Full text
PDF

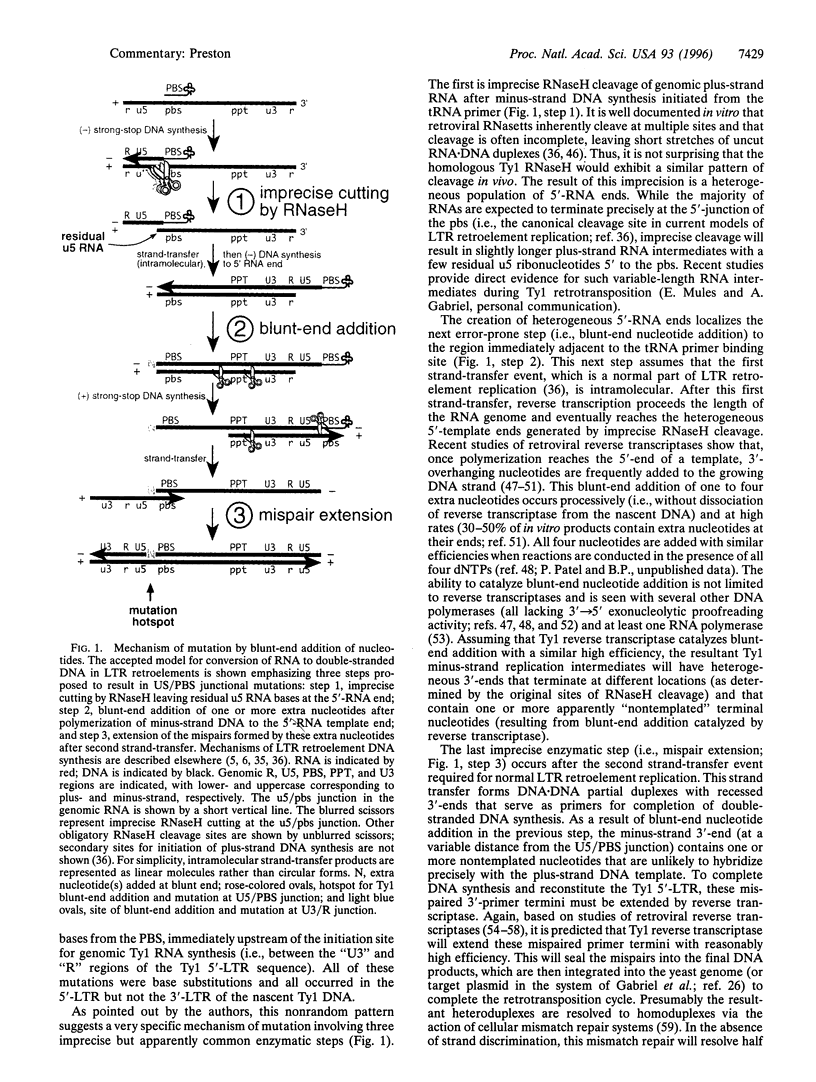


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baltimore D. RNA-dependent DNA polymerase in virions of RNA tumour viruses. Nature. 1970 Jun 27;226(5252):1209–1211. doi: 10.1038/2261209a0. [DOI] [PubMed] [Google Scholar]
- Batschelet E., Domingo E., Weissmann C. The proportion of revertant and mutant phage in a growing population, as a function of mutation and growth rate. Gene. 1976;1(1):27–32. doi: 10.1016/0378-1119(76)90004-4. [DOI] [PubMed] [Google Scholar]
- Bebenek K., Abbotts J., Roberts J. D., Wilson S. H., Kunkel T. A. Specificity and mechanism of error-prone replication by human immunodeficiency virus-1 reverse transcriptase. J Biol Chem. 1989 Oct 5;264(28):16948–16956. [PubMed] [Google Scholar]
- Biessmann H., Champion L. E., O'Hair M., Ikenaga K., Kasravi B., Mason J. M. Frequent transpositions of Drosophila melanogaster HeT-A transposable elements to receding chromosome ends. EMBO J. 1992 Dec;11(12):4459–4469. doi: 10.1002/j.1460-2075.1992.tb05547.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeke J. D., Eichinger D., Castrillon D., Fink G. R. The Saccharomyces cerevisiae genome contains functional and nonfunctional copies of transposon Ty1. Mol Cell Biol. 1988 Apr;8(4):1432–1442. doi: 10.1128/mcb.8.4.1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeke J. D., Garfinkel D. J., Styles C. A., Fink G. R. Ty elements transpose through an RNA intermediate. Cell. 1985 Mar;40(3):491–500. doi: 10.1016/0092-8674(85)90197-7. [DOI] [PubMed] [Google Scholar]
- Boeke J. D., Styles C. A., Fink G. R. Saccharomyces cerevisiae SPT3 gene is required for transposition and transpositional recombination of chromosomal Ty elements. Mol Cell Biol. 1986 Nov;6(11):3575–3581. doi: 10.1128/mcb.6.11.3575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer J. C., Bebenek K., Kunkel T. A. Unequal human immunodeficiency virus type 1 reverse transcriptase error rates with RNA and DNA templates. Proc Natl Acad Sci U S A. 1992 Aug 1;89(15):6919–6923. doi: 10.1073/pnas.89.15.6919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casacuberta J. M., Vernhettes S., Grandbastien M. A. Sequence variability within the tobacco retrotransposon Tnt1 population. EMBO J. 1995 Jun 1;14(11):2670–2678. doi: 10.1002/j.1460-2075.1995.tb07265.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cech T. R. A model for the RNA-catalyzed replication of RNA. Proc Natl Acad Sci U S A. 1986 Jun;83(12):4360–4363. doi: 10.1073/pnas.83.12.4360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalker D. L., Sandmeyer S. B. Ty3 integrates within the region of RNA polymerase III transcription initiation. Genes Dev. 1992 Jan;6(1):117–128. doi: 10.1101/gad.6.1.117. [DOI] [PubMed] [Google Scholar]
