Significance
Gene reporters can be used to track viable cells in vivo and their patterns of gene expression. There have been numerous attempts to develop gene reporters for magnetic resonance imaging (MRI), however these give only modest image contrast and often this is negative, which can be difficult to detect. We describe here a dual-imaging modality reporter that gives intense and positive contrast in magnetic resonance images (up to ∼8× increase in signal), which can also be used with radionuclide imaging, thus combining the sensitivity of radionuclide imaging with the spatial resolution of MRI. The contrast obtained is directly related to the degree of gene expression and is readily reversible, thus allowing longitudinal studies of changes in expression.
Keywords: SPECT, Oatp
Abstract
The ability to track cells and their patterns of gene expression in living organisms can increase our understanding of tissue development and disease. Gene reporters for bioluminescence, fluorescence, radionuclide, and magnetic resonance imaging (MRI) have been described but these suffer variously from limited depth penetration, spatial resolution, and sensitivity. We describe here a gene reporter, based on the organic anion transporting protein Oatp1a1, which mediates uptake of a clinically approved, Gd3+-based, hepatotrophic contrast agent (gadolinium-ethoxybenzyl-diethylenetriamine pentaacetic acid). Cells expressing the reporter showed readily reversible, intense, and positive contrast (up to 7.8-fold signal enhancement) in T1-weighted magnetic resonance images acquired in vivo. The maximum signal enhancement obtained so far is more than double that produced by MRI gene reporters described previously. Exchanging the Gd3+ ion for the radionuclide, 111In, also allowed detection by single-photon emission computed tomography, thus combining the spatial resolution of MRI with the sensitivity of radionuclide imaging.
The development of noninvasive imaging techniques to track cells and their patterns of gene expression in vivo has been a much sought-after goal as it enables imaging of various biological phenomena including stem and immune cell migration, delivery and expression of gene therapy vectors, the genetics of development, and the progression of diseases such as cancer and their response to treatment (1, 2).
Bioluminescent and fluorescent reporters (3, 4), though sensitive, suffer from limited light penetration in whole animal studies and consequently have relatively poor spatial resolution (5). Radionuclide imaging reporters (1) have good sensitivity and depth penetration but lack spatial resolution (5), whereas magnetic resonance imaging (MRI) reporters (6) have good depth penetration and relatively good resolution but currently lack sensitivity and produce poor tissue contrast (7). Table S1 describes the published gene reporters that have been developed for MRI. Apart from the magnetic resonance (MR) spectroscopy reporters, which lack spatial resolution, the majority of MRI gene reporters rely on negative contrast (T2 enhancing agents), which can be difficult to detect in a heterogeneous image. The most promising of these, ferritin (6), gives only modest contrast (Table S1) and has led to calls for reporters with increased sensitivity (8). Deans et al. increased ferritin sensitivity by coexpressing the transferrin receptor and incubating the cells with iron and transferrin in the culture medium (9). However, despite the use of a vector designed to give high level constitutive expression, the R2 relaxation rate (1/T2) increased by only a factor of ∼2 at 7 T and was not significant at 1.5 T. Of the positive contrast agents (T1 enhancing agents), which are easier to detect in vivo, the divalent metal ion transporter (DMT1), which transports Mn2+, and a metabolically biotinylated cell surface protein [biotin acceptor peptide (BAP)-transmembrane (TM)], which was detected using a streptavidin–horseradish peroxidase (SA-HRP) conjugate in conjunction with a peroxidase-sensitive gadolinium agent (10), have so far given the best contrast in vivo. However, DMT1 gave only a 75% increase in image contrast in glioma tumors transfected with a vector expressing DMT1 from a strong constitutive promoter (11). Moreover, there was significant enhancement in surrounding normal tissue and the persistence of the enhancement varied markedly between different tumor types. BAP-TM gave better contrast [3.3× enhancement in a s.c. glioma tumor, where the protein was expressed from the strong cytomegalovirus (CMV) promoter]. However, this first required injection of the macromolecular agent, SA-HRP, which would preclude human use and whose large size may limit tissue access and then, when this had cleared, the gadolinium agent was injected. There was no evidence that this system was quantitative.
An ideal gene reporter would give fast, reversible, quantitative, and intense gene expression-dependent contrast and would work with multiple imaging modalities, thus combining their different advantages. One approach to this has been to create fusion proteins, for example between luciferase, red fluorescent protein (RFP), and thymidine kinase, which allows imaging with bioluminescence, fluorescence, and positron emission tomography (PET), respectively (12). A limitation is that the fusion protein may not be as well expressed or have the same activity as the individual proteins and the large size of the DNA construct may preclude its incorporation into some vectors. An elegant solution to this problem was reported recently by Niers et al. (13), who expressed, from a strong constitutive promoter, a metabolically biotinylated, membrane-bound form of Gaussia luciferase in an implanted glioma tumor. The reporter could be detected using bioluminescence, following injection of coelenterazine, and by fluorescence tomography, MRI, and single-photon emission computed tomography (SPECT), following injection of streptavidin conjugated to a fluorophore, magnetic nanoparticles, and 111In- diethylenetriamine pentaacetic acid (DTPA), respectively. Although the reporter is small (∼40 kDa) the imaging agents are large (the magnetic nanoparticles were ∼45 nm in diameter), the MR image contrast was negative and modest (∼2× increase in R2 in vivo) and there was no evidence that it was reversible, which may limit longitudinal studies of gene expression.
