Abstract
The amino acid Glutamine is converted into Glutamate by a deamidation reaction catalyzed by the enzyme Glutaminase (GLS). Two isoforms of this enzyme have been described, and the GLS2 isoform is regulated by the tumor suppressor gene p53. Here, we show that the p53 family member TAp73 also drives the expression of GLS2. Specifically, we demonstrate that TAp73 regulates GLS2 during retinoic acid-induced terminal neuronal differentiation of neuroblastoma cells, and overexpression or inhibition of GLS2 modulates neuronal differentiation and intracellular levels of ATP. Moreover, inhibition of GLS activity, by removing Glutamine from the growth medium, impairs in vitro differentiation of cortical neurons. Finally, expression of GLS2 increases during mouse cerebellar development. Although, p73 is dispensable for the in vivo expression of GLS2, TAp73 loss affects GABA and Glutamate levels in cortical neurons. Together, these findings suggest a role for GLS2 acting, at least in part, downstream of p73 in neuronal differentiation and highlight a possible role of p73 in regulating neurotransmitter synthesis.
Keywords: p73, GLS2, neuronal differentiation, apoptosis, p53 family, metabolism, neurotransmitter
Introduction
Glutamine (Gln) is an amino acid that plays key roles in many metabolic pathways.1,2 Mammalian glutaminase (GLS) is an enzyme responsible for the hydrolytic deamidation of Gln into Glutamate (Glu) and ammonium ions. Two paralogous genes on separate chromosomes encode distinct GLS isozymes: the kidney-type isozyme, GLS1, and the liver-type isozyme, GLS2.3,4 Although early studies showed that GLS1 is ubiquitously expressed,2 and that GLS2 is mainly expressed in the liver,4 more recent observations indicate that the expression pattern is more complex. Indeed, GLS2 has also been detected in brain,5 pancreas,6 cancer cells,7 and cells of the immune system.8 Both isoforms have been identified in human and mouse brain. At the subcellular level, GLS2 localizes to the inner mitochondrial membrane, while in neurons it has been observed in the nucleus.5 The abundance of a particular glutaminase mRNA species may significantly change depending upon the tissue type and the developmental or metabolic state of the tissue; therefore, each transcript may represent a specific target for different stimuli. However, although GLS distribution has been extensively investigated, the regulation of GLS1/GLS2 expression is poorly understood. Recently, 2 independent groups have shown that GLS2 is a direct target of the tumor suppressor gene p53,9,10 and that p53 drives the expression of GLS2 under both non-stressed and stressed conditions. Increased levels of GLS2 promote Gln metabolism, and the Glu produced by Gln hydrolysis is a precursor in the biosynthesis of Glutathione (GSH) and therefore promotes antioxidant defense through regulating the GSH/GSSG ratio. Moreover, Glu can be further transformed into α-ketoglutarate, which feeds the tricarboxylic acid cycle, resulting in an increase of mitochondrial respiration and ATP production. Therefore, overall, GLS2 plays a key role in energy metabolism and antioxidant defense.
p73 is a pleiotropic protein that belongs to the p53 family.11-15 It is involved in several biological processes, such as cell death,16-19 differentiation,20-22 neuronal stem cell maintenance,23-26 aging,27-29 and metabolism27 through the regulation of gene30-32 and microRNA expression. Mice deficient in p73 display neuronal pathologies, including hydrocephalus and hippocampal dysgenesis, with defects in the CA1-CA3 pyramidal cell layers and the dentate gyrus.33,34 Moreover, they also have a reduction in cortical thickness as a consequence of loss of mature cortical neurons.35 Recently, it has been demonstrated that TAp73 expression increases in parallel with neuronal differentiation, and its ectopic expression induces neurite outgrowth and expression of neuronal markers in neuroblastoma cell lines, suggesting that it has a pro-differentiation role.20 In particular, TAp73 acts, at least in part, through microRNA-34a to regulate neuronal differentiation. Here, we demonstrate that, like p53, p73 is able to drive the expression of GLS2. We show that GLS2 expression itself induces neuronal differentiation in a neuroblastoma (NB) cell line, and that modulation of GLS2 influences retinoic acid (RA)-induced NB differentiation and spontaneous differentiation of cortical neurons in vitro. These data suggest that GLS2 is one mechanism mediating the neuronal effects of p73.
Results
TAp73 regulates the expression of GLS2
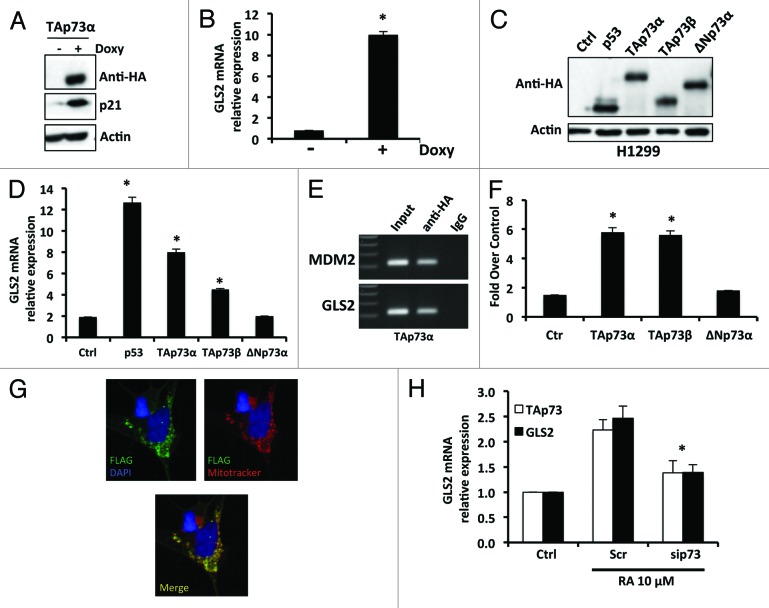
Since both TAp73 and GLS2 are expressed in the brain, and because of the extensive functional similarities between the p53 family of proteins, we asked whether TAp73, like p53, is able to regulate GLS2 expression. We first employed a SAOS2-TAp73α inducible cell line treated with Doxycyclin and extracted total RNA after 24 h induction. As expected, the expression of TAp73α was induced as well as that of p21, a well-known TAp73 target (Fig. 1A). In this cellular context, we also observed a significant upregulation of GLS2 mRNA (Fig. 1B). To confirm that TAp73 is able to regulate the expression of GLS2, we transiently expressed different isoforms of TAp73 in H1299 cells (Fig. 1C). As shown in Figure 1D, GLS2 mRNA was significantly induced (by 6–8-fold) after ectopic expression of TAp73α and β. No induction of GLS2 expression was observed in cells overexpressing ΔNp73α. Next, we asked if GLS2 is directly regulated by TAp73. We first analyzed the human GLS2 promoter and identified a p53 consensus DNA-binding element located at −787 with respect to the transcription start site. In order to verify if TAp73α was able to bind this putative responsive element, we performed a chromatin immunoprecipitation assay (ChIP). As shown in Figure 1E, TAp73 binds the GLS2 promoter containing the p53-responsive element. To confirm that this binding was functionally significant, we transfected TAp73 with a GLS2-luciferase reporter construct. Expression of TAp73α or β resulted in an approximately 6-fold enhancement of luciferase activity (Fig. 1F). To further confirm the ability of TAp73 to modulate the expression of GLS2, we used an in vitro model of neuronal terminal differentiation, in which TAp73 has already been shown to be a positive regulator.20,22 First, we characterized our cellular system for the intracellular localization of GLS2 and expression of the GLS isoforms. In SH-SH5Y cells, GLS2 localizes in the mitochondria (Pearson’s correlation coefficient 0.69) as shown in Figure 1G and Figure S1A. No expression of GLS2 was detected at the nuclear level. Moreover, we have found that GLS1 is also expressed in SH-SY5Y cells (Fig. S1B). Then, we treated SH-SY5Y cells with retinoic acid (RA) in order to induce terminal differentiation and isolated total RNA. As shown in Figure 1H, GLS2 RNA expression was increased after treatment with RA. Moreover, this increase in GLS2 expression induced by RA was abolished when TAp73 expression was inhibited by RNA interference (Fig. 1H).