- Chiang C. C., Kennell J. C., Wanner L. A., Lambowitz A. M. A mitochondrial retroplasmid integrates into mitochondrial DNA by a novel mechanism involving the synthesis of a hybrid cDNA and homologous recombination. Mol Cell Biol. 1994 Oct;14(10):6419–6432. doi: 10.1128/mcb.14.10.6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark J. M. DNA synthesis on discontinuous templates by DNA polymerase I of Escherichia coli. Gene. 1991 Jul 31;104(1):75–80. doi: 10.1016/0378-1119(91)90467-p. [DOI] [PubMed] [Google Scholar]
- Clark J. M., Joyce C. M., Beardsley G. P. Novel blunt-end addition reactions catalyzed by DNA polymerase I of Escherichia coli. J Mol Biol. 1987 Nov 5;198(1):123–127. doi: 10.1016/0022-2836(87)90462-1. [DOI] [PubMed] [Google Scholar]
- Clark J. M. Novel non-templated nucleotide addition reactions catalyzed by procaryotic and eucaryotic DNA polymerases. Nucleic Acids Res. 1988 Oct 25;16(20):9677–9686. doi: 10.1093/nar/16.20.9677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffin J. M. HIV population dynamics in vivo: implications for genetic variation, pathogenesis, and therapy. Science. 1995 Jan 27;267(5197):483–489. doi: 10.1126/science.7824947. [DOI] [PubMed] [Google Scholar]
- Creighton S., Huang M. M., Cai H., Arnheim N., Goodman M. F. Base mispair extension kinetics. Binding of avian myeloblastosis reverse transcriptase to matched and mismatched base pair termini. J Biol Chem. 1992 Feb 5;267(4):2633–2639. [PubMed] [Google Scholar]
- Dombroski B. A., Feng Q., Mathias S. L., Sassaman D. M., Scott A. F., Kazazian H. H., Jr, Boeke J. D. An in vivo assay for the reverse transcriptase of human retrotransposon L1 in Saccharomyces cerevisiae. Mol Cell Biol. 1994 Jul;14(7):4485–4492. doi: 10.1128/mcb.14.7.4485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doolittle R. F., Feng D. F., Johnson M. S., McClure M. A. Origins and evolutionary relationships of retroviruses. Q Rev Biol. 1989 Mar;64(1):1–30. doi: 10.1086/416128. [DOI] [PubMed] [Google Scholar]
- Drake J. W. A constant rate of spontaneous mutation in DNA-based microbes. Proc Natl Acad Sci U S A. 1991 Aug 15;88(16):7160–7164. doi: 10.1073/pnas.88.16.7160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drake J. W. General antimutators are improbable. J Mol Biol. 1993 Jan 5;229(1):8–13. doi: 10.1006/jmbi.1993.1002. [DOI] [PubMed] [Google Scholar]
- Drake J. W. Rates of spontaneous mutation among RNA viruses. Proc Natl Acad Sci U S A. 1993 May 1;90(9):4171–4175. doi: 10.1073/pnas.90.9.4171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drake J. W. Spontaneous mutation. Annu Rev Genet. 1991;25:125–146. doi: 10.1146/annurev.ge.25.120191.001013. [DOI] [PubMed] [Google Scholar]
- Eigen M. Viral quasispecies. Sci Am. 1993 Jul;269(1):42–49. doi: 10.1038/scientificamerican0793-42. [DOI] [PubMed] [Google Scholar]
