Abstract
Bitter taste perception influences human nutrition and health, and the genetic variation underlying this trait may play a role in disease susceptibility. To better understand the genetic architecture and patterns of phenotypic variability of bitter taste perception, we sequenced a 996 bp region, encompassing the coding exon of TAS2R16, a bitter taste receptor gene, in 595 individuals from 74 African populations and in 94 non-Africans from 11 populations. We also performed genotype–phenotype association analyses of threshold levels of sensitivity to salicin, a bitter anti-inflammatory compound, in 296 individuals from Central and East Africa. In addition, we characterized TAS2R16 mutants in vitro to investigate the effects of polymorphic loci identified at this locus on receptor function. Here, we report striking signatures of positive selection, including significant Fay and Wu’s H statistics predominantly in East Africa, indicating strong local adaptation and greater genetic structure among African populations than expected under neutrality. Furthermore, we observed a “star-like” phylogeny for haplotypes with the derived allele at polymorphic site 516 associated with increased bitter taste perception that is consistent with a model of selection for “high-sensitivity” variation. In contrast, haplotypes carrying the “low-sensitivity” ancestral allele at site 516 showed evidence of strong purifying selection. We also demonstrated, for the first time, the functional effect of nonsynonymous variation at site 516 on salicin phenotypic variance in vivo in diverse Africans and showed that most other nonsynonymous substitutions have weak or no effect on cell surface expression in vitro, suggesting that one main polymorphism at TAS2R16 influences salicin recognition. Additionally, we detected geographic differences in levels of bitter taste perception in Africa not previously reported and infer an East African origin for high salicin sensitivity in human populations.
Keywords: selection on standing variation, African genetic diversity, genotype–phenotype association, salicin taste perception
Introduction
Although TAS2R16, a bitter taste receptor gene located on chromosome 7, has traditionally been associated with the detection of toxic β-d-glucopyranosides, such as salicin from willow bark (Bufe et al. 2002; Soranzo et al. 2005; Sakurai, Misaka, Ishiguro et al. 2010; Sakurai, Misaka, Ueno et al. 2010; Greene et al. 2011; Hayes et al. 2011), studies have now discovered a broader range of functions for this locus. Prior research demonstrated that polymorphic variation at TAS2R16 confers a strong differential response to salicin and other bitter ligands in vitro (Bufe et al. 2002; Soranzo et al. 2005; Sakurai, Misaka, Ishiguro et al. 2010; Sakurai, Misaka, Ueno et al. 2010; Greene et al. 2011; Hayes et al. 2011), and this variation has been suggested to influence preference for foods or beverages containing these naturally occurring bitter compounds (Imai et al. 2012). Several studies have also reported that bitter taste receptors are expressed in cell types that are not directly involved in oral sensory perception, such as in the lungs (Shah et al. 2009; Deshpande et al. 2010), sinonasal tissue (Shah et al. 2009; Lee et al. 2012), gastrointestinal tract (Rozengurt and Sternini 2007), and in the testis (Campa et al. 2012). Thus, bitter taste receptors participate in a variety of biological processes, including metabolism and immune response. In addition, nucleotide polymorphisms in or near the coding exon of TAS2R16 have been identified as risk factors for alcohol and nicotine dependence (Hinrichs et al. 2006; Mangold et al. 2008; Hayes et al. 2011) in African Americans and have also been correlated with differences in lifespan and senescence among individuals in an isolated European population from southern Italy (Campa et al. 2012).
Although TAS2R16 is thought to play important roles in nutrition and health, little is known about patterns of genetic and phenotypic variation at this locus in human populations, particularly in Africa. A prior genetic analysis of TAS2R16 found evidence for positive selection, influencing patterns of diversity at this locus, in Africans (consisting of eight populations, four of which share ancestry originating from Bantu-speaking populations) and non-Africans (consisting of 52 populations) (Soranzo et al. 2005), and inferred that functional variation at nucleotide site 516 was the selectively advantageous mutation. A more recent study of global populations (which included nine African populations) detected signals of positive selection at TAS2R16 primarily in Eurasia based on long-range haplotype data and suggested limited evidence for selection at this locus in Africa (Li et al. 2011). However, given the small number of Africans included in these studies and prior evidence for local adaptation in Africa (Tishkoff et al. 2007), we hypothesized that ethnically diverse populations in different geographic regions and/or practicing different subsistence strategies (such as hunting/gathering, pastoralism, agriculture, and agro-pastoralism) in Africa may show distinct patterns of genetic and phenotypic variability and signatures of selection at this gene.
In order to test this hypothesis, we analyzed sequence variation in the intronless coding exon of TAS2R16 in 595 individuals from 74 populations with diverse diets living in West Central, Central, and East Africa and in 94 individuals from 11 populations in the Middle East, Europe, East Asia, and the Americas. We also examined the association between genetic variability at this locus and levels of salicin bitter taste recognition in a subset of 296 individuals from the above African populations. Additionally, we characterized the functional effects of TAS2R16 amino acid substitutions on receptor function in vitro. Here, we report differential signatures of positive selection based on the allele frequency spectrum across Africa, a “star-like” genealogy for haplotypes with the derived T-allele at nucleotide site 516 associated with increased bitter sensitivity, and greater than expected genetic differentiation at this functional polymorphic site consistent with a model of positive selection for “high-sensitivity” variation. In contrast, our analyses indicated that haplotypes carrying the ancestral “low-sensitivity” allele at site 516 have been subject to strong purifying selection. Although a prior in vitro study showed a correlation between nonsynonymous variability at position 516 within TAS2R16 and salicin response (Soranzo et al. 2005), we demonstrated the effect of this nucleotide site on salicin bitter perception in vivo in diverse Africans and suggest a molecular basis for salicin bitter taste recognition using in vitro data. In addition, we observed geographic structuring of bitter taste sensitivity in Africa and infer an East African origin for the salicin “taster” trait in humans. Overall, this study correlates genetic variants that are targets of selection with phenotypic variability, demonstrating the connection between functional variation and local adaptation at a biologically relevant locus in human populations.
Results
Patterns of Nucleotide Diversity
From our analysis of the 876 bp coding exon of TAS2R16 in global populations, including 74 populations from Africa (supplementary table S1, Supplementary Material online), we identified a total of 15 single-nucleotide polymorphisms (SNPs) (supplementary table S2, Supplementary Material online). Overall, African populations had the largest number of polymorphic loci (15) compared to non-Africans (2) (supplementary tables S2 and S3, Supplementary Material online). Of the 15 SNPs identified, 7 were nonsynonymous (A340G, C481T, G516T, C605T, C662T, A665G, and A805G), mostly occurring at low frequency (minor allele frequency [MAF] <1–5.6%) except for G516T and A665G which were found to be more common in Africa (supplementary table S3, Supplementary Material online); the remaining 8 SNPs were synonymous (C300T, C303G, C321G, C333T, T460C, G663T, A846G, and T847C), with MAF ranging from <1% to 20.6% (supplementary table S3, Supplementary Material online). Furthermore, 4 of the 15 SNPs (at positions 300, 303, 516, and 846) had derived alleles at relatively high frequency (23–34%), and we observed strong linkage disequilibrium (LD) among these loci in African populations (supplementary fig. S1, Supplementary Material online). We also identified several novel polymorphisms, namely two nonsynonymous (C605T and A805G) and two synonymous (G663T and T847C) mutations at low frequency (<1–3%) in populations from West Central and Central Africa (supplementary table S3, Supplementary Material online).
Additionally, we observed regional differences in the frequency of common variation at nucleotide site 516 (corresponding to amino acid variability at site 172), previously associated with salicin bitter sensitivity in Africa (Soranzo et al. 2005). Specifically, the derived T-allele (or N amino acid) occurred at relatively high frequency in East Africa (75–94.1%). However, this derived polymorphism was typically present at a lower frequency in populations from West Central and Central Africa (50–78.6%), with the exception of the Fulani where this allele was found at a frequency of 88.6%. In contrast, the derived T-allele was observed at 100% frequency in our non-African samples, suggesting that the ancestral G-allele is rare or absent in populations outside of Africa (supplementary table S3, Supplementary Material online). We also calculated nucleotide diversity statistics, θS and π, in the coding region of our global samples, and we found higher levels of nucleotide diversity for both statistics in African populations compared with non-Africans (supplementary table S4, Supplementary Material online).
