Summary
In the context of a bioassay or an immunoassay, calibration means fitting a curve, usually nonlinear, through the observations collected on a set of samples containing known concentrations of a target substance, and then using the fitted curve and observations collected on samples of interest to predict the concentrations of the target substance in these samples. Recent technological advances have greatly improved our ability to quantify minute amounts of substance from a tiny volume of biological sample. This has in turn led to a need to improve statistical methods for calibration. In this paper, we focus on developing calibration methods robust to dependent outliers. We introduce a novel normal mixture model with dependent error terms to model the experimental noise. In addition, we propose a re-parameterization of the five parameter logistic nonlinear regression model that allows us to better incorporate prior information. We examine the performance of our methods with simulation studies and show that they lead to a substantial increase in performance measured in terms of mean squared error of estimation and a measure of the average prediction accuracy. A real data example from the HIV Vaccine Trials Network Laboratory is used to illustrate the methods.
Keywords: calibration, correlated outliers, 5PL curve, AR(1) process, dose response curve, multiplex bead array
1. Calibration Experiments and Statistical Analyses
Measuring the quantity of a substance in a sample is a fundamental task in many biomedical fields. Due to the difficulty of directly measuring the amount of a biomolecule in a biological/clinical sample, a calibration approach is commonly undertaken. The assay of choice is performed on a set of standard samples which contain known amount of the target analyte at regular intervals. From these, a concentration-response curve is estimated and can be used to predict the analyte concentration in samples of interest which are subjected to the same assay. Most assays fall into two categories: bioassay or immunoassay. A bioassay is based on the effect on living matter by the substance, while immunoassays are performed in an artificial experimental environment such as a test tube and rely on the highly specific binding of antibodies and antigens, hence the name. Two of the most well-known immunoassays to date are the Nobel price-winning radioimmunoassay (RIA, Yalow and Berson, 1960), which produces a radioactive signal in response to a specific binding, and Enzyme-Linked Immunosorbent Assay (ELISA, Engvall and Perlmann, 1971), which produces a safer response, typically a change in color of a chemical substrate. A new type of immunoassay using multiplex bead arrays (MBA) is now gaining popularity. While RIA and ELISA can only measure one analyte at a time, MBA can measure the concentrations of many (up to 100 with the Luminex® 100 IS System) from one biological sample.
As the experimental techniques for calibration experiments evolve, the statistical methods for their analysis have also matured. The shape of the concentration-response curve was a focus from early on (see review by Finney, 1947). D.J. Finney introduced a four parameter logistic (4PL) model in 1970 (according to Rodbard and Frazier, 1975), and it is still widely used today. Generalization of the 4PL model to a five parameter logistic (5PL) model for asymmetrical curves appeared in Prentice (1976), Rodbard et al. (1978) and Finney (1979), while 5PL itself dates back to Richards (1959). More recently, model-robust approaches have been an active research area (Bornkamp and Ickstadt, 2009; Faes et al., 2007; Morales et al., 2006). In this study, we focus on 5PL models, which in many practical settings strike a balance between bias and variance trade-off (Gottschalk and Dunn, 2005).
Statistical modeling of the experimental noise in a calibration experiment also received ample attention (see e.g. Rodbard and Cooper, 1970; Belanger et al., 1996). In particular, the need to model variance robustly was recognized by many. A robust statistical procedure (Box, 1953; Huber and Ronchetti, 2009) is, roughly speaking, one that is less sensitive to the presence of outliers. There are two basic approaches to robust inference: estimating equation-based approach and likelihood-based (or model-based) approach. The former seeks to reduce the influence of outlying observations, examples in calibration include Hamilton (1979), Tiede and Pagano (1979), Miller and Halpern (1980), Normolle (1993), Fomenko et al. (2006), and Wang et al. (2008). The likelihood-based approach aims to provide a better model of the data by replacing normal distributions with more heavy-tailed distributions (Lange et al., 1989; Taplin and Raftery, 1994), examples in assay analysis include Motulsky and Brown (2006). In this study, we are concerned with outliers that appear in a cluster. There have only been a few studies of the so-called dependent outliers or correlated outliers. Grishin and Janczak (2008) presented an algorithm for robustifying tracking algorithms in radars and radio-navigational systems in the presence of correlated outliers. Qiu and Vaswani (2011) proposed an algorithm for solving principal components' analysis problem in the presence of correlated outliers. We will take a likelihood-based approach since it can be extended naturally to model dependent outliers.
Most routine analyses of assay data use frequentist inference to the best of our knowledge. It may be for practical reasons like speed of computation, but it is also because frequentist inference does a reasonably good job in most cases. In several contexts, however, Bayesian inference has been chosen to help improve inference. Racine-Poon (1988) proposed a model to jointly model data from more than one type of bioassay. Gelman et al. (2004) proposed a model for calibration when several different dilutions of an unknown sample were assayed. In this study, a Bayesian approach is adopted to incorporate prior information, without which, the quality of the inference is greatly reduced due to the presence of dependent outliers.
Our study is motivated by the adoption of MBA assay by the HIV Vaccine Trials Network (HVTN) laboratory in the study of immune correlates in the RV144 HIV vaccine trial. The recent RV144 phase III efficacy trial (Rerks-Ngarm et al., 2009) showed a modest 31% efficacy in preventing HIV infection in vaccine compared to placebo recipients. Efforts are now under way to study aspects of vaccine-induced immune responses that may correlate with protection, with the hope of finding clues to feed back into vaccine design in future trials. Because a limited volume of serum sample was collected from trial participants and because we wished to study a wide range of immune responses for possible correlates, the MBA became an important assay in the correlates study. A typical run of the MBA assay is conducted on 96-well plates. Part of the plate, say 20 wells, are used to assay standard samples. The rest of the plate is used to assay unknown samples. If the estimation of the concentration-response curve for one analyte were unduly affected by outliers, the unknown samples would have been wasted with regard to this particular analyte. Thus it is critical that we have a very robust estimation procedure for estimating the concentration-response curves.
