Abstract
The receptor for advanced glycosylation end products (RAGE) is a multiligand receptor involved in diverse cell signaling pathways. Previous studies show that this gene expresses several splice variants in human, mouse, and dog. Alternative splicing (AS) plays an important role in expanding transcriptomic and proteomic diversity, and it has been related to disease. AS is also one of the main evolutionary mechanisms in mammalian genomes. However, limited information is available regarding the AS of RAGE in a wide context of mammalian tissues. In this study, we examined in detail the different RAGE mRNAs generated by AS from six mammals, including two primates (human and monkey), two artiodactyla (cow and pig), and two rodentia (mouse and rat) in 6–18 different tissues including fetal, adult, and tumor. By nested reverse transcription-polymerase chain reaction (RT-PCR) we identified a high number of splice variants including noncoding transcripts and predicted coding ones with different potential protein modifications affecting mainly the transmembrane and ligand-binding domains that could influence their biological function. However, analysis of RNA-seq data enabled detecting only the most abundant splice variants. More than 80% of the detected RT-PCR variants (87 of 101 transcripts) are novel (different exon/intron structure to the previously described ones), and interestingly, 20–60% of the total transcripts (depending on the species) are noncoding ones that present tissue specificity. Our results suggest that RAGE undergoes extensive AS in mammals, with different expression patterns among adult, fetal, and tumor tissues. Moreover, most splice variants seem to be species specific, especially the noncoding variants, with only two (canonical human Tv1-RAGE, and human N-truncated or Tv10-RAGE) conserved among the six different species. This could indicate a special evolution pattern of this gene at mRNA level.
Keywords: RAGE, AGER, alternative splicing, mRNA isoforms, noncoding RNA, comparative genomics
Introduction
Nowadays, it is widely known that alternative splicing (AS) plays an important role in expanding transcriptomic diversity (Blencowe 2006; Nilsen and Graveley 2010; Mercer et al. 2011), as well as in the generation of proteomic variety, which has considerable biological consequences (Xing and Lee 2007; Colak et al. 2013). Recent studies have shown that in addition to proteins, RNA structure is also involved in regulating AS (McManus and Graveley 2011; Yang et al. 2011). Both mechanisms as a whole have tissue- or species-specific functions (Pan et al. 2005; Nurtdinov et al. 2007; Calarco et al. 2011) and have implications in the generation of developmental stages (Blencowe 2006; Xing and Lee 2007; Gingeras 2007; Licatalosi and Darnell 2009). Moreover, AS seems to be involved in several diseases (Villate et al. 2008; Irimia and Blencowe 2012; Colak et al. 2013), because it is a very complex process with several specific splicing site sequences and many RNA-binding proteins, where mutations or misregulation may cause disease (Cooper et al. 2009).
It has also been demonstrated that AS is one of the main evolutionary mechanisms in mammalian genomes (Nurtdinov et al. 2007; Merkin et al. 2012; Barbosa-Morais et al. 2012). Diversity in mammalian transcripts seems to be a consequence of species-specific AS, mainly related to exons conservation among species (Pan et al. 2005). Furthermore, evidences of functional selective pressure for AS events have been previously reported (Xing and Lee 2005). However, because the canonical splice variants carry out critical gene activities, the minority ones may not often have a major impact on physiology (Xing and Lee 2005; Blencowe 2006). Therefore, alternative spliced exons that belong to minority isoforms or species-specific isoforms seem to evolve more rapidly than constitutive ones (Xing and Lee 2005). AS has been described to play an important role in immune system in swine models (Lynch 2004; Sinkora et al. 2005), and in signal transduction and susceptibility to diseases in relation with the evolution of primate species-specific characteristics (Calarco et al. 2007). It should be taken into account that the majority of these kinds of analyses have been based on predictive computational methods and/or in cDNA sequences deposited in different databases (Pan et al. 2005; Xing and Lee 2005; Nurtdinov et al. 2007).
In this study, we focus on the receptor for advanced glycosylation end products (RAGE) as a complex spliced gene in mammals (Yonekura et al. 2003; Hudson et al. 2008; Kalea et al. 2009; Sterenczak et al. 2009). RAGE (also termed AGER) is a member of the immunoglobulin (Ig) superfamily and a cell surface receptor for a broad variety of ligands including advanced glycation end products (Neeper et al. 1992), members of the S100/calgranulin superfamily (Hofmann et al. 1999), amphoterin, also known as HMGB1 (Hori et al. 1995), an amyloidogenic form of serum amyloid A and beta amyloid (Aβ) peptide (Yan et al. 1996). Binding of these ligands to RAGE entails the activation of diverse signal transduction pathways implicated in cell growth and proliferation (Huttunen et al. 1999; Kislinger et al. 1999). The deregulation of this process seems to be related with several diseases (Kalea et al. 2011) including diabetes (Goh and Cooper 2008), immune and inflammatory disorders in arteriosclerosis (Harja et al. 2008), Alzheimer’s disease (Ding and Keller 2005a, 2005b; Emanuele et al. 2005), and invasiveness in various cancers (Hiwatashi et al. 2008; Ghavami et al. 2008).
Several splice variants of RAGE have been previously described in human (Malherbe et al. 1999; Yonekura et al. 2003; Ding and Keller 2005b; Hudson et al. 2008), murine (Kalea et al. 2009), and canine tissues (Sterenczak et al. 2009, 2013). RAGE is composed of three extracellular Ig-like domains: an Ig V-set domain (Ig-V domain) and two Ig C-set domain (Ig-C domain); and a single transmembrane region with a short cytoplasmic tail. Splice variants result in changes that might affect the extracellular ligand-binding domain of RAGE by different modifications of the region encoding the Ig-V domain as well as the removal of the sequence encoding the transmembrane region allowing to generate secreted RAGE (sRAGE) isoforms (Hudson et al. 2008).
Here we report a deep description of RAGE splice variants based on experimental data and compared with RNA-seq data obtained from databases. We have identified by nested reverse transcription-polymerase chain reaction (RT-PCR) numerous novel splice variants of RAGE in 6–18 tissues of six different mammals, and we have performed a wide comparison among them. Furthermore, we have performed a detailed analysis of high-throughput RNA sequences (RNA-seq) from brain and liver samples of five of the six analyzed mammals, which data were obtained from Pan et al. (2008), Barbosa-Morais et al. (2012), Merkin et al. (2012), and ENCODE Project. This is the first comparison of RAGE splice variants among several species within the orders primates, artiodactyla, and rodentia, and our results suggest a special evolution pattern of this gene at the mRNA level. Additionally, we analyzed the relative expression levels of the different splicing variants in the studied tissues and species, as well as the relative abundance of coding and noncoding transcripts showing how the noncoding ones are extensively represented.
Materials and Methods
Analyzed Tissues
For the screening of RAGE splice variants, we analyzed mRNAs from six mammalian species corresponding to different tissues, including adult, embryonic, and cancer tissues, that were obtained from BioChain through one of their European distributor “AMS Biotechnology” as follows: human adult (blood, brain, kidney, liver, lung, pancreas, spleen, and thymus), fetal (brain, heart, kidney, liver, lung, spleen, and thymus), and human cancer tissues (kidney, liver, and lung); rhesus monkey adult tissues (brain, heart, kidney, liver, lung, and pancreas); bovine and swine adult tissues (brain, heart, kidney, liver, lung, and spleen); mouse adult (brain, heart, kidney, liver, lung, and pancreas) and mouse embryonic tissues (11, 15, and 17 days of development); and rat adult (brain, heart, kidney, liver, lung, pancreas, and spleen) and rat embryonic tissues (13 days of development) (for tissue details see supplementary table S1, Supplementary Material online).
