Abstract
Acute promyelocytic leukemia (APL) is initiated by the PML-RARA fusion oncogene and has a characteristic expression profile that includes high levels of the Notch ligand JAG1. In this study, we used a series of bioinformatic, in vitro, and in vivo assays to assess the role of Notch signaling in human APL samples, and in a PML-RARA knockin mouse model of APL (Ctsg-PML-RARA). We identified a Notch expression signature in both human primary APL cells and in Kit+Lin−Sca1+ (KLS) cells from pre-leukemic Ctsg-PML-RARA mice. Both genetic and pharmacologic inhibition of Notch signaling abrogated the enhanced self-renewal seen in hematopoietic stem/progenitor cells (HSPCs) from pre-leukemic Ctsg-PML-RARA mice, but had no influence on cells from age-matched wildtype mice. In addition, 6 of 9 murine APL tumors tested displayed diminished growth in vitro when Notch signaling was inhibited pharmacologically. Finally, we found that genetic inhibition of Notch signaling with a dominant negative MAML protein reduced APL growth in vivo in a subset of tumors. These findings expand the role of Notch signaling in hematopoietic diseases, and further define the mechanistic events important for PML-RARA-mediated leukemogenesis.
Keywords: Notch, Acute Promyelocytic Leukemia, Self-renewal
Introduction
The t(15;17)(q22;q11.2) translocation produces the PML-RARA fusion gene, which has been shown to be the initiating event for acute promyelocytic leukemia (APL, FAB M3) in several mouse models of the disease1-3. The long latency to APL development in these models (frequently over 1 year) suggested the requirement for secondary/cooperating events in leukemogenesis4-8. In our murine model, a human PML-RARA cDNA is knocked into the murine cathepsin G locus (Ctsg-PML-RARA; also referred to as mCG-PR) 3. We and others recently demonstrated that bone marrow cells from young, pre-leukemic Ctsg-PML-RARA mice have increased colony forming and replating ability in vitro, and have a competitive advantage over wild type cells in vivo9-13, implying that PML-RARA expression alone can alter hematopoiesis. Collectively, these results suggest that PML-RARA initially acts in a multipotent progenitor cell to increase self-renewal; the molecular pathways underlying this activity are not yet fully understood.
The Notch signaling cascade is a well-characterized pathway that is important for the self-renewal of several types of stem cells, including HSPCs (reviewed in Sandy et al14). Hematopoietic malignancies frequently demonstrate abnormalities in the Notch cascade, most notably T lymphoblastic leukemias (T-ALLs), where NOTCH1 mutations are found in approximately 60% of cases15. The Notch pathway is also an attractive candidate for involvement in APL, based on several lines of evidence: 1) primary human APL samples overexpress the Notch ligand JAG1, compared to other AML subtypes16,17, to promyelocytes17,18 and to CD34+ cells19, 2) JAG1 mRNA and protein increase after PML-RARA expression is induced in the PR-9 cell line19, 3) JAG1 is rapidly downregulated by all trans retinoic acid (ATRA) treatment of NB4 cells and primary APL blasts 19,20, and 4) PML-RARA expression activates a Hes1 promoter reporter construct, a known target of Notch signaling19. To date, there are no published studies of the role of JAG1 and Notch signaling in APL pathogenesis.
In this report, we show that Notch signaling is important for the pathogenesis of APL. We provide bioinformatic evidence for activation of a known Notch signature in both human APL cells, and in pre-leukemic Kit+Lin−Sca1+ (KLS) cells from Ctsg-PML-RARA mice. Using both pharmacologic and genetic approaches, we also found that Notch blockade abrogates the enhanced self-renewal observed in pre-leukemic cells from Ctsg-PML-RARA mice, but not in bone marrow cells expressing the AML1-ETO fusion gene. Finally, we show that dependence on Notch signaling is retained in a subset of fully transformed murine APL tumors. These findings suggest that Notch signaling is a key downstream effector of PML-RARA, with roles in both early leukemogenesis and in fully transformed cells.
Methods
Expression microarrays and GSEA
Human and murine expression arrays have been previously described elsewhere 17,21,22 and have been deposited in GEO: GSE12662, GSE10358 and GSE24728. Gene set enrichment analysis (GSEA) was performed using phenotype permutation analysis with signal to noise gene ranking (http://www.broad.mit.edu/gsea). Gene sets were considered to be significantly enriched at an FDR < 0.25, according to http://www.broadinstitute.org/gsea/index.jsp.
RNA-seq analysis
RNA-seq data from 176 AML patients with known FAB subtypes were obtained as part of The Cancer Genome Atlas project and analyzed using a custom pipeline based on the Tophat and Cufflinks software packages23. Sequencing reads were mapped with TopHat (version 1.3.1) to the reference genome sequence (GRCh37) using ENSEMBL human gene annotations (release 58). Accepted read alignments were processed with Cufflinks (version 1.1.0) using default parameters to filter alignments to rDNA and mitochondrial sequences and calculate fragments per kilobase of exon per million fragments mapped (FPKM) values for each transcript isoform. An aggregate FPKM value for JAG1 was calculated by summing the values obtained for three annotated JAG1 isoforms.
Cell lines and antibodies
The PR-9 cell line was a kind gift of P. Pelicci of the European Institute of Oncology, Milan, Italy; PML-RARA expression was induced as described 17. OP-9 cells were purchased from ATCC. Cells, including primary APL samples, were lysed directly in SDS sample buffer (final concentration of 0.83% SDS). Antibodies raised against Rara (C-20, Santa Cruz), Jag1 (H-114, Santa Cruz), cleaved-Notch1 Val1744 (Cell Signal Technologies) and actin (C-4, Millipore) were utilized for western blots. Murine APL cells were stained with either FITC- αGr-1 or APC-αc-Kit (eBioscience) for flow cytometry. For intracellular staining, cells were fixed and permeabilized following surface staining, using the FoxP3 Intracellular Staining Buffer Set (eBioscience), and then stained for PEJag-1 (eBioscience).
Mice
The Ctsg-PML-RARA mice have been previously described3, and were back-crossed to the C57BL/6 strain (Taconic) for at least 10 generations. 129SvJ/B6 F1 hybrid animals were generated by mating 129SvJ males with C57BL/6J females (both parental strains obtained from Jackson Laboratory). All animal care and experimental protocols were done in accordance with institutional guidelines and approved by the Animal Studies Committee of Washington University School of Medicine in accordance with current NIH policy.
Retroviral transductions of murine bone marrow cells
The MSCV-DNMAML1-GFP plasmid 24 was provided by Dr. Jon Aster; MSCV-DNMAML1-YFP was generated by replacing GFP with YFP. The MSCV-Jag1 plasmid 25 was a gift from Dr. Rafael Kopan. Dr. Timothy Graubert provided MSCV AML1-ETO ires GFP26. Retrovirus production, transduction of murine bone marrow, and transplants were performed as described previously 8,27. GFP positive or GFP/YFP double positive cells were sorted into liquid media 24-48 hours after spinfection and then plated into methylcellulose media, as previously described 13.
Cryopreserved murine APL samples
Cryopreserved murine APL samples were collected and frozen as described previously28. Cells were rapidly thawed and resuspended in RPMI media supplemented with 10% FBS, IL-3 (6 ng/mL), IL-6 (10 ng/mL) and SCF (100 ng/mL). For experiments with compound E and compound IX (Calbiochem), 200,000 cells were cultured with or without inhibitors for 48 hours. Cells were then plated in methylcellulose media and colony formation was scored at 7 days. Alternatively, cells were thawed and grown on irradiated (2000 cGy) OP9 stromal cells in the presence of murine cytokines TPO (10 ng/ul), SCF (100 ng/ul), IL3 (6 ng/ul) and FLT3-ligand (10 ng/ul) for 24 hrs. Cells were then treated with Compound E (2 μM) for 48 hours. Dead cells were removed by density centrifugation (Histopaque 1083, Sigma) and lysed in SDS sample buffer as described above. For transduction assays, cells were first cultured on OP9 stroma, and non-adherent cells were collected and transduced with MSCV-DN-MAML1-GFP or GFP control retroviral supernatants. Cells were analyzed for GFP positivity after 48 hours, and 1 million cells were engrafted in syngeneic mice via retro-orbital injection. Mice were sacrificed at the first sign of illness (typically 4 weeks).
