Abstract
RNA interference (RNAi) has opened promising avenues to better understand gene function. Though many RNAi screens report on the identification of genes, very few, if any, have been further studied and validated. Data discrepancy is emerging as one of RNAi main pitfalls. We reasoned that a systematic analysis of lethality-based screens, since they score for cell death, would examine the extent of hit discordance at inter-screen level. To this end, we developed a methodology for literature mining and overlap analysis of several screens using both siRNA and shRNA flavors, and obtained 64 gene lists censoring an initial list of 7,430 nominated genes. We further performed a comparative analysis first at a global level followed by hit re-assessment under much more stringent conditions. To our surprise, none of the hits overlapped across the board even for PLK1, which emerged as a strong candidate in siRNA screens; but only marginally in the shRNA ones. Furthermore, EIF5B emerges as the most common hit only in the shRNA screens. A highly unusual and unprecedented result was the observation that 5,269 out of 6,664 nominated genes (~80%) in the shRNA screens were exclusive to the pooled format, raising concerns as to the merits of pooled screens which qualify hits based on relative depletions, possibly due to multiple integrations per cell, data deconvolution or inaccuracies in intracellular processing causing off-target effects. Without golden standards in place, we would encourage the community to pay more attention to RNAi screening data analysis practices, bearing in mind that it is combinatorial in nature and one active siRNA duplex or shRNA hairpin per gene does not suffice credible hit nomination. Finally, we also would like to caution interpretation of pooled shRNA screening outcomes.
Keywords: RNAi, shRNA, siRNA, Gene, screening, bioinformatics, analysis, overlap, lethality, essential, PLK1
INTRODUCTION
RNAi as a screening technology platform has opened the door to functional genomics screen approaches to discover novel, validate current targets, or merely to elucidate gene function within pathways and signaling networks. 1 This has allowed scientists to perform up to genome-scale simultaneous gene knockdowns at an industrial scale against arrayed or pooled RNAi libraries, and with a battery of assay readouts ranging from simple cell metabolic measurements to highly sophisticated deep sequencing methods; as such many mammalian RNAi screens have been conducted to date and reported in various fields with a bias for novel target discovery in cancer biology and host-virus interactions. 2–3
Despite such an endeavor, concerns over hit identification in random RNAi screening have recently been raised; 4–8 mainly due to the continuous lack of reproducibility and follow up on published gene hits and in some cases the unavailability of raw screening data for evaluation post-publication. The problems associated with false discovery are not that uncommon, especially when considering the facts that RNAi is a relatively new screening platform. However, the only unusual thing about it is the total lack of golden standards for the technology; this is a surprise and a growing concern considering its long implementation by a variety of research laboratories yielding ~ 300 published mammalian RNAi screens. In a recent review, Mohr and colleagues tackled the very same issues of false discovery and linked them to problems such as bad instrumentation, poor assay design, noise in large data sets, and/or RNAi reagent design as the main culprits. 9 Though they may well be the culprits in some of reports as they would totally affect RNAi screening outcome, they certainly do not explain false discoveries in cases where despite the use of identical RNAi libraries, same assay technologies, same cell lines, and same screeners, published lists of hit genes were found to be different. 10–13 One potential explanation for such divergent data sets originating from the same screening groups could be data transformation and analysis. 8 For this purpose we developed and implemented the Bhinder-Djaballah Analysis (BDA) method; introducing the H score as a new measure for gene activity as a total active RNAi duplex per gene independent of the assay readout. 14
A higher variability in reported hits from RNAi screens, more so, for the pooled shRNA hairpin screens can be explained at four levels; 1) efficiency of genomic integration at the cellular entry level, 2) multiplicity of integration of plasmids encoding hairpins, 3) efficiency of transcription of the DNA plasmids, and 4) precision of intracellular cleavage of the transcribed oligonucleotide at the hairpin-level processing. The expression-vector based delivery of shRNA hairpins into the cell necessitates their intracellular cleavage by Dicer to yield a functional guide strand, a process similar to miRNA biogenesis. Gu and co-workers have recently demonstrated dicer specificity and cleavage to play a far more crucial role than previously perceived; a northern blot on dicer cleaved strands of exogenous hairpins (with a miR-30 backbone) revealed a high degree of heterogeneity at the cleavage sites, resulting in multiple fragments. 15 An inaccurate Dicer-mediated hairpin processing would shift the guide sequence and consequentially the pivotal seed region from the expected, leading to the prominence of off targeted transcripts. This characteristic is distinctive for the shRNA hairpins screens in comparison to the siRNA duplex screens, intuitively making the former prone to elevated levels of inherent noise in data outputs, questioning the true merits of gene targets identified through this process. Implementation of an activity threshold measure like the H score for hit re-evaluation would facilitate highest levels of stringency allowing for standardized cross study comparisons.
We took advantage of the observations that several published lethality-based RNAi screens have resulted in different active gene sets, to assess the feasibility of identifying genes for cellular viability and to systematically study the extent of their overlap. Our reasoning was simply that these combined efforts must have identified the same essential genes since the readout is cell death in all cases. For this purpose, we surveyed the literature for published RNAi screens and collected a final set of 64 gene lists corresponding to 30 representative studies, comprised of both the siRNA duplex and the shRNA hairpin technologies. We censored an initial hit list of 7,430 nominated genes resulting from an arsenal of different analysis methods utilized by the different groups to score hits in genome-wide as well as focused screening data sets. Considering a conservative estimate of at least 25,000 genes in the human genome, it is rather surprising that 30% of them seem to be required for cellular viability and survival.
In this report, we describe and implement a strategy used for literature mining and systematic overlaps inclusive of a global overview of published data outputs followed by their subjection to hit reassessment through stringent filtering approaches. Before conducting the overlaps, we segregated the gene lists for analysis into siRNA duplex and shRNA hairpin screens, and amongst them, we analyzed the genome-wide and focused screens separately. Surprisingly, none of the genes were found in common across the board and some of the major players of cellular viability were missing. PLK1 was identified as a prominent gene candidate in the systematic analysis of siRNA duplex gene lists, its overlap in the shRNA hairpin gene lists exhibited a marginal presence. A translation factor, EIF5B, dominates the overlap results for lethality from shRNA hairpin screens. An important facet of our findings was the discovery of an unprecedented enrichment of hits exclusively found in pooled shRNA hairpin screens at all stages of the analysis; ~ 80% of the gene candidates at the level of global overlaps and ~ 60% of the gene candidates at the level of stringent overlaps. Our efforts here emphasize dismal reproducibility of RNAi screening data outputs with zero commonality across ~147 distinct lethality gene lists, while showcasing a compelling prominence of gene candidates resulting from relative hairpin depletions in pooled shRNA screens, questioning the merits of this technology in gene discovery and validation.
MATERIALS AND METHODS
Literature mining
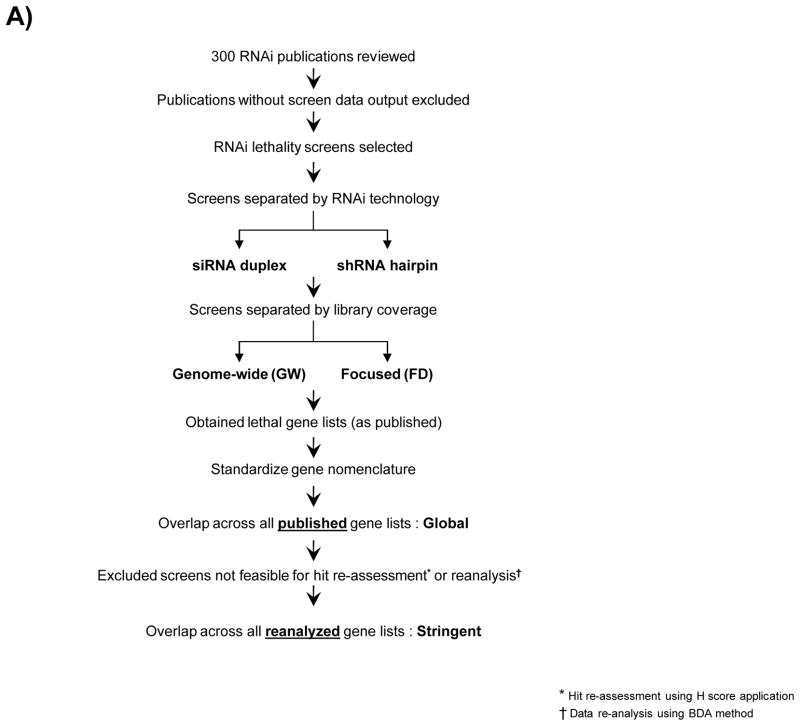
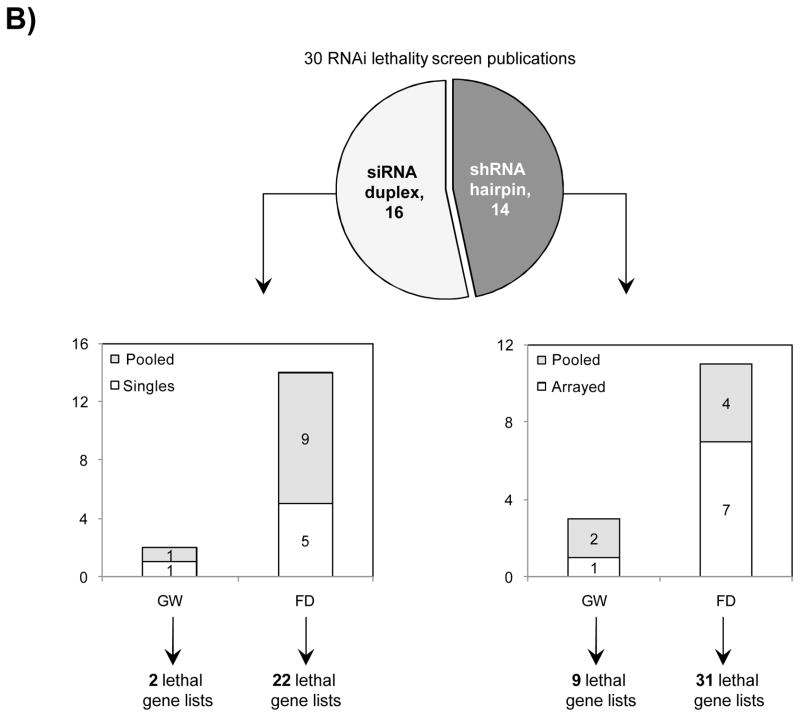
Representative RNAi screening publications reporting genes with a role in cell survival were collected from PubMed (www.ncbi.nlm.nih.gov/pubmed). 30 RNAi publications on cell lethality were selected representing 14 shRNA hairpin and 16 siRNA duplex library screens, performed in arrayed or pooled formats, 5 were genome-wide and 25 were focused library screens (Fig 1A, Suppl Table 1). 10–13, 16–41 The criterion for selection of a publication was based on the availability of data sets pertaining to active gene candidates identified from each RNAi screen. 64 gene lists corresponded to the multiple cell lines screened in the 30 selected publications were obtained.
Figure 1. Workflow of literature mining and data collection from published RNAi screens and overlaps analysis.
