Abstract
The tumor suppressor p21 acts as a cell cycle inhibitor and has also been shown to regulate gene expression by functioning as a transcription corepressor. Here, we identified p21-regulated microRNAs (miRNAs) by sequencing small RNAs from isogenic p21+/+ and p21−/− cells. Three abundant miRNA clusters, miR-200b-200a-429, miR-200c-141, and miR-183-96-182, were downregulated in p21-deficient cells. Consistent with the known function of the miR-200 family and p21 in inhibition of the epithelial-mesenchymal transition (EMT), we observed EMT upon loss of p21 in multiple model systems. To explore a role of the miR-183-96-182 cluster in EMT, we identified its genome-wide targets and found that miR-183 and miR-96 repressed common targets, including SLUG, ZEB1, ITGB1, and KLF4. Reintroduction of miR-200, miR-183, or miR-96 in p21−/− cells inhibited EMT, cell migration, and invasion. Conversely, antagonizing miR-200 and miR-183-96-182 cluster miRNAs in p21+/+ cells increased invasion and elevated the levels of VIM, ZEB1, and SLUG mRNAs. Furthermore, we found that p21 forms a complex with ZEB1 at the miR-183-96-182 cluster promoter to inhibit transcriptional repression of this cluster by ZEB1, suggesting a reciprocal feedback loop.
INTRODUCTION
MicroRNAs (miRNAs) are small noncoding RNAs (∼22 nucleotides [nt]) that function in diverse biological processes, including cellular proliferation, apoptosis, and differentiation (1–4). miRNAs promote mRNA decay and/or reduce translation by base pairing to partially complementary sequences in the 3′ untranslated region (UTR) of target mRNAs (3, 5–9). Altered miRNA expression is frequent in human cancers. Dysregulation of miRNAs in cancer may occur via several mechanisms, including genetic or epigenetic alterations, defects in miRNA processing, or deregulation of miRNA-regulating transcription factors (10). miRNAs, including let-7, miR-34a, and miR-16 (11–15), act as tumor suppressors; others, such as miR-21 and the miR-17∼92 cluster (16, 17), are oncogenic. Some miRNAs (miR-200, miR-31, and miR-10b) regulate metastasis through genes involved in the epithelial-mesenchymal transition (EMT) (18–22). The EMT program is promoted by transcription factors including SNAIL, SLUG, and TWIST and the transcription repressors ZEB1 and ZEB2 (22–24). During EMT, the epithelial marker E-cadherin (CDH1) is downregulated, and the mesenchymal marker vimentin (VIM) is upregulated, resulting in loss of cell-cell contact, increased cell migration, invasion, and metastasis.
p21 (p21Cip1/WAF1) is a cyclin-dependent kinase (CDK) inhibitor that suppresses proliferation by inhibiting CDK2 and CDK1 activity at the G1/S and G2/M transitions (25–28). During the stress response, p21 is transcriptionally upregulated by p53 or by other transcription factors such as E2F1 and MYC (29, 30). p21 acts as a CDK inhibitor, but it can also regulate gene expression. Overexpression of p21 decreases the expression levels of cell cycle progression genes and upregulates senescence-inducing genes (31, 32). Because p21 is a not a bona fide transcription factor, changes in gene expression mediated by p21 can be explained, in part, by its inhibition of CDKs. However, multiple lines of evidence suggest that p21 acts as a transcription cofactor to directly regulate the activity of important transcription factors, including E2F1 and MYC (25, 33).
In addition to its role in cell cycle control, p21 has been shown to inhibit EMT in cell lines and in mouse models (34–36). However, the molecular mechanism(s) by which p21 inhibits EMT is not fully understood. In this study, we identified miRNAs regulated by p21 and show that depletion of p21 results in downregulation of several miRNAs, including miR-200 and the miR-183-96-182 (miR-183) cluster, to inhibit EMT, cell migration, and invasion. These findings expand our knowledge about the cell cycle-independent functions of p21 and indicate a key role of a p21-ZEB1 complex in inhibition of EMT through the miR-183 cluster.
MATERIALS AND METHODS
Cell culture, transfections, and constructs.
The isogenic colorectal cancer cell lines HCT116-p21+/+ and HCT116-p21−/− were provided by Bert Vogelstein and maintained in Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal bovine serum (FBS). MCF10A-p21+/+ and MCF10A-p21−/− cells were previously generated (34) and maintained in DMEM–F-12 medium (Invitrogen) supplemented with insulin (10 μg/ml), epidermal growth factor (EGF) (20 ng/ml), hydrocortisone (500 ng/ml), and cholera toxin (100 ng/ml). All cell lines were maintained in a humidified atmosphere containing 5% CO2 at 37°C and routinely checked for mycoplasma contamination.
On-Target smartPool small interfering RNAs (siRNAs) against p21, p53, and ZEB1 and miR-200a and miR-200b mimics were purchased from Dharmacon. Control (CTL) (cel-miR-67), miR-183, miR-96, and miR-182 mimics were purchased from Sigma. All miRNA mimic and siRNA transfections were performed by reverse transfection at a final concentration of 20 nM using Lipofectamine RNAiMAX (Invitrogen) as directed by the manufacturer. Control antisense oligonucleotide and anti-miRNAs against miR-200a, miR-183, miR-96, and miR-182 were purchased from Ambion and used at a final concentration of 100 nM.
The pGL3-miR-183 promoter luciferase construct was generated by PCR amplification from genomic DNA using gene-specific primers (see Table S8 in the supplemental material), and PCR fragments were digested with NheI and BglII and subcloned into the pGL3-promoter vector (Promega). 3′ UTR luciferase constructs were generated by PCR amplification of ZEB1 and SLUG 3′ UTRs from genomic DNA with gene-specific primers (see Table S9 in the supplemental material). PCR fragments were cloned into the Renilla luciferase 3′ UTR of psiCHECK-2 (Promega).
RNA isolation, RT-qPCR, and miRNA analysis.
Total RNA from cultured cells was isolated by using the miRNeasy isolation kit (Qiagen) as directed by the manufacturer. For quantitative reverse transcription-PCR (RT-qPCR) analysis, 500 ng of total RNA was reverse transcribed by using an iScript RT kit (Bio-Rad), and qPCR was performed by using SYBR green (Applied Biosystems) as described by the manufacturer. Primer sequences are listed in Table S9 in the supplemental material. Mature miRNAs were quantitated by using TaqMan miRNA assays according to the manufacturer's instructions (Applied Biosystems). U6 snRNA was used as an internal control.
Immunoblotting.
Whole-cell lysates were prepared by using radioimmunoprecipitation assay (RIPA) buffer containing protease inhibitors (Roche), and proteins were quantified by using a bicinchoninic acid (BCA) protein quantitation kit (Thermo Scientific). Ten micrograms of whole-cell lysate per lane was used for immunoblotting. The following antibodies were used: anti-CDH1 (Abcam) and anti-VIM (Abcam) at 1:1,000 dilution; anti-p21 (Cell Signaling), anti-ZEB1 (Cell Signaling), and anti-SLUG (Cell Signaling) at 1:2,000 dilution; and antitubulin (Sigma), anti-p53 (DO-1; Santa Cruz), and anti-α-actin (Sigma) at 1:5,000 dilution.
Deep sequencing of small RNAs.
Total RNA was isolated by using the miRNeasy Minikit (Qiagen), and RNA integrity was evaluated with a 2100 BioAnalyzer (Agilent). The small RNA fraction (18 to 30 nt) was enriched from 10 μg total RNA by using 15% denaturing Tris-borate-EDTA (TBE) urea polyacrylamide gel electrophoresis (PAGE). Purified small RNAs were ligated to proprietary 3′ and 5′ RNA adapters (Illumina) and used as templates for cDNA synthesis. PCR amplification was performed by using adapter-specific primers (12 cycles), purified by using TBE-PAGE, and diluted to 8 pM for cluster generation and sequencing on an Illumina GAII instrument. Adapter sequences were clipped and subsequently analyzed by using BWA. Coordinates of mature miRNAs were downloaded from miRBase (v18), and tag densities were counted and averaged over the length of the mature miRNA.
Microarray analysis.
HCT116-p21−/− cells were reverse transfected with CTL (cel-miR-67), miR-183, miR-96, or miR-182 mimics; 48 h later, total RNA was isolated by using an RNeasy minikit (Qiagen). Samples for microarray were labeled by using an IlluminaTotalPrep RNA amplification kit (Applied Biosystems) as recommended by the manufacturer. Microarrays were performed by using a HumanHT-12 v4 Expression BeadChip kit (Illumina) and analyzed by using the R/Bioconductor packages (lumi.limma).
Retrovirus packaging and stable transductions.
Retroviruses expressing short hairpin RNA (shRNA) against p21 (shp21) or luciferase (shCTL) were generated by using an HEK293T (293T) cell-based transfection packaging system. HCT116 cells were transduced with shp21- or shCTL-expressing retroviruses, and 24 h after infection, 1 μg/ml of puromycin (Invitrogen) was added to select stably transduced cells. Stable single clones were generated by using a limitation dilution method and maintained in 0.5 μg/ml puromycin for further analysis.
Cell proliferation, migration, and invasion assays.
Cell proliferation assays were performed after seeding 1,500 cells/well of a 96-well plate and using Cell Counting Kit-8 (catalog number 96992; Fluka). For wound-healing assays, HCT116-p21−/− cells were reverse transfected with CTL, miR-200a, miR-200b, miR-183, miR-96, or miR-182 mimics. Forty-eight hours later, cells were scraped with a p200 pipette tip, and pictures were taken at the indicated time points by using a Zeiss Axiovert 200 microscope. For Transwell migration assays, cells were grown in serum-reduced medium (1% FBS) for 5 h, after which 5 × 104 cells/well were plated onto the top chamber with a noncoated membrane (24-well insert, 8-μm pore size; BD Biosciences). For invasion assays, Matrigel (BD Biosciences) was coated onto Transwell inserts for 5 h and incubated at 37°C. A total of 2 × 105 cells/well were transfected with the indicated miRNA mimics and plated onto the top chamber in medium with 1% FBS. In both assays, the lower chamber was filled with 0.7 ml medium containing 10% FBS, used as a chemoattractant. After 48 h of incubation, the cells that did not migrate or invade through the pores were removed by using a cotton swab. Cells on the lower surface of the membrane were stained with crystal violet, pictures were taken by using a digital camera, and migrated or invaded cells were counted and analyzed by using ImageJ.
Luciferase reporter assays.
For promoter luciferase assays, cells were grown in 24-well plates and transfected by using Lipofectamine 2000. pGL3 or pGL3-183-S was cotransfected with pRL-TK (internal control), and luciferase activity was measured 48 h later by using a dual-luciferase reporter assay (Promega). For siRNA knockdown experiments, HCT116-p21−/− cells were reverse transfected with CTL or ZEB1 siRNAs for 72 h and then with the promoter constructs and pRL-TK. Luciferase activity was measured 48 h after the second transfection. For 3′ UTR luciferase assays, HeLa cells were cotransfected with miRNA mimics and psiCHECK2 constructs; 48 h later, luciferase activity was measured and normalized to firefly luciferase activity (internal control).
Chromatin immunoprecipitation assays.
Chromatin immunoprecipitation (ChIP) was performed by using an Active Motif ChIP assay kit (Active Motif) as directed by the manufacturer. Briefly, HCT116-p21−/− cells were grown in a 10-cm plates. Chromatin was cross-linked with 1% formaldehyde, and cells were lysed and sonicated. Protein-DNA complexes were immunoprecipitated with control IgG or anti-ZEB1 antibody (Cell Signaling). The immunoprecipitation (IP) material was washed and heated at 65°C overnight to reverse the cross-links. The ChIP DNA was column purified (Qiagen), and qPCR was performed by using primers designed to flank the consensus ZEB1 binding sites in the miR-183 cluster promoter (see Table S9 in the supplemental material).
In vitro-biotinylated DNA pulldown assays.
Biotinylated double-stranded DNA (dsDNA) was generated by PCR with biotin-PCR primers (see Table S9 in the supplemental material). PCR products were purified by using a Qiagen PCR purification kit. One hundred twenty nanograms of biotinylated DNA was incubated with 200 μg of whole-cell lysate prepared from 293T cells transfected with hemagglutinin-ZEB1 (HA-ZEB1) or FLAG-p21 for 30 min at room temperature. DNA-protein complexes were then incubated with 20 μl streptavidin Dynabeads for 1 h. Beads were washed 4 times with lysis buffer, and DNA-protein complexes were eluted by the addition of SDS-PAGE sample buffer. Protein samples (input and eluted proteins) were subjected to SDS-PAGE followed by immunoblotting with anti-ZEB1 or anti-p21 antibody.
Immunoprecipitation of Ago2-associated miRNAs.
