Abstract
In B cells, B-cell receptor (BCR) immunoglobulin revision is a common route for modifying unwanted antibody specificities via a mechanism called VH replacement. This in vivo process, mostly affecting heavy-chain rearrangement, involves the replacement of all or part of a previously rearranged IGHV gene with another germline IGHV gene located upstream. Two different mechanisms of IGHV replacement have been reported: type 1, involving the recombination activating genes complex and requiring a framework region 3 internal recombination signal; and type 2, involving an unidentified mechanism different from that of type 1. In the case of light-chain loci, BCR immunoglobulin editing ensures that a second V-J rearrangement occurs. This helps to maintain tolerance, by generating a novel BCR with a new antigenic specificity. We report that human B cells can, surprisingly, undergo type 2 replacement associated with κ light-chain rearrangements. The de novo IGKV-IGKJ products result from the partial replacement of a previously rearranged IGKV gene by a new germline IGKV gene, in-frame and without deletion or addition of nucleotides. There are wrcy/rgyw motifs at the ‘IGKV donor–IGKV recipient chimera junction’ as described for type 2 IGHV replacement, but activation-induced cytidine deaminase (AID) expression was not detected. This unusual mechanism of homologous recombination seems to be a variant of gene conversion-like recombination, which does not require AID. The recombination phenomenon described here provides new insight into immunoglobulin locus recombination and BCR immunoglobulin repertoire diversity.
Keywords: AICDA , B-cell receptor immunoglobulin repertoire diversity, gene conversion-like recombination, immunoglobulin κ, immunoglobulin genes, IGKV replacement
Introduction
Developing B cells generate a vast antibody repertoire, by multistep somatic recombination events involving immunoglobulin genes(IG), also known as V-(D)-J rearrangements. As a consequence, a limitless number of non-self antigens can be recognized. However, this mechanism can also produce autoreactive B cells that, secondarily, may be tolerated by various mechanisms, such as B-cell receptor (BCR) editing.1 During this process, the stimulation of autoreactive BCR induces an initiation signal for secondary V-J rearrangement at light-chain loci. Such secondary rearrangements replace (genotypic editing) or outcompete (phenotypic editing) the primary light-chain rearrangements, thereby modifying autoreactive BCR specificity.2,3 Another process, BCR revision, occurs in mature B cells and involves the modification of self-reactive heavy-chain V-D-J rearrangements by IGHV gene replacement.5–8 Two different types of replacement have been described. The first is mediated by the recombination-activating genes (RAG) complex, using the 3′-recombination signal (3′V-RS) of an incoming germline IGHV (donor) as the substrate and an internal or cryptic RS (cRS), in the opposite orientation, present in the FR3 (framework region 3) of the rearranged VH (recipient).9–15 These type 1 IGHV replacements use the same mechanism as conventional RAG-dependent V-D-J rearrangement and the new junctions formed characteristically display nucleotide deletions and P and N additions. The entire IGHV coding region, except for a short conserved native CDR3 (complementarity-determining region 3) sequence (six to seven nucleotides) downstream from the internal RS, is replaced with a new germline IGHV. This recombination increases the length of the V-D junction and may result in a functional product if the reading frame is conserved. The second type of IGHV replacement was first described by Wilson et al.16 and Itoh et al.17 in normal human peripheral B cells and in B cells from patients with rheumatoid arthritis. Darlow and Stott18–19 recently named this new ‘IGHV chimera’ as type 2 VH replacement. Unlike type 1 replacement, no addition or deletion of nucleotides is observed, and the reading frame is not affected. This phenomenon resembles homologous recombination between a rearranged IGHV gene and a germline IGHV gene. These potentially non-reciprocal homologous recombination events occur at perfectly matched coding regions in the two IGHV genes, such that the exact break point of the recombination cannot be identified. The same resulting sequence signatures are observed in various species, including birds and rabbits, in which gene conversion is used to produce the antibody repertoire and its diversity.20–21 During gene conversion, a segment of DNA is replaced by copying from a cis or trans V sequence, thereby generating a ‘V chimera segment’ without alteration of the donor template.22 This process of gene conversion is initiated by the AICDA gene product, activation-induced cytidine deaminase (AID),23 a key enzyme in somatic hypermutation24–25 and in class switch recombination26–27; its target sequences contain hot spot (a/t)(a/g)c(c/t)/(a/g)g(c/t)(a/t) motifs (wrcy/rgyw).28 These hotspot motifs are found, within the chimera gene, on either side of the perfectly matched sequence corresponding to identical regions in the two fused IGHV genes. Gene conversion, in the context of lymphocyte repertoire diversification, has not been observed in humans, but it has been suggested that type 2 IGHV replacement provides evidence of gene conversion-like events,29–30 involving AID. However, the direct implication of AID in this process has never been proved. Most of the published sequences of type 2 replacement products were obtained from human B cells expressing AICDA, but such products have also been identified in immature B cells (which are theoretically AID-negative).31 It therefore remains unclear whether AID is actually involved in this newly described V gene replacement process. We tried to resolve this issue, by establishing a model of the recombination, de novo, of immunoglobulin V genes in the absence of AICDA. We successfully induced, in vitro, type 2 V replacement in the immunoglobulin κ (IGK) light-chain locus in human pre-B cells, and we report evidence that AICDA is not expressed in this model.
