Abstract
A study of BLM A5 was conducted using a previously isolated library of hairpin DNAs found to bind strongly to metal free BLM. The ability of Fe(II)•BLM to effect cleavage on both the 3' and 5'-arms of the hairpin DNAs was characterized. The strongly bound DNAs were found to be efficient substrates for Fe•BLM A5-mediated hairpin DNA cleavage. Surprisingly, the most prevalent site of BLM-mediated cleavage was found to be the 5′-AT-3′ dinucleotide sequence. This dinucleotide sequence, and other sequences generally not cleaved well by BLM when examined using arbitrarily chosen DNA substrates, were apparent when examining the library of ten hairpin DNAs. In total, 132 sites of DNA cleavage were produced by exposure of the hairpin DNA library to Fe•BLM A5. The existence of multiple sites of cleavage on both the 3′- and 5′-arms of the hairpin DNAs suggested that some of these might be double-strand cleavage events. Accordingly, an assay was developed with which to test the propensity of the hairpin DNAs to undergo double-strand DNA damage. One hairpin DNA was characterized using this method, and gave results consistent with earlier reports of double-strand DNA cleavage, but with a sequence selectivity different from those reported previously.
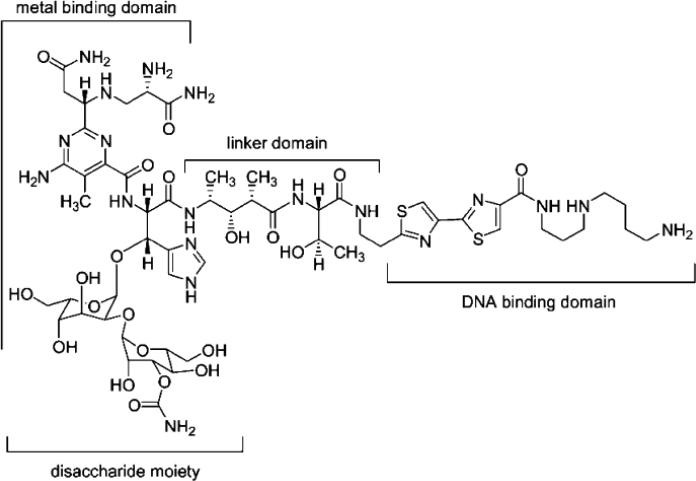
The bleomycins (BLMs) are structurally related antitumor antibiotics exemplified in Figure 1 by BLM A5. They are used in the treatment of certain malignancies, notably squamous cell carcinomas.1,2 The biochemical locus of action of BLM is thought to be DNA2–6 and possibly RNA,7 both of which are bound and oxidatively cleaved by BLM in a reaction requiring both oxygen and a metal ion.8–10 BLM cleaves duplex DNA, producing both single- and double-strand damage, with the latter thought to exert a greater cytotoxic effect.3,11,12 BLM has been reported to cleave DNA sequence selectively at 5′-GPy-3′ sites by abstracting a 4′-H atom from the deoxyribose moiety of the pyrimidine nucleoside.13,14 The ability of BLM to degrade DNA efficiently has been shown to be dependent on the complex structure of the molecule, which is organized into at least four structural domains. These include a metal binding domain, linker domain, DNA binding domain and the disaccharide moiety (Figure 1).15 The metal binding domain contributes importantly to DNA binding16–19 and activates molecular oxygen,16 while the bithiazole and C-terminal substituents are also important in binding DNA efficiently.20–22 The linker region is thought to promote a compact three-dimensional structure for BLM, allowing for efficient cleavage.17,18,23–25 The carbohydrate moiety remains the least well understood domain, but appears to be important for a number of functions including metal binding,26,27 cell surface recognition and cellular uptake,28,29 and DNA cleavage efficiency as well as RNA recognition and cleavage.30,31
Figure 1.
Chemical structure of bleomycin A5.
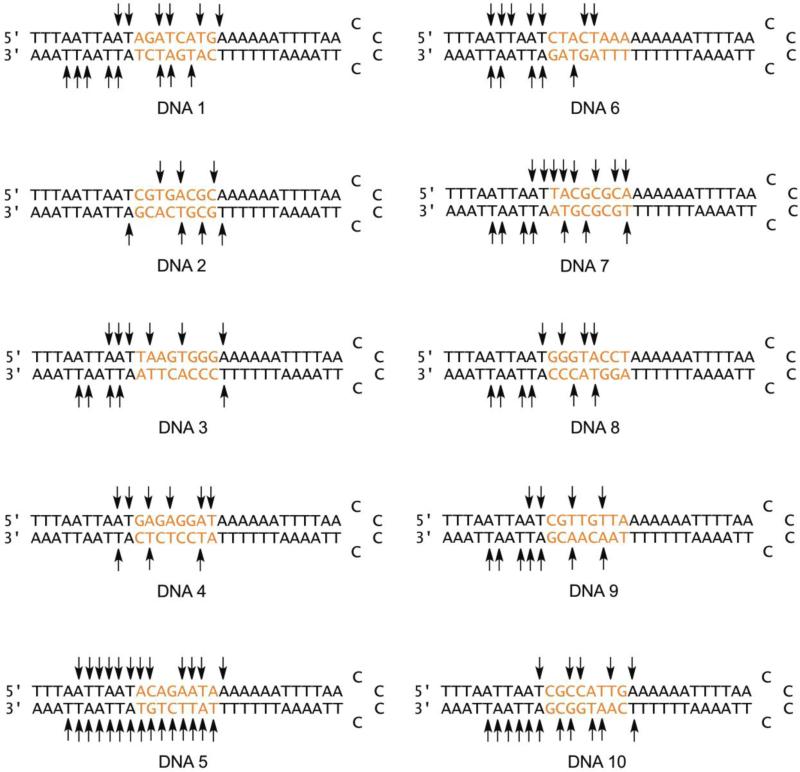
Recently, our laboratory has employed a modified SELEX procedure32 to collect and characterize DNA sequences strongly bound by BLM in order to permit direct examination of the relationship between DNA binding and cleavage.33-35 BLM is used clinically at an atypically low dose (~ 5 μmoles), implying that BLM–DNA binding should be an important determinant of DNA sites that are available for cleavage in a clinical setting. Ten hairpin DNAs were initially identified using this method and their BLM binding specificity was determined using a competition assay, which measured the preference of BLM to bind members of the 64-nt hairpin DNA library (Table 1) when given a choice between that and a previously characterized 16-nt hairpin DNA known to be cleaved stoichiometrically by Fe•BLM.34 The hairpin DNAs were also characterized by high resolution polyacrylamide electrophoresis using [5′-32P]-end labeling.34,36 The length of the [5′-32P]-end labeled hairpin DNAs precluded sequence analysis of the sites at BLM-induced cleavage on the 3′-arms. Accordingly, in the present study, [3′-32P]-end labeled DNAs 1 – 10 were used to characterize the sites of cleavage on the 3′-arms. Because a number of hairpin DNAs had cleavage sites within the variable regions of 5′- and 3′-arms that were in close spatial proximity, it seemed possible that some of these could represent double-strand cleavage events. Accordingly, we developed an assay to identify such events, and demonstrate its utility in characterizing single- and double-strand cleavage events in a representative hairpin DNA.
Table 1.
The 64-nt Hairpin DNAs Isolated Previously
| |||
|---|---|---|---|
| DNA 1 | 5'AGATCATG 3'TCTAGTAC |
DNA 2 | 5'CGTGACGC 3'GCACTGCG |
| DNA 3 | 5'TAAGTGGG 3'ATTCACCC |
DNA 4 | 5'GAGAGGAT 3'CTCTCCTA |
| DNA 5 | 5'ACAGAATA 3'TGTCTTAT |
DNA 6 | 5'CTACTAAA 3'GATGATTT |
| DNA 7 | 5'TACGCGCA 3'ATGCGCGT |
DNA 8 | 5'GGGTACCT 3'CCCATGGA |
| DNA 9 | 5'CGTTGTTA 3'GCAACAAT |
DNA 10 | 5'CGCCATTG 3'GCGGTAAC |
EXPERIMENTAL PROCEDURES
Materials
Terminal deoxynucleotidyl transferase was purchased from Roche Applied Science. T4 polynucleotide kinase was obtained from New England Biolabs. All synthetic oligonucleotides, purified by ion exchange, were purchased from Integrated DNA Technologies. Radiolabeled nucleotides were purchased from Perkin Elmer Life Sciences. BLM A5 solutions were dissolved in water immediately prior to use. Fe(NH4)2(SO4)2·6H2O was purchased from Sigma Aldrich Chemicals and used to prepare fresh Fe2+ solutions immediately prior to use. Chelex 100 was purchased from Sigma Aldrich and used to remove adventitious Fe2+ from solutions prior to experiments.
Methods
Polyacrylamide gel electrophoresis was carried out in 90 mM Tris-borate buffer, pH 8.3, containing 5 mM EDTA. Cleavage sites were confirmed by comparison with the reaction products obtained in a Maxam-Gilbert G lane, Maxam-Gilbert G + A lane and by cytidine specific sequencing protocols.37 Analysis of the polyacrylamide gels was carried out by phosphorimager analysis using a Molecular Dynamics Storm 820 phosphorimager.