Haplotype Variation
We inferred a total of 13 distinct haplotypes (H1–H13) in our global populations (supplementary tables S2 and S5a and b, Supplementary Material online); H1–H9 carry the derived T-allele and H10–H13 contain the ancestral G-allele at nucleotide position 516 within the coding exon of TAS2R16. Among non-Africans, two major haplotypes, H1 and H4, accounted for 100% of sampled chromosomes. In contrast, however, we found higher levels of common haplotype variation among Africans (supplementary table S5a and b, Supplementary Material online). Specifically, in addition to H1 and H4, we also observed common H10 and H12 haplotypes with the ancestral G-allele, and together these four haplotypes accounted for the majority of chromosomes in African populations (88.9–100%). Furthermore, we found population-specific haplotypes (H3, H5, H6, H7, H8, and H11) present at relatively low frequency (<1–5.6%) in different groups across Africa (supplemental table S5a and b, Supplementary Material online). We also observed the presence of region-specific haplotype variation. Specifically, haplotype H2, which carries the derived T-allele at polymorphic site 516 was found only in East Africa, with the highest frequency of this variant occurring in Sandawe hunter-gatherers from Tanzania (13.9 %), followed by the Cushitic Afroasiatic speakers from Tanzania (5.6%) and Kenya (1.9%), and then the Omotic Afroasiatic speakers from Ethiopia (1.2%) (supplementary table S5a, Supplementary Material online).
Genotype–Phenotype Association
Given that salicin, a bitter tasting compound, is a known ligand of the TAS2R16 receptor (Bufe et al. 2005; Mueller et al. 2005; Soranzo et al. 2005; Sakurai, Misaka, Ishiguro et al. 2010; Sakurai, Misaka, Ueno et al. 2010; Greene et al. 2011; Hayes et al. 2011), we examined threshold levels for recognizing salicin bitterness in vivo in 296 individuals originating from Central (N = 161) and East Africa (N = 135). Overall, the distribution of salicin bitter taste sensitivity was continuous in both geographic regions (supplementary fig. S2, Supplementary Material online). However, we did observe regional differences in bitter taste recognition thresholds. In particular, we found a significant difference in the mean salicin score (P = 0.002; two-tailed nonparametric Mann–Whitney test) between East Africa (mean = 5.76 ± 1.886; range = 11; minimum score = 1 [low sensitivity]; maximum score = 12 [high sensitivity]) (supplementary table S6, Supplementary Material online) and Central Africa (mean = 5.06 ± 1.302; range = 8; minimum score = 2; maximum score = 10), indicating geographic structuring of bitter taste sensitivity in Africa.
To test whether or not common nonsynonymous variation at nucleotide 516 influences levels of salicin sensitivity, we compared the mean salicin scores of 1) individuals homozygous for two haplotypes carrying the ancestral G-allele, 2) individuals heterozygous for haplotypes containing the derived T-allele and the ancestral G-allele, and 3) individuals homozygous for haplotypes with the derived T-allele using general linear model (GLM) analysis of variance (ANOVA) adjusting for age and sex in both Central Africa and East Africa. This analysis showed a significant difference in mean salicin sensitivity between genotypic classes (P = 0.035 in Central Africa; P = 0.033 in East Africa) (supplementary table S7, Supplementary Material online), with individuals possessing at least one derived T-allele having a higher mean score than individuals homozygous for the ancestral G-allele, indicating a functional difference between these polymorphisms (supplementary table S7, Supplementary Material online) consistent with in vitro analyses by Soranzo et al. (2005).
Furthermore, to rule out the possibility that other variants may be contributing to salicin phenotypic variance, we also examined the effect of common nonsynonymous variation at polymorphic site 665 (corresponding to amino acid position 222) on haplotypes with the T-allele at polymorphic site 516 by comparing mean salicin score across two genotypic classes: 1) individuals homozygous for the T-allele at site 516 and homozygous for the ancestral A-allele at position 665 and 2) individuals homozygous for the T-allele at site 516 and either homozygous or heterozygous for the derived G-allele at position 665 using a GLM ANOVA adjusting for age and sex. Individuals with the common derived G-allele at position 665 had a lower mean salicin score compared with individuals with two common haplotypes, but the difference in mean score was not statistically significant (P = 0.382 in Central Africa; P = 0.425 in East Africa) (supplementary table S8, Supplementary Material online). Furthermore, we applied the above method to test individuals homozygous or heterozygous for haplotypes containing the low-frequency nonsynonymous variation on haplotypes with the T-allele at nucleotide site 516. This univariate analysis also showed no significant difference (P = 0.933 in Central Africa; P = 0.589 in East Africa) in mean salicin score between groups with and without additional nonsynonymous variants (supplementary table S9, Supplementary Material online).
In Vitro Function of Mutagenized TAS2R16 Receptors
To further understand how polymorphic sites at TAS2R16 influence function in vitro, we constructed multiple TAS2R16 protein receptors with a single amino acid change (wild-type versus derived) at sites where we observed nonsynonymous variants in our sequence data, and we measured receptor surface expression (i.e., the trafficking of TAS2R16 receptor mutants to the cell surface) (table 1). This analysis showed that the N172 TAS2R16 protein sequence with the derived N-variant at amino acid position 172 (corresponding to the T-allele at nucleotide position 516) expresses approximately 1.5- to 2-fold higher than the TAS2R16 receptor protein with the K172 variant (corresponding to the G-allele at site 516), providing a molecular basis for the observed decrease in sensitivity of the K172 variant to salicin in our human population data. In contrast, however, receptor variants at the other sites where we observed nonsynonymous substitutions showed either weak or no effect on receptor cell surface expression (table 1), suggesting that most sites with amino acid changes are not critically involved in receptor function. The one exception was position 114 where the TAS2R16 receptor protein with a derived amino acid variant showed a 2-fold decrease in cell surface trafficking compared with the wild-type receptor with the ancestral amino acid at the same site (table 1).
Table 1.
Surface Expression of TAS2R16 Nonsynonymous Substitutions.
| Nucleotide Site | Amino Acid Site | Ancestral | Derived | Library Mutation | Surface Expression % WT (±Range) | Phenotype of Derived Allele |
|---|---|---|---|---|---|---|
| 340 | 114 | I | V | L | 59 (14.7) | Reduced surface expression and function |
| 481 | 161 | P | S | A | 129 (14.7) | No effect |
| 516 | 172 | K | N | Na | 48 (6.8) | Reduced surface expression of ancestral allele |
| 605 | 202 | S | L | L | 88 (4.0) | No effect |
| 662/3 | 221 | A | V | Vb | 110 (2.2) | No or weak effect on function |
| 665 | 222 | H | R | R | 110 (2.2) | No or weak effect on function |
| 282 | 269 | I | V | V | 110 (5.0) | No effect |
Note.—TAS2R16 mutants (containing nonsynonymous substitutions at a single site) were expressed in HEK293T cells and surface expression was detected by immunostaining against an extracellular FLAG epitope present in all clones. All clones were tested two or three times each and normalized against a wild-type (WT) control set to 100%. The ancestral and derived mutations observed in this study are given for each polymorphic site, as well as the amino acid used in the test of cell surface expression (library mutation) which compares function with the ancestral amino acid (WT). In some cases where the naturally occurring derived mutation and the test mutation are different (namely, amino acid positions 114 and 161), the derived amino acids used in the test of cell surface expression are structurally similar to the naturally occurring derived amino acid. Data are shown as the average expression signal ± range.
aThe WT construct used to make the mutation library contains an Asn (derived N allele) rather than a Lys (ancestral K allele) at residue 172.
bThe clone containing S221L has a second site mutation (Y176N). A clone containing only Y176N shows similar surface expression levels to the double mutant, indicating that the Y176N residue is functionally neutral.
