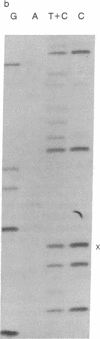
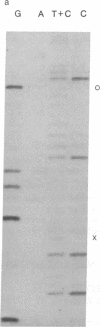
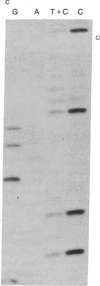
Abstract
The nucleotide sequence of the recognition site for the restriction-modification enzyme of Escherichia coli B (SB site) has been determined. The recognition site is a 15-nucleotide sequence consisting of the trimer 5'TGA3', followed by an 8-nucleotide domain of variable sequence, which in turn is followed by tetramer 5'TGCT3'. The sequence has no 2-fold rotational symmetry. Single base changes in the constant nucleotide domains result in the loss of sensitivity to both restriction and modification. Our data are also consistent with modification occurring by methylation of two adenine residues per SB site: one on the adenine of the trimer 5'TGA3' and the other on the complementary strand on the adenine complementary to the first thymine of the tetramer 5'TGCT3'. All nine independently isolated spontaneous mutants at the SB1 site of bacteriophage f1 are caused by a G-to-T transversion. Mutations at the SB2 site are caused by various single base changes.
Full text
PDF
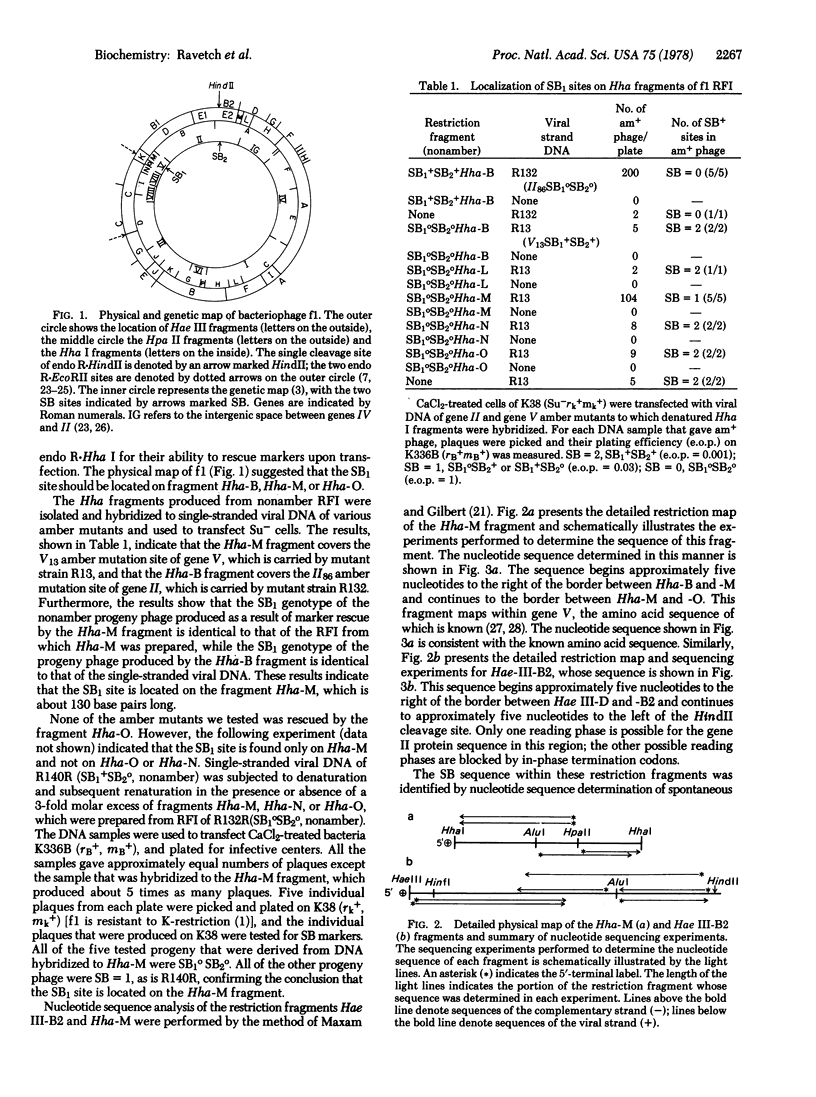



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Air G. M. Amino acid sequences from the gene F (capsid) protein of bacteriophage phiX174. J Mol Biol. 1976 Nov 15;107(4):433–443. doi: 10.1016/s0022-2836(76)80076-9. [DOI] [PubMed] [Google Scholar]
- Air G. M., Blackburn E. H., Sanger F., Coulson A. R. The nucleotide and amino acid sequences of the N (5') terminal region of gene G of bacteriophage phiphiX 174. J Mol Biol. 1975 Aug 25;96(4):703–719. doi: 10.1016/0022-2836(75)90147-3. [DOI] [PubMed] [Google Scholar]
- Arber W. Host specificity of DNA produced by Escherichia coli. 9. Host-controlled modification of bacteriophage fd. J Mol Biol. 1966 Oct;20(3):483–496. doi: 10.1016/0022-2836(66)90004-0. [DOI] [PubMed] [Google Scholar]
- Arber W., Kühnlein U. Mutationeller Verlust B-spezifischer Restriktion des Bakteriophagen fd. Pathol Microbiol (Basel) 1967;30(6):946–952. [PubMed] [Google Scholar]
- Bonner W. M., Laskey R. A. A film detection method for tritium-labelled proteins and nucleic acids in polyacrylamide gels. Eur J Biochem. 1974 Jul 1;46(1):83–88. doi: 10.1111/j.1432-1033.1974.tb03599.x. [DOI] [PubMed] [Google Scholar]
- Boon T., Zinder N. D. Genotypes produced by individual recombination events involving bacteriophage f1. J Mol Biol. 1971 May 28;58(1):133–151. doi: 10.1016/0022-2836(71)90237-3. [DOI] [PubMed] [Google Scholar]
- Cuypers T., van der Ouderaa F. J., de Jong W. W. The amino acid sequence of gene 5 protein of bacteriophage M 13. Biochem Biophys Res Commun. 1974 Jul 24;59(2):557–563. doi: 10.1016/s0006-291x(74)80016-1. [DOI] [PubMed] [Google Scholar]
- Edgell M. H., Hutchison C. A., 3rd, Sclair M. Specific endonuclease R fragments of bacteriophage phiX174 deoxyribonucleic acid. J Virol. 1972 Apr;9(4):574–582. doi: 10.1128/jvi.9.4.574-582.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enea V., Horiuchi K., Turgeon B. G., Zinder N. D. Physical map of defective interfering particles of bacteriophage f1. J Mol Biol. 1977 Apr 25;111(4):395–414. doi: 10.1016/s0022-2836(77)80061-2. [DOI] [PubMed] [Google Scholar]
