Abstract
Lefty expression has been recognized as a stemness marker because Lefty is enriched both in undifferentiated embryonic stem cells (ESCs) and in blastocysts. Here, we examined the function of Lefty1 and Lefty2 in the maintenance of self-renewal and pluripotency of mouse ESCs (mESCs). Suppression of Lefty1 or Lefty2 expression in mESCs did not alter the self-renewal properties of mESCs under nondifferentiating conditions, but suppression of these genes did affect Smad2 phosphorylation and differentiation. Lefty1 knockdown mESCs showed enhanced phosphorylation of Smad2 and increased differentiation potential, whereas Lefty2 knockdown mESCs exhibited reduced phosphorylation of Smad2 and enhanced self-renewal in the presence of a differentiation signal. In vivo, teratomas developed from Lefty2 knockdown mESCs contained massive expansions of immature neuroepithelium, a marker of malignant teratomas. Taken together, these results suggest that optimal expression of Lefty1 and Lefty2 is critical for the balanced differentiation of mESCs into three germ layers.
Introduction
Transforming growth factor (TGF)-β family members are enriched in embryonic stem cells (ESCs), suggesting that these proteins are part of critical signaling pathways that maintain the stemness and pluripotency of these cells [1]. Among inhibitors of TGF-β ligands, only Lefty is enriched both in undifferentiated ESCs and in blastocysts, which indicates that Lefty expression is a marker of stemness [2–4]. Lefty is also highly expressed in the inner cell mass and trophoectoderm [2]. In mouse ESCs (mESCs), Lefty expression is regulated by the binding of a cooperative transcriptional complex composed of Klf4, Oct4, and Sox2 to the proximal element of the Lefty1 promoter [5]. However, unlike ESC self-renewal genes such as Oct4, Lefty expression is not quenched upon differentiation of ESCs. When leukemia inhibitory factor (LIF) is removed from the culture medium of mESCs, the expression of Lefty increases within 48 h of cytokine withdrawal [6]. Similarly, differentiation of ESCs to an embryoid body (EB) leads to increased expression of Lefty [7]. Retinoic acid (RA)–mediated differentiation of ESCs also leads to an increase in Lefty expression in mouse embryonal carcinoma cells [8]. Therefore, Lefty might be important both for the maintenance of self-renewal and the exit from this state that leads to the differentiation of ESCs.
In both humans and mice, the Lefty genes have been localized to chromosome 1 and the locus contains two genes—Lefty1 and Lefty2—with the same transcriptional orientation, and one pseudogene—Lefty3—that has a reverse transcriptional orientation [9,10]. Lefty1 has 91% sequence identity and shares 331 amino acids with Lefty2, indicating that Lefty1 and Lefty2 are closely related to each other. Lefty1 and Lefty2 both block Nodal signaling by binding Nodal and its EGF-CFC coreceptors, such as TDGF-1/Cripto. These interactions prevent the assembly of an active Nodal/Activin receptor complex [11,12].
Even though several findings suggest that TGF-β signaling is required for the maintenance of pluripotency of ESCs [13], the precise role of Lefty1 or Lefty2 in ESCs remains to be elucidated. Recently, our research group reported that the Lefty1-Nodal-Smad2 pathway regulated by Tcea3 is an innate program to determine cell fate choices between self-replication and commitment to differentiation [14]. Here, we studied the function of Lefty isoforms in relation to pluripotency by examining the effect of Lefty1 or Lefty2 suppression on the self-renewal and differentiation of mESCs. Suppression of Lefty1 and Lefty2 produced opposing effects on the differentiation of mESCs. Lefty1 knockdown mESCs (Lefty1 KD) showed enhanced phosphorylation of Smad2 and enhanced differentiation potential, whereas Lefty2 knockdown mESCs (Lefty2 KD) exhibited reduced phosphorylation of Smad2, which might be the result of enhanced expression of Lefty1. In addition, Lefty2 KD mESCs showed enhanced self-renewal and reduced differentiation in response to a differentiation signal. An in vivo teratoma assay showed that Lefty2 KD mESCs formed more malignant tumors that had higher expression of self-renewal factors, such as Oct4 and Sox2. These results suggest that balanced expression of Lefty1 and Lefty2 is critical to maintain the pluripotency of mESCs and that optimal expression of Lefty2 is essential to inhibit carcinogenesis of ESCs.
Materials and Methods
Cell culture, EB formation, and in vitro differentiation of mESCs
J1 mESCs (Cat. No. SCRC-1010) were purchased from ATCC (www.atcc.org) and maintained as described previously [15]. The mESCs were cultured in ESC medium [Dulbecco's modified Eagle's medium (DMEM) supplemented with 15% fetal calf serum (HyClone), 0.1 mM 2-mercaptoethanol (Sigma), 100 U/mL penicillin, 100 μg/mL streptomycin, 2 mM glutamine (Gibco), and 1,000 U/mL LIF (Chemicon)]. To induce mESC differentiation, mESCs were cultured in LIF-deficient ESC medium containing 100 nM all-trans RA. To form EBs, mESCs were trypsinized to achieve a single-cell suspension and, subsequently, cultured on uncoated Petri dishes in ESC medium without LIF. The medium was changed every 2 days for mESC culture or differentiation. Four days after primary EB formation, EBs were collected and dissociated into single cells by trypsinization and trituration. These EB cells were replated into ESC medium without LIF and the efficiency of secondary EB production was assessed after 10 days, to determine the proportion of undifferentiated mESCs present in the primary EBs. Activin-induced mesoendoderm differentiation was performed as previously described [16]. Briefly, ESCs were cultured as a monolayer in gelatinized feeder-free six-well plates with an initial plating density of 1×105 cells/well and the time when 25 ng/mL of Activin was added was counted as day 0. The medium was composed of a 1:1 mixture of DMEM/F12 (Invitrogen) supplemented with N2 supplement (Stem Cell Technologies) and NeuralBasal medium (Invitrogen) supplemented with B27 supplement (Stem Cell Technologies) and β-mercaptoethanol. Cells were harvested at day 4 for gene expression analysis.