[3′-32P] End Labeling and Purification of 64-nt Hairpin DNAs
[3′-32P] end labeling was carried out by combining 10 pmol of the appropriate 64-nt hairpin DNA, 0.06 mCi [α-32P]cordycepin (specific activity 5000 Ci (185 TBq)/mmol) and 400 units of recombinant terminal transferase in 40 μL (total volume) of 25 mM Tris-HCl, pH 6.6, containing 200 mM potassium cacodylate, 2.5 mM CoCl2 and 0.25 mg/mL BSA. The reaction mixture was incubated at 37 °C for 1 h. The [3′-32P] end labeled 64-nt hairpin DNA was purified by 16% polyacrylamide gel electrophoresis at 1800 V for 2.5 h.
[5′-32P] End Labeling and Purification of Hairpin DNA
Ten pmol of 64-nt hairpin DNA was [5′-32P] end labeled by incubation with 20 units of T4 polynucleotide kinase and 0.06 mCi [γ-32P]ATP (specific activity 6000 Ci (222 TBq)/mmol) in 50 μL (total volume) of 70 mM Tris-HCl buffer, pH 7.6, containing 10 mM MgCl2 and 5 mM dithiothrietol. The reaction mixture was incubated at 37 °C for 1 h, followed by heat inactivation of the enzyme at 65 °C for 20 min. The [5′-32P] end labeled 64-nt hairpin DNA was purified by 16% polyacrylamide gel electrophoresis at 1800 V for 2.5 h.
Sequence-Selective Cleavage of Radiolabeled Hairpin DNA by BLM A5
A sample of 5′- or [3′-32P] end labeled hairpin DNA (50,000 cpm) was treated with the appropriate concentrations of Fe2+ and BLM solutions in 5 μL (total volume) of 10 mM Na cacodylate buffer, pH 7.0. Reactions were incubated at 25 °C for 30 min followed by removal of the supernatant under diminished pressure. Ten μL of denaturing gel loading buffer containing 98% formamide, 2 mM EDTA, 0.25% (w/v) bromophenol blue and 0.25% (w/v) xylene cyanol was added to the DNA pellet. The resulting solution was heated at 90 °C for 10 min, followed by chilling on ice. Five microliters of each sample was loaded onto a denaturing gel (16% polyacrylamide, 7 M urea) and run at 50 W for 2.5 h. Gels were visualized using a phosphorimager.
Analysis of Double-Strand DNA Cleavage of [5′-32P]-end and [3′-32P]-end Labeled Hairpin DNAs by Bleomycin A5
Bleomycin cleavage of [5′-32P]-end and [3′-32P]-end labeled hairpin DNAs was performed by incubating hairpin DNA 8 (~30000 cpm) with 5 μM Fe2+ and 5 μM BLM A5 at 25 °C for 30 min in a solution of 10 μL Tris-HCl, pH 8.0. The reactions were quenched using 2 μL of native gel loading buffer containing 0.25 % bromophenol blue, 0.25 % xylene cyanol and 40% D-sucrose and resolved on a 20 % native polyacrylamide gel at 200 V at 4 °C for 16 h. Double strand cleavage sites were confirmed by visualizing co-migrating bands, using a phosphoimager.
Denaturing Gel Electrophoresis of the Double-Strand DNA cleavage products
The relevant [5′-32P]-end and [3′-32P]-end labeled double-strand DNA cleavage bands separated by native gel electrophoresis were excised from gel, isolated by ethanol precipitation and mixed with 5 μL of denaturing loading buffer containing 80% formamide, 2 mM EDTA, 1% bromophenol blue and 1% xylene cyanol, then heated at 90 °C for 10 min. Five μL of the final solutions were chilled on ice and separated on a 16% denaturing polyacrylamide gel containing 16% urea at 50 W for 2.5 h. Two μL of solutions containing [5′-32P]-end labeled and [3′-32P]-end labeled Maxam-Gilbert sequencing markers were used to determine the sequence of cleavage sites. The gels were visualized using a phosphoimager.
RESULTS
Sequence-Selective Degradation of 10 Hairpin DNAs by Fe(II)·BLM A5
Members of the library of hairpin DNAs, of the form 5′-TTTAATTAATXXXXXXXXAAAAAATTTTAACCCCTTAAAATTTTTTYYYYYYYYATTAATTAAA-3′, were characterized by high resolution polyacrylamide electrophoresis and by their ability to suppress cleavage of a previously studied 16-nt hairpin DNA containing a pro-fluorescent nucleoside.34 The hairpin DNAs were selected in what may be regarded as the first step of a SELEX-type procedure32 to provide hairpin DNA sequences that were strongly bound by metal free BLM A5. These hairpin DNAs were found to inhibit cleavage of a stoichiometric amount of the profluorescent 16-nt hairpin DNA to the extent of 76 - 97%. Described below is an analysis of Fe(II)•BLM-mediated cleavage of the 10 hairpin DNAs by high resolution denaturing polyacrylamide gel electrophoresis using both 5'- and [3'-32P] end labeling. This permitted the analysis of every site cleaved by Fe(II)•BLM A5 in the ten hairpin DNAs studied.
DNA 1 was treated with increasing concentrations Fe(II)•BLM A5 and several strong cleavage sites were observed in both the 5' and [3'-32P] end labeled samples. Figure S1A clearly shows the presence of six cleavage sites within the 8-nt randomized region.34,38 These included 5′-GA13-3′, 5′-AT14-3′, 5′-AT17-3′, 5′-AT49-3′, 5′-GA51-3′ and 5′-AT52-3′. None of these sites was a canonical dinucleotide cleavage site for BLM A5 involving -5′-GT-3′ and 5′-GC-3′ sequences. Cleavage sites were also noted within the (invariable) flanking sequences. In spite of the fact that these were identical in all ten hairpin DNAs, BLM-mediated cleavage at different sites on each member of the library (vide infra). For the 5′-32P end labeled hairpin DNA 1, there were several unusual cleavage sites and sequence motifs worthy of note, namely the 5'-PuPu-3' cleavage sites at 5'-AA9-3', 5'-GA13-3' and 5'-GA19-3'. The other nucleobase cleaved on this arm, thymidine, followed the more classic cleavage motif of 5'-PuPy-3'. However, the purine was not the usual guanosine, but rather adenosine. These dinucleotide sequences were 5'-AT10-3', 5'-AT14-3' and 5'-AT17-3'.
The [3'-32P] end labeled DNA cleavage experiment (Figure S1B) for DNA 1 showed very similar results to Figure S1A, as the hairpin contained no classically cleaved dinucleotide sequences for Fe(II)•BLM A5, but did contain several strong sites for cleavage, close to the radiolabel. The sites T49, A51, T52, T56, T57, A59, T60 and T61 were cleaved by BLM A5 on the 3′-arm of the hairpin DNA. The very strong cleavage site at T56 is actually within the invariant flanking region of the hairpin DNA. This is a 5'-AT-3' site, similar to the sites found on the 5'-arm of this hairpin DNA. However, there were also two very unusual 5'-TT-3' cleavage bands (at T57 and T61) which represent cleavage sequences not often noted in studies employing artibrarily chosen DNA sequences.
DNA 2 differed from DNA 1 within the 5'-arm of the hairpin DNA in that it contained two canonical dinucleotide cleavage sequences (5'-GC-3' and 5′-GT-3′). The 5′-GC-3′ was cleaved comparatively weakly relative to the 5'-GT-3' site, which represented the strongest site of cleavage (Figure S2A). DNA 2 also had a binding efficiency of 97% (compared to 82% for 1), which was the highest value determined in the competition assay.34 The cleavage site at A15 is a 5'-PuPu-3′ dinucleotide sequence motif. It was cleaved quite weakly in comparison to both canonical cleavage motifs. The [3′-32P] end labeled hairpin DNA 2 also contained two canonical dinucleotide cleavage sequences (5′-GC48-3′ and 5′GT50-3′). Again, the preferred cleavage site of these two was 5′-GT50-3′ (Figure S2B). These two dinucleotide sequences also represented the strongest sites of cleavage. There were two weak cleavage sites at T46 and A55, the former providing an example of a rarely reported 5′-PyPy-3′ sequence motif. It is of interest that the number of cleavage sites was less on both arms of hairpin DNA 2 compared to hairpin DNA 1. This also included a lesser number of sites in the flanking region, even though these sequences are identical in hairpin DNAs 1 and 2.
Hairpin DNA 3 provided very interesting cleavage results despite its comparatively low binding specificity, reported as 76%.34 This hairpin sequence contains four G:C base pairs, and only one 5′-GPy-3′ site. The latter (5′-GT15-3′) was cleaved, but with an intensity similar to that of the other sites on the 5′-32P end labeled arm (Figure S3). The 5′-arm was cleaved six times at the sites A8, A9, T10, A12, T15 and A19. The number of cleavage sites on the 5′-arm was comparable to DNA 1 and these were distributed throughout both the randomized and flanking regions. In comparison, the 3′-arm of hairpin DNA 3 showed a substantially different result from that of hairpin DNA 1.