- Evans J. P., Palmiter R. D. Retrotransposition of a mouse L1 element. Proc Natl Acad Sci U S A. 1991 Oct 1;88(19):8792–8795. doi: 10.1073/pnas.88.19.8792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furano A. V., Usdin K. DNA "fossils" and phylogenetic analysis. Using L1 (LINE-1, long interspersed repeated) DNA to determine the evolutionary history of mammals. J Biol Chem. 1995 Oct 27;270(43):25301–25304. doi: 10.1074/jbc.270.43.25301. [DOI] [PubMed] [Google Scholar]
- Gabriel A., Willems M., Mules E. H., Boeke J. D. Replication infidelity during a single cycle of Ty1 retrotransposition. Proc Natl Acad Sci U S A. 1996 Jul 23;93(15):7767–7771. doi: 10.1073/pnas.93.15.7767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajjar A. M., Linial M. L. Modification of retroviral RNA by double-stranded RNA adenosine deaminase. J Virol. 1995 Sep;69(9):5878–5882. doi: 10.1128/jvi.69.9.5878-5882.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen S., Heidmann T. An indicator gene for detection of germline retrotransposition in transgenic Drosophila demonstrates RNA-mediated transposition of the LINE I element. EMBO J. 1991 Jul;10(7):1927–1937. doi: 10.1002/j.1460-2075.1991.tb07719.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz R. A., Skalka A. M. Generation of diversity in retroviruses. Annu Rev Genet. 1990;24:409–445. doi: 10.1146/annurev.ge.24.120190.002205. [DOI] [PubMed] [Google Scholar]
- Katz R. A., Skalka A. M. The retroviral enzymes. Annu Rev Biochem. 1994;63:133–173. doi: 10.1146/annurev.bi.63.070194.001025. [DOI] [PubMed] [Google Scholar]
- Kinsey J. A. Transnuclear retrotransposition of the Tad element of Neurospora. Proc Natl Acad Sci U S A. 1993 Oct 15;90(20):9384–9387. doi: 10.1073/pnas.90.20.9384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin H. L., Boeke J. D. Demonstration of retrotransposition of the Tf1 element in fission yeast. EMBO J. 1992 Mar;11(3):1145–1153. doi: 10.1002/j.1460-2075.1992.tb05155.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levis R. W., Ganesan R., Houtchens K., Tolar L. A., Sheen F. M. Transposons in place of telomeric repeats at a Drosophila telomere. Cell. 1993 Dec 17;75(6):1083–1093. doi: 10.1016/0092-8674(93)90318-k. [DOI] [PubMed] [Google Scholar]
- Loeb L. A., Kunkel T. A. Fidelity of DNA synthesis. Annu Rev Biochem. 1982;51:429–457. doi: 10.1146/annurev.bi.51.070182.002241. [DOI] [PubMed] [Google Scholar]
- Luan D. D., Eickbush T. H. RNA template requirements for target DNA-primed reverse transcription by the R2 retrotransposable element. Mol Cell Biol. 1995 Jul;15(7):3882–3891. doi: 10.1128/mcb.15.7.3882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendelman L. V., Petruska J., Goodman M. F. Base mispair extension kinetics. Comparison of DNA polymerase alpha and reverse transcriptase. J Biol Chem. 1990 Feb 5;265(4):2338–2346. [PubMed] [Google Scholar]
