Abstract
Today, most approved therapeutic antibodies are provided as immunoglobulin G (IgG), whereas small recombinant antibody formats are required for in vitro antibody generation and engineering during drug development. Particularly, single chain (sc) antibody fragments like scFv or scFab are well suited for phage display and bacterial expression, but some have been found to lose affinity during conversion into IgG.
In this study, we compared the influence of the antibody format on affinity maturation of the CD30-specific scFv antibody fragment SH313-F9, with the overall objective being improvement of the IgG. The variable genes of SH313-F9 were randomly mutated and then cloned into libraries encoding different recombinant antibody formats, including scFv, Fab, scFabΔC, and FabΔC. All tested antibody formats except Fab allowed functional phage display of the parental antibody SH313-F9, and the corresponding mutated antibody gene libraries allowed isolation of candidates with enhanced CD30 binding. Moreover, scFv and scFabΔC antibody variants retained improved antigen binding after subcloning into the single gene encoded IgG-like formats scFv-Fc or scIgG, but lost affinity after conversion into IgGs. Only affinity maturation using the Fab-like FabΔC format, which does not contain the carboxy terminal cysteines, allowed successful selection of molecules with improved binding that was retained after conversion to IgG. Thus, affinity maturation of IgGs is dependent on the antibody format employed for selection and screening. In this study, only FabΔC resulted in the efficient selection of IgG candidates with higher affinity by combination of Fab-like conformation and improved phage display compared with Fab.
Keywords: phage display, antibody engineering, scFv fragment, scFab fragment, affinity maturation, therapeutic antibody, mutagenesis, CD30, Hodgkin lymphoma, antibody library
Introduction
One lesson learned from the first 20 years of therapeutic antibody development is the almost ubiquitous necessity for improvement of some properties, such as affinity of the original antibody. Affinity maturation by the mammalian immune system can generate antibodies with low picomolar affinity in some cases, but there is also a so-called affinity ceiling effect of B cell responses, which theoretically limits affinity enhancement at 0.1 to 1 nM.1,2 In general, affinity maturation in vitro starts with the diversification of antibody genes, which can be achieved by various methods such as random mutagenesis, targeted mutagenesis or a shuffling approach. Monoclonal antibodies are assumed to require affinities of 1 nM or less to their target antigen to be considered for clinical development; thus, one or several cycles of affinity maturation became part of the standard procedure.3-6 Moreover, humanization of non-human antibody sequences frequently results in some loss of affinity,7,8 so affinity maturation is necessary as an additional step in this process.9,10
Antibody phage display11-14 represents a powerful in vitro selection system for the isolation of human antibody fragments15-17 and can also be employed to improve biochemical properties of antibodies like the affinity or stability18,19 by combining cycles of antibody gene mutation with stringent selection. Other selection processes include the antibody display on ribosomes,20,21 yeast,22,23 Escherichia coli (E. coli),24 and mRNA.25 Mutations can be introduced randomly into the variable regions of antibody genes by error-prone polymerase chain reaction (PCR)26,27 or E. coli mutator strains.28,29 Other methods such as site-directed mutagenesis,30 saturation mutagenesis,31 parsimonious mutagenesis,32 CDR walking33 or look-through mutagenesis34 target certain regions like the CDRs, hence generating limited collections of the specific variants of the parent antibody. Shuffling approaches include DNA shuffling,35 chain shuffling,36-38 or CDR shuffling39,40 to obtain shuffled variants of the parent antibody. Antibody variants with higher affinities can be selected under stringent conditions, for example by extended incubation and intensive washing steps or by competition with soluble antigen.27 These in vitro evolution strategies imitate natural B cell affinity maturation in lymphoid organs, but they usually require small recombinant antibody fragments in order to obtain functional expression in E. coli and display on phage, yeast22 or ribosomes.41 Single chain fragment variable (scFv)42 and single chain Fab (scFab)43 contain a flexible peptide linker combining light (L) and heavy (H) antibody chains into a single polypeptide, which avoids the assembly of two separate polypeptides and makes them less prone to limitations of E. coli protein expression, folding and secretion, and are therefore generally more suitable for in vitro antibody selection using bacteria. The scFv fragment consists only of the variable (V) antibody regions VL and VH, whereas scFab also contains the adjacent constant (C) regions CL and CH1. As previously shown, deletion of the carboxy terminal cysteines (ΔC) of CL and CH1, resulting in the scFabΔC format, improved bacterial expression and phage display.43 Both molecules, scFv and scFabΔC, can also be genetically fused to the Fc moiety to produce single gene encoded bivalent IgG-like scFv-Fc and scIgG formats44,45 that share many properties of IgG, including Fc-mediated effector functions. Nevertheless, IgG is still the most accepted format for therapy. Conversion of in vitro optimized antibody fragments back into IgG can result in similar or even improved antigen binding,46-48 but, without accurate affinity data, apparent effects by increased avidity have to be considered to be responsible for the major part of enhanced antigen binding. The switch from monovalent to bivalent antigen binding can increase the apparent antigen binding several hundred fold.49 Moreover, studies have also demonstrated loss of affinity after conversion of scFv fragments into IgG,50 which can partially abolish the benefit from affinity maturation in the scFv format.19 A variety of strategies try to address issues related to IgG conversion of recombinant antibody fragments, including screening of vast numbers of scFv clones, batch conversion into the IgG format after selection in combination with mammalian expression for screening51,52 or Fab display on yeast.53 Switching to another expression host during screening may overcome restrictions of the E. coli expression system, but it is also associated with greater efforts and potentially creates new biases in the antibody development process due to additional subcloning steps or employment of different expression systems.
In this study, we investigated affinity maturation of the CD30-specific human antibody SH313-F954 in various antibody formats, with a focus on generating IgG variants with higher affinity. CD30 is a member of the tumor necrosis factor receptor superfamily.55 It is overexpressed on Hodgkin lymphomas and some non-Hodgkin lymphoma, whereas normal tissue show only limited expression.55 A number of CD30-specific antibodies have been evaluated as lymphoma treatments, and, in 2011, the antibody-drug conjugate brentuximab vedotin (Adcetris®) was approved by the US Food and Drug Administration for treatment of refractory and relapsed CD30+ hematological malignancies.
The human CD30-specific scFv antibody SH313-F9 was isolated from the universal human naïve scFv library HAL7.17 In this study, random point mutations were introduced into the SH313-F9 scFv gene by consecutive cycles of error-prone PCR. Mutated antibody phage display libraries were constructed in different antibody formats including scFv, scFabΔC, Fab, and FabΔC and their performance was compared.
Results
Phage display vectors for scFv, Fab, FabΔC, and scFabΔC antibody formats
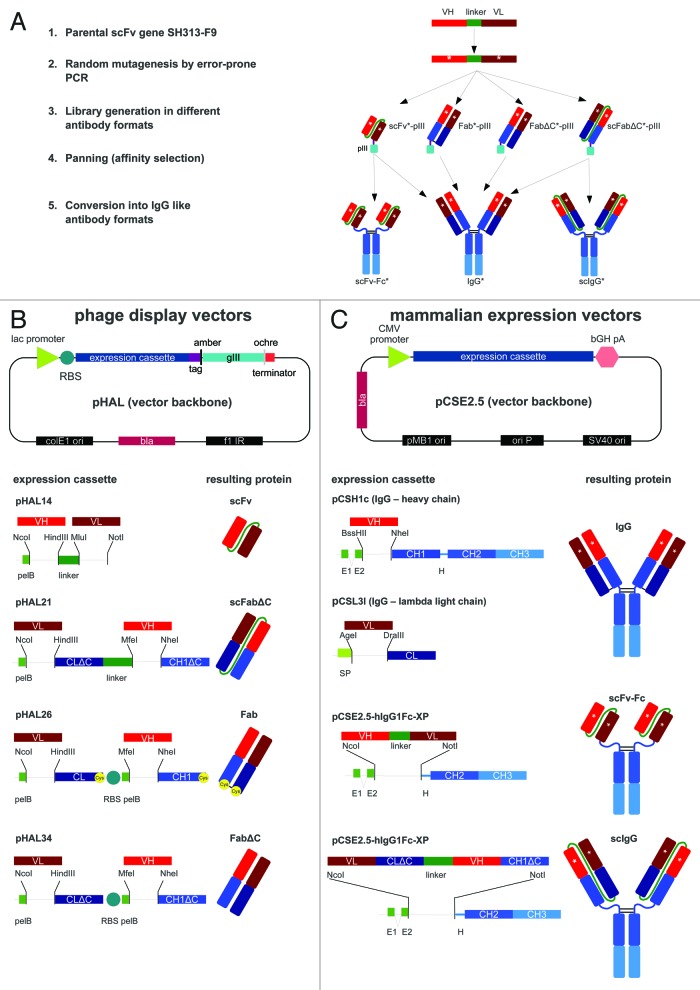
The phage display vector pHAL1417 was used to construct the phagemids pHAL21, pHAL26, and pHAL34 that were used for the generation of antibody gene libraries in scFabΔC, Fab, and FabΔC format, respectively (Fig. 1B). All vectors contained the same vector backbone with gene fragments encoding the secretory signal peptide of the pectate lyase B (pelB) of Erwinia carotovora, the different antibody gene expression cassettes including cloning sites followed by tags for purification and detection, an amber stop codon that is partially translated into the amino acid Q in supE44 E. coli strains, and the filamentous phage gene 3 (gIII) encoding the minor phage surface protein pIII for phage display. The scFabΔC gene expression of the phagemid pHAL21 consisted of gene fragments encoding the PelB leader, the cloning site for VL followed by the human lambda CL without carboxy terminal cysteine (ΔC), a 34 amino acids long G/S linker sequence,43 and the second cloning site for VH and CH1 without carboxy terminal cysteine (ΔC) that was fused to the further downstream elements required for purification, detection and phage display.
