Abstract
Two colistin-susceptible/colistin-resistant (Cols/Colr) pairs of Acinetobacter baumannii strains assigned to international clone 2, which is prevalent worldwide, were sequentially recovered from two patients after prolonged colistin administration. Compared with the respective Cols isolates (Ab248 and Ab299, both having a colistin MIC of 0.5 μg/ml), both Colr isolates (Ab249 and Ab347, with colistin MICs of 128 and 32 μg/ml, respectively) significantly overexpressed pmrCAB genes, had single-amino-acid shifts in the PmrB protein, and exhibited significantly slower growth. The Colr isolate Ab347, tested by proteomic analysis in comparison with its Cols counterpart Ab299, underexpressed the proteins CsuA/B and C from the csu operon (which is necessary for biofilm formation). This isolate also underexpressed aconitase B and different enzymes involved in the oxidative stress response (KatE catalase, superoxide dismutase, and alkyl hydroperoxide reductase), suggesting a reduced response to reactive oxygen species (ROS) and, consequently, impaired colistin-mediated cell death through hydroxyl radical production. Cols isolates that were indistinguishable by macrorestriction analysis from Ab299 caused six sequential bloodstream infections, and isolates indistinguishable from Ab248 caused severe soft tissue infection, while Colr isolates indistinguishable from Ab347 and Ab249 were mainly colonizers. In particular, a Cols isolate identical to Ab299 was still invading the bloodstream 90 days after the colonization of this patient by Colr isolates. These observations indicate considerably lower invasiveness of A. baumannii clinical isolates following the development of colistin resistance.
INTRODUCTION
Acinetobacter baumannii has been an important nosocomial pathogen for the past 30 years, being frequently implicated in ventilator-associated pneumonia and bloodstream, urinary tract, and soft tissue infections (1). The propensity to develop antibiotic resistance makes A. baumannii a difficult-to-treat pathogen (2).
As severe infections caused by multidrug-resistant A. baumannii clinical isolates are increasing worldwide, colistin often constitutes the only active treatment alternative (3). Colistin is rapidly bactericidal for Gram-negative bacteria, affecting the lipid A moiety of lipopolysaccharide (LPS) and thus disorganizing the outer membrane (4). During the last few years, clinical isolates of A. baumannii expressing resistance to colistin have emerged, and outbreaks have been reported (5). The exact mechanism of resistance to colistin still needs to be elucidated, although two unlinked hypotheses have been expressed, involving (i) mutations and overexpression of PmrCAB proteins, leading to LPS modifications (phosphoethanolamine addition on lipid A), or (ii) complete loss of LPS production through inactivation of a lipid A biosynthesis gene (5).
The emergence of colistin resistance has been correlated with the selective pressure exerted by prolonged exposure to this drug (6, 7). There are also preliminary observations in laboratory-derived strains that colistin-resistant A. baumannii may exhibit impaired virulence and in vivo fitness (7). To investigate this hypothesis and also the mechanisms that confer colistin resistance in A. baumannii, we characterized two colistin-susceptible/colistin-resistant (Cols/Colr) pairs of A. baumannii clinical isolates, with each pair to be recovered from the same patient and to include isolates with identical macrorestriction patterns. We report here that compared with the respective Cols isolates, the Colr isolates may exhibit significant growth retardation, impaired virulence, and also considerably lower clinical invasiveness.
MATERIALS AND METHODS
Study isolates and susceptibility testing.
The study included two Cols/Colr pairs of A. baumannii isolates that were recovered consecutively from two intensive-care unit (ICU) patients, as well as the Cols A. baumannii strain ATCC 19606 as a control. The isolates were provisionally identified as belonging to the A. baumannii complex by API 20NE (bioMérieux, Marcy l'Etoile, France) and were all identified as A. baumannii by positive PCR/sequencing that revealed the carriage of a blaOXA-51-like variant gene. Susceptibilities to β-lactams, co-trimoxazole, rifampin, aminoglycosides, and quinolones were determined by Etest (bioMérieux). Colistin MICs were initially determined by Etest and subsequently determined by broth macrodilution (8) using glass tubes (tube dilution [TDS]), which was recently shown to exhibit excellent performance for colistin MIC testing of multidrug-resistant A. baumannii isolates (9).
