Abstract
Actin assembly, filament mechanical properties, and interactions with regulatory proteins depend on the types and concentrations of salts in solution. Salts modulate actin through both nonspecific electrostatic effects and specific binding to discrete sites. Multiple cation-binding site classes spanning a broad range of affinities (nanomolar to millimolar) have been identified on actin monomers and filaments. This review focuses on discrete, low-affinity cation-binding interactions that drive polymerization, regulate filament-bending mechanics, and modulate interactions with regulatory proteins. Cation binding may be perturbed by actin post-translational modifications and linked equilibria. Partial cation occupancy under physiological and commonly used in vitro solution conditions likely contribute to filament mechanical heterogeneity and structural polymorphism. Site-specific cation-binding residues are conserved in Arp2 and Arp3, and may play a role in Arp2/3 complex activation and actin-filament branching activity. Actin-salt interactions demonstrate the relevance of ion-linked equilibria in the operation and regulation of complex biological systems.
Introduction
The assembly of actin monomers into filaments plays fundamental roles in many cellular processes including motility, division, shape maintenance/alteration, and force generation (1–3). Dozens of actin-binding proteins (ABPs) regulate actin (dis)assembly dynamics and organize filaments into highly ordered bundles and networks (2,4,5). Actin assembly, mechanical properties, and interactions with regulatory proteins depend strongly on the solution ionic conditions (6–13).
Multiple cation-binding site classes spanning a broad range of affinities have been identified on actin (6,8,9,14–18). A high-affinity (Kd in the nanomolar range (6,15)) divalent cation—usually Ca2+ or Mg2+ in vitro, and Mg2+ in vivo—associates with bound adenine nucleotide and affects actin monomer conformation, assembly kinetics, and filament mechanical properties (7,11,15,17,19,20). Contributions of the high-affinity nucleotide-associated cation to actin function have been comprehensively reviewed elsewhere (see Carlier (19) and Estes et al. (6)), and will not be discussed further here.
Low-affinity (Kd in the range of micromolar to millimolar (8,10,17,21)) cation-binding sites on actin monomers and filaments have been characterized using biochemical, computational, and structural approaches (16,18,22). At least five classes of saturable sites on monomers (6,8,17,21,22) and two filament-specific classes have been identified (18). Occupancy of these filament-specific cation-binding sites drives actin assembly, modulates filament mechanics, and regulates interactions with ABPs.
The functional consequences of low-affinity actin-cation interactions are the topic of this review. We focus on cation binding to discrete sites on filaments created by conserved amino-acid residues from adjacent filament subunits. Like the high-affinity cation site associated with the actin-bound adenine nucleotide, these discrete sites are specific for cations, but bind different cation species (e.g., Ca2+, Mg2+, K+, H+, etc.) with varying affinities.
Origins of Salt Effects on Biopolymer Function
Elucidating the molecular origins of ion-linked actin filament assembly and mechanics requires consideration of the relative contributions of both general electrostatic screening and ion binding at discrete sites. Charge-charge (Coulombic) interactions are the longest-range physical phenomena that drive molecular interactions (23,24). As such, charged species in aqueous solution interact over long molecular-scale distances as collective many-body systems. Charged molecules display interaction effects that scale with ionic strength-dependent activity coefficients (25), and electrostatically screened mean interaction lengths that are not simple functions of concentration under physiological salt conditions (26).
In the case of charge-dense polymers (i.e., polyelectrolytes; polyampholytes) like actin, three distinct but interdependent phenomena affect biomolecular electrostatic interactions in electrolyte solutions such as the cellular cytoplasm:
-
1.
Ionic strength- and temperature-dependent screening, which scales the electrostatic potential between charges and determines the effective charge-charge interaction length (23,24,27–29);
-
2.
Diffusely bound or condensed counterions, which locally screen charge-charge interactions and (partially) neutralize charge dense segments of polyelectrolytes (30–32); and
-
3.
Discrete ion-specific binding sites, in which individual ions are at least partially desolvated and coordinated by (partially) charged biopolymer groups, including amino-acid side-chain and backbone atoms (33–35).
These various polyelectrolyte effects influence many fundamental and functional properties of actin, including assembly, filament mechanics, and interactions with regulatory ABPs (11,18,36–38). Because actin filaments are densely charged linear polymers (36), both local counterion condensation near the actin filament surface (diffuse ions (30)) and ion binding at discrete sites (chelated ions (30)) contribute to the salt dependence of actin function. We emphasize that site-specific ion binding partially neutralizes charge-dense biopolymer regions, which also modulates localized (mostly/fully solvated) counterion condensation. Accordingly, arbitrarily restricting interpretations of salt-effects on biopolymers to a single type of ion interaction is potentially misleading.
Effects of Salt on Actin Function
Salt effects on actin assembly
Actin polymerization follows a nucleation-elongation mechanism that displays a critical concentration (Cc) for assembly (12) (see Skillman et al. (39) for an interesting exception). Filaments form when the total actin concentration exceeds the Cc for assembly (12). In the case of ADP-actin, the Cc reflects the dissociation constant for reversible equilibrium binding of actin monomers to filament ends (18,40). The Cc of ATP and ADP-Pi actin is the steady-state concentration of free actin monomers in exchange with filament ends (12,40).
The actin Cc depends on the bound nucleotide (ATP, ADP-Pi, or ADP and associated high-affinity divalent cation) and the type and concentration of ions in solution (18,41–43) such that salts lower the Cc value (i.e., filaments become thermodynamically more stable). The salt-dependent Cc has been attributed to ion-dependent neutralization of monomer surface charges (42) as well as specific cation (Mg2+) binding at discrete sites (14,44). Occupancy of (at least) a subset of the low-affinity sites on actin monomers is linked to a conformational change associated with an early step of salt-induced actin polymerization (6,7,16,19,44) and contributes to filament stability and elongation kinetics (7,44–47).
