Summary
AP-1 is a clathrin adaptor complex that sorts cargo between the trans-Golgi network and endosomes. AP-1 recruitment to these compartments requires Arf1-GTP. The crystal structure of the tetrameric core of AP-1 in complex with Arf1-GTP, together with biochemical analyses, shows that Arf1 activates cargo binding by unlocking AP-1. Unlocking is driven by two molecules of Arf1 that bridge two copies of AP-1 at two interaction sites. The GTP-dependent switch I and II regions of Arf1 bind to the N-terminus of the β1 subunit of one AP-1 complex, while the back side of Arf1 binds to the central part of the γ subunit trunk of a second AP-1 complex. A third Arf1 interaction site near the N-terminus of the γ subunit is important for recruitment, but not activation. These observations lead to a model for the recruitment and activation of AP-1 by Arf1.
Introduction
Clathrin-coated vesicles (CCVs) play major roles in intracellular transport of selected cargo molecules from the plasma membrane, trans-Golgi network (TGN) and endosomes (Brodsky et al., 2001; Kirchhausen, 2000). CCV formation starts with the recruitment of adaptor proteins (APs) from the cytosol to the target membranes. The membrane-bound APs interact with sorting signals contained within the cytosolic tails of transmembrane cargo proteins while also inducing the polymerization of clathrin into a polyhedral, lattice-like scaffold. Clathrin-coated membranes curve, eventually leading to the budding of CCVs that contain specific sets of cargo molecules.
The main clathrin APs are two homologous, heterotetrameric complexes named AP-1 (γ-β1-μ1-σ1) and AP-2 (α-β2-μ2-σ2) (subunit composition in parenthesis), which function at the TGN/endosomes and plasma membrane, respectively (Owen et al., 2004; Robinson, 2004). Both complexes are structured as a “core” domain comprising the N-terminal “trunk” portions of γ/α and β1/β2 plus the whole μ1/μ2 and σ1/σ2 subunits, and two “appendage” domains corresponding to the C-terminal portions of γ/α and β1/β2, which are connected to the core by two long, largely unstructured “hinge” sequences. The core domain mediates recruitment to membranes and recognition of sorting signals while the hinge-ear domains interact with clathrin and various accessory proteins. Both AP-1 and AP-2 recognize at least two types of sorting signal: tyrosine-based YXXØ-type signals through binding to the μ1/μ2 subunits (Boll et al., 1996; Ohno et al., 1996; Ohno et al., 1995; Owen and Evans, 1998) and dileucine-based [DE]XXX[LI]-type signals through binding to a site at the interface of the γ-σ1 and α-σ2 subunits (amino acids in single letter code; X is any amino acid and Ø a bulky hydrophobic amino acid) (Chaudhuri et al., 2007; Doray et al., 2007; Janvier et al., 2003; Kelly et al., 2008; Mattera et al., 2011).
The mechanisms of signal recognition and membrane recruitment have been worked out in greatest detail for AP-2. Biochemical and X-ray crystallographic analyses have shown that the AP-2 core occurs in two distinct conformations: a cytosolic, “locked” conformation where binding sites for YXXØ and [DE]XXX[LI] signals are occluded by portions of β2 (Collins et al., 2002), and a membrane-bound, “open” conformation where these binding sites are exposed (Jackson et al., 2010). The AP-2 core also has four clusters of basic residues (one cluster each on α and β2, and two on μ2) that serve as binding sites for the headgroups of membrane phosphatidylinositol-4,5-bisphosphate (PI(4,5)P2) (Collins et al., 2002; Gaidarov et al., 1996; Jackson et al., 2010; Rohde et al., 2002). In the locked conformation, the μ2 C-terminal domain responsible for binding to the YXXØ signal is sequestered in a bowl formed by the trunk domains of the α and β2 subunits. In the open conformation, the two signal-binding sites and four PI(4,5)P2-binding sites become coplanar, enabling simultaneous interactions with cargo proteins and PI(4,5)P2, and thus stabilizing the open conformation of the core (Jackson et al., 2010). The enrichment of PI(4,5)P2 at the plasma membrane (Di Paolo and De Camilli, 2006) ensures that AP-2 is specifically recruited to this compartment.
The structural bases for AP-1 signal recognition and membrane recruitment are less well understood. The AP-1 core also occurs in a locked conformation similar to that of the AP-2 core, as shown by X-ray crystallography (Heldwein et al., 2004). The existence of an open conformation of the AP-1 core has not been demonstrated by structural methods but is supported by other lines of evidence. First, the residues that bind YXXØ and [DE]XXX[LI] signals in AP-2 (Jackson et al., 2010; Kelly et al., 2008; Owen and Evans, 1998) are highly conserved in AP-1 (Heldwein et al., 2004), and mutation of these residues abrogates binding of both types of signal to AP-1 in yeast two- and three-hybrid assays (Carvajal-Gonzalez et al., 2012; Mattera et al., 2011). Second, binding of one type of signal enhances binding of the other type, likely due to stabilization of an open conformation (Lee et al., 2008a).
Whereas the mechanisms of signal recognition by AP-1 and AP-2 appear quite similar, the determinants of recruitment to their corresponding membranes differ significantly. The AP-1 core has a phosphoinositide-binding site with preference for phosphatidylinositol-4-phosphate (PI(4)P) on its γ subunit, at a location similar to that of the PI(4,5)P2-binding site on AP-2 α (Heldwein et al., 2004; Wang et al., 2007). PI(4)P is enriched within domains of the TGN and endosomes (Di Paolo and De Camilli, 2006), consistent with the association of AP-1 to these compartments. In contrast to the case of AP-2, however, phosphoinositides alone are insufficient to recruit AP-1 to its sites of action. Instead, the key determinant of AP-1 targeting to the TGN/endosomes is its interaction with members of the ADP ribosylation factor (Arf) family of small GTPases, particularly Arf1 (Seaman et al., 1996; Stamnes and Rothman, 1993; Traub et al., 1993). Arf1 cooperates with cargo and phosphoinositides such that AP-1 binding to all of these components is thought to be necessary for targeting under normal conditions (Crottet et al., 2002; LeBorgne et al., 1996; Lee et al., 2008a), although enrichment of cargo signals to high levels can override this requirement (Lee et al., 2008b). Arfs cycle between a GDP-bound, inactive cytosolic form and a GTP-bound, active membrane-tethered form (Donaldson and Jackson, 2011). Conversion to the GTP-bound form requires a guanine nucleotide exchange factor (GEF), whereas conversion to the GDP-bound form is catalyzed by a GTPase activating protein (GAP). Loading with GTP causes Arfs to undergo a conformational change, exposing a myristoylated N-terminal amphipathic helix that inserts into the membrane, while reconfiguring its switch I-II and interswitch regions to allow binding of effector proteins (Donaldson and Jackson, 2011). Arf1 has many effectors, including AP-1 as well as the homologous heterotetrameric complexes AP-3 (Ooi et al., 1998), AP-4 (Boehm et al., 2001) and COPI (F subcomplex) (Serafini et al., 1991). Arf1 is not enriched at the plasma membrane, and is not thought to interact with AP-2 in cells. Some studies, however, have suggested that the plasma membrane associated Arf6 could be involved in recruiting AP-2 (Krauss et al., 2003; Montagnac et al., 2011; Paleotti et al., 2005; Poupart et al., 2007). Thus, the question of how Arf family GTPases recognize, recruit, and activate AP complexes has broad implications for intracellular traffic.
Recently, important insight into Arf1 recognition was obtained from the structure of a truncated γζ subcomplex from COPI (Yu et al., 2012). The goal of the present study was to take the next step in understanding whether Arf1 regulates not only the localization, but also the conformation of heterotetrameric sorting complexes. To address this question, we solved the crystal structure of the AP-1 core in complex with GTP-bound Arf1. The most important insight is that Arf1-GTP alone, in the absence of cargo or PI(4)P, can unlock AP-1 and drive it into the open conformation. AP-1 contains two binding sites for the canonical switch I and II surface on Arf1, one on each of the two trunk domains. Both of these Arf1-binding sites are required for high-affinity binding in vitro and for subcellular localization to the TGN/endosomes, but only the site on β1 is important for activation. Moreover, a novel surface on the C-terminal portion (“back side”) of Arf1, distal to switch I and II, was found to be required for full allosteric activation, although it does not contribute to recruitment. Taken together with the dimeric assemblage of AP-1 in the crystal lattice, a model for the allosteric activation mechanism was deduced. Reconstitution of the recruitment of AP-1 to liposomes by Arf1, cargo, and PI(4)P highlights the profound cooperativity between the binding of cargo to AP-1 and the β1 and back side interactions with Arf1.