In Section 2, we introduce a normal mixture model with a dependent error structure to model the experimental noise robustly. In Section 3, we introduce a re-parameterization of the 5PL model, which allows us to incorporate prior information from historical data. We carry out a simulation study in Section 4 and use a real data example from the HVTN lab RV144 study as an illustration in Section 5. We conclude with a discussion in Section 6.
2. A Robust Model for Dependent Experimental Noise
We first describe the structure of a typical set of standard samples from the MBA assay, and establish a Bayesian random effects model for analyses. An experiment often involves multiple plates, each of which has a set of standard samples. Let the plate be indexed by i = 1, ⋯ , I. Each set of standard samples usually comprises two replicates, indexed by j = 1, 2. Each replicate contains one serial dilutions of the standard samples (Blei et al., 2006), in which part of one standard sample is used to prepare the next standard sample in the dilution series. We denote the log concentrations of the standard samples by tijk, for k = 1, ⋯ , Kij. For simplicity, we assume that Kij = K for a certain K, and that tijk = tk, i.e. the concentrations of the standard samples on each plate/replicate combination are identical. Most datasets satisfy these two constraints, and relaxation is straightforward. The outcome in a MBA experiment is median fluorescence intensity (MFI). When untransformed this outcome tends to be more variable when it is higher, but we have found that a log transformation stabilizes the variance to a satisfactory degree. We denote the transformed outcome by Yijk and consider the following nonlinear mixed model (Baharith et al., 2006; Davidian and Giltinan, 1995)
| (1) |
In the above, f5PL (·;θi) denotes the 5PL function with plate-specific parameters θi, εijk is the experimental noise, and bi is the mean-centered plate random effect, which is assumed to follow a normal distribution with precision matrix Ω. We assume conditional conjugate priors on θ0 and Ω. Specifically, θ0 is assumed to follow a normal distribution with mean μ and covariance matrix Σ, and Ω is assumed to follow a Wishart distribution with r degrees of freedom and mean rS.
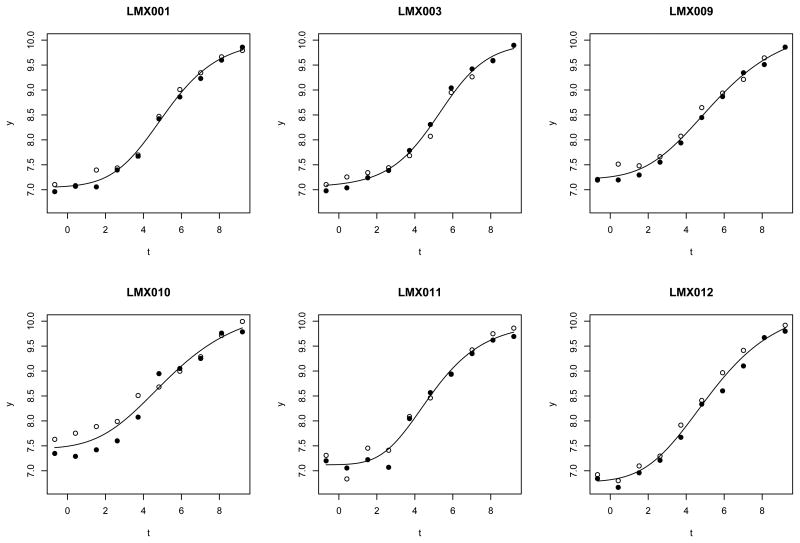
It is common to model the noise terms εijk as independent normal random variables, but this leaves the fit of the curve sensitive to outliers. Sometimes it is possible to manually remove outliers before carrying out statistical analyses, but that is not feasible when there are a number of outliers, in a phenomenon often termed as ‘masking’, i.e. the outliers mask each other from being easily detected. Masking is a complicated effect, but some of it is probably due to the presence of dependent outliers. Figure 1 shows an example of this phenomenon. In plate LMX010, the observations in the first few dilutions from the two replicates are unusually far apart from each other. This pattern can be explained if the observations from one of the replicates in those first few dilutions are outliers. Such clustered occurrences of outliers may be due to certain sequential ordering in the assay protocol. Aside from the structure imposed by serial dilution, in the handling of the standard samples each dilution series is often treated as a unit, e.g. they may be plated in the same column of wells on a plate. While it is possible to come up with experimental protocols to break up this structure, the resulting experimental procedure will be harder to implement by human experimentalists and may lead to decreased overall data quality.
Figure 1.
Motivating dataset. Each panel represents the standard curve data from one plate. The x axis is the log transformed concentration, the y axis is the observed outcome. Two different plotting symbols are used to distinguish the two replicates within a plate. Each replicate comes from one dilution series.
Conceptually we may separate the experimental noise in Figure 1 into two categories. The first category includes noise sources such as detection sensor noise and the stochastic nature of the binding reaction. These sources typically produce small errors, and can be approximately thought of as being independent from observation to observation. The second kind of noise usually leads to larger errors, and may arise due to events that tend to affect several neighboring standard samples in a dilution series. We propose to model both categories with a process of mixture distributions by extending the contaminated normal model (Taplin and Raftery, 1994; Little, 1988). We first describe the process in a generic way. Consider a sequence of K error terms . We say that ε is distributed according to a LAR(1) process if
| (LAR(1)) |
In this model, conditional on π and α, the mixture component indicators follow a multivariate binary distribution with first order autoregressive (AR(1)) correlation (Qaqish, 2003). The marginal mean of is π and the correlation between the kth and k′th indicators is α|k−k′| for k ≠ k′ and |α| < 1. We thus name the above model a Latent first order AutoRegressive (LAR(1)) process. It has four parameters: , , π, and α. It is easy to see that this is a stationary process if |α| < 1. The marginal distribution of a LAR(1) process is a mixture of two normal distributions with mixture coefficient π. A LAR(1) process is not an AR(1) process: the correlation between εk and εk′ is 0 for k ≠ k′. This is because even though are correlated, the normal components of are independent. A LAR(1) process is not even a Markov chain of order 1. To see this, consider the special case of K = 3. We observe that C2|ε2 does not have the same distribution as C2|ε2, ε1. And because ε3 depends on C2, it also depends on ε1. This does not mean that LAR(1) is overly complicated, however. The dependence between εk's is induced by the chain of mixture indicators , and often we can work with the augmented chain , which is an AR(1) process. The proposed model is akin to a hidden Markov model (MacDonald and Zucchini, 1997), which have similar dependence structures in that the observations are not Markov even though the underlying states are.
