Abstract
The influence of DNA duplex structural destabilization introduced by a single base-pair modification was investigated by nanopore measurements. A series of eleven modified base pairs were introduced into the context of an otherwise complementary DNA duplex formed by a 17-mer oligomer and a 65-mer oligomer such that the overhanging ends comprised poly(dT)23 tails, generating a representative set of duplexes that display a range of unzipping mechanistic behaviors and kinetic stabilities. The guanine oxidation products 8-oxo-7,8-dihydroguanine (OG), guanidinohydantoin (Gh), and spiroiminodihydantoin (Sp) were paired with either cytosine (C), adenine (A), or 2,6-diaminopurine (D) to form modified base pairs. The mechanism and kinetic rate constants of duplex dissociation were determined by threading either the 3′ or 5′ overhangs into an α-hemolysin (α-HL) channel under an electrical field and measuring the distributions of unzipping times at constant force. In order of decreasing thermodynamic stability (as measured by duplex melting points), the rate of duplex dissociation increases and the mechanism evolves from a first-order reaction to two sequential first-order reactions. These measurements allow us to rank the kinetic stability of lesion-containing duplexes relative to the canonical G:C base pair, in which the OG:C, Gh:C and Sp:C base pairs are, respectively, 3 to 200 times less stable. The rate constants also depend on whether unzipping was initiated from the 3′ vs. 5′ side of the duplex. The kinetic stability of these duplexes was interpreted in terms of the structural destabilization introduced by the single base pair modification. Specifically, a large distortion of the duplex backbone introduced by the presence of the highly oxidized guanine products Sp and Gh leads to a rapid two-step unzipping. The number of hydrogen bonds in the modified base pair plays a lesser role in determining the kinetics of duplex dissociation.
Keywords: Guanine oxidative lesions, nanopore, kinetic stability
Introduction
In biological cells, DNA nucleotides undergo continuous modification into lesions either spontaneously or by exposure to DNA damaging agents.1 The damage occurs at a surprisingly high rate of tens of thousands of times per cell per day.2 Although the majority of lesions are corrected by repair enzymes, damage beyond the cell’s repair capability may lead to cell death or to cancer, cardiovascular and age-related diseases.3,4 One of the most common forms of DNA damage results from oxidizing free radicals that target the DNA base guanine (G), because it has the lowest redox potential.5 Initial oxidation of G gives rise to 8-oxo-7,8-dihydroguanine (OG), a biomarker for monitoring oxidative stress in the cell. Moreover, OG is prone to further oxidation that yields either spiroiminodihydantoin (Sp) or guanidinohydantoin (Gh), depending on the reaction context.6 The importance of these lesions on the thermodynamic stability of double-stranded DNA is fairly well established;7–10 however, the influence of lesions on the kinetics of unraveling (i.e., unzipping) is less well studied.
The unzipping of duplex DNA has been investigated to gain insight into the process of strand separation during DNA replication and transcription.11–13 Our groups have used the nanopore approach to induce unzipping of lesion-containing dsDNA and to study the kinetic stability of damaged duplexes.14 A preliminary report from our laboratories demonstrated that the force-induced strand dissociation of lesion-containing duplexes occurs by either a first-order reaction or two sequential first-order reactions, depending on the extent of destabilizing effect imparted by the damage.15 Here, the nanopore method is used to investigate the influence of a more extensive series of base modifications on duplex unzipping.
In single-molecule force experiments, the time-dependent trajectory describing the response of a biomolecule to an external force can be used to extract kinetic information concerning structures and interactions of individual molecules.16,17 Kinetic studies using single-molecule force techniques have covered a variety of biological processes, including intramolecular and intermolecular interactions of DNA, RNA, and proteins.18–21 Among them, the nanopore method provides a fast and linker-free approach for molecular manipulation that overcomes some of the drawbacks of optical tweezers and atomic force microscopy (AFM).22–24 In a nanopore experiment using the α-hemolysin (α-HL) nanopore, individual DNA duplexes are driven electrophoretically into the pore with only the overhang, if any, being able to fully thread past the constriction zone into the β-barrel.25 In this experiment, the electrical field imposes a localized rupture force on the duplex, causing strand dissociation to occur.26,27 From the moment of entry to that of rupture, the duplex generates a temporal blockade to the ionic current passing through the channel (carried by the electrolyte ions, K+ and Cl−). The duration of this current blockage is used to extract kinetic information related to strand dissociation.28–30 Typically, the unzipping duration of a double-stranded DNA within the α-HL channel is 2 to 3 orders of magnitude longer than the translocation time of single-stranded DNA (ssDNA) at the same applied voltage.31,32
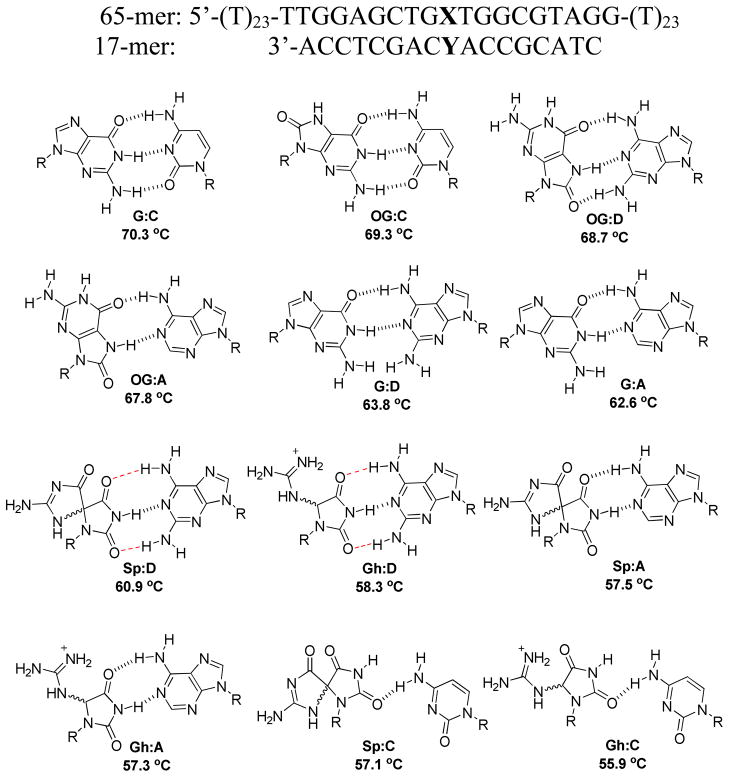
Base pairs for G and its oxidation products OG, Sp and Gh were inserted into a 17-mer duplex DNA (Figure 1). The base-pairing partner was systematically changed to probe the effect on the unzipping kinetics. This allowed us to interrogate the structural features that govern the kinetic stability of lesion-containing base pairs. Each of the base pairs was placed at a mutational hot spot in a portion of the K-ras gene sequence surrounding codon 12, a known location for oxidative damage to G nucleotides.33,34 Eleven base-pair modifications (X:Y) were chosen for study in this work, generating a broad range of variation concerning the extent of destabilizing effects: OG, or one of its second-stage oxidized products, Gh or Sp, was incorporated to introduce destabilization of different levels, while cytosine (C), adenine (A), or 2,6-diaminopurine (D) was paired with the lesion to further adjust the extent of destabilization.35–37 The base D, an analog of A, can increase H-bonding in X:Y compared with A pairing, and can thus be used as a base pair modulator.