Materials and methods
Cell line
The human pre-B cell line 697 was purchased from DSMZ. Cells were cultured at 37°, under a humidified atmosphere containing 5% CO2, in RPMI-1640 medium (Gibco, Paisley, UK) supplemented with 10% heat-inactivated fetal calf serum (PAA Laboratories, Yeovil, UK), 50 U/ml each of penicillin and streptomycin and 2 mm l-glutamine.
Cell treatment
After overnight serum deprivation, 697 cells cultured at a density of 0·5 × 106 cells/ml were induced by incubation for 7 days with 10 ng/ml of the cytokine interleukin-1α (IL-1α) (Tebu-Bio, Le Perray-en-Yvelines, France) or with 1 μg/ml lipopolysaccharide (LPS, Sigma, St Louis, MO). After treatment, a PCR-based test was used to check for the absence of mycoplasma in the cultures.
Genomic DNA extraction
Induced and control cells (∼ 5 × 106) were harvested and washed, and QIAamp spin column technology (Qiagen, Hilden, Germany) was used to extract genomic DNA. The concentration of isolated genomic DNA preparations was measured with a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE).
Multiplex PCR, cloning and sequencing
Genomic IGKV1, IGKV2, IGKV3 to IGKJ5 rearrangements were amplified by multiplex PCR with 500 ng of genomic DNA, using IGKV1f/6, IGKV2f and IGKV3f forward primers and a Jκ5 reverse primer. AmpliTaq Gold and ABI Gold Buffer (Applied Biosystems, Foster City, CA) were used as recommended in the standardized BIOMED-2 PCR protocol.32 PCR amplification conditions were as specified in the BIOMED-2 protocol and PCR was run for 40 cycles. PCR products were subjected to electrophoresis in a 1% agarose gel in 1× TAE buffer for 15 min at 100 V. PCR products were purified with the MinElute gel extraction kit (Qiagen) and cloned with the Topo TA Cloning kit (Invitrogen, Carlsbad, CA). The Nucleospin Plasmid kit (Macherey Nagel, Düren, Germany) was used to purify the plasmids, and the inserts were sequenced with the Big Dye Terminator sequencing kit and subjected to electrophoresis on an Applied Biosystem ABI 3500 sequencer. Both strands were analysed with internal pCR®2.1-TOPO® forward and reverse M13 primers (Invitrogen). Three independent experiments were performed and PCR was carried out three times for each experiment. IMGT®, the international ImMunoGeneTics information system® (http://www.imgt.org)33 and the Basic Local Alignment Search Tool (BLAST) database were used to analyse sequence data for more than 130 clones. Selected IGKV type 2 sequences were submitted to EMBL-EBI (European Nucleotide Archive, http://www.ebi.ac.uk/): Accession numbers: HF674720, HF674721, HF674722, HF674723, HF674724, HF674719 and HF674725.
RNA extraction and RT-PCR
Total cellular RNA was isolated from ∼5 × 106 cells, with RNeasy Mini kits (Qiagen), and treated with DNAse. We reverse-transcribed 1 μg of RNA by the High-Capacity cDNA Reverse Transcription protocol (Applied Biosystems). AmpliTaqGold DNA polymerase (Applied Biosystems) was used for PCR amplification of the AICDA and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) genes from 100 ng of cDNA, and the PCR products were analysed by electrophoresis in 1% agarose gels containing ethidium bromide and visualized with the Chemidoc system (Bio-Rad, Hercules, CA). The primers used were: AICDA forward 5′-cactggactttggttatcttcgc-3′; AICDA reverse 5′-cgtaagtcatcaacctcatacagg-3′; GAPDH forward 5′-tcggagtcaacggatttggtcg-3′; GAPDH reverse 5′-tcagtgtagcccaggatgcct-3′.
Results
Induction of immunoglobulin κ (IGK) locus rearrangement in human B cells
In the mouse B-cell model, immunoglobulin κ (IGK) locus recombination is induced by treatment with LPS34–35 or IL-1.36–37 There are several Toll-like receptor 4-positive and IL1-R1-positive human pre-B-cell lines. We chose to use the RAG1/2-positive 697 cell line for this study. We confirmed that treatment with either IL-1 or LPS successfully induced de novo IGKV-IGKJ rearrangements without altering RAG1/2 mRNA production (data not shown). The 697 cell line was chosen because of its particular configuration of IGK haplotypes: one IGK locus contains a native unmutated IGKV1-8-IGKJ4 rearrangement, and the five IGKJ genes have been deleted from the second locus by IGKV3-7-KDE rearrangement. These particular locus configurations facilitate the exploration of de novo IGKV-IGKJ rearrangements with a single IGKJ5 antisense primer for their detection and for PCR product isolation (Fig. 1). As the single genes of each of the IGKV4, IGKV5 and IGKV7 subgroups (IMGT®; http://www.imgt.org> IMGT Repertoire >Locus representation) have been deleted, de novo IGKV-IGKJ5 rearrangements can only involve genes of the IGKV1, IGKV2 and IGKV3 subgroups. Aliquots of 500 ng of genomic DNA isolated from 105 cells were used for multiplex PCR and electrophoresis of the heterogeneous products of PCR amplification yielded a smear.