Typing assays.
The genetic relationship of all Cols/Colr isolates that were consecutively recovered from the study patients during the study period was tested by pulsed-field gel electrophoresis (PFGE) of ApaI-digested genomic DNA (10); the banding patterns were compared visually using previously proposed criteria (11). The first Cols/Colr isolations of each pair were further tested with the multilocus sequence typing (MLST) scheme developed by the Institute Pasteur (12) and were assigned an MLST (ST) type.
PCR and sequencing.
PCRs for the blaOXA-51-like, blaOXA-58-like, blaOXA-23-like, blaOXA-24-like, blaVIM, blaIMP, blaSIM, pmrA, pmrB, pmrC, lpxA, lpxC, and lpxD genes were performed as described previously (3, 13–16). The nucleotide sequences of both strands of PCR products were determined at Macrogen Inc., Seoul, South Korea, and sequence analysis was carried out with DNAStar software (version 5.07; Lasergene, Madison, WI).
qRT-PCR.
The expression of the pmrA, pmrB, and pmrC genes was tested by quantitative real-time reverse transcription-PCR (qRT-PCR) with the two pairs of clinical isolates in comparison with the A. baumannii control strain ATCC 19606, as described previously (3, 17). In particular, bacteria were grown to the mid-logarithmic phase (as shown in the growth analysis), and total cellular RNA was extracted with an RNeasy minikit (Qiagen, West Sussex, United Kingdom). RNA abundance was quantified spectrophotometrically at 260 nm, and contaminating DNA was removed by DNase I treatment (Promega, Madison, WI, USA). The quantitative RT-PCR was performed with the SuperScript III Platinum SYBR green one-step qRT-PCR kit (Invitrogen Corporation, Carlsbad, CA, USA) with 12 ng of total RNA and previously used primers (3). The 16S rRNA gene was used as an internal control for quantification of relative gene expression (3). Control reactions with untranscribed RNA were also included to detect DNA contamination. The expression of genes was described as the mean values from three independent experiments.
Growth curves.
Growth rates were determined for the two pairs of clinical isolates and the control strain ATCC 19606. Growth curves were performed in triplicate by diluting equal numbers of CFU of each isolate (approximately 5 × 105 CFU/ml) in Muller-Hinton broth, followed by incubation at 37°C under constant shaking. CFU were enumerated after serially 10-fold diluting broth cultures and plating in antibiotic-free medium at each time point from 12 h to 48 h. Growth of isolates at each time point was statistically compared by using the paired t test and Minitab software (version 13.31; Minitab Inc., State College, PA); a P value of <0.05 indicated statistical significance.
Proteomic analysis.
Proteins of bacterial envelopes from the clinical isolates Ab299 and Ab347 were extracted as described previously (18) from cultures with final optical densities at 600 nm (OD600) of 1.5 and 1.7, respectively. Protein concentration, trypsin digestion, and mass spectrometry analyses were performed as described previously (19, 20), except for the injection volume, which was 1 μl. For protein quantification, we used Progenesis LC-MS software (Nonlinear Dynamics) with the same parameters as used previously (19, 20). The merged peak list was searched against the A. baumannii ATCC 17978 database (www.genoscope.cns.fr) using a local version of Mascot (version 2.2; Matrix Science, United Kingdom). Protein fold change was taken into account when it was above 2. At least 3 peptides were used for protein identification and quantification.
RESULTS
First Cols/Colr A. baumannii pair.
A 61-year-old-patient was hospitalized during October 2008 due to severe neurological disease and respiratory distress. The patient was transferred to the intensive-care unit (ICU), underwent tracheostomy and mechanical ventilation, and was given treatment with ampicillin-sulbactam. Blood, urine, and bronchial cultures showed no growth. On hospitalization day 17, an A. baumannii isolate (Ab299) that was resistant to many antibiotic classes and susceptible to colistin (MIC, 0.5 μg/ml) was first recovered from bronchial secretions. During the next 100 days, another 14 A. baumannii isolates with the same susceptibility profile were intermittently grown from bronchial secretions and five isolates from blood. During this period, the patient suffered from bloodstream infection episodes not only due to this A. baumannii strain but also due to Burkholderia cepacia and received multiple antibiotic regimens (including ampicillin-sulbactam, gentamicin, tigecycline, ciprofloxacin, and meropenem for more than 10 days each) and also colistin for a total of 28 days. On day 122, a Colr A. baumannii isolate (Ab347; MIC, 32 μg/ml) was first recovered from bronchial secretions with no signs of infection, and subsequently, another 12 Colr isolates phenotypically similar to Ab347 and indistinguishable from Ab347 by PFGE were grown from bronchial secretions up to day 206. It should be noted that a Cols isolate indistinguishable from Ab299 caused another bloodstream infection episode on day 212, while no Colr isolate was recovered from blood during the whole hospitalization. The patient was discharged on day 224 in a good condition.
