To distribute the protein translation machinery throughout the cytoplasm, polysomes in the fungus Ustilago maydis associate with mobile early endosomes, resulting in long-range motility along microtubules.
Abstract
Early endosomes (EEs) mediate protein sorting, and their cytoskeleton-dependent motility supports long-distance signaling in neurons. Here, we report an unexpected role of EE motility in distributing the translation machinery in a fungal model system. We visualize ribosomal subunit proteins and show that the large subunits diffused slowly throughout the cytoplasm (Dc,60S = 0.311 µm2/s), whereas entire polysomes underwent long-range motility along microtubules. This movement was mediated by “hitchhiking” on kinesin-3 and dynein-driven EEs, where the polysomes appeared to translate EE-associated mRNA into proteins. Modeling indicates that this motor-driven transport is required for even cellular distribution of newly formed ribosomes. Indeed, impaired EE motility in motor mutants, or their inability to bind EEs in mutants lacking the RNA-binding protein Rrm4, reduced ribosome transport and induced ribosome aggregation near the nucleus. As a consequence, cell growth was severely restricted. Collectively, our results indicate that polysomes associate with moving EEs and that “off- and reloading” distributes the protein translation machinery.
Introduction
The mammalian nucleus releases ∼13,000 ribosomal subunits per minute (Kuersten et al., 2001). A subpopulation of these move to sites of local protein translation, either as part of messenger ribonuclear particles (mRNPs; Elvira et al., 2006) or via direct interaction with kinesin motors (Bisbal et al., 2009). However, the majority of ribosomes support cytoplasmic or ER-associated protein translation and thus have to be distributed within the cell. Ribosomal subunits are released from the nucleus by passive diffusion (Politz et al., 2003), which suggests that random motions could distribute them. However, ribosomal subunits are large protein–RNA complexes that are likely to diffuse slowly in the viscoelastic cytoplasm (Luby-Phelps, 2000). Motor-dependent transport processes potentially enhance cellular diffusion rates (Brangwynne et al., 2009); an example here is the transport of nanos mRNA in Drosophila melanogaster (Forrest and Gavis, 2003). However, whether active transport drives ribosome spreading is not known.
The endocytic system comprises several compartments that receive cargo from the plasma membrane for processing, recycling back to the surface or transport to lysosomes for degradation (Seaman, 2008). Key components in the endocytic pathway are early endosomes (EEs), which are characterized by the small GTPase Rab5 that controls biogenesis, membrane fusion, and microtubule (MT)-dependent motility of the EEs (Nielsen et al., 1999; Zerial and McBride, 2001; Zeigerer et al., 2012). Motility of EEs supports sorting, but also participates in long-distance signal transduction within the cell (for review see Miaczynska et al., 2004). In fungi, motile Rab5-positive structures have been described previously (Wedlich-Söldner et al., 2000; Fuchs et al., 2006; Abenza et al., 2009). These were considered putative EEs and were found to be essential for hyphal growth and membrane recycling (Wedlich-Söldner et al., 2000; Fuchs et al., 2006; Lenz et al., 2006). Rapid bidirectional movement of Rab5-positive endosomes is mediated by the molecular motors kinesin-3 and dynein (Wedlich-Söldner et al., 2002b; Lenz et al., 2006; Zhang et al., 2010; Egan et al., 2012b), which frequently turn the transport direction, thereby distributing the moving organelles throughout the hyphal cell (Schuster et al., 2011b). The function of the constant motility of these putative EEs is not understood, but it may mediate long-range signaling from the growing tip to the nucleus, located ∼50 µm behind (Steinberg, 2007). However, recent studies on RNA-binding proteins in Ustilago maydis suggested that the RNA-binding protein Rrm4 binds to the EEs (Baumann et al., 2012), which implies that their motility delivers associated mRNAs from the centrally located nucleus to the cell poles (Becht et al., 2005, 2006; König et al., 2009; Koepke et al., 2011; overview in Vollmeister et al., 2012). Indeed, some EEs travel from the nucleus to the hyphal tip, but the majority undergo much shorter motility and frequently switch direction (Schuster et al., 2011c). Similarly, the ubi1 and rho3 mRNAs undergo bidirectional and short-range movements (König et al., 2009). Such behavior challenges the concept of a role of EEs in long-distance delivery of mRNAs from the nucleus to the cell poles.
Here, we use the model fungus U. maydis to elucidate the mechanism by which ribosomes are transported and distributed in the cell. Surprisingly, we found that bidirectional EE motility randomly distributes entire polysomes. Ribosomes associate with moving EEs via the RNA-binding protein Rrm4, and both are frequently “off-loaded” and “reloaded” from moving EEs. Mutant studies show that motor activity is required to evenly distribute the polysomes and supports polar cell growth. Thus, constant EE motility distributes the translation machinery in the cell.
Results
Ribosomes are evenly distributed within the cell
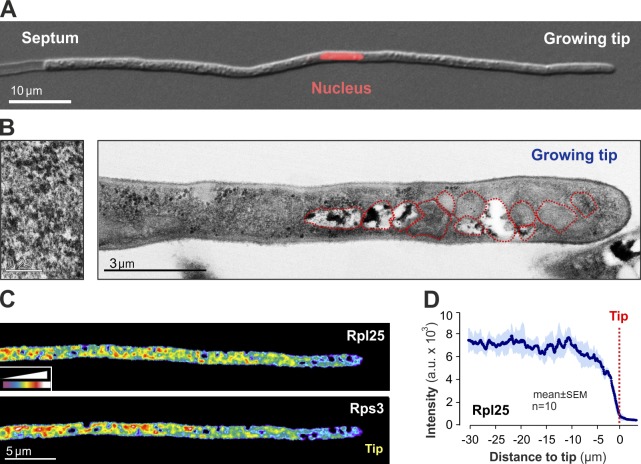
U. maydis hyphal cells are elongated, and their nucleus is positioned ∼50 µm behind the growing tip that produces ribosomal subunits (Fig. 1 A, nucleus labeled with a nucleus-targeted red fluorescent protein; Straube et al., 2005). In electron microscopy images, the apical region of the cell showed a higher concentration of organelles (Fig. 1 B), whereas the cytoplasm is filled with small granules that most likely represent ribosomes (Fig. 1 B, left). To visualize ribosomes in living cells, we identified U. maydis orthologues of the large and small ribosomal subunit proteins Rpl25 and Rps3, respectively (Fig. S1 A). We fused GFP to the end of the endogenous rpl25 gene and a triple red fluorescent mCherry tag to the endogenous rps3 gene (see Table 1 for genotypes of all strains and Table S1 for their usage in this study). This modification did not cause an altered growth phenotype, which suggests that the fusion proteins are biologically active. When coexpressed in the same cell, the large ribosomal subunit marker Rpl25-GFP and the small ribosomal subunit protein Rps3-mCherry3 colocalized with each other (100% colocalization, n = 114 signals from 10 cells; Fig. S1 B), which confirms that both proteins are incorporated into ribosomes. Consistent with the ultrastructural results, both ribosomal proteins were evenly distributed within the cell, but partially excluded from the organelle-rich apical cytoplasm (Fig. 1, C and D).
Figure 1.
Ribosome distribution in U. maydis. (A) Organization of a U. maydis hyphal cell. The cell expands at its apex (“Growing tip”), while vacuolated sections are separated by the septum. The nucleus is located near the center. (B) Electron micrograph of the apical region of a hypha. Left inset shows cytoplasmic ribosomes. Various organelles are encircled by red dotted lines. Bar on the left, 0.25 µm. (C) False-colored images of Rpl25-GFP– and Rps3-mCherry3–expressing hyphal cells. The fluorescent signal is evenly distributed over most parts of the cell, but reduced near the cell tip. The intensity color code is given in the lower left. Images were 2D deconvolved using the software MetaMorph and adjusted in brightness, contrast, and gamma settings. (D) Intensity profile of Rpl25-GFP along hyphal cells. Each data point represents the mean ± SEM (error bars); n = 10 cells from a single representative experiment. Ribosomes are evenly distributed along most parts of the cell, but partially excluded from the organelle rich tip.
Table 1.
Strains and plasmids used in this paper
| Strain or plasmid name | Genotype | Reference |
| AB33nRFP | a2 PnarbW2 PnarbE1, bleR/poNLS3RFP | Schuster et al., 2011b |
| AB33 | a2 PnarbW2 PnarbE1, bleR | Brachmann et al., 2001 |
| AB33R25G | a2 PnarbW2 PnarbE1, Prpl25-rpl25-egfp, bleR, hygR | This paper |
| AB33R3Ch3 | a2 PnarbW2 PnarbE1, Prps3-rps3-3xmcherry, bleR, hygR | This paper |
| AB33R25paG | a2 PnarbW2 PnarbE1, Prpl25-rpl25-pagfp, bleR, hygR | This paper |
| AB33paG2 | a2 PnarbW2 PnarbE1, bleR/popaG2 | This paper |
| AB33R3Ch3_R25G | a2 PnarbW2 PnarbE1, Prps3-rps3-3xmcherry, Prpl25-rpl25-egfp, bleR, hygR, natR | This paper |
| AB33ΔKin3_R25G | a2 PnarbW2 PnarbE1, Δkin3, Prpl25-rpl25-egfp, bleR, natR, hygR | This paper |
| AB5Dyn2ts_R25G | a1 PnarbW2 PnarbE1, Pdyn2-Dyn2ts, Prpl25-rpl25-egfp, bleR, hygR, natR | This paper |
| AB33ΔKin3_Kin3ts_R25G | a2 PnarbW2 PnarbE1, Δkin3, Prpl25-rpl25-egfp, bleR, natR, cbxR/pKin3ts | This paper |
| AB33R25G_ChRab5a_ΔRrm4 | a2 PnarbW2 PnarbE1, Prpl25-rpl25-egfp, Δrrm4, bleR, hygR, cbxR/pomChRab5a | This paper |
| AB33EG | a2 PnarbW2 PnarbE1, bleR/pERGFP | Wedlich-Söldner et al., 2002a |
| AB33ΔKin3_EG_Kin3ts | a2 PnarbW2 PnarbE1, Δkin3, bleR, natR/pERGFP/pKin3ts | This paper |
| AB5Dyn2ts_EG | a1 PnarbW2 PnarbE1, Pdyn2-Dyn2ts, bleR, hygR/pERGFP | This paper |
| AB33R25G_ChRab5a | a2 PnarbW2 PnarbE1, Prpl25-rpl25-egfp, bleR, hygR/pomChRab5a | This paper |
| AB33GRab5a | a2 PnarbW2 PnarbE1, bleR/poGRab5a | Schuster et al., 2011a |
| AB33ChRab5a_PXG | a2 PnarbW2 PnarbE1, bleR/pomChRab5a/poPXG | This paper |
| AB33ChRab5a_GRab4 | a2 PnarbW2 PnarbE1, bleR/pomChRab5a/poGRab4 | This paper |
| AB33ChRab5a_GRab7 | a2 PnarbW2 PnarbE1, bleR/pomChRab5a/poGRab7 | This paper |
| AB33ΔKin3 | a2 PnarbW2 PnarbE1, Δkin3, bleR, natR | This paper |
| FB2N107G | a2 b2 Pnup107-nup107-egfp, bleR | Steinberg et al., 2012 |
| AB33ChRab5a_G3Rho3 | a2 PnarbW2 PnarbE1, bleR/pomChRab5a/poG3Rho3 | This paper |
| AB33Rrm4G | a2 PnarbW2 PnarbE1, Prrm4-rrm4-egfp, bleR, hygR | This paper |
| AB33ChRab5a_Rrm4G | a2 PnarbW2 PnarbE1, Prrm4-rrm4-egfp, bleR, hygR/pomChRab5a | This paper |
| AB33R3Ch3_Rrm4G | a2 PnarbW2 PnarbE1, Prps3-rps3-3xmcherry, Prrm4-rrm4-egfp, bleR, natR, hygR | This paper |
| AB33ΔKin3_Kin3ts_GRab5a | a2 PnarbW2 PnarbE1, Δkin3, bleR, natR/pKin3ts/pCoGRab5a | This paper |
| AB5Dyn2ts_GRab5a | a1 PnarbW2 PnarbE1, Pdyn2-Dyn2ts, bleR, hygR/poGRab5a | Schuster et al., 2011c |
| AB33GRab7 | a2 PnarbW2 PnarbE1, bleR/poGRab7 | This paper |
| AB33Yup1ts_GRab5a | a2 PnarbW2 PnarbE1, Pyup1-yup1ts, bleR, hygR, cbxR/poGRab5a | This paper |
| AB33Yup1ts_R25G | a2 PnarbW2 PnarbE1, Pyup1-yup1ts, Prpl25-rpl25-egfp, bleR, hygR, cbxR | This paper |
| FB2N107R_cG | a2 b2 Pnup107-nup107-mrfp, bleR/pcrgG | This paper |
| AB33R3G | a2 PnarbW2 PnarbE1, Prps3-rps3-egfp, bleR, hygR | This paper |
| AB33G3Dyn2 | a2 PnarbW2 PnarbE1, Pdyn2-3×egfp-dyn2, bleR, hygR | Lenz et al., 2006 |
| AB33Kin3G | a2 PnarbW2 PnarbE1, Pkin3-kin3-egfp, bleR, hygR | Schuster et al., 2011c |
| AB33R25G_ChT | a2 PnarbW2 PnarbE1, Prpl25-rpl25-egfp, bleR, hygR/pNomChTub1 | This paper |
| AB33Kin3G_R3Ch3 | a2 PnarbW2 PnarbE1, Pkin3-kin3-egfp, Prps3-rps3-3xmcherry, bleR, hygR, cbxR | This paper |
| poNLS3RFP | Potef-gal4s-3xmrfp, natR | Schuster et al., 2011b |
| popaG2 | Potef-2×pagfp, cbxR | This paper |
| pKin3ts | Pkin3-kin3ts, hygR | Schuster et al., 2011c |
| pomChRab5a | Potef-mcherry-rab5a, natR | Schuster et al., 2011a |
| pERGFP | Potef-calS-egfp-HDEL, cbxR | Wedlich-Söldner et al., 2002a |
| poGRab5a | Potef-egfp-rab5a, natR | Schuster et al., 2011a |
| poPXG | Potef-PX-egfp, cbxR | This paper |
| poGRab4 | Potef-egfp-rab4, cbxR | This paper |
| poGRab7 | Potef-egfp-rab7, cbxR | This paper |
| poG3Rho3 | Potef-3×egfp-rho3, cbxR | This paper |
| pCoGRab5a | Potef-egfp-rab5a, cbxR | This paper |
| pcrgG | Pcrg-egfp, cbxR | This paper |
| pNomChTub1 | Potef-mcherry-tub1, natR | This paper |
a and b, mating type loci; P, promoter; −, fusion; Δ, deletion; hygR, hygromycin resistance; bleR, phleomycin resistance; natR, nourseothricin resistance; cbxR, carboxin resistance; ts, temperature-sensitive allele; NLS, nuclear localization signal of the GAL-4 DNA-binding domain from pC-ACT1 (Takara Bio Inc.); crg, conditional arabinose-induced promoter; otef, constitutive promoter; /, ectopically integrated; E1, W2, genes of the b mating type locus; egfp, enhanced GFP; pagfp, photoactivatable monomeric GFP; mcherry, monomeric cherry; mrfp, monomeric red fluorescent protein; rpl25, ribosomal large subunit protein; rps3, ribosomal small subunit protein; kin3, kinesin-3; dyn2, C-terminal half of the dynein heavy chain; rrm4, RNA-binding protein; rab4, rab5a, and rab7, small endosomal GTPases; rho3, small GTPase; yup1, endosomal t-SNARE; PX, Phox domain from Yup1 (aa 4–148); calS, signal sequence of calreticulin from rabbit (ntl-51); HDEL, ER retention signal; nup107, nucleoporin; tub1, α tubulin.
