Abstract
The major goal of this study was to create easy-to-use, reusable substrates capable of storing any peptides or bioactive molecules for a desired period of time until cells uptake them without the need for bioactive molecule or peptide-specific techniques. Nanopore arrays of uniform size and distribution were machined into fused silica substrates using femtosecond laser ablation and loaded with peptides by simple adsorption. The nanopore substrates were validated by examining the effect of N-acetyl-seryl-aspartyl-lysyl-proline (Ac-SDKP) loaded nanopores on macrophage phagocytosis and intracellular production of reactive oxygen species (ROS) with and without the pro-inflammatory lipopolysaccharide (LPS). Our results demonstrated that nanopores were generated in a uniform array fashion. Ac-SDKP peptides were stably stored in nanopores and internalized by macrophages. Significant reductions in ROS production and phagocytosis in macrophages were observed over control substrates, even in combination with LPS stimulation, indicating that loading Ac-SDKP peptides in pores significantly improved the anti-inflammatory effects.
Keywords: Nanopore, Ac-SDKP, cellular uptake, anti-inflammation
Background
Surface-mediated or reverse-transfection is a well-established method to deliver genetic information to cells more effectively and efficiently by either simple adsorption or covalent tethering of nucleotide vectors to a culture substrate prior to cell seeding (1). Covalent attachment is the most common surface-mediated protein delivery method but availability to cells, long term stability, and bioactivity are questionable with this approach (1-4). Non-covalent approaches to peptide delivery generally result in burst release of peptide from the surface and are difficult to tailor for longer-term studies (5-7). We hypothesized that loading peptides into nanopore arrays would increase the efficacy of peptide treatment similar to surface-mediated transfection for long-term treatment of cells in vitro. The goal was to create easy-to-use, reusable substrates capable of storing any peptides or bioactive molecules for an extended period of time until cells uptake them without the need for pretreatment or peptide-specific techniques.
The effect of N-acetyl-seryl-aspartyl-lysyl-proline (Ac-SDKP) loaded nanopores on macrophage phagocytosis and intracellular production of reactive oxygen species (ROS) were examined with and without the pro-inflammatory endotoxin lipopolysaccharide (LPS). Macrophages produce ROS in response to inflammatory stimuli (e.g., LPS treatment) to destroy foreign materials and then phagocytose the debris to prevent damage to other cells (8, 9). The tetrapeptide, Ac-SDKP, found in wound fluid, attenuates inflammatory and fibrotic responses by decreasing infiltration and adhesion of macrophages as well as the production of inflammatory cytokines (10, 11).
Methods
Nanopore arrays of uniform size and distribution were machined into fused silica substrates using femtosecond laser ablation with a single 160 fs pulse (12). For in vitro experiments, arrays of 200×200 20μm-spaced nanopores were arranged in a square lattice. Nanopore substrates were characterized by scanning electron microscopy (SEM). Peptides were loaded into the nanopores immediately following laser machining. Briefly, a 50 μL drop of 6 mg/mL Ac-SDKP in ultrapure water was deposited on the pattern or on a flat surface for peptide without pore controls, dried under vacuum overnight, and washed with sterile water prior to use. Confocal microscopy was used to confirm the presence of the fluorescently-labeled peptide FITC-SDKP after 48 hour incubation in culture media.
For in vitro studies, RAW 264.7 cells (ATCC, Manassas, VA) were cultured as suggested by the supplier. Cells were seeded onto each substrate at a density of 6.3×103 cells.cm-2. Cells were allowed to attach for 15 hours, then the media was replaced with normal, LPS (1 mg.mL-1, Sigma-Aldrich, St. Louis, MO), or FITC- SDKP (0.5, 0.25, or 0.125 mg.mL-1) containing media. Endpoint analysis was carried out 72 hours after treatment.
Phagocytosis was measured using a Vybrant® Phagocytosis Assay Kit (V-6694, Life Technologies, Carlsbad, CA) according to the manufacture’s protocol. Intracellular superoxide was stained with 5 μg/mL dihydroethidium (DHE) and counterstained with 5 ug.mL-1 Hoechst. Fluorescence intensities of E. coli particles phagocytized and intracellular superoxide were quantified using a plate reader (Tecan Group, Ltd Männendorf, Switzerland). Results were represented on plots as average ± standard deviation of 45 technical replicates per sample (n=2 for nanopore samples loaded with Ac-SDKP). Statistical analyses were performed by using one-way ANOVA tests followed by one-tailed t-tests for equal variances. For all tests, significance was designated as p < 0.05.
