Abstract
The presence of damage in the transcribed strand (TS) of active genes and its position in relation to nucleosomes influence nucleotide excision repair (NER) efficiency. We examined chromatin structure, transcription and repair at the MET16 gene of wild-type and cbf1Δ Saccharomyces cerevisiae cells under repressing or derepressing conditions. Cbf1p is a sequence-specific DNA binding protein required for MET16 chromatin remodelling. Irrespective of the level of transcription, repair at the MspI restriction fragment of MET16 exhibits periodicity in line with nucleosome positions in both strands of the regulatory region and the non-transcribed strand of the coding region. However, repair in the coding region of the TS is always faster, but exhibits periodicity only when MET16 is repressed. In general, absence of Cbf1p decreased repair in the sequences examined, although the effects were more dramatic in the Cbf1p remodelled area, with repair being reduced to the lowest levels within the nucleosome cores of this region. Our results indicate that repair at the promoter and coding regions of this lowly transcribed gene are dependent on both chromatin structure and the level of transcription. The data are discussed in light of current models relating NER and chromatin structure.
INTRODUCTION
The eukaryotic genomic DNA in vivo is wrapped around octamers of histone proteins in a highly ordered pattern forming the nucleosomes and higher order chromatin structures (1). This packaging is considered to be a general mechanism of silencing genes, preventing the easy access of transcription factors to their binding sites, and it has been shown to influence the activity of important cellular processes like transcription, replication and DNA repair (2,3).
Although the relationship between chromatin structure and DNA repair is not well understood, some reciprocal influences have been detected. Previous studies have shown that in chromatin the rate of formation and repair of DNA lesions depends on the nucleotide sequence and the chromatin environment surrounding the damage (4,5). Furthermore, DNA damage may also alter the structure of the chromatin (6). The rate of nucleotide excision repair (NER) in Saccharomyces cerevisiae depends on the nucleosome structure along the non-transcribed strand (NTS) of the URA3 gene (7,8). Moreover, the wrapping of DNA around a nucleosome has been shown to inhibit the efficiency of NER in vitro (9,10). Recently, we have shown that Cbf1p-dependent changes in nucleosome positioning and/or protein binding affect mainly repair of cyclobutane pyrimidine dimers (CPDs) present downstream of the Cbf1p binding sites in the transcribed strand (TS) of the repressed MET17 gene (11), despite Cbf1p not being essential for transcription of this locus (11,12).
In order to further understand how the level of transcription and the structure of the chromatin influence the rate of CPD removal by NER, here we study these three parameters under conditions that alter the level of transcription and/or nucleosomal conformation of a different Cbf1p-regulated gene, namely the MET16 gene, and where Cbf1p is essential for transcription (12,13).
MET16 encodes the enzyme 3′-phospho 5′-adenylylsulfate reductase of the methionine biosynthetic pathway (14). In conditions where methionine is available in the medium, MET16 is expressed at low levels, i.e. 0.3–0.7 transcripts per cell (15; Mark Gerstein’s laboratory website, http://bioinfo.mbb.yale.edu). MET16 transcription is regulated by two different pathways (12); one specific for methionine, which depends on the binding of a complex of Cbf1p, Met28p and Met4p to the CDE1 site (Fig. 1A), and another through the general control of amino acids that depends on the binding of Gcn4p to the AP-1 site, although it also requires Cbf1p to be fully functional (13). Cbf1p is a non-essential general transcription factor with affinity for the CDE1 sequence present in Saccharomyces cerevisiae centromeres and promoter regions of multiple genes, including most of the genes involved in methionine biosynthesis (16,17). Moreover, Cbf1p has been shown to locally modify the chromatin structure around the CDEI sequence in several Cbf1p-regulated genes, including MET16 and MET17 (11,12,18,19). However, as mentioned earlier, important differences exist with respect to the precise effect that the absence of Cbf1p has on transcription at these loci.
Figure 1.
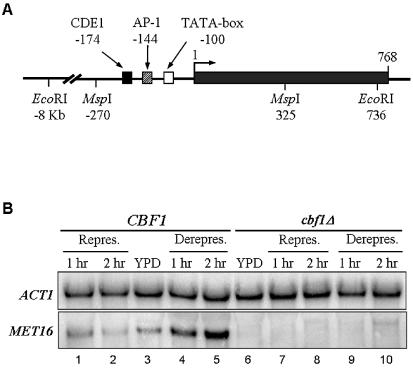
(A) The MET16 gene structure. The three regulatory elements, CDE1, AP-1 and TATA-box, and relevant restriction enzymes sites are shown. Positions are indicated in relation to the first codon of the protein. (B) Representative results of MET16 transcription in CBF1 (lanes 1–5) and cbf1Δ (lanes 6–10) strains. Hybridization with ACT1 probe (top) and with MET16 (bottom) is shown. Lanes 3 and 6 correspond to samples grown in YPD medium. Lanes 1, 2 and 7, 8 correspond to samples grown for 1 and 2 h in repressing conditions, and lanes 4, 5 and 9, 10 to samples grown in derepressing conditions.
We studied how DNA repair in both strands of the promoter and 5′ end of MET16, is influenced by changes in transcription and/or the nucleosome structure. These changes were accomplished by growing yeast cells in high or low concentration of methionine and by deleting CBF1.
MATERIALS AND METHODS
Strains, growing conditions and UV treatment
Two haploid isogenic strains of S.cerevisiae were used: DBY745 (MATα, leu2-3, leu2-112, ura3-52, ade1-110, CBF1) and YAG93 (MATα, leu2-3, leu2-112, ura3-52, ade1-110, cbf1Δ). Cells were grown overnight at 30°C in complete medium (YPD) to 3–4 × 107 cells/ml, washed twice with minimal medium, and resuspended in minimal medium plus either 1 mM methionine (MET16 repressing conditions) or 10 µM methionine (MET16 derepressing conditions) to a final concentration of 2 × 107 cells/ml. After 2 h growing in the conditioning medium, cells were treated with 150 J/m2 of 254 nm UV light as previously described (20) and allowed to repair the damage for a period of 1–4 h in the conditioning medium.
