SUMMARY
The cohesin complex holds together newly-replicated chromatids and is involved in diverse pathways that preserve genome integrity. We show that in budding yeast, cohesin is transiently recruited to active replication origins and it spreads along DNA as forks progress. When DNA synthesis is impeded, cohesin accumulates at replication sites and is critical for the recovery of stalled forks. Cohesin enrichment at replication forks does not depend on γH2A(X) formation, which differs from its loading requirements at DNA double-strand breaks (DSBs). However, cohesin localization is largely reduced in rad50Δ mutants and cells lacking both Mec1 and Tel1 checkpoint kinases. Interestingly, cohesin loading at replication sites depends on the structural features of Rad50 that are important for bridging sister chromatids, including the CXXC hook domain and the length of the coiled-coil extensions. Together, these data reveal a function for cohesin in the maintenance of genome integrity during S phase.
Keywords: Cohesin, Rad50, DNA synthesis, replication stress recovery
INTRODUCTION
Structural maintenance of chromosomes (SMC) proteins are crucial for the condensation and faithful segregation of chromosomes and DNA repair. Failure to execute these functions leads to genomic instability or to cell death. Proteins of the SMC family are characterized by N- and C-terminal globular heads, which are separated by a coiled-coil region with a central hinge domain. The coiled-coil folds back on itself at the hinge, bringing the globular ends together to form an ATPase domain (reviewed in Hirano, 2006). One of the best-characterized SMC complexes is cohesin, which holds sister chromatids together after DNA replication. It is composed of two SMC proteins, Smc1 and Smc3, that associate tightly at the hinge and two non-SMC components, Scc1 and Scc3, which bridge the two head domains. This creates a structure bringing together sister chromatids that are released in metaphase upon cleavage of Scc1 (Uhlmann et al., 1999; Anderson et al., 2002).
Cohesin is loaded at specific sites on chromatin in late G1 phase by the Scc2-Scc4 complex (Ciosk et al., 2000; Tomonaga et al., 2000; Bernard et al., 2006). Sister chromatid cohesion (SCC) is established upon passage of the replication fork and conversion of the cohesin complex to a cohesive state (Lengronne et al., 2006; Rolef Ben-Shahar et al., 2008; Unal et al., 2008). The maintenance of SCC during S phase is essential for accurate chromosome segregation during mitosis. It is also critical for homologous recombination (HR) -mediated repair of DNA double strand breaks (DSBs) in G2 (Sjogren and Nasmyth, 2001).
Incidence of DNA DSBs in G2 provokes “de novo” cohesin loading to the site of damage. This recruitment depends not only on Scc2-Scc4 and Eco1, but also on the Mec1 and Tel1 checkpoint kinases, γH2A(X) and the Mre11/Rad50/Xrs2 (MRX) complex (Strom et al., 2004; Unal et al., 2004; Strom and Sjogren, 2007; Unal et al., 2008; Heidinger-Pauli et al., 2008). The MRX complex can function as a tethering complex between sister chromatids (de Jager et al., 2001; Hopfner et al., 2002; Williams et al., 2007; Williams et al., 2008). It contains a SMC-like protein called Rad50, which is structurally related to Smc1 and Smc3. It has been proposed that Rad50 can dimerize through interactions between hook domains to bridge DNA molecules (Furuse et al., 1998; Usui et al., 1998; Paull and Gellert, 1999). Emphasizing the importance of these structural features, mutations that disrupt the hook interface compromise the tethering function of the complex and render cells sensitive to genotoxic stress (Hopfner et al., 2000; Wiltzius et al., 2005; Hohl et al., 2011).
Replication forks are fragile structures that stall when they encounter DNA lesions or natural replication barriers (Tourriere and Pasero, 2007). A large body of evidence indicates that replication fork restart depends on recombination-mediated mechanisms that are distinct from homologous recombination-dependent DSB repair (Branzei and Foiani, 2007; Allen et al., 2011). Factors involved in the recruitment of cohesin to DSBs, such as Mec1 and the MRX complex, are also involved in the replication stress response (Katou et al., 2003; Osborn and Elledge, 2003; Cobb et al., 2005; Tittel-Elmer et al., 2009). In contrast to DSBs in G2, surprisingly little is known about cohesin dynamics at stalled forks. Moreover, the interplay between fork restart and cohesin function has remained largely unexplored (Blat and Kleckner, 1999; Lengronne et al., 2006). In the current investigation, we performed a genome-wide analysis of cohesin binding to chromatin during an unperturbed S phase and under conditions of replication stress induced by hydroxyurea (HU) or methyl methanesulfonate (MMS). We find that cohesin transiently associates to replication sites and accumulates further when forks pause or stall, presumably to facilitate recovery by maintaining sister chromatids in a conformation that favors recombination-dependent fork restart.
RESULTS
Cohesin accumulates transiently at sites of replication during stress
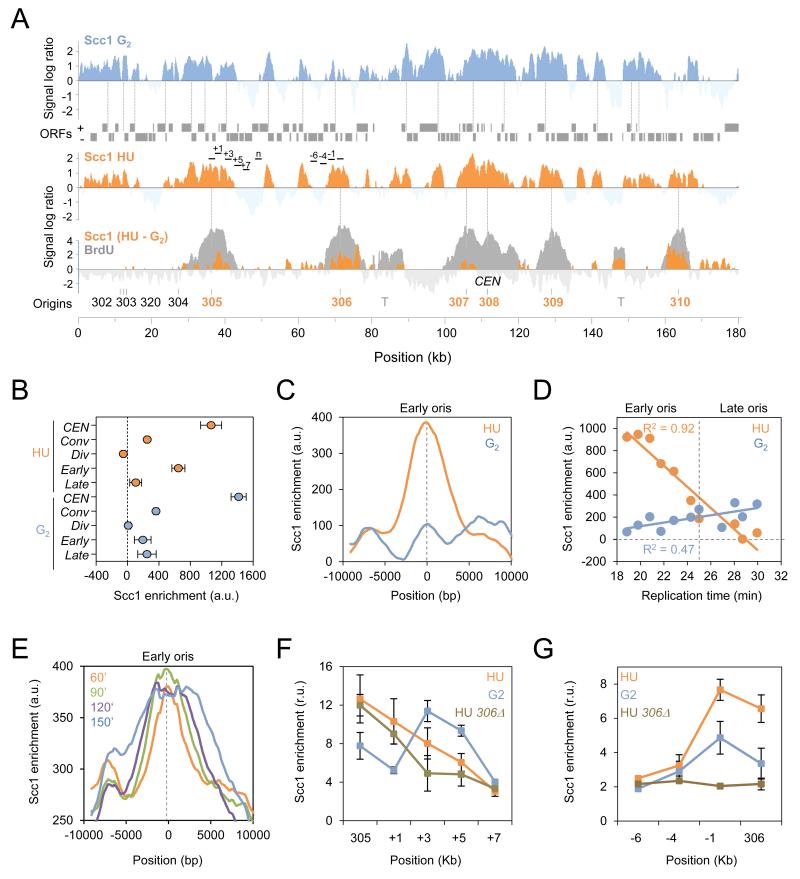
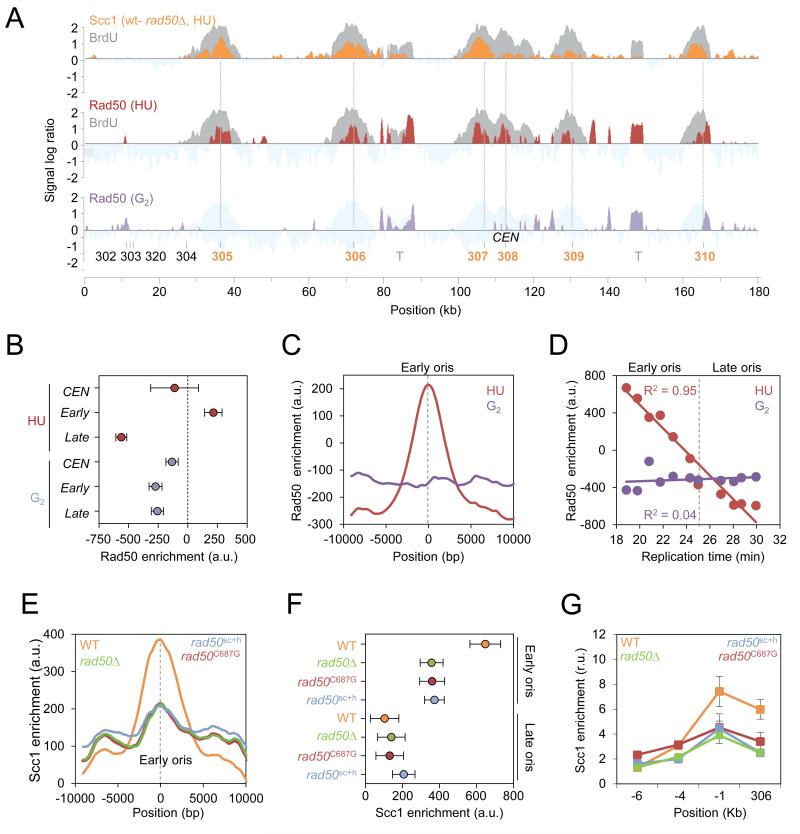
To investigate the pattern of DNA binding by the cohesin complex during replication stress, we compared the genome-wide profiles of Scc1 in cells released synchronously from G1 into S phase for 60 minutes in the presence of 0.2 M hydroxyurea (HU) or in the presence of nocodazole, a microtubule-destabilizing drug blocking cells at the G2/M transition (G2; Figure 1A). In agreement with previous reports (Lengronne et al., 2004), ChIP-chip profiles revealed that Scc1 was enriched between genes that are transcribed in converging directions and at centromeres but not at the promoters of genes with diverging orientation, used here as negative control (Figures 1A, 1B, and S1A). To visualize changes in cohesin binding upon HU exposure, we subtracted the profile of Scc1 in G2 from that of Scc1 in HU and observed that regions of enrichment clustered specifically at active early replication origins, visualized by BrdU incorporation (Crabbe et al., 2010). In contrast, no significant changes were found at centromeres, converging intergenes, repressed late origins or origins proximal to centromeres, which contain high Scc1 levels in G2 (Figures 1A and 1B).
