Abstract
Reduced signaling through the C. elegans insulin/insulin-like growth factor-1-like tyrosine kinase receptor daf-2 and dietary restriction via bacterial dilution are two well-characterized lifespan-extending interventions that operate in parallel or through (partially) independent mechanisms. Using accurate mass and time tag LC-MS/MS quantitative proteomics, we detected that the abundance of a large number of ribosomal subunits is decreased in response to dietary restriction, as well as in the daf-2(e1370) insulin/insulin-like growth factor-1-receptor mutant. In addition, general protein synthesis levels in these long-lived worms are repressed. Surprisingly, ribosomal transcript levels were not correlated to actual protein abundance, suggesting that post-transcriptional regulation determines ribosome content. Proteomics also revealed the increased presence of many structural muscle cell components in long-lived worms, which appeared to result from the prioritized preservation of muscle cell volume in nutrient-poor conditions or low insulin-like signaling. Activation of DAF-16, but not diet restriction, stimulates mRNA expression of muscle-related genes to prevent muscle atrophy. Important daf-2-specific proteome changes include overexpression of aerobic metabolism enzymes and general activation of stress-responsive and immune defense systems, whereas the increased abundance of many protein subunits of the proteasome core complex is a dietary-restriction-specific characteristic.
About two decades ago, it was discovered that mutation in the single gene daf-2, which encodes an insulin/insulin-like growth factor-1 (IGF-1)1 receptor ortholog, approximately doubles the normal lifespan of the small roundworm Caenorhabditis elegans (1, 2). This beneficial effect on lifespan requires changes in gene expression that rely mainly on the activation of DAF-16, a conserved FoxO family transcription factor negatively regulated by DAF-2 (1, 3, 4). Genetic analysis has revealed many processes that are directly or indirectly controlled by this pathway. These include stress and pathogen resistance (5–10), dauer formation (2, 11, 12), autophagy (13–15), and energy metabolism (16–18), all of which seem to be associated to varying extents with the Age phenotype. The evolutionary conservation of the insulin/IGF-1-mediated effect on lifespan in many species, including humans, constitutes the rationale for the intensive ongoing study of the aging process in invertebrate models such as C. elegans (19).
Long before the insulin/IGF-1 regulatory pathway became associated with longevity, it was well known that restricting caloric intake, without causing malnutrition, had a beneficial effect on aging in a wide range of model organisms (20). Although dietary restriction (DR) is a nongenetic intervention, the longevity response to DR requires altered gene expression, controlled by nutrient-sensing pathways such as insulin/IGF-1-like signaling (IIS) (21–23), AMP kinase (24), and target of rapamycin kinase (TOR) signaling (25–29). A number of methods were developed to restrict nutrient uptake and extend the C. elegans lifespan. Importantly, different DR regimes rely on different regulatory pathways to extend the C. elegans lifespan (30). Intermittent fasting or starting DR at mid-life extends lifespan, at least in part, by inhibiting IIS (23, 24), whereas restricting diet throughout an organism's life appears to extend lifespan while bypassing IIS (21, 28, 30–32). However, interaction studies suggest that DR and IIS may act in parallel on common downstream processes (33, 34). One such process is autophagy, which is associated with both IIS- and DR-induced longevity (13, 14). Protein synthesis is another candidate mechanism because in mammals, TOR (35) and insulin signaling (36) regulate global translation levels, and attenuating global translation extends nematode lifespan (27, 37, 38).
Because of the importance of IIS in lifespan extension, a large number of studies have attempted to uncover the transcriptional targets of DAF-16. Potential DAF-16 targets can be identified by the presence of conserved DAF-16 consensus binding sites in the promoter sequence (39, 40). Chromatin profiling techniques such as CHiP (41) and DamID (42) allow the identification of direct regulatory targets of DAF-16. To date, whole-genome transcript profiling based on serial analysis of gene expression or microarrays has been the method of choice for mapping DAF-16-dependent transcriptional changes (43–49). However, cross-platform validation studies have indicated (very) poor concordance between serial analysis of gene expression and microarray technologies (50, 51), and comparisons between candidate DAF-16 target genes from serial analysis of gene expression and microarray studies indeed show little overlap (48, 52). In addition, diverse post-transcriptional and post-translational regulatory processes are responsible for the poor correlation (40% in mammalian cells) between the transcript level and the final protein gene product (53, 54). Therefore, variations in gene expression are always best studied at the proteome level (55).
Dong et al. (2007) published a first profile of the daf-2 proteome involving quantitative 15N stable isotope labeling and identified 86 differentially expressed proteins involved in amino acid biosynthesis, carbohydrate metabolism, and reactive oxygen species detoxification (56). Follow-up genetic analysis identified the protein phosphatase calcineurin A as a novel positive regulator of DAF-2 signaling, antagonizing the daf-2 longevity phenotype (56). Two-dimensional difference gel electrophoresis analysis of the dauer (57, 58) and daf-2 (58) proteome identified a handful of differentially expressed proteins involved in anaerobic metabolism, stress response, and reactive oxygen species detoxification. Recently the first metabolic profiles were generated for daf-2, dauer larvae, and a translation-deficient mutant (18). A recent proteomic analysis of the eat-2 mutant, a genetic model for dietary restriction, indicated a potentially altered metabolism characterized by a subtle shift from carbohydrate to fatty acid metabolism (59).
In this study, we sought to uncover potentially common mechanisms that underlie C. elegans longevity by identifying similar changes in the proteome of long-lived worms either mutated in the daf-2 IIS receptor or fed a restricted diet. We were also interested in proteomic alterations that point to mechanisms unique for each condition. We found that long-lived worms were characterized by an overall decrease in the abundance of ribosomal subunit proteins, along with reduced protein synthesis levels. In addition, we show that changes in ribosome content are not reflected at transcript level and must therefore result from post-transcriptional regulation. Long-lived worms also show a relative increase in striated muscle proteins that is likely the result of maintained muscle integrity even in low-food environments. Condition-specific proteome changes constitute the largest and functionally most diverse group. Prominent categories include the DR-induced expression of proteasome subunits and the daf-2-specific increase in expression of most aerobic metabolism enzymes, together with a general activation of stress and immune responses.
EXPERIMENTAL PROCEDURES
C. elegans Strains
The following strains were used for proteomics and qRT-PCR: GA154 glp-4(bn2ts)I; daf-2(e1370)III (long-lived) and GA153 glp-4(bn2ts)I daf-16(mgDf50)I; daf-2(e1370)III (control strain), which were kindly provided by David Gems at University College London (46). The glp-4(bn2) background allele results in germline-deficient worms when growth occurs at the non-permissive temperature of 24 °C. This allows one to focus on the proteome of aging somatic cells, excluding the gonadal and complex embryonic material (60). The daf-16; daf-2 mutant alleles in our control allow the identification of daf-2 effects mediated by the DAF-16 transcription factor; activation of the latter is required for the daf-2 mutant longevity phenotype. Previous lifespan analyses by McElwee et al. and TeKippe and Aballay demonstrated no increase in lifespan conferred by the presence of the glp-4(bn2) allele in otherwise wild-type or daf-16; daf-2 mutants grown on live E. coli (46, 61). However, glp-4(bn2) causes a small but significant DAF-16-dependent extension of lifespan of daf-2 mutants and nematodes grown on killed E. coli (46, 61). Previous MS proteome profiling of glp-4(bn2) after 15N metabolic labeling revealed only modest changes relative to the N2 wild type (62). The RNAi hypersensitive NL2099 rrf-3(pk1426) II strain was used for RNAi lifespan analyses.
Culturing and Sampling
Large age-synchronized populations were initiated from isolated eggs of alkaline-hypochlorite-bleached gravid adult nematodes (63). Eggs were allowed to hatch overnight in S-buffer (0.1 m NaCl, 0.05 m potassium phosphate, pH 6) without bacteria. First-stage larvae (L1) were seeded onto cholesterol-supplemented (5 mg/ml) nutrient agar plates (Oxoid, Hampshire, UK) containing a lawn of freshly grown E. coli K12 cells. In order to prevent glp-4 daf-16; daf-2 animals from becoming dauer larvae, worms were grown at 16 °C until the third larval stage (L3) and then at 24 °C for the remainder of the experiment. At the fourth larval stage, worms were transferred into Fernbach flasks containing 250 ml S-buffer at densities not exceeding 1500 worms/ml and shaken at 120 rounds per minute. Frozen E. coli K12 cells were added at approximately 3 × 109 cells ml−1 (A550 = 1.8) and approximately 1 × 109 cells ml−1 (A550 = 0.8) for, respectively, fully fed (FF) and DR culturing of nematodes. Although shifting the temperature at the L3 stage rendered glp-4; daf-2 animals sterile, we noticed that by day 2 of adulthood, glp-4 daf-16; daf-2 mutants exhibited gonads with incomplete morphogenesis yet were able to produce 2 to 4 eggs per animal. Therefore, we opted to add 5-fluoro-2′-deoxyuridine (75 μm final concentration; Acros Organics, Geel, Belgium) to all mass cultures in all biological replicates in order to maintain complete sterility. To maintain the initial bacterial concentration, the turbidity at 550 nm (A550) was measured twice each day, and K12 was added as needed. For all experiments, samples were collected at day 2 of adulthood and cleared of dead animals, debris, and bacteria through Percoll (Sigma-Aldrich) washing and floatation on a 60% W/W sucrose solution (64). Animals were immediately flash frozen in liquid nitrogen and stored at −80 °C.
Study Design
Per condition, five biological replicate samples of adult two-day-old C. elegans were used to achieve statistically meaningful results. Each replicate had three pools of FF and DR glp-4 daf-16; daf-2 and FF glp-4; daf-2 mutants, respectively.
Protein Extraction and Tryptic Digestion
For each sample, 40 μl (∼800 worms) of worm suspension in PBS were mixed with 260 μl of denaturing solution consisting of 8 m urea, 1 mm EDTA, and 10 mm DTT in 50 mm TrisHCl buffer (pH 8.0). The samples were immediately frozen in liquid nitrogen and homogenized in random order via pulverization with a hammer, metal pestle, and mortar. Homogenization was followed by recovery of the powder, brief thawing, and sonication for 30 s in a 5510 Branson ultrasonic water bath (Branson Ultrasonics, Soest, The Netherlands). On average, the amount of recovered protein estimated via Coomassie measurement was ∼600 μg. Because the recovery of the powder from the mortar varied significantly, we took a fixed-volume aliquot of 100 μl (∼300 μg) for further processing. The further processing employed a commonly used protocol that is described elsewhere (65). Briefly, the homogenate was incubated for 30 min at 37 °C to allow sufficient time for disulfide bond reduction by 10 mm DTT present in the solution. The sulfhydryl groups of cysteines were protected by alkylation with iodoacetamide, which was added to 40 mm, after which samples were incubated at 37 °C in the dark for 1 h. The mixture was then diluted 4-fold to reduce the urea concentration (to ensure that trypsin was not denatured) and supplemented with 1 mm CaCl2. Trypsin was added at a 1/50 enyzme/protein ratio, which corresponded to 6 μg. After incubation at 37 °C overnight, the resulting tryptic peptides were purified using C18 solid phase extraction (66). The samples were dried down from 500 μl to ∼50 μl via lyophilization to remove the organic solvents. The tryptic peptide samples were reconstituted in 25 mm NH4HCO3, and their concentrations were measured via BCA assay. Concentrations were adjusted to 0.5 μg/μl.
LC/MS analysis
Each individual sample was analyzed with a constant-pressure capillary HPLC system coupled online to an LTQ Orbitrap mass spectrometer (Thermo Fisher, San Jose, CA) using an electrospray ionization interface. The reversed-phase capillary column was prepared by slurry packing 3-μm Jupiter C18 particles (Phenomenex, Torrence, CA) into a 75 μm inner diameter × 65 cm fused silica capillary (Polymicro Technologies, Phoenix, AZ). The mobile phase solvents consisted of (A) 0.2% acetic acid and 0.05% TFA in water and (B) 0.1% TFA in 90% acetonitrile. An exponential gradient was used for the separation, which started with 100% A and gradually increased to 60% B over 100 min. The instrument was operated in a data-dependent mode with an m/z range of 400–2000. The mass spectrometer was operated in the data-dependent mode to automatically switch between MS and MS/MS acquisition. Survey MS spectra (m/z 400–2000) were acquired at 100,000 resolution. The three most intense ions were consecutively selected and fragmented in the linear ion trap using collision-induced dissociation with a normalized collision energy of 30%. Former target ions selected for MS/MS were dynamically excluded for 30 s.