We describe here a gene reporter system in which the dual-modality imaging capability (MRI and SPECT) is invested in a single protein, Oatp1a1 (Slco1a1, Slc21a1, Slc21a3, Oatp1, Oatp1a1, OATP-1). This protein from the SLCO superfamily (14) has the capacity to transport a variety of imageable small molecules across the plasma membrane. These include the clinically approved hepatobillary MRI contrast agent, gadolinium-ethoxybenzyl-DTPA (Gd-EOB-DTPA; gadoxetate or Primovist) (15, 16), which we show produces rapid, intense, and reversible signal enhancement in T1-weighted MR images of cells expressing the reporter in vivo. The reporter can produce more than twice as much signal in vivo as MRI gene reporters described previously (Table S1). Exchanging the gadolinium ion for the radionuclide 111In means, that in the same experiment, the Oatp1 reporter can also be detected using SPECT, thus combining the spatial resolution of MRI with the sensitivity of SPECT.
Results
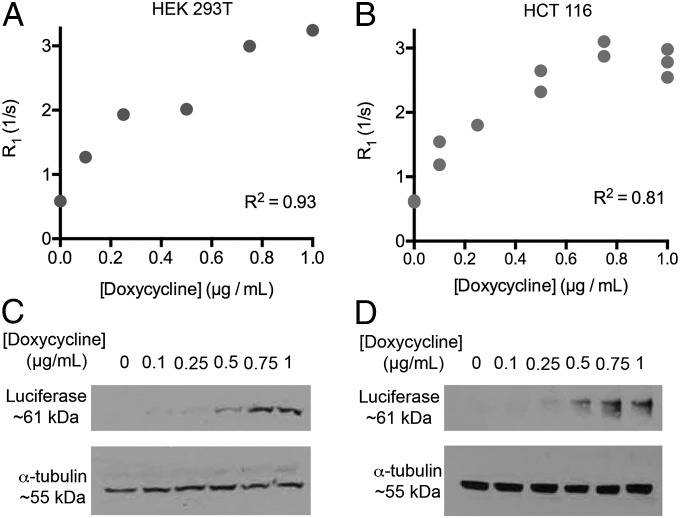
First we demonstrated a quantitative relationship between Oatp1 expression and T1 relaxation enhancement in cells incubated with Gd-EOB-DTPA (Fig. 1). Throughout the majority of the studies reported here, the Oatp1 reporter transgene has either been expressed from the relatively weak PGK promoter (17) and the transgene introduced using lentiviral vectors (which have single sites of integration), or by using a plasmid with the doxycycline-inducible TRE3G promoter (maps of these constructs are shown in Fig. S1). Human embryonic kidney cells (HEK 293T) and human colon adenocarcinoma cells (HCT 116) were transfected with a doxycycline-inducible luciferase (Luc2)–Oatp1 construct (pTRE3G-LO), in which the coding sequences of Luc2 and Oatp1 were separated by an E2A sequence, which results in equimolar production of the two proteins (18). The luciferase provided a marker of transgene expression on Western blots in these experiments. The increase in the relaxation rate (R1 = 1/T1) (Fig. 1 A and B) was correlated with the increase in transgene expression following induction with doxycycline (Fig. 1 C and D). Induction of Oatp1 expression had no effect on the proliferation rate of these cells (Fig. S2 A and B). Transport of hepatocyte-specific MRI contrast agents by Oatp has been shown previously to be specific, saturable, and bidirectional (19, 20). Reversible transport was confirmed here by measurements of Gd-EOB-DTPA uptake and washout in human breast cancer (MCF-7) cells transfected with a vector expressing Oatp1 (Fig. S3). Contrast agent uptake in HEK 293T cells was dependent on the contrast agent concentration and control cells showed no uptake (Fig. S4).
Fig. 1.
Relaxation enhancement rate (R1) correlates with Oatp1 expression. (A) Clonal HEK 293T and (B) HCT 116 cells transfected with the inducible pTRE3G-LO plasmid were induced for 24 h with doxycycline. Cells were then incubated with 0.5 mM (HEK 293T) or 0.25 mM (HCT 116) Gd-EOB-DTPA, washed, pelleted, and imaged with T1 imaging and mapping sequences. Western blots of luciferase in protein extracts from (C) HEK 293T and (D) HCT 116 cells shown in A and B, showing that induction of transgene expression correlated with doxycycline concentration and R1 enhancement.
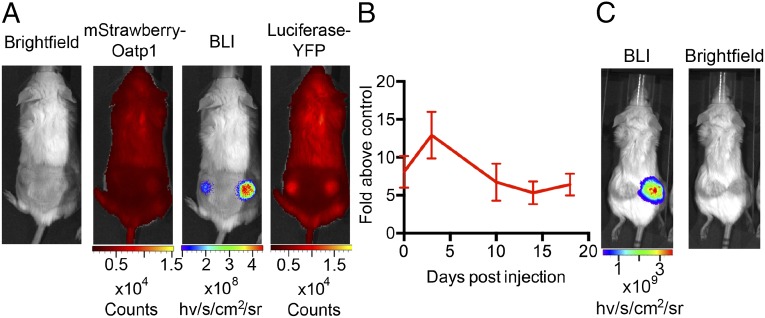
Next we implanted, in the flanks of SCID mice, HEK 293T cells expressing a luciferase–YFP fusion protein, which had been transduced with an mStrawberry- and Oatp1-expressing lentiviral vector (LV-PGK-SO; Fig. S1) and then subcloned. The two coding sequences in this vector are separated by an E2A sequence. The same number of control luciferase–YFP-expressing HEK 293T cells was implanted in the contralateral flanks. During the 16 d following cell implantation, the resulting xenografts expressing Oatp1 showed 5–12× greater light output than the control xenografts (Fig. 2 A and B), suggesting that Oatp1 also mediates transport of luciferin into cells. Luciferase–YFP was equally expressed in the two cell lines, as assessed by the levels of YFP fluorescence. Expression of Oatp1 was confirmed by imaging the red fluorescence from mStrawberry (Fig. 2A) and by immunohistochemical staining for mStrawberry in histological sections obtained postmortem (Fig. S5). These sections also showed that there were low and similar levels of necrosis in these xenografts (Fig. S5). Similar experiments were performed using HCT 116 cells transfected with the doxycycline inducible vector (pTRE3G-LO) that expressed the Luc2 and Oatp transgenes (Fig. 1). There was increased bioluminescence in the transfected cells measured 3 d after addition of doxycycline to the drinking water (Fig. 2). These bioluminescence experiments demonstrated that there was expression of Oatp1 in the HEK 293T and HCT 116 xenografts in vivo and, in the case of the HEK 293T cells, that the expressed transporter was functional. These experiments also showed no evidence for an effect of Oatp1 expression on xenograft growth.