Recent work demonstrates that occupancy of discrete filament-specific cation-binding sites accounts for the salt-dependence of actin polymerization at approximately physiological concentrations (18). Nevertheless, salt concentrations much greater than physiological concentrations (>∼2–3 M for KCl and NaCl) depolymerize actin ((48); see also Pinder et al. (49) for nonphysiological salts). In this regime, charge neutralization, electrostatic screening, and changes in activity coefficients presumably destabilize the largely charged/polar interactions that are required to form stable actin filaments (50), as expected from the interdependence of the various electrostatic interactions discussed above.
Salt effects on filament structure and mechanics
Actins are ∼375-amino-acid-long proteins that fold into a U-shaped structure containing four subdomains (SD1–4, Fig. 1) (22,51,52). SD1 contains both N- and C-termini. SD2 contains a structural element referred to as the DNase I binding loop (DB-loop, residues 36–52), which plays a regulatory role in actin filament structure and interactions with filament binding proteins. SD3 and SD4 form extensive contacts with the base and sugar-ring moieties of the bound nucleotide that sits in the cleft between the SD1–2 and SD3–4 lobes (Fig. 1).
Figure 1.
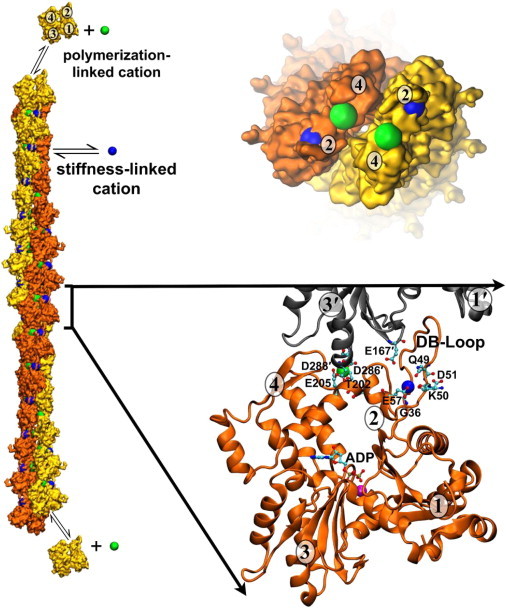
Location of two discrete, actin filament-specific cation-binding sites. The actin filament structure is based on the model of Fujii et al. (50) and includes the predicted cation-binding sites from Kang et al. (18). The barbed end of the filament is toward the bottom of the figure and the pointed end with associated cations is shown following a 90° rotation. (Yellow and orange) Surface rendering of actin monomers; (orange and gray) cartoon rendering of actin monomers. (Numbers in ovals) Actin monomer subdomains. (Green) Polymerization site cation; (blue) stiffness site cation; (magenta) nucleotide-associated cation. (Ball-and-stick representation) ADP nucleotide and specific cation-binding residues. To see this figure in color, go online.
Atomic models of actin filaments generated by x-ray fiber diffraction patterns of oriented filaments (53) or high-resolution cryo-electron microscopy analysis (50) generally agree on many aspects of the filament structure(s). Both models display a flattening of monomers upon incorporation into filaments and favor a very similar relative arrangement of actin subunits (Fig. 1). Although neither model has sufficient resolution to unambiguously place amino-acid side chains, both display encouraging agreement between specific regions that form extensive intersubunit contacts (Table 1) (50,53).
Table 1.
Amino-acid residues that comprise filament intersubunit contacts and discrete, filament-specific cation-binding sites
| Actin filament region | Residuesa | Neighbor subunit residues |
|---|---|---|
| DNase 1 binding loopb | 36–52 | |
| Lateral contactsb | 110–114, 263–273 | 39, 40, 173, 191–197, 202c |
| Longitudinal contactsb | 36–52 | 139, 140, 143, 166–169, 346, 351, 374 |
| Polymerization cation sited | 62, 63, 202–206, 208 | 285–288, 290 |
| Stiffness cation sited | 36–38, 49–54, 57, 58, 61, 64, 65 | 167 |
Italicized residues are common between intersubunit contacts and predicted discrete cation-binding sites.
Residues comprising the DNase I binding loop obtained from Holmes et al. (51). Contacts determined from actin filament models (50,53).
Two potential salt bridges may exist in the lateral intersubunit contacts: one between Glu270 and either Arg39 or Thr202, and another between Lys113 and Glu195.
These sites represent an exhaustive list of residues with at least one nonhydrogen atom within 7.0 Å of predicted bound cation positions (18).
The mechanical properties and large-scale structural dynamics of actin filaments are dictated by the distribution and strength of intersubunit contacts (54,55). Filaments assembled from purified actin in solution can grow several micrometers in length and behave as semiflexible polymers that undergo thermally driven bending (56,57) and twisting (58) motions. Single filament mechanical properties are well described by a bending persistence length (Lp), a torsional persistence length (LT), and a twist-bend coupling persistence length (LTB) (54).
Ionic solution conditions affect actin filament structure, intersubunit contacts, and mechanical properties. SD2 exists in ordered and disordered conformations (59) with a distribution that depends on the type and concentration of salt in solution (9,11). Salts also increase the bending stiffness of most characterized actin filaments (11,18). Because the DB-loop of SD2 participates in longitudinal intersubunit contacts (9,11,53,55,60–63) it has been hypothesized to link salt-dependent filament mechanics and structure (11,59).