A) Workflow for two-step overlaps analysis between gene lists from literature mining and reference screen. B) Breakdown of selected 30 publications based on the RNAi technology used and library coverage.
Reanalysis of published screen data sets
The TRC library performance data for three primary screens was obtained from The Broad Institute website (www.broadinstitute.org); two were pooled shRNA hairpin screens performed by Cheung and co-workers, and Luo and co-workers; while the third was an arrayed shRNA hairpin screen performed by Barbie and co-workers. 17, 25, 11 The data sets for 102 cell lines from Cheung and 19 cell lines from Barbie were analyzed using the BDA method as previously described. 14 For the screens conducted by Luo, data sets were available for 12 cell lines screened as well as the control library DNA plasmid pool in replicates of ten, which were averaged for the purpose of the reanalysis and each cell line was separately analyzed by the BDA method for hit nomination. Briefly, the depletion of individual shRNA hairpins was quantified as fold change (FC) in signal intensity relative to the control pool and active duplexes were identified below a threshold determined at −2 σ from the μ FC of the library hairpins. In the next step, active genes were identified using an H score of ≥ 60 and instances where greater than 5 hairpins targeting a gene were found, a p-value of < 0.05 was considered. This was followed by OTE filtering and re-scoring to obtain a final set of nominated gene candidates for each of the 12 different cell lines. Briefly, the reanalysis of published screen data sets provided us with 133 additional gene lists.
Overlap analysis
To perform a comparison, the 64 gene lists were first divided based on the RNAi technology used- siRNA duplex versus shRNA hairpin. Within these two classes, the gene list were grouped based on the library coverage, broadly categorized as genome-wide or focused and the overlap was conducted separately for the two categories and an overlap between them was also determined towards the end. (Fig 1B). To be included in the overlap analysis, a gene had to be nominated in at least one report meaning all genes reported thus far were included. The overlap was conducted in two steps: First, all genes selected from the literature were consolidated to generate a unique list of genes with a role in cell lethality, therefore global overlap, and second, subset of genes with H score of ≥60, translating into minimal 2 active siRNA duplexes or minimal 3 active shRNA hairpins, were compared, therefore stringent overlap (Fig 1A). 14 H score is defined as the ratio of active duplexes targeting a gene and its corresponding total duplexes in the screening library, multiplied by 100. 14 Reports that did not include the information on individual shRNA hairpins or siRNA duplexes performance were automatically excluded from the stringent overlap analysis. H score was calculated for the gene lists where the information on the total number of hairpins screened was made available. In addition, the BDA method was applied in instances where the raw screen data was publically available and the 133 new gene lists were also included for stringent overlap. For the purpose of active gene candidate nomination in supplementary data provided for 6K and 10K pooled hairpin screens performed by Silva and co-workers 27, the fold change values given in a linear scale were first converted to ratios and numbers < 0 were assigned a negative sign. The fold change values given in log scale were converted to linear fold change values before their conversion to ratios. The hits selection criteria used here is consistent with the one specified by the authors in the report; a fold change of greater than 2 and an false discovery rate of < 10% was considered to determine active hairpins in the screens. These hits were included as such in the global overlap and H score filtering was applied for their inclusion in stringent overlap. Overlap analyses were performed using Perl scripts.
Biological Classifications
Information regarding the functional classes associated with the identified hits in each category were obtained from DAVID (the Database for Annotation, Visualization and Integrated Discovery) Functional Annotation Tool (www.david.abcc.ncifcrf.gov/) 42 and PANTHER (Protein ANalysis THrough Evolutionary Relationships) Classification System. 43 Threshold for statistical level of significance was determined at a p-value of < 0.05. To address the issue of gene synonyms and multiple RefSeq IDs, gene symbols were standardized to the nomenclature used in the TRC1 shRNA hairpin library. RefSeq IDs for the genes not covered in the TRC1 library were obtained from PubMed (www.ncbi.nlm.nih.gov/pubmed) and GeneGo’s Metacore pathway analysis software (www.genego.com/metacore.php). Gene name synonyms and overlapping RefSeq IDs were identified using GeneGo’s Metacore pathway analysis software (www.genego.com/metacore.php). Data on gene names and descriptions was obtained from PubMed (www.ncbi.nlm.nih.gov/pubmed) and UniProt (www.uniprot.org/). 44
RESULTS
RNAi Literature mining to select published gene candidates for cell survival
We reviewed ~ 300 published RNAi screening reports in order to select only those studies screening for genes required for cell survival. In selecting the published reports, we included screens that were not only reflective of genome-wide libraries but also those performed against large-scale and small focused library sets using the arrayed as well as the pooled formats (Fig 1A). After eliminating those publications which did not report on the final list of obtained active genes, the selection was narrowed down to only 30 RNAi reports; 16 of which were performed using siRNA duplex libraries while the remaining 14 where performed using shRNA hairpin libraries comprised of varying collection of genes (Suppl Table 1). 10–13, 16–41 In general, a preference towards using focused screening libraries was observed; 88% of the siRNA duplex screens and 79% of the shRNA hairpin screens were performed using the focused libraries (Fig 1B). Here, we also noticed lack of standardized methods of hit nomination across the board. Majority of the shRNA hairpin pooled screens were analyzed based on fold depletions per hairpin, however the threshold determination was inconsistent across the board. Other commonly used methods for data analysis were found to be growth inhibition, B score; while the remainder used z score, MAD score and RIGER to name a few. 14
For the purpose of the comparative study and hit list consolidation across multiple published RNAi screening data outputs; we first acquired gene lists as provided in the 30 selected publications (Suppl Table 1). Gene lists per each cell line screened from the selected publication were treated as separate for the purpose of a global overlap analysis; yielding a total of 64 individual gene lists from the 30 publications (Fig 1B). Of note, multiple screens provided combined lists of hits for groups of cell lines and therefore such gene lists could not be deconvoluted and were determined to be a single composite gene list in the overlap analysis. Furthermore, a gene reported minimally once in any one of the selected 64 gene lists was considered. Finally, cross gene list overlaps revealed a list of 7,430 unique gene candidates with varying degree of overlap, 62% of which were nominated in a maximum of one gene list and therefore termed as orphan hits. The list of gene candidates was dominated by hits obtained exclusively from the pooled shRNA formats, constituting 73% of the total with a distribution ratio of roughly 1:11 between siRNA duplex and shRNA hairpin gene lists.
We designed and followed a step-by-step workflow to conduct a comparative analysis by first segregating the screens into two broad categories at the level of the RNAi technology used, namely, the siRNA duplex and the shRNA hairpin screens. To eliminate any bias introduced due to the variations in the magnitude of genes screened, we further separated the screens being compared into genome-wide or focused based on the library coverage corresponding to each of the published screen. After these two levels of demarcations, the comparison was conducted separately for each defined category as: 1) Global, to merge all published data so as to gain a comprehensive overview of what has been reported in literature, and 2) Stringent, to re-asses the published hits using a filtering criteria concerning minimal duplex activities so as to obtain a standardized list of common genes with a likely role in cell survival (Fig 1A).
siRNA duplex screens: Global overlaps reveal PLK1 as a top scoring gene candidate
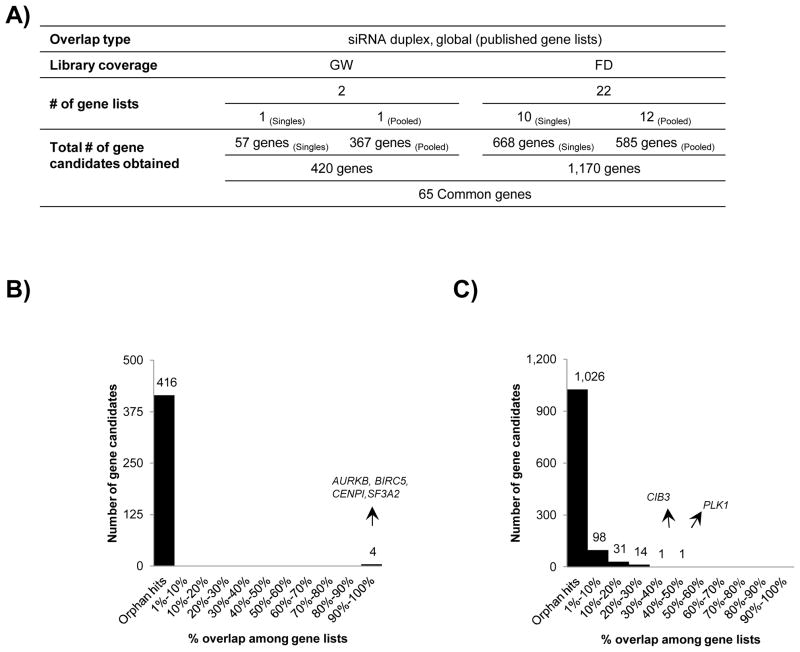
For the purpose of global overlap, we reviewed 16 siRNA duplex screens in total, 2 were genome-wide while the remaining 14 were focused (Fig 1B). As for screening formats, siRNA screens were broadly classified into two categories that of singles, which refer to one duplex per well approach and pooled, which refer to one gene per well approach. In general, the pooled screening formats were comprised of ~ 3 duplexes per well targeting one gene. Of note, 10 out of 16 screens were performed using a pooled approach, reflective of a general preference towards this category (Fig 1B). As a first step of the analysis, we obtained 2 lethal gene lists corresponding to the 2 genome-wide siRNA duplex screens; one screen was conducted in a singles format while the other one was conducted in a pooled format (Fig 2A). We obtained a set of 420 gene candidates in total and found that 88% of the gene candidates originated form pooled siRNA duplex screens (Suppl Table 2). A majority 416 of the gene candidates was associated with only a single gene list and was therefore termed as orphan hits (Fig 2B). Taken together, only 4 gene candidates, namely, AURKB, BIRC5, CENPI, and SF3A2 overlapped among the two gene lists being compared. Interestingly, the first three genes are components of the cell cycle while SF3A2 is an mRNA splicing factor (Table 1).
Figure 2. Global overlap of 24 gene lists procured by literature mining for siRNA duplex screens.
A) Systematic workflow of global overlap for selection of 420 gene candidates from 2 genome-wide and 1,170 gene candidates from 22 focused screen gene lists followed identification of 65 genes in common between the two categories. B) Breakup of number of overlapping genes in genome-wide screens based on their participation in number of gene lists. C) Breakup of number of overlapping genes in focused screens based on their participation in number of gene lists. The degree of overlap is represented as percent of total gene lists selected.
Table 1.