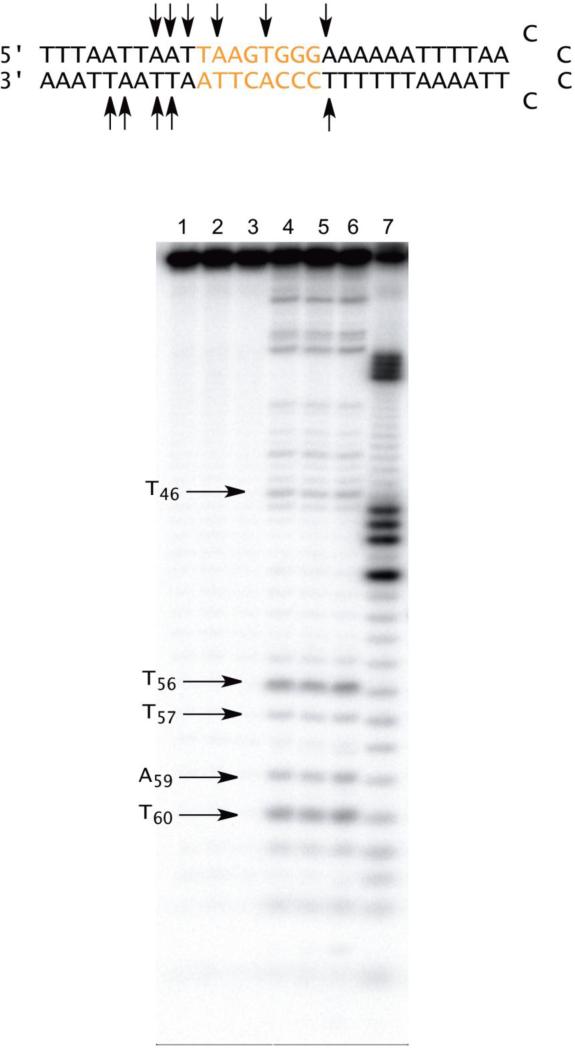
Hairpin DNA 3 was cleaved at T46, T56, T57, A59 and T60 (Figure 2). The strongest cleavages occurred at T56 and T60, both of which are located in the invariant flanking sequence. There was no cleavage within the randomized region, as opposed to the cleavage that appeared within the 5′-arm of hairpin DNA 3, as well as the 5′- and 3′-arms of DNA 1. Hairpin DNA 3 was the only example among the hairpin DNAs tested that exhibited no significant cleavage within the randomized region of the 3′-arm.
Figure 2.
Sequence-selective cleavage of [3′-32P] end labeled 64-nt hairpin DNA 3 by BLM A5. Lane 1, radiolabeled 3 alone; lane 2, 10 μM Fe2+; lane 3, 5 μM BLM A5; lane 4, 1 μM Fe(II)•BLM A5; lane 5, 2.5 μM Fe(II)•BLM A5; lane 6, 5 μM Fe(II)•BLM A5; lane 7, Maxam-Gilbert C lane.
Hairpin DNA 4 (5′-32P end labeled) is shown in Figure S4A. The reported binding specificity of this hairpin DNA was 79%, which is relatively low compared to the other hairpin DNAs.34 It does not contain any canonical sequence cleavage motifs, but showed a comparatively strong cleavage site at 5′-GA12-3′. The other sites of cleavage occurred at A9, T10, A14, A17 and T18. These sites all represent 5′-PuPy-3′ sequence motifs (5′-AT10-3′ and 5′-AT18-3′) or 5′-PuPu-3′ sequence motifs (5′-AA9-3′, 5′-GA12-3′, 5′-GA14-3′ and 5′-GA17-3′). Analogous to hairpin DNA 3, there was less cleavage observed for Fe•BLM A5 on the opposing arm.
The 3′-32P end labeled hairpin DNA 4 showed cleavage at three sites, all of which are thymidines, namely at T48, T53 and T56 (Figure S4B). There was only a single cleavage site in the flanking region, at T56. Two of the three cleavages occurred at 5′-AT-3′ sites, while the third occurred at a 5′-CT-3′ site, which has been observed infrequently.
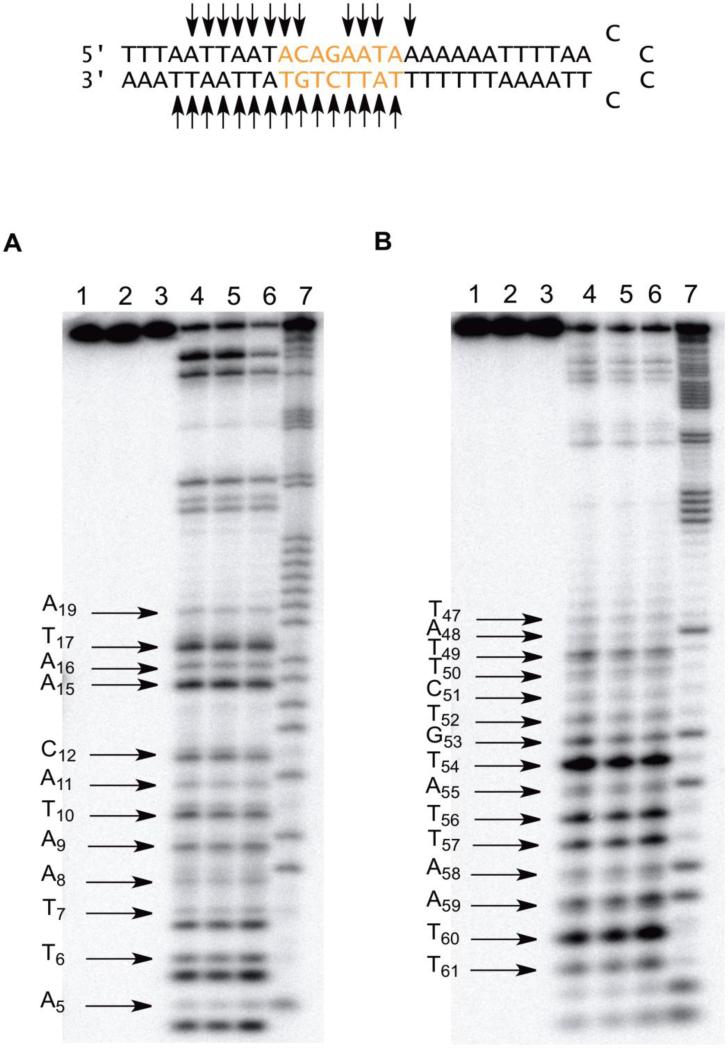
BLM A5 cleaved hairpin DNA 5 at twelve sites on the 5′-arm and did so strongly even when 1 μM Fe(II)•BLM A5 was used. Hairpin DNA 5, remarkably, contains no 5′-GPy-3′ dinucleotide sequences on its 5′–arm (Figure 3A). DNA 5 was cleaved at the most sites of any hairpin DNA in the presently studied library of strongly bound hairpin DNAs, in spite of the fact that the binding specificity (90%) was not the highest measured.34 It was also cleavaged strongly even at the lowest (5 μM) concentration of Fe(II)•BLM A5 tested. The sites of cleavage included 5′-TT7-3′, 5′-AA9-3′ and 5′-AT10-3′ within the flanking region, while relatively strong cleavage occurred at 5′-GA15-3′ in the randomized region of the hairpin. The other sites cleaved by BLM A5 all represent unusual sequence specificities.
Figure 3.
A) Sequence-selective cleavage of [5′-32P]-end labeled 64-nt hairpin DNA 5 by BLM A5. Lane 1, radiolabeled 5 alone; lane 2, 20 μM Fe2+; lane 3, 5 μM BLM A5; lane 4, 5 μM Fe(II)•BLM A5; lane 5, 10 μM Fe(II)•BLM A5; lane 6, 20 μM Fe(II)•BLM A5; lane 7, G+A lane. B) Sequence-selective cleavage of [3′-32P]-end labeled 64-nt hairpin DNA 5 by BLM A5. Lane 1, radiolabeled 5 alone; lane 2, 10 μM Fe2+; lane 3, 5 μM BLM A5; lane 4, 1 μM Fe(II)•BLM A5; lane 5, 5 μM Fe(II)•BLM A5; lane 6, 10 μM Fe(II)•BLM A5; lane 7, Maxam-Gilbert G+A lane.
The 3′-arm of hairpin DNA 5 was also cleaved very extensively. The sequence 5′-GT54-3′ was the preferred site of cleavage, consistent with the expected cleavage pattern for Fe•BLM (Figure 3B). However, fourteen other sites were also cleaved by BLM, albeit some relatively weakly. These included all eight nucleosides in the variable region of the hairpin DNA. A noteworthy site of cleavage, 5′-TG53-3′, represents a 5′-PyPu-3′ sequence cleavage motif. In summary, hairpin DNA 5 was cleaved avidly by Fe•BLM A5 on both arms within the randomized and invariant regions of the hairpin DNA.
Hairpin DNA 6 also lacked any dinucleotide sequences commonly cleaved strongly by Fe•BLM on the 5′-arm. The binding specifically of this DNA (81%) was also lower (Figure S5A).34 Despite this, DNA 6 still underwent cleavage at seven sites on its 5′-arm, including the unusual 5′-AC14-3′ and 5′-CT15-3′ sites in the randomized region of the hairpin. The 5′- flanking region was cleaved at more sites than the randomized region. These sites included 5′-AA5-3′, 5′-AT6-3′ and 5′-TT7-3′. The sites A9 and T10 were also cleaved.
Figure S5B shows the cleavage sites on the 3′-arm of hairpin DNA 6. There were fewer sites cleaved by Fe•BLM on this arm of the hairpin DNA; only one cleavage site was noted in the randomized region of the hairpin DNA (5′-GT52-3′). The remainder of the cleavage occurred in the invariant region at A55, T56, A59 and T60. In summary, this hairpin DNA was cleaved at a total of 12 sites on both arms and most cleavage sites involved a dinucleotide sequence not usually cleaved by Fe•BLM A5 in randomly chosen DNAs. The relative paucity of cleavage sites on this hairpin DNA compared to DNA 5 is notable, and can perhaps be explained by the lower binding specificity of this DNA.