Results
Structure of the AP-1:Arf1 complex
The core of the AP-1 adaptor complex was reconstituted by co-expressing the trunk domains of the murine γ1 (residues 1-595) and human β1 (residues 1-584) subunits with full-length human σ1C and murine α1A subunits in E. coli using a single polycistronic expression plasmid. Human Arf1 bearing the GTPase mutation Q71L and the N-terminal truncation Δ1-16 (Arf1Δ1-16) was loaded with GTP and mixed at a 4:1 excess of Arf1 relative to AP-1. Crystals were obtained that diffracted to 7.0 Å resolution.
The crystal structure of the AP-1:Arf1-GTP complex (Fig. 1A) was determined by the molecular replacement method. Because it was not known a priori whether the crystallized AP-1 core would be in one of the expected locked or open conformations, or in some novel conformation, test searches were run using all of the available crystal structures of AP-1 and AP-2 core complexes. A solution was obtained using as a search model the core of AP-2 in the open conformation (PDB 2XA7) (Jackson et al., 2010). At this stage, clear (Fo-Fc)αcalc difference electron density was visible for the entire Arf1Δ1-16 molecule (Fig. 1B). The 1.6 Å structure of the GTP-bound form of murine Arf1Δ1-17 (Shiba et al., 2003) (PDB 1O3Y) was used as a search model to position the molecule in the unit cell. The clarity of this unbiased difference map, in spite of its low resolution, persuaded us to refine the structure and characterize its functional implications. It is not possible to visualize side-chains at this resolution. However, given the availability of well-refined starting models for substructures, contemporary refinement methodology makes it possible to accurately analyze the overall conformation of large protein complexes and the nature of their interfaces from diffraction data at as low as 7 Å resolution (Brunger et al., 2012).
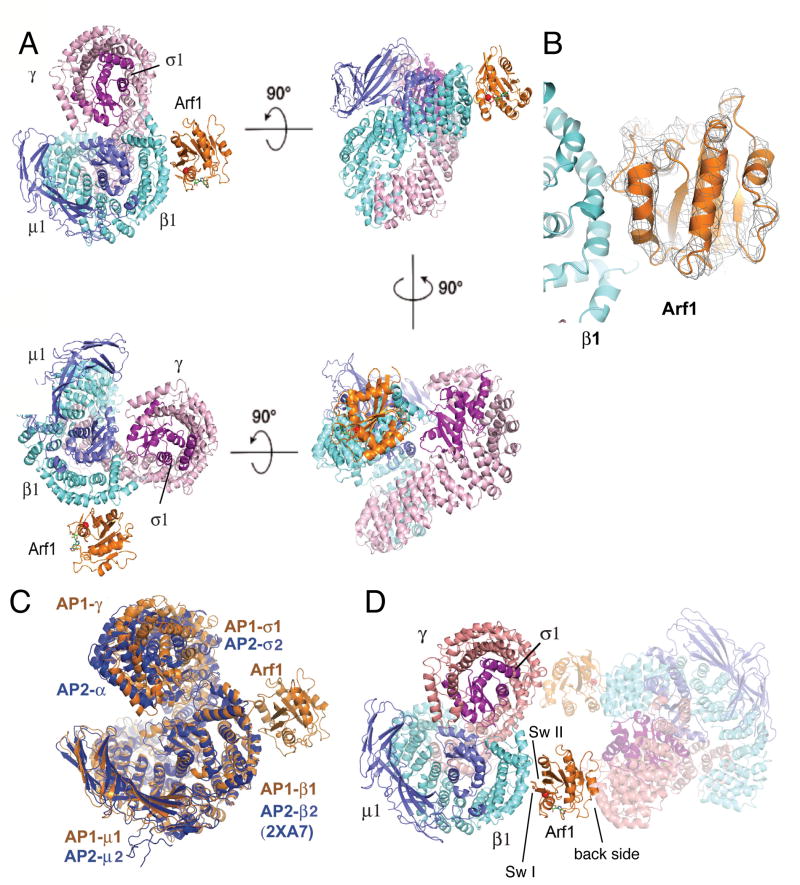
Figure 1. Crystal structure of the AP-1:Arf1 complex.
(A) Views of the overall structure of the AP-1 Arf1 complex, related by rotations about the indicated axes. Colors are γ, light pink; β1, aquamarine; μ1, slate blue; σ1, purple; and Arf1, orange. (B) Unbiased difference density contoured at 2σ around Arf1, which was not present in the search model used to obtain these phases, illustrates the high quality of the molecular replacement phases at 7 Å. (D) Overlay of the YXXØ cargo-bound conformation of AP-2 upon Arf1-GTP-bound AP-1. (E) Overall structure of the crystallographic dimer. See also Table S1.
Refinement of this structure was facilitated by the finding that the Arf1-bound conformation of AP-1 is nearly identical to the open conformation of AP-2, which was refined at 3.1 Å resolution (Jackson et al., 2010). The main-chain of the AP-1 structure was tethered to a model based on the open conformation of AP-2 using the deformable elastic network (DEN) methodology (Schroder et al., 2010). A starting model of AP-1 in this conformation was generated by superimposing the 4.0 Å resolution coordinates of the AP-1 core complex (PDB 1W63) (Heldwein et al., 2004) onto the open conformation of AP-2 on a domain-by-domain basis. The trunk domains of p1 and γ were broken into three fragments for the superposition, and μ1 was broken into its N- and C-terminal domains. All of these domains had excellent fits with the sole exception of the μ1 C-terminal domain. Therefore, the μ1 C-terminal domain model was derived by replacing the side-chains of μ2 in the TGN38-bound structure with their cognates from μ1. Even though side-chains were not visualized, they were included in the refinement in order to account for their contribution to X-ray scattering. Side-chain conformations were allowed to relax in order to accommodate sequence differences with respect to the parent models used for molecular replacement and to avoid steric collisions at Arf1 binding and lattice interfaces. The resulting structure (Fig. 1A) had a free R-factor of 0.25 and excellent stereochemistry (Table S1). Moreover, as described below, the structural interfaces underwent extensive validation on the Arf1 and AP-1 sides, both in solution and in cells.
Open conformation of AP-1
By analogy to AP-2, it was anticipated that AP-1 would be activated through a conformational change and exposure of the YXXØ and [DE]XXX[LI] binding sites. Here, we have visualized the active conformation of AP-1 in the presence of Arf1-GTP but in the absence of cargo tails, phosphoinositides, or soluble phosphoinositide analogs. The overall structure is essentially superimposable on that of the YXXØ-bound AP-2 core (Fig. 1C), which is the structural paradigm of the active conformation. This observation is consistent with the biochemical evidence that Arf1-GTP is a direct allosteric activator of AP-1.
Structures of the AP-1:Arf1 interfaces
The crystals contain one copy each of the AP-1 core and Arf1 (Fig. 1A). Arf1 bridges two copies of the AP-1 core in the crystal lattice, such that Arf1 binds to two sites on AP-1 (Fig. 1D). The larger of the two interfaces (∼720 Å2) buries the switch I and II regions of Arf1 against helices α1, α3, and α5 of the β1 subunit (Fig. 2A, B). The β1 contact is centered on Gln59, Ile85, and Asn89. Arf1 contacts include Ile46, Ile49, Gly50, Phe51, Asn52, and Val53 of switch I, Trp66, Lys73, Ile74, Leu77, His80, Tyr81, and Gln83 of switch II, and Tyr35 of α1 (Fig. 2A, B). Switch I and II are the regions of Arf1 that change conformation upon GTP binding (Goldberg, 1998). The GDP bound conformation of Arf1 (Amor et al., 1994) is not compatible with the β1 structure because of an extensive clash between switch I and β1-α5 (Fig. S1). The involvement of the switch regions in AP-1 binding has been noted (Liang et al., 1997), and is consistent with the GTP requirement for membrane recruitment of AP-1 and its inhibition by the Arf1 GEF inhibitor brefeldin A (Stamnes and Rothman, 1993; Traub et al., 1993). The β1 binding site is in accord with an Arf1 binding site recently predicted to occur on the α4 and α6 helices of the β-COP subunit of COPI (Yu et al., 2012), which correspond to α3 and α5 helices of β1. The direct interaction of the β1 subunit is consistent with the photocross-linking of switch I-labeled Arf1 with the β1 and γ subunits of AP-1(Austin et al., 2002).