Diffusion and active transport distribute ribosomes
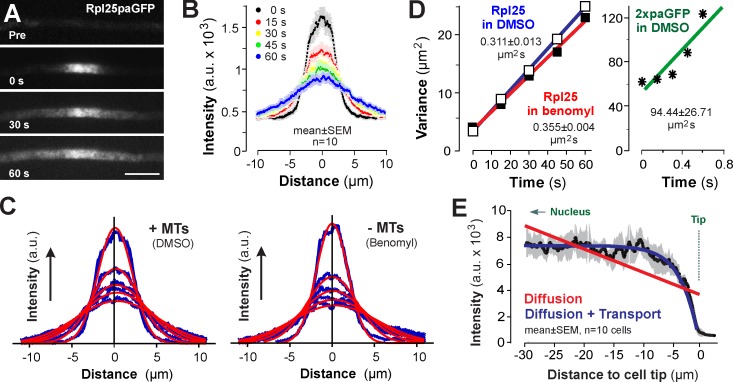
We asked whether thermal fluctuation is sufficient to distribute the Rpl25-GFP–labeled 60 S ribosomal subunit within the cell. To test the diffusion behavior of the ribosomal subunits, we fused photoactivatable GFP (paGFP; Patterson and Lippincott-Schwartz, 2002) to the endogenous rpl25 gene and observed the diffusion of native levels of Rpl25-paGFP after local photoactivation. We found that the fluorescent signal rapidly spreads out (Fig. 2, A and B). The resultant intensity profiles fitted Gaussian distribution curves (Fig. 2 C), from which we obtained variances for each curve. When these were plotted against time, we obtained a cytoplasmic diffusion coefficient of 0.311 µm2/s for Rpl25-paGFP (Fig. 2 D; Rpl25 in DMSO, blue fit; 95% CI = 0.2673–0.338). To test if this slow diffusion represents Rpl25-paGFP incorporated into large ribosomal subunits, we generated a double paGFP, which is of similar size and shape to Rpl25-paGFP (both globular and 54 kD and 43.4 kD, respectively). Activated 2×paGFP spread rapidly, with a diffusion coefficient of 94.44 ± 27.71 µm2/s (95% CI = 12.45–182.4; Fig. 2 D, green fit). Thus, the observed slow spreading of Rpl25-paGFP most likely represents diffusion of the large ribosomal subunit. We used the mean diffusion coefficient to estimate the time that a subunit would need to overcome the ∼50 µm between the nucleus and the hyphal tip and invoked formula t = L2/Dc (t = time in seconds, L = distance in micrometers; Popov and Poo, 1992) to estimate a diffusion time of ∼2 h. We next asked if MT-based motility of organelles can drag the cytoplasm along and increase diffusion rates (Brangwynne et al., 2009). We disrupted the MT cytoskeleton using the inhibitor benomyl, a nocodazole-like, fungal-specific benzimidazole-carbamate that binds to β-tubulin (Davidse and Flach, 1977; Jung et al., 1992), and tested if cytoplasmic diffusion of Rpl25-paGFP is reduced under these conditions. We found that the diffusion was slightly but significantly increased when MTs were absent (Fig. 2, C and D, red fit; P < 0.05; F-test, F = 18.54 with [Fn, Fd] = [1,6]). This demonstrates that streaming of cytoplasm does not foster the diffusion of ribosomal subunits.
Figure 2.
Diffusion of photoactivatable Rpl25-paGFP. (A) Image series showing diffusion of Rpl25-paGFP. The protein becomes visible after activation with a 405-nm laser pulse and spreads rapidly. Time in seconds is indicated. Bar, 5 µm. (B) Diffusion behavior of Rpl25-paGFP. After photoactivation at T = 0 s, the signal spreads with time. Each data point represents the mean ± SEM; n = 10 cells from a single representative experiment. (C) Gaussian fittings (red) to intensity curves (blue) of spreading Rpl25-paGFP signals in cells treated with the MT-disrupting drug benomyl (−MTs) or the solvent DMSO (+MTs). Note that all curves are normally distributed (Shapiro-Wilk test for all curves; P < 0.0001); their R2 values are 0.61–0.86. Intensity profiles are given as the mean of 10 cells. (D) Variances of the fitted Gaussian distributions of Rpl25-paGFP spreading with time. Rpl25 in DMSO: Diffusion of Rpl25-paGFP in the presence of DMSO. Rpl25 in benomyl: Diffusion of Rpl25-paGFP in benomyl-treated cells. 2×paGFP in DMSO: Diffusion of a double-paGFPs in DMSO. Diffusion coefficients are given. Note that the cytoplasmic 2×paGFP diffuses much more rapidly than the slightly smaller Rpl25-paGFP. (E) Intensity profiles predicted by a mathematical model. The red line assumes that only diffusion distributes Rpl25-GFP, whereas the total amount of Rpl25-GFP remains the same. The blue line assumes diffusion and active transport. The latter fits well to the experimentally obtained Rpl25-GFP distribution data (black; data taken from Fig. 1 D).
Next, we asked if diffusion alone is able to support the observed distribution of ribosomal subunits with Rpl25-GFP. To answer this we developed a mathematical model that allowed the inclusion of both diffusion and active transport and that aimed to describe the steady-state distribution shown in Fig. 1 D (see the first paragraph of the Results section). The model assumes that new Rpl25-GFP is constantly generated at the nucleus and that ribosome numbers are reduced at the hyphal tip. Ribosome decay during transport to the tip was excluded, as turnover times are known to be on the order of days (Hirsch and Hiatt, 1966). The model revealed that diffusion alone is not sufficient to distribute the ribosomal subunits. Instead, it predicts accumulation of Rpl25-GFP near the nucleus and a gradual decrease of signal intensity toward the hyphal tip (Fig. 2 E, red fit). However, after inclusion of active transport, the model gave a remarkably good fit for the intensity profile to the experimentally measured Rpl25-GFP distribution data (Fig. 2 E, blue fit; underlying experimental data are taken from Fig. 1 D). This suggests that active motor-dependent transport contributes to the diffusion of ribosomal subunits.
Kinesin-3 and dynein move ribosomes along microtubules
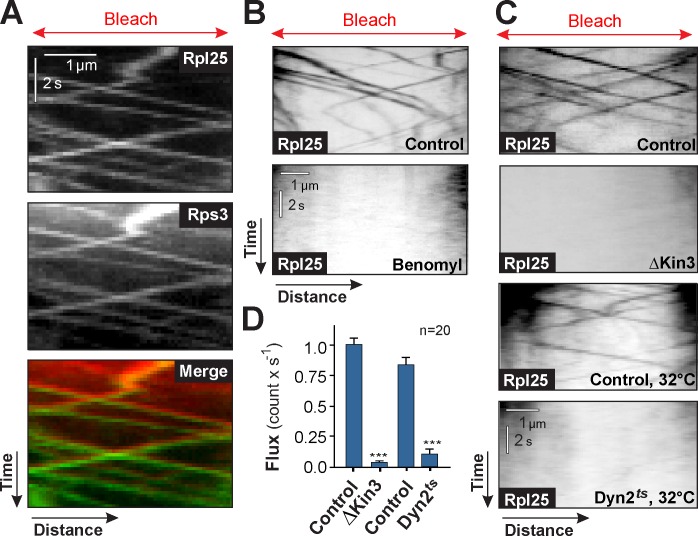
After photoactivation, Rpl25-paGFP showed occasionally directed motility (Fig. S1 C), which suggests that ribosomal subunits are actively transported in the hyphal cell. However, the paGFP signal was relatively weak and disappeared quickly. We therefore observed Rpl25-GFP and/or Rps3-mCherry3 in partially photobleached cells. This treatment did not harm the cells and allowed us to visualize motility of individual dynein motors (Schuster et al., 2011c). We found large and small ribosomal subunits comigrating in all regions of the cell (Fig. 3 A and Video 1). The motility occurred at a mean velocity of 2.01 ± 0.04 µm/s (n = 76) and at a similar frequency along the entire cell (Fig. S1 D). No difference in velocity or frequency was found between anterograde and retrograde motility (velocity, P = 0.32; frequency, P = 0.94; Mann-Whitney test). We next analyzed the mechanism underlying ribosome motility. The rapid and long-range movements suggested that ribosomes are transported along MTs. Indeed, the MT inhibitor benomyl abolished all ribosome motility (Fig. 3 B; motility in the presence of the solvent DMSO) and the large ribosomal subunit moved along mCherry-labeled MTs (Video 2). Collectively, these results suggest that MT-based motors support transport of ribosomal subunits. As motility of organelles in fungi depends largely on kinesin-3 and dynein (Wedlich-Söldner et al., 2002b; Egan et al., 2012a), we tested the involvement of these motors in ribosome motility by introducing the Rpl25-GFP construct into a kinesin-3 deletion mutant (ΔKin3; Schuster et al., 2011b) and a temperature-sensitive dynein mutant (Dyn2ts; Wedlich-Söldner et al., 2002a). We found that ribosome motility was almost abolished in both mutants (Fig. 3, C and D). In addition, the small ribosomal subunit Rps3-mCherry3 colocalized with Kin3-GFP (Video 3). These data strongly suggest that kinesin-3 and dynein transport ribosomes along MTs.
Figure 3.
