Abstract
Summary: High-throughput technologies have led to an explosion of genomic data available for automated analysis. The consequent possibility to simultaneously sample multiple layers of variation along the gene expression flow requires computational methods integrating raw information from different ‘-omics’. It has been recently demonstrated that translational control is a widespread phenomenon, with profound and still underestimated regulation capabilities. Although detecting changes in the levels of total messenger RNAs (mRNAs; the transcriptome), of polysomally loaded mRNAs (the translatome) and of proteins (the proteome) is experimentally feasible in a high-throughput way, the integration of these levels is still far from being robustly approached. Here we introduce tRanslatome, a new R/Bioconductor package, which is a complete platform for the simultaneous pairwise analysis of transcriptome, translatome and proteome data. The package includes most of the available statistical methods developed for the analysis of high-throughput data, allowing the parallel comparison of differentially expressed genes and the corresponding differentially enriched biological themes. Notably, it also enables the prediction of translational regulatory elements on mRNA sequences. The utility of this tool is demonstrated with two case studies.
Availability and implementation: tRanslatome is available in Bioconductor.
Contact: t.tebaldi@unitn.it
Supplementary information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
High-throughput (‘-omics’) measurements of macromolecule variations in the cell offer the possibility to comprehensively understand how the cellular processes are regulated and to reveal how different layers of control are coordinated in producing a physiologically coherent response. These measurements are also invaluable to understand how the loss of this coordination contributes to disease origin. The establishment of high-throughput technologies and the consequent explosion of available data allow us to reach a ‘systems’ understanding of the variations in gene expression only when a parallel evolution of algorithms and data mining techniques is achieved. This eventually enables to suggest and prioritize potential mechanistic processes. Nonetheless, the integration of ‘-omics’ data, ranging from epigenetic chromatin remodeling to the dynamics of transcription, translation and protein activities, still requires considerable experimental and computational developments.
In this context, the low correlation observed between messenger RNA (mRNA) and protein levels is an unsolved issue (Vogel and Marcotte, 2012). Recently we showed that the analysis of the translatome, an intermediate level between the transcriptome and the proteome formed by mRNAs engaged with polysomes, provides substantial and somewhat surprising new information (Tebaldi et al., 2012). This and other examples (Colman et al., 2013; Schwanhäusser et al., 2011; Vogel et al., 2010) show how the integration of ‘-omics’ data can provide a biologically relevant outcome.
Here we present tRanslatome, a new Bioconductor package for the analysis of differential profiles coming from transcriptome, translatome and proteome studies. tRanslatome will help to study mRNA and protein variations in an exhaustive way, providing specific tools for the comparison of polysomal mRNA with total mRNA or protein data.
2 DESCRIPTION AND USAGE
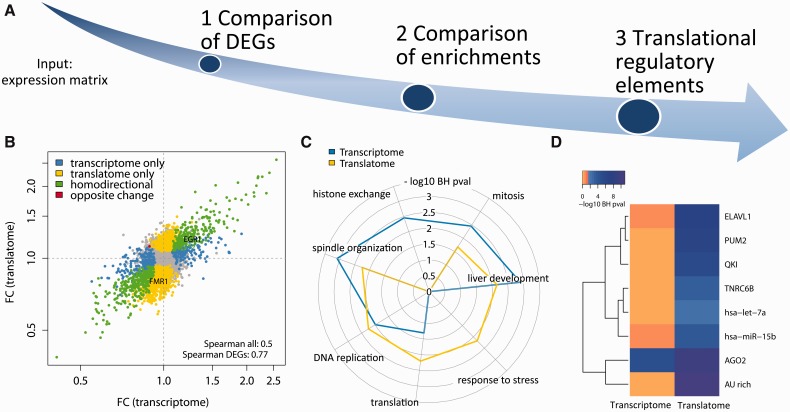
tRanslatome is a complete platform for the analysis and pairwise comparison of two ‘-omics’ levels implemented as a Bioconductor package. It is developed to compare translatome data with transcriptome and/or proteome data. A general overview of the functions offered by tRanslatome is given in Figure 1A. The package is conceptually organized in three modules, described as follows:
Fig. 1.
