Abstract
Although biofilm-based bioprocesses have been increasingly used in various applications, the long-term robust and efficient biofilm performance remains one of the main bottlenecks. In this study, we demonstrated that biofilm cohesiveness and performance of Shewanella oneidensis can be enhanced through disrupting putrescine biosynthesis. Through random transposon mutagenesis library screening, one hyperadherent mutant strain, CP2-1-S1, exhibiting an enhanced capability in biofilm formation, was obtained. Comparative analysis of the performance of biofilms formed by S. oneidensis MR-1 wild type (WT) and CP2-1-S1 in removing dichromate (Cr2O72−), i.e., Cr(VI), from the aqueous phase showed that, compared with the WT biofilms, CP2-1-S1 biofilms displayed a substantially lower rate of cell detachment upon exposure to Cr(VI), suggesting a higher cohesiveness of the mutant biofilms. In addition, the amount of Cr(III) immobilized by CP2-1-S1 biofilms was much larger, indicating an enhanced performance in Cr(VI) bioremediation. We further showed that speF, a putrescine biosynthesis gene, was disrupted in CP2-1-S1 and that the biofilm phenotypes could be restored by both genetic and chemical complementations. Our results also demonstrated an important role of putrescine in mediating matrix disassembly in S. oneidensis biofilms.
INTRODUCTION
In most natural, engineered, and medical settings, bacterial cells often attach onto surfaces or interfaces and develop into sessile communities known as biofilms (1). In biofilms, cells are embedded in a self-produced matrix composed of extracellular polymeric substances (EPS), including proteins, polysaccharides, lipids, and extracellular DNA. Extensive studies have demonstrated a wide range of advantages of the presence of an EPS matrix for the biofilm mode of life (2, 3). For example, compared to their planktonic counterparts, biofilms have been reported to be more tolerant to harsh physicochemical conditions as generated by various toxic contaminants or biocides (1, 4). Because of these advantages, biofilms have been increasingly applied to the efficient removal of toxic contaminants from industrial wastewater (5–8).
Solid substrata, typically suspended plastic pieces or fixed synthetic mesh, have been added to wastewater treatment bioreactors to provide attachment surfaces for biofilms to enhance contaminant removal (9). However, controlling biofilm-based bioprocesses is very challenging because of the intrinsic heterogeneous and dynamic properties of biofilms (10, 11). Previous studies have shown that either unfavorable environmental conditions (e.g., exposure to toxic contaminants) or variations in nutrient supply (e.g., dissolved O2 availability) could cause cell dispersal from biofilms (12–15), resulting in a decrease in the performance of the biofilm-based bioprocesses. To improve biofilm performance, modification of physicochemical properties of substratum surfaces has been applied to enhance biofilm attachment so that the adhesion of biofilms onto substrata becomes stronger, potentially preventing biofilms from being stripped away (16). However, the ability of cells and EPS to stick together in biofilms, i.e., biofilm cohesiveness, could not be improved by enhancing surface attachment through surface modification.
Shewanella oneidensis is a metal-reducing bacterium capable of respiring on metal ions, metal oxides, and solid electrodes (17, 18). Biofilms of S. oneidensis play an important role in many environmental and biotechnological processes, including removing toxic metal contaminants, such as uranium and chromium, from the aqueous phase by transforming them into less soluble species (17, 19–21). Previous studies have suggested that exposure to toxic contaminants, e.g., chromate, could cause dispersal of S. oneidensis biofilms (14). We hypothesize that, through enhancing biofilm cohesiveness, biofilm dispersal triggered by toxic chemicals could be alleviated and biofilm performance could be improved. Here, we used S. oneidensis as a model organism and dichromate as a model contaminant to test this hypothesis.
The specific objectives of this study were to (i) generate an S. oneidensis mutant strain capable of forming highly cohesive biofilms and (ii) evaluate the performance of the cohesive biofilms in Cr(VI) immobilization. Through screening of a random transposon mutagenesis library, we obtained one hyperadherent strain with enhanced biofilm formation capability under both static and hydrodynamic conditions. This mutant showed an enhanced biofilm cohesiveness and performance as indicated by a high viscosity, a low dispersal rate upon exposure to Cr(VI), and an improved Cr(VI) immobilization. Inverse PCR showed the disruption of a putrescine biosynthesis gene, speF, in this mutant, which was confirmed by chemical and genetic complementation experiments. We further elucidated a potential role of putrescine in mediating biofilm matrix disassembly in S. oneidensis.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Stock cultures of S. oneidensis MR-1 (wild type [WT]) and mutants were maintained in lysogeny broth (LB) medium with 20% glycerol at −80°C. Unless otherwise stated, cultures were grown aerobically at 30°C in LB medium or modified M1 defined minimal medium (MM1) (22). The MM1 consisted of 30.00 mM HEPES, 7.50 mM NaOH, 28.04 mM NH4Cl, 1.34 mM KCl, 4.35 mM NaH2PO4, and 0.68 mM CaCl2 supplemented with trace amounts of minerals, vitamins, and amino acids (pH ∼7.0) (23, 24). For bacterial growth in MM1, 20 mM sodium lactate was used as an electron donor and the sole carbon source.
Transposon mutagenesis and mutant selection.
Transposon mutagenesis of S. oneidensis MR-1 was carried out through filter mating with Escherichia coli harboring TnM using the helper strain RK600 according to previously described protocols (25–27). LB medium was used for bacterial growth, and when necessary, the medium was supplemented with 25 μg/ml of kanamycin, 15 μg/ml of tetracycline, 10 μg/ml of gentamicin, and/or 6 μg/ml of chloramphenicol. To select mutants with enhanced biofilm formation capability, 2 μl of overnight (∼16-h) culture of each mutant generated in transposon mutagenesis was dropped onto MM1 agar (1.5% agar in MM1 medium with 20 mM lactate) containing 50 μg/ml Congo red and incubated at 30°C.
Zeta potential measurement and hydrophobicity assay.
The bacterial cell surface charge was assessed by measuring its electrokinetic potential (zeta potential), which characterizes the electrical double-layer potential on the cell surface. For zeta potential measurement, cells from overnight (∼16-h) aerobic cultures of S. oneidensis in MM1 were harvested by centrifugation (6,000 × g, 10 min) and washed three times with 0.9% NaCl. Cells were then resuspended in 10 ml of 0.9% NaCl (pH 7.0) to an optical density at 600 nm (OD600) of ∼0.50, and zeta potential was measured using the Malvern Zetasizer Nano Z dedicated zeta potential analyzer (Nano ZS; Malvern Instruments, Worcestershire, United Kingdom). For the hydrophobicity assay, cells from overnight cultures in MM1 were harvested, washed three times, and resuspended in 2 ml of 0.9% NaCl. Then, 0.4 ml of hexadecane was added to each cell suspension, and the mixtures were vortexed for 15 min followed by standing for 15 min at room temperature for phase separation. Hydrophobicity was calculated as the percentage of cells transferring from aqueous phase to n-hexadecane phase, i.e., (1 − A/A0) × 100%, where A0 and A are OD600s of the aqueous phase before and after mixing with hexadecane. Zeta potential measurement and the hydrophobicity assay were carried out using two independent cultures for each strain with at least three replicates for each culture.
Biofilm formation assay.