We now use the LAR(1) process to complete our model of the assay data. Denote from model (1). For i ∈ {1, ⋯ , I} and j ∈ {1, ⋯ , J}, we assume
The above model implies that the marginally less likely normal component, the contaminating component, has a greater variance than the marginally more likely component. For the hyperparameters, λ = 0.046 is chosen so that the 99% quantile of Exponential (λ) is 100, and α1 = 2, β1 = 0.02 are chosen based on least square 5PL fits (Ritz and Streibig, 2005) of historical data that are apparently free of outliers.
3. Reparameterization of Five Parameter Logistic Model
The intuitive idea behind robust statistics was that “a discordant small minority should never be able to override the evidence of the majority of the observations” (Huber and Ronchetti, 2009). In the example shown in Figure 1, if we only have data from plate LMX010, there is no hope for correcting the influence of outliers, because a good proportion of the observations appear to be outliers. But because we have five other plates, by borrowing information across plates it becomes possible to tease out which observations are more likely to be the outliers in plate LMX010. In our model, the extent to which we can borrow information across plates depends on the prior on the precision matrix of the plate effect prior (1). The HVTN lab has accumulated a large amount of MBA data that we would like to use to inform future inference. It is this need to better capture prior knowledge about the plate random effect from historical data that has led us to propose a new parameterization of the five parameter logistic (5PL) model.
A 5PL model is most commonly parameterized as follows
| (2) |
When f = 1, this model reduces to a four parameter logistic model (4PL). In a 4PL model, three of the parameters have direct interpretation: c and d are the lower and upper asymptotes respectively, and log (e) is the inflection point, i.e. the point at which the curve changes from convex to concave or vice versa, depending on whether the curve is monotonically increasing or decreasing. A 4PL curve is symmetric around the inflection point. The slope at the inflection point, −b(d−c) /4, is proportional to the product of the fourth parameter b and the difference between asymptotes. The 5PL model generalizes the 4PL model to allow fitting of asymmetric concentration-response curves by introducing a fifth parameter f. In a 5PL model as parameterized in (2), c and d retain their interpretation as the asymptotes, but log (e) is no longer the inflection point and the slope at the inflection point is no longer proportional to b (d − c). Without loss of generality, we consider only monotonically increasing curves, thus requiring b < 0 and f > 0. We choose to work with transformed parameters log (−b) and log (f), so that they span the whole real line. We refer to this parameterization as the original parameterization.
One of our historical datasets came from our efforts to validate the MBA assay against the FDA guideline for bioanalytical method. We conducted an experiment that consisted of six plates, and 31 analytes were simultaneously assayed. We use the least square method implemented in the R package drc (Ritz and Streibig, 2005) to fit the 5PL model to each plate/analyte standard curve, in order to obtain parameter estimates. Next, we average the six plates' parameter estimates for each analyte and subtract the averages from the individual parameter estimates to obtain 6×31 = 186 samples of plate random effect. We then carried out Brown-Forsythe tests (Brown and Forsythe, 1974) to test the null hypotheses that the plate effect marginal variances are equal among the analytes. The P values for the five tests on {c, d, log (e) , log(−b) , log(f)} are 0.000, 0.000, 0.000, 0.042, and 0.004.
That the variances of c and d may be different among analytes is not surprising, because we expect to see some mean-variance relationship in these two asymptote parameters. Indeed, as Web Figure 4 shows, as the analyte mean of the lower asymptote c increases from 1 to 4, its plate effect variance tends to decrease. This makes sense because when the MFI is as low as being near exp (1), the background noise is relatively high, which means higher variance on the log(MFI) scale. Similarly, as the analyte mean of the higher asymptote d increases from 9.5 to 11.5, its plate effect variance tends to increase. This is likely because the maximum MFI output is around exp (10), a value of 11 for d has to be extrapolated from the data, hence likely to be subjected to high variability. It is harder to make sense of the test results for b, e and f because they are not easily interpretable.
We propose to re-parameterize 5PL by replacing b and e with the inflection point (denoted by g) and the hill slope at the inflection point (denoted by h). We call this parameterization the g-h parameterization. The reparameterized 5PL function is
| (3) |
Although this is a new parameterization in the calibration literature, it is actually how 5PL was parameterized when it first appeared in Richards (1959) (see also Seber and Wild, 1988, pp. 332-335). We will work with transformed parameter log (h), so that the parameter spans the whole real line. It is worth pointing out that from (3), it is easy to see that the response at the inflection point is only a function of c, d, and f. Using the relationship
| (4) |
we computed estimates of g and log (h) from the historical dataset mentioned before and conducted Brown-Forsythe tests. The P values are 0.266 and 0.184 for g and log (h), respectively. This allows us to pool the plate effect estimates of different analytes to specify the hyperparameters in the priors for these two parameters.