DNA isolation and determination of the rate of NER
The rate of NER at MET16 was determined at nucleotide resolution in the fragment between the MspI restriction sites (Fig. 1A). Samples of cells were taken before the UV treatment (U sample), immediately after treatment (0 sample), and from treated cells allowed to repair the damage for 1, 2, 3 or 4 h (samples 1–4).
Genomic DNA was isolated as previously described (20), and aliquots of ∼30 µg per sample were digested overnight with 120 U of MspI restriction enzyme at 37°C. The DNA was treated with Micrococcus luteus UV endonuclease to cut where CPDs were induced, and both strands were individually isolated and labelled at the 3′-end with [α-32P]dATP using biotinylated probes specific for the MspI fragment of MET16 (see below), as previously described (21). Individual DNA fragments corresponding to strands cut with the UV endonuclease were resolved by electrophoresis in denaturing 6% polyacrylamide gels at 70 W for 2.5 h.
Autoradiographs were scanned in a Storm 860 PhosphorImager (Molecular Dynamics) and the amount of signal present at the undamaged top band and the different pyrimidine tracts was quantified using the ImageQuant 5.0 software. The initial level of damage was calculated as the percentage of radioactivity present in the damaged DNA fragments in relation to the damaged plus undamaged fragments in the 0 sample. CPDs in short pyrimidine tracts were quantified as a single band to simplify presentation and their rate of repair was calculated as the T50% value, the time required for 50% of the lesions present in that band immediately after treatment to be repaired (21). The repair data at nucleotide resolution shown in Results correspond to the average of four to seven different experiments. These data were also used to compare general trends of repair in the upstream regulatory region (from –225 to –38 nt), and the transcribed region (from –38 to +275 nt) of each DNA strand. The statistical comparison of the rate of repair of the group of CPDs present in the same region was carried out by the Student’s t-test for matched-paired samples (H0, mean value of the differences is zero). The statistical comparison of the rate of repair of the group of CPDs present in different DNA regions was carried out by the Mann–Whitney test (H0, both samples are taken from populations with identical median values). Table 1 shows the average T50% of the CPDs present in the promoter, coding region and the whole DNA strands. All data are the average of at least three independent experiments.
Table 1. General CPD repair trends.
| TS | NTS | |||||
|---|---|---|---|---|---|---|
| –225/+275a | –225/–38b | –38/+275c | –225/+275a | –225/–38b | –38/+275c | |
| CBF1-repressing | 3.1 | 4.2 | 2.6 | 3.8 | 3.8 | 3.8 |
| cbf1Δ-repressing | 4.1 | 4.7 | 3.7 | 4.5 | 4.5 | 4.5 |
| CBF1-derepressing | 2.2 | 3.2 | 1.7 | 3.2 | 2.7 | 3.4 |
| cbf1Δ-derepressing | 3.6 | 4.5 | 3.1 | 4.5 | 4.6 | 4.5 |
Rate of CPD repair in the TS and NTS of CBF1 and cbf1Δ cells grown in MET16 transcription repressing and derepressing conditions.
aThe rate of repair is expressed as the average of the T50% values of the CPDs present in the whole MspI fragment.
bThe rate of repair is expressed as the average of the T50% values of the CPDs present in the upstream regulatory region.
cThe rate of repair is expressed as the average of the T50% values of the CPDs present in the transcribed region.
Level of transcription
Total RNA was isolated using the hot-phenol method (22) from samples of 3 × 108 cells, and MET16 transcription was determined by northern blot. Preliminarily, 10 µl of the RNA samples were electrophoresed in 1.5% agarose/0.25 M formaldehyde gels, transferred to nylon membranes (NEN™ Life Science Products) and hybridized overnight with a probe specific for actin (ACT1) to test the integrity and measure the amount of RNA by phosphoimaging. A second set of samples with similar amounts of RNA, as determined in the preliminary experiments, was loaded and hybridized with probes specific for MET16 and ACT1. After quantification, the amount of MET16 transcript was corrected for ACT1 transcription. MET16 transcription in wild-type cells grown in YPD medium was used as a reference (1×). Transcription in the other samples is expressed in relation to the reference value.
Nucleosome mapping
Chromatin mapping at MET16 was carried out at the low and high resolution level by micrococcal nuclease (MNase) digestion of permeabilized yeast cells. The low resolution mapping, which detects MNase-generated double-strand cuts, was performed as described (23). Briefly, 1.8 × 109 cells from each yeast strain were grown as above. Cells were harvested, permeabilized with zymolyase and digested with 5, 10 and 20 U of MNase for 4 min at 37°C. ‘Naked’ DNA samples were prepared from 9 × 108 cells treated as above, but with two phenol/chloroform extractions and an ammonium acetate/isopropanol precipitation before the MNase treatment. Samples of purified naked and chromatin genomic DNA were digested to completion with EcoRI and analysed by Southern blot with a double-strand probe for the MscI–EcoRI fragment (412 to +736 nt).
Nucleosomes at MET16 were mapped at nucleotide resolution by using a biotinylated probe-directed enriching and end-labelling method (24,25), which detects both double- and single-strand cuts. To minimize the effect due to non-specific single-strand breakage induced by MNase digestion, the samples of genomic DNA digested with this enzyme were treated with T4 DNA ligase and T4 polynucleotide kinase for 30 min at 37°C (25). The DNA was then digested to completion with MspI and after further purification the two strands were individually labelled at the 3′-end as for the DNA repair experiments.
Probes and primers used for DNA labelling and RNA probe preparation
The purification and 3′-end [32P]dATP labelling of the MspI fragments of MET16 gene were carried out as previously described for the MFA2 gene (26) using the following probes: MspI-A, 5′-biotin-GATAGCTTTTTT-GGTGGACATCACCT ATTGATTCTAAAT-3′ for the TS and MspI-B, 5′-biotin-GATAGCTTTTTT-GCTTATATACGTGAATGGTTTGATT TTTAG-3′ for the NTS. Sequences in italics correspond to overhang modifications.
DNA probes for MET16 mRNA were prepared from PCR products obtained with MspI-A and -C primers (MspI-C, 5′-CATCCG-GCTTATATACGTGAATGGTTTGATTTTTAG-3′). Probes for ACT1 were similarly prepared using the following primers: 5′-biotin-GCCGGTTTTGCCGGTGACG-3′ and 5′-CCGGCAGATTCCAAACCCAAAA-3′.