The static biofilm formation assay was based on a 96-well plate method described elsewhere with slight modifications (28). One microliter of overnight LB cultures (OD600 of ∼1.0) was mixed with 100 μl of MM1 medium in each well of a 96-well Nunc polystyrene plate (Thermo Scientific) followed by incubation at 30°C. After a 24-h or 48-h incubation, planktonic cells were removed and the surface-associated cells were washed with 0.9% NaCl 10 times. One hundred microliters of a 1% aqueous solution of crystal violet (CV) was added into each well. After removal of excess CV from each well, the plate was air dried at room temperature, followed by the addition of 100 μl of 96% ethanol. The amount of biofilm biomass in each well was indicated by OD590. At least 12 replicates were used to evaluate the biofilm formation capability of each strain.
Flow cell biofilms.
Biofilm formation under hydrodynamic conditions was evaluated using multichannel flow cells (BioCentrum-DTU, Denmark). The dimensions and assembly of the flow cell systems were described elsewhere (29). Each channel of the flow cells was inoculated using 0.4 ml diluted overnight cultures in MM1 medium (OD600 of ∼0.15). After inoculation, medium flow was stopped for initial attachment (∼2 h) onto the glass slide used in the flow cells. Then, air-saturated MM1 medium was continuously supplied with a flow rate of 5.4 ml/h for biofilm growth in each channel. Biofilms grown in the flow cell systems were stained using Syto 9 and propidium iodide (PI) (Invitrogen, Singapore) and imaged using confocal laser scanning microscopy (CLSM) (Carl Zeiss Microscopy LSM 780). The CLSM images were analyzed using IMARIS software (version 7.6.4; Bitplane, Zurich, Switzerland).
Biofilm viscosity.
The viscosity of biofilm samples was measured using a Physica MCR 301 rheometer (Anton Paar). Biofilms of WT and CP2-1-S1 were prepared by dropping 100 μl of overnight cultures (OD600, ∼1.5) onto 0.45-μm filter membranes (diameter, 87 mm; Schleicher & Schuell, Dassel, Germany), placing them on MM1 agar, and incubating them at 30°C for 24 h. The membranes with biofilms were then transferred to the rheometer, and the viscosity of each biofilm was determined as previously described (30, 31).
Biofilms in submerged biofilm reactors.
Milliliter-scale submerged biofilm reactors were set up using 3-ml syringes (Luer-Lok tip, latex free; BD) packed with glass beads (diameter, 2.0 mm) (solid-glass beads; Sigma-Aldrich) for biofilms to attach and grow (see Fig. S1 in the supplemental material). Total surface area for biofilm growth in each reactor was estimated as 5,740 mm2. Each reactor was inoculated using diluted overnight cultures in MM1 medium (OD600 of ∼0.15) followed by a 2-h stop-flow to allow cells to attach onto the glass beads. MM1 medium was then continuously supplied with a flow rate of 2.45 ml/h for biofilm growth. After 120 h, biofilms formed on the glass beads in the biofilm reactors were evaluated. A portion of the glass beads with biofilms was transferred from the reactors to one well of a 96-well plate and stained with Live/Dead stains (Invitrogen) followed by obtaining CLSM images of the biofilms on the beads. To quantify the amount of biofilms in the reactors, a portion of the glass beads was mixed with 0.2 M NaOH and heated at 96°C for 20 min with intermittent vortexing. After cooling to room temperature, protein concentration was determined using a Pierce bicinchoninic acid (BCA) protein assay kit (Thermo Scientific). The amount of biofilms in each reactor was expressed as the amount of proteins per mm2 surface area.
Cr(VI) immobilization in submerged biofilm reactors.
Biofilms were allowed to grow for 120 h in the submerged biofilm reactors before 100 μM potassium dichromate (K2Cr2O7), i.e., 200 μM Cr(VI), was introduced into the growth medium. Effluent samples at the outlet of each reactor were taken periodically to determine cell detachment rate and the amount of Cr(VI) immobilized by the biofilms. Cell detachment rate was quantified using a drop-plate method (32). Briefly, 250 μl of sample was loaded into the first well of each row in a 96-well plate, and 10-fold serial dilutions were made. Then, 6 replicates of 10 μl from each of the selected dilutions were plated onto an LB agar medium. CFU were enumerated after a 24-h incubation at 30°C. The Cr(VI) concentration was determined using a previously reported colorimetric method (33). A 1.6-ml outlet sample was filtered through a 0.22-μm syringe filter and mixed with 0.2 ml of 0.5 g/liter s-diphenylcarbazide in 10% methanol-12.5 mM H2SO4 followed by OD540 measurement after a 20-min incubation. At the end of the Cr(VI) immobilization experiment, a portion of the biofilms in the reactors was analyzed using scanning electron microscopy (SEM) and energy-dispersive X-ray spectroscopy (EDX) (34). The samples were dried and sputtered with Pt. SEM images were taken using a field emission scanning electron microscope (FE-SEM; JSM-7600; JEOL Asia Pte Ltd.) at a voltage of 1.0 to 5.0 kV. The EDX spectrum was obtained using an energy-dispersive X-ray spectrometer (AZtecEnergy; Oxford Instruments, Oxfordshire, United Kingdom).
XANES analysis.
The Cr K-edge (5,989-eV) X-ray absorption near edge structure (XANES) spectra of the biofilm samples from the biofilm reactors were obtained with a transmission mode at room temperature using the XAFCA facility at the Singapore Synchrotron Light Source. A Si(III) double crystal was used to monochromatize the X rays from a 700-MeV electron storage ring. Lyophilized samples were pressed into pellets (∼10 mm in diameter) for measurements. The final spectrum was produced by averaging three scans. Processing of the raw data was done using WinXAS. Cr standards chromium potassium sulfate [Cr(III)] and potassium dichromate [Cr(VI)] were analyzed with Cr K-edge XANES for spectral comparisons with data from Cr immobilized in the biofilms.
iPCR.
The genomic DNA sequences flanking the TnM transposon were amplified by inverse PCR (iPCR) (see Fig. S2 in the supplemental material). Genomic DNA was extracted with the genomic DNA minikit (Invitrogen), and ∼3 μg was digested, respectively, with each of EcoRI, BamHI, SphI, HindIII, and BspHI, which cut at a single site inside the TnM transposon sequence. Following digestion, the DNA was purified with the QIAquick PCR purification kit and ligated overnight at 4°C in 50-μl reaction mixtures with 5 U of T4 DNA ligase (Roche). The circularized DNA sequences flanking the left side or right side of the TnM transposon (approximately 500 ng) were amplified using the MyCycler thermocycler in a total volume of 50 μl containing 350 μM deoxynucleoside triphosphate (dNTP) mixture, 5 μl buffer, 3.75 U enzyme mix (Expand Long Template PCR system; Roche), and 300 nM (each) primer. The primer pairs used for iPCR included TnM-P1/TnM-P2, TnM-P1-2/TnM-P2, and TnM-P3/TnM-P4 (see Table S1). Two-step amplification was used. The first 10 cycles were set as 94°C for 10 s, 60°C for 30 s, and 68°C for 4 min followed by 25 cycles with a 20-s cycle elongation at the 68°C extension step for each successive cycle. iPCR products were purified with the Qiagen QIAquick gel extraction kit and subsequently used as the templates for sequencing analysis. DNA sequencing was carried out using the BigDye Terminator v3.1 cycle sequencing kit (Life Technologies, USA) with the same primers adapted for iPCR, and samples were analyzed in an Applied Biosystems 3730xI DNA sequencer. A BLAST search of the sequences was used to identify the gene in which the TnM transposon was inserted.