Fig. 2.
Bioluminescence measurements demonstrate expression of Oatp1 in vivo. (A) Light output was greater in the bioluminescence image (BLI) from the Oatp1-expressing HEK 293T xenograft (right flank), whereas luciferase–YFP fluorescence was the same in both xenografts. Red fluorescence confirmed expression of the mStrawberry–Oatp1 transgenes. (B) Mean increase in light output from Oatp1-expressing xenografts compared with controls expressing luciferase–YFP alone (n = 5). Points represent the average light output over 20 min following i.p. injection of luciferin. Error bars show SEM. (C) Light output was increased from a representative HCT 116 tumor expressing an inducible luciferase–E2A–Oatp1 construct, 3 d postinduction with doxycycline (10 mg/mL in drinking water), compared with the control tumor (left flank), which was not transfected with the construct.
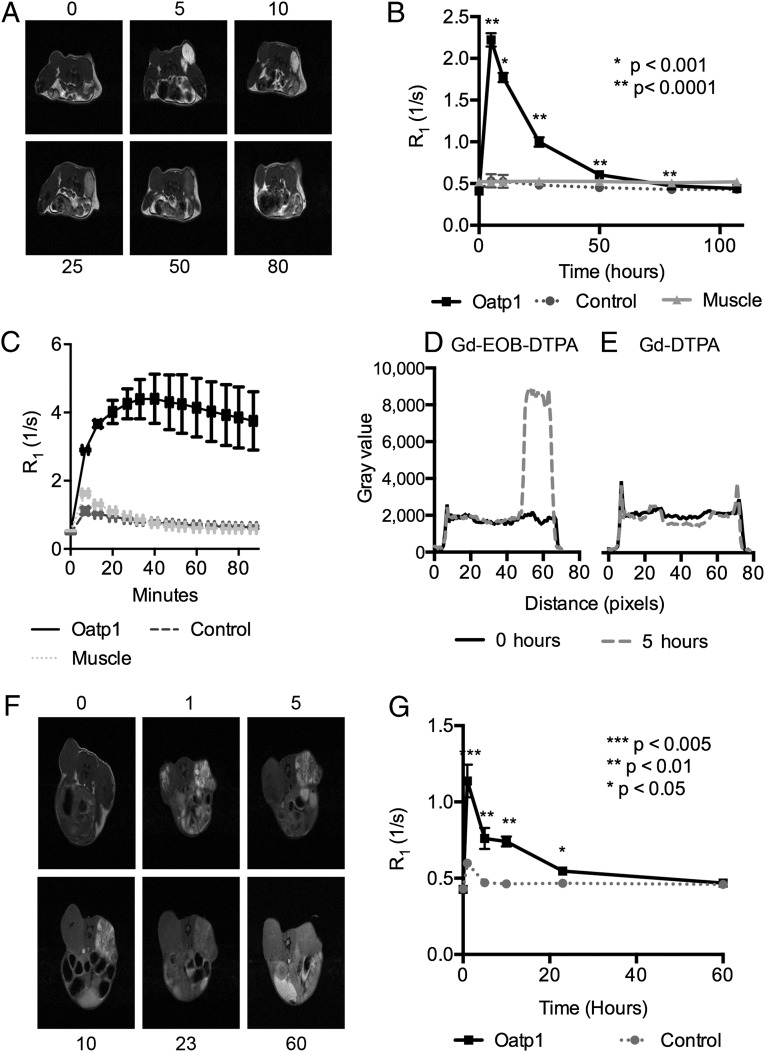
Next we imaged these animals with MRI, using multislice T1-weighted and T1 mapping pulse sequences before and following injection of 0.664 mmoles/kg of Gd-EOB-DTPA. This corresponds to a blood concentration of 8.3 mM, assuming a mouse blood volume of 2 mL (21). There was no difference in T1 (or T2) contrast in the MR images of control or contralateral Oatp1-expressing xenografts before injection of the contrast agent (Fig. 3 A and F), however following injection the spin lattice relaxation rate R1 (1/T1) of Oatp1-expressing xenografts increased by 2–4× (Fig. 3 B and G), which in the HEK 293T xenografts resulted in an increase in pixel intensities of greater than fourfold (Fig. 3 A and D). Assuming a molar relaxivity for Gd-EOB-DTPA of 5.7 mM−1⋅s−1, this corresponds to a tissue concentration in the Oatp1-expressing HEK 293T xenografts of 0.32 mM. There was no significant change in contrast in control xenografts or in muscle (Fig. 3 B and G). Contrast enhancement in the Oatp1-expressing xenografts returned to baseline levels by 107 h for HEK 293T and by 60 h for HCT 116 (Fig. 3 B and G), at which point we injected equimolar amounts of a control contrast agent, Gd-DTPA (Magnevist). This has a similar structure to Gd-EOB-DTPA, but lacks the ethoxybenzene group and is not a substrate for Oatp1. Gd-DTPA produced T1 contrast in both xenografts immediately following injection, and then rapidly washed out, showing no significant signal enhancement in either xenograft after 5 h (Fig. 3E and Fig. S6). Subsequent readministration of Gd-EOB-DTPA showed enhancement of HEK 293T xenografts expressing Oatp1, but not the control xenografts, confirming the ability of the reporter to assess longitudinal changes in gene expression (Fig. 3C). This also showed that a maximum of 6.8-fold increase in signal was obtained in HEK 293T xenografts within 60 min of contrast agent administration. The sensitivity of reporter expression can readily be increased by using higher doses of contrast agent. For example, 2 mmoles/kg of Gd-EOB-DTPA produced 7.8-fold more signal in an HEK 293T xenograft expressing Oatp1 than the control xenograft at 5 h postinjection. The agent has a median lethal dose of 10 mmol/kg when administered i.v. in mice and rats (22). Although the levels of necrosis in these xenografts were relatively low (Fig. S5), some of the early enhancement could be explained by accumulation of contrast agent in necrotic regions, however this had cleared by 5 h after injection (Fig. S7). Contrast agent was also, as expected, taken up by the liver (see whole mouse images in Movie S1), which may preclude imaging of the reporter in and around the liver. However, the degree of contrast enhancement in the liver was comparable to that observed in the HEK 293T xenografts.