Figure 1. (A) Schematic overview of affinity maturation strategies using scFv, scFabΔC, Fab, or FabΔC and subsequent conversion of antigen-binding molecules into scFv-Fc, scIgG, or IgG. (B) Schematic illustration of the vector backbone for phage display of scFv, scFabΔC, Fab, and FabΔC antibody formats including corresponding gene expression cassettes. The pHAL vector backbone of the phagemid pHAL14 was used for generation of scFv gene libraries and for the construction of the vectors pHAL21 for scFabΔC display, pHAL26 for Fab display, and pHAL34 for FabΔC display. Restriction sites used for cloning of antibody gene fragments or V regions are indicated. (C) Schematic illustration of the mammalian expression vector backbone used for production of IgG, scFv-Fc, and scIgG including corresponding gene expression cassettes. The mammalian scFv-Fc expression vector pCSE2.5-hIgG1Fc-XP was also used for expression of scIgG1 and for construction of pSH1c and pSL3l which were used for human IgG1 heavy chain and lambda light chain expression. Co-transfection of pSH1c and pSL3l allowed human IgG1 production in mammalian cells. The vector elements or antibody domains are not drawn to scale. Same type of antibody domains and corresponding gene fragments are indicated by the same color. Abbreviations: amber, amber stop codon; bGH pA, bovine growth hormone poly A signal; bla, ampicillin resistance gene encoding β lactamase; CH1–3 and CL, constant antibody regions of heavy and light chain of IgG1 (blue, dark blue); CMV, cytomegalovirus immediate early promoter; colE1 ori, E. coli origin of replication; f1 IR, F1 intergenic region; gIII, gene encoding for M13 minor phage surface protein pIII; H, IgG1 hinge region; lac, lactose operon promoter; ochre, ochre stop condon; ori P, origin of replication of Epstein-Barr virus (EBV); pelB, secretory pectate lyase B signal peptide sequence; pMB1 ori, a bacterial origin of replication; RBS, ribosomal binding site; tag, hexa-histidine and c-myc tags for purification and detection; SP, secretory mammalian signal peptide; SV40 origin, simian virus (SV) 40 origin of replication; VH and VL, variable antibody regions of light (L) and heavy (H) chain (red, dark red).
The bicistronic antibody expression cassettes of pHAL26 and pHAL34 for Fab and FabΔC display were generated by replacing the linker of pHAL21 with a second ribosomal binding site (RBS) followed by another pelB signal peptide sequence initiating the second cistron. Moreover, an ochre stop codon was introduced at 3′ end of the CL domain to terminate translation of the first cistron. Fab and FabΔC gene expression cassettes only differed in the presence or absence of codons encoding the carboxy terminal cysteines of CL and CH1, respectively.
Expression vectors for expression of scFv-Fc, scIgG and IgG in HEK293–6E cells
The mammalian expression vector pCSE2.5-hIgG1Fc-XP44,45 was used to convert scFvs into scFv-Fc and scFabΔC into scIgG (Fig. 1C). This vector was previously optimized for transient expression in HEK293–6E cells and allows NcoI-NotI single step in-frame cloning of single chain antibody fragments from pHAL14 (scFv) and pHAL21 (scFabΔC) to generate human scFv-Fc and scIgG antibodies, respectively.
The new monocistronic vectors pCSH1c and pCSL3l (Fig. 1C) allow production of human IgG1 lambda antibodies after co-transfection of both plasmids into HEK293–6E cells. Both vectors are based on the pCSE2.5-hIgG1Fc-XP backbone. Human VH gene fragments can be cloned into pCSH1c via BssHII and NheI restriction sites to express the human IgG1 heavy chain, and lambda VL gene fragments can be cloned into pCSL3l via AgeI and DraIII restriction sites to express the lambda light chain without creating any amino acid alteration compared with natural human immunoglobulin sequences.
Evaluation of different antibody formats of SH313-F9 for phage display
The CD30 specific scFv SH313-F954 was converted into the antibody formats scFabΔC,43,56,57 Fab, and FabΔC and cloned into phage vectors pHAL21, pHAL26 and pHAL34, respectively. The different SH313-F9 antibody phage variants were produced using Hyperphage and precipitated before verifying antigen binding to CD30 by titration enzyme linked immunosorbent assay (ELISA). SH313-F9 scFabΔC or FabΔC phage bound less efficient to CD30 than SH313-F9 scFv phage (Fig. 2), which was also visualized in Western immunoblot by a less pronounced antibody::pIII fusion protein band (data not shown). In contrast, SH313-F9 Fab phage bound very poorly to CD30 in the highest phage titers (Fig. 2) and showed a faint antibody::pIII fusion band in Western immunoblot (data not shown). In summary, the parental antibody SH313-F9 showed the best functional display on phage in the scFv format, followed by scFabΔC and FabΔC.
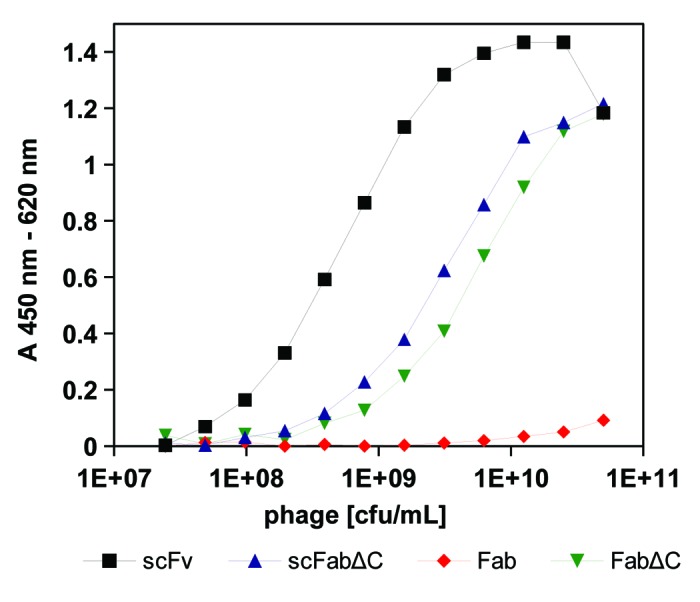
Figure 2. Phage ELISA of scFv, scFabΔC, Fab, and FabΔC antibody formats of SH313-F9. A dilution series of SH313-F9 scFv-, scFabΔC-, Fab- and FabΔC-phage were tested by a monoclonal phage ELISA for binding to CD30. Bound antibody phage were detected with a mouse-α‐M13 antibody HRP conjugate. Background binding to BSA was generally very low (< 0.04) and subtracted from the shown absorption measured for CD30 binding.