Gene complementation.
The speF gene (locus tag SO_0314; GenBank accession number AAN53399) was amplified from S. oneidensis MR-1 genomic DNA using the Expand Long Template PCR system (Roche) with primer pair speF-U2 (5′-AAAGGATCCTCTTCGCGTAGGTAGTAGGCCAATTAAC-3′)/speF-L2 (5′-AAACTCGAGGACTGCAGAGCGGGGTTATTAACTCT-3′). The PCR fragment was purified with the Qiagen QIAquick PCR purification kit, digested with BamHI and XhoI, and then ligated with p15A-Tn5-SoAph at XhoI and BamHI restriction sites to generate a new plasmid, p15A-Tn5-SoAph-speF. Plasmid p15A-Tn5-SoAph-speF (approximately 0.3 to 0.5 μg) containing the speF gene was introduced by electroporation to CP2-1-S1 cells (50 μl) using 0.2-cm Gene Pulser/MicroPulser cuvettes (Bio-Rad) and a Bio-Rad MicroPulser. Subsequently, 0.5 ml 2× yeast extract and tryptone (2YT) medium devoid of antibiotics was added to the cells. Cells were transferred into a sterile culture tube and incubated at 30°C for 2 h under continuous shaking. After incubation, cells were spread onto 2YT agar plates supplemented with 10 μg/ml kanamycin. The plates were incubated in a 30°C incubator overnight. The complementation strain was referred to as CP2-1-S1C.
qPCR.
S. oneidensis MR-1 was cultivated in MM1 at 30°C. About 0.5 ml of the culture taken at 12 h, 16 h, 20 h, and 24 h was mixed with 1 ml of RNAprotect (Qiagen Mini RNA Prep) and centrifuged at 6,000 × g for 10 min. Total RNA content was extracted using a commercially available kit (Qiagen Mini RNA Prep) according to the instructions from the manufacturer. The RNA concentration was determined using a NanoDrop spectrophotometer (Thermo Scientific, DE, USA). Two micrograms of the total RNA was used for first-strand cDNA synthesis by using a commercially available kit (Fermentas Life Sciences). The resultant cDNA was used in quantitative PCRs (qPCRs) on the ABI StepOnePlus system (Life Technologies, CA). Primers (efficiency of >90% for all primers) were obtained from Sigma Life Sciences. PCR was carried out with 1 μl of first-strand cDNA in a total volume of 20 μl containing 0.2 mM (each) primer, 10 μl of PCR Kapa SYBR Fast qPCR Master Mix ABI (2×) Prism, and 0.4 μl ROX (high). Amplification parameters used were initial activation for 3 min at 95°C and then 40 cycles of 3 s at 95°C and 30 s at the respective appropriate annealing temperature. Experiments were performed in triplicate for all the genes. The amount of each target gene was normalized to the 16S rRNA gene and compared to control samples. Data analysis was performed using the threshold cycle (2−ΔΔCT) method, where ΔΔCT = ΔCT (treated sample) − ΔCT (untreated sample), ΔCT = CT (target gene) − CT (translational elongation factor [tsf]), and CT is the threshold cycle value for the amplified gene (35).
EPS extraction and characterization.
Silicon tubing (Masterflex L/S 16; inside diameter [i.d.], 3.2 mm), 20 cm long, was used to grow biofilms for EPS extraction. Peristaltic pumps were used to supply growth medium for biofilm growth. Diluted overnight cultures in MM1 medium (OD600, ∼0.15) were used to inoculate the tubular biofilm reactors. MM1 medium was continuously supplied at a flow rate of 5.40 ml/h. After 96 h, the biofilms were harvested and the EPS was extracted using a cation exchange resin (CER) method (36). Proteins and polysaccharides in the EPS were quantified using the BCA assay and the phenol-sulfuric acid assay (37), respectively.
DLS.
The hydrodynamic diameter of EPS (∼100 μg/ml) in phosphate-buffered saline (PBS) buffer (pH 7.0) was measured using dynamic light scattering (DLS) (ZetaPALS zeta potential analyzer; Brookhaven Instruments) before and after the addition of putrescine (1.0 mM) (Sigma-Aldrich, Singapore). DLS measurements were performed by focusing a vertically polarized light (658 nm) onto the sample and collecting the scattered light with a detector at 90°. Data were collected for 5 min at 30-s intervals, each with a count rate of 50,000 to 300,000 counts/s. The temperature was kept constant at 25°C.
RESULTS AND DISCUSSION
A hyperadherent mutant strain, CP2-1-S1, and its cell surface characteristics.
Congo red, a dye effectively staining cellulose and fimbriae, is often used to screen biofilm mutants because it detects EPS production that impacts biofilm formation (38–40). In this study, S. oneidensis MR-1 random transposon mutants that grew at rates comparable to that of the WT (≤20% variation in OD600 of overnight LB cultures) were incubated on Congo red MM1 agar plates. We found that a number of colonies exhibited morphologies distinct from the WT after a 5-day incubation. One such mutant isolate, designated CP2-1-S1, was found to bind significantly more Congo red dye than the WT did in the colony (see Fig. S3A in the supplemental material). CP2-1-S1 appeared to have a stronger adherent property than did the WT, which was implied by the extensive formation of cell aggregates (cell-cell adhesion) and biofilms (cell-tube wall adhesion) in CP2-1-S1 planktonic cultures (see Fig. S3B and C).
Previous studies demonstrated that cell surface charges and hydrophobicity play an important role in cell adhesion to biotic and abiotic surfaces (25, 41, 42). Hydrophobic interactions have been shown to be an important factor controlling the adhesion of Shewanella cells to amorphous Fe(III) oxide (41). Comparative analyses of the abilities of several different Shewanella strains to adhere to hematite (α-Fe2O3) revealed that strains with more hydrophobic and electronegative cell surfaces exhibited better adhesion (42). In addition, Shewanella cells with a more hydrophobic and electronegative surface have been shown to enhance not only cell adhesion to graphite electrodes but also cell-cell adhesion as evidenced by strong autoaggregation (25). The hyperadherent phenotype of CP2-1-S1 suggests that its cell surface physicochemical properties, such as charge and hydrophobicity, might have been altered. Therefore, cell surface charges and hydrophobicity of the WT and the mutant strain CP2-1-S1 were measured (Table 1). Our results showed that the CP2-1-S1 cell surface was more electronegative and hydrophobic than that of the WT, supporting a stronger adhesion property of CP2-1-S1 cells for attachment surfaces and between cells. We were therefore interested in further characterizing its biofilm formation capability and the biofilm cohesiveness and performance.
TABLE 1.
Surface charge and hydrophobicity of S. oneidensis cellsc
| S. oneidensis strain | Zeta potential (mV)a | Affinity to hexadecane (%)b |
|---|---|---|
| MR-1 WT | −5.68 ± 1.28 | 24.0 ± 8.0 |
| CP2-1-S1 | −8.96 ± 1.36* | 89.0 ± 6.0** |
Measured in 0.9% NaCl (pH 7.0).
Percentage of cells partitioned into hexadecane.
Values are means ± standard deviations (n = 6). Statistical significance of differences (t test): *, P < 0.02; **, P < 0.01.
Biofilm formation by mutant CP2-1-S1.