Second Cols/Colr A. baumannii pair.
A 77-year-old-patient with previous coronary artery bypass surgery was admitted to the hospital in July 2008 due to sternotomy abscess and mediastinitis. The patient was transferred to the ICU; surgical debridement of the sternotomy was conducted, and empirical treatment with imipenem, teicoplanin, and metronidazole was started. On hospitalization day 14, a carbapenem-resistant, Cols A. baumannii isolate (Ab248; MIC, 0.5 μg/ml) was recovered from cultures of drained pus, and treatment with colistin plus tigecycline was administered. On day 37, when the patient's condition was considerably improved, drainage cultures yielded the Colr A. baumannii isolate Ab249 (MIC, 128 μg/ml), and subsequently another three phenotypically similar Colr isolates were recovered, but no antibiotic treatment was given due to the absence of clinical signs of infection. The patient discharged on day 65.
Characteristics of the isolates.
The characteristics of the four A. baumannii study isolates are listed in Table 1. Both pairs of isolates were carbapenem resistant (imipenem and meropenem MICs of >32 μg/ml) and also resistant to most available antibiotics except tigecycline and ampicillin-sulbactam. Ab248, Ab249, and Ab299 were intermediate susceptible, but Ab347 was resistant, to gentamicin and tobramycin, while all four isolates had low rifampin MICs (3 to 6 μg/ml). Colistin MICs determined by TDS were 0.5 μg/ml for the Cols isolates Ab248 and Ab299, while being 128 μg/ml and 32 μg/ml for the Colr isolates Ab249 and Ab347, respectively. It should be noted that both Colr isolates exhibited Etest MICs of 4 μg/ml when tested using the same inoculum with TDS; similar discrepancies between Etest and TDS and other dilution methods for the determination of colistin MICs in multiresistant A. baumannii isolates were also shown recently (9).
TABLE 1.
Characteristics of the A. baumannii study isolates
| Isolate | Date of isolation (mo/day/yr) | Specimen | Colistin TDS MIC (μg/ml)/susceptibility statusa | blaOXA genes | PFGE type | ST | pmrB genotype |
|---|---|---|---|---|---|---|---|
| Ab248 | 8/3/2008 | Pus | 0.5/S | blaOXA-66, blaOXA-58 | Ia | 2 | Wild type |
| Ab249 | 8/26/2008 | Pus | 128/R | blaOXA-66, blaOXA-58 | Ia | 2 | P233S |
| Ab299 | 10/17/2008 | Bronchial secretion | 0.5/S | blaOXA-66, blaOXA-58 | Ib | 2 | Wild type |
| Ab347 | 1/30/2009 | Bronchial secretion | 32/R | blaOXA-66, blaOXA-58 | Ib | 2 | P170L |
S, susceptible; R, resistant.
Typing assays.
All Cols/Colr clinical isolates within each pair had identical PFGE profiles, with the PFGE profile of each pair differing by 2 bands from that of the other pair. All four isolates (Ab299/Ab347 and Ab248/Ab249) tested by MLST belonged to ST2 (international clone 2), which is currently predominant in most regions worldwide (21, 22).
PCR and sequencing.