Figure 1.
Base pairing schemes for X:Y (X = G, OG, Sp or Gh; Y = C, A, or D). The base pairs G:A, G:C, OG:A and OG:C are drawn based on reported structural data, while the hydantoin base pairs are drawn based on predictions in refs. 6 and 38. The duplexes are arranged in order of decreasing melting temperatures (from top down). The D-containing base pairs are predicted based on their H-bonding capabilities as discussed in the text. The melting temperatures are listed under each X:Y-containing duplex and have an average standard deviation of 0.6 °C (See Supporting Information for individual standard deviations in Tm).
Materials and Methods
DNA Preparation and Purification Procedures
The oligodeoxynucleotides were synthesized from commercially available phosphoramidites (Glen Research, Sterling, VA) by the DNA-Peptide Core Facility at the University of Utah. Each oligodeoxynucleotide was cleaved from the synthetic column after synthesis and then deprotected according to the manufacturer’s protocol. The oligonucleotides were purified using an ion-exchange HPLC column with a linear gradient of 25% to 100% B over 30 min while monitoring UV/Vis absorbance at 260 nm (A = 10% CH3CN/90% ddH2O, B = 20 mM NaPi, 1 M NaCl pH 7 in 10% CH3CN/90% ddH2O, flow rate = 1 mL/min). The Gh- and Sp-containing DNAs were synthesized following a previously established protocol from ref 10.
Thermal Denaturation Experiments
Thermal denaturation experiments to determine the Tm values were conducted with the truncated 23-mer strand, 5′-TT TTG GAG CTG XTG GCG TAG GTT-3′, in which X = G, OG, Sp, or Gh. By removing the poly-dT tails of the 65-mer stand, the hyperchromic shift for the transition from dsDNA to ssDNA was more clearly observed. The dsDNA samples were formed by mixing the 23-mer and 17-mer (3′-ACC TCG ACY ACC GCA TC-5′, Y = C, A, or D) in a 1:1 ratio in 1 M KCl, 10 mM PBS, and 1 mM EDTA (pH 7.4) at a concentration of 10 μM. The dsDNAs were annealed in a 90 °C water bath for 5 min, and then slowly cooled to room temperature over 3 h. Thermal denaturation experiments were then conducted on diluted dsDNA samples at 1 μM in the same solution. Samples were thermally equilibrated at 20 °C for 20 min followed by heating to 75 °C at a rate of 0.5 °C/min. As the samples were heated, UV/Vis absorbance readings were recorded at 260 nm every 30 sec. The Tm was determined using a two-point average analysis.
Chemicals and Materials for Nanopore Measurement
Monomer wild-type α-HL (List Biological Laboratories) was dissolved in water at 1 mg/mL and stored at −80 °C. The lipid, 1,2-diphytanoyl-sn-glycero-3-phosphocholine (Avanti Polar Lipids), was dissolved in decane at 10 mg/mL before use. Glass nanopore membranes (GNMs) were fabricated using a bench-top method as described in ref 39 and then chemically modified with 2% (v/v) (3-cyanopropyl) dimethylchlorosilane in acetonitrile to produce a hydrophobic surface as the support structure for the lipid bilayer.40,41 The 65-mer strands (5′-T23-TTG GAG CTG XTG GCG TAG G-T23-3′, X = G, OG, Sp, or Gh) were mixed with the 17-mer strands (3′-ACC TCG ACY ACC GCA TC-5′, Y = C, A or D) at 1:5 mole ratio (65-mer versus 17-mer) to form duplexes in 1 M KCl, 10 mM PBS, and 1 mM EDTA (pH 7.4). The 65-mer and 17-mer mixture was heated in a 90 °C water bath for 5 min and then slowly cooled to room temperature over 3 h.
Duplex Unzipping Current-Time Recording
All current-time recordings were collected using a custom built amplifier and data acquisition system (Electronic BioSciences Inc., San Diego, CA). The same solution to dissolve the DNA (1 M KCl, 10 mM PBS, and 1 mM EDTA at pH 7.4) was used as the electrolyte for ion-channel recordings and to fill up the electrolyte reservoir and the GNM capillary (orifice radius ~500 nm). A voltage of −120 mV (cis vs. trans) was applied across the GNM/bilayer using Ag/AgCl electrodes placed inside and outside of the GNM capillary. A lipid bilayer was formed by painting the lipid/decane solution across the GNM orifice. A positive pressure was applied to the inside of the GNM capillary, allowing the lipid bilayer to be functional for protein channel reconstitution.42 Protein monomers were added to the cis side of the nanopore and self-assembled into the lipid bilayer as the heptamer ion channel. After protein insertion, the duplex DNA sample was added to the electrolyte reservoir at a final concentration of 5 μM. A minimum of 300 unzipping events were collected for each DNA sample. The i-t recordings were filtered at 10 kHz and sampled at 50 kHz.