Motor protein–dependent ribosome motility. (A) Co-motility of Rpl25-GFP (Rpl25; green in Merge) and Rps3-mCherry3 (Rps3; red in Merge). Both subunits travel together in a bidirectional fashion. Brightness, contrast, and gamma settings were adjusted. The photobleached area is indicated by “Bleach” and the red arrows. See Video 1. (B) Motility of Rpl25-GFP in the presence of the MT inhibitor benomyl and the solvent DMSO (Control). Motility is abolished in the absence of MTs. Cells were prebleached to reduce the background (“Bleach”). Images are contrast-inverted, and brightness, contrast, and gamma settings were adjusted. (C) Motility of Rpl25-GFP–labeled ribosomes in hyphal cells grown at 28°C (Control) and 32°C/2 h (Control, 32°C) in kinesin-3–null mutants (ΔKin3), and in temperature-sensitive dynein mutants at 32°C/2 h (Dyn2ts, 32°C). Images are contrast-inverted, and brightness, contrast, and gamma settings were adjusted. (D) Flux rates of Rpl25-GFP in control cells, kinesin-3–null mutants (ΔKin3), and temperature-sensitive dynein mutants (Dyn2ts) after 2 h a at restrictive temperature (32°C). Note that the bars show the combined flux in anterograde and retrograde direction. All bars are given as mean ± SEM (error bars) from a single representative experiment. Sample size is indicated. ***, significant difference to control at P < 0.0001 using a Student’s t test.
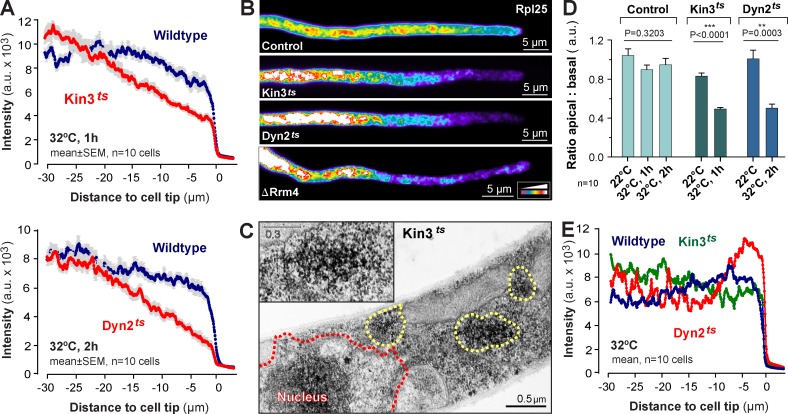
Our model suggests that diffusion and active motor-dependent transport cooperate to distribute ribosomes. We tested this experimentally by observing Rpl25-GFP distribution in the temperature-sensitive kinesin-3 (Kin3ts; Schuster et al., 2011b) and Dyn2ts mutants. At a permissive temperature (22°C), bidirectional EE motility occurred and Rpl25-GFP was normally distributed in both mutant strains (Fig. S2, A and B). At a restrictive temperature (32°C), EE motility was almost abolished and EEs clustered at the cell center (Fig. S2 B, Kin3ts, asterisks) or the tip of the cell (Fig. S2 B, Dyn2ts, asterisks), which indicates that the motors are inactivated under these conditions. This treatment resulted in an accumulation of Rpl25-GFP toward the cell center (Fig. 4, A and B); indeed, electron microscopy revealed that ribosomes formed aggregates close to the nucleus (Fig. 4 C and Fig. S2 C, Dyn2ts). Significantly less Rpl25-GFP signal was found in the apical part of the cells (Fig. 4, A, B, and D; signal intensity in apical region compared with subapical region, P < 0.0001 for Kin3ts and P = 0.0003 for Dyn2ts, Mann-Whitney test). We next tested if changes in the organization of the ER, visualized by the marker GFP-HDEL (Wedlich-Söldner et al., 2002a), account for the ribosome distribution defects in the motor mutants. Indeed, deletion or inactivation of motors slightly altered ER distribution (Fig. 4 E and Fig. S2 D), which suggests that kinesin-3 and dynein participate in ER motility in hyphal cells. However, these changes were minor and thus are not the cause of the altered ribosome distribution. We conclude that kinesin-3 and dynein-driven transport distributes ribosomal subunits within the cytoplasm.
Figure 4.
The role of kinesin-3, dynein, and Rrm4 in distributing ribosomes. (A) Fluorescence intensity profiles of Rpl25-GFP of hyphal wild-type cells (Wildtype) and temperature-sensitive kinesin-3 (Kin3ts) and dynein (Dyn2ts) mutants at 1 h or 2 h at 32°C. Each data point represents the mean ± SEM; n = 10 cells from a single representative experiment. (B) False-colored image of Rpl25-GFP in a control cell and temperature-sensitive kinesin-3 (Kin3ts) and dynein (Dyn2ts) mutants at 32°C, and a Δrrm4 mutant (ΔRrm4). Note that similar ribosome distribution defects are seen in all mutants. Images were 2D-deconvolved and brightness, contrast, and gamma settings were adjusted. The intensity color code is given in the bottom right. (C) Electron micrograph showing ribosomes in kinesin-3ts mutants after 1 h at 32°C. Ribosome clusters (yellow dotted line and inset) appear near the centrally located nucleus (red dotted line, Nucleus). (D) The ratio of Rpl25-GFP fluorescence at the tip (5–10 µm) to the basal region (25–30 µm). ** and ***, statistically significant difference at P = 0.0003 and P < 0.0001, respectively (Student’s t test). No difference was found in control cells at different temperatures (one-way ANOVA test, p-values are indicated). Bars are mean ± SEM (error bars); n = 10 cells from a single representative experiment. (E) Fluorescence intensity profiles of the ER marker GFP-HDEL in wild-type (Wildtype) and temperature-sensitive kinesin-3 (Kin3ts) and dynein (Dyn2ts) mutant cells at 32°C. Note that ER distribution is slightly altered in both mutants, which suggests that the motors participate in ER organization. However, no global reorganization of the network is seen (see also Fig. S2 D). Each data point represents the mean; n = 10 cells from a single representative experiment.
Ribosomes travel on bidirectionally moving EEs
It was shown previously that the RNA-binding protein Rrm4 associates with moving organelles (Baumann et al., 2012; Video 4), which carry the endosomal t-SNARE Yup1 (Wedlich-Söldner et al., 2000) and are transported by dynein and kinesin-3 (Wedlich-Söldner et al., 2002b; Lenz et al., 2006). We therefore speculated that ribosomes travel with Rrm4 in mRNPs. To test this we co-observed Rps3-mCherry3 and Rrm4-GFP and found that ribosomes do, indeed, comigrate with the RNA-binding protein (Fig. S3 A). Interestingly, Rpl25-GFP motility was abolished when rrm4 was deleted (Fig. S3 B), which resulted in subunit clusters near the central nucleus (Fig. S3 C) and partial depletion from apical cell parts (Fig. 4 C, Δrrm4). These results demonstrate a central role of the RNA-binding protein Rrm4 in the association of ribosomes to moving membranes and, consequently, in ribosome transport and distribution.
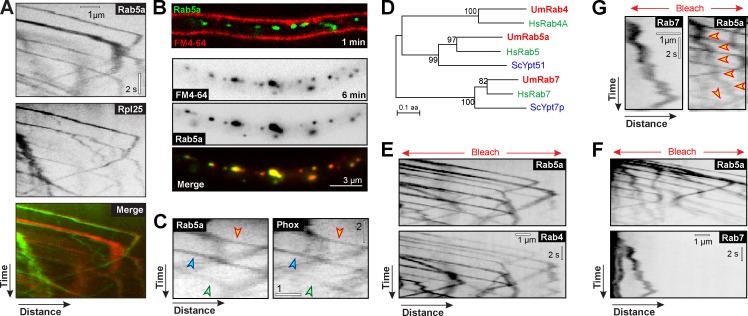
Rrm4 colocalizes with the t-SNARE Yup1 (Baumann et al., 2012), which is localized to membranes of the endosomal pathway (Wedlich-Söldner et al., 2000). To further investigate the nature of these putative endosomes, we coexpressed Rpl25-GFP and the fluorescent EE marker mCherry-Rab5a (Rab5a; Fuchs et al., 2006). Rab5-GTPases specifically bind to EEs (Chavrier et al., 1990). We found that 98% of the large ribosomal subunit protein colocalized with rapidly moving Rab5a signals (Fig. 5 A and Video 5). To gain support for the notion that Rab5a-positive structures are EEs, we tracked the fluorescent endocytic marker FM4-64, which is taken up by endocytosis at the plasma membrane and is delivered through the endocytic pathway to the vacuole/lysosome (Vida and Emr, 1995). 1 min after application, the dye appeared in the plasma membrane, and it reached the Rab5a-positive structures within 6 min (Fig. 5 B and Fig. S3 D). This result confirms that the Rab5a-positive organelles are an early endocytic compartment. EE membranes are enriched in phosphatidylinositol 3-phosphate (PtdIns(3)P; Gillooly et al., 2000), to which PX domains bind (for review see Lemmon, 2003). We fused GFP to the PX domain of Yup1 (Wedlich-Söldner et al., 2000) and found that the fusion protein comigrated with mCherry-Rab5a in 93.4 ± 1.6% (n = 446 signals in 40 cells; Fig. 5 C). This indicates an enrichment of PtdIns(3)P in the Rab5a-positive structures and supports the hypothesis that they are indeed EEs. Finally, we colocalized mCherry-Rab5a and GFP-labeled homologues of the GTPase Rab4, which is known to localize on EEs (Van Der Sluijs et al., 1991), and Rab7, a marker of late endosomes (Chavrier et al., 1990; Fig. 5 D). Rab7-positive structures were labeled with FM4-64 at 15–20 min after dye application (Fig. S3 D). This confirmed that Rab7 labels late endosomes in U. maydis. Consistent with the notion that Rab5a-positive organelles are EEs, we found that Rab4 colocalizes with Rab5a on motile structures (Fig. 5 E), whereas Rab7 was concentrated on largely immobile organelles that only rarely carried Rab5a (Fig. 5, F and G). Collectively, these results strongly indicate that ribosomes travel on rapidly moving EEs.
Figure 5.
Characterization of Rab5a-positive structures. (A) Co-motility of Rpl25-GFP (Rpl25; green in merged image) and the endosomal GTPase mCherry-Rab5a (Rab5a; red in merged image). Ribosomes localize on the bidirectionally moving Rab5a-positive structures. Brightness, contrast, and gamma settings were adjusted. See Video 5. (B) Localization of the endocytic marker FM4-64 at 1 min and 6 min after a pulse and subsequent wash-out. The dye first appears in the plasma membrane (1 min) and concentrates in GFP-Rab5a structures (green) at 6 min. Images are contrast-inverted, and brightness, contrast, and gamma settings were adjusted. (C) Co-motility of Rab5a and a fusion protein of GFP and the PX domain of Yup1 (aa 4–148). Images were contrast-inverted and adjusted with brightness, contrast, and gamma settings. Arrowheads indicate three sets of trajectories. Bars, 2 s and 1 µm. (D) Nearest neighbor tree of Rab4-, Rab5-, and Rab7-GTPases from U. maydis (red) human (green), and budding yeast (blue). Note the absence of Rab4 in yeast. (E) Co-motility of GFP-Rab4 and mCherry-Rab5a. Images were contrast-inverted, and brightness, contrast, and gamma settings were adjusted. (F and G) Colocalization of GFP-Rab7 and mCherry-Rab5a. The late endosome marker Rab7 does not localize to the rapidly moving Rab5a structures. Occasionally, largely immobile Rab7-positive late endosomes carry Rab5a (arrowheads in G). Images were contrast-inverted, and brightness, contrast, and gamma settings were adjusted.
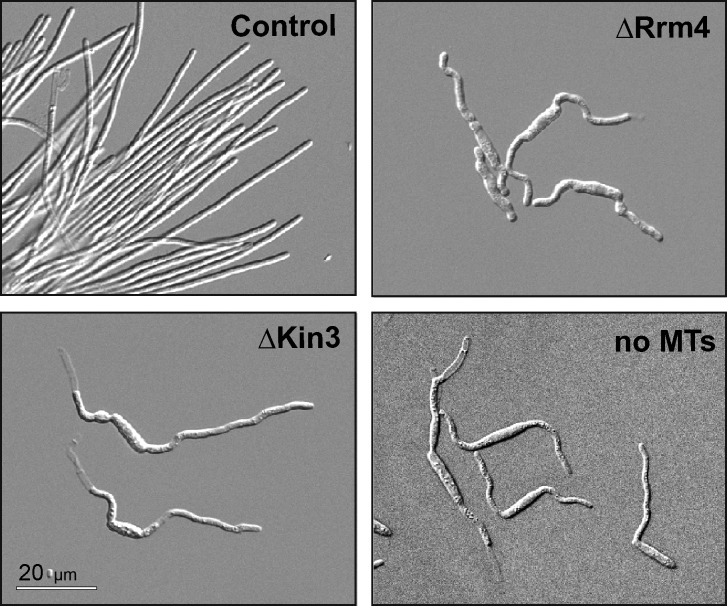
The association of ribosomal subunits with EEs suggests that the observed ribosome distribution defects in kinesin-3 and dynein mutants are caused by defects in EE motility. To test this, we expressed Rpl25-GFP in temperature-sensitive mutants of the endosomal t-SNARE Yup1, where EE numbers decline at restrictive conditions due to a defect in fusion with incoming transport vesicles (Wedlich-Söldner et al., 2000). Indeed, motility of Rpl25-GFP was impaired when Yup1ts mutants were grown at restrictive temperature for 5 h (Fig. S4, A and B). Consequently, the distribution of Rpl25-GFP was affected (Fig. S4, C and D). This distribution defect was not as pronounced as that of the motor mutants, probably because of the higher background motility (∼50% of control when faint signals are included), but this result further supports the idea that EE motility distributes ribosomes in the cell. Finally, we noted that rrm4 and kinesin-3 null mutants share a common morphological phenotype, characterized by short and frequently bipolar hyphae (Fig. 6; Becht et al., 2006; Lenz et al., 2006). A similar phenotype was found when MTs were disrupted by benomyl (Fig. 6, no MTs). Thus, we conclude that EE-based ribosome transport along MTs is most likely essential for hyphal growth.