Top scoring 15 representative gene candidates from global overlap in siRNA duplex screens
| Coverage | Gene | Name | Overlap1 | Biological process |
|---|---|---|---|---|
| GW | AURKB* | Aurora kinase B | 2/2 | Role in chromosome segregation |
| BIRC5 | Baculoviral IAP 2 repeat containing 5 | 2/2 | Role in inhibition of apoptosis | |
| SF3A2 | Splicing factor 3a | 2/2 | mRNA splicing | |
| CENPI | Centromere protein I | 2/2 | Nucleosome assembly at centromere | |
|
| ||||
| FD | PLK1* | Polo-like kinase 1 | 9/22 | Cell cycle regulator important for M phase progression |
| CIB3 | Calcium and integrin binding family member 3 | 7/22 | Exact function unknown | |
| FLJ11149 | Hypothetical Protein | 6/22 | Likely kinase activity | |
| MAP4K1 | Mitogen-activated protein kinase (X4)1 | 6/22 | Activation of JUN kinase activity, stress response | |
| MAPK12 | Mitogen-activated protein kinase 12 | 6/22 | Extracellular signal transduction | |
| UCK1 | Uridine-cytidine kinase 1 | 6/22 | Uridine monophosphate biosynthetic process | |
|
| ||||
| GW & FD | PLK1* | Polo-like kinase 1 | 10/24 | Cell cycle regulator important for M phase progression |
| AURKB* | Aurora kinase B | 3/24 | Role in chromosome segregation | |
| CAMK2B | Calcium/calmodulin-dependent protein kinase II β | 3/24 | Calcium mediated signaling | |
| KIF11 | Kinesin-like protein | 3/24 | Microtubule motor activity, anterograde transport | |
| WEE1 | WEE1 homolog | 3/24 | Cell cycle regulator of G2 to M transition | |
Number of gene lists with nominated gene/total number of gene lists in that category,
IAP; inhibitor of apoptosis,
Genes repeated in top scoring hits in multiple categories, GW; genome-wide, FD; focused.
To perform the global overlap analysis in focused category, we gathered a total of 22 gene lists with a division of 10 gene list originating from the singles and the remaining 12 from pooled formats (Fig 2A). In this part of the analysis, we obtained a total of 1,170 gene candidates from the singles versus pooled screens (Suppl Table 2). Consistent with the observation made in genome-wide overlap, a major portion constituting 88% of the gene candidates were orphan hits (Fig 2C). PLK1 emerged as a top scoring gene candidate in the list with a maximal overlap among 9 out of the 22 gene lists being compared. This was followed by CIB3 (7 gene list), FLJ11149, MAP4K1, MAPK12, and UCK1 (6 gene lists each) (Table 1). It is important to note here that most of the genes topping the overlap list originated from the singles screens. For example, out of the 9 gene lists reporting on PLK1 as a hit, 8 gene lists corresponded to the singles screens. Surprisingly, some of the known gene candidates were not identified as strong candidates like KIF11, and WEE1 exhibited a dismal overlap by being scored among 2 gene lists, while AURKB, UBB, and, UBC were orphan hits.
In order to determine the degree of overlap between the gene candidates obtained from genome-wide and focused screens, we converged the 24 corresponding gene lists to obtain a total of 1,525 gene candidates; out which only a meager 65 genes were identified as common. 23% of the 65 common genes were kinases including PLK1, which once again topped the list owing to its participation in a maximal 10 gene lists (Table 1). The 65 common genes were enriched in functions pertaining to gene expression (21 genes) as well as components of spliceosome (12 genes), translation factors (5 genes), proteasome (4 genes), and ribosome (4 genes).
siRNA duplex screens: PLK1 still a front-runner in stringent overlaps
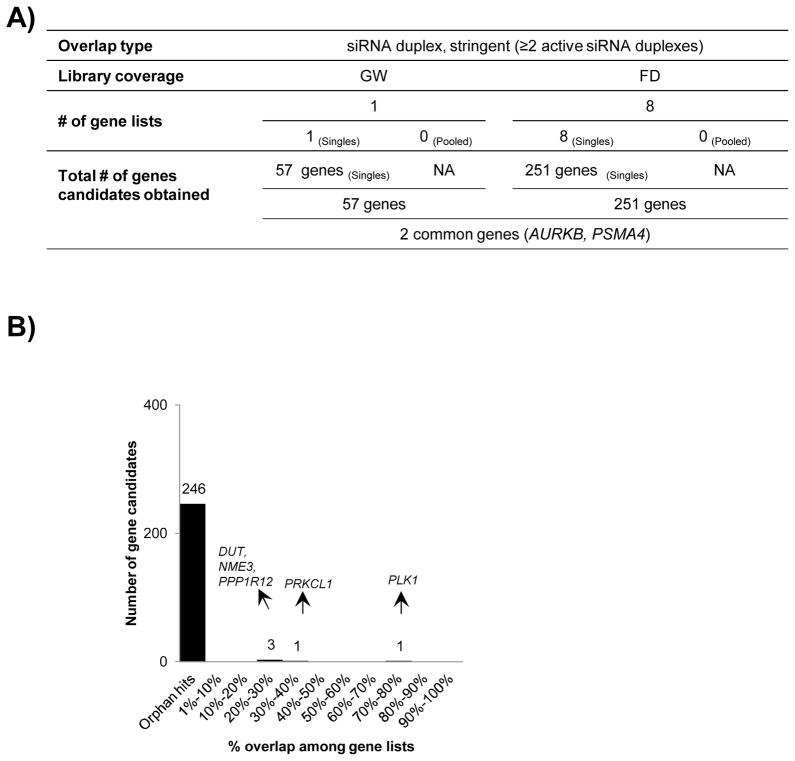
In the first step of global overlap, all gene lists were considered irrespective of the reporting on the number of active duplexes per gene provided in published results. We devised a more conserved filtering strategy for the second step of stringent overlap (Fig 1A). Since RNAi screens are combinatorial in nature, it becomes incumbent to assess gene activity based on its corresponding active relative to inactive duplexes. With this rationale in mind, we opted to review the 24 gene lists for minimal duplex activity based on an H score of ≥ 60 for the second step of stringent overlaps. On a closer scrutiny we found that 15 gene lists got filtered out from further consideration as they did not provide information on the duplex activity per gene or due to unavailability of the raw screening data (Table 2). Of note, all the pooled screens were inadvertently excluded. Since, one of the two gene lists from the genome-wide category got filtered out; a stringent overlap within this category was not feasible (Fig 3A).
Table 2.
Selected 16 siRNA duplex publications highlighting the qualifying criteria for their inclusion in stringent overlaps
| Coverage | Format | Study | Published gene lists | # active duplexes | H score application | Raw data availability | BDA hit nomination | Residual gene lists |
|---|---|---|---|---|---|---|---|---|
| GW | siRNA|Singles | Shum et al. 18 | 1 | Yes | Yes | Yes | Yes | 1 |
|
| ||||||||
| siRNA|Pooled | Moore et al. 19 | 1 | NP | NP | NP | No | - | |
|
| ||||||||
| FD | siRNA|Singles | Aza-Blanc et al.28 | 1 | NP | NP | NP | No | - |
| Henderson et al. 29 | 6 | Yes | NP | NP | No | 6 | ||
| Hu et al. 30 | 1 | Yes | NP | NP | No | 1 | ||
| Sudo et al. 31 | 1 | NP | NP | NP | No | - | ||
| Tiedemann et al. 32 | 1 | Yes | Yes | Yes | No | 1 | ||
|
| ||||||||
| siRNA|Pooled | Giroux et al. 33 | 1 | NP | NP | NP | No | - | |
| MacKeigan et al. 34 | 1 | NP | NP | NP | No | - | ||
| Morgan-Lappe et al. 35 | 1 | NP | NP | NP | No | - | ||
| Sarthy et al. 36 | 1 | NP | NP | NP | No | - | ||
| Sethi et al. 37 | 1 | NP | NP | NP | No | - | ||
| Swanton et al. 38 | 3 | NP | NP | NP | No | - | ||
| Thaker et al. 39 | 1 | NP | NP | NP | No | - | ||
| Thaker et al. 40 | 1 | NP | NP | NP | No | - | ||
| Tyner et al. 41 | 2 | NP | NP | NP | No | - | ||
Figure 3. Stringent overlap of 9 gene lists procured by literature mining for siRNA duplex screens.
A) Systematic workflow of stringent overlap for selection of 57 gene candidates from 1 genome-wide and 251 gene candidates from 8 focused screen gene lists followed identification of 2 genes in common between them. B) Breakup of number of overlapping genes in focused screens based on their participation in number of gene lists. The degree of overlap is represented as percent of total gene lists selected.
We obtained 8 gene lists for focused screens after applying the stringent filtering criteria, with no contribution from the pooled screens at this stage (Table 2, Fig 3A). For the gene lists that did supply active duplex data but mostly defaulted in detailing the total library coverage per gene, we exclusively selected for those gene subset with ≥ 2 independent active siRNA duplexes, as it was difficult to use the H-score as a filter on the published reports. Of note, given a general prevalence of 3 siRNA duplexes in a library, ≥ 2 active siRNA duplexes as an activity cut off translates into an approximate H score value of roughly 60 or greater. Therefore, genes that were targeted by a single siRNA duplex in the library screened were excluded to maintain stringency. Finally, we obtained a total of 251 gene candidates, populated majorly by orphan hits (Fig 3B). Interestingly, PLK1 still topped the list with a participation in 6 out of the 8 gene lists, followed by PRKCL1 (3 gene lists), a protein kinase with a putative role in apoptosis, while only 3 additional genes qualified in > 20% of the gene lists being compared (Table 3). The results from the focused screen overlaps where compared to the single residual gene list from the genome-wide overlaps and after removing for the orphan hits, we were left with only seven genes and only two genes, namely AURKB and PSMA4 were identified as common between one gene lists each of genome-wide and focused categories (Table 3).
Table 3.
Top scoring 7 representative gene candidates from stringent overlap in siRNA duplex screens
| Coverage | Gene | Name | Overlap1 | Biological process |
|---|---|---|---|---|
| GW | N/A | N/A | N/A | N/A |
|
| ||||
| FD | PLK1 | Polo-like kinase 1 | 6/8 | Cell cycle regulator important for M phase progression |
| PRKCL1 | Protein kinase N1 | 3/8 | Signal transduction (apoptosis mediated) | |
| DUT | Deoxyuridine triphosphatase | 2/8 | Essential enzyme of nucleotide metabolism | |
| NME3 | Nucleoside diphosphate kinase 3 | 2/8 | Nucleoside biosynthesis, apoptosis induction | |
| PPP1R12C | Protein phosphatase 1, regulatory subunit 12C | 2/8 | Regulates assembly of actin cytoskeleton | |
|
| ||||
| GW & FD | AURKB | Aurora kinase B | 2/9 | Role in chromosome segregation |
| PSMA4 | Proteasome subunit, α type, 4 | 2/9 | Ubiquitin-dependent peptide cleavage | |
Number of gene lists with nominated gene/total number of gene lists in that category, N/A; Not applicable as only 1 gene lists remain in this category, GW; genome-wide, FD; focused.