RESULTS
MET16 transcription
Discrepancies in the MET16 transcription level obtained by different research groups have been ascribed to small variations in medium composition (12). To avoid this problem and to be able to directly compare the results between different experiments, MET16 transcription was determined under the same growth conditions used to study chromatin structure and repair.
After northern blotting, the amount of MET16 transcription was estimated by normalizing against ACT1 transcription (Fig. 1B). MET16 transcription in the wild-type strain grown in YPD medium (lane 3) was taken as a reference value (1×). Similar levels of transcription were detected in this strain after growth in repressing conditions for either 1 or 2 h (lanes 1 and 2; 1.3× and 1.1×, respectively). However, MET16 transcription was induced 3.2- and 4.2-fold after growth for 1 and 2 h in derepressing conditions (lanes 4 and 5). Conversely, MET16 transcription in the cbf1Δ strain was consistently too low to be detected (lanes 6–10).
Nucleosome mapping at MET16
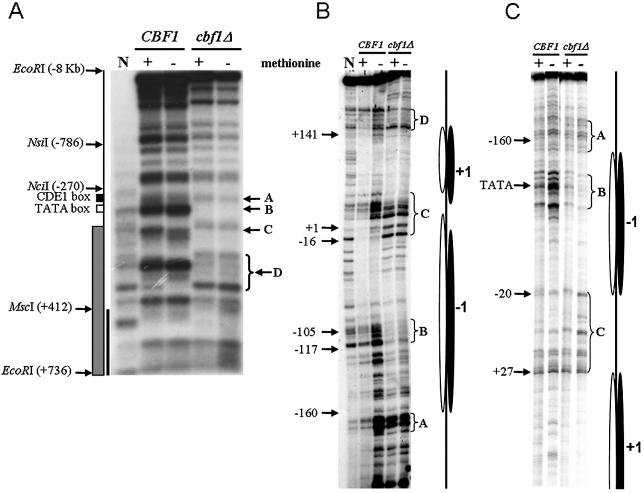
Positions of nucleosomes can be inferred from their ability to protect MNase cleavage sites in naked DNA. Nucleosomes at MET16 were initially mapped at low resolution by MNase digestion of the 9 kb EcoRI restriction fragment that includes this gene (Fig. 2A). These experiments showed some changes in the pattern of accessibility to MNase digestion, mostly related to the presence of Cbf1p, rather than the growth condition. Strain differences started downstream of the Cbf1p binding site (band A) and extended over three nucleosomes into the middle of the coding region (bands D). The main change corresponded to the presence of a new MNase cleavage site around the TATA-box in the wild-type strain (band B) that is absent in cbf1Δ cells. In addition, the positions of the internucleosomal cleavage sites (linkers) are shifted in the wild-type strain compared with the cbf1Δ strain (bands C and D), suggesting small displacements of nucleosomes positions. Two differences dependent on growth conditions were observed in the wild-type strain. Here, an additional MNase cleavage site below band C is evident on gene activation (Fig. 2A, lane 3) and a cleavage site normally present at the end of bands D is missing in low methionine conditions. On the contrary, there were no growth-dependent changes in the cbf1Δ strain.
Figure 2.
Nucleosome mapping at the MET16 locus of naked (N lanes) and chromatin DNA isolated from both strains and growth conditions. Indicated are specific bands/regions as mentioned in the text. (A) Low resolution MNase digestion pattern of the ∼9 kb EcoRI restriction fragment including MET16. Relevant locus components and restriction sites are shown on the left. The black line corresponds to the probe used for the Southern blot. A, B, C and D band(s) indicate the regions more sensitive to MNase within the MspI restriction fragment of MET16. (B) MNase digestion pattern at nucleotide resolution of the MspI restriction fragment of the MET16 TS. (C) MNase digestion pattern at nucleotide resolution of the MspI restriction fragment of the MET16 NTS. Some nucleotide positions are shown for reference. Proposed nucleosomes in the wild-type (white ovals) and mutant strain (black ovals) are shown on the right at their approximate nucleotide positions.
Additionally, the MNase cleavage sites on the MspI restriction fragment were mapped at high resolution to allow a more precise delineation of nucleosome cores and linker regions. Figure 2B shows the TS of the MspI fragment digested with MNase. In the cbf1Δ strain, the region mapping approximately from –160 to –16 (relative to ATG) is resistant to MNase digestion, which is consistent with a positioned nucleosome (–1). This is separated from a second protected region (nucleosome +1), approximately from +20 to +168, by a 30 nt linker region (band C). It is clear that in the wild-type strain the position of band C is shifted by ∼25 nt towards the coding region and as a consequence, nucleosome +1 is also shifted by a similar amount. In the wild-type strain, the presence of the MNase-sensitive region, band B, disrupting the continuous protected region between bands A and C was confirmed, affecting ∼15 nt around the TATA-box. Moreover, in MET16 derepressing conditions the whole region between the CDEI element and the TATA-box is more sensitive to MNase digestion than in repressing conditions. The differences observed in the Southern blot in bands D were not detected at nucleotide resolution. Finally, the region close to the end of the MspI fragment was slightly, but consistently, more prone to MNase digestion in the mutant than in the wild-type strain. Figure 2C shows the equivalent autoradiograph for the NTS. Here, the pattern of bands in the two strains and growth conditions is very similar, but the small displacement of band C detected in the TS is not observed. In the two DNA strands the intensity of the MNase digestion at band B follows the same order, i.e. in the wild-type cells band B is more accessible in derepressing than in repressing conditions, and in these cells the band B is more accessible than in the mutant strain. In summary, the approximate positions of the nucleosomes from these analyses can be seen in Figure 2.
Damage induction and CPD repair at the MspI restriction fragment of MET16
Figure 3 shows typical DNA repair sequencing gels for both strains and transcription conditions of the MET16 TS and NTS. The initial level of damage in the two strains and different growth conditions ranged from 19 to 24% in the TS, and from 14 to 16% in the NTS. This small difference between DNA strands was due to the induction of slightly less damage over the length of the NTS (see Fig. 3). CPD repair rates at the MspI restriction fragment of MET16 are summarized in Figures 4 (average rate of repair at each CPD tract versus the nucleotide position), 5 (relative ratios of T50% for CPD repair between cbf1Δ and CBF1 cells) and 6 (relative ratios of T50% for CPD repair in repressing versus derepressing conditions).