Figure 6.
Morphology of wild-type (Control), rrm4, and kinesin-3 null mutants and cells lacking microtubules. Although the phenotypes may reflect a broad range of defective pathways, their similarity suggests that they are all related to impaired ribosome transport.
Ribosomes are translationally active on moving EEs
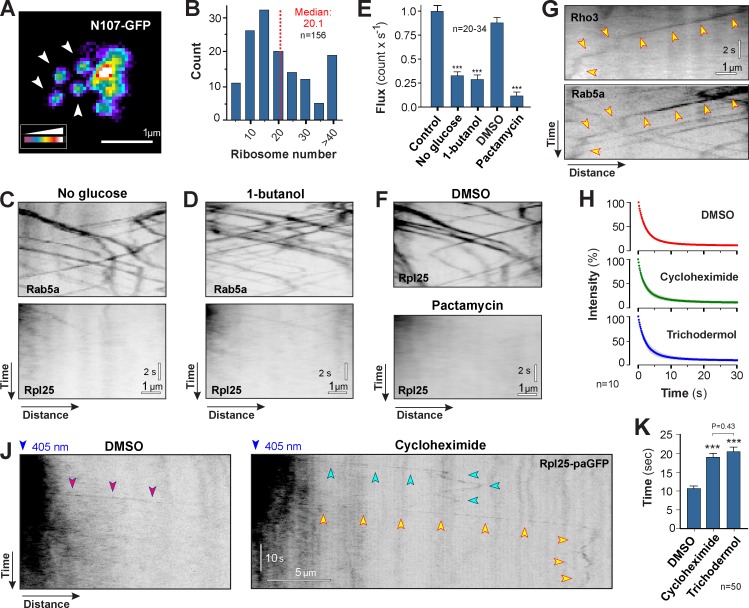
So far, our data show that EE motility distributes ribosomal subunits. Next, we asked how the subunits associate and dissociate with the moving organelles. We noticed that the moving Rpl25-GFP signals varied in fluorescent signal intensity (Video 6), which suggested that variable numbers of ribosomes travel on the EEs. To confirm this, we estimated the number of ribosomes by comparing the intensity of each signal with a fluorescent nucleoporin Nup107-GFP, a method used previously to estimate motor numbers on moving organelles (Schuster et al., 2011a,b). Nup107-GFP fusion proteins are incorporated into single nuclear pores of uniform fluorescent signal intensity (Fig. 7 A, arrows). We determined the intensity of these signals and obtained the mean fluorescent intensity of single pores, which corresponds to 16 GFP molecules (Rabut et al., 2004). When compared with moving Rpl25-GFP signals, we estimated ∼20 ribosomes binding to individual EEs (Fig. 7 B; the median is given, as the dataset was not normally distributed; Shapiro-Wilk test, P < 0.0001; range is 5.65–64.12 ribosomes).
Figure 7.
Association of translationally active polysomes to early endosomes. (A) False-color image of fluorescent nuclear pores in U. maydis. The endogenous copy of Nup107 was fused to GFP (Nup107-GFP). Each nuclear pore contains 16 Nup107-GFP and shows homogenous signal intensity (arrowheads). Images were 2D-deconvolved and false-colored, and brightness, contrast, and gamma settings were adjusted. (B) Number of ribosomes within motile Rpl25-GFP signals. Numbers were estimated using Nup107-GFP as an internal calibration standard. Data are non-normally distributed (Shapiro-Wilk test, P < 0.0001), and median and sample size from a single representative experiment are indicated. (C) Motility of ribosomes (Rpl25-GFP) under glucose depletion stress (no glucose for 10 min), which largely abolished polysome formation on motile EEs (Rab5a). Images are contrast inverted and brightness, contrast, and gamma settings were adjusted. See Video 7. (D) Motility of ribosomes (Rpl25-GFP) under 1-butanol–induced stress (1% [vol/vol] 1-butanol for 10 min). This treatment largely abolished polysome formation on motile EEs (Rab5a). Images are contrast inverted, and brightness, contrast, and gamma settings were adjusted. See Video 7. (E) Frequency of ribosome motility under glucose depletion (No glucose), 1-butanol–induced stress (1-butanol), and block of translation initiation (pactamycin). All bars are given as mean ± SEM (error bars); the sample size for each single representative experiment was between 20 and 34 cells. ***, statistical significance at P < 0.0001 using a Student’s t test. (F) Motility of ribosomes (Rpl25-GFP) in the presence of the solvent DMSO and the translation initiation inhibitor pactamycin. Note that verrucarin A treatment showed the same effect (see Fig. S5, E and F). Images are contrast-inverted and adjusted with brightness, contrast, and gamma settings. See Video 7. (G) Colocalization of nascent GFP3-Rho3 and mCherry-Rab5a–labeled EEs. Note that rho3 mRNA is transported to the septum, where the majority of the Rho3 protein is localized to function (König et al., 2009). Images are contrast inverted and brightness, contrast, and gamma settings were adjusted. Arrowheads indicate one trajectory. (H) Bleaching curves of Rpl25-paGFP in the presence of the solvent DMSO and the translation elongation inhibitors cycloheximide and trichodermol. Each data point represents the mean ± SD (error bars) average signal intensity; n = 10 cells from a single representative experiment. (J) Photoactivated Rpl25-paGFP in DMSO (purple arrowheads) and the inhibitor cycloheximide (green and yellow arrowheads). Note that signals disappear faster in DMSO. Images are contrast inverted and brightness, contrast, and gamma settings were adjusted. (K) Residence time of Rpl25-paGFP on EEs. Bars are given as mean ± SEM (error bars); n = 50 cells from 2–6 experiments. ***, statistical significance at P < 0.0001. The p-value for a nonsignificant pair is given (Mann-Whitney test).
EE association of these ribosomes was dependent on Rrm4 (see Fig. S3 B), which suggests that they could form translationally active polysomes on EE-associated mRNA. To test this further, we exposed cells to mild stress conditions, which have been shown to inhibit protein translation due to polysome disassembly (Ashe et al., 2000, 2001). Indeed, we found that glucose starvation for 10 min and treatment with 1% (vol/vol) 1-butanol reduced Rpl25-GFP association with EEs (Fig. 7, C and D; Rpl25) and significantly reduced ribosome motility (Fig. 7 E), whereas motility of Rab5a-labeled EEs was not affected (Fig. 7, C and D, Rab5a; and Video 7). We further investigated whether EE-associated ribosomes are translationally active by interfering with early steps in protein translation. We chose the inhibitors verrucarin A and pactamycin, which specifically inhibit translation initiation (Macdonald and Goldberg, 1970; Cundliffe et al., 1974). Both prevented the formation of newly formed cytoplasmic GFP when GFP was expressed under an induced promoter (for details see Materials and methods; Fig. S5 A), demonstrating that they inhibit protein synthesis in U. maydis. In contrast, short-term inhibitor treatment (10 min of pactamycin and 1 h of verrucarin A) did not affect the cellular level of Rpl25-GFP or Rps3-GFP, due to the preexisting protein (Fig. S5 B), nor did it inhibit the motility of motors, Rrm4, or EEs (Fig. S5, C and D). Nevertheless, both inhibitors almost abolished Rpl25-GFP motility, which suggests that translation initiation is required to associate ribosomes with EEs (Fig. 7, E and F; Fig. S5, E and F; and Video 7).
Inhibition of polysome formation and translation initiation prevented the association of ribosomes with moving EEs, which suggests that translationally active polysomes “travel” on the organelles. Thus, we considered it possible that polypeptides are formed during mRNA transit to the cell poles. The mRNA of the small GTPase Rho3 is delivered in Rrm4-containing particles to the septum, which suggests local translation (König et al., 2009). We fused triple GFP to the Rho3 N terminus and visualized the nascent GFP3-Rho3 on moving EEs. Indeed, we found faint GFP signals comigrating with 2.76% of the mCherry-Rab5a signals (n = 398 in 40 cells; Fig. 7 G and Fig. S5 G). This result supports the notion that EE-associated polysomes are translationally active. Finally, we set out to investigate the turnover of EE-associated ribosomes. We made use of Rpl25-paGFP, which, after photoactivation, travels on EEs into the nonactivated parts of the cell (see Fig. S1 C). We realized that signals rapidly disappeared, which was likely due to photobleaching of the paGFP. However, if polysomes are translationally active, ribosome turnover due to translation termination would replace fluorescent ribosomal subunits by nonfluorescent subunits. This could enhance fading of the fluorescent signals on EEs. To test this, we observed Rpl25-paGFP motility in the presence of translation elongation inhibitors cycloheximide (Schneider-Poetsch et al., 2010) and trichodermol (Carrasco et al., 1973), which “freeze” ribosomes on the transcript (Cundliffe et al., 1974; Liao et al., 1976). The presence of these inhibitors had no effect on the photobleaching behavior of Rpl25-paGFP (Fig. 7 H). However, both inhibitors significantly increased the time that Rpl25-paGFP signals traveled on EEs (Fig. 7, J and K). As bleaching was the same under all conditions, the change in Rpl25-paGFP signal duration is most likely caused by reduced translation termination, and, consequently, impaired subunit turnover. Collectively, these results indicate that EE-associated ribosomes form active polysomes on Rrm4-anchored mRNA.
Moving EEs off-load polysomes
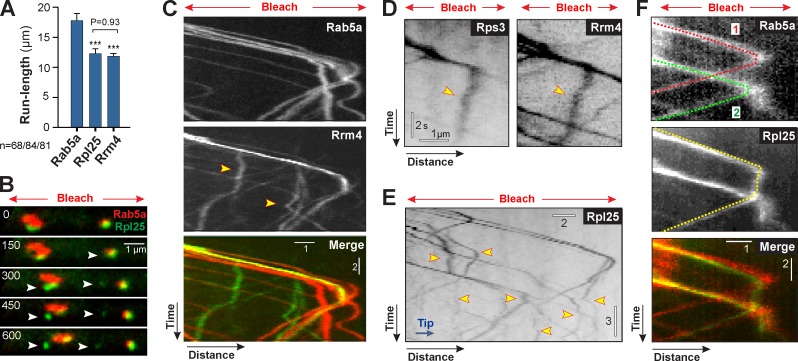
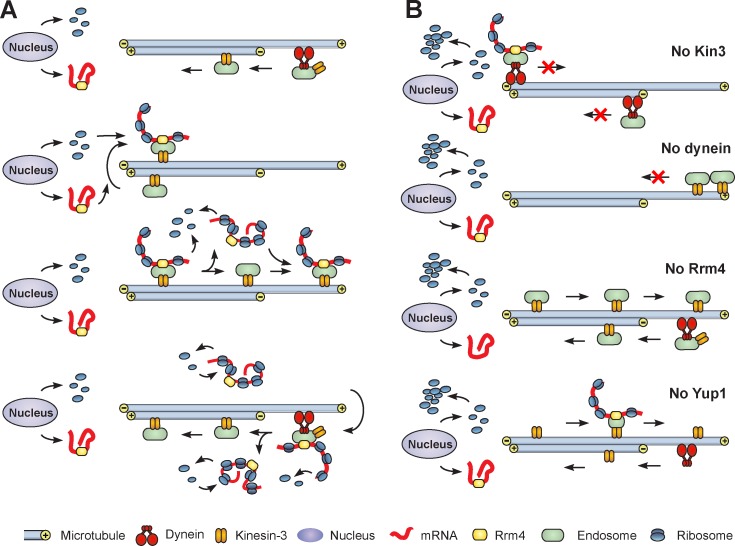
Finally we asked how EE motility ensures an even distribution of ribosomes. We compared the anterograde run length of EEs, starting at the nucleus, with that of polysomes, labeled by Rrm4-GFP and Rpl25-GFP. We found that ribosomes and Rrm4 moved processively to the hyphal tip, with a mean run length of ∼12 µm (Fig. 8 A; Rpl25-GFP: 12.33 ± 0.77 µm, n = 84; Rrm4-GFP: 11.79 ± 0.62 µm, n = 81; not significantly different at P = 0.9299, Mann-Whitney test). In contrast, EEs moved significantly longer distances (Fig. 8 A; Rab5a, 17.77 ± 1.19 µm, n = 68; Rpl25, P = 0.0028; Rrm4, P < 0.0001; Mann-Whitney test), with many runs extending from the nucleus to the hyphal tip. These results indicate that polysomes “fall off” the anterograde moving EEs before they reach the hyphal tip. We visualized this off-loading in cells that expressed Rpl25-GFP and mCherry-Rab5a. We found that Rpl25-GFP signals of various intensity comigrate with EEs into photobleached regions (Fig. 8 B). Occasionally, Rpl25-GFP signals were released from moving EEs (Fig. 8 B, arrowheads; and Video 8). These signals varied in their fluorescent intensity, which suggests that entire polysomes of different sizes are released during EE movement. We tested this possibility further by co-observing the endosomal mCherry-Rab5a and the RNA-binding protein Rrm4-GFP. Again, Rrm4-GFP signals frequently separated from moving EEs (Fig. 8 C, arrowheads). Co-observation of Rrm4-GFP and Rps3-mCherry3 confirmed that ribosomes and the RNA-binding-protein are off-loaded together (seen in 90% of all cases; n = 50; Fig. 8 D). This indicates that entire polysomes dissociate from moving EEs. This off-loading was a frequent process, and polysomes remained relatively stationary after deposition in the cytoplasm (Fig. 8 E, arrowheads). Occasionally, we observed polysomes reattaching to moving EEs (Fig. 8 F). Collectively, these data suggest that repeated and transient interaction with EEs constantly repositions polysomes (Fig. 9 A), thereby ensuring an even distribution of the translation machinery in the cell.