Splice Variants Identification
One microgram of total RNA from each tissue was used for oligo-dT primed cDNA synthesis, which was performed using the ImProm II Reverse Transcription System (Promega) following the manufacturer’s instructions. All cDNAs were checked by amplification of the housekeeping beta-actin gene expression as proof of reliable cDNA template, with the following forward and reverse primers: Actin_F 5′CTTCGCGGGCGACGATGC3′ and Actin_R 5′TGGTGGTGAAGCTGTAGCC3′. The splicing isoforms were amplified by nested PCR from the different tissues cDNAs with GoTaq Green Master Mix (Promega). The PCR conditions were as follows: 95 °C for 5 min followed by 30 cycles of 95 °C for 1 min, 60 °C for 2 min, 72 °C for 2 min, and ending by 72 °C for 5 min. The first round was performed with 1 μl of cDNA in a 20 μl reaction volume and with external forward and reverse primers (extF and extR) as follows: H. sapiens and M. mulatta (RAGE_extF 5′CAGGACCCTGGAAGGAAGCAGG3′ and RAGE_extR 5′CTGGTTGTAGAAGAAAGCTTGGC3′), B. taurus (BtRAGE_extF 5′AGCCTGGGAAGGAAGCAGG3′ and BtRAGE_extR 5′GGTTGTGGAAGAAAGTTTGGG3′), S. scrofa (SsRAGE_extF 5′AGCCTGGGAAGGAAGCAGG3′ and SsRAGE_extR 5′GGGTGAACTGGTCTGGGGCC3′), and R. norvegicus and Mu. musculus (RnRAGE_extF 5′AGCCTGGGAAGGAAGCACC3′ and RnRAGE_extR 5′ATCATGTGGGCTCTGGTTGG3′). Primers were designed to amplify from start to stop codon of the canonical full-length RAGE gene (Hudson et al. 2008). For the second round, we used primers just downstream of the first round ones called internal forward and reverse primers (intF and intR), as follows: H. sapiens and M. mulatta (RAGE_intF 5′AGCCGGAACAGCAGTTGGAGCC3′ and RAGE_intR 5′GCCCTCCAGTACTACTCTCGCC3′), B. taurus (BtRAGE_intF 5′GGGGCAGTGGTCGGAGCC3′ and BtRAGE_intR 5′CTCCTGTGCTGCTCTCTGCC3′), S. scrofa (SsRAGE_intF 5′GCAGCAGGGACAACGGCC3′ and SsRAGE_intR 5′GCTCCTGCACTGCTCTCAGCC3′), and R. norvegicus and Mu. musculus (RnRAGE_intF 5′ACAGCAGCTAGAGCCTGGG3′ and RnRAGE_intR 5′CCCGGCACCATTCTCTGGC3′). The second round was performed with 1 μl of a 1:10 dilution of the first round product as template on a final volume of 20 μl and only 20 cycles. To verify product size and experimental reproducibility, two independent PCR experiments were performed and loaded on a 2% agarose gel. The splicing isoforms products were gel purified as single bands, except from human and rhesus monkey where 1–1.5 kb bands were extracted at the same time, using the Wizard SV Gel and PCR Clean up System (Promega), and then cloned into the pGEM-T Easy vector (Promega). The resultant plasmids were purified by Wizard Plus SV Minipreps DNA Purification Systems (Promega) and sequenced using T7 (5′TAATACGACTCACTATAGGG3′) and Sp6 (5′GATTTAGGTGACACTATAGAATAC3′) primers by the Genomics Unit at the Parque Científico de Madrid (http://www.fpcm.es/, last accessed December 2, 2013). Relative abundance of each splice variant was measured as the result of the gel intensity for each single band using ImageJ software (http://rsb.info.nih.gov/ij/index.html, last accessed December 2, 2013). In the case of 1–1.5 kb bands, their relative abundance was related to the number of colonies found from the 48 total colonies checked in each tissue and measure as follows: lower than 20% of the total colonies was considered as low expression, 20–40% as moderate expression, 40–50% as moderate–high expression, and 50% of the total colonies or higher was considered as high expression levels.
Bioinformatics
General Analysis
Sequence data were annotated using the BioEdit software (http://www.mbio.ncsu.edu/BioEdit/bioedit.html, last accessed December 2, 2013) and aligned to Tv1-RAGE mRNA (cDNA) and to genomic RAGE DNA in each species using EBI Pairwise Alignment (http://www.ebi.ac.uk/Tools/psa/, last accessed December 2, 2013) to obtain the exonic composition of each isoform and to compare it to the canonical one. The translation of each splice variant to protein was performed using ORF Finder (www.ncbi.nlm.nih.gov/gorf/, last accessed December 2, 2013). Pfam (http://pfam.sanger.ac.uk/, last accessed December 2, 2013) was used for general domain prediction and SignalP4.0 Server (http://www.cbs.dtu.dk/services/SignalP/, last accessed December 2, 2013) for signal peptide prediction. Splice variants with less than one protein domain and one or more premature stop codons (PSC) were considered as noncoding RNA (ncRNA) variants. mRNA splice variants were named as Sp_RAGE_VN (where species was abbreviated by Gsss = first letter of genus and the three first letters of the species name, VN = mRNA variant number). The sequence data from this study have been submitted to GenBank with the accession numbers: H. sapiens (GU139370–84), M. mulatta (GU139408–22), B. taurus (GU139385–407), S. scrofa (GU164706–14), Mu. musculus (GU164727–39) and R. norvegicus (GU164715–26) (For details see supplementary fig. S1, Supplementary Material online).
RNA-Seq Data Analyses
RNA-seq data from brain and liver tissues of different species were obtained from the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA) database, single reads of SRP007412 (Barbosa-Morais et al. 2012) and paired reads of SRP016501 (Merkin et al. 2012). Paired reads of human Monocytes-CD14+ from ENCODE project (SRX179434) were also analyzed. Reads were mapped to the genome of each species, obtained from the Ensembl Database (H. sapiens: GRCh37, M. mulatta: MMUL_1, B. taurus: UMD3.1, Mu. musculus: GRCm38, R. norvegicus: Rnor_5.0), using the default parameters of TopHat and Bowtie (Trapnell et al. 2009; Langmead et al. 2009). Junction reads of the BED files (TopHat outfile) were normalized (per million of mapped reads) to compare splice events among samples. Samtools software (Li et al. 2009) was used for indexing BAM files (TopHat outfile), conversing alignment formats, and sorting the gene region of interest. The resulting RAGE-aligned reads were used to assemble them into transcripts using Cufflinks (Trapnell et al. 2010) by both default (-F 0.1 and -j 0.15) and modified (-F 0 and -j 0.5) parameters. Additionally, the experimentally described transcripts were provided to Cufflinks by a GTF file, and their relative abundances were estimated. The expression of each transcript was measured in Fragments Per Kilobase of exon model per Million mapped fragments. The IGV browser (Robinson et al. 2011) was used for visualization of mapped reads and assembled transcripts to its genome context.
Results
Primates RAGE Splice Variants
Identification and Characterization of Human RAGE Splice Variants
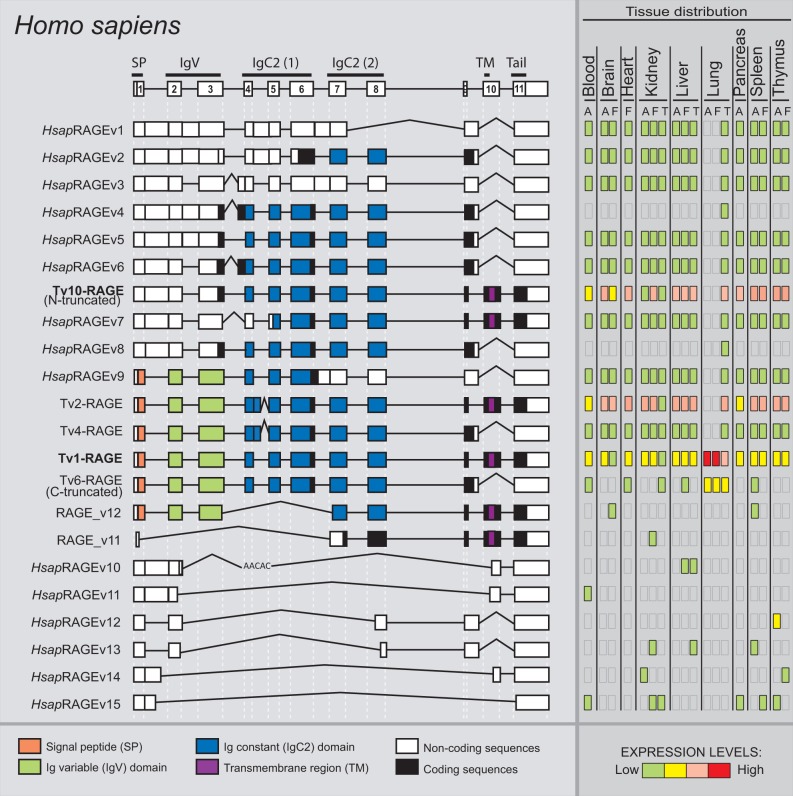
To fully characterize H. sapiens RAGE splice variants, we performed specific nested RT-PCR assays in 18 different tissues. Detailed analyses of these results showed a total of 22 different variants obtained from eight adult, seven fetal, and three tumoral tissues (fig. 1). Interestingly, 15 of these isoforms were novel RAGE splice variants (different exon/intron structure to the previously described ones) named HsapRAGEv1 to v15 (detailed information regarding these novel variants and the previously described ones is available in supplementary fig. S1, Supplementary Material online). Transcript variant 1 (also known as full-length RAGE, and hereafter Tv1-RAGE) was identified in this study as one of the most abundant splice variants (fig. 1), in agreement to previous descriptions (Hudson et al. 2008). This isoform is composed by a signal peptide encoded within exon 1, an Ig-V domain within exons 2 and 3, two Ig-C domains within exons 4–6 and 7–8, respectively, and a single transmembrane region within exon 10 followed by a short cytoplasmic tail within exon 11 (Hudson et al. 2008) (fig. 1).
Fig. 1.—
RAGE transcript variants in Homo sapiens. Representation of human RAGE splice variants and their relative expression levels in different adult (A), fetal (F), and tumor (T) tissues. Potential protein domains are shown with different colors, and noncoding sequences are shown in white. Junctions from the canonical variant are shown as straight lines, whereas the alternative junctions are shown as open triangles. Splice variants are arranged by length.