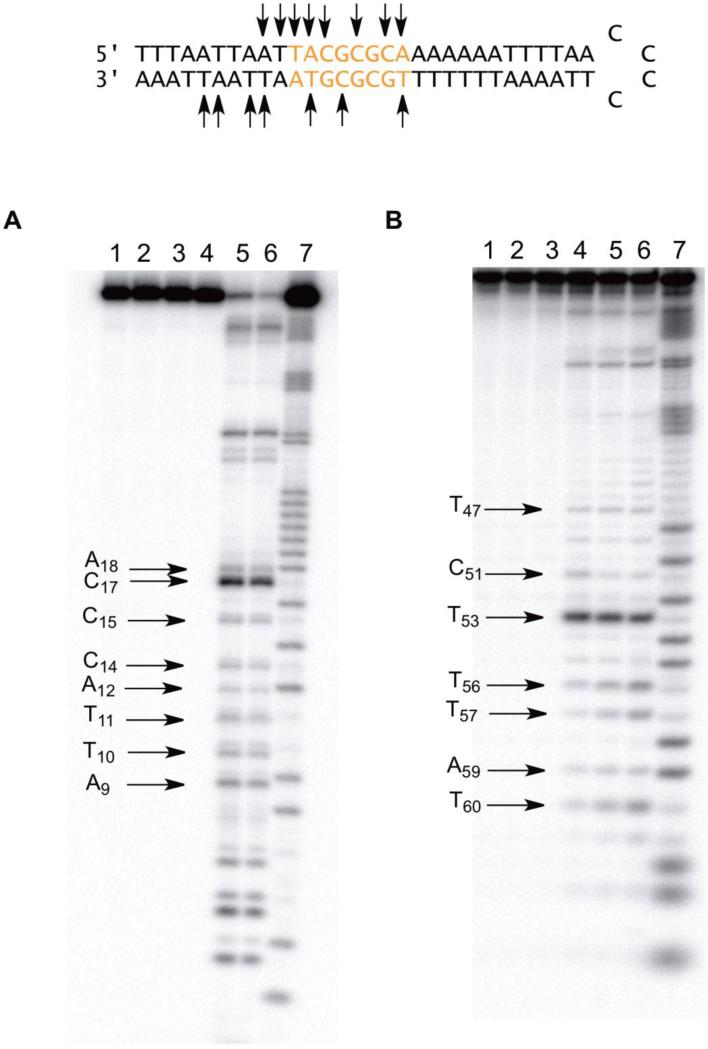
The sequence selective cleavage of DNA 7 within the 5′-arm is presented in Figure 4A. Hairpin DNA 7 was bound exceptionally well by BLM A5, with a measured binding specificity value of 97%, the highest recorded value for members of the library.34 Except for two cleavage sites at A9 and T10, the hairpin 7 was cleaved within the randomized portion of the DNA. The only nucleotides not cleaved were G14 and G16. The 5′-arm of this hairpin DNA contains two 5′-GC-3′ dinucleotide sequences. The 5′-GC17-3′ dinucleotide sequence was the preferred site of cleavage on the 5′-arm of hairpin DNA 7. Remarkably, the 5′-GC15-3′ site was not cleaved to an extent greater than that of the other sites of cleavage.
Figure 4.
A) Sequence-selective cleavage of [5′-32P]-end labeled 64-nt hairpin DNA 7 by BLM A5. Lane 1, radiolabeled 7 alone; lane 2, 5 μM Fe2+; lane 3, 5 μM BLM A5; lane 4, 1 μM Fe(II)•BLM A5; lane 5, 2.5 μM Fe(II)•BLM A5; lane 6, 5 μM Fe(II)•BLM A5; lane 7, G+A lane. B) Sequence-selective cleavage of [3′-32P]-end labeled 64-nt hairpin DNA 7 by BLM A5. Lane 1, radiolabeled 7 alone; lane 2, 10 μM Fe2+; lane 3, 5 μM BLM A5; lane 4, 1 μM Fe(II)•BLM A5; lane 5, 2.5 μM Fe(II)•BLM A5; lane 6, 5 μM Fe(II)•BLM A5; lane 7, Maxam-Gilbert G+A lane.
The 3′-arm of hairpin DNA 7 differed from the 5′-arm with respect to distribution of cleavage sites; four of the seven cleavage sites occurred in the flanking region of the DNA(Figure 4B). This preference is especially interesting in light of the presence of three 5′-GPy-3′ cleavage motifs within the randomized region. Of the three sites, two were cleaved: 5′-GC51-3′ and 5′-GT53-3′. The lack of significant cleavage at 5′-GC49-3′ is notable. The 5′-GT53-3′ site was the most efficiently cleaved site within the 3′-arm of this hairpin. In summary, BLM A5 cleaved DNA 7 at 15 sites and had the highest binding efficiency measured for any of the DNAs studied (97%). Interestingly, BLM A5 preferred certain 5′-GPy-3′ dinucleotide sites over others, with 5′-GT-3′ being the most efficiently cleaved, and certain other sites (e.g. 5′-GC49-3′) not undergoing significant cleavage.
The binding specificity of hairpin DNA 8 for BLM was 92%.34 The 5′-arm contained four cleavage sites, including a strong site at 5′-GT14-3′ (Figure S6A). The other sites were cleaved weakly in comparison, including sites at dinucleotide sequences 5′-AT10-3′, 5′-GG12-3′ and 5′-TA15-3′. The 5′-GG12-3′ site is notable, representing one of the few sites of cleavage at a G residue and one of only two recorded 5′-GG-3′ cleavage sites within the ten hairpin DNAs studied. The 3′-arm of the hairpin DNA had a strong cleavage site at 5′-GT50-3′, but the 3′-arm also contained several cleavage sites in the flanking region of the hairpin DNA.
Hairpin DNA 8 was cleaved at six sites on the 3′-arm of the hairpin DNA (Figure S6B). As noted, the strongest site of cleavage was at the 5′-GT50-3′ dinucleotide sequence. Weaker sites included a site in the randomized region at 5′-AC52-3′, and several sites in the invariant region: 5′-AT56-3′, 5′-TT57-3′, 5′-AA59-3′ and 5′-AT60-3′. This hairpin DNA contains a 5′-GT-3′ dinucleotide sequence in each arm, and they both represented the strongest sites of cleavage within the individual arms of the hairpin DNA. The high binding specificity (97%) and relative paucity of cleavage sites represent a notable example of binding and cleavage preference for BLM A5.
The BLM-mediated cleavage sites resolved by [5′-32P] end labeling of hairpin DNA 9 are presented in Figure S7A. There were two cleaved dinucleotide sequences in the randomized region, both following the preferred sequence composition of BLM. The cleavage sites occurred at 5′-GT13-3′ and 5′-GT16-3′, with the former cleaved more extensively. Two other cleavage sites occurred in the invariant region at A9 and T10.
The randomized region of the 3′-arm of the hairpin DNA contained no canonical dinucleotide sequences for cleavage by BLM (Figure S7B). The strongest site of cleavage was the 5′-GA55-3′ sequence. The 3′-arm of the hairpin DNA also showed stronger cleavage in the randomized region of the hairpin DNA (5′AA49-3′ and 5′-AA52-3′) than the 5′-arm. In summary, hairpin DNA 9 was bound with a specificity of 89% and cleaved at a total of 11 sites, including sites within both 3′ and 5′-arms of the hairpin DNA.
The site specificity of cleavage of 5′-32P end labeled DNA 10 is presented in Figure S8A. Fe•BLM utilized five sites for cleavage on the 5′-arm of hairpin DNA 10 including 5′-AT10-3′, 5′-GC13-3′, an interesting 5′-CC14-3′ dinucleotide sequence, as well as a 5′-TT17-3′ and 5′-GA19-3′. Cleavage of the 5′-CC-3′ dinucleotide sequence was not observed for any other hairpin DNA in this library, but it was cleaved inefficiently. The 5′-GC13-3′ dinucleotide sequence represented the dominant cleavage site, as has been seen regularly on the hairpin DNAs that contain a canonical Fe•BLM cleavage site. Little cleavage was observed in the flanking region of this arm of the DNA, compared to other DNAs such as DNA 3 and 5, which exhibited a number of cleavage sites in the flanking regions compared to that within the randomized nucleotide sequence. Hairpin DNA 10 also had a binding specificity not much different than DNA 5 (86 vs 90%), but did not have as many sites subject to cleavage by BLM. The paucity of cleavage sites on the 5′-arm of the hairpin is also in contrast to the eleven sites of cleavage observed within the 3′-arm.
Similar to the 5′-arm, the 3′-arm had one canonical BLM cleavage site at 5′-GC53-3′ (Figure S8B). This site and a variety of other sequence motifs were cleaved with comparable efficiency, including cleavage at 5′-GG52-3′. There were four cleavage sites in the randomized region, but seven cleavage sites were observed for hairpin DNA 10 in the invariant region. For many of the hairpin DNAs treated with Fe•BLM in this study, there was a significant difference between the number of cleavage sites on the 5′- and 3′-arms.