Figure 3.
Representative autoradiographs at nucleotide resolution showing the repair of CPDs in the TS and NTS of the MspI fragment of MET16 in wild-type and cbf1Δ cells grown in 10 µM (derepressing) and 1 mM methionine conditions (repressing). Sanger A+G and T+C sequencing ladders are included to determine the position of the CPDs induced. U, untreated cells; 0, cells treated with UV light no repair; (1–4), UV-treated cells allowed to repair the damage for 1–4 h. The intense top band corresponds to the undamaged MspI fragment of MET16, the bands below represent DNA fragments with a CPD lesion that was cut by the CPD-specific endonuclease. Proposed nucleosomes in CBF1 (white ovals) and cbf1Δ cells (black ovals) are shown at their approximate nucleotide positions.
Figure 4.
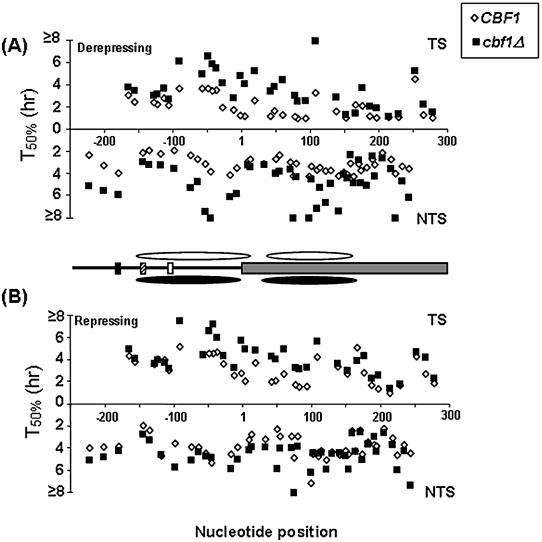
Repair of individual CPDs at MET16 versus the nucleotide position in (A) derepressing and (B) repressing conditions. The rate of repair in CBF1 (diamonds) and cbf1Δ cells (squares) is expressed as the T50% for CPDs in the TS (top), and NTS (bottom). Position 1 corresponds to the start of the coding region. Cbf1p binding site (black box), Gcn4p binding site (striped box), TATA-box (white box) and coding region (grey box). Approximate nucleosome positions according to the high resolution mapping of the MET16 TS are shown for the CBF1 (white ovals) and cbf1Δ strain (black ovals).
Figure 5.
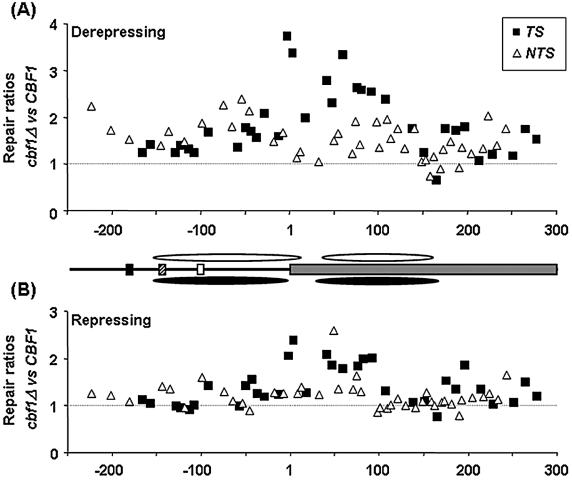
Relative ratios of T50% values for CPD repair between CBF1 and cbf1Δ cells in the MET16 TS (squares) and NTS (triangles) in (A) derepressing and (B) repressing conditions. Values above 1 indicate faster repair in the CBF1 strain and those below 1 faster repair in the cbf1Δ mutant. Position 1 corresponds to the start of the coding region. Cbf1p binding site (black box), Gcn4p binding site (striped box), TATA-box (white box) and coding region (grey box). Approximate nucleosome positions according to the high resolution mapping of the MET16 TS are shown for the CBF1 (white ovals) and cbf1Δ strain (black ovals).
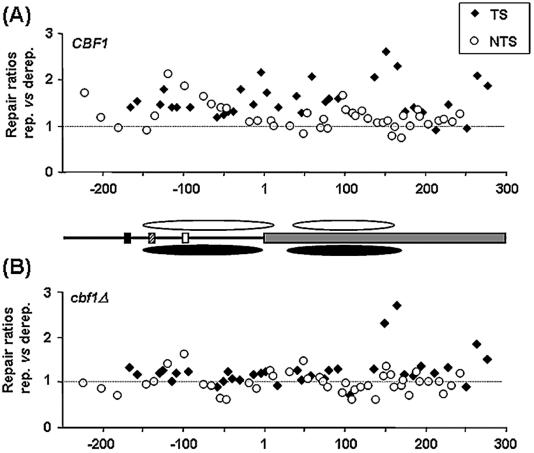
Figure 6.
Relative ratios of T50% values for CPD repair between MET16 derepressing and repressing conditions in the TS (diamonds) and NTS (circles) of (A) CBF1 cells and (B) cbf1Δ cells. Values above 1 indicate faster repair when transcription is derepressed, and values below 1, faster repair with transcription repressed. Position 1 corresponds to the start of the coding region. Cbf1p binding site (black box), Gcn4p binding site (striped box), TATA-box (white box) and coding region (grey box). Approximate nucleosome positions according to the high resolution mapping of the MET16 TS are shown for the CBF1 (white ovals) and cbf1Δ strain (black ovals).
Cbf1p influences CPD repair in derepressing conditions
In either yeast strain the group of CPDs within the coding region of the TS (Fig. 4A) was repaired faster than those in the respective promoter region of the TS or the NTS overall (P ≤ 0.05). In addition, Figures 4A and 5A clearly illustrate that, in general, repair in the cbf1Δ strain was less efficient than in the wild-type strain. Moreover, the statistical analysis showed that repair of specific CPDs within the same region was faster in the wild-type strain (P < 0.001) in all the different regions.