Affinity maturation of SH313-F9 in the scFv format
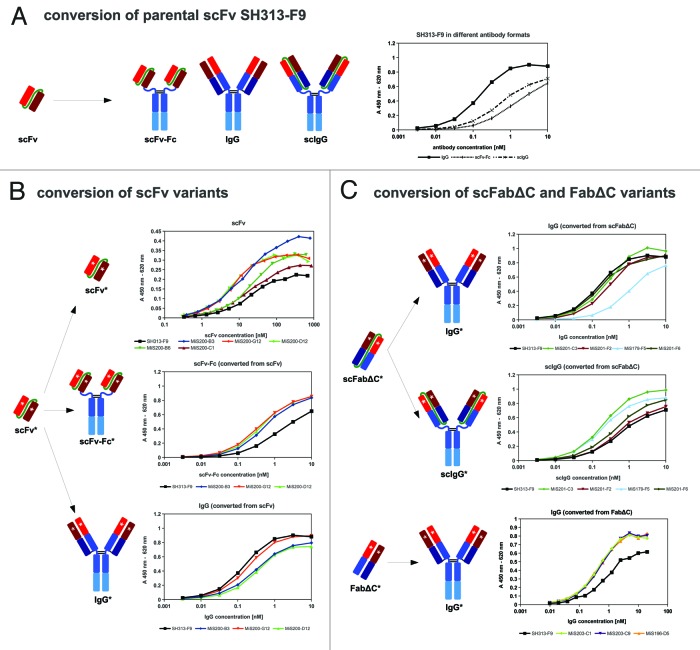
Error-prone PCR was used to introduce random point mutations into the SH313-F9 scFv antibody gene. Amplified DNA derived from the third round of consecutive error-prone PCRs was cloned into the phage display vector pHAL1458-61 (Fig. 1B). After transformation, a mutated SH313-F9 scFv gene library was obtained comprising 1 × 107 independent clones with a content of full size scFv inserts of 73% (Table 1). Sequence analysis of randomly picked clones revealed 9 to 22 (average 14) point mutations per scFv gene corresponding to 7 to 11 (average 8) amino acid substitutions encoded by each clone. The mutated SH313-F9 scFv gene library was packaged with Hyperphage for oligovalent display. Display of mutated SH313-F9 scFv antibody fragments on phage was demonstrated by Western immunoblot analysis as shown by the clear scFv::pIII fusion protein band corresponding to a molecular mass of about 90 kDa (Fig. 3). Another aliquot of this mutated scFv gene library was again packaged with M13K07 for monovalent display and used for panning against CD30 antigen in immunoplates in the presence of soluble antigen for competition. After soluble expression in E. coli, a total of 92 scFv clones were analyzed by ELISA. Five of these scFv clones (MiS200-B3, -B6, -C1, -G12, -D12) demonstrated higher CD30 specific ELISA signals compared with the parental SH313-F9 scFv (Table 1). Further validation of these scFv clones by titration ELISA (Fig. Four B) confirmed enhanced binding to CD30.
Table 1. Mutant antibody gene libraries of SH313-F9 in scFv, scFabΔC, Fab and FabΔC format and antibody candidates obtained after affinity maturation.
| Antibody format | Library size | Full size antibody genes | Phage titer | Number of antibody clones | |||
|---|---|---|---|---|---|---|---|
| FabΔC | 2.1x107 | 100 (n = 8) | 1.7 × 1013 | 2.0 × 1011 | 184 | 3 | 3 |
1 Packaging with M13K07;2packaging with Hyperphage;3tested in titration ELISA on CD30.
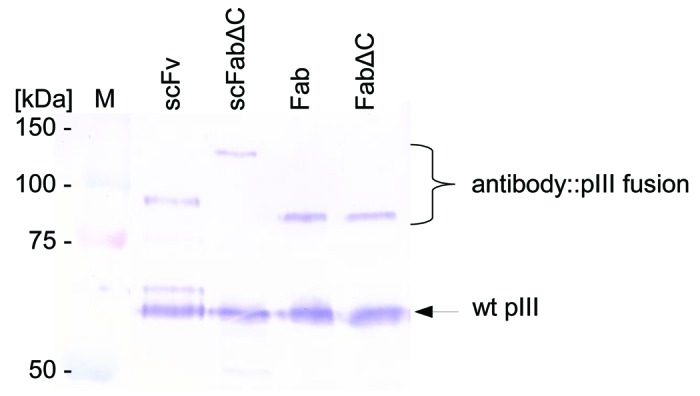
Figure 3. Display of mutated SH313-F9 scFv, scFabΔC, Fab, and FabΔC antibody fragments on phage. Mutated libraries of SH313-F9 in different formats were packaged with Hyperphage. After phage production, each library was analyzed by SDS-PAGE and western blot including immunostaining with a pIII antibody.
Conversion of affinity enhanced SH313-F9 scFv clones into the scFv-Fc and IgG format
The scFv antibodies MiS200-B3, MiS200-G12 and MiS200-D12 showing best antigen binding in ELISA were converted into the bivalent scFv-Fc and IgG formats (Fig. 1A) as described above. IgGs and scFv-Fc antibodies were transiently produced in HEK293–6E cells and purified by protein A affinity chromatography. Yields of scFv-Fc antibodies were between 19 to 31 mg/L, and the corresponding IgGs were less efficiently produced (3–16 mg/L; Table 2). The scFv-Fc generated from the parental scFv SH313-F9 showed the highest yield, whereas the corresponding IgG achieved the lowest yield, illustrating that scFv conversion into different formats can strongly influence production levels in mammalian cells.
Table 2. Yields of different IgG and IgG-like formats of SH313-F9 variants produced in HEK293–6E cells.
| Antibody | Format | Initial format | Yield 10 mL scale [mg/L] |
Yield 50–100 mL scale [mg/L] |
|---|---|---|---|---|
| SH313-F9 | IgG | scFv | 2.7 | 8 |
| MiS200-B3 | IgG | scFv | 7.0 | |
| MiS200-G12 | IgG | scFv | 6.1 | |
| MiS200-D12 | IgG | scFv | 16.0 | |
| MiS201-C3 | IgG | scFabΔC | 10.3 | |
| MiS201-F2 | IgG | scFabΔC | 4.0 | |
| MiS179-F5 | IgG | scFabΔC | 8.7 | |
| MiS201-F6 | IgG | scFabΔC | 0.2 | |
| MiS203-C1 | IgG | FabΔC | 0.1 | 11 |
| MiS203-C9 | IgG | FabΔC | 0.1 | 7 |
| MiS196-D5 | IgG | FabΔC | 0.4 | 5 |
| SH313-F9 | scFv-Fc | scFv | 31.1 | 303 |
| MiS200-B3 | scFv-Fc | scFv | 20.4 | |
| MiS200-G12 | scFv-Fc | scFv | 19.3 | |
| MiS200-D12 | scFv-Fc | scFv | 27.4 | |
| SH313-F9 | scIgG | scFv | 9.8 | 106 |
| MiS201-C3 | scIgG | scFabΔC | 5.3 | |
| MiS201-F2 | scIgG | scFabΔC | 25.0 | |
| MiS179-F5 | scIgG | scFabΔC | 10.3 | |
| MiS201-F6 | scIgG | scFabΔC | 14.7 |
CD30 binding of these scFv-Fc and IgG antibodies was confirmed by titration ELISA (Fig. 4B). In the scFv-Fc format, all affinity matured scFv antibodies showed stronger antigen binding compared with the parental antibody SH313-F9, whereas in the IgG format the situation was opposite, i.e., the increased affinity of the scFv affinity mutants could not be transferred to the IgG format. In contrast, the parent SH313-F9 IgG version bound stronger than the scFv-Fc, when determined by titration ELISA (Fig. 4A). Therefore, affinity maturation of SH313-F9 in the scFv format did not lead to IgGs with enhanced affinity.