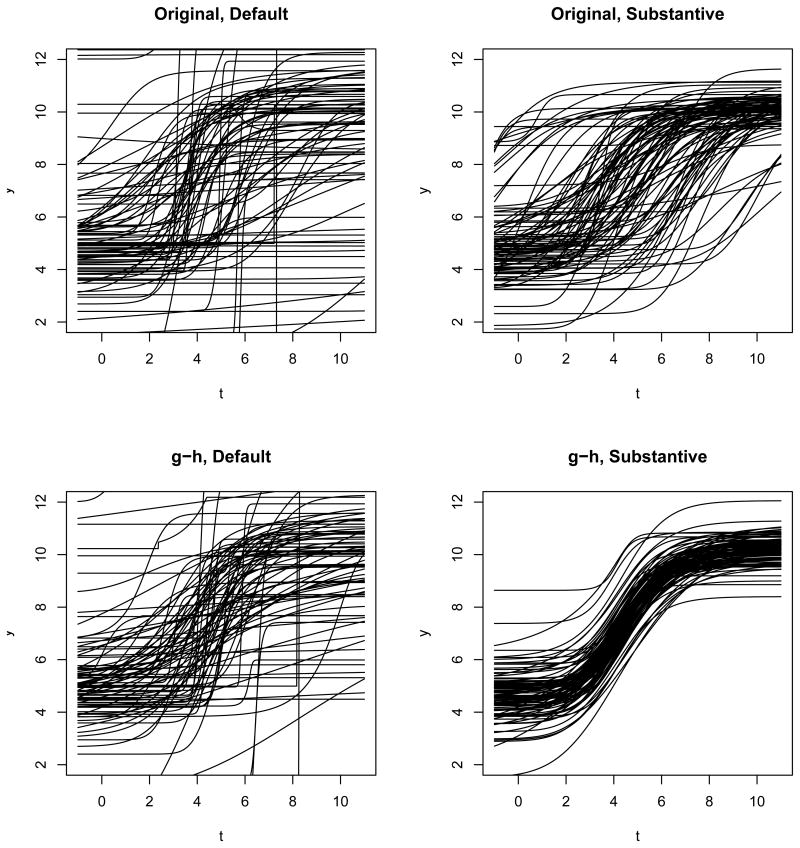
Because of the mean-variance relationship of the random effects, it is difficult to reliably extract information about the non-diagonal elements of S from the historical dataset. Thus we assume S to be diagonal. Note this does not imply Ω is diagonal. We specify the diagonal elements of S following the procedure in Fong et al. (2010). Let Sm be the hyperparameter corresponding to an arbitrary parameter m. Integrating out the precision parameter Ω, the plate effect bi is distributed as a multivariate Student's t distribution. Assuming that all marginal distributions of the plate effect have four degrees of freedom, we find r = 8. Then , where is the 97.5th quantile of Student's t-distribution with 4 degrees of freedom. For g and log (h), we let (−Rm, Rm) be the range of 95% quantile of the estimated random effects from the historical dataset. For other parameters, since we cannot pool information from different analytes, we resort to letting Rm be the maximum absolute plate effect in the whole historical dataset. Under the original parameterization the hyperparameter is S{c,d,log(e),log(−b),log(f)} = diag (0.46, 2.34, 0.07, 2.69, 0.5); under the g-h parameterization the hyperparameter is S{c,d,g,log(h),log(f)} = diag (0.46, 2.34, 23,172, 0.5). For comparison, we also work with a default hyperparameter: r = 5 and S = diag (1, 1, 1, 1, 1). To visualize the different plate effect variance component priors, we simulate random plate effects, add them to a common θ0, and plot the curves in Figure 2. Under either parameterization, the default prior results in a bigger spread than the substantive prior derived from the historical data. In addition, the substantive prior under the g-h parameterization appears to be a stronger prior than the substantive prior under the original parameterization.
Figure 2.
Comparison of plate random effect priors. 100 samples are simulated from each prior. All random effects are added to a common θ0 = {c = 4.45, d = 10.3, log (e) = 3.30, log (−b) = −0.178, log (f) = 0.758, g = 4.26, log (h) = 0.358}.
We also use the historical dataset to specify the hyperparameters μ and Σ in (1). For each analyte, we average the parameter estimates from the six plates, and call this θ̄. We take the average of θ̄ to be μ. We assume Σ to be a diagonal matrix, and let the diagonal be the empirical variance of θ̄ (across analytes) multiplied by 4. For a default prior, we use μ = 0, Σ = diag (10−4, 10−4, 10−4, 10−4, 10−4).
4. Simulation Study
To investigate the operating characteristic of the proposed methods, we carry out a simulation study. Nine different estimating procedures are compared, three of them sensitive and six of them robust to outliers. The outlier-sensitive methods include ‘drc’ (Ritz and Streibig, 2005), which produces nonlinear least square estimates for each plate, and two Bayesian random effects models, both labeled ‘original, norm’ but differing in the priors used. These two models both use the original parameterization and a variant of the Bayesian random effects model laid out in Section 2 with π set to be 0, i.e. the error terms of a dilution series are assumed to be independent and normally distributed errors conditional on . The difference between the two is that one uses default hyperparameters and the other uses substantive hyperparameters. For robust estimating procedures, we examine four models: ‘original norm mix’ uses the original parameterization and a variant of the error model from Section 2 with α set to be 0, i.e. each error term is assumed to be independently distributed as a normal mixture of two components; ‘original LAR(1)’ uses the original parameterization and assumes the error model from Section 2; ‘original t-AR(1)’ uses the original parameterization and assumes that the error terms follow a Student's t4 AR(1) process (Heracleous and Spanos, 2006); ‘g-h LAR(1)’ uses the g-h parameterization and assumes the error model from Section 2. These four models are paired with both default hyperparameters and substantive hyperparameters. Furthermore, we investigate a reference model under the g-h parameterization that allows a replicate-level random effect to be nested within plate-level random effect. It differs from (1) in that Yijk = f5PL(tk;θij) + εijk and θij = θ0+bi+sij. For estimating concentrations of the unknown samples, we need to have one curve per plate, and we use (θi1 + θi2) /2 for that. We assume sij |W ∼iid N (0, precision=W) and W ∼ Wishart5 {5, diag (1, 1, 1, 1, 1)}. Inference for the Bayesian models are carried out using JAGS (Plummer, 2003).
We simulate 1000 datasets. Each simulation dataset is generated from Yijk = f5PL (tk; θ0,i)+εijk, where i = 1, ⋯ , 6, j = 1, 2, k = 1, ⋯ , 10, are equally spaced between log (0.51) and log (103), and are the estimated parameters for an arbitrarily chosen analyte from our historical dataset described in Section 3. The 5PL curves corresponding to are plotted in Web Figure 2.
Two sets of criteria are used to compare the nine estimating procedures. The first set is the mean squared errors for parameters c, d, g, h, f and is denoted by AVE(θ̄ − θ0)2. For each of the parameters, AVE(θ̄ − θ0)2 is defined to be the average of the squared difference between the posterior median of the parameter and the truth over six plates and 1000 simulation datasets. The reason we have chosen the posterior median as a summary of the posterior distribution over the posterior mean is that the posterior distributions tend to be highly skewed.