Data Analysis
Unzipping events were extracted using QuB (version 1.5.0.31). According to previously reported duplex unzipping experiments in ref 15, the blockades in the i-t traces that lasted longer than 0.5 ms were analyzed as duplex unzipping events. Event durations shorter than 0.5 ms represent the translocation of the excess 17-mer strands (due to the 1:5 mixing ratio of 65-mer:17-mer) and are not discussed in this work. The duplex unzipping process was described by either a first-order reaction or two sequential first-order reactions, depending on the specific duplex sequence. The shape of the unzipping duration (t) histogram was used to determine which kinetic model best describes the duplex unzipping. The unzipping rate constants (k for Type I, or k1 and k2 for Type II) were obtained based on fit of the t histogram using the corresponding kinetic equations.
The kinetic equation for Type I histograms is (equation derivation in ref 15):
| (1) |
where C/T is event counts in a time increment t centered at time t divided by the total counts. In Model I, the unzipping time constant τ = k−1.
The kinetic equation for Type II histograms is (equation derivation in ref 15):
| (2) |
where k1 and k2 correspond to the rate constants for the first and second unzipping steps. For Type II, the time constants for the first and second steps τ1 and τ2 can be calculated from τ1 = k1−1 and τ2 = k2−1. Because k1 and k2 are interchangeable in eq 2, the fit of this equation to the histograms does not allow assignment of the extracted rate constant values to step 1 or 2. Unzipping experiments using duplexes in which either the 5′ or 3′ poly(dT)23 tail of the 65-mer oligomer is removed are necessary in order to identify k1 and k2 with the individual steps (the details of this analysis are presented in ref 15).
Histograms were plotted and fit using OriginPro (version 8.5.1). Density plots were generated using the data analysis program provided by Electronic Biosciences, San Diego, CA.
Results and Discussion
In this study, the 65-mer target strand containing a guanine derivative of interest X (X = G, OG, Sp, or Gh) was hybridized with a 17-mer Y-containing (Y = C, A, or D) strand, forming a duplex with the base pair X opposite to Y (Figure 1). The resultant duplex is fully complementary except for the modified base pair X:Y. For the sake of brevity, the duplex is hereafter denoted simply as X:Y.
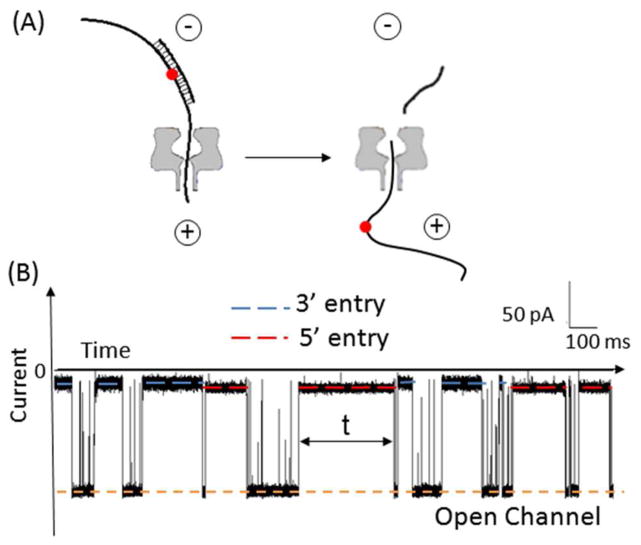
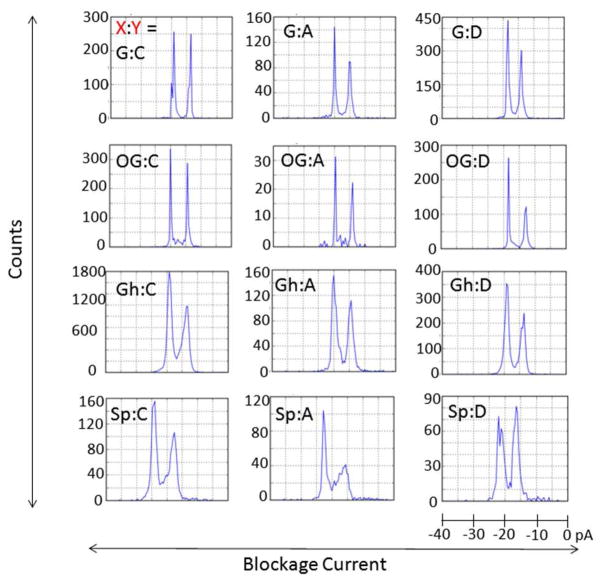
Ion-channel measurements were performed to record the unzipping of single molecules of the X:Y duplexes within the α-HL channel under the influence of an electrical field (Figure 2). The blockage events due to occupation of the channel by the duplex yielded two distinct current levels, depending on whether the 3′-or the 5′-poly(dT)23 overhang initiated threading into the channel; we have previously determined that the more blocking current level is associated with 3′ entry of the duplex and the less blocking level with 5′ entry (the 3′ and 5′ directions refer to the ends of the 65-mer target strand).15 As shown in Figure 3, histograms of the blockage current extracted from unzipping events for each double-overhang duplex do not display a general preference of entry direction; between ~40 and 60% of events correspond to 3′ entry. This result is in contrast to the reported biased entry preferred at the 3′ end for ssDNA and hairpins.43,44 The histograms shown in Figure 3 also indicate that the current blockage levels are nearly independent of the identity of X:Y for G-, OG-, and Gh-containing duplexes. The Sp-containing duplexes, on the other hand, always generated shallower current blockages than the other duplexes by 3 to 5 pA, regardless of the pairing base or the entry direction. Broader current distributions were observed for all Gh- and Sp-containing duplexes, as expected based on the conformational variations that originate from coexistence of diastereomers for both Gh and Sp.36,45
Figure 2.