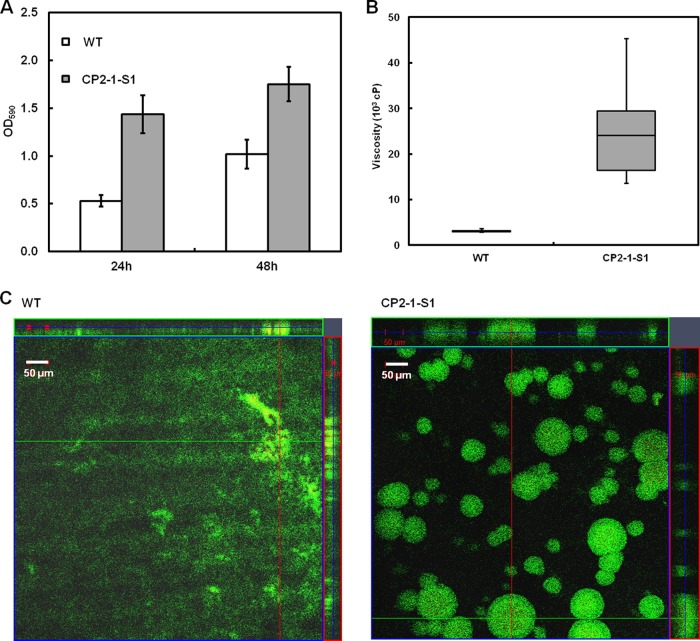
Quantitative analysis of biofilm formation by CP2-1-S1 during growth under static conditions was performed in polystyrene microtiter plates. After 24 h and 48 h, the amounts of biofilms formed by CP2-1-S1 were ∼3.0- and ∼1.7-fold larger than those formed by the WT, respectively (Fig. 1A). The results confirmed the hyperadherent phenotype of CP2-1-S1. Further, biofilms formed by the hyperadherent mutant CP2-1-S1 were substantially more cohesive than that of the WT, as evidenced by a significantly higher viscosity for the mutant biofilms (Fig. 1B).
FIG 1.
(A) Quantitative analysis of biofilm formation by S. oneidensis MR-1 WT and CP2-1-S1 during growth in MM1 medium in microtiter plates; values are means ± standard deviations (n = 12). (B) Box-and-whisker chart of biofilm viscosity for WT and CP2-1-S1 (n = 10). (C) CLSM images of biofilm structures formed under flow conditions by WT and CP2-1-S1 after 72 h of growth. Live cells were stained green and dead cells were stained red and yellow with the Live/Dead stain.
Biofilm development under hydrodynamic conditions in flow chambers was also analyzed using CLSM. After growing for 3 days, biofilms of the mutant strain CP2-1-S1 developed into three-dimensional (3-D) structures dominated by towering, mushroom-like protrusions (∼50 μm), while the biofilms of the WT were relatively flat and unstructured with a thickness of ∼20 μm (Fig. 1C).
In organisms such as Pseudomonas aeruginosa, it has been shown that the biofilms dominated by 3-D tower structures are significantly more tolerant than are flat and unstructured biofilms formed by certain mutants to biocides, including detergents and antibiotics (43, 44). Hence, it is expected that under unfavorable conditions the biofilms of CP2-1-S1 could be more robust than those of the WT. To confirm this, the performances of biofilms of CP2-1-S1 and the WT in Cr(VI) immobilization in submerged biofilm reactors were compared.
Immobilization of Cr(VI) by CP2-1-S1 biofilms.
To compare the performance of Cr(VI) immobilization by biofilms of CP2-1-S1 and the WT, both biofilms were grown in submerged biofilm reactors for 5 days before Cr(VI) was introduced into the reactors. The 5-day biofilms of CP2-1-S1 on the glass beads in the reactors were significantly thicker (∼50 versus 20 μm) than those of the WT (P = 0.02) (see Fig. S4A in the supplemental material), consistent with our observations in the above-mentioned flow cell experiments. As a quantitative indicator of the biofilms formed on the glass beads, total amount of proteins per unit surface area or surface density of total proteins was determined for each strain. The results showed that the surface density of total proteins for CP2-1-S1 biofilms was ∼6.7-fold that of the WT in submerged biofilm reactors (see Fig. S4B). We further quantified cell density in the WT and CP2-1-S1 biofilms and found that the biofilms contained about 4.0 × 108 to 8.0 × 108 CFU/mg biomass, and no significant difference was observed for the WT and the mutant (P > 0.05).
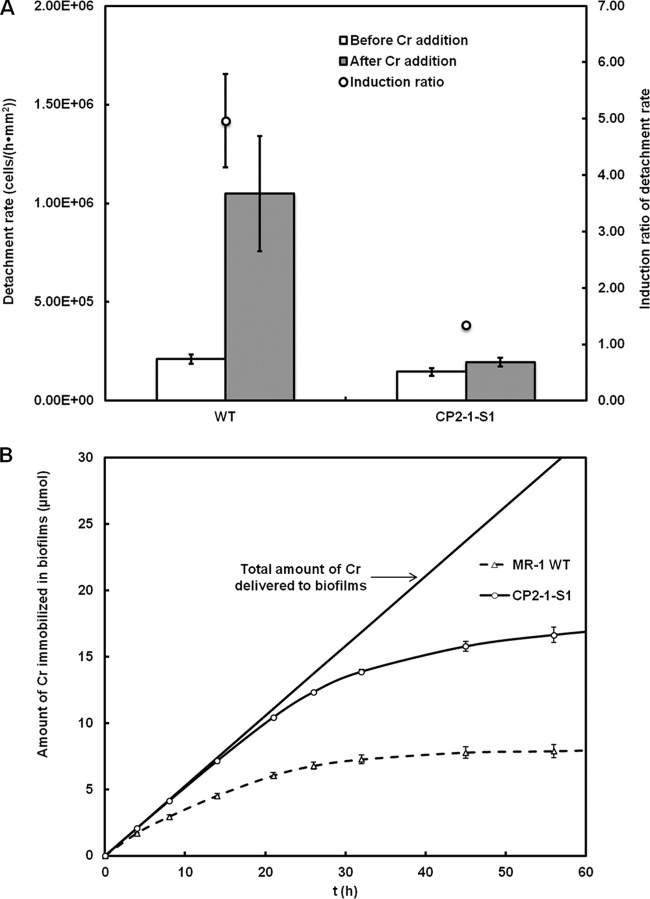
To the 5-day biofilms formed in the reactors, MM1 medium containing 200 μM Cr(VI) was introduced continuously at a flow rate the same as that used for biofilm growth. Previous studies have shown that the respiration of S. oneidensis biofilms was inhibited upon the exposure to Cr(VI) due to the toxicity of Cr(VI) and Cr(III) produced during Cr(VI) reduction (14). Inhibited energy metabolism could lead to cell dispersal from the biofilms, because biofilm maintenance has been shown to be metabolic energy dependent (45). In fact, horizontal channels with high diffusion coefficients of water in S. oneidensis biofilms exposed to Cr(VI) have been observed, which were followed by biofilm detachment (14). Here in this study, we quantified the cell detachment rate for both CP2-1-S1 and the WT before and after Cr(VI) exposure, and we found that, before Cr(VI) exposure, cell detachment rates for the WT and the CP2-1-S1 were only marginally different (2.1 × 105 ± 0.2 × 105 versus 1.4 × 105 ± 0.2 × 105 cells · h−1 · mm−2); however, after a 12-h exposure to Cr(VI), the cell detachment rate for the WT was ∼5-fold that for CP2-1-S1 (1.0 × 106 ± 0.3 × 106 versus 1.9 × 105 ± 0.2 × 105 cells · h−1 · mm−2) (Fig. 2A). Intriguingly, the exposure to Cr(VI) caused an ∼5.0-fold induction of detachment from the WT biofilms; in contrast, only ∼1.3-fold induction was observed for CP2-1-S1 biofilms, suggesting that, in the presence of toxic chemicals, CP2-1-S1 biofilms were more cohesive than those formed by the WT. Higher cohesiveness may prevent the biofilms from being disassembled, and hence, a higher performance of the biofilms would be expected. The amount of Cr immobilized by the biofilms over time is shown in Fig. 2B.
