Abstract
In normal tissues or tumors, cells have extensive opportunities for adhesion to their neighbors. This state is mimicked by dense cell cultures. In this review, we integrate some recent findings on a key signal transducer, STAT3 (signal transducer and activator of transcription-3), whose activity is dramatically increased following cadherin-mediated cell to cell adhesion. Cadherin engagement, favored in dense cell cultures, causes a dramatic increase in total Rac/Cdc42 protein levels through inhibition of proteasomal degradation, which is followed by activation of IL-6 and STAT3. The cadherin/Rac/IL-6/STAT3 axis offers a potent survival signal that is a prerequisite for neoplastic transformation, as well as normal tissue function.
Keywords: signal transducer and activator of transcription-3, cadherins, apoptosis, interleukin-6, Rac, neoplastic transformation, YAP/TAZ, Erk1/2
Introduction
In the past 30 years, the signal transduction pathways initiated by growth factors or oncogenes have been the focus of intense research and the major ones have been characterized. It also became apparent that the three-dimensional tissue architecture and mechanical forces applied upon a cell in a tissue are important signals informing fundamental cell decisions such as proliferation, differentiation, and apoptosis (reviewed in ref. 1).
In normal tissues or tumors, cells have extensive opportunities for adhesion to their neighbors, unlike sparsely growing, cultured cells. Dense cell cultures however, although two-dimensional, may in part mimic some of the physiological stress signals present in tissues. In this review, we integrate some recent findings on a common signal transducer, STAT3 (signal transducer and activator of transcription-3), whose activity was demonstrated to be dramatically increased through cadherin-mediated, cell-to-cell contact, thus contributing substantially to cellular survival. Apoptosis inhibition is, in fact, a prerequisite for neoplastic transformation, as well as normal tissue function.
Activation of STAT3
Classical pathways of STAT3 activation
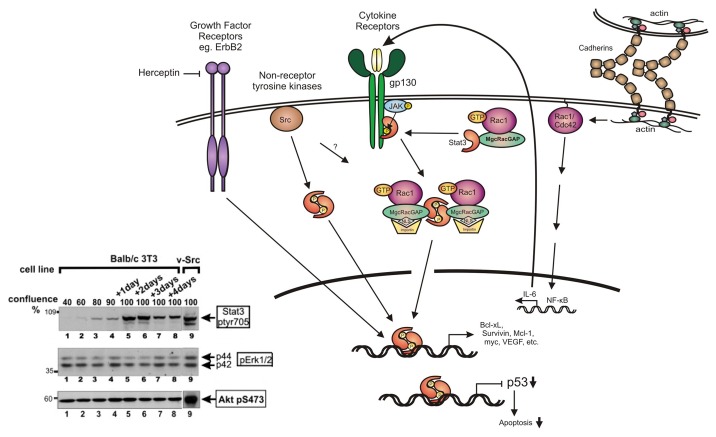
STAT3 is a member of the STAT family of transcription factors (STAT1 to STAT6), that mediate a variety of responses in mammalian cells. STAT3 is activated by cytokine receptors especially of the IL-6 family, receptor tyrosine kinases, such as the EGFR (epidermal growth factor receptor) family including Her2/neu, as well as members of the non-receptor tyrosine kinase Src family (Fig. 1). A number of studies have shown that in quiescent cells STAT3 is inactive in the cytoplasm. Following receptor stimulation, STAT3 is phosphorylated at the critical tyr-705 by the associated JAK or Src kinases (STAT3-ptyr705). This leads to STAT3 dimerization through reciprocal Src homology 2 (SH2)-ptyr interactions, which are facilitated by the MgcRacGAP (male germ cell RacGAP) molecule, bound to activated Rac1. The complex is then targeted to the nucleus, driven by the NLS (nuclear localization signal) of MgcRacGAP (reviewed in ref. 2). The phosphorylated STAT3 dimer then binds a 9 bp sequence (TTCNNNGAA) in the regulatory region of target genes, thereby activating the transcription of specific genes involved in cell division and survival, such as myc, bcl-xL, mcl-1, and survivin while it downregulates the tumor suppressor p53,3 thus protecting tumor cells from apoptosis.4,5 In addition, STAT3 monomers were shown to regulate transcription by binding to NFκB.6
Figure 1. STAT3 activation. Following activation of receptors such as cytokine receptors of the IL-6 family or growth factor receptors such as Her2/ErbB2 (that can be inhibited by drugs such as herceptin), or expression of activated forms of non-receptor tyrosine kinases such as Src, STAT3 is activated by phosphorylation at tyr705 by the receptor itself, or the associated Src or JAK kinases. STAT3 phosphorylation by the IL-6/JAK complex or Src is facilitated through binding to activated Rac1-GTP in a complex with MgcRacGAP. This results in targeting of the complex to the nuclear envelope, driven by the NLS (nuclear localization signal) of MgcRacGAP. The STAT3 dimer then binds specific DNA sequences to initiate transcription of STAT3-responsive genes, or downregulation of other genes such as the p53 tumor suppressor. In addition to this mechanism, cadherin engagement was shown to cause a dramatic increase in the levels of Rac1 and Cdc42 proteins and activity, which results in a transcriptional activation of IL-6 through NFκB, hence STAT3 activation. At the same time, cadherin engagement suppresses Erk1/2 activation by IL-6 (not shown). Inset: Lysates from mouse Balb/c3T3 fibroblasts were grown to different densities as indicated and probed for STAT3-ptyr705, pErk1/2 or active Akt-pser473 (lanes 1–8). Lane 9: Balb/c3T3 cells transformed by activated Src. Note that STAT3-ptyr705 is dramatically increased with density, while levels of p-Erk and Akt-pS473 remain unchanged. Similar results were obtained by plating cells on surfaces coated with E-cadherin or cadherin-11 fragments.19,20 (Modified and adapted from refs. 2 and 20).
In addition to these functions, it has recently emerged that STAT3 can also inhibit apoptosis by affecting the cellular metabolism. STAT3-ptyr705 transcriptionally activates the oxygen sensor HIF1α (hypoxia inducible factor-1α) transcription factor, and this leads to increased aerobic glycolysis.7 At the same time, STAT3-ptyr705 downregulates mRNAs of mitochondrial genes, thereby reducing oxidative phosphorylation and ROS (reactive oxygen species) production with apoptosis reduction as a result.8 STAT3 is also phosphorylated on ser727 downstream of a number of stimuli that trigger MAP kinase activation, including Ras signaling or stress.8,9 Interestingly, contrary to STAT3-ptyr705, which is acting in the nucleus, STAT3-ser727 localizes to the mitochondria where it acts in a transcription-independent manner to enhance the activity of ETC (electrotransfer chain) complexes and glycolysis, thus promoting survival as well. Moreover, STAT3-ptyr727 opposes the mitochondrial permeability transition pore, thereby inhibiting apoptosis further.7,10,11
STAT3 is found to be overactive in a number of human cancers and to be required for transformation by a number of oncogenes.5 Persistent activation of STAT3 occurs mainly downstream of pro-oncogenic tyrosine kinases and rather infrequently through a direct STAT3 mutation.12 In any event, a constitutively active form of STAT3 (STAT3C) alone is sufficient to induce neoplastic transformation of cultured mouse fibroblasts,13 which points to an etiological role for STAT3 in neoplasia.