shRNA hairpin screens: Global overlaps identify KRAS as an genes candidate
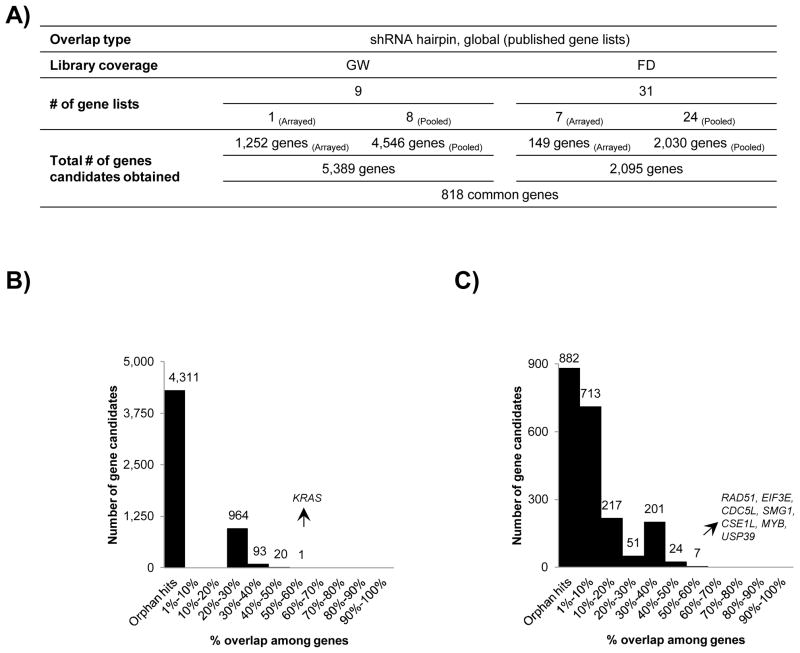
Among the 14 selected publications for shRNA hairpin screens, 3 screens were genome-wide libraries and 11 screens were focused yielding a total of 40 lethality gene lists (Fig 1B). Contrary to the standard arrayed formats used in siRNA duplex screening, an arrayed format in shRNA hairpin screen refers to a systematic one hairpin per well knockdown while pooled format refers to an amalgamation of up to the entire library hairpins, which could be as many as a million in one pool, targeting thousands of genes collectively. We conducted a global overlap using the 9 gene lists corresponding to the three genome-wide screens (Fig 4A). A preference towards pooled screening approach was observed in the genome-wide shRNA hairpin screens, perhaps due to the ease of screening and lower initial screening costs involved. In the genome-wide overlaps, we included our recently published shRNA hairpin screen, which till date is the only genome-wide shRNA hairpin screen to identify gene candidates with putative role in cell survival performed in an arrayed format. 16 The remaining 8 genome-wide screens were conducted in pooled formats only (Fig 4B). Taken together, we obtained a total of 5,389 gene candidates, 80% of which were orphan hits. Topping the list was a known oncogene, KRAS due to its nomination in 5 separate gene lists, followed by 20 other genes scored in 4 out of 9 gene lists (Table 4, Suppl Table 3).
Figure 4. Global overlap of 40 gene lists procured by literature mining for shRNA hairpin screens.
A) Systematic workflow of global overlap for selection of 5,389 candidates from 9 genome-wide and 2,095 gene candidates from 31 focused screen gene lists followed identification of 818 genes in common between them. B) Breakup of number of overlapping genes in genome-wide screens based on their participation in number of gene lists. C) Breakup of number of overlapping genes in focused screens based on their participation in number of gene lists. The degree of overlap is represented as percent of total gene lists selected.
Table 4.
Top scoring 23 representative gene candidates from global overlap in shRNA hairpin screens
| Coverage | Gene | Name | Overlap1 | Biological process |
|---|---|---|---|---|
| GW | KRAS* | v-Ki-ras2 Kirsten rat sarcoma viral oncogene homolog | 5/9 | Ras protein signal transduction (oncogene) |
| MCM6 | Minichromosome maintenance complex component 6 | 4/9 | Essential for replication inititation | |
| SELS | Selenoprotein S | 4/9 | ER overload response2, degradation of misfolded proteins | |
| SENP5 | Sentrin-specific protease 5 | 4/9 | Essential in SUMO pathway, required for cell division | |
| SNRPC | Small nuclear ribonucleoprotein polypeptide C | 4/9 | Spliceosomal snRNP assembly | |
|
| ||||
| FD | RAD51* | DNA repair protein RAD51 homolog 1 | 18/31 | DNA recombination and repair (oncogene) |
| EIF3E* | Eukaryotic translation initiation factor 3, subunit E | 18/31 | Regulation of translational initiation | |
| CDC5L* | Cell division cycle 5-like | 18/31 | Cell cycle regulator important for G2/M transition | |
| SMG1* | Phosphatidylinositol 3-kinase-related kinase | 18/31 | Nonsense-mediated mRNA decay | |
| CSE1L* | Cellular apoptosis susceptibility protein | 16/31 | Protein transport | |
| MYB | Myeloblastosis viral oncogene homolog | 16/31 | Transcription factor | |
| USP39 | Ubiquitin specific peptidase 39 | 16/31 | mRNA splicing | |
| CDC2L1 | Cyclin-dependent kinase 11B | 15/31 | Cell cycle control | |
| KRAS | v-Ki-ras2 Kirsten rat sarcoma viral oncogene homolog | 15/31 | Ras protein signal transduction (oncogene) | |
| NR2F1 | Nuclear receptor subfamily 2, group F, member 1 | 15/31 | Transcription initiation | |
| PIK4CA | Phosphatidylinositol 4-kinase | 15/31 | Biosynthesis of phosphatidylinositol 4,5-bisphosphate | |
| TAF10 | TATA box binding protein (TBP)-associated factor | 15/31 | DNA-dependent transcription initiation | |
|
| ||||
| GW & FG | KRAS* | v-Ki-ras2 Kirsten rat sarcoma viral oncogene homolog | 20/40 | Ras protein signal transduction (oncogene) |
| EIF3E* | Eukaryotic translation initiation factor 3, subunit E | 20/40 | Regulation of translational initiation | |
| CDC5L* | Cell division cycle 5-like | 19/40 | Cell cycle regulator important for G2/M transition | |
| RAD51* | DNA repair protein RAD51 homolog 1 | 19/40 | DNA recombination and repair (oncogene) | |
| SMG1* | Phosphatidylinositol 3-kinase-related kinase | 18/40 | Nonsense-mediated mRNA decay | |
| CSE1L* | Cellular apoptosis susceptibility protein | 18/40 | Protein transport | |
Number of gene lists with nominated gene/total number of gene lists in that category,
ER; endoplasmic reticulum,
Genes repeated in top scoring hits in multiple categories, GW; genome-wide, FD; focused.
For the purpose of performing the overlap in the focused category, we analyzed 31 gene lists obtained from the 11 focused library screens, we encountered a similar trend with regards to prevalence of hits nominated from pooled screens (24 gene lists) versus the arrayed screens (7 gene lists). We collected a total of 2,095 genes at this stage, and a rather surprising result was that 1,946 hits (93%) corresponded exclusively to pooled shRNA formats (Fig 4A). RAD51, EIF3E, SMG1, and CDC5L ranked highest in this category making them the only four genes common in close to 60% of the gene lists being compared (Fig 4C, Table 4). In the next step of comparative analysis, we merged the hits obtained from 40 gene lists corresponding to the genome-wide and focused screens taken together and collected a total of 6,664 genes out which 4,284 were orphans and therefore ignored, and amongst the remainder, 818 gene candidates were identified in common (Fig 4A, Suppl Table 3). KRAS, the top scoring gene candidate from genome-wide category once again emerged as strong hits in the merged results from focused library screens with preponderance in 20 gene lists. EIF3E also scored in 20 gene lists, however, is given lesser significance that KRAS owing to its identification in only pooled formats when compared to that of KRAS which was identified in all categories of genome-wide and focused screens (Suppl Table 3). PLK1 showed a marginal performance with an overlap among 11 out of the 40 gene lists with its dominance in 33% of genome-wide and 26% of focused gene lists (Suppl Table 3). Among the 818 common genes, 78 genes were associated with pathways in cancer (hsa05200), 60 genes with a role in MAPK signaling pathway (hsa04010), and ~ 40 genes each associated with cell cycle and ubiquitin mediated proteolysis.
shRNA hairpin: Stringent overlaps reveal EIF5B as a top gene candidate
Similar to the methodology adopted for stringent overlap in siRNA duplex screens, we filtered out the gene lists where hit re-assessment was not feasible (Table 5). Surprisingly, 11 gene lists were deemed incompetent for participation in the stringent analysis. In the next step towards re-evaluating the gene lists, we attempted to re-analyze the raw data from the published screens but were posed by major handicap of unavailability of primary screen raw data. For the purpose of selecting high-confidence hits in shRNA screening data for instances where the library information was not provided, we set a selection threshold of ≥ 3 active hairpins targeting a gene; which ideally is representative of an H score of ≥ 60 conforming to the fairly standard practice of including 5 hairpins targeting per gene on an average in a shRNA hairpin library.
Table 5.
Selected 14 shRNA hairpin publications highlighting the qualifying criteria for their inclusion in stringent overlap
| Coverage | Format | Study | Published gene lists | # active duplexes | H score application | Raw data availability | BDA hit nomination | Residual gene lists |
|---|---|---|---|---|---|---|---|---|
| GW | shRNA|Arrayed | Bhinder et al. 16 | 1 | Yes | Yes | Yes | Yes | 1 |
|
| ||||||||
| shRNA|Pooled | Cheung et al. 17 | 6 | NP | NP | Yes | Yes | 102 | |
| Luo et al. 12 | 2 | Yes | NP | NP | No | 2 | ||
|
| ||||||||
| FD | shRNA|Arrayed | Barbie et al. 11 | 1 | NP | NP | Yes | Yes | 19 |
| Duan et al. 20 | 1 | NP | NP | NP | No | - | ||
| Giodts et al. 21 | 1 | NP | NP | NP | No | - | ||
| Scholl et al. 10 | 1 | Yes | Yes | NP | No | 1 | ||
| Whitworth et al. 22 | 1 | NP | NP | NP | No | - | ||
| Yang et al. 23 | 1 | NP | NP | NP | No | - | ||
| Yeh et al. 24 | 1 | Yes | NP | NP | No | 1 | ||
|
| ||||||||
| shRNA|Pooled | Luo et al. 25 | 12 | Yes | NP | Yes | Yes | 12 | |
| Ngo et al. 26 | 1 | Yes | NP | NP | No | 1 | ||
| Schlabach et al. 13 | 4 | Yes | Yes | NP | No | 4 | ||
| Silva et al. 27 | 7 | Yes | NP | NP | No | 4 | ||
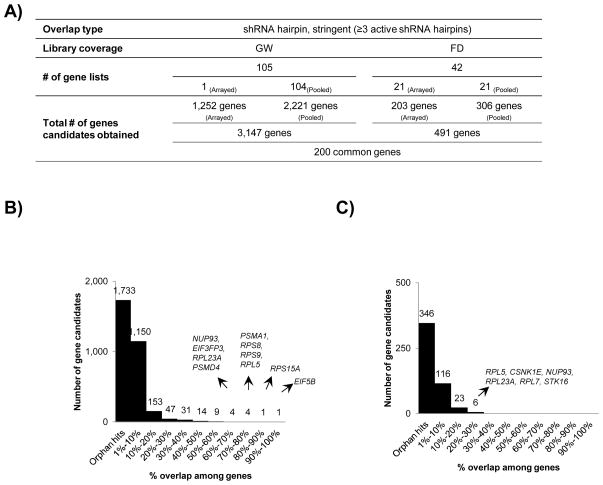
For the genome-wide overlaps, we collected the residual 3 gene lists; one form arrayed and two from pooled formats. In addition, we took advantage of the raw shRNA hairpin screening data provided by the Broad Institute for the genome-wide shRNA hairpin screen published by Cheung and co-workers 17 in pooled format conducted in 102 cancer cell lines. We applied the entire BDA workflow for hit candidate nomination using the BDA method. 14 We merged the additional gene lists thus obtained, expanding our hits lists under consideration to cover 105 cell lines (Fig 5A). We compiled the results of stringent overlaps from filtered published hits as well as re-analyzed published screen data, and an overall hit breakup per gene list was performed, once again highlighting the preponderance of orphan hits at a 55% (Fig 5B). More so, the hits appeared to be more enriched in exclusively pooled shRNA hairpin screens at 60% in the data output; such an observation is not surprising given the inherent noise of the system (Suppl Table 4). As a manifestation of the stringent filtering majority of the top scoring gene candidates from the global overlaps exhibited a decline in their prevalence and EIF5B emerged as the strong hit in the stringent overlap (Table 6).