As illustrated in Figure 5, each of the ten hairpin DNA differed in specific cleavage sites induced by Fe•BLM. However, quantitative analysis of cleavage sites mediated by BLM on the hairpin DNAs in the library showed that, in the aggregate, the 3′ and 5′-arms had a similar number of cleavage sites. On the 5′ arms, these occurred between A5 and A19. This 15-nt range represented the extreme positions of BLM-mediated cleavage on the hairpin DNAs when [5′-32P] end labeled. Table 2 shows the number of cleavage sites and the dinucleotide sequences at which the cleavages occurred on the 5′-arm. Thus, 16 AT nucleotide sequences in the 5′-arms of DNAs 1 – 10 underwent significant cleavage by Fe•BLM, as did 11 AA sequences. The dinucleotide sequence GA was cleaved 9 times, while the canonical cleavage sequences GT and GC were cleaved only 5 and 4 times, respectively. These sites represented 45 of the 61 cleavage sites observed on the 5′-arms of hairpin DNAs 1 – 10. However, when the frequency of cleavage is considered (Table 3), only 64% of the AT sequences and 39% of the AA sequences actually present were cleaved to a significant extent. In comparison, all of the GA, GC and GT dinucleotides present in the 5′-arms were cleaved by Fe•BLM.
Figure 5.
Site of BLM-mediated damage for the ten hairpin DNAs studied.
Table 2.
Dinucleotide Sequences Cleaved on the 5′-Arm by BLM A5a
| 5′-XX-3′ | Hairpin DNA | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | totalb | |
| AA | 1 | 1 | 1 | 4 | 2 | 1 | 1 | 11 | |||
| AC | 1 | 1 | 1 | 3 | |||||||
| AG | 0 | ||||||||||
| AT | 3 | 1 | 2 | 3 | 2 | 1 | 1 | 1 | 2 | 16 | |
| CA | 1 | 1 | |||||||||
| CC | 1 | 1 | |||||||||
| CG | 0 | ||||||||||
| CT | 1 | 1 | |||||||||
| GA | 2 | 1 | 1 | 3 | 1 | 1 | 9 | ||||
| GC | 1 | 2 | 1 | 4 | |||||||
| GG | 1 | 1 | |||||||||
| GT | 1 | 1 | 1 | 2 | 5 | ||||||
| TA | 2 | 2 | 1 | 1 | 6 | ||||||
| TC | 0 | ||||||||||
| TG | 0 | ||||||||||
| TT | 1 | 1 | 1 | 3 |
|||||||
| 61 | |||||||||||
Assayed by the use of 5'-32P-end labeled DNAs as substrates.
Total number of cleavages at each dinucleotide sequence in the 5'-arm of hairpin DNAs 1 – 10.
Table 3.
Frequency of Dinucleotide Sequence Appearance Within the 5′-Arm of a Hairpin DNA between A5 and A19, and Analysis of Frequency of Cleavage at Each Dinucleotide
| 5′-XX-3′ | Hairpin DNA | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | Occurrences | Cleavages | Percent cleaved | |
| AA | 2 | 2 | 3 | 2 | 4 | 5 | 3 | 2 | 3 | 2 | 28 | 11 | 39% |
| AC | 1 | 1 | 1 | 1 | 1 | 5 | 3 | 60% | |||||
| AG | 1 | 1 | 2 | 1 | 5 | 0 | 0% | ||||||
| AT | 4 | 2 | 2 | 3 | 3 | 2 | 2 | 2 | 2 | 3 | 25 | 16 | 64% |
| CA | 1 | 1 | 1 | 1 | 1 | 5 | 1 | 20% | |||||
| CC | 1 | 1 | 2 | 1 | 50% | ||||||||
| CG | 2 | 2 | 1 | 1 | 6 | 0 | 0% | ||||||
| CT | 2 | 1 | 3 | 1 | 33% | ||||||||
| GA | 2 | 1 | 1 | 3 | 1 | 1 | 9 | 9 | 100% | ||||
| GC | 1 | 2 | 1 | 4 | 4 | 100% | |||||||
| GG | 2 | 1 | 2 | 5 | 1 | 20% | |||||||
| GT | 1 | 1 | 1 | 2 | 5 | 5 | 100% | ||||||
| TA | 2 | 1 | 2 | 2 | 3 | 3 | 2 | 3 | 2 | 1 | 21 | 6 | 29% |
| TC | 1 | 1 | 1 | 1 | 1 | 5 | 0 | 0% | |||||
| TG | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 7 | 0 | 0% | |||
| TT | 1 | 1 | 2 | 1 | 1 | 1 | 2 | 1 | 3 | 2 | 15 | 3 | 20% |
On the 3′-arm of the hairpin DNA library, the range of Fe•BLM mediated cleavage sites spanned nucleotides T46 to T61, inclusively (Tables 4 and 5). A total of 71 BLM-mediated cleavage sites were noted. BLM mediated cleavage on the 3′-arms showed more cleavage at 5′-AT-3′ dinucleotide sequences (21 sites), as well as cleavage at 15 5′-TT-3′ and 11 5′-AA-3′ dinucleotide sequences. Relative to the total number of such sequences present, the efficiencies of cleavage of these dinucleotides sequences were only 81%, 41% and 73%, respectively. The dinucleotide sequence 5′-GC-3′ appeared five times, but was only cleaved by Fe•BLM at three of those sites (60% cleavage efficiency). The dinucleotide sequences 5′-GT-3′ and 5′-GA-3′, each approved five times in the 3′-arms, and were cleaved each time they were present in the hairpin DNA.
Table 4.
Dinucleotide Sequences Cleaved on the 3′-Arm by BLM A5a
| 5′-XX-3′ | Hairpin DNA | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | Totalb | |
| AA | 1 | 1 | 1 | 1 | 1 | 1 | 3 | 2 | 11 | ||
| AC | 1 | 1 | |||||||||
| AG | 0 | ||||||||||
| AT | 4 | 2 | 1 | 3 | 2 | 2 | 2 | 2 | 3 | 21 | |
| CA | 0 | ||||||||||
| CC | 0 | ||||||||||
| CG | 0 | ||||||||||
| CT | 2 | 1 | 3 | ||||||||
| GA | 1 | 1 | 1 | 1 | 1 | 5 | |||||
| GC | 1 | 1 | 1 | 3 | |||||||
| GG | 1 | 1 | |||||||||
| GT | 1 | 1 | 1 | 1 | 1 | 5 | |||||
| TA | 3 | 1 | 4 | ||||||||
| TC | 1 | 1 | |||||||||
| TG | 1 | 1 | |||||||||
| TT | 2 | 1 | 2 | 4 | 2 | 1 | 1 | 2 | 15 |
||
| 71 | |||||||||||
Assayed by the use of 3'-32P-end labeled DNAs as substrates.
Total number of cleavages at each dinucleotide sequence in the 3'-arm of hairpin DNAs 1 – 10.
Table 5.
Frequency of Dinucleotide Sequence Appear Once within the 3′-Arm of a Hairpin DNA between T46 and T61, and Analysis of Frequency of Cleavage at Each Dinucleotide
| 5′-XX-3′ | Hairpin DNA | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | occurrences | cleavages | percent cleaved | |
| AA | 1 | 1 | 2 | 1 | 1 | 1 | 2 | 1 | 3 | 2 | 15 | 11 | 73% |
| AC | 1 | 1 | 1 | 2 | 5 | 1 | 20% | ||||||
| AG | 2 | 1 | 3 | 0 | 0% | ||||||||
| AT | 4 | 2 | 2 | 3 | 4 | 2 | 2 | 2 | 2 | 3 | 26 | 21 | 81% |
| CA | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 7 | 0 | 0% | |||
| CC | 2 | 1 | 2 | 5 | 0 | 0% | |||||||
| CG | 2 | 2 | 1 | 1 | 6 | 0 | 0% | ||||||
| CT | 1 | 1 | 2 | 1 | 5 | 4 | 80% | ||||||
| GA | 1 | 1 | 1 | 1 | 1 | 5 | 5 | 100% | |||||
| GC | 1 | 2 | 1 | 5 | 3 | 60% | |||||||
| GG | 1 | 1 | 2 | 1 | 50% | ||||||||
| GT | 1 | 1 | 1 | 1 | 1 | 5 | 5 | 100% | |||||
| TA | 2 | 1 | 2 | 2 | 3 | 3 | 2 | 3 | 2 | 1 | 21 | 4 | 19% |
| TC | 2 | 1 | 1 | 3 | 1 | 1 | 9 | 1 | 11% | ||||
| TG | 1 | 1 | 1 | 1 | 1 | 5 | 1 | 20% | |||||
| TT | 3 | 3 | 4 | 3 | 4 | 6 | 4 | 3 | 4 | 3 | 37 | 15 | 41% |
In summary, the 5′-AT-3′ dinucleotide was the sequence cleaved the most often on both the 3′- and 5′-arms of the hairpin DNAs. This sequence is generally not preferred for cleavage by BLM within randomly chosen DNAs, but this library of hairpin DNAs contains many AT sequences, which BLM cleaved roughly 73% of the time the dinucleotide sequence appeared. There were 132 sites of cleavage on these 10 hairpin DNAs, almost 30% of which were 5′-AT-3′ dinucleotide sequences.