Cytoplasmic extracts were prepared from HCT116 cells as described previously (14). Ago2 IPs were performed with cytoplasmic extracts by using a monoclonal Ago2 antibody, provided by Zissimos Mourelators. Briefly, HCT116-p21−/− cells were transfected with miRNA mimics (20 nM) for 48 h, and cytoplasmic extracts containing protease inhibitors (Roche) and RNaseOUT (Invitrogen) were used directly for immunoprecipitation (IP). Ago2-miRNA complexes were subjected to IP with a control IgG or Ago2 antibody for 4 h at 4°C. RNA bound to the beads was isolated by phenol-chloroform extraction (Ambion) followed by ethanol precipitation, as previously described (14). Ago2-bound miRNAs were detected by TaqMan RT-qPCR analysis.
TGF-β treatment.
MCF10A-p21+/+ and MCF10A-p21−/− cells were cultured in MEGM medium (Lonza) containing 5% horse serum. For transforming growth factor β (TGF-β) treatment, cells were seeded into a 6-well plate (about 50% confluence) with MEBM basal medium containing 5% horse serum for 24 h and then treated with TGF-β (5 ng/ml) for 6 days. At day 3, the medium was changed, and fresh TGF-β was added. Pictures were taken by using a Zeiss Axiovert 200 microscope, and RNA was isolated after 6 days of TGF-β treatment.
Mutant MRE constructs.
ZEB1 3′ UTR and SLUG 3′ UTR miRNA recognition element (MRE) mutants were generated by GeneScript. Briefly, using full-length ZEB1 3′ UTR or SLUG 3′ UTR constructs in psiCHECK2 as the template, PCR products containing the designed mutant MRE sequences were restriction enzyme digested and inserted into psiCHECK2. Mutant 3′ UTRs were confirmed by DNA sequencing.
RESULTS
Loss of p21 alters the levels of several miRNAs, including the miR-200 and miR-183 clusters.
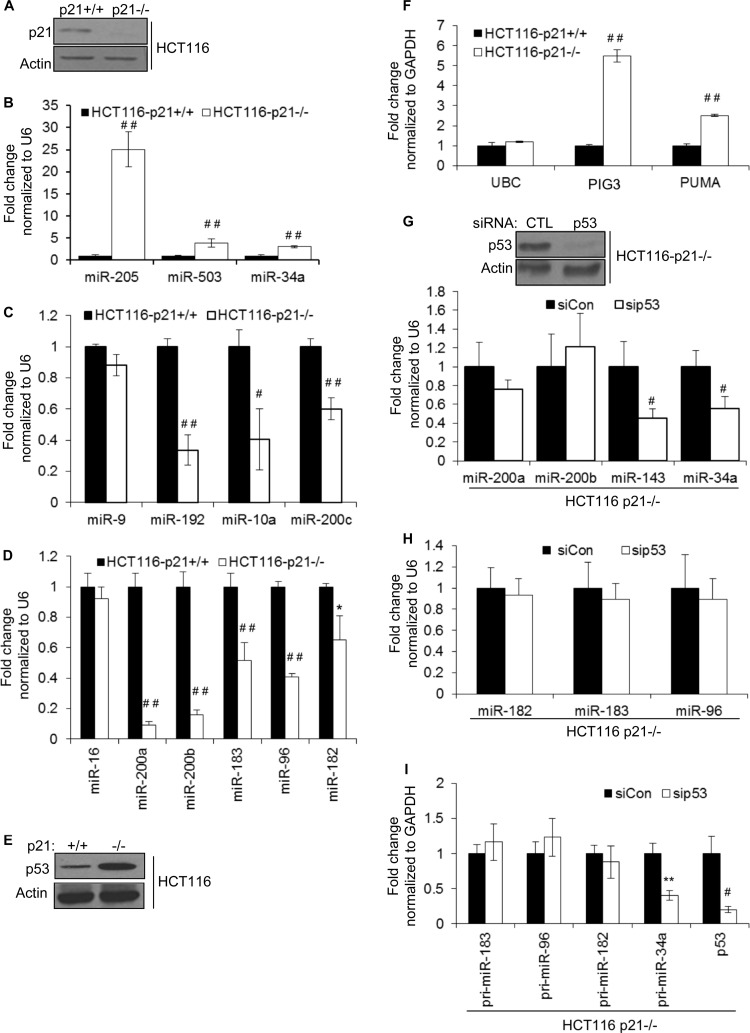
To identify miRNAs regulated by p21, we sequenced small RNAs from the colorectal cancer cell line HCT116 (HCT116-p21+/+) and from isogenic HCT116-p21−/− cells (28). As expected, p21 was expressed only in HCT116-p21+/+ cells (Fig. 1A). To identify p21-regulated miRNAs, we used an arbitrary cutoff of 1.50-fold (≥500 reads) and found 29 upregulated and 36 downregulated miRNAs in p21−/− cells (see Tables S1 and S2 in the supplemental material). We performed TaqMan miRNA RT-qPCR for 13 of these candidate miRNAs for follow-up validation (Table 1). In HCT116-p21−/− cells, levels of miR-34a, miR-205, and miR-503 were consistently higher (∼3- to 25-fold) (Fig. 1B). Significant downregulation (Fig. 1C and D) was observed for 8 out of 9 miRNAs (miR-200a, miR-200b, miR-200c, miR-183, miR-96, miR-182, miR-9, miR-192, and miR-10a). miR-9 was the only miRNA that could not be validated. miR-16 levels did not change significantly, and therefore, miR-16 was used as an internal control (Table 1).
FIG 1.
Depletion of p21 alters the expression levels of several miRNAs. (A) p21 protein levels in isogenic HCT116-p21+/+ and HCT116-p21−/− cells were measured by immunoblotting using actin as loading control. (B to D) Deep sequencing results for 8 miRNAs were validated by RT-qPCR. (E) Levels of p53 protein in isogenic HCT116-p21+/+ and HCT116-p21−/− cells were measured by immunoblotting using actin as a loading control. (F) The mRNA levels of the p53 target genes PIG3 and PUMA in HCT116-p21+/+ and HCT116-p21−/− cells were measured by RT-qPCR. The housekeeping gene UBC was used as a negative control. GAPDH, glyceraldehyde-3-phosphate dehydrogenase. (G) HCT116-p21−/− cells were transfected with a control siRNA (CTL) or p53 siRNAs for 48 h and p53 knockdown was measured by immunoblotting (top), and the effect of silencing of p53 on the p53-regulated miRNAs miR-34a and miR-143 was measured by RT-qPCR (bottom). (H and I) The effect of silencing of p53 in HCT116-p21−/− cells on the levels of mature and primary transcripts of p21-regulated miRNAs was measured by RT-qPCR. Error bars represent means ± standard deviations of three independent experiments. *, P < 0.05; #, P < 0.01; **, P < 0.005; ##, P < 0.001.
TABLE 1.
Numbers of normalized reads of 12 candidate p21-regulated miRNAs and the control miRNA miR-16
| miRNA | No. of normalized readsa |
Fold change (p21KO/WT) | |
|---|---|---|---|
| WT | p21KO | ||
| miR-200a | 13,609 | 1,444 | 0.11 |
| miR-200b | 23,964 | 2,550 | 0.11 |
| miR-200c | 61,071 | 47,718 | 0.78 |
| miR-141 | 173,843 | 88,230 | 0.51 |
| miR-429 | 6,652 | 856 | 0.13 |
| miR-183 | 10,520 | 7,668 | 0.73 |
| miR-96 | 31,469 | 17,298 | 0.55 |
| miR-182 | 34,607 | 16,777 | 0.48 |
| miR-10a | 8,648 | 2,799 | 0.32 |
| miR-34a | 1,628 | 6,078 | 3.73 |
| miR-205 | 3 | 528 | 175.93 |
| miR-503 | 165 | 976 | 5.91 |
| miR-16 | 53,426 | 48,345 | 0.90 |
WT, wild type; p21KO, p21 knockout.
Among the above-mentioned p21-responsive miRNAs, miR-34a, miR-200 family, and miR-183 cluster miRNAs were shown recently to be p53 targets (11, 37, 38). It is known that p53 is upregulated in HCT116-p21−/− cells due to downregulation of MDM2 (32, 39). Indeed, we found elevated levels of p53 and its targets PUMA and PIG3 in HCT116-p21−/− cells (Fig. 1E and F). Although silencing of p53 in HCT116-p21−/− cells with siRNAs reduced levels of the p53-regulated miR-34a and miR-143, the abundance of miR-200 and the miR-183 cluster did not change significantly (Fig. 1G and H). In this experiment, pri-miR-34a was downregulated, but the primary transcript for the miR-183 cluster, as assessed by 3 sets of primers that span the primary transcript region corresponding to miR-183 (pri-miR-183), miR-96 (pri-miR-96), and miR-182 (pri-miR-182), did not change, suggesting that p53 does not regulate miR-200 and miR-183 cluster miRNAs in p21−/− cells (Fig. 1I).
In this study, we focused on the miR-200 family and the miR-183 clusters for two main reasons. First, these miRNAs were abundant (∼6,600 to 173,000 reads) in parental HCT116 cells (Table 1) and significantly downregulated in HCT116-p21−/− cells, suggesting that they may contribute to p21-regulated gene expression. Second, the miR-200 family and miR-183 clusters encode 8 miRNAs (from 3 clusters) that can potentially regulate common targets that contribute to the biological function of p21. The miR-200 family consists of 2 clusters that have similar seed sequences: one cluster encodes miR-200a, miR-200b, and miR-429, and the other cluster expresses miR-200c and miR-141. The miR-183 cluster encodes miR-183, miR-96, and miR-182, which have similar seed sequences and may therefore target an overlapping set of genes.
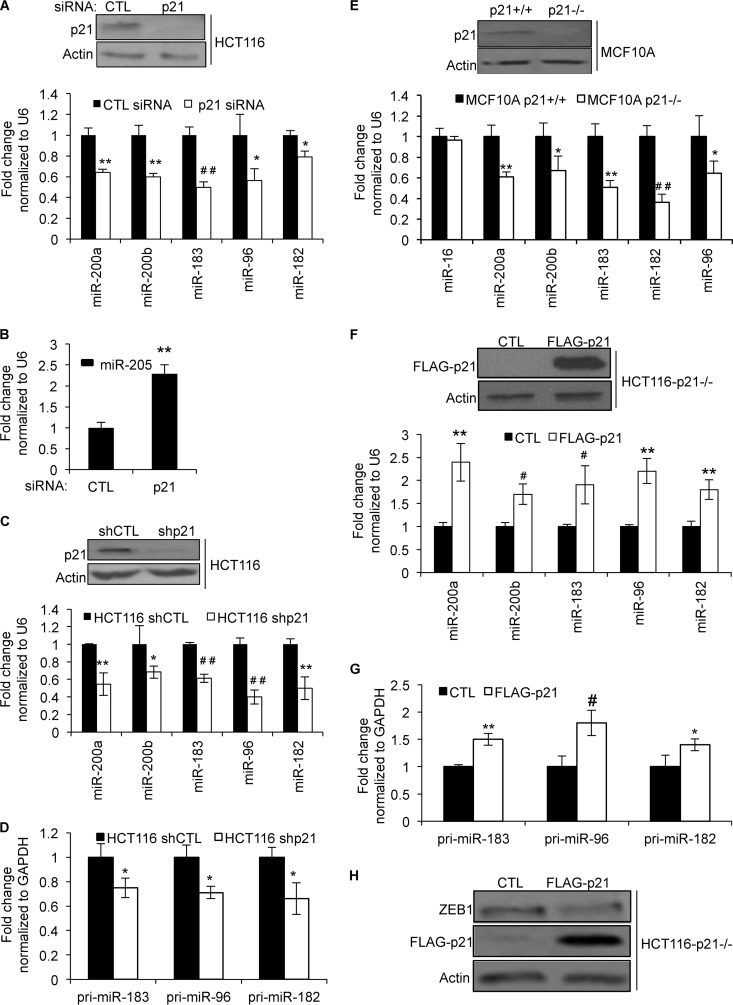
To further validate the sequencing results, we silenced p21 by transfecting HCT116 cells with control (CTL) or p21 siRNAs and measured miRNA expression after 48 h. Silencing of p21 significantly downregulated p21, miR-200a, miR-200b, miR-183, miR-96, and miR-182 (Fig. 2A), and miR-205 was upregulated (Fig. 2B). Moreover, downregulation of these miRNAs and the miR-183 cluster primary transcript was also observed upon the stable knockdown of p21 with a p21 shRNA in HCT116 cells (Fig. 2C and D). This result also suggested that the loss of p21 decreases miR-183 cluster transcription. Furthermore, regulation of these miRNAs by p21 was not cell type specific because downregulation of miR-200 and the miR-183 cluster was also observed upon the targeted deletion of p21 in MCF10A cells (mammary epithelial cell line) (34) (Fig. 2E). Reintroduction of p21 by transient transfection of HCT116-p21−/− cells with a p21-expressing construct significantly elevated miR-200a, miR-200b, miR-183, miR-96, and miR-182 levels and the expression level of the miR-183 cluster primary transcript (Fig. 2F and G). Because the miR-200 family has been shown to regulate ZEB1, a master regulator of EMT, we next looked at the effect of p21 overexpression on ZEB1 protein levels. As shown in Fig. 2H, overexpression of p21 in HCT116-p21−/− cells decreased ZEB1 protein levels, indicating that increased expression of miR-200 upon p21 overexpression may downregulate ZEB1 expression. These results suggest that p21 plays a role in regulating the expression of multiple miRNAs, including the miR-200 and miR-183 clusters.