Figure 5. Stringent overlap of 147 gene lists procured by literature mining for shRNA hairpin screens.
A) Systematic workflow of stringent overlap for selection of 3,147 gene candidates from 105 genome-wide and 491 gene candidates from 42 focused screen gene lists followed identification of 200 genes in common between the two categories. B) Breakup of number of overlapping genes in genome-wide screens based on their participation in number of gene lists. C) Breakup of number of overlapping genes in focused screens based on their participation in number of gene lists. The degree of overlap is represented as percent of total gene lists selected.
Table 6.
Top scoring 32 representative gene candidates from stringent overlap in shRNA hairpin screens
| Coverage | Gene | Name | Overlap1 | Biological process |
|---|---|---|---|---|
| GW | EIF5B* | Eukaryotic translation initiation factor 5B | 99/105 | Translation initiation |
| RPS15A* | Ribosomal protein S15a | 90/105 | Ribosomal (40S) biogenesis, translation | |
| PSMA1* | Proteasome subunit, α type, 1 | 83/105 | Ubiquitin-dependent peptide cleavage | |
| RPS8 | Ribosomal protein S8 | 78/105 | Ribosomal (40S) biogenesis, translation | |
| RPS9 | Ribosomal protein S9 | 78/105 | Ribosomal (60S) biogenesis, translation | |
| RPL5* | Ribosomal protein L5 | 77/105 | Ribosomal (60S) biogenesis, rRNA maturation | |
| EIF3FP3 | Eukaryotic translation initiation factor 3 | 72/105 | Pseudogene | |
| NUP93* | Nucleoporin | 72/105 | Required for nuclear pore assembly, protein transport | |
| PSMD4 | Proteasome 26S subunit, non-ATPase, 4 | 64/105 | Ubiquitin-dependent peptide cleavage | |
| RPL23A* | Ribosomal protein L23a | 64/105 | Ribosomal (60S) biogenesis, translation | |
| RPS15 | Ribosomal protein S15 | 61/105 | Ribosomal (40S) biogenesis, translation | |
| TUBB | Tubulin β | 61/105 | Major constituent of microtubules | |
| NACA | Nascent polypeptide-associated complex α subunit | 58/105 | Blocks nascent polypeptides interactions with SRP2 | |
| DDX21 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 21 | 56/105 | Helicase, foldase (RNA processing) | |
| DYNC1H1 | Dynein cytoplasmic 1 heavy chain 1 | 56/105 | Microtubule-based movement | |
| SRSF2 | Serine/arginine-rich splicing factor 2 | 56/105 | Necessary for the splicing of pre-mRNA | |
| LSM4 | U6 small nuclear RNA associated | 55/105 | Role in mRNA splicing | |
| RPL7 | Ribosomal protein L7 | 53/105 | Ribosomal (60S) biogenesis, translation | |
| USP39 | Ubiquitin specific peptidase 39 | 53/105 | Likely role in mRNA splicing | |
| EIF2B3 | Eukaryotic translation initiation factor 2B | 52/105 | Regulation of translation inititation | |
| RPS25 | Ribosomal protein S25 | 52/105 | Ribosomal (40S) biogenesis, translation | |
|
| ||||
| FD | RPL5* | Ribosomal protein L5 | 12/42 | Ribosomal (60S) biogenesis, rRNA maturation |
| NUP93* | Nucleoporin | 11/42 | Required for nuclear pore assembly, protein transport | |
| RPL7* | Ribosomal protein L7 | 10/42 | Ribosomal (60S) biogenesis, translation | |
| CSNK1E | Casein kinase 1 | 9/42 | DNA replication and repair | |
| CHEK1 | Checkpoint kinase 1 | 7/42 | DNA damage response | |
| RPA2 | Replication protein A2 | 7/42 | DNA replication and repair | |
|
| ||||
| GW & FD | EIF5B* | Eukaryotic translation initiation factor 5B | 104/147 | Translation inititation |
| RPS15A* | Ribosomal protein S15a | 92/147 | Ribosomal (40S) biogenesis, translation | |
| RPL5* | Ribosomal protein L5 | 89/147 | rRNA maturation, formation of 60S ribosomal subunits | |
| PSMA1* | Proteasome subunit, α type, 1 | 88/147 | Ubiquitin-dependent peptide cleavage | |
| NUP93* | Nucleoporin | 83/147 | Required for nuclear pore assembly, protein transport | |
Number of gene lists with nominated gene/total number of gene lists in that category,
SRP; signal recognition particle,
Genes repeated in top scoring hits in multiple categories, GW; genome-wide, FD; focused.
To define the gene candidates for the residual gene lists in focused category, we also included the 19 gene lists from an arrayed shRNA hairpin screen performed by Barbie and co-workers 11 and the data re-analyzed using the BDA method as previously described. 14 Similarly, we also reanalyzed data sets available at The Broad Institute website for shRNA hairpin screen performed in a pooled format by Luo and co-workers and obtained 12 corresponding gene lists. 25 Signal intensity values were obtained for the 12 cell lines screened as well as the control library DNA plasmid pool in replicates of ten and BDA method was applied for the purpose of hit nomination. FC of library shRNA hairpins relative to the control pools were computed to determine the relative depletion and the thresholds for FC were determined at a −2 σ from the average FC per each cell line. Following through the steps of stringent filtering, we finally obtained a total of 42 gene lists with varying degree of overlaps; 59% of the total hits were obtained exclusively from pooled shRNA hairpin screens (Fig 5A). The degree of overlap was not very strong across the board with RPL5 topping the lists with its prevalence in only 29% of the gene lists (Fig 5C, Table 6).
Convergence of the two categories led to identification of 3,438 gene candidates in total with only 200 genes found in common between the two categories (Fig 5A, Suppl Table 4). Amongst them were genes known to play role in cancer progression; for example, leukemia (FLT3, and KRAS), and colorectal cancer (ATM, CHEK1, DCC, EGFR, HRAS, KRAS, MDM2, and PTEN) to name a few. Although, we did find PLK1 among the 200 common genes, it showed a marginal enrichment of only 3 gene lists in total. As far as the biological classification of the 200 genes is concerned, the highest enriched functional GO categories were found to be cell communication (GO:0007154; 37%), cell cycle (GO:0007049; 29%), developmental process (GO:0032502; 20%), transport (GO:0006810; 16%), immune system process (GO:0002376; 13%), and apoptosis (GO:0006915; 12%). Also dominating the list were 57 transferases, 50 kinases, 17 ribosomal proteins, 18 genes associated with ubiquitin dependent proteolysis, 16 genes associated with RNA processing including mRNA splicing factors and 5 genes associated with mRNA transport & localization.
Evaluation of hit preponderance: enrichment bias from pooled shRNA hairpin screens
We reviewed the overall hit distributions so as to gain insights into and to identify a bias, if any, in the nominated hits towards a specific RNAi technology or screening format. We first reviewed the 24 published siRNA duplex gene lists, and found that 53% of the hits originated exclusively from 13 pooled screens while 39% originated exclusively from 11 singles screens and the remaining 8% were commonly identified in both formats (Fig 6A). Next, we reviewed the 40 published shRNA hairpin gene lists and observed that a drastic 79% of the gene candidates were exclusively associated with pooled shRNA hairpin screens accounting for up to 5,269 out of 6,664 gene candidates, majority of which were orphan hits and therefore associated with maximal 1 gene list (Fig 6B). Furthermore, a technology specific breakup revealed that even amongst the pooled screening hits, shRNA hairpin screens were highly enriched; 63% of the total hits obtained from all literature taken together where from pooled shRNA screens (Fig 6C). Taking a broader overview of global overlap inclusive of both siRNA duplex and shRNA hairpin screens, we looked at a total of 7,430 gene candidates collected from 64 published gene lists and observed a significant dominance of candidates obtained from pooled formats (Fig 6C). A similar trend was observed in the stringent overlaps for shRNA hairpins screens, 1,993 gene candidates out of 3,438 total gene candidates were obtained exclusively from pooled format screens (Fig 6D). Despite such enrichment in stringent overlaps, major players of cellular viability either displayed a marginal overlap (e.g., PLK1) or where completely missed (e.g., AURKA). The top scoring gene candidates for shRNA hairpin screens from stringent overlaps were over-shadowed by a bias towards pooled screening formats, for example, the top scoring gene candidate, EIF5B was active in 104 gene lists in total, 103 out of which were associated with screens conducted in pooled formats while only one gene list corresponded to the screens performed in arrayed formats.