Salt-dependent filament bundle formation and ABP interactions
Actin filaments are linear polyampholytes that display polyelectrolyte behavior (36). Accordingly, actin-filament bundling and ABP interactions are expected to be mediated by both global electrostatic screening effects and specific ion binding at discrete sites. Multivalent cation concentrations greater than required for polymerization (e.g., [Mg2+] > 10 mM) promote bundle- (17,36) or angle-layered aggregate (64) formation. Salt-induced actin filament bundling has been compared to that of DNA condensation, and interpreted according to the theory of linear polyelectrolytes (27,36,65). Interestingly, the actin bundling (crosslinking) protein smooth muscle calponin has been suggested to bundle filaments via nonspecific electrostatic interactions (36,37) similar to the process of counterion-mediated DNA condensation (12,66). However, the cation species- and concentration-dependence of bundle formation suggests that discrete and specific cation-binding interactions also contribute to salt-mediated filament bundling (17).
Interactions of nonbundling ABPs with actin filaments also display strong salt dependencies (e.g., cofilin, myosin, tropomyosin (38,67,68)), typically becoming weaker with salt. Established actin purification protocols utilize high salt concentrations to dissociate contaminating regulatory proteins (69). Although the salt-dependence of ABP binding is usually referred to as an ionic strength effect, cations can weaken ABP binding via direct competition or through conformationally linked equilibria. For example, binding of the filament-severing protein cofilin is linked to net cation release (∼1 Mg2+ per bound cofilin) (38).
Discrete Filament-Specific Cation-Binding Sites have Distinct Effects on Actin
Although we emphasize the interdependence of different electrolyte-driven phenomena in solution, at or near-physiological conditions, the salt-dependence of actin filament thermodynamic stability (Cc) and bending mechanics (Lp) are best described by discrete cation-binding interactions rather than general electrostatic screening effects (18). In other words, both screening and specific binding occur at physiological concentration ranges, but the latter dominates the salt-dependent actin filament assembly and mechanics. Filament Cc and Lp are both salt-dependent, but display sensitivities over divalent cation concentration regimes that differ by approximately an order of magnitude (Fig. 2). This behavior indicates that (at least) two distinct classes of cation-binding/interaction sites exist on actin: one class responds to Mg2+ at submillimolar concentration and drives polymerization, whereas the second class responds to millimolar concentration and stiffens filaments.
Figure 2.
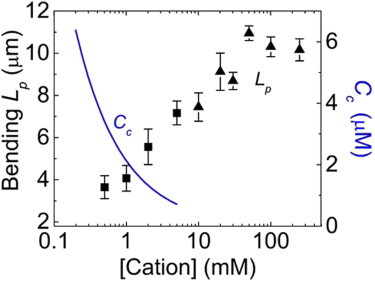
Two distinct classes of cation interaction sites exist on actin. One class responds to submillimolar cation concentrations and lowers Cc (blue; data shown for Mg2+). The second class responds to millimolar concentration and stiffens filaments as measured by Lp (black; squares represent Mg2+, triangles represent K+). Figure adapted from Kang et al. (18). To see this figure in color, go online.
Two discrete, filament-specific cation-binding sites were predicted by structural bioinformatics and verified with site-specific mutagenesis (18). Residues from two adjacent filament subunits form both binding sites, such that cations are coordinated by neighboring subunits (Fig. 1, Table 1). The two cation-binding sites are referred to as polymerization and stiffness sites according to their roles in actin assembly and filament bending mechanics, respectively (18).
Polymerization cation-binding site
The polymerization cation-binding site is positioned between SD3 and SD4 of longitudinally adjacent filament subunits (Fig. 1). Residues comprising the polymerization site (Fig.1, Table 1) are conserved among the vast majority of actins (70). Certain mutations within this predicted site are lethal in yeast (71), and other mutations at this specific subunit interface antagonize polymerization of Drosophila cytoplasmic actin (98.7% identical to human γ-cytoplasmic actin) (72). Mutation of a polymerization site residue (T203C) shifts the Mg2+-dependence of yeast actin Cc, consistent with a weaker Mg2+ binding affinity (18).
Cation binding to the polymerization site and subsequent filament assembly could potentially be modulated by posttranslational modifications and actin-binding proteins that perturb the site geometry or stability. For example, phosphorylation of the polymerization site at Thr202 and Thr203 (Figs. 1 and 3) dramatically disrupts actin polymerization (73–75), as would be expected if phosphorylation compromises cation-binding site integrity at the subunit interface. Accordingly, regulatory proteins or other factors that displace or weaken cation binding to the polymerization site, either through direct competition or reorganization of the binding site, are predicted to inhibit polymerization (Fig. 3).
Figure 3.
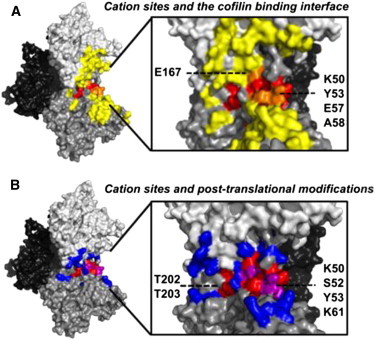
Actin regulatory protein-binding interfaces and posttranslational modification sites overlap with filament-specific cation sites. (A) Residues participating in the actin-cofilin interface (yellow) (78); (inset) 90° turn and zoom to cation sites where site residues in the cofilin binding interface are highlighted (orange). (B) Acetylation, ADP-ribosylation, arginylation, carbonylation, malonylation, methylation, nitrosylation, oxidation, phosphorylation, and ubiquitylation sites (83) are shown (blue) for longitudinal subunits; (inset) 90° turn and zoom to cation sites where modifiable site residues are highlighted (purple). To see this figure in color, go online.