Figure 1. TAp73 drives the expression of GLS2. (A) SAOS-2-TAp73α inducible cell lines were treated with Doxycyclin (Doxy) for 24 h in order to overexpress the human TAp73α protein, and endogenous levels of GLS2 were assessed by real-time PCR (B). Induction of TAp73α led to a significant (P < 0.05) increase of GLS2 expression, as evaluated by real-time PCR. (C) H1299 cells were transfected with the indicated plasmids and expression of GLS2 was evaluated by real-time PCR as in (D). (E) TAp73 binds to the promoter of GLS2 as shown by ChIP. (F) TAp73 activates the GLS2 promoter as evaluated by luciferase activity. Co-transfection of a Renilla luciferase control plasmid was used to normalize the transfection efficiency. (G) Exogenous GLS2 localize in the mitochondria. SH-SY5Y were transfected with FLAG-GLS2 expressing vector and after 24 h stained with MitoTracker® Red CMXRos and antibody against FLAG epitope as described in “Materials and Methods”. A representative micrograph is shown. Magnification 40×. (H) TAp73 regulates GLS2 expression during neuronal terminal differentiation of neuroblastoma cells. SH-SY5Y cells were treated with 10 μM retinoic acid in order to induce differentiation. Retinoic acid (RA) treatment induces the expression of GLS2. Inhibition of TAp73 expression induced by retinoic acid prevents the upregulation of GLS2. Real-time PCR data are normalized to the housekeeping gene GAPDH and relative to control (Ctrl) Data represent mean ± s.d. of 3 different experiments. *P < 0.05
Together these results demonstrate that TAp73 directly regulates the expression of GLS2 and suggest a potential role for GLS2 in the regulation of RA-induced terminal differentiation of neuroblastoma cells.
GLS2 expression contributes to the terminal neuronal differentiation of neuroblastoma cell lines
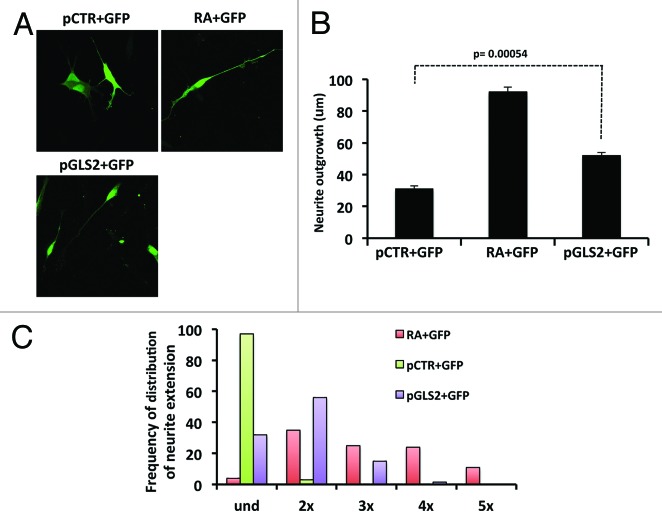
Since we have shown that RA induces the expression of GLS2 via a TAp73-dependent mechanism, we asked whether GLS2 could itself play a role in the terminal neuronal differentiation of NB cells. To assess this, we overexpressed GLS2 together with GFP in SH-SY5Y cells (Fig. 2A) and, after 24 h, evaluated neurite outgrowth. Figure 2B shows that overexpression of GLS2 (pGLS2) significantly increases neurite outgrowth though not to the same extent as RA. Transfection of GFP (C+GFP) or GFP+empty vector (pCTR+GFP) had no effect. Moreover, as shown in Figure 2C, cells overexpressing GLS2 show a high frequency of neurites twice, and to a lesser extent, 3 times the length of the soma. GFP+empty vector transfected cells remained in an undifferentiated state (und).
Figure 2. Ectopic expression of GLS2 increases neurite outhgrowth. (A) Representative images of neurite outgrowth in the SHSY-5Y cell line. Cells were transfected with the indicated plasmids and either left untreated or treated with RA. A representative micrograph is shown. Magnification 40× (B) After 48 h cells were fixed and analyzed for neurite extension as described in “Materials and Methods”. (C) SH-SY5Y cells overexpressing GLS2 (violet columns) have neurites 2–3 times longer than the cell body. Data represent mean ± s.d. of 3 different experiments.