Fig. 3.
MRI of Oatp1 expression in vivo. (A) T1-weighted images before (0 h) and 5, 10, 25, 50, and 80 h after i.v. injection of Gd-EOB-DTPA, from a representative animal bearing an HEK 293T xenograft (as shown in Fig. 2). Control xenografts (left flank) expressed luciferase–YFP and the xenografts on the right flank also expressed Oatp1. (B) Xenograft relaxation rates (R1) before (0 h) and following i.v. injection of Gd-EOB-DTPA (0.664 mmoles/kg) were significantly greater compared with controls [*P < 0.001, **P < 0.0001, n = 5 (n = 4 at 107 h)]. (C) R1 increased in control and Oatp1-expressing xenografts after repeat i.v. injection with Gd-EOB-DTPA (0.664 mmoles/kg), following washout of the original dose in B and injection and washout of Gd-DTPA (E) (n = 2). Error bars show SEM. (D) Pixel intensities along a line bisecting the control and Oatp1-expressing xenografts shown in A, obtained before (0 h) and 5 h after Gd-EOB-DTPA injection. (E) The same as in D, but before (0 h) and 5 h after injection of Gd-DTPA. (F) T1-weighted images before (0 h) and 1, 5, 10, 23, and 60 h after i.v. injection of Gd-EOB-DTPA (0.664 mmoles/kg) from a representative mouse bearing a control HCT 116 xenograft (left flank) and a xenograft expressing the pTRE3G-LO construct (right flank). Animals were induced with 10 mg/mL doxycycline in the drinking water for 3 d before imaging. (G) Relaxation rates (R1) before (0 h) and following i.v. injection of Gd-EOB-DTPA. R1 was significantly greater in xenografts expressing Oatp1 compared with controls (one-tailed paired t test, *P < 0.05, **P < 0.01, ***P < 0.005, n = 4 at 0–24 h, n = 3 at 60 h). Error bars show the SEM. Some error bars are obscured by the data points.
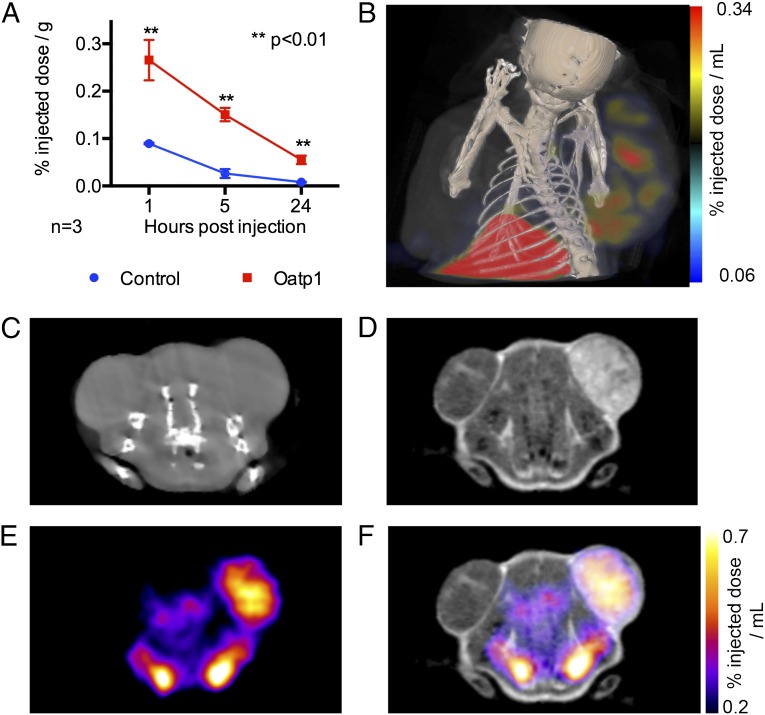
To demonstrate that Oatp1 could be used as a dual reporter for radionuclide imaging, we replaced Gd3+ in the MRI contrast agent with 111In (111In-EOB-DTPA). Mice with control and Oatp1-expressing HEK 293T xenografts, as used in the MRI experiments (Fig. 3), were injected with 50–70 MBq of 111In-EOB-DTPA and imaged using SPECT (Fig. 4). Xenografts expressing Oatp1 contained 3.1-fold more of the injected dose per gram tissue compared with control tumors at 1 h postinjection, 7.8-fold more at 5 h, and 6.9-fold at 24 h (Fig. 4A). SPECT-CT imaging at 1 and 5 h postinjection showed enhancement in the Oatp1-expressing xenografts in comparison with control xenografts, see Fig. 4 B and F. Enhancement of Oatp1-expressing xenografts in both MR and SPECT images was demonstrated, following sequential injections of Gd- EOB-DTPA and 111In-EOB-DTPA, see Fig. 4 D–F. Background enhancement of vasculature was visible on SPECT images at 1 h postinjection, however this had cleared at the 5-h imaging time point. Autoradiography of frozen tissue sections, which had been excised at 1 and 5-h post-111In-EOB-DTPA injection, demonstrated a close correlation between tissue viability and 111In-EOB-DTPA uptake in Oatp1-expressing tissue. There was 16.6-fold more 111In-EOB-DTPA in viable regions of the Oatp1-expressing xenografts than in viable regions of control tumors at 5-h postinjection (Fig. S7).