Results
JAG1 is dysregulated in APL cells
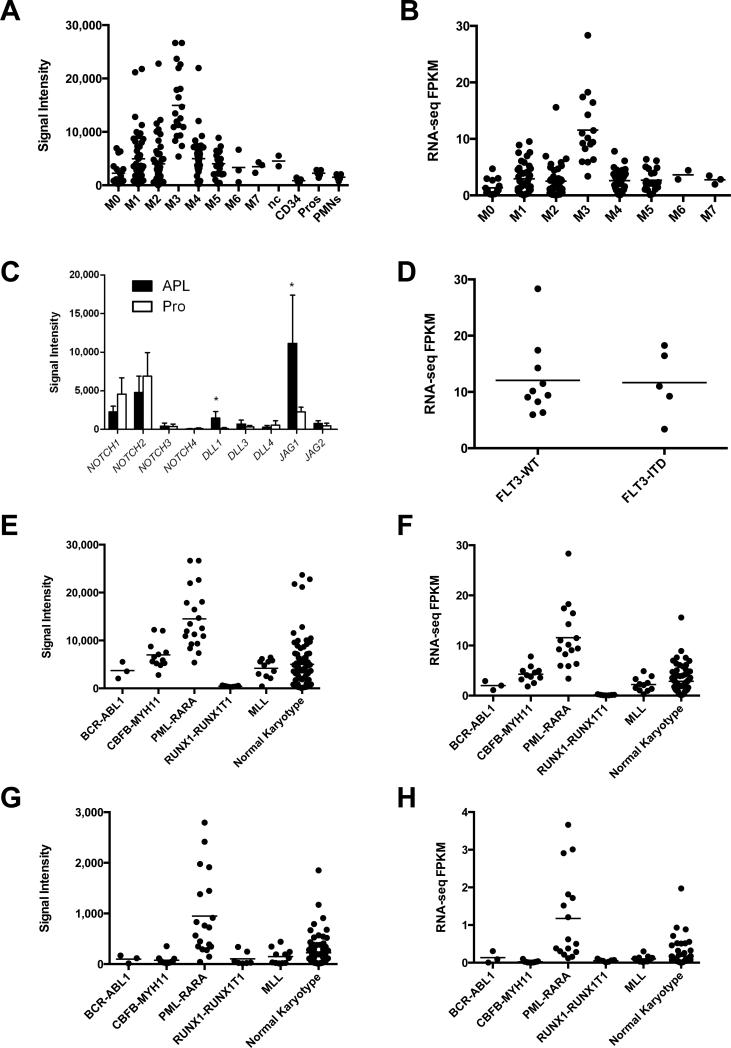
We previously reported a signature of genes with altered expression in APL cells; the Notch ligand Jagged-1 (JAG1) was among this set17. Using gene expression profiling, we examined the expression of JAG1 in bone marrow samples collected from a set of 180 de novo AML patients21 and in purified normal myeloid populations (CD34+ cells, promyelocytes, and neutrophils) from 5 normal human bone marrow samples17. JAG1 expression is somewhat variable in AML samples, but is expressed at significantly higher levels in FAB M3/APL samples compared to all other FAB subtypes, as well as normal myeloid populations (Figure 1A and data not shown). This pattern of JAG1 expression was validated by RT-PCR in a subset of the patients (Figure S1) and by using RNA-seq data from 176 AML patients (that completely overlap with the patient cohort with microarray expression studies) with known FAB subtypes that were part of The Cancer Genome Atlas (TCGA) project on AML (Figure 1B). Further validation was also performed using an independent set of de novo AML samples from the Cancer and Leukemia Group B (CALGB) Cooperative group (Figure S1). In addition, genes encoding the components of Notch activation, including the Notch receptors and various genes involved in processing and transcriptional activation are also expressed in APL cells, indicating that the essential components of Notch signaling are present in APL cells (Figures 1C and S2).
Figure 1. Expression of JAG1 and Notch signaling components in APL, other AML and normal myeloid cells.
A). Expression array-derived data for JAG1 in a set of 180 de novo AML samples, and flow sorted normal CD34+ cells, promyelocytes, and neutrophils. B). JAG1 expression data from 178 de novo AML samples with known FAB subtypes obtained from The Cancer Genome Atlas RNA-Seq database for AML. Each data point in A and B represents one patient sample or one normal sample. C). Expression of Notch receptors and ligands in APL (closed bars) and normal promyelocytes (open bars). D). Expression of JAG1 by RNA-seq in APL cases separated by FLT3-ITD status. E and F). Expression of JAG1 in fusion-driven AML samples by expression arrays (E) and RNA-seq (F); G and H). Expression of DLL1 in fusion-driven AMLs by expression arrays (G) and RNA-seq (H). For panels A, B and E-H, mean expression in M3/PML-RARA cases is statistically different than all other comparison groups as determined by one-way ANOVA with Tukey's multiple comparison test (alpha 0.05). For panel C, *, p<0.05.
Although an association between FLT3-ITD and JAG1 expression has been noted in other studies29,30, there was no difference in JAG1 expression within APL cases when segregated by FLT3-ITD status (Figure 1D). Using both expression platforms (microarray and RNA-seq) we also found that JAG1 was consistently over-expressed in APL cells relative to other fusion oncogene-driven AML cells (Figures 1E and 1F). Similar findings were observed for another Notch ligand, DLL1, although the levels of expression are much lower than that observed for JAG1 (Figures 1G and 1H). These results are similar to multiple other AML gene expression profiling studies 16,18,19,29-31, and strongly suggest that overexpression of JAG1 (and DLL1) is a characteristic of APL. Because JAG1 is a well-characterized Notch ligand, and the dominant Notch ligand in APL cells, we decided to investigate the role of JAG1 and Notch signaling in APL.
Bioinformatic evidence that Notch signaling is present in APL cells
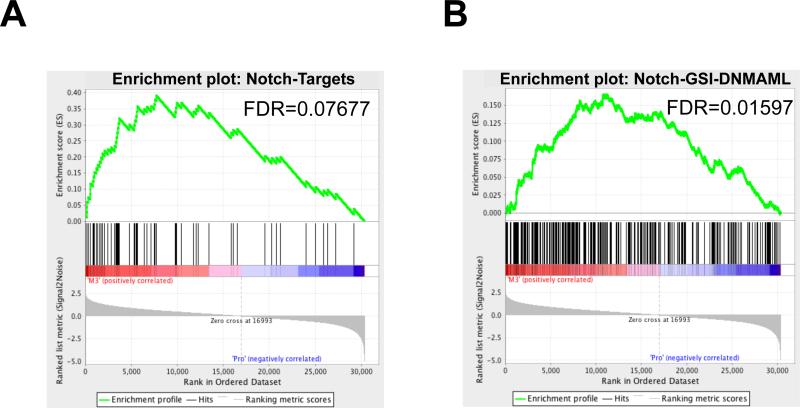
Increased Notch signaling is a major component of T-ALL due to activating mutations in NOTCH1 15, and several studies have reported dysregulated gene expression due to this aberrant Notch signaling in T-ALL cells 32-34. We used gene set enrichment analysis (GSEA) with three Notch signatures identified in T-ALL, including 1) ‘GSI-Notch’ (comprised of genes whose expression changes in T-ALL cells upon treatment with gamma secretase inhibitors 32), 2) ‘Notch-Targets’ (comprised of genes previously reported to be transcriptional targets of NOTCH1 33), and 3) ‘Notch-GSIDNMAML’ (comprised of transcriptional targets that are inhibited by both GSI treatment and DNMAML expression 34). All these signatures are enriched in APL cells compared to normal promyelocytes (Figures 2 and S3), providing strong bioinformatic evidence that Notch signaling is activated in human APL cells.
Figure 2. Notch target gene signatures are enriched in human APL cells.