Role of endogenous GLS2 in neuronal differentiation
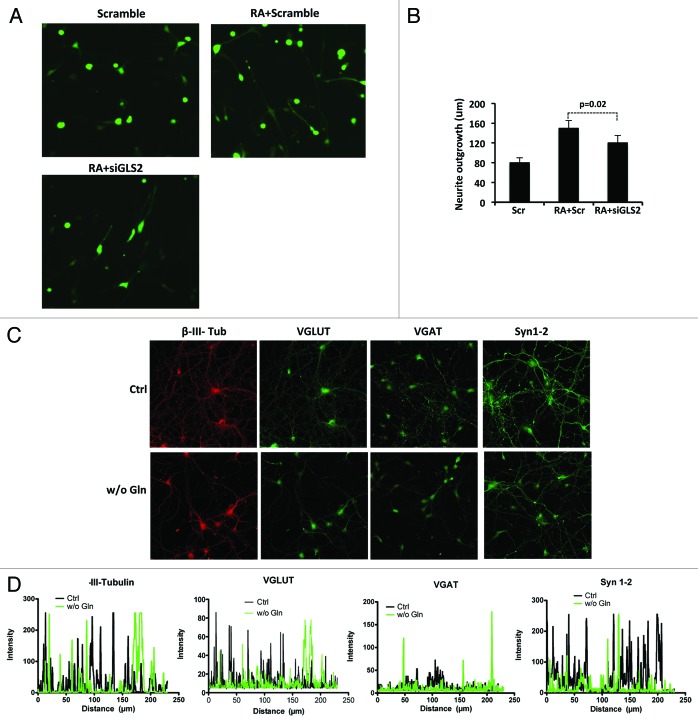
To assess whether endogenous GLS2 also contributes to neuronal differentiation, we knocked down GLS2 expression in SH-SY5Y cells treated with RA (Fig. 3A; Fig. S1C). Figure 3B shows that the inhibition of GLS2 expression resulted in a modest but significant (P = 0.02) reduction of the neurite extension induced by RA at 24 h. This small effect may be due to the compensatory effect of the concomitant expression of the GLS1 isoform in SH-SY5Y cells as mentioned above. To better understand, in a more physiological context, the role of GLS isoforms in neuronal differentiation, we functionally inhibited GLS activity by Gln withdrawal during in vitro differentiation of cortical neurons. Primary cortical neurons were plated, and after 24 h the complete medium was replaced with medium without Gln. After 6 days cortical neurons with or without Gln were processed for immunocytochemistry. As shown in Figure 3C and D, deprivation of Gln results in a reduction in the number of neurites, as shown by β-III-Tubulin staining. This reduction was confirmed by staining the cortical neurons for the specific synaptic proteins VGAT, VGLUT, and Synapsin 1/2. No significant differences in apoptosis were detected between cortical neurons in normal medium and in medium without Gln (Fig. S2).36
Figure 3. Inhibition of endogenous levels of GLS2 reduce neurite outgrowth induced by RA. (A) Representative images of neurite outgrowth in the SHSY-5Y cell line. Cells were transfected with scramble + GFP or siGLS2 + GFP and untreated or treated with retinoic acid (RA). (B) After 48 h cells were fixed and analyzed for neurite extension as described in Methods. Data represent mean ± s.d. of 3 different experiments. Glutamine withdrawal impairs in vitro terminal differentiation of cortical neurons. (C) Cortical neurons were cultured either in the presence or absence of glutamine and fixed and stained for the indicated proteins after 7 days. Magnification 40×. (D) Quantification of fluorescence intensity of the neuronal marker β-III-Tubulin and of the indicated synaptic proteins was performed using Zeiss LSM 510 software analysis.
Taken together, these observations indicate that GLS2 participates in the regulation of neuronal differentiation in both RA-treated NB cells and in cortical neurons.
Metabolic function of GLS2 in NB cells
It is well known that GLS2 plays a key role in energy metabolism and antioxidant defense. Indeed, it regulates ATP levels and the GSH/GSSG ratio in cells.9,10,37 Hence, we asked whether GLS2 could also exert the same functions in SH-SY5Y cells. First, we overexpressed GLS2 (Fig. 4A) and measured the intracellular levels of ATP. As shown in Figure 4C (left hand bars), the ectopic expression of GLS2 led to a significant increase (about +40%) of ATP intracellular levels. In contrast, when we knocked down GLS2 expression by siRNA (Fig. 4B), we observed a significant reduction (about −20%) of ATP levels. The same results were obtained when we inhibited the activity of GLS2 with the glutamine antagonist DON (Fig. 4C). However, the data in Figure 4D show that neither overexpression nor knockdown of GLS2 significantly effects the GSH/GSSG ratio.
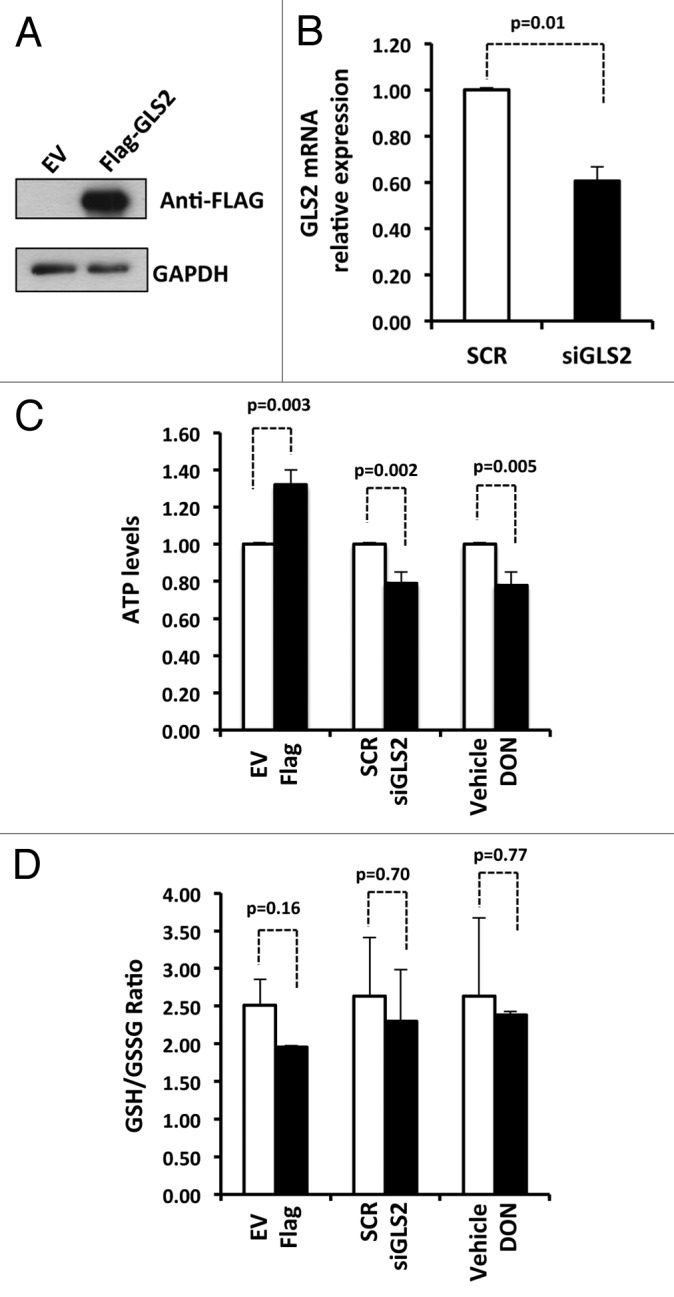
Figure 4. GLS2 regulates intracellular levels of ATP. (A) A representative western blot showing expression of GLS2 after transfection of SHSY-5Y cells with empty vector (EV) or FLAG-GLS2. (B) Real-time PCR showing inhibition of GLS2 expression after transfection of SHSY-5Y cells with short interfering RNA against GLS2 (siGLS2). Scramble (SCR). Real-time PCR data are normalized to the housekeeping gene GAPDH relative to scramble (SCR). Data represent mean ± s.d. of 3 different experiments. (C) Intracellular levels of ATP in SHSY-5Y transfected or treated as indicated were measured as described in “Materials and Methods”. The inhibitor of Glutaminase DON (6-Diazo-5-oxo-L-norleucine) was used at 200 μM. (D) The intracellular levels of GSH and GSSG are not affected by GLS2 in SHSY-5Y cells. Ratio of GSH/GSSG was measured as described in “Materials and Methods”. Data represent mean ± s.d. of 3 different experiments.