Results
SEM images showed that nanopore substrates were highly uniform in space, distribution, and pore size. Nanopores had roughly-elliptical openings with average major and minor axes of 2.95 and 2.56 μm (FIGURE 1A-B). Below the surface the pore diameter reduces to around 540 nm. Confocal microscopy of empty nanopores indicated that nanopores could be as much as 40 μm deep (Figure 1C). Confocal microscopy of FITC-SDKP loaded nanopores showed that nanopore substrates remained loaded with FITC-SDKP after 48 hours of incubation in culture media (FIGURE 1D, FIGURE 2H). Confocal microscopy also confirmed that nanopore fluorescence is not due to auto-fluorescence of the pore structure. FITC-labeled SDKP peptides were visible inside macrophages after 4 day culture on FITC-SDKP loaded nanopores, whereas peptide delivered in solution was only internalized at the highest concentration (FIGURE 2).
FIGURE 1.
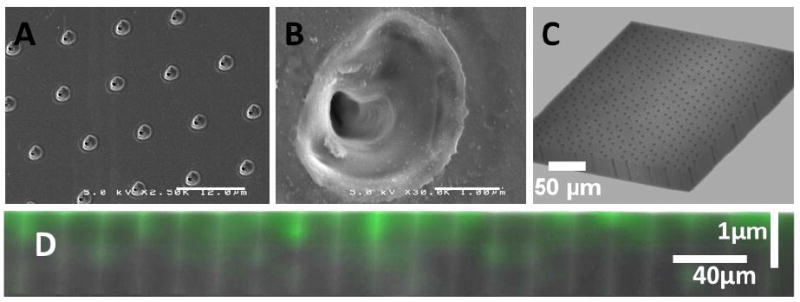
(A) SEM of 1 million nanopore array showing uniformity of pore size, shape, and distribution (B) SEM of a single nanopore showing pore morphology (C) Z-stack confocal micrograph of empty nanopore array (D) Z section of FITC-SDKP loaded nanopore array after two days of incubation in culture media.
FIGURE 2.
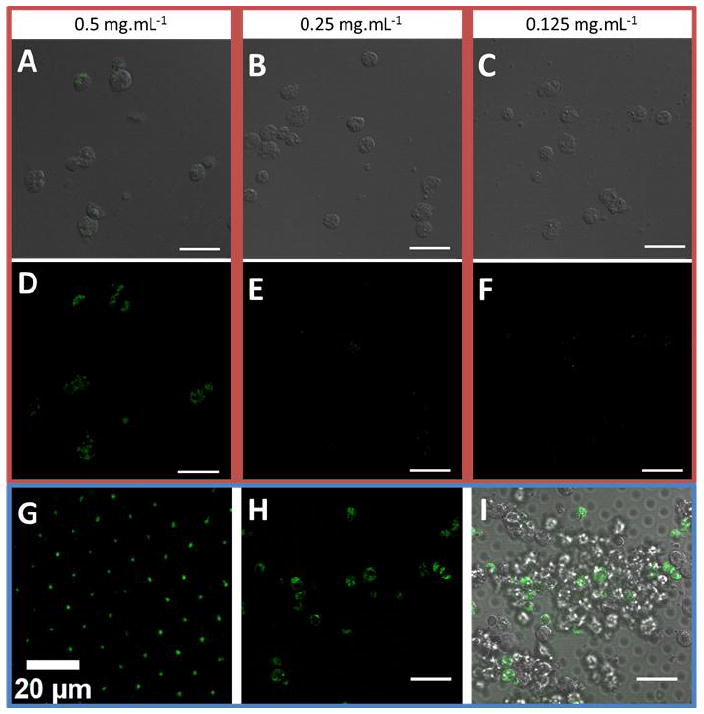
Uptake of FITC-SDKP by Raw-264.7 from solution and from nanopore substrates demonstrated enhanced cellular uptake from nanopores. Merged fluorescent and phase contrast of Raw 264.7 cells cultured with FITC-SDKP dissolved in solution at 0.5 (A), 0.25 (B), and 0.125 (C) mg.mL-1. Fluorescence images of Raw 264.7 cells cultured with FITC-SDKP in solution 0.5 (A), 0.25 (B), and 0.125 (C) mg.mL-1. (G) FITC-SDKP visualized in a nanopore array after two days of incubation in culture media (H) Fluorescent image of Raw 264.7 cells cultured on FITC-SDKP loaded nanopore array (I) Merged fluorescent and phase contrast image of Raw 264.7 cells cultured on FITC-SDKP loaded nanopore array.
Ac-SDKP release significantly reduced intracellular superoxide production and phagocytosis over control substrates, even in the case of LPS stimulation (FIGURE 3). When loaded in pores, the Ac-SDKP effects were improved significantly compared to peptide loaded on unpatterned substrates.
FIGURE 3.