GSEA plots of 22 APL samples compared to 5 normal promyelocyte (Pro) samples demonstrate significant enrichment in APL of A) a previously described set of Notch transcriptional targets in T-ALL33 and B) a previously published set of transcriptional targets of Notch whose expression is inhibited by both gamma secretase inhibitor treatment and DNMAML expression34. Normalized enrichment scores-A: 1.6523; B: 1.1921.
Notch signaling is present in APL cell lines
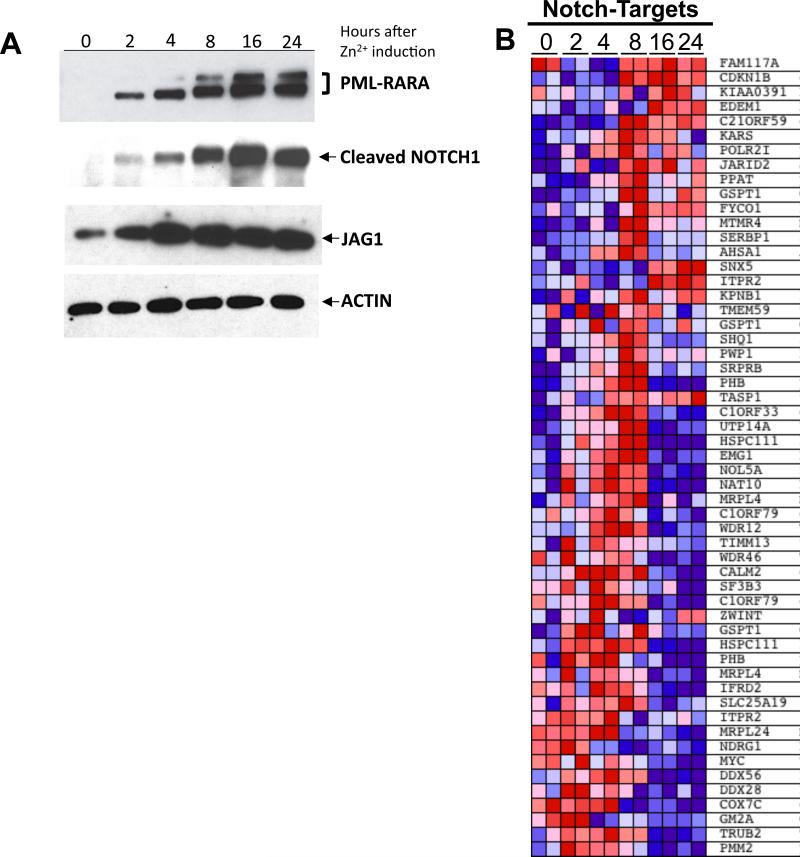
The PR-9 cell line, which contains a zinc inducible PML-RARA cDNA, is frequently used to study early events following PML-RARA expression. Consistent with previous reports19, JAG1 protein levels increased following induction of PML-RARA; JAG1 protein was detected by intracellular flow cytometry but not by conventional extracellular staining (Figure S4). JAG1 levels decreased following treatment with ATRA (data not shown), which has been reported previously 20. Cleaved Notch-1 protein levels peaked soon after JAG1 protein levels reached a maximum (Figure 3A), suggesting that Notch activation is a direct result of JAG1 upregulation. To determine whether downstream transcriptional targets of Notch signaling are induced with PML-RARA expression, we examined gene expression data from resting and ZnSO4-treated PR-9 cells. We found that the Notch signatures enriched in primary APL cells (Figure 2) have overall increased expression at 8-16 hours after zinc induction of PML-RARA expression (Figure 3B and Figure S5). Several known Notch targets (HSPC111, TASP1, PHB, GSPT1) in T-ALL 32-34, as well as JAG1, showed increased expression over time (Figure S6). These results demonstrate that activation of Notch signaling occurs as a consequence of PML-RARA induction in PR-9 cells, where it activates a similar transcriptional program to that found in primary APL cells.
Figure 3. Increased JAG1 and activation of Notch signaling are found in induced PR-9 cells.
A). Western blots showing protein levels of PML-RARA (using an anti-RARA antibody), JAG1, cleaved Notch-1 (ICN1), and actin in PR-9 cells at 0, 2, 4, 8, 16 and 24 hours after Zn2+ induction. Data are representative of three independent inductions. B) Heat map showing induction of the Notch-Targets33 signature in PR-9 cells 0, 2, 4, 8, 16 or 24 hours after Zn2+ induction of PML-RARA expression. Panels A and B were generated from different induction experiments.
Jag1 overexpression and Notch signaling are found in a murine model of APL
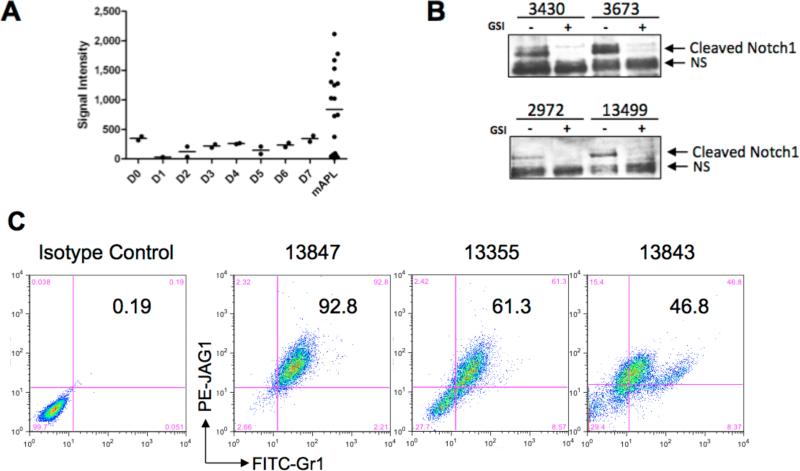
We next examined Notch signaling in the previously described Ctsg-PML-RARA mouse model of APL3, which produces a lethal leukemia that responds to ATRA both in vitro and in vivo and which has a similar gene expression signature as human APL 17,35. We examined the expression of Jag1 using previously published gene expression profiles of 21 murine APL samples and wildtype Lin−Sca+ (LS) progenitor cells undergoing 7 days of G-CSF induced myeloid differentiation (Figure 4A) 35. Jag1 expression was detectable in the majority of tumors and was higher than that of Lin−/Sca+ cells (d0) or promyelocytes (d2). Additionally, Jag1 mRNA levels remained below a signal intensity of 500 for the entire 7 day in vitro differentiation, suggesting that Jag1 is not significantly expressed at any stage of normal myeloid development, similar to the expression data obtained with normal human myeloid cells (Figure 1A-B). We then tested for expression of Jag1 protein and activated Notch1 in primary murine APL tumor samples. Cleaved Notch1 was detected by western blotting and the activated Notch signal in these tumors was sensitive to GSI inhibition (Figure 4B). Further, Jag1 protein was detected by flow cytometry in all tumors tested, with a range of 19% to greater than 90% of the cells containing Jag1 (Figure 4C and Supplemental Table 1). Similar to induced PR-9 cells, Jag1 protein was detectable only by intracellular (and not extracellular) flow cytometry (Figure S7). Therefore, like human APL and APL cell lines, murine APL cells both overexpress Jag1 and have activated Notch signaling, providing a rationale for utilizing the Ctsg-PML-RARA model to investigate the role of Notch signaling in leukemogenesis.
Figure 4. Jag1 and activated Notch signaling are found in murine APL samples.
A). Microarray expression data for Jag1 in murine Lin−Sca+ cells undergoing in vitro differentiation with G-CSF, and in 22 murine APL samples. Promyelocytes are highly enriched on days 2 and 3 of in vitro differentiation33 B) Western blot showing cleaved Notch1 in murine APL tumors treated for 48 hrs with either DMSO vehicle, or 2 μM compound E. The faster migrating band is non-specific (NS) and not sensitive to GSI treatment. 13499 and 3430 are GSI-sensitive tumors (Figure 7, Supplemental Table 1); 2972 and 3673 are insensitive. C). Intracellular flow cytometry detection of Jag1 in representative murine APL samples. Numbers correspond to percentage of cells co-expressing Gr-1 and Jag1.