Cadherin engagement also activates STAT3
Classical cadherins activate STAT3 through Rac/Cdc42, NFκB, and IL-6
Recent results from a number of labs indicated that besides growth factors and oncogenes, confluence of cultured cells induces a dramatic surge in STAT3 and ptyr705 phosphorylation and activity in a number of cell lines14-20 (reviewed in ref. 21). The fact that STAT3 activity is actually increased in growth-arrested cells cultured to high densities was an unexpected finding, since STAT3 was generally accepted to have a positive role in cell proliferation. It was further demonstrated that this STAT3 increase requires calcium, which pointed to a role for the cadherins, calcium-dependent, cell-to-cell adhesion proteins.
Cadherins are a superfamily of adhesive receptors that control the specificity, organization and dynamics of intercellular recognition and junction formation, which is crucial for the development, stability, and homeostasis of tissue architecture and function in all multicellular metazoans.22-24 Classical cadherins are glycoproteins that consist of an extracellular (EC) domain of five cadherin modules (EC1 to EC5), a single-pass transmembrane domain and a highly conserved intracellular part, which interacts with the cytoskeleton via the catenin family of proteins.25,26 Upon calcium binding, the EC domains change conformation to form adhesive structures between adjacent cells, named adherens junctions.23,27,28 Such adhesive structures participate in cell polarity and differentiation, and provide local reinforcement against mechanical stress. Classical cadherins are subdivided into type I (E, N, M, P, R, and C cadherin) and type II (VE, 6–12, 18–20, 22, 24) based on the structure of their EC domains.26 Interestingly, it has been demonstrated that differences observed in the adhesion efficiency between cells expressing type I and type II cadherins could be due to differences in the structure of the adhesive interface.29-33
The first discovered and best characterized classical type I cadherin is the E (epithelial)-cadherin, which is involved in the formation and maintenance of epithelial structures and is abundant in cultured cells of epithelial origin.24,34,35 Early results showed that continued plasma membrane expression of E-cadherin is required for cells to remain tightly associated within the epithelium, so that loss of E-cadherin function, including mislocalization to the cytoplasm, is associated with metastasis of breast cancer.22 Other cadherins however, such as N-cadherin and cadherin-11, recently defined as “mesenchymal”,36 have been positively implicated in cancer as putative proto-oncogenic proteins and are often upregulated in tumor tissues of epithelial origin. N-cadherin overexpression and engagement has been reported to be associated with a highly invasive phenotype and motility in mammary cell lines.22,37 Cadherin-11 (classical type II) was originally identified in mouse osteoblasts, but it was later found to be constitutively expressed in a variety of normal tissues of mesodermal origin,38 as well as in cultured fibroblasts.20 Cadherin-11 correlates with metastasis and a poor prognosis in cancers such as cancer of the breast and prostate.39,40
Cadherin engagement was previously shown to regulate the activity of the Rho family of small GTPases,41 which, in turn, regulate actin organization.42 Examination of the mechanism of STAT3 activation in confluent cultures later indicated that the engagement of E-cadherin in HC11, mouse breast epithelial cells induces a dramatic increase in total protein levels of the Rac and Cdc42, small GTPases through inhibition of proteasomal degradation, and this could account for the increase in Rac/Cdc42 activity observed (Fig. 1). Activated Rac leads to a surge in secretion of cytokines of the IL-6 family through the transcription factor NFκB and JAK kinases, and this, in turn activates STAT3, which constitutes a potent survival signal19 (reviewed in ref. 21). Downregulation of gp130, the common subunit of the family, did reduce STAT3-ptyr705 substantially,19,20 indicating that the IL-6 family is a significant contributor to the density-induced, STAT3 upregulation. Other cytokines such as the TNF family may also be involved, however their levels varied with the cell line, indicating that the additional pathways may not be well preserved between different cells and tissues.19,20 Interestingly, although JAK activity increased with cell density,14 Src activity remained unchanged.43,44 In addition, fibroblasts from mice where cSrc, as well as the related Fyn and Yes genes had been genetically ablated displayed the same increase in STAT3-ptyr705 with cell density, and treatment with the Src inhibitors Dasatinib or PD180970 did not reduce the density-dependent STAT3 activation in a number of lines.43 These data indicate that, despite the fact that it has been implicated in cell adhesion signaling, Src is not involved in the density-mediated STAT3 activation.14,15 Similarly, cells from Fer or IGF-R-knockout mice displayed the same increase in STAT3 activity with cell density, indicating that these receptor kinases are unlikely to be involved.14
Recent findings revealed that cadherin engagement can directly activate STAT3, in the absence of cell to cell contact. Plating noneoplastic, mouse breast epithelial HC11 cells on surfaces coated with a cadherin fragment encompassing the two outermost modules of E-cadherin, resulted in a dramatic increase in STAT3, tyr705 phosphorylation and activity. These data conclusively demonstrated that the cell density-dependent, STAT3 activation is directly due to cadherin engagement on the functionalized surface.19 As anticipated, this activation was mediated through an increase in total Rac/Cdc42 protein, followed by a transcriptional upregulation of IL-6 family cytokines, indicating that cadherin engagement is sufficient to initiate this cellular signal, in the absence of cell to cell contact.19 Interestingly, examination of E-cadherin and STAT3-ptyr705 levels in normal mouse breast tissues by immunohistochemistry staining, demonstrated the presence of constitutively activated STAT3 in the breast luminal epithelial cells where E-cadherin is engaged, i.e., the corresponding normal breast tissue of origin of HC11 cells. This finding reveals a distinct correlation in the distribution and presumably the function of these two molecular markers observed in HC11 cells growing at high, but not low densities in culture, with the same type of cell in vivo.19
Recent results demonstrated that the “mesenchymal” cadherins, N-cadherin and cadherin-11, which belong in the type I and type II classical cadherins, respectively, and whose expression in epithelial cells, unlike E-cadherin, often correlates with metastasis,39 can also trigger a dramatic surge in STAT3 activity in mouse Balb/c3T3 fibroblasts.20 This activation occurs through upregulation of Rac and members of the IL-6 family of cytokines as well, and this is necessary for cell survival, proliferation, and migration. A cadherin-mediated Rac activation was also observed for C-45 and M-cadherin,46 strongly suggesting that this may be a feature common for classical cadherins. Taken together, the above data indicate that the cadherin/Rac/IL-6/STAT3 axis may be a pathway used by multiple cadherins of widely divergent functions.