Figure 1. Scc1 accumulates at early origins during a HU-induced replication stress.
(A) Genome-wide localization of Scc1-PK9 in wild type (JC-1315) cells arrested in G2 by nocodazole treatment for 120 minutes (blue) or released from G1 into S phase for 60 minutes in the presence of 0.2M HU (orange). Scc1 enrichment in the immunoprecipitated fraction relative to input genomic DNA is shown for part of chromosome 3 and is expressed as the log2 of the IP to input ratio. Gray boxes represent open reading frames and dashed lines represent converging intergenes. Early and late origins are indicated in orange and black, respectively; centromere (CEN); transposons (T). The BrdU profile of cells released from G1 in S phase for 60 minutes in the presence of 0.2M HU (Gray) is overlaid with the subtraction profile of Scc1 binding sites in HU - G2 (orange).
(B-D) Quantitation of Scc1 enrichment in HU-treated (orange) and G2-arrested cells (blue). Enrichment is calculated from ChIP-chip data as described in Materials and Methods and is expressed in arbitrary units (a. u.). (B) Mean Scc1 enrichment at centromeres (CEN) n=16, converging intergenes (conv) n=1757, diverging genes (div) n=1496, early origins (early) n=146 and late origins (late) n=85. Error bars indicate 95% CI. (C) Average distribution of Scc1 enrichment at early origins +/− 10 kb (n=112). (D) Scc1 enrichment at replication origins expressed as a function of replication time (Yabuki et al., 2002). Each dot represents a set of 20 origins. R2=0.92 and R2=0.47 correspond to the linear regression calculated for HU- and G2-arrested cells, respectively.
(E) Average profiles of Scc1 enrichment at early origins and regions +/− 10 kb in wild type cells released from G1 into S phase + 0.2M HU for 60 (orange), 90 (green), 120 (purple), and 150 (blue) minutes (n=112).
(F-G) ChIP followed by qPCR was performed on Scc1-PK9 cells released from G1 into S phase + 0.2M HU for 60 minutes (orange) or cells arrested in G2 with 7.5 μg/ml nocodazole for 120 minutes (blue) in WT and ars306Δ (JC-1969; brown) cells. Probes correspond to (F) ARS305 and regions downstream +1kb, +3kb, +5kb, and +7kb or (G) ARS306 and regions upstream −1kb, −4kb, and −6kb. Data were normalized to a region devoid of Scc1 (n) located 11 kb downstream of ARS305 and are expressed in relative units (r. u.). Error bars represent s.d. of fold enrichment of multiple runs and three individual experiments. All assessed loci are shown in Figure 1A.
To expand our analysis, Scc1 enrichment profiles were averaged at early origins (Figure 1C) and at converging intergenes (Figure S1B) in HU- and G2-arrested cells. We observed a clear HU-specific increase of Scc1 at early origins compared to G2 (Figure 1C), which was not recapitulated at converging intergenes (Figure S1B) or other Scc1-enriched domains (Figure S1A). Scc1 enrichment at active origins was also plotted as a function of BrdU incorporation and of the median time of replication, as previously determined (Yabuki et al., 2002). There was considerable Scc1 enrichment at sites of BrdU incorporation, which correlated with replication timing (R2=0.92; Figures 1D and S1C). Moreover, we also observed an expansion of cohesin around early origins as time progressed in the presence of 0.2 M HU (Figure 1E), reflecting the slow advancement of the replisome in low dNTP conditions (Poli et al., 2012).
In order to quantify the HU-induced accumulation of Scc1 at replication sites, we performed ChIP-qPCR and compared Scc1 enrichment near two early origins, ARS305 and ARS306. Upon HU treatment, Scc1 levels at both origins increased by 50% over levels in G2, but no significant differences were seen −4 kb or −6 kb upstream of ARS306 (Figures 1F and 1G, probe sets are shown in Figure 1A). Scc1 enrichment was observed in G2 samples at regions +3 kb and +5 kb of ARS305 (Figure 1F), which is a region of gene convergence (Figure 1A). Lastly, deletion of ARS306 abrogated Scc1 enrichment specifically at this locus and adjacent sites, but not at ARS305 (Figures 1F and 1G).
To determine if Scc1 accumulation at early origins during HU-induced stress represented a ‘permanent’ redistribution of cohesin until mitosis we analyzed the profile of Scc1 binding in G2 cells after HU treatment (HU/G2) by ChIP-chip. The Scc1 enrichment profile in HU/G2 was compared either to the profile of Scc1 in HU or in G2 arrested cells. The distribution at early origins in HU/G2 treated cells correlated more closely with cells arrested in G2 (R2=0.83) than HU (R2=0.44), suggesting that cohesin localization during replication stress was relatively transient (Figures S1D and S1E). The data revealed a greater than 2-fold increase in cohesin at 12 origins in HU/G2 vs. G2 (Figure S1F), however the implications of this distinction remain unclear.