Genetic modification of mESCs
shRNA plasmids that target mouse Lefty1 and Lefty2 were purchased (RMM4534-EG13590 and RMM4534-EG320202; Open Biosystems) to generate knockdown cell lines of mESCs. Plasmids were transfected into mESCs with Lipofectamine 2000 (Invitrogen) and stably transfected lines were established according to the manufacturer's instructions. Five Lefty1 KD and five Lefty2 KD cell lines were established, and two different cell lines of each knockdown were used to study the effect of suppression of Lefty1 or Lefty2 in mESCs.
Alkaline phosphatase activity
Alkaline phosphatase activity was measured using the AnaSpec kit (No. 71230; AnaSpec, www.anaspec.com) according to the manufacturer's instructions.
Cell cycle analysis by flow cytometry
Samples (1×106 cells) were washed with 0.5 mL phosphate-buffered saline (PBS) and fixed with ice-cold 70% (v/v) ethanol at 4°C for at least 1 h. After washing in PBS again, cells were incubated with staining buffer containing 50 μg/mL propidium iodide (Sigma), 0.2 mg/mL RNase A (Sigma), and 0.1% (v/v) Triton X-100 in PBS in the dark for 40 min at 37°C. The DNA content was analyzed using an FACSCalibur flow cytometer (BD Biosciences) and the data were analyzed using Cell Quest software (BD Biosciences).
RNA extraction and real-time reverse transcriptase (RT)-PCR
Total RNA from mESCs and teratomas was extracted using TRIzol (Invitrogen), and 2–5 μg of total RNA was reverse-transcribed using the SuperScriptII™ First-Strand Synthesis System (Invitrogen) according to the manufacturer's instructions. The cDNA was treated with 2 U of RNase H (Invitrogen) for 20 min at 37°C. Real-time RT-PCR was carried out using cDNAs with the Quantitect SYBR Green PCR kit (Qiagen). Reactions were carried out in triplicate using an Exicycler™ 96 (Bioneer). For quantification, target genes were normalized against glyceraldehyde-3-phosphate dehydrogenase (Gapdh). PCR primers used in this study are listed in Supplementary Table S1 (Supplementary Data are available online at www.liebertpub.com/scd).
Immunoblot analysis
For immunoblotting assays, cells were washed twice with cold PBS, lysed with tissue lysis buffer [20 mM Tris-base (pH 7.4), 137 mM NaCl, 2 mM EDTA, 1% Triton X-100, 25 mM β-glycerophosphate, 2 mM sodium pyrophosphate, 10% glycerol, 1 mM sodium orthovanadate, 1 mM phenylmethylsulfonyl fluoride, and 1 mM benzamidine], and clarified by centrifugation at 12,000 g for 10 min. Whole-cell extracts were prepared and 20–50 μg of proteins was resolved by sodium dodecyl sulfate polyacrylamide gel electrophoresis, transferred to polyvinylidene difluoride membranes (Perkin Elmer Life Sciences), and probed using antibodies against Oct4 (sc-9081; Santa Cruz), Sox2 (sc-20088; Santa Cruz), pStat3 (Tyr-705) (No. 9131; Cell Signaling Technology), Stat3 (sc-482; Santa Cruz), pSmad2 (No. 3101; Cell Signaling Technology), Smad2/3 (sc-8332; Santa Cruz), or β-actin (sc-47778; Santa Cruz). Immunoreactivity was detected by enhanced chemiluminescence (ECL; Amersham). For quantitative analysis, the mean density of each band was measured using Multi Gauge V3.0 software.
Teratoma formation
For teratoma formation assays, cells were trypsinized, and 5×105 cells were suspended in a DMEM/Matrigel solution [BD Biosciences, Inc.; 1:1 ratio (v/v)]. The cell/Matrigel suspension was then injected subcutaneously into NOD/SCID mice (Charles River Laboratories). Teratoma formation of Lefty1 KD and Lefty2 KD was examined at 6 and 4 weeks after injection, respectively. The experiments were reviewed and approved by the Institutional Animal Care and Use Committee of CHA University. All procedures were performed in accordance with the Guidelines for the Care and Use of Laboratory Animals published by the U.S. National Institutes of Health (NIH publication No. 85-23, revised 1996).
Statistical analysis
Graphical data are presented as mean±SD. Each experiment was performed at least three times and subjected to statistical analysis. Significant differences between two groups were determined using Student's t-test. P<0.05 was considered significant. P<0.05 and P<0.01 were marked as ** and *. Statistical analysis was performed using the SAS statistical package v.9.13 (SAS, Inc.).
