Abstract
Myxobacteria exhibit complex social traits during which large populations of cells coordinate their behaviors. An iconic example is their response to starvation: thousands of cells move by gliding motility to build a fruiting body in which vegetative cells differentiate into spores. Here we review mechanisms that the model species Myxococcus xanthus uses for cell—cell interactions, with a focus on developmental signaling and social gliding motility. We also discuss a newly discovered cell—cell interaction whereby myxobacteria exchange their outer membrane (OM) proteins and lipids. The mechanism of OM transfer requires physical contact between aligned cells on a hard surface and is apparently mediated by OM fusion. The TraA and TraB proteins are required in both donor and recipient cells for transfer, suggesting bidirectional exchange, and TraA is thought to serve as a cell surface adhesin. OM exchange results in phenotypic changes that can alter gliding motility and development and is proposed to represent a novel microbial interacting platform to coordinate multicellular activities.
Keywords: Cell—cell signaling, Fruiting body, Type IV pilus, Membrane fusion, Protein transfer, Surface receptor
1. Introduction
Myxobacteria are true socialites; they exhibit complex and cooperative social behaviors that are unparalleled in the bacterial kingdom. Because of these behaviors, they were originally misidentified as eukaryotes (Kaiser, 1993). A hallmark myxobacterial behavior is their ability to organize the movements of thousands of cells to build a fruiting body in which many of the cells differentiate into environmentally resistant spores (Fig. 1A). Myxobacteria are also noted for other multicellular behaviors such as swarming, rippling, elasticotaxis and predation (Whitworth, 2008). The goal of this review is to provide an overview of known cellecell interaction mechanisms in the model species Myxococcus xanthus, with an emphasis on recent developments concerning how they exchange their outer membrane (OM) components.
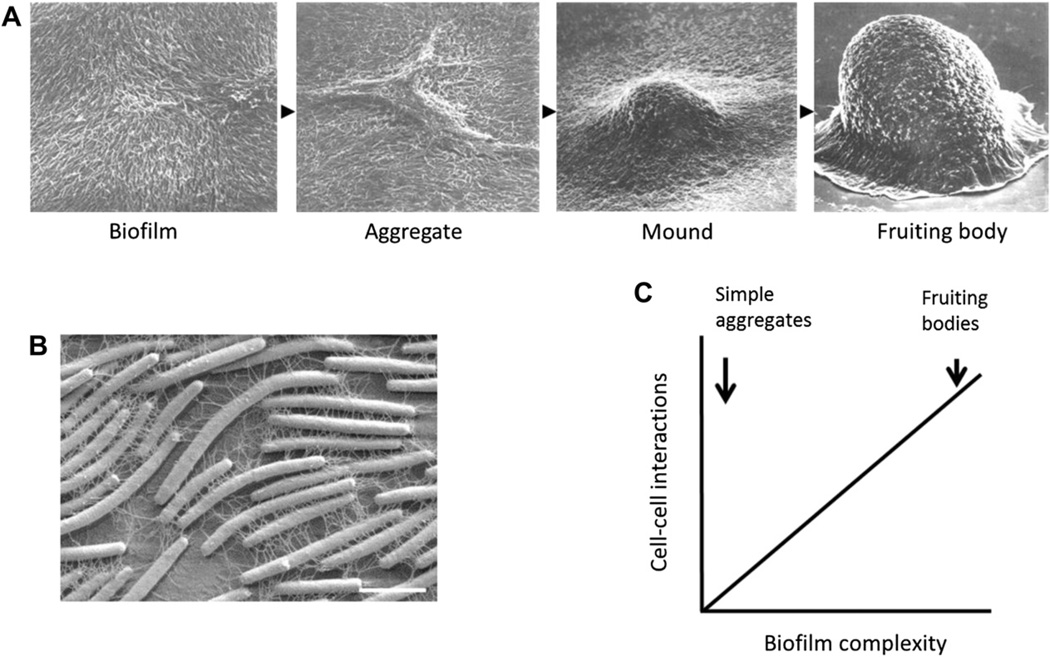
Fig. 1.
Cellecell interactions within biofilms. A) SEMs that reconstruct temporal steps in M. xanthus fruiting body formation. Left-most panel shows a biofilm that has formed on a plastic surface when the bacteria were grown in a rich medium. Upon medium replacement with starvation buffer, development ensued. These developmental events occurred over 3 days. Fig. was adapted with permission from Kuner and Kaiser (1982). B) SEM of a M. xanthus biofilm showing the fibrils that connect individual cells. Fig. was adapted with permission from Kearns and Shimkets (2001). Scale bar, 2 µm. C) Graph depicts an idealized sliding scale of cell–cell interactions between bacteria in biofilms. At one extreme, cells only aggregate into a biofilm; there is no cell–cell signaling. At the other extreme are complex cell–cell interactions exemplified by myxobacteria and discussed in the text.
Fig. 1A shows a series of scanning electron micrographs (SEMs) that reconstruct the temporal steps in fruiting body development. M. xanthus cells do not swim; instead these terrestrial microbes attach to surfaces and move by gliding (Fig. 1A and B). When these cells are grown in a static (not shaking) cell culture they form a biofilm on a plastic surface. Upon replacement of nutrient-rich media with starvation buffer, the cells aggregate and then erect a mound that develops into a fruit (Fig. 1A). From these micrographs it is apparent that cell behaviors are synchronized and coordinated to allow successful completion of their predetermined developmental program.
M. xanthus has evolved intercellular signaling mechanisms to govern their multicellular behaviors and sporulation decision. Cellecell signaling helps ensure that the population makes an informed decision before it engages in the energetically costly process of development. By using diffusible or quorum signals, individual cells can ‘vote’ to create an integrated group decision about how best to proceed (Kaiser, 1996). The ability of cells to produce erect multicellular fruits facilitates their dispersion by wind, water and/or animals to richer feeding grounds. Dispersion of a fruit also ensures that their social network remains intact upon germination.
Central to their multicellular behaviors is gliding motility, which powers and organizes cells on solid surfaces. M. xanthus has two separate motility systems or motors that drive gliding. They are called A (adventurous) and S (social) motility. Fibrils and type IV pili are the extracellular structures that are associated with S-motility and physically link cells together. Fig. 1B is a SEM of a M. xanthus swarm that reveals an extensive fibril network that connects cells and creates an extracellular fabric. Fibrils have been purified, and their molecular composition is roughly half protein and half polysaccharide (Dworkin, 1999). Fibril polysaccharides are frequently called exopolysaccharides (EPS), and for convenience this term is adopted here. It should, however, be noted that M. xanthus produces other forms of EPS that are not associated with fibrils (Ducret et al., 2012; Muller et al., 2012). Fibrils play a key role in cell–cell adhesion (a.k.a. agglutination or clumping), recognition, motility and development (Dana and Shimkets, 1993; Li et al., 2003). Polarly localized type IV pili function as the motor that powers S-motility; their retraction from the ‘front end’ of the rod-shaped cell pulls the cell forward (Skerker and Berg, 2001; Wall and Kaiser, 1999). As a recognition anchor for retraction, pili bind to EPS deposited in the extracellular matrix (ECM) (Li et al., 2003).
A-motility is associated with polar extracellular ‘slime’ filaments, which are thought to be composed of an as-yet-to-be-characterized polysaccharide (Ducret et al., 2012; Wolgemuth et al., 2002; Yu and Kaiser, 2007). The A-motility motor appears to be powered by mobile cell surface adhesins. Thus, this mechanism of motility might be analogous to how tank or bulldozer tracks propel those vehicles (Nan and Zusman, 2011; Sun et al., 2011) It is also striking that the A-motility motor appears to be functionally analogous to Flavobacterium johnsoniae gliding motility (Jarrell and McBride, 2008), yet the proteins involved in these machines are not homologous, suggesting that the motors evolved independently.
Myxobacterial behaviors are usually conceptualized in the context of a biofilm, although the term biofilm was only recently adopted by the field (O’Toole et al., 2000). In a broader context, many microbial species form biofilms, and those cells interact by a variety of mechanisms. Of these many species, however, some exhibit only limited cellecell interactions, perhaps simply consisting of cells sticking together without cell–cell signaling. Fig. 1C illustrates the point that the complexity of cellular biofilm interactions is likely to vary over a wide range, and we suggest that myxobacteria represent an extreme group that has evolved numerous cell–cell interactions, for which the known mechanisms are described here.
The complexity of myxobacterial social interactions is mirrored by their genomic complexity; these species contain some of the largest known bacterial genomes. For example, the model species M. xanthus contains a moderately sized myxobacterial genome at 9.14 Mb (Goldman et al., 2006). With ~7454 predicted genes, M. xanthus has ~20% more genes than the model eukaryote Saccharomyces cerevisiae. Without the intracellular organelle complexity found in yeasts, much of the large genome of myxobacteria contributes to their multicellular life cycle. Although the function of most M. xanthus genes remains to be determined, bioinformatic analysis clearly reveals complex signaling and regulatory networks. For instance, the M. xanthus genome encodes 53 σ54 enhancer binding proteins, 38 extracytoplasmic function sigma factors, 97 serine threonine protein kinases and 272 two-component signal transduction proteins (Goldman et al., 2006). Many of these regulatory proteins are likely to function in pathways governing interactions with other cells and their environment, and future work needs to elucidate their roles. Myxobacterial genetic complexity also leads a geneticist to pause; numerous cellular processes are likely to involve redundant pathways, making the identification of phenotypes for particular genes especially daunting. Given this, genetic analysis has nevertheless served as a fruitful discovery tool.
The primary approach used to identify myxobacterial genes involved in cell–cell interactions is the isolation of mutants followed by extracellular complementation tests (Hagen et al., 1978; Hodgkin and Kaiser, 1977). In particular, determining whether mutants that are defective in development or motility are rescued upon mixing with wild-type cells or mutants that belong to different complementation groups has been useful. The principal idea behind this approach is that a signalingdefective mutant should be rescued when co-cultured with a strain that produces the missing signal. The results of such efforts are discussed below.
2. Developmental signaling
A paper published 50 years ago provided the first evidence for cell–cell signaling in M. xanthus development (McVittie et al., 1962). This paper showed that certain fruiting body mutants were rescued when mixed with another mutant class. A more comprehensive study was subsequently conducted (Hagen et al., 1978). Based on strain-mixing experiments from this latter study, mutants were placed into four extracellular complementation groups designated as A, B, C and D. A fifth group (E) was discovered independently (Downard et al., 1993). Thus, when an A mutant and a C mutant are mixed, they provide the missing functions (signals) to allow the other strain to develop. These findings led to the idea that M. xanthus produces five developmental signals. To date, A- and C-signals are the best understood (Kaiser, 2004; Konovalova et al., 2012b; Shimkets, 1999).
2.1. A-signal
Upon starvation, M. xanthus initiates their developmental program (Fig. 1A). An A-signal is proposed to serve as a nutritional and quorum sensor, which monitors population density to ensure that a minimum number of cells are present and that those cells are starving (Kuspa et al., 1992b; Singer and Kaiser, 1995). Once a threshold concentration (10–50 µM) of A-signaling is reached, the starving population triggers the expression of A-signal-dependent genes, and the developmental program proceeds. Based on gene expression profiling, an A-signal is required for the expression of genes 2 h or later after starvation (Kroos and Kaiser, 1987; Kroos et al., 1986). To date, five genes involved in A-factor production are known. asgA and asgD encode hybrid histidine protein kinases (Cho and Zusman, 1999; Plamann et al., 1995); asgB encodes a putative DNA-binding protein (Plamann et al., 1994); asgC encodes the major sigma factor, RpoD (Davis et al., 1995); and asgE has sequence homology with amidohydrolases and contains putative membrane-spanning helices (Garza et al., 2000). Mutations in any of these asg genes decrease A-factor production, suggesting that these proteins function in the same or related pathways.