Cohesin accumulates transiently at sites of replication during an unperturbed S phase
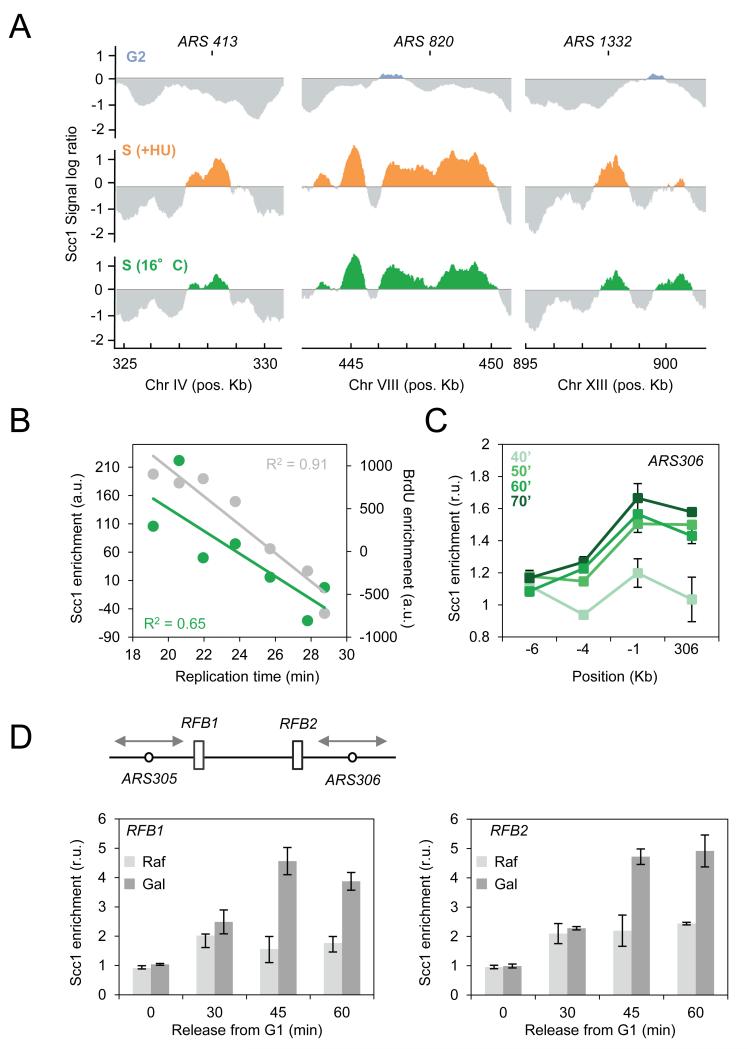
Our data indicate that cohesin is recruited to paused forks in HU-treated cells. We next checked whether cohesin is also present at replication sites in normal growth conditions. To this end, we determined the genome-wide binding profiles of Scc1 in cells released from G1 for 70 minutes at 16°C. At this temperature, the kinetics are slow enough to capture replisome components at early origins in the absence of HU (Cobb et al., 2003). Early origins such as ARS413, 820, 1332, which have no Scc1 in G2, were instrumental in visualizing enrichment during unperturbed replication. The Scc1 pattern at 16°C was similar to that of HU-arrested cells (Figure 2A). At the genome-wide level, Scc1 binding at origins correlated with replication timing (R2=0.65) and BrdU incorporation (R2=0.91; Figure 2B). Moreover, we performed ChIP-qPCR at ARS306 and adjacent regions to quantify levels during a normal S phase. Scc1 enrichment at ARS306 increased significantly by 50 minutes (Figure 2C), but remained below the levels observed in HU-arrested cells (Figure 1G), presumably because forks persist in the vicinity of ARS306 longer in the presence of HU.
Figure 2. Scc1 associates to replication sites during an unperturbed S phase.
(A) Cells containing Scc1-Pk9 were arrested in G2 by nocodazole treatment for 120 minutes (blue) or released from G1 into S phase for 60 minutes in the presence of 0.2M HU (orange) or released from G1 into S phase for 70 minutes at 16°C (green). Enrichment in the immunoprecipitat ed fraction relative to input DNA is shown at ARS413, ARS820 and ARS1332. The y-axis scale is log2.
(B) Scc1 (green) and BrdU (gray) enrichment were expressed as a function of replication timing of cells released from G1 into S phase for 70 minutes at 16°C. Each dot represents the median enrichment of 30 origins. R2=0.65 and R2=0.91 correspond to the linear regression calculated for Scc1 and BrdU respectively.
(C) ChIP followed by qPCR was performed on Scc1-PK9 (JC-1315) cells released synchronously from G1 into S phase for 40, 50, 60 or 70 minutes at 16°C. Probes corresponding to ARS306 and upstream regions were used as described in Figure 1G.
(D) ChIP followed by qPCR was performed on Scc1-PK9 (PP-1678) in S phase cells first synchronized in G1 before Gal-FOB1 induction for 45 min. Scc1 enrichment was quantified in cells with Fob1 expression (Gal, dark gray) or not (Raf, light gray) using probes corresponding to RFB1 and RFB2 (Calzada et al., 2005). Data were normalized to a region free of Scc1 at position 182’000 on chromosome 3. Error bars represent s.d. of fold enrichment of multiple runs and three individual experiments.
To better understand the relationship between fork progression and cohesin enrichment, we used two natural replication fork barriers (RFB1 and RFB2) integrated between ARS305 and ARS306 (Figure 2D). RFB activity at ectopic sites depends on the fork blocking protein Fob1, whose expression is under control of the Gal1,10 promoter (Calzada et al., 2005). As cells traverse S phase, Scc1 levels were 2-3 fold higher in Fob1-bound RFBs (Gal) (Figure 2D). These data indicate that similar to HU-arrested forks, cohesin accumulates at natural replication pause sites.
Cohesin is critical for replication fork restart
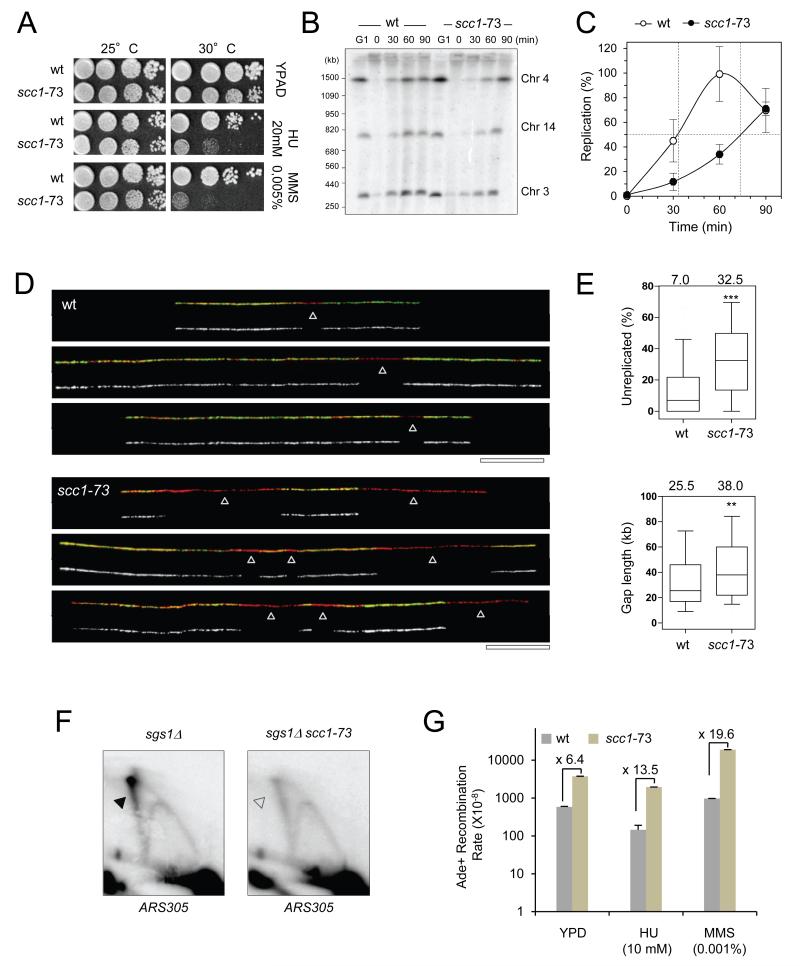
Cohesin is recruited to DSBs in G2 to promote HR-mediated repair, using the sister chromatid as a template (reviewed in Wu and Yu, 2012). HR-related mechanisms are also involved in replication fork restart (Vanoli et al., 2010; Allen et al., 2011), yet the contribution of cohesin function in this process has not been demonstrated. To determine if cohesin promotes fork recovery after replication stress, we exposed cells carrying the SCC1 thermo-sensitive (ts) allele scc1-73 (Michaelis et al., 1997) to sub-lethal doses of HU or MMS. As shown in Figure 3A, scc1-73 cells are exquisitely sensitive to both drugs at a semi-permissive temperature of 30°C.
Figure 3. Cohesin promotes recovery after replication fork arrest.
(A) Drop assays on exponentially growing wild type (JC-470) and scc1-73 (JC-1339) cells. 1:10 serial dilutions were grown on YPD + 0.2M HU or 0.005% MMS at 25 or 30°C.
(B-C) Scc1 is required for recovery of MMS-induced forks stalling. WT (PP-872) and scc1-73 (PP-1186) cells were grown at 25°C and synchroni zed in G1 with α-factor 30°C then released into S phase with 400 μg/ml BrdU to label newly-replicated DNA and 0.033% MMS for 60 minutes at 30°C. Cells were then washed and resuspended in fresh medium at 30°C for the indicated time after MMS release (t=0). Samples were subjected to PFGE analysis. (B) Southern blot probed for chromosomes 3, 4 and 14. (C) Quantification of chromosome mobility in wild-type (open circles) and scc1-73 (black circles). Median values and standard deviation for the three probes are shown.