Results
Lefty1 critically controls the differentiation potential of mESCs
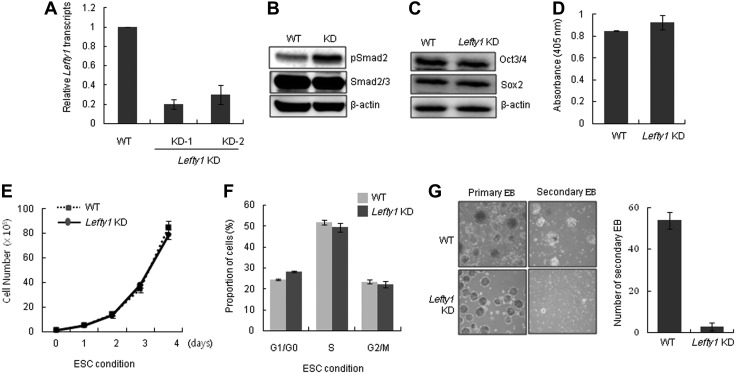
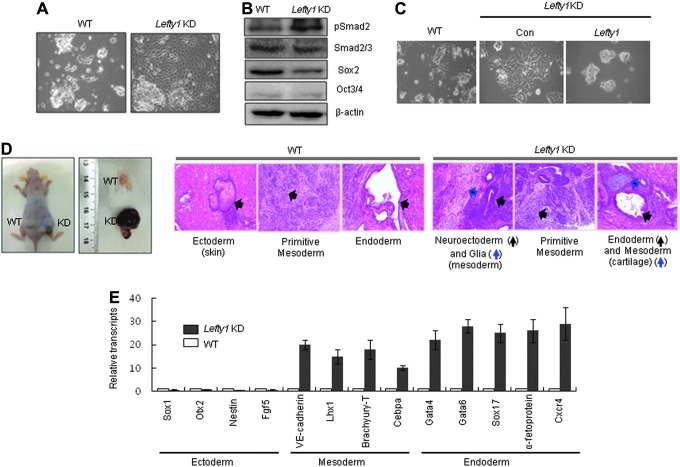
To study the role of Lefty1 and Lefty2 in mESCs, we first generated mESC lines that stably expressed shRNA for Lefty1 or Lefty2 transcripts. The relative transcript levels of Lefty1 in the Lefty1 KD were <50% of wild type (WT) (Fig. 1A). Because Lefty inhibits phosphorylation of Smad2/3 by blocking Nodal/Activin signaling, we examined the phosphorylation of Smad2 in Lefty1 KD cell lines. As expected, knockdown of Lefty1 expression resulted in an increase in Smad2 phosphorylation in ESCs (Fig. 1B). The expression of self-renewal factors, such as Oct3/4 and Sox2, was not affected by the suppression of Lefty1 (Fig. 1C). In addition, the level of alkaline phosphatase activity in Lefty1 KD cells was comparable with that in the WT, indicating that changes in the level of Lefty1 expression did not affect the self-renewal of mESCs (Fig. 1D). The cell proliferation and cell cycle profiles of Lefty1 KD cells were similar to those of WT cells (Fig. 1E, F). However, the efficiency of secondary EB formation by Lefty1 KD cells was much lower than that of WT cells (Fig. 1G). Secondary EB formation reflects the ability of mESCs to maintain their self-renewal capacity [17]. Previously, we reported that Lefty1-Nodal-pSmad2 signals regulated by Tcea3 form a critical pathway that regulates cell fate choices between self-renewal and commitment to differentiation [14]. As Lefty1 is a negative regulator of Nodal signaling and Smad2-mediated Activin/Nodal signaling is required for proper differentiation of mESCs toward the mesoendoderm lineages [16], we hypothesized that suppression of Lefty1 expression would facilitate differentiation of mESCs. As expected, upon removal of LIF and addition of RA, Lefty1 KD cells rapidly developed epithelial-like outgrowths by day 2 whereas WT mESCs still maintained ESC-like colonies (Fig. 2A). Consistent with the differentiating cell morphology, the expression of lineage markers was markedly increased in differentiating Lefty1 KD cells compared with that in the WT (Supplementary Fig. S1). Despite the facilitated differentiation of Lefty1 KD cells, cell proliferation and the cell cycle profiles of Lefty1 KD cells were similar to those of the WT (Supplementary Fig. S2A, B). Smad2 phosphorylation in the differentiating Lefty1 KD cells was maintained at a higher level than that in WT cells, whereas Sox2 levels rapidly decreased in differentiating Lefty1 KD cells (Fig. 2B). When a Lefty1-expressing plasmid was transiently introduced, the differentiation properties of Lefty1 KD cells became similar to WT (Fig. 2C). Consistent with the in vitro differentiation properties, Lefty1 KD cells developed teratomas more rapidly in vivo than WT (Fig. 2D, left panel). When examined by histological staining, teratomas harvested 4 weeks after transplantation of Lefty1 KD cells showed well-developed and differentiated glia (mesoderm) and cartilage (mesoderm), indicating that Lefty1 KD cells had a tendency to differentiate into cells of the mesodermal lineage (Fig. 2D, right panel).
FIG. 1.
Suppression of Lefty1 expression does not affect the self-renewal of mESCs. (A) Expression level of Lefty1 in two Lefty1 KD (KD-1 and KD-2) cell lines was analyzed by real-time reverse transcriptase (RT)-PCR. (B) Phospho-Smad2 of Lefty1 KD cells under self-renewal culture conditions was analyzed by immunoblot analysis. (C) Expression of Oct4 and Sox2 by Lefty1 KD cells under ESC culture conditions was analyzed by immunoblot analysis. (D) Alkaline phosphatase activity of Lefty1 KD cells was compared with that in WT cells. (E) The proliferation of Lefty1 KD cells was compared with that of WT cells at the indicated times. (F) The cell cycle distribution of Lefty1 KD cells was compared with that of WT cells by flow cytometry. (G) Primary EBs and secondary EBs formed by Lefty1 KD and WT cells were examined by light microscopy after 4 and 10 days in culture, respectively (left panel). The number of secondary EBs was compared between WT and Lefty2 KD mESCs (right panel). All values are mean±SD from at least three independent experiments. Lefty1 KD, stable Lefty1-knocked-down mESC line; WT, wild-type mESCs; EB, embryoid body; mESCs, mouse embryonic stem cells.
FIG. 2.