- Milligan J. F., Uhlenbeck O. C. Synthesis of small RNAs using T7 RNA polymerase. Methods Enzymol. 1989;180:51–62. doi: 10.1016/0076-6879(89)80091-6. [DOI] [PubMed] [Google Scholar]
- Modrich P. Mechanisms and biological effects of mismatch repair. Annu Rev Genet. 1991;25:229–253. doi: 10.1146/annurev.ge.25.120191.001305. [DOI] [PubMed] [Google Scholar]
- Nowak M. A., McMichael A. J. How HIV defeats the immune system. Sci Am. 1995 Aug;273(2):58–65. doi: 10.1038/scientificamerican0895-58. [DOI] [PubMed] [Google Scholar]
- Parthasarathi S., Varela-Echavarría A., Ron Y., Preston B. D., Dougherty J. P. Genetic rearrangements occurring during a single cycle of murine leukemia virus vector replication: characterization and implications. J Virol. 1995 Dec;69(12):7991–8000. doi: 10.1128/jvi.69.12.7991-8000.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel P. H., Preston B. D. Marked infidelity of human immunodeficiency virus type 1 reverse transcriptase at RNA and DNA template ends. Proc Natl Acad Sci U S A. 1994 Jan 18;91(2):549–553. doi: 10.1073/pnas.91.2.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peliska J. A., Balasubramanian S., Giedroc D. P., Benkovic S. J. Recombinant HIV-1 nucleocapsid protein accelerates HIV-1 reverse transcriptase catalyzed DNA strand transfer reactions and modulates RNase H activity. Biochemistry. 1994 Nov 22;33(46):13817–13823. doi: 10.1021/bi00250a036. [DOI] [PubMed] [Google Scholar]
- Peliska J. A., Benkovic S. J. Mechanism of DNA strand transfer reactions catalyzed by HIV-1 reverse transcriptase. Science. 1992 Nov 13;258(5085):1112–1118. doi: 10.1126/science.1279806. [DOI] [PubMed] [Google Scholar]
- Perrino F. W., Preston B. D., Sandell L. L., Loeb L. A. Extension of mismatched 3' termini of DNA is a major determinant of the infidelity of human immunodeficiency virus type 1 reverse transcriptase. Proc Natl Acad Sci U S A. 1989 Nov;86(21):8343–8347. doi: 10.1073/pnas.86.21.8343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preston B. D., Dougherty J. P. Mechanisms of retroviral mutation. Trends Microbiol. 1996 Jan;4(1):16–21. doi: 10.1016/0966-842x(96)81500-9. [DOI] [PubMed] [Google Scholar]
- Pulsinelli G. A., Temin H. M. High rate of mismatch extension during reverse transcription in a single round of retrovirus replication. Proc Natl Acad Sci U S A. 1994 Sep 27;91(20):9490–9494. doi: 10.1073/pnas.91.20.9490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pélisson A., Finnegan D. J., Bucheton A. Evidence for retrotransposition of the I factor, a LINE element of Drosophila melanogaster. Proc Natl Acad Sci U S A. 1991 Jun 1;88(11):4907–4910. doi: 10.1073/pnas.88.11.4907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts J. D., Preston B. D., Johnston L. A., Soni A., Loeb L. A., Kunkel T. A. Fidelity of two retroviral reverse transcriptases during DNA-dependent DNA synthesis in vitro. Mol Cell Biol. 1989 Feb;9(2):469–476. doi: 10.1128/mcb.9.2.