Other found alternative splice variants show changes in these domains. For instance, intron 1 retention in Tv10-RAGE (previously named N-truncated) and HsapRAGEv1 to v8 splice variants results in a PSC, but there is an alternative downstream potential start codon located at exon 3 excluding the signal peptide (which could affect to the functionality of the protein) and the Ig-V domain from their potential encoded isoforms (fig. 1). Tv10-RAGE was recently defined as a noncoding variant by NCBI, although several authors consider it as a coding isoform that lacks the Ig-V domain (Yonekura et al. 2003; Sterenczak et al. 2013). Tv10-RAGE variant only presents this splice event, whereas HsapRAGEv1 to v8 present a combination of other additional AS events (fig. 1). Another common splicing event is the inclusion of part of intron 9 and skipping of exon 10 (Tv6-RAGE or C-truncated, Tv4-RAGE, HsapRAGEv1 to v6, v8, and v9 splice variants) that results in the loss of the transmembrane region and cytoplasmic tail and, therefore, generating potential sRAGE protein isoforms. We have also found other different splicing events, such as the use of a different 5′-donor splice site in exon 4 (Tv2-RAGE and Tv4-RAGE, previously described by Hudson et al. 2008), that would cause variations in the region encoding the corresponding Ig-C domain and therefore, this could modify the ligand specificity and/or the binding affinity. In addition, we found several predicted noncoding short variants (RAGE_v11, HsapRAGEv10 to v15 splice variants), which present skipping of several exons and use of different acceptor splice sites (fig. 1).
We have also assessed the relative abundance of each splice variant showing differences in expression levels among all the analyzed tissues. Tv1-RAGE, Tv2-RAGE, and Tv10-RAGE variants were the most prevalent of the detected transcripts in the majority of the analyzed tissues. Moreover, large splice variants with different intron retentions show low but uniform expression among the different tissues. By contrast, small splice variants also show low expression levels but with a very specific distribution on particular tissues, even among embryonic or tumoral states within the equivalent tissue. All the analyzed tissues show this similar pattern of expression levels, but interestingly we have observed that adult and fetal lung only express Tv1-RAGE and Tv6-RAGE (C-truncated) variants at high levels, whereas tumor lung expresses all the different large splice variants but at low levels. Nearly 40% of the total analyzed variants in human are noncoding (fig. 2).
Fig. 2.—
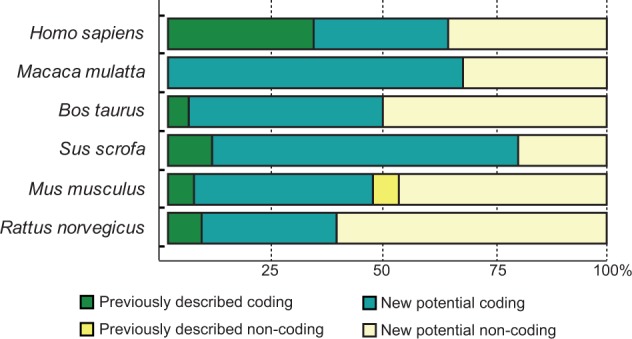
Relative abundance of coding versus noncoding transcript analyzed. Stacked percentage bars reflect the portion of previously described variants and the variants that we found in our study (coding and noncoding) present in different analyzed species.
Identification and Characterization of Rhesus Macaque RAGE Splice Variants
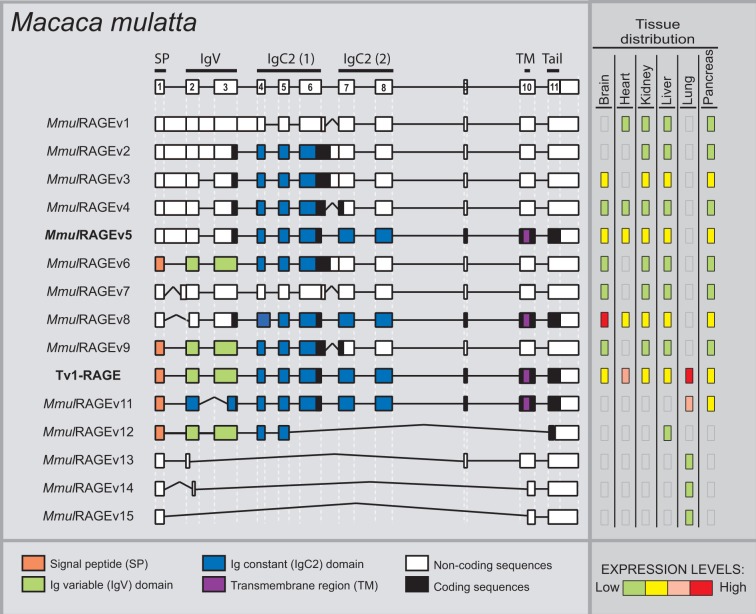
In M. mulatta, we detected in total 15 novel variants of RAGE within six different adult tissues (fig. 3), and one of them was the predicted canonical Tv1-RAGE (NM_001205117–GU139417). We have named these splice variants as MmulRAGEv1 to v15. Only Tv1-RAGE was previously annotated on databases based on homology to human, but we show here the first biological evidence for this isoform. MmulRAGEv5 and Tv1-RAGE from M. mulatta are directly related to the exon/intron structure of Tv10-RAGE and Tv1-RAGE human splice variants, respectively. Furthermore, we found an isoform (MmulRAGEv3) that retains introns 1 and 6 and is related to an isoform (NtRAGEΔ) previously described in humans (supplementary fig. S1, Supplementary Material online) and not detected by us, involved in Alzheimer’s disease (Ding and Keller 2005a, 2005b). None of the remainder macaque variants have a human homologous variant (table 1).
Fig. 3.—
RAGE transcript variants in Macaca mulatta. Representation of monkey RAGE splice variants and their relative expression levels in different adult tissues. See figure 1 for details. MmulRAGEv5 (bold) is related to the human Tv10-RAGE.
Table 1.
Conserved RAGE Variants Based on Exon/Intron Composition across Mammals
 |
Inclusion of intron 1 is shown in MmulRAGEv1 to v5, in which MmulRAGEv1 is predicted as ncRNAs due to the presence of different PSC in introns 1 and 6 or exon 7. The whole inclusion of intron 6 in MmulRAGEv6 and its partial inclusion in MmulRAGEv9 resulted in the appearance of PSCs. The potential encoded isoforms of these two variants could be new sRAGE isoforms lacking also the last IgC domain (fig. 3). We found changes in the amino acid sequence of the Ig-V domain in MmulRAGEv11 which turns it into an Ig-C domain as a result of the use of a 5′-donor alternative splice site in exon 3. Moreover, an alternative 3′-acceptor splice site in exon 2 detected in MmulRAGEv8 transcript implies changes in the reading phase and, due to that a PSC appears, although an isoform could be generated from a downstream start codon in exon 3 (fig. 3). In addition, we found four smaller isoforms (MmulRAGEv12 to v15), which present skipping of large number of exons, one of them could be a potential coding variant (MmulRAGEv12), whereas the other three are predicted as ncRNA. Noncoding variants form around 30% of the total monkey RAGE isoforms (fig. 2).
Macaque tissues show different expression levels of each variant (fig. 3). Tv1-RAGE, MmulRAGEv8, MmulRGEv5, and MmulRAGEv3 were the most abundant variants among different tissues. MmulRAGEv8 is overrepresented in brain, whereas macaque Tv1-RAGE is overrepresented in lung tissue, as it happens with its homologous human variant. As MmulRAGEv8 lacks the IgV domain in relation to Tv1-RAGE, the overexpression in brain could indicate that MmulRAGEv8 is probably binding to another ligand. However, we have to consider that it would not encode a signal peptide. Small noncoding variants show low expression levels and are only present in lung tissue, with the exception of the potential coding MmulRAGEv12 that it is only expressed in liver.