Expression of GLS2 during neuronal development
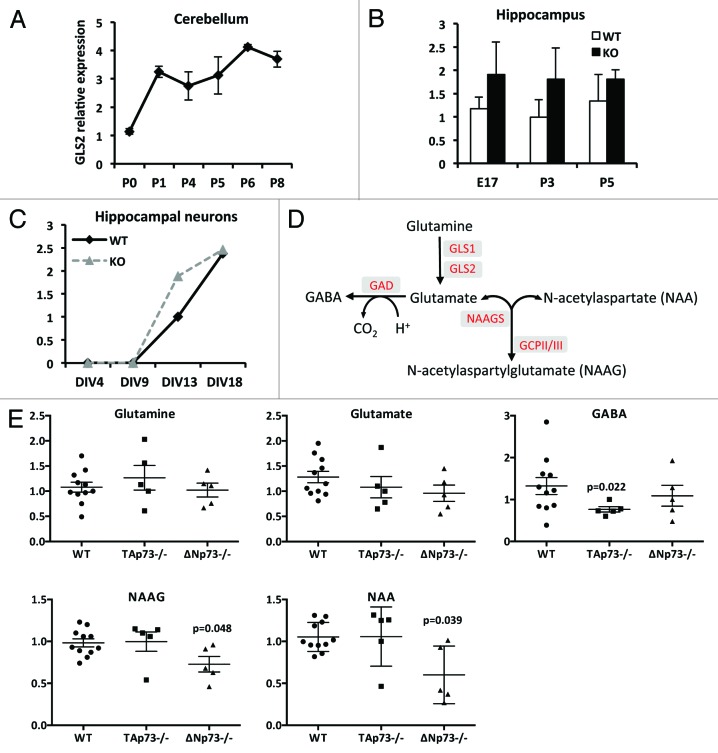
To further investigate whether GLS2 is involved in neuronal differentiation in vivo, we monitored its expression during cerebellar development. We collected mouse cerebellum at different stages (from postnatal day [P] 0 to P8), extracted total RNA, and used qPCR to assess the level of GLS2 expression. Figure 5A shows that the expression of GLS2 increases from P0 to P1 and then remains constant at least until P8.
Figure 5. In vivo expression of GLS2. (A) GLS2 expression increases during mouse cerebellar development. The cerebellum was isolated from wild-type mice (n = 4 per each point) and expression of GLS2 was evaluated during cerebellar development by real-time PCR at birth (P0) and subsequent postnatal (P) days. (B) GLS2 expression in the hippocampus dissected from wild-type and p73−/− mice at the indicated age (E, embryo; P, postnatal). (C) GLS2 expression during in vitro terminal differentiation of hippocampal neurons derived from wild-type and p73−/− mice. Real-time data are normalized to the housekeeping gene GAPDH relative. Data represent mean ± s.d. of 3 different experiments. (D) Schematic representation of neurotransmitters biosynthesis. In red the enzymes involved in each reaction. GLS, Glutaminase; GAD, Glutamic acid decarboxylase; NAGGS, N-acetylglutamate synthase; GCP, glutamate carboxypeptidase. (E) Effect of p73 deficiency on intracellular levels of neurotransmitters. Cortical neurons derived from wild-type (WT), TAp73−/− and ΔNp73−/− were harvested after 7 days of culture and subjected to mass spectrometry analysis as described in “Materials and Methods”.
Our in vitro data prompted us to explore whether TAp73 also regulated the expression of GLS2 in this in vivo context. To do this, we first isolated RNA from hippocampus obtained from wild-type (WT) and p73−/− mice at different ages and monitored the expression of GLS2. As shown in Figure 5B, no differences in the expression of GLS2 mRNA were found between WT and p73−/− mice. The same results were obtained when we compared the expression of GLS2 during the in vitro differentiation of hippocampal neurons. Indeed, as shown in Figure 5C, GLS2 expression appears to increase during the differentiation of p73−/− hippocampal neurons, at least at some time points.
Although our results suggest that p73 is not required for the in vivo expression of GLS2, we sought to investigate whether the absence of p73 could affect glutamate metabolism (Fig. 5D). Cortical neurons derived from WT, TAp73−/−, and ΔNp73−/− mice were plated, harvested after 7 days, and subjected to mass spectrometry analysis. Interestingly, as shown in Figure 5E, cortical neurons from the TAp73−/− appeared to have reduced levels of GABA, which may be driven by slightly lower levels of glutamate. However, aspartate, which is also transported by glutamine transporters, and levels of N-acetylaspartylglutamate (NAAG), are unchanged in the TAp73−/− neurons. Thus, reduction of GABA in these neurons is more likely to be a result of altered glutamic acid decarboxylase (GAD) activity. Conversely, NAAG and its precursor N-acetylaspartate (NAA), as well as the inhibitory neurotransmitter glycine (Fig. S3A), were reduced in ΔNp73−/− neurons at DIV 7. Decreased levels of these molecules in p73−/− neurons could result in altered neuronal function or may reflect a delay in neuronal differentiation during in vitro culture.
Glutamate is one of the precursor amino acids in the biosynthesis of GSH. We observed that cortical neurons derived from TAp73−/− mice show a reduction in GSH levels and in the GSH/GSSG ratio (Fig. S3B). However, these differences fail to reach statistical significance.
Discussion
The p53 family is involved in many aspects of development38-41 and, in particular, p73 regulates neuronal differentiation. First, p73 deletion results in neuronal defects, including hydrocephalus and hippocampal dysgenesis, with abnormalities in the CA1-CA3 pyramidal cell layers and in the dentate gyrus.33,34 Second, TAp73 regulates neuronal differentiation of cortical neurons at least in part throughout the transcription of the microRNA, miR-34a.20,42 Third, TAp73 is implicated in the terminal differentiation of NB cells induced by retinoic acid (RA) and which is associated with an increased expression of TAp73. In addition, ectopic expression of TAp73 itself induces terminal neuronal differentiation.22 Recently, we have also demonstrated that TAp73 regulates RA-induced differentiation of NB cells through miR-34a, since RA-induced differentiation of NB cells is inhibited both by knockdown of TAp73 and by antagomiR-34a.20 In this report we show that TAp73 is able to directly regulate the expression of GLS2. Our results suggest that GLS2 is under the control of TAp73 during neuronal differentiation of NB cells induced by retinoic acid, and that GLS2 itself can modulate neuronal differentiation.