Stiffness cation-binding site
The stiffness cation-binding site is comprised predominantly by residues within the DB-loop of SD2 but also includes glutamate at position 167 (Glu167) within SD3 of a neighboring subunit (Fig. 1, Table 1) (18). Most actins have Glu167 in the stiffness site, and in cases examined display salt-dependent filament-bending stiffness (18). Saccharomyces cerevisiae (yeast) actin has alanine at this position (Ala167 (70)), and filaments display a salt-independent bending stiffness that is more flexible than filaments with Glu167 (e.g., vertebrate actin) (76). Substitution of yeast Ala167 with glutamate confers filaments with salt-dependent stiffness (18), consistent with occupancy of the stiffness site dominating the observed salt effects on filament mechanics. These stiffening effects are thought to arise from stabilization of the SD2 conformation and DB-loop interactions with both the SD1–3 nonpolar cleft and the adjacent subunit loop containing Glu167 (18). Mutations adjacent to the stiffness site (Y166C) of human actin have been linked to hypertrophic cardiomyopathy (77). Although the biochemical and functional consequences of the Y166C mutation have not been evaluated, the observed phenotype raises the possibility that cation binding at the stiffness site plays a critical role in actin physiology, perhaps through modulation of mechanics and/or assembly.
Outlook: Cation Site Occupancy as a Tunable Actin-Regulatory Mechanism
ABP-linked cation binding
Many regulatory proteins bind filaments at the SD2-SD1/3 interface of adjacent subunits where the stiffness site is located (Fig. 1 and Table 1). Stiffness-site cation release can occur if considerable actin reorganization is linked to ABP binding (and/or if direct contacts are made with stiffness site residues). For example, the cofilin binding interface overlaps with the stiffness site (Fig. 3 A) (78) and binding is associated with net release of a single divalent cation (38). It has been suggested (18) that cofilin-mediated dissociation of the stiffness site cation is coupled to DB-loop reorganization (63,78,79), which lowers the radial distribution and density of filament intersubunit contacts (55,61), thereby enhancing filament fluctuations thought to promote severing (76,80–82). We emphasize that effects on filament mechanics resulting from stiffness-site cation dissociation and DB-loop reorganization may be compensated by stiffening effects when ABPs stably interact with one or more filament subunits.
Actin posttranslational modifications and cation binding
Actin posttranslational modifications (PTMs) potentially tune cation occupancy at the stiffness site, as inferred for phosphorylation of the polymerization site (see Polymerization Cation-Binding Site, above). Residues 50, 52, 53, and 61 of SD2 located at or near the stiffness site (Fig. 3 B) are targets of modification (83), including acetylation, phosphorylation, and ubiquitylation. Changes in filament mechanics and interactions with regulatory proteins are expected if PTMs disrupt site geometry. Evaluating the consequences of stiffness site PTMs on actin properties and the frequency with which PTMs vary among cell types in response to cellular stimuli may therefore reveal an additional cellular mechanism for spatial and temporal regulation of actin.
Cation-site occupancy and actin structural polymorphism
Actin filaments adopt multiple distinct structural states, and the models used to describe actin filaments represent only a subset of these states (84). For example, filaments in solution display variable twist and subunit tilt distributions that can vary with ABP occupancy (58,78,85). In addition, the conformation of the DB-loop in SD2 is also highly variable (60,61,62), and is linked to ABP interactions (61,78,79). This heterogeneity and plasticity in actin filament structure has been referred to as “actin structural polymorphism”.
The divalent cation-binding affinity at the stiffness site is in the low millimolar range (Fig. 2 (18)), comparable to physiological and commonly-used in vitro concentrations. Filament stiffness sites are partially saturated with divalent cation under these conditions, and filaments exist as a mixed population—some stiffness sites are (on average) occupied with divalent cation, whereas others are vacant. Physiological K+ concentrations and other cations provide additional stiffness-site occupancy in mixed salt solutions, such as cellular cytoplasm.
Structural heterogeneity among individual actin filaments, filament segments, and subunits could reflect partial cation occupancy of the stiffness site. ABP binding can influence the distribution of actin filament structures and actin-salt interactions, consistent with contributions from stiffness-site cation-linked equilibria. As such, it is important to assess actin structural polymorphism in the presence of stiffness-site-saturating divalent and monovalent cation concentrations.
Cation-binding and filament nucleation by Arp2/3
We put forth the hypothesis that the polymerization and stiffness cation-binding sites are conserved features utilized by the Arp2/3 complex to help template the daughter filament in Arp2/3-mediated actin filament nucleation (86,87). Using the web-based tool FATCAT (88), we generated an optimal structural alignment between the Bos taurus Arp2/3 actin-like subunits, Arp2 and Arp3 (86,87), and a subunit of vertebrate muscle actin (Fig. 4) in the F-actin conformation (50). Although Arp2 shares <50% sequence identity with actin, three barbed-end-facing residues defining the actin polymerization and stiffness cation-binding sites (Glu167, Asp286, Asp288) align almost perfectly with corresponding Arp2 residues (Fig. 4). Arp3 shares <40% sequence identity with actin, yet also aligns with two of the three conserved residues (Glu167 and Asp288; Fig. 4). Imperfect actin filament templating at the barbed end of inactive Arp3 (Pro286 instead of Asp) may be a feature allowing for regulation of Arp2/3 activation (see Fig. 4).
Figure 4.