FIG 2.
(A) Rates of cell detachment from the biofilms of S. oneidensis MR-1 WT and strain CP2-1-S1 before Cr(VI) addition and after 12-h exposure to Cr(VI). (B) Amounts of Cr accumulated in the biofilms over time. Data are presented as means ± standard deviations (n = 3).
Although biofilms of both the WT and CP2-1-S1 could continuously remove Cr(VI) from the aqueous phase, as evidenced by the increasing amount of immobilized Cr in the biofilms over time, CP2-1-S1 biofilms showed a significantly higher capability than the WT biofilms: after a 56-h exposure to Cr(VI), ∼56% of the total amount of Cr(VI) delivered to the reactors was immobilized by the CP2-1-S1 biofilms, while only ∼27% was immobilized by the WT biofilms (Fig. 2B).
SEM-EDX analysis showed that the biofilms collected at the end of the immobilization experiments were coated with Cr precipitates (see Fig. S5 in the supplemental material). As it has been reported that, under similar conditions, S. oneidensis MR-1 reduces dichromate to Cr(III) (19, 46), we obtained the XANES spectra of Cr in the biofilms of both WT and CP2-1-S1 to examine the Cr oxidation states (Fig. 3). Cr in biofilms of both WT and CP2-1-S1 had very similar XANES spectra, and the two exhibited the same absorption edge energy (5.995 keV) as did the Cr(III) standard, suggesting that the valence of Cr in the biofilms was +3. In addition, the feature of a prepeak of Cr(VI) was not found in the XANES spectra of Cr in the biofilms, indicating that there was no or an undetectable amount of Cr(VI) in the biofilms. Taken together, our results showed that, compared to the MR-1 WT, the mutant strain CP2-1-S1 formed stronger biofilms with a significantly lower cell detachment rate in the presence of Cr(VI) and removed Cr(VI) more efficiently from the aqueous phase by reducing Cr(VI) and immobilizing Cr(III) in the biofilms.
FIG 3.
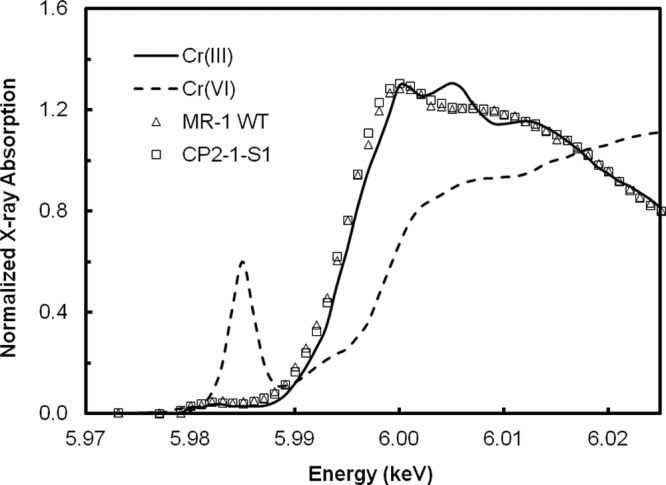
Cr K-edge XANES spectra of Cr-immobilized biofilms of S. oneidensis MR-1 WT and CP2-1-S1 from the submerged biofilm reactors. The spectra were plotted with Cr(III) and Cr(VI) standards for comparisons.
Genetic analysis of CP2-1-S1.
We carried out inverse PCR and sequence analysis to identify the transposon insertion site in strain CP2-1-S1 and found that the transposon had inserted into the gene speF (SO_0314), which encodes ornithine decarboxylase (ODC), an enzyme responsible for the conversion of l-ornithine to putrescine. Putrescine is one of the most common bacterial polyamines, a group of positively charged organic polycations (47). Polyamines in several model organisms such as E. coli have been reported to play a critical role in many biological processes, including binding to nucleic acids, stabilizing outer membranes, and protecting cells from toxic effects of reactive oxygen species and acid stresses (48–52). Polyamines have also been suggested to influence biofilm formation, and the impacts of different polyamines vary in different organisms. In Yersinia pestis, putrescine has been reported to be essential for the biofilm formation (53). Norspermidine has been suggested to enhance biofilm formation of Vibrio cholerae, while spermidine reduces its biofilm formation (54, 55). In Bacillus subtilis, norspermidine, but not closely related polyamines, has been reported to be capable of disassembling biofilms through interacting with extracellular polysaccharides (56).
Here, we show that the disruption of speF, a gene involved in biosynthesis of putrescine, in S. oneidensis led to a hyperadherent phenotype. The phenotype could be reversed by chemical complementation (via addition of exogenous putrescine into the mutant cultures) or genetic complementation (via introduction of WT speF gene into the mutant strain) (Fig. 4). These data ruled out polar effects of the transposon insertion and confirmed an important role of putrescine in mediating cell adhesion and biofilm formation in S. oneidensis.
FIG 4.
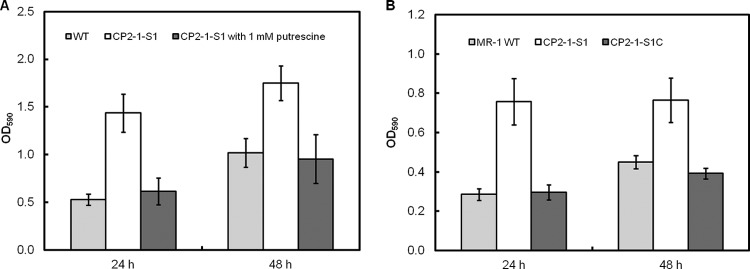
Chemical and genetic complementation. (A) Effect of putrescine on biofilm formation by the mutant strain CP2-1-S1 (speF::TnM). (B) Biofilm formation of the complementation strain CP2-1-S1C (speF::TnM, p15A-speF). The assays were conducted using MM1 medium. Values are means ± standard deviations (n = 12).
Effects of disruption of putrescine biosynthesis on biofilm matrix.
How does putrescine influence biofilm formation of S. oneidensis? We examined the effects of putrescine on established biofilms (24 h) of the mutant strain CP2-1-S1 and found that the addition of putrescine triggered disassembly of the biofilms (see Fig. S6A in the supplemental material). Further, quantitative analysis of gene expression using qPCR revealed that the gene speF was highly expressed in the stationary growth phase (see Fig. S6B). These results suggest that the production of putrescine could be a strategy employed by the bacteria for active dispersal from biofilms to find new niches. Hence, we hypothesize that putrescine affects the stability of biofilm matrix and that the disruption of putrescine biosynthesis in CP2-1-S1 helps the retention of EPS in the biofilms. We therefore quantitatively compared the EPS contents in mature biofilms of the WT and the mutant CP2-1-S1. The biofilms of CP2-1-S1 were found to contain a substantially higher EPS content than that of the WT biofilms. EPS extracted from 4-day WT biofilms was ∼0.74% of the dry weight of the biofilms, while the value for 4-day biofilms of CP2-1-S1 was ∼1.13% (Fig. 5). There was no significant difference (P = 0.09) in the relative amounts of protein and polysaccharide (i.e., protein/polysaccharide ratio) in the biofilms (0.73 ± 0.14 for WT versus 0.98 ± 0.14 for CP2-1-S1) (Fig. 5).