PCR for β-lactamase genes was positive for the blaOXA-51-like and blaOXA-58-like genes and negative for the blaOXA-23-like, blaOXA-24-like, and class B carbapenemase genes. By nucleotide sequencing, all Cols/Colr study isolates were shown to carry the oxacillinase alleles blaOXA-66 and blaOXA-58 and did not have any mutations in genes pmrA, pmrC, lpxA, lpxC, and lpxD. Compared with the Cols isolate Ab299, the Colr isolate Ab347 harbored one nucleotide shift in pmrB, leading to the amino acid replacement P170L in the PmrB protein, which is involved in colistin resistance via LPS modifications (23). Similarly, the Colr isolate Ab249 harbored the amino acid replacement P233S in the PmrB protein compared with the Cols isolate Ab248.
qRT-PCR.
The Colr isolates Ab249 and Ab347 had increased expression of genes pmrA (3.6- and 10.9-fold, respectively) and pmrB (5.7- and 23.7-fold, respectively) compared with that of their Cols counterparts Ab248 and Ab299, respectively, whereas they exhibited smaller increases in pmrC expression (1.6- and 2.4-fold, respectively). The relative gene expression represents the mean from three independent experiments. The differences in pmrCAB gene expression were significant in both pairs of isolates (P < 0.05 for all genes).
Growth analysis.
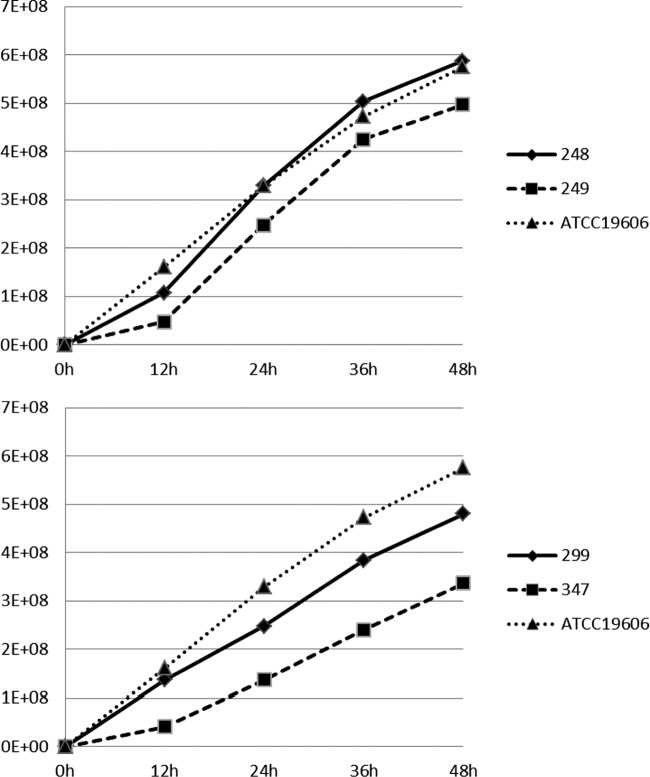
The growth curves of the two pairs of Cols/Colr clinical isolates and the ATCC 19606 control are shown in Fig. 1. The growth of the Colr isolates was considerably slower than that of the respective Cols isolates, with the difference being significant (P < 0.05) at all time points for the pair Ab299/Ab347 and at 24 to 48 h for the pair Ab248/Ab249. It should be noted that the colonies grown from the growth analysis tubes of the Colr isolate Ab347 were considerably smaller than those of the Cols isolate Ab299.
FIG 1.
Growth curves of the study and control isolates. y axis, CFU/ml from broth cultures; x axis, time of growth (hours).
Proteomic analysis.
Interestingly, the proteomic comparison between the envelopes of Cols/Colr isolates Ab299/Ab347 showed a significant underexpression of the CsuA/B and CsuC proteins in Ab347 (Table 2). These proteins are part of the chaperone-usher pilus assembly system that produces pili necessary for biofilm formation. This underexpression supported the impaired biofilm formation that has already been observed by Fernandez-Reyes et al. (24) for Colr strains. The Colr isolate also underexpressed different enzymes involved in the oxidative stress response, i.e., the KatE catalase, the superoxide dismutase, and the alkyl hydroperoxide reductase, suggesting a reduced capacity or necessity to respond to reactive oxygen species (ROS). Furthermore, of note, the aconitate hydratase (aconitase B) enzyme of the tricarboxylic acid (TCA) cycle is underexpressed in the Colr isolate, suggesting that the production of NADH may be reduced and that the rapid cell death caused by colistin through hydroxyl radical production may be impaired (25). As already observed by Fernandez-Reyes et al. (24) in an A. baumannii reference strain where colistin resistance was induced, the carbapenem resistance-associated protein CarO was also underexpressed. Finally, among proteins associated with antibiotic resistance, the carbapenem-hydrolyzing oxacillinase OXA-66 was significantly underexpressed and the putative RND-type efflux pump AdeT, which is involved in aminoglycoside resistance, was significantly overexpressed in the Colr isolate. It should be noted that Ab299 and other indistinguishable Cols isolates were resistant to amikacin and netilmicin and intermediate to gentamicin and tobramycin (MICs of 6 and 8 μg/ml, respectively), while Ab347 and indistinguishable Colr isolates exhibited considerably elevated MICs to gentamicin and tobramycin (32 and 64 μg/ml, respectively).