Haplotype Relationships
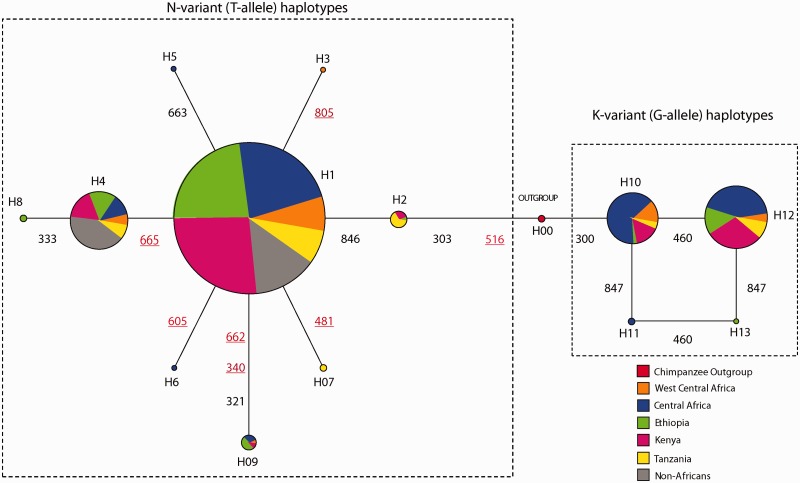
We constructed a median-joining network which showed that haplotype variation grouped into two major clusters, each defined by the presence of either the derived T- or ancestral G-allele at nucleotide position 516 (corresponding to the N or K amino acid variant, respectively, at position 172). Notably, the N-bearing H2 haplotype, specific to East Africa, is located closest to the root of the network, implying that this is the oldest haplotype associated with increased sensitivity to salicin (fig. 1). Common haplotypes were also observed at varying frequencies in Africa. For example, we found a higher proportion of common K-variant haplotypes (H10 and H12) in West Central and Central Africa and a lower proportion of these haplotypes in East Africa, indicating a geographic structuring of haplotype variation associated with bitter taste sensitivity across the continent. Additionally, populations practicing different subsistence strategies (such as hunting/gathering, pastoralism, agriculture, and agro-pastoralism) had relatively similar frequencies of common N- or K-variant haplotypes (supplementary tables S5a and b, Supplementary Material online), suggesting that patterns of haplotype variation are not likely correlated with diet.
Fig. 1.
Inferred haplotype relationships for global populations. Haplotype relationships were constructed using Network 4.15 (Bandelt et al. 1999). The circles represent haplotypes and the size of the circles is proportional to the number of chromosomes with a given haplotype. Colors within each haplotype represent the proportion of chromosomes with that haplotype within a population or geographic region. The alpha/numerical label beside each haplotype is the designated name of each haplotype. The numbers and lines between haplotypes represent the nucleotide position of mutations that differentiate haplotypes.
The low-frequency haplotypes, defined by polymorphic sites in addition to the functional SNP at position 516, also showed a star-like pattern of variation radiating from the common H1 high-sensitivity haplotype in the N-variant clade consistent with the genetic pattern expected under a model of positive selection (fig. 1). Moreover, nonsynonymous polymorphisms, including derived mutations, occurred only on haplotypes with the N-variant (153 chromosomes out of a total of 1,378 chromosomes), whereas nonsynonymous mutations were not observed on any haplotype with the low-sensitivity K-variant (273 chromosomes out of a total of 1,378 chromosomes), a significant difference based on χ2 analysis (χ2 = 42.5; df = 1; P < 0.001) (supplementary table S14, Supplementary Material online).
Population Differentiation
To investigate patterns of genetic differentiation, we generated multidimensional scaling (MDS) plots of genetic relationships among African populations based on estimates of FST derived from (A) randomly selected genome-wide SNPs and (B) TAS2R16 sequence data (fig. 2 and supplementary fig. S3, Supplementary Material online). Our analysis using genome-wide SNPs showed that African populations generally clustered on the basis of genetic similarity and/or linguistic properties consistent with previous studies of autosomal loci (Tishkoff et al. 2007; Scheinfeldt et al. 2010). For example, the Luo from Kenya, who speak a Western Nilotic language, grouped more closely with Niger-Kordofanian Bantu speakers with whom the Luo share high levels of ancestry. In contrast, however, MDS analysis of our sequence data from TAS2R16 revealed a different pattern of genetic relationships among populations (fig. 2B and supplementary fig. S3, Supplementary Material online). Here, we found that, in most cases, populations from the same geographic region generally clustered together, irrespective of shared ancestry (fig. 2B). For example, most Afroasiatic-, Nilo-Saharan-, and Niger-Kordofanian-speaking populations from Central Africa formed a distinct cluster. Similarly, East African populations clustered together as a separate group, with the exception of the Niger-Kordofanian and Nilo-Saharan speakers from Kenya (fig. 2B). Another notable exception were the Fulani from Cameroon (West Central Africa) who grouped more closely with East African populations rather than with other Niger-Kordofanian speakers from West Central Africa with whom they share recent ancestry as in figure 2A (fig. 2B).
Fig. 2.
MDS plots of allele frequencies for genome-wide and TAS2R16 data in African populations. The MDS plots indicate the spatial configuration of diverse African populations, represented by different symbols, based on pairwise FST calculated (A) from genome-wide SNP data genotyped using the Illumina 1 M Duo platform and (B) from SNPs ascertained through resequencing of the TAS2R16 gene in similar sets of African populations. The circles encompassing populations represent groups of populations that clustered together based on pairwise FST estimates. For the genome-wide SNP data set in (A), geographic region from which populations originated is given in parentheses, whereas for the TAS2R16 data set in (B), linguistic affiliation is indicated in parentheses.
Our analyses of population differentiation also revealed that the observed FST value among Africans (FST = 0.076) at the common functional nonsynonymous SNP at position 516 was an outlier in the empirical distribution of FST values derived from our genome-wide SNP data, falling in the top one percentile of values. Furthermore, the FST value at position 516 was significantly different from the mean FST for our genome-wide SNP data (P = 0.000001; one-tailed test) based on the one-sample t-test. Together, the unusually high FST for the common functional variant at TAS2R16 and the extensive population structure among Africans based on sequence data are consistent with a model of local adaptation at this locus in Africa.
Estimated Age of TAS2R16 Variants
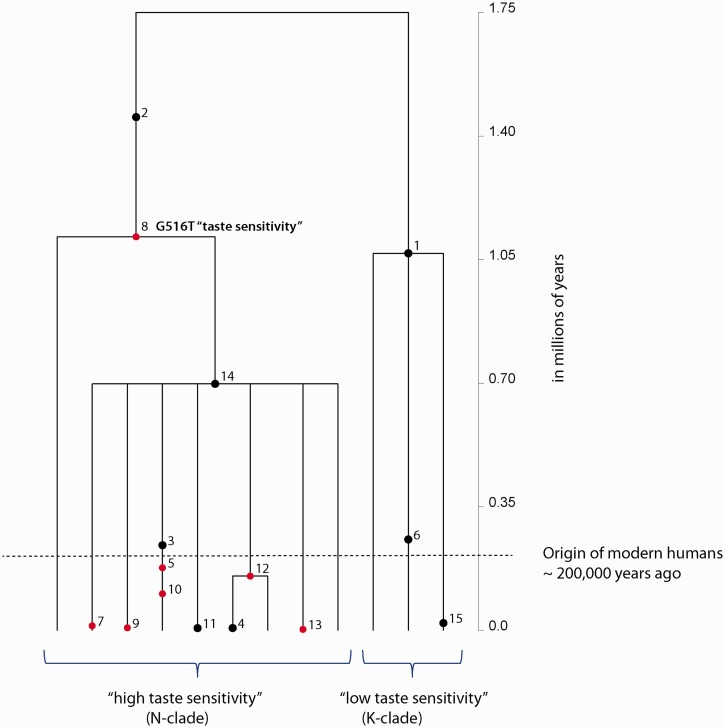
We constructed a gene tree showing the mutation history of global variation in the coding region of TAS2R16 and estimated the time since the most recent common ancestor (TMRCA) of the tree, as well as ages of nucleotide polymorphisms (Griffiths 2007) (fig. 3 and supplementary table S11, Supplementary Material online). Like the haplotype network, the shape of the gene tree indicated that variation at this locus clustered into two major clades, each defined by the presence of either the ancestral K or derived N amino acid variant (corresponding to the G- or T-allele, respectively, at nucleotide site 516). Our analysis yielded a deep coalescence time for the origin of variation at TAS2R16 (mean TMRCA was ∼1.75 Ma ± 745,743 years) and an ancient age for the derived nonsynonymous allele at position 516 associated with increased salicin sensitivity (mean age was ∼1.1 Ma ± 440,115 years) (fig. 3 and supplementary table S11, Supplementary Material online). Our genealogy also showed the relatively more recent emergence of other nonsynonymous mutations on haplotypes carrying the derived “high-sensitivity” allele, suggesting ongoing diversification of “high-sensitivity” haplotype variation.
Fig. 3.
Inferred TMRCA of global variation and individual ages of mutations at TAS2R16. A gene tree describing the mutation history or genealogy of polymorphisms in the coding region of TAS2R16 was constructed using a coalescent-based method implemented in GENETREE (Griffiths 2007). The TMRCA and ages of polymorphisms are mean estimates with standard deviations listed in supplementary table S10, Supplementary Material online. The scale (in millions of years) is along the vertical axis of this figure. Dots represent mutations, and the numbers beside each dot are the designated identifier for each polymorphism given by the GENETREE program. The red dots indicate nonsynonymous variants, while black dots signify synonymous variation. We also indicated the polymorphic variation associated with salicin bitter taste (G516T). The dashed horizontal line represents the origin of anatomically modern humans ∼200,000 years ago.