Characterization of Bleomycin-induced Double-strand Cleavage of DNA 4
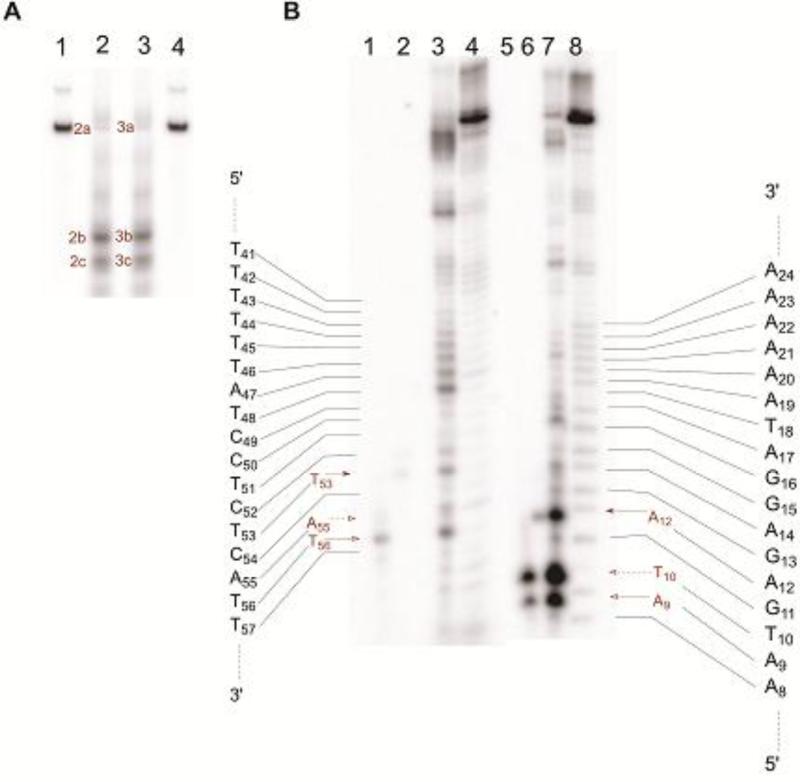
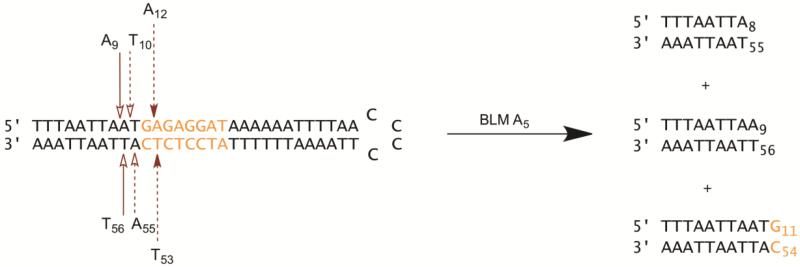
Treatment of hairpin DNAs with Fe(II)•BLM produced strand breaks on both arms of most hairpin DNAs studied (Figure 5). The proximity of the breaks on opposing arms of the DNA suggested that if both occurred within a single hairpin DNA molecule, they might effectively constitute double-strand breaks. In order to study this possibility, hairpin DNA 4, which contained neither 5′-GT-3′ nor 5′-GC-3′ sequences and had six cleavage sites within the region of the DNA that had been randomized in the initial library, was analyzed for possible double strand cleavage. This was done by radiolabeling samples of this DNA with 32P alternatively at the 5′- and 3′-ends. Equal amounts of these labeled DNAs, having roughly the same specific activity, were then treated with 5 μM Fe(II)•BLM A5. The products were analyzed on a native polyacrylamide gel. As shown in Figure 6A, both 32P-end labeled DNAs produced two relatively strong bands which co-migrated on the native gel (2b/3b and2c/3c). The co-migration (and similar intensities) of the bands in lanes 2 and 3 argues that each was formed by cleavage at the same sites on opposite strands. Bands 2b/3b each constituted about 31% of the total radioactivity in the lane in which it appeared, while bands 2c/3c each constituted about 22% of the total radioactivity. There were also weaker bands apparent, such as 2a/3a (12%), which comprise mixtures of full-length hairpin DNA and single nicked hairpin DNAs. These bands were excised from the native gel, purified and electrophoresed on a denaturing gel, using Maxam-Gilbert sequencing ladders to permit characterization of the sites of double strand DNA cleavage. The denaturing gel in Figure 6B shows that DNA 4 was cleaved by Fe(II)•BLM at A12 and T53 (lanes 6 and lane 2, respectively), which were the samples from gel bands 2b/3b in Figure 6A. Analysis of gel bands 2c/3c in Figure 6A revealed that these bands actually contained a mixture of two DNA duplexes, one resulting from cleavage at T10 and A55, the other from cleavage at A9 and T56. Interestingly, cleavage at A55 was not especially strong in the sequencing gels (Figure S4). Likewise, no strong double strand cleavage band resulted from the single strand cleavages at A17 and T48, although some weak bands are apparent in Figure 6A. Thus this hairpin DNA underwent double strand cleavage at three sites, as summarized in Figure 7.
Figure 6.
Analysis of bleomycin-induced double strand cleavage sites on DNA 4. (A) Double strand cleavage of [5′-32P]-end labeled (lane 2) and [3′-32P]-end labeled (lane 3) 64-nucleotide hairpin DNA 4 by Fe(II)•bleomycin A5. Lane 1, [5′-32P]-end labeled DNA alone; lane 2, [5′-32P]-end labeled DNA + 5 μM Fe(II)•BLM A5; lane 3, [3′-32P]-end labeled DNA + 5 μM Fe(II)•BLM A5; lane 4, [3′-32P]-end labeled DNA alone. (B) Sequencing gel analysis of bleomycin induced double strand cleavage products of [3′-32P]-end labeled DNA 4 (lanes 1-4) and [5′-32P]-end labeled (lanes 5-8) DNA 4. Each lane (except lanes 4 and 8) corresponds to a numbered cleavage band, isolated from the gel shown in Figure 4A. Lane1, band 3c; lane 2, band 3b; lane 3, band 3a; lane 4, Maxam-Gilbert G+A sequencing lane for the [3′-32P]-end labeled DNA 4; lane 5, band 2c; lane 6, band 2b; lane 7, band 2a; lane 8, Maxam-Gilbert G+A sequencing lane for the [5′-32P]-end labeled DNA 4.
Figure 7.
Summary of bleomycin-induced double strand cleavage sites on hairpin DNA 4. Similar arrows on opposite strands indicate paired cleavages. The double strand products resulting from cleavage at A9-T56 and A10-T55 both migrated in bands 2c and 3c of the gel in Figure 6A. The product resulting from cleavage at A12 and T53 migrated in bands 2b and 2c. The length of the arrows reflect the relative intensities of the cleavage bands 2b/3b and 2c/3c.
DISCUSSION
The bleomycins have been studied extensively for several decades, and have been in clinical use for nearly as long. Nonetheless, many facets of the action of BLM are incompletely understood, as evidenced by the present study and related earlier efforts.34-36,38 Presently, the relationship between DNA binding of BLM and its cleavage of DNA was examined using a library of 64-nt hairpin DNAs. The members of this library of ten hairpin DNAs were chosen arbitrarily from a larger random hairpin DNA library on the basis of their ability to bind tightly to metal-free BLM. The initial library provided a sequence space of 65,536 possible combinations.34 The present work differs from previous studies in this and other laboratories, which have used arbitrarily chosen DNA substrates in reactions with BLM that have employed high BLM:DNA ratios. The experimental conditions applied in such studies differ from those present during clinical administration of the drug. When used clinically, the concentration of DNA present must be greatly in excess of the concentration of BLM.
The present study has characterized 10 of these hairpin DNAs using both 3′- and [5′ -32P] end labeling to permit measurement of the Fe•BLM cleavage sites within both arms of hairpin DNAs 1 - 10. The goal of the study was to identify DNA sequence elements preferred for cleavage in substrates bound strongly by BLM, gain inferences into the obligatory binding step that occurs before C-4′ H abstraction, which has been reported to be rate-limiting for DNA degradation.39 Study of the hairpin DNAs through BLM cleavage site analysis has shown that they are all substrates for BLM cleavage, although some were cleaved more efficiently than others. The hairpin DNAs also showed sites of cleavage that have not traditionally been observed in DNA cleavage reactions with BLM, including cleavage in the AT-rich invariant regions of the hairpin DNA.
Overall, the 10 DNAs contained 132 sites that were cleaved by BLM, while the number of cleavage sites per DNA molecule varied from seven sites on DNA 2 (Figure S2) to 27 on DNA 5 (Figure 3). Hairpin DNAs 3 (Figure 2) and 4 (Figure S4) differed significantly in the number and relative intensities of cleavage sites on their 5′ and 3′-arms. Hairpin DNA 3 offered perhaps the most interesting results: the 5′-arm contained six cleavage sites (Figure 2). In comparison, the 3′-arm had no cleavage sites within the randomized region and minimal cleavage at sites not usually cleaved by Fe•BLM in the invariant flanking AT-rich sequence regions. The latter were apparent only at a high (20 μM) concentration of Fe(II)•BLM A5. Considering the low binding specificity for this DNA (76%), the lack of cleavage in the randomized region suggests that the efficiency of BLM–DNA binding can be an important determinant of cleavage efficiency.
DNA 4 afforded a similar result. The 5′-arm of the hairpin DNA, having a G-rich sequence but no canonical BLM cleavage motifs, underwent cleavage at six sites but only at 20 μM Fe(II)•BLM A5 concentration (Figure S4). Nonetheless, this DNA was cleaved more strongly and at more sites than on the 3′-arm, which is pyrimidine rich. The lack of 3′-arm cleavage sites for hairpin DNAs 3 and 4 may simply reflect the fact that both have pyrimidine-rich 3′-arms. However, the propensity for DNA cleavage on a given strand cannot be accommodated in all cases by an analysis this straightforward, as noted below for hairpin DNA 5.