Outline of tRanslatome workflow and graphical outputs. (A) General overview of the three modules provided by tRanslatome. (B) Scatterplot of fold changes. Each gene is mapped according to the fold change in the transcriptome and the translatome. Different classes of DEGs are color labeled. The Spearman correlation coefficients are displayed for all genes and for all DEGs. (C) Radar plot of the top enriched GO biological process terms for the transcriptome and the translatome DEGs. (D) Heatmap of the top enriched post-transcriptional regulators for the transcriptome and the translatome DEGs. The color scale is based on the -log10 of the enrichment P-value, calculated with a Fisher test
2.1 DEGs detection
The only input required by tRanslatome is an expression matrix containing either read counts (from next-generation sequencing data) or normalized signals (from microarray or proteome experiments). To select differentially expressed genes (DEGs), the package offers, in the same computational environment, the integrative analysis of established and emerging statistical methods: (i) DEseq (Anders and Huber, 2010) and edgeR (Robinson et al., 2010), specifically implemented for the analysis of next-generation sequencing data; (ii) significance analysis of microarrays (SAM) (Tusher et al., 2001), developed for the analysis of microarray data; (iii) t-test, RankProd (Breitling et al., 2004), linear models and moderated t-test (Smyth, 2004), suitable to deal with general quantitative data; and (iv) methods dealing specifically with the comparison of translatome and transcriptome data, e.g. ANOTA (Larsson et al., 2011) and translational efficiency, derived from the ratio of polysomal and subpolysomal signals (Powley et al., 2009) or the ratio of ribosome protected fragments and RNA-seq reads (Ingolia et al., 2011). These techniques are described more exhaustively in the documentation and in the Supplementary Material.
To study all the relevant differences arising from the two ‘-omics’ levels, tRanslatome offers a variety of graphical outputs, helping the quality assessment and interpretation of the results. The minus-average plots aid the identification of intensity-dependent patterns, whereas the standard deviation plot can help the selection of the best method for DEGs identification (see Supplementary Figs 4 and 5). The graphics also include scatterplots, displaying changes in the expression of genes in terms of fold changes at both levels (Fig. 1B), and histograms, showing a detailed representation of all the DEGs classes (Supplementary Figs 2 and 3).
2.2 Gene Ontology enrichment comparison
One of the most frequent applications of the Gene Ontology (GO) is enrichment analysis, i.e. the identification of significantly overrepresented GO terms in a given gene set (Ashburner et al., 2000). tRanslatome includes the detection and comparison of GO terms, resuming information about cellular components, molecular functions and biological processes associated to DEGs detected from the two ‘-omics’ levels. Multiple choices are offered for the overrepresentation test, which exploits the GO ‘tree’ structure by means of the Bioconductor package ‘topGO’: these choices can satisfy the need for either more general or more specific biological themes. To simplify the inspection of the results and to effectively represent the differences in the enrichment of ontological terms, the corresponding radar plots and heatmaps can be produced (Fig. 1C and Supplementary Figs 7 and 8). tRanslatome also provides methods for a sensitive comparison of the similarity between enriched GO terms, including the semantic similarity scores (Wang et al., 2007) between terms at each level, and the global similarity score between the two levels.
2.3 Enrichment analysis of post-transcriptional regulatory elements
As tRanslatome focuses on the study of global translational controls, enrichment analysis of RNA binding proteins and microRNA binding sites or other RNA regulatory motifs (e.g. AU-rich elements) can be performed on the lists of DEGs. This analysis allows the user to identify possible regulatory factors responsible for the translational regulation of genes in the experiment under consideration (Fig. 1D). The list of genes regulated by each post-transcriptional element is obtained from the recently established Atlas of UTR Regulatory Activity (AURA) database (Dassi et al., 2012). The method computes a Fisher test P-value indicating whether binding sites for each regulator are significantly enriched in the DEGs lists. The annotations from AURA will be updated on every release of tRanslatome. Users can also specify a custom annotation file in place of the one provided by default.