In the TS, the maximum differences in repair for individual CPDs between both strains were obtained around the initial ATG codon, i.e. 3.7- and 3.4-fold faster repair in the CBF1 strain for the CPDs at –3/TTTT/+1 and +3/CTT/+5 (Fig. 5A). These differences decreased over the length of the coding region, reaching the minimum in a small region towards the end of the MspI fragment, namely for CPDs at 213, 228 and 251 nt positions.
Inspection of the rate of repair over the length of the NTS suggested the presence of a wave-like pattern in the two strains (Fig. 4A), although this pattern was less obvious in the wild-type strain. To confirm the presence of this pattern, data from Figure 4A were examined using curve analysis software (CurveExpert 1.3). For both strains, the best fit for the rate of CPD repair along the TS and NTS corresponded to a 7–9 degree polynomial function, supporting the existence of the wave pattern (data not shown).
In general, repair in the NTS was faster in the wild-type than in the cbf1Δ strain (Fig. 4A) and less differences occurred in regions that were repaired rapidly (Fig. 5A). These include the region between the Gcn4p binding site and the TATA-box, the first 50–70 nt of the coding region and the area between +160 to +210 nt. However, in regions where repair was slow, differences in the T50% of up to 2.4-fold were seen (Fig. 5A). The position of areas subject to fast repair corresponds primarily to MNase cleavage sites in chromatin, whereas the slow repairing ones correspond to the regions contained within nucleosome cores.
Cbf1p influences CPD repair in repressing conditions
Since the absence of a detectable MET16 transcript in the cbf1Δ strain could be responsible for the differences in repair observed in derepressing conditions by reducing transcription coupled repair (TCR), we have also determined the repair rates for individual CPDs under repressing conditions (Fig. 4B).
As was the case for derepressing conditions, in both yeast strains repair in the coding region of the TS was faster than in the respective promoter region of the TS or the whole NTS (P ≤ 0.05). Interestingly, repair of the group of CPDs within the same region was still statistically faster (P ≤ 0.05) in the wild-type than in the mutant strain (Figs 4B and 5B).
In this growing condition, repair of the group of six CPDs present in the TS between the CDE1 element and the TATA-box was statistically homogeneous in the two strains (Figs 4B and 5B). Immediately downstream of the TATA-box, repair rates started to diverge, i.e. 5.2 ± 0.9 h in CBF1 versus 7.5 ± 0.8 h in the cbf1Δ strain for the CPDs at –92/TTTTTTTC/–86 (T50% ± SE). The maximum differences in this strand were obtained around the initial ATG codon, 2.1- and 2.4-fold difference for the CPDs at –3/TTTT/+1 and +3/CTT/+5, respectively, decreasing towards the end of the coding region (Fig. 5B).
Differences in CPD repair in the NTS were less pronounced than in derepressing conditions (Fig. 5B). The largest differences corresponded to the CPDs at +49/TT/+50 (2.6-fold), +74/TC/+75 (1.7-fold) and around the TATA-box –99/TT/–98 (1.6-fold). Finally, the rate of repair in the NTS was also characterized by a wave pattern, which was supported by curve analysis (data not shown) as in derepressing conditions. In this condition, the three regions of fast repair matched the Gcn4p binding site, the first 50–70 nt of the coding region, and the fragment between +160 to +210 nt positions (Fig. 4B).
Transcription influences CPD repair at the MET16 NTS and TS
The influence of the transcriptional status on NER was studied by comparing for each yeast strain the rate of CPD repair in the two growth conditions.
In the wild-type strain (Fig. 6A) the CPDs induced in the TS, taken as a group, were repaired faster in low methionine than in high methionine (P < 0.001). In general, repair was 1.6-fold faster in low methionine, with the exception of a small region from +200 to +250 nt with similar rates of repair (Fig. 6A). Similarly, the CPDs in the NTS, as a group, were also repaired faster in derepressing conditions (P < 0.01). The biggest differences in repair were observed in a fragment of ∼80 nt around the TATA-box, and in a small region between the +100 to +125 nt positions, both of them lying within the core of the proposed nucleosomes.
In the cbf1Δ strain, a smaller difference in the rate of repair was detected between growth conditions (Fig. 6B). In general, cells grown in derepressing conditions repaired the group of CPDs induced in the TS faster than those grown in high methionine (P ≤ 0.05), on average a 1.3-fold difference. However, no significant differences between the two growth conditions were detected in repair of the group of CPDs on the NTS, either taking them as a whole or for the promoter or coding regions individually. In this strand the biggest difference in the rate of repair for individual CPDs corresponded to those close to the TATA-box (1.6-fold difference), which were faster repaired in derepressing conditions.
DISCUSSION
Cbf1p influences MET16 transcription and chromatin structure
The results from the transcription experiments show a profound impairment of MET16 transcription in the absence of Cbf1p and a modest (4-fold) increase of MET16 transcription in wild-type cells grown in derepressing conditions. This is similar to the induction previously reported for this gene and in agreement with the importance of Cbf1p in MET16 regulation (12,13).
In addition, Cbf1p causes, either directly or indirectly, the remodelling of MET16 chromatin structure at promoter and coding regions. The different methods used in this work for the nucleosome mapping experiments showed small discrepancies in the pattern of MNase accessibility. This is probably due to a combination of several factors, i.e. the detection in the low resolution mapping of non-specific single-strand cuts, the presence of the DNA-histone contacts at different positions in the TS and NTS (27,28), and the effect of transcription factors bound to one or the other strand.
