org/10.1083/jcb.201308001.dv.
Supplementary Material
Acknowledgments
We thank the Nikon imaging center at Harvard Medical School, specifically Jennifer Waters and Josh Rosenberg, for invaluable help with imaging. We thank Aaron Groen, Phuong Nguyen, and Keisuke Ishihara for help with X. laevis extracts and MT preparation, and Julie Huang for advice on tubulin purification. We are grateful to Robin Klemm, Alex Stein, and Tom Shemesh for suggestions and to Adrian Salic and Robin Klemm for critical reading of the manuscript.
T.A. Rapoport is a Howard Hughes Medical Institute Investigator. T.J. Mitchison was supported by a grant from the National Institutes of Health (GM 39565).
Footnotes
Abbreviations used in this paper:
- ATL
- atlastin
- CC1
- coiled-coil region 1
- CSF
- cytostatic factor
- MT
- microtubule
- MTOC
- MT-organizing center
- NE
- nuclear envelope
References
- Allan V. 1995. Protein phosphatase 1 regulates the cytoplasmic dynein-driven formation of endoplasmic reticulum networks in vitro. J. Cell Biol. 128:879–891 10.1083/jcb.128.5.879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allan V.J., Vale R.D. 1991. Cell cycle control of microtubule-based membrane transport and tubule formation in vitro. J. Cell Biol. 113:347–359 10.1083/jcb.113.2.347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson D.J., Hetzer M.W. 2007. Nuclear envelope formation by chromatin-mediated reorganization of the endoplasmic reticulum. Nat. Cell Biol. 9:1160–1166 10.1038/ncb1636 [DOI] [PubMed] [Google Scholar]
- Audhya A., Desai A., Oegema K. 2007. A role for Rab5 in structuring the endoplasmic reticulum. J. Cell Biol. 178:43–56 10.1083/jcb.200701139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baur T., Ramadan K., Schlundt A., Kartenbeck J., Meyer H.H. 2007. NSF- and SNARE-mediated membrane fusion is required for nuclear envelope formation and completion of nuclear pore complex assembly in Xenopus laevis egg extracts. J. Cell Sci. 120:2895–2903 10.1242/jcs.010181 [DOI] [PubMed] [Google Scholar]
- Bian X., Klemm R.W., Liu T.Y., Zhang M., Sun S., Sui X., Liu X., Rapoport T.A., Hu J. 2011. Structures of the atlastin GTPase provide insight into homotypic fusion of endoplasmic reticulum membranes. Proc. Natl. Acad. Sci. USA. 108:3976–3981 10.1073/pnas.1101643108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrnes L.J., Sondermann H. 2011. Structural basis for the nucleotide-dependent dimerization of the large G protein atlastin-1/SPG3A. Proc. Natl. Acad. Sci. USA. 108:2216–2221 10.1073/pnas.1012792108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan R.C., Forbes D.I. 2006. In vitro study of nuclear assembly and nuclear import using Xenopus egg extracts. Methods Mol. Biol. 322:289–300 10.1007/978-1-59745-000-3_20 [DOI] [PubMed] [Google Scholar]
- Chen S., Novick P., Ferro-Novick S. 2013. ER structure and function. Curr. Opin. Cell Biol. 25:428–433 10.1016/j.ceb.2013.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collas P., Poccia D. 1996. Distinct egg membrane vesicles differing in binding and fusion properties contribute to sea urchin male pronuclear envelopes formed in vitro. J. Cell Sci. 109:1275–1283 [DOI] [PubMed] [Google Scholar]
- Desai A., Murray A., Mitchison T.J., Walczak C.E. 1999. The use of Xenopus egg extracts to study mitotic spindle assembly and function in vitro. Methods Cell Biol. 61:385–412 10.1016/S0091-679X(08)61991-3 [DOI] [PubMed] [Google Scholar]
- Dreier L., Rapoport T.A. 2000. In vitro formation of the endoplasmic reticulum occurs independently of microtubules by a controlled fusion reaction. J. Cell Biol. 148:883–898 10.1083/jcb.148.5.883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Y., Ferro-Novick S., Novick P. 2004. Dynamics and inheritance of the endoplasmic reticulum. J. Cell Sci. 117:2871–2878 10.1242/jcs.01286 [DOI] [PubMed] [Google Scholar]