TABLE 2.
Proteins related to antibiotic resistance and virulence differentially expressed in Colr strain Ab347
| Accession no. | No. of peptidesa | Scoreb | ANOVAc (P) | Fold changed | Description |
|---|---|---|---|---|---|
| ABYAL1639|katE | 39 | 2,919 | 1.78 × 10−13 | −35.39 | Catalase hydroperoxidase II |
| ABYAL3026 | 4 | 207 | 5.00 × 10−15 | −28.34 | Putative porin protein associated with imipenem resistance, CarO |
| ABYAL2670 | 6 | 549 | 3.27 × 10−13 | −27.38 | Putative protein CsuA/B |
| ABYAL2667 | 3 | 130 | 1.24 × 10−6 | −14.64 | Putative protein CsuC |
| ABYAL2566|acnB | 9 | 410 | 1.64 × 10−9 | −3.13 | Fragment of aconitate hydratase 2 (part 3) |
| ABYAL2567|acnB | 7 | 332 | 1.43 × 10−7 | −2.46 | Fragment of aconitate hydratase 2 (part 2) |
| ABYAL3715|sodC | 7 | 376 | 1.41 × 10−4 | −2.15 | Superoxide dismutase precursor (Cu-Zn) |
| ABYAL1416|ahpC | 14 | 881 | 1.81 × 10−8 | −2.13 | Alkyl hydroperoxide reductase (detoxification of hydroperoxides) |
| ABYAL2568|acnB | 7 | 291 | 3.17 × 10−6 | −2.12 | Fragment of aconitate hydratase 2 (part 1) |
| ABYAL1790|blaOXA-66 | 3 | 104 | 2.17 × 10−7 | −2.03 | Carbapenem-hydrolyzing oxacillinase OXA-66 |
| ABYAL0009 | 9 | 578 | 1.08 × 10−9 | +3.15 | Putative RND-type efflux pump involved in aminoglycoside resistance (AdeT) |
Number of peptides for identification and quantification.
Confidence score for identification by Mascot software.
ANOVA, analysis of variance.
Negative values indicate underexpression in Colr strain Ab347.
DISCUSSION
The growing worldwide issue of antibiotic resistance among A. baumannii isolates often necessitates the use of colistin for the treatment of severe infections, particularly in ICUs (3). However, the widespread administration of colistin in hospital settings has exerted selective pressure for the development of resistant A. baumannii clinical isolates, which are increasingly reported from many regions (5, 26).
The exact colistin resistance mechanism has not yet been elucidated, although two unrelated resistance mechanisms affecting lipid A, the target molecule of colistin, have been described in A. baumannii isolates from different locations. The first hypothesis suggested the complete loss of LPS through inactivation of lipid A biosynthesis gene lpxA, lpxC, or lpxD (16). The second proposed mechanism showed mutations in the genes pmrA and pmrB leading to phosphoethanolamine addition to hepta-acylated lipid A to be linked to colistin resistance in A. baumannii, along with increased expression of the PmrCAB system (3, 23, 27). In the present study, the underlying mechanisms of colistin resistance development were mutations in PmrB protein along with pmrCAB upregulation, while the lpxA, lpxC, and lpxD genes were not affected. It should be noted that in most of the previous clinical or laboratory reports of colistin resistance mechanisms in A. baumannii, resistance development was linked to exposure to colistin (3, 5, 6), similarly to our clinical cases.