Taking into account the results from the low and high resolution mapping experiments, the pattern of MNase digestion of cbf1Δ cells suggests the presence of two nucleosomes in the MspI fragment of MET16, one covering the area between the CDEI element and the beginning of the coding region (–1), and the other between bands C and D inside the transcribed region (+1). These positions are similar to those previously described in a different cbf1Δ strain employing a low resolution approach (12), i.e. nucleosome –1 would correspond to that between the MNase cleavage sites E′ and G, and nucleosome +1 to the one between G and H. Results from that study suggested that in the presence of Cbf1p, nucleosome –1 would be completely displaced from the DNA. However, two facts indicate that this is not happening in our experiments. In CBF1 cells we observe a MNase-sensitive region over the TATA-box, leaving a protected region between bands B and C of only 90 bp, which is too short to accommodate a nucleosome. Yet, the bands observed in the naked DNA between bands B and C do not appear in the CBF1 background. In addition, the MNase-sensitive area C, a possible linker region, is displaced in the TS towards the coding region, but is still partly overlapping the equivalent area in the cbf1Δ strain. These facts suggest that in the CBF1 strain nucleosome –1 would still be present over the TATA-box, but covering a slightly different region. This poses the interesting question as to how a positioned nucleosome can coexist with the presence of an internal MNase-sensitive region. According to previous chromatin studies, this coexistence could result from the partial disruption of some DNA-histone contacts (27,28) by the RNA polymerase, chromatin remodelling factors or histone acetyltransferases (29–34), leaving the TATA-box region in a small intranucleosomal loop. This kind of structure would be consistent with the presence of the transcriptional intermediates detected in vitro with the SP6 RNA polymerase (32) or the stepwise uncoiling of individual nucleosomes obtained by applying tension to the end of DNA wrapped around a nucleosomal array (35). Alternatively, it has been proposed that the displacement or destabilization of one H2A/H2B dimer would facilitate the access of transcription factors to nucleosomal DNA and transcription (33,36–38), which would also be compatible with the appearance of the new MNase-sensitive region and the reduced size of the protected region between bands B and C.
The effect of Cbf1p on the level of transcription and chromatin structure is different at MET16 than in the a priori similar MET17 gene, i.e. Cbf1p is essential for MET16 transcription but not for MET17 (11–13; this work); at MET17 Cbf1p changes the position of two nucleosomes maintaining the TATA-box in a nucleosome-free region, whereas at MET16 the nucleosome over the TATA-box is preserved basically at the same position in the presence of Cbf1p but in an altered state, thus leaving the TATA-box in a small unattached region (11,12,18,19; this work). The reasons behind this different response are not exactly known. It may depend on the presence of two Cbf1p binding sites at MET17 instead of only the one present at MET16, and where the sequence is slightly different. It is also worthy to note that MET16 is considerably less transcribed than MET17 either for basal or induced levels of transcription (15; Mark Gerstein’s laboratory website, http://bioinfo.mbb.yale.edu), so the way in which transcription factors influence these loci cannot be identical.
CPD repair at MET16 depends both on chromatin structure and transcription extent
Our results show a significant influence of methionine concentration on the rate of CPD repair at the MET16 locus, although this effect is more marked in the wild-type than in the cbf1Δ strain, which is consistent with its bigger influence on MET16 transcription in the wild-type strain. Interestingly, many of the CPDs in the cbf1Δ NTS were repaired slightly faster in repressing than in derepressing conditions. Since no transcription of MET16 is detected in the cbf1Δ strain and repair in the NTS is greatly dependent on chromatin structure (7,39), we assume this effect is due to local changes in chromatin structure that increase the accessibility or functionality of the NER machinery in repressing conditions.
Despite the substantial differences detected in MET16 transcription, the group of CPDs in the coding region of the TS is always repaired faster than those in the corresponding non-transcribed regions. This fast repair always starts ∼30–40 nt upstream of the start of translation, and reaches the maximum efficiency around the initial ATG codon. Although when transcription occurs this can be attributed to TCR, the presence of fast repair in the coding region even when transcription is absent or highly suppressed (cbf1Δ strain) supports the idea that at a primary level, efficient repair of some lowly transcribed genes, such as MET16, may depend on chromatin characteristics more than on transcription per se enhancing TCR (40).
The efficiency of NER in the NTS of the URA3 gene has been shown to be modulated by nucleosome positioning (7,8). Accordingly, the rate of repair of individual CPDs in the MET16 NTS follows a wave pattern with a periodicity approximately equal to nucleosome spacing. However, this pattern is somehow less obvious in the wild-type strain grown in derepressing conditions. The peaks and troughs of this wave occur basically in the same regions in both strains, supporting that the nucleosomes occupy similar positions irrespective of Cbf1p status. Furthermore, the more efficient repair of the four CPDs present in the NTS between the Gcn4p binding site and the TATA-box detected in MET16 derepressing conditions correlates with a higher MNase sensitivity in this region.
Although this pattern of nucleosome-dependent repair is not obvious in the TS, in both strains and growth conditions the areas of fast and slow repair in the region covered by nucleosome –1 are concordant with those in the NTS. Moreover, contrary to that seen with the URA3 gene (7,8), a small patch of slow repair coincident with the core of the nucleosome over the coding region of the TS (+1) can be detected in cells grown in repressing conditions. This is likely because with lowly transcribed genes, such as MET16, TCR is not operating to repair all CPDs in the TS. Thus, GGR would repair many lesions in the TS when transcription levels are low. With respect to URA3, transcription levels are much higher, so the role of GGR in repairing the TS would be much diminished, and TCR would operate more effectively, explaining why no wave pattern was seen for repair of the TS at this locus.
Deletion of the CBF1 gene, particularly when transcription is derepressed, decreases the efficiency of CPD removal in both strands of MET16, although its influence is not homogeneous along the MspI fragment. In both transcription conditions, the highest differences in CPD repair detected in the TS correspond to the region between the TATA-box and the +100 nt position, which includes the main area remodelled in the presence of Cbf1p. Absence of Cbf1p has a small, but significant, effect on CPD repair along the NTS when transcription is repressed, while its influence under derepressing conditions, especially in the promoter region, is more marked. Here, although the ratio of repair between strains is similar in core and linker regions, CBF1 deletion reduced NER efficiency to the lowest levels at the CPDs within the nucleosome cores. The differences detected in the rate of repair at MET16 due to Cbf1p absence are probably the consequence of a local effect because the overall efficiency of NER in cbf1Δ cells is indistinguishable from wild-type cells when the rate of CPD removal from total genomic DNA is measured by CPD immunoblotting (data not shown).