FIG 2.
Loss of p21 downregulates miR-200 and miR-183 cluster miRNAs in HCT116 and MCF10A cells. (A) HCT116 cells were transfected with CTL or p21 siRNAs for 48 h. p21 knockdown was confirmed by immunoblotting (top), and downregulation of p21-regulated miRNAs was assessed by RT-qPCR (bottom). (B) Increased miR-205 expression upon p21 knockdown (A) was validated by RT-qPCR. (C) HCT116 cells were transduced with a control shRNA (shCTL) or a p21 shRNA (shp21). p21 silencing was confirmed by immunoblotting (top), and expression of p21-regulated miRNAs was assessed by RT-qPCR (bottom). (D) The effect of stable knockdown of p21 on the miR-183 cluster primary transcript in HCT116 cells was assessed by RT-qPCR. (E) The expression of p21 protein in MCF10A-p21+/+ and MCF10A-p21−/− cells was measured by immunoblotting (top), and downregulation of p21-regulated miRNAs was measured by RT-qPCR (bottom). (F and G) HCT116-p21−/− cells were transfected with an empty vector (CTL) or a FLAG-p21-expressing construct for 48 h. p21 overexpression was measured by immunoblotting (F, top), and the effect on p21-regulated miRNAs (F, bottom) and the miR-183 cluster primary transcript (G) was assessed by RT-qPCR. (H) The effect of p21 overexpression on ZEB1 protein expression was assessed by immunoblotting. Error bars represent means ± standard deviations of three independent experiments. *, P < 0.05; #, P < 0.01; **, P < 0.005; ##, P < 0.001.
ZEB1 inhibits miR-183 cluster transcription.
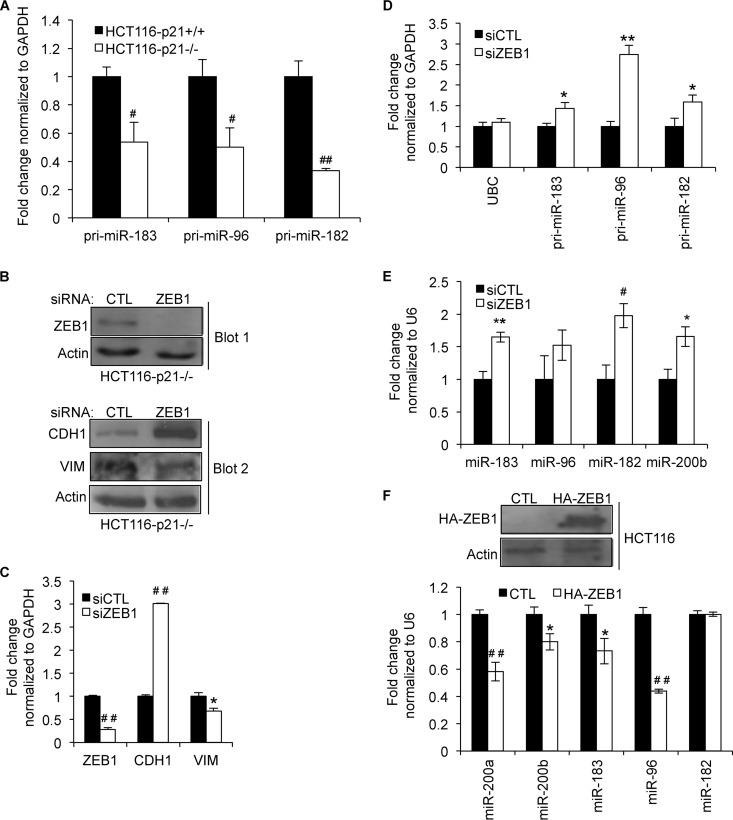
p21 can regulate gene expression by (i) indirectly inhibiting E2F activity through regulation of retinoblastoma (Rb) phosphorylation or (ii) functioning as a transcription corepressor to directly inhibit the activity of transcription factors such as STAT3, MYC, and E2F (25, 33). The loss of p21 may therefore activate these transcription factors to enhance miRNA transcription. Because the miR-183 cluster was downregulated upon the loss of p21, we hypothesized that p21 may instead inhibit a transcription repressor to directly inhibit transcription of the miR-183 cluster. It has been shown that the miR-200 family is directly repressed by ZEB1, a transcription repressor that has been shown to play an important role in EMT. We therefore hypothesized that ZEB1 represses miR-183 cluster transcription in HCT116-p21−/− cells. Indeed, significant downregulation (∼2-fold) of the miR-183 cluster primary transcript was observed in HCT116-p21−/− cells, suggesting that transcription of the miR-183 cluster was repressed in p21−/− cells (Fig. 3A). To investigate a potential role of ZEB1 in regulation of the miR-183 cluster, we transfected HCT116-p21−/− cells with CTL or ZEB1 siRNAs for 48 h. ZEB1 knockdown was confirmed by immunoblotting, and as expected, CDH1 protein was upregulated, whereas VIM was downregulated upon ZEB1 silencing (Fig. 3B). Changes in CDH1 and VIM protein expression levels were associated with increased CDH1 mRNA (3-fold) and decreased VIM mRNA (∼30%) levels (Fig. 3C). Moreover, silencing of ZEB1 significantly elevated the levels of miR-183 cluster primary transcripts and upregulated mature miR-183 and miR-182 levels (Fig. 3D and E). Conversely, overexpression of HA-ZEB1 by transient transfection of HCT116 cells for 72 h enhanced the levels of miR-200a, miR-200b, miR-183, and miR-96 (Fig. 3F). Surprisingly, miR-182 levels did not change under these conditions, and this needs further investigation. Taken together, these results suggest that ZEB1 inhibits transcription of the miR-183 cluster.
FIG 3.
ZEB1 inhibits transcription of the miR-183 cluster. (A) The effect of targeted deletion of p21 on the transcription of the miR-183 cluster was evaluated by measuring the primary transcript levels spanning miR-183 (pri-miR-183), miR-96 (pri-miR-96), and miR-182 (pri-miR-182) by RT-qPCR in HCT116-p21+/+ and HCT116-p21−/− cells. (B, top) Immunoblotting was performed to confirm ZEB1 knockdown 48 h after transfection of HCT116-p21−/− cells with CTL or ZEB1 siRNAs. (Bottom) The effect of ZEB1 knockdown on the EMT markers CDH1 and VIM was assessed by immunoblotting. (B to E) HCT116-p21−/− cells were transfected with CTL or ZEB1 siRNAs, and 48 h later, RT-qPCR was performed to determine the effect of ZEB1 silencing on ZEB1, CDH1, and VIM mRNAs (C), 3 segments of the miR-183 cluster primary transcript (D), and mature miR-183, miR-96, miR-182, and miR-200b levels (E). (F) HA-ZEB1 was transiently transfected into HCT116 cells. Immunoblotting was used to validate HA-ZEB1 expression (top), and the effect of HA-ZEB1 overexpression on p21-regulated miRNAs in HCT116 cells was measured by RT-qPCR (bottom). Error bars represent means ± standard deviations of three independent experiments. *, P < 0.05; #, P < 0.01; **, P < 0.005; ##, P < 0.001.
p21 associates with ZEB1 to inhibit ZEB1-mediated transcriptional repression of the miR-183 cluster.
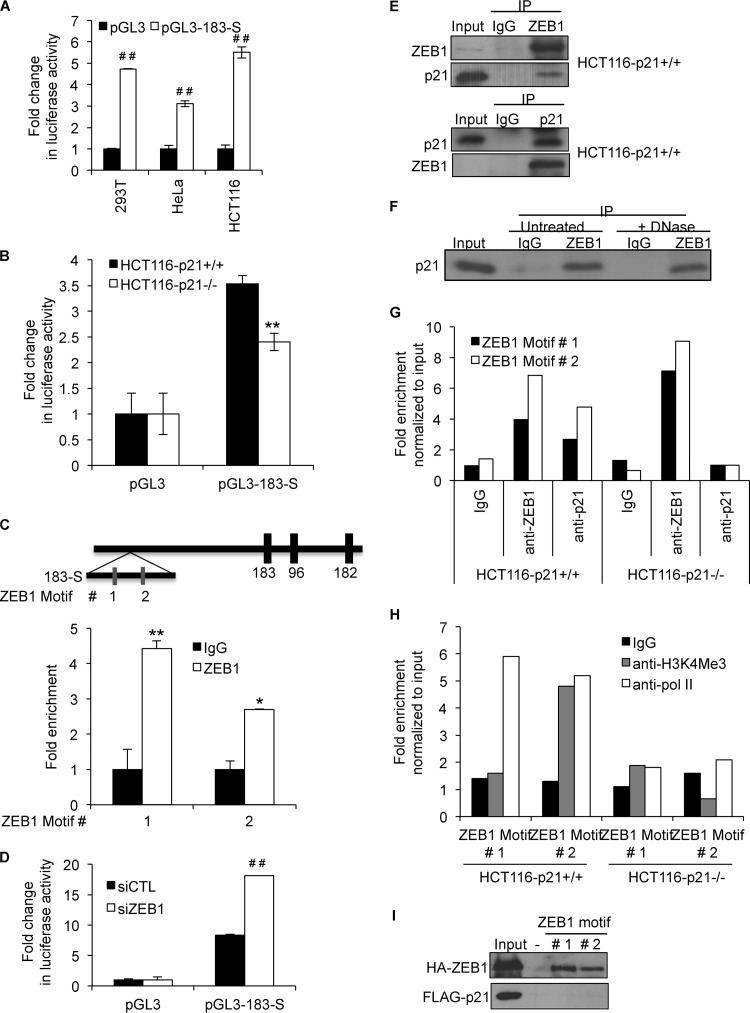
The human miR-183 cluster is intergenic and located on chromosome 7. Although its transcription start site has not been identified, we found several histone acetylation marks (indicative of active chromatin) in a 1-kb DNA region upstream of miR-183 (bp 4470 to 5470) using the UCSC genome browser (data not shown). When this 1-kb DNA region (183-S) was inserted into the pGL3 promoter vector and luciferase assays were performed, we consistently observed increased luciferase activity in 293T, HeLa, and HCT116 cells upon transfection with the pGL3-183-S reporter for 48 h (Fig. 4A). Moreover, the luciferase expression level driven by 183-S was significantly higher in HCT116-p21+/+ than in HCT116-p21−/− cells, indicating that the 183-S promoter region of the miR-183 cluster is transcriptionally repressed in p21−/− cells (Fig. 4B). We also found two ZEB1 binding motifs (22) in the 183-S DNA fragment, located 3,800 and 5,100 bases upstream of miR-183 (Fig. 4C). Both ZEB1 motifs were significantly enriched in chromatin immunoprecipitation (ChIP) assays with HCT116-p21−/− cells by using an anti-ZEB1 antibody (Fig. 4C). Furthermore, knockdown of ZEB1 in HCT116-p21−/− cells enhanced luciferase expression driven by the 183-S DNA fragment, suggesting that the 183-S region is transcriptionally repressed by ZEB1 in p21−/− cells (Fig. 4D). These results suggest that ZEB1 binds to the miR-183 cluster promoter and may repress its transcription in p21−/− cells.