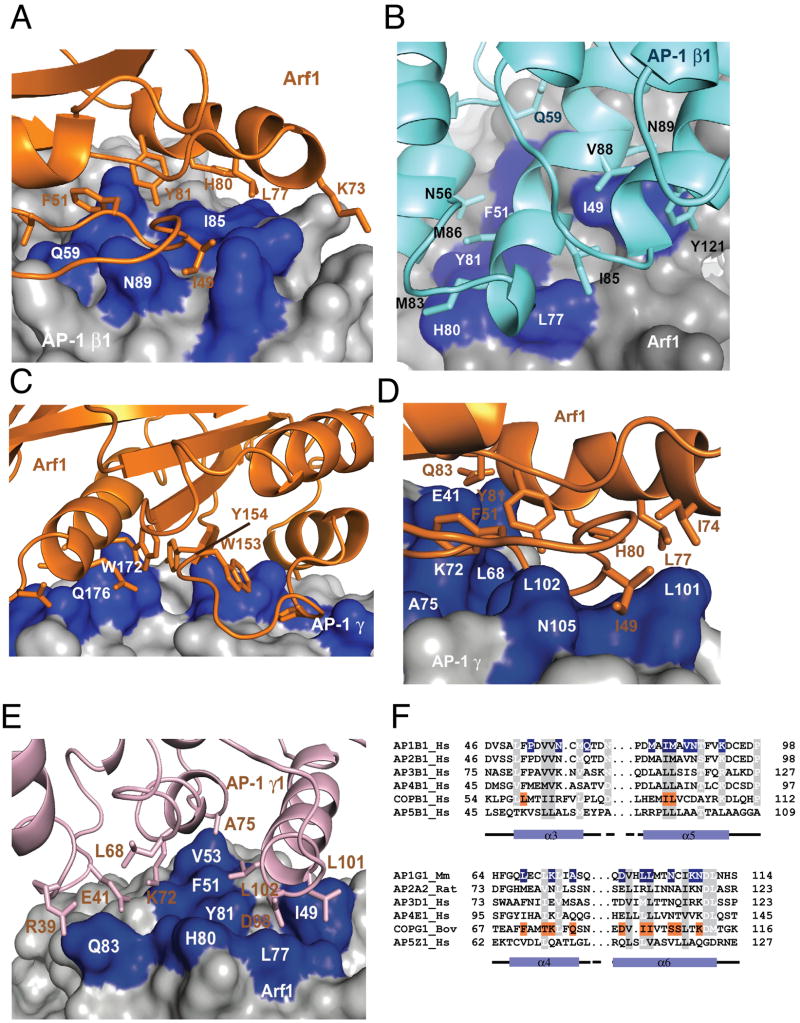
Figure 2. Arf1 interfaces with AP-1 subunits.
(A) Arf1 is shown in a ribbon model (orange) as it interacts with the surface of the β1 subunit. Functionally important residues of β1 are highlighted in blue. (B) The same interface shown in (A) is represented with a ribbon model of the β1 subunit and a surface representation of Arf1, with key switch I and II residues of Arf1 highlighted in blue. (C) The back side of Arf1 distal to switch I and II is shown in a ribbon model (orange) as it interacts with the surface of the γ subunit in the crystal. Interacting residues of γ are highlighted in blue. (D, E) Two views of the functional Arf1 interface with the γ subunit. This interface is not present in this crystal structure, but is modeled on the basis of the β1 interface and by analogy to the COPI complex (Yu et al., 2012), represented as in (A, B). (F) Structure-based alignment of key Arf1 switch I and II-binding helices of the β1 and γ subunits of AP-1 with corresponding subunits of other AP complexes and COPI. See also Fig. S1.
The smaller of the two interfaces (∼690 Å2) buries the C-terminal α4, β6, and α5 of the back side of Arf1, which is on the opposite face from switch I and II, against the region of helices α12-α16 of the γ subunit (Fig. 2C). A cluster of large hydrophobic residues from the back side of Arf1 participate in this interface: Trp153, Tyr154, and Trp172. At the periphery of this hydrophobic cluster, Ala136, Ala137, and Gln176 also make contacts with γ. Confidence in the identity of residues on the γ subunit is limited by the resolution of the structure. Thus far it has not been possible for us to corroborate the residues on this face of the site by mutagenesis. As described below, mutational analysis of the interaction suggested this interface is involved in allosteric activation, but not in recruitment.
Several considerations led us to localize a functional Arf1 switch I and II binding site near the N-terminus of γ (Fig. 2D, E). First, the lattice contact between γ and the back side of Arf1 did not explain the observation of cross-linking between switch I and γ (Austin et al., 2002). Second, the crystal structure of a γζ-COP complex containing a 15-helix fragment of the y-COP trunk was recently determined in complex with Arf1 (Yu et al., 2012). This structure showed that helices α4 and α6 of γ-COP bind to Arf1 switch I and II. The Arf1-binding residues of γ-COP are partially conserved in the γ subunit of AP-1 (Fig. 2F). Finally, the trunk domains of the β1 and γ subunits are structurally homologous to each other, another line of suggestion that the γ subunit might possess an Arf1 switch I binding site similar to the one found near the N-terminus of β1. The β1 and γ subunits were overlaid on one another, and used to generate a provisional model for Arf1 bound to γ via switch I and II (Fig. 2D, E). The putative binding site is centered on Leu68 and Leu71 of helix α4 and Leu102 of helix α6 of γ. This model is consistent with the results of overlaying the γ-COP and AP-1 γ structures, and was subsequently validated by mutational dissection.
The Arf1 switch I and II binding sites on β1 and γ are both important for subcellular targeting, as described below, therefore we refer to them as recruitment sites. The character of these two recruitment sites is well-conserved in other Arf1-dependent APs, including AP-3, AP-4, and COPI. The Arf1 binding site on the β1 subunit also appears to be conserved in the AP-2 subunit β2 (Fig. 2F). Switch I and II residues are highly conserved among Arf family GTPases, thus this finding is consistent with the possibility that Arf6 is a direct activator of AP-2. The cognate of the γ subunit Arf1 binding site on the AP-2 α subunit is less clearly conserved, in that the key hydrophobic Leu101 of γ is replaced by an Arg (Fig. 2F). It remains to be determined if this or other nearby changes render the AP-2 α subunit unable to bind Arf family members. Each of the Arf1 binding sites comprises ∼700 Å of buried surface area, which taken individually would amount to a low-affinity interaction. The modest amount of surface area buried in each site explains why, as described below, neither one of the sites by itself can support high-affinity binding in vitro or TGN/endosomal localization in cells.