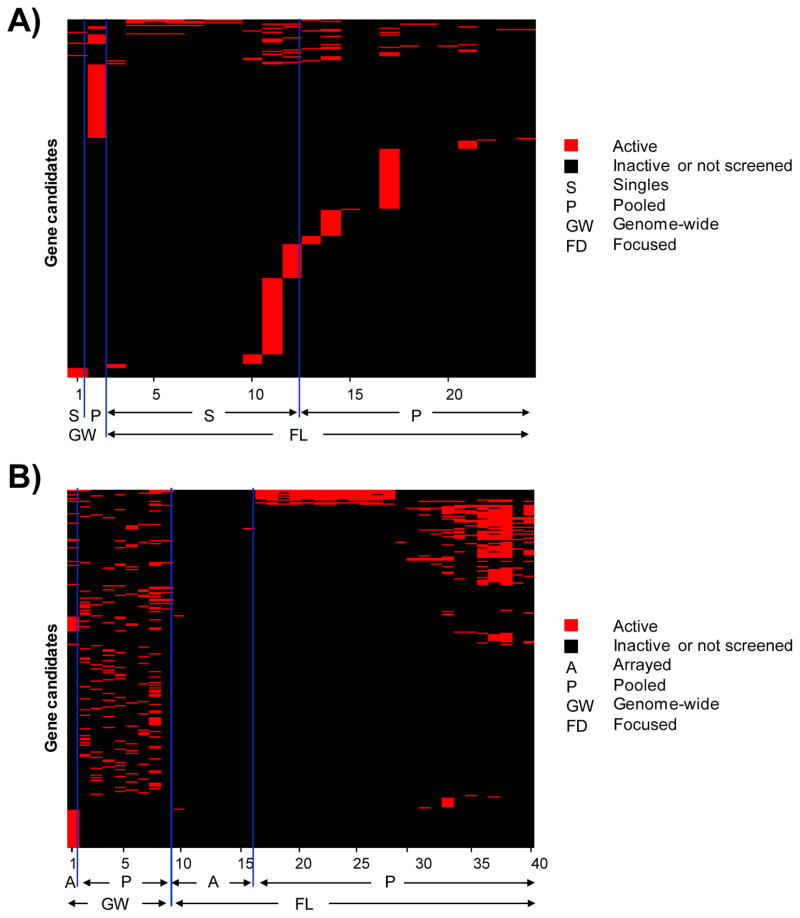
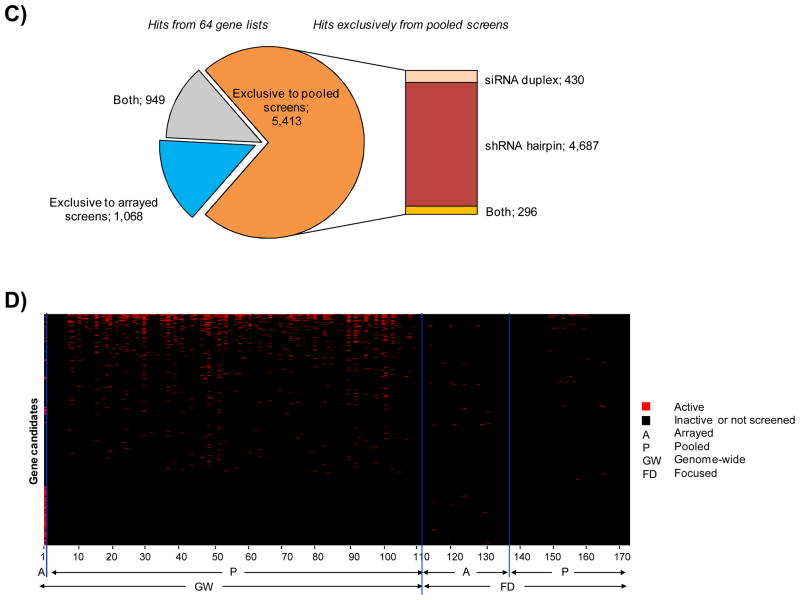
Figure 6. Visualization of hit preponderance in siRNA duplex versus shRNA hairpin screens.
A) Heat map plot representation of global overlap among 1,525 genes in siRNA duplex screens. B) Heat map plot representation of global overlap among 6,664 genes showing prominence of hits from pooled shRNA hairpin screens. C) Pie chart illustrating the breakup of gene candidates obtained from each category at the stage of global overlaps. A strong enrichment of hits originating exclusively form shRNA hairpin pooled formats can be observed. D) Heat map plot representation of stringent overlap among 3,644 genes showing prominence of hits from pooled shRNA hairpin screens.
The gene lists are ordered based on the screening format and are sorted by numbers corresponding to the publication index (Suppl Table 6).
DISCUSSION
RNAi screening technology has been viewed as a promising tool over the past decade, enabling researchers to perform up to genome-scale knockdowns in mammalian cells, to elucidate gene function. 1 Approximately 300 RNAi screens have been published using siRNA duplex or shRNA hairpin libraries in both arrayed and pooled formats to yield a multitude of promising targets. It wasn’t until 2009 that the field faced a major criticism with regards to hit reproducibly and poor overlap across different studies, and has brought into question the true therapeutic potential of the high value targets identified from RNAi screening. 5–6 As we become aware of the major pitfalls of the technology in terms of its sensitivity and specificity, it has become all the more important to consolidate results of published RNAi screens so as to gain insights into the degree of hit discordance and get a perspective on the differential practices which might lead to such an outcome. Our reasoning was to simply analyze published lethality screening data with the hope of finding a high degree of gene commonality across the board as one would intuitively expect. With this objective, we selected 30 RNAi lethality screens to profile gene candidates required for cell survival.
A variety of lethality RNAi screens studying genes with a role in cell survival have been reported in literature thus far, and it is intuitive to expect a high overlap among the data outputs from such screens. With an aim to review gene-level overlaps in these RNAi lethality screens, we first obtained 64 gene lists from 30 published studies, to include both siRNA duplex and shRNA hairpin screens performed in arrayed as well as pooled formats. To our surprise, we found a lack of a general consensus in terms of hit nomination in RNAi screens, which is rather surprising especially in comparison to chemical screening, where data analysis practices are fairly standardized. To gain a general perspective, we merged the results from the 64 lists to procure a set of 7,430 gene candidates. Strikingly, the global analysis captured about 30% of the human genome to play a role in cell survival with a substantial number of hits being reported only once among the gene lists, perhaps indicative of a generic preponderance of off-target mediated false positives in RNAi results.
An overview of the global overlap from the 64 gene lists unveiled that a relatively higher number of genes were obtained from shRNA hairpin screens when compared to the siRNA screens; 6,664 gene candidates were acquired from 40 shRNA hairpin gene lists while only 1,525 gene candidates were acquired from 24 siRNA duplex gene lists. This could partly be explained by the fundamental difference between the two technologies, which lies at the basal level of their processing into active entities; the siRNA duplexes are introduced into the cell as double stranded RNA molecules comprised of a guide and a passenger strand whereas the shRNA hairpins are introduced into the cell as an expression vector construct, and therefore have to undergo sequential steps of genomic integration, transcription of the DNA insert and intracellular processing of the transcribed hairpin to finally yield a guide and passenger strand. Consequentially, the accuracy of intracellular shRNA hairpin processing becomes a principal determinant of a hairpin’s target specificity; which has become questionable in wake of recent findings with regards to heterogeneous intracellular cleavage of hairpins. 15, 45–46
In the siRNA duplex screens, we observed that a higher fraction of genes were obtained from the focused library screens; 68% additional gene candidates were obtained from focused libraries relative to the genome-wide. Since focused libraries are smaller in scale, smaller number of hits would be expected from the data outputs. It can be argued that, small-scale screens with lesser number of genes screened are likely to have lower standard deviations when compared to large-scale screens, therefore enabling identification of weaker hits in the screen. This observation was however not consistent with the results from shRNA hairpin screens, where majority of the hits corresponded to the genome-wide screens, specifically to those performed in pooled formats, perhaps indicative of the higher noise in shRNA hairpin screens.
Our systematic analysis reveals an unprecedented enrichment in hits from pooled shRNA hairpin screens; ~80% of the total hits obtained from the global overlaps and ~60% of the total hits obtained from stringent filtering were an outcome of pooled shRNA hairpin screens. We divided the gene lists into the categories of genome-wide and focused during the course of the analysis, and observed a similar trend in both cases. In fact, exclusion of the pooled shRNA hairpin screens would have reduced our global overlap set of gene candidates to mere 27%. It is important to note that an arrayed screening format has a comparative advantage over the pooled screening counterpart in its enhanced initial assay sensitivity as it allows for one gene one duplex knockdowns while obviating the necessity for shRNA hairpins deconvolution by microarray hybridization or deep sequencing. In addition, the sheer magnitude of distinct hairpins in a pool might also elevate the likelihood of multiple integrations per cell.
Furthermore, a striking bias in pooled formats as observed in the shRNA hairpin screens, was not encountered in the siRNA screen data outputs; 53% hits were obtained exclusively from pooled and 39% of the hits were obtained exclusively from arrayed formats in siRNA duplex screens. Here it is important to note that there is an innate difference in the common practices pertaining to pooled methodologies in siRNA duplexes versus shRNA hairpins; siRNA pools are generally comprised of 2–4 siRNA duplexes targeting a single gene whereas an shRNA pool maybe comprised of up to ~50,000 hairpins targeting several thousand genes collectively and probably to yield a synergistic phenotype. Taken together, these findings are suggestive of a higher inherent noise associated with pooled shRNA hairpin screens and perhaps the uncertainty associated with specific target knockdowns.
RNAi screening is combinatorial in nature and therefore, it is crucial to score a phenotype based on the maximal hairpin activity corresponding to a gene taking into account the active duplexes relative to the total targeting duplexes in the library screened. A closer scrutiny of the selected gene lists used in the global overlaps revealed that almost none of the published screens employed a strategy to incorporate minimal duplex activity corresponding to an H score of 60 and OTE filtering. 14 Accordingly, we re-assessed the reported hits from 30 published RNAi screening studies as refined stringent overlaps by applying the activity metric, H score of ≥ 60 or the BDA method where applicable. The unavailability of raw primary screen data and screening library posed as two major handicaps in the endeavor. Surprisingly, 16 out of 30 studies merely reported list of active genes and not the corresponding number of active duplexes per gene and therefore had to be eliminated from the stringent level of comparative analysis. Interestingly, application of stringent filtering on the global data outputs revealed a significant decline in gene prevalence across the board; 14% gene candidates were retained in the genome-wide category and 21% in the focused category for the siRNA duplex screens while 58% gene candidates were retained in the genome-wide and 23% in the focused category for the shRNA hairpin screens. Of note, 133 additional reanalyzed gene lists were added to the stringent overlap in shRNA hairpin screens. Irrespective, such a decline in gene candidates is strongly suggestive of the fact that most of the genes from global overlap did not qualify the minimal H score criteria of 60, thereby accentuating the need to employ stringency in hit nomination for RNAi screens in general.
RNAi literature mining based gene signature analysis reported here revealed that none of the genes overlapped across all cell lines and showed poor overlaps within similar cell lines (Suppl Table 5) while top scoring published targets deemed high value were surprisingly missing, for example STK33 and TBK1. 10–11 Of interest was the differential performance of PLK1, a routinely used control in RNAi screens and perhaps a strong candidate for gene lethality. PLK1 was identified as a top scoring gene candidate in siRNA duplex screens at both global as well as stringent filtering overlaps but was not a prevalent hit in the shRNA hairpin screens. Although PLK1 was found to be nominated as a hit in 11 published shRNA screening reports, application of stringent filtering threshold dropped its prevalence to a mere 3 gene lists; one of which was arrayed16 while the other two were from a pooled hairpin screen.13 Of note, the pooled gene list reporting on identification of PLK1 only had maximal 3 targeting hairpins in the library screened. We have previously reported on a marginal performance of the shRNA hairpins targeting PLK1 in an arrayed screen performed in HeLa cell line; only four out of the 20 independently validated hairpins were scored as active. 16 Of note, the shRNA hairpins for PLK1 were validated in MCF7 cell line. Since PLK1 is believed to be an important gene in cellular viability, the failure to score it as a hit is suggestive of the differential perturbation produced by shRNA hairpins perhaps due to inefficiencies in intracellular processing or maybe due to cell line specificity.