FIG 4.
p21 interacts with ZEB1 and inhibits ZEB1 binding to the miR-183 cluster promoter. (A and B) Luciferase assays were performed with 293T, HeLa, and HCT116 cells (A) or HCT116-p21+/+ and HCT116-p21−/− cells (B) 48 h after transfection of the cells with a control vector (pGL3) or pGL3 containing the 1-kb region upstream of miR-183 (pGL3-183-S). (C) Schematic diagram (top) showing a 1-kb region (183-S) upstream of miR-183 that has several histone acetylation marks. Two ZEB1 motifs in the 183-S DNA fragment are shown. ChIP assays were performed to pull down ZEB1-DNA complexes from HCT116-p21−/− cells using an anti-ZEB1 antibody, and fold enrichment of the two ZEB1 motifs (motifs 1 and 2) was determined by qPCR (bottom). (D) HCT116 p21−/− cells were transfected with CTL or ZEB1 siRNAs; 72 h later, the cells were transfected with pGL3 or pGL3-183-S, and luciferase assays were performed after 48 h. (E) Association of p21 with ZEB1 was determined by immunoprecipitating ZEB1 with anti-ZEB1 (top) and p21 with anti-p21 (bottom) from whole-cell lysates prepared from HCT116-p21+/+ cells. (F) Whole-cell lysates from HCT116 cells were prepared and divided into 2 parts. One part was treated with DNase for 30 min. ZEB1 was pulled down from these lysates, and immunoblotting was performed to determine whether the p21-ZEB1 interaction was DNA independent. (G and H) Enrichment of p21 and ZEB1 (G) and Pol II and H3K4Me3 (H) at the ZEB1 motifs in the miR-183 promoter region (183-S) was assessed by ChIP-qPCR. (I) 293T cells were transfected with a construct expressing HA-ZEB1 or FLAG-p21, and binding of biotin–183-S DNA motif 1 or 2 was assessed by incubating the lysates with beads only (−) and by immunoblotting after streptavidin pulldowns. Error bars represent means ± standard deviations of three independent experiments. *, P < 0.05; #, P < 0.01; **, P < 0.005; ##, P < 0.001.
p21 is not a transcription factor per se, but it can bind to select transcription factors to inhibit their activity (25). We therefore hypothesized that p21 may directly regulate miR-183 cluster transcription by interacting with ZEB1 to inhibit its activity. To test this, we immunoprecipitated ZEB1 from whole-cell lysates prepared from HCT116-p21+/+ cells. As shown in Fig. 4E, we observed enrichment of p21 in ZEB1 IPs, suggesting that p21 interacts with ZEB1. Conversely, we were able to pull down ZEB1 in p21 IPs, further confirming this interaction (Fig. 4E). Furthermore, DNase treatment did not affect the association of p21 and ZEB1, indicating that p21 may interact directly with ZEB1 (Fig. 4F). To determine whether the p21-ZEB1 complex binds to the miR-183 promoter, we performed ChIP-qPCR assays with HCT116-p21+/+ and HCT116-p21−/− cells. As shown by the results from a representative ChIP-PCR experiment, both ZEB1 motifs were significantly enriched in the p21 and ZEB1 ChIPs from HCT116-p21+/+ cells (Fig. 4G). As expected, p21 did not bind to the miR-183 promoter in HCT116-p21−/− cells. The level of binding of ZEB1 to the miR-183 promoter was significantly higher in p21−/− cells than in p21+/+ cells, suggesting that p21 inhibits ZEB1 promoter occupancy on the miR-183 promoter (Fig. 4G). Moreover, binding of RNA polymerase II (Pol II) and H3K4Me3 to these motifs was significantly reduced upon the loss of p21, suggesting that miR-183 cluster transcription is repressed in p21−/− cells (Fig. 4H).
To determine whether ZEB1 and/or p21 may bind directly to the ZEB1 motifs, we performed in vitro DNA-protein binding assays as previously described (40). Whole-cell lysates prepared from 293T cells transfected with constructs expressing HA-ZEB1 or FLAG-p21 were incubated with biotinylated miR-183 DNA motifs (ZEB1 motifs 1 and 2) generated by PCR, and biotinylated DNA-protein complexes were pulled down by using streptavidin beads. Binding of FLAG-p21 or HA-ZEB1 to the ZEB1 motifs was determined by immunoblotting with the eluted material. As shown in Fig. 4I, we were able to detect specific binding of HA-ZEB1 to both biotin–miR-183 motifs, whereas FLAG-p21 did not bind. Taken together, these results indicate that p21 associates with the miR-183 cluster promoter through ZEB1 and that the interaction of p21 with ZEB1 inhibits ZEB1 binding to the miR-183 cluster promoter.
Depletion of p21 induces EMT to enhance cell migration and invasion.
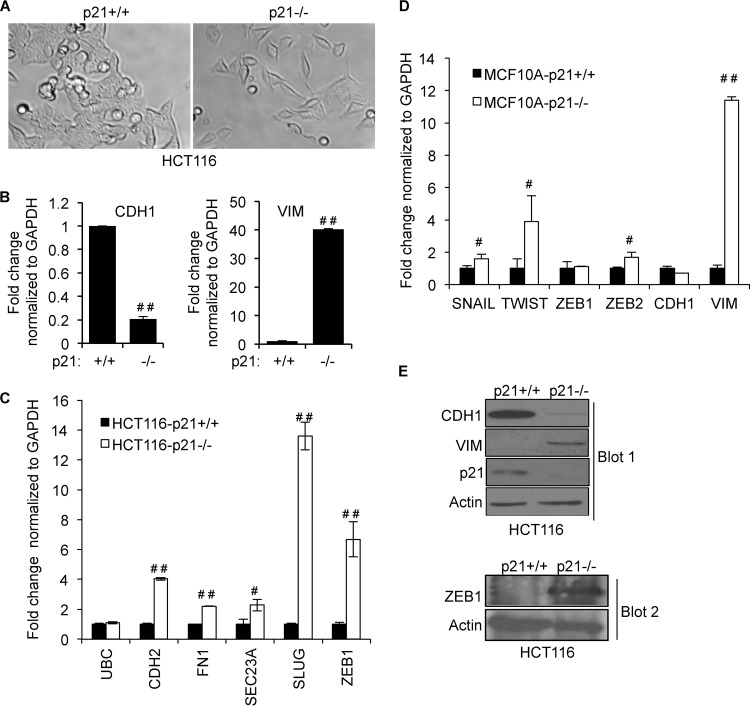
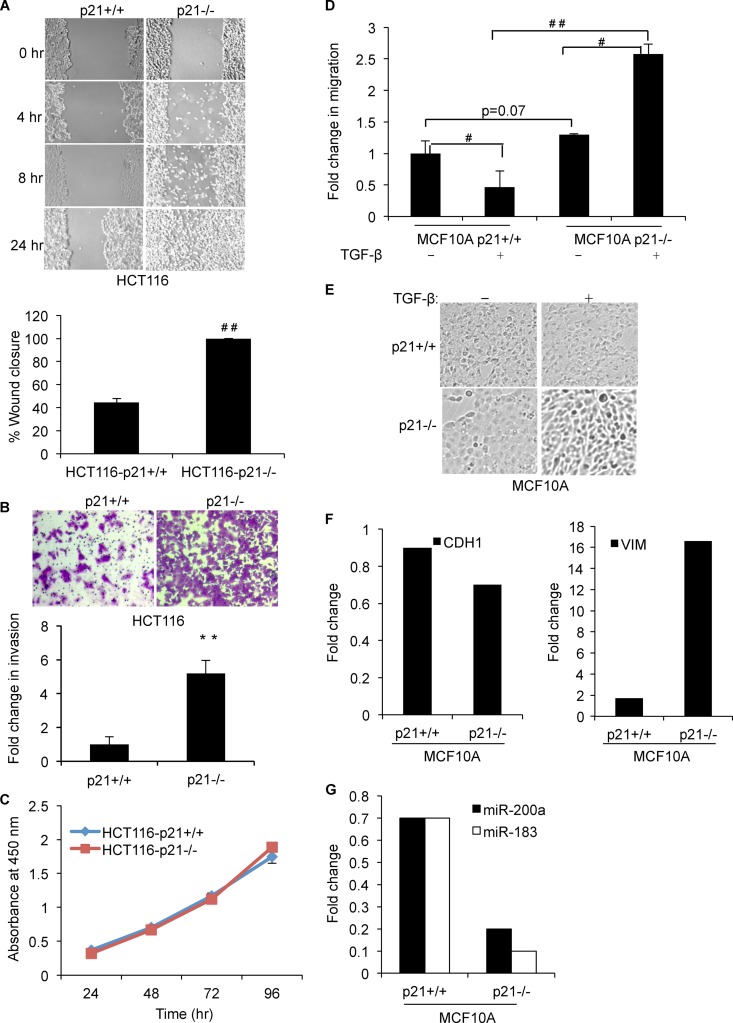
We and others have shown that p21 inhibits EMT (34–36). Consistent with data reported in the literature, we observed morphological changes reminiscent of EMT upon depletion of p21 in HCT116 cells (Fig. 5A). The loss of p21 resulted in a more fibroblast-like morphology with a loss of cell-cell contact. Depletion of p21 upregulated the mRNA levels of multiple mesenchymal genes, including VIM, CDH2, ZEB1, and SLUG, and downregulated the epithelial cell marker CDH1 in HCT116 and MCF10A cells (Fig. 5B to D). Downregulation of CDH1 and upregulation of VIM and ZEB1 proteins upon the loss of p21 were validated by immunoblotting (Fig. 5E). To determine whether induction of EMT upon p21 depletion resulted in increased cell migration and invasion, we performed wound-healing assays with HCT116-p21+/+ and HCT116-p21−/− cells. Strikingly, unlike HCT116-p21+/+ cells, HCT116-p21−/− cells migrated toward the wound within 4 h, and complete wound closure was observed in 24 h (Fig. 6A). Increased cell migration was also associated with enhanced invasion (∼5-fold), as assessed by Matrigel invasion assays (Fig. 6B), and these phenotypes were not due to altered proliferation (Fig. 6C). Next, we performed Transwell migration assays with isogenic MCF10A-p21+/+ and MCF10A-p21−/− cells after treatment of the cells with TGF-β for 24 h. Consistent with the known tumor-suppressive function of TGF-β, migration of MCF10A-p21+/+ cells was inhibited upon TGF-β treatment. Loss of p21 significantly enhanced (∼2.5-fold) the migration of MCF10A cells in response to TGF-β (Fig. 6D), further suggesting a function of p21 in inhibiting cell migration.
FIG 5.
Targeted deletion of p21 induces EMT. (A) Phase-contrast microscopy shows that loss of p21 in HCT116 cells leads to changes in cellular morphology similar to induction of EMT. (B) The mRNA levels of CDH1 (left) and VIM (right) in isogenic HCT116 cells (p21+/+ or p21−/−) were measured by RT-qPCR. (C and D) Expression levels of EMT-associated genes in HCT116-p21+/+, HCT116-p21−/−, MCF10A-p21+/+, and MCF10A-p21−/− cells were determined by RT-qPCR. (E) The effect of targeted deletion of p21 in HCT116 cells on CDH1 or VIM protein levels was assessed by immunoblotting. Error bars represent means ± standard deviations of three independent experiments. #, P < 0.01; ##, P < 0.001.
FIG 6.
Targeted deletion of p21 increases cell migration and invasion. (A) Wound-healing assays were performed with HCT116 -p21+/+ or -p21−/− cells at the indicated time points. Results from a representative experiment are shown (top), and results from 3 independent wound-healing assays (24 h) with HCT116 -p21+/+ or -p21−/− cells were quantified by using ImageJ (bottom). (B, top) Representative picture of invading cells at the 48-h time point in Matrigel invasion assays of HCT116-p21+/+ and HCT116-p21−/− cells. (Bottom) Fold change in invasion from 3 independent experiments quantified by using ImageJ. (C) Rates of proliferation of HCT116-p21+/+ and HCT116-p21−/− cells were measured by using Cell Counting Kit-8. (D) The effect of p21 loss on migration of MCF10A -p21+/+ or -p21−/− cells was assessed by Transwell migration assays in the presence and absence of TGF-β (24 h). (E and F) MCF10A -p21+/+ or -p21−/− cells were treated with TGF-β (6 days), and induction of EMT in p21−/− cells was observed by morphological changes (E) and decreased CDH1 and increased VIM mRNA levels (F). RT-qPCR results from two independent experiments are shown. (G) Induction of EMT in MCF10A-p21−/− cells 6 days after TGF-β treatment was associated with decreased miR-200a and miR-183 levels, as measured by RT-qPCR. Error bars represent means ± standard deviations of three independent experiments. #, P < 0.01; **, P < 0.005; ##, P < 0.001.
To examine the role of p21-regulated miRNAs in EMT in a more physiological setting, we looked at changes in expression levels of EMT markers (CDH1 and VIM) and p21-regulated miRNAs in the MCF10A isogenic cell lines after treatment of the cells with TGF-β. These experiments were performed with MCF10A cells instead of HCT116 cells because HCT116 cells have mutations in the TGF-β receptor (TGFBR2) and, as a result, do not respond to TGF-β. As shown in Fig. 6E, we observed EMT in MCF10A-p21−/− cells as early as 6 days after TGF-β treatment, and this morphological change was also associated with decreased CDH1 and increased VIM mRNA levels, as measured by RT-qPCR (Fig. 6F). Induction of EMT in MCF10A-p21−/− cells was also associated with substantial decreases (>80%) in miR-200a and miR-183 levels (Fig. 6G). Taken together, these results suggest an important physiological function of p21-regulated miRNAs in the inhibition of EMT.
Overexpression of miR-200 or miR-183 cluster miRNAs in p21-deficient cells inhibits cell migration and invasion.