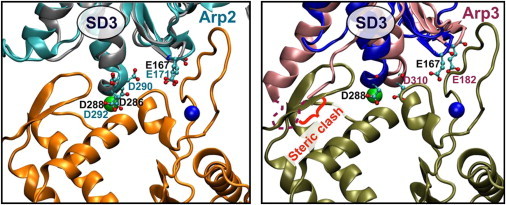
Conserved cation-binding residues between Arp2/3 and actin suggest salt-dependent regulation of Arp2/3 activation. Arp2 and Arp3 are overlaid with an actin subunit (best alignment calculated by the software FATCAT (88)) and interact with the next longitudinally neighboring subunit at the barbed-end face of Arp2/3. Actin subunits at the bottom of each panel represent the first two actin monomers that associate with Arp2/3 to nucleate the daughter filament at a branch point. Arp2 aligns very well with actin, especially the putative cation-binding residues E171, D290, and D292, which help form the polymerization and stiffness cation-binding sites with the incoming actin monomer. Arp3 does not align as well in the inactive crystal conformation. Arp2/3 activation is thought to require WASP/Scar-dependent conformational rearrangement of Arp3. We hypothesize that this rearrangement relieves a steric clash with the incoming actin monomer while forming a better cation-binding geometry at both the polymerization and stiffness cation-binding sites shared with the incoming daughter filament subunit. In this figure, pivoting of the Arp3 SD3 to the left would both alleviate the steric clash and place E182 and D310 into the proper position to bind interfacial cations. To see this figure in color, go online.
Cation binding at the Arp/actin interface could be coupled to Arp2/3 complex activation. An Arp3 conformational change is required to activate the Arp2/3 complex and nucleate a daughter filament (86). The resulting binding surface formed on the Arp3 barbed-end must rearrange relative to Arp2 to accommodate the first two daughter filament subunits. Discrete cation binding at the Arp/actin interface could stabilize activated Arp2/3 complex with incoming actin subunits in a manner similar to native actin filaments. Such a mechanism predicts that the Arp2/3-mediated actin nucleation and/or branching rates depend strongly on both the concentration and type of cations in solution.
Acknowledgments
This work was supported by National Institutes of Health grant No. RO1-GM097348 and American Heart Association Established Investigator Award No. 0655849T awarded to E.M.D.L.C.
Footnotes
Hyeran Kang and Michael J. Bradley contributed equally to this work.
References
- 1.Korn E.D., Carlier M.F., Pantaloni D. Actin polymerization and ATP hydrolysis. Science. 1987;238:638–644. doi: 10.1126/science.3672117. [DOI] [PubMed] [Google Scholar]
- 2.Pollard T.D., Borisy G.G. Cellular motility driven by assembly and disassembly of actin filaments. Cell. 2003;112:453–465. doi: 10.1016/s0092-8674(03)00120-x. [DOI] [PubMed] [Google Scholar]
- 3.Pollard T.D., Cooper J.A. Actin, a central player in cell shape and movement. Science. 2009;326:1208–1212. doi: 10.1126/science.1175862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pollard T.D., Blanchoin L., Mullins R.D. Molecular mechanisms controlling actin filament dynamics in nonmuscle cells. Annu. Rev. Biophys. Biomol. Struct. 2000;29:545–576. doi: 10.1146/annurev.biophys.29.1.545. [DOI] [PubMed] [Google Scholar]
- 5.Ditlev J.A., Mayer B.J., Loew L.M. There is more than one way to model an elephant. Experiment-driven modeling of the actin cytoskeleton. Biophys. J. 2013;104:520–532. doi: 10.1016/j.bpj.2012.12.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Estes J.E., Selden L.A., Gershman L.C. Tightly-bound divalent cation of actin. J. Muscle Res. Cell Motil. 1992;13:272–284. doi: 10.1007/BF01766455. [DOI] [PubMed] [Google Scholar]
- 7.Frieden C. The Mg2+-induced conformational change in rabbit skeletal muscle G-actin. J. Biol. Chem. 1982;257:2882–2886. [PubMed] [Google Scholar]
- 8.Strzelecka-Gołaszewska H., Pròchniewicz E., Drabikowski W. Interaction of actin with divalent cations. 2. Characterization of protein-metal complexes. Eur. J. Biochem. 1978;88:229–237. doi: 10.1111/j.1432-1033.1978.tb12442.x. [DOI] [PubMed] [Google Scholar]
- 9.Strzelecka-Golaszewska H., Wozniak A., Lindberg U. Effects of the type of divalent cation, Ca2+ or Mg2+, bound at the high-affinity site and of the ionic composition of the solution on the structure of F-actin. Biochem. J. 1996;316:713–721. doi: 10.1042/bj3160713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zimmerle C.T., Patane K., Frieden C. Divalent cation binding to the high- and low-affinity sites on G-actin. Biochemistry. 1987;26:6545–6552. doi: 10.1021/bi00394a039. [DOI] [PubMed] [Google Scholar]
- 11.Orlova A., Egelman E.H. A conformational change in the actin subunit can change the flexibility of the actin filament. J. Mol. Biol. 1993;232:334–341. doi: 10.1006/jmbi.1993.1393. [DOI] [PubMed] [Google Scholar]
- 12.Oosawa F., Kasai M. A theory of linear and helical aggregations of macromolecules. J. Mol. Biol. 1962;4:10–21. doi: 10.1016/s0022-2836(62)80112-0. [DOI] [PubMed] [Google Scholar]
- 13.Straub F.B., Feuer G. Adenosinetriphosphate the functional group of actin. Biochim. Biophys. Acta. 1950;4:455–470. [PubMed] [Google Scholar]