FIG 5.
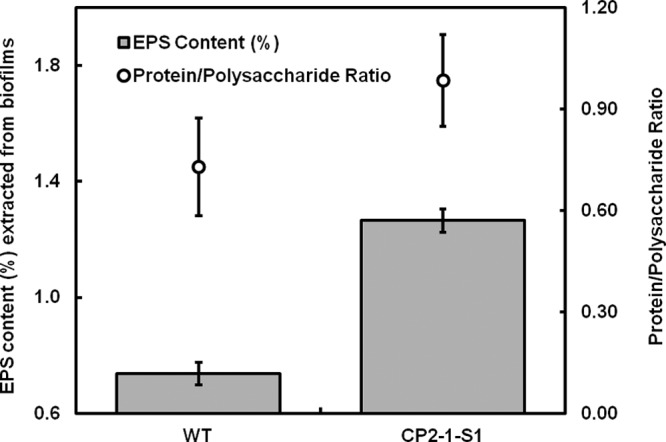
EPS content extracted from 4-day biofilms using a CER-based extraction method and protein-to-polysaccharide ratios in the extracted EPS. The content is expressed as a percentage in total dried biofilm biomass. Data are presented as means ± standard deviations (n = 3).
Although the biofilms of CP2-1-S1 contained a higher EPS content than did the WT biofilms, a comparable protein/polysaccharide ratio was observed, which implies that putrescine may disassemble the biofilm matrix through influencing the interaction between proteins and polysaccharides. To attempt to detect such an influence, we measured the hydrodynamic diameter of the EPS in solution as impacted by putrescine. The extracted EPS from CP2-1-S1 biofilms exhibited an average diameter of 686 ± 60 nm at pH 7.0, while the addition of putrescine reduced the average diameter to 575 ± 25 nm (Fig. 6). A change in hydrodynamic radius of polymers in solution often indicates a structural reconfiguration of the polymers (57). Here, a decrease in the hydrodynamic diameter of the EPS suggests that putrescine can cause configurational changes of EPS structures, which may negatively influence the interaction between proteins and polysaccharides. Detailed molecular interactions among putrescine and EPS components warrant further investigation.
FIG 6.
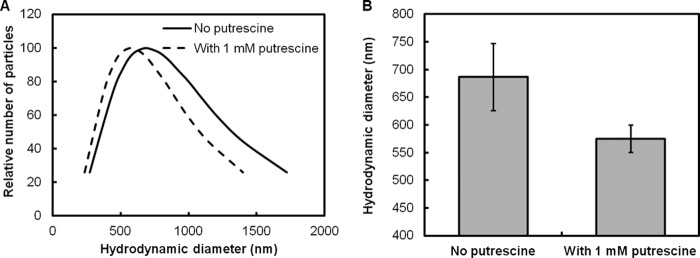
Effect of putrescine on the hydrodynamic diameter of EPS extracted from CP2-1-S1 biofilms (100 μg/ml in PBS buffer, pH 7.0). (A) Distribution of hydrodynamic diameter. (B) Average hydrodynamic diameter. Data are presented as means ± standard deviations (n = 10).
In summary, through random transposon mutagenesis library screening, we obtained one hyperadherent S. oneidensis mutant strain CP2-1-S1, in which a putrescine biosynthesis gene, speF, was disrupted. The mutant exhibited an enhanced capability in biofilm formation under both static and hydrodynamic conditions. Biofilms of the WT and the mutant strain CP2-1-S1 were used to remove Cr(VI) from the aqueous phase in submerged biofilm reactors. We found that, (i) upon exposure to Cr(VI), a significantly lower cell detachment rate in the CP2-1-S1 biofilms could be achieved, suggesting a higher cohesiveness of the mutant biofilms, and (ii) a significantly larger amount of Cr(III) was immobilized in the CP2-1-S1 biofilms, indicating an enhanced performance of the mutant biofilms in Cr(VI) bioremediation. Further, an important role of putrescine in mediating biofilm matrix disassembly through influencing the structural configuration of EPS was implied in S. oneidensis. Our work demonstrates, for the first time, that biofilm cohesiveness and performance can be improved through manipulating polyamine biosynthesis, providing a novel strategy to engineer biofilms for a better performance in biofilm-based bioprocesses. The engineered biofilms may be used in a wide range of environmental and biotechnological applications, such as treatment of contaminated water and production of high-value chemicals.
Supplementary Material
ACKNOWLEDGMENTS
We thank Simon Goh and Rong Xu for their assistance in DLS measurement, Jegan Roy for viscosity measurement, Yingdan Zhang for quantifying cell detachment rates, and Anee Mohanty for qPCR analysis. We also thank Staffan Kjelleberg and Yehuda Cohen for their useful discussion and advice.
This research is supported by the National Research Foundation and Ministry of Education Singapore under its Research Centre of Excellence Programme, Singapore Centre on Environmental Life Sciences Engineering (SCELSE) (M4330005.C70), and a start-up grant (M4080847.030) from the College of Engineering, Nanyang Technological University, Singapore.
Footnotes
Published ahead of print 20 December 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.03461-13.