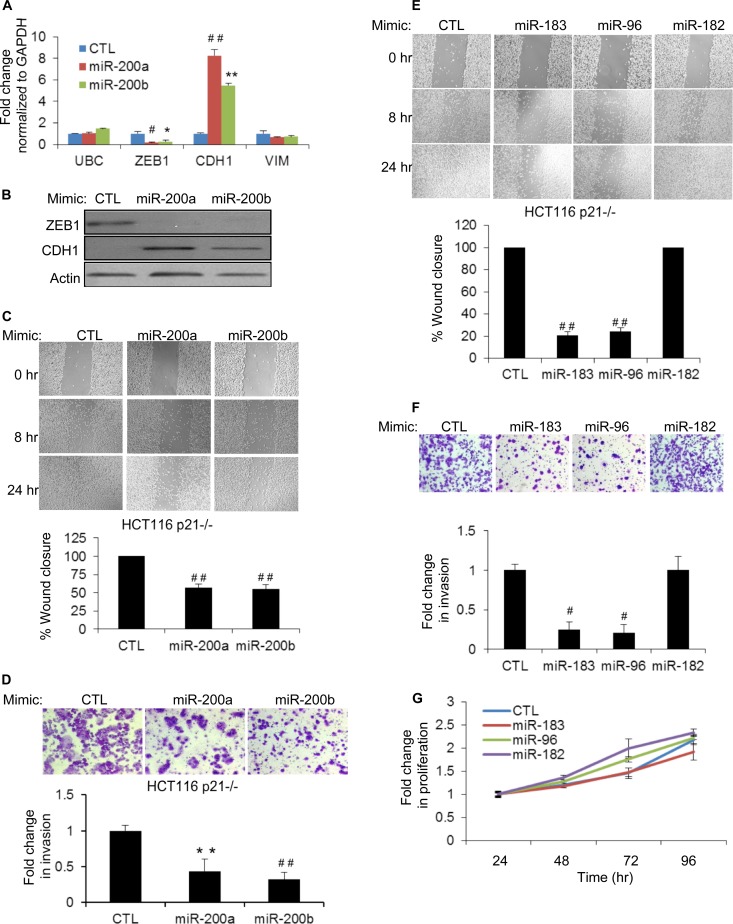
We next investigated whether the induction of EMT upon the loss of p21 could be mediated, in part, by downregulation of the miR-200 family. HCT116-p21−/− cells were transfected with CTL, miR-200a, or miR-200b mimics, and the abundance of ZEB1 mRNA was examined by RT-qPCR after 48 h. Consistent with previous reports (18, 22, 41), miR-200a or miR-200b overexpression downregulated ZEB1 (>5-fold) and upregulated CDH1 (>5-fold) (a transcriptional target of ZEB1) mRNAs and proteins (Fig. 7A and B). miR-200 also had a modest but reproducible effect on VIM mRNA levels. To determine whether enhanced migration and invasion of p21−/− cells could be mediated partly by downregulation of miR-200, we transfected HCT116-p21−/− cells with CTL, miR-200a, or miR-200b mimics and performed wound-healing and Matrigel invasion assays after 48 h. The introduction of miR-200a or miR-200b significantly inhibited migration (Fig. 7C) and invasion (Fig. 7D) of HCT116-p21−/− cells, and these phenotypes were not due to reduced proliferation or altered cell viability (data not shown). These results suggest that downregulation of the miR-200 family may play a role in inducing EMT upon the loss of p21.
FIG 7.
Overexpression of miR-200, miR-183, or miR-96 inhibits migration and invasion in p21 knockout cells. (A) The effect of miR-200a or miR-200b overexpression in HCT116-p21−/− cells on the levels of ZEB1, CDH1, and VIM mRNAs was assessed by RT-qPCR after transfecting CTL, miR-200a, or miR-200b mimics for 48 h. The housekeeping gene UBC was used as a negative control. (B) Immunoblotting of ZEB1 and CDH1 after 48 h of transfection of CTL, miR-200a, or miR-200b mimics in HCT116-p21−/− cells. (C) Wound healing was monitored at the indicated time points in HCT116-p21−/− cells transfected 48 h earlier with CTL, miR-200a, or miR-200b mimics (top), and results from 3 experiments were quantitated by using ImageJ (bottom). (D, top) Representative pictures of invading HCT116-p21−/− cells 48 h after transfection with CTL, miR-200a, or miR-200b mimics. (Bottom) Results from 3 invasion assays were quantitated by using the ImageJ program. (E and F) Forty-eight hours after transfection of HCT116-p21−/− cells with CTL, miR-183, miR-96, or miR-182 mimics, the effect on cell migration (E) or invasion (F) was examined at the indicated time points by wound-healing assays (E) or Matrigel invasion assays (F). (G) The effect of introducing miR-183 cluster miRNAs on cell proliferation was determined by using Cell Counting Kit-8 from HCT116-p21−/− cells transfected with CTL, miR-183, miR-96, or miR-182 mimics. Error bars represent means ± standard deviations of three independent experiments. *, P < 0.05; #, P < 0.01; **, P < 0.005; ##, P < 0.001.
The role of the miR-183 cluster in EMT is not fully understood. This miRNA cluster has been shown to inhibit or promote EMT depending on the cell type (42–45). Therefore, we next examined the functional consequences of downregulation of the miR-183 cluster upon the loss of p21. We transfected HCT116-p21−/− cells with CTL, miR-183, miR-96, or miR-182 mimics (20 nM) for 48 h and performed wound-healing and Matrigel invasion assays. Reintroduction of miR-183 or miR-96 but not miR-182 significantly inhibited cell migration (Fig. 7E) and invasion (Fig. 7F), and these phenotypes were not caused by reduced proliferation (Fig. 7G). These results suggest that decreased miR-183 and miR-96 levels in HCT116-p21−/− cells may play a role in enhancing cell migration and invasion.
Concurrent knockdown of miR-200 and the miR-183 cluster upregulates expression of mesenchymal markers and enhances invasion of p21-proficient cells.
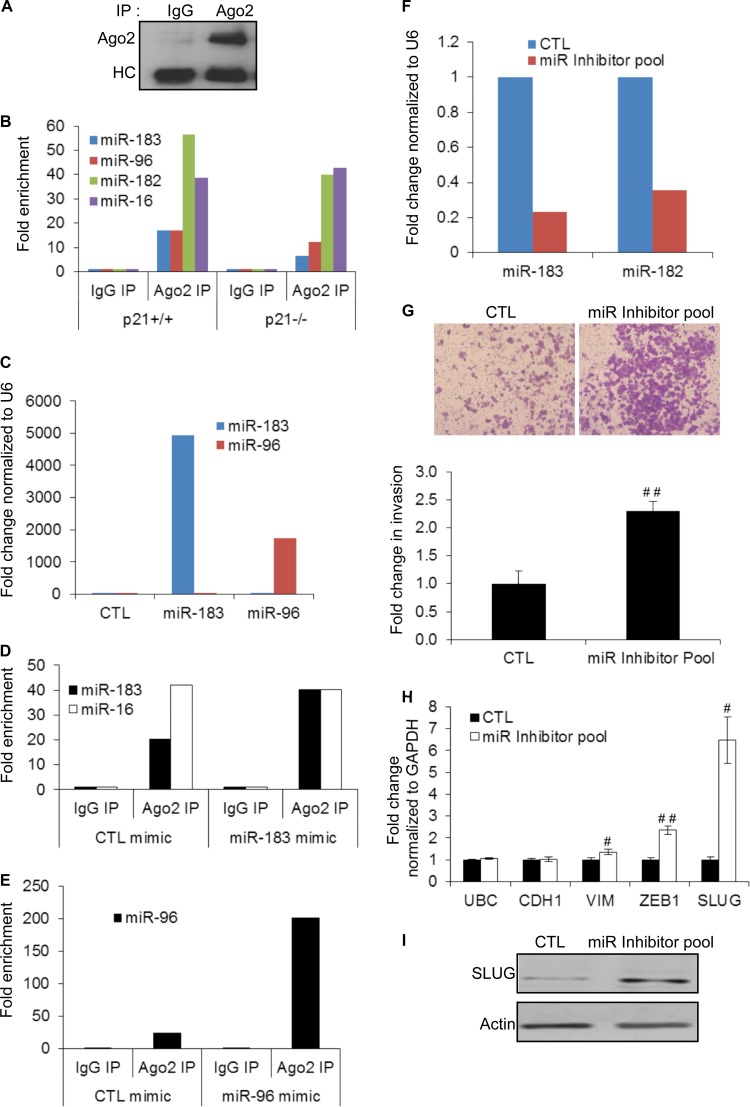
Although miRNA mimics are commonly used for miRNA overexpression experiments, they can result in dramatic increases in miRNA expression levels and may therefore raise concerns regarding the physiological significance of the results. To determine the proportion of the transfected miRNA that is functional, we first immunoprecipitated Ago2 from HCT116-p21−/− cytoplasmic extracts using an antibody that specifically recognizes Ago2 (46) (Fig. 8A). The abundance of miR-183 cluster miRNAs in the IP material from isogenic HCT116 cells was assessed by RT-qPCR. Consistent with the downregulation of the miR-183 cluster in p21−/− cells, we observed decreased enrichment of miR-183 cluster miRNAs in p21−/− cells compared to p21+/+ cells (Fig. 8B). As a negative control, the association of Ago2 to miR-16, a miRNA whose expression level did not change upon the loss of p21 (Table 1), was unchanged. We next transfected HCT116-p21−/− cells with CTL, miR-183, or miR-96 mimics (20 nM) and performed Ago2 IPs after 48 h. Despite the fact that the mimics resulted in >1,000-fold increases in total miR-96 or miR-183 levels (Fig. 8C), the enrichment of miR-183 increased by only 2-fold (from ∼20-fold to ∼40-fold) (Fig. 8D). Likewise, the enrichment of miR-96 in the Ago2 IPs increased by only ∼8-fold (from ∼25-fold to ∼200-fold) (Fig. 8E). These results therefore suggest that although transfection of HCT116-p21−/− cells with miR-183 or miR-96 mimics increases the abundance of these miRNAs to very high levels, the incorporation into Argonaute is within physiological levels and does not significantly influence the levels of the negative-control miRNA miR-16 in the Ago2 IPs. Similar results for miRNA mimic overexpression have been reported recently for multiple miRNAs by using the same Ago2 antibody (47).
FIG 8.
Concurrent knockdown of miR-200 and miR-183 cluster miRNAs increases invasion. (A) Immunoblotting from IPs performed from HCT116-p21−/− cytoplasmic extracts shows specific enrichment of Ago2 in the Ago2 IP. HC corresponds to the antibody heavy chain. (B) Enrichment of miR-183 cluster miRNAs and miR-16 (a control miRNA) in Ago2 IPs performed in duplicate from cytoplasmic extracts of HCT116 -p21+/+ and -p21−/− cells was measured by RT-qPCR of pulldown RNA. IgG IP was used as a negative control. (C) HCT116-p21−/− cells were transfected with CTL, miR-183, or miR-96 mimics for 48 h, and miR-183 and miR-96 levels were measured by RT-qPCR. (D) HCT116-p21−/− cells were transfected in duplicate with CTL or miR-183 mimics for 48 h, and enrichment of miR-183 and miR-16 in Ago2 IPs from cytoplasmic extracts was measured by RT-qPCR normalized to U6. (E) HCT116-p21−/− cells were transfected with CTL or miR-96 mimics in duplicate for 48 h, and enrichment of miR-96 in Ago2 IPs from cytoplasmic extracts was measured by RT-qPCR normalized to U6. IgG IP was used as a negative control. (F) Parental HCT116 cells were transfected in duplicate for 48 h with a control antisense oligonucleotide or a pool of antisense oligonucleotides (miR inhibitor pool) targeting miR-200a, miR-183, miR-96, and miR-182, and knockdown efficiency of miR-183 and miR-182 was assessed by RT-qPCR. (G) Parental HCT116 cells were transfected with a control antisense oligonucleotide or the miR inhibitor pool targeting miR-200a, miR-183, miR-96, and miR-182 for 48 h, and the effect on invasion was assessed by Matrigel invasion assays. (H) RT-qPCR was performed with HCT116 cells transfected with the miR inhibitor pool to determine changes in mRNA levels of EMT markers. The housekeeping gene UBC was used as a negative control. (I) Immunoblotting was performed with HCT116 cells transfected with the miR inhibitor pool to determine changes in SLUG protein levels. Error bars represent means ± standard deviations of three independent experiments. #, P < 0.01; ##, P < 0.001.