Artiodactyla RAGE Splice Variants
Identification and Characterization of Bovine RAGE Splice Variants
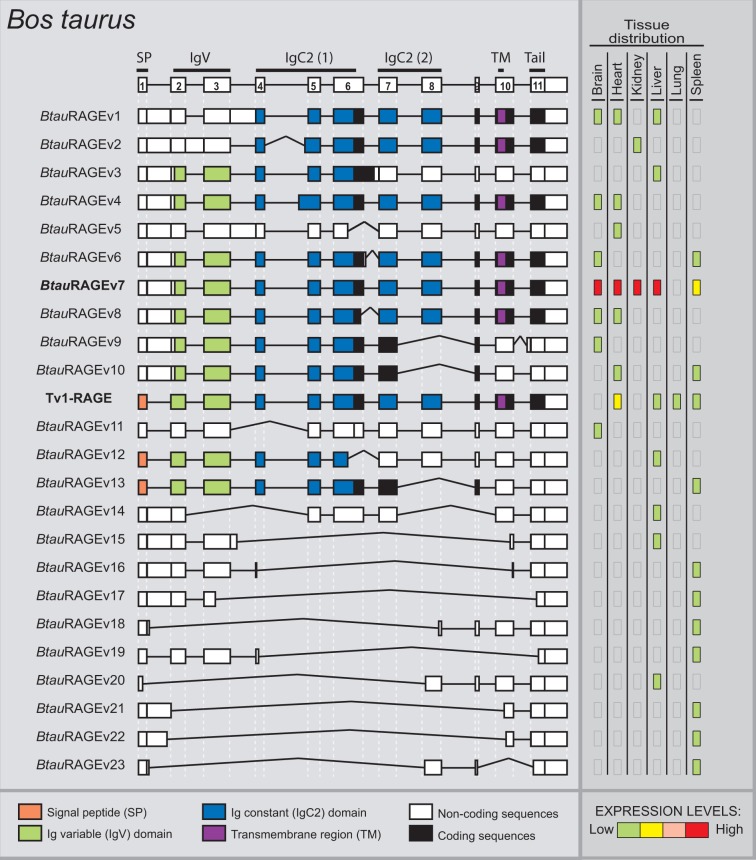
In B. taurus we have detected a total of 24 alternative splice variants of RAGE within six different adult tissues (fig. 4), 23 of them were novel alternative splice variants (named as BtauRAGEv1 to v23), Tv1-RAGE being the only one previously described by Neeper et al. (1992) (NM_173982) (supplementary fig. S1, Supplementary Material online). Tv1-RAGE and BtauRAGEv7 isoforms are related structurally to the similar human and macaque variants (table 1). However, BtauRAGEv7 splice variant leads to several changes at protein level when compared with the primates counterparts, due to an alternative downstream start codon located at exon 2, instead of exon 3. Only BtauRAGEv1 and BtauRAGEv2 could generate N-truncated proteins similar to the human one but, in this case, due to the retention of introns 1 and 3 or introns 1 and 2, respectively. BtauRAGEv22 is similar to HsapRAGEv14, being predicted as ncRNAs, and BtauRAGEv3 and v6 have homologs in macaque (table 1). Furthermore, sRAGE isoforms could be generated by exon 8 skipping (BtauRAGEv9, v10, and v13), using alternative donor sites (BtauRAGEv12), or by intron retention (BtauRAGEv3), but in all these cases lacking the second Ig-C domain. In addition, several noncoding splice variants are found with large “deletions” (BtauRAGEv14 to v23), or small ones and intron retentions (BtauRAGEv5 and v11), which represent around 50% of whole transcripts (fig. 2).
Fig. 4.—
RAGE transcript variants in Bos taurus. Representation of bovine RAGE splice variants and their relative expression levels in different adult tissues. See figure 1 for details. BtauRAGEv7 (bold) is related to the human Tv10-RAGE.
In relation to relative abundance of each splice variant, figure 4 shows high and wide expression levels of BtauRAGEv7 splice variant, whereas the rest of splice variants show in general low and tissue-specific expression levels. In contrast to human, bovine Tv1-RAGE presents low expression levels in the lung tissue.
Identification and Characterization of Swine RAGE Splice Variants
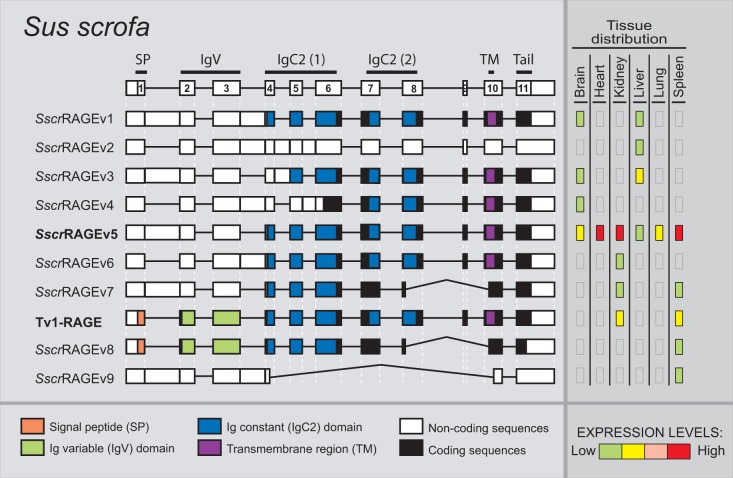
Sus scrofa only shows 10 alternative splice variants of RAGE within the six different analyzed adult tissues (fig. 5), becoming the species with fewer variants, even when comparing the same number and type of tissues. Databases searches only show the predicted Tv1-RAGE isoform, whereas the rest are novel alternative splice variants (named SscrRAGEv1 to v9). Tv1-RAGE and SscrRAGEv5 isoforms are again the only two variants similar to human splice variants (Tv1-RAGE and Tv10-RAGE, respectively, table 1), showing the same domains as the humans counterparts. Retentions of intron 1 and/or 3 are common events in swine RAGE (SscrRAGEv1 to v7 and v9). These events result in a PSC and a downstream start codon located at exon 4, instead of exon 3 in primates or in exon 2 in B. taurus, which could encode proteins that lack the Ig-V domain and the signal peptide. SscrRAGEv1 isoform is related to bovine BtauRAGEv1 isoform with similar protein domains (table 1). Furthermore, soluble isoforms could also be present in swine RAGE, as a result of the use of an alternative 3′-acceptor splice site in exon 10 (SscrRAGEv7 and v8) and therefore lacking the transmembrane region. On the other hand, swine RAGE only shows two predicted noncoding splice variants (SscrRAGEv2 and v9), representing 20% of all variants detected (fig. 2).
Fig. 5.—
RAGE transcript variants in Sus scrofa. Representation of porcine RAGE splice variants and their relative expression levels in different adult tissues. See figure 1 for details. Splice variants are arranged by length. SscrRAGEv5 (bold) is related to the human Tv10-RAGE.
Expression levels of the different splice variants in swine (fig. 5) are quite different compared with human ones. In swine, SscrRAGEv5 isoform, which is equivalent to human Tv10-RAGE, is also highly and widely expressed in all the analyzed tissues, even in lung tissue where the human homolog is not expressed. However, Tv1-RAGE expression is moderate and only in two tissues (kidney and spleen), remarkably, there is no Tv1-RAGE expression in lung (fig. 5).
Rodents RAGE Splice Variants
Identification and Characterization of Mouse RAGE Splice Variants
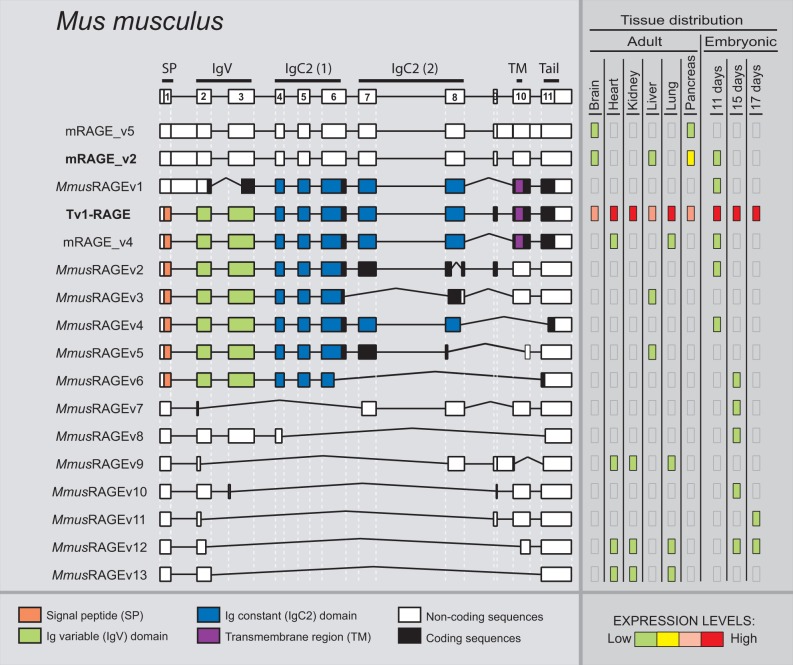
We detected a total of 17 alternative splice variants of RAGE from Mu. musculus within different adult and embryonic tissues (fig. 6). Tv1-RAGE was already in databases, and mRAGE_v2, v4, and v5 were previously described (Kalea et al. 2009) (supplementary fig. S1, Supplementary Material online). The other 13 splice variants are novel and named as MmusRAGEv1 to v13 (fig. 6). Only two of these variants, Tv1-RAGE and mRAGE_v2 are related, in terms of exon/intron structure, to the equivalent splice variants in other analyzed mammalian species (table 1). However, mRAGE_v2 presents another PSC in exon 5, not present in the other homologs, and as a result, mRAGE_v2 is a noncoding variant (fig. 6). There is a potential soluble murine splice variant (MmusRAGEv5) related to a swine one (SscrRAGEv8) (table 1). In addition, other potential soluble variants containing the signal peptide and the first two Ig domains could be generated (MmusRAGEv2 to v6) (fig. 6), and three by the presence of stop codons before exon 10 (MmusRAGEv2, v3 and v5). Harashima et al. (2006) had previously described an isoform (esRAGE) that could generate soluble proteins (by intron 9 retention), but we have not been able to identify it (supplementary fig. S1, Supplementary Material online). Another event that could produce domain alterations is present in MmusRAGEv1 due to the use of an alternative 3′-acceptor splice site in exon 3 and, therefore, removing the Ig-V domain. We found several noncoding splice variants of which mRAGE_v5 and mRAGE_v2 were previously described, and MmusRAGEv7 to v13 being novel from this study (supplementary fig. S1, Supplementary Material online). In total, they represent more than 50% of murine splice variants (fig. 2).