The second criterion measures the average absolute prediction error. Denoted by S̄ (θ̄, θ0), it is defined as the area between two 5PL curves parameterized by the truth θ0,i and the posterior median θ̄i (shaded area between curves in Web Figure 1), averaged over plates and 1000 simulation datasets. To see that it measures the average prediction accuracy, suppose the black and gray lines in Web Figure 1 correspond to θ0,i and θ̄i, separately. The area between the two curves is then approximately the Riemann sum of the absolute difference between the true concentration of an unknown sample and the concentration predicted based on a perfectly measured outcome. The approximation becomes exact if the predicted concentration is defined to be the lowest/highest concentration of the standard samples when the outcome is below/above the estimated asymptote.
We first assess the performance of using robust estimating procedures when the data do not have outliers. For this purpose, are simulated independently from a mean 0 normal distribution with standard deviation 0.3. Results are summarized in Table 1. We first focus on comparing area between curves. Using either default or substantive hyperparameters, the robust Bayesian random effect models show slightly higher area between curves than the non-robust models. For example, S̄ increases from 0.377 for ‘original norm’ to 0.378 for ‘original LAR(1)’ under default priors, and increases from 0.362 to 0.365 under substantive priors. So the price we pay for guarding against outliers is almost negligible. It is also worth noting that the robust Bayesian random effects models perform worse than ‘drc’ when using default priors, and better than ‘drc’ when using substantive priors, suggesting that the gain from using substantive priors can outweigh the loss from using long tailed error distributions. The ‘g-h, nested r.e.’ model also shows a slightly higher area between curves than the non-robust models. Comparing the mean squared errors of the parameters, we see mostly the same trend as for the area between curves.
Table 1.
Performance comparison in the absence of outliers. S̄ (θ̄, θ0) is the area between the curves parameterized by θ̄ and θ0 averaged over plates and simulation datasets.
| AVE (θ̄ − θ0)2 | S̄ (θ̄, θ0) | |||||
|---|---|---|---|---|---|---|
| c | d | g | log (h) | log (f) | ||
| drc | 0.0041 | 0.0061 | 0.014 | 0.0023 | 2.16 | 0.377 |
| default priors | ||||||
| original, norm | 0.0036 | 0.0058 | 0.014 | 0.0022 | 1.27 | 0.378 |
| original, norm mix | 0.0036 | 0.0060 | 0.015 | 0.0022 | 1.55 | 0.382 |
| original, t-AR(1) | 0.0039 | 0.0063 | 0.016 | 0.0024 | 1.62 | 0.397 |
| original, LAR(1) | 0.0037 | 0.0060 | 0.015 | 0.0022 | 2.15 | 0.382 |
| g-h, LAR(1) | 0.0035 | 0.0058 | 0.013 | 0.0023 | 1.48 | 0.374 |
| g-h, nested r.e. | 0.0035 | 0.0066 | 0.013 | 0.0024 | 2.30 | 0.392 |
| substantive priors | ||||||
| original, norm | 0.0035 | 0.0039 | 0.014 | 0.0017 | 1.17 | 0.362 |
| original, norm mix | 0.0035 | 0.0040 | 0.015 | 0.0018 | 1.17 | 0.365 |
| original, t-AR(1) | 0.0038 | 0.0041 | 0.015 | 0.0019 | 1.21 | 0.377 |
| original, LAR(1) | 0.0035 | 0.0040 | 0.015 | 0.0018 | 1.28 | 0.364 |
| g-h, LAR(1) | 0.0035 | 0.0040 | 0.013 | 0.0014 | 1.05 | 0.350 |
| g-h, nested r.e. | 0.0034 | 0.0058 | 0.012 | 0.0022 | 1.02 | 0.383 |
We now assess the performance of these estimation procedures in the presence of outliers. We use the same simulation setup as in the previous study, except now εijk are simulated according to a modified , where forms an AR(1) process with correlation coefficient 0.9. Table 2 shows the results. Comparing area between curves, we see that with either the default or the substantive hyperparameters, successive refinement of the noise model from normal to normal mixture to LAR(1) process leads to better and better performance. In addition, switching to the g-h parameterization from the original parameterization offers additional improvement. The t-AR(1) models, although somewhat robust, do not perform well. The ‘g-h, nested r.e.’ model does not perform better than the non-robust models. We conjecture that this might be due to the symmetry between the replicate-level random effects si1 and si2. Comparing the mean squared errors of the parameters, we see mostly the same trend.
Table 2.
Performance comparison in the presence of outliers. S̄ (θ̄, θ0) is the area between the curves parameterized by θ̄ and θ0 averaged over plates and simulation datasets.