Mutational analysis of the AP-1:Arf1 interaction
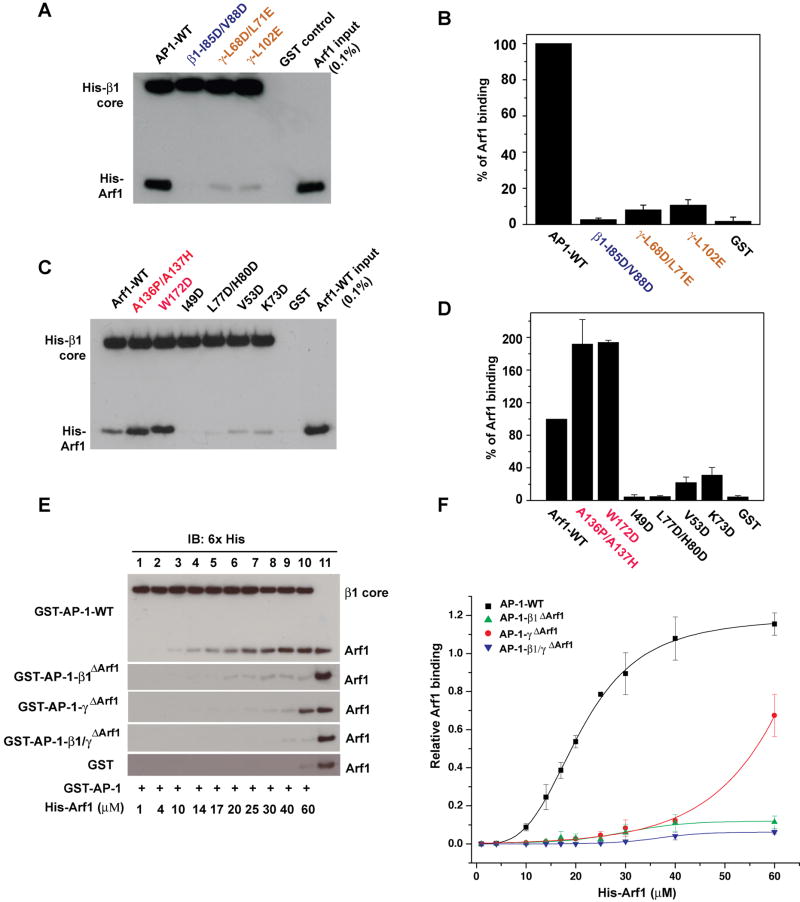
The β1 binding site and both the crystallographic and modeled γ binding sites for Arf1 were assessed by mutational disruption. The mutations β1I85D/V88D, γL68D/L71E, and γL102E were constructed in the context of the recombinant AP-1 core and purified from E. coli as GST fusions. Purification yields and subunit stoichiometries were essentially identical to wild-type for all mutants (Fig. S2A). Wild-type and mutant AP-1 cores tagged with GST were immobilized on glutathione-Sepharose beads, and their ability to bind to 5 μM His6-Arf1Δ1-16-GTP was determined. The β1I85D/V88D mutant was the only mutant that had, within experimental error, no binding of Arf1 above the GST control (Fig. 3A, B). Thus, mutation of the crystallographic binding site on β1 completely eliminated binding. Two mutants designed to disrupt the modeled γ α4-α6 binding site, γL68D/L71E and γL102E, reduced binding to ∼10 % of wild-type levels (Fig. 3A, B). This indicates that the γ α4-α6 binding site is functional, but that its affinity for Arf1 is less than that of the β1 binding site. We also tested the effect of mutations in the switch I and II (Arf1I49D, Arf1V53D, Arf1K73D and Arf1L77D/H80D) and back side regions (Arf1A136P/A137H and Arf1W172D) of Arf1 on binding to AP-1. Purification yields for all Arf1 mutants were similar to wild-type (Fig. S2B). Switch I and II mutants sharply reduced or eliminated binding, whereas back side mutants actually enhanced binding (Fig. 3C, D). These results are consistent with the crystallographic observations at the β1 interface and with the proposed model for the functional γ interface.
Figure 3. Mutational analysis of Arf1 binding sites.
(A, B) Representative gel (A) and quantification (B) of GST pull-down assays to assess binding of the immobilized GST-AP-1 core and its mutants to Arf1-GTP. GST-AP-1 core proteins (15μg) were immobilized on glutathione-Sepharose beads and incubated with His-Arf1Δ16-Q71L (5 μM). The AP-1-bound Arf1 was detected by anti-His-tag antibody using Western blotting. (C, D) The same procedures as in (A) were used to determine the effects of switch I (I49D, V53D), switch II (K73D, L77D/H80D), and back side (A136P/A137H, W172D) mutations in Arf1 on binding to the wild-type AP-1 core. (E, F) Arf1-AP1 binding curves derived from quantitative immunoblotting. Lane 11 in (E) represents 1.6 pmol of His-Arf1 input, which was used for normalization. Relative Arf1 binding was quantified and fitted to the Hill equation in (F). See also Fig. S2 and S3. Error bars represent the standard deviation of three measurements.
In order to probe the function of the β1 and γ interfaces in cellular function, multiple mutations were constructed at each site. The β1I85D/V88D mutant is hereafter referred to as “β1ΔArf1”, and the triple mutant γL68D/L71E/L102E is hereafter “γΔArf1”. To verify that these mutants did not cause a loss in thermal stability, differential scanning fluorimetry of wild-type and the β1ΔArf1 and γΔArf1 mutant cores was carried out. Melting temperatures Tm of 56 to 58 °C were measured (Fig. S3), and no sign of melting was observed for any of the constructs at the temperatures T = 25 or T = 37 °C at which the in vitro and biological experiments were carried out.
Binding curves were obtained for wild-type and each mutant. Binding of the wild-type AP-1 core His-Arf1 could be fit to the Hill equation with Kd = 20 ± 0.6 μM and a Hill coefficient of nH=3.3. The β1I85D/V88D mutant (hereafter “β1ΔArf1”, Fig. 3E, F) nearly eliminated binding as compared to wild-type. The triple mutant γL68D/L71E/L102E (hereafter “γΔArf1”, Fig 3E, F) showed residual binding and its curve retained a sigmoidal character, consistent with the presence of an intact β1 interface functioning in the context of an AP-1 multimer. A β1/γΔArf1 construct was prepared by combining β1ΔArf1 and γΔArf1 in the same complex, and it was found that this construct completely eliminated binding (Fig. 3E, F).
Roles of Arf1 binding sites on β1 and γ in TGN/endosomal localization
Having established the presence of two Arf1 recruitment sites in vitro, we examined the effect of disrupting the Arf1-binding sites of β1 and γ on the recruitment of these proteins to the TGN/endosomes in whole cells. GFP-tagged forms of β1ΔArf1 and γΔArf1 were incorporated into AP-1 complexes as efficiently as their wild-type counterparts when expressed by transfection into cells (Fig. 4A, B), consistent with previous observations (Farias et al., 2012; Huang et al., 2001). GFP-tagged β1 (β1WT-GFP) and mCherry-tagged γ (γWT-mCh) co-localized with endogenous γ and transgenic μ1A-GFP, respectively, to a juxtanuclear structure characteristic of the TGN/endosomes (Fig. 4, C-E and I-K). In contrast, the mutant β1ΔArf1-GFP and γΔArf1-mCh were largely cytosolic (Fig. 4, F-H and L-N). These observations indicated that the Arf1 recruitment sites on both β1 and γ are required for targeting of AP-1 to the TGN/endosomes within cells.
Figure 4. Arf1-binding sites on β1 and γ are required for association of AP-1 to the TGN/endosomes.
(A, B) MDCK-μ1A-HA cells transfected with plasmids encoding β1WT-GFP or β1ΔArf1-GFP (A) and γWT-GFP or γΔArf1-GFP (B) or GFP (A, B) were subjected to immunoprecipitation (IP) with antibody to GFP followed by SDS-PAGE and immunoblotting with HRP-conjugated antibodies to the HA epitope and GFP. The position of molecular mass markers (in kDa) are indicated on the left. Loading was adjusted to normalize for β1 and γ expression. Assembly of β1 and γ mutants with μ1A-HA was 99±6% and 97±5% of the corresponding wild-type proteins (n=3). (C-H) HeLa cells transfected with plasmids encoding β1WT-GFP (C-E) or β1ΔArf1-GFP (F-H) were immunostained for endogenous γ. (I-N) HeLa cells were co-transfected with plasmids encoding γWT-mCh (I-K) or γΔArf1-mCh (L-N) together with μ1A-GFP. Nuclei were stained with DAPI. Images were obtained by confocal microscopy. The third image in each row is a merge of images in the green, red and blue channels. Scale bar: 10 μm.
Roles of Arf1 binding sites on β1 and γ in the allosteric activation of AP-1
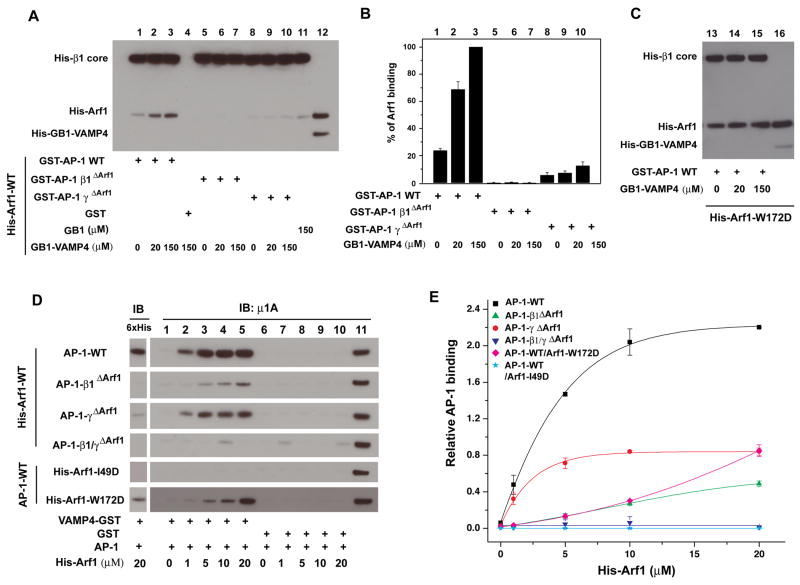
Binding of cargo peptides bearing either tyrosine- or dileucine-based sorting signals strongly promotes the binding of Arf1 to AP-1 in solution (Lee et al., 2008a). We applied this principle to map the involvement of the individual Arf1-binding sites on β1 and γ in this conformational change. The addition of 20 μM of a dileucine-containing peptide from VAMP4 (Peden et al., 2001) led to a 4-fold increase in the amount of Arf1 bound to the AP-1 core (Fig. 5A, B), similar to previous results for the binding of full-length AP-1 to Arf1 in the presence of a dileucine-containing peptide from the cation-independent mannose 6-phosphate receptor (Lee et al., 2008a). As shown above in Fig. 3 (E, F), β1ΔArf1 has essentially no interaction with Arf1 in the absence of peptide. Addition of peptide failed to rescue Arf1 binding to AP-1-β1ΔArf1 (Fig. 5). The γΔArf1 form of the AP-1 complex binds weakly to Arf1 in the absence of peptide (Fig. 5A, B). When the peptide concentration was increased to 150 μM, a 4-fold enhancement was seen relative to the absence of peptide. This suggests that the γ subunit recruitment site is not necessary for allosteric coupling between the Arf1 and cargo.