To further confirm the role of miR-200 and the miR-183 cluster in inhibition of invasion, we performed Matrigel invasion assays with HCT116-p21+/+ cells after knocking down individual members of this cluster using antisense oligonucleotides. Although knockdown of individual members of the miR-183 cluster or miR-200a alone did not affect invasion, significant increases in invasion (Fig. 8F and G) and mRNA expression levels of the EMT markers VIM, ZEB1, and SLUG (Fig. 8H) were observed when parental HCT116 cells were transfected with a pool of miRNA inhibitors targeting miR-200a, miR-183, miR-96, and miR-182 (miR inhibitor pool). Immunoblotting from whole-cell lysates transfected with the CTL or miR inhibitor pool confirmed significant upregulation of SLUG protein upon the concurrent knockdown of miR-200 and miR-183 cluster miRNAs (Fig. 8I). The effect on ZEB1 protein could not be evaluated because ZEB1 was undetectable in HCT116-p21+/+ cells (Fig. 5E). These results are consistent with the known mechanism by which miRNAs function and indicate functional redundancy between miR-200 and miR-183 cluster miRNAs.
miR-183 and miR-96 repress common targets, including the 3′ UTRs of SLUG and ZEB1.
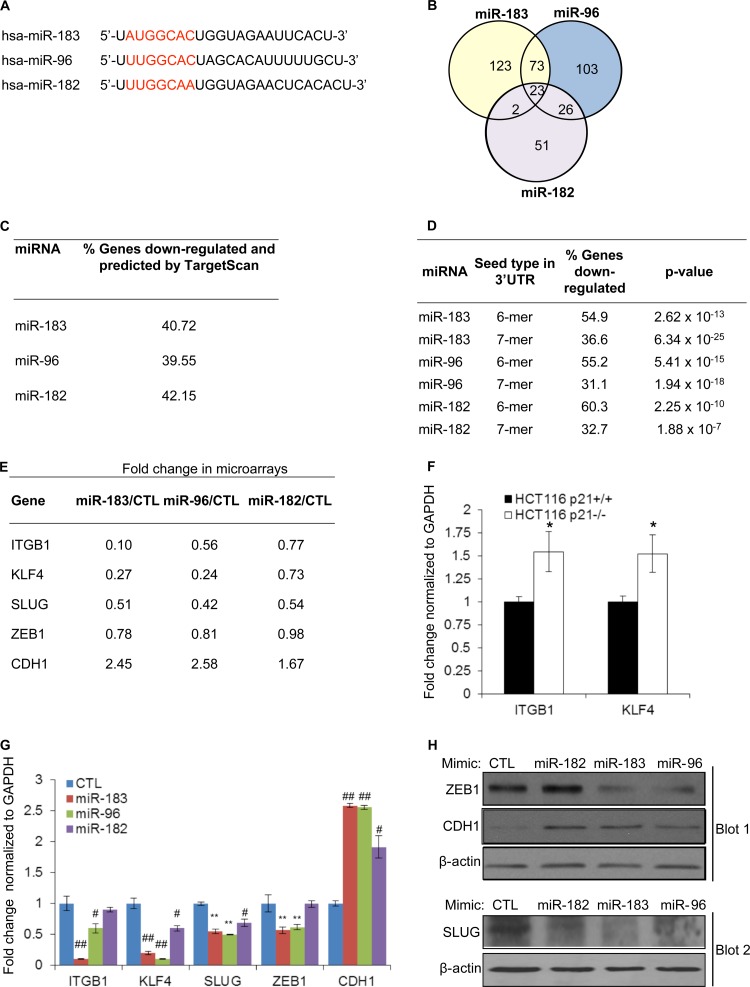
To understand the mechanism(s) by which reintroduction of the miR-183 cluster in p21−/− cells inhibited migration and invasion, we identified the genome-wide mRNAs downregulated by each miRNA of this cluster using microarrays (Fig. 9A and B). Although genes regulated exclusively at the level of mRNA translation will be missed by using this approach, we and others have successfully employed this strategy to identify a majority of targets of specific miRNAs (11, 48, 49). Therefore, we transfected HCT116-p21−/− cells with CTL, miR-183, miR-96, or miR-182 mimics and performed microarrays after 48 h. Using a cutoff of 2-fold (P < 0.01) (Fig. 9B; see also Tables S3 to S5 in the supplemental material) or 1.5-fold (P < 0.01), miR-182 repressed less than half as many genes as miR-183 or miR-96, indicating that miR-96 and miR-183 may inhibit the expression of an overlapping set of genes to exert common functions. Moreover, a substantial proportion of the genes downregulated by the miR-183 cluster miRNAs were also predicted by TargetScan (Fig. 9C), and their 3′ UTRs were significantly enriched for an exact match to the hexamer or heptamer sequence complementary to the seed region of the miR-183 cluster miRNAs, suggesting that many genes downregulated by the miR-183 cluster miRNAs are direct targets (Fig. 9D).
FIG 9.
miR-183 and miR-96 downregulate common targets involved in EMT. (A) Sequence of mature miRNAs from the miR-183 cluster. The heptamer seed sequence is shown in red. (B) Venn diagram showing the overlap between genes downregulated ≥2-fold (P value of <0.01) in the microarrays (see Table S3 to S5 in the supplemental material) for HCT116-p21−/− cells 48 h after transfection with CTL, miR-183, miR-96, or miR-182 mimics. (C) Table showing the overlap between TargetScan predictions and the genes downregulated by the miR-183 cluster miRNAs. (D) Table showing the enrichment of the sequence complementary to the hexamer region (nt 2 to 7) and heptamer (nt 2 to 8) of miR-183, miR-96, or miR-182 in the 3′ UTR of the transcripts that were downregulated ≥2-fold (P value of <0.01) in the microarrays (see Tables S3 to S5 in the supplemental material). (E) Table showing fold changes in expression of CDH1 and select miR-183 cluster target mRNAs (involved in cell migration and invasion) in the microarrays. (F) Effect of p21 deletion on the expression of ITGB1 and KLF4 mRNAs was measured by RT-qPCR for HCT116-p21+/+ and HCT116-p21−/− cells. (G) HCT116-p21−/− cells were transfected with CTL, miR-183, miR-96, or miR-182 mimics for 48 h, and RT-qPCR was performed to measure changes in levels of miR-183 cluster-regulated and CDH1 mRNAs. (H) The effect of miR-183 cluster miRNAs on ZEB1, CDH1, and SLUG protein levels was assessed by immunoblotting from HCT116-p21−/− cells 48 h after transfection with CTL, miR-183, miR-96, or miR-182 mimics. Error bars represent means ± standard deviations of three independent experiments. *, P < 0.05; #, P < 0.01; **, P < 0.005; ##, P < 0.001.
Our microarray results suggested that the miR-183 cluster miRNAs strongly reduced the expression levels of key genes involved in regulating migration, invasion, and metastasis. In particular, miR-183 and miR-96 significantly downregulated the mRNAs encoding the fibronectin receptor integrin β-1 (ITGB1) and the transcription factor KLF4 (Fig. 9E). ITGB1 is a known target of miR-183 (42) and plays a crucial role in tumor invasion (50). KLF4 is a direct regulator of p21 transcription (51) and functions as an oncogene or a tumor suppressor depending on the p21 status (52). These mRNAs were also upregulated in HCT116-p21−/− cells (Fig. 9F). In agreement with the microarray results, overexpression of miR-183 or miR-96 in HCT116-p21−/− cells downregulated ITGB1 and KLF4 mRNAs (∼2- to 10-fold), as assessed by RT-qPCR (Fig. 9G), indicating that downregulation of the miR-183 cluster in p21−/− cells may contribute to the upregulation of these mRNAs upon the loss of p21.
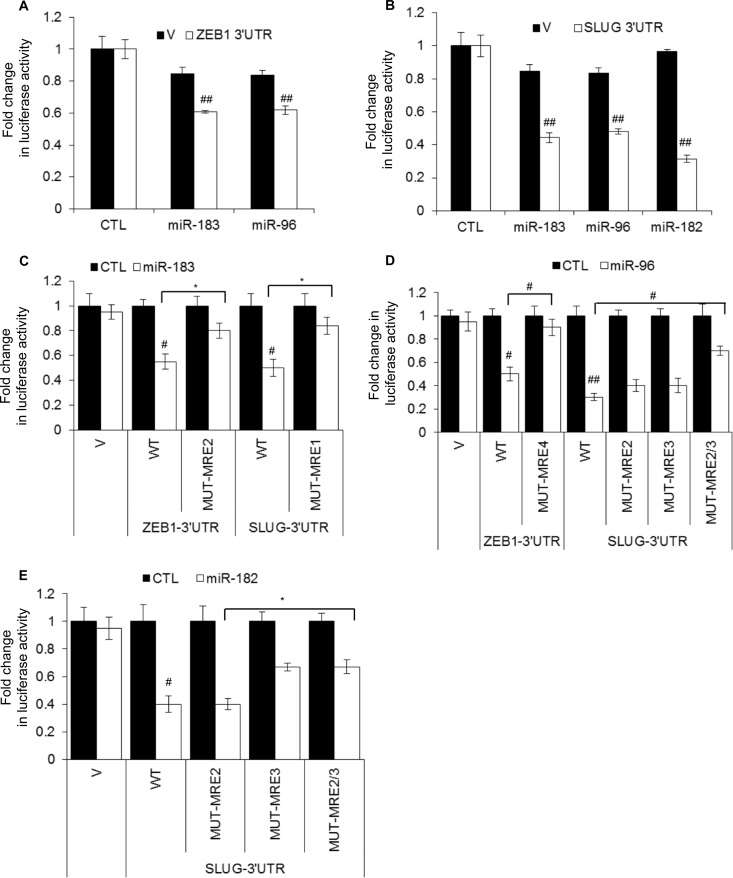
According to our microarray results, miR-183 and miR-96 significantly increased (∼2.5-fold) CDH1 mRNA levels (Fig. 9E; see also Tables S3 and S4 in the supplemental material). We therefore searched for alterations in the expression levels of genes that regulate CDH1 transcription and found significantly decreased (∼2-fold) SNAI2 (SLUG) mRNA levels in the microarrays (Fig. 9E), a result that was confirmed by RT-qPCR (Fig. 9G). Although ZEB1, a known repressor of CDH1 transcription, was not identified as a miR-183 cluster target in our microarrays (Fig. 9E), ZEB1 mRNA and protein were consistently downregulated by miR-183 or miR-96, as measured by RT-qPCR (Fig. 9G) and immunoblotting (Fig. 9H), suggesting that downregulation of the miR-183 cluster upon the loss of p21 may play a role in enhancing the expression of these genes. 3′ UTR luciferase reporter assays suggested that ZEB1 and SLUG 3′ UTRs were repressed by miR-183 and miR-96 (Fig. 10A and B). Consistent with the downregulation of SLUG mRNA and protein by miR-182 (Fig. 9G and H), miR-182 repressed the 3′ UTR of SLUG mRNA (Fig. 10B). To identify the miRNA recognition elements (MREs) for the miR-183 cluster miRNAs in the 3′ UTR of ZEB1 and SLUG mRNAs, we used PITA, as previously described (48) (see Table S7 in the supplemental material). To determine if these MREs were responsive to the miR-183 cluster miRNAs, we inserted the putative MREs into the 3′ UTR of Renilla luciferase of psiCHECK2 and performed luciferase assays after cotransfecting the reporters with mimics corresponding to the miR-183 cluster miRNAs. This approach identified one ZEB1 MRE each for miR-183 and miR-96 (ZEB1-MRE2 and -MRE4), one MRE for miR-183 in the SLUG 3′ UTR (SLUG-MRE1), and 2 MREs for miR-96 and miR-182 in the SLUG 3′UTR (SLUG-MRE2 and -MRE3) (data not shown). Point mutations in the seed region of these MREs in the 3′ UTRs of ZEB1 and SLUG mRNAs decreased the repression by the miR-183 cluster miRNAs (Fig. 10C to E; see also Table S7 in the supplemental material). These results further confirm that ZEB1 is a direct target of miR-183 and miR-96, whereas SLUG is a direct target of miR-183, miR-96, and miR-182.
FIG 10.
miR-183 and miR-96 directly repress the 3′ UTRs of SLUG and ZEB1 mRNAs. Luciferase assays were performed with HeLa cells 48 h after cotransfection of the cells with psiCHECK2 (V) or psiCHECK2 containing the wild-type (WT) or mutant (MUT) 3′ UTR of ZEB1 or SLUG mRNAs and CTL, miR-183, miR-96, or miR-182 mimics. Sequences of wild-type and mutant MREs are shown in Table S7 in the supplemental material.
The results from this study together with those of our previous studies where we demonstrated a feedback loop between ZEB1 and miR-200 (22, 53) indicate that p21 forms a complex with ZEB1 to regulate the expression of the miR-183 cluster and miR-200 family to modulate the expression of SLUG, ZEB1, ITGB1, and KLF4 mRNAs to inhibit EMT, cell migration, and invasion.