Unzipping of a DNA duplex using an α-HL nanopore under an electrical field. (A) dsDNA strand dissociation within an α-HL. The red dot in the target strand defines the relative position of X (X = G, OG, Sp, or Gh) opposite to Y (Y = C, A, or D) in the duplex. The + and − signs show the polarity of the electrodes. (B) Example current-time trace (X:Y = OG:C) at an applied voltage of −120 mV (cis versus trans). The blockades that last up to hundreds of milliseconds correspond to unzipping events of duplexes (unzipping duration shown as t). Events with duration shorter than 0.5 ms correspond to translocation of excess 17-mer ssDNA (the 65-mer and 17-mer strands are present in a 1:5 mole ratio). The red and blue dashed lines show the current blockage levels associated with 5′ and 3′ entry, respectively.
Figure 3.
Histograms of blockage current for the duplex formed by hybridizing the X-containing 65-mer with the Y-containing 17-mer at −120 mV. X:Y represents the modified base pair.
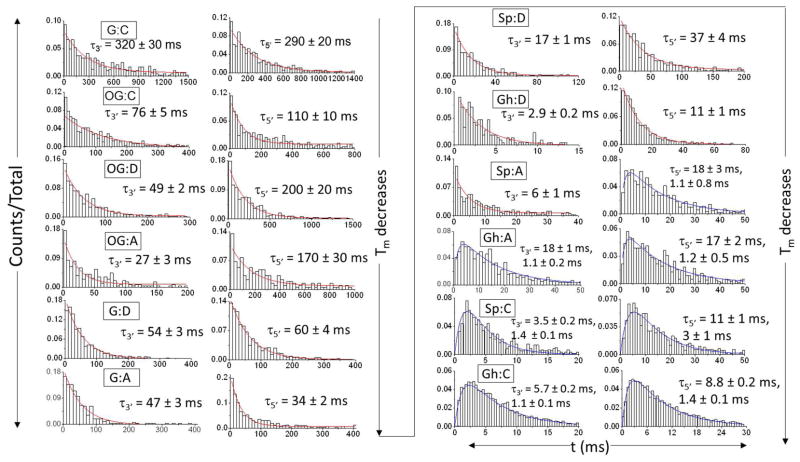
Unzipping durations (t) sorted by entry direction (3′ vs. 5′) were used to investigate the mechanism and kinetics of strand separation for each X:Y duplex. The histograms are consistent with either a first-order reaction path (Type I) or a path comprising two sequential first-order reactions (Type II). In Figure 4, histograms of unzipping durations for the X:Y duplexes (X = G, OG, Sp, or Gh, and Y = C, or A) show a transition from the Type I exponential shape to the Type II peak shape as the Tm decreases, with the less stable duplexes tending to unzip more rapidly and in two steps. In cases where the strand dissociation follows a two-step model, the unzipping time constants are presented in Figure 4 without assignment of the first and second steps. Additional experiments using truncated duplexes that place X:Y asymmetrically in the sequence as demonstrated in ref 15 are necessary to determine the unzipping time constants for each step.
Figure 4.
Histograms of unzipping time (t) via 3′ entry (left column) and 5′ entry (right column) at −120 mV for the duplex that contains X:Y (where X = G, OG, Gh or Sp, and Y = C, A, D). The unzipping time constants (τ) were obtained based on the fit (red or blue curves) of the t histograms using the corresponding kinetic model, either a first-order reaction (Type I, red) or two sequential first-order reactions (Type II, blue). If the strand dissociation follows the Type II model, the unzipping time constants for each step are presented without assignment of the first and second steps. The histograms are plotted on different time scales in order to emphasize the shape as a determinant for the model type.
Well-defined Type II shapes of duration histograms were observed for Gh:C and Sp:C. Among the twelve base pair duplexes investigated here, Gh:C and Sp:C have the lowest Tm, suggesting that these duplexes are most capable of generating a pronounced energy well for the existence of an intermediate state.23 In contrast, unzipping of the OG-containing duplex follows the Type I model, indicating that it is relatively stable despite the presence of the slightly destabilizing base pair. The histograms for Sp:A/D and Gh:A/D, whose stability is intermediate between the above cases, display shapes of either the Type I or the Type II model, consistent with the tendency of model evolution in terms of stability.
Tm values of the duplexes were used to order the histograms in Figure 4 to indicate the general correlation of thermodynamic and kinetic stabilities (decreasing from top to bottom). However, we cannot precisely predict the unzipping model or kinetic rates based merely on Tm, especially for duplexes of moderate stability.46–49 Thus, instead of explaining the unzipping results using Tm values, we attempt to compare the kinetic data in terms of two features of local duplex structures: (1) known duplexdistortion induced by the X:Y base pair, and (2) the number of hydrogen bonds between the modified base pair.