Overexpression of Jag1 does not lead to increased self-renewal in vitro or AML in in vivo
We next sought to determine if overexpression of Jag1 is sufficient for transformation by transducing wildtype C57BL/6 bone marrow cells with retroviruses expressing either a murine Jag1 cDNA 25 or a GFP cDNA (Figure 5A). We found that Jag1 overexpression did not lead to increased colony forming ability compared to GFP or mock-transduced controls in serial plating experiments (Figure 5B), and the percentage of GFP-expressing cells was similar in cultures of Jag1 and GFP transduced marrow (Figure 5C). We transplanted irradiated syngeneic host animals with either Jag1 or GFP transduced bone marrow. GFP positivity in the peripheral blood was similar in the Jag1 and GFP control groups over time (Figure 5D-E). After 300 or more days post-transplant, none of the mice had developed leukocytosis (Figure 5F-G), anemia, thrombocytopenia or splenomegaly (data not shown). Collectively, these results suggest that while Jag1 overexpression and Notch signaling are present in APL cells, Jag1 overexpression in wildtype cells is not sufficient to induce self-renewal or initiate APL.
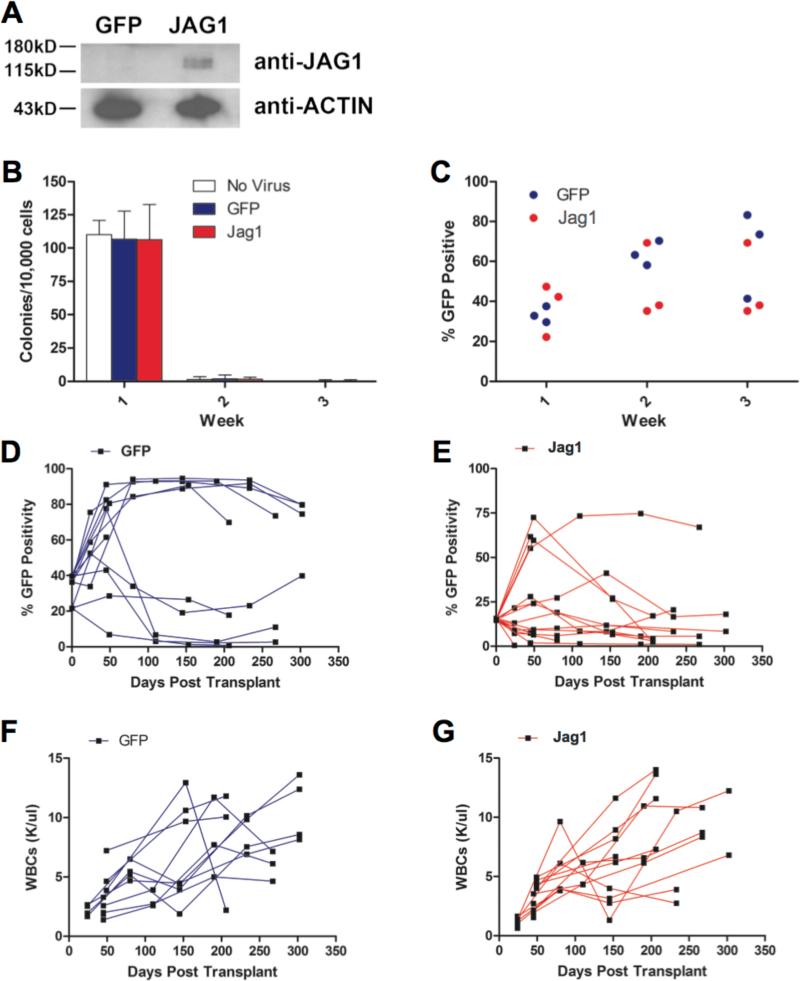
Figure 5. Jag1 overexpression does not lead to changes in self-renewal in vitro nor cause AML in vivo.
A) Western blot analysis of GFP sorted bone marrow transduced with either control MSCV-IRES-GFP virus (GFP) or Jag1 expressing virus (Jag1), expanded for 1 week on OP-9 stroma and sorted for GFP positivity, demonstrating overexpression of Jag1 in the transduced cells. B) Colony formation for wildtype C57BL/6 marrow that was mock transduced, or transduced with either MSCV-Jag1-IRES-GFP or MSCV-IRES-GFP control, and plated in methylcellulose media containing IL-3, IL-6 and SCF. Data shown are means +/− standard deviations. C) GFP positivity in marrow transduced with Jag1 virus or GFP vector control during 3 weeks of growth in methylcellulose media. Each data point represents the average of three plates. There was no statistically significant difference in % GFP+ cells at any time point. (D, E) GFP positivity in the peripheral blood of mice transplanted with whole marrow transduced with either the GFP control (panel D) or the Jag1 vector (panel E). (F, G) Peripheral blood WBC counts in animals transplanted with the GFP control (panel F) or the Jag1 vector (panel G) transduced marrow.
Inhibition of Notch signaling reduces self-renewal in marrow cells from Ctsg-PML-RARA mice
We next investigated the role of Notch signaling in PML-RARA induced leukemogenesis. Marrow cells from pre-leukemic Ctsg-PML-RARA animals have increased colony forming ability in vitro and a competitive advantage over wildtype cells in vivo 9-13. We and others10,22 have shown that PML-RARA is expressed in the KLS cells of these mice. To determine whether Notch signaling is activated in Ctsg-PML-RARA KLS cells, we performed GSEA on KLS cells from young (6-8 week) pre-leukemic Ctsg-PML-RARA (PR) mice and wildtype (WT) controls; the Notch target signature 33 that was enriched in human APL (Figure 2) and induced PR-9 cells (Figure 3) was also present in KLS cells derived from Ctsg-PML-RARA animals (Figure 6A and Figure S8).
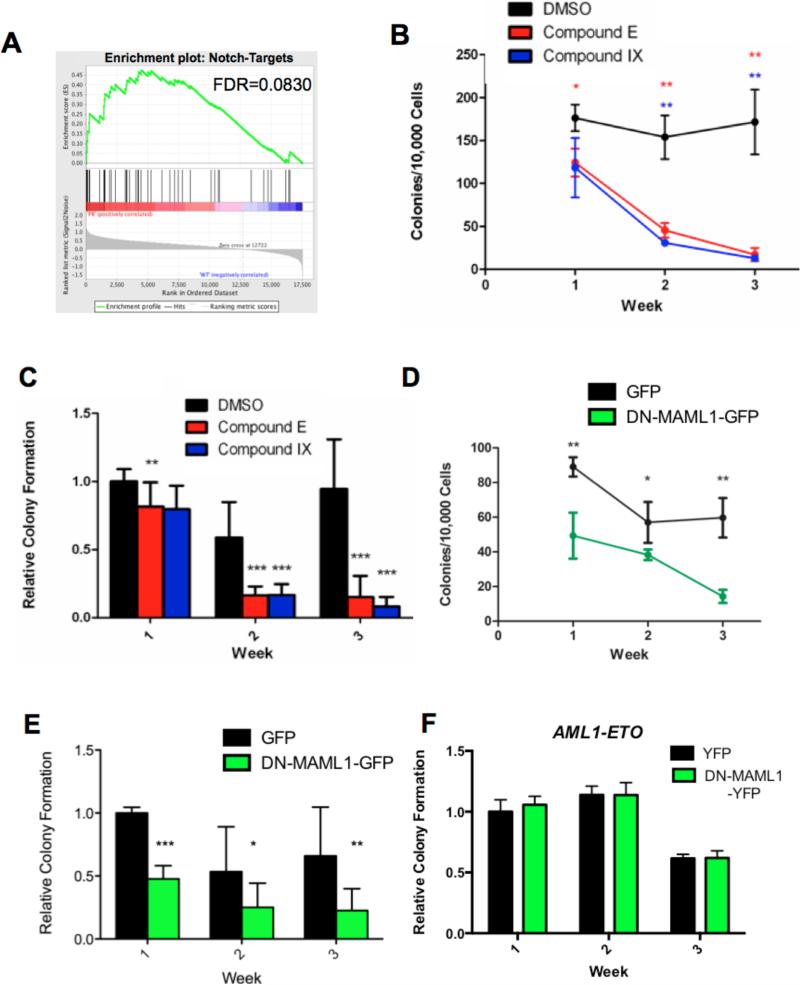
Figure 6. Notch signaling in pre-leukemic progenitor cells from Ctsg-PML-RARA mice.