GLS2 has recently attracted attention because it is a target of the p53 tumor suppressor gene.9,10 In particular, GLS2 regulates energy metabolism and antioxidant defense in cancer cells. Indeed our results indicate and confirm that GLS2 is a key player in the regulation of, at least, intracellular ATP levels. However, a role in cell differentiation has also been attributed to GLS2.7 Our findings reinforce this possible role of GLS2 in cell differentiation, particularly neuronal differentiation. Thus, GLS2 gain- and loss-of-function experiments result in an increase and decrease of neuronal differentiation, respectively.
Therefore, the role of TAp73 in neuronal differentiation, as revealed by the TAp73-knockout mouse phenotype, is complex and involves several downstream pathways. The demonstration in this report that GLS2, in addition to miR-34a, is a TAp73 target involved in the differentiation of NB cells, and, in particular, that direct manipulation of GLS2 expression itself modulates NB differentiation, and that Gln deprivation influences the differentiation of cortical neurons in vitro, suggests that the neuronal effects of TAp73 are partly due to its effects on metabolism, since p53 family members, including TAp73, have been reported to have metabolic effects43-45 in addition to regulating Gln metabolism. Despite the fact that TAp73 is not essential for the in vivo regulation of GLS2 expression, our results suggest that TAp73 loss affects glutamate metabolism. Indeed, cortical neurons derived from TAp73−/− mice show a reduction in the levels of the neurotransmitters glutamate and GABA. Because neuronal differentiation is a complex biological process that is regulated by intrinsic pathways46-51 and extrinsic signals,52,53 it will be of interest to see whether these other metabolic effects of TAp73 also affect this biological process.
Materials and Methods
Mice
Mice were bred and subjected to listed procedures under the Project License released from the Home Office. TAp73−/− and ΔNp73−/− mice were generated as previously described.40,42
Cell culture and transfection
TAp73α-SAOS-2-Tet-On cells were grown in RPMI, 250 μM L-glutamine (Gibco), penicillin/streptomycin 1 U/ml (Gibco), and 10% Tet-free FCS (Clontech); HEK 293E cells were grown in DMEM high glucose, 10% FBS, penicillin (100 U), streptomycin (100 μg) (Invitrogen); SH-SY5Y cells were maintained in DMEM 10% FCS, penicillin/streptomycin 1 U/ml (Gibco), and H1299 cells were grown in RPMI, 250 μM L-glutamine (Gibco), penicillin/streptomycin 1 U/ml (Gibco), 10% FCS (Invitrogen). For differentiation, SH-SY5Y cells were cultured in DMEM supplemented with 1% FCS and 10 μM RA.
Cells were transfected by Lipofectamine 2000 according to the manufacturer’s protocols (Invitrogen). Human GLS2-expressing vector was kindly provided by Dr Zhaohui Feng (Cancer Institute of New Jersey). Primary cortical neuronal cultures were prepared from E17.5 embryos mouse as previously described.42
RNA extraction and real-time PCR
Total RNA from cells or tissues was isolated using Trizol (Invitrogen) according to the manufacturer's instructions. Total RNA (3 μg) was reverse transcribed using RevertAid H Minus Reverse Transcriptase and oligo(dT) (Thermo Scientific). qRT-PCR was performed in an ABI PRISM 7000 Sequence Detection System (Applied Biosystem) with SYBR green ready mix (Applied Biosystem) and specific primers. The expression of each gene was defined from the threshold cycle (Ct), and relative expression levels were calculated by using the 2−ΔΔCt method after normalization with reference to expression of the housekeeping gene GAPDH. The following primers were used: mouse GAPDH Fwd 5′-CAATGAATAC GGCTACAGCA AC-3′ and mouse GAPDH Rev 5′-AGGGAGATGC TCAGTGTTGG-3′; mouse GLS2 Fwd 5′-AGCGTATCCC TATCCACAAG TTCA-3′ and mouse GLS2 Rev 5′-GCAGTCCAGT GGCCTTCAGA G-3′; human TAp73 Fwd 5′-CTCTGGAGCT CTCTGGAACC A-3′ and human TAp73 Rev 5′-CGCCCACCAC CTCATTATTC-3′; human GLS2 Fwd 5′- TGCCTATAGT GGCGATGTCT CA -3′ and human GLS2 Rev 5′-GTTCCATATC CATGGCTGAC AA -3′; human GAPDH Fwd 5′-AGCCACATCG CTCAGACAC-3′ and human GAPDH Rev 5′-GCCCAATACG ACCAAATCC-3′; human GLS1 Fwd 5′-GCTGTGCTCC ATTGAAGTGA CT-3′; and human GLS1 Rev 5′-TTGGGCAGAA ACCACCATTA G-3′.
Chromatin immunoprecipitation
TAp73α-SaOS-2-inducible cells were crosslinked for 10 min in a solution containing 1% formaldehyde and ChIP assays were performed using a MAGnify ChIP system (Invitrogen), as specified in the manufacturer’s instructions. Cell lysates were sonicated in order to obtain chromatin fragments of ~700 bp. The immune complex was immunoprecipitated using an anti-HA specific antibody (16B12, Covance) and nonspecific IgG as a control. Collected DNA fragments were tested by PCR. The following oligos were used for amplifying the p53 putative responsive elements (RE) found in the GLS2 promoter: Fwd 5′-GGCCTCCCAA GTCACCAGTT CA-3′ Rev 5′-TGTTTTTGCT TGTTTTCGCC TTCT-3′. The p53 RE located on hMDM2, used as a positive control, was amplified with one set of primers: Fwd 5′-GGTTGACTCA GCTTTTCCTC TTG-3′ and Rev 5′-GGAAAATGCA TGGTTTAAAT AGCC-3′ (119 bp).
Luciferase assay
HEK 293E cells were plated in 12-well plates (1 × 105 and 1.5 × 105 per well, respectively). After 24 h, pGL3control vectors (200 ng) were cotransfected with TAp73α, β, and ΔNp73α or empty vector and Renilla luciferase pRL-CMV vector (10 ng), using Lipofectamine 2000. Luciferase activities were measured 24 h after transfection using a Dual Luciferase Reporter Assay System (Promega); light emission was measured over 10 s using an OPTOCOMP I luminometer. Efficiency of transfection was normalized using Renilla luciferase activity.
Western blot
Proteins were extracted with RIPA buffer containing an inhibitor cocktail (Roche) and protein concentration was determined using a Bradford dye-based assay (Biorad). Total protein (30 μg) was subjected to SDS-PAGE followed by immunoblotting with appropriate antibodies at the recommended dilutions. The blots were then incubated with peroxidase-linked secondary antibodies followed by enhanced chemiluminescent detection using the Super Signal chemiluminescence kit (Thermo scientific). Antibodies: HA-HRP (1:5000; Sigma), p21 (1:1000, Santa Cruz), FLAG M2 clone (1:5000; Sigma-Aldrich), and GAPDH (1:10000; Sigma-Aldrich).