Figure 8.
Off-loading of polysomes from moving early endosomes. (A) Mean run-length of EEs (Rab5a), the large ribosomal subunit (Rpl25), and the RNA-binding protein Rrm4. Bars are given as mean ± SEM (error bars); sample size n is 68, 84, and 81, respectively, from 2–4 experiments. ***, statistical significance at P ≤ 0.0004 (Mann-Whitney test). Run length of Rrm4 and Rpl25 was not different; P = 0.90991 (Mann-Whitney test). (B) Image series showing off-loading of ribosomes (arrowheads), labeled with Rpl25-GFP (green), from moving EEs, labeled with mCherry-Rab5a (red). The photobleached area is indicated by “Bleach” and the red arrows. See also Video 8. Brightness, contrast, and gamma settings were adjusted. Time is indicated in milliseconds. (C) Off-loading of Rrm4-GFP (arrowheads; green in Merge) from moving EEs (Rab5a; red in Merge). Brightness, contrast, and gamma settings were adjusted. The photobleached area is indicated by “Bleach” and the red arrows. Bars, 2 s and 1 µm. (D) Off-loading of Rrm4-GFP (Rrm4) and Rps3-mCherry3 (Rps3). Both markers are deposited together (arrowheads), which indicates that entire polysomes are released from the EEs. Images are contrast inverted, and brightness, contrast, and gamma settings were adjusted. The photobleached area is indicated by “Bleach” and the red arrows. (E) Anterograde motility of Rpl25-GFP subunits in a photobleached region (red arrows). Ribosome subunits are off-loaded before they reach the hyphal tip (yellow arrowheads). Images are contrast inverted, and brightness, contrast, and gamma settings were adjusted. The photobleached area is indicated by “Bleach” and the red arrows. Bars, 3 s and 2 µm. (F) “Off- and re-loading” of Rpl25-GFP onto EEs. The ribosome is indicated by a yellow dotted line (Rpl25); the two EEs involved in transport are indicated by a red (1, Rab5a) and green dotted line (2, Rab5a). The photobleached area is indicated by “Bleach” and the red arrows. Bars, 2 s and 1 µm.
Figure 9.
Model showing the role of EE motility in polysome distribution. (A) Ribosomal subunits are released from the nucleus, form polysomes on Rrm4-bound mRNA, and get loaded onto EEs that travel through the ribosome subunit–rich region by the activity of kinesin-3 and dynein. Off- and reloading of polysomes from the moving EEs distributes the entire translation machinery in the cell. (B) Ribosome distribution defects in the mutants used in this study. Absence of bidirectional EE motility (No kin3, No dynein), the inability to bind to EEs (No Rrm4), or reduced EE numbers (No Yup1) results in formation of central ribosome subunit clusters.
Discussion
EEs motility is a general feature of the eukaryotic cell and is thought to support the sorting of endocytosed cargo, thereby controlling numerous cellular processes (Lemmon and Traub, 2000; Seaman, 2008). The discovery of additional roles in signaling over long distances (Sorkin and Von Zastrow, 2002; Howe and Mobley, 2004; Miaczynska et al., 2004), cytokinesis (Carlton and Martin-Serrano, 2007; Schiel et al., 2013), and cell polarity and migration (Emery and Knoblich, 2006) suggests that motile EEs serve as a platform for the assembly and function of key molecular machines (Gould and Lippincott-Schwartz, 2009; Pálfy et al., 2012). In this study we provide evidence for a new and unexpected role of EEs that is in their fostering of the distribution of polysomes.
Ribosomes “hitchhike” on moving EEs
The hyphal cells of filamentous fungi share organizational features with neurons (Steinberg and Perez-Martin, 2008). In particular, in the fungus U. maydis, the motors kinesin-3 and dynein move Rab5a-carrying organelles in a bidirectional fashion along a bipolar MT array (Lenz et al., 2006; Schuster et al., 2011c). Several arguments support the notion that these Rab5-positive structures are EEs: (1) In animal cells, Rab4- and Rab5-like GTPases bind to EEs, whereas Rab7 localizes to late endosomes (Chavrier et al., 1990; Van Der Sluijs et al., 1991). We find Rab5a and Rab4, but not Rab7, on the moving organelles in U. maydis; (2) shortly after internalization, the endocytic marker FM4-64 appears in Rab5a-positive organelles, which suggests that these organelles are the first endocytic compartments; and (3) EEs are enriched in the lipid PtdIns(3)P (Gillooly et al., 2000), which binds PX domains (Lemmon, 2003). The Rab5a-carrying organelles bind the Yup1 PX domain, which suggests that they are rich in PtdIns(3)P. Collectively, these results support the notion that the Rab5a-carrying organelles are EEs. Previous work demonstrates that the tSNARE Yup1 binds to these EEs (Fuchs et al., 2006) and that RNA-binding protein Rrm4 (Becht et al., 2005; Becht et al., 2006) anchors mRNA to these organelles (König et al., 2009). Here, we report that Rrm4 anchors polysomes to the EEs, indicating that entire mRNPs “travel” along MTs in U. maydis. Animal mRNPs are also transported along MTs (Ainger et al., 1993; Knowles et al., 1996). However, while animal mRNPs directly bind to MT motors for transport along the cytoskeleton (Kanai et al., 2004; Dienstbier et al., 2009), the fungal Rrm4-containing mRNPs associate with EEs and use the endosome transport machinery to hitchhike through the cell.
How widespread this mechanism of mRNP transport by EEs is remains unknown. However, the RNA-binding protein Rrm4 is pivotal to this process, as it anchors mRNA and attaches polysomes to EEs. Rrm4 was initially identified as an RNA recognition motif 1–containing protein, which shows similarity to human and plant RNA-binding proteins (Becht et al., 2005). Subsequently, Rrm4 was reported to interact with >50 mRNAs, including that of the small G protein Rho3, which is delivered to the distal cell pole for possible local translation (König et al., 2009). Thus, if the process of EE-associated mRNP movement is conserved, Rrm4 homologues should be found in other systems. However, the closest Rrm4 homologues in human, worms, and fruit flies have much higher similarity with another putative RNA-binding protein in U. maydis (31.8% identical amino acids; accession no. XP_759641.1). Furthermore, database searches with the C-terminal part of Rrm4 (293 aa), which, on its own, travels on MTs (König et al., 2009) and thus contains the EE-binding region, revealed that Rrm4 homologues are restricted to the basidiomycete fungi (e.g., XP_003027868 in Schizophyllum commune). Thus, we consider it unlikely that Rrm4-mediated mRNP movement on EEs is widely conserved. However, RNA-binding proteins are generally known to anchor mRNAs to membranes (for review see Cohen, 2005), and unknown factors could link mRNPs to moving EEs or other membranes in higher eukaryotes. Indeed, the principle link of mRNA transport to membrane trafficking may be more general, as Saccharomyces cerevisiae and Xenopus laevis mRNAs “piggyback” on motile ER (Cohen, 2005; Schmid et al., 2006; Paquin and Chartrand, 2008).
EE-associated ribosomes are most likely translationally active
We suggest here that the mechanism by which ribosomes associate with EEs involves active translation of Rrm4-anchored mRNA. Several lines of evidence support this: (1) ∼20 ribosomes move in a single Rrm4-containing mRNP, and their association requires the RNA-binding protein Rrm4, suggesting that ribosomes form polysomes on Rrm4-bound mRNA; (2) association of ribosomes with EEs requires the initiation of translation, again suggesting protein synthesis on EEs; (3) nascent GFP3-Rho3 protein co-migrating with EEs during delivery of rho3 mRNA to the septum, where the GTPase is thought to function; and (4) turnover of ribosomal subunits on moving EEs is decreased when translation elongation is inhibited and polysomes are “frozen.” Whether EE-associated translation distributes polypeptides is currently not clear. We consider it likely that translation initiation might be a simple and efficient way of assembling ribosomes on Rrm4-anchored mRNA for long-distance transport on moving EEs.
Bidirectional EE motility supports ribosome and mRNA diffusion
The association of mRNPs with moving EEs has been suggested to deliver mRNAs to the cell poles for local translation (Zarnack and Feldbrügge, 2010). However, fungal EEs and associated polysomes move bidirectionally (Wedlich-Söldner et al., 2000; Schuster et al., 2011b; this study), and we found no net flux of mRNPs toward the cell poles. Furthermore, we show that most polysomes do not reach the growth region of the cell tip, but fall off the moving EEs before they reach the cell pole. This behavior is reminiscent of the bidirectional motility of ubi1 and rho3 mRNAs, which on average travel only 5 µm in one direction (König et al., 2009), whereas EEs have a mean run length of 18 µm (Schuster et al., 2011c; this study). The limited run length and the bidirectional motility of the mRNPs challenge a role of the EE-based motility in delivering mRNAs to the cell ends for local translation. Instead, our results suggest that EE motility has a much more fundamental role in distributing polysomes throughout the cell. This mechanism is based on three assumptions and findings: (1) Newly synthesized ribosomes are constantly released from the nucleus; (2) cellular diffusion is not sufficient to distribute the subunits efficiently within the elongated cell; and (3) ribosomes associate and dissociate transiently from moving EEs that pass the nucleus, thereby picking up the newly formed subunits (Fig. 9 A). The phenotype of the tested motor, yup1ts, and Δrrm4 mutants support such a model. Inactivation of dynein or kinesin-3 causes an imbalance in EE motility and ultimately traps the EEs at the hyphal tip or the cell center, respectively (Lenz et al., 2006; this study). Consequently, EEs do not pass through the ribosome-rich nuclear area and do not “pick up” ribosomes for long-distance transport (Fig. 9 B, No Kin3, No Dynein). This leads to ribosome depletion near the cell poles, whereas the constant production of ribosomal subunits in the nucleus results in central ribosome clusters. A similar phenotype is seen in Δrrm4 mutants, where EEs still move, but have lost the ability to capture ribosomes (Fig. 9 B, no Rrm4). In yup1ts mutants, the number of EEs is reduced due to a defect in membrane fusion. Consequently, fewer moving EEs are present to distribute ribosomes, again resulting in ribosome accumulation around the nucleus and reduced abundance near the cell poles (Fig. 9 B, no Yup1). We conclude that transient association and release of polysomes from kinesin-3 and dynein-driven EEs is of central importance for the constant reorganization and cellular mixing of the translation machinery within the viscous cytoplasm, and defects in this process eventually result in aberrant cell growth of motor and Δrrm4 mutants. This phenotype may be consequent upon numerous cellular defects, making it difficult to draw specific functional conclusions. However, it does demonstrate the importance of EE-associated polysome transport.
Conclusion
Increasing evidence suggests that EEs are “multipurpose platforms” that participate in numerous signaling pathways (Gould and Lippincott-Schwartz, 2009). The MT-based motility of EEs enables rapid delivery signaling complexes into specific cellular locations, which is of particular importance in elongated cells, where passive diffusion is not sufficient to overcome large distances (Miaczynska et al., 2004). Here, we provide evidence for a novel role of EE motility in delivering the translation machinery. The core components of this, the ribosomal subunits, are constantly formed at the nucleus and need to be distributed within the cell. The large size of these protein–RNA complexes and the structured viscoelastic nature of the cytoplasm make passive diffusion unsuitable for transport of ribosomes to the distant regions of the cell. In U. maydis, this limitation is overcome by transient association of entire polysomes to rapidly moving EEs. This active long-range delivery of ribosomes, mRNA-binding proteins, and mRNAs complements short-range passive diffusion. Thus, MT-based EE motility supports Brownian motion in a process named “active cytoplasmic diffusion,” which was suggested to increase motility of molecules and vesicles in cells (Brangwynne et al., 2009).