| AVE (θ̄ − θ0)2 | S̄ (θ̄, θ0) | |||||
|---|---|---|---|---|---|---|
| c | d | g | log (h) | log (f) | ||
| drc | 0.0085 | 0.0116 | 0.022 | 0.0046 | 2.98 | 0.520 |
| default priors | ||||||
| original, norm | 0.0069 | 0.0105 | 0.019 | 0.0043 | 3.24 | 0.519 |
| original, norm mix | 0.0060 | 0.0091 | 0.019 | 0.0037 | 3.23 | 0.483 |
| original, t-AR(1) | 0.0062 | 0.0093 | 0.019 | 0.0038 | 5.25 | 0.500 |
| original, LAR(1) | 0.0059 | 0.0089 | 0.018 | 0.0035 | 2.60 | 0.472 |
| g-h, LAR(1) | 0.0058 | 0.0087 | 0.016 | 0.0035 | 5.43 | 0.469 |
| g-h, nested r.e. | 0.0071 | 0.0116 | 0.019 | 0.0045 | 8.60 | 0.533 |
| substantive priors | ||||||
| original, norm | 0.0066 | 0.0062 | 0.018 | 0.0033 | 1.31 | 0.490 |
| original, norm mix | 0.0056 | 0.0052 | 0.018 | 0.0027 | 1.48 | 0.452 |
| original, t-AR(1) | 0.0057 | 0.0049 | 0.017 | 0.0028 | 1.25 | 0.462 |
| original, LAR(1) | 0.0055 | 0.0050 | 0.017 | 0.0026 | 1.25 | 0.439 |
| g-h, LAR(1) | 0.0053 | 0.0052 | 0.014 | 0.0018 | 1.15 | 0.413 |
| g-h, nested r.e. | 0.0069 | 0.0098 | 0.016 | 0.0042 | 1.34 | 0.517 |
To get a better picture of the efficiency gain, we divide the simulated data into overlapping strata with strata defined by the number of the Ck indicators that are consecutively equal to 1. For each stratum, we measure the relative efficiency versus the non-robust model ‘original, norm’ as measured by the percent reduction in area between curves: 100 × (1 − S̄/S̄original norms). The results are shown in Table 3. We first look at substantive priors. With the noise modeled as normal mixture random variables, the efficiency gain increases from 8% to 13% when the minimum run length increases from 0 to 2, stays at 13% until the run length reaches 4, and then drops to 9% when the run length is 5. With the noise modeled as LAR(1) process, the trend is different. For both parameterizations, the efficiency gain keeps increasing until the minimum run length reaches 4 and stays there when the minimum run length is 5. Looking across different robust estimating procedures for a minimum run length of 4, we see that under the original parameterization, assuming the normal mixture noise model leads to a 12% reduction; assuming LAR(1) noise model brings an additional 8% reduction; switching to the g-h parameterization leads to a further 8% reduction, bringing the total reduction to twice that of the normal mixture model with original parameterization. Under default priors, we see similar trends, except that the difference between ‘original LAR(1)’ and ‘g-h LAR(1)’ is much less. This makes sense because the main advantage of the g-h parameterization is in choosing substantive hyperparameters.
Table 3.
Percent efficiency gain over non-robust Bayesian models, stratified by minimum run length.
5. Examples
We now apply the proposed methods to the dataset shown in Figure 1 to illustrate the impact that different noise models and different parameterizations have on the estimation of the concentration-response curves. As mentioned before, the dataset was collected as part of the RV144 immune correlates study, where we were interested in measuring the cytokine production profile of peripheral blood mononuclear cells taken from the study participants after incubation with HIV-specific peptides. The particular cytokine assayed in this example was interleukin 3, which is a protein that is part of the signaling pathways in an immune response. We fit the four Bayesian models using the substantive hyperparameters described in Section 4.
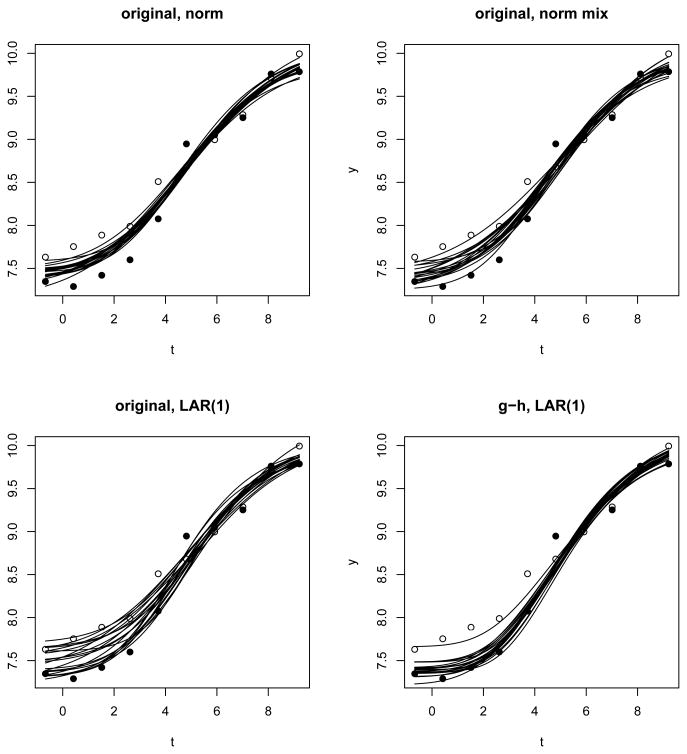
Web Figure 3 shows estimated concentration response curves for the six plates by three of the models: ‘original, norm’, ‘g-h, nested r.e.’ and ‘g-h, LAR(1)’. The plate LMX010 exhibits the most difference between these models (S̄ (θ̄, θ0) = 0:866 between ‘original, norm’ and ‘g-h, LAR(1)’) and is examined in more detail in Figure 3. Here, in each panel, we show 20 concentration-response curves using parameters from the posterior distribution. Under ‘original, norm’, the curves tend to go through the middle of the two replicates in the first four concentrations. Under ‘original, norm mix’, we start to see two modes in the posterior distribution, one close to each replicate. Under ‘original, LAR(1)’, we see a clearer split of the posterior samples, and a ‘vacuum’ of posterior density is visible where the posterior samples from the ‘original, norm’ model mostly reside. This transition from normal errors to normal mixture errors to LAR(1) errors is exactly what we hope to see under such circumstances. Because one of the replicates seems to be an outlier in each of the first four concentrations, the ‘true’ concentration-response curve should be near one of the replicates, and not in between. In light of the fitted curves from the other plates (Web Figure 3), if the ‘true’ curve were close to the replicate plotted in empty circles, it would be a lot flatter than other curves. The result of the ‘g-h, LAR(1)’ fit shows that given our belief about the plate random effect variance the ‘true’ curve is much more likely to be close to the replicate plotted in solid circles.
Figure 3.
Posterior distributions of the concentration-response curve for plate LMX010. All use substantive priors. Two replicates in the data are distinguished by two different plotting symbols.