Suppression of Lefty1 expression enhances differentiation potential of mESCs. (A) Lefty1 KD cells spontaneously differentiate when leukemia inhibitory factor (LIF) is removed and RA is added for 3 days. Cell morphology was examined by light microscopy. (B) Expression of phospho-Smad2 and self-renewal markers, including Oct4 and Sox2, by differentiating Lefty1 KD cells was analyzed by immunoblotting. (C) Lefty1 KD cells were transfected with control (Con) or a Lefty1 expression plasmid and then spontaneously differentiated for 3 days. Differentiating cells were examined by light microscopy. (D) Lefty1 KD and control mESCs were injected into NOD/SCID mice and the size of the teratomas was examined 4 weeks after injection. Teratomas were stained with hematoxylin-eosin (H&E) and the skin, primitive mesoderm, glia, and cartilage in the teratomas were identified by examination under a light microscope. (E) Lineage-specific marker expression in Lefty1 KD teratomas was compared with that of WT by real-time RT-PCR. All values are mean±SD from at least three independent experiments. Lefty1 KD, stable Lefty1-knocked-down mESC line; WT, wild-type mESCs; RA, retinoic acid.
Markedly enhanced expression of mesoderm and endoderm markers in 4-week-old Lefty1 KD teratomas suggested that larger teratoma formation was an in vivo indicator of differentiation potential (Fig. 2E). To investigate whether the activation of TGF-β signaling in the Lefty1 KD cells leads to the increased differentiation of mESCs, we examined the effects of a chemical inhibitor of TGF-β signaling upon the differentiation of Lefty1 KD cells. Treatment with SB431542 compromised the morphology of rapidly differentiating mESC colonies and the increased expression of lineage markers in the differentiating Lefty1 KD cells (Supplementary Fig. S3A, B). These results indicate that Lefty1 critically controls the differentiation potential of mESCs via a TGF-β signaling pathway.
Suppression of Lefty2 impairs differentiation of mESCs
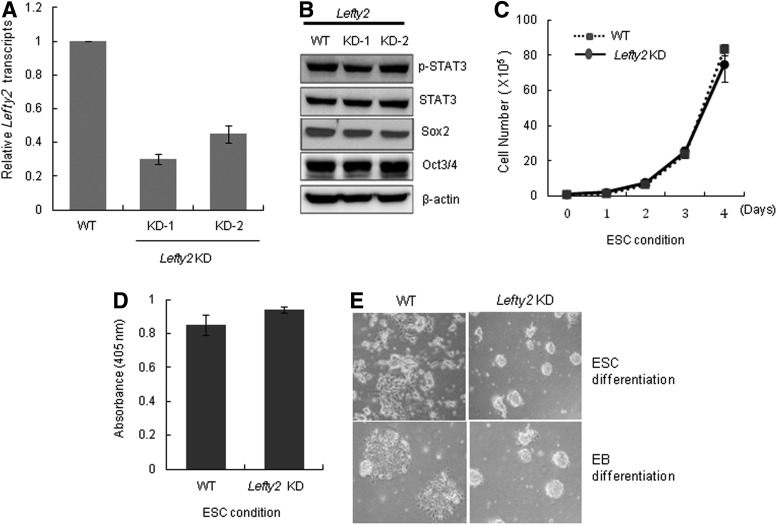
We next examined the function of Lefty2 in mESCs by generating Lefty2 KD cell lines using an shRNA targeting Lefty2 (Fig. 3A). Suppression of Lefty2 expression did not affect the expression of self-renewal markers, such as phosphorylated Stat3 (p-Stat3), Sox2, and Oct4 (Fig. 3B). The cell proliferation and cell cycle profiles of the Lefty2 KD cell populations were similar to those of the WT both under self-renewal and differentiation culture conditions (Fig. 3C and Supplementary Fig. S4). In addition, the alkaline phosphatase activity of Lefty2 KD cells was similar to that of WT cells (Fig. 3D). These results suggest that alteration of Lefty2 expression does not have any effect on the self-renewal of mESCs. We next investigated whether Lefty2 has a similar function to Lefty1 in regulating pluripotent differentiation of mESCs. In contrast to Lefty1 KD, Lefty2 KD cells maintained undifferentiated colony morphology under RA-induced differentiation conditions (Fig. 3E, upper panel). To confirm the effect of Lefty2 suppression on the differentiation of mESCs, we cultured mESCs under mesoendoderm differentiation conditions. Consistent with the RA-induced spontaneous differentiation pattern, Lefty2 KD cells did not respond to differentiation stimuli (Fig. 3E, lower panel). Based on these results, we concluded that suppression of Lefty2 significantly impairs the pluripotent differentiation potential of mESCs.
FIG. 3.
Lefty2 KD mESCs have defective differentiation properties. (A) Expression level of Lefty2 in two Lefty2 KD (KD-1 and KD-2) cell lines was analyzed by real-time RT-PCR. (B) The expression of phospho-Stat3, Sox2, and Oct3/4 by Lefty2 KD cells under ESC culture conditions was analyzed by immunoblotting. (C) Proliferation of Lefty2 KD cells in ESCs was compared with that of WT cells at the indicated times. (D) Alkaline phosphatase activity of Lefty2 KD cells was compared with that in the WT. (E) Lefty2 KD cells spontaneously differentiated (upper) or differentiated into the mesoendoderm lineage (bottom) for 3 days and cell morphology was examined by light microscopy. All values are mean±SD from at least three independent experiments. Lefty2 KD, stable Lefty2-knocked-down mESC line; WT, wild-type mESCs.