(D-E) Representative DNA fibers of WT or scc1-73 cells exponentially grown at permissive temperature (25°C), arrested in G1 with α-factor at semi-permissive (30°C) and released synchronously into S phase in t he presence of 0.033% MMS for 60 minutes at 30°C. Green: BrdU, red: DNA, arro ws: unreplicated DNA. Bar: 50 kb. (E) Box plots depict the percentage of unreplicated DNA per DNA fiber and the length of unreplicated gaps. Box and whiskers indicate 25-75 and 10-90 percentiles, respectively.
(F) 2D gel analysis of replication intermediates in sgs1Δ (PP-35) and sgs1Δ scc1-73 (PP-1688) cells shifted at restrictive temperature (37°C) upon G1 arrest and released synchronously into S phase in the presence of 0.033% MMS for 30 minutes at 37°C. Genomic DNA was extracted and dige sted with EcoRI and the replication intermediates were analyzed by southern blot with a probe recognizing the ARS305 region.
(G) Rates of interhomologous recombination were measured as described (Mozlin et al., 2008) in diploid ade2-I/ade2-n WT (JC-2596) and scc1-73 (JC-2597) cells. Error bars correspond to s.d. of 9 diploids per condition in each experiment. Frequencies were determined at least twice in separate experiments giving comparable results at 30°C after overnight i ncubation in YPD + 10 mM HU or 0.001% MMS.
To investigate if the increased sensitivity is due to fork recovery defects, we monitored the completion of replication after stress. Since HU slows down DNA synthesis but does not actually block fork progression (Poli et al., 2012), we focused on MMS, an alkylating agent that induces fork stalling at a high frequency. As with HU, the binding pattern of Scc1 in MMS showed enrichment at early origins with levels proportionally to replication timing and BrdU incorporation (Figures S2A-D). Pulsed field gel electrophoresis (PFGE) was utilized to measure the kinetics of replication completion after acute MMS treatment. Chromosomes from wild type and scc1-73 cells were separated by PFGE and the replication status of three chromosomes was quantified by Southern blot analysis (Figures 3B and 3C). Wild type cells completed replication 60 minutes after MMS removal, however chromosome reentry into the gel was significantly delayed by at least 40 minutes in scc1-73 mutants. Notably, wild type and scc1-73 cells progressed through an unperturbed S phase with indistinguishable kinetics (Figure S2E), suggesting cohesin deficiency correlates with defects in replication primarily during times of stress.
To further characterize replication in scc1-73 cells upon MMS treatment, we monitored individual molecules by DNA combing. DNA fibers were purified and stretched along silanized coverslips as described (Poli et al., 2012). The length of BrdU tracks (green) and unreplicated gaps (red) were measured for more than 25 Mb of genomic DNA (Figures 3D and S2F). This analysis revealed a 4- to 5-fold increase in the percentage of unreplicated DNA in scc1-73 mutants relative to wild type cells (p<0.0001; Figure 3E) and a proportional reduction in the number of fully-replicated DNA fibers in these mutants (Figures 3E and S2G). Unreplicated gaps were also significantly larger in scc1-73 mutants than in wild type cells (38.0 kb versus 25.5 kb, p<0.0001; Figure 3E). Together, these data indicate that cohesin-deficient cells fail to complete DNA replication when exposed to agents inducing fork stalling.
Upon MMS treatment, stalled forks can be rescued by an HR-related process called template switching. To determine if cohesin is required for this we analyzed replication intermediates at ARS305 by 2D gel electrophoresis. Template switching generates branched molecules that are rapidly resolved through the combined action of Sgs1 and Top3 (Branzei and Foiani, 2007). In sgs1Δ cells, these structures accumulate and migrate as an X-spike on 2D gels (Vanoli et al., 2010). Consistent with this, 2D gel analysis of cells released into S phase with MMS revealed a prominent accumulation of X-molecules in sgs1Δ cells (Figure 3F). Remarkably, the X-spike intensity was reduced by 50% in sgs1Δ/scc1-73 mutants (Figures 3F, S2H and S2I), indicating that recombination intermediates formed less efficiently, or were possibly less stable when cohesin is compromised.
Finally, we reasoned that if cohesin promoted recombination between sister chromatids at stalled forks, it should also restrain interactions between homologs. To address this we measured mitotic heteroallelic recombination in cohesincompromised cells at 30°C in diploids containing th e ade2 heteroalleles, ade2-I and ade2-n, where recombination restores a functional ADE2 gene (Huang and Symington, 1994; Mozlin et al., 2008). The spontaneous rate in scc1-73 mutants was 6.4-fold over that of wild type and this increased to 13.5 and 19.6 fold upon HU and MMS treatment respectively (Figure 3G). Although we cannot formally exclude the possibility that cohesin located at a distance from stalled forks contribute to fork recovery, these data support a role for cohesin at stalled forks to promote completion of DNA replication and to prevent genomic instability.
Cohesin accumulation at sites of replication depends on Rad50 but not on Mec1 alone or γH2A(X)
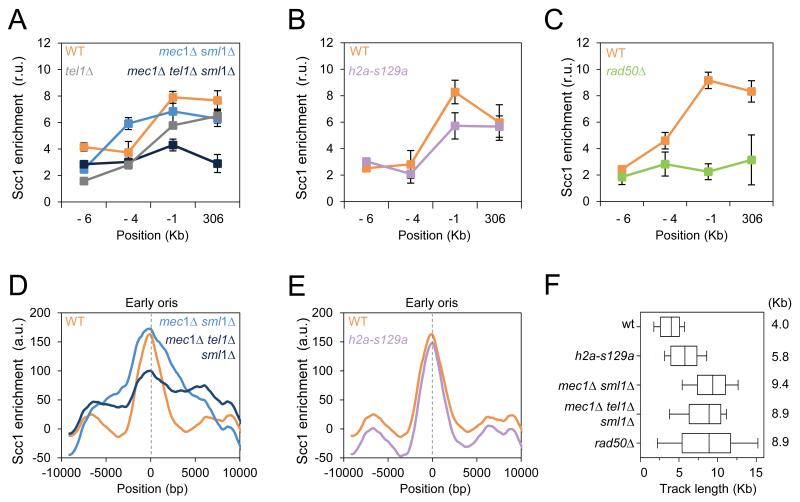
It is established that cohesin loading at DSBs depends on the MRX complex and on the checkpoint kinases Mec1 and Tel1, through the phosphorylation of histone H2A to form γ-H2A(X) domains (Unal et al., 2004; Strom et al., 2007). This prompted us to dissect the mechanism of Scc1 accumulation at sites of replication. We first monitored cohesin enrichment at ARS306 as described in Figure 1G in cells lacking either Mec1, Tel1 or Rad50 and in cells carrying the h2a-S129A allele, encoding an unphosphorylatable form of H2A. Scc1 was recovered to wild type levels in HU-arrested mec1Δ/sml1Δ, tel1Δ and h2a-S129A mutants (Figures 4A and 4B). In contrast, rad50Δ and mec1Δ/tel1Δ/sml1Δ mutants showed a significant decrease in Scc1 enrichment at ARS306 and adjacent regions (Figures 4A and 4C). To further characterize these observations, we performed ChIP-chip analysis of Scc1 in the above-mentioned mutants, as described in Figure 1C. The Scc1 profile at sites of replication in h2a-S129A mutants was nearly identical to the profile in wild type cells (Figure 4E). In mec1Δ/sml1Δ mutants, Scc1 enrichment was comparable to wild type, but Scc1 extended over a larger region (Figure 4D). This reflects the fact that forks progress further in this mutant (Figures 4F and S3). Consistent with ChIP-qPCR profiles, Scc1 was specifically reduced at early origins, but not at sites of converging intergenes in the absence of both Mec1 and Tel1 (Figure 4D). Nonetheless, Scc1 enrichment at replication sites does not depend on γH2A(X), therefore having distinct requirements compared to cohesin loading at DSBs. Scc1 enrichment also depends on Rad50, prompting us to further investigate the role of this SMC-like protein in the accumulation of cohesin at sites of replication.
Figure 4. Rad50 is required for cohesin localization at early origins.
(A-E) ChIP followed by qPCR was performed on Scc1-PK9 as described in Figure 1G (A-C), and the average profiles of Scc1 distribution at early origins as in Figure 1C (D-E) compare wild type (JC-1315), mec1Δ/sml1Δ (JC-1890), tel1Δ (JC-2378), mec1Δ/tel1Δ/sml1Δ (JC-2377), h2a-S129A (JC-1597) and rad50Δ (JC-1314).