Neurite outgrowth assay
SHSY-5Y cells were transfected as indicated using Lipofctamine 2000 and then treated with RA (10 μM) and analyzed 24 h or 48 h later. The projection images were semi-automatically traced with NIH ImageJ using the NeuronJ plugin. The neurite length of each individual GFP positive cell was analyzed.
ATP and GSH/GSSG measurements
Intracellular levels of ATP were measured by using the ELITEN ATP Assay System Bioluminescence Detection Kit (Promega). GSH and GSSG levels were measured by using the GSH/GSSG-Glo Assay (Promega). Briefly, cells were transfected either with FLAG-GLS2 or siGLS2 and after 24 h or 48 h, respectively, harvested and processed according to the manufacturer’s instructions.
Immunofluorescence
SH-SY5Y cells were transfected with human-FLAG-GLS2 and after 24 h were incubated for 30 min at 37 °C with MitoTracker® Red CMXRos (Invitrogen). Then cells were washed twice with warm medium, fixed, and processed for immunofluorescence. SHSY-5Y cells were stained with anti-FLAG (1:1000, Sigma-Aldrich) antibody. Cortical neurons cultured with or without glutamine were fixed with 3% paraformaldehyde in PBS followed by treatment with 0.1% Triton X-100 for 5 min and 10% normal goat serum in PBS for 1 h at RT. Then, cells were stained at 4 °C overnight with the following primary antibodies: neuronal-specific anti-β-III-Tubulin (1:2000; Promega), anti-VGAT (1:500; Synaptic System), anti-VGLUT1 (1:500; Synaptic System), and anti-Synapsin1–2 (1:500; Synaptic System). After 3 washes (10 min in PBS), the cells were incubated with Alexa Fluor conjugated secondary antibody (1:1000; Jackson Immuno Research Laboratories). After 3 washes (10 min, PBS), nuclei were stained with DAPI (1:10 000) for 10 min; this step was followed by a further wash (10 min in PBS). The coverslips were then mounted with Aquapolymount antifading solution (Polysciences) onto glass slides and observed under a confocal microscope (Zeiss LSM 510). Colocalization analysis was performed with Volocity 3D image analysis software version 6.3 (Perkin Elmer).
Metabolomic analysis of cortical neurons
DIV7 cortical neurons were harvested and immediately stored at −80°C and samples were processed as previously described.54,55 Briefly, the sample preparation process was performed using the automated MicroLab STAR® system from the Hamilton Company. Recovery standards were added prior to the first step in the extraction process for QC purposes. Sample preparation was conducted using a proprietary series of organic and aqueous extractions to remove the protein fraction while allowing maximum recovery of small molecules. The resulting extract was divided into 2 fractions: one for analysis by LC and one for analysis by GC. Samples were placed briefly on a TurboVap® (Zymark) to remove the organic solvent. Each sample was then frozen and dried under vacuum. Samples were then prepared for the appropriate instrument either LC/MS or GC/MS. Compounds were identified by comparison to library entries of purified standards or recurrent unknown entities. Identification of known chemical entities was based on comparison to metabolomic library entries of purified standards.
Statistical analysis
All results are expressed as means ± s.d. P < 0.05 was considered significant. For metabolomics analysis we perform Welch 2-sample t tests.
Supplementary Material
Acknowledgments
We thank David J Read for technical support. This work has been supported by the Medical Research Council, UK; grants from “Alleanza contro il Cancro” (ACC12), MIUR/PRIN (20078P7T3K_001)/FIRB (RBIP06LCA9_0023, RBIP06LCA9_0C), AIRC (2011-IG11955), AIRC 5xmille (#9979), Telethon Grant GGPO9133, Min. Salute (Ricerca oncologica 26/07), and IDI-IRCCS (RF06 c.73, RF07 c.57, RF08 c.15, RF07 c.57) to GM.
Glossary
Abbreviations:
- TAp73
transcriptionally active p73
- ΔNp73
amino truncated p73
- Gln
glutamine
- Glu
glutamate
- RA
retinoic acid
- NB
neuroblastoma
- P
postnatal day
- ChIP
chromatin immunoprecipitation
- GFP
green fluorescent protien
- GABA
gamma-aminobutyric acid
- VGAT
vesicular GABA transporter
- VGLUT
vesicular glutamate transporter
- DON
6-diazo-5-oxo-L-norleucine
- DMEM
Dulbecco minimal essential medium
- FBS
foetal bovine serum
- Ct
cycle
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplemental Materials
Supplemental materials may be found here: www.landesbioscience.com/journals/cc/article/26771
Footnotes
Previously published online: www.landesbioscience.com/journals/cc/article/26771
Reference
- 1.Matés JM, Segura JA, Campos-Sandoval JA, Lobo C, Alonso L, Alonso FJ, Márquez J. Glutamine homeostasis and mitochondrial dynamics. Int J Biochem Cell Biol. 2009;41:2051–61. doi: 10.1016/j.biocel.2009.03.003. [DOI] [PubMed] [Google Scholar]
- 2.Kovacevic Z, McGivan JD. Mitochondrial metabolism of glutamine and glutamate and its physiological significance. Physiol Rev. 1983;63:547–605. doi: 10.1152/physrev.1983.63.2.547. [DOI] [PubMed] [Google Scholar]
- 3.Shapiro RA, Farrell L, Srinivasan M, Curthoys NP. Isolation, characterization, and in vitro expression of a cDNA that encodes the kidney isoenzyme of the mitochondrial glutaminase. J Biol Chem. 1991;266:18792–6. [PubMed] [Google Scholar]
- 4.Smith EM, Watford M. Molecular cloning of a cDNA for rat hepatic glutaminase. Sequence similarity to kidney-type glutaminase. J Biol Chem. 1990;265:10631–6. [PubMed] [Google Scholar]
- 5.Olalla L, Gutiérrez A, Campos JA, Khan ZU, Alonso FJ, Segura JA, Márquez J, Aledo JC. Nuclear localization of L-type glutaminase in mammalian brain. J Biol Chem. 2002;277:38939–44. doi: 10.1074/jbc.C200373200. [DOI] [PubMed] [Google Scholar]
- 6.Gómez-Fabre PM, Aledo JC, Del Castillo-Olivares A, Alonso FJ, Núñez De Castro I, Campos JA, Márquez J. Molecular cloning, sequencing and expression studies of the human breast cancer cell glutaminase. Biochem J. 2000;345:365–75. doi: 10.1042/0264-6021:3450365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pérez-Gómez C, Campos-Sandoval JA, Alonso FJ, Segura JA, Manzanares E, Ruiz-Sánchez P, González ME, Márquez J, Matés JM. Co-expression of glutaminase K and L isoenzymes in human tumour cells. Biochem J. 2005;386:535–42. doi: 10.1042/BJ20040996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Castell L, Vance C, Abbott R, Marquez J, Eggleton P. Granule localization of glutaminase in human neutrophils and the consequence of glutamine utilization for neutrophil activity. J Biol Chem. 2004;279:13305–10. doi: 10.1074/jbc.M309520200. [DOI] [PubMed] [Google Scholar]