Materials and methods
Strains and plasmids
The U. maydis strains (AB33nRFP, AB33, AB33EG, AB33GRab5a, FB2N107G, AB5Dyn2ts_GRab5a, AB33G3Dyn2, and AB33Kin3G) were described previously (Brachmann et al., 2001; Wedlich-Söldner et al., 2002a; Lenz et al., 2006; Schuster et al., 2011a,b,c; Steinberg et al., 2012). The genotype and the experimental usage of all strains and plasmids in this study are summarized in Table 1 and Table S1, respectively. The following plasmids were generated through the standard cloning method or in vivo recombination in the S. cerevisiae strain DS94.
pHrpl25G.
To obtain an S. cerevisiae– Escherichia coli shuttle vector, a 2,680-bp fragment containing the S. cerevisiae URA3 marker, the 2 µm ori, the ampicillin resistance cassette and an E. coli origin of replication, and a 3,823-bp fragment containing egfp and the hygromycin phosphotransferase gene resistance cassette (hygR) were obtained from pKin3G_H (Schuster et al., 2011c) digested with BamHI and SacI. An 864-bp fragment, including the rpl25 ORF but excluding the stop codon, and 1,000 bp downstream of rpl25 ORF were amplified by PCR with 30-bp homology to the upstream and downstream of the sequence stretch of vectors.
pCrpl25G and pNrpl25G.
To replace hygR with carboxin (cbxR) or nourseothricin resistance cassette (natR), pHrpl25G was digested with both SalI and XhoI, and cbxR or natR was amplified by PCR with 30 bp homology sequences of the vector for pCrpl25G or pNrpl25G, respectively.
pHrps3mCh3.
pKin3G_H was digested with both BamHI and SacI to obtain the yeast recombination vector. A 1,000-bp fragment served as a left flank, including an intron sequence and the last 726 bp of the rps3 ORF (not including the stop codon). The right flank consisted of 1,000 bp downstream of rps3 ORF. Both flanks, the mcherry and hygR resistance genes, were amplified by PCR from U. maydis genomic DNA, pomChTub1 (Schuster et al., 2011b) and pKin3G_H, respectively, thereby introducing 30 bp overlapped sequences to allow recombination in the budding yeast. The fragments were transformed into yeast, and two more copies of mcherry were introduced by standard ligation into the resulting plasmid after digestion with BsrGI.
pCrps3mCh3 and pNrps3mCh3.
To replace hygR with cbxR or natR, pHrps3mCh3 was digested with both EcoRI and SalI, and cbxR or natR was amplified by PCR with 30-bp homology sequences of the vector for pCrps3mCh3 or pNrps3mCh3, respectively.
prpl25paG.
To replace egfp with pagfp, pHrpl25G was digested with both BsrGI and SfoI. An 864-bp fragment, including the rpl25 ORF but excluding the stop codon, and pagfp were amplified by PCR from popaGRab5a (Schuster et al., 2011b) with 30-bp homology sequences.
prrm4G.
pHrpl25G was digested with SphI, and both a 1,018-bp fragment of rrm4 ORF, excluding the stop codon, and 984 bp downstream of rrm4 ORF were amplified by PCR with 30 bp homology sequences.
pΔrrm4.
pHrpl25G was digested with SphI, and both 980-bp and 984-bp fragments upstream and downstream of rrm4 ORF were amplified by PCR with 30-bp homology sequences, respectively.
prps3G.
pHrpl25G was digested with SphI, and both a 1,000-bp fragment, including an intron sequence and the last 726 bp of the rps3 ORF, excluding the stop codon, and a 1,000-bp fragment downstream of rps3 ORF were amplified by PCR with 30-bp homology sequences.
pcrgG.
To replace the otef promoter of p123 (Aichinger et al., 2003) with the crg promoter, p123 was digested with NdeI and NcoI and introduced with the crg promoter.
pCoGRab5a.
For visualization of EE, natR of poGRab5a (Schuster et al., 2011a) was replaced with cbxR.
pNomChTub1.
For visualization of MT, cbxR of pomChTub1 (Schuster et al., 2011b) was replaced with natR.
popaG2.
For observation of diffusion of cytoplasmic 2×paGFP, egfp of p123 was replaced with 2×pagfp.
poGRab4.
Both ORF and a 394-bp downstream region of tub1 of pomChTub1 were replaced with both ORF and a 721-bp downstream region of rab4.
poGRab7.
Both ORF and a 394 bp downstream region of tub1 of pomChTub1 were replaced with ORF of rab7.
poPXG.
The otef promoter, egfp, and a 438-bp fragment encoding the sequence of the PX domain (aa 4–148) from the endosomal t-SNARE Yup1 (Wedlich-Söldner et al., 2000) were inserted into pNEBcbx yeast (Schuster et al., 2011b).
poG3Rho3.
Two additional copies of egfp were introduced into the BsrGI site of p123. The resultant plasmid poG3 was inserted with both ORF and a 500-bp downstream region of rho3 under the sequence of 3×egfp.
Growth conditions
All cultures of U. maydis strains, with the exception of temperature-sensitive mutants, were grown overnight in complete medium (CM), containing 1% (wt/vol) glucose (CMglc), shaking at 200 rpm at 28°C. For induction of the nitrase reductase promoter to form hypha, cells were grown in CMglc, transferred into nitrate minimal medium (NM) supplemented with 1% (wt/vol) glucose (NMglc), and incubated for 8–14 h at 28°C, shaking at 200 rpm. All temperature-sensitive strains were grown overnight in CMglc, and hyphal formation was induced in NMglc at a permissive temperature (22°C). To inactivate Kin3, Dyn2, and Yup1, cells in the respective strains were shifted to a restrictive temperature (32°C for Kin3ts and Dyn2ts; 34°C for Yup1ts) for 1, 2, and 5 h, respectively. For induction of regulatable crg promoter in the strain FB2N107R_cG, cells were grown overnight in CMglc, transferred into CM containing 1% (wt/vol) arabinose as the sole carbon source (CMara), and incubated for the indicated times at 28°C with or without inhibitor, shaking at 200 rpm.
Sequence analysis
Sequences were retrieved from the NCBI database. Accession numbers are as follows. Ribosome subunit proteins: UmRpl25, XP_762145.1; Ce23A1, NP_508808.1; ScRpl25p, NP_014514.1; DmL23A, AAF46914.1; mouse L23a-like, NP_997406.1; AtL23a, AAB87692.1; UmRps3, XP_759103.1; ScRps3p, NP_014221.3; CeRps-3, NP_498349.1; MmS3, NP_036182.1; DmS3, NP_476632.1; AtRps3, NP_085481.1. Rab proteins: UmRab4, XP_757882.1; UmRab5a, XP_758632.1; UmRab7, XP_761658.1; HsRab4, P20338.3; HsRab5, AAB08927.1; HsRab7, AAA86640.1; ScYpt51, P36017; ScYpt7p, P32939.1. Evolutionary trees were generated using MEGA 5.1 (Tamura et al., 2011).
Laser-based epifluorescence microscopy
Microscopy of cells in culture medium was performed as described previously (Schuster et al., 2011a). In brief, cells were placed on a 2% agarose cushion, covered with a coverslip, and immediately observed using a motorized inverted microscope (IX81; Olympus) with Plan-Apochromat 100×/1.45 NA oil total internal reflection fluorescence microscopy or UPlan-SApochromat 60×/1.35 NA oil objective lenses (Olympus) and a VS-LMS4 Laser Merge System (Visitron) with 70-mW observation solid-state lasers at 488 and 561 nm. Photoactivation and photobleaching experiments were performed using a 2D FRAP system (Visitron). For temperature-dependent experiments, the objective lenses were cooled or heated using a metal hull connected to a water bath (Huber). Images were captured using a charge-coupled device camera (Photometric CoolSNAP HQ2; Roper Scientific). All parts of the system were under the control of the software package MetaMorph (Molecular Devices), which was also used for fluorescence measurements and image processing. For FRAP analyses, a region of variable length was photobleached by a 150-ms light pulse using a solid-state 405-nm laser at 100% laser power with a beam diameter of 30 pixels. Subsequently, 30–100 frames were acquired using the 488-nm and/or 561-nm lasers at exposure times of 150 or 250 ms. Kymographs were generated from the acquired image series using MetaMorph. For analyzing the colocalization of green and red fluorescent proteins, a region 10 µm in length from 10 µm behind the tip or a region of 10 µm from the tip was photobleached and immediately observed. The 488-nm laser at 40%, 40%, 100%, and 50% and the 561 nm laser at 100%, 10%, 10%, and 10% output power at an exposure time of 150 ms were used for strains AB33R3Ch3_Rrm4G, AB33ChRab5a_Rrm4G, AB33R25G_ChRab5a, and AB33ChRab5a_G3Rho3, respectively. To analyze ribosome and/or EE motility under glucose starvation or 1-butanol stresses, culture in NMglc was shifted to NM or added with 1% (vol/vol) 1-butanol for 10 min with shaking, respectively, and observed immediately. The endocytic pathway was investigated using pulse/chase analysis of FM4-64 at 16 µM (stock: 16 mM in water; Molecular Probes), and vacuolar staining was performed using CellTracker blue CMAC at 10 µM (stock: 10 mM in DMSO; Molecular Probes) according to previously published protocols (Wedlich-Söldner et al., 2000).
Quantitative motility analysis
For flux measurement, image series were taken with 75–100 frames at 150 or 100 ms for Rpl25-GFP or GFP-Rab5a, respectively. The flux of Rpl25-GFP motility was measured in kymographs using MetaMorph and determined by counting signals that crossed a line in 10 µm of the FRAP region for the first 5 s to minimize the bleaching effect of fluorescence. All experiments were done at least twice; the second experiment was usually nonquantitative.
Quantitative fluorescent intensity measurement analysis
Quantitative analysis of fluorescent intensity of moving Rpl25-GFP signals was performed as described previously (Schuster et al., 2011a). In brief, for the standard of intensity measurement, images of Nup107-GFP were taken with a 150-ms exposure time. All measurements were corrected for the background in the nuclear envelope. All corrected values were plotted and the mean integrated intensity value for a single Nup107-GFP was calculated. The number of ribosomes was estimated by comparing the corrected integrated intensity values of Rpl25-GFP with the mean value of the background-corrected integrated intensity values of a single Nup107-GFP. All experiments were done at least twice; the second experiment was usually nonquantitative.
Measurement of ribosome and ER distribution
To measure ribosome or ER distribution, images of Rpl25-GFP or GFP-HDEL fluorescence were analyzed using the linescan function in MetaMorph. The fluorescence profile was determined by averaging the individual profiles of 10 cells. Fluorescence intensity ratio of ribosome distribution was calculated by dividing the mean intensity of a region from 5–10 µm from the hyphal tip by that from 25–30 µm.
Inhibitor experiments
For inhibitor experiments, 500-µl cultures were incubated at 200 rpm in a 2-ml tube at 28°C with either benomyl at 30 µM (stock: 30 mM in DMSO; Fluka; Sigma-Aldrich), verrucarin A at 10 µg/ml (stock: 10 mg/ml in DMSO; Sigma-Aldrich), pactamycin at 100 µM (stock: 100 mM in DMSO; Sigma-Aldrich), cycloheximide at 100 µg/ml (stock: 100 mg/ml in DMSO; Sigma-Aldrich), or 100 µg/ml trichodermol (stock: 100 mg/ml in DMSO; provided by J. Zhao, Zhejiang University, China). For control experiments to analyze cytoplasmic GFP production driven under a crg promoter, cells were treated with verrucarin A, pactamycin, cycloheximide, or trichodermol for 3 h. For ribosome motility experiments, cells were cultured with benomyl for 30 min, verrucarin A for 1 h, or pactamycin for 10 min. Control cells were treated with the respective amount of solvent DMSO. Cells were placed onto a 2% agar cushion containing the respective inhibitor and immediately observed. For measurement of Rpl25-paGFP running time, cells were put onto a 2% agar cushion containing cycloheximide or trichodermol for 5 min and subsequently observed for 10 min. All experiments were done at least twice; the second experiment was usually nonquantitative.
Electron microscopy
Hyphal strains were prepared and processed as described previously (Steinberg, 2012). In brief, the cells were fixed in paraformaldehyde/glutaraldehyde phosphate buffer (2% [vol/vol] paraformaldehyde plus 2.5% [vol/vol] glutaraldehyde in 0.1 M phosphate buffer, pH 7.4), and cells were sedimented by centrifugation. After treatment with 1% OsO4 for 1 h, the cells were furthermore stained with uranyl acetate 1% (wt/vol) overnight at 4°C to enhance protein staining. Subsequently, cells were embedded in Spurr resin (TAAB), and then ultrathin sections were cut and placed on nickel grids (Agar Scientific), as previously reported (Steinberg, 2012), followed by staining with lead citrate. Samples were investigated using a transmission electron microscope (JEM 1400; JEOL, Ltd.).