(F) Distribution of BrdU tracks length in cells released from G1 into S phase + 0.2M HU for 60 minutes. Boxes and whiskers indicate 25-75 and 10-90 percentiles, respectively. Median BrdU track length is indicated in kb; Wild type (JC-1315), (PP-1323), (PP-1325), h2a-S129A (PP-1028) and )
Rad50 localizes to early firing origins during HU-induced replication stress.
First, we assessed genome-wide differences in Scc1 enrichment between WT and rad50Δ cells after HU exposure using the subtraction profile of Scc1 between WT and rad50Δ (Figures 5A and S4A). This demonstrated that regions of Rad50-dependent Scc1 binding corresponded to sites of BrdU incorporation (Figures 5A and S4E). To refine our analysis, Scc1 enrichment was averaged at early and late-firing origins, centromeres and converging intergenes in rad50Δ cells and compared to WT. The defect in Scc1 accumulation in cells lacking Rad50 was unique to early-firing origins as no significant changes were found at centromeres, late origins or converging intergenes (Figures 5A, S4B and S4F). Highlighting the defect at early-firing origins, there was a poor correlation between Scc1 and BrdU incorporation (R2=0.12; Figure S4C) and a significant reduction of Scc1 recruitment when plotted as a function of replication timing in rad50Δ cells (Figure S4D). Importantly, we verified that cohesin association deficiency in rad50Δ cells is not an indirect consequence of massive replication fork defects by monitoring fork progression in this mutant using BrdU-IP-chip. This analysis demonstrated that the distribution of BrdU track lengths is comparable between rad50Δ and mec1Δ mutants, which accumulate Scc1 at arrested forks (Figure 4F).
Figure 5. Structural features of Rad50 mediate Scc1 accumulation at replication sites.
(A) Upper panel: Rad50-dependent Scc1 enrichment in HU-arrested cells. WT and rad50Δ cells were synchronized in G1 and released into S phase + 0.2M HU for 60 minutes. Scc1 signal log ratio from rad50Δ cells was subtracted to signal from WT cells (orange). A representative region on chromosome 3 is shown. BrdU-IP-chip profile from WT cells (gray) is overlaid with the Scc1 profile. Middle-lower panels: Genome wide localization of Rad50-HA in wild type cells (JC-1300) arrested in G2 with 7.5 μg/ml nocodazole for 120 minutes (purple) or cells released from G1 into S phase + 0.2M HU for 60 minutes (red). The BrdU-IP-chip profile from HU-arrested cells is shown in gray.
(B-D) Quantitation of Rad50 enrichment in HU-treated cells (red) or in G2-arrested cells (purple). (B) Mean Rad50 enrichment at centromeres (CEN; n=16), early origins (early; n=146) and late origins (late; n=85). Error bars indicate 95% CI. (C) Average distribution of Rad50 at early origins (n=112). (D) Rad50 enrichment expressed as a function of replication timing. Each dot represents the median enrichment for 20 origins. R2=0.95 and R2=0.04 correspond to the linear regression calculated for HU-arrested and G2 arrested cells, respectively.
(E-G) Average distribution of Scc1 at replication origins as in Figures 1B and 1C and ChIP followed by qPCR as in Figure 1G were performed on Scc1-PK9 in WT (orange; JC-1315), rad50Δ (green; JC-1314), rad50C687G (red; JC-1988) and rad50sch (blue; JC-1967).
We had previously shown that Rad50 was recruited to the early-firing origin ARS607 in the presence of HU-induced replication stress during S phase (Tittel-Elmer et al., 2009). This, together with our observations that Scc1 levels decreased specifically at early origins in the absence of Rad50, led us to investigate the localization of MRX relative to sites of HU-specific cohesin accumulation. To obtain a genome-wide picture of MRX binding, ChIP-chip was performed on Rad50-HA in HU- (red) and G2-arrested (purple) cells (Figure 5A). Rad50 profiles showed clear peaks of enrichment at early origins upon HU-induced replication stress and no enrichment was observed in G2 arrested cells (Figures 5A-D). Moreover, sites of Rad50 enrichment in the presence of HU correlated with sites where Scc1 accumulation was lost in rad50Δ cells (Figure 5A). At the genome-wide level, there was a clear enrichment of Rad50 at early origins but not at late origins or centromeres (Figure 5B). As for Scc1, this enrichment was specific to HU-arrested cells and was not detected in G2 (Figures 5B and 5C). It also strongly correlated to replication timing (R2=0.95; Figure 5D). Taken together, these data suggest that the MRX complex contributes to the accumulation of Scc1 at early origins during replication stress.
Structural features of Rad50 mediate cohesin accumulation in HU-treated cells
To gain an understanding for how the MRX complex could mediate cohesin accumulation at stalled forks, we considered the pertinent structural features of the complex and its previously reported role in replisome stability as being independent of Mre11 nuclease activity (Hopfner et al., 2002; Wiltzius et al., 2005; Williams et al., 2008; Tittel-Elmer et al., 2009). We reasoned that the structural attributes intrinsic to the MRX complex might promote cohesin recruitment to paused forks. To investigate this we utilized the rad50sc and rad50C687G alleles, which are altered in the CXXC domain compromising hook-hook interactions, and the rad50sc+h allele, which abridges the length of the coiled-coil domain shortening the length of the ‘molecular tether’ (Hopfner et al., 2002; Hohl et al., 2011).
Consistent with previous reports (Hohl et al., 2011), the MRX complex remained intact in rad50sc+h and rad50sc mutants and was still proficiently recruited to early origins (Figures S5A-C). In addition, disruptions in the hook region resulted in phenotypes comparable to those of rad50 null mutants (Hohl et al., 2011), particularly for survival after HU exposure (Figure S5D). Strikingly, BrdU track lengths for the rad50C687G and rad50sc+h alleles were undistinguishable from rad50Δ mutant cells (Figure S5F), providing evidence that structural features of the MRX complex are important for its functionality during replication HU stress.
To investigate the structural role of Rad50 in Scc1 accumulation at early origins, we performed a genome-wide analysis of Scc1 levels at origins in rad50sc+h and rad50C687G mutants and compared these levels to those measured in rad50Δ cells (Figure 5E). Defective Scc1 enrichment was observed for both rad50sc+h and rad50C687G mutants at early origins with level indistinguishable from those of rad50Δ cells (Figures 5E and 5F). In order to quantify Scc1 enrichment in these rad50 alleles, ChIP-qPCR showed that Scc1 levels at ARS306 and regions upstream were diminished in all rad50 mutants (Figures 5G and S5E) corroborating the global loss of Scc1 at early origins. Together, these data underscore the importance of the hook and coiled-coil regions of Rad50 and the tether capability of the MRX complex for cohesin accumulation to forks during replication stress.
DISCUSSION
The cohesin complex functions in many aspects of chromosome organization and DNA metabolism processes, including DNA replication and repair (Nasmyth and Haering, 2009). We assessed the binding pattern of cohesin during replication stress and determined that cohesin accumulates at sites of replication by a mechanism that depends on emerging forks and correlates with replication timing. This profile is distinct from cohesin binding at centromeres and converging intergenes, which does not change from late G1 to mitosis (Lengronne et al., 2004). Our data are consistent with a model where cohesin associates transiently to replication sites during S phase and accumulates on newly-replicated chromatin if fork progression is inhibited, in order to facilitate fork restart.
During an unperturbed S phase, cohesin levels at replication sites are significantly lower than under replication stress conditions. This could either reflect the presence of low levels of cohesin at moving forks or its accumulation at spontaneous replication pause sites. In any case, this accumulation could result from different mechanisms, including the redistribution of already bound cohesin, de novo loading of additional cohesin or the retention of otherwise transitory cohesin interactions. In support of the former possibility, there is precedence for cohesin mobility as the complex has been shown to translocate along DNA and to relocalize in a transcription-dependent manner (Lengronne et al., 2004). Thus, it is plausible that inhibiting fork progression could transiently alter cohesin distribution patterns, driving enrichment where DNA synthesis stalls in a manner analogous to transcription.