We have found gene clusters among the overlapping genes corresponding to ribosomal proteins, proteasome and mRNA splicing. As for some of the genes deemed important for cell survival, we observed a varying prevalence of common genes between global and stringent overlaps. For example, AURKA was found to be common in 7 gene lists pertaining to shRNA hairpin screens and 1 gene list pertaining to siRNA duplex screen, but was missed in the stringent filtering, because of either being targeted by utmost single duplex in the screen or unavailability of screen data. Similarly, WEE1 was found in a total of 7 gene lists but dropped down to a mere 1 gene list after application of stringent filtering. KIF11 was found in a total of 5 gene lists in the global overlaps but after addition of reanalyzed additional gene-lists, it was found in 12 gene lists in the stringent overlap. Interesting, KRAS was identified only in shRNA hairpins screens; predominantly in those performed in the pooled formats; 20 gene lists in the global overlap and 41 gene lists in the stringent overlap. Similarly, the ubiquitin protein, UBB, was identified in 12 gene lists in the global overlaps, out of which 10 gene lists corresponded to shRNA screens, and showed a prominence in 34 gene lists at the stage of stringent overlap.
Comparative analysis reports in the past have explored commonalities at a pathway and functional level at a small scale, to harmonize distinct hits lists identified by various RNAi screening groups. 3 In lieu of such inferences, we had hypothesized that a true positive must consistently exhibit a strong phenotype across maximal screening data outputs irrespective of the technology used. To this end, we undertook the first systematic analysis approach to combine data outputs from variety of RNAi lethality screens, reviewed their overlaps at gene level and asked a simple question as to the feasibility of identifying a set of gene candidates with a role in cell survival that was found in all RNAi lethality screens. Collaborative attempts have being made in the past towards creating repositories for RNAi screen data integration. As an example, GenomeRNAi database contains a collection of ~ 297 published RNAi screens performed in Drosophila and mammalian cells with an aim to centralize gene perturbation information for different phenotypes from multiple sources. 47 However, it would now be beneficial to take this endeavor forward by merging multiple RNAi data outputs and assessing the hit reproducibility; as has been attempted in this report.
In summary, we have reported the first large-scale systematic analysis for 30 RNAi lethality reports and surprisingly observed zero gene commonality across all the selected gene lists; none of the genes displayed a consistently lethal phenotype across 64 gene lists considered for global overlap and 147 gene lists re-evaluated for stringent overlap. This is of great concern if this RNAi technology cannot identify similar genes in lethality screens. Our results, also, report an unprecedented enrichment in active gene candidates obtained exclusively from pooled shRNA hairpin screens, identified via either microarray hybridization or deep sequencing. This finding question the merits of performing pooled shRNA screens using either deep sequencing and/or microarray hybridization as readout; could the observed phenotype be merely a consequence of synergistic silencing due to multiple cellular integrations and off-targeted effects due to inaccurate hairpin cleavage? Our findings unfortunately concur with the current pitfalls of the technology, especially as to the abuse recently reported by Kaelin. 7 Without any golden standards in place, we would encourage the community to pay more attention to RNAi screening data analysis practices, bearing in mind that the important aspect of this technology is its combinatorial knockdowns. Finally, we also would like to convene cautionary measures while interpreting outcomes pertaining to pooled shRNA hairpin screens.
Supplementary Material
Suppl Table 1. Selected 30 publications for identification of genes associated with cell survival
TRC; The RNAi Consortium, H&E; Hannon and Elledge, NKI; Netherlands Cancer Institute
Suppl Table 2. List of 197 gene candidates obtained from selected publications in global overlaps for siRNA duplex screens, excluding orphan hits
GW; genome-wide, FD; focused, GL; gene list
Suppl Table 3. List of 1,046 gene candidates obtained from selected publications in global overlaps for shRNA hairpin screens, excluding hits from ≤ 2 GLs
GW; genome-wide, FD; focused, GL; gene list
Suppl Table 4. List of 1,014 gene candidates obtained from selected publications in stringent overlaps for shRNA hairpin screens, excluding hits from ≤ 2 GLs
GW; genome-wide, FD; focused, GL; gene list
Suppl Table 5. List of 6 representative cell line specific overlaps for global and stringent levels of comparative analysis
GL; gene list
Suppl Table 6. List of gene lists used for global and stringent levels of comparative analysis in siRNA duplex and shRNA hairpin screens
Acknowledgments
The HTS Core Facility is partially supported by Mr. William H. Goodwin and Mrs. Alice Goodwin and the Commonwealth Foundation for Cancer Research, the Experimental Therapeutics Center of the Memorial Sloan-Kettering Cancer Center, the William Randolph Hearst Fund in Experimental Therapeutics, the Lillian S. Wells Foundation, and by a NIH/NCI Cancer Center Support Grant 5 P30 CA008748-44.
Abbreviations
- RNAi
RNA interference
- OTE
off-target effect
- shRNA
short hairpin RNA
- siRNA
small interfering RNA
- BDA
Bhinder-Djaballah Analysis
- H score
hit rate per gene score
- NUCL
nuclei count
- TRC
The RNAi Consortium
- FC
fold change
Footnotes
DISCLOSURE STATEMENT
The authors declare no competing financial interests.
LIST OF REFERENCES
- 1.Mohr SE, Bakal C, Perrimon N. Genomic screening with RNAi: Results and challenges. Annu Rev Biochem. 2010;79:37–64. doi: 10.1146/annurev-biochem-060408-092949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Simpson KJ, Davis GM, Boag PR. Comparative high-throughput RNAi screening methodologies in C. elegans and mammalian cells. Nat Biotechnol. 2012;29(4):459–470. doi: 10.1016/j.nbt.2012.01.003. [DOI] [PubMed] [Google Scholar]
- 3.Bushman FD, Malani N, Fernandes J, D’Orso I, Cagney G, Diamond TL, Zhou H, Hazuda DJ, Espeseth AS, König R, Bandyopadhyay S, Ideker T, Goff SP, Krogan NJ, Frankel AD, Young JA, Chanda SK. Host Cell Factors in HIV Replication: Meta-Analysis of Genome-Wide Studies. PLoS Pathog. 2009;5(5):e1000437. doi: 10.1371/journal.ppat.1000437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cherry S. What have RNAi screens taught us about viral-host interactions? Curr Opin Microbiol. 2009;12(4):446–452. doi: 10.1016/j.mib.2009.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Babij C, Zhang Y, Kurzeja RJ, Munzli A, Shehabeldin A, Fernando M, Quon K, Kassner PD, Ruefli-Brasse AA, Watson VJ, Fajardo F, Jackson A, Zondlo J, Sun Y, Ellison AR, Plewa CA, San MT, Robinson J, McCarter J, Schwandner R, Judd T, Carnahan J, Dussault I. STK33 kinase activity is nonessential in KRAS-dependent cancer cells. Cancer Res. 2011;71(17):5818–5826. doi: 10.1158/0008-5472.CAN-11-0778. [DOI] [PubMed] [Google Scholar]
- 6.Naik G. Scientists’ Elusive Goal: Reproducing Study Results. Wall Street Journal. 2011 [Google Scholar]
- 7.Kaelin WG., Jr Use and Abuse of RNAi to Study Mammalian Gene Function. Science. 2012;337(6093):421–422. doi: 10.1126/science.1225787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Djaballah H. Random RNAi Screening Data Analysis: A Call for Standardization. Comb Chem High Throughput Screen. 2012;15(9):685. doi: 10.2174/138620712803519725. [DOI] [PubMed] [Google Scholar]
- 9.Mohr SE, Perrimon N. RNAi screening: new approaches, understanding, and organisms. Wiley Interdiscip Rev RNA. 2012;3(2):145–158. doi: 10.1002/wrna.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Scholl C, Fröhling S, Dunn IF, Schinzel AC, Barbie DA, Kim SY, Silver SJ, Tamayo P, Wadlow RC, Ramaswamy S, Döhner K, Bullinger L, Sandy P, Boehm JS, Root DE, Jacks T, Hahn WC, Gilliland DG. Synthetic lethal interaction between oncogenic KRAS dependency and STK33 suppression in human cancer cells. Cell. 2009;137(5):821–834. doi: 10.1016/j.cell.2009.03.017. [DOI] [PubMed] [Google Scholar]
- 11.Barbie DA, Tamayo P, Boehm JS, Kim SY, Moody SE, Dunn IF, Schinzel AC, Sandy P, Meylan E, Scholl C, Fröhling S, Chan EM, Sos ML, Michel K, Mermel C, Silver SJ, Weir BA, Reiling JH, Sheng Q, Gupta PB, Wadlow RC, Le H, Hoersch S, Wittner BS, Ramaswamy S, Livingston DM, Sabatini DM, Meyerson M, Thomas RK, Lander ES, Mesirov JP, Root DE, Gilliland DG, Jacks T, Hahn WC. Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature. 2009;462(7269):108–112. doi: 10.1038/nature08460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Luo J, Emanuele MJ, Li D, Creighton CJ, Schlabach MR, Westbrook TF, Wong KK, Elledge SJ. Genome-wide RNAi screen identifies multiple synthetic lethal interactions with the Ras oncogene. Cell. 2009;137(5):835–848. doi: 10.1016/j.cell.2009.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schlabach MR, Luo J, Solimini NL, Hu G, Xu Q, Li MZ, Zhao Z, Smogorzewska A, Sowa ME, Ang XL, Westbrook TF, Liang AC, Chang K, Hackett JA, Harper JW, Hannon GJ, Elledge SJ. Cancer proliferation gene discovery through functional genomics. Science. 2008;319(5863):620–624. doi: 10.1126/science.1149200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bhinder B, Djaballah H. A simple method for analyzing actives in random RNAi screens: Introducing the “H score” for gene nomination and prioritization. Comb Chem High Throughput Screen. 2012;15(9):686–704. doi: 10.2174/138620712803519671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gu S, Jin L, Zhang Y, Huang Y, Zhang F, Valdmanis PN, Kay MA. The Loop Position of shRNAs and Pre-miRNAs Is Critical for the Accuracy of Dicer Processing In Vivo. Cell. 2012;151(4):900–911. doi: 10.1016/j.cell.2012.09.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bhinder B, Antczak C, Ramirez CN, Shum D, Liu-Sullivan N, Radu C, Frattini MG, Djaballah H. An Arrayed Genome Scale Lentiviral Enabled shRNA Screen Identifies Lethal & Rescuer Gene Candidates. Assay Drug Dev Technol. 2013;11(3):173–190. doi: 10.1089/adt.2012.475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cheung HW, Cowley GS, Weir BA, Boehm JS, Rusin S, Scott JA, East A, Ali LD, Lizotte PH, Wong TC, Jiang G, Hsiao J, Mermel CH, Getz G, Barretina J, Gopal S, Tamayo P, Gould J, Tsherniak A, Stransky N, Luo B, Ren Y, Drapkin R, Bhatia SN, Mesirov JP, Garraway LA, Meyerson M, Lander ES, Root DE, Hahn WC. Systematic investigation of genetic vulnerabilities across cancer cell lines reveals lineage-specific dependencies in ovarian cancer. Proc Natl Acad Sci USA. 2011;108(30):12372–12377. doi: 10.1073/pnas.1109363108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shum D, Bhinder B, Ramirez CN, Radu C, Calder PA, Beauchamp L, Farazi T, Landthaler M, Tuschi T, Magdaleno S, Djaballah H. An Arrayed RNA Interference Genome-Wide Screen Identifies Candidate Genes Involved in the MicroRNA 21 Biogenesis Pathway. Assay Drug Dev Technol. 2013;11(3):191–205. doi: 10.1089/adt.2012.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moore MJ, Wang Q, Kennedy CJ, Silver PA. An alternative splicing network links cell-cycle control to apoptosis. Cell. 2010;142(4):625–636. doi: 10.1016/j.cell.2010.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Duan Z, Ji D, Weinstein EJ, Liu X, Susa M, Choy E, Yang C, Mankin H, Hornicek FJ. Lentiviral shRNA screen of human kinases identifies PLK1 as a potential therapeutic target for osteosarcoma. Cancer Lett. 2010;293(2):220–229. doi: 10.1016/j.canlet.2010.01.014. [DOI] [PubMed] [Google Scholar]