Western blot analysis
Cells cultured with or without inhibitor were harvested and suspended with 1 ml of buffer B (50 mM Tris-HCl, 50 mM NaCl, and 1 mM DTT, pH 7.5) supplemented with complete mini protease inhibitor cocktail (Roche), and kept on ice. Cell suspension was lysed with glass beads, and supernatant was collected after centrifugation and used for SDS-PAGE with 10% (wt/vol) polyacrylamide gels. To detect GFP-labeled ribosomal proteins or α-tubulin for loading controls, monoclonal anti-GFP (1:5,000; Roche) or anti–α-tubulin (1:5,000; Oncogene Research Products) antibodies were used, respectively. In each case, an anti–mouse immunoglobulin G (H+L) horseradish peroxidase-conjugated antibody (1:5,000; Promega) was used for secondary antibody reaction.
Mathematical modeling
Suppose that r(x,t) is the linear density of ribosomes (in units of particles per unit length) at location x < 0 along the cell, where x = 0 represents the tip, and at time t. We assume that the ribosomes propagate away from the nucleus boundary located at x = −x0, through a combination of uniform advection (equal to active transport) at effective velocity v toward the tip and diffusion at effective diffusion rate D. The model does not consider ribosome turnover, as it is at a much slower time scale than the processes consider here (Hirsch and Hiatt, 1966). Then r evolves according to the following: The J comprises an advective and a diffusive flux J = vr − D dr/dx. Concentrating now on the steady (statistical equilibrium) state R(x) = r(x,t) we have
Solving this for v ≠ 0, we have
whereas for v = 0 (i.e., diffusion only), then R(x) = A + Bx, where in both cases the constants A and B are determined by the boundary conditions at x = x_0 < 0 and x = 0. From experimental data on the equilibrium density of ribosomes we can measure A, B, and v/D as parameters that we fit to the collected data using the nonlinear curve fitting procedure of the software Prism 4 (GraphPad Software). If we assume that ribosomes are produced at a rate α at the nucleus, there is a boundary condition on the flux J(x_0) = α, and so A = α/v. The model suggests that ribosomes are consumed at x = 0; there is a “ribosome sink” at x = 0. In terms of the flux, the expression above becomes
By nonlinear curve fitting of A + B exp(Kx) with parameters A, B, and K to the experimental curves of fluorescent intensity we found estimates for α/v = A, ξA = B, and v/D = K. The parameter ξ is a dimensionless parameter that characterizes the relative importance of advection and diffusion near the sink, and is determined by the boundary condition at x = 0.
Estimation of diffusion rates for ribosomal subunits
For monitoring paGFP with Rpl25 and cytoplasmic 2×paGFP, a small region ∼15 µm from the tip was irradiated for 150 ms using 10% output power of the 405-nm laser, followed by acquiring pictures at 100-ms exposure time for Rpl25-paGFP or at 150-ms exposure time for 2×paGFP using the 488-nm laser at 20% power. The diffusion rate was measured by fitting to a spreading Gaussian pattern, using the nonlinear curve fitting procedures of Prism 4. After subtracting the background intensity, the best fit Gaussian profiles are shown in Fig. 2 C. The growth in standard deviation σ of the Gaussian at times t = 0, 15, 30, 45, and 60 s (for Rpl25-paGFP in DMSO and benomyl) and t = 0, 0.15, 0.30, 0.45, and 0.6 s (2×paGFP in DMSO) was then plotted and fitted to a square root temporal growth where D is the diffusion rate (in square micrometers per second) and t the time (in seconds).
Statistical analysis
The Shapiro-Wilk testing for normality, Student’s t test, one-way analysis of variance (ANOVA) testing, F-testing for model comparison, and nonparametric Mann-Whitney testing were performed using Prism 4 the software.
Online supplemental material
Fig. S1 shows the position of Rpl25 and Rps3 in a phylogenetic tree, colocalization of both ribosome proteins, long-range motility of Rpl25-paGFP, and flux of Rpl25-GFP–labeled ribosome subunits near the septum and the tip. Fig. S2 shows fluorescence intensity profiles of Rpl25-GFP in Kin3ts and Dyn2ts mutants at 22°C, EE motility in Kin3ts and Dyn2ts mutants at 22°C and 32°C,ibosome clusters in Dyn2ts mutants at 32°C, and ER organization in motor mutants at restrictive conditions. Fig. S3 shows comigration of Rps3 and the RNA-binding protein Rrm4, motility of EEs and Rpl25, as well as ribosome clusters in rrm4-null mutants, and the endocytic pathway, visualized with the FM4-64. Fig. S4 shows motility of EEs and Rpl25, as well as Rpl25-GFP distribution defects in control and Yup1ts mutants. Fig. S5 shows the effect of translation inhibitors on the synthesis of cytoplasmic GFP, the expression of Rpl25-GFP and Rps3-GFP and intracellular motility of EEs and motors in the presence of the translation initiation inhibitors verrucarin A and pactamycin, Rpl25-GFP flux in the presence of verrucarin A, and colocalization of Rho3-GFP and EEs. Table S1 shows experimental usage of strains. Video 1 shows bidirectional motility of Rpl25-GFP. Video 2 shows motility of Rpl25-GFP along MTs. Video 3 shows comigration of ribosomes and kinesin-3. Video 4 shows colocalization of Rrm4-GFP and mCherry-Rab5a on EEs. Video 5 shows colocalization of Rpl25-GFP and mCherry-Rab5a on EEs. Video 6 shows moving Rpl25-GFP signals with variable fluorescent intensity. Video 7 shows motility of Rpl25-GFP under stress conditions and in the presence of translation inhibitors. Video 8 shows off-loading of ribosomes from moving EEs. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201307164/DC1.
Supplementary Material
Acknowledgments
We are grateful to Drs. Martin Schuster and Massimo Micaroni, Bioimaging Centre Exeter, for technical help with light and electron microscopy, respectively. We also thank to Drs. Ewa Bielska for providing poPXG and the strains AB33ΔKin3, AB33ChRab5a_GRab4, and AB33ChRab5a_PXG; Isabel Schuchardt for pCoGRab5a; Christoph Hemetsberger for pcrgG and the strain FB2N107R_cG; Uta Fuchs for poGRab4; and Anastassia Eskova for poGRab7. We acknowledge to Dr. Jinhao Zhao (Zhejiang University, China) for providing trichodermol and Prof. Sarah J. Gurr for critically reading the manuscript.
This work was supported by Wellcome Trust (097835/Z/11/Z) and the Biotechnology and Biological Sciences Research Council (BB/H019774/1).
Author contributions: Y. Higuchi performed experiments, generated strains, and analyzed data; P. Ashwin analyzed Rpl25-paGFP diffusion behavior and developed the mathematical model; Y. Roger generated strains and analyzed data; G. Steinberg analyzed data, conceived the project, and wrote the manuscript.
Footnotes
Abbreviations used in this paper:
- ANOVA
- analysis of variance
- EE
- early endosome
- mRNP
- messenger ribonuclear particles
- MT
- microtubule
- paGFP
- photoactivatable GFP
- PtdIns(3)P
- phosphatidylinositol 3-phosphate
References
- Abenza J.F., Pantazopoulou A., Rodríguez J.M., Galindo A., Peñalva M.A. 2009. Long-distance movement of Aspergillus nidulans early endosomes on microtubule tracks. Traffic. 10:57–75 10.1111/j.1600-0854.2008.00848.x [DOI] [PubMed] [Google Scholar]
- Aichinger C., Hansson K., Eichhorn H., Lessing F., Mannhaupt G., Mewes W., Kahmann R. 2003. Identification of plant-regulated genes in Ustilago maydis by enhancer-trapping mutagenesis. Mol. Genet. Genomics. 270:303–314 10.1007/s00438-003-0926-z [DOI] [PubMed] [Google Scholar]
- Ainger K., Avossa D., Morgan F., Hill S.J., Barry C., Barbarese E., Carson J.H. 1993. Transport and localization of exogenous myelin basic protein mRNA microinjected into oligodendrocytes. J. Cell Biol. 123:431–441 10.1083/jcb.123.2.431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashe M.P., De Long S.K., Sachs A.B. 2000. Glucose depletion rapidly inhibits translation initiation in yeast. Mol. Biol. Cell. 11:833–848 10.1091/mbc.11.3.833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashe M.P., Slaven J.W., De Long S.K., Ibrahimo S., Sachs A.B. 2001. A novel eIF2B-dependent mechanism of translational control in yeast as a response to fusel alcohols. EMBO J. 20:6464–6474 10.1093/emboj/20.22.6464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann S., Pohlmann T., Jungbluth M., Brachmann A., Feldbrügge M. 2012. Kinesin-3 and dynein mediate microtubule-dependent co-transport of mRNPs and endosomes. J. Cell Sci. 125:2740–2752 10.1242/jcs.101212 [DOI] [PubMed] [Google Scholar]
- Becht P., Vollmeister E., Feldbrügge M. 2005. Role for RNA-binding proteins implicated in pathogenic development of Ustilago maydis. Eukaryot. Cell. 4:121–133 10.1128/EC.4.1.121-133.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becht P., König J., Feldbrügge M. 2006. The RNA-binding protein Rrm4 is essential for polarity in Ustilago maydis and shuttles along microtubules. J. Cell Sci. 119:4964–4973 10.1242/jcs.03287 [DOI] [PubMed] [Google Scholar]
- Bisbal M., Wojnacki J., Peretti D., Ropolo A., Sesma J., Jausoro I., Cáceres A. 2009. KIF4 mediates anterograde translocation and positioning of ribosomal constituents to axons. J. Biol. Chem. 284:9489–9497 10.1074/jbc.M808586200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brachmann A., Weinzierl G., Kämper J., Kahmann R. 2001. Identification of genes in the bW/bE regulatory cascade in Ustilago maydis. Mol. Microbiol. 42:1047–1063 10.1046/j.1365-2958.2001.02699.x [DOI] [PubMed] [Google Scholar]
- Brangwynne C.P., Koenderink G.H., MacKintosh F.C., Weitz D.A. 2009. Intracellular transport by active diffusion. Trends Cell Biol. 19:423–427 10.1016/j.tcb.2009.04.004 [DOI] [PubMed] [Google Scholar]
- Carlton J.G., Martin-Serrano J. 2007. Parallels between cytokinesis and retroviral budding: a role for the ESCRT machinery. Science. 316:1908–1912 10.1126/science.1143422 [DOI] [PubMed] [Google Scholar]
- Carrasco L., Barbacid M., Vazquez D. 1973. The trichodermin group of antibiotics, inhibitors of peptide bond formation by eukaryotic ribosomes. Biochim. Biophys. Acta. 312:368–376 10.1016/0005-2787(73)90381-X [DOI] [PubMed] [Google Scholar]
- Chavrier P., Parton R.G., Hauri H.P., Simons K., Zerial M. 1990. Localization of low molecular weight GTP binding proteins to exocytic and endocytic compartments. Cell. 62:317–329 10.1016/0092-8674(90)90369-P [DOI] [PubMed] [Google Scholar]
- Cohen R.S. 2005. The role of membranes and membrane trafficking in RNA localization. Biol. Cell. 97:5–18 10.1042/BC20040056 [DOI] [PubMed] [Google Scholar]
- Cundliffe E., Cannon M., Davies J. 1974. Mechanism of inhibition of eukaryotic protein synthesis by trichothecene fungal toxins. Proc. Natl. Acad. Sci. USA. 71:30–34 10.1073/pnas.71.1.30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidse L.C., Flach W. 1977. Differential binding of methyl benzimidazol-2-yl carbamate to fungal tubulin as a mechanism of resistance to this antimitotic agent in mutant strains of Aspergillus nidulans. J. Cell Biol. 72:174–193 10.1083/jcb.72.1.174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dienstbier M., Boehl F., Li X., Bullock S.L. 2009. Egalitarian is a selective RNA-binding protein linking mRNA localization signals to the dynein motor. Genes Dev. 23:1546–1558 10.1101/gad.531009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan M.J., McClintock M.A., Reck-Peterson S.L. 2012a. Microtubule-based transport in filamentous fungi. Curr. Opin. Microbiol. 15:637–645 10.1016/j.mib.2012.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan M.J., Tan K., Reck-Peterson S.L. 2012b. Lis1 is an initiation factor for dynein-driven organelle transport. J. Cell Biol. 197:971–982 10.1083/jcb.201112101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elvira G., Wasiak S., Blandford V., Tong X.K., Serrano A., Fan X., del Rayo Sánchez-Carbente M., Servant F., Bell A.W., Boismenu D., et al. 2006. Characterization of an RNA granule from developing brain. Mol. Cell. Proteomics. 5:635–651 10.1074/mcp.M500255-MCP200 [DOI] [PubMed] [Google Scholar]