Figure 5. Allosteric coupling between the Arf1 and dileucine binding sites.
(A) Pull-down of Arf1 with GST-AP-1 cores in the presence of VAMP4 peptide. Wild-type or mutant GST-AP-1 core (100 nM) was immobilized, and incubated with His-Arf1-Q71L (4 μM) and 2 mM GTP and the indicated concentrations of His-GB1 tagged VAMP4 (20-28, the dileucine motif). AP-1-bound Arf1 was analyzed by the western blot using anti-polyHis antibody, followed by quantification (B) Lane 12 and lane 16 represent 0.1% of the input from the reaction containing 4 μM His-tagged Arf1 and 150 mM His-GB1-VAMP4 peptide. (C) Arf1-W172D tightly bound to GST-AP1-A core independent of the VAMP4 peptide. (D). Recruitment of AP-1 cores to VAMP4-GST was activated by Arf1, dependent on and intact AP-1 β1 Arf1 binding site. In each reaction, VAMP4 (1-51)-GST (100 nM) was immobilized and incubated with the indicated AP-1 core (0.5 μM) and His-Arf1. The bound fraction was immunoblotted using anti-polyHis to detect Arf1 (left panel) and anti-μ1 to detect AP-1 (right panel). Lane 11 represents 0.1 pmol of AP-1 core input. The relative AP-1 binding was quantified (E) to plot with the function of His-Arf1 input concentration. The active affinity of AP-1 core WT binding to his-Arf1 in the presence of VAMP4-GST was 4.0 ± 0.3 μM. Error bars represent the standard deviation of three measurements. See also Movie S1.
To provide a second view of conformational coupling between Arf1 and VAMP4, the VAMP4 sequence was immobilized, and the binding of AP-1 was measured in the presence of increasing concentrations of Arf1 (Fig. 5D, E). We observed that Arf1 enhanced binding of wild-type AP-1 to VAMP4 following a hyperbolic curve with an effective activation constant Kact = 4.0 ± 0.3 μM. As compared to the Arf1 binding curve in the absence of VAMP4 (Fig. 3F), the activation curve is shifted sharply to the left and the apparent cooperativity is absent. Thus, the presence of VAMP4 sharply increases the affinity of AP-1 for Arf1, consistent with its promotion of the open conformation. The defect in β1ΔArf1 activation is much greater than that for γΔArf1 (Fig. 5C, D). Indeed, γΔArf1 activates at a slightly lower concentration, with Kact = 2.4 ± 0.6 μM. The γ recruitment interface is thus much less important for activation than for binding. The Arf1 switch I mutant I49D completely loses its ability to activate AP-1, consistent with its lack of binding to AP-1. Strikingly, Arf1 back side mutant W172D shows a sharp decrease in AP-1 activation (Fig. 5C, D). Since the W172D mutation actually increases Arf1 binding to AP-1 (Fig. 3C, D), the loss of activation cannot be ascribed to a loss of overall affinity. We hypothesize that, by bridging the AP-1 dimer, the back side of Arf1 couples binding to the conformational change in AP-1. By decoupling binding from the conformational change, the energetic cost of the conformational change is avoided, and the affinity increases. The phenotype of Arf1 W172D connects the mechanism inferred from the crystal structure to the activation of cargo binding as seen in solution (Movie S1).
Reconstitution of synergistic recruitment by Arf1 and PI(4)P
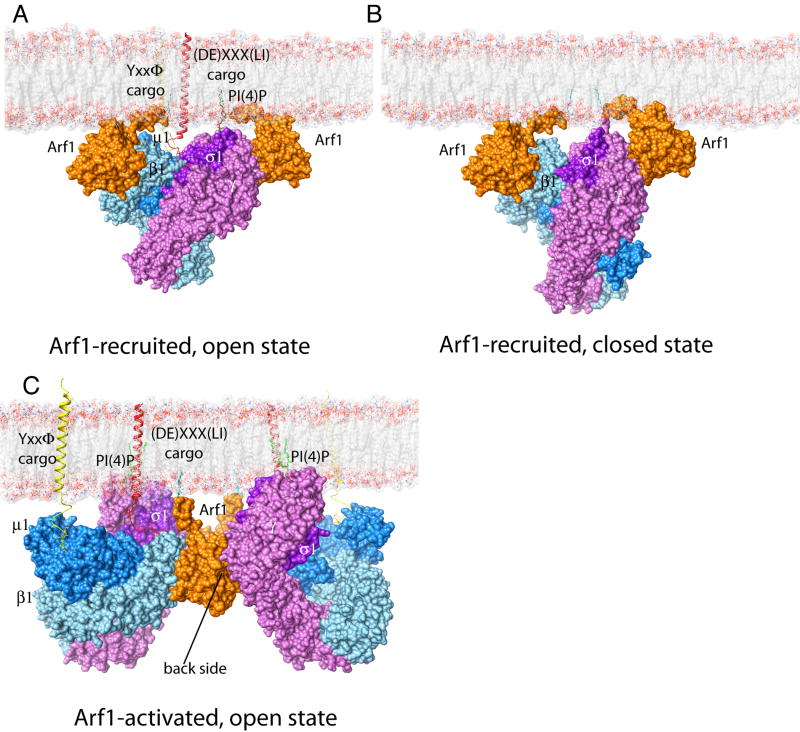
Models for Arf1 recruitment of AP-1 were generated for the open conformation as bound to two copies of full-length myristoylated Arf1-GTP, one copy each of a tyrosine and dileucine signal-bearing cargo, and one molecule of PI(4)P (Fig. 6A), and for the closed state in the absence of cargo (Fig. 6B). The modeling suggests that Arf1, cargo and PI(4)P function synergistically in promoting binding. The models also indicate that simultaneous binding of both recruitment sites is sterically compatible with membrane binding by either the closed or open states. A third model was constructed based on the activated crystallographic dimer (Fig. 6C), which suggested that the γ recruitment site might not be sterically compatible with the membrane-bound activated dimer (Fig. S4).
Figure 6. AP-1 recruitment and activation at the membrane.