DISCUSSION
p21 functions as a downstream effector of several tumor suppressors, including p53 (26–28). Our present findings suggest that in addition to its role in cell cycle control during the stress response, loss of p21 induces EMT, an effect partly mediated by downregulation of the miR-200 family and the miR-183 cluster through activation of ZEB1. Identification of miR-183 cluster targets suggested that miR-183 and miR-96 repressed an overlapping set of genes that control cell migration, invasion, or EMT, including ITGB1, KLF4, SLUG, and ZEB1. In addition, our results indicate that transcription of the miR-183 cluster is repressed directly by ZEB1 in p21 knockout cells, suggesting the existence of a reciprocal feedback loop.
p21 has been implicated in regulation of gene expression (25, 32, 33, 54) by inhibiting transcription factors such as E2F1 (55), STAT3 (56), MYC (57), NRF2 (58), and p300 (59). Because these transcription factors also regulate miRNA expression (60–62), we sought to identify miRNAs under p21 control. We identified several p21-regulated miRNAs and showed that activation of p53 (32, 39) is responsible for the upregulation of miR-34a (11) and miR-143 (63) in p21−/− cells. Because p21 is known to inhibit E2F1, MYC, and STAT3, it is possible that in addition to p53, the expression of a subset of the upregulated miRNAs is attributable to the activation of these transcription factors in the absence of p21. Our results also identified a novel interaction between p21 and ZEB1. However, further studies with recombinant proteins are necessary to determine whether p21 interacts directly with ZEB1 or whether this interaction is mediated via other proteins. Although our paper focused on regulation of the miR-183 cluster through a p21-ZEB1 complex, it is possible that the p21-ZEB1 complex regulates the expression of other miRNAs.
In this study, we focused on understanding how the miR-200 and miR-183 clusters were downregulated in p21−/− cells and the downstream consequences. Consistent with recent studies establishing a role of the miR-200 family and p21 in inhibition of EMT (22, 34–36, 64, 65), we observed induction of EMT upon the depletion of p21. The expression of many EMT regulators, including CDH1, ZEB1, and SLUG, was altered upon the depletion of p21 in both HCT116 and MCF10A cells. Induction of EMT in p21 knockout HCT116 and MCF10A cells enhanced cell migration and invasion, indicating that p21 inhibits cell migration and invasion by suppressing EMT. Our results support a role for p21 in inhibiting EMT in breast and colorectal cancer and are consistent with previous studies that demonstrated a role of p21 in inhibition of EMT (34–36).
Among the 3 miRNAs of the miR-183 cluster, miR-183 and miR-96 but not miR-182 inhibited cell migration and invasion when these miRNAs were reintroduced into HCT116-p21−/− cells. Consistent with these phenotypic effects, we found that miR-183 and miR-96 share more targets with each other than with miR-182. Interestingly, miR-182 and miR-96 have the same hexamer seed sequence (UGCCAA), whereas the seed sequence of miR-183 (UGCCAU) differs by only one nucleotide. Therefore, sequences beyond the hexamer seed region may play a role in gene silencing by these miRNAs. In addition to the miR-183 cluster, reintroduction of miR-200a or miR-200b in p21−/− cells downregulated ZEB1 (a known target of miR-200) and increased CDH1 expression levels to reduce migration and invasion. Our miRNA knockdown experiments suggest that the enhanced migration and invasion upon p21 loss are mediated, in part, by downregulation of the miR-200 family and the miR-183 cluster. Functional redundancy between these 3 miRNA clusters may ensure that aberrant expression of one of these clusters does not deregulate their targets to inhibit EMT and prevent tumor progression.
Our results are consistent with recent evidences which indicate that miR-183 or miR-96 inhibits migration, invasion, and metastasis (42, 43, 66, 67). However, some studies have shown that miR-183 or miR-96 promotes migration and invasion (44, 45). Likewise, the miR-200 family has also been shown to function as a tumor suppressor by inhibiting EMT (22, 38, 65, 68), but it can also exert oncogenic effects by promoting metastatic colonization (41, 69). These findings indicate that the miR-200 family and the miR-183 cluster miRNAs have context-dependent functions. In summary, our findings are indicative of a mechanism by which p21 inhibits EMT through the miR-200 and miR-183 clusters. It remains to be seen if p21-regulated miRNAs also play a role in other phenotypes regulated by p21.
Supplementary Material
ACKNOWLEDGMENTS
Ashish Lal was supported by the Intramural Research Program of the National Institutes of Health, National Cancer Institute, Center for Cancer Research. Thomas Brabletz was supported by the DFG (BR 1399/6-1, SFB 850/B2, and SFB992/C6) and the Deutsche Krebshilfe (grant no. 109430). Sudha Sharma was supported by the National Institute of General Medical Sciences of the National Institutes of Health under award number SC1GM093999.
We thank K. Prasanth (University of Illinois at Urbana-Champaign) and Marshall Thomas (Harvard University) for their comments on the manuscript. The HCT116 isogenic cell lines were a kind gift from Bert Vogelstein (Johns Hopkins University), and the Ago2 antibody was a kind gift from Zissimos Mourelators (University of Pennsylvania).
Footnotes
Published ahead of print 25 November 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MCB.01043-13.
REFERENCES
- 1.Ambros V. 2004. The functions of animal microRNAs. Nature 431:350–355. 10.1038/nature02871 [DOI] [PubMed] [Google Scholar]
- 2.Bartel DP. 2004. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116:281–297. 10.1016/S0092-8674(04)00045-5 [DOI] [PubMed] [Google Scholar]
- 3.Bartel DP. 2009. MicroRNAs: target recognition and regulatory functions. Cell 136:215–233. 10.1016/j.cell.2009.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berezikov E. 2011. Evolution of microRNA diversity and regulation in animals. Nat. Rev. Genet. 12:846–860. 10.1038/nrg3079 [DOI] [PubMed] [Google Scholar]
- 5.Baek D, Villen J, Shin C, Camargo FD, Gygi SP, Bartel DP. 2008. The impact of microRNAs on protein output. Nature 455:64–71. 10.1038/nature07242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Djuranovic S, Nahvi A, Green R. 2012. miRNA-mediated gene silencing by translational repression followed by mRNA deadenylation and decay. Science 336:237–240. 10.1126/science.1215691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fabian MR, Sonenberg N. 2012. The mechanics of miRNA-mediated gene silencing: a look under the hood of miRISC. Nat. Struct. Mol. Biol. 19:586–593. 10.1038/nsmb.2296 [DOI] [PubMed] [Google Scholar]
- 8.Selbach M, Schwanhausser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. 2008. Widespread changes in protein synthesis induced by microRNAs. Nature 455:58–63. 10.1038/nature07228 [DOI] [PubMed] [Google Scholar]
- 9.Thomas M, Lieberman J, Lal A. 2010. Desperately seeking microRNA targets. Nat. Struct. Mol. Biol. 17:1169–1174. 10.1038/nsmb.1921 [DOI] [PubMed] [Google Scholar]
- 10.Croce CM. 2009. Causes and consequences of microRNA dysregulation in cancer. Nat. Rev. Genet. 10:704–714. 10.1038/nrg2634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chang TC, Wentzel EA, Kent OA, Ramachandran K, Mullendore M, Lee KH, Feldmann G, Yamakuchi M, Ferlito M, Lowenstein CJ, Arking DE, Beer MA, Maitra A, Mendell JT. 2007. Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. Mol. Cell 26:745–752. 10.1016/j.molcel.2007.05.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cimmino A, Calin GA, Fabbri M, Iorio MV, Ferracin M, Shimizu M, Wojcik SE, Aqeilan RI, Zupo S, Dono M, Rassenti L, Alder H, Volinia S, Liu CG, Kipps TJ, Negrini M, Croce CM. 2005. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc. Natl. Acad. Sci. U. S. A. 102:13944–13949. 10.1073/pnas.0506654102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Johnson CD, Esquela-Kerscher A, Stefani G, Byrom M, Kelnar K, Ovcharenko D, Wilson M, Wang X, Shelton J, Shingara J, Chin L, Brown D, Slack FJ. 2007. The let-7 microRNA represses cell proliferation pathways in human cells. Cancer Res. 67:7713–7722. 10.1158/0008-5472.CAN-07-1083 [DOI] [PubMed] [Google Scholar]
- 14.Lal A, Thomas MP, Altschuler G, Navarro F, O'Day E, Li XL, Concepcion C, Han YC, Thiery J, Rajani DK, Deutsch A, Hofmann O, Ventura A, Hide W, Lieberman J. 2011. Capture of microRNA-bound mRNAs identifies the tumor suppressor miR-34a as a regulator of growth factor signaling. PLoS Genet. 7:e1002363. 10.1371/journal.pgen.1002363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yu F, Yao H, Zhu P, Zhang X, Pan Q, Gong C, Huang Y, Hu X, Su F, Lieberman J, Song E. 2007. let-7 regulates self renewal and tumorigenicity of breast cancer cells. Cell 131:1109–1123. 10.1016/j.cell.2007.10.054 [DOI] [PubMed] [Google Scholar]
- 16.Medina PP, Nolde M, Slack FJ. 2010. OncomiR addiction in an in vivo model of microRNA-21-induced pre-B-cell lymphoma. Nature 467:86–90. 10.1038/nature09284 [DOI] [PubMed] [Google Scholar]
- 17.Mu P, Han YC, Betel D, Yao E, Squatrito M, Ogrodowski P, de Stanchina E, D'Andrea A, Sander C, Ventura A. 2009. Genetic dissection of the miR-17∼92 cluster of microRNAs in Myc-induced B-cell lymphomas. Genes Dev. 23:2806–2811. 10.1101/gad.1872909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gregory PA, Bert AG, Paterson EL, Barry SC, Tsykin A, Farshid G, Vadas MA, Khew-Goodall Y, Goodall GJ. 2008. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat. Cell Biol. 10:593–601. 10.1038/ncb1722 [DOI] [PubMed] [Google Scholar]
- 19.Ma L, Teruya-Feldstein J, Weinberg RA. 2007. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature 449:682–688. 10.1038/nature06174 [DOI] [PubMed] [Google Scholar]
- 20.Valastyan S, Chang A, Benaich N, Reinhardt F, Weinberg RA. 2011. Activation of miR-31 function in already-established metastases elicits metastatic regression. Genes Dev. 25:646–659. 10.1101/gad.2004211 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 21.Valastyan S, Reinhardt F, Benaich N, Calogrias D, Szasz AM, Wang ZC, Brock JE, Richardson AL, Weinberg RA. 2009. A pleiotropically acting microRNA, miR-31, inhibits breast cancer metastasis. Cell 137:1032–1046. 10.1016/j.cell.2009.03.047 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 22.Wellner U, Schubert J, Burk UC, Schmalhofer O, Zhu F, Sonntag A, Waldvogel B, Vannier C, Darling D, zur Hausen A, Brunton VG, Morton J, Sansom O, Schuler J, Stemmler MP, Herzberger C, Hopt U, Keck T, Brabletz S, Brabletz T. 2009. The EMT-activator ZEB1 promotes tumorigenicity by repressing stemness-inhibiting microRNAs. Nat. Cell Biol. 11:1487–1495. 10.1038/ncb1998 [DOI] [PubMed] [Google Scholar]
- 23.Yang J, Mani SA, Donaher JL, Ramaswamy S, Itzykson RA, Come C, Savagner P, Gitelman I, Richardson A, Weinberg RA. 2004. Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell 117:927–939. 10.1016/j.cell.2004.06.006 [DOI] [PubMed] [Google Scholar]
- 24.Yang J, Weinberg RA. 2008. Epithelial-mesenchymal transition: at the crossroads of development and tumor metastasis. Dev. Cell 14:818–829. 10.1016/j.devcel.2008.05.009 [DOI] [PubMed] [Google Scholar]
- 25.Abbas T, Dutta A. 2009. p21 in cancer: intricate networks and multiple activities. Nat. Rev. Cancer 9:400–414. 10.1038/nrc2657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Deng C, Zhang P, Harper JW, Elledge SJ, Leder P. 1995. Mice lacking p21CIP1/WAF1 undergo normal development, but are defective in G1 checkpoint control. Cell 82:675–684. 10.1016/0092-8674(95)90039-X [DOI] [PubMed] [Google Scholar]