- 9.Suzuki S, Tanaka T, Poyurovsky MV, Nagano H, Mayama T, Ohkubo S, Lokshin M, Hosokawa H, Nakayama T, Suzuki Y, et al. Phosphate-activated glutaminase (GLS2), a p53-inducible regulator of glutamine metabolism and reactive oxygen species. Proc Natl Acad Sci U S A. 2010;107:7461–6. doi: 10.1073/pnas.1002459107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hu W, Zhang C, Wu R, Sun Y, Levine A, Feng Z. Glutaminase 2, a novel p53 target gene regulating energy metabolism and antioxidant function. Proc Natl Acad Sci U S A. 2010;107:7455–60. doi: 10.1073/pnas.1001006107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dötsch V, Bernassola F, Coutandin D, Candi E, Melino G. p63 and p73, the ancestors of p53. Cold Spring Harb Perspect Biol. 2010;2:a004887. doi: 10.1101/cshperspect.a004887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rufini A, Agostini M, Grespi F, Tomasini R, Sayan BS, Niklison-Chirou MV, Conforti F, Velletri T, Mastino A, Mak TW, et al. p73 in Cancer. Genes Cancer. 2011;2:491–502. doi: 10.1177/1947601911408890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Killick R, Niklison-Chirou M, Tomasini R, Bano D, Rufini A, Grespi F, Velletri T, Tucci P, Sayan BS, Conforti F, et al. p73: a multifunctional protein in neurobiology. Mol Neurobiol. 2011;43:139–46. doi: 10.1007/s12035-011-8172-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McCubrey JA, Demidenko ZN. Recent discoveries in the cycling, growing and aging of the p53 field. Aging (Albany NY) 2012;4:887–93. doi: 10.18632/aging.100529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Amelio I, Grespi F, Annicchiarico-Petruzzelli M, Melino G. p63 the guardian of human reproduction. Cell Cycle. 2012;11:4545–51. doi: 10.4161/cc.22819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gong JG, Costanzo A, Yang HQ, Melino G, Kaelin WG, Jr., Levrero M, Wang JY. The tyrosine kinase c-Abl regulates p73 in apoptotic response to cisplatin-induced DNA damage. Nature. 1999;399:806–9. doi: 10.1038/21690. [DOI] [PubMed] [Google Scholar]
- 17.Muppani N, Nyman U, Joseph B. TAp73alpha protects small cell lung carcinoma cells from caspase-2 induced mitochondrial mediated apoptotic cell death. Oncotarget. 2011;2:1145–54. doi: 10.18632/oncotarget.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Al-Bahlani S, Fraser M, Wong AY, Sayan BS, Bergeron R, Melino G, Tsang BK. P73 regulates cisplatin-induced apoptosis in ovarian cancer cells via a calcium/calpain-dependent mechanism. Oncogene. 2011;30:4219–30. doi: 10.1038/onc.2011.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grespi F, Amelio I, Tucci P, Annicchiarico-Petruzzelli M, Melino G. Tissue-specific expression of p73 C-terminal isoforms in mice. Cell Cycle. 2012;11:4474–83. doi: 10.4161/cc.22787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Agostini M, Tucci P, Killick R, Candi E, Sayan BS, Rivetti di Val Cervo P, Nicotera P, McKeon F, Knight RA, Mak TW, et al. Neuronal differentiation by TAp73 is mediated by microRNA-34a regulation of synaptic protein targets. Proc Natl Acad Sci U S A. 2011;108:21093–8. doi: 10.1073/pnas.1112061109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Billon N, Terrinoni A, Jolicoeur C, McCarthy A, Richardson WD, Melino G, Raff M. Roles for p53 and p73 during oligodendrocyte development. Development. 2004;131:1211–20. doi: 10.1242/dev.01035. [DOI] [PubMed] [Google Scholar]
- 22.De Laurenzi V, Raschellá G, Barcaroli D, Annicchiarico-Petruzzelli M, Ranalli M, Catani MV, Tanno B, Costanzo A, Levrero M, Melino G. Induction of neuronal differentiation by p73 in a neuroblastoma cell line. J Biol Chem. 2000;275:15226–31. doi: 10.1074/jbc.275.20.15226. [DOI] [PubMed] [Google Scholar]
- 23.Agostini M, Tucci P, Chen H, Knight RA, Bano D, Nicotera P, McKeon F, Melino G. p73 regulates maintenance of neural stem cell. Biochem Biophys Res Commun. 2010;403:13–7. doi: 10.1016/j.bbrc.2010.10.087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Talos F, Abraham A, Vaseva AV, Holembowski L, Tsirka SE, Scheel A, Bode D, Dobbelstein M, Brück W, Moll UM. p73 is an essential regulator of neural stem cell maintenance in embryonal and adult CNS neurogenesis. Cell Death Differ. 2010;17:1816–29. doi: 10.1038/cdd.2010.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gonzalez-Cano L, Herreros-Villanueva M, Fernandez-Alonso R, Ayuso-Sacido A, Meyer G, Garcia-Verdugo JM, Silva A, Marques MM, Marin MC. p73 deficiency results in impaired self renewal and premature neuronal differentiation of mouse neural progenitors independently of p53. Cell Death Dis. 2010;1:e109. doi: 10.1038/cddis.2010.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Flores ER. p73 is critical for the persistence of memory. Cell Death Differ. 2011;18:381–2. doi: 10.1038/cdd.2010.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rufini A, Niklison-Chirou MV, Inoue S, Tomasini R, Harris IS, Marino A, Federici M, Dinsdale D, Knight RA, Melino G, et al. TAp73 depletion accelerates aging through metabolic dysregulation. Genes Dev. 2012;26:2009–14. doi: 10.1101/gad.197640.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grespi F, Melino G. P73 and age-related diseases: is there any link with Parkinson Disease? Aging (Albany NY) 2012;4:923–31. doi: 10.18632/aging.100515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guglielmino MR, Santonocito M, Vento M, Ragusa M, Barbagallo D, Borzì P, Casciano I, Banelli B, Barbieri O, Astigiano S, et al. TAp73 is downregulated in oocytes from women of advanced reproductive age. Cell Cycle. 2011;10:3253–6. doi: 10.4161/cc.10.19.17585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Melino G, Bernassola F, Ranalli M, Yee K, Zong WX, Corazzari M, Knight RA, Green DR, Thompson C, Vousden KH. p73 Induces apoptosis via PUMA transactivation and Bax mitochondrial translocation. J Biol Chem. 2004;279:8076–83. doi: 10.1074/jbc.M307469200. [DOI] [PubMed] [Google Scholar]