Suppression of Lefty2 results in enhanced expression of Lefty1
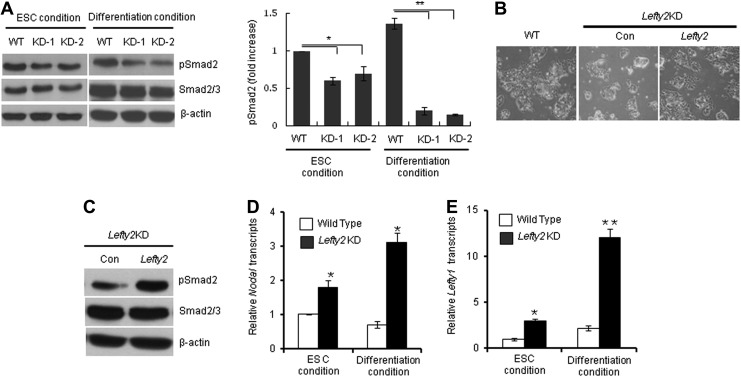
Because Lefty2 functions as a negative regulator of Nodal signaling and active Nodal/Activin signaling induces phosphorylation of the effectors Smad2/3 [18], we investigated whether suppression of Lefty2 leads to phosphorylation of Smad2. Surprisingly, the levels of phosphorylated Smad2 (p-Smad2) in Lefty2 KD cells were lower than those in the WT, and were further reduced when cells were differentiated for 3 days under RA-mediated differentiation conditions (Fig. 4A). This result suggests that Smad2-mediated Activin/Nodal signaling, which is required for the proper differentiation of mESCs toward mesoendoderm lineages, was impaired in the Lefty2 KD cell lines. To confirm that the defective differentiation phenotype of Lefty2 KD cell lines was caused by suppression of Lefty2 expression, we rescued Lefty2 expression by introducing a Lefty2 expression plasmid into Lefty2 KD and then examined whether increased Lefty2 expression could rescue the defective differentiation of Lefty2 KD. The differentiation pattern of Lefty2 KD became similar to WT as a result of re-expression of Lefty2 (Fig. 4B). When Lefty2 was re-expressed in Lefty2 KD mESCs, phosphorylation of Smad2 increased, consistent with our hypothesis that the low level of p-Smad2 in the differentiating Lefty2 KD mESCs was indeed caused by suppression of Lefty2 expression (Fig. 4C).
FIG. 4.
Smad2 phosphorylation and Lefty1 expression in Lefty2 KD cells. (A) Phospho-Smad2 in two Lefty2 KD mESC lines under self-renewal or differentiation culture conditions was analyzed by immunoblot analysis (left panel). The band intensity of p-Smad2 was quantified by normalization against β-actin (right panel). Cells were differentiated by removing LIF and adding RA for 3 days (RA-d3). (B) Lefty2 KD was transfected with a control (Con) or a Lefty2 expression plasmid and then spontaneously differentiated for 3 days. Differentiating cells were examined by light microscopy. (C) Lefty2 KD mESCs transfected with a control (Con) or a Lefty2 expression plasmid were analyzed for phospho-Smad2 levels by immunoblot analysis. (D) Nodal expression in Lefty2 KD and control ESCs was analyzed by real-time RT-PCR. (E) Lefty1 expression in Lefty2 KD and control ESCs was analyzed by real-time RT-PCR. All values are mean±SD from at least three independent experiments. *P<0.05 and **P<0.01 based on Student's t-test analysis. Lefty2KD, stable Lefty2-knocked-down mESC line; Con, Lefty2KD transfected with control plasmid.
To understand the underlying mechanism of dephosphorylation of Smad2 and the impaired differentiation of Lefty2 KD cells, we examined whether the inhibition of Lefty2 expression in mESCs results in the suppression of Nodal expression. However, Nodal expression was increased in Lefty2 KD cells both in ESC culture and in differentiation conditions (Fig. 4D). We then analyzed the expression level of Lefty1, another negative regulator of Nodal-mediated Smad2 phosphorylation. Interestingly, the expression of Lefty1 was markedly increased in both ESCs and differentiating Lefty2 KD cells (Fig. 4E). These results suggest that abnormal overexpression of Lefty1 is the main cause of reduced phosphorylation of Smad2 and the impaired differentiation of Lefty2 KD cells. The inhibitory effect of Lefty1 overexpression on the differentiation of ESCs was further supported by the observation that transient transfection of a Lefty1-expressing plasmid inhibited RA-mediated differentiation of mESCs (Supplementary Fig. S5).
Self-renewal is enhanced in the differentiating Lefty2 KD mESCs
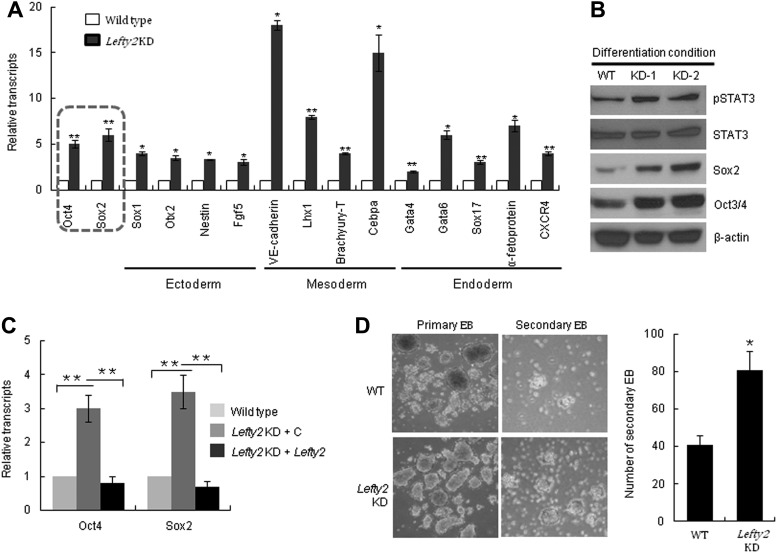
To further investigate the mechanism of impaired differentiation of Lefty2 KD cells, we analyzed the expression of self-renewal and differentiation markers in the differentiating Lefty2 KD mESCs. Unexpectedly, both self-renewal and differentiation marker genes were highly expressed in differentiating Lefty2 KD cells (Fig. 5A). This result indicates that the expression of self-renewal genes in Lefty2 KD mESCs was increased upon exposure to differentiation stimuli, which could result in the enhancement of self-renewal of mESCs under differentiation conditions. Immunoblot analysis of self-renewal markers, including phospho-Stat3, Sox2, and Oct3/4, further supports the possibility that self-renewal of Lefty2 KD was not decreased during RA-mediated differentiation (Fig. 5B). To confirm that the expression of self-renewal genes in differentiating Lefty2 KD was caused by the suppression of Lefty2 expression, we analyzed Oct4 and Sox2 expression levels in differentiating Lefty2 KD that were transfected with a Lefty2-expressing plasmid. The expression of Oct4 and Sox2 in differentiating Lefty2 KD cells was decreased to that of WT when Lefty2 expression was rescued (Fig. 5C). The increase in secondary EB formation by Lefty2 KD cells further supports the hypothesis that self-renewal of Lefty2 KD is enhanced under differentiating culture condition (Fig. 5D).