All asg mutants except asgD block the expression of the early development reporter Ω4521, which was identified by a Tn5lac insertion (Kroos et al., 1986). This observation was exploited to develop an Ω4521 bioassay to biochemically identify an A-factor that restored reporter gene expression to an asgB mutant (Kuspa et al., 1992b). To do this, conditioned medium was prepared from starved wild-type cells and was fractionated. Active material was found to contain heat-stable components, primarily consisting of six amino acids (Tyr, Pro, Phe, Trp, Leu and Ile) and peptide fragments (Fig. 2), and heat-labile activity identified as proteases, which presumably produce the A-factor amino acids (Kuspa et al., 1992a). The addition of A-factor components was able to rescue developmental gene expression, aggregation and sporulation to various degrees in asg mutants (Kuspa et al., 1992b). These results are interesting in the context of M. xanthus physiology, as their primary carbon/energy source is proteins/amino acids, not carbohydrates, and thus the A-signal seems to link nutrition with developmental signaling.
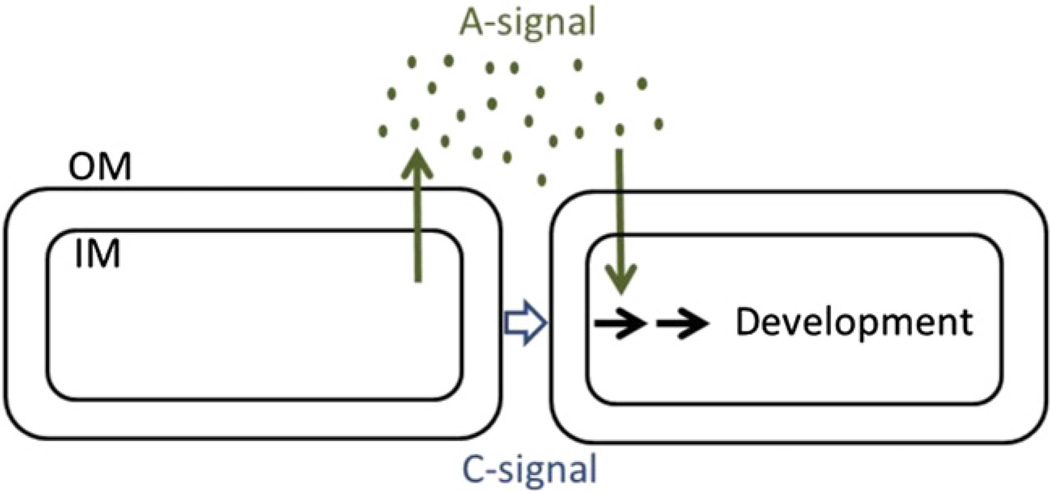
Fig. 2.
Signaling pathways during M. xanthus development. Cells secrete an A-signal into the extracellular milieu that is taken up by responder cells and by themselves early (~2 h) during development. The C-signal requires cellecell contact and triggers developmental gene expression after 6 h of starvation. Cells produce and respond to developmental signals in a similar manner (i.e., bidirectionally); however, for simplicity the signal transduction pathways are depicted unidirectionally.
No receptor for A-factor has been identified. However, to help address how the A-signal is perceived and processed, suppressors of asgB and asgA mutants were isolated and designated sasB and sasA, respectively. The sasB locus encodes three putative regulatory proteins named sasS, sasR and sasN (Kaplan et al., 1991). The sasB mutation restores expression of Ω4521, but not fruiting body formation or sporulation, in asgA or asgB mutants (Yang and Kaplan, 1997). SasS is a histidine kinase required for A-signal reception, and SasR is a NtrC-like activator that, together with SasS, positively regulates Ω4521 expression (Kaplan, 2003). SasN, which contains five of the independent sasB mutations, has no sequence similarity with other proteins and acts as a negative regulator, because sasN null mutations increase Ω4521 expression (Xu et al., 1998). This led to the proposal that SasN monitors intracellular A-signal levels, and, when these amino acid levels are low, SasN becomes inactive, which in turn activates SasSR to induce Ω4521 expression. A second locus, designated sasA, resides in a gene cluster involved in lipopolysaccharide (LPS) biosynthesis (Kaplan, 2003; Kaplan et al., 1991). The sasA suppressors bypass the A-signal requirement for Ω4521 expression; however, these mutants still cannot fruit or sporulate. The isolation of sasA suppressors shows a link between the OM and developmental gene expression. Additionally, a null mutation in rodK, which encodes a hybrid histidine protein kinase, increases A-signal production (Rasmussen et al., 2005). The rodK mutant also forms abnormal fruiting bodies and uncouples sporulation from fruit formation, indicating that RodK regulates temporal and spatial processes during development. Strikingly, a rodK null mutation suppresses fruiting body and sporulation defects of a csgA mutant, suggesting that RodK may link A- and C-signaling.
2.2. C-signal
csgA mutants are unable to ripple, aggregate or sporulate and exhibit abnormal developmental gene expression after 6 h of starvation (Kroos and Kaiser, 1987; Shimkets et al., 1983). csgA developmental defects are restored by extracellular complementation with wild-type cells or asg, bsg, dsg or esg mutants (Downard et al., 1993; Hagen et al., 1978) or, importantly, by the addition of purified CsgA, either as a recombinant fusion protein or as endogenously made protein (Kim and Kaiser, 1990a; Lee et al., 1995). These and other data indicate that C-factor acts as a short-range morphogen (Fig. 2) (Kaiser, 2004; Konovalova et al., 2010; Shimkets, 1999). Several lines of evidence suggest that the C-signal works in a dose-dependent manner to induce rippling, aggregation, developmental gene expression and sporulation. In vivo manipulation of csgA expression found that low C-signaling levels induce rippling, whereas intermediate levels induce aggregation and high levels are required for sporulation (Li et al., 1992). Similarly, the addition of an intermediate amount of purified C-factor to starving csgA cells rescues early C-signal-dependent genes, and the addition of a higher concentration induces late C-signal-dependent gene expression and sporulation (Kim and Kaiser, 1991). Finally, CsgA overexpression prematurely induces aggregation, sporulation and skips the rippling stage (Kruse et al., 2001).
Early observations showed a link between motility and C-signaling. First, non-motile and csgA mutants exhibit a similar temporal block in developmental gene expression and fruiting (Kroos et al., 1988). Second, C-signal transmission requires cell motility in both donor and recipient cells (Kim and Kaiser, 1990c). Strikingly, the requirement for cell motility is bypassed by artificial cell alignment in microscopic grooves that enrich end-to-end cell contacts or by the addition of C-signal at a high concentration (Kim and Kaiser, 1990b). These results imply that cell motility facilitates C-signaling by allowing proper cell–cell contacts. Details of the downstream C-signaling pathway have been reviewed (Kroos, 2007).
Full-length csgA encodes a 25-kDa protein that shows homology to small-chain alcohol dehydrogenase (SCAD) (Lee et al., 1995). SCAD contains two conserved sequence motifs, an N-terminal NAD(P)+ coenzyme binding pocket and a C-terminal catalytic domain, both of which are encoded by csgA. Consistent with this, affinity-purified MalE-CsgA binds to NAD+, and CsgA amino acid substitutions in either the putative NAD+ binding pocket (T6A or R10A) or catalytic site (S135T or T155R) did not rescue the aggregation and sporulation defects in csgA mutant cells in vivo or by exogenous addition of purified mutant proteins (Lee et al., 1995). To help elucidate the C-signaling pathway – and similar to what was discovered with rodK – an early study isolated csgA bypass suppressors (Rhie and Shimkets, 1989). One suppressor proved to be informative: SocA overexpression, a CsgA and SCAD homolog, bypassed the developmental requirement for csgA (Lee and Shimkets, 1994). More recently, lysophosphatidylethanolamine was identified as a lipid substrate for SocA, thus demonstrating enzymatic activity (Avadhani et al., 2006). Taken together, these data suggest that CsgA functions as an enzyme to produce the C-signal. Such a signaling mechanism might resemble sterol signaling in eukaryotes (Edwards and Ericsson, 1999).
An alternative model proposes that a processed form of CsgA serves as a cell surface morphogen (Kim and Kaiser, 1990a; Lobedanz and Sogaard-Andersen, 2003). C-factor activity, as assessed with a bioassay, can be purified by detergent extraction from starved cells and is found to be associated with a 17-kDa protein derived from CsgA (Kim and Kaiser, 1990a,d). Immunoblot analysis during development detects CsgA in two forms, full-length protein (25 kDa) and a minor C-terminal processed 17-kDa form (Lobedanz and Sogaard-Andersen, 2003). Importantly, a purified engineered MalE recombinant fusion containing C-terminal CsgA (p17), which lacks the NAD+ binding pocket, when added to a csgA mutant, elicits similar C-factor-specific activity as does full-length p25 (Lobedanz and Sogaard-Andersen, 2003). Thus, surprisingly, and contrary to an earlier observation (Lee et al., 1995), this finding argues that the CsgA NAD+ binding pocket is dispensable. Recently, PopC was identified as the protease that cleaves p25 to produce p17 (Rolbetzki et al., 2008). In vegetative cells, PopC accumulates in the cytoplasm and forms a complex with PopD that prevents its secretion (Konovalova et al., 2012a). Secretion of PopC is activated in response to starvation by a RelA-dependent pathway that activates FtsHD, which in turn results in PopD degradation. Removal of PopD then allows PopC to be slowly secreted by an unknown pathway that results in the gradual build-up of p17.
To date, both C-signaling models are incomplete. Central to both models is the elucidation of how C-factor is detected by cells, which requires the identification of its receptor. According to the models, the receptor is expected to be either a cell surface protein that binds p17 or a protein that binds the CsgA enzymatic product. For the enzyme model, it is also critical to identify the CsgA substrate and reaction product, which presumably is a lipid. For the p17 cell surface ligand model, there are questions about how CsgA is transported and localized to the cell surface, as it does not encode signaling or sorting sequences. The proposed subcellular localization of CsgA also varies. An initial study by immunogold electron microscopy found CsgA in the ECM, but it was subsequently shown that fibrils are not required for C-signal transmission (Li and Shimkets, 1993; Shimkets and Rafiee, 1990). More recently, two different cell fractionation methods yielded different results, with CsgA fractionating to either the OM or inner membrane (IM) (Lobedanz and Sogaard-Andersen, 2003; Simunovic et al., 2003). Part of the technical challenge in determining protein localization is that, during development, M. xanthus cells become fragile and lyse. The use of modern fluorescent fusion reporters or immunofluorescent methods may help to clarify CsgA subcellular localization.
2.3. B-, D- and E-signals
The roles that the bsg, dsg and esg gene products play in development are less clear and, consequently, they are considered to be inferred cellecell signals (Kaiser, 2004). bsgA and dsg mutants are defective in aggregation and sporulation and exhibit early developmental arrest around 2 and 5 h after starvation, respectively (Kroos and Kaiser, 1987; Cheng and Kaiser, 1989). As defined by this mutant class, the developmental defects of bsgA and dsg mutants can be complemented by strain mixing, albeit poorly (Hagen et al., 1978). The bsgA locus encodes an ATP-dependent LonD-protease (Gill et al., 1993) and dsg encodes the translation initiation factor IF3 (Cheng et al., 1994). The mechanism behind bsgA and dsg extracellular complementation remains to be discovered.
Morphologically, an esg mutant exhibits early developmental arrest and is rescued when co-developed with esg+ cells (Downard et al., 1993). The esg locus encodes E1α decarbox-ylase of the branched-chain keto acid dehydrogenase (BCKDH) (Toal et al., 1995). This enzyme contains two subunits (E1α and E1β) and catalyzes the conversion of branched-chain amino acids into coenzyme A derivatives, which serve as precursors for the synthesis of long branched-chain fatty acids and polyketides (Mahmud et al., 2002). Although esg mutants completely lack BCKDH activity, they still produce about half the wild-type level of iso-branched fatty acids from an alternative pathway (Bode et al., 2006). Thus, apparently one or more branched-chain fatty acids are required for development (Downard and Toal, 1995). Importantly, the esg fruiting body and sporulation defect are rescued by addition of certain short branched-chain fatty acids to the medium (Ring et al., 2006; Toal et al., 1995). These and other findings suggest that lipids are exchanged between the cells. To date, it is not clear whether esg extracellular complementation is the result of intercellular signaling (lipid derivative) or by metabolic cross-feeding (Stacy et al., 2012), as fatty acids may serve as important metabolites to power development.