Fig. 4.
Oatp1 expression was detectable in HEK 293T xenografts using MRI and SPECT after sequential injections of Gd-EOB-DTPA and 111In-EOB-DTPA, respectively. (A) Measurements on extracted tissue showed that 111In-EOB-DTPA was taken up by Oatp1-expressing xenografts (n = 3, two-tailed t test, **P < 0.01). (B) SPECT-CT image obtained 5 h after injection of 111In-EOB-DTPA. The xenograft on the left flank was a control and the xenograft on the right flank expressed Oatp1. (C) CT image showing control (left flank) and Oatp1-expressing (right flank) xenografts. (D) T1-weighted MR image of the section shown in C, 1 h postinjection of Gd-EOB-DTPA. (E) SPECT image of the cross section shown in C, 1 h postinjection of 111In-EOB-DTPA. (F) MRI and SPECT images shown in D and E combined.
Discussion
We have described a gene reporter that can be used with MRI and radionuclide (SPECT) imaging. Exchanging Gd3+ for 68Ga would also allow imaging with PET (23). As an MRI gene reporter the construct described here has a number of advantages over those described previously. Detection is based on a Gd3+ contrast agent, which gives large positive contrast in T1-weighted images and is easier to detect than the negative contrast generated by T2-agents, such as superparamagnetic iron oxide (SPIO) nanoparticles (24) magA (25), and ferritin (26, 27). The contrast agent is small, thus readily exiting the vasculature to gain access to Oatp1-expressing cells, and because Oatps show restricted tissue expression (28) background uptake in most tissues is expected to be low (see a whole mouse with a Gd-EOB-DTPA–enhanced xenograft in Movie S1). The observed relationship between R1 and induction of Oatp1 expression indicates that the kinetics of contrast agent uptake should give quantitative estimates of gene expression levels. Finally, because Gd3+-based hepatocyte-specific contrast agents have been approved for human use (15), and human OATP1B3 has been shown to take up this contrast agent (16), this could facilitate transfer of this technology to the clinic in the longer term.
The reporter may be particularly useful for tracking implanted stem cells, because reporter detection should be unaffected by cell division or exocytosis, which in the case of SPIO-based labeling causes dilution of the label (24, 29), and for interrogating the differentiation state of the cells by incorporating tissue-specific promoters into the construct (30). The reporter could also be used for monitoring the integration and expression of gene therapy vectors (1). The combination of MRI and SPECT detection with Gd-EOB-DTPA and 111In-EOB-DTPA, respectively, may allow whole-body screening with SPECT, for detection of labeled cell deposits, followed by more high-resolution and targeted imaging with MRI.
Methods
Vector Construction.
An Oatp1-expressing plasmid was constructed by cloning a cDNA encoding Oatp1a1 (kindly provided by Allan Wolkoff, Department of Medicine, and Department of Anatomy and Structural Biology, Albert Einstein College of Medicine, Bronx, NY, National Center for Biotechnology Information, Nucleotide Database, reference sequence NM_017111.1) into pEF6-V5/His A (Invitrogen). A 3,729-bp sequence encoding mStrawberry (4), E2A (QCTNYALLKLAGDVESNPGP) (18), and Oatp1, with a stop codon at the end of Oatp1, were cloned into the lentiviral transfer plasmid pBOBI (Inder M. Verma laboratory, Salk Institute, La Jolla, CA) 3′ to a 500-bp PGK promoter (17) (referred to as “LV-PGK-SO”). Replication-defective vesicular stomatitis virus glycoprotein-pseudotyped lentiviral vectors were produced (31), and purified (32) according to published protocols, using the transfer plasmids and three packaging plasmids (Inder M. Verma laboratory). Inducible plasmids were produced using a doxycycline responsive promoter (TRE3G): one with the mStrawberry–E2A–Oatp sequence described above and one with mStrawberry replaced by the luciferase (Luc2)-coding sequence (referred to as “pTRE3G-SO” and “pTRE3G-LO”).
Animal Experiments.
Female SCID mice were obtained from Charles River at ∼20 g (2–3 mo) and used within a few months of acquisition. Animals were injected s.c. with 1 × 107 viable cells per flank and anesthetized using 2% Isofluorane in 75% air/25% O2 (vol/vol; flow rate, 2 L/min) for bioluminescence, fluorescence, radionuclide, and MRI experiments. Animal experiments were carried out under the authority of project and personal licenses issued by the Home Office of the United Kingdom and had been subject to ethical review by the Cancer Research UK, Cambridge Institute Animal Welfare and Ethical Review Body.
Cell Culture.
HEK 293T cells were acquired from Thermo Scientific, breast adenocarcinoma cells (MCF-7) from the European Collection of Animal Cell Cultures (Teddington, UK) and HCT 116 cells from LGC Promochem. All cells were grown in DMEM (Invitrogen), supplemented with 10% (vol/vol) FBS, 2 mM l-glutamine, and 0.1 mM MEM nonessential amino acids, except HCT 116 cells, which were grown in McCoy’s 5A GlutaMAX media (Invitrogen), supplemented with 10% (vol/vol) FBS. Cells were mycoplasma tested upon receipt. Additionally, before experimentation, HEK 293T and HCT 116 cells were genotyped using AmpFISTR profiling (Invitrogen), with HCT 116 cells having 94% identity with the American Type Culture Collection profile, and HEK 293T cells having 88% identity.