It has been recently demonstrated that the mechanism of bacterial killing by colistin could be via the production of hydroxyl radicals, leading to rapid cell death (25). This mechanism remained active in A. baumannii multidrug-resistant strains, but hydroxyl radical production might be abolished when colistin resistance appeared (25). In this study, the proteomic comparison of envelopes from isolates Ab299/Ab347 showed an underexpression of the aconitase B protein, a component of the TCA cycle, in the Colr isolate Ab347. Therefore, in accordance with the model and studies described by Kohanski et al. (28), this underexpression should reduce the production of NADH, decrease superoxide generation (and then decrease ferrous iron availability for the Fenton reaction), and finally increase the bacterial resistance to colistin. In accordance with a reduced cell death following colistin exposure, the large underexpression in the Colr isolate Ab347 of enzymes such as catalase, alkyl hydroperoxide reductase, and superoxide dismutase may be linked to a reduced necessity to counteract ROS production.
Colr A. baumannii laboratory strains or, very recently, clinical isolates have been associated with reduced virulence relative to that of their Cols counterparts, as reflected by reduced experimental fitness and mortality in animals (7, 29). In the current study, the two Colr isolates, compared with their respective Cols clinical isolates, exhibited significantly slower growth, indicating a reduced fitness. Further, the Ab347 Colr isolate underexpressed the Csu system, which is necessary for biofilm formation, the outer membrane protein CarO (30), and antioxidant proteins that could protect it from ROS toxic effects generated by macrophages (31), overall possibly indicating a lower virulence of the Colr isolates and supporting the previous observations (7, 29). Furthermore, it had been reported that a Colr isolate was associated with a prolonged clinical carriage without signs of infection, in contrast to the case for its Cols counterpart causing bloodstream infections (6). In our case patients, of particular importance, the Colr isolates probably had a reduced ability to cause invasive infection, as the patients remained carriers for prolonged time periods. Furthermore, their Cols counterparts repeatedly caused severe clinical infections, including multiple bloodstream infections, while a Cols isolate from case patient 1 was still evading the bloodstream long after the emergence of Colr isolates. These observations could suggest that the changes contributing to colistin resistance development confer a considerable fitness cost and affect the capacity to produce clinical infections.
ACKNOWLEDGMENT
This study was funded by internal funding.
Footnotes
Published ahead of print 18 November 2013
REFERENCES
- 1.Fournier PE, Richet H. 2006. The epidemiology and control of Acinetobacter baumannii in health care facilities. Clin. Infect. Dis. 42:692–699. 10.1086/500202 [DOI] [PubMed] [Google Scholar]
- 2.Bonomo RA, Szabo D. 2006. Mechanisms of multidrug resistance in Acinetobacter species and Pseudomonas aeruginosa. Clin. Infect. Dis. 43(Suppl 2):S49–S56. 10.1086/504477 [DOI] [PubMed] [Google Scholar]
- 3.Beceiro A, Llobet E, Aranda J, Bengoechea JA, Doumith M, Hornsey M, Dhanji H, Chart H, Bou G, Livermore DM, Woodford N. 2011. Phosphoethanolamine modification of lipid A in colistin-resistant variants of Acinetobacter baumannii mediated by the pmrAB two-component regulatory system. Antimicrob. Agents Chemother. 55:3370–3379. 10.1128/AAC.00079-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hancock RE. 1997. Peptide antibiotics. Lancet 349:418–422. 10.1016/S0140-6736(97)80051-7 [DOI] [PubMed] [Google Scholar]