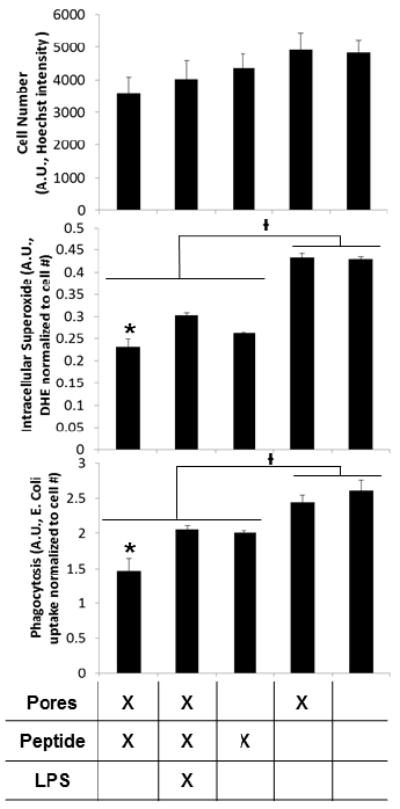
Ac-SDKP loaded nanopores significantly reduced inflammatory activation of RAW 264.7. Intracellular superoxide as measured by DHE fluorescence and phagocytosis as measured by fluorescent E. Coli particle uptake (both normalized to cell number) were significantly lower with Ac-SDKP treatments. In addition, treatment with Ac-SDKP loaded pores significantly reduced intracellular superoxide and phagocytosis compared to other conditions of Ac-SDKP treatment. Ɨ p<0.05 between groups, * indicates p<0.05 within groups).
Discussion
The nanopore delivery system developed in this study provides a simple and efficient means to store and deliver bioactive molecules in vitro. This flexible technique is capable of producing user-defined patterns with nanometer resolution (13). In addition, we demonstrated that peptide-loaded nanopore substrates were stable enough to be shipped and stored at room temperature.
Fluorescent peptide (FITC-SDKP) was shown to persist in nanopore arrays after two days of incubation in culture media but was not detectable in the supernatant medium during release studies (data not shown). Although peptide release was not measureable, the substrates produced a significant biological effect on cultured macrophages by cellular uptake of peptide (FIGURE 2, 3). Ac-SDKP-loaded nanopores significantly limited the inflammatory activation of macrophages, as measured by ROS production and phagocytosis assays. This indicates that the peptide is constrained to the pores but can be sequestered by cultured cells and elicit a biological response. In addition, the total amount of peptide used to load nanopore substrates was 0.3 mg and most of this was washed away prior to cell seeding. However, cellular uptake of FITC-SDKP was greater from nanopore substrates than 0.25mg/mL FITC-SDKP delivered in solution. This indicates the potential to use less reagent, while eliciting a greater biological response. Macrophages were not influenced by the presence of empty nanopores compared to flat substrates, indicating this pore feature has minimal effects on macrophages in vitro.
Future studies will utilize nanopore substrates to release bioactive molecules with spatial gradient patterns and expand on the utility of these substrates to store and deliver different bioactive molecules. This study demonstrated that femtosecond laser-patterned nanopore substrates are a practical tool for studying the effects of bioactive molecules on cultured cells and increases the efficacy of treatment akin to surface-mediated cellular uptake.
Supplementary Material
Acknowledgments
This study was supported by NIH HL091465, NIH 1UH2 TR000491, NSF DMR 1006558, and NSF CAREER CBET 1056046. The authors would like to acknowledge the use of optical instruments in Vanderbilt Institute of Nanoscale Science and Engineering (VINSE), a facility renovated under NSF ARI-R2 DMR-0963361. Support for LC and WHH as well as femtosecond laser fabrication was provided by the Tennessee Higher Education Commission through a grant to the Center for Laser Applications at the University of Tennessee Space Institute. Confocal images were performed in part through the use of the VUMC Cell Imaging Shared Resource (supported by NIH grants CA68485, DK20593, DK58404, HD15052, DK59637 and EY08126). The authors would also like to acknowledge Dr. Xintong Wang for assistance with operating the HPLC, and Mr. Brian Evans for providing the reagents for the TNF-α ELISA.
Footnotes
The authors have no conflicts of interest. This manuscript has not been published nor submitted for publication elsewhere in whole or in part.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Angela L. Zachman, Vanderbilt University, Department of Biomedical Engineering, VU Station B 351631, Nashville, TN 37235, angela.l.zachman@vanderbilt.edu.
Lucas H. Hofmeister, Vanderbilt University, Department of Biomedical Engineering, VU Station B 351631, Nashville, TN 37235, lucas.h.hofmeister@vanderbilt.edu.
Lino Costa, University of Tennessee Space Institute, Center for Laser Applications, 411 B.H. Goethert Parkway, MS 35, Tullahoma, TN 37388-9700, lcosta@utsi.edu.