3. Gliding motility structures and cell–cell interactions
Early work recognized that cell–cell interactions are important in gliding motility, as swarm expansion rates increase at higher cell densities (Kaiser and Crosby, 1983). In addition, S-motility itself requires cell–cell contact for cell movement (Rodriguez and Spormann, 1999). These effects are explained in part by the extracellular fibrils that form a mesh, holding cells together and provides essential ligands for type IV pili (Fig. 1B) (Li et al., 2003). The pilus motor consists of a PilA homopolymer, where PilA is initially synthesized as prepilin, which is processed by the PilD peptidase to form mature pilin. The assembly of pili requires the function of the PilB, PilC, PilM, PilN, PilO, PilP and PilQ proteins (Wall and Kaiser, 1999). PilQ forms an OM channel (secretin) through which pilus filaments protrude, and the Tgl lipoprotein is required for PilQ assembly (Nudleman et al., 2005, 2006; Wall and Kaiser, 1999). Assembled pili can extend 10 µm or farther from the cell surface. Recently, PilA was shown to bind fibril EPS directly on neighboring cells or in the ECM (Hu et al., 2012). Mutants that are defective in fibril EPS production not only are defective in S-motility but also are hyperpiliated, suggesting that EPS controls the abundance of pili by presumably triggering retraction (Li et al., 2003; Wu et al., 1997). Pilus retraction is powered by the ATPase activity of PilT, which disassembles pilin subunits into the IM (Fig. 3) (Jakovljevic et al., 2008; Skerker and Berg, 2001; Wall and Kaiser, 1999). Pili also connect adjacent cells and help coordinate social behaviors.
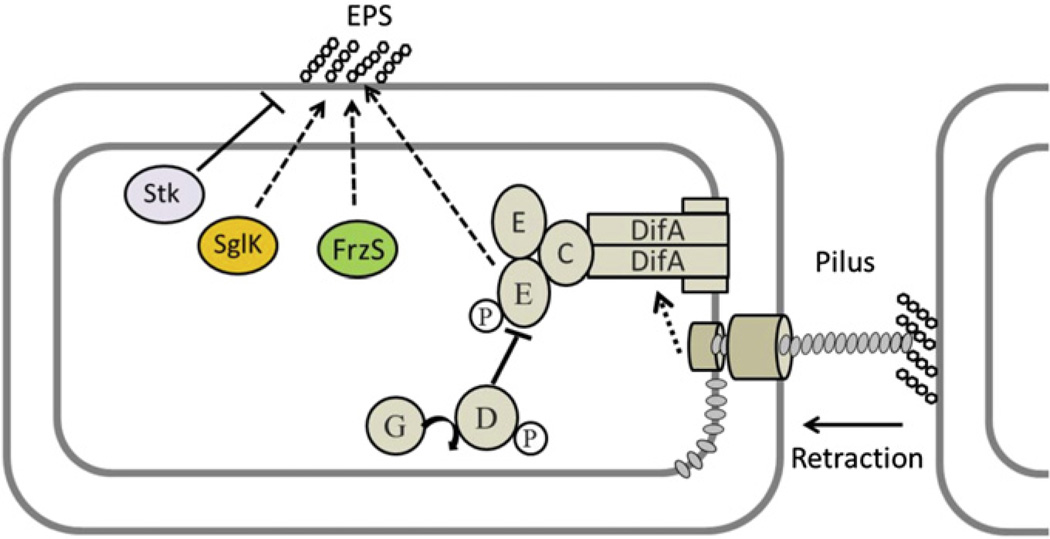
Fig. 3.
Type IV pili and fibril EPS interactions. A pilus binds EPS on a neighboring cell, which triggers its retraction, pulling the cell forward. The pilus is also proposed to function as a sensor to activate the Dif signal transduction pathway, which results in EPS production. Other proteins known to influence EPS production are also shown and described in the text. Arrows with dashed lines indicate positive regulation, and bars indicate negative regulation. Circles with P represent a phosphate group; gray shaded ovals represent the pilin subunits and Dif proteins are labeled with letters. For details, see text and Black et al. (2006).
In an apparent circular regulatory network, the binding of pili to EPS also stimulates EPS production, as pili-deficient mutants produce less EPS. The Dif proteins mediate pili-dependent EPS biosynthesis and were originally identified as mutants that are defective in fruiting-body formation and S-motility (Fig. 3) (Black et al., 2006; Yang et al., 1998). The dif genes are homologous to the well-characterized chemotaxis genes of Escherichia coli. Subsequently, the dif genes were found to be allelic to the dsp (dispersed growth) genes required for fibril production (Lancero et al., 2002; Shimkets, 1986). Similar to rescue of certain developmental mutants described above, developmental defects in fruit formation, gene expression and sporulation of dsp/dif mutants can also be rescued by the addition of purified fibrils or by co-culture with dsp+ cells (Chang and Dworkin, 1994; Li and Shimkets, 1993). For this reason, dsp/dif genes are sometimes described as the sixth developmental complementation group (Shimkets, 1999).
A regulatory pathway for pilus retraction and EPS biosynthesis is summarized in Fig. 3. Binding of pili to fibril EPS on neighboring cells triggers retraction. Pili are also proposed to act as a sensor to transduce a ‘signal’ to DifA, a membrane receptor homologous to methyl-accepting chemotaxis proteins (MCPs). A signal is then transduced through the Dif pathway to activate EPS production (Black et al., 2010; Campodonico and Zusman, 2010). Unpolymerized PilA subunits localized in the IM constitute part of the regulatory signal (Yang et al., 2010). EPS production is further regulated by two DnaK paralogs, SglK and Stk, and by FrzS (Fig. 3) (Berleman et al., 2011; Dana and Shimkets, 1993; Konovalova et al., 2010; Weimer et al., 1998). The EPS biosynthetic gene clusters, eps and eas, have been identified (Lu et al., 2005; Youderian and Hartzell, 2006) and await future studies including an analysis of their tran-scriptional control.
LPS plays an important role in S-motility and development. Although LPS constituents lipid A and core oligosaccharide are likely to be essential for growth, O-antigen mutants were isolated and exhibit S-motility and developmental defects (Bowden and Kaplan, 1998; Youderian and Hartzell, 2006). Because pili, EPS, A-motility and cell adhesion appear normal, O-antigen mutants do not cause general pleiotropic defects. As mentioned above, certain LPS (O-antigen) mutants also suppress A-signaling defects (Bowden and Kaplan, 1998).
Interestingly, Zusman and colleagues recently uncovered a new cell-contact-dependent interaction that reorganizes intracellular localization of the FrzCD protein (Mauriello et al., 2009). The Frz chemosensory pathway controls the gliding motility reversal frequency (Zusman et al., 2007). Specifically, these investigators fused a fluorescent protein to FrzCD, which is a MCP homolog but lacks the transmembrane/periplasmic domains typically associated with MCPs. FrzCD–GFP forms random and dynamic clusters in the cell. However, when motility leads to side-by-side cell contacts, the FrzCD–GFP clusters become aligned between cells. The transient FrzCD–GFP alignments are typically followed by one of the cells reversing its direction of movement. The alignment of FrzCD clusters indicates that cells sense and respond to contact with one another. Although the mechanism remains unknown, those authors and others have speculated that FrzCD cluster alignments help coordinate population movements by regulating or synchronizing a postulated Frz biochemical clock that governs reversal frequencies (Igoshin et al., 2004; Mauriello et al., 2009).
4. Outer membrane exchange as a new cell-interaction platform
Recently, we described a process whereby myxobacteria in a structured biofilm fuse their OMs and exchange contents (Pathak et al., 2012; Wei et al., 2011). To date this process is unique to myxobacteria, although other bacteria may conduct a similar behavior that simply is awaiting discovery. The transfer of DNA, protein, small molecules and other cellular components occurs in other bacterial systems, although those processes are mechanistically different than the described myxobacterial system (Konovalova and Sogaard-Andersen, 2011).
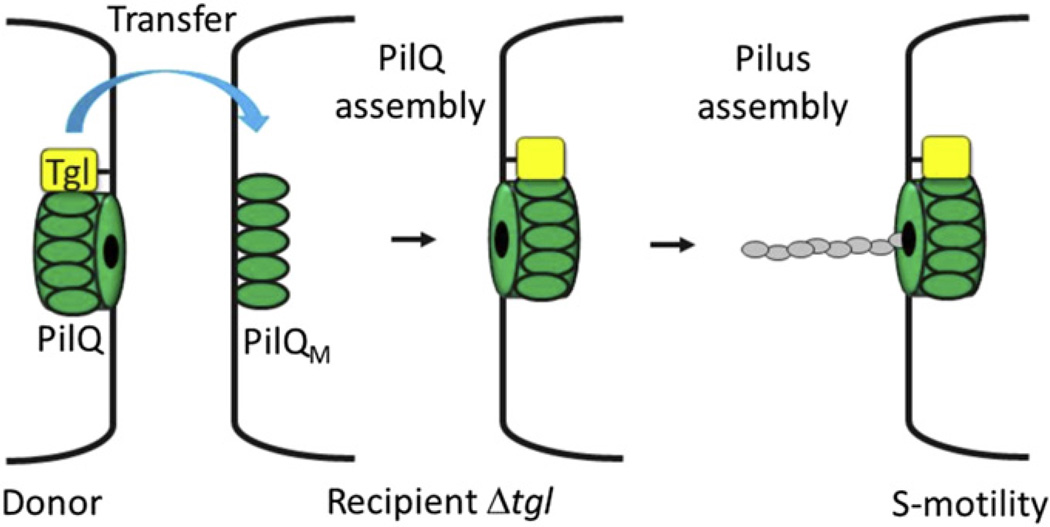
The path to OM exchange discovery again originated from extracellular complementation (stimulation) studies. Early work found that a small subset of motility mutants is transiently rescued when mixed with different strains (Hodgkin and Kaiser, 1977). Importantly, stimulation requires that the ‘donor’ strain contain the wild-type gene that is mutated in the ‘recipient.’ Stimulation is transient, meaning that the mutants again become non-motile over time, thus indicating that DNA is not transferred. The mechanism of stimulation is by the transfer of missing protein function from donor to recipient cells (Nudleman et al., 2005). Fig. 4 shows an overview of how a particular motility mutant is stimulated. Here a Δtgl mutant lacks S-motility because the Tgl lipoprotein is required for functional assembly of the PilQ secretin. As illustrated, when a Dtgl mutant makes physical contact with a Tgl+ donor cell, the Tgl protein is efficiently transferred (Nudleman et al., 2005). Upon receipt of Tgl, the mutant assembles stable and functional PilQ secretin, thus allowing pili assembly. In the S-motility system tgl is the only mutant known to be rescued by stimulation.
Fig. 4.
Model for Tgl transfer and stimulation of S-motility. Juxtaposed cell poles of a Tgl+ donor (left) and a Δtgl recipient are shown. A tgl mutant expresses PilQ monomer (PilQM), but these monomers fail to assemble into a stable multimeric OM channel. Following physical contact, the Tgl protein is transferred from donor to recipient, where Tgl facilitates assembly of the PilQ secretin (Nudleman et al., 2005). Once PilQ is assembled, type IV pili are made at the cell pole to power S-motility.
In A-motility there are five complementation groups corresponding to five genes called cglB/C/D/E/F (Hodgkin and Kaiser, 1977; Pathak and Wall, 2012; Rodriguez and Spormann, 1999). All of the Cgl proteins encode type I or II signal sequences, suggesting envelope localization, and some of these proteins are associated with an A-motility macro-molecular complex (Luciano et al., 2011; Nan et al., 2010). The mechanism of stimulation for each cgl mutant has either been demonstrated or is inferred to involve cell-to-cell protein transfer (Nudleman et al., 2005; Pathak and Wall, 2012).