Figure 4. (A) SH313-F9 was converted into scFv-Fc, scIgG and IgG, produced in mammalian cells and tested by titration ELISA for binding to CD30. (B) Affinity maturation of SH313-F9 in the scFv format resulted in molecules with enhanced antigen binding compared with the parental scFv SH313-F9. These candidates were tested as scFv, scFv-Fc and IgG by titration ELISA for binding to CD30. (B) Affinity maturation of SH313-F9 in the scFabΔC or FabΔC formats resulted in molecules with enhanced antigen binding compared with the parental antibody. These candidates were tested as scIgG (only scFabΔC) and IgG by titration ELISA for binding to CD30. Background binding to BSA was generally very low (< 0.02) and subtracted from the shown absorption measured for CD30 binding.
Affinity maturation of the SH313-F9 antibody in Fab, scFabΔC, and FabΔC formats
The same template of randomly mutated SH313-F9 scFv gene fragments was also used for the construction of three antibody gene phage display libraries in other formats: scFabΔC (vector: pHAL21), Fab (vector: pHAL26) and FabΔC (vector: pHAL34). The resulting libraries comprised 1.9 × 107 to 5.7 × 107 independent clones (Table 1). The quality of the library was tested by colony PCR of randomly selected single clones, revealing 100% full size antibody inserts for the scFabΔC and FabΔC libraries and 75% for the Fab library (Table 1). All libraries were packaged with either Hyperphage or M13K07. Phage titers after phage production with helperphage M13K07 and precipitation were between 4.0 × 1012 to 4.6 × 1013 cfu/mL. All mutated SH313-F9 antibody gene libraries, including the mutant Fab library packaged with Hyperphage, showed antibody-pIII fusion proteins in Western immunoblot (Fig. 3).
The selection of high affinity mutant antibody fragments was performed as described for the scFv library. CD30-specific molecules with elevated signals in ELISA compared with the corresponding parental SH313-F9 antibody fragment were identified from mutated scFabΔC and FabΔC libraries, but not from the corresponding Fab library (Table 1).
Sequence analysis of affinity enhanced antigen-binding molecules from scFv, scFabΔC, and FabΔC libraries
DNA sequencing revealed that the molecules with higher affinity were unique, reflecting an appropriate diversity of antibody gene mutations in the library. The encoded amino acid sequences of 21 individual antibody clones were compared (Fig. 5). Some regions within the SH313-F9 antibody sequence were not affected by any amino acid exchange, such as framework (FR) 2 and FR4 of VH and the complementary-determining region (CDR) 2 and FR4 of VL. In contrast, the first amino acid in CDR3 of VH was identified as a mutation hot spot, showing the same amino acid exchange in 11 out of 21 cases. Moreover, some mutation hot spots seem to depend on the antibody format that was employed for affinity maturation. For example, all three FabΔC antibody mutants with higher affinity than the parental antibody contained the same amino acid exchange at the third amino acid in FR3 of VL (data not shown), thus indicating that affinity maturation results in a format-dependent selection of antibody variants.
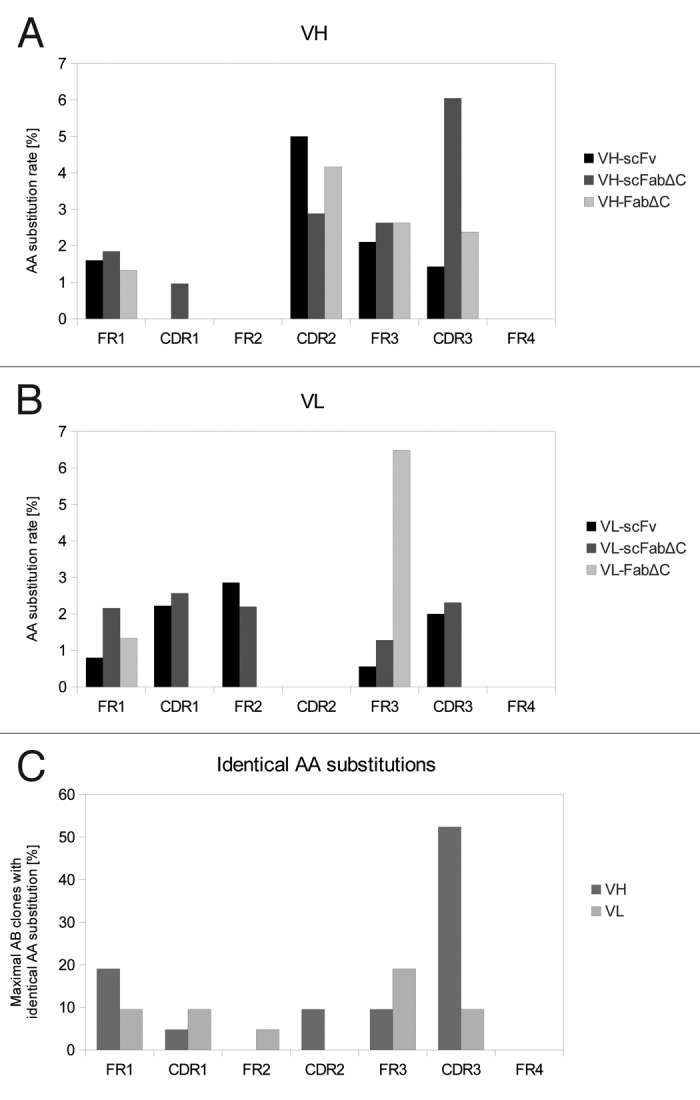
Figure 5. Distribution of AA substitutions in SH313-F9 variants after affinity maturation. Amino acid substitutions in VH (A) and VL (B) of SH313-F9 variants with higher affinity obtained from affinity maturation in different antibody formats (scFv, scFabΔC, FabΔC). The amino acid substitution rate was calculated for each region (FRs and CDRs) in respect to its length. The total number of AA of each region was set 100%. (C) The maximal percentage of antibody clones per region carrying the identical amino acid substitution was calculated in order to indicate mutation hot spots.
Conversion of affinity enhanced scFabΔC and FabΔC clones into IgG
Affinity improved scFabΔC (MiS201-C3, -F2, -F6 and MiS179-F5) and FabΔC (MiS196-D5, MiS203-C1, -C9) antibody clones were converted into the IgG format. In addition, the scFabΔC variants were also cloned into the bivalent IgG-like scIgG format (Fig. One A). IgGs and scIgGs were produced in HEK293–6E cells in 10 mL scale. For further analysis, transient production of the IgGs SH313-F9, MiS203-C1, -C9, and MiS196-D5 was repeated in HEK293–6E cells in 100 mL scale. In addition, SH313-F9 was also produced as scIgG in HEK293–6E cells in 50 mL scale. All antibodies were purified by protein A affinity chromatography for further validation. IgG production yields of 100 mL scale productions were between 4 to 11 mg/L, which was significantly higher than those obtained by the small scale production (Table 2). The yields of scIgGs were between 5 to 25 mg/L in 10 mL small scale productions, which is generally higher than the yields of the corresponding IgGs. Production in shake flasks with a volume of 50 mL achieved volumetric yields of up to 106 mg/L for the parental antibody SH313-F9 scIgG (Table 2).