Fig. 6.—
RAGE transcript variants in Mus musculus. Representation of mouse RAGE splice variants and their relative expression levels in different adult and embryonic tissues. See figure 1 for details. mRAGE_v2 (bold) is related to the human Tv10-RAGE.
Relative abundance of each splice variant shows differences in expression levels among tissues (fig. 6) as well as compared with other mammals analyzed in this study. Tv1-RAGE presents, as in humans, high and wide expression, either in embryonic or adult tissues. Nevertheless, the noncoding mRAGE_v2 (equivalent in structure to potential coding human TV10-RAGE) shows low expression levels in brain, liver, and 11 days embryonic tissue, and only shows moderate expression levels in pancreas adult tissue, in contrast to the human homolog which is highly expressed. Other splice variants show low expression levels but with a specific distribution among tissues and state of development.
Identification and Characterization of Rat RAGE Splice Variants
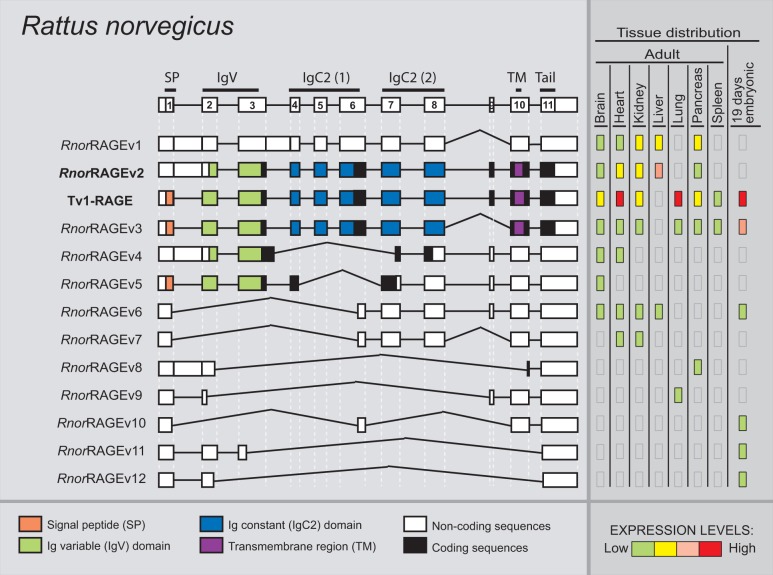
We detected a total of 13 alternative splice variants of RAGE from R. norvegicus within different adult and embryonic tissues (fig. 7). Tv1-RAGE sequence was previously found based on homology, whereas the other 12 were novel alternative splice variants named as RnorRAGEv1 to v12 (see supplementary fig. S1, Supplementary Material online). Tv1-RAGE and RnorRAGEv2 isoforms are related to human splice variants, as well as in the other analyzed species (table 1). Rat RnorRAGEv2, although related structurally to human TV10-RAGE, shows changes at protein level being similar to bovine BtauRAGEv7 isoform, with the Ig-V domain present (fig. 7).
Fig. 7.—
RAGE transcript variants in Rattus norvegicus. Representation of rat RAGE splice variants and their relative expression levels in different adult and an embryonic tissue. See figure 1 for details. RnorRAGEv2 (bold) is related to the human Tv10-RAGE.
Intron 1 retention is present in some splice variants (RnorRAGEv1, v2, v4, and v8) resulting in PSC on the retained intron, with an alternative downstream start codon located at exon 2 that conforms potential coding splice variants, and noncoding splice variant when intron 3 is also retained (RnorRAGEv1) or due to several exon skipping events (RnorRAGEv8). Some rat RAGE splice variants are related to murine RAGE splice variants (table 1), of which RnorRAGEv3 (related to mRAGE_v4) is broadly expressed and quite similar in protein structure to TV1-RAGE as it only lacks the short region between the second IgC. Noncoding splice variants (RnorRAGEv1 and RnorRAGEv6 to v12) represent more than 60% of ncRNA in R. norvegicus (fig. 2).
Relative abundance of each rat splice variant shows differences in expression levels among tissues, especially noncoding variants that show specific distribution around tissues and embryonic developmental stages. Tv1-RAGE presents the highest expression levels in all tissues, with the exception of the liver where it is not expressed. Lung tissue shows similar expression levels to human ones, with high levels of Tv1-RAGE and low levels of RnorRAGEv3 and smaller noncoding isoforms. RnorRAGEv1 and RnorRAGEv2 variants show moderate and moderate–high levels in adult tissues, the last one in agreement with the human homolog TV10-RAGE. In embryonic tissue, Tv1-RAGE and RnorRAGEv3 are the most abundant transcripts (fig. 7).
Identification of RAGE Splice Variants by High-Throughput Technologies
Nowadays, RNA-seq data are a powerful tool to perform transcriptional studies, from description of new isoforms to quantification of all transcripts. For this reason, we decided to carry out high-throughput RNA analyses of sequencing data deposited in the SRA database (Pan et al. 2008; Barbosa-Morais et al. 2012; Merkin et al. 2012) to compare them with our nested RT-PCR results. We mapped these publically available RNA-seq reads from brain and liver tissues to each genome of reference (human, monkey, bovine, rat, and mouse), and afterward, relative abundances of RT-PCR detected RAGE transcripts were estimated using Cufflinks. The analyses confirmed that RAGE is a very lowly expressed gene due to a minimum or null coverage (supplementary table S2, Supplementary Material online). In human, Tv1-RAGE shows moderate expression in brain and Tv10_RAGE in liver, in accordance with our RT-PCR results. In addition, macaque MmulRAGEv2 and MmulRAGEv6 variants are the mainly expressed in brain and liver followed, in brain, by Tv1-RAGE, which is not expressed in liver. These RNA-seq results are not in agreement with the RT-PCR ones where MmulRAGEv8 is more expressed in all tissues followed by Tv1-RAGE. In the bovine analyses, BtauRAGEv1, v2, and v4 are highly expressed in brain and BtauRAGEv1 and v3 in liver (with some correlation to our PCR results). Moreover, bovine Tv1-RAGE is moderately expressed in brain but not expressed in liver, whereas BtauRAGEv7, which is very highly detected by PCR, shows no expression in RNA-seq samples. In mouse RNA-seq samples, the mainly expressed variants are Tv1-RAGE (as by our PCR results) and mRAGE_v5 in both tissues. In addition, we observe similar expression results between both technologies in Tv1-RAGE, RnorRAGEv1, and v2 rat variants. Nevertheless, it is important to observe that all expression levels show extremely low values.
Furthermore, we analyzed data from a human sample (monocytes) from the ENCODE project with a very high number of reads. Using default parameters, Cufflinks was only able to assemble three transcripts (supplementary fig. S2, Supplementary Material online) which do not correlate with those we found by nested RT-PCR, even the canonical transcript could not be assembled. As Cufflinks suppresses the output of smaller fractions of splice variants, we approached this analysis altering the parameters which modify the minimal isoform fraction and the pre-mRNA fraction. As a result, we were able to assemble Tv1-RAGE, Tv10-RAGE, and HsapRAGEv8 (supplementary fig. S2, Supplementary Material online) in the human monocytes sample. We extended this approach to the previously analyzed brain and liver samples in all the different species. Tv1-RAGE was detected in only one brain human sample, and N-truncated-related variants were assembled in macaque brain (MmulRAGEv5) and both bovine tissues (BtauRAGEv7). MmulRAGEv6 was also assembled in macaque brain. (supplementary fig. S2, Supplementary Material online). No variants could be assembled in the rodent samples.
Observing that Cufflinks analyses were not able to assemble all the isoforms detected by RT-PCR, not even the mainly expressed ones, we decided to analyze the number of splicing events detected by TopHat, by using the reported junctions. We were able to detect most of the canonical splice events but only in those samples which have high read coverage. It is interesting to notice that the splice event between exons 1 and 2 is not present in any bovine sample, in accordance to our results obtained by RT-PCR. For that reason, we were not able to assemble the Tv1-RAGE isoform using Cufflinks, and the equivalent to the human N-truncated variant was the only isoform assembled in these samples. Moreover, a few alternative spliced events were also detected in all the species (supplementary fig. S3, Supplementary Material online) but all of them with extremely low expression values. Some of these events showed values between 0.004 and 0.008 junction reads per million of mapped reads, which correspond to just one or two junctions found in the total sample. These events show a similar tissue expression pattern as our experimental results. For instance, partial retention of intron 6 in rhesus monkey is lowly detected in brain, as the variants which contain this splice event (MmulRAGEv5, v7, and v9). The same consistent results are shown in the skipping of exon 9 event in rat samples, with low levels in brain observed by both approaches.