Cohesin at sites of replication stress could also result from de novo loading. Indeed, Mec1 and Tel1 promote Scc1 recruitment through the phosphorylation of H2A at DSBs in G2 (Unal et al., 2004). However, at forks there was a dramatic difference between the cohesin profiles of h2a-S129A and mec1Δ tel1Δ double mutants arguing against such a mechanism at stalled forks. Defective Scc1 loading at stalled forks in mec1Δ tel1Δ cells might be attributed to the loss of checkpoint activation yet this too seems unlikely because RFB arrested forks do not activate the checkpoint (Calzada et al., 2005) and we measured robust cohesin enrichment at the sites of fork blockage. Taken together, these data indicate that checkpoint activation in S phase per se is not the key determinant for cohesin accumulation at sites of replication stress.
The mechanism by which Rad50 promotes the accumulation of cohesin on newly-replicated chromatin is currently unclear. Since the MRX/MRN complex acts at stalled or damaged replication forks (Costanzo et al., 2001; Tittel-Elmer et al., 2009), an attractive possibility could be that MRX stabilizes transiently-bound cohesin at replication sites upon fork arrest. The fact that Rad50 colocalizes with Scc1 during HU-induced stress is consistent with this view and implicates MRX as an integral factor in the maintenance of fork-associated cohesin. Hook-mediated bridging is essential for cohesin localization at stalled forks, presumably by maintaining sister chromatids in a conformation compatible with cohesin accumulation. It is important to note that we detected no physical interaction between cohesin and MRX and observed no difference in Rad50 binding in scc1-73 mutants (data not shown), indicating MRX functions independently or upstream of cohesin.
What is the physiological role of cohesin at stalled forks? The cohesin complex maintains the positioning of sister chromatids relative to each other and is therefore critical to repair one-ended DSBs occurring when the replisome encounters nicks or ssDNA gaps (Cortes-Ledesma and Aguilera, 2006). However, it is worth noting that most fork-blocking lesions, such as those induced by MMS, do not induce DSBs (Lundin et al., 2005; Groth et al., 2010), suggesting that cohesin has a role at stalled forks that is distinct from DSB repair. This view is supported by a recent synthetic gene array (SGA) analysis indicating that cohesin mutants show negative genetic interactions with genes involved in fork stability, but not with genes involved in DSB repair (McLellan et al., 2012). Moreover, it is consistent with our observation that cohesin contributes to HR-mediated template switching, a process contributing to fork restart (Vanoli et al., 2010; Allen et al., 2011). Importantly, defective fork restart in scc1-73 mutants is not a consequence of altered fork progression as fork speed is normal in scc1-73 mutants (AL, unpublished results), unlike in cohesin compromised human cells (Terret et al., 2009).
A more speculative model could be extended to include a need for more MRX and cohesin-dependent ‘constraining support’ as replicated regions grows in length and precatenane torsional stress increases (Bermejo et al., 2007). Consistent with this reasoning, we detected a strong correlation between the timing of replication initiation and the abundance of Scc1 and Rad50 at sites of replication. Interestingly, the accumulation of torsional stress also leads to fork reversal through a process mediated by poly(ADP-ribose) polymerases to promote fork recovery (Ray Chaudhuri et al., 2012). Taken together, these data suggest that events at the fork contribute significantly to the synthetic lethal interactions detected between cohesin mutations and PARP inhibitors in human cells (McLellan et al., 2012). Finally, the functional relationship between MRX/MRN and cohesin at stalled forks may explain why individuals with NBS1, ATLD and cohesinopathy disorders develop overlapping types of cancer when either complex is disrupted (Dzikiewicz-Krawczyk, 2008; Iwaizumi et al., 2009; Xu et al., 2011a; Xu et al., 2011b).
EXPERIMENTAL PROCEDURES
Yeast strains
All strains used in this study are listed in Table S1. They were derived from W303 and corrected for the rad5-535 mutation (RAD5+).
ChIP-qPCR and ChIP-chip
ChIP-qPCR was performed as described in (Katou et al., 2003; Tittel-Elmer et al., 2009) for Rad50-HA, Mre11-Myc, and Scc1-PK. Antibodies used were α-Pk clone SV5-Pk1 (Serotec), α-HA clone F-7 (Santa-Cruz), and α-Myc clone 9E10 coupled to α-mouse (Invitrogen,110.31) or α-Protein A Dynabeads (Invitrogen, 100.02). To synchronize cultures in G1, cells were incubated with α-factor for 2.5 hours and released by washes or by addition of 50 g/ml Pronase. For quantitative PCR (qPCR), uncoupled Dynabeads were used for background controls. DNA was quantified by real-time PCR using an ABI 7900 Sequence Detector System as described in (Tittel-Elmer et al., 2009). Primer sequences are listed in Table S3. For ChIP-chip, DNA was amplified, labeled and hybridized as described in (Crabbe et al., 2010). Immunoprecipitation of BrdU-labeled DNA was performed as described in (Crabbe et al., 2010).
DNA combing, 2D and PFGE gel electrophoresis
DNA combing and two-dimensional gel analysis of replication intermediates were done as described (Tourriere et al., 2005). ssDNA was detected using anti-mouse MAB 3034 (Millipore) and goat anti-mouse coupled to Alexa 546; BrdU was detected with a rat monoclonal antibody (Abcys, clone BU1/75) and goat anti-rat coupled to Alexa 488 (Invitrogen). Images were recorded on a Leica DM6000 microscope. 2D gel were done in triplicate and the intensity of X-spike was quantitated with a PhosphorImager (Typhoon, GE) after Southern blotting, normalized to the amount of linear DNA fragments and expressed relative to sgs1Δ levels. PFGE gel electrophoresis was performed as described (Lengronne et al., 2001). Briefly, yeast cells were embedded in low-melting agarose plugs (5.107 cells/ml) and genomic DNA was extracted. Chromosomes were separated on a Gene Navigator system (GE) for 32 h at 6 V/cm with 60 s pulses, stained with ethidium bromide and transferred to Hybond XL (GE Healthcare). Quantitation of chromosome intensity was performed by Southern blotting and hybridization using radioactive probes specific for 3 representative chromosomes (3, 4 and 14), using a PhosphorImager (Typhoon Trio, GE).
Supplementary Material
Highlights.
-
-
Cohesin is present at replication sites and promotes restart of stalled forks.
-
-
Cohesin accumulation at forks is distinct from its loading at DNA breaks.
-
-
Rad50 structural integrity is essential for cohesin enrichment at replication sites.
Acknowledgements
We thank D. Branzei (IFOM, Milan) for communicating unpublished data. We thank L.S. Symington (Columbia University) and S.M. Gasser (Friedrich Miescher Institute for Biomedical Research) for plasmids and yeast strains. We thank V. Pantesco for the microarray hybridization and scanning. We thank E. Schwob and the DNA combing facility of Montpellier for providing silanized coverslips and the Montpellier RIO Imaging microscopy facility. MTE thanks EMBO for support (EMBO STF 36-2012). Work in the Pasero laboratory is supported by FRM (Equipe FRM) and ANR. Work in the Petrini laboratory is supported by GM56888. JC receives support from AIHS and the Cobb laboratory is funded by CIHR (# MOP-82736) and ACF (# 23575).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
ACCESSION NUMBERS
Microarray data can be obtained from Gene Expression Omnibus with the accession numbers GSE39078, GSE33686 and GSE21014.
SUPPLEMENTAL INFORMATION Supplemental Information includes five figures and three tables.