Peptide Identification
The analyses of quantitative LC-MS datasets were performed as previously described in detail (65). LC-MS feature detection was performed by deisotoping of individual MS scans using Decon2LS (version 1.0.3464.28342; parameters are all defaults except PeakBackgroundRatio = 1.3 and SignalToNoiseThreshold = 2) (67) followed by grouping of monoisotopic peaks constituting the LC profile and assignment of LC-MS features to peptides using VIPER (version 3.48; all defaults except mass and elution time measurement error tolerances as described below) (68). The latter was done by matching the elution time and monoisotopic mass to the database of peptides previously identified using the MS/MS fragmentation approach with tolerances of 2% of normalized elution time and 2 ppm, respectively. False peptide identifications are likely to be present only in one or a few samples. Therefore, to get a highly confident list of identifications, we discarded all the peptides that were not fully present (five out of five) in any of three experimental conditions. The false discovery rate of peptide identification for the accurate mass and time approach or the number of false peptide identifications among all identifications that passed the mass and elution time tolerance thresholds was estimated using the 11-Da shift approach (69) and was contained just below 1%. Identified peptides are accessible through PeptideAtlas (70) (supplemental File S1).
Quantification of Changes in Protein Abundance
Peptide arbitrary abundance was measured using a label-free approach as the sum of the intensities of all monoisotopic ions constituting the peptide's elution profile or the LC-MS feature. To minimize systematic errors in quantitation, we applied a normalization procedure that estimates sample-specific biases and corrects the intensities of a corresponding peptide for each sample (69, 71). For the purpose of statistical testing, the missing values were inputted using the K-NN approach (72) with the Spearman rank correlation as a distance metric between the abundance profiles of two peptides (missing values were assigned the lowest rank within the peptide abundance profile). In order to avoid a misleading interpretation of the quantitative data, we retained only those peptides that unambiguously matched proteins within the WS210 WormBase freeze of the protein sequences. Relative protein abundances were estimated as the sum of abundances of constituting peptides. Differentially abundant proteins were identified using a t test followed by correction for multiplicity of testing.
Functional Analysis
The normalized dataset was visualized and analyzed with MultiExperiment Viewer, part of the TM4 microarray software suit (73, 74). Multivariate analyses (principal component analysis (PCA) and hierarchical clustering analysis (HCA)) were performed using the MultiExperiment Viewer package. PCA was used to discover principal patterns in protein expression (PCA on proteins) and also to identify prevalent components among samples (PCA on conditions) and how they separated after analysis (75). HCA of samples was used to quantify the degree of similarity between samples visualized as sets of nested clusters organized as a tree (dendrogram), with Pearson correlation as the distance metric and average pairwise distance as the Linkage method (76). Pavlidis template matching (77) was used to generate gene lists conforming to an expression pattern of interest. The Pavlidis template matching algorithm allows a dataset to be searched for proteins with abundance profiles that match a user-defined template profile, based on the Pearson correlation between the template and the proteins in the dataset (77). To further aid in the identification of important altered biological processes, we performed functional gene annotation clustering using the DAVID Functional Annotation Tool with a modified Fisher's exact p value (EASE score) of 0.05 (78, 79). Gene set enrichment analysis (GSEA) was used to determine whether predefined sets of proteins derived from annotation clustering showed statistically significant differential expression as described in detail in Ref. 80. In short, a ranked list of proteins is generated according to their differential abundance level between two experimental groups. An enrichment score test statistic is calculated that reflects the degree to which a defined set of proteins is overrepresented at one of the extremes of the entire ranked list. Positive enrichment score values correspond to enrichment for enzymes ranked as having higher expression levels in daf-2 mutants, and vice versa. Gene sets eligible for GSEA are reported with their nominal p value and false discovery rate (FDR), calculated from 1000 permutations of the experimental group labels. The nominal p value estimates the statistical significance of the enrichment score for a single set. The FDR is the estimated probability that an enriched set represents a false positive finding. Subramanian et al. have suggested an FDR cutoff of 25% as appropriate to signify biologically significant results (80). The GSEA-P software used for GSEA can be found at the Broad Institute website.
35S Protein Incorporation Assay
35S-labeled bacteria were obtained by growing E. coli K12 overnight at 37 °C in low-sulfate medium (44 mm Na2HPO4, 22 mm KH2PO4, 85 mm NaCl, 20 mm NH4Cl, 1.25 mg/l thiamine, 0.1% (w/v) glucose, 2 mm MgCl2) (161) supplemented with LB medium (1% final concentration) and 5 μCi/ml [35S]sulfate (PerkinElmer Life Sciences, Waltham, MA). The bacterial concentration was determined by measuring the optical density at 550 nm. 35S-bacteria were fed to worms cultured in 10 ml S-buffer (approximately 1000 worms/ml). FF or DR nematodes were fed 35S-bacteria at concentrations corresponding to their dietary status (FF: A550 = 1.8; DR: A550 = 0.8). Samples of two-day-old adult worms were taken at regular time intervals and worms were washed five times in S-buffer supplemented with nonradioactive E. coli K12 to purge the intestines of undigested 35S-labeled bacteria. To isolate proteins, worms were first boiled for 15 min in 50% Tris-SDS buffer (25 mm Tris, 250 mm NaCl, 5% SDS, pH 7.4) and samples were centrifuged for 5 min at 14,000 rpm. To precipitate proteins, trichloroacetic acid (TCA) (final concentration of 9.3%) was added to the supernatant, and the mixture was allowed to incubate at room temperature for 1 h. Precipitated proteins were centrifuged at 14,000 rpm for 5 min and washed once with 1 ml of 10% TCA. The protein pellet was dissolved in 150 μl 350 mm NaOH for at least 1 h at room temperature. 100 μl was added to 5 ml Ultima Gold LSC mixture (PerkinElmer Life Sciences) for liquid scintillation counting in a Tri-Carb 2800TR liquid scintillation counter (PerkinElmer Life Sciences). Counts per minute were normalized to the total protein concentration as determined using a BCA Protein Assay Kit (Thermo Scientific, Rockford, IL). The protein synthesis rate was expressed as the slope of 35S protein incorporation over time.
Quantitative Real-time RT-PCR
Total RNA was isolated using the RNeasy Midi Kit (Qiagen, Venlo, The Netherlands) according to manufacturer's instructions. DNase I (Zymo Research, Orange, CA) was added to all samples. RNA concentration and purity were determined using a NanoDrop ND 1000 spectrophotometer (NanoDrop, Wilmington, DE). First-strand cDNA of ribosomal and muscle-related genes was synthesized from 2 μg RNA using an oligo(dT) primer and Moloney murine leukemia virus reverse transcriptase (Fermentas, Vilnius, Lithuania) at 42 °C for 1 h. Full-length gene sequences were obtained from WormBase (release WS170), and primers were designed using Primer3 software according to Ref. 81. Primer binding specificity was tested using UCSC In-Silico PCR. Primer binding sites were evaluated for the absence of secondary structures by means of MFOLD analysis using default settings, 50 mm Na+, 3 mm Mg2+, and a temperature of 60 °C. Primers were purchased from Invitrogen (Carlsbad, CA). Quantitative RT-PCR was performed using a Rotor-Gene 2000 centrifugal real-time cycler (Corbett Research, Mortlake, Australia) using the Platinum SYBR Green qPCR SuperMix-UDG (Invitrogen) as described previously (82). The identity of each PCR product was confirmed by a single melt peak for each reaction. For each assay and for every primer pair, a no-template control was included. For each biological replicate (four in total), all measurements were performed in duplicate to account for variation. The threshold cycle (Ct) values obtained from the Rotor-Gene software (version 6, Corbett Research, Bath, UK) were exported to qBasePLUS software (version 2.2, Biogazelle, Zwijnaarde, Belgium) for real-time PCR data analysis as described in detail elsewhere (82). Three stable reference genes (cdc-42, pmp-3, and eif-3.C) were identified by GeNorm 3.4 software out of 10 previously selected candidate genes (82). An unpaired two-tailed Student's t test was used to compare differences in expression levels between strains.
Transmission Electron Microscopy and Image Analysis
Three day-2 young adult glp-4(bn2) daf-16(mgDf50); daf-2(e1370) and glp-4(bn2); daf-2(e1370) C. elegans nematodes were fixed and transverse sections were prepared according to the procedure described by Fonderie et al. (83). Electron microscopy was done using a Jeol JEM 1010 (Jeol, Tokyo, Japan) operating at 60 kV. Images were digitized using a DITABIS system (Pforzheim, Germany). KS 400 imaging software (Carl Zeiss, Zaventem, Belgium) was used to calculate surface areas. Relative differences in striated body-wall muscle transverse area were tested for significance with analysis of variance and Fisher's least significant difference post hoc test.
Quantitation of Branched Chain Amino Acids
The determination of amino acid concentrations via HPLC was performed as described elsewhere (84). In short, free amino acids were extracted by treating worm homogenates with 15% TCA and taking the supernatant. After filtration over a 0.45-μm polytetrafluoroethylene syringe filter (Grace, Lokeren, Belgium), amino acids were derivatized with o-phthaldialdehyde in the injector of the Agilent 1100 HPLC system (Agilent Technologies, Santa Clara, CA). Labeled amino acids were then separated on a Zorbax Eclipse Amino Acid Analysis HPLC column (Agilent) and quantized via fluorometry (340ex./450em. nm). For the quantification, norvaline and sarcosine were used as internal standards.
Lifespan Analysis
Worms were subjected to RNAi either during their adult life only or from hatching (L1 stage). For adult-only lifespan analysis, L1 worms were added to OP50-seeded NGM plates and pre-fertile adult worms were transferred to NGM plates seeded with gene-specific RNAi bacteria. 5-fluoro-2′-deoxyuridine (final concentration of 50 μm) was added to the plates to prevent the development of progeny. Lifespan analyses were started at day 0 (pre-fertile adults), and worms were kept at 20 °C throughout the experiment. Nematodes were transferred to new RNAi plates every two to three days until they lost most of their capacity to move freely. The online application OASIS was used for Kaplan–Meier-based survival analysis and generation of the survival plots (85).
RESULTS
Quantitative Proteomics Analysis of Long-lived daf-2 and DR C. elegans
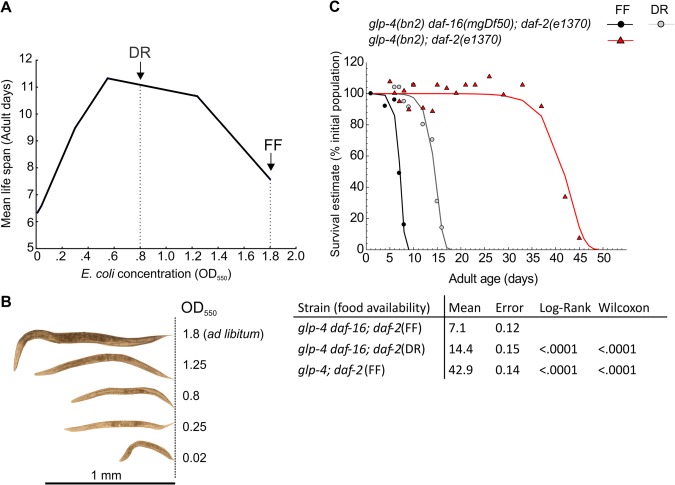
Quantitative proteome profiles of FF and DR C. elegans nematodes were generated using the long-lived glp-4(bn2); daf-2(e1370) IIS receptor mutant and the glp-4(bn2) daf-16(mgDf50); daf-2(e1370) triple mutant as a control. A full list of all quantified proteins can be found in supplemental File S2. In the remainder of this article we refer to these strains as, respectively, daf-16; daf-2 and daf-2. Using serial dilution of bacterial food (Fig. 1A), a level of bacterial dilution was chosen that resulted in a robust increase of mean and maximum lifespans of the daf-16; daf-2 control strain (p < 0.0001; log-rank test; Fig. 1C). Besides lifespan extension, bacterial dilution also resulted in nematodes with smaller body size and increased transparency and mobility, similar to that seen in feeding defective mutants (86), whereas worms fed ad libitum had a larger body size and a darker intestine (Fig. 1B).
Fig. 1.
Lifespan analysis of IIS mutants and DR regime. L1 juvenile worms were grown on plates at 17 °C for 48 h (L3 stage), and the temperature was switched to 24 °C for 24 h before the worms were transferred into liquid medium (S-buffer) for mass culture. Worms were kept at 24 °C for the remainder of the lifespan analysis, and survival was determined daily by counting the number of worms in three independent subsamples of the total population. A, serial dilution of bacterial food results in a local maximum in mean lifespan of glp-4 daf-16; daf-2, and further dilution results in starvation, causing a reduction in mean lifespan. Arrows indicate bacterial concentrations used for the proteomics analysis of worms fed a full (FF) or restricted (DR) diet. B, bacterial dilution resulted in an increasingly smaller body size, higher transparency, and elevated mobility (not shown) of daf-16; daf-2 nematodes. C, survival curves of IIS strains fed either a full or a restricted diet.