Discussion
In this study, we have deeply characterized a high number of alternative splice variants of RAGE in different mammals and tissues, and we have compared their relative expression levels. A diversity of functions of RAGE has been previously described as a result of AS events (Yonekura et al. 2003; Hudson et al. 2008; Kalea et al. 2009; Sterenczak et al. 2009), which can modify protein domains. Previous studies focused on splice variants of RAGE in human (Hudson et al. 2008) and mouse (Kalea et al. 2009) showed similar results in expression levels of each splice variant. In human, RAGE had only been studied in detail in lung and aortic smooth muscle cells (AoSMCs) (Hudson et al. 2008). Comparison of adult lung in previous (Hudson et al. 2008) and our studies show similar expression levels for Tv1-RAGE (80% and high expression levels, respectively) and different for Tv6-RAGE (C-truncated) (7% and moderate expression levels, respectively). Interestingly, in our study adult and fetal lung were the only two tissues showing these two isoforms, whereas tumor lung presents a high number of different splice variants.
Previous studies related to RAGE splice variants in different mouse tissues have been performed (Harashima et al. 2006; Kalea et al. 2009). Harashima et al. (2006) identified a soluble isoform on brain using nested PCR, and the relative abundance among other tissues and isoforms was quantified using a small PCR (exons 9–11) so these cannot be compared with our results. In that work, they also describe a soluble isoform (esRAGE) not found in our study, but they generated it with a primer in the second round of PCR specific to the intron 9 retained. In the study by Kalea et al. (2009), murine lung tissue showed both Tv1-RAGE (53%) and mRAGE_v4 (41%) expressions, and we have found high expression and low expression, respectively. The smaller isoforms they found are different from those described here, but they also detected them at very low expression levels (1% in each splice variant). In general, our results are somewhat consistent with previous studies (Hudson et al. 2008; Kalea et al. 2009) as percentages higher than 50% are related to our high expression levels, 50–40% to moderate–high, and 40–20% to low, in most cases. Lower than 20% is sometimes not detected in our study, and, on the other hand, low-expression variants detected by us were not detected by them. In both human and mouse previous studies (Hudson et al. 2008; Kalea et al. 2009), the splice variants were analyzed by RT-PCR and restriction enzyme recognition sequences, whereas we separated the RT-PCR product electrophoretically, cloned and verified by sequencing. Both approaches use nested PCR, by reason of the extremely low expression levels of RAGE, which makes it impossible to approach the study in the dynamic PCR range. Therefore, this methodology could slightly influence the findings limiting the power to differentiate splice variants expression and their relative abundance. For this reason, low-expression variants, which are the most difficult to detect, sometimes showed differences among these studies. Indeed, the optimal approach for determining the relative expression of each transcript should be a quantitative RT-PCR (qRT-PCR). However, because of the presence of multiple splice events common to several isoforms is completely impossible to design a qRT-PCR assay to discriminate one against each other. Nevertheless, this technique was used in canine RAGE (Sterenczak et al. 2009) to describe splice variants in healthy and tumor tissues, but these analyses were performed only with the intron 1 retention event. They detected high expression levels of intron 1 retained variants in neoplastic samples. On the other hand, intron 1 skipping was appointed to healthy transcript variants and, therefore, with lower expression levels of the intron 1 retention event in healthy tissues (Sterenczak et al. 2009). This assay was performed as a global effect of the intron 1, not trying to discriminate among the different variants. The wide diversity of intron 1 retention splice variants that we found cannot be differentiated against each other with that assay and, for this reason, in our study, we find splice variants with intron 1 retention that are highly expressed in healthy adult and fetal tissues, and intron 1 skipping splice variants also present in tumor tissues with different expression levels.
Nowadays, the high-throughput sequencing technology is being employed to describe the complete transcriptome of several organisms (Barbosa-Morais et al. 2012; Merkin et al. 2012; Rastrojo et al. 2013). This technique provides information on differential expression of genes, posttranscriptional mutations or edition, and gene fusions, among others. Furthermore, differences in transcript expression levels should also be observed, but sufficient coverage is required. In the case of RAGE, both low expression and high transcriptomic complexity made extremely difficult to approach the transcriptomic study by this technology. Indeed, in the sample used with high coverage (human monocytes, with 200 millions of paired reads), we were able to detect only three of the splice variants detected by nested PCR, and only when Cufflinks parameters were altered. For these reasons, particular detailed transcriptional studies of low expression genes such as RAGE could be misinterpreted by high-throughput sequencing technologies, and it would be better to study them by other techniques like the one we use in this study.
The transcript variants found and their common splice events could have potential regulatory functions about the overall role of the Tv1-RAGE isoform, and could be involved in the development of several diseases including cancer or Alzheimer’s disease, among others. For instance, low levels of circulating sRAGE have been associated with an increased risk of Alzheimer’s disease (Emanuele et al. 2005), diabetic complications (Grossin et al. 2008), cardiovascular disease (Falcone et al. 2005), and hypertension (Geroldi et al. 2005) in humans. Removal of the transmembrane region allows the generation of sRAGE isoforms by different biological processes. These soluble variants (sRAGE) found in humans are due to either alternative splicing (esRAGE) or proteolytic cleavage (Maillard-Lefebvre et al. 2009) of carboxyl-terminal truncation of membrane-bound isoforms (cRAGE) (Hanford et al. 2004) by the sheddase ADAM10 (Raucci et al. 2008). Adult rat sRAGE shows high protein expression levels, 2-fold higher compared with neonatal rat (Lizotte et al. 2007) and then, proteolytic cleavage of the membrane-bound isoforms seems to be the main way of sRAGE generation (Hanford et al. 2004). To conclude, any of these biological events could generate soluble receptors of RAGE which could bind to ligands before they find membrane-bound receptor RAGE, and thereby attenuate intracellular RAGE signaling. We have been able to detect potential soluble isoforms in all the analyzed species, although only in human (Tv4-RAGE and Tv6-RAGE) and mouse (MmusRAGEv4) they contain the three Ig domains and a signal peptide. Interestingly, all species except rat present soluble isoforms with only the first two Ig domains, which are the ones involved in ligand binding.
Furthermore, we have found splicing events resulting in changes in the extracellular domain that may affect the ligand binding domain, by insertion, deletion, or removal of partial Ig-V domain of RAGE. Binding of AGEs (Grossin et al. 2009) or Aβ peptides (Ding and Keller 2005a) to the Ig-V domain in the membrane-bound receptor RAGE has been suggested to contribute to the pathogenesis of diabetic vascular complications and Alzheimer’s disease, respectively. Ig-V domain modification could generate changes in ligand affinity of RAGE, and splice variants with these domains modifications could be important factors in disease modulation. Indeed, there are several variants that lack the signal peptide, and hence, their specific intracellular transport and structure could be altered. This sort of regulation could modify the final location of each isoform and, therefore, its specific function. Furthermore, most of these splice variants have a PSC and they should be degraded quickly by nonsense-mediated decay. However, they seem to be stable, as it has been shown by other studies (Hudson et al. 2008; Kalea et al. 2009) and our case, finding them to be relatively abundant. The only two variants that are found in all the analyzed species are the equivalent isoforms to human Tv1-RAGE and Tv10-RAGE (table 1), which are also the most abundant ones. In fact, Tv10-RAGE presents a PSC but is even more expressed than Tv1-RAGE in some species (Artiodactyla group). This is very interesting from the evolutionary point of view of the conservation of the highly and widely expressed Tv10-RAGE variant in tissues and species, which can be related to generation of a new function. In human, monkey, and pig samples, the predicted protein lacks the first IgV domain, whereas in cow and rat samples, it would also encode the first IgV domain, although it has to be considered that they do not contain a signal peptide. In addition, this Tv10-RAGE is a predicted noncoding variant in mouse.
On the other hand, other splice variants of RAGE were shown in previous studies, such as dominant-negative RAGE (dnRAGE or RAGEΔ), which only lacks 16 amino acids of the intracellular domain (Ding and Keller 2005a). These splice variants can bind to Aβ ligand and thereby also prevent RAGE signaling. But neither our study nor previous related studies (Yonekura et al. 2003; Hudson et al. 2008; Kalea et al. 2009; Sterenczak et al. 2009) have found these dnRAGE variants, therefore dnRAGE could be tissue or disease specific. We have not detected any other alterations on the cytoplasmic tail in any of the analyzed mammals.