Models. (A) A model of AP-1 recruited by two myristoylated Arf1-GTP molecules, in cooperation with PI(4)P on a membrane. The Y- and LL-bearing cargos further bind and stabilize AP-1. (B) The closed conformation of AP-1 is sterically compatible with the simultaneous binding of Arf1 to β1 and to the recruitment site on γ. Therefore, the docking of AP-1 to a membrane via the simultaneous binding to these two Arf1 molecules does not, by itself, appear to be sufficient for activation. (C) The crystallized AP-1 Arf1 dimer can be docked onto a cargo bearing membrane such that the Arf1 myristate moieties, the ends of transmembrane helices of cargo proteins, and PI(4)P all lie in the plane of the membrane surface. See supplmental figure 4
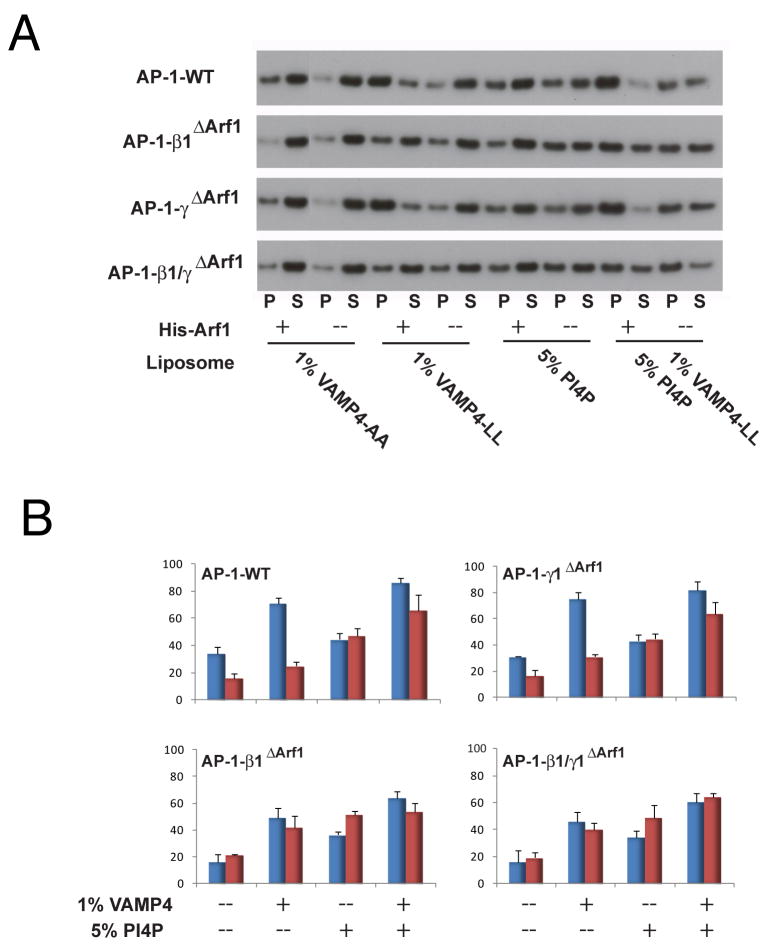
To test whether there is synergy in membrane binding in vitro, PC:PE liposomes with and without PI(4)P were decorated with Arf1 via a His6-Ni2+-NTA linkage. Peptidoliposomes were prepared by chemically conjugating a VAMP4 tail construct to lipids. A VAMP4 LL->AA mutant was constructed as a control. Lipid-peptide conjugates were incorporated into liposomes at 1 mol %. In the absence of Arf1 and PI(4)P, AP-1 bound minimally (16 ± 3%) to PC:PE:VAMP4-AA liposomes (Fig. 7A, B). A moderate increase in binding (24 ± 3%) was seen when wild-type VAMP4 was incorporated in place of VAMP4-AA. In the presence of 50 nM Arf1, however, the majority (71 ± 4%) of AP-1 bound to VAMP4 liposomes. Binding to liposomes containing 5 mol% PI(4)P was significant at 47 ± 6%, but showed little dependence on Arf1. The incorporation of PI(4)P into liposomes bearing VAMP4 and Arf1 drove binding essentially to completion at 86 ± 4%, (Fig. 7A, B). All of the mutant complexes behave like wild-type in the absence of Arf1 (Fig. 7A, B). However, β1ΔArf1 and β1/γΔArf1 are completely insensitive to the presence of Arf1 (Fig. 7A, B), consistent with their complete or nearly complete loss of activation by Arf1. In contrast, γArf1 behaves like wild-type in both the presence and absence of Arf1 (Fig. 7A, B), consistent with the concept that the γ recruitment site does not function in the activation step.
Figure 7. Reconstitution of membrane recruitment and activation by Arf1 and cargo.
(A) Recruitment of the AP-1 core to peptidoliposomes by lipid sedimentation assay. Liposomes were made of 5% DOGS-NTA:POPC:POPE, 1% VAMP4-LL/AA lipopeptide, 1% VAMP4 (1-51) lipopeptide, 5% PI(4)P, or both PI(4)P and VAMP4 lipopeptide. AP-1 cores (20 nM) were incubated with or without His-Arf1-GTP (50 nM) ultracentrifuged to separate the pellet (P) and supernatant fractions (S). Fractions were immunoblotted with anti-μ1 (A), and quantified using ImageJ. The AP-1 membrane binding percentage was calculated according to the formula (P/P+S) × 100% (B) Quantification of sedimentation data. Assays containing Arf1 are colored code in blue and without Arf1 in red. The error bars represent the standard deviation of three replicates.
Discussion
The discovery that AP-1 is recruited to the TGN/endosomes by Arf1-GTP dates back nearly 20 years (Stamnes and Rothman, 1993; Traub et al., 1993). This recruitment event is the prototype for a larger class of heterotetrameric sorting adaptor complex, comprising AP-1, -3, and -4, and COPI. If the Arf GTPase family is considered more broadly to include Arf6, this event might apply to AP-2 as well. The structural basis for Arf1 recognition by this class of sorting adaptor began to emerge with the structure determination of a fragment of γ-COP bound to Arf1 (Yu et al., 2012). Here we have extended these findings by directly visualizing the recognition of Arf1 by β1-adaptin, which we find is the primary binding site for Arf1 on the AP-1 complex. We confirm the prediction that the mode of Arf1 binding described for γ-COP is conserved in γ-adaptin and serves as a second important, albeit lower affinity, binding site for Arf1 on AP-1. Finally, we discover an unexpected role for the back side of Arf1 in allosterically activating AP-1 via a contact with the central part of the γ trunk domain.
Here, we have visualized the active form of the intact AP-1 core in crystals, in the absence of membranes or sorting signals. The activation mechanism is derived from mutational analysis of the coupling between the binding of Arf1 and the dileucine signal peptide of VAMP4 in solution, taken together with the structural analysis. The linchpin of the activation mechanism is the Arf1: β1 interface, the highest affinity Arf1-binding site on AP-1. A molecular pathway for activation was inferred in which formation of contacts between switch I and II of Arf1 and β1, and the back side of Arf1 and γ, pivots the trunk domains and drives their opening. This model requires that at least one additional Arf1-binding site must act as a fulcrum. The ability of the two Arf1-binding sites to open AP-1, and their synergism in high-affinity binding, suggest that there is cross-talk between the two sites. In the crystal, we were able to visualize how a 2:2 arrangement of Arf1 and AP-1 molecules led to activation (Movie S1).
Taken together, the crystallography, biochemical and mutational analyses, and modeling provide a picture of one of the most complex membrane-associated allosteric pathways elucidated to date. The activation of AP-1 within a 2:2 Arf1:AP-1 assembly provides a more complicated contrast to the activation of AP-2 by PI(4,5)P2 (Collins et al., 2002). AP-2 activation by PI(4,5)P2 appears to be fully explained by events occurring within the context of a single AP-2 complex (Collins et al., 2002). The multiplicity of activation and recruitment sites was another surprise. The biochemical analysis shows that the γ recruitment site does not function to any great extent in activation, and modeling suggests that occupancy of this site is sterically incompatible with membrane binding by the dimeric assembly. This raises the intriguing possiblity the initial targeting of AP-1 to the trans-Golgi network might occur through binding of one AP-1 complex in the closed state to two copies of Arf1 via the two recruitment sites (Fig. 6B). Once on the membrane, the presence of cargo could shift the AP-1 equilibrium towards the open state, as shown in Fig. 6A. The high local concentration of AP-1 complexes would then promote dimerization via formation of the Arf1 back side contact (Fig. 6C), stabilizing the open form. The γ recruitment site on Arf1 would then have to dissociate concommitant with AP-1 unlocking in order to allow membrane docking in the cargo-bound conformation.
Very recently, the crystal structure of the HIV-1 Nef in complex with the cytosolic tail of MHC-I and the C-terminal domain of μ1 was determined (Jia et al., 2012). When docked onto the open AP-1 core in the orientation like the one shown in Fig. 6C, Nef presents its myristoyl group to the membrane (Jia et al., 2012). The concepts outlined here suggest that Nef binding is compatible with the dimeric Arf1-activated open state, and it is possible that it could promote activation in addition to its accepted function as an adaptor that links MHC-I to AP-1.
In conclusion, the structural and biochemical details of AP-1 membrane recruitment by two molecules of Arf1 have been elucidated. The most important finding in the study is that Arf1 is capable of activating AP-1 by promoting the open conformation, independent of its role in targeting AP-1 to membranes. A remarkable and unexpected structural pathway for activation has been elucidated. The principles of targeting described for AP-1 appear to extend to the activation of AP-3, -4, and COPI, and perhaps to a lesser extent to Arf6 activation of AP-2. However, it remains to be explored whether activation via dimerization and the Arf1 back side contact occurs in these other systems. The stage is now set for a holistic structural and biophysical understanding of the interplay of Arf GTPases, phosphoinositides, and other elements in CCV biogenesis in the recruitment and activation of AP complexes.