References
- Allen C, Ashley AK, Hromas R, Nickoloff JA. More forks on the road to replication stress recovery. J Mol Cell Biol. 2011;3:4–12. doi: 10.1093/jmcb/mjq049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson DE, Losada A, Erickson HP, Hirano T. Condensin and cohesin display different arm conformations with characteristic hinge angles. J Cell Biol. 2002;156:419–424. doi: 10.1083/jcb.200111002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bermejo R, Doksani Y, Capra T, Katou YM, Tanaka H, Shirahige K, Foiani M. Top1- and Top2-mediated topological transitions at replication forks ensure fork progression and stability and prevent DNA damage checkpoint activation. Genes Dev. 2007;21:1921–1936. doi: 10.1101/gad.432107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard P, Drogat J, Maure JF, Dheur S, Vaur S, Genier S, Javerzat JP. A screen for cohesion mutants uncovers Ssl3, the fission yeast counterpart of the cohesin loading factor Scc4. Curr Biol. 2006;16:875–881. doi: 10.1016/j.cub.2006.03.037. [DOI] [PubMed] [Google Scholar]
- Blat Y, Kleckner N. Cohesins bind to preferential sites along yeast chromosome III, with differential regulation along arms versus the centric region. Cell. 1999;98:249–259. doi: 10.1016/s0092-8674(00)81019-3. [DOI] [PubMed] [Google Scholar]
- Branzei D, Foiani M. Template switching: from replication fork repair to genome rearrangements. Cell. 2007;131:1228–1230. doi: 10.1016/j.cell.2007.12.007. [DOI] [PubMed] [Google Scholar]
- Calzada A, Hodgson B, Kanemaki M, Bueno A, Labib K. Molecular anatomy and regulation of a stable replisome at a paused eukaryotic DNA replication fork. Genes Dev. 2005;19:1905–1919. doi: 10.1101/gad.337205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciosk R, Shirayama M, Shevchenko A, Tanaka T, Toth A, Nasmyth K. Cohesin’s binding to chromosomes depends on a separate complex consisting of Scc2 and Scc4 proteins. Mol Cell. 2000;5:243–254. doi: 10.1016/s1097-2765(00)80420-7. [DOI] [PubMed] [Google Scholar]
- Cobb JA, Bjergbaek L, Shimada K, Frei C, Gasser SM. DNA polymerase stabilization at stalled replication forks requires Mec1 and the RecQ helicase Sgs1. EMBO J. 2003;22:4325–4336. doi: 10.1093/emboj/cdg391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobb JA, Schleker T, Rojas V, Bjergbaek L, Tercero JA, Gasser SM. Replisome instability, fork collapse, and gross chromosomal rearrangements arise synergistically from Mec1 kinase and RecQ helicase mutations. Genes Dev. 2005;19:3055–3069. doi: 10.1101/gad.361805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortes-Ledesma F, Aguilera A. Double-strand breaks arising by replication through a nick are repaired by cohesin-dependent sister-chromatid exchange. EMBO Rep. 2006;7:919–926. doi: 10.1038/sj.embor.7400774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzo V, Robertson K, Bibikova M, Kim E, Grieco D, Gottesman M, Carroll D, Gautier J. Mre11 protein complex prevents double-strand break accumulation during chromosomal DNA replication. Mol Cell. 2001;8:137–147. doi: 10.1016/s1097-2765(01)00294-5. [DOI] [PubMed] [Google Scholar]
- Crabbe L, Thomas A, Pantesco V, De Vos J, Pasero P, Lengronne A. Analysis of replication profiles reveals key role of RFC-Ctf18 in yeast replication stress response. Nat Struct Mol Biol. 2010;17:1391–1397. doi: 10.1038/nsmb.1932. [DOI] [PubMed] [Google Scholar]
- D’Amours D, Jackson SP. The yeast Xrs2 complex functions in S phase checkpoint regulation. Genes Dev. 2001;15:2238–2249. doi: 10.1101/gad.208701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jager M, Dronkert ML, Modesti M, Beerens CE, Kanaar R, van Gent DC. DNA-binding and strand-annealing activities of human Mre11: implications for its roles in DNA double-strand break repair pathways. Nucleic Acids Res. 2001;29:1317–1325. doi: 10.1093/nar/29.6.1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dzikiewicz-Krawczyk A. The importance of making ends meet: mutations in genes and altered expression of proteins of the MRN complex and cancer. Mutat Res. 2008;659:262–273. doi: 10.1016/j.mrrev.2008.05.005. [DOI] [PubMed] [Google Scholar]
- Furuse M, Nagase Y, Tsubouchi H, Murakami-Murofushi K, Shibata T, Ohta K. Distinct roles of two separable in vitro activities of yeast Mre11 in mitotic and meiotic recombination. EMBO J. 1998;17:6412–6425. doi: 10.1093/emboj/17.21.6412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groth P, Auslander S, Majumder MM, Schultz N, Johansson F, Petermann E, Helleday T. Methylated DNA causes a physical block to replication forks independently of damage signalling, O(6)-methylguanine or DNA single-strand breaks and results in DNA damage. J Mol Biol. 2010;402:70–82. doi: 10.1016/j.jmb.2010.07.010. [DOI] [PubMed] [Google Scholar]
- Heidinger-Pauli JM, Unal E, Guacci V, Koshland D. The kleisin subunit of cohesin dictates damage-induced cohesion. Mol Cell. 2008;31:47–56. doi: 10.1016/j.molcel.2008.06.005. [DOI] [PubMed] [Google Scholar]
- Hirano T. At the heart of the chromosome: SMC proteins in action. Nat Rev Mol Cell Biol. 2006;7:311–322. doi: 10.1038/nrm1909. [DOI] [PubMed] [Google Scholar]
- Hohl M, Kwon Y, Galvan SM, Xue X, Tous C, Aguilera A, Sung P, Petrini JH. The Rad50 coiled-coil domain is indispensable for Mre11 complex functions. Nat Struct Mol Biol. 2011;18:1124–1131. doi: 10.1038/nsmb.2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopfner KP, Craig L, Moncalian G, Zinkel RA, Usui T, Owen BA, Karcher A, Henderson B, Bodmer JL, McMurray CT, et al. The Rad50 zinc-hook is a structure joining Mre11 complexes in DNA recombination and repair. Nature. 2002;418:562–566. doi: 10.1038/nature00922. [DOI] [PubMed] [Google Scholar]
- Hopfner KP, Karcher A, Shin DS, Craig L, Arthur LM, Carney JP, Tainer JA. Structural biology of Rad50 ATPase: ATP-driven conformational control in DNA double-strand break repair and the ABC-ATPase superfamily. Cell. 2000;101:789–800. doi: 10.1016/s0092-8674(00)80890-9. [DOI] [PubMed] [Google Scholar]
- Huang KN, Symington LS. Mutation of the gene encoding protein kinase C 1 stimulates mitotic recombination in Saccharomyces cerevisiae. Mol Cell Biol. 1994;14:6039–6045. doi: 10.1128/mcb.14.9.6039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwaizumi M, Shinmura K, Mori H, Yamada H, Suzuki M, Kitayama Y, Igarashi H, Nakamura T, Suzuki H, Watanabe Y, et al. Human Sgo1 downregulation leads to chromosomal instability in colorectal cancer. Gut. 2009;58:249–260. doi: 10.1136/gut.2008.149468. [DOI] [PubMed] [Google Scholar]
- Katou Y, Kanoh Y, Bando M, Noguchi H, Tanaka H, Ashikari T, Sugimoto K, Shirahige K. S-phase checkpoint proteins Tof1 and Mrc1 form a stable replication-pausing complex. Nature. 2003;424:1078–1083. doi: 10.1038/nature01900. [DOI] [PubMed] [Google Scholar]
- Lengronne A, Katou Y, Mori S, Yokobayashi S, Kelly GP, Itoh T, Watanabe Y, Shirahige K, Uhlmann F. Cohesin relocation from sites of chromosomal loading to places of convergent transcription. Nature. 2004;430:573–578. doi: 10.1038/nature02742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengronne A, McIntyre J, Katou Y, Kanoh Y, Hopfner KP, Shirahige K, Uhlmann F. Establishment of sister chromatid cohesion at the S. cerevisiae replication fork. Mol Cell. 2006;23:787–799. doi: 10.1016/j.molcel.2006.08.018. [DOI] [PubMed] [Google Scholar]