Binding studies of IgG converted antibodies
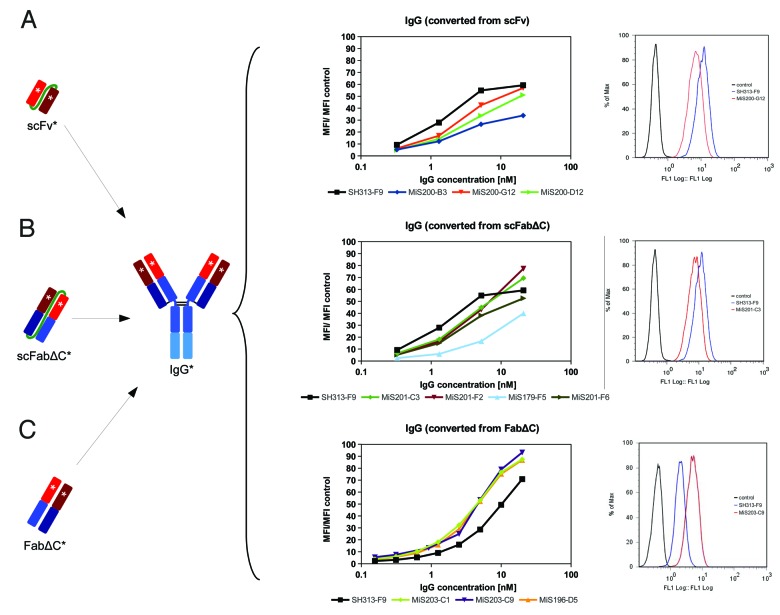
The IgGs and scIgGs generated from affinity matured scFabΔC (MiS201-C3, -F2, -F6 and MiS179-F5) and FabΔC clones (MiS196-D5, MiS203-C1, -C9) were compared by titration ELISA regarding binding to CD30 (Fig. 4B). The scIgGs originating from scFabΔC mutants showed elevated binding compared with the parental SH313-F9 in the scIgG format (Fig. 4B). CD30 binding of SH313-F9 IgG in titration ELISA was elevated compared with SH313-F9 scIgG (Fig. Four A), but, after conversion of the affinity matured scFabΔC clones into IgG, antigen binding was the same or reduced compared with parental SH313-F9 IgG (Fig. 4C). On the contrary, all IgGs generated from the FabΔC clones MiS196-D5, MiS203-C1, and MiS203-C9 showed increased CD30 binding in ELISA compared with parental SH313-F9 IgG (Fig. 4C), indicating that affinity maturation in the FabΔC format was beneficial. Moreover, these FabΔC-derived IgGs (MiS196-D5, MiS203-C1, -C9) also showed increased binding on the CD30+ lymphoma cell line Karpas299 in flow cytometry compared with the parental SH313-F9 IgG (Fig. 6C), whereas all IgGs generated from affinity matured scFv or scFabΔC antibody candidates showed lower binding to CD30+ cells (Fig. 6A and B).
Figure 6. Cell binding of SH313-F9 variants. CD30+ Karpas299 cells were stained with a dilution series of IgGs which were converted from affinity matured scFv (A), scFabΔC (B), or FabΔC (C) candidates. The mean fluorescence intensities are summarized in the diagrams shown in the middle column. In addition, overlay histogram plots (right column) of CD30+ Karpas299 cells stained with 0.6 nM (C) or 1.3 nM (A and B) of representative affinity matured IgG candidates (red line) are shown in comparison to the parental SH313-F9 IgG (blue line). Cells only stained with secondary detection antibody conjugate are shown as negative control (black line).
Surface plasmon resonance-based affinity ranking of IgGs
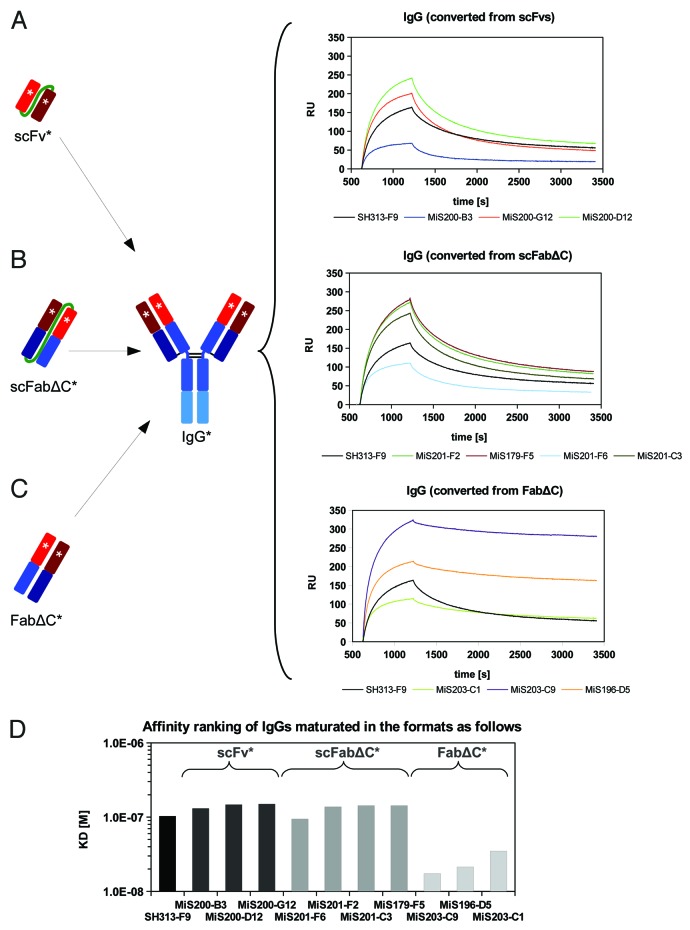
In addition, IgGs converted from different antibody formats obtained by affinity maturation were analyzed by surface plasmon resonance (SPR) for affinity ranking. Each IgG variant was captured by a monoclonal mouse anti-human IgG coupled to a CM5 chip. Then, binding kinetics of monomeric CD30 were measured and analyzed (Fig. 7). All IgGs converted from scFvs had lower affinities compared with SH313-F9 IgG (Fig. 7A and D). The four IgGs converted from the scFabΔC format showed slightly lower or similar affinities compared with the parental SH313-F9 IgG (Fig. 7B and D). In contrast, all three FabΔC candidates that showed improved antigen binding in ELISA and flow cytometry also demonstrated higher affinity after IgG conversion compared with the parental SH313-F9 IgG (Fig. 7C and D).
Figure 7. Surface plasmon resonance (SPR) based affinity ranking for IgGs converted from affinity matured SH313-F9 variants. IgGs converted from scFv (A), scFabΔC (B), or FabΔC (C) affinity matured candidates were captured on a CM5 chip modified with a mouse anti-IgG Fc specific capture antibody and probed with monomeric CD30. (D) Summary of apparent affinity constants.
SPR analysis for determination of kinetic binding constants kon, koff, and KD
IgGs derived from the FabΔC clones MiS196-D5, MiS203-C1 and MiS203-C9 showed 8- to 12-fold higher affinities (lower KDs) compared with the parental SH313-F9 IgG (Table 3). The MiS203-C9 IgG had the lowest KD (1.32 × 10−8 M). The affinity enhancement was due to a combination of faster association and slower dissociation rates, with the latter having a greater effect on the affinity enhancement. Moreover, the parental SH313-F9 antibody was also tested in different IgG-like formats, including scFv-Fc, scIgG and IgG. Surprisingly, the IgG generated from the parental scFv SH313-F9 had a 2.5- and 3.3-fold lower KD value (1.59 × 10−7 M) than the corresponding scFv-Fc and scIgG, respectively.
Table 3. Affinity determined by SPR.
| Antibody | kon [1/Ms] | koff [1/s] | KD [M] | χ2 | Affinity compared with SH313-F9 IgG |
|---|---|---|---|---|---|
| SH313-F9 IgG | 1.38 × 104 | 2.20 × 10−3 | 1.59 × 10−7 | 2.2 | 1.0 |
| SH313-F9 scFv-Fc | 7.18 × 103 | 3.30 × 10−3 | 4.18 × 10−7 | 5.3 | 0.4 |
| SH313-F9 scIgG | 7.18 × 103 | 3.28 × 10−3 | 4.56 × 10−7 | 8.7 | 0.3 |
| MiS203-C1 IgG | 3,03 × 104 | 6.11 × 10−4 | 2.02 × 10−8 | 0.3 | 7.9 |
| MiS203-C9 IgG | 3,96 × 104 | 5.22 × 10−4 | 1.32 × 10−8 | 1.2 | 12.0 |
| MiS196-D5 IgG | 3,75 × 104 | 5.42 × 10−4 | 1.45 × 10−8 | 1.0 | 11.0 |
kon, off-rate; koff, off-rate; KD dissociation equilibrium constant (affinity); χ2, chi square value (curve fitting)
Analysis of aggregation and multimerization by SEC
The IgGs SH313-F9, MiS203-C1, -C9 and MiS196-D5, as well as scFv-Fc and scIgG formats of the parental antibody SH313-F9, were analyzed by size-exclusion chromatography (SEC) to study aggregation and multimerization. None of the IgGs tended to form dimers or larger aggregates, whereas scFv-Fc and scIgG showed an additional small population, indicating the formation of dimers (Fig. 8).