In considering the influence of duplex distortion on stability, we find that the unzipping duration is very sensitive to even a slight displacement of the backbone relative to the perfectly matched G:C duplex. The unzipping duration decreased by a factor of 3 to 4 as a result of substituting G:C with OG:C (For G:C, τ3′ = 320 ± 30 ms, τ5′ = 290 ± 20 ms. For OG:C, τ3′ = 76 ± 5 ms, τ5′ = 110 ± 10 ms.); in the oxidized structure, the Tm decreased by 1 °C due to a subtle distortion in the sugar-phosphate backbone originating from the repulsion between the C8 oxo group of the base and the C4′ oxygen of the ribose.50, In terms of discriminating a G:C and OG:C, the nanopore method outrivals thermal denaturation methods, in which a modest 1 °C Tm (Figure 1) difference exists between these two base pairs translates to a 3–4 fold increase in the rate of unzipping (Figure 4). In the OG:A base pair the Tm decreased by 2.5 °C (Figure 1) relative to the G:C base pair while the unzipping rate was highly directionally dependent and increased by 2–12 fold (Figure 4). From OG:C to OG:D, the kinetic stability did not change unambiguously based on either 3′ and 5′ entry. Although no structural details are available, we propose that OG adopts the syn conformation when paired with D forming three H-bonds (Figure 1). 51 Consequently, for OG:D, the tension between the C8 oxo group and the phosphate backbone is eased by rotating the C8 oxo group into the groove, but not to the degree of a full recovery based on the ~2 °C decrease in Tm observed for the OG:D base pair relative to the G:C base pair (Figure 1). Further, by substituting OG:D/A with G:D/A, the decrease in durations were expected, due to destabilization from widening of the phosphate backbone at the G:D/A mismatch site by ~3 Å that is reflected in a ~6–7 °C decrease in Tm relative to a G:C base pair (Figure 1).52 For all Gh- and Sp-containing duplexes, experimental and theoretical studies have demonstrated that these nonplanar lesions cause serious distortions to the phosphate backbone as well as interruption of base stacking and hydrogen bonding, making them unzip much faster than G- and OG-containing duplexes.38,53
In cases where backbone distortion remains similar, the number of H-bonds appears to determine the rate of duplex unzipping. The decrease in unzipping duration for OG:A versus OG:D, G:A versus G:D, and Sp:A versus Sp:D can be explained by reduction in the number of hydrogen bonds. Nevertheless, an exception occurs for Gh:A versus Gh:D (Figure 1). With fewer hydrogen bonds, Gh:A unzips slower than Gh:D, though the two-step model for Gh:A indicates that it is more destabilized than Gh:D. For Sp:C and Gh:C, in addition to forming poor H-bonds between Gh/Sp and C, the incorporation of these two base pairs dramatically alters the duplex backbone, resulting in the most severe destabilizing effect observed. In view of the influence of backbone distortions and hydrogen bonding, backbone distortion that generates a large-scale structural perturbation (i.e., nonplanar lesions such as Sp and Gh) should facilitate unzipping more proficiently than removal of H-bonds whose effect is constrained mostly within the base pair. Thus, higher weight in determining the kinetic stability was assigned to the influence of distorting lesions over the number of hydrogen bonds in X:Y.
The results in Figure 4 confirmed that the sequence-dependent unzipping duration is also influenced by the unzipping direction. In Figure 4, for the duplexes in which the unzipping time constants for 3′ and 5′ entry are significantly different, unzipping from the 3′ end always proceeds faster than unzipping from the 5′ end. Further, we found that the oxidized lesions destabilize the duplex in the 3′-orientation to a larger degree than the 5′-orientation, as evidenced by the more notable change of duration at 3′ entry for modified duplexes versus G:C. For example, by replacing G:C with the modified base pair OG:D, the unzipping duration decreased by a factor of 6.5 at 3′ entry and a factor of 1.5 at 5′ entry (For G:C, τ3′ = 320 ± 30 ms, τ5′ = 290 ± 20 ms. For OG:D, τ3′ = 49 ± 2 ms, τ5′ = 200 ± 20 ms).
In addition to studying the kinetics of strand separation in the nanopore, the results of unzipping duration also provide insight into detection of damaged DNA products.54–56 The mutagenic potentials of OG, Sp, and Gh make it important to detect them for the purpose of disease diagnosis and management.57–61 We are able to differentiate target strands containing X = G, OG, Sp, or Gh using three different 17-mer probes, each containing Y = C, A, or D. The C probe destabilizes the target strand that contains oxidative lesions relative to the undamaged strand and causes the unzipping duration to decrease by 4 times for OG versus G and up to 60 times for Sp/Gh versus G. The stabilizing effect of D on OG relative to G enhances the detection selectivity toward OG and increases the unzipping duration for G:D relative to OG:D by a factor of 3 at 5′ entry. Specifically, Sp can be distinguished from G, OG, and Gh by inspecting the blockage current that results from its corresponding duplex.
Conclusion
The present nanopore-based experiments have demonstrated how the structural destabilization introduced by lesions influences the kinetic stability of dsDNA. A progression of the kinetic mechanism and rate has been observed from a single-step path of first-order kinetics to a path of two sequential first-order reactions, with the duplex containing the more destabilizing base pairs X:Y being prone to unravel in a two-step fashion. The kinetic properties of unzipping have been interpreted in terms of the degree of backbone distortion induced by the modified base pair, X:Y, as well as the number of hydrogen bonds within X:Y. Among the two factors mentioned here, the backbone distortion plays a major role in determining the stability of the duplex in α-HL. This study on duplexes with a broad range of structures leads us toward a better understanding of the physical models for the strand-separation process, as well as providing new information about DNA local structures and interactions, which are often concealed in ensemble-averaged measurements. In addition, the wide selection of bases for pairing opposite a lesion, including synthetic heterocycles, allows us multiple options for tuning the properties of DNA and RNA assemblies in nucleic acids research. This work also sheds light on single-molecule damage detection by the ion channel method using the unzipping concept in which one interrogates a specific sequence of a target strand with a customized probe.
Supplementary Material
Acknowledgments
Funding Sources
This work was supported by a research grant from the National Institutes of Health (GM093099).
Instruments and software were donated by Electronic BioSciences Inc., San Diego. We thank Dr. J. G. Muller (University of Utah) for assistance with mass spectrometry.