Figure 1.
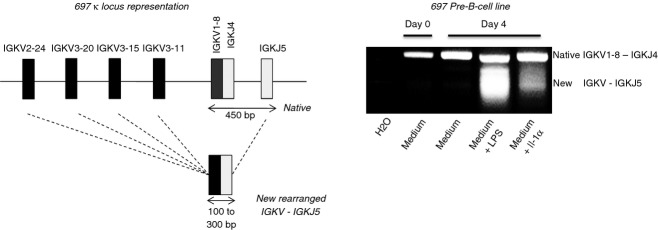
IGKV-IGKJ5 multiplex PCR with DNA from the human pre-B cell line 697 after induction with lipopolysaccharide (LPS) or interleukin-1 (IL-1). Genomic DNA was isolated and IGKV-IGKJ rearrangements were amplified by PCR with the IGKV1f/6, IGKV2f and IGKV3f forward primers and the Jk5 reverse primer.32 A schematic diagram of the organization of the native haplotype carrying the IGKV1-8-IGKJ4 rearrangement in the 697 cell line is shown on the left (EMBL accession number HF674726). New IGKV-IGKJ5 rearrangements result in a smear migrating between 120 and 300 bp (right). After longer periods of induction, the signal for the native IGKV1-8-IGKJ4 disappeared and the signal from the new IGKV3-IGKJ5 became more intense (see Supplementary material, Fig. S1).
V replacement in κ locus rearrangement
After cloning and sequencing, we analysed 220 sequences obtained from several independent experiments, with IMGT® and Ig-BLAST tools and associated databases. In total, 130 of the 220 clones analysed had different sequences, and 28 of these clones were mutated. Comparisons of the sequences we obtained with the IMGT/V-QUEST38 tool indicated that about 22% of the individual IGKV-IGKJ5 rearrangements induced de novo carried somatic mutations, including 18 with at least two mutations or about 14% of the independent sequences analysed. Comparisons between IGKV-IGKJ5 sequences carrying a single mutation and germline IGKV genes were not considered in this study. Alignments of apparently multiple mutated rearrangements with germline reference IGKV sequences revealed an ‘IGKV chimera gene’ and led to the first identification of IGKV-IGKJ sequences with type 2 IGKV replacement (Fig. 2). We chose seven chimera sequences with clear characteristics of IGKV type 2 replacement for further study. As shown in Fig. 2, IGKV replacement can take place at various sites in the V-REGION, from the FR3-IMGT (e.g. HF674720) to the CDR3-IMGT (HF674724). The use of an IGKV gene as both recipient and donor sequence is illustrated by the HF674719 and HF674720 sequences. An example of internal IGKV replacement is provided by the HF674725 sequence. A comprehensive alignment of full sequences is provided in the Supplementary material (Fig. S3). Type 2 replacement was observed in de novo IGKV-IGKJ5 rearrangements, irrespective of the IGKV subgroup involved (IGKV1, IGKV2 or IGKV3), although IGKV2-IGKJ5 recombinations were found preferentially. IGKV replacement occurred only between donor and recipient IGKV genes from the same subgroup. Within the same subgroup, no difference was observed between IGKV genes: IGKV2-30 could be replaced with IGKV2-24 and vice versa. No cryptic RS in the IGKV recipient gene was identified to account for the localization of replacement sites. Following type 2 replacement, the chimera sequences encode new Vκ domains displaying amino acid changes when aligned with the donor sequence or the recipient sequence (Fig. 3). Unlike type 1 IGHV replacement, in which the 3′V-RS of the IGHV donor gene rearranges with the internal RS in the FR3 of the IGHV recipient gene with a resulting longer sequence, no change in the length of type 2 V chimera IGKV genes was observed. The ‘IGKV chimera genes’ were all in-frame, without the addition or deletion of nucleotides. The alignment of IGKV-IGKJ5 sequences with IGKV germline sequences, using IMGT/V- QUEST38 made it possible to assign particular regions to specific IGKV genes accurately (Fig. 2). However, at the junctions of these assigned regions, there are short sequences of 10–20 nucleotides, the origin of which is unclear because they are identical to the sequences of both the IGKV receiver gene and the IGKV donor gene. These short unassigned regions correspond to stretches of strict identity between the donor and recipient genes. As found for type 2 IGHV replacements, AID motifs are present at sites flanking these strictly identical sequences.
Figure 2.