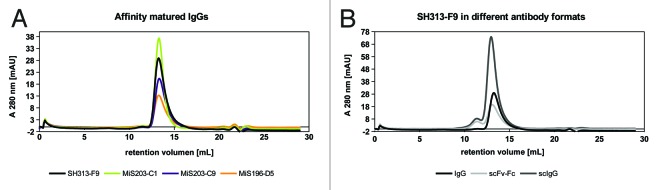
Figure 8. Analytical SEC of IgGs SH313-F9, MiS196-D5, MiS203-C1 and MiS203-C9 (A) as well as of scFv-Fc, scIgG, and IgG generated from the parental antibody SH313-F9 (B).
Discussion
In this study, we used three consecutive steps of error-prone PCR to introduce random point mutations into the SH313-F9 scFv54 gene, which correlated with about eight amino acid substitutions per mutant on the translational level. The resulting mutated scFv gene fragments were used as templates to generate four phage display libraries encoding different antibody formats including scFv, scFabΔC, Fab, and FabΔC. All libraries showed similar properties like high full-length antibody insert rates, comparable phage titers and clearly detectable levels of antibody::pIII fusion protein after packaging with Hyperphage. After a single in vitro selection step under identical stringent conditions, all mutated libraries except the Fab library allowed the selection of molecules with improved antigen binding properties compared with the parental SH313-F9 antibody in the same antibody formats.
On examination of the distribution of the mutations, it is evident that mutations were not evenly distributed with preference in some regions including both, FRs and CDRs. Some mutations were clearly dependent on the antibody format used for affinity maturation, for example FR3 mutations were enriched in the VL of the FabΔC format. Moreover, hotspots of single amino acid substitutions, e.g., in CDR3 of VH, were observed in all antibody formats and seem to be important for increasing the affinity to the antigen, whereas other amino acid substitutions did only occur in a specific antibody format. A detailed understanding of structural effects would require crystallization. However, the enrichment of large numbers of FR mutations indicates the importance of introducing random mutations into the entire antibody V genes compared with approaches that are only restricted to CDRs.
The conversion of the parental scFv SH313-F9 into Fab already revealed poor CD30 antigen binding of antibody phage, which indicated that this Fab is not well-produced in E. coli and not functionally displayed on phage. This changed after eliminating both of the carboxy terminal cysteines that form the interchain disulfide bridge between light chain and Fd fragment in Fab fragments. This so-called FabΔC format provided functional antibody phage of the parental SH313-F9 antibody with similar performance compared with the SH313-F9 scFabΔC. In a previous study, scFabΔC also showed elevated bacterial expression levels and display on phage than scFab.43 Unpaired cysteines have been shown before to have a negative impact on bacterial expression.62
In contrast to Fab, the scFv, scFabΔC, and FabΔC libraries of mutated SH313-F9 allowed the isolation of CD30-specific molecules, and some of them showed improved antigen binding compared with the parental antibody SH313-F9. However, all affinity enhanced antibody clones originating from scFv and scFabΔC lost affinity after conversion into IgG. In contrast to scFv and scFabΔC, the three molecules obtained from affinity maturation performed in the FabΔC format retained their increased affinity after conversion into IgG. The FabΔC format only differs from scFabΔC by the absence of the linker. Affinity maturation in the scFabΔC format may promote structural alterations of the antigen binding surface away from Fab or IgG due to the presence of the linker, an effect that would also explain the results of affinity maturation in scFv format. We noted that the parental scFv SH313-F9, obtained from the human naïve antibody gene library HAL7, showed a 2.5- to 3.3-fold increased affinity as IgG compared with scFv-Fc and scIgG. This can be explained by the fact that all antibody genes in the library HAL7 were isolated from peripheral B lymphocytes and have already undergone a process of natural selection of functional B cell receptors and IgG antibodies in vivo. Other studies have demonstrated that conversion of scFvs derived from an antibody gene library also being constructed using B lymphocytes to IgGs can improve affinity due to increased stability in IgG.63 However, introduction of new mutations during affinity maturation into single chain antibodies can result in variants that favor structural changes requiring the presence of the peptide linker. This effect was clearly observed in this study. Most mutations enriched by affinity maturation in the scFv and scFabΔC formats did not enhance but rather reduced affinity of the corresponding IgG.
The IgG derived from the FabΔC MiS203-C9 showed the highest affinity in this study, with a KD of 13 nM, which is a 12-fold improvement compared with the parental SH313-F9 IgG. Other groups described three to more than 1000-fold enhanced antigen binding after random mutagenesis of CDRs or entire antibody V genes in combination with phage display selection when compared in the antibody formats used for affinity maturation.28,29,64 Because these studies did not test antigen binding in the Fab or IgG format, it remains unclear if subsequent IgG conversion of these antibody fragments would result in loss of affinity, as shown in this and other previous studies.19 In some reports, scFv antibody fragments were also converted into IgG, but antigen binding kinetics were not measured47,48 or in other cases apparent binding effects by different avidity were not adequately considered.66 The increase of avidity can elevate antigen binding several hundred fold, particularly when the antibody format switches from monovalent to bivalent binding.49
During panning and screening, single chain antibody formats may have an increased apparent affinity due to formation of dimers or multimers that compete with antibody clones with increased affinity. This can be avoided by employing monomeric antibody fragments like FabΔC (or Fab). In this study, avidity effects did not play an important role because neither scFv-Fc nor scIgG of the parental antibody SH313-F9 showed any advantage in affinity compared with IgG, even though both IgG equivalent single chain formats showed some degree of dimer formation.
It should be noted that affinity maturation employing the scFv format were successfully used to generate high affinity IgGs. The most prominent example is adalimumab (D2E7, Humira®), which was humanized and engineered by an scFv-based guided selection strategy using a murine antibody as template and by iterative rounds of mutagenesis and chain shuffling. Generally, higher efforts have to be expected for screening and validation to generate antibody fragments that retain high affinity after conversion into IgG. Moreover, there is an intrinsic risk for a bias during scFv selection, which suppresses optimal IgG variants. Such bias against optimal IgG variants has also to be expected for Fab-based affinity maturation due to higher selection pressure by the bacterial expression and secretion system compared with FabΔC. Therefore, the Fab format should only be considered for affinity maturation of antibodies that allow functional Fab display on phage and expression in E. coli and only in addition to FabΔC.
To summarize, the choice of the antibody format during affinity maturation has a substantial effect on the development of IgGs with improved affinity. Affinity maturation in single chain antibody formats like scFv or scFabΔC bears the risk of generating a bias for affinity-enhanced molecules with altered conformation whose improved antigen binding properties cannot be transferred to IgG. The two chain FabΔC format, rather than the Fab format, represented the best compromise, preserving the functional conformation of the antigen binding site while still allowing sufficient bacterial expression for phage display.
Materials and Methods
Construction of phage display vectors
Phage display vectors pHAL21 (scFabΔC), pHAL26 (Fab) and pHAL34 (FabΔC) are based on the phagemid backbone of pHAL14 (scFv).58-61 All modifications compared with pHAL14 are shown in Figure 1B. For the construction of the scFabΔC phage vector pHAL21, the entire scFv gene expression cassette of pHAL14 was modified by introducing gene fragments encoding CL, a 32 amino acid linker (SGGG)2-(SEGGG)4-SGGGSG and the CH1. The 3′ codon of CL and CH1 encoding the carboxy terminal cysteines were deleted (ΔC). The 5′ end of both CLΔC and CH1ΔC was endowed with appropriate restriction cloning sites for cloning of VL and VH, respectively.
The bicistronic Fab and FabΔC phagemids pHAL26 and pHAL34 were constructed from pHAL 21 by replacing the linker with a 3′ ochre stop codon after the CL (or CLΔC), followed by a non-coding spacer with a second RBS and pelB leader sequence and the VH cloning site to enable bicistronic expression of the light chain and heavy chain Fd fragment, respectively. The CH1ΔC gene fragment was replaced in pHAL26 by the complete CH1 to obtain both cysteine residues for the interchain disulfide bond in the Fab.
Cloning, production and purification of the antigen CD30
The gene of the extracellular domain of CD30 was cloned into the eukaryotic expression vector pCSE2.5-His-XP, which was generated by replacing the Fc gene fragment of vector pCSE2.5-hIgG1Fc-XP by a hexa-histidine tag (data not shown). The monomeric his-tagged CD30 protein and a dimeric CD30-Fc fusion protein54 were transiently produced in HEK293–6E cells66,67 and purified by immobilized metal ion (Ni2+) or protein A affinity chromatography, respectively, using the ProfiniaTM Affinity Chromatography Protein Purification System (Bio-Rad) according to the manufacturer's description.