Utilizing accurate mass and time tag LC-MS/MS proteomics (87), we were able to quantitatively determine 829 proteins, of which 101 (12.2%) were differentially abundant in both daf-2 and DR nematodes (41 up- and 60 down-regulated; p value < 0.05 after Benjamini–Hochberg adjustment for multiplicity of testing, which is equivalent to FDR (88)). In addition, 263 (31.7%; 178 up- and 85 down-regulated) and 155 (18.7%; 76 up- and 79 down-regulated) proteins where uniquely identified as differentially abundant in daf-2 mutants and DR worms, respectively. These are discussed in more detail in the supplemental data. In total, over 60% of all detected proteins showed a consistently altered expression pattern in the long-lived worm cohorts relative to the normal-lived controls. A complete list of significantly altered proteins is available as supplemental File S3. Previously, quantitative proteomics of glp-4(bn2ts); daf-2(e1370) by the Yates group identified 86 differentially expressed proteins, of which 53 could be identified in our dataset (56). Of these 53 proteins, 37 also showed significant differential expression in the daf-2 mutant, with only 2 proteins showing conflicting expression patterns between datasets (Fig. 2, supplemental File S4). Therefore, a high level of conformity exists between these studies. We attribute the fact that this study detected substantially more differentially abundant proteins to our superior instrumentation and study design involving five biological replicates.
Fig. 2.
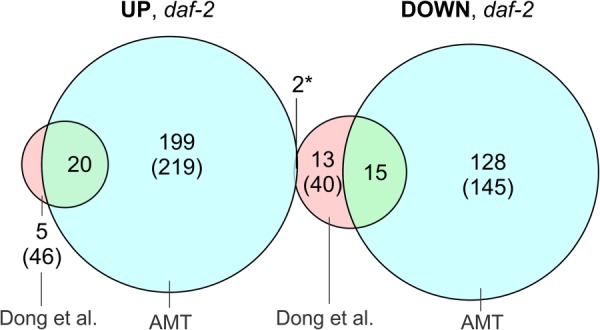
Venn diagram showing the degree of overlap between all differentially expressed proteins in the glp-4(bn2ts); daf-2(e1370) adult found in the here-presented accurate mass and time proteomics dataset and the dataset previously published by Dong et al. (56). Numbers between brackets denote the total number of differentially expressed proteins in each dataset. *Y38F1A.6 and T22B11.5.
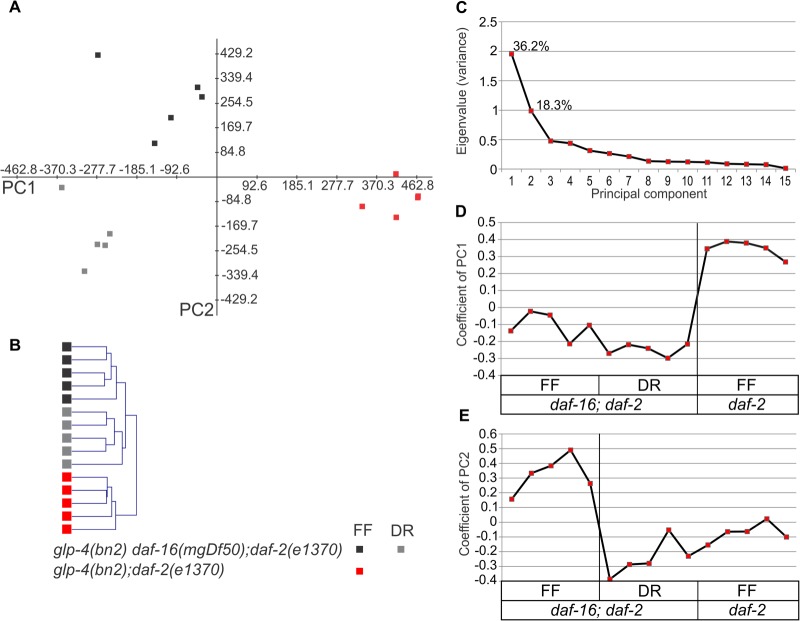
PCA and HCA of the accurate mass and time proteome profiles unambiguously grouped the experimental replicas (Figs. 3A and 3B). This indicates high reproducibility among the five biological replicates and serves as quality assurance of our dataset. The first two principal components (PCs) explained over 50% of the variation in abundance levels among samples (Fig. 3C). PC1 separated samples with high DAF-16 activity (daf-2) from those with no DAF-16 activity (daf-16; daf-2), whereas PC2 clearly separated the long-lived from the control strain samples. Therefore, PC2 reflected proteome changes associated with longevity (Figs. 3D and 3E).
Fig. 3.
Principal component and hierarchical clustering analysis. A, PCA plot of the first two principal components (PC1 and PC2) generated from the proteome profiles of young adult nematodes either fed a full E. coli diet or dietarily restricted, and with or without reduced IIS. Squares represent individual biological replicates. B, HCA separates biological replicate samples into three non-overlapping clusters. C, the eigenvalue of each PC is plotted. The higher the eigenvalue, the more variance can be accounted for by the component. PC1 and PC2 account for, respectively, 36.2% and 18.3% of the total variation. D, E, plot of the eigenvector coefficients of the first two PCs. Sign transitions are a measure of change in the expression along the different experimental groups.
Reduced Protein Synthesis Is Characteristic for Increased Longevity
Functional annotation clustering (78) was used to subdivide proteins that were significantly altered in both daf-2 and DR worms (Table I). Note that broad and functional complex gene classes (e.g. GO:OO18988) with highly varying protein expression profiles can result in the annotation term being enriched in both up- and down-regulated classes. Separate gene annotation clustering analysis for either DR or daf-2 conditions can be found in supplemental Table S1.
Table I. Gene ontology information from DAVID Bioinformatics Resource 6.7 of proteins that are respectively down- or up-regulated in both daf-2 and diet-restricted nematodes. Reported p value is the modified Fisher's exact p value or EASE score. Only clusters with enrichment scores greater than 1.3 are shown.
| Representative term (GO, INTERPRO, KEGG pathway) | Enrichment score | p value |
|---|---|---|
| Common down-regulated | ||
| GO:0005840—ribosome | 22.45 | 1.51E-43 |
| GO:0045735—nutrient reservoir activity, IPR015816: vitellogenin | 4.75 | 2.01E-07 |
| GO:0008340—determination of adult life span GO:0007568—aging | 3.94 | 1.14E-04 |
| GO:0018988—molting cycle, protein-based cuticle | 3.88 | 1.31E-04 |
| GO:0004190—aspartic-type endopeptidase activity | 2.40 | 2.93E-04 |
| Common up-regulated | ||
| IPR002048:calcium-binding EF-hand | 2.72 | 4.99E-05 |
| GO:0055114—oxidation reduction | 2.58 | 1.73E-06 |
| GO:0045333—cellular respiration | 2.58 | 4.58E-04 |
| GO:0006084—acetyl-CoA metabolic process | 2.31 | 2.97E-03 |
| GO:0016459—myosin complex | 2.20 | 1.31E-03 |
| GO:0018988—molting cycle, protein-based cuticle | 1.71 | 1.10E-02 |
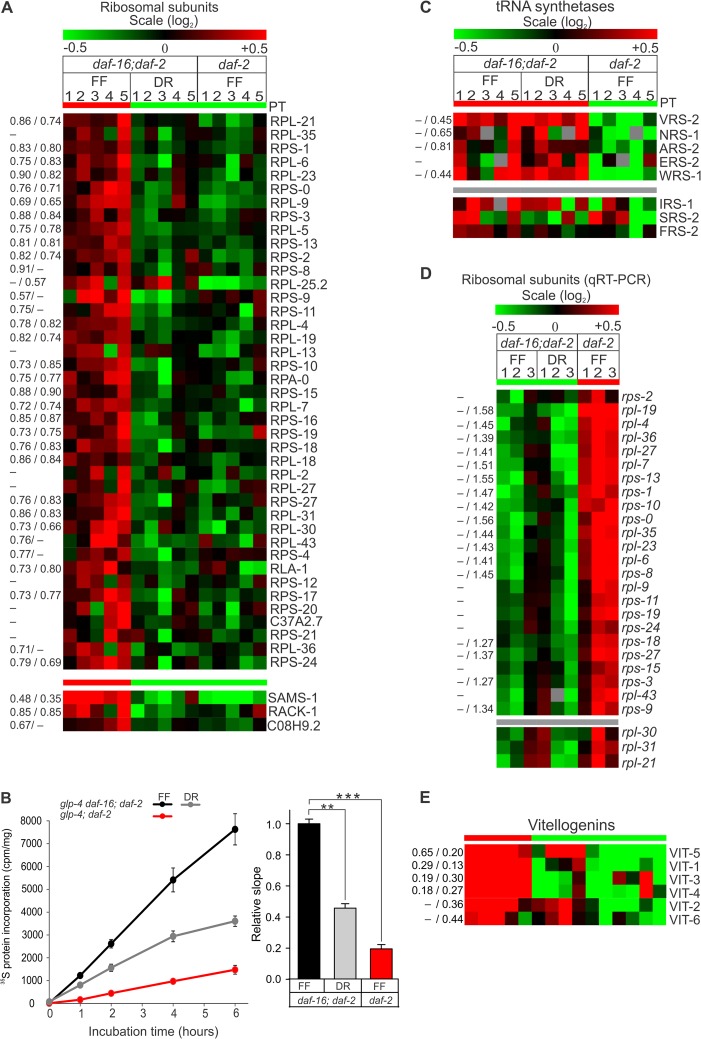
By far the most striking pattern in the proteome fingerprints of both long-lived IIS mutant and DR worms was the significant overall decrease in abundance of a large number of ribosomal proteins (RPs) of both the large (60S) and small (40S) subunits (Fig. 4A). In addition, mutation in daf-2 worms, but not DR, also resulted in lower levels of several tRNA synthetases (Fig. 4C). GSEA allows one to assess whether a group of proteins (e.g. the set of all ribosomal subunit proteins in our dataset) shows significant differential expression (see “Experimental Procedures” for details, as well as Ref. 80). For the set of ribosomal proteins, a significant negative normalized enrichment score (NES) was found for both DR and daf-2 worms (NESDR = −2.67, p < 0.001, FDR < 0.001; NESdaf-2 = −2.69, p < 0.001, FDR < 0.001). tRNA synthetases had a significant negative enrichment score for the daf-2 samples (NESdaf-2 = −1.59, p = 0.028, FDR = 0.039). These findings suggest protein synthesis may be reduced in daf-2 mutants and DR nematodes. We therefore measured protein synthesis rates by feeding worms with radiolabeled bacteria and confirmed that overall protein synthesis rates were significantly lower in the long-lived cohorts than in the FF control population (Fig. 4B). Reduced protein synthesis was also confirmed in daf-2 mutants that did not carry the glp-4 background mutation and in mutants carrying the less pleiotrophic daf-2(m577) allele (supplemental Fig. S1) (89). Thus, the reduction in protein synthesis is independent of any interaction between daf-2 and glp-4 and is likely a common feature in reduced IIS receptor mutants. We then asked whether the reduced expression of RP subunits in these worms is a direct consequence of attenuated RP gene transcription. We found that mRNA levels of ribosomal subunit genes were unchanged in DR worms and, surprisingly, even increased in daf-2 mutants (Fig. 4D, supplemental Table S2). Therefore, post-transcriptional mechanisms are an important factor in determining RP content in C. elegans.
Fig. 4.
Repression of global protein synthesis is a common mechanism in DAF-16 and DR-induced longevity. A, heat map of ribosomal subunit proteins (base-2 logarithmic scale). Each column represents one biological replicate. Red and green colors indicate a relative increase or decrease in protein content for a particular protein, respectively (row). Proteins are grouped according to whether or not a protein expression profile matches a preset template as defined by the Pavlidis template (PT) matching algorithm (see text and Ref. 77 for details). Green and red PT bars indicate, respectively, lower and higher protein levels between conditions. Gray PT bars indicate protein profiles that do not match any obvious expression pattern. The relative expression level is given on the left-hand side of the heatmap for DR worms (left) and daf-2 mutants (right). Values are separated by a forward slash; a dash or no value indicates that the ratio was not significantly different from 1 (no change in expression level relative to control at the 5% significance level with two-tailed t test). B, young adult worms were fed 35S-labeled bacteria, and the incorporation of 35S into newly formed proteins was monitored and normalized to the total protein content. Bars represent relative slope (± S.E.) to control condition glp-4 daf-16; daf-2 from three independent experiments. Protein synthesis was significantly lower in both daf-2 and DR worms relative to the control strain fed ad libitum as determined via two-sided, unpaired t test (p < 0.001 and p < 0.0005, respectively). C, heat map of tRNA synthetase protein abundance levels. D, heat map representation of mRNA levels of genes encoding ribosomal subunit proteins as determined by qRT-PCR. On average, a 2-fold increase in the transcript level of ribosomal genes was observed in daf-2 mutants, but not in DR worms. E, heat map of vitellogenin protein abundance levels.