Hybrid IGKV sequences from multiplex PCR IGKV-IGKJ5. Examples of sequences from the cloned PCR products obtained from interleukin-1α (IL-1α) or lipopolysaccharide (LPS) -treated human pre-B 697 cells. Seven chimeric sequences were selected to illustrate different examples of IGKV type 2 replacement in terms of localization, in particular: from the FR to CDR targeted regions, internal replacement and IGKV gene usage as either a recipient or donor sequence. IMGT/V-QUEST38–33 was used for alignment with IGKV germline genes. IGKV-IGKJ5 rearrangements were identified as follow: on the alphabetic sequences of the cloned PCR products, dark green corresponds to IGKV genes, IGKJ5 sequences are shown in blue, and P- and N-diversity are shown by orange and red letters, respectively. The figures in brackets indicate the number of nucleotides deleted during the initial IGKV-IGKJ rearrangement. The lower aligned reference sequences are represented as dashes in cases of a perfect match and as letters in cases of discordance. The IGKV genes initially rearranged with the IGKJ5 gene (recipient IGKV gene) are shown in green and the donor IGKV genes replacing the initial rearranged recipient IGKV gene are shown in blue. Superimposed blue and green sequences indicate regions of sequence identity (not assigned). Potential AID hotspots (WRCY/RGYW motif with W:A/T; R:A/G; Y:C/T) are underlined and in bold.
Figure 3.
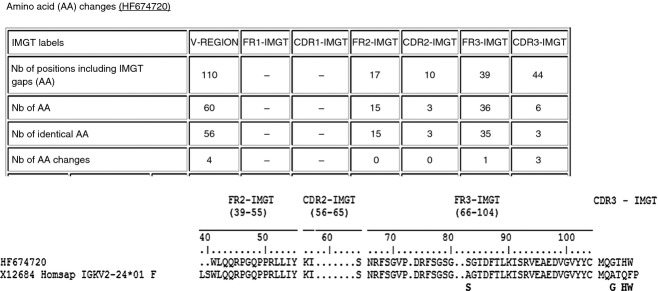
V-region protein display from a hybrid sequence. Amino-acid sequence of the HF674720 clone from lipopolysaccharide (LPS) -treated human pre-B cell line 697. HF674720, a chimeric sequence, is shown as an example of amino-acid changes resulting from IGKV type 2 replacement. IMGT/V-QUEST38–33 was used and provides a V-REGION alignment together with the number of amino acid changes. Sequence alignment with the IGKV2-24 donor gene is shown, together with the amino acid changes resulting from IGKV2-30 replacement.
AICDA expression after LPS or IL-1 induction
We investigated the expression of AICDA in 697 pre-B cells after stimulation. Cultures of 697 pre-B cells were treated with LPS or IL-1. Genomic DNA was extracted for the analysis of IGKV-IGKJ5 rearrangements and total RNA was isolated and tested for AICDA expression. The AID-positive Ramos cell line was used as a positive control. No AICDA mRNA was detected in 697 cells, after either 2 hr or 4 days of induction or at intermediate times. Moreover, the treatment of 697 cells with three AICDA inducers – the synthetic analogue of dsRNA, Poly(I:C), which acts as a ligand of Toll-like receptor 3; the Toll-like receptor 9 agonist CpG and the CD40 ligand – did not induce AICDA expression, as assessed by RT-qPCR (data not shown). Neither LPS nor IL-1 induced AICDA gene expression (Fig. 4). Consequently, this enzyme cannot be considered to be an inducer of, or even be involved in, the type 2 IGKV replacement observed in these cells.
Figure 4.
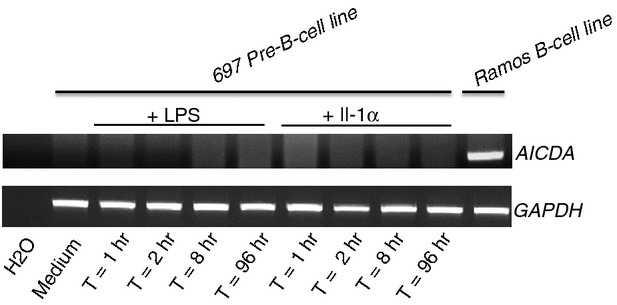
AICDA expression in the human pre-B cell line 697 after induction with lipopolysaccharide (LPS) or interleukin-1 (IL-1). Total RNA was reverse-transcribed to generate cDNA and AICDA expression was analysed by PCR. The controls used were: GAPDH as a positive control for RT-PCR and the Ramos mature B-cell line as a positive control for AICDA expression. The results shown are representative of three independent experiments.