4.1. Requirements for transfer
Protein transfer requires that cells be in physical contact on a hard surface (Wall and Kaiser, 1998; Wei et al., 2011). Transfer does not occur in liquid or even on a soft agar surface, even when cells are densely packed. These and other findings argue that the Tgl/Cgl proteins are not transferred by secretion/diffusion into the extracellular milieu with subsequent recipient cell uptake. Furthermore, cell motility has a profound indirect effect on protein transfer (Wall and Kaiser, 1998; Wei et al., 2011). In other words, only the donor or recipient or even a third-party cell, not involved in exchange, needs to be motile to facilitate transfer. Motility derived from either A or S systems works equally well; there is no requirement for a particular system. The role of gliding motility in this process is not simply to create local areas of high cell densities, as concentrating non-motile cells by centrifugation does not alleviate the motility requirement (X. Wei and D. Wall; unpublished data). From these studies, the role of motility is interpreted to facilitate proper cell–cell contacts or alignments that lead to exchange (Wall and Kaiser, 1998; Wei et al., 2011). It is also striking to recall that the transmission of C-signal requires motility (Kim and Kaiser, 1990c; Kroos et al., 1988), although a connection between these processes is not yet established.
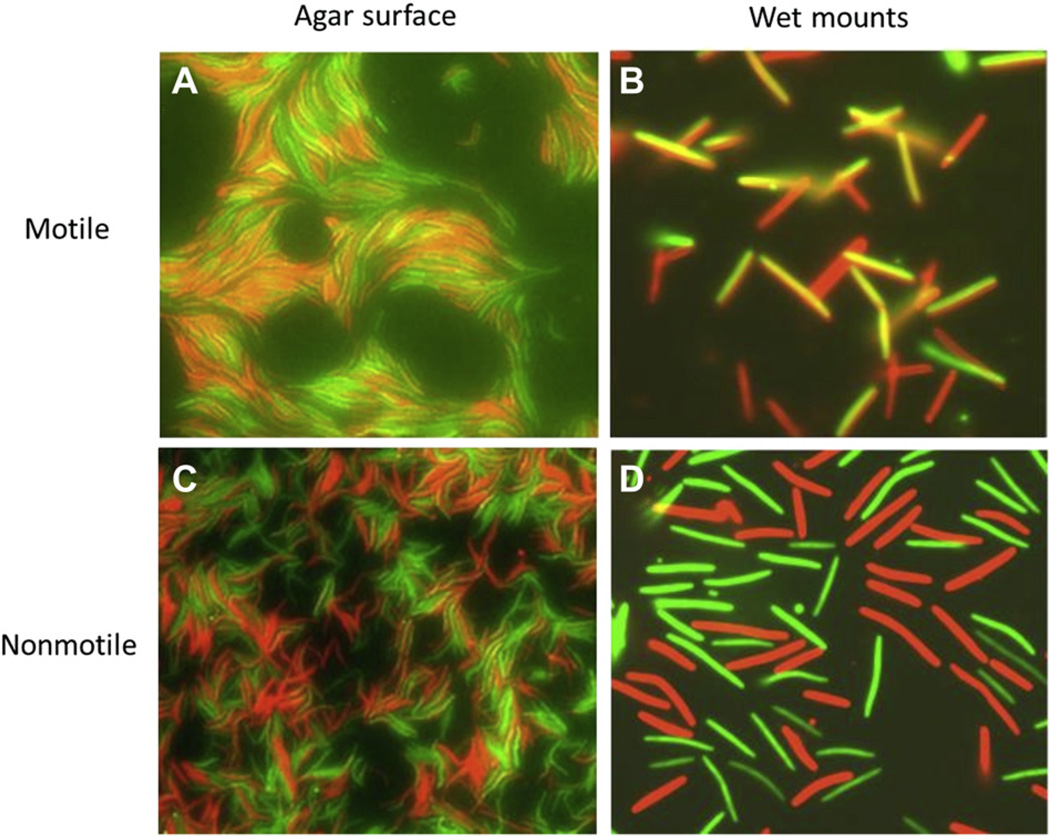
The effect that motility has on the anatomy of a biofilm is readily visualized by microscopic examination (Fig. 5). In these experiments, non-motile cells were fluorescently labeled (red, donors; green, recipients) and placed on hard agar pads, where they were randomly positioned, analogous to how wood matches are oriented after being dropped on the floor (Fig. 5C). In contrast, when a third-party motile, but non-fluorescent, strain was mixed with the same non-motile cells, the non-motile fluorescent cells became highly aligned and were enriched in end-to-end and side-by-side contacts (Fig. 5A). Thus, motility creates a structured biofilm that facilitates reporter transfer (compare Fig. 5B to D), which we hypothesize is required for key cell–cell interactions between juxtaposed cells (Wei et al., 2011). The friction caused by cells ‘bumping’ into each other might also help engage cell surface receptors and lead to OM stress for fusion.
Fig. 5.
Cell motility aligns cells for OM exchange. A) Merged fluorescent micrograph on a hard agar surface of non-motile fluorescently-labeled donor (red, SSOM-mCherry) and recipient (green, GFP) cells mixed with a third-party strain that is motile and non-fluorescent (1:1:1 ratio). B) Same cells as in A, except cells were harvested and placed on a glass slide for clear analysis of protein transfer; green recipients become yellow/orange in merged micrograph. C) Same non-motile labeled donor/recipients cells as in A, except without the motile strain. Cells are not aligned. D) Same cells as in C, except harvested cells are placed on a glass slide, showing that the non-motile cells do not transfer, as red and green cells remain distinct in color in the merged micrograph. For details, see text and Wei et al. (2011).
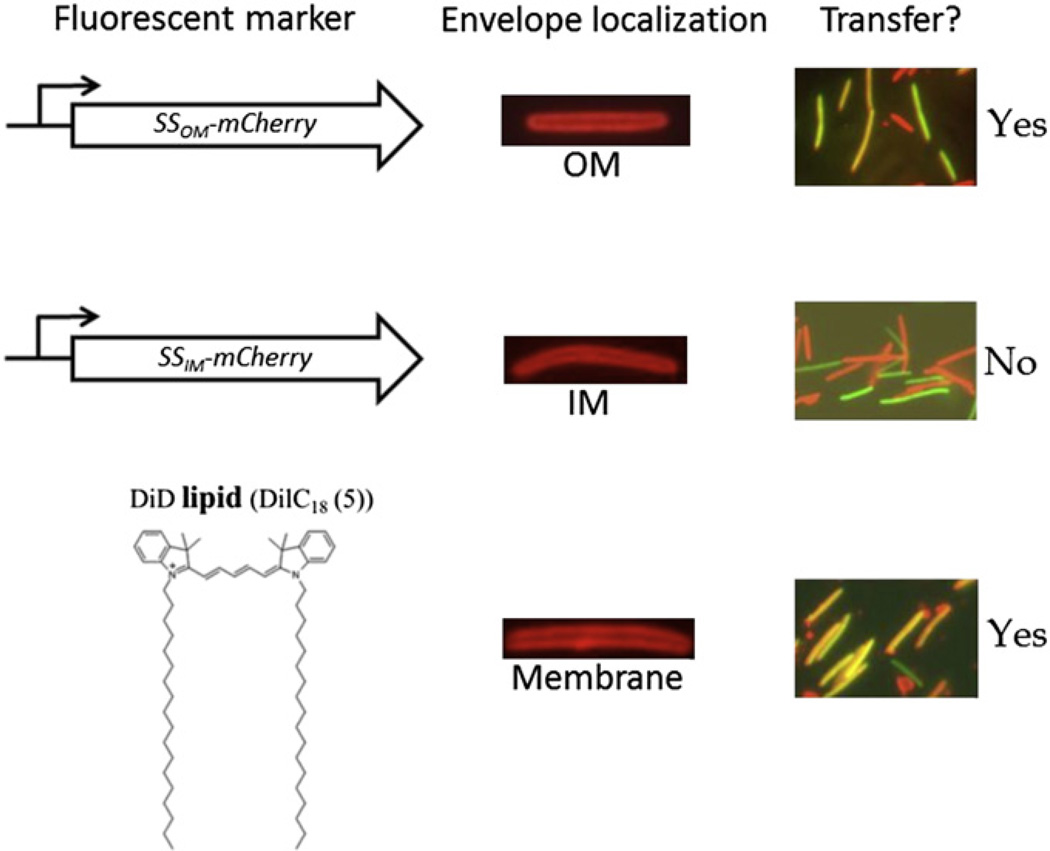
Several fluorescent probes were used to determine what cellular material is actually transferred. Because Tgl and the majority of the Cgl proteins contain type II signal sequences for lipoprotein biogenesis (Pathak and Wall, 2012), we tested whether a type II signal sequence fused to a heterologous protein reporter would mediate transfer. As shown in Fig. 6, fusion of a type II signal sequence to mCherry indeed allowed for transfer when it coded for OM localization (SSOM-mCherry), but not when the type II signal sequence coded for IM localization (SSIM-mCherry; Fig. 6) (Wei et al., 2011). As cytoplasmic GFP does not transfer, this reporter was used to identify recipients in a live cell transfer assay by testing whether green cells receive mCherry from donors and thus green cells gain red fluorescence (Wei et al., 2011). This assay is particularly useful and now serves as our primary method to detect transfer (compare Fig. 5B and D). Importantly, these results show that a type II signal sequence coding for OM localization is necessary and sufficient for heterologous protein transfer among motile M. xanthus cells placed on a hard surface. These findings also imply that many OM lipoproteins are shuttled between M. xanthus cells, as their genome contain an expansive lipoprotein processing pathway and encodes >400 predicted lipoproteins (Goldman et al., 2006; Xiao et al., 2012). Recently, the cglE and cglF genes were identified and found to encode type I signal sequences, which target proteins for transport across the cytoplasmic membrane without lipid modification (Pathak and Wall, 2012). This finding suggests that non-lipoproteins are also transferred, although CglE and CglF may be associated with the OM. Thus the reservoir of cargo proteins is apparently large and may not be restricted to OM lipoproteins.
Fig. 6.
Fluorescent markers used to monitor cell envelope exchange. The ability of markers to label specific cell envelope compartments and to be transferred is shown (Pathak et al., 2012; Wei et al., 2011). DiD is a lipid fluorescent dye, which based on our experience, becomes irreversible imbedded in the M. xanthus OM.
Because OM protein transfer occurs efficiently and involves a form of bulk transfer, we investigated whether OMs fuse by testing for lipid transfer. This hypothesis was corroborated by the finding that lipophilic fluorescent dyes are indeed exchanged (Fig. 6) (Pathak et al., 2012). Importantly, the transfer of lipophilic dyes, and hence membrane lipids, has the same stringent requirements as does protein transfer. That is, dye transfer is concurrent with protein transfer and does not occur by diffusion into the extracellular milieu or by diffusible OM vesicles (Pathak et al., 2012; Wei et al., 2011). Instead, lipophilic dye transfer requires cell motility and a solid surface. In this context it is interesting to recall, as discussed above, that the E-signal likely involves fatty acid exchange. In addition, transmission of the C-signal, either as a derivative from a CsgA lipid substrate or as the p17 protein itself, might be mediated by OM exchange. Future experiments will need to test these possibilities.
4.2. Genetic determinants for transfer
We have recently identified mutants that are universally defective in cgl/tgl stimulation and protein transfer; the associated gene products are named TraA and TraB (Pathak et al., 2012). In a tra− mutant, the defect in protein transfer is absolute; under no condition is stimulation/transfer detected. Importantly, the TraAB proteins are also required for lipophilic dye transfer, thus cementing the assertion that protein and lipid transfer are coupled. Furthermore, TraAB functions are required in both donors and recipients. Thus, unlike other bacterial secretion systems, where the transfer apparatus is only required in donors, in this system the transfer machinery is required in both cells. Taken together, these findings imply that transfer occurs bidirectionally, leading to the exchange or sharing of OM resources between cells (Pathak et al., 2012).