Cell Transfection and Transduction.
pEF6-Oatp1a1 and pEF6-V5/His A were linearized with Fsp I and transfected into MCF-7 cells seeded at a density of 1 × 104 cells per cm2 using FuGene (Roche). Transfected cells were selected using Blasticidin (15 μg/mL) and cloned. HEK 293T cells were transduced with a feline immunodeficiency virus-based vector (33) encoding a Luc2–YFP fusion protein under a CMV promoter. Transgene-expressing clones were selected using hygromycin. Stable clones from lentivirus-infected cells were expanded from single cells, following plating out at low seeding densities. Expression of the transgenes was confirmed by Western blot and by flow cytometry (FACSCalibur II) or fluorescence imaging (IVIS 200 series camera; Caliper Life Sciences) or by microscopy. The doxycycline-inducible luciferase–E2A–Oatp1 and mStrawberry–E2A–Oatp1 plasmids used for MRI and radionuclide experiments in vivo and the MRI in vitro, were constructed using a modified TRE3G vector (Clontech) containing a puromycin resistance cassette for selection of stable clones.
Cell Uptake Experiments.
MCF-7 cells were seeded at 1.2 × 105 cells per cm2 and after ∼24 h were washed with transport buffer (142 mM NaCl, 5 mM KCl, 1 mM KH2PO4, 1.2 mM MgSO4, 1.5 mM CaCl2, 5 mM d-glucose, 12.2 mM Hepes, pH 7.3) and incubated with 5 mM Gd-EOB-DTPA (Bayer) at 37 °C, 5% CO2. After incubation, cells were washed and lysed with Nonidet P-40. HEK 293T and HCT 116 cells expressing luciferase–YFP and cells expressing mStrawberry–OATP, were grown to ∼80% confluence and incubated with or without Gd-EOB-DTPA (0.25 mM, 0.5 mM) in the growth media for 90 min. The media was then removed and the cells washed twice in PBS. Cells were then trypsinized, pelleted in 1.5-mL Eppendorf centrifuge tubes, the supernatant removed, and T1 of the pellets determined using a T1 imaging experiment at 7 T.
Western Blots.
Cells were extracted using M-Per reagent (Thermo Scientific) with Complete Mini Protease inhibitors (Roche), and protein concentration determined using the Bradford assay. Extracts were run on an SDS PAGE gel (NuPAGE, Invitrogen), transferred to a nitrocellulose membrane and luciferase–YFP detected using polyclonal goat anti-luciferase antibody (g475A, Promega), at 1 in 25,000 dilution with incubation for 1 h at room temperature. For α-tubulin staining, a monoclonal mouse anti–α-tubulin (T1799, Sigma-Aldrich) was used at 1 in 1,000 dilution with overnight incubation at 4 °C. For mStrawberry staining, rabbit polyclonal anti-RFP (ab34771, AbCam) was used at 1 in 10,000 dilution with overnight incubation at 4 °C. Horseradish peroxidase-conjugated anti-rabbit (111-035-003), anti-goat (705-035-003-JIR), and anti-mouse (115-035-006-JIR) antibodies (Jackson Immunoresearch) were used as the secondary antibodies at 1 in 10,000 dilution with an incubation time of 45 min at room temperature. All antibodies were diluted with 5% (wt/vol) milk powder and 0.1% Tween.
Fluorescence and Bioluminescence Imaging.
Images were acquired using an IVIS 200 series camera (Perkin-Elmer) with an F-stop of 1 and exposure of 60 s. RFP imaging was performed with excitation wavelengths between 580 and 610 nm and emission wavelengths between 615 and 665 nm, and YFP imaging with excitations between 480 and 520 nm and emission between 525 and 560 nm. Before fluorescence imaging, mice were shaved around the site of cell implantation. Regions of interest were analyzed using Living Image software (Perkin-Elmer).
MRI.
Images were acquired using a 7-T horizontal system and VnmrJ software (Agilent). T1-weighted spin-echo images (14 slices 1.5 mm thick) were acquired with an echo time of 8 ms, repetition time of 400 ms, 50 × 50 mm2 field of view (FOV), 256 × 128 resolution, and four averages per increment. T1 maps were acquired using an inversion recovery fast low angle shot pulse sequence with eight inversion times between 0.05 s and 6.4 s, repetition time (TR) 4.46 ms, echo time (TE) 2.1 ms, 12-s delay between images, resolution 128 × 64, FOV of 60 × 60 mm, slice thickness of 2 mm, two averages per increment, and a flip angle of 10°. Gd3+-chelate concentration in cell lysates was determined by measuring the T1 relaxation time at 9.4 T using an inversion recovery pulse sequence with seven inversion times between 0.25 s and 16 s. Whole-body acquisitions were performed using a 3D gradient-echo pulse sequence (TR 20 ms, TE 3 ms, flip angle 20°, FOV 76.8 × 38.4 × 25.6 mm, data matrix 384 × 192 × 128).
SPECT Imaging.
SPECT imaging was performed using a NanoSPECT system (Bioscan Inc.), and CT was performed using a microtomography system (NanoPET/CT; Mediso). SPECT apertures of 2 mm were used with a scan time of 1 h. Images were reconstructed and data analyzed using Vivoquant 1.42 software (InviCRO).
Histology.