As indicated above, transcription regulation and chromatin remodelling at the MET16 and MET17 genes show important differences. Concurrently, some differences in CPD repair at the two genes can be seen (11; this work). First of all, fast repair of CPDs in the coding region of the TS is detected in the two strains and growth conditions at MET16, while this only happens under inducing conditions at MET17. Moreover, while fast repair at the MET16 TS starts 30–40 nt before the initial ATG codon, at MET17 it starts ∼200 nt upstream of the coding region. On average, repair rates in the NTS at both genes are quite similar. The most striking difference between MET16 and MET17 corresponds to repair of CPDs in the TS. Here, while cbf1Δ cells repair CPDs with a more or less similar efficiency at either gene, the wild-type cells repair CPDs more efficiently at the MET16 TS when transcription is activated. This is surprising because the amount of transcription from MET16 is ∼10-fold less than that from MET17. Finally, repair at the MET16 gene shows a stronger correlation with nucleosome positions than repair at the MET17 gene. Together, these differences indicate the difficulty in predicting how efficiently repair at the nucleotide level is going to operate at different genes in relation to transcription, the binding of regulatory proteins and nucleosome positions.
In summary, the rate of CPD repair via NER at the MET16 gene, can be modulated by a number of different parameters. Repair is dependent on the nucleotide sequence of the damage-containing region (4,5). It can depend on the location of the damage in relation to nucleosome positioning and it can be further influenced by the binding of specific chromatin remodelling factors governing nucleosome position and/or conformation (7,8; this work). Finally, the actual rate of repair for each CPD position in the transcribed portion of the TS would depend on the transcriptional status and the amount of transcription of the gene in question.
Lastly, given the fact that NER is faster in the TS of the lowly expressed MET16 when compared with that seen at the more highly expressed MET17 (15; Mark Gerstein’s laboratory website, bioinfo.mbb.yale.edu), simple extrapolations concerning rates of repair versus rates of transcription are not possible for these loci. This may well be a reflection of the complexity of factors governing NER rates throughout the genome and suggests caution in generalizing from NER effects seen at specific loci.
Acknowledgments
ACKNOWLEDGEMENTS
This work was supported by The Wellcome Trust (056922/A/99/Z/BS/DC) and the MRC (G99 00118). J. A. Ferreiro was funded by FICYT (Spain) and an EU Marie Curie fellowship (QLK6-CT-1999-51401).
REFERENCES
- 1.Wolffe A. (1998) Chromatin: Structure and Function. Academic Press, San Diego, USA. [Google Scholar]
- 2.Meijer M. and Smerdon,M.J. (1999) Accessing DNA damage in chromatin: insights from transcription. BioEssays, 21, 596–603. [DOI] [PubMed] [Google Scholar]
- 3.Meyer P. (2001) Chromatin remodelling. Curr. Opin. Plant Biol., 4, 457–462. [DOI] [PubMed] [Google Scholar]
- 4.Pfeifer G.P. (1997) Formation and processing of UV photoproducts: effects of DNA sequence and chromatin environment. Photochem. Photobiol., 65, 270–283. [DOI] [PubMed] [Google Scholar]
- 5.Smerdon M.J. and Conconi,A. (1999) Modulation of DNA damage and DNA repair in chromatin. Prog. Nucleic Acid Res. Mol. Biol., 62, 227–255. [DOI] [PubMed] [Google Scholar]
- 6.Mann D.B., Springer,D.L. and Smerdon,M.J. (1997) DNA damage can alter the stability of nucleosomes: effects are dependent on damage type. Proc. Natl Acad. Sci. USA, 94, 2215–2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wellinger R.E. and Thoma,F. (1997) Nucleosome structure and positioning modulate nucleotide excision repair in the non-transcribed strand of an active gene. EMBO J., 16, 5046–5056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tijsterman M., De Pril,R., Tasseron-de Jong,J.G. and Brouwer,J. (1999) RNA polymerase II transcription suppresses nucleosomal modulation of UV-induced (6–4) photoproduct and cyclobutane pyrimidine dimer repair in yeast. Mol. Cell. Biol., 19, 934–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hara R., Mo,J. and Sancar,A. (2000) DNA damage in the nucleosome core is refractory to repair by human excision nuclease. Mol. Cell. Biol., 20, 9173–9181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ura K., Araki,M., Saeki,H., Masutani,C., Ito,T., Iwai,S., Mizukoshi,T., Kaneda,Y. and Hanaoka,F. (2001) ATP-dependent chromatin remodeling facilitates nucleotide excision repair of UV-induced DNA lesions in synthetic dinucleosomes. EMBO J., 20, 2004–2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Powell N.G., Ferreiro,J., Karabetsou,N., Mellor,J. and Waters,R. (2003) Transcription, nucleosome positioning and protein binding modulate nucleotide excision repair of the Saccharomyces cerevisiae MET17 promoter. DNA Repair (Amst.), 2, 375–386. [DOI] [PubMed] [Google Scholar]
- 12.O’Connell K.F., Surdin-Kerjan,Y. and Baker,R.E. (1995) Role of the Saccharomyces cerevisiae general regulatory factor CP1 in methionine biosynthetic gene transcription. Mol. Cell. Biol., 15, 1879–1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kuras L., Chérest,H., Surdin-Kerjan,Y. and Thomas,D. (1996) A heteromeric complex containing the centromere binding factor 1 and two basic leucine zipper factors, Met4 and Met28, mediates the transcription activation of yeast sulfur metabolism. EMBO J., 15, 2519–2529. [PMC free article] [PubMed] [Google Scholar]
- 14.Thomas D., Barbey,R. and Surdin-Kerjan,Y. (1990) Gene-enzyme relationship in the sulfate assimilation pathway of Saccharomyces cerevisiae. Study of the 3′-phosphoadenylylsulfate reductase structural gene. J. Biol. Chem., 265, 15518–15524. [PubMed] [Google Scholar]
- 15.Wang Y., Liu,C.L., Storey,J.D., Tibshirani,R.J., Herschlag,D. and Brown,P.O. (2002) Precision and functional specificity in mRNA decay. Proc. Natl Acad. Sci. USA, 99, 5860–5865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cai M. and Davis,R.W. (1990) Yeast centromere binding protein CBF1, of the helix-loop-helix protein family, is required for chromosome stability and methionine prototrophy. Cell, 61, 437–446. [DOI] [PubMed] [Google Scholar]