Intriguingly, we also found a significant reduction in the abundance of C. elegans S-adenosyl methionine synthase-1 (SAMS-1) in both DR and daf-2 mutant worms (p < 0.001) (Fig. 4A), which is consistent with previous proteomics (56) and transcriptomics (46) analyses. RNAi knockdown of sams-1 expression was shown to repress global protein translation by as much as 40% (90). Moreover, SAMS-1 was identified as part of the DR longevity response in eat-2 mutants, possibly acting downstream of the TOR-kinase signaling pathway (90, 91). Therefore, we hypothesized that decreased levels of SAMS-1 could be involved in the observed DAF-16-dependent reduction in protein synthesis rate. C08H9.2 in Fig. 4A encodes the C. elegans orthologs of the mammalian 80S ribosomal RNA binding protein vigilin (92), and RACK-1 the ortholog of the receptor for activated kinase-1, a core component of the 40S subunit eukaryotic ribosome (93–95). Vigilin binds 80S ribosomes and is involved in the nucleocytoplasmic translocation of tRNA and their subsequent association with actively translating ribosomes. In yeast, RACK1 acts as a scaffold protein for several signaling molecules, such as protein kinase C, which is thought to phosphorylate translation initiation factors regulating translation (96, 97). In S. cerevisiae, RACK1 is also required for the binding of Vigilin to ribosomes (98). RACK1 thus allows direct regulation of translation in response to distinct signaling pathways. In sum, the reduced expression of regulatory components of the translation machinery, such as vigilin and RACK-1, adds further support for a general repression of mRNA translation in IIS mutants.
Vitellogenins are the protein components of yolk, which is produced in great abundance in the intestine and translocated to the gonads to serve as an energy source for developing oocytes (99). The continued (over)production of yolk in post-reproductive animals likely contributes to mortality with age (44, 100). Although it is already known that yolk synthesis is greatly reduced in daf-2 mutants (101), our proteomics suggests yolk levels in the DR nematode are at least equally low (Fig. 4E). We hypothesize that decreased yolk production in these long-lived animals is a (beneficial) side effect of overall reduced protein synthesis.
Muscle Mass Is Preserved in Long-lived Worms
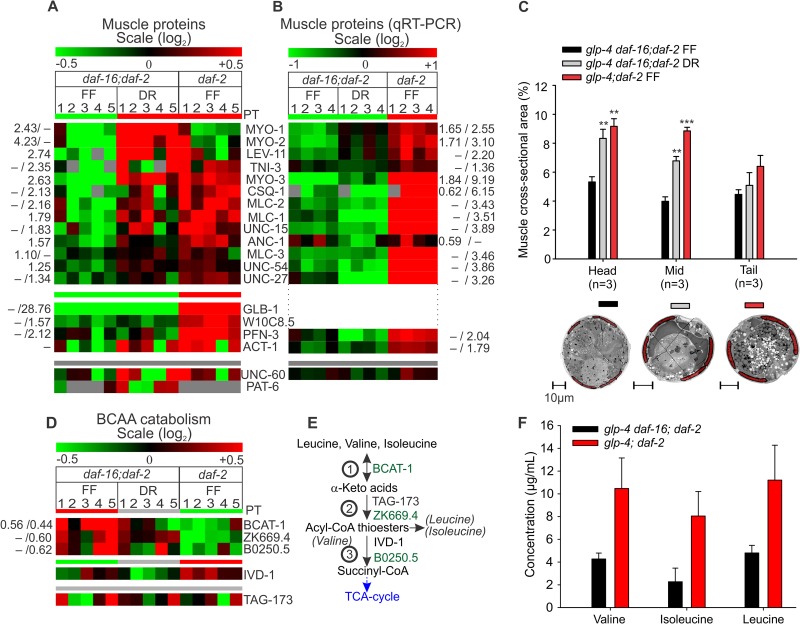
We were intrigued to find myosin complex (GO:0016459) among the ontology terms of proteins up-regulated in the long-lived strains (Table I). A heat map of all muscle-related proteins found in our dataset is given in Fig. 5A. Although the increased abundance of muscle proteins was not always significant in both daf-2 and DR worms at the individual protein level, both Pavlidis template matching and GSEA confirmed enrichment of the set of muscle proteins in the long-lived samples (NESDR = 1.90, p < 0.002, FDR < 0.001; NESdaf-2 = 1.86, p < 0.001, FDR < 0.003). Among these were many essential proteins that constitute the contractile sarcomeres of muscle cells in C. elegans. These include actin isoform ACT-1, the actin binding tropomyosin (LEV-11) and troponin I orthologs (TNI-3, UNC-27), all four isoforms of the myosin class II heavy chain (MYO-1, MYO-2, MYO-3, and UNC-54) and myosin class II light chain proteins (MLC-1, MLC-2, and MLC-3), and the invertebrate-specific paramyosin (UNC-15), which interacts with the myosin heavy chain proteins. Note that MYO-1 and MYO-2 constitute pharynx-specific myosins, whereas MYO-3 and UNC-54 are present in all other muscles (102). Profilin is an actin-binding protein that controls actin polymerization (103). Out of the three profilin isoforms in our dataset (PFN-1, -2, and -3), only the body-wall muscle-specific PFN-3 (PFN-1 and -2 are expressed in nonmuscle cells) was up-regulated in long-lived daf-2 nematodes (103). ANC-1 is a structural protein that binds the actin cytoskeleton in cytoplasm but is also expressed in body-wall muscles (104). It is reported that after a short heat shock, ANC-1 completely co-localizes with actin in body wall muscles. Other muscle-related proteins that are not part of the contractile apparatus but are relevant to muscle physiology include a putative mitochondrial creatine kinase (W10C8.5), the putative C. elegans myoglobin GLB-1, and calsequestrin (CSQ-1), all of which show strong DAF-16-dependent up-regulation.
Fig. 5.
Muscle biomass is preserved in diet-restricted and IIS-defective worms. A, increased abundance of muscle-related proteins in long-lived nematodes (see legend of Fig. 1 for details). B, heatmap representation of mRNA levels of genes encoding muscle proteins as determined by qRT-PCR. C, comparison of the relative surface area of body-wall muscle cross-sections in head (intestine posterior to the pharyngeal terminal bulb), mid-, and tail (hindgut anterior to rectum) regions of two-day-old worms with the total worm cross-section surface. TEM images in the lower part illustrate the relative share of body-wall muscle in total surface area (midsection area). Body-wall muscles are overlain in red. D, decreased expression of BCAA catabolic enzymes in daf-2(e1370). E, overview of the BCAA catabolic process: 1, branched chain aminotransferase; 2, branched chain α-keto acid decarboxylase complex; 3, IVD-1: isovaryl-CoA dehydrogenase, B0250.5: 3-hydroxyisobutyrate dehydrogenase. F, BCAA levels showed a borderline-significant increase in daf-2 mutants (valine, p = 0.086; isoleucine, p = 0.079; leucine, p = 0.11) as determined by two-sided, unpaired t tests. Bars represent mean BCAA concentration from three separate experiments.
Creatine kinase catalyzes the generation of the high-energy compound phosphocreatine in mitochondria via reversible phosphorylation of creatine from mitochondrially derived ATP (105). In accordance, we found that W10C8.5 is predicted (106) to be directed to mitochondria with 81% probability by MitoProt II. Phosphocreatine exported from the mitochondria into the cytosol acts in turn as a readily available temporal energy buffer in active muscle cells, maintaining a constant ATP/ADP ratio (105). The globin GLB-1 shows an approximately 30-fold change in protein expression upon DAF-16 activation, consistent with the transcriptional activation of glb-1 in daf-2 reported earlier by our group (107). A translational reporter construct of GLB-1 shows high expression in (all) muscle tissues.2 In addition, GLB-1 exhibits high O2 ligand affinity in vitro (108). These findings have led to the conclusion that GLB-1 is very likely to act as the C. elegans myoglobin. Finally, calsequestrin binds and buffers Ca2+ in the sarcoplasmic reticulum of body-wall and vulval muscle cells (109). The high abundance of proteins involved in sarcomere structures indicates that C. elegans maintains muscle integrity when confronted with low food availability.
We wondered whether the increased abundance of muscle proteins had any observable effect on muscle biomass. To investigate this, we generated cross-sectional transmission electron microscopy images of the head, mid-, and tail regions of our worms and measured the absolute and relative cross-sectional volumes of striated body-wall muscles. We found that the relative, but not absolute, cross-sectional area of body-wall muscle was increased in the head and mid-section of both daf-2 and DR worms, implying a relative increase in muscle biomass (Fig. 5C). Thus, the increased abundance levels of muscle-related proteins appear to be the result of muscle preservation in daf-2 and DR worms, relative to an overall decrease in their whole-body volume. We asked whether this maintenance of muscle integrity is, at least partly, supported by an increased transcription of muscle-protein-encoding genes. qRT-PCR analysis revealed that this is indeed the case for daf-2 mutants, which show substantially higher mRNA expression of most muscle genes (Fig. 5B, supplemental Table S3). Such a general up-regulation is not seen in DR daf-16; daf-2 nematodes, although a moderate increase in expression is seen for pharyngeal MYO-1 and MYO-2 and body-muscle myosin heavy-chain MYO-3, possibly as a result of the relatively increased muscle biomass. Our results thus indictate that the activation of DAF-16 prevents muscle atrophy, at least in part, by stimulating the expression of muscle-related genes, but that complementary mechanisms (e.g. selective inhibition of structural muscle protein degradation or tissue-specific suppression of protein synthesis) are likely at play under DR.
Reduced Catabolism of Branched Chain Amino Acids in daf-2 Mutants
In mammals, the branched chain amino acids (BCAAs) valine, isoleucine, and leucine stimulate protein synthesis via mTOR activation in skeletal muscle (110, 111). Recently, metabolic profiling has revealed increased levels of BCAAs in several daf-2 mutants and the translation-defective ife-2 mutant (18). Therefore, it is tempting to hypothesize that also in C. elegans, BCAAs might be involved in preserving muscle-specific protein synthesis in daf-2, while global mRNA translation is attenuated. Because C. elegans lacks the enzymes required for the de novo synthesis of these amino acids (112), it was suggested that elevated levels of BCAAs are the result of their decreased degradation (18). BCAAs share the first two enzymatic steps in catabolism: reversible transamination by the branched chain aminotransferase enzyme (BCAT) to the corresponding α-keto acid, and rate-limiting oxidative decarboxylation to the corresponding acyl-CoA by the branched chain α-keto acid dehydrogenase complex (Fig. 5E) (113–115). In line with previous transcription-based evidence (18), we found reduced concentrations of C. elegans orthologs for both mitochondrial BCAT (BCAT-1) and the E2 subunit (dihydrolipoamide acyltransferase) of the branched chain α-keto acid dehydrogenase complex (ZK669.4), and B0250.5, a 3-hydroxyisobutyrate dehydrogenase involved in the catabolism of valine, in the daf-2 mutant (Fig. 5D). The level of BCAT-1 is also significantly lowered in response to DR. We were able to detect increased BCAA levels in daf-2 homogenates, albeit with only borderline significance (Fig. 5F). Possibly, changes in BCAA catabolism are restricted to muscle tissue, partly masking their increase.
Testing Candidate Genes for Lifespan Extension
The list of differentially expressed proteins in both daf-2 and DR worms contains several novel candidate protein determinants of C. elegans lifespan. Therefore, we screened 17 genes with decreased protein abundance in our long-lived strains for their capacity to influence lifespan by RNAi knockdown in the RNAi sensitive rrf-3(pk1426) strain (supplemental File S5). Disappointingly, all genes tested either shortened rrf-3 lifespan, such as the BCAA catabolic gene bcat-1, or had no consistent significant effect on longevity. This could indicate that although these genes are regulated similarly under DR and reduced IIS, they have no connection with the Age phenotype. Alternatively, in vivo regulation of these genes is subtler or requires a context (e.g. tissue specificity) that is not met when these genes are knocked down individually by RNAi.
Other Significant daf-2- or DR-specific Functional Categories
Many functional annotation clusters are uniquely identified for either daf-2 or DR worms; further discussion of this can be found in supplemental File S6. A notable DR-specific annotation term is the proteasome core complex. We found that several α and β subunits of the 20S core and 19S regulator complex of the proteasome were increased significantly in DR worms, which was also confirmed by GSEA (NESDR = 2.08, p < 0.001, FDR < 0.003). Transthyretin-like proteins were significantly up-regulated in a DAF-16-dependent manner (NESdaf-2 = 1.73, p = 0.003, FDR = 0.017). Many proteins with variable daf-2-specific expression can be categorized under the broad denominator of stress-responsive enzymes. These include phase-2 detoxification enzymes (glutathione S-transferases, UDP-glucoronyl transferases, lipid binding and cadmium responsive proteins), reactive oxygen species neutralizing enzymes (catalases, superoxide dismutases), chaperones and heat-shock proteins, and proteins associated with innate immunity (lysozymes, C-type lectins, aspartyl proteases). Annotation clustering of daf-2-specific alterations in protein concentrations reveals many ontology terms linked to intermediary metabolism (carbohydrate, lipid, and oxidative metabolism). The extensive DAF-16-driven restructuring of intermediary metabolism in daf-2 mutants will be presented and discussed in detail elsewhere.
DISCUSSION
Although bacterial dilution-induced DR and reduced insulin-like signaling extend lifespan through parallel signaling pathways, we reasoned that common alterations in biological processes that are possibly required for longevity might emerge in the proteome profiles of these long-lived worms. In this study, the attenuation of protein synthesis and maintenance of muscle integrity were identified as such processes. A large number of gene categories were also identified with unique differential expression either in the daf-2 mutant or under dietary restriction. A discussion of these categories and comparison with key transcriptomics-derived gene categories is provided in supplemental File S6.