Footnotes
Supporting Information. Melting temperatures for the duplexes, histogram of current blockage levels for a duplex containing both 3′- and 5′-overhangs, and unzipping time constants as a function of melting temperature. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Loeb LA, Harris CC. Advances in chemical carcinogenesis: a historical review and prospective. Cancer Res. 2008;68:6863–6872. doi: 10.1158/0008-5472.CAN-08-2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mangal D, Vudathala D, Park JH, Lee SH, Penning TM, Blair IA. Analysis of 7, 8-dihydro-8-oxo-2′-deoxyguanosine in cellular DNA during oxidative stress. Chem Res Toxicol. 2009;22:788–797. doi: 10.1021/tx800343c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sohal RS, Weindruch R. Oxidative stress, caloric restriction, and aging. Science. 1996;273:59–63. doi: 10.1126/science.273.5271.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zawia NH, Lahiri DK, Cardozo-Pelaez F. Epigenetics, oxidative stress, and Alzheimer disease. Free Radic Biol Med. 2009;46:1241–1249. doi: 10.1016/j.freeradbiomed.2009.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Steenken S, Jovanovic SV. How easily oxidizable is DNA? One-electron reduction potentials of adenosine and guanosine radicals in aqueous solution. J Am Chem Soc. 1997;119:617–618. [Google Scholar]
- 6.Fleming AM, Muller JG, Dlouhy AC, Burrows CJ. Structural context effects in the oxidation of 8-oxo-7, 8-dihydro-2′-deoxyguanosine to hydantoin products: electrostatics, base stacking, and base pairing. J Am Chem Soc. 2012;134:15091–15102. doi: 10.1021/ja306077b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Plum GE, Grollman AP, Johnson F, Breslauer KJ. Influence of the oxidatively damaged adduct 8-oxodeoxyguanosine on the conformation, energetics, and thermodynamic stability of a DNA duplex. Biochemistry. 1995;34:16148–16160. doi: 10.1021/bi00049a030. [DOI] [PubMed] [Google Scholar]
- 8.Khutsishvili I, Zhang N, Marky LA, Crean C, Patel DJ, Geacintov NE, Shafirovich V. Thermodynamic profiles and nuclear magnetic resonance studies of oligonucleotide duplexes containing single diastereomeric spiroiminodihydantoin lesions. Biochemistry. 2013;52:1354–1363. doi: 10.1021/bi301566v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yennie CJ, Delaney S. Thermodynamic consequences of the hyperoxidized guanine lesion guanidinohydantoin in duplex DNA. Chem Res Toxicol. 2012;25:1732–1739. doi: 10.1021/tx300190a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kornyushyna O, Berges AM, Muller JG, Burrows CJ. In vitro nucleotide misinsertion opposite the oxidized guanosine lesions spiroiminodihydantoin and guanidinohydantoin and DNA synthesis past the lesions using Escherichia coli DNA polymerase I (Klenow fragment) Biochemistry. 2002;41:15304–16314. doi: 10.1021/bi0264925. [DOI] [PubMed] [Google Scholar]
- 11.Bockelmann U, Thomen P, Essevaz-Roulet B, Viasnoff V, Heslet F. Unzipping DNA with optical tweezers: high sequence sensitivity and force flips. Biophys J. 2002;82:1537–1553. doi: 10.1016/S0006-3495(02)75506-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Larson MH, Greenleaf WJ, Landick R, Block SM. Applied force reveals mechanistic and energetic details of transcription termination. Cell. 2008;132:971–982. doi: 10.1016/j.cell.2008.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cocco S, Monasson R, Marko JF. Force and kinetic barriers to unzipping of the DNA double helix. Proc Natl Acad Sci. 2001;98:8608–8613. doi: 10.1073/pnas.151257598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schibel AEP, Fleming AM, Jin Q, An N, Liu J, Blakemore CP, White HS, Burrows CJ. Sequence-specific single-molecule analysis of 8-oxo-7,8-dihydroguanine lesions in DNA based on unzipping kinetics of complementary probes in ion channel recordings. J Am Chem Soc. 2011;133:14778–14784. doi: 10.1021/ja205653v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jin Q, Fleming AM, Burrows CJ, White HS. Unzipping kinetics of duplex DNA containing oxidized lesions in an α-hemolysin nanopore. J Am Chem Soc. 2012;134:11006–11010. doi: 10.1021/ja304169n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rief M, Gautel M, Oesterhelt F, Fernandez JM, Gaub HE. Reversible unfolding of individual titin immunoglobulin domains by AFM. Science. 1997;276:1109–1112. doi: 10.1126/science.276.5315.1109. [DOI] [PubMed] [Google Scholar]
- 17.Bennink ML, Leuba SH, Leno GH, Zlatanova J, de Grooth BG, Greve J. Unfolding individual nucleosomes by stretching single chromatin fibers with optical tweezers. Nat Struct Mol Biol. 2001;8:606–610. doi: 10.1038/89646. [DOI] [PubMed] [Google Scholar]
- 18.Wang MD, Yin H, Landick R, Gelles H, Block SM. Stretching DNA with optical tweezers. Biophys J. 1997;71:1335–1346. doi: 10.1016/S0006-3495(97)78780-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li PTX, Bustamante C, Tinoco I. Real-time control of the energy landscape by force directs the folding of RNA molecules. Proc Natl Acad Sci USA. 2007;104:7039–7044. doi: 10.1073/pnas.0702137104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cecconi C, Shank EA, Bustamante C, Marqusee S. Direct observation of the three-state folding of a single protein molecule. Science. 2005;309:2057–2060. doi: 10.1126/science.1116702. [DOI] [PubMed] [Google Scholar]