Timothy C. Boire, Vanderbilt University, Department of Biomedical Engineering, VU Station B 351631, Nashville, TN 37235, timothy.c.boire@vanderbilt.edu.
Yu-Shik Hwang, Kyung Hee University, Department of Maxillofacial Biomedical Engineering, 1 Hoegidong, Dongdaemun-gu, Seoul 130-701, South Korea, yshwang@khu.ac.kr.
William H. Hofmeister, University of Tennessee Space Institute, Center for Laser Applications, 411 B.H. Goethert Parkway, MS 35, Tullahoma, TN 37388-9700, hof@utsi.edu.
Hak-Joon Sung, Vanderbilt University, Department of Biomedical Engineering, VU Station B 351631, Nashville, TN 37235, hak-joon.sung@vanderbilt.edu.
References
- 1.MacBeath G, Schreiber SL. Printing proteins as microarrays for high-throughput function determination. Science. 2000;289(5485):1760–3. doi: 10.1126/science.289.5485.1760. [DOI] [PubMed] [Google Scholar]
- 2.Ziauddin J, Sabatini DM. Microarrays of cells expressing defined cDNAs. Nature. 2001;411(6833):107–10. doi: 10.1038/35075114. [DOI] [PubMed] [Google Scholar]
- 3.Bengali Z, Rea JC, Shea LD. Gene expression and internalization following vector adsorption to immobilized proteins: dependence on protein identity and density. J Gene Med. 2007;9(8):668–78. doi: 10.1002/jgm.1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang H, Yang RH, Yang L, Tan WH. Nucleic Acid Conjugated Nanomaterials for Enhanced Molecular Recognition. Acs Nano. 2009;3(9):2451–60. doi: 10.1021/nn9006303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Habraken WJEM, Boerman OC, Wolke JGC, Mikos AG, Jansen JA. In vitro growth factor release from injectable calcium phosphate cements containing gelatin microspheres. J Biomed Mater Res A. 2009;91A(2):614–22. doi: 10.1002/jbm.a.32263. [DOI] [PubMed] [Google Scholar]
- 6.Gombotz WR, Wee SF. Protein release from alginate matrices. Adv Drug Deliver Rev. 2012;64:194–205. doi: 10.1016/s0169-409x(97)00124-5. [DOI] [PubMed] [Google Scholar]
- 7.Huang X, Brazel CS. On the importance and mechanisms of burst release in matrix-controlled drug delivery systems. J Control Release. 2001;73(2-3):121–36. doi: 10.1016/s0168-3659(01)00248-6. [DOI] [PubMed] [Google Scholar]
- 8.Zhang X, Shan P, Sasidhar M, Chupp GL, Flavell RA, Choi AMK, Lee PJ. Reactive Oxygen Species and Extracellular Signal-Regulated Kinase 1/2 Mitogen-Activated Protein Kinase Mediate Hyperoxia-Induced Cell Death in Lung Epithelium. American Journal of Respiratory Cell and Molecular Biology. 2003;28(3):305–15. doi: 10.1165/rcmb.2002-0156OC. [DOI] [PubMed] [Google Scholar]
- 9.Maderna P, Godson C. Phagocytosis of apoptotic cells and the resolution of inflammation. Biochim Biophys Acta-Mol Basis Dis. 2003;1639(3):141–51. doi: 10.1016/j.bbadis.2003.09.004. [DOI] [PubMed] [Google Scholar]
- 10.Yang F, Yang XP, Liu YH, Xu J, Cingolani O, Rhaleb NE, Carretero OA. Ac-SDKP reverses inflammation and fibrosis in rats with heart failure after myocardial infarction. Hypertension. 2004;43(2):229–36. doi: 10.1161/01.HYP.0000107777.91185.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sharma U, Rhaleb N-E, Pokharel S, Harding P, Rasoul S, Peng H, Carretero OA. Novel anti-inflammatory mechanisms of N-Acetyl-Ser-Asp-Lys-Pro in hypertension-induced target organ damage. American Journal of Physiology - Heart and Circulatory Physiology. 2008;294(3):H1226–H32. doi: 10.1152/ajpheart.00305.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zalloum OHY, Parrish M, Terekhov A, Hofmeister W. An amplified femtosecond laser system for material micro-/nanostructuring with an integrated Raman microscope. Review of Scientific Instruments. 2010;81(5) doi: 10.1063/1.3430073. [DOI] [PubMed] [Google Scholar]
- 13.Rajput D, Crowder SW, Hofmeister L, Costa L, Sung HJ, Hofmeister W. Cell interaction study method using novel 3D silica nanoneedle gradient arrays. Colloids Surf B Biointerfaces. 2013;102:111–6. doi: 10.1016/j.colsurfb.2012.07.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