Cadherin engagement does not allow Erk1/2 activation by IL-6
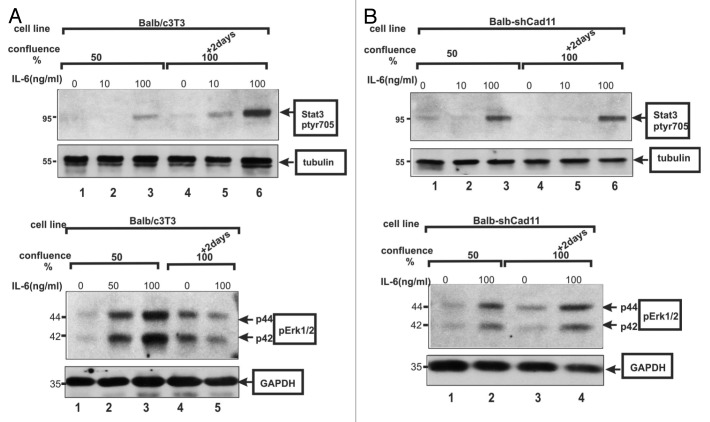
Besides STAT3, IL-6 stimulation was shown to activate the Erk1/2 (Erk) kinase by triggering its phosphorylation at a TEY sequence.47 Surprisingly however, despite the IL-6 secretion by the confluent cells, p-Erk1/2 levels remained the same at high cell densities, or by direct cadherin engagement, at the time when STAT3-ptyr705 levels were dramatically increased. This was found to hold true for both E-cadherin19 and cadherin-11.20 It was subsequently demonstrated that IL-6 addition to confluent, HC11 or Balb/c3T3 cells was unable to activate Erk1/2, hinting at the possibility of a profound effect of confluence on the response of Balb/c3T3 cells to IL-6 addition. To investigate whether this might be due to cadherin function per se, the ability of IL-6 to activate Erk in Balb/c3T3 cells rendered deficient in cadherin-11 was examined. The results demonstrated that, in sharp contrast to the parental Balb/c3T3 cells, IL-6 does stimulate Erk in densely growing, cadherin-11-deficient, Balb-shCad11 cells, clearly indicating that it is indeed cadherin-11 that prevents Erk activation by IL-6 (Fig. 2).
Figure 2. (A) IL-6 activates STAT3, but not Erk, at high densities in Balb/c3T3 cells. IL-6 was added at 0, 10, 50, or 100 ng/mL for 15 min to Balb/c3T3 cells grown to 50% confluence (lanes 1–3), or 2 d postconfluence as indicated. Cell extracts were probed for STAT3-ptyr705 (upper panel) or Erk1/2 (lower panel), with tubulin or GAPDH as loading controls. Note the absence of Erk activation at high densities (lower panel, lanes 4 and 5). (B) IL-6 activates STAT3 and Erk in the absence of cadherin-11. Same as above, cadherin-11-deficient, Balb-shCad11 cells. Note the Erk activation at high densities (lower panel, lanes 3 and 4), same as STAT3, even at high densities (upper panel, 4 vs 6). (From ref. 20, reproduced with permission.)
The reasons for the STAT3 specificity of cadherin engagement are at present unclear. The fact that other growth factors such as EGF or PDGF can activate Erk in densely growing, mouse fibroblasts,48 argues against a blanket Erk inhibition by cadherin engagement, for example, through the activation of Erk-specific phosphatases such as Cdc25A.49 It is tempting to speculate that cadherin engagement stimulates IL-6 secretion in dense tissues in order to counteract apoptotic death signals through STAT3 activation. On the other hand, Erk would promote mainly cell division, which is absent in contact-inhibited cells. In any event, taken together, these findings, obtained with three different type I and type II cadherins demonstrate a specific STAT3 response of cells to cadherin engagement, despite the fact that the two pathways, Erk and STAT3, have been reported to be coordinately regulated by cytokine receptors.
Consequences of STAT3 Activation
The three cadherins described were found to activate STAT3 via the same Rac/NFκB/IL-6/gp130/JAK/STAT3 pathway. The fact that the “mesenchymal” cadherins, cadherin-11 and N-cadherin, actually activate STAT3, although, contrary to the epithelial E-cadherin, they generally promote metastasis in cells of epithelial origin,22,36,39,40 may point to STAT3 as a central survival, rather than metastasis, factor, whose function is important in both tumor and normal tissues. Following are possible mechanisms:
Survival function in neoplastic transformation
Since the initial discovery that STAT3 is required for transformation by activated Src,50,51 it became evident that STAT3 inhibition in Src-transformed cells induces apoptosis, not simply reversion of the cell to a normal phenotype. This initial observation provided the first clue as to the important role of STAT3 in the survival of tumor cells.
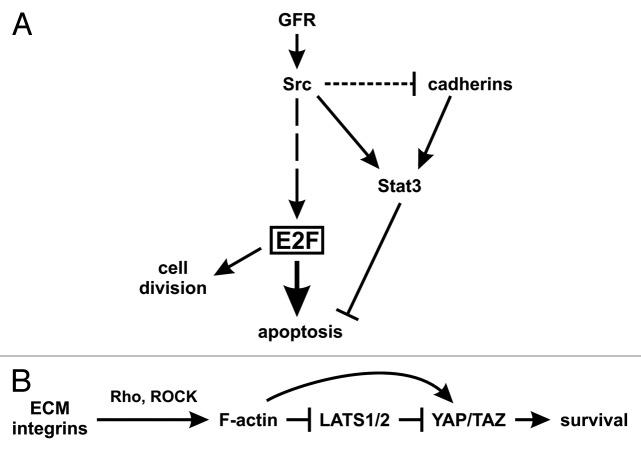
A variety of receptor and non-receptor tyrosine kinase oncogenes affect the phosphorylation status of the Rb (retinoblastoma susceptibility) family of nuclear phosphoproteins, through a number of upstream regulators (G1 cyclins, cyclin-dependent kinases, and their inhibitors). Rb phosphorylation reduces its ability to bind to and regulate their best characterized targets, the E2F family of transcription factors (the “activating” E2Fs, E2F1–3a), which are important cell cycle controllers.52 A detailed examination of E2F-activated genes by microarray analysis indicated that they have many targets including genes involved in DNA synthesis, as well as growth factor and receptor genes.53 Interestingly, at the same time E2F activation leads to apoptosis, as shown in vivo in Rb knockout animals.54 Moreover, ectopic E2F expression has been demonstrated to induce apoptosis under conditions where serum growth factors, which normally impart survival signals, are limiting. Apoptosis is induced through expression of p19ARF by E2F, which negatively regulates the Mdm2, p53-specific ubiquitin ligase, with a surge in p53 levels as a result.55 Furthermore, E2F can also induce apoptosis through p53-independent mechanisms, such as activation of the tumor suppressor p73,56 and induction of Apaf1 expression which, in combination with cytosolic cytochrome c and caspase-9 forms the apoptosome, which activates the final apoptosis effectors57 (reviewed in ref. 58). Therefore, E2F activation, which was shown to occur in the majority of tumors, also induces apoptosis, through both p53-dependent and -independent mechanisms. Certainly, apoptosis is normally prevented, due to activation of PI3 kinase or STAT3 by tyrosine kinase receptors induced by E2F, or directly by the oncogenes which had activated E2F in the first place, so that transformation does occur. Upon inhibition of STAT3 activity however, tumor cells, having high E2F levels, may succumb to apoptosis.15 Most importantly, inhibition of cadherin-11 or N-cadherin would induce apoptosis (through STAT3 inhibition) in metastatic tumor cells specifically, since normal cells, even cells with cadherin-11, would have low E2F activity hence would be spared (Fig. 3A; ref. 43; Raptis, Guy, and Geletu, in preparation).