- 14.Estes J.E., Selden L.A., Gershman L.C. Tight binding of divalent cations to monomeric actin. Binding kinetics support a simplified model. J. Biol. Chem. 1987;262:4952–4957. [PubMed] [Google Scholar]
- 15.Gershman L.C., Selden L.A., Estes J.E. High affinity binding of divalent cation to actin monomer is much stronger than previously reported. Biochem. Biophys. Res. Commun. 1986;135:607–614. doi: 10.1016/0006-291x(86)90036-7. [DOI] [PubMed] [Google Scholar]
- 16.Rich S.A., Estes J.E. Detection of conformational changes in actin by proteolytic digestion: evidence for a new monomeric species. J. Mol. Biol. 1976;104:777–792. doi: 10.1016/0022-2836(76)90181-9. [DOI] [PubMed] [Google Scholar]
- 17.Strzelecka-Gołaszewska H., Pròchniewicz E., Drabikowski W. Interaction of actin with divalent cations. 1. The effect of various cations on the physical state of actin. Eur. J. Biochem. 1978;88:219–227. doi: 10.1111/j.1432-1033.1978.tb12441.x. [DOI] [PubMed] [Google Scholar]
- 18.Kang H., Bradley M.J., De La Cruz E.M. Identification of cation-binding sites on actin that drive polymerization and modulate bending stiffness. Proc. Natl. Acad. Sci. USA. 2012;109:16923–16927. doi: 10.1073/pnas.1211078109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carlier M.F. Actin: protein structure and filament dynamics. J. Biol. Chem. 1991;266:1–4. [PubMed] [Google Scholar]
- 20.Selden L.A., Estes J.E., Gershman L.C. High affinity divalent cation binding to actin. Effect of low affinity salt binding. J. Biol. Chem. 1989;264:9271–9277. [PubMed] [Google Scholar]
- 21.Martonosii A., Molino C.M., Gergely J. The binding of divalent cations to actin. J. Biol. Chem. 1964;239:1057–1064. [PubMed] [Google Scholar]
- 22.Otterbein L.R., Graceffa P., Dominguez R. The crystal structure of uncomplexed actin in the ADP state. Science. 2001;293:708–711. doi: 10.1126/science.1059700. [DOI] [PubMed] [Google Scholar]
- 23.Dill K.A., Bromberg S., Stigter D. Garland Science; New York: 2011. Molecular Driving Forces: Statistical Thermodynamics in Biology, Chemistry, Physics, and Nanoscience. [Google Scholar]
- 24.Phillips R., Kondev J., Chasan B. 2nd Ed. Garland Science; New York: 2012. Physical Biology of the Cell. [Google Scholar]
- 25.Robinson R.A., Stokes R.H. Butterworths; London, UK: 1959. Electrolyte Solutions. [Google Scholar]
- 26.Israelachvili J.N. 3rd Ed. Academic Press; Waltham, MA: 2011. Intermolecular and Surface Forces. [Google Scholar]
- 27.Manning G.S. The molecular theory of polyelectrolyte solutions with applications to the electrostatic properties of polynucleotides. Q. Rev. Biophys. 1978;11:179–246. doi: 10.1017/s0033583500002031. [DOI] [PubMed] [Google Scholar]
- 28.Oosawa F. Marcel Dekker; New York: 1971. Polyelectrolytes. [Google Scholar]
- 29.Atkins P.W. Oxford University Press; Oxford, UK: 1982. Physical Chemistry. [Google Scholar]
- 30.Draper D.E. A guide to ions and RNA structure. RNA. 2004;10:335–343. doi: 10.1261/rna.5205404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Record M.T., Jr., Lohman M.L., De Haseth P.L. Ion effects on ligand-nucleic acid interactions. J. Mol. Biol. 1976;107:145–158. doi: 10.1016/s0022-2836(76)80023-x. [DOI] [PubMed] [Google Scholar]
- 32.Record M.T., Jr., Anderson C.F., Lohman T.M. Thermodynamic analysis of ion effects on the binding and conformational equilibria of proteins and nucleic acids: the roles of ion association or release, screening, and ion effects on water activity. Q. Rev. Biophys. 1978;11:103–178. doi: 10.1017/s003358350000202x. [DOI] [PubMed] [Google Scholar]
- 33.Liu T., Altman R.B. Prediction of calcium-binding sites by combining loop-modeling with machine learning. BMC Struct. Biol. 2009;9:72. doi: 10.1186/1472-6807-9-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nayal M., Di Cera E. Predicting Ca2+-binding sites in proteins. Proc. Natl. Acad. Sci. USA. 1994;91:817–821. doi: 10.1073/pnas.91.2.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zheng H., Chruszcz M., Minor W. Data mining of metal ion environments present in protein structures. J. Inorg. Biochem. 2008;102:1765–1776. doi: 10.1016/j.jinorgbio.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tang J.X., Janmey P.A. The polyelectrolyte nature of F-actin and the mechanism of actin bundle formation. J. Biol. Chem. 1996;271:8556–8563. doi: 10.1074/jbc.271.15.8556. [DOI] [PubMed] [Google Scholar]
- 37.Tang J.X., Szymanski P.T., Tao T. Electrostatic effects of smooth muscle calponin on actin assembly. Eur. J. Biochem. 1997;247:432–440. doi: 10.1111/j.1432-1033.1997.00432.x. [DOI] [PubMed] [Google Scholar]
- 38.Cao W.X., Goodarzi J.P., De La Cruz E.M. Energetics and kinetics of cooperative cofilin-actin filament interactions. J. Mol. Biol. 2006;361:257–267. doi: 10.1016/j.jmb.2006.06.019. [DOI] [PubMed] [Google Scholar]
- 39.Skillman K.M., Ma C.I., Sibley L.D. The unusual dynamics of parasite actin result from isodesmic polymerization. Nat. Commun. 2013;4:2285. doi: 10.1038/ncomms3285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yarmola E.G., Bubb M.R. Profilin: emerging concepts and lingering misconceptions. Trends Biochem. Sci. 2006;31:197–205. doi: 10.1016/j.tibs.2006.02.006. [DOI] [PubMed] [Google Scholar]