REFERENCES
- 1.Hall-Stoodley L, Costerton JW, Stoodley P. 2004. Bacterial biofilms: from the natural environment to infectious diseases. Nat. Rev. Microbiol. 2:95–108. 10.1038/nrmicro821 [DOI] [PubMed] [Google Scholar]
- 2.Flemming H, Wingender J. 2010. The biofilm matrix. Nat. Rev. Microbiol. 8:623–633. 10.1038/nrmicro2415 [DOI] [PubMed] [Google Scholar]
- 3.Flemming H. 2011. The perfect slime. Colloids Surf. B Biointerfaces 86:251–259. 10.1016/j.colsurfb.2011.04.025 [DOI] [PubMed] [Google Scholar]
- 4.Harrison J, Ceri H, Turner A. 2007. Multimetal resistance and tolerance in microbial biofilms. Nat. Rev. Microbiol. 5:928–938. 10.1038/nrmicro1774 [DOI] [PubMed] [Google Scholar]
- 5.Cao B, Ahmed B, Beyenal H. 2010. Immobilization of uranium in groundwater using biofilms, p 1–34 In Shah V. (ed), Emerging environmental technologies, vol II Springer Science+Business Media BV, Dordrecht, Netherlands [Google Scholar]
- 6.Martin KJ, Nerenberg R. 2012. The membrane biofilm reactor (MBfR) for water and wastewater treatment: principles, applications, and recent developments. Bioresour. Technol. 122:83–94. 10.1016/j.biortech.2012.02.110 [DOI] [PubMed] [Google Scholar]
- 7.Singh R, Paul D, Jain RK. 2006. Biofilms: implications in bioremediation. Trends Microbiol. 14:389–397. 10.1016/j.tim.2006.07.001 [DOI] [PubMed] [Google Scholar]
- 8.Bishop PL. 2007. The role of biofilms in water reclamation and reuse. Water Sci. Technol. 55(1–2):19–26. 10.2166/wst.2007.005 [DOI] [PubMed] [Google Scholar]
- 9.Matsumoto S, Ohtaki A, Hori K. 2012. Carbon fiber as an excellent support material for wastewater treatment biofilms. Environ. Sci. Technol. 46:10175–10181. 10.1021/es3020502 [DOI] [PubMed] [Google Scholar]
- 10.Halan B, Schmid A, Buchler K. 2010. Maximizing the productivity of catalytic biofilms on solid supports in membrane aerated reactors. Biotechnol. Bioeng. 106:516–527. 10.1002/bit.22732 [DOI] [PubMed] [Google Scholar]
- 11.Rosche B, Li XZ, Hauer B, Schmid A, Buehler K. 2009. Microbial biofilms: a concept for industrial catalysis? Trends Biotechnol. 27:636–643. 10.1016/j.tibtech.2009.08.001 [DOI] [PubMed] [Google Scholar]
- 12.Sawyer LK, Hermanowicz SW. 1998. Detachment of biofilm bacteria due to variations in nutrient supply. Water Sci. Technol. 37:211–214. 10.1016/S0273-1223(98)00108-5 [DOI] [Google Scholar]
- 13.Hunt SM, Werner EM, Huang BC, Hamilton MA, Stewart PS. 2004. Hypothesis for the role of nutrient starvation in biofilm detachment. Appl. Environ. Microbiol. 70:7418–7425. 10.1128/AEM.70.12.7418-7425.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cao B, Majors PD, Ahmed B, Renslow RS, Silvia CP, Shi L, Kjelleberg S, Fredrickson JK, Beyenal H. 2012. Biofilm shows spatially stratified metabolic responses to contaminant exposure. Environ. Microbiol. 14:2901–2910. 10.1111/j.1462-2920.2012.02850.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thormann KM, Saville RM, Shukla S, Spormann AM. 2005. Induction of rapid detachment in Shewanella oneidensis MR-1 biofilms. J. Bacteriol. 187:1014–1021. 10.1128/JB.187.3.1014-1021.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Khan MM, Chapman T, Cochran K, Schuler AJ. 2013. Attachment surface energy effects on nitrification and estrogen removal rates by biofilms for improved wastewater treatment. Water Res. 47:2190–2198. 10.1016/j.watres.2013.01.036 [DOI] [PubMed] [Google Scholar]
- 17.Fredrickson JK, Romine MF, Beliaev AS, Auchtung JM, Driscoll ME, Gardner TS, Nealson KH, Osterman AL, Pinchuk G, Reed JL, Rodionov DA, Rodrigues JLM, Saffarini DA, Serres MH, Spormann AM, Zhulin IB, Tiedje JM. 2008. Towards environmental systems biology of Shewanella. Nat. Rev. Microbiol. 6:592–603. 10.1038/nrmicro1947 [DOI] [PubMed] [Google Scholar]
- 18.Fredrickson JK, Zachara JM. 2008. Electron transfer at the microbe-mineral interface: a grand challenge in biogeochemistry. Geobiology 6:245–253. 10.1111/j.1472-4669.2008.00146.x [DOI] [PubMed] [Google Scholar]
- 19.Belchik SM, Kennedy DW, Dohnalkova AC, Wang Y, Sevinc PC, Wu H, Lin Y, Lu HP, Fredrickson JK, Shi L. 2011. Extracellular reduction of hexavalent chromium by cytochromes MtrC and OmcA of Shewanella oneidensis MR-1. Appl. Environ. Microbiol. 77:4035–4041. 10.1128/AEM.02463-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cao B, Ahmed B, Kennedy DW, Wang Z, Shi L, Marshall MJ, Fredrickson JK, Isern NG, Majors PD, Beyenal H. 2011. Contribution of extracellular polymeric substances from Shewanella sp. HRCR-1 biofilms to U(VI) immobilization. Environ. Sci. Technol. 45:5483–5490. 10.1021/es200095j [DOI] [PubMed] [Google Scholar]
- 21.Marshall MJ, Beliaev AS, Dohnalkova AC, Kennedy DW, Shi L, Wang Z, Boyanov MI, Lai B, Kemner KM, McLean JS, Reed SB, Culley DE, Bailey VL, Simonson CJ, Saffarini DA, Romine MF, Zachara JM, Fredrickson JK. 2006. c-type cytochrome-dependent formation of U(IV) nanoparticles by Shewanella oneidensis. PLoS Biol. 4:e268. 10.1371/journal.pbio.0040268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zachara JM, Cowan CE, Schmidt RL, Ainsworth CC. 1988. Chromate adsorption by kaolinite. Clays Clay Miner. 36:317–326. 10.1346/CCMN.1988.0360405 [DOI] [Google Scholar]
- 23.Cao B, Shi LA, Brown RN, Xiong YJ, Fredrickson JK, Romine MF, Marshall MJ, Lipton MS, Beyenal H. 2011. Extracellular polymeric substances from Shewanella sp. HRCR-1 biofilms: characterization by infrared spectroscopy and proteomics. Environ. Microbiol. 13:1018–1031. 10.1111/j.1462-2920.2010.02407.x [DOI] [PubMed] [Google Scholar]
- 24.Zachara JM, Fredrickson JK, Li SM, Kennedy DW, Smith SC, Gassman PL. 1998. Bacterial reduction of crystalline Fe3+ oxides in single phase suspensions and subsurface materials. Am. Miner. 83:1426–1443 [Google Scholar]
- 25.Kouzuma A, Meng XY, Kimura N, Hashimoto K, Watanabe K. 2010. Disruption of the putative cell surface polysaccharide biosynthesis gene SO3177 in Shewanella oneidensis MR-1 enhances adhesion to electrodes and current generation in microbial fuel cells. Appl. Environ. Microbiol. 76:4151–4157. 10.1128/AEM.00117-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rizzi A, Pontiroli A, Brusetti L, Borin S, Sorlini C, Abruzzese A, Sacchi GA, Vogel TM, Simonet P, Bazzicalupo M, Nielsen KM, Monier JM, Daffonchio D. 2008. Strategy for in situ detection of natural transformation-based horizontal gene transfer events. Appl. Environ. Microbiol. 74:1250–1254. 10.1128/AEM.02185-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thormann KM, Saville RM, Shukla S, Pelletier DA, Spormann AM. 2004. Initial phases of biofilm formation in Shewanella oneidensis MR-1. J. Bacteriol. 186:8096–8104. 10.1128/JB.186.23.8096-8104.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Merritt J, Kadouri D, O'Toole GA. 2005. Growing and analyzing static biofilms. Curr. Protoc. Microbiol. Chapter 1:Unit 1B.1. 10.1002/9780471729259.mc01b01s00 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Crusz S, Popat R, Rybtke M, Camara M, Givskov M, Tolker-Nielsen T, Diggle S, Williams P. 2012. Bursting the bubble on bacterial biofilms: a flow cell methodology. Biofouling 28:835–842. 10.1080/08927014.2012.716044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Powell LC, Sowedan A, Khan S, Wright CJ, Hawkins K, Onsoyen E, Myrvold R, Hill KE, Thomas DW. 2013. The effect of alginate oligosaccharides on the mechanical properties of Gram-negative biofilms. Biofouling 29:413–421. 10.1080/08927014.2013.777954 [DOI] [PubMed] [Google Scholar]