FIG. 5.
Self-renewal is not decreased in the differentiating Lefty2 KD cells. (A) Lefty2 KD cells were spontaneously differentiated for 3 days and expression of the indicated genes was analyzed by real-time RT-PCR. (B) Lefty2 KD cells were spontaneously differentiated for 3 days and expression of phospho-Stat3, Sox2, and Oct4 was analyzed by immunoblot. (C) Oct4 and Sox2 expression in differentiating Lefty2 KD mESCs transfected with a control or a Lefty2 expression plasmid was compared with differentiating WT mESCs by real-time RT-PCR. mESCs were spontaneously differentiated for 3 days. (D) Primary and secondary EBs were formed for 4 days and examined by light microscopy (left panel). The number of secondary EBs was compared between WT and Lefty2 KD mESCs (right panel). All values are mean±SD from at least three independent experiments. *P<0.05 and **P<0.01 based on Student's t-test analysis. Lefty2 KD, stable Lefty2-knocked-down mESC line; WT, wild-type mESCs.
Lefty2 KD teratomas show a massive expansion of immature malignant tissue
We next analyzed the impact of Lefty2 on the differentiation of mESCs in xenografts. Similar to Lefty1 KD cells, Lefty2 KD cells showed an obvious increase in teratoma growth compared with WT mESCs (Fig. 6A). Because teratomas are an in vivo indicator of pluripotency and in vitro differentiation properties, the knockdown of Lefty2 suggested that Lefty2 suppression leads to a defect in the pluripotent differentiation of mESCs. The development of larger teratomas from Lefty2 KD cells was an unexpected result. As teratomas generated from mESCs contain a small proportion of malignant, undifferentiated carcinoma tissue [19], we hypothesized that the enlarged size of Lefty2 KD teratomas is due to a large proportion of malignant tissue. Unlike other epithelial cells (eg, squamous epithelial cells or gut-epithelium), immature neuroepithelium that resembles a neural tube was embedded in the glial fibrillary background. The areas of immature neuroepithelium were composed of stratified columnar cells that formed rosettes. Fine eosinophilic glial fibrils of neuroglia without a collagen matrix provided a supporting framework for the neuroepithelium, which was confirmed by immunohistochemistry for glial fibrillary acidic protein (data not shown).
FIG. 6.
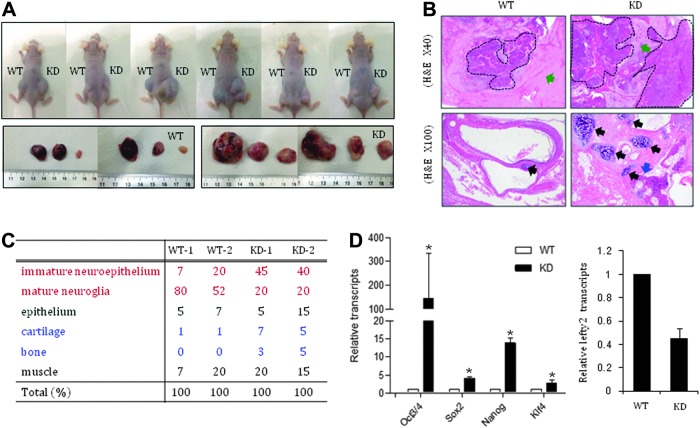
Lefty2 KD cells develop immature malignant teratomas. (A) Lefty2 KD and control mESCs were injected into NOD/SCID mice and teratoma development was examined. The size of the teratomas was examined 6 weeks after injection. (B) H&E staining and data analysis of teratoma sections were performed by a pathologist. Immature neuroepithelium is indicated by dotted lines and mature neuroglia is indicated by green arrows (upper panel). Cartilage and bone in the teratoma are indicated by black and blue arrows, respectively (lower panel). (C) Quantification of immature neuroepithelium, mature neuroglia, epithelium, cartilage, bone, and muscle was performed using serially sectioned teratomas of two WT and two Lefty2 KD teratomas. (D) Oct4, Sox2, Nanog, Klf4, and Lefty2 expression in the teratoma tissue was analyzed by real-time RT-PCR. All values are mean±SD from at least three independent experiments. *P<0.05 based on Student's t-test analysis. KD, stable Lefty2-knocked-down mESC line; WT, wild-type mESCs.
In teratomas formed by Lefty2 KD cells, massive numbers of immature neuroepithelial cells, which represent malignant immature teratogenic cells, were present and the proportion of mature neuroglia was significantly reduced (Fig. 6B upper panel, C). Cartilage and bone tissues were easily detected by their blue-gray matrix or dense calcification, as well as by the presence of lacunar chondrocytes and bony osteocytes. Mesoderm-derived cartilage and/or bone were highly developed in Lefty2 KD teratomas compared with WT (Fig. 6B lower panel, C). High expression levels of the pluripotency genes Oct4, Sox2, Nanog, and Klf4 in the bulk teratoma tissues derived from Lefty2 KD cells suggest that prolonged self-renewal might be the main cause of malignant and immature teratoma development (Fig. 6D). Together, these data indicate that suppression of Lefty2 leads to the malignant transformation of differentiating mESCs.