A) GSEA plot of 6 KLS samples sorted from pre-leukemic Ctsg-PML-RARA (PR) mice compared to 4 wildtype C57BL/6 KLS (WT) samples. The data demonstrates significant enrichment of the previously described Notch-Targets (normalized enrichment score 1.5395)33 signature in Ctsg-PML-RARA KLS cells. B) Colony formation from total bone marrow cells from a representative Ctsg-PML-RARA mouse plated in methylcellulose media containing IL-3, IL-6 and SCF and supplemented with either DMSO, 2 μM compound E, or 25 μM compound IX. Colonies were counted and cells replated weekly for 3 weeks. Data shown are means +/− standard deviation. C) Summarized colony formation data from 5 independent Ctsg-PML-RARA marrow samples treated as shown in C. Data shown are means +/- standard deviation, and are normalized to the week 1 DMSO treated control colony counts. D). Colony formation from total bone marrow from a representative Ctsg-PML-RARA animal transduced with either MSCV-DN-MAML1-GFP or a GFP control virus, sorted for GFP+ cells, and plated in methylcellulose media supplemented with IL-3, IL-6 and SCF. E) Summarized colony formation data from 4 independent Ctsg-PML-RARA marrow samples treated as shown in Panel D. Data shown are means +/− standard deviation and are normalized to the week 1 GFP control. In all experiments shown, cells from each animal were plated in triplicate for each condition. Summarized data are representative of 3-5 animals per group. F. Relative colony formation from total bone marrow cells transduced with AML1-ETO and either YFP or DN-MAML1-YFP. GFP/YFP double positive cells were sorted and plated in methylcellulose media supplemented with IL-3, IL-6 and SCF. A representative example from 3 independent transductions is shown. In all graphs, one asterisk (*) indicates p<0.05, two asterisks (**) indicates p<0.01, and three asterisks (***) indicates p<0.001.
To examine the functional role of Notch signaling in early leukemogenesis, we cultured bone marrow cells from young (6-8 week old) pre-leukemic Ctsg-PML-RARA mice (or wildtype C57BL/6 mice) in methylcellulose media supplemented with IL3, IL6, and SCF, and assessed colony formation in the presence of GSIs (compound E or compound IX) or DMSO control. As expected, marrow derived from wildtype C57BL/6 animals did not serially replate; this was not influenced by either compound 36 (Figure S9). In contrast, after 1 week in culture, colony formation by Ctsg-PML-RARA bone marrow grown in media containing GSIs was significantly reduced (Figure 6B and 6C). This effect was further enhanced after 2 rounds of replating. These data suggest that Notch signaling may be partially responsible for the abnormal replating phenotype observed with Ctsg-PML-RARA progenitor cells.
We further validated the role of Notch signaling in serial replating with a dominant-negative fragment of Mastermind-like 1 (MAML1) fused to GFP; this portion of MAML contains the domains necessary to interact with cleaved Notch, but lacks the domains needed to recruit transcriptional machinery 24,37,38. We retrovirally transduced wildtype C57BL/6 or Ctsg-PML-RARA bone marrow with either DNMAML-GFP or GFP control virus, sorted GFP+ cells to >95% purity, and plated them in methylcellulose media. DNMAML-transduced Ctsg-PML-RARA bone marrow significantly reduced colony-forming activity at weeks 1, 2 and 3, similar to that observed with pharmacological inhibition of Notch signaling (Figure 6D-E). In contrast, expression of DN-MAML1 had no impact on the growth and colony forming activity of the RUNX1-RUNX1T1/AML1-ETO fusion in mouse bone marrow cells (Figure 6F), suggesting that the ability of Notch to regulate self-renewal is not a shared feature among all AML-initiating oncogenic fusion proteins. As previously shown36, genetic inhibition of Notch signaling did not impair colony formation by wildtype cells (Figure S9).
Inhibition of Notch signaling reduces colony formation by primary murine APL cells
To investigate the role of Notch signaling in murine APL tumors, we cultured cells in the presence of DMSO, ATRA, compound E or compound IX for 48 hours in liquid culture, followed by 7 days in methylcellulose without inhibitors (Figure 7A). As a control, 129/B6 F1 total bone marrow was subjected to the same assay. Of 13 tumors evaluated, 9 formed colonies using vehicle control conditions (Supplemental Table 1). As expected, ATRA exposure resulted in significantly fewer colonies in 9/9 tumors (Figures 7B and S10 for individual tumors and Figure 7C for summarized data). Six of the 9 assayed tumors formed significantly fewer colonies after exposure to GSIs, although neither compound E nor compound IX significantly altered colony formation by wild type marrow cells (Figure S9). In many of the GSI responsive tumors, the reduction in colony formation was similar to that observed with ATRA treatment. On average, exposure to compound E or compound IX resulted in colony formation that was 62.2% (SD 35.0%) or 45.2% (SD 24.4%) of the DMSO controls, respectively. In addition, the degree of inhibition of colony formation by compound E and compound IX were significantly correlated (p<0.01 for compound E, and p<0.001 for compound IX, Figure 7D), which suggests that Notch signaling plays a role in the survival or self-renewal of some APL tumors in vitro. Exposure of four different primary human APL samples to compound IX (using a recently characterized in vitro stromal co-culture system 39) also resulted in a significant dose-dependent inhibition of growth in 2 of 4 samples (Figure S11).
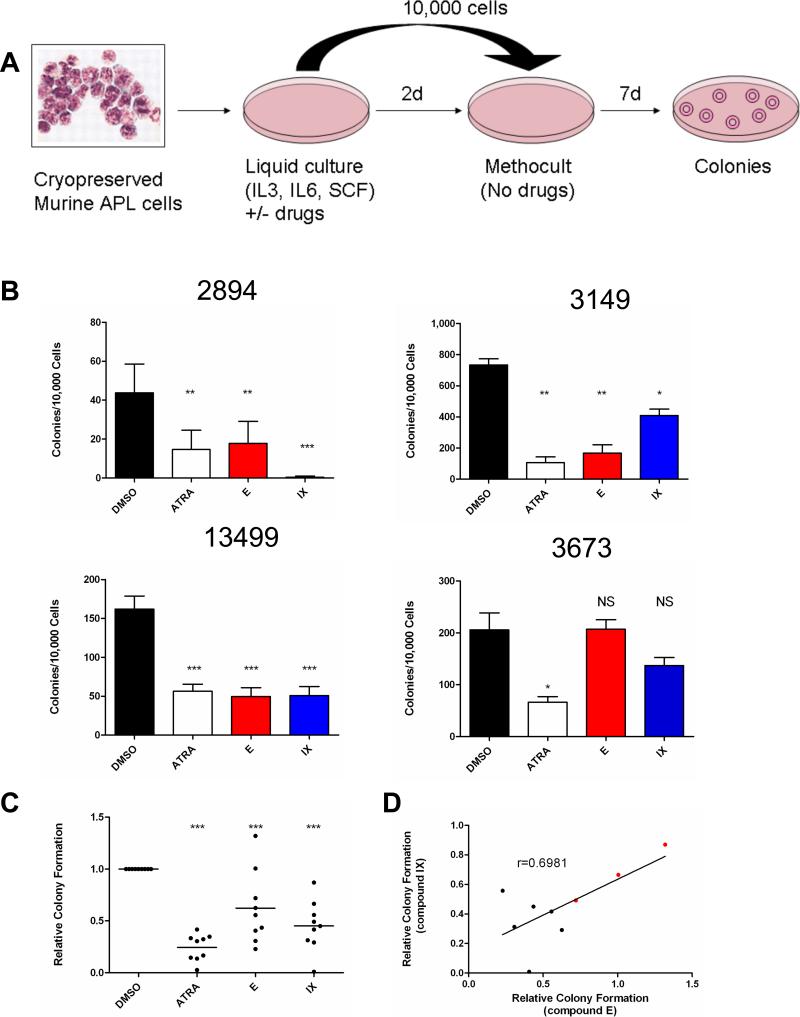
Figure 7. Inhibition of Notch signaling in murine APL cells in vitro and in vivo.