Figure 3. Model of STAT3 as a survival signal. (A) In transformed cells. Growth factor receptors or Src activate the transcription factor E2F1–3a, through a number of steps.58 E2F, in turn, can induce apoptosis through p53-dependent and -independent pathways, while Src or activated receptors also activate STAT3 which blocks apoptosis, so that transformation can occur. However, cadherin engagement may also be necessary to increase STAT3 activity further, even in cells expressing STAT3 activating oncogenes such as Src, and despite the fact that Src may also trigger cadherin downregulation (Guy, Raptis, et al., in preparation). (B) In normal tissues or cells grown to high densities. When cells are spread on the extracellular matrix (ECM), the forces exerted through integrin-ECM adhesion promote the formation of contractile F-actin structures with myosin molecules, regulated by the Rho GTPase. F-actin filaments oppose YAP/TAZ phosphorylation and degradation through inhibition of the LATS kinases and other mechanisms yet to be identified. In confluent cultures or tissues on the other hand, mechanical constraints reduce integrin-ECM adhesion and F-actin formation, thus allowing LATS activation, hence downregulation of YAP/TAZ and apoptosis.1 At the same time, cadherins activate STAT3 to achieve cell survival. As a result, STAT3 inhibition would cause apoptosis in confluent cultures. (Not shown: Cadherin engagement may also lead to the formation of contractile F-actin.)
Survival function in normal cells and tissues
In tumor cells, the high E2F levels would necessitate the action of a survival agent, such as STAT3, in order to block apoptosis and allow transformation to occur (Fig. 3A). Still, the fact that STAT3 inhibition in cultured, non-malignant, densely growing cells induces apoptosis14,15,19,59 points to the possibility that even in normal cells the role of STAT3 at high cell densities, which may be the densities with relevance in vivo, is not simply to promote proliferation, but to provide a potent survival signal as well.
What could be the apoptotic signal in normal cells grown to high densities? Recent evidence has implicated the cytoskeletal changes occurring as a result of cell shape constraints.
Cell shape is modulated by the biomechanical properties of the cellular environment. Cells can feel the environment via two types of adhesive junctions, namely (a) junctions between the cell and the extracellular matrix (ECM) and (b) junctions between cells. Due to their transmembrane structure, both types of adhesive receptors can establish bridges with cytoskeletal filaments.60-62 These interactions are subject to mechanical forces, and the receptors’ behavior under force is an important parameter of cytoskeletal organization and signal regulation.1,63-65
Cell-ECM adhesion is mediated by the integrins, a family of receptors clustered into focal adhesions. Integrins sense the mechanical properties of the ECM by changing their conformation, avidity, and clustering, and this, in turn, regulates signal transduction.66,67 In fact, it is the degree of cell spreading itself, forced by the actin cytoskeleton, rather than the extent of cell-ECM contact or signaling through the ECM, which is a fundamental, dose-dependent factor of proliferation and differentiation.64,68-70 Integrins promote the bundling of actin filaments64,70 and in a feedback loop, the rate of actin polymerization as well as actin regulatory molecules affect integrin function.65 Integrin adhesion triggers the development of contractile filamentous F-actin structures with myosin molecules, a process regulated by the Rho family of small GTPases.71
Cell-cell interactions are also key players in cell and tissue architecture and cytoskeletal dynamics. At the adherens junctions, cadherins generate, receive, and integrate contractile forces.72,73 At the molecular level, cadherin interactions, mediated through the extracellular domain, are very dynamic and sensitive to force, while their organization into clusters is complex, with several pathways under investigation25,29-31,74,75 (Bron and Feracci, in preparation). Cell to cell contact maturation includes intracellular strengthening by recruitment of the actomyosin cytoskeleton, as well as actin-associated proteins.72
It has recently emerged that key mediators of the biological effects observed in response to cell shape and actin cytoskeletal changes are the transcription coactivators YAP (yes-associated protein) and TAZ (transcriptional co-activator with PDZ-binding motif) which have partly overlapping functions.76,77 The YAP and TAZ shuttle between the cytoplasm and nucleus where they associate with promoter-specific transcription factors, such as TEA domain (TEAD) family members to regulate gene expression.78,79 As a result, high YAP/TAZ levels promote proliferation, while low YAP/TAZ shifts cell responses toward apoptosis or growth arrest.1 The action of YAP and TAZ is kept in check by the LATS1 and 2 (large tumor suppressor-1 and 2) kinases, which phosphorylate YAP/TAZ, thereby promoting their cytoplasmic localization or degradation.78,79 In extended cells, YAP/TAZ are active and localized in the nucleus, while in cells grown on a small adhesive area they are excluded from the nucleus and inactive. It was further demonstrated that the activity of YAP/TAZ determines the response of the cell to mechanical cues; knockdown of YAP/TAZ in cells grown in large adhesive areas produced a phenotype typical of cells grown in small adhesive areas. Conversely, YAP/TAZ overexpression was sufficient to induce an extended cell response.76 Moreover, it was recently shown that contractile F-actin cytoskeletal structures can sustain YAP and TAZ nuclear localization and activity, and oppose YAP and TAZ phosphorylation and degradation through inhibition of the LATS kinases.76
Interestingly, high cell density (hence cadherin engagement) was in fact shown to induce LATS1/2 activity, thereby inhibiting YAP/TAZ target gene expression.80 Therefore, it is tempting to speculate that, at least in certain systems,81 the mechanically constrained cell shape hence low YAP/TAZ levels in confluent cultures is the apoptosis trigger, and this would need to be overcome by cadherin engagement and STAT3, to confer cell and tissue survival.