Discussion
The importance of Activin/Nodal signaling in the earliest cell fate decisions during embryogenesis has been well recognized. Nodal induces mesoderm and endoderm, patterns the nervous system, and determines left-right (L-R) asymmetry in vertebrates. Smad2 mediates Activin/Nodal signaling functions to orchestrate mesoendoderm lineage commitment of mESCs through direct modulation of the expression of corresponding developmental regulators [18]. Therefore, Lefty is a critical regulator of balanced lineage differentiation of cell fate determinants of ESCs. The present study revealed that Lefty1 functions to regulate mesoendoderm lineage differentiation commitments in response to external differentiation signals. However, in sharp contrast to the differentiation properties of Lefty1 KD, self-renewal of Lefty2 KD was not decreased under differentiation culture medium. Lefty1 is abnormally overexpressed and Smad2 is dephosphorylated in the differentiating Lefty2 KD, implying that proper expression of Lefty2 is crucial to maintain optimal Lefty1 and phospho-Smad2 levels in the differentiating mESCs. In addition, Oct4 and Sox2 are highly expressed in the differentiating Lefty2 KD. The Lefty1 promoter contains an ESC-specific enhancer that contains binding sites for Oct4 and Sox2 [5], indicating that overexpression of Lefty1 in the differentiating Lefty2 KD is mainly caused by enhanced expression of Oct4 and Sox2. Similar to Lefty2 KD cells, phospho-Smad2 levels in mESCs that overexpress Lefty1 (Lefty1 OE) were lower than those in WT cells (Supplementary Fig. S6A). However, Lefty1 OE cells developed smaller teratomas, which showed impaired differentiation of the three germ layers (Supplementary Fig. S6B). This result is opposite to that obtained for Lefty2 KD cells (Fig. 6A). Consistent with the teratoma size, the expression of several lineage markers in Lefty1 OE cells was lower than that in WT cells (Supplementary Fig. S6C). These results suggest that the Smad2 dephosphorylation in Lefty2 KD cells was caused by the activation of Lefty1, whereas the development of malignant and immature teratomas from Lefty2 KD cells was caused by a distinct mechanism, such as the increased self-renewal properties of Lefty2 KD mESCs under differentiation conditions. Contrary to the results obtained with Lefty2 KD cells, the expression of Nodal or Lefty2 was not altered in Lefty1 KD cells (Supplementary Fig. S7). This result implies that Nodal signaling is not regulated by reciprocal interactions of Lefty1 and Lefty2, at least in mESCs.
An in vivo teratoma assay revealed that Lefty2 KD mESCs formed enlarged teratomas with an enormous expansion of immature tissues. Recent studies demonstrated that LeftyA in conditioned medium from human liver stem cells (MSCs) has antitumor activity [20]. It would be important to elucidate whether Lefty2 has tumor-suppressive activity that could inhibit the malignant transformation of ESCs by repressing self-renewal properties in the differentiating mESCs. It is interesting to note that cartilage and bone were more developed in Lefty2 KD teratomas compared with WT. The reinforced differentiation of Lefty2 KD mESCs, at least into mesodermal lineages, was further supported by the enhanced expression of differentiation marker genes in the differentiating Lefty2 KD cells in vitro (Fig. 5A). This result indicates that suppression of Lefty2 also enhances the mesodermal differentiation potential of Lefty2 KD as well as the malignancy of mESCs. In addition, these results also suggest that enhanced self-renewal in the differentiating mESCs could transform mESCs into malignant carcinoma tissues regardless of the activation of a commitment to differentiation.
Contrary to Lefty2 KD, Lefty1 KD mESCs exhibited enhanced phospho-Smad2 levels both in self-renewal and in differentiating cells. In addition, suppression of Lefty1 facilitates differentiation of mESCs. These results suggest that, unlike Lefty1, the function of Lefty2 in mESCs is more than a simple Nodal inhibitor. Indeed, inhibition of Lefty in Xenopus results in Nodal overexpression [21], indicating that the regulatory network of Nodal signaling and Lefty expression is more complicated than we understand. Overexpression of Lefty1 in the differentiating Lefty2 KD explains that Lefty2 functions to adjust the intensity of Nodal signaling at the crossroads of self-renewal and differentiation of mESCs. Even though enhanced expression of Oct4 and Sox2 is thought to be the main cause of Lefty1 overexpression in the differentiating Lefty2 KD cells, we cannot exclude the presence of additional mechanisms that coordinate the expression of Lefty1 and Lefty2 during the differentiation of mESCs. Further studies are necessary to uncover the mechanism by which Lefty2 regulates the differentiation commitment of mESCs and regulates Lefty1 expression. Regardless of Smad2 phosphorylation and the differentiation patterns of Lefty1 KD and Lefty2 KD cells, phosphorylation of Smad1 and 5, which are pivotal intracellular effectors of the bone morphogenetic protein (BMP), was suppressed in differentiating Lefty1 KD and Lefty2 KD cells (Supplementary Fig. S8). The intracellular TGF-β signaling network, which is finely tuned by ligands such as nodal and BMP (as well as by Lefty1/2 inhibitors) during the differentiation of mESCs, remains to be elucidated.
The study of Lefty1 or Lefty2 function in human ESCs also remains to be elucidated to understand the evolutionary conservation of Nodal signaling and Lefty activity.
Supplementary Material
Acknowledgments
This research was supported by grants from the National Research Foundation of Korea funded by the Ministry of Science, ICT & Future Planning (2012-M3A9C6050367, 2011-0014084) and by the Priority Research Centers Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science and Technology (2012-0006679).