Experimental Procedures
Crystallization and crystallographic analysis
The S-carboxymethylated AP-1 core protein was mixed with Arf1Δ16-Q71L at 1:4 molar ratio in 20 mM Tris pH 7.4, 200 mM NaCl, 0.3 mM TCEP, 5 mM MgCl2 and 2 mM GTP (Axxora). Crystals were grown in 3-5 days at 288 K by hanging drop vapor diffusion against a reservoir containing 0.2 M lithium sulfate, 0.1 M Tris pH 8.5, 0.2 M lithium nitrate, 0.7 M ammonium sulphate, 1 mM TCEP. The final crystal of the complex used for data collection was obtained by micro-seeding at 6 mg/ml AP-1 concentration. Crystals were cryoprotected in the reservoir solution supplemented with 30% glycerol and frozen in liquid nitrogen.
Native data were collected from a single frozen crystal using a MAR CCD detector at beamline 22-ID, Advanced Photon Source. All data were processed and scaled using HKL2000 (HKL research). The crystal diffracted to 7.0 Å resolution, and belonged to space group P64 with unit cell dimensions a=b=267.49 Å, c=191.41 Å, α=β=90°, γ=120°. A molecular replacement solution was found using the AP-2 core/TGN38 peptide structure (PDB: 2XA7) as a search model with Phaser (McCoy et al., 2007). Model building and refinement was carried out with Coot (Emsley et al., 2010) and CNS 1.3 using the DEN method (Schroder et al., 2010) (Table S1). In DEN refinement, it is standard practice to allow several final cycles of refinement that are not constrained by the elastic network. In view of the lower resolution of this data set, these extra cycles were suppressed. Only one B-factor per subunit (or two for μ1 N- and C-terminal domains) was refined. Structural figures were generated with PyMol (W Delano; http://pymol.sourceforge.net/).
Supplementary Material
Highlights.
Crystal structure of the AP-1 clathrin adaptor complex bound to Arf1
Arf1 promotes the open conformation and cargo binding by AP-1
AP-1 is recruited via canonical Arf1 contacts with β and γ subunits
AP-1 is activated by dimerizing via the back side of Arf1
Acknowledgments
We thank G. Mardones for assistance with constructs and W. Yang for critically reading the manuscript. Crystallographic data were collected at Southeast Regional Collaborative Access Team 22-ID beamline at the Advanced Photon Source, Argonne National Laboratory. Use of the Advanced Photon Source was supported by the U. S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract No. W-31-109-Eng-38. This research was supported by the Intramural Programs of NICHD (J.S.B.) and NIDDK (J.H.H.), NIH.
Footnotes
Other methods are described in the extended experimental procedures online.
Accession Numbers: Crystallographic coordinates are being deposited in the RCSB data bank with accession code 4HMY.
Supplemental Information: Supplemental information includes extended experimental procedures, three supplemental figures, two supplemental tables, and one movie, which can be found online at:
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Amor JC, Harrison DH, Kahn RA, Ringe D. Structure of the human ADP-ribosylation factor-1 complexed with GDP. Nature. 1994;372:704–708. doi: 10.1038/372704a0. [DOI] [PubMed] [Google Scholar]
- Austin C, Boehm M, Tooze SA. Site-specific cross-linking reveals a differential direct interaction of class 1, 2, and 3 ADP-ribosylation factors with adaptor protein complexes 1 and 3. Biochemistry. 2002;41:4669–4677. doi: 10.1021/bi016064j. [DOI] [PubMed] [Google Scholar]
- Boehm M, Aguilar RC, Bonifacino JS. Functional and physical interactions of the adaptor protein complex AP-4 with ADP-ribosylation factors (ARFs) EMBO J. 2001;20:6265–6276. doi: 10.1093/emboj/20.22.6265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boll W, Ohno H, Zhou SY, Rapoport I, Cantley LC, Bonifacino JS, Kirchhausen T. Sequence requirements for the recognition of tyrosine-based endocytic signals by clathrin AP-2 complexes. EMBO J. 1996;15:5789–5795. [PMC free article] [PubMed] [Google Scholar]
- Brodsky FM, Chen CY, Knuehl C, Towler MC, Wakeham DE. Biological basket weaving: Formation and function of clathrin- coated vesicles. Annu Rev Cell Dev Biol. 2001;17:517–568. doi: 10.1146/annurev.cellbio.17.1.517. [DOI] [PubMed] [Google Scholar]
- Brunger AT, Adams PD, Fromme P, Fromme R, Levitt M, Schroder GF. Improving the accuracy of macromolecular structure refinement at 7 Å resolution. Structure. 2012;20:957–966. doi: 10.1016/j.str.2012.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvajal-Gonzalez JM, Gravotta D, Mattera R, Diaz F, Bay AP, Roman AC, Schreiner RP, Thuenauer R, Bonifacino JS, Rodriguez-Boulan E. Basolateral sorting of the coxsackie and adenovirus receptor through interaction of a canonical YXX Phi motif with the clathrin adaptors AP-1A and AP-1B. Proc Natl Acad Sci U S A. 2012;109:3820–3825. doi: 10.1073/pnas.1117949109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhuri R, Lindwasser OW, Smith WJ, Hurley JH, Bonifacino JS. Downregulation of CD4 by human immunodeficiency virus type 1 Nef is dependent on clathrin and involves direct interaction of Nef with the AP2 clathrin adaptor. J Virol. 2007;81:3877–3890. doi: 10.1128/JVI.02725-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins BM, McCoy AJ, Kent HM, Evans PR, Owen DJ. Molecular architecture and functional model of the endocytic AP2 complex. Cell. 2002;109:523–535. doi: 10.1016/s0092-8674(02)00735-3. [DOI] [PubMed] [Google Scholar]
- Crottet P, Meyer DM, Rohrer J, Spiess M. ARF1 center dot GTP, tyrosine-based signals, and phosphatidylinositol 4,5-bisphosphate constitute a minimal machinery to recruit the AP-1 clathrin adaptor to membranes. Molecular Biology of the Cell. 2002;13:3672–3682. doi: 10.1091/mbc.E02-05-0309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Paolo G, De Camilli P. Phosphoinositides in cell regulation and membrane dynamics. Nature. 2006;443:651–657. doi: 10.1038/nature05185. [DOI] [PubMed] [Google Scholar]
- Donaldson JG, Jackson CL. ARF family G proteins and their regulators: roles in membrane transport, development and disease. Nature Reviews Molecular Cell Biology. 2011;12:362–375. doi: 10.1038/nrm3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doray B, Lee I, Knisely J, Bu GJ, Kornfeld S. The gamma/sigma 1 and alpha/sigma 2 hemicomplexes of clathrin adaptors AP-1 and AP-2 harbor the dileucine recognition site. Molecular Biology of the Cell. 2007;18:1887–1896. doi: 10.1091/mbc.E07-01-0012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr Sect D-Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farias GG, Cuitino L, Guo X, Ren X, Jarnik M, Mattera R, Bonifacino JS. Signal-Mediated, AP-1/Clathrin-Dependent Sorting of Transmembrane Receptors to the Somatodendritic Domain of Hippocampal Neurons. Neuron. 2012;75:810–823. doi: 10.1016/j.neuron.2012.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaidarov I, Chen Q, Falck JR, Reddy KK, Keen JH. A functional phosphatidylinositol 3,4,5-trisphosphate/phosphoinositide binding domain in the clathrin adaptor AP-2 ‡ subunit. Implications for the endocytic pathway. J Biol Chem. 1996;271:20922–20929. doi: 10.1074/jbc.271.34.20922. [DOI] [PubMed] [Google Scholar]
- Goldberg J. Structural basis for activation of ARF GTPase: mechanisms of guanine nucleotide exchange and GTP-myristoyl switching. Cell. 1998;95:237–248. doi: 10.1016/s0092-8674(00)81754-7. [DOI] [PubMed] [Google Scholar]
- Heldwein EE, Macia E, Jing W, Yin HL, Kirchhausen T, Harrison SC. Crystal structure of the clathrin adaptor protein 1 core. Proc Natl Acad Sci U S A. 2004;101:14108–14113. doi: 10.1073/pnas.0406102101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang FT, Nesterov A, Carter RE, Sorkin A. Trafficking of yellow-fluorescent-protein-tagged mu 1 subunit of clathrin adaptor AP-1 complex in living cells. Traffic. 2001;2:345–357. doi: 10.1034/j.1600-0854.2001.25020506.x. [DOI] [PubMed] [Google Scholar]