- 17.Mellor J., Jiang,W., Funk,M., Rathjen,J., Barnes,C.A., Hinz,T., Hegemann,J.H. and Philippsen,P. (1990) CPF1, a yeast protein which functions in centromeres and promoters. EMBO J., 9, 4017–4026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McKenzie E.A., Kent,N.A., Dowell,S.J., Moreno,F., Bird,L.E. and Mellor,J. (1993) The centromere and promoter factor 1, CPF1, of Saccharomyces cerevisiae modulates gene activity through a family of factors including SPT21, RPD1 (SIN3), RPD3 and CCR4. Mol. Gen. Genet., 240, 374–386. [DOI] [PubMed] [Google Scholar]
- 19.Kent N.A., Tsang,J.S., Crowther,D.J. and Mellor,J. (1994) Chromatin structure modulation in Saccharomyces cerevisiae by centromere and promoter factor 1. Mol. Cell. Biol., 14, 5229–5241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reed S.H., Boiteux,S. and Waters,R. (1996) UV-induced endonuclease III-sensitive sites at the mating type loci in Saccharomyces cerevisiae are repaired by nucleotide excision repair: RAD7 and RAD16 are not required for their removal from HML alpha. Mol. Gen. Genet., 250, 505–514. [DOI] [PubMed] [Google Scholar]
- 21.Teng Y., Li,S., Waters,R. and Reed,S.H. (1997) Excision repair at the level of the nucleotide in the Saccharomyces cerevisiae MFA2 gene: mapping of where enhanced repair in the transcribed strand begins or ends and identification of only a partial rad16 requisite for repairing upstream control sequences. J. Mol. Biol., 267, 324–337. [DOI] [PubMed] [Google Scholar]
- 22.Ausubel F.M. (1989) Current Protocols in Molecular Biology. Wiley, New York, NY. [Google Scholar]
- 23.Kent N.A. and Mellor,J. (1995) Chromatin structure snap-shots: rapid nuclease digestion of chromatin in yeast. Nucleic Acids Res., 23, 3786–3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Teng Y., Yu,S. and Waters,R. (2001) The mapping of nucleosomes and regulatory protein binding sites at the Saccharomyces cerevisiae MFA2 gene: a high resolution approach. Nucleic Acids Res., 29, e64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morillon A., Karabetsou,N., O’Sullivan,J., Kent,N., Proudfoot,N. and Mellor,J. (2003) Isw1 chromatin remodeling ATPase coordinates transcription elongation and termination by RNA Polymerase II. Cell, 115, 425–435. [DOI] [PubMed] [Google Scholar]
- 26.Teng Y. and Waters,R. (2000) Excision repair at the level of the nucleotide in the upstream control region, the coding sequence and in the region where transcription terminates of the Saccharomyces cerevisiae MFA2 gene and the role of RAD26. Nucleic Acids Res., 28, 1114–1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dutnall R.N. and Ramakrishnan,V. (1997) Twists and turns of the nucleosome: tails without ends. Structure, 5, 1255–1259. [DOI] [PubMed] [Google Scholar]
- 28.Luger K., Mader,A.W., Richmond,R.K., Sargent,D.F. and Richmond,T.J. (1997) Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature, 389, 231–233. [DOI] [PubMed] [Google Scholar]
- 29.Studitsky V.M., Kassavetis,G.A., Geiduschek,E.P. and Felsenfeld,G. (1997) Mechanism of transcription through the nucleosome by eukaryotic RNA polymerase. Science, 278, 1960–1963. [DOI] [PubMed] [Google Scholar]
- 30.Cote J., Peterson,C.L. and Workman,J.L. (1998) Perturbation of nucleosome core structure by the SWI/SNF complex persists after its detachment, enhancing subsequent transcription factor binding. Proc. Natl Acad. Sci. USA, 95, 4947–4952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tse C., Sera,T., Wolffe,A.P. and Hansen,J.C. (1998) Disruption of higher-order folding by core histone acetylation dramatically enhances transcription of nucleosomal arrays by RNA polymerase III. Mol. Cell. Biol., 18, 4629–4638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bednar J., Studitsky,V.M., Grigoryev,S.A., Felsenfeld,G. and Woodcock,C.L. (1999) The nature of the nucleosomal barrier to transcription: direct observation of paused intermediates by electron cryomicroscopy. Mol. Cell, 4, 377–386. [DOI] [PubMed] [Google Scholar]
- 33.Wolffe A.P. and Hayes,J.J. (1999) Chromatin disruption and modification. Nucleic Acids Res., 27, 711–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Workman J.L. and Kingston,R.E. (1998) Alteration of nucleosome structure as a mechanism of transcriptional regulation. Annu. Rev. Biochem., 67, 545–579. [DOI] [PubMed] [Google Scholar]
- 35.Brower-Toland B.D., Smith,C.L., Yeh,R.C., Lis,J.T., Peterson,C.L. and Wang,M.D. (2002) Mechanical disruption of individual nucleosomes reveals a reversible multistage release of DNA. Proc. Natl Acad. Sci. USA, 99, 1960–1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hayes J.J. and Wolffe,A.P. (1992) Histones H2A/H2B inhibit the interaction of transcription factor IIIA with the Xenopus borealis somatic 5S RNA gene in a nucleosome. Proc. Natl Acad. Sci. USA, 89, 1229–1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hansen J.C. and Wolffe,A.P. (1994) A role for histones H2A/H2B in chromatin folding and transcriptional repression. Proc. Natl Acad. Sci. USA, 91, 2339–2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Spangenberg C., Eisfeld,K., Stunkel,W., Luger,K., Flaus,A., Richmond,T.J., Truss,M. and Beato,M. (1998) cAMP-dependent phosphorylation sites and macroscopic activity of recombinant cardiac L-type calcium channels. J. Mol. Biol., 278, 725–739. [DOI] [PubMed] [Google Scholar]
- 39.Hu W., Feng,Z., Chasin,L.A. and Tang,M. (2002) Transcription-coupled and transcription-independent repair of cyclobutane pyrimidine dimers in the dihydrofolate reductase gene. J. Biol. Chem., 277, 38305–38310. [DOI] [PubMed] [Google Scholar]
- 40.Zheng Y., Pao,A., Adair,G.M. and Tang,M. (2001) Cyclobutane pyrimidine dimers and bulky chemical DNA adducts are efficiently repaired in both strands of either a transcriptionally active or promoter-deleted APRT gene. J. Biol. Chem., 276, 16786–16796. [DOI] [PubMed] [Google Scholar]