In C. elegans, partial inhibition of protein synthesis by RNAi against any of a large number of RPs or genes involved in ribosome biogenesis, translation initiation, or elongation, as well as several tRNA synthetases, was previously shown to increase nematode lifespan and stress resistance (27, 37, 38, 116–120). We have shown here that a general decrease in mRNA translation could in fact be part of a strategy that allows worms to be long-lived.
In mammalian systems, the insulin/IGF-1 signaling pathways and the energy sensing TOR pathway are the major regulators of protein synthesis, and their inhibition leads to attenuation of global translation (25, 26, 35, 36, 121–124). Although there is extensive cross-talk between the IIS and TOR pathways, our results (as well as others) imply that the two pathways can act in parallel and converge on one or more common mediators of ribosomal protein expression, downstream of FoxO/DAF-16, to promote longevity (27, 123, 125–129). Recently, knock-down of sams-1 was found to significantly attenuate total protein synthesis in a pathway downstream of TOR, but in parallel to S6K and pha-4 (90, 91). In line with these results, we observed a strong reduction in SAMS-1 protein levels in both DR and daf-2 animals, suggesting that in C. elegans, SAMS-1 could be such a common mediator of protein synthesis. 7′methyl guanosine capping of the 5′ ends of mRNA by RNA (guanine-7)-methyltransferase is essential for efficient translation of the majority of genes (162). Given that the activity of this enzyme is strongly dependent on the level of S-adenosylmethionine, which is in turn determined by S-adenosylmethionine synthase, reduced 5′ capping of mRNA is one way SAMS-1 could control mRNA translation levels (130). S-adenosylmethionine-dependent methylation of rRNA molecules is required for ribosome assembly and function (131). Therefore, a possible alternative mechanism for SAMS-1-mediated translation is interference with ribosome assembly. In fact, knock-down of two evolutionarily conserved S-adenosylmethionine-requiring RNA methyltransferases, T07A9.8 and nol-1/W07E6.1, both predicted to be involved in ribosome formation, was shown to extend the lifespan of wild-type C. elegans (118). Additionally, RNAi knockdown of Nop2-like SKIP interacting protein, a recently identified putative S-adenosylmethionine-dependent RNA methyltransferase, was shown to interfere with 80S ribosome maturation in HeLa cells and to extend the lifespan of both C. elegans and D. melanogaster.3
The lesser abundance of ribosomal subunit proteins in our long-lived worms was not reflected at the mRNA level. daf-2 mutants even showed an increase in ribosomal transcripts. In yeast, the expression of ribosomal genes can be repressed at many stages, most of which are post-transcriptional, such as decreased translation initiation and translation efficiency, inhibition of splicing, and turnover of excess ribosomal subunit proteins (132–137). For example, 40% of all genes showing a significant reduction in translation efficiency after acute amino acid starvation are involved in ribosome biogenesis (137). Therefore, it is conceivable that similar post-transcriptional mechanisms were also responsible for altered ribosome abundance in our long-lived worms. In fact, increased thermotolerance in daf-2(e1370) was found to require changes in the translation efficiency of individual mRNAs, indicating that post-transcriptional regulatory mechanisms could be an important contributing factor in altered gene expression in daf-2 mutants (138).
Several hypotheses have been proposed to explain why reduced mRNA translation promotes longevity (139). One possibility is that the attenuation of excess biosynthetic and reproductive pathways, such as RP translation and vitellogenesis, could save valuable energy during times of low food availability or channel more resources toward specific cell maintenance processes (38, 101, 140–142). In the same vein, reduced global translation might also lessen the burden on protein repair and degradation machinery, facilitating the maintenance of protein homeostasis. This hypothesis is plausible, as 30% to 50% of nascent polypeptides have been estimated to be degraded co-translationally by the ubiquitin proteasome system as a result of errors in translation and protein folding (143, 144).
On the other hand, evidence from C. elegans also supports a model in which the perturbation of protein synthesis activates specific stress-regulatory pathways involving the DAF-16 and SKN-1 transcription factors to convey increased stress resistance and lifespan (27, 38, 120, 129). Besides this transcriptional response, reduced overall protein synthesis in C. elegans also induces extensive shifts in the ribosomal loading of individual mRNAs (i.e. post-transcriptional expression regulation), with larger transcripts (associated with stress-related genes) being preferentially translated over shorter transcripts (associated with biosynthetic pathways) (145), similar to what has been observed in yeast (137, 146) and Drosophila (147).
The proteome profile of long-lived worms was also characterized by the increased abundance of many muscle-related proteins, which we showed is likely the result of a relative increase in the biomass of (body-wall) muscle tissue in these worms. daf-2 mutants, but not DR worms, also show increased mRNA expression of muscle-protein-coding genes. Our results thus indicate that in C. elegans, some mechanism exists that protects muscles from atrophy when the organism is confronted with reduced nutrient availability or reduced IIS. Increased expression of a large number of muscle proteins was also reported for dauer larvae relative to mixed-stage wild types (148). We note that at first sight our results seem to contrast with reports that mutation in daf-2 or acute starvation induces the degradation of a small cytosolic soluble N-terminal fragment of UNC-54, a muscle myosin class II heavy chain (149, 150). However, full-length UNC-54 incorporated into myofibrils was reported by the same group to be resistant to starvation-induced proteolysis, suggesting preferential catabolism of soluble muscle proteins over proteins that constitute the contractile fibers (149). We reason that these results support the notion that in C. elegans, excessive catabolism of muscle tissue is prevented under conditions of low food or reduced IIS in order to preserve muscle function and sustain locomotion. Previous work has demonstrated that undisturbed daf-2(e1370) mutants reside in a mostly locomotory inactive state, yet these animals retain the capability for high mobility in response to external stimuli, bearing much similarity to dauer behavior (151, 152). For the dauer larva, retaining the ability to move freely even when in a dormant state is evolutionarily advantageous, as it allows for active scouting for other, more nutrient-rich environments. Alternatively, retained muscle function allows for nictation behavior in which a worm stands up straight on its tail and waves its head in order to attach itself to invertebrate carriers for dispersal (153). In mammalian systems, low insulin/IGF-1 signaling and activation of FoxO transcription factors is usually associated with muscle atrophy and a loss of muscle protein (154). We acknowledge that it is therefore surprising that we found increased muscle protein expression in IIS-defective daf-2 mutants. However, in a recent study of mouse myoblast cells, FoxO1 was shown to induce a transition from slow oxidative to fast glycolytic muscle fibers, suggesting a role for FoxO1 in myofiber specification (155). This finding is intriguing in that it suggests the possibility that in C. elegans, DAF-16 promotes a similar remodeling of muscle fibers, consistent with their locomotory behavior (156). Finally, daf-2 mutants show increased expression of the putative mitochondrial creatine kinase W10C8.5 and the putative myoglobin GLB-1, as well as higher concentrations of muscle-growth-stimulating BCAAs (110). All our findings therefore point to increased muscle physiology in daf-2 mutants.
Fuchs et al. were the first to observe a DAF-16-dependent increase in BCAA levels, but the role of BCAA in daf-2 longevity remains to be determined (18). In mammals, BCAAs have a well-established role in stimulating protein synthesis and decreasing protein degradation specifically in skeletal muscle tissue (110, 111). In addition, supplemental dietary administration of BCAAs was demonstrated to extend the average and chronological lifespan in, respectively, middle-aged mice and Saccharomyces cerevisiae (157, 158). Thus BCAAs serve as survival-promoting agents in these animals. The increase in the lifespan of aging mice was accompanied by markedly improved motor coordination and muscle preservation and increased mitochondrial biogenesis in cardiac and skeletal muscles (158). These results from other model organisms thus strongly suggest that the increased BCAA levels are linked with the here-observed maintenance of muscle integrity in daf-2 worms. Increased preservation of muscle structural integrity and function could potentially, like in mice, add significantly to daf-2's long life (159). The BCAA catabolic enzyme bcat-1 is required during embryonic development (160), and we found that it is also required for the normal adult lifespan. Perhaps altering BCAA levels through careful tissue-specific manipulation of bcat-1 expression will be more successful for conferring longevity.
In conclusion, using accurate mass and time tag LC-MS technology, we established the most extensive dataset of changes in protein abundance resulting from induced IIS and diet restriction in C. elegans to date. This database has been made publically available and will thus serve as a valuable resource for researchers working with daf-2 mutants or on dietary restriction in C. elegans. Given the unpredictable relation between measured transcript and final protein product, as was demonstrated in this study, proteomics datasets capture biologically important agents more accurately than transcriptomics data. As further endeavors are continuously made to increase detection sensitivity, proteome analyses will become increasingly important for understanding biological phenomena and disease conditions, including aging.
Supplementary Material
Acknowledgments
The strains GA154 glp-4(bn2ts)I; daf-2(e1370)III and GA153 glp-4(bn2ts)I daf-16(mgDf50)I; daf-2(e1370)III were kindly provided by David Gems. We are grateful to Renata Coopman for her assistance in culturing and sampling worm cohorts. We also thank Myriam Claeys and Ineke Dhondt for assisting with TEM imaging. Proteomic analyses were performed in the Environmental Molecular Sciences Laboratory, a U.S. Department of Energy (DOE) national scientific user facility located at the Pacific Northwest National Laboratory in Richland, WA. PNNL is a multi-program national laboratory operated by Battelle Memorial Institute for the U.S. Department of Energy under Contract DE-AC05–76RL01830. HPLC analysis was performed by the NutriFOODchem unit (Department of Food Safety and Food Quality) at Ghent University.
Footnotes
* This work was supported by a grant from the Fund for Scientific Research-Flanders (G.04371.0N) to B.P.B. and the NIH National Center for Research Resources (RR18522 to R.D.S.). G.D. was supported by a BOF project of Ghent University (01J04208).
 This article contains supplemental material.
This article contains supplemental material.
2 S. De Henau, personal communication.
3 M. Schosserer and E. Fuchs, personal communication.
1 The abbreviations used are:
- BCAA
- branched chain amino acid
- BCAT
- branched chain aminotransferase
- C. elegans
- Caenorhabditis elegans
- DR
- dietary restriction
- FDR
- false discovery rate
- FF
- fully fed
- FoxO
- forkhead box-O
- GSEA
- gene set enrichment analysis
- HCA
- hierarchical clustering analysis
- IGF-1
- insulin-like growth factor-1
- IIS
- insulin/insulin-like growth factor-1 signaling
- NES
- normalized enrichment score
- NGM
- nematode growth medium
- PC
- principal component
- PCA
- principal component analysis
- qRT-PCR
- quantitative reverse-transcription polymerase chain reaction
- RP
- ribosomal protein
- SAMS-1
- S-adenosyl methionine synthase-1
- TEM
- transmission electron microscopy
- TOR
- target of rapamycin kinase.