- 5.Cai Y, Chai D, Wang R, Liang B, Bai N. 2012. Colistin resistance of Acinetobacter baumannii: clinical reports, mechanisms and antimicrobial strategies. J. Antimicrob. Chemother. 67:1607–1615. 10.1093/jac/dks084 [DOI] [PubMed] [Google Scholar]
- 6.Rolain JM, Roch A, Castanier M, Papazian L, Raoult D. 2011. Acinetobacter baumannii resistant to colistin with impaired virulence: a case report from France. J. Infect. Dis. 204:1146–1147. 10.1093/infdis/jir475 [DOI] [PubMed] [Google Scholar]
- 7.Lopez-Rojas R, Dominguez-Herrera J, McConnell MJ, Docobo-Perez F, Smani Y, Fernandez-Reyes M, Rivas L, Pachon J. 2011. Impaired virulence and in vivo fitness of colistin-resistant Acinetobacter baumannii. J. Infect. Dis. 203:545–548. 10.1093/infdis/jiq086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clinical and Laboratory Standards Institute 2010. Performance standards for antimicrobial susceptibility testing, 20th informational supplement. M100-S20. Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 9.Hindler JA, Humphries RM. 2013. Colistin MIC variability by method for contemporary clinical isolates of multidrug-resistant gram-negative bacilli. J. Clin. Microbiol. 51:1678–1684. 10.1128/JCM.03385-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ikonomidis A, Neou E, Gogou V, Vrioni G, Tsakris A, Pournaras S. 2009. Heteroresistance to meropenem in carbapenem-susceptible Acinetobacter baumannii. J. Clin. Microbiol. 47:4055–4059. 10.1128/JCM.00959-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tenover FC, Arbeit RD, Goering RV, Mickelsen PA, Murray BE, Persing DH, Swaminathan B. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233–2239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Diancourt L, Passet V, Nemec A, Dijkshoorn L, Brisse S. 2010. The population structure of Acinetobacter baumannii: expanding multiresistant clones from an ancestral susceptible genetic pool. PLoS One 5:e10034. 10.1371/journal.pone.0010034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pournaras S, Markogiannakis A, Ikonomidis A, Kondyli L, Bethimouti K, Maniatis AN, Legakis NJ, Tsakris A. 2006. Outbreak of multiple clones of imipenem-resistant Acinetobacter baumannii isolates expressing OXA-58 carbapenemase in an intensive care unit. J. Antimicrob. Chemother. 57:557–561. 10.1093/jac/dkl004 [DOI] [PubMed] [Google Scholar]
- 14.Lee K, Yum JH, Yong D, Lee HM, Kim HD, Docquier JD, Rossolini GM, Chong Y. 2005. Novel acquired metallo-β-lactamase gene, blaSIM-1, in a class 1 integron from Acinetobacter baumannii clinical isolates from Korea. Antimicrob. Agents Chemother. 49:4485–4491. 10.1128/AAC.49.11.4485-4491.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Oh EJ, Lee S, Park YJ, Park JJ, Park K, Kim SI, Kang MW, Kim BK. 2003. Prevalence of metallo-β-lactamase among Pseudomonas aeruginosa and Acinetobacter baumannii in a Korean university hospital and comparison of screening methods for detecting metallo-β-lactamase. J. Microbiol. Methods 54:411–418. 10.1016/S0167-7012(03)00090-3 [DOI] [PubMed] [Google Scholar]
- 16.Moffatt JH, Harper M, Harrison P, Hale JD, Vinogradov E, Seemann T, Henry R, Crane B, St Michael F, Cox AD, Adler B, Nation RL, Li J, Boyce JD. 2010. Colistin resistance in Acinetobacter baumannii is mediated by complete loss of lipopolysaccharide production. Antimicrob. Agents Chemother. 54:4971–4977. 10.1128/AAC.00834-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Higgins PG, Wisplinghoff H, Stefanik D, Seifert H. 2004. Selection of topoisomerase mutations and overexpression of adeB mRNA transcripts during an outbreak of Acinetobacter baumannii. J. Antimicrob. Chemother. 54:821–823. 10.1093/jac/dkh427 [DOI] [PubMed] [Google Scholar]