- Enea V., Vovis G. F., Zinder N. D. Genetic studies with heteroduplex DNA of bacteriophage fl. Asymmetric segregation, base correction and implications for the mechanism of genetic recombination. J Mol Biol. 1975 Aug 15;96(3):495–509. doi: 10.1016/0022-2836(75)90175-8. [DOI] [PubMed] [Google Scholar]
- Enea V., Zinder N. D. Heteroduplex DNA: a recombinational intermediate in bacteriophage f1. J Mol Biol. 1976 Feb 15;101(1):25–38. doi: 10.1016/0022-2836(76)90064-4. [DOI] [PubMed] [Google Scholar]
- Haberman A., Heywood J., Meselson M. DNA modification methylase activity of Escherichia coli restriction endonucleases K and P. Proc Natl Acad Sci U S A. 1972 Nov;69(11):3138–3141. doi: 10.1073/pnas.69.11.3138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horiuchi K., Vovis G. F., Enea V., Zinder N. D. Cleavage map of bacteriophage f1: location of the Escherichia coli B-specific modification sites. J Mol Biol. 1975 Jun 25;95(2):147–165. doi: 10.1016/0022-2836(75)90388-5. [DOI] [PubMed] [Google Scholar]
- Horiuchi K., Vovis G. F., Zinder N. D. Effect of deoxyribonucleic acid length on the adenosine triphosphatase activity of Escherichia coli restriction endonuclease B. J Biol Chem. 1974 Jan 25;249(2):543–552. [PubMed] [Google Scholar]
- Horiuchi K., Zinder N. D. Cleavage of bacteriophage fl DNA by the restriction enzyme of Escherichia coli B. Proc Natl Acad Sci U S A. 1972 Nov;69(11):3220–3224. doi: 10.1073/pnas.69.11.3220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kühnlein U., Arber W. Host specificity of DNA produced by Escherichia coli. XV. The role of nucleotide methylation in in vitro B-specific modification. J Mol Biol. 1972 Jan 14;63(1):9–19. doi: 10.1016/0022-2836(72)90518-9. [DOI] [PubMed] [Google Scholar]
- Laskey R. A., Mills A. D. Quantitative film detection of 3H and 14C in polyacrylamide gels by fluorography. Eur J Biochem. 1975 Aug 15;56(2):335–341. doi: 10.1111/j.1432-1033.1975.tb02238.x. [DOI] [PubMed] [Google Scholar]
- Lautenberger J. A., Kan N. C., Lackey D., Linn S., Edgell M. H., Hutchison C. A., 3rd Recognition site of Escherichia coli B restriction enzyme on phi XsB1 and simian virus 40 DNAs: an interrupted sequence. Proc Natl Acad Sci U S A. 1978 May;75(5):2271–2275. doi: 10.1073/pnas.75.5.2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lautenberger J. A., Linn S. The deoxyribonucleic acid modification and restriction enzymes of Escherichia coli B. I. Purification, subunit structure, and catalytic properties of the modification methylase. J Biol Chem. 1972 Oct 10;247(19):6176–6182. [PubMed] [Google Scholar]
- Lyons L. B., Zinder N. D. The genetic map of the filamentous bacteriophage f1. Virology. 1972 Jul;49(1):45–60. doi: 10.1016/s0042-6822(72)80006-0. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci U S A. 1977 Feb;74(2):560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meselson M., Yuan R. DNA restriction enzyme from E. coli. Nature. 1968 Mar 23;217(5134):1110–1114. doi: 10.1038/2171110a0. [DOI] [PubMed] [Google Scholar]
- Model P., Zinder N. D. In vitro synthesis of bacteriophage f1 proteins. J Mol Biol. 1974 Feb 25;83(2):231–251. doi: 10.1016/0022-2836(74)90389-1. [DOI] [PubMed] [Google Scholar]