Following MRI, tissues were excised and fixed in 10% formalin in PBS for 24 h, then changed into ethanol, and paraffin embedded. Sections 6 μm thick were cut and stained with hematoxylin/eosin (H&E). RFP was stained with rabbit polyclonal anti-RFP (ab34771, AbCam), at 1 in 500 dilution at 4 °C overnight and developed using ABC Vectastain and DAB peroxidase kits according to the manufacturer’s instructions (Vectorlabs). Slides were scanned using a Scanscope XT (Aperio). Following 111In-EOB-DTPA injection, excised xenografts were frozen on dry ice and sectioned (10 µm) at −20 °C in a cryostat (CM3050S Cryostat; Leica). Sections were thaw mounted onto microscope slides and air dried before placing next to a storage phosphor screen for 1 wk (BAS-IP MS 2040; GE Healthcare). Slides were then stained with H&E. Phosphor screens were imaged on a Typhoon Trio+ (GE Healthcare) at pixel size of 25 µm. Images were analyzed using ImageJ software (National Institutes of Health).
Statistical Analyses.
Student’s t test and linear regression were performed using Excel (Microsoft) and Graphpad Prism (Graphpad Software Inc.) software. All t tests were calculated to find an effect size of 3.5 (Cohen’s d) between populations with a P ≤ 0.05 using three biological replicates per condition, with a power of 80%, assuming a normal distribution. Unequal variance t tests were used when variance between groups was not similar. No blinding or randomization were used in any of the experiments, and no animals were excluded from analyses.
Production of 111In-EOB-DTPA.
Primovist (Bayer) was diluted to 0.1 M in a 0.4-M solution of oxalate dihydrate. The precipitate was removed by centrifugation and the supernatant passed through a syringe column (Oasis HLB, 225-mg resin; Waters), which had been preflushed with 1 mL ethanol and 10 mL water. Excess oxalate was eluted with 10 mL water, then the gadolinium-free chelate was eluted with 5 mL 10% ethanol and vacuum dried. Identity of the chelate and labeling with cold indium were confirmed by mass spectrometry. Labeling was performed in 0.1 M ammonium acetate buffer (pH 7), in which the chelate was dissolved at 1.5 mg/mL and mixed with 111In at a ratio of 2:1, reducing the pH to 6. Labeling was assessed at 98%, using HPLC and TLC, after 10 min incubation at room temperature.
Supplementary Material
Acknowledgments
We thank Andre Neves for help with purifying the 111In-labelled chelate, Yelena Wainman for mass spectrometric analysis of the complex, and Piotr Dzien for help with genotyping. This work was supported by the Medical Research Council and Cancer Research UK (CRUK) doctoral training grants (to P.S.P.), a Biotechnology and Biological Sciences Research Council studentship (to J.H.), a University of Cambridge Overseas Research Studentship (to L.L.), a Yousef Jameel studentship (to S.-S.T.), and by the European Commission 7th Framework ENCITE Integrated project. T.B.R. was in receipt of Intra-European Marie Curie (FP7-PEOPLE-2009-IEF, Imaging Lymphoma) and Long-Term European Molecular Biology Organization (EMBO-ALT-1145-2009) fellowships. This work was also supported by a CRUK Programme Grant (to K.M.B.).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1319000111/-/DCSupplemental.
References
- 1.Brader P, Serganova I, Blasberg RG. Noninvasive molecular imaging using reporter genes. J Nucl Med. 2013;54(2):167–172. doi: 10.2967/jnumed.111.099788. [DOI] [PubMed] [Google Scholar]
- 2.Lyons SK, Patrick PS, Brindle KM. 2013. Imaging mouse cancer models in vivo using reporter transgenes. Cold Spring Harb Protoc 2013(8):685–699.
- 3.Contag CH, Bachmann MH. Advances in in vivo bioluminescence imaging of gene expression. Annu Rev Biomed Eng. 2002;4:235–260. doi: 10.1146/annurev.bioeng.4.111901.093336. [DOI] [PubMed] [Google Scholar]
- 4.Shaner NC, et al. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat Biotechnol. 2004;22(12):1567–1572. doi: 10.1038/nbt1037. [DOI] [PubMed] [Google Scholar]
- 5.James ML, Gambhir SS. A molecular imaging primer: Modalities, imaging agents, and applications. Physiol Rev. 2012;92(2):897–965. doi: 10.1152/physrev.00049.2010. [DOI] [PubMed] [Google Scholar]
- 6.Vandsburger MH, Radoul M, Cohen B, Neeman M. MRI reporter genes: Applications for imaging of cell survival, proliferation, migration and differentiation. NMR Biomed. 2013;26(7):872–884. doi: 10.1002/nbm.2869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gilad AA, Winnard PT, Jr, van Zijl PC, Bulte JW. Developing MR reporter genes: Promises and pitfalls. NMR Biomed. 2007;20(3):275–290. doi: 10.1002/nbm.1134. [DOI] [PubMed] [Google Scholar]
- 8.Vande Velde G, et al. Quantitative evaluation of MRI-based tracking of ferritin-labeled endogenous neural stem cell progeny in rodent brain. Neuroimage. 2012;62(1):367–380. doi: 10.1016/j.neuroimage.2012.04.040. [DOI] [PubMed] [Google Scholar]
- 9.Deans AE, et al. Cellular MRI contrast via coexpression of transferrin receptor and ferritin. Magn Reson Med. 2006;56(1):51–59. doi: 10.1002/mrm.20914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tannous BA, et al. Metabolic biotinylation of cell surface receptors for in vivo imaging. Nat Methods. 2006;3(5):391–396. doi: 10.1038/nmeth875. [DOI] [PubMed] [Google Scholar]