Discussion
In this study, we demonstrated that type 2 V replacements, previously described in IGH rearrangements, also occur in V-J rearrangements of the κ locus in human pre-B-cell lines. We also showed that they could be induced in vitro. We focused our study on a human B-cell line able to generate new IGKV-IGKJ rearrangements after LPS or IL-1 treatment. Due to the particular configuration of the two κ haplotypes, pre-B cells of the 697 line can generate de novo IGKV-IGKJ rearrangements only from a single haplotype already harbouring a native IGKV1-8-IGKJ4 rearrangement. As all IGKJ genes other than IGKJ5 have been deleted (by IGKV1-8-IGKJ4 rearrangement), any subsequent de novo IGKV-IGKJ rearrangements must use IGKJ5 as the junction gene for IGK recombination. Alignments of sequences corresponding to new recombination products showed that most IGKV-IGKJ5 rearrangements were not mutated, but that a minority (about 10–15%) carried more than two mutations. We considered only the IGKV-IGKJ5 sequences carrying two or more mutations in subsequent analyses of type 2 IGKV replacements. These IGKV replacements, at the IGKV-IGKJ5 rearrangements induced de novo, occurred in the 5′ upstream region of the IGKV recipient, resulting in a new, in-frame sequence for the antigen-binding Vκ domain. Indeed the V chimera domain generated could carry, for example, the IGKV donor sequence from the FR1 to CDR2 and the native sequence from the FR3 to CDR3, so contributing to the diversity of the BCR immunoglobulin repertoire. Remarkably, the length of the replaced sequences was identical to that of potential germline IGKV sequences. Hence, the upstream replacements of the rearranged IGKV genes were not the result of the repair of RAG-induced double-strand breaks at a putative cryptic RS,39 as described in type 1 replacement. The replacement of the inner region of the de novo rearranged IGKV segments, as observed for the HF674725 sequence, could be explained by a mechanism of homologous recombination from a cis or trans donor sequence with double cross-overs affecting a very short sequence. However, homologous recombination does not seem to be a plausible mechanism for such short regions because double cross-overs generally occur in conditions in which the sequence exchanged is longer than 3 kb.22 The observation of inner replacements, as described for HF674725, also confirms that the type 2 IGKV replacements observed in this study were not PCR artefacts. As IGKV2-24 was deleted during IGKV2-30-IGKJ5 recombination, it is possible that IGKV genes from the excision circle, are used as the template for type 2 replacements either while still in circular form, or after the reintegration of the excision circle into the genome.40 This possibility is supported by one of the interesting features of this model: type 2 replacement was not detected in any of the native IGKV1-8-IGKJ4 sequences after IL-1 or LPS stimulation (see Supplementary material, Fig. S2), suggesting that type 2 replacement and V-J rearrangement may be chronologically linked. As a more conventional model, in the case of reintegration of the excision circle, it is possible that, at the time of V-J rearrangement, the spatial chromatin conformation promotes the juxtaposition of donor and recipient genes, thereby facilitating the exchange of DNA sequences. We cannot exclude the possibility of the use of IGKV genes from the second allele for κ type 2 replacement, by interchromosomal recombination. Indeed, all of the 30 or so sequences with a type 2 replacement analysed displayed the use, as the donor, of an IGKV gene and allele present on both loci. Furthermore, both IGK haplotypes in the 697 cell line contain exactly the same IGKV genes, with the exception of the IGKV3-7 gene, which is present in the second haplotype only. Hence, whatever the IGKV gene used as the donor, it is present simultaneously on both strands (in both cis and trans). The 697 cell line is therefore not the most appropriate model for distinguishing between cis (intra-allelic) and trans (inter-allelic) exchanges of DNA. However, it could be argued that the absence of IGKV replacement in the IGKV1-8-IGKJ4 native rearrangement may be a result of the chromatin conformation and epigenetic organization of the IGK haplotype, which remains inaccessible to the DNA replacement machinery in non-rearranging loci but acquires a permissive conformation in the locus undergoing de novo IGKV-IGKJ rearrangements,41 which may promote DNA replacement events. The hypothesis that inter-allelic DNA exchange is responsible for type 2 replacement is theoretically possible, but this would require the simultaneous availability of both κ loci, which would be incompatible with the phenomenon of allelic exclusion.
As previously described for type 2 VH replacements, (a/t)(a/g)c(c/t)/(a/g)g(c/t)(a/t) hotspot motifs (wrcy/rgyw) were identified on either side of the short, unassigned region showing strict identity between the donor and recipient IGKV genes. However, many other AID motifs42 can also be identified throughout the IGKV-IGKJ5 sequences, both within and outside the unassigned region. The short unassigned region may ensure matching between recipient and donor IGKV sequences. After matching, the flanking AID hotspots or other DNA motifs22–43 may then trigger double-strand breaks and the process of type 2 IGKV replacement. In species using gene conversion for the generation of their immunoglobulin repertoire diversity, such AID motifs are essential for homologous recombination. As previously suggested,16–19 type 2 replacement may result from a mechanism related to the ‘gene conversion-like’ mechanism involving AID. Surprisingly, no AICDA expression was detected in our pre-B-cell model either before or after treatment with IL-1 or LPS, even after 8 hr of stimulation, the time-point corresponding to the beginning of the rearrangement process, or after 4 days, corresponding to the end of stimulation. By contrast, previous studies have shown that LPS treatment greatly increases AICDA expression in mature B cells.44–45 However, in this study, we used κ recombination inducers (IL-1 and LPS) that can cause type 2 IGKV replacements in precursor B cells in the absence of AICDA expression. Hence, AID is not required for type 2 recombination processes in this context.