- 21.Koch SJ, Shundrovsky A, Jantzen BC, Wang MD. Probing protein-DNA interactions by unzipping a single DNA double helix. Biophys J. 2002;83:1098–1105. doi: 10.1016/S0006-3495(02)75233-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Howorka S, Movileanu L, Braha O, Bayley H. Kinetics of duplex formation for individual DNA strands within a single protein nanopore. Proc Natl Acad Sci. 2001;98:12996–13001. doi: 10.1073/pnas.231434698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dudko OK, Mathé J, Szabo A, Meller A. Nanopore force spectroscopy tools for analyzing single biomolecular complexes. Methods Enzymol. 2010;475:565–589. doi: 10.1016/S0076-6879(10)75021-7. [DOI] [PubMed] [Google Scholar]
- 24.Dudko OK, Mathé J, Szabo A, Meller A, Hummer G. Extracting kinetics from single-molecule force spectroscopy: nanopore unzipping of DNA hairpins. Biophys J. 2007;92:4188–4195. doi: 10.1529/biophysj.106.102855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Song L, Hobaugh MR, Shustak C, Cheley S, Bayley H, Gouax JE. Structure of staphylococcal alpha-hemolysin, a heptameric transmembrane pore. Science. 1996;274:1859–1865. doi: 10.1126/science.274.5294.1859. [DOI] [PubMed] [Google Scholar]
- 26.Wanunu M, Sutin J, Meller A. DNA profiling using solid-state nanopores: detection of DNA-binding molecules. Nano Lett. 2009;9:3498–3502. doi: 10.1021/nl901691v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Howorka S, Cheley S, Bayley H. Sequence-specific detection of individual DNA strands using engineered nanopores. Nat Biotechnol. 2001;19:636–639. doi: 10.1038/90236. [DOI] [PubMed] [Google Scholar]
- 28.Vercoutere W, Winters-Hilt S, Olsen H, Deamer D, Haussler D, Akeson M. Rapid discrimination among individual DNA hairpin molecules at single-nucleotide resolution using an ion channel. Nat Biotechnol. 2001;19:248–252. doi: 10.1038/85696. [DOI] [PubMed] [Google Scholar]
- 29.Vercoutere WA, Winters-Hilt S, DeGuzman VS, Deamer D, Ridino SE, Rodgers JT, Olsen HE, Marziali A, Akeson M. Discrimination among individual Watson–Crick base pairs at the termini of single DNA hairpin molecules. Nucleic Acids Res. 2003;31:1311–1318. doi: 10.1093/nar/gkg218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mathè J, Visram H, Viasnoff V, Rabin Y, Meller A. Nanopore unzipping of individual DNA hairpin molecules. Biophys J. 2004;87:3205–3212. doi: 10.1529/biophysj.104.047274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kasianowicz J, Brandin E, Branton D, Deamer D. Characterization of individual polynucleotide molecules using a membrane channel. Proc Natl Acad Sci USA. 1996;93:13770–13773. doi: 10.1073/pnas.93.24.13770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Akeson M, Branton D, Kasianowicz J, Brandin E, Deamer D. Microsecond timescale discrimination among polycytidylic acid, poly-adenylic acid, and polyuridylic acid as homopolymers or as segments within single RNA molecules. Biophys J. 1999;77:3227–3233. doi: 10.1016/S0006-3495(99)77153-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xie Y, Yang H, Cunanan C, Okamoto K, Shibata D, Pan J, Barnes DE, Lindahl T, McIlhantton M, Fishel R, Miller JH. Deficiencies in mouse Myh and Ogg1 result in tumor predisposition and G to T mutations in codon 12 of the K-ras oncogene in lung tumors. Cancer Res. 2004;64:3096–3102. doi: 10.1158/0008-5472.can-03-3834. [DOI] [PubMed] [Google Scholar]
- 34.Husgafvel-Pursiainen K, Hackman P, Ridanpaa M, Anttila S, Karjalainen A, Partanen T, Taikina-Aho O, Heikkila L, Vainio H. K-ras mutations in human adenocarcinoma of the lung: association with smoking and occupational exposure to asbestos. Int J Cancer. 1993;53:250–256. doi: 10.1002/ijc.2910530213. [DOI] [PubMed] [Google Scholar]
- 35.Luo W, Muller JG, Burrows CJ. The pH-dependent role of superoxide in riboflavin-catalyzed photooxidation of 8-oxo-7,8-dihydroguanosine. Org Lett. 2001;3:2801–2804. doi: 10.1021/ol0161763. [DOI] [PubMed] [Google Scholar]
- 36.Luo W, Muller JG, Rachlin EM, Burrows CJ. Characterization of spiroiminodihydantoin as a product of one-electron oxidation of 8-oxo-7,8-dihydroguanosine. Org Lett. 2000;2:613–616. doi: 10.1021/ol9913643. [DOI] [PubMed] [Google Scholar]
- 37.Luo W, Muller JG, Rachlin EM, Burrows CJ. Characterization of hydantoin products from one-electron oxidation of 8-oxo-7,8-dihydroguanosine in a nucleoside model. Chem Res Toxicol. 2001;14:927–938. doi: 10.1021/tx010072j. [DOI] [PubMed] [Google Scholar]
- 38.Jia L, Shafirovich V, Shapiro R, Geacintov NE, Broyde S. Structural and thermodynamic features of spiroiminodihydantoin damaged DNA duplexes. Biochemistry. 2005;44:13342–13353. doi: 10.1021/bi050790v. [DOI] [PubMed] [Google Scholar]
- 39.Zhang B, Galusha J, Shiozawa PG, Wang G, Bergren AJ, Jones RM, White RJ, Ervin EN, Cauley C, White HS. Bench-top method for fabricating glass-sealed nanodisk electrodes, glass nanopore electrodes, and glass nanopore membranes of controlled size. Anal Chem. 2007;79:4778–4787. doi: 10.1021/ac070609j. [DOI] [PubMed] [Google Scholar]
- 40.White RJ, Ervin EN, Yang T, Chem X, Daniel S, Cremer PS, White HS. Single ion-channel recordings using glass nanopore membranes. J Am Chem Soc. 2007;129:11766–11775. doi: 10.1021/ja073174q. [DOI] [PubMed] [Google Scholar]