- 27.el-Deiry WS, Tokino T, Velculescu VE, Levy DB, Parsons R, Trent JM, Lin D, Mercer WE, Kinzler KW, Vogelstein B. 1993. WAF1, a potential mediator of p53 tumor suppression. Cell 75:817–825. 10.1016/0092-8674(93)90500-P [DOI] [PubMed] [Google Scholar]
- 28.Waldman T, Kinzler KW, Vogelstein B. 1995. p21 is necessary for the p53-mediated G1 arrest in human cancer cells. Cancer Res. 55:5187–5190 [PubMed] [Google Scholar]
- 29.Gartel AL, Radhakrishnan SK. 2005. Lost in transcription: p21 repression, mechanisms, and consequences. Cancer Res. 65:3980–3985. 10.1158/0008-5472.CAN-04-3995 [DOI] [PubMed] [Google Scholar]
- 30.Starostina NG, Kipreos ET. 2012. Multiple degradation pathways regulate versatile CIP/KIP CDK inhibitors. Trends Cell Biol. 22:33–41. 10.1016/j.tcb.2011.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chang BD, Swift ME, Shen M, Fang J, Broude EV, Roninson IB. 2002. Molecular determinants of terminal growth arrest induced in tumor cells by a chemotherapeutic agent. Proc. Natl. Acad. Sci. U. S. A. 99:389–394. 10.1073/pnas.012602599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chang BD, Watanabe K, Broude EV, Fang J, Poole JC, Kalinichenko TV, Roninson IB. 2000. Effects of p21Waf1/Cip1/Sdi1 on cellular gene expression: implications for carcinogenesis, senescence, and age-related diseases. Proc. Natl. Acad. Sci. U. S. A. 97:4291–4296. 10.1073/pnas.97.8.4291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Perkins ND. 2002. Not just a CDK inhibitor: regulation of transcription by p21(WAF1/CIP1/SDI1). Cell Cycle 1:39–41. 10.4161/cc.1.1.98 [DOI] [PubMed] [Google Scholar]
- 34.Bachman KE, Blair BG, Brenner K, Bardelli A, Arena S, Zhou S, Hicks J, De Marzo AM, Argani P, Park BH. 2004. p21(WAF1/CIP1) mediates the growth response to TGF-beta in human epithelial cells. Cancer Biol. Ther. 3:221–225. 10.4161/cbt.3.2.666 [DOI] [PubMed] [Google Scholar]
- 35.Liu M, Casimiro MC, Wang C, Shirley LA, Jiao X, Katiyar S, Ju X, Li Z, Yu Z, Zhou J, Johnson M, Fortina P, Hyslop T, Windle JJ, Pestell RG. 2009. p21CIP1 attenuates Ras- and c-Myc-dependent breast tumor epithelial mesenchymal transition and cancer stem cell-like gene expression in vivo. Proc. Natl. Acad. Sci. U. S. A. 106:19035–19039. 10.1073/pnas.0910009106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang Y, Yan W, Jung YS, Chen X. 2013. PUMA cooperates with p21 to regulate mammary epithelial morphogenesis and epithelial-to-mesenchymal transition. PLoS One 8:e66464. 10.1371/journal.pone.0066464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chang CJ, Chao CH, Xia W, Yang JY, Xiong Y, Li CW, Yu WH, Rehman SK, Hsu JL, Lee HH, Liu M, Chen CT, Yu D, Hung MC. 2011. p53 regulates epithelial-mesenchymal transition and stem cell properties through modulating miRNAs. Nat. Cell Biol. 13:317–323. 10.1038/ncb2173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim T, Veronese A, Pichiorri F, Lee TJ, Jeon YJ, Volinia S, Pineau P, Marchio A, Palatini J, Suh SS, Alder H, Liu CG, Dejean A, Croce CM. 2011. p53 regulates epithelial-mesenchymal transition through microRNAs targeting ZEB1 and ZEB2. J. Exp. Med. 208:875–883. 10.1084/jem.20110235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Javelaud D, Besancon F. 2002. Inactivation of p21WAF1 sensitizes cells to apoptosis via an increase of both p14ARF and p53 levels and an alteration of the Bax/Bcl-2 ratio. J. Biol. Chem. 277:37949–37954. 10.1074/jbc.M204497200 [DOI] [PubMed] [Google Scholar]
- 40.Parvathaneni S, Stortchevoi A, Sommers JA, Brosh RM, Jr, Sharma S. 2013. Human RECQ1 interacts with Ku70/80 and modulates DNA end-joining of double-strand breaks. PLoS One 8:e62481. 10.1371/journal.pone.0062481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dykxhoorn DM, Wu Y, Xie H, Yu F, Lal A, Petrocca F, Martinvalet D, Song E, Lim B, Lieberman J. 2009. miR-200 enhances mouse breast cancer cell colonization to form distant metastases. PLoS One 4:e7181. 10.1371/journal.pone.0007181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li G, Luna C, Qiu J, Epstein DL, Gonzalez P. 2010. Targeting of integrin beta1 and kinesin 2alpha by microRNA 183. J. Biol. Chem. 285:5461–5471. 10.1074/jbc.M109.037127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lowery AJ, Miller N, Dwyer RM, Kerin MJ. 2010. Dysregulated miR-183 inhibits migration in breast cancer cells. BMC Cancer 10:502. 10.1186/1471-2407-10-502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sarver AL, Li L, Subramanian S. 2010. MicroRNA miR-183 functions as an oncogene by targeting the transcription factor EGR1 and promoting tumor cell migration. Cancer Res. 70:9570–9580. 10.1158/0008-5472.CAN-10-2074 [DOI] [PubMed] [Google Scholar]
- 45.Weeraratne SD, Amani V, Teider N, Pierre-Francois J, Winter D, Kye MJ, Sengupta S, Archer T, Remke M, Bai AH, Warren P, Pfister SM, Steen JA, Pomeroy SL, Cho YJ. 2012. Pleiotropic effects of miR-183∼96∼182 converge to regulate cell survival, proliferation and migration in medulloblastoma. Acta Neuropathol. 123:539–552. 10.1007/s00401-012-0969-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nelson PT, De Planell-Saguer M, Lamprinaki S, Kiriakidou M, Zhang P, O'Doherty U, Mourelatos Z. 2007. A novel monoclonal antibody against human Argonaute proteins reveals unexpected characteristics of miRNAs in human blood cells. RNA 13:1787–1792. 10.1261/rna.646007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thomson DW, Bracken CP, Szubert JM, Goodall GJ. 2013. On measuring miRNAs after transient transfection of mimics or antisense inhibitors. PLoS One 8:e55214. 10.1371/journal.pone.0055214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lal A, Navarro F, Maher CA, Maliszewski LE, Yan N, O'Day E, Chowdhury D, Dykxhoorn DM, Tsai P, Hofmann O, Becker KG, Gorospe M, Hide W, Lieberman J. 2009. miR-24 inhibits cell proliferation by targeting E2F2, MYC, and other cell-cycle genes via binding to “seedless” 3′UTR microRNA recognition elements. Mol. Cell 35:610–625. 10.1016/j.molcel.2009.08.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM. 2005. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature 433:769–773. 10.1038/nature03315 [DOI] [PubMed] [Google Scholar]
- 50.White DE, Kurpios NA, Zuo D, Hassell JA, Blaess S, Mueller U, Muller WJ. 2004. Targeted disruption of beta1-integrin in a transgenic mouse model of human breast cancer reveals an essential role in mammary tumor induction. Cancer Cell 6:159–170. 10.1016/j.ccr.2004.06.025 [DOI] [PubMed] [Google Scholar]
- 51.Zhang W, Geiman DE, Shields JM, Dang DT, Mahatan CS, Kaestner KH, Biggs JR, Kraft AS, Yang VW. 2000. The gut-enriched Kruppel-like factor (Kruppel-like factor 4) mediates the transactivating effect of p53 on the p21WAF1/Cip1 promoter. J. Biol. Chem. 275:18391–18398. 10.1074/jbc.C000062200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rowland BD, Peeper DS. 2006. KLF4, p21 and context-dependent opposing forces in cancer. Nat. Rev. Cancer 6:11–23. 10.1038/nrc1780 [DOI] [PubMed] [Google Scholar]
- 53.Burk U, Schubert J, Wellner U, Schmalhofer O, Vincan E, Spaderna S, Brabletz T. 2008. A reciprocal repression between ZEB1 and members of the miR-200 family promotes EMT and invasion in cancer cells. EMBO Rep. 9:582–589. 10.1038/embor.2008.74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ferrandiz N, Caraballo JM, Garcia-Gutierrez L, Devgan V, Rodriguez-Paredes M, Lafita MC, Bretones G, Quintanilla A, Munoz-Alonso MJ, Blanco R, Reyes JC, Agell N, Delgado MD, Dotto GP, Leon J. 2012. p21 as a transcriptional co-repressor of S-phase and mitotic control genes. PLoS One 7:e37759. 10.1371/journal.pone.0037759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Delavaine L, La Thangue NB. 1999. Control of E2F activity by p21Waf1/Cip1. Oncogene 18:5381–5392. 10.1038/sj.onc.1202923 [DOI] [PubMed] [Google Scholar]
- 56.Coqueret O, Gascan H. 2000. Functional interaction of STAT3 transcription factor with the cell cycle inhibitor p21WAF1/CIP1/SDI1. J. Biol. Chem. 275:18794–18800. 10.1074/jbc.M001601200 [DOI] [PubMed] [Google Scholar]
- 57.Kitaura H, Shinshi M, Uchikoshi Y, Ono T, Iguchi-Ariga SM, Ariga H. 2000. Reciprocal regulation via protein-protein interaction between c-Myc and p21(cip1/waf1/sdi1) in DNA replication and transcription. J. Biol. Chem. 275:10477–10483. 10.1074/jbc.275.14.10477 [DOI] [PubMed] [Google Scholar]
- 58.Chen W, Sun Z, Wang XJ, Jiang T, Huang Z, Fang D, Zhang DD. 2009. Direct interaction between Nrf2 and p21(Cip1/WAF1) upregulates the Nrf2-mediated antioxidant response. Mol. Cell 34:663–673. 10.1016/j.molcel.2009.04.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Snowden AW, Anderson LA, Webster GA, Perkins ND. 2000. A novel transcriptional repression domain mediates p21(WAF1/CIP1) induction of p300 transactivation. Mol. Cell. Biol. 20:2676–2686. 10.1128/MCB.20.8.2676-2686.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chang TC, Yu D, Lee YS, Wentzel EA, Arking DE, West KM, Dang CV, Thomas-Tikhonenko A, Mendell JT. 2008. Widespread microRNA repression by Myc contributes to tumorigenesis. Nat. Genet. 40:43–50. 10.1038/ng.2007.30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.O'Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT. 2005. c-Myc-regulated microRNAs modulate E2F1 expression. Nature 435:839–843. 10.1038/nature03677 [DOI] [PubMed] [Google Scholar]
- 62.Petrocca F, Visone R, Onelli MR, Shah MH, Nicoloso MS, de Martino I, Iliopoulos D, Pilozzi E, Liu CG, Negrini M, Cavazzini L, Volinia S, Alder H, Ruco LP, Baldassarre G, Croce CM, Vecchione A. 2008. E2F1-regulated microRNAs impair TGFbeta-dependent cell-cycle arrest and apoptosis in gastric cancer. Cancer Cell 13:272–286. 10.1016/j.ccr.2008.02.013 [DOI] [PubMed] [Google Scholar]
- 63.Suzuki HI, Yamagata K, Sugimoto K, Iwamoto T, Kato S, Miyazono K. 2009. Modulation of microRNA processing by p53. Nature 460:529–533. 10.1038/nature08199 [DOI] [PubMed] [Google Scholar]
- 64.Brabletz S, Brabletz T. 2010. The ZEB/miR-200 feedback loop—a motor of cellular plasticity in development and cancer? EMBO Rep. 11:670–677. 10.1038/embor.2010.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Park SM, Gaur AB, Lengyel E, Peter ME. 2008. The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev. 22:894–907. 10.1101/gad.1640608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhao H, Guo M, Zhao G, Ma Q, Ma B, Qiu X, Fan Q. 2012. miR-183 inhibits the metastasis of osteosarcoma via downregulation of the expression of Ezrin in F5M2 cells. Int. J. Mol. Med. 30:1013–1020. 10.3892/ijmm.2012.1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhu J, Feng Y, Ke Z, Yang Z, Zhou J, Huang X, Wang L. 2012. Down-regulation of miR-183 promotes migration and invasion of osteosarcoma by targeting Ezrin. Am. J. Pathol. 180:2440–2451. 10.1016/j.ajpath.2012.02.023 [DOI] [PubMed] [Google Scholar]
- 68.Liu YN, Yin JJ, Abou-Kheir W, Hynes PG, Casey OM, Fang L, Yi M, Stephens RM, Seng V, Sheppard-Tillman H, Martin P, Kelly K. 2012. MiR-1 and miR-200 inhibit EMT via Slug-dependent and tumorigenesis via Slug-independent mechanisms. Oncogene 32:296–306. 10.1038/onc.2012.58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Korpal M, Ell BJ, Buffa FM, Ibrahim T, Blanco MA, Celia-Terrassa T, Mercatali L, Khan Z, Goodarzi H, Hua Y, Wei Y, Hu G, Garcia BA, Ragoussis J, Amadori D, Harris AL, Kang Y. 2011. Direct targeting of Sec23a by miR-200s influences cancer cell secretome and promotes metastatic colonization. Nat. Med. 17:1101–1108. 10.1038/nm.2401 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.