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux L., Simon A. E., Holland J. J. Effects of defective interfering viruses on virus replication and pathogenesis in vitro and in vivo. Adv Virus Res. 1991;40:181–211. doi: 10.1016/S0065-3527(08)60279-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheen F. M., Levis R. W. Transposition of the LINE-like retrotransposon TART to Drosophila chromosome termini. Proc Natl Acad Sci U S A. 1994 Dec 20;91(26):12510–12514. doi: 10.1073/pnas.91.26.12510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimamoto T., Inouye M., Inouye S. The formation of the 2',5'-phosphodiester linkage in the cDNA priming reaction by bacterial reverse transcriptase in a cell-free system. J Biol Chem. 1995 Jan 13;270(2):581–588. doi: 10.1074/jbc.270.2.581. [DOI] [PubMed] [Google Scholar]
- Singer M. F. Unusual reverse transcriptases. J Biol Chem. 1995 Oct 20;270(42):24623–24626. doi: 10.1074/jbc.270.42.24623. [DOI] [PubMed] [Google Scholar]
- TEMIN H. M. The control of cellular morphology in embryonic cells infected with rous sarcoma virus in vitro. Virology. 1960 Feb;10:182–197. doi: 10.1016/0042-6822(60)90038-6. [DOI] [PubMed] [Google Scholar]
- Temin H. M., Mizutani S. RNA-dependent DNA polymerase in virions of Rous sarcoma virus. Nature. 1970 Jun 27;226(5252):1211–1213. doi: 10.1038/2261211a0. [DOI] [PubMed] [Google Scholar]
- Wain-Hobson S. Human immunodeficiency virus type 1 quasispecies in vivo and ex vivo. Curr Top Microbiol Immunol. 1992;176:181–193. doi: 10.1007/978-3-642-77011-1_12. [DOI] [PubMed] [Google Scholar]
- Ward C. D., Stokes M. A., Flanegan J. B. Direct measurement of the poliovirus RNA polymerase error frequency in vitro. J Virol. 1988 Feb;62(2):558–562. doi: 10.1128/jvi.62.2.558-562.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams K. J., Loeb L. A. Retroviral reverse transcriptases: error frequencies and mutagenesis. Curr Top Microbiol Immunol. 1992;176:165–180. doi: 10.1007/978-3-642-77011-1_11. [DOI] [PubMed] [Google Scholar]
- Xu H., Boeke J. D. High-frequency deletion between homologous sequences during retrotransposition of Ty elements in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1987 Dec;84(23):8553–8557. doi: 10.1073/pnas.84.23.8553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinnen S., Hsieh J. C., Modrich P. Misincorporation and mispaired primer extension by human immunodeficiency virus reverse transcriptase. J Biol Chem. 1994 Sep 30;269(39):24195–24202. [PubMed] [Google Scholar]
- de Mercoyrol L., Corda Y., Job C., Job D. Accuracy of wheat-germ RNA polymerase II. General enzymatic properties and effect of template conformational transition from right-handed B-DNA to left-handed Z-DNA. Eur J Biochem. 1992 May 15;206(1):49–58. doi: 10.1111/j.1432-1033.1992.tb16900.x. [DOI] [PubMed] [Google Scholar]