Conclusions
The three cadherins described so far, E-cadherin,19 N-cadherin, and cadherin-11,20 in various combinations are present in essentially all tissues. The fact that all activate the same STAT3 pathway, points to a central importance of this pathway in cellular survival. This may explain the presence of at least one cadherin in all cells of the organism during embryonic development and homeostasis. It is interesting to note that a close correspondence was found in genes expressed specifically in the prostate carcinoma line, LNCaP, cultured to high, but not low, densities with genes associated with prostate cancer in vivo, which further stresses the relevance of cell interactions observed in densely growing cultured cells to the in vivo situation.82
The need for STAT3 to maintain survival, in both tumor and normal cells may be an indication that survival signals are in limited supply; in tumor cells this may be because of the high E2F-apoptosis inducers, while in normal tissues due to the low YAP/TAZ levels, caused by mechanical constraints. It is interesting to note that no increase in active Akt-p473, the effector of the PI3 kinase survival pathway, was noted in confluent cultures (Fig. 1, inset). Therefore, the fact that cells have evolved to call up from dormancy the STAT3 pathway rather than PI3k to overcome apoptosis, may be an indication that STAT3 may be a more potent inhibitor of apoptosis in both normal and tumor tissues.
Since the discovery of the STAT3 activity surge upon cadherin engagement, other genes have been shown to be affected by cell density to different degrees.83,84 This includes “housekeeping” genes such as GAPDH (glyceraldehyde 3-phosphate dehydrogenase), whose function has long been considered to be constitutively needed. Whether cadherins or STAT3 affect the expression levels of housekeeping genes remains to be determined.
Engagement of cadherins is known to affect cell differentiation. As such, STAT3 is emerging as an oncogene which, unlike activated receptor or non-receptor kinases, it is promoting rather than inhibit the differentiation of epithelial MDCK cells,16 as well as adipocytes (Raptis, Guy, and Geletu, in preparation). Cadherins are also required for gap junction formation and it was recently demonstrated that, contrary to other oncogenes, STAT3 can increase, rather than decrease intercellular, gap junctional communication.83,84 In any event, the cadherin/STAT3 axis is emerging as a central survival factor, which would be especially critical to tumor cells. Most importantly, since cadherin-11 promotes metastasis, inhibition of cadherin-11 would promote apoptosis of metastatic tumor cells specifically (through STAT3 inhibition), a finding which could have important therapeutic implications.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We would like to thank Dr Xiaolong Yang of Queen’s University for valuable suggestions and a critical reading of the manuscript. The financial assistance of the Canadian Institutes of Health Research, the Canadian Breast Cancer Foundation (Ontario Chapter), the Natural Sciences and Engineering Research Council of Canada (NSERC), the Canadian Breast Cancer Research Alliance, the Ontario Centers of Excellence, the Breast Cancer Action Kingston, and the Clare Nelson bequest fund through grants to L Raptis is gratefully acknowledged. S Guy was supported by an NSERC studentship and a Queen’s University Graduate Award. M Geletu was supported by a postdoctoral fellowship from the US Army Breast Cancer Program, the Ministry of Research and Innovation of the Province of Ontario, and the Advisory Research Committee of Queen’s University.
Footnotes
Previously published online: www.landesbioscience.com/journals/jak-stat/article/27363
References
- 1.Halder G, Dupont S, Piccolo S. Transduction of mechanical and cytoskeletal cues by YAP and TAZ. Nat Rev Mol Cell Biol. 2012;13:591–600. doi: 10.1038/nrm3416. [DOI] [PubMed] [Google Scholar]
- 2.Raptis L, Arulanandam R, Geletu M, Turkson J. The R(h)oads to Stat3: Stat3 activation by the Rho GTPases. Exp Cell Res. 2011;317:1787–95. doi: 10.1016/j.yexcr.2011.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Niu G, Wright KL, Ma Y, Wright GM, Huang M, Irby R, Briggs J, Karras J, Cress WD, Pardoll D, et al. Role of Stat3 in regulating p53 expression and function. Mol Cell Biol. 2005;25:7432–40. doi: 10.1128/MCB.25.17.7432-7440.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gritsko T, Williams A, Turkson J, Kaneko S, Bowman T, Huang M, Nam S, Eweis I, Diaz N, Sullivan D, et al. Persistent activation of stat3 signaling induces survivin gene expression and confers resistance to apoptosis in human breast cancer cells. Clin Cancer Res. 2006;12:11–9. doi: 10.1158/1078-0432.CCR-04-1752. [DOI] [PubMed] [Google Scholar]
- 5.Yu H, Pardoll D, Jove R. STATs in cancer inflammation and immunity: a leading role for STAT3. Nat Rev Cancer. 2009;9:798–809. doi: 10.1038/nrc2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang J, Liao X, Agarwal MK, Barnes L, Auron PE, Stark GR. Unphosphorylated STAT3 accumulates in response to IL-6 and activates transcription by binding to NFkappaB. Genes Dev. 2007;21:1396–408. doi: 10.1101/gad.1553707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Demaria M, Poli V. From the nucleus to the mitochondria and back: the odyssey of a multitask STAT3. Cell Cycle. 2011;10:3221–2. doi: 10.4161/cc.10.19.17379. [DOI] [PubMed] [Google Scholar]