The traA and traB genes form a bicistronic operon in which their open reading frames overlap by four bases (Pathak et al., 2012). Bioinformatic analysis revealed that TraA and TraB contain type I signal sequences, indicating cell envelope localization. The N-terminal region of TraA includes a distantly related PA14 domain, a domain broadly distributed across the tree of life and implicated in glycan binding (Rigden et al., 2004). However, the distant sequence relationship of the TraA PA14 domain to other family members hints that it is not necessarily a lectin. C-terminal to the distant PA14 domain of TraA is a cysteine-rich region, which includes nine repeat elements, each of which consists of 25 amino acids. Because traA encodes an unusually high number of cysteines (79 total) for a secreted protein, it is likely to contain an inordinate number of disulfide bonds. At the C terminus, TraA contains a MYXO-CTERM motif predicted to function as a protein-sorting tag for cell surface localization. The domain architecture of TraA thus suggests that it functions as a cell surface receptor/adhesin. This prediction is supported by the finding that TraAB overexpression dramatically increases cell—cell adhesion (Pathak et al., 2012). The domain architecture of TraA also shows striking similarity to the S. cerevisiae FLO1 and FLO5 cell surface adhesins involved in yeast flocculation and kin recognition (Rigden et al., 2004; Smukalla et al., 2008; Veelders et al., 2010). The C-terminal region of TraB contains an OmpA-like domain that may mediate peptidoglycan binding. The rest of TraB shows no homology to other database domains. Given that these genes are adjacent and have identical mutant phenotypes, we conclude that TraAB function in the same pathway.
4.3. Transfer mechanism
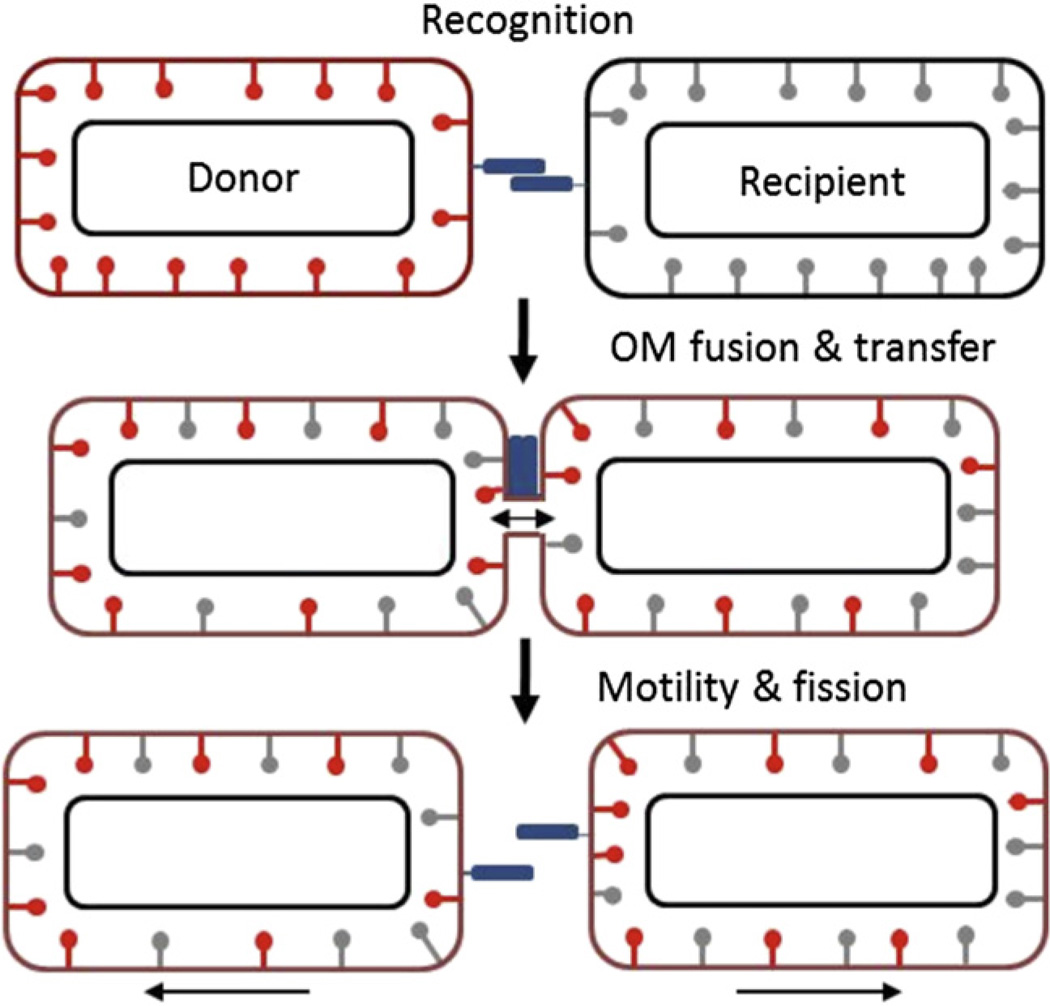
A cartoon depicting TraAB-dependent OM exchange is shown in Fig. 7. The model proposes that TraA serves as a cell surface receptor/adhesin (Pathak et al., 2012). As discussed above, TraA could bind to glycans on neighboring cells, for example LPS, EPS or itself. A key component of the model requires reciprocal TraA binding between cells. Furthermore, we have evidence that TraA functions as a homophilic receptor for self-recognition (D. Pathak, X. Wei, A. Dey and D. Wall, unpublished). The function of TraB is unknown and is not depicted, but it might bind TraA and/or facilitate its subcellular localization. Once cellecell binding occurs, the OMs are proposed to fuse, which then would lead to component exchange by membrane diffusion. In Borrelia, OM fusion has been suggested to occur between motile cells on a hard surface based on fluorescence and electron microscopy observations (Kudryashev et al., 2011). During fusion, the cell walls may act as barriers to prevent IM fusion and restrict the transfer of other envelope components. In Fig. 7, the left cell is labeled with red lollipops symbolizing SSOM-mCherry lipoproteins, whereas the right cell shows endogenous lipoproteins (gray lollipops). OM fusion thus triggers the exchange of lipoproteins. Although the mechanism for fusion is unknown, tight cellecell binding between cell poles, where membrane curvature is highest and leads to its destabilization, may help catalyze fusion (Martens and McMahon, 2008; Wickner and Schekman, 2008). Gliding motility on hard surfaces is likely to facilitate proper cell–cell contacts and perhaps contributes to membrane stress once cells are adhered to one another. Subsequently, gliding motility is likely to power fission between OMs.
Fig. 7.
A working model of OM exchange. The blue rectangles represent TraA, and the red and gray lollipops represent OM lipoproteins. See text for details.
To date, M. xanthus represents the clearest example of OM fusion during which protein exchange with phenotypic consequences has been demonstrated. Gram-negative bacteria, including M. xanthus, secrete diffusible OM vesicles that potentially can fuse with other cells, including eukaryotic cells (Kulp and Kuehn, 2010; Mashburn-Warren and Whiteley, 2006), yet the process described here is mechanistically distinct. Based on a bioinformatic analysis, TraAB orthologs are restricted to myxobacteria, although other bacterial groups may have functional analogs and may carry out similar behavior (Kudryashev et al., 2011).
4.4. Social and ecological perspectives
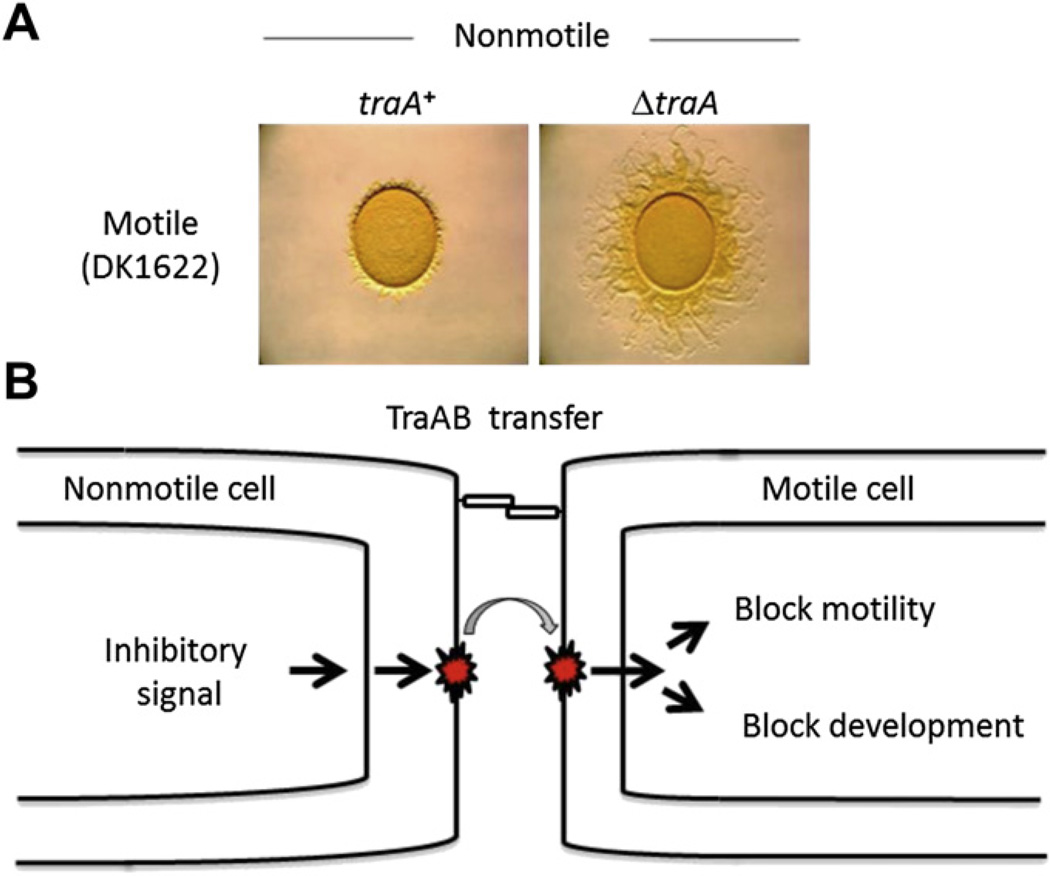
We postulate that myxobacteria use OM exchange to help regulate their complex social behaviors. Although TraAB are required for stimulation of certain motility mutants, these proteins are not required for A- or S-motility (Pathak et al., 2012). Insights into the physiological significance of OM exchange were found in strain-mixing experiments (Pathak et al., 2012). OM exchange is able to regulate swarming and development behaviors between strains. That is, the swarm expansion of a motile strain is (partially) blocked in a Tra-dependent manner when it is mixed with a non-motile strain (Fig. 8A). As was found for protein/lipid transfer, the ‘relief of swarm inhibition’ occurs when a tra mutation is introduced into either the non-motile or motile strains (Fig. 8A; traA mutation shown only in non-motile donor) (Pathak et al., 2012). These studies suggest that non-motile cells produce a signal that can be transferred via TraAB to motile cells that blocks their motility (Fig. 8B). In a similar set of experiments, a non-motile strain inhibits development of a motile strain, again by a Tra-dependent mechanism (Pathak et al., 2012). Taken together, these results support the idea that OM exchange serves as a cell–cell signaling mechanism. As opposed to diffusible signaling molecules, cell contact signaling allows unambiguous identification of neighbors. In a general context, myxobacteria use signaling by cell contact as a major tool to coordinate multicellular behaviors (Kaiser et al., 2010; Konovalova et al., 2010).
Fig. 8.
OM exchange regulates swarming and development. A) Motile and non-motile strains mixed at a 1:1 ratio and incubated for two days. Swarming is inhibited by a Tra-dependent mechanism. B) Non-motile cells are proposed to produce a signal(s) that is transferred to motile cells via TraAB that blocks swarming and development.