Tests of Neutrality and Extended Haplotype Homozygosity
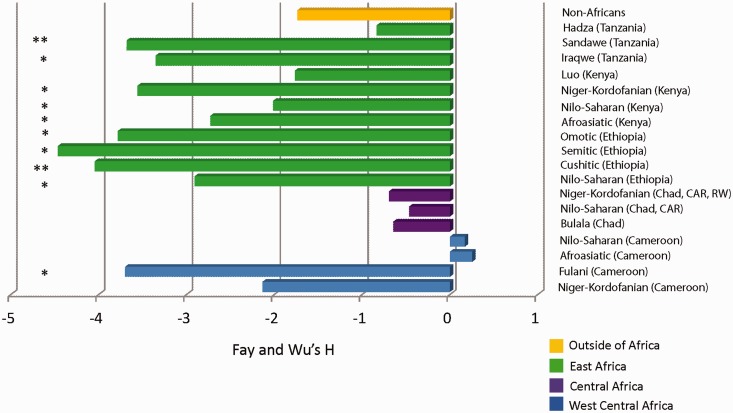
Given the presence of high-frequency derived variation at this locus (supplementary table S3, Supplementary Material online), we applied Fay and Wu’s H statistic (Fay and Wu 2000; Jensen et al. 2005), which measures departures from neutrality based on high-frequency and intermediate-frequency alleles (Fay and Wu 2000), to sequence data in the coding region of TAS2R16 in African and non-African populations. Our results showed that most populations in East Africa exhibited significantly negative Fay and Wu’s H statistics (under an assumption of constant population size), indicating an excess of high-frequency derived alleles within this geographic region. However, H values did not significantly deviate from neutral expectation in populations from West Central and Central Africa, with the exception of the Fulani population (H = −3.699; P < 0.05) (fig. 4 and supplementary table S12, Supplementary Material online). Estimates of Tajima’s D (DT) were also calculated for the coding region in our global samples (supplementary table S12, Supplementary Material online), but we did not detect significant deviation from neutrality using this statistic. Given that DT does not distinguish between ancestral and derived states and is influenced mostly by intermediate- and low-frequency variation (Fay and Wu 2000; Jensen et al. 2005), this statistical test may not be sensitive enough to detect selection based on the observed allele frequency distribution at TAS2R16. Because population growth can skew the observed allele frequency spectrum, we also reestimated the significance of Fay and Wu’s H under different models of population growth using coalescent simulations (Hudson et al. 1992). Our results showed that H values in East Africa and the Fulani from West Central Africa were significantly more negative than expected (P < 0.025) under scenarios of low growth, with the strongest departures from neutrality occurring in Ethiopian populations (supplementary table S13, Supplementary Material online). We also applied the McDonald–Kreitman test (McDonald and Kreitman 1991) to our African and non-African populations using the common chimpanzee as an outgroup. This analysis of the relative number of polymorphic and fixed synonymous and nonsynonymous variation was not significant (P > 0.05; supplementary table S14, Supplementary Material online) in our samples. However, we did observe low levels of fixed nonsynonymous variation between species (i.e., humans and chimpanzee) at this locus indicative of strong purifying selection (supplementary table S14, Supplementary Material online).
Fig. 4.
A plot of observed Fay and Wu’s H values. Bars in the plot represent Fay and Wu’s H statistics, based on the site frequency spectrum, in the intronless coding region of TAS2R16 in global populations using DnaSP (Librado and Rozas 2009). Populations are listed in close proximity to each bar, and colors of the bars denote the geographic region from where our population samples originated. Within parentheses, CAR and RW represent Central African Republic and Rwanda, respectively. The * and ** symbols indicate statistical significance at P < 0.05 and P ≤ 0.01, respectively, under an assumption of constant population size.
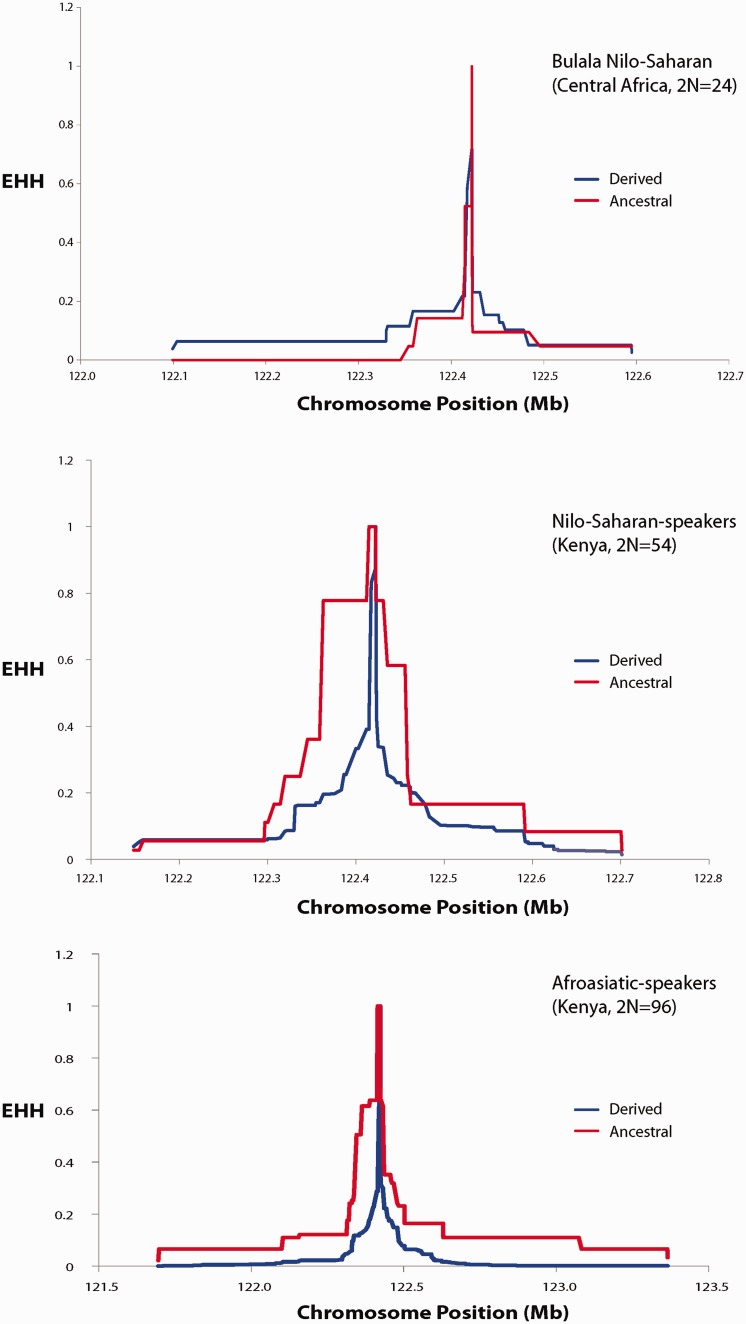
To further explore signatures of selection at TAS2R16, we integrated SNPs ascertained through resequencing of this locus with ∼60,000 SNPs genotyped across an ∼158 Mb region on chromosome 7 for a subset of 165 individuals from Africa. We plotted the decay of haplotype homozygosity (extended haplotype homozygosity [EHH] statistics) with increasing distance from the core SNP (represented by nucleotide site 516) (fig. 5 and supplementary fig. S4, Supplementary Material online). However, we did not observe long-range haplotype homozygosity on chromosomes carrying the derived T-allele relative to chromosomes carrying the ancestral G-allele in any African population using this statistic (fig. 5 and supplementary fig. S4, Supplementary Material online).
Fig. 5.
EHH plots for selected African populations. These plots show the decay of haplotype homozygosity with increasing distance from the core SNP represented here as the derived and ancestral alleles at nucleotide position 516 in the TAS2R16 gene. The x axis is the chromosome position of SNPs spanning chromosome 7 and the y axis is the probability that two chromosomes are homozygous at all SNPs for the entire interval from the core region to distance x. EHH = 0 means all extended haplotypes are different, while EHH = 1 indicates that all extended haplotypes are the same. In our plot, the red line represents the decay of homozygosity of chromosomes carrying the derived allele at the core, while the blue line signifies the decay of homozygosity on chromosomes bearing the ancestral core allele.