- 41.Oda T., Makino K., Maéda Y. Distinct structural changes detected by x-ray fiber diffraction in stabilization of F-actin by lowering pH and increasing ionic strength. Biophys. J. 2001;80:841–851. doi: 10.1016/S0006-3495(01)76063-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rouayrenc J.F., Travers F. The first step in the polymerization of actin. Eur. J. Biochem. 1981;116:73–77. doi: 10.1111/j.1432-1033.1981.tb05302.x. [DOI] [PubMed] [Google Scholar]
- 43.Tellam R. Mechanism of CaCl2-induced actin polymerization. Biochemistry. 1985;24:4455–4460. doi: 10.1021/bi00337a029. [DOI] [PubMed] [Google Scholar]
- 44.Frieden C. Polymerization of actin: mechanism of the Mg2+-induced process at pH 8 and 20°C. Proc. Natl. Acad. Sci. USA. 1983;80:6513–6517. doi: 10.1073/pnas.80.21.6513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cooper J.A., Pollard T.D. Effect of capping protein on the kinetics of actin polymerization. Biochemistry. 1985;24:793–799. doi: 10.1021/bi00324a039. [DOI] [PubMed] [Google Scholar]
- 46.Vavylonis D., Yang Q., O’Shaughnessy B. Actin polymerization kinetics, cap structure, and fluctuations. Proc. Natl. Acad. Sci. USA. 2005;102:8543–8548. doi: 10.1073/pnas.0501435102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Doi Y., Frieden C. Actin polymerization. The effect of brevin on filament size and rate of polymerization. J. Biol. Chem. 1984;259:11868–11875. [PubMed] [Google Scholar]
- 48.Nagy B., Jencks W.P. Depolymerization of F-actin by concentrated solutions of salts and denaturing agents. J. Am. Chem. Soc. 1965;87:2480–2488. doi: 10.1021/ja01089a030. [DOI] [PubMed] [Google Scholar]
- 49.Pinder J.C., Sleep J.A., Gratzer W.B. Concentrated Tris solutions for the preparation, depolymerization, and assay of actin: application to erythroid actin. Anal. Biochem. 1995;225:291–295. doi: 10.1006/abio.1995.1157. [DOI] [PubMed] [Google Scholar]
- 50.Fujii T., Iwane A.H., Namba K. Direct visualization of secondary structures of F-actin by electron cryomicroscopy. Nature. 2010;467:724–728. doi: 10.1038/nature09372. [DOI] [PubMed] [Google Scholar]
- 51.Holmes K.C., Popp D., Kabsch W. Atomic model of the actin filament. Nature. 1990;347:44–49. doi: 10.1038/347044a0. [DOI] [PubMed] [Google Scholar]
- 52.Dominguez R., Holmes K.C. Actin structure and function. Annu. Rev. Biophys. 2011;40:169–186. doi: 10.1146/annurev-biophys-042910-155359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Oda T., Iwasa M., Narita A. The nature of the globular- to fibrous-actin transition. Nature. 2009;457:441–445. doi: 10.1038/nature07685. [DOI] [PubMed] [Google Scholar]
- 54.De La Cruz E.M., Roland J., Martiel J.L. Origin of twist-bend coupling in actin filaments. Biophys. J. 2010;99:1852–1860. doi: 10.1016/j.bpj.2010.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fan J., Saunders M.G., Voth G.A. Molecular origins of cofilin-linked changes in actin filament mechanics. J. Mol. Biol. 2013;425:1225–1240. doi: 10.1016/j.jmb.2013.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Oosawa F. The flexibility of F-actin. Biophys. Chem. 1980;11:443–446. doi: 10.1016/0301-4622(80)87021-9. [DOI] [PubMed] [Google Scholar]
- 57.Yanagida T., Nakase M., Oosawa F. Direct observation of motion of single F-actin filaments in the presence of myosin. Nature. 1984;307:58–60. doi: 10.1038/307058a0. [DOI] [PubMed] [Google Scholar]
- 58.Prochniewicz E., Zhang Q., Thomas D.D. Cooperativity in F-actin: binding of gelsolin at the barbed end affects structure and dynamics of the whole filament. J. Mol. Biol. 1996;260:756–766. doi: 10.1006/jmbi.1996.0435. [DOI] [PubMed] [Google Scholar]
- 59.Durer Z.A.O., Kudryashov D.S., Reisler E. Structural states and dynamics of the D-loop in actin. Biophys. J. 2012;103:930–939. doi: 10.1016/j.bpj.2012.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chu J.W., Voth G.A. Coarse-grained modeling of the actin filament derived from atomistic-scale simulations. Biophys. J. 2006;90:1572–1582. doi: 10.1529/biophysj.105.073924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pfaendtner J., De La Cruz E.M., Voth G.A. Actin filament remodeling by actin depolymerization factor/cofilin. Proc. Natl. Acad. Sci. USA. 2010;107:7299–7304. doi: 10.1073/pnas.0911675107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Saunders M.G., Voth G.A. Comparison between actin filament models: coarse-graining reveals essential differences. Structure. 2012;20:641–653. doi: 10.1016/j.str.2012.02.008. [DOI] [PubMed] [Google Scholar]
- 63.Bobkov A.A., Muhlrad A., Reisler E. Structural effects of cofilin on longitudinal contacts in F-actin. J. Mol. Biol. 2002;323:739–750. doi: 10.1016/s0022-2836(02)01008-2. [DOI] [PubMed] [Google Scholar]