- 8.Gao X, Wang H, Yang JJ, Liu X, Liu ZR. Pyruvate kinase M2 regulates gene transcription by acting as a protein kinase. Mol Cell. 2012;45:598–609. doi: 10.1016/j.molcel.2012.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Turkson J, Bowman T, Adnane J, Zhang Y, Djeu JY, Sekharam M, Frank DA, Holzman LB, Wu J, Sebti S, et al. Requirement for Ras/Rac1-mediated p38 and c-Jun N-terminal kinase signaling in Stat3 transcriptional activity induced by the Src oncoprotein. Mol Cell Biol. 1999;19:7519–28. doi: 10.1128/mcb.19.11.7519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wegrzyn J, Potla R, Chwae YJ, Sepuri NB, Zhang Q, Koeck T, Derecka M, Szczepanek K, Szelag M, Gornicka A, et al. Function of mitochondrial Stat3 in cellular respiration. Science. 2009;323:793–7. doi: 10.1126/science.1164551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gough DJ, Corlett A, Schlessinger K, Wegrzyn J, Larner AC, Levy DE. Mitochondrial STAT3 supports Ras-dependent oncogenic transformation. Science. 2009;324:1713–6. doi: 10.1126/science.1171721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pilati C, Amessou M, Bihl MP, Balabaud C, Nhieu JT, Paradis V, Nault JC, Izard T, Bioulac-Sage P, Couchy G, et al. Somatic mutations activating STAT3 in human inflammatory hepatocellular adenomas. J Exp Med. 2011;208:1359–66. doi: 10.1084/jem.20110283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bromberg JF, Wrzeszczynska MH, Devgan G, Zhao Y, Pestell RG, Albanese C, Darnell JE., Jr. Stat3 as an oncogene. Cell. 1999;98:295–303. doi: 10.1016/S0092-8674(00)81959-5. [DOI] [PubMed] [Google Scholar]
- 14.Vultur A, Cao J, Arulanandam R, Turkson J, Jove R, Greer P, Craig A, Elliott B, Raptis L. Cell-to-cell adhesion modulates Stat3 activity in normal and breast carcinoma cells. Oncogene. 2004;23:2600–16. doi: 10.1038/sj.onc.1207378. [DOI] [PubMed] [Google Scholar]
- 15.Vultur A, Arulanandam R, Turkson J, Niu G, Jove R, Raptis L. Stat3 is required for full neoplastic transformation by the Simian Virus 40 large tumor antigen. Mol Biol Cell. 2005;16:3832–46. doi: 10.1091/mbc.E04-12-1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Su HW, Yeh HH, Wang SW, Shen MR, Chen TL, Kiela PR, Ghishan FK, Tang MJ. Cell confluence-induced activation of signal transducer and activator of transcription-3 (Stat3) triggers epithelial dome formation via augmentation of sodium hydrogen exchanger-3 (NHE3) expression. J Biol Chem. 2007;282:9883–94. doi: 10.1074/jbc.M606754200. [DOI] [PubMed] [Google Scholar]
- 17.Kreis S, Munz GA, Haan S, Heinrich PC, Behrmann I. Cell density dependent increase of constitutive signal transducers and activators of transcription 3 activity in melanoma cells is mediated by Janus kinases. Mol Cancer Res. 2007;5:1331–41. doi: 10.1158/1541-7786.MCR-07-0317. [DOI] [PubMed] [Google Scholar]
- 18.Onishi A, Chen Q, Humtsoe JO, Kramer RH. STAT3 signaling is induced by intercellular adhesion in squamous cell carcinoma cells. Exp Cell Res. 2008;314:377–86. doi: 10.1016/j.yexcr.2007.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Arulanandam R, Vultur A, Cao J, Carefoot E, Elliott BE, Truesdell PF, Larue L, Feracci H, Raptis L. Cadherin-cadherin engagement promotes cell survival via Rac1/Cdc42 and signal transducer and activator of transcription-3. Mol Cancer Res. 2009;7:1310–27. doi: 10.1158/1541-7786.MCR-08-0469. [DOI] [PubMed] [Google Scholar]
- 20.Geletu M, Arulanandam R, Chevalier S, Saez B, Larue L, Feracci H, Raptis L. Classical cadherins control survival through the gp130/Stat3 axis. Biochim Biophys Acta. 2013;1833:1947–59. doi: 10.1016/j.bbamcr.2013.03.014. [DOI] [PubMed] [Google Scholar]
- 21.Raptis L, Arulanandam R, Vultur A, Geletu M, Chevalier S, Feracci H. Beyond structure, to survival: activation of Stat3 by cadherin engagement. Biochem Cell Biol. 2009;87:835–43. doi: 10.1139/O09-061. [DOI] [PubMed] [Google Scholar]
- 22.Wheelock MJ, Shintani Y, Maeda M, Fukumoto Y, Johnson KR. Cadherin switching. J Cell Sci. 2008;121:727–35. doi: 10.1242/jcs.000455. [DOI] [PubMed] [Google Scholar]
- 23.Halbleib JM, Nelson WJ. Cadherins in development: cell adhesion, sorting, and tissue morphogenesis. Genes Dev. 2006;20:3199–214. doi: 10.1101/gad.1486806. [DOI] [PubMed] [Google Scholar]
- 24.Larue L, Antos C, Butz S, Huber O, Delmas V, Dominis M, Kemler R. A role for cadherins in tissue formation. Development. 1996;122:3185–94. doi: 10.1242/dev.122.10.3185. [DOI] [PubMed] [Google Scholar]
- 25.Patel SD, Chen CP, Bahna F, Honig B, Shapiro L. Cadherin-mediated cell-cell adhesion: sticking together as a family. Curr Opin Struct Biol. 2003;13:690–8. doi: 10.1016/j.sbi.2003.10.007. [DOI] [PubMed] [Google Scholar]
- 26.Nollet F, Kools P, van Roy F. Phylogenetic analysis of the cadherin superfamily allows identification of six major subfamilies besides several solitary members. J Mol Biol. 2000;299:551–72. doi: 10.1006/jmbi.2000.3777. [DOI] [PubMed] [Google Scholar]
- 27.Koch AW, Pokutta S, Lustig A, Engel J. Calcium binding and homoassociation of E-cadherin domains. Biochemistry. 1997;36:7697–705. doi: 10.1021/bi9705624. [DOI] [PubMed] [Google Scholar]
- 28.Courjean O, Chevreux G, Perret E, Morel A, Sanglier S, Potier N, Engel J, van Dorsselaer A, Feracci H. Modulation of E-cadherin monomer folding by cooperative binding of calcium ions. Biochemistry. 2008;47:2339–49. doi: 10.1021/bi701340d. [DOI] [PubMed] [Google Scholar]
- 29.Pierres A, Feracci H, Delmas V, Benoliel AM, Thiery JP, Bongrand P. Experimental study of the interaction range and association rate of surface-attached cadherin 11. Proc Natl Acad Sci U S A. 1998;95:9256–61. doi: 10.1073/pnas.95.16.9256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Perret E, Benoliel AM, Nassoy P, Pierres A, Delmas V, Thiery JP, Bongrand P, Feracci H. Fast dissociation kinetics between individual E-cadherin fragments revealed by flow chamber analysis. EMBO J. 2002;21:2537–46. doi: 10.1093/emboj/21.11.2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Patel SD, Ciatto C, Chen CP, Bahna F, Rajebhosale M, Arkus N, Schieren I, Jessell TM, Honig B, Price SR, et al. Type II cadherin ectodomain structures: implications for classical cadherin specificity. Cell. 2006;124:1255–68. doi: 10.1016/j.cell.2005.12.046. [DOI] [PubMed] [Google Scholar]
- 32.Thiery JP, Engl W, Viasnoff V, Dufour S. Biochemical and biophysical origins of cadherin selectivity and adhesion strength. Curr Opin Cell Biol. 2012;24:614–9. doi: 10.1016/j.ceb.2012.06.007. [DOI] [PubMed] [Google Scholar]