- 41.Lan WJ, White HS. Diffusional motion of a particle translocating through a nanopore. ACS Nano. 2012;6:1757–1765. doi: 10.1021/nn2047636. [DOI] [PubMed] [Google Scholar]
- 42.Schibel AEP, Heider EC, Harris JM, White HS. Fluorescence microscopy of the pressure-dependent structure of lipid bilayers suspended across conical nanopores. J Am Chem Soc. 2011;133:7810–7815. doi: 10.1021/ja1117182. [DOI] [PubMed] [Google Scholar]
- 43.Mathé J, Aksimentiev A, Nelson DR, Schulten K, Meller A. Orientation discrimination of single-stranded DNA inside the α-hemolysin membrane channel. Proc Natl Acad Sci USA. 2005;102:12377–12382. doi: 10.1073/pnas.0502947102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Butler TZ, Gundlach JH, Troll M. Ionic current blockages from DNA and RNA molecules in the α-hemolysin nanopore. Biophys J. 2007;93:3229–3240. doi: 10.1529/biophysj.107.107003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ye Y, Muller JG, Luo W, Mayne CL, Shallop AJ, Jones RA, Burrows CJ. Formation of 13C-, 15N-, and 18O-labeled guanidinohydantoin from guanosine oxidation with singlet oxygen. Implications for structure and mechanism. J Am Chem Soc. 2003;125:13926–13927. doi: 10.1021/ja0378660. [DOI] [PubMed] [Google Scholar]
- 46.McNally B, Wanunu M, Meller A. Electromechanical unzipping of individual DNA molecules using synthetic sub-2 nm pores. Nano Lett. 2008;8:3418–3422. doi: 10.1021/nl802218f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Viasnoff V, Chiaruttini N, Bockelmann U. Probing DNA base pairing energy profiles using a nanopore. Eur Biophys J. 2009;38:263–269. doi: 10.1007/s00249-008-0372-2. [DOI] [PubMed] [Google Scholar]
- 48.Renner S, Bessonov A, Gerland U, Simmel FC. Sequence-dependent unfolding kinetics of DNA hairpins studied by nanopore force spectroscopy. J Phys: Condens Matter. 2010;22:454119. doi: 10.1088/0953-8984/22/45/454119. [DOI] [PubMed] [Google Scholar]
- 49.Viasnoff V, Chiaruttini N, Muzard J, Bockelmann U. Force fluctuations assist nanopore unzipping of DNA. J Phys: Condens Matter. 2010;22:454122. doi: 10.1088/0953-8984/22/45/454122. [DOI] [PubMed] [Google Scholar]
- 50.Lipscomb LA, Peek ME, Morningstar ML, Verghis SM, Miller EM, Rich A, Essigmann JM, Williams LD. X-ray structure of a DNA decamer containing 7,8-dihydro-8-oxoguanine. Proc Natl Acad Sci USA. 1995;92:719–723. doi: 10.1073/pnas.92.3.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cheong C, Tinoco I, Jr, Chollet A. Thermodynamic studies of base pairing involving 2,6-diaminopurine. Nucleic Acids Res. 1988;16:5115–5122. doi: 10.1093/nar/16.11.5115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Berashevich J, Chakraborty T. Thermodynamics of G·A mispairs in DNA: Continuum electrostatic model. J Chem Phys. 2009;130:015101. doi: 10.1063/1.3050107. [DOI] [PubMed] [Google Scholar]
- 53.Aller P, Ye Y, Wallace SS, Burrows CJ, Doublié S. Crystal structure of a replicative DNA polymerase bound to the oxidized guanine lesion guanidinohydantoin. Biochemistry. 2010;49:2502–2509. doi: 10.1021/bi902195p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Howorka S, Siwy Z. Nanopore analytics: sensing of single molecules. Chem Soc Rev. 2009;38:2360–2384. doi: 10.1039/b813796j. [DOI] [PubMed] [Google Scholar]
- 55.Wanunu M, Cohen-Karni D, Johnson RR, Fields L, Benner J, Peterman N, Zheng Y, Klein ML, Drndic M. Discrimination of methylcytosine from hydroxymethylcytosine in DNA molecules. J Am Chem Soc. 2010;133:486–492. doi: 10.1021/ja107836t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schibel AEP, An N, Jin Q, Fleming AM, Burrows CJ, White HS. Nanopore detection of 8-oxo-7,8-dihydro-2′-deoxyguanosine in immobilized single-stranded DNA via adduct formation to the DNA damage site. J Am Chem Soc. 2010;132:17992–17995. doi: 10.1021/ja109501x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Duarte V, Muller JG, Burrows CJ. Insertion of dGMP and dAMP during in vitro DNA synthesis opposite an oxidized form of 7,8-dihydro-8-oxoguanine. Nucleic Acids Res. 1999;27:496–502. doi: 10.1093/nar/27.2.496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hailer MK, Slade PG, Martin BD, Sugden KD. Nei deficient Escherichia coli are sensitive to chromate and accumulate the oxidized guanine lesion spiroiminodihydantoin. Chem Res Toxicol. 2005;18:1378–1383. doi: 10.1021/tx0501379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Delaney S, Neeley WL, Delaney JC, Essigmann JM. The substrate specificity of MutY for hyperoxidized guanine lesions in vivo. Biochemistry. 2007;46:1448–1455. doi: 10.1021/bi061174h. [DOI] [PubMed] [Google Scholar]
- 60.Henderson PT, Delaney JC, Muller JG, Neeley WL, Tannenbaum SR, Burrows CJ, Essigmann JM. The hydantoin lesions formed from oxidation of 7,8-dihydro-8-oxoguanine are potent sources of replication errors in vivo. Biochemistry. 2003;42:9257–9262. doi: 10.1021/bi0347252. [DOI] [PubMed] [Google Scholar]
- 61.Mangerich A, Knutson CG, Parry NM, Muthupalani S, Ye W, Prestwich E, Gui L, McFaline JL, Mobley M, Ge Z, Taghizadeh K, Wishnok JS, Wogan GN, Fox JG, Tannenbaum SR, Dedon PC. Infection-induced colitis in mice causes dynamic and tissue-specific changed in stress response and DNA damage leading to colon cancer. Proc Natl Acad Sci USA. 2012;109:E1820–1829. doi: 10.1073/pnas.1207829109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.