- English A.R., Voeltz G.K. 2013a. Endoplasmic reticulum structure and interconnections with other organelles. Cold Spring Harb. Perspect. Biol. 5:a013227 10.1101/cshperspect.a013227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- English A.R., Voeltz G.K. 2013b. Rab10 GTPase regulates ER dynamics and morphology. Nat. Cell Biol. 15:169–178 10.1038/ncb2647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman J.R., Webster B.M., Mastronarde D.N., Verhey K.J., Voeltz G.K. 2010. ER sliding dynamics and ER-mitochondrial contacts occur on acetylated microtubules. J. Cell Biol. 190:363–375 10.1083/jcb.200911024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goyal U., Blackstone C. 2013. Untangling the web: mechanisms underlying ER network formation. Biochim. Biophys. Acta. 1833:2492–2498 10.1016/j.bbamcr.2013.04.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigoriev I., Gouveia S.M., van der Vaart B., Demmers J., Smyth J.T., Honnappa S., Splinter D., Steinmetz M.O., Putney J.W., Jr, Hoogenraad C.C., Akhmanova A. 2008. STIM1 is a MT-plus-end-tracking protein involved in remodeling of the ER. Curr. Biol. 18:177–182 10.1016/j.cub.2007.12.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetzer M., Bilbao-Cortés D., Walther T.C., Gruss O.J., Mattaj I.W. 2000. GTP hydrolysis by Ran is required for nuclear envelope assembly. Mol. Cell. 5:1013–1024 10.1016/S1097-2765(00)80266-X [DOI] [PubMed] [Google Scholar]
- Hu J., Shibata Y., Zhu P.P., Voss C., Rismanchi N., Prinz W.A., Rapoport T.A., Blackstone C. 2009. A class of dynamin-like GTPases involved in the generation of the tubular ER network. Cell. 138:549–561 10.1016/j.cell.2009.05.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyman A., Drechsel D., Kellogg D., Salser S., Sawin K., Steffen P., Wordeman L., Mitchison T. 1991. Preparation of modified tubulins. Methods Enzymol. 196:478–485 10.1016/0076-6879(91)96041-O [DOI] [PubMed] [Google Scholar]
- Juwana J.P., Henderikx P., Mischo A., Wadle A., Fadle N., Gerlach K., Arends J.W., Hoogenboom H., Pfreundschuh M., Renner C. 1999. EB/RP gene family encodes tubulin binding proteins. Int. J. Cancer. 81:275–284 [DOI] [PubMed] [Google Scholar]
- King S.J., Brown C.L., Maier K.C., Quintyne N.J., Schroer T.A. 2003. Analysis of the dynein-dynactin interaction in vitro and in vivo. Mol. Biol. Cell. 14:5089–5097 10.1091/mbc.E03-01-0025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogh A., Larsson B., von Heijne G., Sonnhammer E.L. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305:567–580 10.1006/jmbi.2000.4315 [DOI] [PubMed] [Google Scholar]
- Lane J.D., Allan V.J. 1999. Microtubule-based endoplasmic reticulum motility in Xenopus laevis: activation of membrane-associated kinesin during development. Mol. Biol. Cell. 10:1909–1922 10.1091/mbc.10.6.1909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T.Y., Bian X., Sun S., Hu X., Klemm R.W., Prinz W.A., Rapoport T.A., Hu J. 2012. Lipid interaction of the C terminus and association of the transmembrane segments facilitate atlastin-mediated homotypic endoplasmic reticulum fusion. Proc. Natl. Acad. Sci. USA. 109:E2146–E2154 10.1073/pnas.1208385109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lourim D., Krohne G. 1993. Membrane-associated lamins in Xenopus egg extracts: identification of two vesicle populations. J. Cell Biol. 123:501–512 10.1083/jcb.123.3.501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu L., Ladinsky M.S., Kirchhausen T. 2009. Cisternal organization of the endoplasmic reticulum during mitosis. Mol. Biol. Cell. 20:3471–3480 10.1091/mbc.E09-04-0327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu L., Ladinsky M.S., Kirchhausen T. 2011. Formation of the postmitotic nuclear envelope from extended ER cisternae precedes nuclear pore assembly. J. Cell Biol. 194:425–440 10.1083/jcb.201012063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maresca T.J., Heald R. 2006. Methods for studying spindle assembly and chromosome condensation in Xenopus egg extracts. Methods Mol. Biol. 322:459–474 10.1007/978-1-59745-000-3_33 [DOI] [PubMed] [Google Scholar]