Correlation between Allele Frequency at TAS2R16 and Malaria Endemicity
It has been suggested that the “lower sensitivity” ancestral G-allele at polymorphic site 516 occurs at relatively high frequency in regions of Africa with endemic malaria and may be protective against this disease (Soranzo et al. 2005). To explore this hypothesis, we measured the strength of association between ancestral G-allele frequency at nucleotide site 516 and levels of malaria endemicity for several African populations from West Central, Central, and East Africa for which we had endemicity data using Spearman’s correlation coefficient (ρ) (supplementary table S15, Supplementary Material online). We did not, however, find a significant association (P = 0.317) between ancestral allele frequency at 516 and malaria endemicity in Africans (supplementary table S15, Supplementary Material online).
Discussion
Genotype–Phenotype Association in Africa
Our genotype–phenotype analysis in vivo showed that individuals with at least one derived T-allele at polymorphic site 516 have a higher sensitivity to salicin bitterness compared with individuals homozygous for the ancestral G-allele. Our in vitro data also supported this finding and further showed that the T-allele (coding for the N amino acid at residue 172) increases trafficking of the receptor to the cell membrane. While a prior study also reported a functional difference between the T- and G-alleles in vitro (Soranzo et al. 2005), we confirm the effect of allelic variability at site 516 on levels of salicin sensitivity in vivo in diverse Africans and suggest, based on in vitro data, that differences in cell surface expression of TAS2R16 receptor variants may contribute to differences in salicin bitter taste perception. In addition, our data showed that most other nonsynonymous substitutions at TAS2R16 do not have a strong effect on receptor trafficking, suggesting that a very limited number of amino acid polymorphisms at this locus result in differential response to salicin bitterness in human populations. However, it remains possible that these other nonsynonymous variants at TAS2R16 could play a role in the detection of other bitter compounds or that they may participate in a number of physiological processes beyond gustatory function. Alternatively, low-frequency nonsynonymous variants on haplotypes containing the derived T-allele could be tolerated due to weak purifying selection, whereas strong purifying selection acting on haplotypes containing the G-allele has constrained accumulation of nonsynonymous variation.
In addition, we observed higher levels of salicin sensitivity (higher mean and range of salicin scores) in East Africa relative to West Central and Central Africa. Similarly, genetic variability associated with salicin sensitivity was geographically structured, with salicin “taster” variation occurring at higher frequency in East Africa than in West Central and Central Africa, which had been reported by Soranzo et al. (2005) for a smaller number of populations. Previous studies have also identified a strong correlation between decreased salicin sensitivity and medically relevant behavioral traits, such as nicotine and alcohol dependence (Hinrichs et al. 2006; Mangold et al. 2008) in African American populations. Our observation of a relatively high frequency of the G-allele at position 516 associated with decreased salicin sensitivity in West Central and Central Africa could explain the high frequency of this risk allele in African Americans who originated mainly from these geographic regions in Africa (Tishkoff et al. 2009).
Selection for Salicin Bitter Taste Sensitivity in Africa
We identified striking differential patterns of positive selection at TAS2R16 in diverse African populations based on the allele frequency spectrum. Specifically, we detected significantly negative Fay and Wu’s H statistics predominantly in East African populations, indicating an excess of high-frequency derived alleles, whereas Fay and Wu’s H did not reach statistical significance in most populations from West Central and Central Africa. The star-like phylogeny observed for the derived N-clade haplotypes also indicates that haplotypes with the derived allele at polymorphic site 516 have undergone expansion due to selection for “high-sensitivity” variation, which could also explain the prevalence of low-frequency synonymous and nonsynonymous variants on these haplotypes. In addition, we observed extensive population structure among African populations based on total TAS2R16 sequence variation, as well as a high level of genetic differentiation at the common SNP associated with salicin bitter taste sensitivity. Overall, these patterns of diversity are congruent with a model of positive selection operating at TAS2R16 in Africa, and we also suggest that the derived “high-sensitivity” T-allele at site 516 is the likely target of selection in Africa, as was proposed by Soranzo et al. (2005).
Our GENETREE analysis showed that both “taster” and “non-taster” alleles have existed for a relatively long period of time in Africa (up to a million years). However, it appears that the “taster” T-allele rose to high frequency primarily in East Africa on multiple haplotype backgrounds consistent with a model of positive selection from standing variation (a soft sweep). This model of selection typically yields a relatively deep time of origin for selected mutations within a coalescent framework and is distinct from a classic selective sweep where the age of advantageous mutations is generally more recent (Bamshad and Wooding 2003; Barrett and Schluter 2008; Coop et al. 2008; Kelley 2012; Peter et al. 2012; Fu and Akey 2013). Nevertheless, the global distribution of the derived T-allele at site 516 suggests that this variant at least predates the origin of anatomically modern humans. Furthermore, the presence of the taster haplotypes at relatively high frequency in populations with divergent diets in East Africa indicates that diet is not likely the selective agent driving signals of selection. Indeed, given the diverse functions of TAS2R16, the pressures that have shaped patterns of genetic variation could be non-gustatory in nature. A similar hypothesis has also been proposed for TAS2R38, another bitter taste receptor gene (Campbell et al. 2012). Although we cannot identify the selective pressures affecting the geographic distribution of phenotypic and genetic variability, it is clear that positive selection on standing variation has maintained high-sensitivity haplotypes in East African populations. In addition, the fixation of the derived T-allele outside of Africa suggests that this mutation may have either risen to high frequency due to a selective sweep or may have resulted from the population bottleneck during the geographic expansion of modern humans from Africa ∼40,000–80,000 years ago.
The absence of nonsynonymous mutations on haplotypes with the “low-sensitivity” allele in the K-clade may be due to strong pressure to maintain the ancestral trait of salicin insensitivity in West Central and Central African populations (i.e., strong purifying selection), consistent with the low number of fixed nonsynonymous variants in the McDonald–Kreitman test. These findings raise the possibility that ancestral K-variant haplotypes without amino acid changes are functionally important, resulting in this pattern of evolutionary constraint on “low-sensitivity” haplotypes. Although it has been suggested that decreased bitter taste sensitivity may be selectively advantageous against malaria susceptibility, possibly explaining the relatively high frequency of the “low taste sensitivity” allele in populations from West Central and Central Africa (Soranzo et al. 2005), we did not observe a significant association (Spearman’s rank correlation, P = 0.317) between variation at 516 and malaria endemicity in Africans (supplementary table S13, Supplementary Material online). Furthermore, we did not detect signatures of positive selection, based on Fay and Wu’s H statistic, in most populations from West Central and Central Africa, suggesting that the elevated frequency of low taste sensitivity variation within these geographic regions is not likely due to recent selection for malaria resistance.
Of particular interest, however, is the fact that the Fulani from Cameroon had the highest frequency of the derived “high taste sensitivity” allele and the most significantly negative Fay and Wu’s H value in West Central Africa. Based on previous data on African population structure (Tishkoff et al. 2007; Scheinfeldt et al. 2010), it is possible that the high-sensitivity allele was introduced into the Fulani by migration and admixture with other populations from East Africa and/or outside of Africa. However, we cannot distinguish whether the above pattern of variation in the Fulani was due to strong selection after the mutation was introduced into the population and/or whether a selective event had first occurred in other populations who then admixed with the Fulani.
Timing and Spread of Salicin Bitter Taste Sensitivity in Africa
Typically, when a mutation is advantageous, it can rapidly increase in frequency in populations, together with linked variants, resulting in EHH around the selected mutation (Sabeti et al. 2005; Grossman et al. 2013). Over time, mutations arise and recombination breaks up the genomic region containing the advantageous variant, leading to shorter blocks of EHH in the genomic regions around the beneficial allele under selection. In this study, we compared patterns of EHH flanking the derived and ancestral allele at the core SNP (nucleotide position 516) in African populations. However, we did not find evidence for extensive EHH on chromosomes with the derived allele in Africa, in general agreement with a recent analysis of long-range haplotype (LRH) structure around TAS2R16 in global populations (Li et al. 2011). While this prior study inferred selection for “high taste sensitivity” variation in only a single African population, we argue that “high taste sensitivity” haplotypes have likely existed in humans for a long period of time, in light of the ancient age of variability at nucleotide 516, decreasing the likelihood of observing long-range LD in Africans. Indeed, given that the LRH and EHH tests are aimed at detecting signals of recent sweeps (Sabeti et al. 2005), these methods will not have sufficient power for identifying nonclassical selective events (Przeworski et al. 2005; Voight et al. 2006; Fu and Akey 2013), such as a soft sweep of variation at TAS2R16 which may have occurred at a deeper point in modern human history.