Type 2 IGKV and IGHV replacements cannot both be due to a phenomenon of AID-dependent gene conversion.46 Instead, they may both involve a new unidentified mechanism of homologous recombination (gene conversion-like recombination). Presumably, proteins other than AID are involved in this mechanism: possible candidates include error-prone DNA polymerases with spontaneous mutagenic activity on undamaged DNA47–48 and other members of the APOBEC family.49–50 As native IGKV1-8-IGKJ4 does not undergo type 2 IGKV replacements, this phenomenon of homologous recombination seems to be tightly associated with the V-J rearrangements process. These new IGKV and IGHV replacements independently contribute to the broadening of B-cell repertoire diversity, by taking part in BCR immunoglobulin editing/revision in an AID-independent manner. We are currently investigating the relationships between this new replacement process, the stimulation of κ enhancers and B-cell transcription factors.
Acknowledgments
We thank Prof. J. Rochette from the Genetics Laboratory of Amiens University Hospital for assistance with the Genetic Analyzer. This work was supported by funds from the Amiens University Hospital, INSERM and University of Picardie Jules Verne. We thank, in particular, the CIEL Association and its president, Christophe Lacombe, for their initial contribution, without which this study would not have been initiated. HOH and HG performed the experiments with the help of AR, DH and ST; HOH and BG designed the study; HOH, BG, MPL and JPM wrote the paper.
Disclosure
The authors declare that they have no conflict of interest.
Supporting Information
Additional Supporting Information may be found in the online version of this article:
The native rearrangement of 697 cells disappears after a long period of induction with interleukin-1.
Identical sequence for the all native 450 bp PCR products from treated 697 cells.
Comprehensive alignments of full hybrid IGKV sequences.
References
- Wang YH, Diamond B. B cell receptor revision diminishes the autoreactive B cell response after antigen activation in mice. J Clin Invest. 2008;118:2896–907. doi: 10.1172/JCI35618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gay D, Saunders T, Camper S, Weigert M. Receptor editing: an approach by autoreactive B cells to escape tolerance. J Exp Med. 1993;177:999–1008. doi: 10.1084/jem.177.4.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yachimovich N, Mostoslavsky G, Yarkoni Y, Verbovetski I, Eilat D. The efficiency of B cell receptor (BCR) editing is dependent on BCR light chain rearrangement status. Eur J Immunol. 2002;32:1164–74. doi: 10.1002/1521-4141(200204)32:4<1164::AID-IMMU1164>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- Tiegs SL, Russell DM, Nemazee D. Receptor editing in self-reactive bone marrow B cells. J Exp Med. 1993;177:1009–20. doi: 10.1084/jem.177.4.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reth M, Gehrmann P, Petrac E, Wiese P. A novel VH to VHDJH joining mechanism in heavy-chain-negative (null) pre-B cells results in heavy-chain production. Nature. 1986;322:840–2. doi: 10.1038/322840a0. [DOI] [PubMed] [Google Scholar]
- Nemazee D, Weigert M. Revising B cell receptors. J Exp Med. 2000;191:1813–7. doi: 10.1084/jem.191.11.1813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Zemlin M, Wang YH, et al. Contribution of Vh gene replacement to the primary B cell repertoire. Immunity. 2003;19:21–31. doi: 10.1016/s1074-7613(03)00170-5. [DOI] [PubMed] [Google Scholar]
- Liu J, Lange MD, Hong SY, et al. Regulation of VH replacement by B cell receptor-mediated signaling in human immature B cells. J Immunol. 2013;190:5559–66. doi: 10.4049/jimmunol.1102503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covey LR, Ferrier P, Alt FW. VH to VHDJH rearrangement is mediated by the internal VH heptamer. Int Immunol. 1990;2:579–83. doi: 10.1093/intimm/2.6.579. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Burrows PD, Cooper MD. The molecular basis and biological significance of VH replacement. Immunol Rev. 2004;197:231–42. doi: 10.1111/j.0105-2896.2004.0107.x. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Wang YH, Zemlin M, et al. Molecular mechanism of serial VH gene replacement. Ann N Y Acad Sci. 2003;987:270–3. doi: 10.1111/j.1749-6632.2003.tb06060.x. [DOI] [PubMed] [Google Scholar]
- Chen C, Nagy Z, Prak EL, Weigert M. Immunoglobulin heavy chain gene replacement: a mechanism of receptor editing. Immunity. 1995;3:747–55. doi: 10.1016/1074-7613(95)90064-0. [DOI] [PubMed] [Google Scholar]