- Emery G., Knoblich J.A. 2006. Endosome dynamics during development. Curr. Opin. Cell Biol. 18:407–415 10.1016/j.ceb.2006.06.009 [DOI] [PubMed] [Google Scholar]
- Forrest K.M., Gavis E.R. 2003. Live imaging of endogenous RNA reveals a diffusion and entrapment mechanism for nanos mRNA localization in Drosophila. Curr. Biol. 13:1159–1168 10.1016/S0960-9822(03)00451-2 [DOI] [PubMed] [Google Scholar]
- Fuchs U., Hause G., Schuchardt I., Steinberg G. 2006. Endocytosis is essential for pathogenic development in the corn smut fungus Ustilago maydis. Plant Cell. 18:2066–2081 10.1105/tpc.105.039388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillooly D.J., Morrow I.C., Lindsay M., Gould R., Bryant N.J., Gaullier J.M., Parton R.G., Stenmark H. 2000. Localization of phosphatidylinositol 3-phosphate in yeast and mammalian cells. EMBO J. 19:4577–4588 10.1093/emboj/19.17.4577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould G.W., Lippincott-Schwartz J. 2009. New roles for endosomes: from vesicular carriers to multi-purpose platforms. Nat. Rev. Mol. Cell Biol. 10:287–292 10.1038/nrm2652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch C.A., Hiatt H.H. 1966. Turnover of liver ribosomes in fed and in fasted rats. J. Biol. Chem. 241:5936–5940 [PubMed] [Google Scholar]
- Howe C.L., Mobley W.C. 2004. Signaling endosome hypothesis: A cellular mechanism for long distance communication. J. Neurobiol. 58:207–216 10.1002/neu.10323 [DOI] [PubMed] [Google Scholar]
- Jung M.K., Wilder I.B., Oakley B.R. 1992. Amino acid alterations in the benA (β-tubulin) gene of Aspergillus nidulans that confer benomyl resistance. Cell Motil. Cytoskeleton. 22:170–174 10.1002/cm.970220304 [DOI] [PubMed] [Google Scholar]
- Kanai Y., Dohmae N., Hirokawa N. 2004. Kinesin transports RNA: isolation and characterization of an RNA-transporting granule. Neuron. 43:513–525 10.1016/j.neuron.2004.07.022 [DOI] [PubMed] [Google Scholar]
- Knowles R.B., Sabry J.H., Martone M.E., Deerinck T.J., Ellisman M.H., Bassell G.J., Kosik K.S. 1996. Translocation of RNA granules in living neurons. J. Neurosci. 16:7812–7820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koepke J., Kaffarnik F., Haag C., Zarnack K., Luscombe N.M., König J., Ule J., Kellner R., Begerow D., Feldbrügge M. 2011. The RNA-binding protein Rrm4 is essential for efficient secretion of endochitinase Cts1. Mol. Cell. Proteomics. 10:M111.011213 10.1074/mcp.M111.011213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- König J., Baumann S., Koepke J., Pohlmann T., Zarnack K., Feldbrügge M. 2009. The fungal RNA-binding protein Rrm4 mediates long-distance transport of ubi1 and rho3 mRNAs. EMBO J. 28:1855–1866 10.1038/emboj.2009.145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuersten S., Ohno M., Mattaj I.W. 2001. Nucleocytoplasmic transport: Ran, beta and beyond. Trends Cell Biol. 11:497–503 10.1016/S0962-8924(01)02144-4 [DOI] [PubMed] [Google Scholar]
- Lemmon M.A. 2003. Phosphoinositide recognition domains. Traffic. 4:201–213 10.1034/j.1600-0854.2004.00071.x [DOI] [PubMed] [Google Scholar]
- Lemmon S.K., Traub L.M. 2000. Sorting in the endosomal system in yeast and animal cells. Curr. Opin. Cell Biol. 12:457–466 10.1016/S0955-0674(00)00117-4 [DOI] [PubMed] [Google Scholar]
- Lenz J.H., Schuchardt I., Straube A., Steinberg G. 2006. A dynein loading zone for retrograde endosome motility at microtubule plus-ends. EMBO J. 25:2275–2286 10.1038/sj.emboj.7601119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao L.L., Grollman A.P., Horwitz S.B. 1976. Mechanism of action of the 12,13-epoxytrichothecene, anguidine, an inhibitor of protein synthesis. Biochim. Biophys. Acta. 454:273–284 10.1016/0005-2787(76)90230-6 [DOI] [PubMed] [Google Scholar]
- Luby-Phelps K. 2000. Cytoarchitecture and physical properties of cytoplasm: volume, viscosity, diffusion, intracellular surface area. Int. Rev. Cytol. 192:189–221 [DOI] [PubMed] [Google Scholar]
- Macdonald J.S., Goldberg I.H. 1970. An effect of pactamycin on the initiation of protein synthesis in reticulocytes. Biochem. Biophys. Res. Commun. 41:1–8 10.1016/0006-291X(70)90460-2 [DOI] [PubMed] [Google Scholar]
- Miaczynska M., Pelkmans L., Zerial M. 2004. Not just a sink: endosomes in control of signal transduction. Curr. Opin. Cell Biol. 16:400–406 10.1016/j.ceb.2004.06.005 [DOI] [PubMed] [Google Scholar]
- Nielsen E., Severin F., Backer J.M., Hyman A.A., Zerial M. 1999. Rab5 regulates motility of early endosomes on microtubules. Nat. Cell Biol. 1:376–382 10.1038/14075 [DOI] [PubMed] [Google Scholar]
- Pálfy M., Reményi A., Korcsmáros T. 2012. Endosomal crosstalk: meeting points for signaling pathways. Trends Cell Biol. 22:447–456 10.1016/j.tcb.2012.06.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paquin N., Chartrand P. 2008. Local regulation of mRNA translation: new insights from the bud. Trends Cell Biol. 18:105–111 10.1016/j.tcb.2007.12.004 [DOI] [PubMed] [Google Scholar]
- Patterson G.H., Lippincott-Schwartz J. 2002. A photoactivatable GFP for selective photolabeling of proteins and cells. Science. 297:1873–1877 10.1126/science.1074952 [DOI] [PubMed] [Google Scholar]
- Politz J.C., Tuft R.A., Pederson T. 2003. Diffusion-based transport of nascent ribosomes in the nucleus. Mol. Biol. Cell. 14:4805–4812 10.1091/mbc.E03-06-0395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popov S., Poo M.M. 1992. Diffusional transport of macromolecules in developing nerve processes. J. Neurosci. 12:77–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabut G., Doye V., Ellenberg J. 2004. Mapping the dynamic organization of the nuclear pore complex inside single living cells. Nat. Cell Biol. 6:1114–1121 10.1038/ncb1184 [DOI] [PubMed] [Google Scholar]
- Schiel J.A., Childs C., Prekeris R. 2013. Endocytic transport and cytokinesis: from regulation of the cytoskeleton to midbody inheritance. Trends Cell Biol. 23:319–327 10.1016/j.tcb.2013.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid M., Jaedicke A., Du T.G., Jansen R.P. 2006. Coordination of endoplasmic reticulum and mRNA localization to the yeast bud. Curr. Biol. 16:1538–1543 10.1016/j.cub.2006.06.025 [DOI] [PubMed] [Google Scholar]
- Schneider-Poetsch T., Ju J., Eyler D.E., Dang Y., Bhat S., Merrick W.C., Green R., Shen B., Liu J.O. 2010. Inhibition of eukaryotic translation elongation by cycloheximide and lactimidomycin. Nat. Chem. Biol. 6:209–217 10.1038/nchembio.304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster M., Kilaru S., Ashwin P., Lin C., Severs N.J., Steinberg G. 2011a. Controlled and stochastic retention concentrates dynein at microtubule ends to keep endosomes on track. EMBO J. 30:652–664 10.1038/emboj.2010.360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster M., Kilaru S., Fink G., Collemare J., Roger Y., Steinberg G. 2011b. Kinesin-3 and dynein cooperate in long-range retrograde endosome motility along a nonuniform microtubule array. Mol. Biol. Cell. 22:3645–3657 10.1091/mbc.E11-03-0217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster M., Lipowsky R., Assmann M.A., Lenz P., Steinberg G. 2011c. Transient binding of dynein controls bidirectional long-range motility of early endosomes. Proc. Natl. Acad. Sci. USA. 108:3618–3623 10.1073/pnas.1015839108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seaman M.N. 2008. Endosome protein sorting: motifs and machinery. Cell. Mol. Life Sci. 65:2842–2858 10.1007/s00018-008-8354-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorkin A., Von Zastrow M. 2002. Signal transduction and endocytosis: close encounters of many kinds. Nat. Rev. Mol. Cell Biol. 3:600–614 10.1038/nrm883 [DOI] [PubMed] [Google Scholar]
- Steinberg G. 2007. On the move: endosomes in fungal growth and pathogenicity. Nat. Rev. Microbiol. 5:309–316 10.1038/nrmicro1618 [DOI] [PubMed] [Google Scholar]
- Steinberg G. 2012. Cytoplasmic fungal lipases release fungicides from ultra-deformable vesicular drug carriers. PLoS ONE. 7:e38181 10.1371/journal.pone.0038181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinberg G., Perez-Martin J. 2008. Ustilago maydis, a new fungal model system for cell biology. Trends Cell Biol. 18:61–67 10.1016/j.tcb.2007.11.008 [DOI] [PubMed] [Google Scholar]
- Steinberg G., Schuster M., Theisen U., Kilaru S., Forge A., Martin-Urdiroz M. 2012. Motor-driven motility of fungal nuclear pores organizes chromosomes and fosters nucleocytoplasmic transport. J. Cell Biol. 198:343–355 10.1083/jcb.201201087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straube A., Weber I., Steinberg G. 2005. A novel mechanism of nuclear envelope break-down in a fungus: nuclear migration strips off the envelope. EMBO J. 24:1674–1685 10.1038/sj.emboj.7600644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28:2731–2739 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Der Sluijs P., Hull M., Zahraoui A., Tavitian A., Goud B., Mellman I. 1991. The small GTP-binding protein rab4 is associated with early endosomes. Proc. Natl. Acad. Sci. USA. 88:6313–6317 10.1073/pnas.88.14.6313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vida T.A., Emr S.D. 1995. A new vital stain for visualizing vacuolar membrane dynamics and endocytosis in yeast. J. Cell Biol. 128:779–792 10.1083/jcb.128.5.779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vollmeister E., Schipper K., Baumann S., Haag C., Pohlmann T., Stock J., Feldbrügge M. 2012. Fungal development of the plant pathogen Ustilago maydis. FEMS Microbiol. Rev. 36:59–77 10.1111/j.1574-6976.2011.00296.x [DOI] [PubMed] [Google Scholar]
- Wedlich-Söldner R., Bölker M., Kahmann R., Steinberg G. 2000. A putative endosomal t-SNARE links exo- and endocytosis in the phytopathogenic fungus Ustilago maydis. EMBO J. 19:1974–1986 10.1093/emboj/19.9.1974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wedlich-Söldner R., Schulz I., Straube A., Steinberg G. 2002a. Dynein supports motility of endoplasmic reticulum in the fungus Ustilago maydis. Mol. Biol. Cell. 13:965–977 10.1091/mbc.01-10-0475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wedlich-Söldner R., Straube A., Friedrich M.W., Steinberg G. 2002b. A balance of KIF1A-like kinesin and dynein organizes early endosomes in the fungus Ustilago maydis. EMBO J. 21:2946–2957 10.1093/emboj/cdf296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarnack K., Feldbrügge M. 2010. Microtubule-dependent mRNA transport in fungi. Eukaryot. Cell. 9:982–990 10.1128/EC.00030-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeigerer A., Gilleron J., Bogorad R.L., Marsico G., Nonaka H., Seifert S., Epstein-Barash H., Kuchimanchi S., Peng C.G., Ruda V.M., et al. 2012. Rab5 is necessary for the biogenesis of the endolysosomal system in vivo. Nature. 485:465–470 10.1038/nature11133 [DOI] [PubMed] [Google Scholar]
- Zerial M., McBride H. 2001. Rab proteins as membrane organizers. Nat. Rev. Mol. Cell Biol. 2:107–117 10.1038/35052055 [DOI] [PubMed] [Google Scholar]
- Zhang J., Zhuang L., Lee Y., Abenza J.F., Peñalva M.A., Xiang X. 2010. The microtubule plus-end localization of Aspergillus dynein is important for dynein-early-endosome interaction but not for dynein ATPase activation. J. Cell Sci. 123:3596–3604 10.1242/jcs.075259 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.