Finally, we observed the highest frequency of the oldest haplotype with the derived T-allele (H2), in the Sandawe- and Cushitic-speaking populations from Tanzania. Previous autosomal studies have indicated that the Sandawe hunter-gatherers have admixed considerably with neighboring groups, particularly with southern Cushitic speakers who migrated to Tanzania from a southern Ethiopian homeland (Tishkoff et al. 2009; Campbell and Tishkoff, 2010). Additional genetic and archaeological evidence also suggested a long history of gene flow between Cushitic speakers and other East African populations (Tishkoff et al. 2009; Campbell and Tishkoff 2010). Ethiopia is also thought to be the likely site of origin of migration of modern humans out of Africa. It is also interesting to note that East Africans have the highest frequency of the derived variant and we also observed fixation of the derived allele outside of Africa. We, therefore, propose an early origin of the derived high-sensitivity trait in Cushitic speakers originating from Ethiopia that spread to other populations in East Africa through admixture, and subsequently increased in frequency due to positive selection within this geographic region, which is also the likely source for the “high taste sensitivity” variant outside of Africa.
Though the field of TAS2R receptor research has advanced over the last several years, further investigation is needed in order to understand the genetic and environmental components influencing patterns of phenotypic variation and human adaptation. In particular, additional resequencing data on regulatory regions of the TAS2R genes and genome-wide association analyses of bitter taste perception traits will be important for discovering novel mutations contributing to phenotypic differences between populations. Furthermore, as we learn more about the diverse physiological functions of bitter taste receptors, we will have a better idea of the selective pressures that have been acting on the TAS2R loci. In the end, the integration of genetic, phenotypic, and functional data will be critical for identifying the key factors underlying the evolution of these important but relatively understudied genes.
Material and Methods
Population Samples
We sequenced 595 African individuals from 74 populations originating from West Central (Cameroon), Central (Chad, Central African Republic, and Rwanda), and East Africa (Ethiopia, Kenya, and Tanzania) practicing diverse modes of subsistence (such as hunting/gathering, pastoralism, agriculture, and agro-pastoralism), and a comparative set of 94 individuals from 11 populations in the Middle East, Europe, East Asia, and the Americas (a total of 1,378 chromosomes) (supplementary table S1, Supplementary Material online). African populations with shared genetic similarity as well as cultural and/or linguistic properties (e.g., Khoesan-speaking hunter-gatherers and Niger-Kordofanian speakers) were pooled together for analyses as described in Tishkoff et al. (2009).
Institutional Review Board (IRB) approval for this project was obtained from the University of Maryland at College Park and the University of Pennsylvania, and written informed consent was obtained from all African participants. IRB approval and permits were also obtained from the following institutions prior to sample collection: The Commission for Science and Technology and the National Institute for Medical Research in Dar es Salaam, Tanzania; the Kenya Medical Research Institute in Nairobi, Kenya; the Ministry of Health and National Committee of Ethics; the Federal Democratic Republic of Ethiopia Ministry of Science and Technology National Health Research Ethics Review Committee and the University of Addis Ababa; the Ministry of Health and National Committee of Ethics, Cameroon. Individuals from the Central African Republic, Chad, and Rwanda were recent immigrants to Cameroon, and these participants gave informed consent under our Cameroon IRB.
DNA Sequencing
A 996 bp region, encompassing the 876 bp coding exon of TAS2R16, was amplified by polymerase chain reaction (PCR). All nucleotide sequence data were generated using the 3730xl automated sequencer (Applied Biosystems). DNA sequences were imported into Sequencher 4.0 (Gene Codes Corporation, Ann Arbor, MI). All reported polymorphisms were then visually inspected for accurate base calls. Consensus sequences were exported and aligned by the MEGA program which was also used to confirm the occurrence of nucleotide polymorphisms.
Haplotype Analysis
The program PHASE v.2.1, which implements a Bayesian statistical method, was used (Stephens et al. 2001) to reconstruct multisite haplotypes from sequence data for subsequent analyses. The genealogical relationships among inferred haplotypes were constructed using the median-joining algorithm implemented in the Network 4.5 program (Bandelt et al. 1999). The resulting topology was the tree with the minimum number of changes among all possible maximum parsimony trees from a given data set. Haplotypes within this phylogeny were classified as ancestral or derived based on the presence of either the ancestral G- or derived T-allele at polymorphic site 516.
To further investigate patterns of haplotype variation, we used the chi-square (χ2) statistic to test whether or not there is a relationship between two categorical variables: 1) haplotype classification (i.e., ancestral or derived haplotype defined by nucleotide position 516) and 2) the number of haplotypes with or without nonsynonymous polymorphisms (at sites outside of nucleotide position 516 in the coding region) in a 2 × 2 contingency table. The χ2 statistic measures the divergence of observed data from values that are expected under the null hypothesis of no association between variables. Statistical significance was assessed at P < 0.05, representing the probability of the observed data occurring by chance alone.
Phenotype Determination
Levels of salicin bitter taste sensitivity were measured using a modification of the quality recognition threshold method described by Harris and Kalmus (1951) in 296 individuals from Central (Chad, Central African Republic, and Rwanda) and East Africa (Kenya and Tanzania). During this test, subjects were asked to taste increasingly concentrated solutions of salicin in bottles labeled 13 (containing the most dilute solution, 2 × 10−6 M) to 1 (containing concentrated stock solution, 1.64 × 10−2 M), until they detected the bitter taste of salicin. Subjects were given a raw phenotype score, corresponding to the bottle number at which they recognized bitter taste. For example, if an individual detected salicin bitterness at bottle number 12, that individual was assigned a salicin score of 12.
Genotype–Phenotype Association
To ascertain the functional effect of genetic variability at TAS2R16 on phenotype, we examined mean salicin score across different genotypic classes in individuals from Central (N = 161) and East (N = 135) Africa using a GLM ANOVA adjusting for age and sex in SPSS. In this univariate analysis, log-transformed salicin scores were entered as the dependent variable and genotypes were included as fixed factors; age and sex were also entered as covariates.
TAS2R16 Receptor Analysis
Mutations in TAS2R16 were introduced by random PCR mutagenesis using a Diversity Mutagenesis kit (Clontech Laboratories, Inc., Mountain View, CA) as part of a larger library covering the entire TAS2R16 receptor, and plasmids were used for functional analysis, as described previously in Greene et al. (2011). Briefly, HEK-293T cells were transfected with individual TAS2R16 plasmid mutations and a plasmid expressing a Gα16 chimera containing the last 44 amino acids of rat gustducin (Gα16gust44) and incubated for 22 h at 37 °C. To measure cell surface expression, cells were washed with PBS (HyClone), fixed with 4% paraformaldehyde (Electron Microscopy Sciences, Hatfield, PA), and stained for 1 h with anti-FLAG MAb M2 (1:500; Stratagene) in phosphate buffered saline (PBS) plus calcium and magnesium (PBS++). After washing, cells were incubated with goat anti-mouse Cy3-conjugated secondary antibody (1:500; Jackson Laboratories). After final washing, secondary antibody fluorescence was measured from a minimum of 500 cells by flow cytometry on an IntelliCyt HTFC screening system. Raw data obtained from surface expression and flux experiments were normalized to wild-type TAS2R16 values. Data sets were analyzed and represented as % over baseline signal in the wild-type TAS2R16, using Prism 5.0 software, and values shown represent the average of two or three independent measurements. In some cases where the derived amino acid used in the test of cell surface expression in a TAS2R16 clone was different from the naturally occurring derived amino acid at the same site, the derived amino acid used in the test was structurally similar to the naturally occurring derived amino acid (e.g., if the naturally occurring amino acid was hydrophobic, the replacement amino acid used in the test of surface expression was also hydrophobic and of similar size). Data are shown as the average expression signal ± range.
LD Analysis
Haploview version 4.2 (Barrett et al. 2005) was used to construct pairwise comparisons of LD among SNP loci. LD was determined based on the frequency of each SNP and the frequency with which a given SNP was associated with another nucleotide variant. Pairwise LD plots were constructed for pooled populations from West Central, Central, and East Africa, separately. LD plots were also constructed using all identified SNP variation.