- Shirasawa T, Miyazoe I, Hagiwara S, et al. Heavy chain variable (VH) region diversity generated by VH gene replacement in the progeny of a single precursor cell transformed with a temperature-sensitive mutant of Abelson murine leukemia virus. J Exp Med. 1992;176:1209–14. doi: 10.1084/jem.176.4.1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenze D, Greiner A, Knorr C, Anagnostopoulos I, Stein H, Hummel M. Receptor revision of immunoglobulin heavy chain genes in human MALT lymphomas. Mol Pathol. 2003;56:249–55. doi: 10.1136/mp.56.5.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L, Lange MD, Yu Y, Li S, Su K, Zhang Z. Contribution of V(H) replacement products in mouse antibody repertoire. PLoS ONE. 2013;8:e57877. doi: 10.1371/journal.pone.0057877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson PC, Wilson K, Liu YJ, Banchereau J, Pascual V, Capra JD. Receptor revision of immunoglobulin heavy chain variable region genes in normal human B lymphocytes. J Exp Med. 2000;191:1881–94. doi: 10.1084/jem.191.11.1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh K, Meffre E, Albesiano E, et al. Immunoglobulin heavy chain variable region gene replacement As a mechanism for receptor revision in rheumatoid arthritis synovial tissue B lymphocytes. J Exp Med. 2000;192:1151–64. doi: 10.1084/jem.192.8.1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darlow JM, Stott DI. Gene conversion in human rearranged immunoglobulin genes. Immunogenetics. 2006;58:511–22. doi: 10.1007/s00251-006-0113-6. [DOI] [PubMed] [Google Scholar]
- Darlow JM, Stott DI. V(H) replacement in rearranged immunoglobulin genes. Immunology. 2005;114:155–65. doi: 10.1111/j.1365-2567.2004.02084.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinstein PD, Anderson AO, Mage RG. Rabbit IgH sequences in appendix germinal centers: VH diversification by gene conversion-like and hypermutation mechanisms. Immunity. 1994;1:647–59. doi: 10.1016/1074-7613(94)90036-1. [DOI] [PubMed] [Google Scholar]
- McCormack WT, Tjoelker LW, Thompson CB. Avian B-cell development: generation of an immunoglobulin repertoire by gene conversion. Annu Rev Immunol. 1991;9:219–41. doi: 10.1146/annurev.iy.09.040191.001251. [DOI] [PubMed] [Google Scholar]
- Chen JM, Cooper DN, Chuzhanova N, Ferec C, Patrinos GP. Gene conversion: mechanisms, evolution and human disease. Nat Rev Genet. 2007;8:762–75. doi: 10.1038/nrg2193. [DOI] [PubMed] [Google Scholar]
- Arakawa H, Hauschild J, Buerstedde JM. Requirement of the activation-induced deaminase (AID) gene for immunoglobulin gene conversion. Science. 2002;295:1301–6. doi: 10.1126/science.1067308. [DOI] [PubMed] [Google Scholar]
- Muramatsu M, Kinoshita K, Fagarasan S, Yamada S, Shinkai Y, Honjo T. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 2000;102:553–63. doi: 10.1016/s0092-8674(00)00078-7. [DOI] [PubMed] [Google Scholar]
- Dickerson SK, Market E, Besmer E, Papavasiliou FN. AID mediates hypermutation by deaminating single stranded DNA. J Exp Med. 2003;197:1291–6. doi: 10.1084/jem.20030481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honjo T, Kinoshita K, Muramatsu M. Molecular mechanism of class switch recombination: linkage with somatic hypermutation. Annu Rev Immunol. 2002;20:165–96. doi: 10.1146/annurev.immunol.20.090501.112049. [DOI] [PubMed] [Google Scholar]
- Durandy A. Activation-induced cytidine deaminase: a dual role in class-switch recombination and somatic hypermutation. Eur J Immunol. 2003;33:2069–73. doi: 10.1002/eji.200324133. [DOI] [PubMed] [Google Scholar]
- Foster SJ, Dorner T, Lipsky PE. Somatic hypermutation of VκJκ rearrangements: targeting of RGYW motifs on both DNA strands and preferential selection of mutated codons within RGYW motifs. Eur J Immunol. 1999;29:4011–21. doi: 10.1002/(SICI)1521-4141(199912)29:12<4011::AID-IMMU4011>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- Duvvuri B, Wu GE. Gene conversion-like events in the diversification of human rearranged IGHV3-23*01 gene sequences. Front Immunol. 2012;3:158. doi: 10.3389/fimmu.2012.00158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krawinkel U, Zoebelein G, Bruggemann M, Radbruch A, Rajewsky K. Recombination between antibody heavy chain variable-region genes: evidence for gene conversion. Proc Natl Acad Sci USA. 1983;80:4997–5001. doi: 10.1073/pnas.80.16.4997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolar GR, Capra JD. Immunoglobulin heavy-chain receptor editing is observed in the NOD/SCID model of human B-cell development. Scand J Immunol. 2004;60:108–11. doi: 10.1111/j.0300-9475.2004.01467.x. [DOI] [PubMed] [Google Scholar]
- van Dongen JJ, Langerak AW, Bruggemann M, et al. Design and standardization of PCR primers and protocols for detection of clonal immunoglobulin and T-cell receptor gene recombinations in suspect lymphoproliferations: report of the BIOMED-2 Concerted Action BMH4-CT98-3936. Leukemia. 2003;17:2257–317. doi: 10.1038/sj.leu.2403202. [DOI] [PubMed] [Google Scholar]