There are also novel transcripts of unknown function, which are mainly long and small ncRNAs. Intron retention is the common splice event in potential long ncRNAs, whereas large exon skipping generates the potential small ones. Previous studies related to ncRNAs show noncoding genes, such as XIST (Duret et al. 2006) or HOTAIR (Rinn et al. 2007), but not much is known about coding genes with alternative noncoding splice variants, as we show in this study. This event could significantly increase the number of ncRNAs among wide RNAs. Moreover, these ncRNAs may underlie vital intracellular functions, such as transcriptional regulation or epigenetics among others (Kikuchi et al. 2009; Marques and Ponting 2009; Ponting et al. 2009), being generated from individual transcript or multiple rounds of cleavage of a unique loci (Tuck and Tollervey 2011). In this particular case, the presence of these different noncoding variants could be to regulate the full-length RAGE isoform and/or its functions in each specific tissue. Because of that, long and small noncoding RAGE variants seem to be involved in different processes, since long ncRNAs show wide expression among the different tissues and, by contrast, small ncRNAs only had low expression and very specific distribution. Furthermore, we have also observed that most of the ncRNAs detected in our study seem to be species-specific splice variants and only a few of them are conserved among the analyzed mammals. These nonconserved variants could be due to transcriptional noise corresponding to the phenomenon described as noisy splicing (Baek and Green 2005; Zhang et al. 2007, 2009; Melamud and Moult 2009; Pickrell et al. 2010) as it is associated to lowly expressed genes and splice junctions not conserved in different tissues nor in species. Some authors have mentioned the implication of noisy splicing as an important property of genome evolution (Pickrell et al. 2010). Nevertheless, to further characterize this, it would require increasing the number of tissues analyzed in each species to fully determine the species specificity of each splice variant. Either different distribution or species-specific noncoding RAGE variants could generate a wide diversity of functional roles that remain still unknown.
Conclusions
In conclusion, we provide evidence that RAGE undergoes extensive AS in the six analyzed mammalian species, generating different isoforms which are distributed among tissues (fetal, adult, and tumor samples) and species (human, monkey, cow, pig, mouse, and rat). No such detailed analysis, which compares a complex alternative spliced gene in different species and tissues, has been previously performed. These transcripts have been detected through direct RT-PCR amplifications and sequencing of the products but not by RNA-seq using default Cufflinks parameters although a very few can be detected after altering them. In summary, we can classify these transcripts in: highly expressed conserved splice variants; low but stable expressed, less or not conserved variants; and lowly expressed, in only one or two conditions, species-specific variants. Comparison between the splice variants in these mammals reveals only two totally conserved ones (human TV1-RAGE and TV10-RAGE and their homologs) indicating a special evolution pattern at the mRNA level. This is interesting from the point of view of different biological functions of one particular gene among different species and, in particular, when they are used as animal models. Furthermore, a large majority of these variants are predicted as noncoding isoforms with unknown function, more than previously known. In fact, Tv10-RAGE and homologs could be a noncoding variant that is highly and broadly expressed. After this analysis, we have observed an even more complex and tissue-specific AS variants than expected among mammals, including a great variability of coding and noncoding variants. For this reason, further extensive studies of functionally relevance of each splice variant are necessary to understand the overall role of RAGE biology, and their possible implication among diseases and evolution. Indeed, similar studies should be performed with other genes to evaluate whether complex AS patterns are common.
Supplementary Material
Supplementary figures S1–S3 and tables S1 and S2 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
R.L.-D. carried out the experiments and bioinformatic analyses presented in this article and drafted the manuscript. A.R. participated in the sequence alignments, data collection, and interpretation. O.V. participated at the initial bases of the project. B.A. conceived the study, participated in its design and interpretation of date, and on the writing of the manuscript. All authors read, helped with the writing, and approved the final manuscript. The authors are grateful to the Genomics Service at CBMSO, and in particular to Fernando Carrasco, for their technical expertise. This work was supported by grants from the Ministerio de Educación y Ciencia (BFU2005-03683), Ministerio de Ciencia e Innovación (BFU2008-03126), Comunidad de Madrid (GR/SAL/0670/2004, and 200620M078), Fundación Ramón Areces. R.L.-D. held a postgraduate fellowship from the Universidad Autónoma de Madrid, A.R. held a postgraduate fellowship (FPU) from the Ministerio de Educación y Ciencia, O.V. held a postgraduate fellowship (FPI) from the Comunidad de Madrid, and B.A. held a Programa Ramón y Cajal contract and an Amarouto (Comunidad Madrid-Fundación Severo Ochoa) contract. The CBMSO receives an institutional grant from Fundación Ramón Areces.
Literature Cited
- Baek D, Green P. Sequence conservation, relative isoform frequencies, and nonsense-mediated decay in evolutionarily conserved alternative splicing. Proc Natl Acad Sci U S A. 2005;102:12813–12818. doi: 10.1073/pnas.0506139102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbosa-Morais NL, et al. The evolutionary landscape of alternative splicing in vertebrate species. Science. 2012;338:1587–1593. doi: 10.1126/science.1230612. [DOI] [PubMed] [Google Scholar]
- Blencowe BJ. Alternative splicing: new insights from global analyses. Cell. 2006;126:37–47. doi: 10.1016/j.cell.2006.06.023. [DOI] [PubMed] [Google Scholar]
- Calarco JA, Zhen M, Blencowe BJ. Networking in a global world: establishing functional connections between neural splicing regulators and their target transcripts. RNA. 2011;17:775–791. doi: 10.1261/rna.2603911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calarco JA, et al. Global analysis of alternative splicing differences between humans and chimpanzees. Genes Dev. 2007;21:2963–2975. doi: 10.1101/gad.1606907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colak R, et al. Distinct types of disorder in the human proteome: functional implications for alternative splicing. PLoS Comput Biol. 2013;9:e1003030. doi: 10.1371/journal.pcbi.1003030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper TA, Wan L, Dreyfuss G. RNA and disease. Cell. 2009;136:777–793. doi: 10.1016/j.cell.2009.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Q, Keller JN. Evaluation of rage isoforms, ligands, and signaling in the brain. Biochim Biophys Acta. 2005a;1746:18–27. doi: 10.1016/j.bbamcr.2005.08.006. [DOI] [PubMed] [Google Scholar]
- Ding Q, Keller JN. Splice variants of the receptor for advanced glycosylation end products (RAGE) in human brain. Neurosci Lett. 2005b;373:67–72. doi: 10.1016/j.neulet.2004.09.059. [DOI] [PubMed] [Google Scholar]
- Duret L, Chureau C, Samain S, Weissenbach J, Avner P. The Xist RNA gene evolved in eutherians by pseudogenization of a protein-coding gene. Science. 2006;312:1653–1655. doi: 10.1126/science.1126316. [DOI] [PubMed] [Google Scholar]
- Emanuele E, et al. Circulating levels of soluble receptor for advanced glycation end products in Alzheimer disease and vascular dementia. Arch Neurol. 2005;62:1734–1736. doi: 10.1001/archneur.62.11.1734. [DOI] [PubMed] [Google Scholar]
- Falcone C, et al. Plasma levels of soluble receptor for advanced glycation end products and coronary artery disease in nondiabetic men. Arterioscler Thromb Vasc Biol. 2005;25:1032–1037. doi: 10.1161/01.ATV.0000160342.20342.00. [DOI] [PubMed] [Google Scholar]
- Geroldi D, et al. Decreased plasma levels of soluble receptor for advanced glycation end-products in patients with essential hypertension. J Hypertens. 2005;23:1725–1729. doi: 10.1097/01.hjh.0000177535.45785.64. [DOI] [PubMed] [Google Scholar]
- Ghavami S, et al. S100A8/A9 at low concentration promotes tumor cell growth via RAGE ligation and MAP kinase-dependent pathway. J Leukoc Biol. 2008;83:1484–1492. doi: 10.1189/jlb.0607397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingeras TR. Origin of phenotypes: genes and transcripts. Genome Res. 2007;17:682–690. doi: 10.1101/gr.6525007. [DOI] [PubMed] [Google Scholar]
- Goh SY, Cooper ME. Clinical review: the role of advanced glycation end products in progression and complications of diabetes. J Clin Endocrinol Metab. 2008;93:1143–1152. doi: 10.1210/jc.2007-1817. [DOI] [PubMed] [Google Scholar]
- Grossin N, Wautier MP, Picot J, Stern DM, Wautier JL. Differential effect of plasma or erythrocyte AGE-ligands of RAGE on expression of transcripts for receptor isoforms. Diabetes Metab. 2009;35:410–417. doi: 10.1016/j.diabet.2009.04.009. [DOI] [PubMed] [Google Scholar]
- Grossin N, et al. Severity of diabetic microvascular complications is associated with a low soluble RAGE level. Diabetes Metab. 2008;34:392–395. doi: 10.1016/j.diabet.2008.04.003. [DOI] [PubMed] [Google Scholar]