In this example, the ‘g-h, LAR(1)’ model fit differs markedly from the ‘original, LAR(1)’ model fit (area between curves 1.017). Since this is mainly due to the amount of prior information encoded in the respective substantial priors (see Figure 2), we hereby examine the sensitivity of the ‘g-h, LAR(1)’ fit to the strength of the prior. We do this by reducing each diagonal element of S by 4 or 16 fold, which corresponds to doubling or quadrupling the 95% quantiles of the corresponding random effects used in the specification of the hyperparameters. The left-hand panel of Web Figure 5 shows the results from reducing Sg and Slog(h) by 4 or 16 fold. The posterior medians move towards the default prior fit noticeably, but gradually. This confirms the importance of using information from the historical dataset in a proper way, and it also shows the posterior inference is reasonably robust to the quantile information extracted from the historical dataset. The right-hand panel of Web Figure 5 shows the results from reducing Sc, Sd and Slog(f) by 4 or 16 fold. Results, presented in the right panel of Web Figure 5, show that the lower asymptote of the posterior median curve moves up a little bit at 4 fold, and stabilizes at 16 fold. It does not change, however, the feature that distinguishes the ‘g-h, LAR(1)’ fit from the ‘original, LAR(1)’ fit, namely the fitted curve is much closer to the solid circle data points than the empty circle data points. These results also suggest that under the g-h parameterization the posterior fit is more sensitive to the priors on g and log (h) than to the priors on c, d, and log (f), presumably because there is more information in the data with regard to the latter group of parameters.
6. Discussion
In this paper we introduce a novel normal mixture model with a dependent error structure which we call the latent first order autoregressive process (LAR(1)). We use LAR(1) to model the experimental noise in a calibration experiment where dependent outliers may arise due to an intrinsic order in the data generating process. In addition, we propose a reparameterization of the five parameter logistic model, the g-h parameterization. The reparameterization appears to reduce the mean-variance dependence in the prior distribution of the plate effect variance component parameters, thus allowing us to elicit priors from the historical data we have. The g-h parameterization also offers better mixing properties in MCMC (data not shown) under both the default and the substantive priors. We show through simulation experiments and a real example that the proposed method leads to substantial gain in prediction performance in the presence of dependent gross outliers. We implement our methods in an R package, which is available at http://labs.fhcrc.org/fong/Ruminex/index.html.
D.J. Finney once wrote that “many features of bioassay are outstandingly good for concentrating the mind on important parts of biometric practice and statistical inference … because some requirements are more sharply defined” (Finney, 1979). These words still ring true today. Our encounter with dependent outliers in the bioassay data has motivated the LAR(1) process. This process differs from previous work on correlated residuals in non-linear regression (e.g. Glasbey, 1980) in that it allows the distribution of each residual to be heavy-tailed. On the other hand, it is set apart from other dependent heavy-tailed distributions such as AR(1) process with Student's t distribution in that the latent mixture indicators are modeled as an AR(1) process. This allows the flexibility that one or both components of the normal mixture at different time points be independent. The posteriors on the latent mixture indicators can also indicate which points are outliers. It would be interesting to see whether LAR(1) process can be used to model other dependent heavy-tailed data, such as financial time series data, for which AR(1) process with heavy-tailed distributions have been used in the modeling (Heracleous and Spanos, 2006). We speculate that our LAR(1) model could also be useful in pharmacokinetic and pharmacodynamic modeling (when the latter involves a continuous response), see Davidian and Giltinan (1995) for examples.
Supplementary Material
Acknowledgments
We thank Olivier Defawe and members of the HVTN laboratory for performance of the assays. We thank Xuesong Yu for helpful discussion on the simulation study. We also thank the Editor, Associate Editor and referee for insightful comments.
Footnotes
Supplementary Materials: Web Figures referenced in Sections 3-5 are available are available with this paper at the Biometrics website on Wiley Online Library.
Contributor Information
Y. Fong, Email: yfong@fhcrc.org, Vaccine and Infectious Disease Division, Fred Hutchinson Cancer Research Institute, WA 98109, USA.
J. Wakefield, Email: jonno@uw.edu, Department of Statistics, University of Washington, Seattle 98105, USA.
S. De Rosa, Email: sderosa@fhcrc.org, Vaccine and Infectious Disease Division, Fred Hutchinson Cancer Research Institute, WA 98109, USA.
N. Frahm, Email: nfrahm@fhcrc.org, Vaccine and Infectious Disease Division, Fred Hutchinson Cancer Research Institute, WA 98109, USA.
References
- Baharith L, Al-Khouli A, Raab G. Cytotoxic assays for screening anticancer agents. Statistics in Medicine. 2006;25:2323–2339. doi: 10.1002/sim.2272. [DOI] [PubMed] [Google Scholar]
- Belanger B, Davidian M, Giltinan D. The effect of variance function estimation on nonlinear calibration inference in immunoassay data. Biometrics. 1996;52:158–175. [PubMed] [Google Scholar]
- Blei I, Odian G, Selfe S. General, Organic, and Biochemistry Lab Manual. WH Freeman & Co; 2006. [Google Scholar]
- Bornkamp B, Ickstadt K. Bayesian nonparametric estimation of continuous monotone functions with applications to dose–response analysis. Biometrics. 2009;65:198–205. doi: 10.1111/j.1541-0420.2008.01060.x. [DOI] [PubMed] [Google Scholar]
- Box G. Non-normality and tests on variances. Biometrika. 1953;40:318–335. [Google Scholar]
- Brown M, Forsythe A. Robust tests for the equality of variances. Journal of the American Statistical Association. 1974;69:364–367. [Google Scholar]
- Davidian M, Giltinan D. Nonlinear models for repeated measurement data. Vol. 62. Chapman & Hall/CRC; 1995. [Google Scholar]