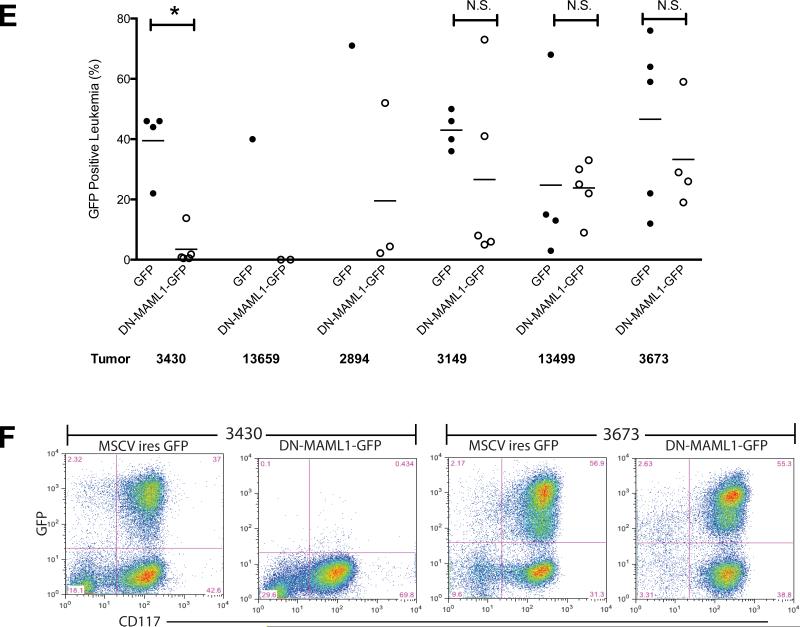
A). Schematic diagram of experimental protocol. B). Representative colony formation data for 4 murine APL samples treated for 48 hours with 1 μM ATRA, 2 μM compound E, 25 μM compound IX, or DMSO vehicle control, and then plated in methylcellulose media supplemented with IL-3, IL-6 and SCF in triplicate, as described in Panel A. Data bars are the mean numbers of colonies per 10,000 cells plated. Error bars represent standard deviations. C). Summarized data for 9 independent tumors treated as described in Panels A and B. Each data point represents the average of three plates for a single tumor, normalized to the average colony numbers for DMSO vehicle controls for that tumor. In all graphs, one asterisk (*) indicates p<0.05, two asterisks (**) indicates p<0.01 and three asterisks (***) indicates p<0.001. D) Relative colony numbers with compound E plotted against relative colony numbers with compound IX for the 9 tumors shown in Panel C. Each data point represents a single tumor, and colony formation was normalized to DMSO vehicle controls. The three GSI-resistant tumors are highlighted in red. E). Leukemia cells derived from 6 Ctsg-PML-RARA tumors were transduced with MSCV DN-MAML1-GFP or MSCV-ires-GFP control virus, and engrafted in C57BL/6 × 129SvJ F1 mice. After 4 weeks, total splenic cells were analyzed for GFP and CD117 (c-Kit) positivity by flow cytometry. Data shown are the percentage of cells positive for both GFP and CD117. *, p<0.05; N.S., not significant. Data points represent individual recipient animals. F) Flow cytometry plots from tumors 3430 and 3673, showing GFP and CD117 positivity in the spleens of animals transplanted with either DNMAML-GFP or GFP control transduced tumors.
Finally we investigated whether Notch signaling is required for growth of APL cells in vivo. We transduced 6 tumors (including samples that were either susceptible or resistant to GSIs) with MSCV-DNMAML1-GFP or a GFP control virus, and engrafted 1 million transduced cells into syngeneic 129/B6 host animals 48 hrs after transduction (Figure 7E-F). At the time of engraftment, the tumors had similar degrees of GFP positivity (DNMAML1-GFP, mean, 37.8%, range, 27%-47.8%; vs. GFP control, mean, 28.5%, range, 18%-48.6%). GFP positivity in the CD117+ splenic tumor population was then assessed when the animals became moribund, typically at about 4 weeks. In some animals engrafted with DNMAML1-GFP tumors, very few of the recovered tumor cells were GFP positive, despite having high GFP levels at the time of engraftment. These results suggest that inhibition of Notch signaling with DNMAML1 reduced the ability of some tumors to expand in mice, which could be caused by reduced engraftment, diminished growth, and/or reduced self-renewal of cells expressing this inhibitor.
Discussion
In this report we extended the observation that the Notch ligand JAG1 is overexpressed in APL cells 16,18,19,29-31 by performing a functional study of the role of Notch signaling in both human and murine APL samples. Not only do human APL cells express the core components of the Notch pathway, but they also show enrichment in Notch signaling, and are sometimes sensitive to gamma secretase inhibitors. Pharmacological and genetic inhibition of Notch signaling resulted in a loss of serial replating by Ctsg-PML-RARA progenitor cells, demonstrating that the Notch pathway is necessary for the increased self-renewal found in this mouse model. Finally, most murine APL samples retain sensitivity to Notch inhibition in vitro and in vivo, suggesting that Notch signaling is also relevant for the growth or survival of some fully transformed APL cells.
Our data suggest that JAG1 overexpression is characteristic of APL, but not absolutely restricted to this AML subtype (Figure 1A), suggesting that there may be multiple distinct pathways leading to high JAG1 expression. Despite the data presented here and elsewhere 19,20, the mechanism by which PML-RARA regulates JAG1 expression and Notch activation is unclear. Three independent whole genome chromatin immunoprecipitation studies failed to find evidence of PML-RARA binding in the JAG1 promoter in either cell lines or primary APL cells 40-42, despite the presence of an inverted repeat and combined direct repeat-PU.1 motif sites similar to known PMLRARA binding sites 20,40 in the proximal JAG1 promoter. In addition, cotransfection of a JAG1 promoter-reporter construct (1.5 kb of 5’ flanking sequence) with PML-RARA did not result in JAG1 promoter activation (data not shown). Clearly, the relationship between PML-RARA expression and JAG1 activation is indirect, and additional experiments will be required to define this pathway.
We initially hypothesized that overexpression of Jag1 alone would be leukemogenic in wildtype marrow, since other studies have found that Jag1 overexpression in stromal cells results in myeloproliferation 43,44. Surprisingly, we found no detectable hematopoietic alterations in mice transplanted with Jag1 overexpressing marrow cells, nor did we observe increased colony formation nor replating of Jag1 transduced cells in vitro. Importantly, over-expression of Jag1 in our study was confined to hematopoietic cells rather than the bone marrow stroma. Further, in both of the stromal overexpression studies, increased Jag1 expression was caused indirectly by another genetic event35,43.
Our work adds to the increasing body of literature demonstrating that the cellular context, as well as the level of Notch activation, can influence the resulting phenotype14,45-48. Klinakis et al 49 demonstrated that loss of Notch signaling results in an expansion of GMPs and eventual development of a CMML-like myeloproliferative disease, which can be rescued by reacquisition of Notch signaling. However, the leukemia-initiating cell in these CMML models may be a more differentiated progenitor than that of Ctsg-PMLRARA mice, where PML-RARA is first expressed in the KLS compartment 10,22. Our findings suggest that Notch signaling induced by PML-RARA can promote the self-renewal of myeloid progenitors without affecting the downstream commitment to the myeloid lineage, since preleukemic Ctsg-PML-RARA animals initially have increased self-renewal in the absence of any block in myeloid differentiation9-13. The results presented here also suggest that canonical Notch activation and high JAG1 expression may be specific for PML-RARA, since these findings were not observed in cells expressing AML1-ETO (Figs 1 and 6). In addition, recent studies have suggested that the Notch pathway may behave as a tumor suppressor in some myeloid disorders, including those driven by MLL-AF950. Long-term experiments designed to determine the consequences of genetic inhibition (or promotion) of Notch signaling on leukemogenesis in Ctsg-PML-RARA mice are currently in progress, and should further clarify the role of this pathway for leukemic transformation.