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Seuntjens E, Umans L, Zwijsen A, Sampaolesi M, Verfaillie CM. and Huylebroeck D. (2009). Transforming growth factor type beta and Smad family signaling in stem cell function. Cytokine Growth Factor Rev 20:449–458 [DOI] [PubMed] [Google Scholar]
- 2.Adjaye J, Huntriss J, Herwig R, BenKahla A, Brink TC, Wierling C, Hultschig C, Groth D, Yaspo ML, et al. (2005). Primary differentiation in the human blastocyst: comparative molecular portraits of inner cell mass and trophectoderm cells. Stem Cells 23:1514–1525 [DOI] [PubMed] [Google Scholar]
- 3.Richards M SP, Tan JH, Tan WK, Chan and Bongso A. (2004). The transcriptome profile of human embryonic stem cells as defined by SAGE. Stem Cells 22:51–64 [DOI] [PubMed] [Google Scholar]
- 4.Sperger JM, Chen X, Draper JS, Antosiewicz JE, Chon CH, Jones SB, Brooks JD, Andrews PW, Brown PO. and Thomson JA. (2003). Gene expression patterns in human embryonic stem cells and human pluripotent germ cell tumors. Proc Natl Acad Sci U S A 100:13350–13355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nakatake Y, Fukui N, Iwamatsu Y, Masui S, Takahashi K, Yagi R, Yagi K, Miyazaki J, Matoba R, Ko MS. and Niwa H. (2006). Klf4 cooperates with Oct3/4 and Sox2 to activate the Lefty1 core promoter in embryonic stem cells. Mol Cell Biol 26:7772–7782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sekkai D, Gruel G, Herry M, Moucadel V, Constantinescu SN, Albagli O, Tronik-Le Roux D, Vainchenker W. and Bennaceur-Griscelli A. (2005). Microarray analysis of LIF/Stat3 transcriptional targets in embryonic stem cells. Stem Cells 23:1634–1642 [DOI] [PubMed] [Google Scholar]
- 7.Dvash T, Mayshar Y, Darr H, McElhaney M, Barker D, Yanuka O, Kotkow KJ, Rubin LL, Benvenisty N. and Eiges R. (2004). Temporal gene expression during differentiation of human embryonic stem cells and embryoid bodies. Hum Reprod 19:2875–2883 [DOI] [PubMed] [Google Scholar]
- 8.Oulad-Abdelghani M, Chazaud C, Bouillet P, Mattei MG, Dolle P. and Chambon P. (1998). Stra3/lefty, a retinoic acid-inducible novel member of the transforming growth factor-beta superfamily. Int J Dev Biol 42:23–32 [PubMed] [Google Scholar]
- 9.Kosaki K, Bassi MT, Kosaki R, Lewin M, Belmont J, Schauer G. and Casey B. (1999). Characterization and mutation analysis of human LEFTY A and LEFTY B, homologues of murine genes implicated in left-right axis development. Am J Hum Genet 64:712–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yashiro K, Saijoh Y, Sakuma R, Tada M, Tomita N, Amano K, Matsuda Y, Monden M, Okada S. and Hamada H. (2000). Distinct transcriptional regulation and phylogenetic divergence of human LEFTY genes. Genes Cells 5:343–357 [DOI] [PubMed] [Google Scholar]
- 11.Schier AF. and MM Shen. (2000). Nodal signalling in vertebrate development. Nature 403:385–389 [DOI] [PubMed] [Google Scholar]
- 12.Cheng SK, Olale F, Brivanlou AH. and Schier AF. (2004). Lefty blocks a subset of TGFbeta signals by antagonizing EGF-CFC coreceptors. PLoS Biol 2:E30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.James D, Levine AJ, Besser D. and Hemmati-Brivanlou A. (2005). TGFbeta/activin/nodal signaling is necessary for the maintenance of pluripotency in human embryonic stem cells. Development 132:1273–1282 [DOI] [PubMed] [Google Scholar]
- 14.Park KS, Cha Y, Kim CH, Ahn HJ, Kim D, Ko S, Kim KH, Chang MY, JH Ko, et al. (2013). Transcription elongation factor Tcea3 regulates the pluripotent differentiation potential of mouse embryonic stem cells via the Lefty1-Nodal-Smad2 pathway. Stem Cells 31:282–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jirmanova L, Afanassieff M, Gobert-Gosse S, Markossian S. and Savatier P. (2002). Differential contributions of ERK and PI3-kinase to the regulation of cyclin D1 expression and to the control of the G1/S transition in mouse embryonic stem cells. Oncogene 21:5515–5528 [DOI] [PubMed] [Google Scholar]
- 16.Fei T, Zhu S, Xia K, Zhang J, Li Z, Han JD. and Chen YG. (2010). Smad2 mediates Activin/Nodal signaling in mesendoderm differentiation of mouse embryonic stem cells. Cell Res 20:1306–1318 [DOI] [PubMed] [Google Scholar]
- 17.Chan RJ, Johnson SA, Li Y, Yoder MC. and Feng GS. (2003). A definitive role of Shp-2 tyrosine phosphatase in mediating embryonic stem cell differentiation and hematopoiesis. Blood 102:2074–2080 [DOI] [PubMed] [Google Scholar]
- 18.Shiratori H, Sakuma R, Watanabe M, Hashiguchi H, Mochida K, Sakai Y, Nishino J, Saijoh Y, Whitman M. and Hamada H. (2001). Two-step regulation of left-right asymmetric expression of Pitx2: initiation by nodal signaling and maintenance by Nkx2. Mol Cell 7:137–149 [DOI] [PubMed] [Google Scholar]
- 19.Blum B, Bar-Nur O, Golan-Lev T. and Benvenisty N. (2009). The anti-apoptotic gene survivin contributes to teratoma formation by human embryonic stem cells. Nat Biotechnol 27:281–287 [DOI] [PubMed] [Google Scholar]
- 20.Cavallari C, Fonsato V, Herrera MB, Bruno S, Tetta C. and Camussi G. (2013). Role of Lefty in the antitumor activity of human adult liver stem cells. Oncogene 32:819–826 [DOI] [PubMed] [Google Scholar]
- 21.Branford WW. and HJ Yost. (2002). Lefty-dependent inhibition of Nodal- and Wnt-responsive organizer gene expression is essential for normal gastrulation. Curr Biol 12:2136–2141 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.