Production and purification of antibody phage
A total of 50 mL 2xTY,68 100 µg/mL ampicillin, 100 mM glucose was inoculated with an overnight culture of phagemid bearing E. coli XL1-Blue MRF' (Agilent Technologies) and grown at O.D.600 0.4–0.5 at 37 °C and 250 rpm. A total of 2 mL of the bacteria was infected with 2 × 1010 pfu M13K07 or hyperphage69 for 30 min at 37 °C, followed by incubation for 30 min at 37 °C and 250 rpm. Bacteria were pelleted at 3,220 xg for 10 min at RT. The pellet was resuspended in 30 mL 2 xTY supplemented with 100 µg/mL ampicillin and 50 µg/mL kanamycin and incubated for about 20 h at 30 °C and 250 rpm. Bacteria were centrifuged at 3,220 xg for 10 min and 4 °C and phage particles in the supernatant were precipitated with 1/5 volume 20% (w/v) polyethylene glycol (PEG) 6000 + 2.5 M NaCl for 1 h at 4 °C, followed by centrifugation at 3220 xg for 1 h at 4 °C. The pellet was resuspended in phage dilution buffer (10 mM TRIS-HCl pH 7.5, 20 mM NaCl, 2 mM EDTA) and centrifuged again at 16,000xg for 1 min to pellet cell debris. The phage containing supernatant was stored at 4 °C.
Construction of mutation libraries
A total of 0.05 ng template DNA of SH313-F9 scFv was amplified using a random mutagenesis PCR kit (GeneMorphII, Stratagene) and 0.2 µM oligonucleotides (MHColE1: 5′ cccaatacgcaaaccgcc 3′; MhgIII_r: 5′ ctaaagttttgtcgtctttcc 3′) in a total volume of 25 µL. The first denaturating step was done for 120 s at 95 °C, followed by 35 cycles (95 °C 60 s, 65 °C 60 s, 72 °C 70 s) and a final synthesis step for 10 min at 72 °C. After agarose gel electrophoresis the PCR product was purified via Nucleospin Extract 2 Kit (Macherey- Nagel, Düren, Germany) and used as template for the next round of error-prone PCR with nested primers (MHLacZ-Pro_f: 5′ ggctcgtatgttgtgtgg 3′; HT-gIII-Beginn-1-rev: 5′ taaacaactttcaacagtttcagct 3′). This step was repeated as third round of error-prone PCR followed by an additional amplification step using the Phusion® high-fidelity DNA polymerase (Thermo Fisher Scientific) with 40 nM dNTPs and 0.1 µM desoxyoligonucleotides llB6-Aff-2-fwd 5′ gccgctggcttgctgctgctggcagctcagccggccatgg 3′ and llB6-Aff-2-rev 5′ gttctgcggccccgtgatggtgatgatgatgagcggccgc. The amplified PCR product was purified and cloned using the restriction sites NcoI and NotI into the phagemid pHAL14. For the antibody formats scFabΔC, Fab and FabΔC amplification and cloning was done as follows: For amplification of VL the desoxyoligonucleotides PaD_F9_NcoI_f 5′ ATCTCCATGGCCCAGCCTGGGCTGACTC 3′ and PaD_F9_Hind_r 5′ ACTGAAGCTTGGTCCCTCCGCCGAATACC 3′ were used and the PCR product was cloned via NcoI and HindIII into the phagemids pHAL21 (scFabΔC), pHAL34 (Fab) and pHAL34 (FabΔC). For the amplification of VH, the oligonucleotides PaD_F9_MfeI_f 5′ ATCTCAATTGGTGCAGTCTGGGTCTG 3′ and PaD_B5F9_NheI_r 5′ ATCGGCTAGCTGAAGAGACGGTGACC 3′ were used and the PCR product was cloned via MfeI and NheI into the VL containing phagemids pHAL21-VL (scFabΔC), pHAL26-VL (Fab) and pHAL34-VL (FabΔC). The ligation was performed in a 100 µL reaction using 3 units T4 DNA ligase (Promega, Fitchburg, USA) for 16 °C and 12 h with a vector:insert ratio of 1:3. The ligation was stopped at 65 °C for 10 min. E. coli XL1-Blue MRF' were transformed by electroporation and packaged with M13K07 as described.43
Selection of affinity matured antibody fragments
A total of 100 ng CD30-Fc was coated in PBS68 into Nunc Maxisorp stripes (Nunc, Langenselbold, Germany) for 1 h at RT. Free binding sites were blocked with 1% (w/v) BSA in M-PBST (PBS supplemented with 0.1% Tween 20 and 2% skim milk powder) for 1 h at RT. The wells were washed with PBST using an ELISA washer (Tecan Columbus, Tecan, Crailsheim, Germany). Additionally, Nunc Maxisorp stripes without antigen were also blocked with 1% (w/v) BSA in M-PBST. A total of 1010 phage particles of each mutation library were diluted in M-PBST and incubated in separate wells prepared without antigen for 1 h at RT to remove unspecific binding antibody phage particles. The non-bound antibody phage particles were transferred into wells containing CD30-Fc antigen. After 1 h at RT, the wells were washed 10 × with a stringent bottom wash program using the ELISA washer (COLUMBUS, Tecan). A total 150 µL of 1 nM soluble CD30-Fc antigen was added for competition with the immobilized antigen. After 1 h incubation at RT the wells were washed again under stringent conditions. Bound phage particles were eluted with 200 µL/well 10 µg/mL trypsin for 30 min at 37 °C. Eluted antibody phage were used for infection of 50 µL E. coli XL1-Blue MRF' (O.D.600 0.4–0.5). A dilution series of these infected bacteria was plated on 2xTY agar plates68 supplemented with 100 µg/mL ampicillin and 100 mM glucose and incubated over night at 37 °C.
Production of antibody fragments in microtiter plates (MTPs)
After panning, colonies of bacteria infected with eluted antibody phage were isolated and inoculated in microtiter plates (MTP) to produce soluble antibody fragments as described previously.70 These antibody fragments were analyzed by ELISA on CD30-Fc and compared with parent SH313-F9 in the corresponding antibody format.
Enzyme linked immunosorbent assays for different antibody formats and antibody phage
A number of ELISAs were performed at different steps in this study, but in all cases a total of 50 ng CD30-Fc or CD30-His antigen was coated in 96-well MTPs (High Binding, Costar) in PBS over night at 4 °C and subsequent blocking was performed with 2% M-PBST (2% (w/v) skim milk powder in PBST) for 1 h at RT. Analogously, wells were prepared with BSA as control antigen. After three washing steps using an ELISA washer (Columbus, TECAN), antibody containing supernatants, purified antibodies or antibody phage were incubated for 1 h at RT. All antibody or antibody phage incubations, including all detection steps, were performed for 1 h at RT, including a subsequent set of three washing steps using an ELSA washer. Finally, substrate TMB (3,3′,5,5′-tetramethylbenzidine) was added and the color reaction was stopped by adding 100 µL 1 N sulfuric acid. Absorbances were measured at 450 nm (with 620 nm reference wavelength) using an ELISA reader (SUNRISE, Tecan).
Screening ELISA after affinity maturation was performed with 50 µL production supernatant containing soluble antibody fragments (scFv, Fab, scFabΔC, or FabΔC) as described previously.70 Detection of bound antibody fragments was performed with the c-myc tag specific IgG 9E10 followed by goat anti-mouse Fc-specific secondary antibody horseradish peroxidase (HRP) conjugate (Sigma).
A total of 100 µL/well of a dilution series of antibody-phage in different SH313-F9 formats (scFv, Fab, scFabΔC, or FabΔC) were prepared in 2% M-PBST and incubated on antigen-coated wells. Detection was performed with a mouse anti-M13 antibody HRP conjugate (GE Healthcare) followed by goat anti-mouse Fc specific secondary antibody horseradish peroxidase (HRP) conjugate (Sigma).