Not only is Notch signaling active in pre-leukemic Ctsg-PML-RARA stem/progenitor cells, but some fully transformed APL samples (both human and murine) are sensitive to Notch inhibition. Surprisingly, murine tumors that were either sensitive (3430 and 13499) or resistant (3673 and 2972) to GSIs in our replating assays (Figure 7 and Supplemental Table 1) had constitutive cleavage of Notch1 that was blocked by GSI treatment (Figure 4B). There was no correlation between Jag1 expression and GSI sensitivity in these tumors, suggesting that some APL tumors rely on Notch1-independent mechanisms for their progression. The Myc gene is a known target of Notch signaling that is required for PML-RARA induced APL7 ; it is possible that some APL tumors utilize the Notch pathway to activate Myc, while others find alternative pathways (e.g. gene amplification). In addition, our previous studies have identified significant genetic heterogeneity among APL8 tumors, with different mutations cooperating with PML-RARA to cause APL in each tumor.
In summary, we have demonstrated a role for Notch signaling in the pathogenesis of APL. The expression of JAG1 and activation of Notch signaling are common events in the development of both murine and human APL. Collectively, our results demonstrate that activation of Notch signaling is at least partially responsible for increased self-renewal in progenitors that express PML-RARA. We also show that dependence on Notch signaling is retained by some tumors in vitro and in vivo. Further investigation will be necessary to demonstrate the precise requirements for Notch signaling during PML-RARA induced leukemogenesis. Regardless, the demonstration that this pathway is active in some APL tumors may result in new approaches for treating patients with relapsed or refractory APL.
Supplementary Material
Acknowledgments
We thank Drs. Raphael Kopan, Daniel Link, Matthew Walter, Jacqueline Payton, David H. Spencer, and John Welch for assistance and/or helpful discussions. Mieke Hoock, Dan George, and Nick Protopsaltis provided invaluable animal husbandry assistance, and Erin Wehmeyer provided excellent technical support. We are grateful to the TCGA for access to the RNA-Seq data for JAG1. We thank the High Speed Cell Sorter Core, the Laboratory of Clinical Genomics, the Tissue Procurement Core, and the Bioinformatics Core of the Siteman Cancer Center at Washington University for their assistance with this study. This work was supported by NIH R01 CA083962, NIH PO1 CA101937, and the Barnes Jewish Hospital Foundation (all to T.J.L.), NIH T32 HL007088-35 (N.R.G.) and the Doris Duke Charitable Foundation to Washington University (S.M.S, Clinical Research Fellow).
Footnotes
Authorship:
N.R.G., J.M.K, and T.J.L. designed the research, analyzed data and wrote the paper; T.L. A.M.V, L.D.W. and S.M.S. performed experiments.
Conflict of interest. The authors declare no conflict of interest.
REFERENCES
- 1.Brown D, Kogan S, Lagasse E, et al. A PMLRARalpha transgene initiates murine acute promyelocytic leukemia. Proceedings of the National Academy of Sciences of the United States of America. 1997;94(6):2551–2556. doi: 10.1073/pnas.94.6.2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Grisolano JL, Wesselschmidt RL, Pelicci PG, Ley TJ. Altered myeloid development and acute leukemia in transgenic mice expressing PML-RAR alpha under control of cathepsin G regulatory sequences. Blood. 1997;89(2):376–387. [PubMed] [Google Scholar]
- 3.Westervelt P, Lane AA, Pollock JL, et al. High-penetrance mouse model of acute promyelocytic leukemia with very low levels of PML-RARalpha expression. Blood. 2003;102(5):1857–1865. doi: 10.1182/blood-2002-12-3779. [DOI] [PubMed] [Google Scholar]
- 4.Kelly LM, Kutok JL, Williams IR, et al. PML/RARalpha and FLT3-ITD induce an APL-like disease in a mouse model. Proceedings of the National Academy of Sciences of the United States of America. 2002;99(12):8283–8288. doi: 10.1073/pnas.122233699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kogan SC, Brown DE, Shultz DB, et al. BCL-2 cooperates with promyelocytic leukemia retinoic acid receptor alpha chimeric protein (PMLRARalpha) to block neutrophil differentiation and initiate acute leukemia. The Journal of experimental medicine. 2001;193(4):531–543. doi: 10.1084/jem.193.4.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Walter MJ, Park JS, Ries RE, et al. Reduced PU.1 expression causes myeloid progenitor expansion and increased leukemia penetrance in mice expressing PML RARalpha. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(35):12513–12518. doi: 10.1073/pnas.0504247102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jones L, Wei G, Sevcikova S, et al. Gain of MYC underlies recurrent trisomy of the MYC chromosome in acute promyelocytic leukemia. The Journal of experimental medicine. 2010;207(12):2581–2594. doi: 10.1084/jem.20091071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wartman LD, Larson DE, Xiang Z, et al. Sequencing a mouse acute promyelocytic leukemia genome reveals genetic events relevant for disease progression. The Journal of clinical investigation. 2011;121(4):1445–1455. doi: 10.1172/JCI45284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Viale A, De Franco F, Orleth A, et al. Cell-cycle restriction limits DNA damage and maintains self-renewal of leukaemia stem cells. Nature. 2009;457(7225):51–56. doi: 10.1038/nature07618. [DOI] [PubMed] [Google Scholar]
- 10.Wojiski S, Guibal FC, Kindler T, et al. PML-RARalpha initiates leukemia by conferring properties of self-renewal to committed promyelocytic progenitors. Leukemia : official journal of the Leukemia Society of America, Leukemia Research Fund, UK. 2009;23(8):1462–1471. doi: 10.1038/leu.2009.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Uy GL, Lane AA, Welch JS, Grieselhuber NR, Payton JE, Ley TJ. A protease-resistant PML-RAR{alpha} has increased leukemogenic potential in a murine model of acute promyelocytic leukemia. Blood. 2010;116(18):3604–3610. doi: 10.1182/blood-2008-11-189282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Welch JS, Klco JM, Varghese N, Nagarajan R, Ley TJ. Rara haploinsufficiency modestly influences the phenotype of acute promyelocytic leukemia in mice. Blood. 2011;117(8):2460–2468. doi: 10.1182/blood-2010-08-300087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Welch JS, Yuan W, Ley TJ. PML-RARA can increase hematopoietic self-renewal without causing a myeloproliferative disease in mice. The Journal of clinical investigation. 2011;121(4):1636–1645. doi: 10.1172/JCI42953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sandy AR, Jones M, Maillard I. Notch signaling and development of the hematopoietic system. Advances in experimental medicine and biology. 2012;727:71–88. doi: 10.1007/978-1-4614-0899-4_6. [DOI] [PubMed] [Google Scholar]
- 15.Weng AP, Ferrando AA, Lee W, et al. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science. 2004;306(5694):269–271. doi: 10.1126/science.1102160. [DOI] [PubMed] [Google Scholar]
- 16.Ross ME, Mahfouz R, Onciu M, et al. Gene expression profiling of pediatric acute myelogenous leukemia. Blood. 2004;104(12):3679–3687. doi: 10.1182/blood-2004-03-1154. [DOI] [PubMed] [Google Scholar]
- 17.Payton JE, Grieselhuber NR, Chang LW, et al. High throughput digital quantification of mRNA abundance in primary human acute myeloid leukemia samples. The Journal of clinical investigation. 2009;119(6):1714–1726. doi: 10.1172/JCI38248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Casorelli I, Tenedini E, Tagliafico E, et al. Identification of a molecular signature for leukemic promyelocytes and their normal counterparts: Focus on DNA repair genes. Leukemia : official journal of the Leukemia Society of America, Leukemia Research Fund, UK. 2006;20(11):1978–1988. doi: 10.1038/sj.leu.2404376. [DOI] [PubMed] [Google Scholar]