- 31.Conforti F, Sayan AE, Sreekumar R, Sayan BS. Regulation of p73 activity by post-translational modifications. Cell Death Dis. 2012;3:e285. doi: 10.1038/cddis.2012.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Graupner V, Alexander E, Overkamp T, Rothfuss O, De Laurenzi V, Gillissen BF, Daniel PT, Schulze-Osthoff K, Essmann F. Differential regulation of the proapoptotic multidomain protein Bak by p53 and p73 at the promoter level. Cell Death Differ. 2011;18:1130–9. doi: 10.1038/cdd.2010.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tomasini R, Tsuchihara K, Wilhelm M, Fujitani M, Rufini A, Cheung CC, Khan F, Itie-Youten A, Wakeham A, Tsao MS, et al. TAp73 knockout shows genomic instability with infertility and tumor suppressor functions. Genes Dev. 2008;22:2677–91. doi: 10.1101/gad.1695308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang A, Walker N, Bronson R, Kaghad M, Oosterwegel M, Bonnin J, Vagner C, Bonnet H, Dikkes P, Sharpe A, et al. p73-deficient mice have neurological, pheromonal and inflammatory defects but lack spontaneous tumours. Nature. 2000;404:99–103. doi: 10.1038/35003607. [DOI] [PubMed] [Google Scholar]
- 35.Wilhelm MT, Rufini A, Wetzel MK, Tsuchihara K, Inoue S, Tomasini R, Itie-Youten A, Wakeham A, Arsenian-Henriksson M, Melino G, et al. Isoform-specific p73 knockout mice reveal a novel role for delta Np73 in the DNA damage response pathway. Genes Dev. 2010;24:549–60. doi: 10.1101/gad.1873910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen J, Herrup K. Glutamine acts as a neuroprotectant against DNA damage, beta-amyloid and H2O2-induced stress. PLoS One. 2012;7:e33177. doi: 10.1371/journal.pone.0033177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Giacobbe A, Bongiorno-Borbone L, Bernassola F, Terrinoni A, Markert EK, Levine AJ, Feng Z, Agostini M, Zolla L, Agrò AF, et al. p63 regulates glutaminase 2 expression. Cell Cycle. 2013;12:1395–405. doi: 10.4161/cc.24478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Danilova N, Sakamoto KM, Lin S. p53 family in development. Mech Dev. 2008;125:919–31. doi: 10.1016/j.mod.2008.09.003. [DOI] [PubMed] [Google Scholar]
- 39.Moll UM, Slade N. p63 and p73: roles in development and tumor formation. Mol Cancer Res. 2004;2:371–86. [PubMed] [Google Scholar]
- 40.Shalom-Feuerstein R, Lena AM, Zhou H, De La Forest Divonne S, Van Bokhoven H, Candi E, Melino G, Aberdam D. ΔNp63 is an ectodermal gatekeeper of epidermal morphogenesis. Cell Death Differ. 2011;18:887–96. doi: 10.1038/cdd.2010.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Paris M, Rouleau M, Pucéat M, Aberdam D. Regulation of skin aging and heart development by TAp63. Cell Death Differ. 2012;19:186–93. doi: 10.1038/cdd.2011.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Agostini M, Tucci P, Steinert JR, Shalom-Feuerstein R, Rouleau M, Aberdam D, Forsythe ID, Young KW, Ventura A, Concepcion CP, et al. microRNA-34a regulates neurite outgrowth, spinal morphology, and function. Proc Natl Acad Sci U S A. 2011;108:21099–104. doi: 10.1073/pnas.1112063108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vousden KH, Ryan KM. p53 and metabolism. Nat Rev Cancer. 2009;9:691–700. doi: 10.1038/nrc2715. [DOI] [PubMed] [Google Scholar]
- 44.Thotala DK, Hallahan DE, Yazlovitskaya EM. Glycogen synthase kinase 3β inhibitors protect hippocampal neurons from radiation-induced apoptosis by regulating MDM2-p53 pathway. Cell Death Differ. 2012;19:387–96. doi: 10.1038/cdd.2011.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Madan E, Gogna R, Bhatt M, Pati U, Kuppusamy P, Mahdi AA. Regulation of glucose metabolism by p53: emerging new roles for the tumor suppressor. Oncotarget. 2011;2:948–57. doi: 10.18632/oncotarget.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ninkovic J, Götz M. Signaling in adult neurogenesis: from stem cell niche to neuronal networks. Curr Opin Neurobiol. 2007;17:338–44. doi: 10.1016/j.conb.2007.04.006. [DOI] [PubMed] [Google Scholar]
- 47.Zimmer B, Kuegler PB, Baudis B, Genewsky A, Tanavde V, Koh W, Tan B, Waldmann T, Kadereit S, Leist M. Coordinated waves of gene expression during neuronal differentiation of embryonic stem cells as basis for novel approaches to developmental neurotoxicity testing. Cell Death Differ. 2011;18:383–95. doi: 10.1038/cdd.2010.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Volvert ML, Rogister F, Moonen G, Malgrange B, Nguyen L. MicroRNAs tune cerebral cortical neurogenesis. Cell Death Differ. 2012;19:1573–81. doi: 10.1038/cdd.2012.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Brett JO, Renault VM, Rafalski VA, Webb AE, Brunet A. The microRNA cluster miR-106b~25 regulates adult neural stem/progenitor cell proliferation and neuronal differentiation. Aging (Albany NY) 2011;3:108–24. doi: 10.18632/aging.100285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nigro A, Menon R, Bergamaschi A, Clovis YM, Baldi A, Ehrmann M, Comi G, De Pietri Tonelli D, Farina C, Martino G, et al. MiR-30e and miR-181d control radial glia cell proliferation via HtrA1 modulation. Cell Death Dis. 2012;3:e360. doi: 10.1038/cddis.2012.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Frank CL, Tsai LH. Alternative functions of core cell cycle regulators in neuronal migration, neuronal maturation, and synaptic plasticity. Neuron. 2009;62:312–26. doi: 10.1016/j.neuron.2009.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen N, Napoli JL. All-trans-retinoic acid stimulates translation and induces spine formation in hippocampal neurons through a membrane-associated RARalpha. FASEB J. 2008;22:236–45. doi: 10.1096/fj.07-8739com. [DOI] [PubMed] [Google Scholar]
- 53.Aoto J, Nam CI, Poon MM, Ting P, Chen L. Synaptic signaling by all-trans retinoic acid in homeostatic synaptic plasticity. Neuron. 2008;60:308–20. doi: 10.1016/j.neuron.2008.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tucci P, Porta G, Agostini M, Dinsdale D, Iavicoli I, Cain K, Finazzi-Agró A, Melino G, Willis A. Metabolic effects of TiO2 nanoparticles, a common component of sunscreens and cosmetics, on human keratinocytes. Cell Death Dis. 2013;4:e549. doi: 10.1038/cddis.2013.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tucci P, Porta G, Agostini M, Antonov A, Garabadgiu AV, Melino G, Willis AE. Rapamycin regulates biochemical metabolites. Cell Cycle. 2013;12:2454–67. doi: 10.4161/cc.25450. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.