- Mitchison T.J., Nguyen P., Coughlin M., Groen A.C. 2013. Self-organization of stabilized microtubules by both spindle and midzone mechanisms in Xenopus egg cytosol. Mol. Biol. Cell. 24:1559–1573 10.1091/mbc.E12-12-0850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison E.E., Wardleworth B.N., Askham J.M., Markham A.F., Meredith D.M. 1998. EB1, a protein which interacts with the APC tumour suppressor, is associated with the microtubule cytoskeleton throughout the cell cycle. Oncogene. 17:3471–3477 10.1038/sj.onc.1202247 [DOI] [PubMed] [Google Scholar]
- Murray A.W. 1991. Cell cycle extracts. Methods Cell Biol. 36:581–605 10.1016/S0091-679X(08)60298-8 [DOI] [PubMed] [Google Scholar]
- Murray A.W., Solomon M.J., Kirschner M.W. 1989. The role of cyclin synthesis and degradation in the control of maturation promoting factor activity. Nature. 339:280–286 10.1038/339280a0 [DOI] [PubMed] [Google Scholar]
- Newmeyer D.D., Wilson K.L. 1991. Egg extracts for nuclear import and nuclear assembly reactions. Methods Cell Biol. 36:607–634 10.1016/S0091-679X(08)60299-X [DOI] [PubMed] [Google Scholar]
- Niclas J., Allan V.J., Vale R.D. 1996. Cell cycle regulation of dynein association with membranes modulates microtubule-based organelle transport. J. Cell Biol. 133:585–593 10.1083/jcb.133.3.585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niethammer P., Kueh H.Y., Mitchison T.J. 2008. Spatial patterning of metabolism by mitochondria, oxygen, and energy sinks in a model cytoplasm. Curr. Biol. 18:586–591 10.1016/j.cub.2008.03.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orso G., Pendin D., Liu S., Tosetto J., Moss T.J., Faust J.E., Micaroni M., Egorova A., Martinuzzi A., McNew J.A., Daga A. 2009. Homotypic fusion of ER membranes requires the dynamin-like GTPase atlastin. Nature. 460:978–983 10.1038/nature08280 [DOI] [PubMed] [Google Scholar]
- Petry S., Pugieux C., Nédélec F.J., Vale R.D. 2011. Augmin promotes meiotic spindle formation and bipolarity in Xenopus egg extracts. Proc. Natl. Acad. Sci. USA. 108:14473–14478 10.1073/pnas.1110412108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poteryaev D., Squirrell J.M., Campbell J.M., White J.G., Spang A. 2005. Involvement of the actin cytoskeleton and homotypic membrane fusion in ER dynamics in Caenorhabditis elegans. Mol. Biol. Cell. 16:2139–2153 10.1091/mbc.E04-08-0726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puhka M., Vihinen H., Joensuu M., Jokitalo E. 2007. Endoplasmic reticulum remains continuous and undergoes sheet-to-tubule transformation during cell division in mammalian cells. J. Cell Biol. 179:895–909 10.1083/jcb.200705112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puhka M., Joensuu M., Vihinen H., Belevich I., Jokitalo E. 2012. Progressive sheet-to-tubule transformation is a general mechanism for endoplasmic reticulum partitioning in dividing mammalian cells. Mol. Biol. Cell. 23:2424–2432 10.1091/mbc.E10-12-0950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reck-Peterson S.L., Yildiz A., Carter A.P., Gennerich A., Zhang N., Vale R.D. 2006. Single-molecule analysis of dynein processivity and stepping behavior. Cell. 126:335–348 10.1016/j.cell.2006.05.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross J.L., Wallace K., Shuman H., Goldman Y.E., Holzbaur E.L. 2006. Processive bidirectional motion of dynein-dynactin complexes in vitro. Nat. Cell Biol. 8:562–570 10.1038/ncb1421 [DOI] [PubMed] [Google Scholar]