- 19.Alcalay M, Meani N, Gelmetti V, et al. Acute myeloid leukemia fusion proteins deregulate genes involved in stem cell maintenance and DNA repair. The Journal of clinical investigation. 2003;112(11):1751–1761. doi: 10.1172/JCI17595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meani N, Minardi S, Licciulli S, et al. Molecular signature of retinoic acid treatment in acute promyelocytic leukemia. Oncogene. 2005;24(20):3358–3368. doi: 10.1038/sj.onc.1208498. [DOI] [PubMed] [Google Scholar]
- 21.Tomasson MH, Xiang Z, Walgren R, et al. Somatic mutations and germline sequence variants in the expressed tyrosine kinase genes of patients with de novo acute myeloid leukemia. Blood. 2008;111(9):4797–4808. doi: 10.1182/blood-2007-09-113027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wartman LD, Welch JS, Uy GL, et al. Expression and function of PML-RARA in the multipotent hematopoietic progenitor cells of Ctsg-PML-RARA mice. PLoS One. 2012 doi: 10.1371/journal.pone.0046529. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Trapnell C, Roberts A, Goff L, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nature protocols. 2012;7(3):562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weng AP, Nam Y, Wolfe MS, et al. Growth suppression of pre-T acute lymphoblastic leukemia cells by inhibition of notch signaling. Molecular and cellular biology. 2003;23(2):655–664. doi: 10.1128/MCB.23.2.655-664.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ong CT, Sedy JR, Murphy KM, Kopan R. Notch and presenilin regulate cellular expansion and cytokine secretion but cannot instruct Th1/Th2 fate acquisition. PloS one. 2008;3(7):e2823. doi: 10.1371/journal.pone.0002823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fortier JM, Payton JE, Cahan P, Ley TJ, Walter MJ, Graubert TA. POU4F1 is associated with t(8;21) acute myeloid leukemia and contributes directly to its unique transcriptional signature. Leukemia : official journal of the Leukemia Society of America, Leukemia Research Fund, UK. 2010;24(5):950–957. doi: 10.1038/leu.2010.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Luo H, Li Q, O'Neal J, Kreisel F, Le Beau MM, Tomasson MH. c-Myc rapidly induces acute myeloid leukemia in mice without evidence of lymphoma-associated antiapoptotic mutations. Blood. 2005;106(7):2452–2461. doi: 10.1182/blood-2005-02-0734. [DOI] [PubMed] [Google Scholar]
- 28.Pollock JL, Westervelt P, Kurichety AK, Pelicci PG, Grisolano JL, Ley TJ. A bcr-3 isoform of RARalpha-PML potentiates the development of PML-RARalpha-driven acute promyelocytic leukemia. Proceedings of the National Academy of Sciences of the United States of America. 1999;96(26):15103–15108. doi: 10.1073/pnas.96.26.15103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Valk PJ, Verhaak RG, Beijen MA, et al. Prognostically useful gene-expression profiles in acute myeloid leukemia. The New England journal of medicine. 2004;350(16):1617–1628. doi: 10.1056/NEJMoa040465. [DOI] [PubMed] [Google Scholar]
- 30.Verhaak RG, Wouters BJ, Erpelinck CA, et al. Prediction of molecular subtypes in acute myeloid leukemia based on gene expression profiling. Haematologica. 2009;94(1):131–134. doi: 10.3324/haematol.13299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Alcalay M, Tiacci E, Bergomas R, et al. Acute myeloid leukemia bearing cytoplasmic nucleophosmin (NPMc+ AML) shows a distinct gene expression profile characterized by up-regulation of genes involved in stem-cell maintenance. Blood. 2005;106(3):899–902. doi: 10.1182/blood-2005-02-0560. [DOI] [PubMed] [Google Scholar]
- 32.Moellering RE, Cornejo M, Davis TN, et al. Direct inhibition of the NOTCH transcription factor complex. Nature. 2009;462(7270):182–188. doi: 10.1038/nature08543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Palomero T, Lim WK, Odom DT, et al. NOTCH1 directly regulates c-MYC and activates a feed-forward-loop transcriptional network promoting leukemic cell growth. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(48):18261–18266. doi: 10.1073/pnas.0606108103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang H, Zou J, Zhao B, et al. Genome-wide analysis reveals conserved and divergent features of Notch1/RBPJ binding in human and murine T-lymphoblastic leukemia cells. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(36):14908–14913. doi: 10.1073/pnas.1109023108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yuan W, Payton JE, Holt MS, et al. Commonly dysregulated genes in murine APL cells. Blood. 2007;109(3):961–970. doi: 10.1182/blood-2006-07-036640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maillard I, Koch U, Dumortier A, et al. Canonical notch signaling is dispensable for the maintenance of adult hematopoietic stem cells. Cell stem cell. 2008;2(4):356–366. doi: 10.1016/j.stem.2008.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nam Y, Sliz P, Song L, Aster JC, Blacklow SC. Structural basis for cooperativity in recruitment of MAML coactivators to Notch transcription complexes. Cell. 2006;124(5):973–983. doi: 10.1016/j.cell.2005.12.037. [DOI] [PubMed] [Google Scholar]
- 38.Maillard I, Weng AP, Carpenter AC, et al. Mastermind critically regulates Notch-mediated lymphoid cell fate decisions. Blood. 2004;104(6):1696–1702. doi: 10.1182/blood-2004-02-0514. [DOI] [PubMed] [Google Scholar]
- 39.Klco JM, Spencer DH, Lamprecht TL, et al. Genomic impact of transient low-dose decitabine treatment on primary AML cells. Blood. 2012 doi: 10.1182/blood-2012-09-459313. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang K, Wang P, Shi J, et al. PML/RARalpha targets promoter regions containing PU.1 consensus and RARE half sites in acute promyelocytic leukemia. Cancer cell. 2010;17(2):186–197. doi: 10.1016/j.ccr.2009.12.045. [DOI] [PubMed] [Google Scholar]
- 41.Martens JH, Brinkman AB, Simmer F, et al. PML-RARalpha/RXR Alters the Epigenetic Landscape in Acute Promyelocytic Leukemia. Cancer cell. 2010;17(2):173–185. doi: 10.1016/j.ccr.2009.12.042. [DOI] [PubMed] [Google Scholar]
- 42.Hoemme C, Peerzada A, Behre G, et al. Chromatin modifications induced by PML-RARalpha repress critical targets in leukemogenesis as analyzed by ChIP-Chip. Blood. 2008;111(5):2887–2895. doi: 10.1182/blood-2007-03-079921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Calvi LM, Adams GB, Weibrecht KW, et al. Osteoblastic cells regulate the haematopoietic stem cell niche. Nature. 2003;425(6960):841–846. doi: 10.1038/nature02040. [DOI] [PubMed] [Google Scholar]
- 44.Rupec RA, Jundt F, Rebholz B, et al. Stroma-mediated dysregulation of myelopoiesis in mice lacking I kappa B alpha. Immunity. 2005;22(4):479–491. doi: 10.1016/j.immuni.2005.02.009. [DOI] [PubMed] [Google Scholar]
- 45.Chiang MY, Shestova O, Xu L, Aster JC, Pear WS. Divergent effects of supraphysiological Notch signals on leukemia stem cells and hematopoietic stem cells. Blood. 2012 doi: 10.1182/blood-2012-03-416503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lobry C, Oh P, Aifantis I. Oncogenic and tumor suppressor functions of Notch in cancer: it's NOTCH what you think. The Journal of experimental medicine. 2011;208(10):1931–1935. doi: 10.1084/jem.20111855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mercher T, Cornejo MG, Sears C, et al. Notch signaling specifies megakaryocyte development from hematopoietic stem cells. Cell stem cell. 2008;3(3):314–326. doi: 10.1016/j.stem.2008.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Santos MA, Sarmento LM, Rebelo M, et al. Notch1 engagement by Delta-like-1 promotes differentiation of B lymphocytes to antibody-secreting cells. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(39):15454–15459. doi: 10.1073/pnas.0702891104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Klinakis A, Lobry C, Abdel-Wahab O, et al. A novel tumour-suppressor function for the Notch pathway in myeloid leukaemia. Nature. 2011;473(7346):230–233. doi: 10.1038/nature09999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lobry C, Ntziachristos P, Ndiaye-Lobry D, et al. Notch pathway activation targets AML-initiating cell homeostasis and differentiation. The Journal of experimental medicine. 2013;210(2):301–319. doi: 10.1084/jem.20121484. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.