- 33.Leckband D, Sivasankar S. Cadherin recognition and adhesion. Curr Opin Cell Biol. 2012;24:620–7. doi: 10.1016/j.ceb.2012.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Takeichi M. Morphogenetic roles of classic cadherins. Curr Opin Cell Biol. 1995;7:619–27. doi: 10.1016/0955-0674(95)80102-2. [DOI] [PubMed] [Google Scholar]
- 35.Jeanes A, Gottardi CJ, Yap AS. Cadherins and cancer: how does cadherin dysfunction promote tumor progression? Oncogene. 2008;27:6920–9. doi: 10.1038/onc.2008.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Berx G, van Roy F. Involvement of members of the cadherin superfamily in cancer. Cold Spring Harb Perspect Biol. 2009;1:a003129. doi: 10.1101/cshperspect.a003129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hazan RB, Qiao R, Keren R, Badano I, Suyama K. Cadherin switch in tumor progression. Ann N Y Acad Sci. 2004;1014:155–63. doi: 10.1196/annals.1294.016. [DOI] [PubMed] [Google Scholar]
- 38.Hoffmann I, Balling R. Cloning and expression analysis of a novel mesodermally expressed cadherin. Dev Biol. 1995;169:337–46. doi: 10.1006/dbio.1995.1148. [DOI] [PubMed] [Google Scholar]
- 39.Pishvaian MJ, Feltes CM, Thompson P, Bussemakers MJ, Schalken JA, Byers SW. Cadherin-11 is expressed in invasive breast cancer cell lines. Cancer Res. 1999;59:947–52. [PubMed] [Google Scholar]
- 40.Chu K, Cheng CJ, Ye X, Lee YC, Zurita AJ, Chen DT, Yu-Lee LY, Zhang S, Yeh ET, Hu MC, et al. Cadherin-11 promotes the metastasis of prostate cancer cells to bone. Mol Cancer Res. 2008;6:1259–67. doi: 10.1158/1541-7786.MCR-08-0077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yamada S, Nelson WJ. Synapses: sites of cell recognition, adhesion, and functional specification. Annu Rev Biochem. 2007;76:267–94. doi: 10.1146/annurev.biochem.75.103004.142811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fukata M, Kaibuchi K. Rho-family GTPases in cadherin-mediated cell-cell adhesion. Nat Rev Mol Cell Biol. 2001;2:887–97. doi: 10.1038/35103068. [DOI] [PubMed] [Google Scholar]
- 43.Geletu M, Guy S, Raptis L. Effects of SRC and STAT3 upon gap junctional, intercellular communication in lung cancer lines. Anticancer Res. 2013;33:4401–10. [PubMed] [Google Scholar]
- 44.Arulanandam R, Geletu M, Raptis L. The simian virus 40 large tumor antigen activates cSrc and requires cSrc for full neoplastic transformation. Anticancer Res. 2010;30:47–53. [PubMed] [Google Scholar]
- 45.Noren NK, Niessen CM, Gumbiner BM, Burridge K. Cadherin engagement regulates Rho family GTPases. J Biol Chem. 2001;276:33305–8. doi: 10.1074/jbc.C100306200. [DOI] [PubMed] [Google Scholar]
- 46.Charrasse S, Comunale F, Fortier M, Portales-Casamar E, Debant A, Gauthier-Rouvière C. M-cadherin activates Rac1 GTPase through the Rho-GEF trio during myoblast fusion. Mol Biol Cell. 2007;18:1734–43. doi: 10.1091/mbc.E06-08-0766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fischer P, Hilfiker-Kleiner D. Role of gp130-mediated signalling pathways in the heart and its impact on potential therapeutic aspects. Br J Pharmacol. 2008;153(Suppl 1):S414–27. doi: 10.1038/bjp.2008.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Raptis L, Brownell HL, Vultur AM, Ross GM, Tremblay E, Elliott BE. Specific inhibition of growth factor-stimulated extracellular signal-regulated kinase 1 and 2 activation in intact cells by electroporation of a growth factor receptor-binding protein 2-Src homology 2 binding peptide. Cell Growth Differ. 2000;11:293–303. [PubMed] [Google Scholar]
- 49.Lazo JS, Nemoto K, Pestell KE, Cooley K, Southwick EC, Mitchell for Cdc25 dual specificity phosphatase inhibitors. Mol Pharmacol. 2002;61:720–8. doi: 10.1124/mol.61.4.720. [DOI] [PubMed] [Google Scholar]
- 50.Turkson J, Bowman T, Garcia R, Caldenhoven E, De Groot RP, Jove R. Stat3 activation by Src induces specific gene regulation and is required for cell transformation. Mol Cell Biol. 1998;18:2545–52. doi: 10.1128/mcb.18.5.2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bromberg JF, Horvath CM, Besser D, Lathem WW, Darnell JE., Jr. Stat3 activation is required for cellular transformation by v-src. Mol Cell Biol. 1998;18:2553–8. doi: 10.1128/mcb.18.5.2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wong JV, Dong P, Nevins JR, Mathey-Prevot B, You L. Network calisthenics: control of E2F dynamics in cell cycle entry. Cell Cycle. 2011;10:3086–94. doi: 10.4161/cc.10.18.17350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Young AP, Nagarajan R, Longmore GD. Mechanisms of transcriptional regulation by Rb-E2F segregate by biological pathway. Oncogene. 2003;22:7209–17. doi: 10.1038/sj.onc.1206804. [DOI] [PubMed] [Google Scholar]
- 54.Tsai KY, Hu Y, Macleod KF, Crowley D, Yamasaki L, Jacks T. Mutation of E2f-1 suppresses apoptosis and inappropriate S phase entry and extends survival of Rb-deficient mouse embryos. Mol Cell. 1998;2:293–304. doi: 10.1016/S1097-2765(00)80274-9. [DOI] [PubMed] [Google Scholar]
- 55.Leone G, DeGregori J, Sears R, Jakoi L, Nevins JR. Myc and Ras collaborate in inducing accumulation of active cyclin E/Cdk2 and E2F. Nature. 1997;387:422–6. doi: 10.1038/387422a0. [DOI] [PubMed] [Google Scholar]
- 56.Irwin M, Marin MC, Phillips AC, Seelan RS, Smith DI, Liu W, Flores ER, Tsai KY, Jacks T, Vousden KH, et al. Role for the p53 homologue p73 in E2F-1-induced apoptosis. Nature. 2000;407:645–8. doi: 10.1038/35036614. [DOI] [PubMed] [Google Scholar]
- 57.Moroni MC, Hickman ES, Lazzerini Denchi E, Caprara G, Colli E, Cecconi F, Müller H, Helin K. Apaf-1 is a transcriptional target for E2F and p53. Nat Cell Biol. 2001;3:552–8. doi: 10.1038/35078527. [DOI] [PubMed] [Google Scholar]
- 58.Sears RC, Nevins JR. Signaling networks that link cell proliferation and cell fate. J Biol Chem. 2002;277:11617–20. doi: 10.1074/jbc.R100063200. [DOI] [PubMed] [Google Scholar]
- 59.Anagnostopoulou A, Vultur A, Arulanandam R, Cao J, Turkson J, Jove R, Kim JS, Glenn M, Hamilton AD, Raptis L. Differential effects of Stat3 inhibition in sparse vs confluent normal and breast cancer cells. Cancer Lett. 2006;242:120–32. doi: 10.1016/j.canlet.2005.10.047. [DOI] [PubMed] [Google Scholar]