- 31.Lieleg O, Caldara M, Baumgartel R, Ribbeck K. 2011. Mechanical robustness of Pseudomonas aeruginosa biofilms. Soft Matter 7:3307–3314. 10.1039/c0sm01467b [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen C-Y, Nace G, Irwin P. 2003. A 6x6 drop plate method for simultaneous colony counting and MPN enumeration of Campylobacter jejuni, Listeria monocytogenes, and Escherichia coli. J. Microbiol. Methods 55:475–479. 10.1016/S0167-7012(03)00194-5 [DOI] [PubMed] [Google Scholar]
- 33.Guha H, Jayachandran K, Maurrasse F. 2001. Kinetics of chromium (VI) reduction by a type strain Shewanella alga under different growth conditions. Environ. Pollut. 115:209–218. 10.1016/S0269-7491(01)00108-7 [DOI] [PubMed] [Google Scholar]
- 34.Ng C, Sivakumar K, Liu X, Madhaiyan M, Ji L, Yang L, Tang C, Song H, Kjelleberg S, Cao B. 2013. Influence of outer membrane c-type cytochromes on particle size and activity of extracellular polymeric substances produced by Shewanella oneidensis. Biotechnol. Bioeng. 110:1831–1837. 10.1002/bit.24856 [DOI] [PubMed] [Google Scholar]
- 35.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25:402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- 36.Jahn A, Nielsen PH. 1995. Extraction of extracellular polymeric substances (EPS) from biofilms using a cation exchange resin. Water Sci. Technol. 32:157–164. 10.1016/0273-1223(96)00020-0 [DOI] [Google Scholar]
- 37.Fournier E. 2001. Colorimetric quantification of carbohydrates. Curr. Protoc. Food Anal. Chem. Chapter E1:Unit E1.1. 10.1002/0471142913.fae0101s00 [DOI] [Google Scholar]
- 38.Friedman L, Kolter R. 2004. Genes involved in matrix formation in Pseudomonas aeruginosa PA14 biofilms. Mol. Microbiol. 51:675–690. 10.1046/j.1365-2958.2003.03877.x [DOI] [PubMed] [Google Scholar]
- 39.Hickman JW, Tifrea DF, Harwood CS. 2005. A chemosensory system that regulates biofilm formation through modulation of cyclic diguanylate levels. Proc. Natl. Acad. Sci. U. S. A. 102:14422–14427. 10.1073/pnas.0507170102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ueda A, Wood TK. 2009. Connecting quorum sensing, c-di-GMP, Pel polysaccharide, and biofilm formation in Pseudomonas aeruginosa through tyrosine phosphatase TpbA (PA3885). PLoS Pathog. 5:e1000483. 10.1371/journal.ppat.1000483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Caccavo F, Schamberger PC, Keiding K, Nielsen PH. 1997. Role of hydrophobicity in adhesion of the dissimilatory Fe(III)-reducing bacterium Shewanella alga to amorphous Fe(III) oxide. Appl. Environ. Microbiol. 63:3837–3843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Korenevsky A, Beveridge TJ. 2007. The surface physicochemistry and adhesiveness of Shewanella are affected by their surface polysaccharides. Microbiology 153:1872–1883. 10.1099/mic.0.2006/003814-0 [DOI] [PubMed] [Google Scholar]
- 43.Haagensen JAJ, Klausen M, Ernst RK, Miller SI, Folkesson A, Tolker-Nielsen T, Molin S. 2007. Differentiation and distribution of colistin- and sodium dodecyl sulfate-tolerant cells in Pseudomonas aeruginosa biofilms. J. Bacteriol. 189:28–37. 10.1128/JB.00720-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Parsek MR, Tolker-Nielsen T. 2008. Pattern formation in Pseudomonas aeruginosa biofilms. Curr. Opin. Microbiol. 11:560–566. 10.1016/j.mib.2008.09.015 [DOI] [PubMed] [Google Scholar]
- 45.Saville R, Rakshe S, Haagensen J, Shukla S, Spormann A. 2011. Energy-dependent stability of Shewanella oneidensis MR-1 biofilms. J. Bacteriol. 193:3257–3264. 10.1128/JB.00251-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang YM, Sevinc PC, Belchik SM, Fredrickson J, Shi L, Lu HP. 2013. Single-cell imaging and spectroscopic analyses of Cr(VI) reduction on the surface of bacterial cells. Langmuir 29:950–956. 10.1021/la303779y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shah P, Swiatlo E. 2008. A multifaceted role for polyamines in bacterial pathogens. Mol. Microbiol. 68:4–16. 10.1111/j.1365-2958.2008.06126.x [DOI] [PubMed] [Google Scholar]
- 48.Igarashi K, Sugawara K, Izumi I, Nagayama C, Hirose S. 1974. Effect of polyamines on polyphenylalanine synthesis by Escherichia coli and rat-liver ribosomes. Eur. J. Biochem. 48:495–502. 10.1111/j.1432-1033.1974.tb03790.x [DOI] [PubMed] [Google Scholar]
- 49.Dela Vega AL, Delcour AH. 1996. Polyamines decrease Escherichia coli outer membrane permeability. J. Bacteriol. 178:3715–3721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Khan AU, Di Mascio P, Medeiros MHG, Wilson T. 1992. Spermine and spermidine protection of plasmid DNA against single-strand breaks induced by singlet oxygen. Proc. Natl. Acad. Sci. U. S. A. 89:11428–11430. 10.1073/pnas.89.23.11428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chattopadhyay MK, Tabor CW, Tabor H. 2003. Polyamines protect Escherichia coli cells from the toxic effect of oxygen. Proc. Natl. Acad. Sci. U. S. A. 100:2261–2265. 10.1073/pnas.2627990100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jung IL, Kim IG. 2003. Polyamines and glutamate decarboxylase-based acid resistance in Escherichia coli. J. Biol. Chem. 278:22846–22852. 10.1074/jbc.M212055200 [DOI] [PubMed] [Google Scholar]
- 53.Patel CN, Wortham BW, Lines JL, Fetherston JD, Perry RD, Oliveira MA. 2006. Polyamines are essential for the formation of plague biofilm. J. Bacteriol. 188:2355–2363. 10.1128/JB.188.7.2355-2363.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McGinnis MW, Parker ZM, Walter NE, Rutkovsky AC, Cartaya-Marin C, Karatan E. 2009. Spermidine regulates Vibrio cholerae biofilm formation via transport and signaling pathways. FEMS Microbiol. Lett. 299:166–174. 10.1111/j.1574-6968.2009.01744.x [DOI] [PubMed] [Google Scholar]
- 55.Karatan E, Duncan TR, Watnick PI. 2005. NspS, a predicted polyamine sensor, mediates activation of Vibrio cholerae biofilm formation by norspermidine. J. Bacteriol. 187:7434–7443. 10.1128/JB.187.21.7434-7443.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kolodkin-Gal I, Cao SG, Chai L, Bottcher T, Kolter R, Clardy J, Losick R. 2012. A self-produced trigger for biofilm disassembly that targets exopolysaccharide. Cell 149:684–692. 10.1016/j.cell.2012.02.055 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 57.Orgad O, Oren Y, Walker SL, Herzberg M. 2011. The role of alginate in Pseudomonas aeruginosa EPS adherence, viscoelastic properties and cell attachment. Biofouling 27:787–798. 10.1080/08927014.2011.603145 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.