- Jackson LP, Kelly BT, McCoy AJ, Gaffry T, James LC, Collins BM, Honing S, Evans PR, Owen DJ. A Large-Scale Conformational Change Couples Membrane Recruitment to Cargo Binding in the AP2 Clathrin Adaptor Complex. Cell. 2010;141:1220–1229. doi: 10.1016/j.cell.2010.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janvier K, Kato Y, Boehm M, Rose JR, Martina JA, Kim BY, Venkatesan S, Bonifacino JS. Recognition of dileucine-based sorting signals from HIV-1 Nef and LIMP-II by the AP-1 gamma-sigma 1 and AP-3 delta-sigma 3 hemicomplexes. J Cell Biol. 2003;163:1281–1290. doi: 10.1083/jcb.200307157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia X, Singh R, Homann R, Yang H, Guatelli J, Xiong X. Structural basis of evasion of cellular adaptive immunity by HIV-1 Nef. Nat Struct Mol Biol. 2012;19:701–706. doi: 10.1038/nsmb.2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly BT, McCoy AJ, Spate K, Miller SE, Evans PR, Honing S, Owen DJ. A structural explanation for the binding of endocytic dileucine motifs by the AP2 complex. Nature. 2008;456:976–U981. doi: 10.1038/nature07422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchhausen T. Clathrin. Annu Rev Biochem. 2000;69:699–727. doi: 10.1146/annurev.biochem.69.1.699. [DOI] [PubMed] [Google Scholar]
- Krauss M, Kinuta M, Wenk MR, De Camilli P, Takei K, Haucke V. ARF6 stimulates clathrin/AP-2 recruitment to synaptic membranes by activating phosphatidylinositol phosphate kinase type Igamma. J Cell Biol. 2003;162:113–124. doi: 10.1083/jcb.200301006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeBorgne R, Griffiths G, Hoflack B. Mannose 6-phosphate receptors and ADP-ribosylation factors cooperate for high affinity interaction of the AP-1 Golgi assembly proteins with membranes. J Biol Chem. 1996;271:2162–2170. doi: 10.1074/jbc.271.4.2162. [DOI] [PubMed] [Google Scholar]
- Lee I, Doray B, Govero J, Kornfeld S. Binding of cargo sorting signals to AP-1 enhances its association with ADP ribosylation factor 1-GTP. J Cell Biol. 2008a;180:467–472. doi: 10.1083/jcb.200709037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee I, Drake MT, Traub LM, Kornfeld S. Cargo-sorting signals promote polymerization of adaptor protein-1 in an Arf-1.GTP-independent manner. Archives of Biochemistry & Biophysics. 2008b;479:63–68. doi: 10.1016/j.abb.2008.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang JO, Sung TC, Morris AJ, Frohman MA, Kornfeld S. Different domains of mammalian ADP-ribosylation factor 1 mediate interaction with selected target proteins. J Biol Chem. 1997;272:33001–33008. doi: 10.1074/jbc.272.52.33001. [DOI] [PubMed] [Google Scholar]
- Mattera R, Boehm M, Chaudhuri R, Prabhu Y, Bonifacino JS. Conservation and Diversification of Dileucine Signal Recognition by Adaptor Protein (AP) Complex Variants. J Biol Chem. 2011;286:2022–2030. doi: 10.1074/jbc.M110.197178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montagnac G, de Forges H, Smythe E, Gueudry C, Romao M, Salamero J, Chavrier P. Decoupling of Activation and Effector Binding Underlies ARF6 Priming of Fast Endocytic Recycling. Curr Biol. 2011;21:574–579. doi: 10.1016/j.cub.2011.02.034. [DOI] [PubMed] [Google Scholar]
- Ohno H, Fournier MC, Poy G, Bonifacino JS. Structural determinants of interaction of tyrosine-based sorting signals with the adaptor medium chains. J Biol Chem. 1996;271:29009–29015. doi: 10.1074/jbc.271.46.29009. [DOI] [PubMed] [Google Scholar]
- Ohno H, Stewart J, Fournier MC, Bosshart H, Rhee I, Miyatake S, Saito T, Gallusser A, Kirchhausen T, Bonifacino JS. Interaction of tyrosine-based sorting signals with clathrin-associated proteins. Science. 1995;269:1872–1875. doi: 10.1126/science.7569928. [DOI] [PubMed] [Google Scholar]
- Ooi CE, Dell'Angelica EC, Bonifacino JS. ADP-ribosylation factor 1 (ARF1) regulates recruitment of the AP- 3 adaptor complex to membranes. J Cell Biol. 1998;142:391–402. doi: 10.1083/jcb.142.2.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen DJ, Collins BM, Evans PR. Adaptors for clathrin coats: Structure and function. Annu Rev Cell Dev Biol. 2004;20:153–191. doi: 10.1146/annurev.cellbio.20.010403.104543. [DOI] [PubMed] [Google Scholar]
- Owen DJ, Evans PR. A structural explanation for the recognition of tyrosine-based endocytotic signals. Science. 1998;282:1327–1332. doi: 10.1126/science.282.5392.1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paleotti O, Macia E, Luton F, Klein S, Partisani M, Chardin P, Kirchhausen T, Franco M. The small G-protein Arf6(GTP) recruits the AP-2 adaptor complex to membranes. J Biol Chem. 2005;280:21661–21666. doi: 10.1074/jbc.M503099200. [DOI] [PubMed] [Google Scholar]
- Poupart ME, Fessart D, Cotton M, Laporte SA, Claing A. ARF6 regulates angiotensin II type 1 receptor endocytosis by controlling the recruitment of AP-2 and clathrin. Cell Signal. 2007;19:2370–2378. doi: 10.1016/j.cellsig.2007.07.015. [DOI] [PubMed] [Google Scholar]
- Robinson MS. Adaptable adaptors for coated vesicles. Trends Cell Biol. 2004;14:167–174. doi: 10.1016/j.tcb.2004.02.002. [DOI] [PubMed] [Google Scholar]
- Rohde G, Wenzel D, Haucke V. A phosphatidylinositol (4,5)-bisphosphate binding site within mu 2-adaptin regulates clathrin-mediated endocytosis. J Cell Biol. 2002;158:209–214. doi: 10.1083/jcb.200203103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroder GF, Levitt M, Brunger AT. Super-resolution biomolecular crystallography with low-resolution data. Nature. 2010;464:1218–1222. doi: 10.1038/nature08892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seaman MNJ, Sowerby PJ, Robinson MS. Cytosolic and membrane-associated proteins involved in the recruitment of AP-1 adaptors onto the trans-Golgi network. J Biol Chem. 1996;271:25446–25451. doi: 10.1074/jbc.271.41.25446. [DOI] [PubMed] [Google Scholar]
- Serafini T, Orci L, Amherdt M, Brunner M, Kahn RA, Rothman JE. ADP-ribosylation factor is a subunit of the coat of Golgi- derived COP-coated vesicles: a novel role for a GTP-binding protein. Cell. 1991;67:239–253. doi: 10.1016/0092-8674(91)90176-y. [DOI] [PubMed] [Google Scholar]
- Shiba T, Kawasaki M, Takatsu H, Nogi T, Matsugaki N, Igarashi N, Suzuki M, Kato R, Nakayama K, Wakatsuki S. Molecular mechanism of membrane recruitment of GGA by ARF in lysosomal protein transport. Nat Struct Biol. 2003;10:386–393. doi: 10.1038/nsb920. [DOI] [PubMed] [Google Scholar]
- Stamnes MA, Rothman JE. The binding of AP-1 clathrin adaptor particles to Golgi membranes requires ADP-ribosylation factor, a small GTP-binding protein. Cell. 1993;73:999–1005. doi: 10.1016/0092-8674(93)90277-w. [DOI] [PubMed] [Google Scholar]
- Traub LM, Ostrom JA, Kornfeld S. Biochemical dissection of AP-1 recruitment onto Golgi membranes. J Cell Biol. 1993;123:561–573. doi: 10.1083/jcb.123.3.561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Sun HQ, Macia E, Kirchhausen T, Watson H, Bonifacino JS, Yin HL. PIP promotes the recruitment of the GGA adaptor proteins to the trans-Golgi TS recognition of the ubiquitin network and regulates their sorting signal. Molecular Biology of the Cell. 2007;18:2646–2655. doi: 10.1091/mbc.E06-10-0897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu XC, Breitman M, Goldberg J. A Structure-Based Mechanism for Arf1-Dependent Recruitment of Coatomer to Membranes. Cell. 2012;148:530–542. doi: 10.1016/j.cell.2012.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.