REFERENCES
- 1. Kenyon C., Chang J., Gensch E., Rudner A., Tabtiang R. (1993) A C. elegans mutant that lives twice as long as wild type. Nature 366, 461–464 [DOI] [PubMed] [Google Scholar]
- 2. Kimura K. D., Tissenbaum H. A., Liu Y., Ruvkun G. (1997) daf-2, an insulin receptor-like gene that regulates longevity and diapause in Caenorhabditis elegans. Science 277, 942–946 [DOI] [PubMed] [Google Scholar]
- 3. Lin K., Dorman J. B., Rodan A., Kenyon C. (1997) daf-16: an HNF-3/forkhead family member that can function to double the life-span of Caenorhabditis elegans. Science 278, 1319–1322 [DOI] [PubMed] [Google Scholar]
- 4. Ogg S., Paradis S., Gottlieb S., Patterson G. I., Lee L., Tissenbaum H. A., Ruvkun G. (1997) The Fork head transcription factor DAF-16 transduces insulin-like metabolic and longevity signals in C. elegans. Nature 389, 994–999 [DOI] [PubMed] [Google Scholar]
- 5. Lithgow G. J., White T. M., Melov S., Johnson T. E. (1995) Thermotolerance and extended life-span conferred by single-gene mutations and induced by thermal stress. Proc. Natl. Acad. Sci. U.S.A. 92, 7540–7544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Murakami S., Johnson T. E. (1996) A genetic pathway conferring life extension and resistance to UV stress in Caenorhabditis elegans. Genetics 143, 1207–1218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Barsyte D., Lovejoy D. A., Lithgow G. J. (2001) Longevity and heavy metal resistance in daf-2 and age-1 long-lived mutants of Caenorhabditis elegans. FASEB J. 15, 627–634 [DOI] [PubMed] [Google Scholar]
- 8. Scott B. A., Avidan M. S., Crowder C. M. (2002) Regulation of hypoxic death in C. elegans by the insulin/IGF receptor homolog DAF-2. Science 296, 2388–2391 [DOI] [PubMed] [Google Scholar]
- 9. Garsin D. A., Villanueva J. M., Begun J., Kim D. H., Sifri C. D., Calderwood S. B., Ruvkun G., Ausubel F. M. (2003) Long-lived C. elegans daf-2 mutants are resistant to bacterial pathogens. Science 300, 1921. [DOI] [PubMed] [Google Scholar]
- 10. Walker G. A., Lithgow G. J. (2003) Lifespan extension in C. elegans by a molecular chaperone dependent upon insulin-like signals. Aging Cell 2, 131–139 [DOI] [PubMed] [Google Scholar]
- 11. Gottlieb S., Ruvkun G. (1994) daf-2, daf-16 and daf-23: genetically interacting genes controlling dauer formation in Caenorhabditis elegans. Genetics 137, 107–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Tissenbaum H. A., Ruvkun G. (1998) An insulin-like signaling pathway affects both longevity and reproduction in Caenorhabditis elegans. Genetics 148, 703–717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Melendez A., Talloczy Z., Seaman M., Eskelinen E. L., Hall D. H., Levine B. (2003) Autophagy genes are essential for dauer development and life-span extension in C. elegans. Science 301, 1387–1391 [DOI] [PubMed] [Google Scholar]
- 14. Hansen M., Chandra A., Mitic L. L., Onken B., Driscoll M., Kenyon C. (2008) A role for autophagy in the extension of lifespan by dietary restriction in C. elegans. PLoS Genet. 4, e24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Jia K., Thomas C., Akbar M., Sun Q., Adams-Huet B., Gilpin C., Levine B. (2009) Autophagy genes protect against Salmonella typhimurium infection and mediate insulin signaling-regulated pathogen resistance. Proc. Natl. Acad. Sci. U.S.A. 106, 14564–14569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lee S. S., Kennedy S., Tolonen A. C., Ruvkun G. (2003) DAF-16 target genes that control C. elegans life-span and metabolism. Science 300, 644–647 [DOI] [PubMed] [Google Scholar]
- 17. Houthoofd K., Braeckman B. P., Lenaerts I., Brys K., Matthijssens F., De Vreese A., Van Eygen S., Vanfleteren J. R. (2005) DAF-2 pathway mutations and food restriction in aging Caenorhabditis elegans differentially affect metabolism. Neurobiol. Aging 26, 689–696 [DOI] [PubMed] [Google Scholar]
- 18. Fuchs S., Bundy J. G., Davies S. K., Viney J. M., Swire J. S., Leroi A. M. (2010) A metabolic signature of long life in Caenorhabditis elegans. BMC Biol. 8, 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kenyon C. J. (2010) The genetics of ageing. Nature 464, 504–512 [DOI] [PubMed] [Google Scholar]
- 20. McDonald R. B., Ramsey J. J. (2010) Honoring Clive McCay and 75 years of calorie restriction research. J. Nutr. 140, 1205–1210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bishop N. A., Guarente L. (2007) Two neurons mediate diet-restriction-induced longevity in C. elegans. Nature 447, 545–549 [DOI] [PubMed] [Google Scholar]
- 22. Arum O., Bonkowski M. S., Rocha J. S., Bartke A. (2009) The growth hormone receptor gene-disrupted mouse fails to respond to an intermittent fasting diet. Aging Cell 8, 756–760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Honjoh S., Yamamoto T., Uno M., Nishida E. (2009) Signalling through RHEB-1 mediates intermittent fasting-induced longevity in C. elegans. Nature 457, 726–730 [DOI] [PubMed] [Google Scholar]
- 24. Greer E. L., Dowlatshahi D., Banko M. R., Villen J., Hoang K., Blanchard D., Gygi S. P., Brunet A. (2007) An AMPK-FOXO pathway mediates longevity induced by a novel method of dietary restriction in C. elegans. Curr. Biol. 17, 1646–1656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kapahi P., Zid B. M., Harper T., Koslover D., Sapin V., Benzer S. (2004) Regulation of lifespan in Drosophila by modulation of genes in the TOR signaling pathway. Curr. Biol. 14, 885–890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kaeberlein M., Powers R. W., Steffen K. K., Westman E. A., Hu D., Dang N., Kerr E. O., Kirkland K. T., Fields S., Kennedy B. K. (2005) Regulation of yeast replicative life span by TOR and Sch9 in response to nutrients. Science 310, 1193–1196 [DOI] [PubMed] [Google Scholar]
- 27. Hansen M., Taubert S., Crawford D., Libina N., Lee S. J., Kenyon C. (2007) Lifespan extension by conditions that inhibit translation in Caenorhabditis elegans. Aging Cell 6, 95–110 [DOI] [PubMed] [Google Scholar]
- 28. Panowski S. H., Wolff S., Aguilaniu H., Durieux J., Dillin A. (2007) PHA-4/Foxa mediates diet-restriction-induced longevity of C. elegans. Nature 447, 550–555 [DOI] [PubMed] [Google Scholar]
- 29. Sheaffer K. L., Updike D. L., Mango S. E. (2008) The Target of Rapamycin pathway antagonizes pha-4/FoxA to control development and aging. Curr. Biol. 18, 1355–1364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Greer E. L., Brunet A. (2009) Different dietary restriction regimens extend lifespan by both independent and overlapping genetic pathways in C. elegans. Aging Cell 8, 113–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lakowski B., Hekimi S. (1998) The genetics of caloric restriction in Caenorhabditis elegans. Proc. Natl. Acad. Sci. U.S.A. 95, 13091–13096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Houthoofd K., Braeckman B. P., Johnson T. E., Vanfleteren J. R. (2003) Life extension via dietary restriction is independent of the Ins/IGF-1 signalling pathway in Caenorhabditis elegans. Exp. Gerontol. 38, 947–954 [DOI] [PubMed] [Google Scholar]
- 33. Walker G., Houthoofd K., Vanfleteren J. R., Gems D. (2005) Dietary restriction in C. elegans: from rate-of-living effects to nutrient sensing pathways. Mech. Ageing Dev. 126, 929–937 [DOI] [PubMed] [Google Scholar]
- 34. Yen K., Mobbs C. V. (2010) Evidence for only two independent pathways for decreasing senescence in Caenorhabditis elegans. Age 32, 39–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sharp Z. D., Bartke A. (2005) Evidence for down-regulation of phosphoinositide 3-kinase/Akt/mammalian target of rapamycin (PI3K/Akt/mTOR)-dependent translation regulatory signaling pathways in ames dwarf mice. J. Gerontol. A Biol. Sci. Med. Sci. 60, 293–300 [DOI] [PubMed] [Google Scholar]
- 36. Hsieh C. C., Papaconstantinou J. (2004) Akt/PKB and p38 MAPK signaling, translational initiation and longevity in Snell dwarf mouse livers. Mech. Ageing Dev. 125, 785–798 [DOI] [PubMed] [Google Scholar]
- 37. Pan K. Z., Palter J. E., Rogers A. N., Olsen A., Chen D., Lithgow G. J., Kapahi P. (2007) Inhibition of mRNA translation extends lifespan in Caenorhabditis elegans. Aging Cell 6, 111–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Syntichaki P., Troulinaki K., Tavernarakis N. (2007) eIF4E function in somatic cells modulates ageing in Caenorhabditis elegans. Nature 445, 922–926 [DOI] [PubMed] [Google Scholar]
- 39. Furuyama T., Nakazawa T., Nakano I., Mori N. (2000) Identification of the differential distribution patterns of mRNAs and consensus binding sequences for mouse DAF-16 homologues. Biochem. J. 349, 629–634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ookuma S., Fukuda M., Nishida E. (2003) Identification of a DAF-16 transcriptional target gene, scl-1, that regulates longevity and stress resistance in Caenorhabditis elegans. Curr. Biol. 13, 427–431 [DOI] [PubMed] [Google Scholar]
- 41. Oh S. W., Mukhopadhyay A., Dixit B. L., Raha T., Green M. R., Tissenbaum H. A. (2006) Identification of direct DAF-16 targets controlling longevity, metabolism and diapause by chromatin immunoprecipitation. Nat. Genet. 38, 251–257 [DOI] [PubMed] [Google Scholar]
- 42. Schuster E., McElwee J. J., Tullet J. M., Doonan R., Matthijssens F., Reece-Hoyes J. S., Hope I. A., Vanfleteren J. R., Thornton J. M., Gems D. (2010) DamID in C. elegans reveals longevity-associated targets of DAF-16/FoxO. Mol. Syst. Biol. 6, 399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. McElwee J., Bubb K., Thomas J. H. (2003) Transcriptional outputs of the Caenorhabditis elegans forkhead protein DAF-16. Aging Cell 2, 111–121 [DOI] [PubMed] [Google Scholar]
- 44. Murphy C. T., McCarroll S. A., Bargmann C. I., Fraser A., Kamath R. S., Ahringer J., Li H., Kenyon C. (2003) Genes that act downstream of DAF-16 to influence the lifespan of Caenorhabditis elegans. Nature 424, 277–283 [DOI] [PubMed] [Google Scholar]
- 45. Golden T. R., Melov S. (2004) Microarray analysis of gene expression with age in individual nematodes. Aging Cell 3, 111–124 [DOI] [PubMed] [Google Scholar]
- 46. McElwee J. J., Schuster E., Blanc E., Thomas J. H., Gems D. (2004) Shared transcriptional signature in Caenorhabditis elegans dauer larvae and long-lived daf-2 mutants implicates detoxification system in longevity assurance. J. Biol. Chem. 279, 44533–44543 [DOI] [PubMed] [Google Scholar]
- 47. Halaschek-Wiener J., Khattra J. S., McKay S., Pouzyrev A., Stott J. M., Yang G. S., Holt R. A., Jones S. J. M., Marra M. A., Brooks-Wilson A. R., Riddle D. L. (2005) Analysis of long-lived C. elegans daf-2 mutants using serial analysis of gene expression. Genome Res. 15, 603–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ruzanov P., Riddle D. L., Marra M. A., McKay S. J., Jones S. M. (2007) Genes that may modulate longevity in C. elegans in both dauer larvae and long-lived daf-2 adults. Exp. Gerontol. 42, 825–839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ruzanov P., Riddle D. L. (2010) Deep SAGE analysis of the Caenorhabditis elegans transcriptome. Nucleic Acids Res. 38, 3252–3262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Griffith O. L., Pleasance E. D., Fulton D. L., Oveisi M., Ester M., Siddiqui A. S., Jones S. J. (2005) Assessment and integration of publicly available SAGE, cDNA microarray, and oligonucleotide microarray expression data for global coexpression analyses. Genomics 86, 476–488 [DOI] [PubMed] [Google Scholar]
- 51. van Ruissen F., Ruijter J. M., Schaaf G. J., Asgharnegad L., Zwijnenburg D. A., Kool M., Baas F. (2005) Evaluation of the similarity of gene expression data estimated with SAGE and Affymetrix GeneChips. BMC Genomics 6, 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wang J., Kim S. K. (2003) Global analysis of dauer gene expression in Caenorhabditis elegans. Development 130, 1621–1634 [DOI] [PubMed] [Google Scholar]
- 53. Tian Q., Stepaniants S. B., Mao M., Weng L., Feetham M. C., Doyle M. J., Yi E. C., Dai H. Y., Thorsson V., Eng J., Goodlett D., Berger J. P., Gunter B., Linseley P. S., Stoughton R. B., Aebersold R., Collins S. J., Hanlon W. A., Hood L. E. (2004) Integrated genomic and proteomic analyses of gene expression in mammalian cells. Mol. Cell. Proteomics 3, 960–969 [DOI] [PubMed] [Google Scholar]
- 54. Maier T., Guell M., Serrano L. (2009) Correlation of mRNA and protein in complex biological samples. FEBS Lett. 583, 3966–3973 [DOI] [PubMed] [Google Scholar]