- 11.Bartelle BB, Szulc KU, Suero-Abreu GA, Rodriguez JJ, Turnbull DH. 2013. Divalent metal transporter, DMT1: A novel MRI reporter protein. Magn Reson Med 70(3):842–850. [DOI] [PMC free article] [PubMed]
- 12.Ray P, De A, Min JJ, Tsien RY, Gambhir SS. Imaging tri-fusion multimodality reporter gene expression in living subjects. Cancer Res. 2004;64(4):1323–1330. doi: 10.1158/0008-5472.can-03-1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Niers JM, et al. Single reporter for targeted multimodal in vivo imaging. J Am Chem Soc. 2012;134(11):5149–5156. doi: 10.1021/ja209868g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jacquemin E, Hagenbuch B, Stieger B, Wolkoff AW, Meier PJ. Expression cloning of a rat liver Na(+)-independent organic anion transporter. Proc Natl Acad Sci USA. 1994;91(1):133–137. doi: 10.1073/pnas.91.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van Montfoort JE, et al. Hepatic uptake of the magnetic resonance imaging contrast agent gadoxetate by the organic anion transporting polypeptide Oatp1. J Pharmacol Exp Ther. 1999;290(1):153–157. [PubMed] [Google Scholar]
- 16.Leonhardt M, et al. Hepatic uptake of the magnetic resonance imaging contrast agent Gd-EOB-DTPA: Role of human organic anion transporters. Drug Metab Dispos. 2010;38(7):1024–1028. doi: 10.1124/dmd.110.032862. [DOI] [PubMed] [Google Scholar]
- 17.Adra CN, Boer PH, McBurney MW. Cloning and expression of the mouse pgk-1 gene and the nucleotide sequence of its promoter. Gene. 1987;60(1):65–74. doi: 10.1016/0378-1119(87)90214-9. [DOI] [PubMed] [Google Scholar]
- 18.Szymczak AL, et al. Correction of multi-gene deficiency in vivo using a single ‘self-cleaving’ 2A peptide-based retroviral vector. Nat Biotechnol. 2004;22(5):589–594. doi: 10.1038/nbt957. [DOI] [PubMed] [Google Scholar]
- 19.Pascolo L, et al. Molecular mechanisms for the hepatic uptake of magnetic resonance imaging contrast agents. Biochem Biophys Res Commun. 1999;257(3):746–752. doi: 10.1006/bbrc.1999.0454. [DOI] [PubMed] [Google Scholar]
- 20.Shi X, et al. Stable inducible expression of a functional rat liver organic anion transport protein in HeLa cells. J Biol Chem. 1995;270(43):25591–25595. doi: 10.1074/jbc.270.43.25591. [DOI] [PubMed] [Google Scholar]
- 21.Riches AC, Sharp JG, Thomas DB, Smith SV. Blood volume determination in the mouse. J Physiol. 1973;228(2):279–284. doi: 10.1113/jphysiol.1973.sp010086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schuhmann-Giampieri G, et al. Preclinical evaluation of Gd-EOB-DTPA as a contrast agent in MR imaging of the hepatobiliary system. Radiology. 1992;183(1):59–64. doi: 10.1148/radiology.183.1.1549695. [DOI] [PubMed] [Google Scholar]
- 23.Wadas TJ, Wong EH, Weisman GR, Anderson CJ. Coordinating radiometals of copper, gallium, indium, yttrium, and zirconium for PET and SPECT imaging of disease. Chem Rev. 2010;110(5):2858–2902. doi: 10.1021/cr900325h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rogers WJ, Meyer CH, Kramer CM. Technology insight: In vivo cell tracking by use of MRI. Nat Clin Pract Cardiovasc Med. 2006;3(10):554–562. doi: 10.1038/ncpcardio0659. [DOI] [PubMed] [Google Scholar]
- 25.Zurkiya O, Chan AW, Hu X. MagA is sufficient for producing magnetic nanoparticles in mammalian cells, making it an MRI reporter. Magn Reson Med. 2008;59(6):1225–1231. doi: 10.1002/mrm.21606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cohen B, Dafni H, Meir G, Harmelin A, Neeman M. Ferritin as an endogenous MRI reporter for noninvasive imaging of gene expression in C6 glioma tumors. Neoplasia. 2005;7(2):109–117. doi: 10.1593/neo.04436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Genove G, DeMarco U, Xu H, Goins WF, Ahrens ET. A new transgene reporter for in vivo magnetic resonance imaging. Nat Med. 2005;11(4):450–454. doi: 10.1038/nm1208. [DOI] [PubMed] [Google Scholar]
- 28.Tamai I, et al. Molecular identification and characterization of novel members of the human organic anion transporter (OATP) family. Biochem Biophys Res Commun. 2000;273(1):251–260. doi: 10.1006/bbrc.2000.2922. [DOI] [PubMed] [Google Scholar]
- 29.Cromer Berman SM, et al. Cell motility of neural stem cells is reduced after SPIO-labeling, which is mitigated after exocytosis. Magn Reson Med. 2013;69(1):255–262. doi: 10.1002/mrm.24216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Agostini S, Recchia FA, Lionetti V. Molecular advances in reporter genes: The need to witness the function of stem cells in failing heart in vivo. Stem Cell Rev. 2012;8(2):503–512. doi: 10.1007/s12015-011-9296-9. [DOI] [PubMed] [Google Scholar]
- 31.Tiscornia G, Singer O, Verma IM. Production and purification of lentiviral vectors. Nat Protoc. 2006;1(1):241–245. doi: 10.1038/nprot.2006.37. [DOI] [PubMed] [Google Scholar]
- 32.Kutner RH, Zhang XY, Reiser J. Production, concentration and titration of pseudotyped HIV-1-based lentiviral vectors. Nat Protoc. 2009;4(4):495–505. doi: 10.1038/nprot.2009.22. [DOI] [PubMed] [Google Scholar]
- 33.Curran MA, Kaiser SM, Achacoso PL, Nolan GP. Efficient transduction of nondividing cells by optimized feline immunodeficiency virus vectors. Mol Ther. 2000;1(1):31–38. doi: 10.1006/mthe.1999.0007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.