- 60.Chu YS, Thomas WA, Eder O, Pincet F, Perez E, Thiery JP, Dufour S. Force measurements in E-cadherin-mediated cell doublets reveal rapid adhesion strengthened by actin cytoskeleton remodeling through Rac and Cdc42. J Cell Biol. 2004;167:1183–94. doi: 10.1083/jcb.200403043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gao M, Sotomayor M, Villa E, Lee EH, Schulten K. Molecular mechanisms of cellular mechanics. Phys Chem Chem Phys. 2006;8:3692–706. doi: 10.1039/b606019f. [DOI] [PubMed] [Google Scholar]
- 62.Burute M, Thery M. Spatial segregation between cell-cell and cell-matrix adhesions. Curr Opin Cell Biol. 2012;24:628–36. doi: 10.1016/j.ceb.2012.07.003. [DOI] [PubMed] [Google Scholar]
- 63.Dufrêne YF, Evans E, Engel A, Helenius J, Gaub HE, Müller DJ. Five challenges to bringing single-molecule force spectroscopy into living cells. Nat Methods. 2011;8:123–7. doi: 10.1038/nmeth0211-123. [DOI] [PubMed] [Google Scholar]
- 64.Vogel V, Sheetz M. Local force and geometry sensing regulate cell functions. Nat Rev Mol Cell Biol. 2006;7:265–75. doi: 10.1038/nrm1890. [DOI] [PubMed] [Google Scholar]
- 65.Gardel ML, Schneider IC, Aratyn-Schaus Y, Waterman CM. Mechanical integration of actin and adhesion dynamics in cell migration. Annu Rev Cell Dev Biol. 2010;26:315–33. doi: 10.1146/annurev.cellbio.011209.122036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Paszek MJ, Zahir N, Johnson KR, Lakins JN, Rozenberg GI, Gefen A, Reinhart-King CA, Margulies SS, Dembo M, Boettiger D, et al. Tensional homeostasis and the malignant phenotype. Cancer Cell. 2005;8:241–54. doi: 10.1016/j.ccr.2005.08.010. [DOI] [PubMed] [Google Scholar]
- 67.Levental KR, Yu H, Kass L, Lakins JN, Egeblad M, Erler JT, Fong SF, Csiszar K, Giaccia A, Weninger W, et al. Matrix crosslinking forces tumor progression by enhancing integrin signaling. Cell. 2009;139:891–906. doi: 10.1016/j.cell.2009.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Singhvi R, Kumar A, Lopez GP, Stephanopoulos GN, Wang DI, Whitesides GM, Ingber DE. Engineering cell shape and function. Science. 1994;264:696–8. doi: 10.1126/science.8171320. [DOI] [PubMed] [Google Scholar]
- 69.Chen CS, Mrksich M, Huang S, Whitesides GM, Ingber DE. Geometric control of cell life and death. Science. 1997;276:1425–8. doi: 10.1126/science.276.5317.1425. [DOI] [PubMed] [Google Scholar]
- 70.Schwartz MA. Integrins and extracellular matrix in mechanotransduction. Cold Spring Harb Perspect Biol. 2010;2:a005066. doi: 10.1101/cshperspect.a005066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mammoto A, Ingber DE. Cytoskeletal control of growth and cell fate switching. Curr Opin Cell Biol. 2009;21:864–70. doi: 10.1016/j.ceb.2009.08.001. [DOI] [PubMed] [Google Scholar]
- 72.Gomez GA, McLachlan RW, Yap AS. Productive tension: force-sensing and homeostasis of cell-cell junctions. Trends Cell Biol. 2011;21:499–505. doi: 10.1016/j.tcb.2011.05.006. [DOI] [PubMed] [Google Scholar]
- 73.Maître JL, Heisenberg CP. Three functions of cadherins in cell adhesion. Curr Biol. 2013;23:R626–33. doi: 10.1016/j.cub.2013.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Perret E, Leung A, Feracci H, Evans E. Trans-bonded pairs of E-cadherin exhibit a remarkable hierarchy of mechanical strengths. Proc Natl Acad Sci U S A. 2004;101:16472–7. doi: 10.1073/pnas.0402085101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Harrison OJ, Jin X, Hong S, Bahna F, Ahlsen G, Brasch J, Wu Y, Vendome J, Felsovalyi K, Hampton CM, et al. The extracellular architecture of adherens junctions revealed by crystal structures of type I cadherins. Structure. 2011;19:244–56. doi: 10.1016/j.str.2010.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Dupont S, Morsut L, Aragona M, Enzo E, Giulitti S, Cordenonsi M, Zanconato F, Le Digabel J, Forcato M, Bicciato S, et al. Role of YAP/TAZ in mechanotransduction. Nature. 2011;474:179–83. doi: 10.1038/nature10137. [DOI] [PubMed] [Google Scholar]
- 77.Wada K, Itoga K, Okano T, Yonemura S, Sasaki H. Hippo pathway regulation by cell morphology and stress fibers. Development. 2011;138:3907–14. doi: 10.1242/dev.070987. [DOI] [PubMed] [Google Scholar]
- 78.Zhao B, Li L, Lei Q, Guan KL. The Hippo-YAP pathway in organ size control and tumorigenesis: an updated version. Genes Dev. 2010;24:862–74. doi: 10.1101/gad.1909210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Halder G, Johnson RL. Hippo signaling: growth control and beyond. Development. 2011;138:9–22. doi: 10.1242/dev.045500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhao B, Wei X, Li W, Udan RS, Yang Q, Kim J, Xie J, Ikenoue T, Yu J, Li L, et al. Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control. Genes Dev. 2007;21:2747–61. doi: 10.1101/gad.1602907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kim NG, Koh E, Chen X, Gumbiner BM. E-cadherin mediates contact inhibition of proliferation through Hippo signaling-pathway components. Proc Natl Acad Sci U S A. 2011;108:11930–5. doi: 10.1073/pnas.1103345108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chen Q, Watson JT, Marengo SR, Decker KS, Coleman I, Nelson PS, Sikes RA. Gene expression in the LNCaP human prostate cancer progression model: progression associated expression in vitro corresponds to expression changes associated with prostate cancer progression in vivo. Cancer Lett. 2006;244:274–88. doi: 10.1016/j.canlet.2005.12.027. [DOI] [PubMed] [Google Scholar]
- 83.Geletu M, Chaize C, Arulanandam R, Vultur A, Kowolik C, Anagnostopoulou A, Jove R, Raptis L. Stat3 activity is required for gap junctional permeability in normal rat liver epithelial cells. DNA Cell Biol. 2009;28:319–27. doi: 10.1089/dna.2008.0833. [DOI] [PubMed] [Google Scholar]
- 84.Geletu M, Arulanandam R, Greer S, Trotman-Grant A, Tomai E, Raptis L. Stat3 is a positive regulator of gap junctional intercellular communication in cultured, human lung carcinoma cells. BMC Cancer. 2012;12:605. doi: 10.1186/1471-2407-12-605. [DOI] [PMC free article] [PubMed] [Google Scholar]