- 55. Schrimpf S. P., Weiss M., Reiter L., Ahrens C. H., Jovanovic M., Malmstrom J., Brunner E., Mohanty S., Lercher M. J., Hunziker P. E., Aebersold R., von Mering C., Hengartner M. O. (2009) Comparative functional analysis of the Caenorhabditis elegans and Drosophila melanogaster proteomes. PLoS Biol. 7, 616–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Dong M. Q., Venable J. D., Au N., Xu T., Park S. K., Cociorva D., Johnson J. R., Dillin A., Yates J. R., 3rd (2007) Quantitative mass spectrometry identifies insulin signaling targets in C. elegans. Science 317, 660–663 [DOI] [PubMed] [Google Scholar]
- 57. Madi A., Mikkat S., Koy C., Ringel B., Thiesen H. J., Glocker M. O. (2008) Mass spectrometric proteome analysis suggests anaerobic shift in metabolism of Dauer larvae of Caenorhabditis elegans. Biochim. Biophys. Acta 1784, 1763–1770 [DOI] [PubMed] [Google Scholar]
- 58. Jones L. M., Staffa K., Perally S., LaCourse E. J., Brophy P. M., Hamilton J. V. (2010) Proteomic analyses of Caenorhabditis elegans dauer larvae and long-lived daf-2 mutants implicates a shared detoxification system in longevity assurance. J. Proteome Res. 9, 2871–2881 [DOI] [PubMed] [Google Scholar]
- 59. Yuan Y., Kadiyala C. S., Ching T. T., Hakimi P., Saha S., Xu H., Yuan C., Mullangi V., Wang L., Fivenson E., Hanson R. W., Ewing R., Hsu A. L., Miyagi M., Feng Z. (2012) Enhanced energy metabolism contributes to the extended life span of calorie-restricted Caenorhabditis elegans. J. Biol. Chem. 287, 31414–31426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Beanan M. J., Strome S. (1992) Characterization of a germ-line proliferation mutation in C. elegans. Development 116, 755–766 [DOI] [PubMed] [Google Scholar]
- 61. TeKippe M., Aballay A. (2010) C. elegans germline-deficient mutants respond to pathogen infection using shared and distinct mechanisms. PLoS One 5, e11777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Krijgsveld J., Ketting R. F., Mahmoudi T., Johansen J., Artal-Sanz M., Verrijzer C. P., Plasterk R. H., Heck A. J. (2003) Metabolic labeling of C. elegans and D. melanogaster for quantitative proteomics. Nat. Biotechnol. 21, 927–931 [DOI] [PubMed] [Google Scholar]
- 63. Wood W. B. (1988) The Nematode Caenorhabditis elegans, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 64. Fabian T. J., Johnson T. E. (1994) Production of age-synchronous mass cultures of Caenorhabditis elegans. J. Gerontol. 49, B145–B156 [DOI] [PubMed] [Google Scholar]
- 65. Petyuk V. A., Qian W. J., Smith R. D., Smith D. J. (2010) Mapping protein abundance patterns in the brain using voxelation combined with liquid chromatography and mass spectrometry. Methods 50, 77–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Delahunty C., Yates J. R., 3rd (2003) Identification of proteins in complex mixtures using liquid chromatography and mass spectrometry. Curr. Protoc. Cell Biol. Chapter 5, Unit 5.6 [DOI] [PubMed] [Google Scholar]
- 67. Jaitly N., Mayampurath A., Littlefield K., Adkins J. N., Anderson G. A., Smith R. D. (2009) Decon2LS: an open-source software package for automated processing and visualization of high resolution mass spectrometry data. BMC Bioinformatics 10, 87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Monroe M. E., Tolic N., Jaitly N., Shaw J. L., Adkins J. N., Smith R. D. (2007) VIPER: an advanced software package to support high-throughput LC-MS peptide identification. Bioinformatics 23, 2021–2023 [DOI] [PubMed] [Google Scholar]
- 69. Petyuk V. A., Qian W. J., Chin M. H., Wang H., Livesay E. A., Monroe M. E., Adkins J. N., Jaitly N., Anderson D. J., Camp D. G., 2nd, Smith D. J., Smith R. D. (2007) Spatial mapping of protein abundances in the mouse brain by voxelation integrated with high-throughput liquid chromatography-mass spectrometry. Genome Res. 17, 328–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Desiere F., Deutsch E. W., King N. L., Nesvizhskii A. I., Mallick P., Eng J., Chen S., Eddes J., Loevenich S. N., Aebersold R. (2006) The PeptideAtlas project. Nucleic Acids Res. 34, D655–D658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Polpitiya A. D., Qian W. J., Jaitly N., Petyuk V. A., Adkins J. N., Camp D. G., 2nd, Anderson G. A., Smith R. D. (2008) DAnTE: a statistical tool for quantitative analysis of -omics data. Bioinformatics 24, 1556–1558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Troyanskaya O., Cantor M., Sherlock G., Brown P., Hastie T., Tibshirani R., Botstein D., Altman R. B. (2001) Missing value estimation methods for DNA microarrays. Bioinformatics 17, 520–525 [DOI] [PubMed] [Google Scholar]
- 73. Saeed A. I., Bhagabati N. K., Braisted J. C., Liang W., Sharov V., Howe E. A., Li J., Thiagarajan M., White J. A., Quackenbush J. (2006) TM4 microarray software suite. Methods Enzymol. 411, 134–193 [DOI] [PubMed] [Google Scholar]
- 74. Saeed A. I., Sharov V., White J., Li J., Liang W., Bhagabati N., Braisted J., Klapa M., Currier T., Thiagarajan M., Sturn A., Snuffin M., Rezantsev A., Popov D., Ryltsov A., Kostukovich E., Borisovsky I., Liu Z., Vinsavich A., Trush V., Quackenbush J. (2003) TM4: a free, open-source system for microarray data management and analysis. BioTechniques 34, 374–378 [DOI] [PubMed] [Google Scholar]
- 75. Raychaudhuri S., Stuart J. M., Altman R. B. (2000) Principal components analysis to summarize microarray experiments: application to sporulation time series. Pac. Symp. Biocomput. 2000, 455–466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Eisen M. B., Spellman P. T., Brown P. O., Botstein D. (1998) Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. U.S.A. 95, 14863–14868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Pavlidis P., Noble W. S. (2001) Analysis of strain and regional variation in gene expression in mouse brain. Genome Biol. 2, RESEARCH0042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Huang da W., Sherman B. T., Lempicki R. A. (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 [DOI] [PubMed] [Google Scholar]
- 79. Dennis G., Jr., Sherman B. T., Hosack D. A., Yang J., Gao W., Lane H. C., Lempicki R. A. (2003) DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 4, P3. [PubMed] [Google Scholar]
- 80. Subramanian A., Tamayo P., Mootha V. K., Mukherjee S., Ebert B. L., Gillette M. A., Paulovich A., Pomeroy S. L., Golub T. R., Lander E. S., Mesirov J. P. (2005) Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A. 102, 15545–15550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Hoebeeck J., Speleman F., Vandesompele J. (2007) Real-time quantitative PCR as an alternative to Southern blot or fluorescence in situ hybridization for detection of gene copy number changes. Methods Mol. Biol. 353, 205–226 [DOI] [PubMed] [Google Scholar]
- 82. Hoogewijs D., Houthoofd K., Matthijssens F., Vandesompele J., Vanfleteren J. R. (2008) Selection and validation of a set of reliable reference genes for quantitative sod gene expression analysis in C. elegans. BMC Mol. Biol. 9, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Fonderie P., Willems M., Bert W., Houthoofd W., Steel H., Claeys M., Borgonie G. (2009) Intestine ultrastructure of the facultative parasite Halicephalobus gingivalis (Nematoda: Panagrolaimidae). Nematology 11, 859–868 [Google Scholar]
- 84. Kerkaert B., Mestdagh F., Cucu T., Shrestha K., Van Camp J., De Meulenaer B. (2011) The impact of photo-induced molecular changes of dairy proteins on their ACE-inhibitory peptides and activity. Amino Acids 43, 951–962 [DOI] [PubMed] [Google Scholar]
- 85. Yang J. S., Nam H. J., Seo M., Han S. K., Choi Y., Nam H. G., Lee S. J., Kim S. (2011) OASIS: online application for the survival analysis of lifespan assays performed in aging research. PLoS One 6, e23525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Morck C., Pilon M. (2006) C. elegans feeding defective mutants have shorter body lengths and increased autophagy. BMC Dev. Biol. 6, 39–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Pasa-Tolic L., Masselon C., Barry R. C., Shen Y., Smith R. D. (2004) Proteomic analyses using an accurate mass and time tag strategy. Biotechniques 37, 621–624, 626,–633, 636 passim [DOI] [PubMed] [Google Scholar]
- 88. Benjamini Y., Hochberg Y. (1995) Controlling the false discovery rate—a practical and powerful approach to multiple testing. J. R. Stat. Soc. Series B Stat. Methodol. 57, 289–300 [Google Scholar]
- 89. Gems D., Sutton A. J., Sundermeyer M. L., Albert P. S., King K. V., Edgley M. L., Larsen P. L., Riddle D. L. (1998) Two pleiotropic classes of daf-2 mutation affect larval arrest, adult behavior, reproduction and longevity in Caenorhabditis elegans. Genetics 150, 129–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Ching T. T., Paal A. B., Mehta A., Zhong L., Hsu A. L. (2010) drr-2 encodes an eIF4H that acts downstream of TOR in diet-restriction-induced longevity of C. elegans. Aging Cell 9, 545–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Hansen M., Hsu A. L., Dillin A., Kenyon C. (2005) New genes tied to endocrine, metabolic, and dietary regulation of lifespan from a Caenorhabditis elegans genomic RNAi screen. PLoS Genet. 1, 119–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Vollbrandt T., Willkomm D., Stossberg H., Kruse C. (2004) Vigilin is co-localized with 80S ribosomes and binds to the ribosomal complex through its C-terminal domain. Int. J. Biochem. Cell Biol. 36, 1306–1318 [DOI] [PubMed] [Google Scholar]
- 93. Link A. J., Eng J., Schieltz D. M., Carmack E., Mize G. J., Morris D. R., Garvik B. M., Yates J. R., 3rd (1999) Direct analysis of protein complexes using mass spectrometry. Nat. Biotechnol. 17, 676–682 [DOI] [PubMed] [Google Scholar]
- 94. Gerbasi V. R., Weaver C. M., Hill S., Friedman D. B., Link A. J. (2004) Yeast Asc1p and mammalian RACK1 are functionally orthologous core 40S ribosomal proteins that repress gene expression. Mol. Cell. Biol. 24, 8276–8287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Nilsson J., Sengupta J., Frank J., Nissen P. (2004) Regulation of eukaryotic translation by the RACK1 protein: a platform for signalling molecules on the ribosome. EMBO Rep. 5, 1137–1141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. McCahill A., Warwicker J., Bolger G. B., Houslay M. D., Yarwood S. J. (2002) The RACK1 scaffold protein: a dynamic cog in cell response mechanisms. Mol. Pharmacol. 62, 1261–1273 [DOI] [PubMed] [Google Scholar]
- 97. Ceci M., Gaviraghi C., Gorrini C., Sala L. A., Offenhauser N., Marchisio P. C., Biffo S. (2003) Release of eIF6 (p27BBP) from the 60S subunit allows 80S ribosome assembly. Nature 426, 579–584 [DOI] [PubMed] [Google Scholar]
- 98. Baum S., Bittins M., Frey S., Seedorf M. (2004) Asc1p, a WD40-domain containing adaptor protein, is required for the interaction of the RNA-binding protein Scp160p with polysomes. Biochem. J. 380, 823–830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Kimble J., Sharrock W. J. (1983) Tissue-specific synthesis of yolk proteins in Caenorhabditis elegans. Dev. Biol. 96, 189–196 [DOI] [PubMed] [Google Scholar]
- 100. Gems D. H., de la Guardia Y. I. (2012) Alternative perspectives on aging in C. elegans: reactive oxygen species or hyperfunction? Antioxid. Redox. Signal. 19, 321–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. DePina A. S., Iser W. B., Park S. S., Maudsley S., Wilson M. A., Wolkow C. A. (2011) Regulation of Caenorhabditis elegans vitellogenesis by DAF-2/IIS through separable transcriptional and posttranscriptional mechanisms. BMC Physiol. 11, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Riddle D. (1997) C. elegans II, 2nd ed., Cold Spring Harbor Laboratory Press, Plainview, NY: [PubMed] [Google Scholar]
- 103. Polet D., Lambrechts A., Ono K., Mah A., Peelman F., Vandekerckhove J., Baillie D. L., Ampe C., Ono S. (2006) Caenorhabditis elegans expresses three functional profilins in a tissue-specific manner. Cell Motil. Cytoskeleton 63, 14–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Starr D. A., Han M. (2002) Role of ANC-1 in tethering nuclei to the actin cytoskeleton. Science 298, 406–409 [DOI] [PubMed] [Google Scholar]
- 105. Wallimann T., Tokarska-Schlattner M., Schlattner U. (2011) The creatine kinase system and pleiotropic effects of creatine. Amino Acids 40, 1271–1296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Claros M. G., Vincens P. (1996) Computational method to predict mitochondrially imported proteins and their targeting sequences. Eur. J. Biochem. 241, 779–786 [DOI] [PubMed] [Google Scholar]