DNA 5 provided a stark contrast to the other DNAs studied. It contains only a single canonical 5′-GPy-3′ cleavage site but was still cleaved by Fe•BLM 12 times on the 5′-arm (Figure 3A) and 15 times on the 3′-arm (Figure 3B). The BLM-mediated cleavages occurred in both the randomized and invariant regions and were readily apparent even at low Fe•BLM concentrations. The binding specificity of DNA 5 was 90%, which is relatively high, but not the greatest measured. The coincidence of a high binding specificity and a large number of cleavage sites could indicate that BLM responds to the tertiary structure assumed by this hairpin DNA in a fashion which enables it to make many contacts with the molecule. Unlike DNA 2 (Figure S2), which had comparatively few sites of cleavage and a higher binding specificity, DNA 5 (Figure 3) may bind less tightly, allowing for interaction sufficient to support cleavage activity, but sufficiently indiscriminate that BLM binding and cleavage can occur at many points on the DNA molecule.
In this context, it is instructive to consider the results of a recent study in which we analyzed the dynamics of BLM interaction with some of these hairpin DNAs using surface plasmon resonance (SPR).35 Two important observations were made during this study. First, each of the hairpin DNAs studied, including 2, 4 and 5, formed a single, strongly bound complex with Fe(III)•BLM but at least one, and likely multiple, more weakly bound and more transient complexes. Nonetheless, all of these DNAs had more than one site cleaved strongly by Fe(II)•BLM, arguing that efficient cleavage at a given site need not result from persistent binding at that site. The second observation was that for strongly bound hairpin DNAs, even when BLM was not bound to the DNA as judged by SPR analysis, it was not available to bind to a second hairpin DNA known to be an efficient substrate for cleavage by Fe•BLM. The picture that emerges from the present study, as inferred by the recent study of the dynamic interaction of BLM with the hairpin DNAs, is that the binding interactions of BLM with the individual hairpin DNAs must be driven primarily by one, or a small number of specific interactions, but that these binding interactions are far from irreversible, and permit sampling of numerous other sites on the same DNA, some of which may lead more readily to DNA cleavage.
The selection and characterization of the present library through the study of its sites and facility of cleavage by Fe•BLM represents an important step in delineating the relationship between DNA binding and cleavage by BLM. These results, in concert with an understanding of double-strand DNA damage mediated by BLM may well provide key insights into the way in which BLM acts as a therapeutic agent in vivo. The exact DNA structures and precise motifs that BLM prefers, however, are still elusive and worthy of further study. Possible strategies include the use of a SELEX-type procedure that permits iterative selections to be employed, along with a larger randomized region for binding by BLM and metallobleomycins.
The characterization of double-strand DNA damage has been an important priority in BLM studies due to the belief that it may represent the mechanism through which BLM exerts its antitumor effects. While double-strand cleavage is undoubtedly sequence dependent, it has been estimated to represent about 20% of all DNA strand breaks mediated by Fe•BLM.40 The Povirk laboratory pioneered the study of the characteristics and mechanism of double-strand DNA breaks mediated by bleomycin, and was the first to define specific sequence patterns associated with this double-strand cleavage.41-43 Other laboratories subsequently provided important mechanistic observations concerning double-strand cleavage.44-46 Given the large number of closely spaced DNA cleavage sites on opposite strands of the hairpin DNAs studied here, it seemed logical to anticipate that some of these might constitute sites of double-strand DNA cleavage. In order to characterize this hairpin DNA library for its propensity to undergo double-strand DNA cleavage by Fe(II)•BLM A5, we developed a new strategy for analyzing double strand cleavage, exemplified here for hairpin DNA 4. The strategy involved alternatively 32P-labeling samples of the DNA on the 5′ and 3′-ends. Following Fe•BLM-mediated DNA cleavage, the samples were analyzed by native polyacrylamide gel analysis. Double strand cleavage should result in 5′ and [3′-32P] end labeled DNA duplexes that co-migrate, as actually observed in Figure 6A. Recovery of the individual co-migrating bands, followed by sequencing gel analysis of each, then permitted the sites of double-strand cleavage to be determined (Figure 6B). When hairpin DNA 4 was subjected to this method of characterization three individual sites of double strand cleavage were identified (Figures 6 and 7). The A12-T53 site was located in the variable region of the hairpin DNA whereas the other two sites were in the flanking region of the hairpin DNA common to all members of the library. It may be noted that none of these three sites conforms to the patterns of double-strand cleavage noted previously by the Povirk laboratory. 41-43 The presence of large numbers of closely spaced DNA cleavage sites on both strands of the hairpin DNA library suggests that numerous double-strand cleavage sites may be present. While the extent of double-strand cleavage of randomly chosen DNAs occurs at a level of frequency well beyond can be accounted for by the random accumulation of single strand breaks,40,41 the hairpin DNAs studied here, which were selected for tight binding to BLM, have recently been shown to sequester Fe•BLM,35 such that double-strand cleavage might plausibly result from two independent cleavage events more frequently than for randomly chosen DNAs.
The methods developed for studying the damage inflicted by BLM A5 on hairpin DNAs now extends to the identification of sites of double-strand cleavage. The use of the newly developed method for identifying double-strand cleavage sites will no doubt provide additional insights into the mechanism of BLM action when it is applied to a number of strongly bound hairpin DNAs. For example, the finding of strongly bound hairpin DNAs subject to DNA damage at multiple sites might suggest critical loci in DNA particularly susceptible to BLM-mediated cleavage, and which constitute sites at which BLM exerts its therapeutic effects.
Supplementary Material
Footnotes
Supporting Information
High resolution polyacrylamide electrophoresis gels for several hairpin DNAs are available in the supporting information. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.de Wit R, Stoter G, Kaye SB, Sleifer DT, Jones WG, Huinink W. W. t. B., Rea LA, Collette L, Sylvester R. Importance of bleomycin in combination chemotherapy for good-prognosis testicular nonseminoma: a randomized study of the European Organization for Research and Treatment of Cancer Genitourinary Tract Cancer Cooperative Group. J. Clin. Oncol. 1997;15:1837–1843. doi: 10.1200/JCO.1997.15.5.1837. [DOI] [PubMed] [Google Scholar]
- 2.Hecht SM. Bleomycin Group Antitumor Agents. In: Cragg GM, Kingston DGI, Newman DJ, editors. Anticancer Agents from Natural Products. 2nd ed. CRC Press; Florida: 2012. pp. 451–478. [Google Scholar]
- 3.Sausville EA, Peisach J, Horwitz SB. Inhibition of SV40 DNA synthesis by camptothecin and neocarzinostatin. Biochemistry. 1978;17:2740–2746. [PubMed] [Google Scholar]
- 4.Sausville EA, Stein RWP, Horwitz SB. Properties and products of the degradation of DNA by bleomycin and iron(II). Biochemistry. 1978;17:2746–2754. doi: 10.1021/bi00607a008. [DOI] [PubMed] [Google Scholar]
- 5.Hecht SM. The chemistry of activated bleomycin. Acc. Chem. Res. 1986;19:383–391. [Google Scholar]
- 6.Burger RM. Cleavage of nucleic acids by bleomycin. Chem. Rev. 1998;98:1153–1170. doi: 10.1021/cr960438a. [DOI] [PubMed] [Google Scholar]
- 7.Carter BJ, de Vroom E, Long EC, van der Marel GA, van Boom JH, Hecht SM. Site-specific cleavage of RNA by Fe(II)•bleomycin. Proc. Natl. Acad. Sci. U.S.A. 1990;87:9373–9377. doi: 10.1073/pnas.87.23.9373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kuramochi H, Takahashi K, Takita T, Umezawa H. An active intermediate formed in the reaction of bleomycin-Fe(II) complex with oxygen. J. Antibiot. 1981;34:576–582. doi: 10.7164/antibiotics.34.576. [DOI] [PubMed] [Google Scholar]
- 9.Burger RM, Peisach J, Horwitz SB. Activated bleomycin. A transient complex of drug, iron, and oxygen that degrades DNA. J. Biol. Chem. 1981;256:11636–11644. [PubMed] [Google Scholar]