- Giudicelli V, Brochet X, Lefranc MP. IMGT/V-QUEST: IMGT standardized analysis of the immunoglobulin (IG) and T cell receptor (TR) nucleotide sequences. Cold Spring Harb Protoc. 2011;2011:695–715. doi: 10.1101/pdb.prot5633. [DOI] [PubMed] [Google Scholar]
- Schlissel MS, Baltimore D. Activation of immunoglobulin κ gene rearrangement correlates with induction of germline κ gene transcription. Cell. 1989;58:1001–7. doi: 10.1016/0092-8674(89)90951-3. [DOI] [PubMed] [Google Scholar]
- Feddersen RM, Van Ness BG. Corrective recombination of mouse immunoglobulin κ alleles in Abelson murine leukemia virus-transformed pre-B cells. Mol Cell Biol. 1990;10:569–76. doi: 10.1128/mcb.10.2.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giri JG, Kincade PW, Mizel SB. Interleukin 1-mediated induction of κ-light chain synthesis and surface immunoglobulin expression on pre-B cells. J Immunol. 1984;132:223–8. [PubMed] [Google Scholar]
- Bomsztyk K, Toivola B, Emery DW, et al. Role of cAMP in interleukin-1-induced κ light chain gene expression in murine B cell line. J Biol Chem. 1990;265:9413–7. [PubMed] [Google Scholar]
- Alamyar E, Duroux P, Lefranc MP, Giudicelli V. IMGT((R)) tools for the nucleotide analysis of immunoglobulin (IG) and T cell receptor (TR) V-(D)-J repertoires, polymorphisms, and IG mutations: IMGT/V-QUEST and IMGT/HighV-QUEST for NGS. Methods Mol Biol. 2012;882:569–604. doi: 10.1007/978-1-61779-842-9_32. [DOI] [PubMed] [Google Scholar]
- Davila M, Liu F, Cowell LG, et al. Multiple, conserved cryptic recombination signals in VH gene segments: detection of cleavage products only in pro B cells. J Exp Med. 2007;204:3195–208. doi: 10.1084/jem.20071224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanura K, Montpellier B, Le T, et al. In vivo reinsertion of excised episomes by the V(D)J recombinase: a potential threat to genomic stability. PLoS Biol. 2007;5:e43. doi: 10.1371/journal.pbio.0050043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vettermann C, Schlissel MS. Allelic exclusion of immunoglobulin genes: models and mechanisms. Immunol Rev. 2010;237:22–42. doi: 10.1111/j.1600-065X.2010.00935.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longo NS, Satorius CL, Plebani A, Durandy A, Lipsky PE. Characterization of Ig gene somatic hypermutation in the absence of activation-induced cytidine deaminase. J Immunol. 2008;181:1299–306. doi: 10.4049/jimmunol.181.2.1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuzhanova N, Chen JM, Bacolla A, et al. Gene conversion causing human inherited disease: evidence for involvement of non-B-DNA-forming sequences and recombination-promoting motifs in DNA breakage and repair. Hum Mutat. 2009;30:1189–98. doi: 10.1002/humu.21020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pone EJ, Zhang J, Mai T, et al. BCR-signalling synergizes with TLR-signalling for induction of AID and immunoglobulin class-switching through the non-canonical NF-κB pathway. Nat Commun. 2012;3:767. doi: 10.1038/ncomms1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Pone EJ, Al-Qahtani A, Park SR, Zan H, Casali P. Regulation of AICDA expression and AID activity: relevance to somatic hypermutation and class switch DNA recombination. Crit Rev Immunol. 2007;27:367–97. doi: 10.1615/critrevimmunol.v27.i4.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longo NS, Grundy GJ, Lee J, Gellert M, Lipsky PE. An activation-induced cytidine deaminase-independent mechanism of secondary VH gene rearrangement in preimmune human B cells. J Immunol. 2008;181:7825–34. doi: 10.4049/jimmunol.181.11.7825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gearhart PJ, Wood RD. Emerging links between hypermutation of antibody genes and DNA polymerases. Nat Rev Immunol. 2001;1:187–92. doi: 10.1038/35105009. [DOI] [PubMed] [Google Scholar]
- Ohashi E, Bebenek K, Matsuda T, et al. Fidelity and processivity of DNA synthesis by DNA polymerase κ, the product of the human DINB1 gene. J Biol Chem. 2000;275:39678–84. doi: 10.1074/jbc.M005309200. [DOI] [PubMed] [Google Scholar]
- Conticello SG. The AID/APOBEC family of nucleic acid mutators. Genome Biol. 2008;9:229. doi: 10.1186/gb-2008-9-6-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conticello SG, Langlois MA, Yang Z, Neuberger MS. DNA deamination in immunity: AID in the context of its APOBEC relatives. Adv Immunol. 2007;94:37–73. doi: 10.1016/S0065-2776(06)94002-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The native rearrangement of 697 cells disappears after a long period of induction with interleukin-1.
Identical sequence for the all native 450 bp PCR products from treated 697 cells.
Comprehensive alignments of full hybrid IGKV sequences.