- 107. Hoogewijs D., Geuens E., Dewilde S., Vierstraete A., Moens L., Vinogradov S., Vanfleteren J. R. (2007) Wide diversity in structure and expression profiles among members of the Caenorhabditis elegans globin protein family. BMC Genomics 8, 356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Geuens E., Hoogewijs D., Nardini M., Vinck E., Pesce A., Kiger L., Fago A., Tilleman L., De Henau S., Marden M. C., Weber R. E., Van Doorslaer S., Vanfleteren J., Moens L., Bolognesi M., Dewilde S. (2010) Globin-like proteins in Caenorhabditis elegans: in vivo localization, ligand binding and structural properties. BMC Biochem. 11, 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Cho J. H., Oh Y. S., Park K. W., Yu J., Choi K. Y., Shin J. Y., Kim D. H., Park W. J., Hamada T., Kagawa H., Maryon E. B., Bandyopadhyay J., Ahnn J. (2000) Calsequestrin, a calcium sequestering protein localized at the sarcoplasmic reticulum, is not essential for body-wall muscle function in Caenorhabditis elegans. J. Cell Sci. 113(Pt 22), 3947–3958 [DOI] [PubMed] [Google Scholar]
- 110. Blomstrand E., Eliasson J., Karlsson H. K., Kohnke R. (2006) Branched-chain amino acids activate key enzymes in protein synthesis after physical exercise. J. Nutr. 136, 269S–273S [DOI] [PubMed] [Google Scholar]
- 111. Kimball S. R., Jefferson L. S. (2006) Signaling pathways and molecular mechanisms through which branched-chain amino acids mediate translational control of protein synthesis. J. Nutr. 136, 227S–231S [DOI] [PubMed] [Google Scholar]
- 112. Payne S. H., Loomis W. F. (2006) Retention and loss of amino acid biosynthetic pathways based on analysis of whole-genome sequences. Eukaryot. Cell 5, 272–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Patel M. S., Harris R. A. (1995) Mammalian alpha-keto acid dehydrogenase complexes: gene regulation and genetic defects. FASEB J. 9, 1164–1172 [DOI] [PubMed] [Google Scholar]
- 114. Hutson S. (2001) Structure and function of branched chain aminotransferases. Prog. Nucleic Acids Res. Mol. Biol. 70, 175–206 [DOI] [PubMed] [Google Scholar]
- 115. Brosnan J. T., Brosnan M. E. (2006) Branched-chain amino acids: enzyme and substrate regulation. J. Nutr. 136, 207S–211S [DOI] [PubMed] [Google Scholar]
- 116. Hamilton B., Dong Y., Shindo M., Liu W., Odell I., Ruvkun G., Lee S. S. (2005) A systematic RNAi screen for longevity genes in C. elegans. Genes Dev. 19, 1544–1555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Chen D., Pan K. Z., Palter J. E., Kapahi P. (2007) Longevity determined by developmental arrest genes in Caenorhabditis elegans. Aging Cell 6, 525–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Curran S. P., Ruvkun G. (2007) Lifespan regulation by evolutionarily conserved genes essential for viability. PLoS Genet. 3, 479–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Tohyama D., Yamaguchi A., Yamashita T. (2008) Inhibition of a eukaryotic initiation factor (eIF2Bdelta/F11A3.2) during adulthood extends lifespan in Caenorhabditis elegans. FASEB J. 22, 4327–4337 [DOI] [PubMed] [Google Scholar]
- 120. Wang J. L., Robida-Stubbs S., Tullet J. M. A., Rual J. F., Vidal M., Blackwell T. K. (2010) RNAi screening implicates a SKN-1-dependent transcriptional response in stress resistance and longevity deriving from translation inhibition. PLoS Genet. 6, 1–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Hayashi A. A., Proud C. G. (2007) The rapid activation of protein synthesis by growth hormone requires signaling through mTOR. Am. J. Physiol. Endocrinol. Metab. 292, E1647–E1655 [DOI] [PubMed] [Google Scholar]
- 122. Selman C., Tullet J. M. A., Wieser D., Irvine E., Lingard S. J., Choudhury A. I., Claret M., Al-Qassab H., Carmignac D., Ramadani F., Woods A., Robinson I. C. A., Schuster E., Batterham R. L., Kozma S. C., Thomas G., Carling D., Okkenhaug K., Thornton J. M., Partridge L., Gems D., Withers D. J. (2009) Ribosomal protein S6 kinase 1 signaling regulates mammalian life span. Science 326, 140–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Bjedov I., Toivonen J. M., Kerr F., Slack C., Jacobson J., Foley A., Partridge L. (2010) Mechanisms of life span extension by rapamycin in the fruit fly Drosophila melanogaster. Cell Metab. 11, 35–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Bjedov I., Partridge L. (2011) A longer and healthier life with TOR down-regulation: genetics and drugs. Biochem. Soc. Trans. 39, 460–465 [DOI] [PubMed] [Google Scholar]
- 125. Grewal S. S., Li L., Orian A., Eisenman R. N., Edgar B. A. (2005) Myc-dependent regulation of ribosomal RNA synthesis during Drosophila development. Nat. Cell Biol. 7, 295–U109 [DOI] [PubMed] [Google Scholar]
- 126. Wullschleger S., Loewith R., Hall M. N. (2006) TOR signaling in growth and metabolism. Cell 124, 471–484 [DOI] [PubMed] [Google Scholar]
- 127. Teleman A. A., Hietakangas V., Sayadian A. C., Cohen S. M. (2008) Nutritional control of protein biosynthetic capacity by insulin via Myc in Drosophila. Cell Metab. 7, 21–32 [DOI] [PubMed] [Google Scholar]
- 128. Li L., Edgar B. A., Grewal S. S. (2010) Nutritional control of gene expression in Drosophila larvae via TOR, Myc and a novel cisregulatory element. BMC Cell Biol. 11, 7–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Robida-Stubbs S., Glover-Cutter K., Lamming D. W., Mizunuma M., Narasimhan S. D., Neumann-Haefelin E., Sabatini D. M., Blackwell T. K. (2012) TOR signaling and rapamycin influence longevity by regulating SKN-1/Nrf and DAF-16/FoxO. Cell Metab. 15, 713–724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Schwer B., Saha N., Mao X., Chen H. W., Shuman S. (2000) Structure-function analysis of yeast mRNA cap methyltransferase and high-copy suppression of conditional mutants by AdoMet synthase and the ubiquitin conjugating enzyme Cdc34p. Genetics 155, 1561–1576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Chiang P. K., Gordon R. K., Tal J., Zeng G. C., Doctor B. P., Pardhasaradhi K., McCann P. P. (1996) S-adenosylmethionine and methylation. FASEB J. 10, 471–480 [PubMed] [Google Scholar]
- 132. Warner J. R., Mitra G., Schwindinger W. F., Studeny M., Fried H. M. (1985) Saccharomyces cerevisiae coordinates accumulation of yeast ribosomal proteins by modulating mRNA splicing, translational initiation, and protein turnover. Mol. Cell. Biol. 5, 1512–1521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Tsay Y. F., Thompson J. R., Rotenberg M. O., Larkin J. C., Woolford J. L., Jr. (1988) Ribosomal protein synthesis is not regulated at the translational level in Saccharomyces cerevisiae: balanced accumulation of ribosomal proteins L16 and rp59 is mediated by turnover of excess protein. Genes Dev. 2, 664–676 [DOI] [PubMed] [Google Scholar]
- 134. Jorgensen P., Rupes I., Sharom J. R., Schneper L., Broach J. R., Tyers M. (2004) A dynamic transcriptional network communicates growth potential to ribosome synthesis and critical cell size. Genes Dev. 18, 2491–2505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Hinnebusch A. G. (2005) Translational regulation of GCN4 and the general amino acid control of yeast. Annu. Rev. Microbiol. 59, 407–450 [DOI] [PubMed] [Google Scholar]
- 136. Pleiss J. A., Whitworth G. B., Bergkessel M., Guthrie C. (2007) Rapid, transcript-specific changes in splicing in response to environmental stress. Mol. Cell 27, 928–937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Ingolia N. T., Ghaemmaghami S., Newman J. R. S., Weissman J. S. (2009) Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science 324, 218–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. McColl G., Rogers A. N., Alavez S., Hubbard A. E., Melov S., Link C. D., Bush A. I., Kapahi P., Lithgow G. J. (2010) Insulin-like signaling determines survival during stress via posttranscriptional mechanisms in C. elegans. Cell Metab. 12, 260–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Mehta R., Chandler-Brown D., Ramos F. J., Shamieh L. S., Kaeberlein M. (2010) Regulation of mRNA translation as a conserved mechanism of longevity control. Adv. Exp. Med. Biol. 694, 14–29 [DOI] [PubMed] [Google Scholar]
- 140. Warner J. R. (1999) The economics of ribosome biosynthesis in yeast. Trends Biochem. Sci. 24, 437–440 [DOI] [PubMed] [Google Scholar]
- 141. Moss T., Stefanovsky V. Y. (2002) At the center of eukaryotic life. Cell 109, 545–548 [DOI] [PubMed] [Google Scholar]
- 142. Lam Y. W., Lamond A. I., Mann M., Andersen J. S. (2007) Analysis of nucleolar protein dynamics reveals the nuclear degradation of ribosomal proteins. Curr. Biol. 17, 749–760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Schubert U., Anton L. C., Gibbs J., Norbury C. C., Yewdell J. W., Bennink J. R. (2000) Rapid degradation of a large fraction of newly synthesized proteins by proteasomes. Nature 404, 770–774 [DOI] [PubMed] [Google Scholar]
- 144. Turner G. C., Varshavsky A. (2000) Detecting and measuring cotranslational protein degradation in vivo. Science 289, 2117–2120 [DOI] [PubMed] [Google Scholar]
- 145. Rogers A. N., Chen D., McColl G., Czerwieniec G., Felkey K., Gibson B. W., Hubbard A., Melov S., Lithgow G. J., Kapahi P. (2011) Life span extension via eIF4G inhibition is mediated by posttranscriptional remodeling of stress response gene expression in C. elegans. Cell Metab. 14, 55–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Steffen K. K., MacKay V. L., Kerr E. O., Tsuchiya M., Hu D., Fox L. A., Dang N., Johnston E. D., Oakes J. A., Tchao B. N., Pak D. N., Fields S., Kennedy B. K., Kaeberlein M. (2008) Yeast life span extension by depletion of 60S ribosomal subunits is mediated by Gcn4. Cell 133, 292–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Zid B. M., Rogers A. N., Katewa S. D., Vargas M. A., Kolipinski M. C., Lu T. A., Benzer S., Kapahi P. (2009) 4E-BP extends lifespan upon dietary restriction by enhancing mitochondrial activity in Drosophila. Cell 139, 149–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Jeong P. Y., Na K., Jeong M. J., Chitwood D., Shim Y. H., Paik Y. K. (2009) Proteomic analysis of Caenorhabditis elegans. Methods Mol. Biol. 519, 145–169 [DOI] [PubMed] [Google Scholar]
- 149. Fostel J. L., Benner Coste L., Jacobson L. A. (2003) Degradation of transgene-coded and endogenous proteins in the muscles of Caenorhabditis elegans. Biochem. Biophys. Res. Commun. 312, 173–177 [DOI] [PubMed] [Google Scholar]
- 150. Szewczyk N. J., Peterson B. K., Barmada S. J., Parkinson L. P., Jacobson L. A. (2007) Opposed growth factor signals control protein degradation in muscles of Caenorhabditis elegans. EMBO J. 26, 935–943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Gaglia M. M., Kenyon C. (2009) Stimulation of movement in a quiescent, hibernation-like form of Caenorhabditis elegans by dopamine signaling. J. Neurosci. 29, 7302–7314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Hsu A. L., Feng Z., Hsieh M. Y., Xu X. Z. (2009) Identification by machine vision of the rate of motor activity decline as a lifespan predictor in C. elegans. Neurobiol. Aging 30, 1498–1503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Felix M. A., Braendle C. (2010) The natural history of Caenorhabditis elegans. Curr. Biol. 20, R965–R969 [DOI] [PubMed] [Google Scholar]
- 154. Sandri M., Sandri C., Gilbert A., Skurk C., Calabria E., Picard A., Walsh K., Schiaffino S., Lecker S. H., Goldberg A. L. (2004) Foxo transcription factors induce the atrophy-related ubiquitin ligase atrogin-1 and cause skeletal muscle atrophy. Cell 117, 399–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Yuan Y., Shi X. E., Liu Y. G., Yang G. S. (2011) FoxO1 regulates muscle fiber-type specification and inhibits calcineurin signaling during C2C12 myoblast differentiation. Mol. Cell. Biochem. 348, 77–87 [DOI] [PubMed] [Google Scholar]
- 156. Burnell A. M., Houthoofd K., O'Hanlon K., Vanfleteren J. R. (2005) Alternate metabolism during the dauer stage of the nematode Caenorhabditis elegans. Exp. Gerontol. 40, 850–856 [DOI] [PubMed] [Google Scholar]
- 157. Alvers A. L., Fishwick L. K., Wood M. S., Hu D., Chung H. S., Dunn W. A., Jr., Aris J. P. (2009) Autophagy and amino acid homeostasis are required for chronological longevity in Saccharomyces cerevisiae. Aging Cell 8, 353–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. D'Antona G., Ragni M., Cardile A., Tedesco L., Dossena M., Bruttini F., Caliaro F., Corsetti G., Bottinelli R., Carruba M. O., Valerio A., Nisoli E. (2010) Branched-chain amino acid supplementation promotes survival and supports cardiac and skeletal muscle mitochondrial biogenesis in middle-aged mice. Cell Metab. 12, 362–372 [DOI] [PubMed] [Google Scholar]
- 159. Herndon L. A., Schmeissner P. J., Dudaronek J. M., Brown P. A., Listner K. M., Sakano Y., Paupard M. C., Hall D. H., Driscoll M. (2002) Stochastic and genetic factors influence tissue-specific decline in ageing C. elegans. Nature 419, 808–814 [DOI] [PubMed] [Google Scholar]
- 160. Fraser A. G., Kamath R. S., Zipperlen P., Martinez-Campos M., Sohrmann M., Ahringer J. (2000) Functional genomic analysis of C. elegans chromosome I by systematic RNA interference. Nature 408, 325–330 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.