- 21.Goidts V, Bageritz J, Puccio L, Nakata S, Zapatka M, Barbus S, Toedt G, Campos B, Korshunov A, Momma S, Van Schaftingen E, Reifenberger G, Herold-Mende C, Lichter P, Radlwimmer B. RNAi screening in glioma stem-like cells identifies PFKFB4 as a key molecule important for cancer cell survival. Oncogene. 2012;31(27):3235–3243. doi: 10.1038/onc.2011.490. [DOI] [PubMed] [Google Scholar]
- 22.Whitworth H, Bhadel S, Ivey M, Conaway M, Spencer A, Hernan R, Holemon H, Gioeli D. Identification of kinases regulating prostate cancer cell growth using an RNAi phenotypic screen. PLoS One. 2012;7(6):e38950. doi: 10.1371/journal.pone.0038950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang C, Ji D, Weinstein EJ, Choy E, Hornicek FJ, Wood KB, Liu X, Mankin H, Duan Z. The kinase Mirk is a potential therapeutic target in osteosarcoma. Carcinogenesis. 2010;31(4):552–558. doi: 10.1093/carcin/bgp330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yeh MH, Tsai TC, Kuo HP, Chang NW, Lee MR, Chung JG, Tsai MH, Liu JY, Kao MC. Lentiviral short hairpin RNA screen of human kinases and phosphatases to identify potential biomarkers in oral squamous cancer cells. Int J Oncol. 2011;39(5):1221–1231. doi: 10.3892/ijo.2011.1100. [DOI] [PubMed] [Google Scholar]
- 25.Luo B, Cheung HW, Subramanian A, Sharifnia T, Okamoto M, Yang X, Hinkle G, Boehm JS, Beroukhim R, Weir BA, Mermel C, Barbie DA, Awad T, Zhou X, Nguyen T, Piqani B, Li C, Golub TR, Meyerson M, Hacohen N, Hahn WC, Lander ES, Sabatini DM, Root DE. Highly parallel identification of essential genes in cancer cells. Proc Natl Acad Sci USA. 2008;105(51):20380–20385. doi: 10.1073/pnas.0810485105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ngo VN, Davis RE, Lamy L, Yu X, Zhao H, Lenz G, Lam LT, Dave S, Yang L, Powell J, Staudt LM. A loss-of-function RNA interference screen for molecular targets in cancer. Nature. 2006;441(7089):106–110. doi: 10.1038/nature04687. [DOI] [PubMed] [Google Scholar]
- 27.Silva JM, Marran K, Parker JS, Silva J, Golding M, Schlabach MR, Elledge SJ, Hannon GJ, Chang K. Profiling essential genes in human mammary cells by multiplex RNAi screening. Science. 2008;319(5863):617–620. doi: 10.1126/science.1149185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aza-Blanc P, Cooper CL, Wagner K, Batalov S, Deveraux QL, Cooke MP. Identification of modulators of TRAIL-induced apoptosis via RNAi-based phenotypic screening. Mol Cell. 2003;12(3):627–637. doi: 10.1016/s1097-2765(03)00348-4. [DOI] [PubMed] [Google Scholar]
- 29.Henderson MC, Gonzales IM, Arora S, Choudhary A, Trent JM, Von Hoff DD, Mousses S, Azorsa DO. High-throughput RNAi screening identifies a role for TNK1 in growth and survival of pancreatic cancer cells. Mol Cancer Res. 2011;9(6):724–732. doi: 10.1158/1541-7786.MCR-10-0436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hu K, Lee C, Qiu D, Fotovati A, Davies A, Abu-Ali S, Wai D, Lawlor ER, Triche TJ, Pallen CJ, Dunn SE. Small interfering RNA library screen of human kinases and phosphatases identifies polo-like kinase 1 as a promising new target for the treatment of pediatric rhabdomyosarcomas. Mol Cancer Ther. 2009;8(11):3024–3035. doi: 10.1158/1535-7163.MCT-09-0365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sudo H, Tsuji AB, Sugyo A, Kohda M, Sogawa C, Yoshida C, Harada YN, Hino O, Saga T. Knockdown of COPA identified by loss-of-function screen induces apoptosis and suppresses tumor growth in mesothelioma mouse model. Genomics. 2010;95(4):210–216. doi: 10.1016/j.ygeno.2010.02.002. [DOI] [PubMed] [Google Scholar]
- 32.Tiedemann RE, Zhu YX, Schmidt J, Shi CX, Sereduk C, Yin H, Mousses S, Stewart AK. Identification of molecular vulnerabilities in human multiple myeloma cells by RNA interference lethality screening of the druggable genome. Cancer Res. 2012;72(3):757–768. doi: 10.1158/0008-5472.CAN-11-2781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Giroux V, Iovanna J, Dagorn JC. Probing the human kinome for kinases involved in pancreatic cancer cell survival and gemcitabine resistance. FASEB J. 2006;20(12):1982–1991. doi: 10.1096/fj.06-6239com. [DOI] [PubMed] [Google Scholar]
- 34.MacKeigan JP, Murphy LO, Blenis J. Sensitized RNAi screen of human kinases and phosphatases identifies new regulators of apoptosis and chemoresistance. Nat Cell Biol. 2005;7(6):591–600. doi: 10.1038/ncb1258. [DOI] [PubMed] [Google Scholar]
- 35.Morgan-Lappe SE, Tucker LA, Huang X, Zhang Q, Sarthy AV, Zakula D, Vernetti L, Schurdak M, Wang J, Fesik SW. Identification of Ras-related nuclear protein, targeting protein for xenopus kinesin-like protein 2, and stearoyl-CoA desaturase 1 as promising cancer targets from an RNAi-based screen. Cancer Res. 2007;67(9):4390–4398. doi: 10.1158/0008-5472.CAN-06-4132. [DOI] [PubMed] [Google Scholar]
- 36.Sarthy AV, Morgan-Lappe SE, Zakula D, Vernetti L, Schurdak M, Packer JC, Anderson MG, Shirasawa S, Sasazuki T, Fesik SW. Survivin depletion preferentially reduces the survival of activated K-Ras-transformed cells. Mol Cancer Ther. 2007;6(1):269–276. doi: 10.1158/1535-7163.MCT-06-0560. [DOI] [PubMed] [Google Scholar]
- 37.Sethi G, Pathak HB, Zhang H, Zhou Y, Einarson MB, Vathipadiekal V, Gunewardena S, Birrer MJ, Godwin AK. An RNA interference lethality screen of the human druggable genome to identify molecular vulnerabilities in epithelial ovarian cancer. PLoS One. 2012;7(10):e47086. doi: 10.1371/journal.pone.0047086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Swanton C, Marani M, Pardo O, Warne PH, Kelly G, Sahai E, Elustondo F, Chang J, Temple J, Ahmed AA, Brenton JD, Downward J, Nicke B. Regulators of mitotic arrest and ceramide metabolism are determinants of sensitivity to paclitaxel and other chemotherapeutic drugs. Cancer Cell. 2007;11(6):498–512. doi: 10.1016/j.ccr.2007.04.011. [DOI] [PubMed] [Google Scholar]
- 39.Thaker NG, McDonald PR, Zhang F, Kitchens CA, Shun TY, Pollack IF, Lazo JS. Designing, optimizing, and implementing high-throughput siRNA genomic screening with glioma cells for the discovery of survival genes and novel drug targets. J Neurosci Methods. 2010;185(2):204–212. doi: 10.1016/j.jneumeth.2009.09.023. [DOI] [PubMed] [Google Scholar]
- 40.Thaker NG, Zhang F, McDonald PR, Shun TY, Lewen MD, Pollack IF, Lazo JS. Identification of survival genes in human glioblastoma cells by small interfering RNA screening. Mol Pharmacol. 2009;76(6):1246–1255. doi: 10.1124/mol.109.058024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tyner JW, Walters DK, Willis SG, Luttropp M, Oost J, Loriaux M, Erickson H, Corbin AS, O’Hare T, Heinrich MC, Deininger MW, Druker BJ. RNAi screening of the tyrosine kinome identifies therapeutic targets in acute myeloid leukemia. Blood. 2008;111(4):2238–2245. doi: 10.1182/blood-2007-06-097253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID Bioinformatics Resources. Nature Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 43.Thomas PD, Kejariwal A, Campbell MJ, Mi H, Diemer K, Guo N, Ladunga I, Ulitsky-Lazareva B, Muruganujan A, Rabkin S, Vandergriff JA, Doremieux O. PANTHER: a browsable database of gene products organized by biological function, using curated protein family and subfamily classification. Nucleic Acids Res. 2003;31(1):334–341. doi: 10.1093/nar/gkg115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.UniProt Consortium. Reorganizing the protein space at the Universal Protein Resource (UniProt) Nucleic Acids Res. 2012;40:D71–D75. doi: 10.1093/nar/gkr981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Park JE, Heo I, Tian Y, Simanshu DK, Chang H, Jee D, Patel DJ, Kim VN. Dicer recognizes the 5′ end of RNA for efficient and accurate processing. Nature. 2011;475(7355):201–205. doi: 10.1038/nature10198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vermeulen A, Behlen L, Reynolds A, Wolfson A, Marshall WS, Karpilow J, Khvorova A. The contributions of dsRNA structure to Dicer specificity and efficiency. RNA. 2005;11(5):674–682. doi: 10.1261/rna.7272305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gilsdorf M, Horn T, Arziman Z, Pelz O, Kiner E, Boutros M. GenomeRNAi: a database for cell-based RNAi phenotypes. 2009 update. Nucleic Acids Res. 2010;38:D448–D452. doi: 10.1093/nar/gkp1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Suppl Table 1. Selected 30 publications for identification of genes associated with cell survival
TRC; The RNAi Consortium, H&E; Hannon and Elledge, NKI; Netherlands Cancer Institute
Suppl Table 2. List of 197 gene candidates obtained from selected publications in global overlaps for siRNA duplex screens, excluding orphan hits
GW; genome-wide, FD; focused, GL; gene list
Suppl Table 3. List of 1,046 gene candidates obtained from selected publications in global overlaps for shRNA hairpin screens, excluding hits from ≤ 2 GLs
GW; genome-wide, FD; focused, GL; gene list
Suppl Table 4. List of 1,014 gene candidates obtained from selected publications in stringent overlaps for shRNA hairpin screens, excluding hits from ≤ 2 GLs
GW; genome-wide, FD; focused, GL; gene list
Suppl Table 5. List of 6 representative cell line specific overlaps for global and stringent levels of comparative analysis
GL; gene list
Suppl Table 6. List of gene lists used for global and stringent levels of comparative analysis in siRNA duplex and shRNA hairpin screens