As noted, myxobacteria are rather unusual. Like many bacteria they live as unicellular entities and grow by binary fission. Their vegetative life is social – the cells swarm in tightly associated packs and at times single cells will venture away from the pack. Their developmental program touches on a fundamental biological question of how single cells recognize related or kin individuals to coalesce and cooperate to build a multicellular tissue. Central to this process is an appreciation that myxobacteria do not exist as a prototypic eukaryote, in which sibling cells are derived from clonal expansion of a fertilized egg. In some cases, populations are likely to be enriched toward homogeneity by clonal expansion, for example, by inoculation of a single myxobacteria cell and its subsequent growth on a rich nutrient source, e.g., a dung pellet. More often, however, myxobacteria exist in heterogeneous populations, consisting of assorted microbial species and various myxobacterial species and subspecies (Vos and Velicer, 2009). Moreover, Velicer and colleagues showed that fruiting bodies derived from soil contain genetically similar, though not necessarily identical siblings (Kraemer and Velicer, 2011). Their work suggests that kin recognition and selection may occur during development.
Another noted property of myxobacteria is that they are highly antagonistic toward other related species and subspecies (Smith and Dworkin, 1994; Vos and Velicer, 2009). Given the heterogeneous and antagonistic nature of myxobacteria, how they are able to recognize kin to elicit cooperative behaviors is an intriguing problem. A complete answer is not known, but it is plausible to speculate that OM exchange represents one layer for self-recognition. Indeed, our recent work has suggested that OM exchange does involve kin recognition (D. Pathak, X. Wei, A. Dey & D. Wall, unpublished). Future work will need to elucidate if OM exchange and kin recognition play a role in cell selection during multicellular development. Kin recognition is probably relevant in environmental niches as there can be a high degree of diversity; for example, 1 g of soil can contain >30,000 microbial species (Mendes et al., 2011). Other microbes, such as Dictyostelium, are confronted with similar problems of kin recognition during multicellular development, and they have indeed devised self-recognition mechanisms (Strassmann et al., 2011).
OM exchange may also represent a form of cooperative behavior whereby cells share resources. Kin selection ensures that ‘cheater’ (non-kin) cells do not exploit kin resources (Velicer et al., 2000). OM exchange would thus represent an extension of uncontrolled resource sharing whereby M. xanthus cells cooperatively secrete hydrolytic enzymes and antibiotics into the extracellular milieu to kill, digest and feed on prey as microbial ‘wolf-packs’ (Berleman and Kirby, 2009; Rosenberg et al., 1977; Xiao et al., 2011). Sharing resources also provides a mechanism to reduce physiological heterogeneity in the population, which inevitably leads to some cells being less fit and introduces physiological noise. Thus, we hypothesize that sharing their OM proteome allows the population to establish envelope homeostasis and allows individuals to gain fitness. Homeostasis in turn creates improved population fitness by normalizing intercellular signal output and reception, thus helping to coordinate cell behaviors.
Acknowledgments
This work was support by NSF grant MCB-848141 and NIH grant GM101449 to D.W.
Biographies
Darshankumar Pathak received his Bachelor’s as well as Master’s degrees in Microbiology from Sardar Patel University in India. He is currently pursuing a Ph.D. degree in the Molecular and Cellular Life Science program with Dr. Daniel Wall at the University of Wyoming. He uses molecular, genetic and microscopy methods to understand a novel mechanism whereby myxobacteria distinguish self from non-self to fuse and exchange outer membrane components.
Dr. Xueming Wei received his Ph.D. degree from The Ohio State University studying rhizobial physiology and ecology. He then conducted a postdoctoral fellowship at Oregon State University using molecular genetics and genomics to study nitrification in Nitrosomonas europaea. He then joined the laboratory of Dr. Daniel Wall at University of Wyoming to study social interactions in myxobacteria.
Dr. Daniel Wall received his Bachelor’s degree in Biology from Sonoma State University. He then earned his Ph.D. degree from the University of Utah with Dr. Costa Georgopoulos studying molecular chaperones and the heat-shock response in E. coli. His postdoctoral fellowship was conducted with Dr. Dale Kaiser at Stanford University investigating gliding motility and extracellular complementation in myxobacteria. After working on antibiotic drug discovery at two different San Diego, CA, biotechnology companies he joined the Molecular Biology Department at the University of Wyoming in 2007. His lab focuses on social interactions and antibiotics produced by myxobacteria.
Contributor Information
Darshankumar T. Pathak, Email: dpathak@uwyo.edu.
Xueming Wei, Email: xwei@uwyo.edu.
References
- Avadhani M, Geyer R, White DC, Shimkets LJ. Lysophosphati-dylethanolamine is a substrate for the short-chain alcohol dehydrogenase SocA from Myxococcus xanthus. J. Bacteriol. 2006;188:8543–8550. doi: 10.1128/JB.01047-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berleman JE, Kirby JR. Deciphering the hunting strategy of a bacterial wolfpack. FEMS Microbiol. Rev. 2009;33:942–957. doi: 10.1111/j.1574-6976.2009.00185.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berleman JE, Vicente JJ, Davis AE, Jiang SY, Seo YE, Zusman DR. FrzS regulates social motility in Myxococcus xanthus by controlling exopolysaccharide production. PLoS One. 2011;6:e23920. doi: 10.1371/journal.pone.0023920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black WP, Schubot FD, Li Z, Yang Z. Phosphorylation and dephosphorylation among Dif chemosensory proteins essential for exo-polysaccharide regulation in Myxococcus xanthus. J. Bacteriol. 2010;192:4267–4274. doi: 10.1128/JB.00403-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black WP, Xu Q, Yang Z. Type IV pili function upstream of the Dif chemotaxis pathway in Myxococcus xanthus EPS regulation. Mol. Microbiol. 2006;61:447–456. doi: 10.1111/j.1365-2958.2006.05230.x. [DOI] [PubMed] [Google Scholar]
- Bode HB, Ring MW, Kaiser D, David AC, Kroppenstedt RM, Schwar G. Straight-chain fatty acids are dispensable in the myx-obacterium Myxococcus xanthus for vegetative growth and fruiting body formation. J. Bacteriol. 2006;188:5632–5634. doi: 10.1128/JB.00438-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowden MG, Kaplan HB. The Myxococcus xanthus lipopolysac-charide O-antigen is required for social motility and multicellular development. Mol. Microbiol. 1998;30:275–284. doi: 10.1046/j.1365-2958.1998.01060.x. [DOI] [PubMed] [Google Scholar]
- Campodonico EM, Zusman DR. Developments in defining Dif. J. Bacteriol. 2010;192:4264–4266. doi: 10.1128/JB.00700-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang BY, Dworkin M. Isolated fibrils rescue cohesion and development in the Dsp mutant of Myxococcus xanthus. J. Bacteriol. 1994;176:7190–7196. doi: 10.1128/jb.176.23.7190-7196.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y, Kaiser D. dsg, a gene required for cellecell interaction early in Myxococcus development. J. Bacteriol. 1989;171:3719–3726. doi: 10.1128/jb.171.7.3719-3726.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng YL, Kalman LV, Kaiser D. The dsg gene of Myxococcus xanthus encodes a protein similar to translation initiation factor IF3. J. Bacteriol. 1994;176:1427–1433. doi: 10.1128/jb.176.5.1427-1433.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho K, Zusman DR. AsgD, a new two-component regulator required for A-signalling and nutrient sensing during early development of Myxococcus xanthus. Mol. Microbiol. 1999;34:268–281. doi: 10.1046/j.1365-2958.1999.01594.x. [DOI] [PubMed] [Google Scholar]
- Dana JR, Shimkets LJ. Regulation of cohesion-dependent cell interactions in Myxococcus xanthus. J. Bacteriol. 1993;175:3636–3647. doi: 10.1128/jb.175.11.3636-3647.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis JM, Mayor J, Plamann L. A missense mutation in rpoD results in an A-signalling defect in Myxococcus xanthus. Mol. Microbiol. 1995;18:943–952. doi: 10.1111/j.1365-2958.1995.18050943.x. [DOI] [PubMed] [Google Scholar]
- Downard J, Ramaswamy SV, Kil KS. Identification of esg, a genetic locus involved in cellecell signaling during Myxococcus xanthus development. J. Bacteriol. 1993;175:7762–7770. doi: 10.1128/jb.175.24.7762-7770.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downard J, Toal D. Branched-chain fatty acids: the case for a novel form of cellecell signalling during Myxococcus xanthus development. Mol. Microbiol. 1995;16:171–175. doi: 10.1111/j.1365-2958.1995.tb02290.x. [DOI] [PubMed] [Google Scholar]
- Ducret A, Valignat MP, Mouhamar F, Mignot T, Theodoly O. Wet-surface-enhanced ellipsometric contrast microscopy identifies slime as a major adhesion factor during bacterial surface motility. Proc. Natl. Acad. Sci. U.S.A. 2012;109:10036–10041. doi: 10.1073/pnas.1120979109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dworkin M. Fibrils as extracellular appendages of bacteria: their role in contact-mediated cellecell interactions in Myxococcus xanthus. Bio-essays. 1999;21:590–595. doi: 10.1002/(SICI)1521-1878(199907)21:7<590::AID-BIES7>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- Edwards PA, Ericsson J. Sterols and isoprenoids: signaling molecules derived from the cholesterol biosynthetic pathway. Annu. Rev. Biochem. 1999;68:157–185. doi: 10.1146/annurev.biochem.68.1.157. [DOI] [PubMed] [Google Scholar]
- Garza AG, Harris BZ, Pollack JS, Singer M. The asgE locus is required for cellecell signalling during Myxococcus xanthus development. Mol. Microbiol. 2000;35:812–824. doi: 10.1046/j.1365-2958.2000.01753.x. [DOI] [PubMed] [Google Scholar]
- Gill RE, Karlok M, Benton D. Myxococcus xanthus encodes an ATP-dependent protease which is required for developmental gene transcription and intercellular signaling. J. Bacteriol. 1993;175:4538–4544. doi: 10.1128/jb.175.14.4538-4544.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman BS, Nierman WC, Kaiser D, Slater SC, Durkin AS, Eisen JA, Ronning CM, Barbazuk WB, et al. Evolution of sensory complexity recorded in a myxobacterial genome. Proc. Natl. Acad. Sci. U.S.A. 2006;103:15200–15205. doi: 10.1073/pnas.0607335103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagen DC, Bretscher AP, Kaiser D. Synergism between morpho-genetic mutants of Myxococcus xanthus. Dev. Biol. 1978;64:284–296. doi: 10.1016/0012-1606(78)90079-9. [DOI] [PubMed] [Google Scholar]