- Hanford LE, et al. Purification and characterization of mouse soluble receptor for advanced glycation end products (sRAGE) J Biol Chem. 2004;279:50019–50024. doi: 10.1074/jbc.M409782200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harashima A, et al. Identification of mouse orthologue of endogenous secretory receptor for advanced glycation end-products: structure, function and expression. Biochem J. 2006;396:109–115. doi: 10.1042/BJ20051573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harja E, et al. Vascular and inflammatory stresses mediate atherosclerosis via RAGE and its ligands in apoE-/- mice. J Clin Invest. 2008;118:183–194. doi: 10.1172/JCI32703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiwatashi K, et al. A novel function of the receptor for advanced glycation end-products (RAGE) in association with tumorigenesis and tumor differentiation of HCC. Ann Surg Oncol. 2008;15:923–933. doi: 10.1245/s10434-007-9698-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann MA, et al. RAGE mediates a novel proinflammatory axis: a central cell surface receptor for S100/calgranulin polypeptides. Cell. 1999;97:889–901. doi: 10.1016/s0092-8674(00)80801-6. [DOI] [PubMed] [Google Scholar]
- Hori O, et al. The receptor for advanced glycation end products (RAGE) is a cellular binding site for amphoterin. Mediation of neurite outgrowth and co-expression of rage and amphoterin in the developing nervous system. J Biol Chem. 1995;270:25752–25761. doi: 10.1074/jbc.270.43.25752. [DOI] [PubMed] [Google Scholar]
- Hudson BI, et al. Identification, classification, and expression of RAGE gene splice variants. FASEB J. 2008;22:1572–1580. doi: 10.1096/fj.07-9909com. [DOI] [PubMed] [Google Scholar]
- Huttunen HJ, Fages C, Rauvala H. Receptor for advanced glycation end products (RAGE)-mediated neurite outgrowth and activation of NF-kappaB require the cytoplasmic domain of the receptor but different downstream signaling pathways. J Biol Chem. 1999;274:19919–19924. doi: 10.1074/jbc.274.28.19919. [DOI] [PubMed] [Google Scholar]
- Irimia M, Blencowe BJ. Alternative splicing: decoding an expansive regulatory layer. Curr Opin Cell Biol. 2012;24:323–332. doi: 10.1016/j.ceb.2012.03.005. [DOI] [PubMed] [Google Scholar]
- Kalea AZ, Schmidt AM, Hudson BI. Alternative splicing of RAGE: roles in biology and disease. Front Biosci. 2011;17:2756–2770. doi: 10.2741/3884. [DOI] [PubMed] [Google Scholar]
- Kalea AZ, et al. Alternative splicing of the murine receptor for advanced glycation end-products (RAGE) gene. FASEB J. 2009;23:1766–1774. doi: 10.1096/fj.08-117739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi K, et al. Transcripts of unknown function in multiple-signaling pathways involved in human stem cell differentiation. Nucleic Acids Res. 2009;37:4987–5000. doi: 10.1093/nar/gkp426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kislinger T, et al. N(epsilon)-(carboxymethyl)lysine adducts of proteins are ligands for receptor for advanced glycation end products that activate cell signaling pathways and modulate gene expression. J Biol Chem. 1999;274:31740–31749. doi: 10.1074/jbc.274.44.31740. [DOI] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licatalosi DD, Darnell RB. RNA processing and its regulation: global insights into biological networks. Nat Rev Genet. 2009;11:75–87. doi: 10.1038/nrg2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lizotte PP, et al. Developmental expression of the receptor for advanced glycation end-products (RAGE) and its response to hyperoxia in the neonatal rat lung. BMC Dev Biol. 2007;7:15. doi: 10.1186/1471-213X-7-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch KW. Consequences of regulated pre-mRNA splicing in the immune system. Nat Rev Immunol. 2004;4:931–940. doi: 10.1038/nri1497. [DOI] [PubMed] [Google Scholar]
- Maillard-Lefebvre H, et al. Soluble receptor for advanced glycation end products: a new biomarker in diagnosis and prognosis of chronic inflammatory diseases. Rheumatology. 2009;48:1190–1196. doi: 10.1093/rheumatology/kep199. [DOI] [PubMed] [Google Scholar]
- Malherbe P, et al. cDNA cloning of a novel secreted isoform of the human receptor for advanced glycation end products and characterization of cells co-expressing cell-surface scavenger receptors and Swedish mutant amyloid precursor protein. Brain Res Mol Brain Res. 1999;71:159–170. doi: 10.1016/s0169-328x(99)00174-6. [DOI] [PubMed] [Google Scholar]
- Marques AC, Ponting CP. Catalogues of mammalian long noncoding RNAs: modest conservation and incompleteness. Genome Biol. 2009;10:R124. doi: 10.1186/gb-2009-10-11-r124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McManus CJ, Graveley BR. RNA structure and the mechanisms of alternative splicing. Curr Opin Genet Dev. 2011;21:373–379. doi: 10.1016/j.gde.2011.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melamud E, Moult J. Stochastic noise in splicing machinery. Nucleic Acids Res. 2009;37:4873–4886. doi: 10.1093/nar/gkp471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer TR, et al. Targeted RNA sequencing reveals the deep complexity of the human transcriptome. Nat Biotechnol. 2011;30:99–104. doi: 10.1038/nbt.2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkin J, Russell C, Chen P, Burge CB. Evolutionary dynamics of gene and isoform regulation in Mammalian tissues. Science. 2012;338:1593–1599. doi: 10.1126/science.1228186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neeper M, et al. Cloning and expression of a cell surface receptor for advanced glycosylation end products of proteins. J Biol Chem. 1992;267:14998–15004. [PubMed] [Google Scholar]
- Nilsen TW, Graveley BR. Expansion of the eukaryotic proteome by alternative splicing. Nature. 2010;463:457–463. doi: 10.1038/nature08909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurtdinov RN, Neverov AD, Favorov AV, Mironov AA, Gelfand MS. Conserved and species-specific alternative splicing in mammalian genomes. BMC Evol Biol. 2007;7:249. doi: 10.1186/1471-2148-7-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet. 2008;40:1413–1415. doi: 10.1038/ng.259. [DOI] [PubMed] [Google Scholar]
- Pan Q, et al. Alternative splicing of conserved exons is frequently species-specific in human and mouse. Trends Genet. 2005;21:73–77. doi: 10.1016/j.tig.2004.12.004. [DOI] [PubMed] [Google Scholar]
- Pickrell JK, Pai AA, Gilad Y, Pritchard JK. Noisy splicing drives mRNA isoform diversity in human cells. PLoS Genet. 2010;6:e1001236. doi: 10.1371/journal.pgen.1001236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629–641. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- Rastrojo A, et al. The transcriptome of Leishmania major in the axenic promastigote stage: transcript annotation and relative expression levels by RNA-seq. BMC Genomics. 2013;14:223. doi: 10.1186/1471-2164-14-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raucci A, et al. A soluble form of the receptor for advanced glycation endproducts (RAGE) is produced by proteolytic cleavage of the membrane-bound form by the sheddase a disintegrin and metalloprotease 10 (ADAM10) FASEB J. 2008;22:3716–3727. doi: 10.1096/fj.08-109033. [DOI] [PubMed] [Google Scholar]
- Rinn JL, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JT, et al. Integrative genomics viewer. Nat Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinkora M, Butler JE, Holtmeier W, Sinkorova J. Lymphocyte development in fetal piglets: facts and surprises. Vet Immunol Immunopathol. 2005;108:177–184. doi: 10.1016/j.vetimm.2005.08.013. [DOI] [PubMed] [Google Scholar]
- Sterenczak KA, Nolte I, Murua Escobar H. RAGE splicing variants in mammals. Methods Mol Biol. 2013;963:265–276. doi: 10.1007/978-1-62703-230-8_16. [DOI] [PubMed] [Google Scholar]
- Sterenczak KA, et al. Cloning, characterisation, and comparative quantitative expression analyses of receptor for advanced glycation end products (RAGE) transcript forms. Gene. 2009;434:35–42. doi: 10.1016/j.gene.2008.10.027. [DOI] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuck AC, Tollervey D. RNA in pieces. Trends Genet. 2011;27:422–432. doi: 10.1016/j.tig.2011.06.001. [DOI] [PubMed] [Google Scholar]
- Villate O, Rastrojo A, Lopez-Diez R, Hernandez-Torres F, Aguado B. Differential splicing, disease and drug targets. Infect Disord Drug Targets. 2008;8:241–251. doi: 10.2174/187152608786734188. [DOI] [PubMed] [Google Scholar]
- Xing Y, Lee C. Evidence of functional selection pressure for alternative splicing events that accelerate evolution of protein subsequences. Proc Natl Acad Sci U S A. 2005;102:13526–13531. doi: 10.1073/pnas.0501213102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing Y, Lee C. Relating alternative splicing to proteome complexity and genome evolution. Adv Exp Med Biol. 2007;623:36–49. doi: 10.1007/978-0-387-77374-2_3. [DOI] [PubMed] [Google Scholar]
- Yan SD, et al. RAGE and amyloid-beta peptide neurotoxicity in Alzheimer's disease. Nature. 1996;382:685–691. doi: 10.1038/382685a0. [DOI] [PubMed] [Google Scholar]
- Yang Y, et al. RNA secondary structure in mutually exclusive splicing. Nat Struct Mol Biol. 2011;18:159–168. doi: 10.1038/nsmb.1959. [DOI] [PubMed] [Google Scholar]
- Yonekura H, et al. Novel splice variants of the receptor for advanced glycation end-products expressed in human vascular endothelial cells and pericytes, and their putative roles in diabetes-induced vascular injury. Biochem J. 2003;370:1097–1109. doi: 10.1042/BJ20021371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Krainer AR, Zhang MQ. Evolutionary impact of limited splicing fidelity in mammalian genes. Trends Genet. 2007;23:484–488. doi: 10.1016/j.tig.2007.08.001. [DOI] [PubMed] [Google Scholar]
- Zhang Z, et al. Noisy splicing, more than expression regulation, explains why some exons are subject to nonsense-mediated mRNA decay. BMC Biol. 2009;7:23. doi: 10.1186/1741-7007-7-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.