- Nakashima Y., Dunker A. K., Marvin D. A., Konigsberg W. The amino acid sequence of a DNA binding protein, the gene 5 product of fd filamentous bacteriophage. FEBS Lett. 1974 Apr 1;40(2):290–292. doi: 10.1016/0014-5793(74)80246-2. [DOI] [PubMed] [Google Scholar]
- Ravetch J. V., Horiuchi K., Zinder N. D. Nucleotide sequences near the origin of replication of bacteriophage f1. Proc Natl Acad Sci U S A. 1977 Oct;74(10):4219–4222. doi: 10.1073/pnas.74.10.4219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravetch J., Horiuchi K., Model P. Mapping of bacteriophage f1 ribosome binding sites to their cognate genes. Virology. 1977 Sep;81(2):341–351. doi: 10.1016/0042-6822(77)90150-7. [DOI] [PubMed] [Google Scholar]
- Roberts R. J. Restriction endonucleases. CRC Crit Rev Biochem. 1976 Nov;4(2):123–164. doi: 10.3109/10409237609105456. [DOI] [PubMed] [Google Scholar]
- Sanger F., Air G. M., Barrell B. G., Brown N. L., Coulson A. R., Fiddes C. A., Hutchison C. A., Slocombe P. M., Smith M. Nucleotide sequence of bacteriophage phi X174 DNA. Nature. 1977 Feb 24;265(5596):687–695. doi: 10.1038/265687a0. [DOI] [PubMed] [Google Scholar]
- Schnegg B., Hofschneider P. H. Mutant of phi X174 accessible to host-controlled modification. J Virol. 1969 May;3(5):541–542. doi: 10.1128/jvi.3.5.541-542.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp P. A., Sugden B., Sambrook J. Detection of two restriction endonuclease activities in Haemophilus parainfluenzae using analytical agarose--ethidium bromide electrophoresis. Biochemistry. 1973 Jul 31;12(16):3055–3063. doi: 10.1021/bi00740a018. [DOI] [PubMed] [Google Scholar]
- Smith J. D., Arber W., Kühnlein U. Host specificity of DNA produced by Escherichia coli. XIV. The role of nucleotide methylation in in vivo B-specific modification. J Mol Biol. 1972 Jan 14;63(1):1–8. doi: 10.1016/0022-2836(72)90517-7. [DOI] [PubMed] [Google Scholar]
- Vovis G. F., Horiuchi K., Hartman N., Zinder N. D. Restriction endonuclease B and f1 heteroduplex DNA. Nat New Biol. 1973 Nov 7;246(149):13–16. doi: 10.1038/newbio246013a0. [DOI] [PubMed] [Google Scholar]
- Vovis G. F., Horiuchi K., Zinder N. D. Endonuclease R-EcoRII restriction of bacteriophage f1 DNA in vitro: ordering of genes V and VII, location of an RNA promotor for gene VIII. J Virol. 1975 Sep;16(3):674–684. doi: 10.1128/jvi.16.3.674-684.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vovis G. F., Horiuchi K., Zinder N. D. Kinetics of methylation of DNA by a restriction endonuclease from Escherichia coli B. Proc Natl Acad Sci U S A. 1974 Oct;71(10):3810–3813. doi: 10.1073/pnas.71.10.3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vovis G. F., Zinder N. D. Methylation of f1 DNA by a restriction endonuclease from escherichia coli B. J Mol Biol. 1975 Jul 15;95(4):557–568. doi: 10.1016/0022-2836(75)90317-4. [DOI] [PubMed] [Google Scholar]
- WATSON J. D., CRICK F. H. Molecular structure of nucleic acids; a structure for deoxyribose nucleic acid. Nature. 1953 Apr 25;171(4356):737–738. doi: 10.1038/171737a0. [DOI] [PubMed] [Google Scholar]
- van Ormondt H., Lautenberger J. A., Linn S., de Waard A. Methylated oligonucleotides derived from bacteriophage fd RF-DNA modified in vitro by E. coli B modification methylase. FEBS Lett. 1973 Jul 1;33(2):177–180. doi: 10.1016/0014-5793(73)80186-3. [DOI] [PubMed] [Google Scholar]