- Hodgkin J, Kaiser D. Cell-to-cell stimulation of movement in nonmotile mutants of Myxococcus. Proc. Natl. Acad. Sci. U.S.A. 1977;74:2938–2942. doi: 10.1073/pnas.74.7.2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W, Yang Z, Lux R, Zhao M, Wang J, He X, Shi W. Direct visualization of the interaction between pilin and exopolysaccharides of Myxococcus xanthus with eGFP-fused PilA protein. FEMS Microbiol. Lett. 2012;326:23–30. doi: 10.1111/j.1574-6968.2011.02430.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igoshin OA, Goldbeter A, Kaiser D, Oster G. A biochemical oscillator explains several aspects of Myxococcus xanthus behavior during development. Proc. Natl. Acad. Sci. U S A. 2004;101:15760–15765. doi: 10.1073/pnas.0407111101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakovljevic V, Leonardy S, Hoppert M, Sogaard-Andersen L. PilB and PilT are ATPases acting antagonistically in type IV pilus function in Myxococcus xanthus. J. Bacteriol. 2008;190:2411–2421. doi: 10.1128/JB.01793-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarrell KF, McBride MJ. The surprisingly diverse ways that prokaryotes move. Nat. Rev. Microbiol. 2008;6:466–476. doi: 10.1038/nrmicro1900. [DOI] [PubMed] [Google Scholar]
- Kaiser D. Roland Thaxter’s legacy and the origins of multicellular development. Gene. 1993;135:249–254. doi: 10.1093/genetics/135.2.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser D. Bacteria also vote. Science. 1996;272:1598–1599. doi: 10.1126/science.272.5268.1598. [DOI] [PubMed] [Google Scholar]
- Kaiser D. Signaling in myxobacteria. Annu. Rev. Microbiol. 2004;58:75–98. doi: 10.1146/annurev.micro.58.030603.123620. [DOI] [PubMed] [Google Scholar]
- Kaiser D, Crosby C. Cell movements and its coordination in swarms of Myxococcus xanthus. Cell Motil. 1983;3:227–245. [Google Scholar]
- Kaiser D, Robinson M, Kroos L. Myxobacteria, polarity, and multicellular morphogenesis. Cold Spring Harb. Perspect. Biol. 2010;2:a000380. doi: 10.1101/cshperspect.a000380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan HB. Multicellular development and gliding motility in Myxococcus xanthus. Curr. Opin. Microbiol. 2003;6:572–577. doi: 10.1016/j.mib.2003.10.006. [DOI] [PubMed] [Google Scholar]
- Kaplan HB, Kuspa A, Kaiser D. Suppressors that permit A-signal-independent developmental gene expression in Myxococcus xanthus. J. Bacteriol. 1991;173:1460–1470. doi: 10.1128/jb.173.4.1460-1470.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearns DB, Shimkets LJ. Lipid chemotaxis and signal transduction in Myxococcus xanthus. Trends Microbiol. 2001;9:126–129. doi: 10.1016/s0966-842x(01)01948-5. [DOI] [PubMed] [Google Scholar]
- Kim SK, Kaiser D. C-factor: a cellecell signaling protein required for fruiting body morphogenesis of M. xanthus. Cell. 1990a;61:19–26. doi: 10.1016/0092-8674(90)90211-v. [DOI] [PubMed] [Google Scholar]
- Kim SK, Kaiser D. Cell alignment required in differentiation of Myxococcus xanthus. Science. 1990b;249:926–928. doi: 10.1126/science.2118274. [DOI] [PubMed] [Google Scholar]
- Kim SK, Kaiser D. Cell motility is required for the transmission of C-factor, an intercellular signal that coordinates fruiting body morphogenesis of Myxococcus xanthus. Genes Dev. 1990c;4:896–904. doi: 10.1101/gad.4.6.896. [DOI] [PubMed] [Google Scholar]
- Kim SK, Kaiser D. Purification and properties of Myxococcus xanthus C-factor, an intercellular signaling protein. Proc. Natl. Acad. Sci. U S A. 1990d;87:3635–3639. doi: 10.1073/pnas.87.10.3635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SK, Kaiser D. C-factor has distinct aggregation and sporulation thresholds during Myxococcus development. J. Bacteriol. 1991;173:1722–1728. doi: 10.1128/jb.173.5.1722-1728.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konovalova A, Lobach S, Sogaard-Andersen L. A RelA-dependent two-tiered regulated proteolysis cascade controls synthesis of a contact-dependent intercellular signal in Myxococcus xanthus. Mol. Microbiol. 2012a;84:260–275. doi: 10.1111/j.1365-2958.2012.08020.x. [DOI] [PubMed] [Google Scholar]
- Konovalova A, Petters T, Sogaard-Andersen L. Extracellular biology of Myxococcus xanthus. FEMS Microbiol. Rev. 2010;34:89–106. doi: 10.1111/j.1574-6976.2009.00194.x. [DOI] [PubMed] [Google Scholar]
- Konovalova A, Sogaard-Andersen L. Close encounters: contact-dependent interactions in bacteria. Mol. Microbiol. 2011;81:297–301. doi: 10.1111/j.1365-2958.2011.07711.x. [DOI] [PubMed] [Google Scholar]
- Konovalova A, Wegener-Feldbrugge S, Sogaard-Andersen L. Two intercellular signals required for fruiting body formation in Myxococcus xanthus act sequentially but non-hierarchically. Mol. Microbiol. 2012b;86:65–81. doi: 10.1111/j.1365-2958.2012.08173.x. [DOI] [PubMed] [Google Scholar]
- Kraemer SA, Velicer GJ. Endemic social diversity within natural kin groups of a cooperative bacterium. Proc. Natl. Acad. Sci. U S A. 2011;108:10823–10830. doi: 10.1073/pnas.1100307108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroos L. The Bacillus and Myxococcus developmental networks and their transcriptional regulators. Annu. Rev. Genet. 2007;41:13–39. doi: 10.1146/annurev.genet.41.110306.130400. [DOI] [PubMed] [Google Scholar]
- Kroos L, Hartzell P, Stephens K, Kaiser D. A link between cell movement and gene expression argues that motility is required for cellecell signaling during fruiting body development. Genes Dev. 1988;2:1677–1685. doi: 10.1101/gad.2.12a.1677. [DOI] [PubMed] [Google Scholar]
- Kroos L, Kaiser D. Expression of many developmentally regulated genes in Myxococcus depends on a sequence of cell interactions. Genes Dev. 1987;1:840–854. doi: 10.1101/gad.1.8.840. [DOI] [PubMed] [Google Scholar]
- Kroos L, Kuspa A, Kaiser D. A global analysis of developmentally regulated genes in Myxococcus xanthus. Dev. Biol. 1986;117:252–266. doi: 10.1016/0012-1606(86)90368-4. [DOI] [PubMed] [Google Scholar]
- Kruse T, Lobedanz S, Berthelsen NM, Sogaard-Andersen L. C-signal: a cell surface-associated morphogen that induces and coordinates multicellular fruiting body morphogenesis and sporulation in Myxococcus xanthus. Mol. Microbiol. 2001;40:156–168. doi: 10.1046/j.1365-2958.2001.02365.x. [DOI] [PubMed] [Google Scholar]
- Kudryashev M, Cyrklaff M, Alex B, Lemgruber L, Baumeister W, Wallich R, Frischknecht F. Evidence of direct cellecell fusion in Borrelia by cryogenic electron tomography. Cell. Microbiol. 2011;13:731–741. doi: 10.1111/j.1462-5822.2011.01571.x. [DOI] [PubMed] [Google Scholar]
- Kulp A, Kuehn MJ. Biological functions and biogenesis of secreted bacterial outer membrane vesicles. Annu. Rev. Microbiol. 2010;64:163–184. doi: 10.1146/annurev.micro.091208.073413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuner JM, Kaiser D. Fruiting body morphogenesis in submerged cultures of Myxococcus xanthus. J. Bacteriol. 1982;151:458–461. doi: 10.1128/jb.151.1.458-461.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuspa A, Plamann L, Kaiser D. Identification of heat-stable A-factor from Myxococcus xanthus. J. Bacteriol. 1992a;174:3319–3326. doi: 10.1128/jb.174.10.3319-3326.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuspa A, Plamann L, Kaiser D. A-signalling and the cell density requirement for Myxococcus xanthus development. J. Bacteriol. 1992b;174:7360–7369. doi: 10.1128/jb.174.22.7360-7369.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancero H, Brofft JE, Downard J, Birren BW, Nusbaum C, Naylor J, Shi W, Shimkets LJ. Mapping of Myxococcus xanthus social motility dsp mutations to the dif genes. J. Bacteriol. 2002;184:1462–1465. doi: 10.1128/JB.184.5.1462-1465.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee BU, Lee K, Mendez J, Shimkets LJ. A tactile sensory system of Myxococcus xanthus involves an extracellular NAD(P)(+)-containing protein. Genes Dev. 1995;9:2964–2973. doi: 10.1101/gad.9.23.2964. [DOI] [PubMed] [Google Scholar]
- Lee K, Shimkets LJ. Cloning and characterization of the socA locus which restores development to Myxococcus xanthus C-signaling mutants. J. Bacteriol. 1994;176:2200–2209. doi: 10.1128/jb.176.8.2200-2209.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Lee BU, Shimkets LJ. csgA expression entrains Myxococcus xanthus development. Genes Dev. 1992;6:401–410. doi: 10.1101/gad.6.3.401. [DOI] [PubMed] [Google Scholar]
- Li SF, Shimkets LJ. Effect of dsp mutations on the cell-to-cell transmission of CsgA in Myxococcus xanthus. J. Bacteriol. 1993;175:3648–3652. doi: 10.1128/jb.175.11.3648-3652.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Sun H, Ma X, Lu A, Lux R, Zusman D, Shi W. Extracellular polysaccharides mediate pilus retraction during social motility of Myxococcus xanthus. Proc. Natl. Acad. Sci. U S A. 2003;100:5443–5448. doi: 10.1073/pnas.0836639100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobedanz S, Sogaard-Andersen L. Identification of the C-signal, a contact-dependent morphogen coordinating multiple developmental responses in Myxococcus xanthus. Genes Dev. 2003;17:2151–2161. doi: 10.1101/gad.274203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu A, Cho K, Black WP, Duan XY, Lux R, Yang Z, Kaplan HB, Zusman DR, et al. Exopolysaccharide biosynthesis genes required for social motility in Myxococcus xanthus. Mol. Microbiol. 2005;55:206–220. doi: 10.1111/j.1365-2958.2004.04369.x. [DOI] [PubMed] [Google Scholar]
- Luciano J, Agrebi R, Le Gall AV, Wartel M, Fiegna F, Ducret A, Brochier-Armanet C, Mignot T. Emergence and modular evolution of a novel motility machinery in bacteria. PLoS Genet. 2011;7:e1002268. doi: 10.1371/journal.pgen.1002268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahmud T, Bode HB, Silakowski B, Kroppenstedt RM, Xu M, Nordhoff S, Hofle G, Muller R. A novel biosynthetic pathway providing precursors for fatty acid biosynthesis and secondary metabolite formation in myxobacteria. J. Biol. Chem. 2002;277:32768–32774. doi: 10.1074/jbc.M205222200. [DOI] [PubMed] [Google Scholar]
- Martens S, McMahon HT. Mechanisms of membrane fusion: disparate players and common principles. Nat. Rev. Mol. Cell. Biol. 2008;9:543–556. doi: 10.1038/nrm2417. [DOI] [PubMed] [Google Scholar]
- Mashburn-Warren LM, Whiteley M. Special delivery: vesicle trafficking in prokaryotes. Mol. Microbiol. 2006;61:839–846. doi: 10.1111/j.1365-2958.2006.05272.x. [DOI] [PubMed] [Google Scholar]
- Mauriello EM, Astling DP, Sliusarenko O, Zusman DR. Localization of a bacterial cytoplasmic receptor is dynamic and changes with cellecell contacts. Proc. Natl. Acad. Sci. U.S.A. 2009;106:4852–4857. doi: 10.1073/pnas.0810583106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McVittie A, Messik F, Zahler SA. Developmental biology of Myxococcus. J. Bacteriol. 1962;84:546–551. doi: 10.1128/jb.84.3.546-551.1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendes R, Kruijt M, de Bruijn I, Dekkers E, van der Voort M, Schneider JH, Piceno YM, DeSantis TZ, et al. Deciphering the rhizosphere microbiome for disease-suppressive bacteria. Science. 2011;332:1097–1100. doi: 10.1126/science.1203980. [DOI] [PubMed] [Google Scholar]