Population Differentiation
Metric MDS analyses based on estimates of FST 1) derived from 1,069 genome-wide SNPs randomly selected from the Illumina 1M-Duo SNP array and 2) calculated from TAS2R16 sequence data were performed in similar sets of African populations in XLSTAT (http://www.xlstat.com/en/download.html, last accessed November 26, 2013). This method created a mapping of objects (in this case, populations) from a proximities matrix (i.e., genetic distances based on FST) between objects. This analysis converts the proximity matrix between N objects to coordinates of these same objects in a p-dimensional space; p was generally fixed at two dimensions so that the objects may be easily visualized. The 1,069 SNPs were randomly selected from across the genome using the “– thin” option implemented in the PLINK program (Purcell et al. 2007).
Average FST, derived from sequence data, was calculated for the common functional SNP at polymorphic site 516 using the program FSTAT 2.9.3.2 (Goudet 2002) (supplementary table S3, Supplementary Material online). This observed FST estimate was then compared with the empirical distribution of FST values derived from the 1,069 genome-wide SNPs. We also performed a one-sample t-test comparing the average FST value for the functional common SNP at nucleotide site 516 with average FST for all of the 1,069 SNPs using the R statistical package.
Age Estimates of Mutations
We used a maximum-likelihood approach to estimate the expected TMRCA for the coding region polymorphisms and the expected ages of individual mutations implemented in GENETREE (Griffiths 2007). This method is based on the coalescent process and assumes an infinite alleles model and no recombination. Haplotypes and sites that were not compatible with the condition of no recombination (which was one singleton haplotype) were removed from the analysis. Each simulation produced an ordering of coalescent and mutation events. The age of mutations and the TMRCA of the gene tree were also estimated for each run. The expected age was estimated from the weighted average of the simulated ages over different independent runs (Griffiths 2007).
Statistical Tests of Neutrality
To detect departures from neutral evolution at TAS2R16, Fay and Wu’s H and Tajima’s D statistics were calculated for global populations. Fay and Wu’s H compares estimates of θH (derived from high-frequency alleles) and π (derived from intermediate-frequency alleles) (Fay and Wu 2000); Tajima’s D (DT) compares estimates of θW (the number of segregating sites) and θπ (pairwise nucleotide divergence) (Tajima 1989). Under neutral evolution, θ and π are expected to be equal assuming constant population size (Fay and Wu 2000). The TAS2R16 sequence from Pan troglodytes (common chimpanzee) was downloaded from the National Center for Biotechnology Information (NCBI; http://www.ncbi.nlm.nih.gov, last accessed November 26, 2013) database and used as an outgroup in the Fay and Wu’s H analysis. Significance (P value) was determined with coalescence simulations assuming constant population size. Using the ms program (Hudson 1990), statistical significance of observed Fay and Wu’s H values was also assessed under a range of simple demographic models of population growth in coalescent simulations. For Africans, we simulated a 2-, 4-, 8-, and 10-fold increase in population size (from an initial population size of 10,000 individuals) starting at 70 kya until the present (Marth et al. 2004; Voight et al. 2005).
In addition, we compared the ratio of nonsynonymous and synonymous mutations within and between species for the coding region in African and non-African populations using the McDonald–Kreitman test (McDonald and Kreitman 1991). For the between-species comparison in this analysis, we used the TAS2R16 sequence from P. troglodytes downloaded from NCBI.
EHH Analyses
We applied the EHH statistic (Sabeti et al. 2005) to our data to identify long-range homozygosity on chromosomes carrying the functional derived T-allele at TAS2R16. We performed this analysis in a subset of 165 sequenced African samples for which we also had Illumina 1M-Duo genotype data. We integrated our sequencing SNP data from 165 individuals with ∼60,000 SNPs, encompassing an ∼158 Mb region of chromosome 7 flanking the TAS2R16 gene. SNPs were phased using fastPHASE (v1.4) (Scheet and Stephens 2006). We then measured the decay of LD for a given core SNP by calculating EHH for the core SNP and the surrounding SNPs on chromosome 7 in the order of increasing distances, starting with the core SNP (EHH = 1) followed by the next closest neighboring SNPs on either side of the core SNP. The estimated EHH values of the core and neighboring SNPs were plotted against their chromosomal position for both the derived and ancestral alleles present at the core site (Sabeti et al. 2005). Alleles were inferred to be ancestral or derived using the chimpanzee as an outgroup.
Correlation between Allele Frequency at TAS2R16 and Malaria Endemicity
To examine the relationship between genetic variability at TA2R16 and malaria endemicity, we calculated Spearman’s rank correlation coefficient (ρ) between the frequency of the ancestral G-allele at polymorphic site 516 and levels of malaria endemicity, measured by the Plasmodium falciparum parasite rate (PfPr), in a subset of the sampled African populations (supplementary table S13, Supplementary Material online). PfPr for each population was determined by locating the latitude and longitude coordinates of a sampling site for a given population on the country-level map of the spatial distribution of P. falciparum malaria endemicity in Africa created by the Malaria Atlas Project (Hay et al. 2009). Significance was determined by testing whether or not the observed value of ρ was significantly different from zero.
Supplementary Material
Supplementary tables S1–S15 and figures S1–S4 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
The authors thank Fathya Abdo, Eva Aluvalla, Birhanu Mekaunintie, Alemayehu Moges, Hussein Musa, Lilian Nyindodo, and Solomon Taye who helped with the field work, as well as the various institutions in West Central, Central, and East Africa for their generous support. They thank Edgar Davidson, Felicia Gomez, Dongbo Hu, Wen-Ya Ko, Joseph Lachance, Laura Scheinfeldt, Sameer Soi, and Anu Thomas for data collection and/or useful discussions. They also express our deepest appreciation to the many Africans who generously donated their DNA and time so that we could learn more about the origins of salicin bitter taste sensitivity in Africa. This work was supported by the US National Science Foundation (grant numbers BCS-0552486, BCS-0827436 to S.A.T.), US National Institutes of Health (grant numbers R01GM076637, 5DP1ES022577 05 to S.A.T.); US National Institutes of Health (grant number RO1 DC02995 to P.A.S.B.); and US National Institutes of Health (grant number DC010105 to J.B.R.).