- 64.Egelman E.H., Francis N., DeRosier D.J. Helical disorder and the filament structure of F-actin are elucidated by the angle-layered aggregate. J. Mol. Biol. 1983;166:605–629. doi: 10.1016/s0022-2836(83)80286-1. [DOI] [PubMed] [Google Scholar]
- 65.Manning G.S. Limiting laws and counterion condensation in polyelectrolyte solutions I. Colligative properties. J. Chem. Phys. 1969;51:924–933. [Google Scholar]
- 66.Manning G.S. Counterion binding in polyelectrolyte theory. Acc. Chem. Res. 1979;12:443–449. [Google Scholar]
- 67.Eaton B.L., Kominz D.R., Eisenberg E. Correlation between the inhibition of the acto-heavy meromyosin ATPase and the binding of tropomyosin to F-actin: effects of Mg2+ ion, potassium chloride troponin I, and troponin C. Biochemistry. 1975;14:2718–2725. doi: 10.1021/bi00683a025. [DOI] [PubMed] [Google Scholar]
- 68.Tonomura Y., Tokura S., Sekiya K. Binding of myosin A to F-actin. J. Biol. Chem. 1962;237:1074–1081. [PubMed] [Google Scholar]
- 69.Pardee J.D., Spudich J.A. Purification of muscle actin. Methods Cell Biol. 1982;24:271–289. doi: 10.1016/s0091-679x(08)60661-5. [DOI] [PubMed] [Google Scholar]
- 70.Sheterline P., Clayton J., Sparrow J. Academic Press; London, UK: 1996. Actins. [Google Scholar]
- 71.Wertman K.F., Drubin D.G., Botstein D. Systematic mutational analysis of the yeast ACT1 gene. Genetics. 1992;132:337–350. doi: 10.1093/genetics/132.2.337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rould M.A., Wan Q., Trybus K.M. Crystal structures of expressed non-polymerizable monomeric actin in the ADP and ATP states. J. Biol. Chem. 2006;281:31909–31919. doi: 10.1074/jbc.M601973200. [DOI] [PubMed] [Google Scholar]
- 73.Sonobe S., Takahashi S., Kuroda K. Phosphorylation of Amoeba G-actin and its effect on actin polymerization. J. Biol. Chem. 1986;261:14837–14843. [PubMed] [Google Scholar]
- 74.Furuhashi K., Hatano S., Inagaki M. Phosphorylation by actin kinase of the pointed end domain on the actin molecule. J. Biol. Chem. 1992;267:9326–9330. [PubMed] [Google Scholar]
- 75.Gettemans J., De Ville Y., Waelkens E. Physarum actin is phosphorylated as the actin-fragmin complex at residues Thr203 and Thr202 by a specific 80 kDa kinase. EMBO J. 1992;11:3185–3191. doi: 10.1002/j.1460-2075.1992.tb05395.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.McCullough B.R., Grintsevich E.E., De La Cruz E.M. Cofilin-linked changes in actin filament flexibility promote severing. Biophys. J. 2011;101:151–159. doi: 10.1016/j.bpj.2011.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Müller M., Mazur A.J., Mannherz H.G. Functional characterization of the human α-cardiac actin mutations Y166C and M305L involved in hypertrophic cardiomyopathy. Cell. Mol. Life Sci. 2012;69:3457–3479. doi: 10.1007/s00018-012-1030-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Galkin V.E., Orlova A., Egelman E.H. Remodeling of actin filaments by ADF/cofilin proteins. Proc. Natl. Acad. Sci. USA. 2011;108:20568–20572. doi: 10.1073/pnas.1110109108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Muhlrad A., Kudryashov D., Reisler E. Cofilin induced conformational changes in F-actin expose subdomain 2 to proteolysis. J. Mol. Biol. 2004;342:1559–1567. doi: 10.1016/j.jmb.2004.08.010. [DOI] [PubMed] [Google Scholar]
- 80.McCullough B.R., Blanchoin L., De la Cruz E.M. Cofilin increases the bending flexibility of actin filaments: implications for severing and cell mechanics. J. Mol. Biol. 2008;381:550–558. doi: 10.1016/j.jmb.2008.05.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.De La Cruz E.M. How cofilin severs an actin filament. Biophys. Rev. 2009;1:51–59. doi: 10.1007/s12551-009-0008-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Elam W.A., Kang H., De La Cruz E.M. Biophysics of actin filament severing by cofilin. FEBS Lett. 2013;587:1215–1219. doi: 10.1016/j.febslet.2013.01.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Terman J.R., Kashina A. Post-translational modification and regulation of actin. Curr. Opin. Cell Biol. 2013;25:30–38. doi: 10.1016/j.ceb.2012.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Galkin V.E., Orlova A., Egelman E.H. Structural polymorphism in F-actin. Nat. Struct. Mol. Biol. 2010;17:1318–1323. doi: 10.1038/nsmb.1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Prochniewicz E., Janson N., De La Cruz E.M. Cofilin increases the torsional flexibility and dynamics of actin filaments. J. Mol. Biol. 2005;353:990–1000. doi: 10.1016/j.jmb.2005.09.021. [DOI] [PubMed] [Google Scholar]
- 86.Nolen B.J., Pollard T.D. Insights into the influence of nucleotides on actin family proteins from seven structures of Arp2/3 complex. Mol. Cell. 2007;26:449–457. doi: 10.1016/j.molcel.2007.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Robinson R.C., Turbedsky K., Pollard T.D. Crystal structure of Arp2/3 complex. Science. 2001;294:1679–1684. doi: 10.1126/science.1066333. [DOI] [PubMed] [Google Scholar]
- 88.Ye Y., Godzik A. Flexible structure alignment by chaining aligned fragment pairs allowing twists. Bioinformatics. 2003;19(Suppl 2):ii246–ii255. doi: 10.1093/bioinformatics/btg1086. [DOI] [PubMed] [Google Scholar]