- Lengronne A, Pasero P, Bensimon A, Schwob E. Monitoring S phase progression globally and locally using BrdU incorporation in TK(+) yeast strains. Nucleic Acids Res. 2001;29:1433–1442. doi: 10.1093/nar/29.7.1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundin C, North M, Erixon K, Walters K, Jenssen D, Goldman AS, Helleday T. Methyl methanesulfonate (MMS) produces heat-labile DNA damage but no detectable in vivo DNA double-strand breaks. Nucleic Acids Res. 2005;33:3799–3811. doi: 10.1093/nar/gki681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLellan JL, O’Neil NJ, Barrett I, Ferree E, van Pel DM, Ushey K, Sipahimalani P, Bryan J, Rose AM, Hieter P. Synthetic lethality of cohesins with PARPs and replication fork mediators. PLoS Genet. 2012;8:e1002574. doi: 10.1371/journal.pgen.1002574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaelis C, Ciosk R, Nasmyth K. Cohesins: chromosomal proteins that prevent premature separation of sister chromatids. Cell. 1997;91:35–45. doi: 10.1016/s0092-8674(01)80007-6. [DOI] [PubMed] [Google Scholar]
- Mozlin AM, Fung CW, Symington LS. Role of the Saccharomyces cerevisiae Rad51 paralogs in sister chromatid recombination. Genetics. 2008;178:113–126. doi: 10.1534/genetics.107.082677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasmyth K, Haering CH. Cohesin: its roles and mechanisms. Annu Rev Genet. 2009;43:525–558. doi: 10.1146/annurev-genet-102108-134233. [DOI] [PubMed] [Google Scholar]
- Osborn AJ, Elledge SJ. Mrc1 is a replication fork component whose phosphorylation in response to DNA replication stress activates Rad53. Genes Dev. 2003;17:1755–1767. doi: 10.1101/gad.1098303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paull TT, Gellert M. Nbs1 potentiates ATP-driven DNA unwinding and endonuclease cleavage by the Mre11/Rad50 complex. Genes Dev. 1999;13:1276–1288. doi: 10.1101/gad.13.10.1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poli J, Tsaponina O, Crabbe L, Keszthelyi A, Pantesco V, Chabes A, Lengronne A, Pasero P. dNTP pools determine fork progression and origin usage under replication stress. EMBO J. 2012;31:883–894. doi: 10.1038/emboj.2011.470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray Chaudhuri A, Hashimoto Y, Herrador R, Neelsen KJ, Fachinetti D, Bermejo R, Cocito A, Costanzo V, Lopes M. Topoisomerase I poisoning results in PARP-mediated replication fork reversal. Nat Struct Mol Biol. 2012;19:417–423. doi: 10.1038/nsmb.2258. [DOI] [PubMed] [Google Scholar]
- Rolef Ben-Shahar T, Heeger S, Lehane C, East P, Flynn H, Skehel M, Uhlmann F. Eco1-dependent cohesin acetylation during establishment of sister chromatid cohesion. Science. 2008;321:563–566. doi: 10.1126/science.1157774. [DOI] [PubMed] [Google Scholar]
- Sjogren C, Nasmyth K. Sister chromatid cohesion is required for postreplicative double-strand break repair in Saccharomyces cerevisiae. Curr Biol. 2001;11:991–995. doi: 10.1016/s0960-9822(01)00271-8. [DOI] [PubMed] [Google Scholar]
- Strom L, Karlsson C, Lindroos HB, Wedahl S, Katou Y, Shirahige K, Sjogren C. Postreplicative formation of cohesion is required for repair and induced by a single DNA break. Science. 2007;317:242–245. doi: 10.1126/science.1140649. [DOI] [PubMed] [Google Scholar]
- Strom L, Lindroos HB, Shirahige K, Sjogren C. Postreplicative recruitment of cohesin to double-strand breaks is required for DNA repair. Mol Cell. 2004;16:1003–1015. doi: 10.1016/j.molcel.2004.11.026. [DOI] [PubMed] [Google Scholar]
- Strom L, Sjogren C. Chromosome segregation and double-strand break repair - a complex connection. Curr Opin Cell Biol. 2007;19:344–349. doi: 10.1016/j.ceb.2007.04.003. [DOI] [PubMed] [Google Scholar]
- Terret ME, Sherwood R, Rahman S, Qin J, Jallepalli PV. Cohesin acetylation speeds the replication fork. Nature. 2009;462:231–234. doi: 10.1038/nature08550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tittel-Elmer M, Alabert C, Pasero P, Cobb JA. The MRX complex stabilizes the replisome independently of the S phase checkpoint during replication stress. EMBO J. 2009;28:1142–1156. doi: 10.1038/emboj.2009.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomonaga T, Nagao K, Kawasaki Y, Furuya K, Murakami A, Morishita J, Yuasa T, Sutani T, Kearsey SE, Uhlmann F, et al. Characterization of fission yeast cohesin: essential anaphase proteolysis of Rad21 phosphorylated in the S phase. Genes Dev. 2000;14:2757–2770. doi: 10.1101/gad.832000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tourriere H, Pasero P. Maintenance of fork integrity at damaged DNA and natural pause sites. DNA Repair (Amst) 2007;6:900–913. doi: 10.1016/j.dnarep.2007.02.004. [DOI] [PubMed] [Google Scholar]
- Tourriere H, Versini G, Cordon-Preciado V, Alabert C, Pasero P. Mrc1 and Tof1 promote replication fork progression and recovery independently of Rad53. Mol Cell. 2005;19:699–706. doi: 10.1016/j.molcel.2005.07.028. [DOI] [PubMed] [Google Scholar]
- Uhlmann F, Lottspeich F, Nasmyth K. Sister-chromatid separation at anaphase onset is promoted by cleavage of the cohesin subunit Scc1. Nature. 1999;400:37–42. doi: 10.1038/21831. [DOI] [PubMed] [Google Scholar]
- Unal E, Arbel-Eden A, Sattler U, Shroff R, Lichten M, Haber JE, Koshland D. DNA damage response pathway uses histone modification to assemble a double-strand break-specific cohesin domain. Mol Cell. 2004;16:991–1002. doi: 10.1016/j.molcel.2004.11.027. [DOI] [PubMed] [Google Scholar]
- Unal E, Heidinger-Pauli JM, Koshland D. DNA double-strand breaks trigger genome-wide sister-chromatid cohesion through Eco1 (Ctf7) Science. 2007;317:245–248. doi: 10.1126/science.1140637. [DOI] [PubMed] [Google Scholar]
- Unal E, Heidinger-Pauli JM, Kim W, Guacci V, Onn I, Gygi SP, Koshland DE. A molecular determinant for the establishment of sister chromatid cohesion. Science. 2008;321:566–569. doi: 10.1126/science.1157880. [DOI] [PubMed] [Google Scholar]
- Usui T, Ohta T, Oshiumi H, Tomizawa J, Ogawa H, Ogawa T. Complex formation and functional versatility of Mre11 of budding yeast in recombination. Cell. 1998;95:705–716. doi: 10.1016/s0092-8674(00)81640-2. [DOI] [PubMed] [Google Scholar]
- Vanoli F, Fumasoni M, Szakal B, Maloisel L, Branzei D. Replication and recombination factors contributing to recombination-dependent bypass of DNA lesions by template switch. PLoS Genet. 2010;6:e1001205. doi: 10.1371/journal.pgen.1001205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Versini G, Comet I, Wu M, Hoopes L, Schwob E, Pasero P. The yeast Sgs1 helicase is differentially required for genomic and ribosomal DNA replication. EMBO J. 2003;22:1939–1949. doi: 10.1093/emboj/cdg180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams RS, Moncalian G, Williams JS, Yamada Y, Limbo O, Shin DS, Groocock LM, Cahill D, Hitomi C, Guenther G, et al. Mre11 dimers coordinate DNA end bridging and nuclease processing in double-strand-break repair. Cell. 2008;135:97–109. doi: 10.1016/j.cell.2008.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams RS, Williams JS, Tainer JA. Mre11-Rad50-Nbs1 is a keystone complex connecting DNA repair machinery, double-strand break signaling, and the chromatin template. Biochem Cell Biol. 2007;85:509–520. doi: 10.1139/O07-069. [DOI] [PubMed] [Google Scholar]
- Wiltzius JJ, Hohl M, Fleming JC, Petrini JH. The Rad50 hook domain is a critical determinant of Mre11 complex functions. Nat Struct Mol Biol. 2005;12:403–407. doi: 10.1038/nsmb928. [DOI] [PubMed] [Google Scholar]
- Wu N, Yu H. The Smc complexes in DNA damage response. Cell Biosci. 2012;2:5. doi: 10.1186/2045-3701-2-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Tomaszewski JM, McKay MJ. Can corruption of chromosome cohesion create a conduit to cancer? Nat Rev Cancer. 2011a;11:199–210. doi: 10.1038/nrc3018. [DOI] [PubMed] [Google Scholar]
- Xu H, Yan M, Patra J, Natrajan R, Yan Y, Swagemakers S, Tomaszewski JM, Verschoor S, Millar EK, van der Spek P, et al. Enhanced RAD21 cohesin expression confers poor prognosis and resistance to chemotherapy in high grade luminal, basal and HER2 breast cancers. Breast Cancer Res. 2011b;13:R9. doi: 10.1186/bcr2814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yabuki N, Terashima H, Kitada K. Mapping of early firing origins on a replication profile of budding yeast. Genes Cells. 2002;7:781–789. doi: 10.1046/j.1365-2443.2002.00559.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.