- Shibata Y., Voss C., Rist J.M., Hu J., Rapoport T.A., Prinz W.A., Voeltz G.K. 2008. The reticulon and DP1/Yop1p proteins form immobile oligomers in the tubular endoplasmic reticulum. J. Biol. Chem. 283:18892–18904 10.1074/jbc.M800986200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata Y., Hu J., Kozlov M.M., Rapoport T.A. 2009. Mechanisms shaping the membranes of cellular organelles. Annu. Rev. Cell Dev. Biol. 25:329–354 10.1146/annurev.cellbio.042308.113324 [DOI] [PubMed] [Google Scholar]
- Shibata Y., Shemesh T., Prinz W.A., Palazzo A.F., Kozlov M.M., Rapoport T.A. 2010. Mechanisms determining the morphology of the peripheral ER. Cell. 143:774–788 10.1016/j.cell.2010.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon I., Zerial M., Goody R.S. 1996. Kinetics of interaction of Rab5 and Rab7 with nucleotides and magnesium ions. J. Biol. Chem. 271:20470–20478 10.1074/jbc.271.34.20470 [DOI] [PubMed] [Google Scholar]
- Smyth J.T., Beg A.M., Wu S., Putney J.W., Jr, Rusan N.M. 2012. Phosphoregulation of STIM1 leads to exclusion of the endoplasmic reticulum from the mitotic spindle. Curr. Biol. 22:1487–1493 10.1016/j.cub.2012.05.057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffen W., Karki S., Vaughan K.T., Vallee R.B., Holzbaur E.L., Weiss D.G., Kuznetsov S.A. 1997. The involvement of the intermediate chain of cytoplasmic dynein in binding the motor complex to membranous organelles of Xenopus oocytes. Mol. Biol. Cell. 8:2077–2088 10.1091/mbc.8.10.2077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terasaki M., Chen L.B., Fujiwara K. 1986. Microtubules and the endoplasmic reticulum are highly interdependent structures. J. Cell Biol. 103:1557–1568 10.1083/jcb.103.4.1557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirnauer J.S., O’Toole E., Berrueta L., Bierer B.E., Pellman D. 1999. Yeast Bim1p promotes the G1-specific dynamics of microtubules. J. Cell Biol. 145:993–1007 10.1083/jcb.145.5.993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voeltz G.K., Prinz W.A., Shibata Y., Rist J.M., Rapoport T.A. 2006. A class of membrane proteins shaping the tubular endoplasmic reticulum. Cell. 124:573–586 10.1016/j.cell.2005.11.047 [DOI] [PubMed] [Google Scholar]
- Waterman-Storer C.M., Salmon E.D. 1998. Endoplasmic reticulum membrane tubules are distributed by microtubules in living cells using three distinct mechanisms. Curr. Biol. 8:798–806 10.1016/S0960-9822(98)70321-5 [DOI] [PubMed] [Google Scholar]
- Waterman-Storer C.M., Gregory J., Parsons S.F., Salmon E.D. 1995. Membrane/microtubule tip attachment complexes (TACs) allow the assembly dynamics of plus ends to push and pull membranes into tubulovesicular networks in interphase Xenopus egg extracts. J. Cell Biol. 130:1161–1169 10.1083/jcb.130.5.1161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams R.C., Jr, Lee J.C. 1982. Preparation of tubulin from brain. Methods Enzymol. 85:376–385 10.1016/0076-6879(82)85038-6 [DOI] [PubMed] [Google Scholar]
- Wedlich-Söldner R., Schulz I., Straube A., Steinberg G. 2002. Dynein supports motility of endoplasmic reticulum in the fungus Ustilago maydis. Mol. Biol. Cell. 13:965–977 10.1091/mbc.01-10-0475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiese C., Goldberg M.W., Allen T.D., Wilson K.L. 1997. Nuclear envelope assembly in Xenopus extracts visualized by scanning EM reveals a transport-dependent ‘envelope smoothing’ event. J. Cell Sci. 110:1489–1502 [DOI] [PubMed] [Google Scholar]
- Woźniak M.J., Bola B., Brownhill K., Yang Y.C., Levakova V., Allan V.J. 2009. Role of kinesin-1 and cytoplasmic dynein in endoplasmic reticulum movement in VERO cells. J. Cell Sci. 122:1979–1989 10.1242/jcs.041962 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.