- Muller FD, Schink CW, Hoiczyk E, Cserti E, Higgs PI. Spore formation in Myxococcus xanthus is tied to cytoskeleton functions and polysaccharide spore coat deposition. Mol. Microbiol. 2012;83:486–505. doi: 10.1111/j.1365-2958.2011.07944.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nan B, Mauriello EM, Sun IH, Wong A, Zusman DR. A multi-protein complex from Myxococcus xanthus required for bacterial gliding motility. Mol. Microbiol. 2010;76:1539–1554. doi: 10.1111/j.1365-2958.2010.07184.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nan B, Zusman DR. Uncovering the mystery of gliding motility in the myxobacteria. Annu. Rev. Genet. 2011;45:21–39. doi: 10.1146/annurev-genet-110410-132547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nudleman E, Wall D, Kaiser D. Cell-to-cell transfer of bacterial outer membrane lipoproteins. Science. 2005;309:125–127. doi: 10.1126/science.1112440. [DOI] [PubMed] [Google Scholar]
- Nudleman E, Wall D, Kaiser D. Polar assembly of the type IV pilus secretin in Myxococcus xanthus. Mol. Microbiol. 2006;60:16–29. doi: 10.1111/j.1365-2958.2006.05095.x. [DOI] [PubMed] [Google Scholar]
- O’Toole G, Kaplan HB, Kolter R. Biofilm formation as microbial development. Annu. Rev. Microbiol. 2000;54:49–79. doi: 10.1146/annurev.micro.54.1.49. [DOI] [PubMed] [Google Scholar]
- Pathak DT, Wall D. Identification of the cglC, cglD, cglE, and cglF genes and their role in cell contact-dependent gliding motility in Myxococcus xanthus. J. Bacteriol. 2012;194:1940–1949. doi: 10.1128/JB.00055-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pathak DT, Wei X, Bucuvalas A, Haft DH, Gerloff DL, Wall D. Cell contact-dependent outer membrane exchange in myxobacteria: genetic determinants and mechanism. PLoS Genet. 2012;8:e1002626. doi: 10.1371/journal.pgen.1002626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plamann L, Davis JM, Cantwell B, Mayor J. Evidence that asgB encodes a DNA-binding protein essential for growth and development of Myxococcus xanthus. J. Bacteriol. 1994;176:2013–2020. doi: 10.1128/jb.176.7.2013-2020.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plamann L, Li Y, Cantwell B, Mayor J. The Myxococcus xanthus asgA gene encodes a novel signal transduction protein required for multicellular development. J. Bacteriol. 1995;177:2014–2020. doi: 10.1128/jb.177.8.2014-2020.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen AA, Porter SL, Armitage JP, Sogaard-Andersen L. Coupling of multicellular morphogenesis and cellular differentiation by an unusual hybrid histidine protein kinase in Myxococcus xanthus. Mol. Microbiol. 2005;56:1358–1372. doi: 10.1111/j.1365-2958.2005.04629.x. [DOI] [PubMed] [Google Scholar]
- Rhie HG, Shimkets LJ. Developmental bypass suppression of Myxococcus xanthus csgA mutations. J. Bacteriol. 1989;171:3268–3276. doi: 10.1128/jb.171.6.3268-3276.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigden DJ, Mello LV, Galperin MY. The PA14 domain, a conserved all-beta domain in bacterial toxins, enzymes, adhesins and signaling molecules. Trends Biochem. Sci. 2004;29:335–339. doi: 10.1016/j.tibs.2004.05.002. [DOI] [PubMed] [Google Scholar]
- Ring MW, Schwar G, Thiel V, Dickschat JS, Kroppenstedt RM, Schulz S, Bode HB. Novel iso-branched ether lipids as specific markers of developmental sporulation in the myxobacterium Myxococcus xanthus. J. Biol. Chem. 2006;281:36691–36700. doi: 10.1074/jbc.M607616200. [DOI] [PubMed] [Google Scholar]
- Rodriguez AM, Spormann AM. Genetic and molecular analysis of cglB, a gene essential for single-cell gliding in Myxococcus xanthus. J. Bacteriol. 1999;181:4381–4390. doi: 10.1128/jb.181.14.4381-4390.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolbetzki A, Ammon M, Jakovljevic V, Konovalova A, Sogaard-Andersen L. Regulated secretion of a protease activates intercellular signaling during fruiting body formation in M. xanthus. Dev. Cell. 2008;15:627–634. doi: 10.1016/j.devcel.2008.08.002. [DOI] [PubMed] [Google Scholar]
- Rosenberg E, Keller KH, Dworkin M. Cell density-dependent growth of Myxococcus xanthus on casein. J. Bacteriol. 1977;129:770–777. doi: 10.1128/jb.129.2.770-777.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimkets LJ. Correlation of energy-dependent cell cohesion with social motility in Myxococcus xanthus. J. Bacteriol. 1986;166:837–841. doi: 10.1128/jb.166.3.837-841.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimkets LJ. Intercellular signaling during fruiting-body development of Myxococcus xanthus. Annu. Rev. Microbiol. 1999;53:525–549. doi: 10.1146/annurev.micro.53.1.525. [DOI] [PubMed] [Google Scholar]
- Shimkets LJ, Gill RE, Kaiser D. Developmental cell interactions in Myxococcus xanthus and the spoC locus. Proc. Natl. Acad. Sci. U.S.A. 1983;80:1406–1410. doi: 10.1073/pnas.80.5.1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimkets LJ, Rafiee H. CsgA, an extracellular protein essential for Myxococcus xanthus development. J. Bacteriol. 1990;172:5299–5306. doi: 10.1128/jb.172.9.5299-5306.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simunovic V, Gherardini FC, Shimkets LJ. Membrane localization of motility, signaling, and polyketide synthetase proteins in Myxococcus xanthus. J. Bacteriol. 2003;185:5066–5075. doi: 10.1128/JB.185.17.5066-5075.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer M, Kaiser D. Ectopic production of guanosine penta- and tetraphosphate can initiate early developmental gene expression in Myxococcus xanthus. Genes Dev. 1995;9:1633–1644. doi: 10.1101/gad.9.13.1633. [DOI] [PubMed] [Google Scholar]
- Skerker JM, Berg HC. Direct observation of extension and retraction of type IV pili. Proc. Natl. Acad. Sci. U.S.A. 2001;98:6901–6904. doi: 10.1073/pnas.121171698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DR, Dworkin M. Territorial interactions between two Myxococcus Species. J. Bacteriol. 1994;176:1201–1205. doi: 10.1128/jb.176.4.1201-1205.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smukalla S, Caldara M, Pochet N, Beauvais A, Guadagnini S, Yan C, Vinces MD, Jansen A, et al. FLO1 is a variable green beard gene that drives biofilm-like cooperation in budding yeast. Cell. 2008;135:726–737. doi: 10.1016/j.cell.2008.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stacy AR, Diggle SP, Whiteley M. Rules of engagement: defining bacterial communication. Curr. Opin. Microbiol. 2012;15:155–161. doi: 10.1016/j.mib.2011.11.007. [DOI] [PubMed] [Google Scholar]
- Strassmann JE, Gilbert OM, Queller DC. Kin discrimination and cooperation in microbes. Annu. Rev. Microbiol. 2011;65:349–367. doi: 10.1146/annurev.micro.112408.134109. [DOI] [PubMed] [Google Scholar]
- Sun M, Wartel M, Cascales E, Shaevitz JW, Mignot T. Motor-driven intracellular transport powers bacterial gliding motility. Proc. Natl. Acad. Sci. U.S.A. 2011;108:7559–7564. doi: 10.1073/pnas.1101101108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toal DR, Clifton SW, Roe BA, Downard J. The esg locus of Myxococcus xanthus encodes the E1 alpha and E1 beta subunits of a branched-chain keto acid dehydrogenase. Mol. Microbiol. 1995;16:177–189. doi: 10.1111/j.1365-2958.1995.tb02291.x. [DOI] [PubMed] [Google Scholar]
- Veelders M, Bruckner S, Ott D, Unverzagt C, Mosch HU, Essen LO. Structural basis of flocculin-mediated social behavior in yeast. Proc. Natl. Acad. Sci. U.S.A. 2010;107:22511–22516. doi: 10.1073/pnas.1013210108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velicer GJ, Kroos L, Lenski RE. Developmental cheating in the social bacterium Myxococcus xanthus. Nature. 2000;404:598–601. doi: 10.1038/35007066. [DOI] [PubMed] [Google Scholar]
- Vos M, Velicer GJ. Social conflict in centimeter-and global-scale populations of the bacterium Myxococcus xanthus. Curr. Biol. 2009;19:1763–1767. doi: 10.1016/j.cub.2009.08.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall D, Kaiser D. Alignment enhances the cell-to-cell transfer of pilus phenotype. Proc. Natl. Acad. Sci. U.S.A. 1998;95:3054–3058. doi: 10.1073/pnas.95.6.3054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall D, Kaiser D. Type IV pili and cell motility. Mol. Microbiol. 1999;32:1–10. doi: 10.1046/j.1365-2958.1999.01339.x. [DOI] [PubMed] [Google Scholar]
- Wei X, Pathak DT, Wall D. Heterologous protein transfer within structured myxobacteria biofilms. Mol. Microbiol. 2011;81:315–326. doi: 10.1111/j.1365-2958.2011.07710.x. [DOI] [PubMed] [Google Scholar]
- Weimer RM, Creighton C, Stassinopoulos A, Youderian P, Hartzell PL. A chaperone in the HSP70 family controls production of extracellular fibrils in Myxococcus xanthus. J. Bacteriol. 1998;180:5357–5368. doi: 10.1128/jb.180.20.5357-5368.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitworth DE. Myxobacteria Multicellularity and Differentiation. ASM Press: Washington DC; 2008. [Google Scholar]
- Wickner W, Schekman R. Membrane fusion. Nat. Struct. Mol. Biol. 2008;15:658–664. doi: 10.1038/nsmb.1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolgemuth C, Hoiczyk E, Kaiser D, Oster G. How myxobacteria glide. Curr. Biol. 2002;12:369–377. doi: 10.1016/s0960-9822(02)00716-9. [DOI] [PubMed] [Google Scholar]
- Wu SS, Wu J, Kaiser D. The Myxococcus xanthus pilT locus is required for social gliding motility although pili are still produced. Mol. Microbiol. 1997;23:109–121. doi: 10.1046/j.1365-2958.1997.1791550.x. [DOI] [PubMed] [Google Scholar]
- Xiao Y, Gerth K, Muller R, Wall D. Myxobacterium-produced antibiotic TA (myxovirescin) inhibits type II signal peptidase. Antimicrobial. Agents Chemother. 2012;56:2014–2021. doi: 10.1128/AAC.06148-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Y, Wei X, Ebright R, Wall D. Antibiotic production by myxobacteria plays a role in predation. J. Bacteriol. 2011;193:4626–4633. doi: 10.1128/JB.05052-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D, Yang C, Kaplan HB. Myxococcus xanthus sasN encodes a regulator that prevents developmental gene expression during growth. J. Bacteriol. 1998;180:6215–6223. doi: 10.1128/jb.180.23.6215-6223.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C, Kaplan HB. Myxococcus xanthus sasS encodes a sensor histidine kinase required for early developmental gene expression. J. Bacteriol. 1997;179:7759–7767. doi: 10.1128/jb.179.24.7759-7767.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Geng Y, Xu D, Kaplan HB, Shi W. A new set of chemotaxis homologues is essential for Myxococcus xanthus social motility. Mol. Microbiol. 1998;30:1123–1130. doi: 10.1046/j.1365-2958.1998.01160.x. [DOI] [PubMed] [Google Scholar]
- Yang Z, Lux R, Hu W, Hu C, Shi W. PilA localization affects extracellular polysaccharide production and fruiting body formation in Myxococcus xanthus. Mol. Microbiol. 2010;76:1500–1513. doi: 10.1111/j.1365-2958.2010.07180.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youderian P, Hartzell PL. Transposon insertions of magellan-4 that impair social gliding motility in Myxococcus xanthus. Genetics. 2006;172:1397–1410. doi: 10.1534/genetics.105.050542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu R, Kaiser D. Gliding motility and polarized slime secretion. Mol. Microbiol. 2007;63:454–467. doi: 10.1111/j.1365-2958.2006.05536.x. [DOI] [PubMed] [Google Scholar]
- Zusman DR, Scott AE, Yang Z, Kirby JR. Chemosensory pathways, motility and development in Myxococcus xanthus. Nat. Rev. Microbiol. 2007;5:862–872. doi: 10.1038/nrmicro1770. [DOI] [PubMed] [Google Scholar]