- 18.Marti S, Nait Chabane Y, Alexandre S, Coquet L, Vila J, Jouenne T, Dé E. 2011. Growth of Acinetobacter baumannii in pellicle enhanced the expression of potential virulence factors. PLoS One 6:e26030. 10.1371/journal.pone.0026030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Van Oudenhove L, Devreese B. 2013. A review on recent developments in mass spectrometry instrumentation and quantitative tools advancing bacterial proteomics. Appl. Microbiol. Biotechnol. 97:4749–4762. 10.1007/s00253-013-4897-7 [DOI] [PubMed] [Google Scholar]
- 20.Michaux C, Martini C, Shioya K, Ahmed Lecheheb S, Budin-Verneuil A, Cosette P, Sanguinetti M, Hartke A, Verneuil N, Giard JC. 2012. CspR, a cold shock RNA-binding protein involved in the long-term survival and the virulence of Enterococcus faecalis. J. Bacteriol. 194:6900–6908. 10.1128/JB.01673-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Higgins PG, Dammhayn C, Hackel M, Seifert H. 2010. Global spread of carbapenem-resistant Acinetobacter baumannii. J. Antimicrob. Chemother. 65:233–238. 10.1093/jac/dkp428 [DOI] [PubMed] [Google Scholar]
- 22.Zarrilli R, Pournaras S, Giannouli M, Tsakris A. 2013. Global evolution of multidrug-resistant Acinetobacter baumannii clonal lineages. Int. J. Antimicrob. Agents 41:11–19. 10.1016/j.ijantimicag.2012.09.008 [DOI] [PubMed] [Google Scholar]
- 23.Adams MD, Nickel GC, Bajaksouzian S, Lavender H, Murthy AR, Jacobs MR, Bonomo RA. 2009. Resistance to colistin in Acinetobacter baumannii associated with mutations in the PmrAB two-component system. Antimicrob. Agents Chemother. 53:3628–3634. 10.1128/AAC.00284-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fernández-Reyes M, Rodríguez-Falcón M, Chiva C, Pachón J, Andreu D, Rivas L. 2009. The cost of resistance to colistin in Acinetobacter baumannii: a proteomic perspective. Proteomics 9:1632–1645. 10.1002/pmic.200800434 [DOI] [PubMed] [Google Scholar]
- 25.Sampson TR, Liu X, Schroeder MR, Kraft CS, Burd EM, Weiss DS. 2012. Rapid killing of Acinetobacter baumannii by polymyxins is mediated by a hydroxyl radical death pathway. Antimicrob. Agents Chemother. 56:5642–5649. 10.1128/AAC.00756-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Valencia R, Arroyo LA, Conde M, Aldana JM, Torres MJ, Fernández-Cuenca F, Garnacho-Montero J, Cisneros JM, Ortíz C, Pachón J, Aznar J. 2009. Nosocomial outbreak of infection with pan-drug-resistant Acinetobacter baumannii in a tertiary care university hospital. Infect. Control Hosp. Epidemiol. 30:257–263. 10.1086/595977 [DOI] [PubMed] [Google Scholar]
- 27.Park YK, Choi JY, Shin D, Ko KS. 2011. Correlation between overexpression and amino acid substitution of the PmrAB locus and colistin resistance in Acinetobacter baumannii. Int. J. Antimicrob. Agents 37:525–530. 10.1016/j.ijantimicag.2011.02.008 [DOI] [PubMed] [Google Scholar]
- 28.Kohanski MA, Dwyer DJ, Hayete B, Lawrence CA, Collins JJ. 2007. A common mechanism of cellular death induced by bactericidal antibiotics. Cell 130:797–810. 10.1016/j.cell.2007.06.049 [DOI] [PubMed] [Google Scholar]
- 29.López-Rojas R, McConnell MJ, Jiménez-Mejías ME, Domínguez-Herrera J, Fernández-Cuenca F, Pachón J. 2013. Colistin resistance in a clinical Acinetobacter baumannii strain appearing after colistin treatment: effect on virulence and bacterial fitness. Antimicrob. Agents Chemother. 57:4587–4589. 10.1128/AAC.00543-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fernández-Cuenca F, Smani Y, Gómez-Sánchez MC, Docobo-Pérez F, Caballero-Moyano FJ, Domínguez-Herrera J, Pascual A, Pachón J. 2011. Attenuated virulence of a slow-growing pandrug-resistant Acinetobacter baumannii is associated with decreased expression of genes encoding the porins CarO and OprD-like. Int. J. Antimicrob. Agents 38:548–549. 10.1016/j.ijantimicag.2011.08.002 [DOI] [PubMed] [Google Scholar]
- 31.Mendez JA, Soares NC, Mateos J, Gayoso C, Rumbo C, Aranda J, Tomas M, Bou G. 2012. Extracellular proteome of a highly invasive multidrug-resistant clinical strain of Acinetobacter baumannii. J. Proteome Res. 11:5678–5694. 10.1021/pr300496c [DOI] [PubMed] [Google Scholar]