- Engvall E, Perlmann P. Enzyme-linked immunosorbent assay (ELISA). Quantitative assay of immunoglobulin G. Immunochemistry. 1971;8:871–874. doi: 10.1016/0019-2791(71)90454-x. [DOI] [PubMed] [Google Scholar]
- Faes C, Aerts M, Geys H, Molenberghs G. Model averaging using fractional polynomials to estimate a safe level of exposure. Risk Analysis. 2007;27:111–123. doi: 10.1111/j.1539-6924.2006.00863.x. [DOI] [PubMed] [Google Scholar]
- Finney D. The principles of biological assay. Supplement to the Journal of the Royal Statistical Society. 1947;9:46–91. [Google Scholar]
- Finney D. Bioassay and the practice of statistical inference. International Statistical Review/Revue Internationale de Statistique. 1979;47:1–12. [Google Scholar]
- Fomenko I, Durst M, Balaban D. Robust regression for high throughput drug screening. Computer Methods and Programs in Biomedicine. 2006;82:31–37. doi: 10.1016/j.cmpb.2006.01.008. [DOI] [PubMed] [Google Scholar]
- Fong Y, Rue H, Wakefield J. Bayesian Inference for Generalized Linear Mixed Models. Biostatistics. 2010;11:397–412. doi: 10.1093/biostatistics/kxp053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelman A, Chew G, Shnaidman M. Bayesian analysis of serial dilution assays. Biometrics. 2004;60:407–417. doi: 10.1111/j.0006-341X.2004.00185.x. [DOI] [PubMed] [Google Scholar]
- Glasbey C. Nonlinear regression with autoregressive time series errors. Biometrics. 1980:135–139. [Google Scholar]
- Gottschalk P, Dunn J. The five-parameter logistic: a characterization and comparison with the four-parameter logistic. Analytical Biochemistry. 2005;343:54–65. doi: 10.1016/j.ab.2005.04.035. [DOI] [PubMed] [Google Scholar]
- Grishin Y, Janczak D. A robust traclang fixed-lag smoothing algorithm in the presence of correlated outliers. Radar Symposium, 2008 International; IEEE. 2008. pp. 1–4. [Google Scholar]
- Hamilton M. Robust estimates of the ED50. Journal of the American Statistical Association. 1979:344–354. [Google Scholar]
- Heracleous M, Spanos A. The Student's t dynamic linear regression: re-examining volatility modeling. Advances in Econometrics. 2006;20:289–319. [Google Scholar]
- Huber P, Ronchetti E. Robust Statistics. John Wiley & Sons Inc; 2009. [Google Scholar]
- Lange K, Little R, Taylor J. Robust statistical modeling using the t distribution. Journal of the American Statistical Association. 1989:881–896. [Google Scholar]
- Little R. Robust estimation of the mean and covariance matrix from data with missing values. Applied Statistics. 1988;37:23–38. [Google Scholar]
- MacDonald I, Zucchini W. Monographs on statistics and applied probability. Chapman & Hall; 1997. Hidden Markov and other models for discrete-valued time series. [Google Scholar]
- Miller R, Halpern J. Robust estimators for quantal bioassay. Biometrika. 1980;67:103–110. [Google Scholar]
- Morales K, Ibrahim J, Chen C, Ryan L. Bayesian model averaging with applications to benchmark dose estimation for arsenic in drinking water. Journal of the American Statistical Association. 2006;101:9–17. [Google Scholar]
- Motulsky H, Brown R. Detecting outliers when fitting data with nonlinear regression–a new method based on robust nonlinear regression and the false discovery rate. BMC bioinformatics. 2006;7:123. doi: 10.1186/1471-2105-7-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Normolle D. AN algorithm for robust non-linear analysis of radioimmunoassay and other bioassays. Statistics in Medicine. 1993;12:2025–2042. doi: 10.1002/sim.4780122106. [DOI] [PubMed] [Google Scholar]
- Plummer M. JAGS: A program for analysis of Bayesian graphical models using Gibbs sampling. Proceedings of the 3rd International Workshop on Distributed Statistical Computing; 2003. Mar, pp. 20–22. [Google Scholar]
- Prentice R. A generalization of the probit and logit methods for dose response curves. Biometrics. 1976;32:761–768. [PubMed] [Google Scholar]
- Qaqish B. A family of multivariate binary distributions for simulating correlated binary variables with specified marginal means and correlations. Biometrika. 2003;90:455–463. [Google Scholar]
- Qiu C, Vaswani N. Support-Predicted Modified-CS for recursive robust principal components' Pursuit. Information Theory Proceedings (ISIT), 2011 IEEE International Symposium on; IEEE. 2011. pp. 668–672. [Google Scholar]
- Racine-Poon A. A Bayesian approach to nonlinear calibration problems. Journal of the American Statistical Association. 1988:650–656. [Google Scholar]
- Rerks-Ngarm S, Pitisuttithum P, Nitayaphan S, Kaewkungwal J, Chiu J, Paris R, Premsri N, Namwat C, de Souza M, Adams E, et al. Vaccination with ALVAC and AIDSVAX to prevent HIV-1 infection in Thailand. New England Journal of Medicine. 2009;361:2209–2220. doi: 10.1056/NEJMoa0908492. [DOI] [PubMed] [Google Scholar]
- Richards F. A flexible growth function for empirical use. Journal of Experimental Botany. 1959;10:290. [Google Scholar]
- Ritz C, Streibig J. Bioassay analysis using R. Journal of Statistical Software. 2005;12:1–22. [Google Scholar]
- Rodbard D, Cooper J. A model for prediction of confidence limits in radioimmunoassays and competitive protein binding assays. Vienna International Atomic Energy Agency; 1970. [Google Scholar]
- Rodbard D, Frazier G. Statistical analysis of radioligand assay data. Methods in Enzymology. 1975;37:3–22. doi: 10.1016/s0076-6879(75)37003-1. [DOI] [PubMed] [Google Scholar]
- Rodbard D, Munson P, DeLean A. Improved curve-fitting, parallelism testing, characterization of sensitivity and specificity, validation, and optimization for radioligand assays. Radioimmunoassay and Related Procedures in Medicine. 1978;1:469–504. [Google Scholar]
- Seber G, Wild C. Nonlinear regression. New York, NY (EUA): J. Wiley; 1988. [Google Scholar]
- Taplin R, Raftery A. Analysis of agricultural field trials in the presence of outliers and fertility jumps. Biometrics. 1994;50:764–781. [Google Scholar]
- Tiede J, Pagano M. The application of robust calibration to radioimmunoassay. Biometrics. 1979;35:567–574. [PubMed] [Google Scholar]
- Wang D, Burton R, Nahm M, Soong S. A four-parameter logistic model for estimating titers of functional multiplexed pneumococcal opsonophagocytic killing assay. Journal of Biopharmaceutical Statistics. 2008;18:307–325. doi: 10.1080/10543400701697182. [DOI] [PubMed] [Google Scholar]
- Yalow R, Berson S. Immunoassay of endogenous plasma insulin in man. Journal of Clinical Investigation. 1960;39:1157–1175. doi: 10.1172/JCI104130. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.