3 EXAMPLES
A worked example, derived from data on differentiated versus undifferentiated human hepatocytes (Parent et al., 2007) is used to generate the panels contained in Figure 1. Detailed explanations of this example, along with a second example dealing with the comparison of the proteome and the transcriptome between two human cell lines (Stevens and Brown, 2013), are provided in the Supplementary Material.
4 CONCLUSION
tRanslatome allows a user-friendly comparison and integration of data generated from two ‘-omics’ measurements, empowering the discovery of regulatory mechanisms underlying the uncoupling processes among the transcriptome, the translatome and the proteome.
Funding: This work is supported by the University and Scientific Research Services of the Autonomous Province of Trento.
Conflicts of interest: none declared.
Supplementary Material
REFERENCES
- Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitling R, et al. Rank products: a simple, yet powerful, new method to detect differentially regulated genes in replicated microarray experiments. FEBS Lett. 2004;573:83–92. doi: 10.1016/j.febslet.2004.07.055. [DOI] [PubMed] [Google Scholar]
- Colman H, et al. Genome-wide analysis of host mRNA translation during hepatitis C virus infection. J. Virol. 2013;87:6668–6677. doi: 10.1128/JVI.00538-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dassi E, et al. AURA: Atlas of UTR Regulatory Activity. Bioinformatics. 2012;28:142–144. doi: 10.1093/bioinformatics/btr608. [DOI] [PubMed] [Google Scholar]
- Ingolia NT, et al. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell. 2011;147:789–802. doi: 10.1016/j.cell.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson O, et al. anota: Analysis of differential translation in genome-wide studies. Bioinformatics. 2011;27:1440–1441. doi: 10.1093/bioinformatics/btr146. [DOI] [PubMed] [Google Scholar]
- Parent R, et al. Mammalian target of rapamycin activation impairs hepatocytic differentiation and targets genes moderating lipid homeostasis and hepatocellular growth. Cancer Res. 2007;67:4337–4345. doi: 10.1158/0008-5472.CAN-06-3640. [DOI] [PubMed] [Google Scholar]
- Powley IR, et al. Translational reprogramming following UVB irradiation is mediated by DNA-PKcs and allows selective recruitment to the polysomes of mRNAs encoding DNA repair enzymes. Genes Dev. 2009;23:1207–1220. doi: 10.1101/gad.516509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, et al. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwanhäusser B, et al. Global quantification of mammalian gene expression control. Nature. 2011;473:337–342. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 2004;3:Article3. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- Stevens SG, Brown CM. In silico estimation of translation efficiency in human cell lines: potential evidence for widespread translational control. PLoS One. 2013;8:e57625. doi: 10.1371/journal.pone.0057625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tebaldi T, et al. Widespread uncoupling between transcriptome and translatome variations after a stimulus in mammalian cells. BMC Genomics. 2012;13:220. doi: 10.1186/1471-2164-13-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tusher VG, et al. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. USA. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel C, Marcotte EM. Insights into the regulation of protein abundance from proteomic and transcriptomic analyses. Nat. Rev. Genet. 2012;13:227–232. doi: 10.1038/nrg3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel C, et al. Sequence signatures and mRNA concentration can explain two-thirds of protein abundance variation in a human cell line. Mol. Syst. Biol. 2010;6:400. doi: 10.1038/msb.2010.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JZ, et al. A new method to measure the semantic similarity of GO terms. Bioinformatics. 2007;23:1274–1281. doi: 10.1093/bioinformatics/btm087. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.