References
- Bamshad M, Wooding SP. Signatures of natural selection in the human genome. Nat Rev Genet. 2003;4:99–111. doi: 10.1038/nrg999. [DOI] [PubMed] [Google Scholar]
- Bandelt HJ, Forster P, Rohl A. Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol. 1999;16:37–48. doi: 10.1093/oxfordjournals.molbev.a026036. [DOI] [PubMed] [Google Scholar]
- Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Barrett RD, Schluter D. Adaptation from standing genetic variation. Trends Ecol Evol. 2008;23:38–44. doi: 10.1016/j.tree.2007.09.008. [DOI] [PubMed] [Google Scholar]
- Bufe B, Breslin PAS, Kuhn C, Reed DR, Tharp CD, Slack JP, Kim UK, Drayna D, Meyerhof W. The molecular basis of individual differences in phenylthiocarbamide and propylthiouracil bitterness perception. Curr Biol. 2005;15:322–327. doi: 10.1016/j.cub.2005.01.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bufe B, Hofmann T, Krautwurst D, Raguse JD, Meyerhof W. The human TAS2R16 receptor mediates bitter taste in response to beta-glucopyranosides. Nat Genet. 2002;32:397–401. doi: 10.1038/ng1014. [DOI] [PubMed] [Google Scholar]
- Campa D, De Rango F, Carrai M, Crocco P, Montesanto A, Canzian F, Rose G, Rizzato C, Passarino G, Barale R. Bitter taste receptor polymorphisms and human aging. PLoS One. 2012;7:e45232. doi: 10.1371/journal.pone.0045232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell MC, Ranciaro A, Froment A, et al. (12 co-authors) Evolution of functionally diverse alleles associated with PTC bitter taste sensitivity in Africa. Mol Biol Evol. 2012;29:1141–1153. doi: 10.1093/molbev/msr293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell MC, Tishkoff SA. The evolution of human genetic and phenotypic variation in Africa. Curr Biol. 2010;20:R166–R173. doi: 10.1016/j.cub.2009.11.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coop G, Bullaughey K, Luca F, Przeworski M. The timing of selection at the human FOXP2 gene. Mol Biol Evol. 2008;25:1257–1259. doi: 10.1093/molbev/msn091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshpande DA, Wang WC, McIlmoyle EL, Robinett KS, Schillinger RM, An SS, Sham JS, Liggett SB. Bitter taste receptors on airway smooth muscle bronchodilate by localized calcium signaling and reverse obstruction. Nat Med. 2010;16:1299–1304. doi: 10.1038/nm.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fay JC, Wu CI. Hitchhiking under positive Darwinian selection. Genetics. 2000;155:1405–1413. doi: 10.1093/genetics/155.3.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu W, Akey JM. Selection and adaptation in the human genome. Annu Rev Genomics Hum Genet. 2013;14:467–489. doi: 10.1146/annurev-genom-091212-153509. [DOI] [PubMed] [Google Scholar]
- Goudet J. Fstat 2.9.3.2. 2002 [updated 2005 Aug 23; cited 2013 Nov 26]. Available from: http://www2.unil.ch/popgen/softwares/fstat.htm. [Google Scholar]
- Greene TA, Alarcon S, Thomas A, Berdougo E, Doranz BJ, Breslin PAS, Rucker JB. Probenecid inhibits the human bitter taste receptor TAS2R16 and suppresses bitter perception of salicin. PLoS One. 2011;6:e20123. doi: 10.1371/journal.pone.0020123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths RC. Genetree v. 9.0. 2007 [cited 2013 Nov 26]. Available from: http://www.stats.ox.ac.uk/∼griff/software.html. [Google Scholar]
- Grossman SR, Andersen KG, Shlyakhter I, et al. (19 co-authors) Identifying recent adaptations in large-scale genomic data. Cell. 2013;152:703–713. doi: 10.1016/j.cell.2013.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris H, Kalmus H. The distribution of taste thresholds for phenylthiourea of 384 sib pairs. Ann Eugen. 1951;16:226–230. doi: 10.1111/j.1469-1809.1951.tb02475.x. [DOI] [PubMed] [Google Scholar]
- Hay SI, Guerra CA, Gething PW, et al. (13 co-authors) A world malaria map: Plasmodium falciparum endemicity in 2007. PLoS Med. 2009;6:e1000048. doi: 10.1371/journal.pmed.1000048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes JE, Wallace MR, Knopik VS, Herbstman DM, Bartoshuk LM, Duffy VB. Allelic variation in TAS2R bitter receptor genes associates with variation in sensations from and ingestive behaviors toward common bitter beverages in adults. Chem Senses. 2011;36:311–319. doi: 10.1093/chemse/bjq132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinrichs AL, Wang JC, Bufe B, et al. (24 co-authors) Functional variant in a bitter-taste receptor (hTAS2R16) influences risk of alcohol dependence. Am J Hum Genet. 2006;78:103–111. doi: 10.1086/499253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson RR. Gene genealogies and the coalescent process. Oxford: Oxford University Press; 1990. [Google Scholar]
- Hudson RR, Slatkin M, Maddison WP. Estimation of levels of gene flow from DNA sequence data. Genetics. 1992;132:583–589. doi: 10.1093/genetics/132.2.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai H, Suzuki N, Ishimaru Y, Sakurai T, Yin L, Pan W, Abe K, Misaka T, Hirai H. Functional diversity of bitter taste receptor TAS2R16 in primates. Biol Lett. 2012;8:652–656. doi: 10.1098/rsbl.2011.1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen JD, Kim Y, DuMont VB, Aquadro CF, Bustamante CD. Distinguishing between selective sweeps and demography using DNA polymorphism data. Genetics. 2005;170:1401–1410. doi: 10.1534/genetics.104.038224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley JL. Systematic underestimation of the age of selected alleles. Front Genet. 2012;3:165. doi: 10.3389/fgene.2012.00165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee RJ, Xiong G, Kofonow JM, (17 co-authors) T2R38 taste receptor polymorphisms underlie susceptibility to upper respiratory infection. J Clin Invest. 2012;22:4145–4159. doi: 10.1172/JCI64240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Pakstis AJ, Kidd JR, Kidd KK. Selection on the human bitter taste gene, TAS2R16, in Eurasian populations. Hum Biol. 2011;83:363–377. doi: 10.3378/027.083.0303. [DOI] [PubMed] [Google Scholar]
- Librado P, Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- Mangold JE, Payne TJ, Ma JZ, Chen G, Li MD. Bitter taste receptor gene polymorphisms are an important factor in the development of nicotine dependence in African Americans. J Med Genet. 2008;45:578–582. doi: 10.1136/jmg.2008.057844. [DOI] [PubMed] [Google Scholar]
- Marth GT, Czabarka E, Murvai J, Sherry ST. The allele frequency spectrum in genome-wide human variation data reveals signals of differential demographic history in three large world populations. Genetics. 2004;166:351–372. doi: 10.1534/genetics.166.1.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald JH, Kreitman M. Adaptive protein evolution at the Adh locus in Drosophila. Nature. 1991;351:652–654. doi: 10.1038/351652a0. [DOI] [PubMed] [Google Scholar]
- Mueller KL, Hoon MA, Erlenbach I, Chandrashekar J, Zuker CS, Ryba NJ. The receptors and coding logic for bitter taste. Nature. 2005;434:225–229. doi: 10.1038/nature03352. [DOI] [PubMed] [Google Scholar]
- Peter BM, Huerta-Sanchez E, Nielsen R. Distinguishing between selective sweeps from standing variation and from a de novo mutation. PLoS Genet. 2012;8:e1003011. doi: 10.1371/journal.pgen.1003011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Przeworski M, Coop G, Wall JD. The signature of positive selection on standing genetic variation. Evolution. 2005;59:2312–2323. [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, et al. (11 co-authors) PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozengurt E, Sternini C. Taste receptor signaling in the mammalian gut. Curr Opin Pharmacol. 2007;7:557–562. doi: 10.1016/j.coph.2007.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabeti PC, Walsh E, Schaffner SF, et al. (15 co-authors) The case for selection at CCR5-Delta32. PLoS Biol. 2005;3:e378. doi: 10.1371/journal.pbio.0030378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakurai T, Misaka T, Ishiguro M, et al. (11 co-authors) Characterization of the beta-d-glucopyranoside binding site of the human bitter taste receptor hTAS2R16. J Biol Chem. 2010;285:28373–28378. doi: 10.1074/jbc.M110.144444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakurai T, Misaka T, Ueno Y, Ishiguro M, Matsuo S, Ishimaru Y, Asakura T, Abe K. The human bitter taste receptor, hTAS2R16, discriminates slight differences in the configuration of disaccharides. Biochem Biophys Res Commun. 2010;402:595–601. doi: 10.1016/j.bbrc.2010.10.059. [DOI] [PubMed] [Google Scholar]
- Scheet P, Stephens M. A fast and flexible statistical model for large-scale population genotype data: applications to inferring missing genotypes and haplotypic phase. Am J Hum Genet. 2006;78:629–644. doi: 10.1086/502802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheinfeldt LB, Soi S, Tishkoff SA. Colloquium paper: working toward a synthesis of archaeological, linguistic, and genetic data for inferring African population history. Proc Natl Acad Sci U S A. 2010;107(Suppl 2):8931–8938. doi: 10.1073/pnas.1002563107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah AS, Ben-Shahar Y, Moninger TO, Kline JN, Welsh MJ. Motile cilia of human airway epithelia are chemosensory. Science. 2009;325:1131–1134. doi: 10.1126/science.1173869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soranzo N, Bufe B, Sabeti PC, Wilson JF, Weale ME, Marguerie R, Meyerhof W, Goldstein DB. Positive selection on a high-sensitivity allele of the human bitter-taste receptor TAS2R16. Curr Biol. 2005;15:1257–1265. doi: 10.1016/j.cub.2005.06.042. [DOI] [PubMed] [Google Scholar]
- Stephens M, Smith NJ, Donnelly P. A new statistical method for haplotype reconstruction from population data. Am J Hum Genet. 2001;68:978–989. doi: 10.1086/319501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tishkoff SA, Reed FA, Friedlaender FR, et al. (25 co-authors) The genetic structure and history of Africans and African Americans. Science. 2009;324:1035–1044. doi: 10.1126/science.1172257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tishkoff SA, Reed FA, Ranciaro A, et al. (19 co-authors) Convergent adaptation of human lactase persistence in Africa and Europe. Nat Genet. 2007;39:31–40. doi: 10.1038/ng1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voight BF, Adams AM, Frisse LA, Qian Y, Hudson RR, Di Rienzo A. Interrogating multiple aspects of variation in a full resequencing data set to infer human population size changes. Proc Natl Acad Sci U S A. 2005;102:18508–18513. doi: 10.1073/pnas.0507325102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voight BF, Kudaravalli S, Wen X, Pritchard JK. A map of recent positive selection in the human genome. PLoS Biol. 2006;4:e72. doi: 10.1371/journal.pbio.0040072. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.