- 10.Burger RM, Kent TA, Horwitz SB, Munck E, Peisach J. Mössbauer study of iron bleomycin and its activation intermediates. J. Biol. Chem. 1983;258:1559–1564. [PubMed] [Google Scholar]
- 11.Hecht SM. DNA strand scission by activated bleomycin group antibiotics. Fed. Proc. 1986;45:2784–2791. [PubMed] [Google Scholar]
- 12.Steighner RJ, Povirk LF. Bleomycin-induced DNA lesions at mutational hot spots: Implications for the mechanism of double-strand Cleavage. Proc. Natl. Acad. Sci. U.S.A. 1990;87:8350–8354. doi: 10.1073/pnas.87.21.8350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu JC, Kozarich JW, Stubbe J. Mechanism of bleomycin: Evidence for a rate-determining 4'-hydrogen abstraction from poly(dA-dU) associated with the formation of both free base and base propenal. Biochemistry. 1985;24:7562–7568. doi: 10.1021/bi00347a009. [DOI] [PubMed] [Google Scholar]
- 14.Kross J, Henner WD, Hecht SM, Haseltine WA. Specificity of deoxyribonucleic acid cleavage by bleomycin, phleomycin, and tallysomycin. Biochemistry. 1982;21:4310–4318. doi: 10.1021/bi00261a021. [DOI] [PubMed] [Google Scholar]
- 15.Hecht SM. Bleomycin: New perspectives on the mechanism of action. J. Nat. Prod. 2000;63:158–168. doi: 10.1021/np990549f. [DOI] [PubMed] [Google Scholar]
- 16.Kilkuskie RES, Hecht SM. Oxygen transfer by bleomycin analogs dysfunctional in DNA cleavage. J. Am. Chem. Soc. 1985;107:260–261. [Google Scholar]
- 17.Carter BJ, Murty VS, Reddy KS, Wang SN, Hecht SM. A role for the metal binding domain in determining the DNA sequence selectivity of Fe-bleomycin. J. Biol. Chem. 1990;265:4193–4196. [PubMed] [Google Scholar]
- 18.Tan JD, Farinas ET, David SS, Mascharak PK. NMR evidence of sequence specific DNA binding by a cobalt(III)-bleomycin analog with tethered acridine. J. Inorg. Chem. 1994;33:4295–4308. [Google Scholar]
- 19.Wu W, Vanderwall DE, Turner CJ, Kozarich JW, Stubbe J. Solution structure of Co•bleomycin A2 green complexed with d(CCAGGCCTGG). J. Am. Chem. Soc. 1996;118:1281–1294. [Google Scholar]
- 20.Povirk LF, Hogan M, Dattagupta N. Binding of bleomycin to DNA: intercalation of the bithiazole rings. Biochemistry. 1979;18:96–101. doi: 10.1021/bi00568a015. [DOI] [PubMed] [Google Scholar]
- 21.Sucheck SJ, Ellena JF, Hecht SM. Characterization of Zn(II)•deglycobleomycin A2 and interaction with d(CGCTAGCG)2: direct evidence for minor groove binding of the bithiazole moiety. J. Am. Chem. Soc. 1998;120:7450–7460. [Google Scholar]
- 22.Abraham AT, Zhou X, Hecht SM. Metallobleomycin-mediated cleavage of DNA not involving a threading-intercalation mechanism. J. Am. Chem. Soc. 2001;123:5167–5175. doi: 10.1021/ja002460y. [DOI] [PubMed] [Google Scholar]
- 23.Wu W, Vanderwall DE, Lui SM, Tang X-J, Turner CJ, Kozarich JW, Stubbe J. Studies of Co·bleomycin A2 green: its detailed structural characterization by NMR and molecular modeling and its sequence-specific interaction with DNA oligonucleotides. J. Am. Chem. Soc. 1996;118:1268–1280. [Google Scholar]
- 24.Boger DL, Ramsey TM, Cai H, Hoehn ST, Stubbe J. A systematic evaluation of the bleomycin A2 L-threonine side chain: its role in preorganization of a compact conformation implicated in sequence-selective DNA cleavage. Nucleic Acids Res. 1998;120:9139–9148. [Google Scholar]
- 25.Rishel MJ, Thomas CJ, Tao ZF, Vialas C, Leitheiser CJ, Hecht SM. Conformationally constrained analogues of bleomycin A5. J. Am. Chem. Soc. 2003;125:10194–10205. doi: 10.1021/ja030057w. [DOI] [PubMed] [Google Scholar]
- 26.Oppenheimer NJ, Rodriguez LO, Hecht SM. Structural studies of “active complex” of bleomycin: assignment of ligands to the ferrous ion in a ferrous-bleomycin-carbon monoxide complex. Proc. Natl. Acad. Sci. U.S.A. 1979;76:5616–5620. doi: 10.1073/pnas.76.11.5616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oppenheimer NJ, Chang C, Chang LH, Ehrenfeld G, Rodriguez LO, Hecht SM. Deglyco-bleomycin. Degradation of DNA and formation of a structurally unique Fe(II)•CO complex. J. Biol. Chem. 1982;257:1606–1609. [PubMed] [Google Scholar]
- 28.Chapuis JC, Schmaltz RM, Tsosie KS, Belohlavek M, Hecht SM. Carbohydrate dependent targeting of cancer cells by bleomycin-microbubble conjugates. J. Am. Chem. Soc. 2009;131:2438–2439. doi: 10.1021/ja8091104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yu Z, Schmaltz RM, Bozeman TC, Paul R, Rishel MJ, Tsosie KS, Hecht SM. Selective tumor cell targeting by the disaccharide moiety of bleomycin. J. Am. Chem. Soc. 2013;135:2883–2886. doi: 10.1021/ja311090e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thomas CJ, Chizhov AO, Leitheiser CJ, Rishel MJ, Konishi K, Tao Z-F, Hecht SM. Solid phase synthesis of bleomycin A5 and three monosaccharide analogues. Exploring the role of the carbohydrate moiety in RNA cleavage. J. Am. Chem. Soc. 2002;124:12926–12927. doi: 10.1021/ja0208916. [DOI] [PubMed] [Google Scholar]
- 31.Leitheiser CJ, Smith KL, Rishel MJ, Hashimoto S, Konishi K, Thomas CJ, Li C, McCormick MM, Hecht SM. Solid phase synthesis of bleomycin group antibiotics. Construction of a 108-member deglycobleomycin library. J. Am. Chem. Soc. 2003;125:8218–8227. doi: 10.1021/ja021388w. [DOI] [PubMed] [Google Scholar]
- 32.Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 33.Akiyama Y, Ma Q, Edgar E, Laikhter A, Hecht SM. A novel DNA hairpin substrate for bleomycin. Org. Lett. 2008;10:2127–2130. doi: 10.1021/ol800445x. [DOI] [PubMed] [Google Scholar]
- 34.Ma Q, Akiyama Y, Xu Z, Konishi K, Hecht SM. Identification and cleavage site analysis of DNA sequences bound strongly by bleomycin. J. Am. Chem. Soc. 2009;131:2013–2022. doi: 10.1021/ja808629s. [DOI] [PubMed] [Google Scholar]
- 35.Bozeman TC, Nanjunda R, Tang C, Liu Y, Segerman ZJ, Zaleski PA, Wilson WD, Hecht SM. Dynamics of bleomycin interaction with a strongly bound hairpin DNA substrate, and implications for cleavage of the bound DNA. J. Am. Chem. Soc. 2012;134:17842–17845. doi: 10.1021/ja306233e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Giroux RA, Hecht SM. Characterization of bleomycin cleavage sites in strongly bound hairpin DNAs. J. Am. Chem. Soc. 2010;132:16987–16996. doi: 10.1021/ja107228c. [DOI] [PubMed] [Google Scholar]
- 37.Maxam AM, Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65:499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- 38.Akiyama Y, Ma Q, Edgar E, Laikhter A, Hecht SM. Identification of strong DNA binding motifs for bleomycin. J. Am. Chem. Soc. 2008;130:9650–9651. doi: 10.1021/ja802905g. [DOI] [PubMed] [Google Scholar]
- 39.Kozarich JW, Worth L, Jr., Frank BL, Christner DF, Vanderwall DE, Stubbe J. Sequence-specific isotope effects on the cleavage of DNA by bleomycin. Science. 2013;243:1396–1399. doi: 10.1126/science.2476851. [DOI] [PubMed] [Google Scholar]
- 40.Boger DL, Honda T, Menezes RF, Colletti SL. Total synthesis of bleomycin A2 and related agents. 3. Synthesis and comparative evaluation of deglycobleomycin A2 , epideglycobleomycin A2, deglycobleomycin A1, and desacetamido-, descarboxamido-, desmethyl-, and desimidazolyldeglycobleomycin A2. J. Am. Chem. Soc. 1994;116:5631–5646. [Google Scholar]
- 41.Povirk LF, Houlgrave CW. Effect of apurinic/apyrimidinic endonucleases and polyamines on DNA treated with bleomycin and neocarzinostatin: specific formation and cleavage of closely opposed lesions in complementary strands. Biochemistry. 1988;27:3850–3857. doi: 10.1021/bi00410a049. [DOI] [PubMed] [Google Scholar]
- 42.Povirk LF, Han YH, Steighner RJ. Structure of bleomycin-induced DNA double-strand breaks: predominance of blunt ends and single-base 5' extensions. Biochemistry. 1989;28:5808–5814. doi: 10.1021/bi00440a016. [DOI] [PubMed] [Google Scholar]
- 43.Steighner RR, Povirk LF. Bleomycin-induced DNA lesions at mutational hot spots: implications for the mechanism of double-strand cleavage. Proc. Natl. Acad. Sci. U.S.A. 1990;87:8350–8354. doi: 10.1073/pnas.87.21.8350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Absalon MJ, Kozarich JW, Stubbe J. Sequence-specific double-strand cleavage of DNA by Fe-bleomycin. 1. The detection of sequence-specific double-strand breaks using hairpin oligonucleotides. Biochemistry. 1989;34:2065–2074. doi: 10.1021/bi00006a029. [DOI] [PubMed] [Google Scholar]
- 45.Absalon MJ, Wu W, Kozarich JW, Stubbe J. Sequence-specific double-strand cleavage of DNA by Fe-bleomycin. 2. Mechanism and dynamics. Biochemistry. 1995;34:2076–2086. doi: 10.1021/bi00006a030. [DOI] [PubMed] [Google Scholar]
- 46.Keck MV, Manderville RA, Hecht SM. Chemical and structural characterization of the interaction of bleomycin A2 with d(CGCGAATTCGCG)2. Efficient, double-strand DNA cleavage accessible without structural reorganization. J. Am. Chem. Soc. 2001;123:8690–8700. doi: 10.1021/ja003795i. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.