A total of 100 µL of a concentration series of scFv-Fc, scIgG, and IgG antibodies were diluted in 2% M-PBST and incubated on antigen coated wells. Detection was done with goat anti-human Fc specific antibody HRP conjugate (Sigma)
SDS-PAGE and Immunoblot
Samples of antibody phage packaged with Hyperphage were run on a reducing 10% SDS-PAGE gel and blotted onto polyvinylidene fluoride (PVDF) membrane using a semidry electroblotting system (BioRad, Hercules, USA). Then, the membrane was blocked for 1 h at room temperature in 2% MPBS (2% skim milk powder in PBS). Monoclonal mouse anti-pIII antibody (1:2000, MoBiTec) was incubated for 1 h at RT. After three washing steps with PBST the membrane was incubated with goat anti-mouse IgG alkaline phosphatase (AP) conjugate (1:10 000, Sigma) for 1 h at RT. After two washing steps with PBST, a third washing step with substrate buffer was performed (100 mM TRIS-HCl, 0.5 mM MgCl2, pH 9.5). Substrate buffer with 1% (v/v) 5-Brom-4-chlor-3-indolylphosphate (BCIP) (Applichem) and 1% (v/v) nitro-blue tetrazolium (Applichem) was incubated for visualization. This reaction was stopped by washing with water.
Construction of mammalian expression vectors
Monocistronic IgG expression vectors pCSH1c, pCSL3l (Fig. 1C) and pCSL3k were generated using the backbone of pCSE2.5-hIgG1Fc-XP44,45 and light and heavy immunoglobulin chains of the heavy and light chain IgG expression vectors pSH1 and pSL1, respectively.71
For the generation of the heavy chain expression vector pCSH1c, the entire expression cassette, including the CMV promoter, the mouse IgG heavy chain signal peptide and the constant regions of the human IgG1 heavy chain of pSH1, was cloned into the vector backbone of pCSE2.5-hIgG1Fc-XP via EcoRI and XbaI. Cloning of VH can be done by using BssHII and NheI restriction sites. To obtain the light chain expression vectors with cloning cassettes that allow the introduction of VL gene fragments without altering any codon compared with human immunoglobulin sequences, restriction sites were generated by silent mutations that were introduced into the gene sequences of human kappa signal peptide and C-kappa of vector pSL1 by several PCRs. First, the DNA sequence comprising CMV promoter and signal peptide was amplified from pSL1 by using desoxyoligonucleotide primers CMV_EcorRI_f2 (cct agc gaa ttc aca ttg att att gag tag) and AF_k_Leader-AgeI_SacI_r (cct agc gag ctc acc ggt gct ttc tgg gag cca g), with the latter introducing the AgeI site at the 3′ end of leader sequence (underlined). This PCR fragment was cloned into the light chain expression vector pSL1 using EcoRI and SacI. In a second step, the desoxyoligonucleotide primers AF_CLk_XbaI_r (cct agc tct aga act aac act ctc ccc) and AF_CLk_HindIII_BsiWI_f (cct agc aag ctt cgt acg gtg gct gca cca tct gtg) were used to amplify C-kappa from pSL1 and to introduce a BsiWI site to its 5′ end. This PCR fragment was cloned into the pSL1 vector containing the modified leader sequence using HindIII and XbaI. Finally, the entire expression cassette was inserted into the pCSE2.5 vector backbone to obtain the kappa light chain expression vector pCSL3k. The vector pCSL3k was not used in this study.
Lambda light chain 2 (C2) was amplified using the desoxyoligonucleotide primers AF_Cl_HindIII_DraIII_f (cta aaa gct tac cgt cct agg tca gcc caa ggc tgc acc caa gtg tca ctc tg) and AF_Cl_XbaI_r (cga gct tct aga gtt taa act cac tat gaa cat tct gta ggg g) with the first primer introducing DraIII-site at the 5′ end of the C2 gene fragment (underlined in the primer sequence). The PCR product was finally cloned into pCSL3k to replace the C-kappa sequence with the lambda C2 gene fragment using HindIII and XbaI, resulting in the lambda light chain expression vector pCSL3l.
Cloning, Production and purification of IgGs, scFv-Fcs, and IgGs
For IgG format VH and VL of the antibody fragments were recloned into the heavy chain vector pCSH1c and the light chain vector pCSL3l using the restriction sites BssHII/NheI and AgeI/DraIII, respectively. Therefore, VH and VL were amplified using the oligonucleotides SH313-B5/F9-VH_for (5′ gatcgcgcgcactcccaggtgcagctggtgcag 3′), AF110.1_B12_HC_NheI_r (5′ ccgagcgctagctgaagagacggtgaccattgtccc 3′) and SH313-F9_VL_for (5′ gatccaccggtcagcctgggctgactcag 3′), AF_pCSL3_DraIII-rev (5′ AGTGACACTTGGTGCAGCCTTGGGCTGACC 3′), respectively.
For the conversion of scFvs in scFv-Fcs and scFabΔC into scIgGs, the antibody fragments were recloned into the vector pCSE2.5-hIgG1-Fc-XP using the restriction sites NcoI/NotI.
Production and purification of the antibodies was done as described above for CD30-hIgG1-Fc.
Flow cytometry
Cells were washed and resuspended in FACS buffer (PBS supplemented with 2% (v/v) fetal calf serum and 2 mM EDTA). A total of 105 cells were incubated with the anti-CD30 IgGs at 4 °C for 15 min. The cells were washed with FACS buffer and incubated with anti-human IgG Fc gamma specific conjugated with FITC (1:100, Dianova) at 4 °C for 15 min. Cells were washed again and resuspended in FACS buffer containing 1 µg/mL propidium iodide and analyzed by flow cytometry using a FC500 (emission at 488 nm, Beckman Coulter). For each sample, 10,000 events were measured and analyzed using FlowJo7.2.5 (Tree Star). Debris and dead cells were excluded during analysis.
Surface plasmon resonance
SPR was performed using Biacore 2000 (GE Healthcare) according to the manufacturer's manual. The human Antibody Capture Kit (GE Healthcare) was used to couple monoclonal mouse anti-human IgG Fc specific capture antibody to a CM5 sensor chip (GE Healthcare) according to manufacturer's protocol. Following, 7 µg/mL IgGs were captured at a flow rate of 10 µL/min. The binding to different dilutions of monomeric CD30-His (0.01–1.5 µM) was measured at a flow rate of 25 µL/min. Regeneration was done with 3 M MgCl2 at a flow rate of 100 µL/min. For reference, a flow cell was activated with 1-ethyl-3-[3-dimethylaminopropyl]carbodiimide hydrochloride / N-hydroxysuccinimide and then blocked with ethanolamine. Data fitting was performed using 1:1 Langmuir binding algorithm with drifting baseline and local Rmax of the BIAevaluation software.
Size exclusion chromatography
Superdex-200 10/300 GL (GE Healthcare) on ÄKTA purifier (GE Healthcare) with UnicornTM 5.10 software (GE Healthcare) was employed for analytical SEC of IgG, scFv-Fc, and scIgG. A total of 500 µL of each sample was applied on the column using a flow rate of 0.5 mL/min and PBS as running buffer. Absorbance at 280 nm was plotted against the retention volume to identify the aggregation behavior of the antibodies. Molecular masses were calculated according to calibration standard proteins (GE Healthcare).
Disclosure of Potential Conflicts of Interest
The authors declare that they have no conflicting interests.
Acknowledgments
We acknowledge the financial support by the EU FP7 collaborative projects “Affinity Proteome” (contract 222635) and “Affinomics” (contract 241481).
Footnotes
Previously published online: www.landesbioscience.com/journals/mabs/article/27227
Reference
- 1.Foote J, Eisen HN. Kinetic and affinity limits on antibodies produced during immune responses. Proc Natl Acad Sci U S A. 1995;92:1254–6. doi: 10.1073/pnas.92.5.1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Batista FD, Neuberger MS. Affinity dependence of the B cell response to antigen: a threshold, a ceiling, and the importance of off-rate. Immunity. 1998;8:751–9. doi: 10.1016/S1074-7613(00)80580-4. [DOI] [PubMed] [Google Scholar]
- 3.Johnson S, Griego SD, Pfarr DS, Doyle ML, Woods R, Carlin D, Prince GA, Koenig S, Young JF, Dillon SB. A direct comparison of the activities of two humanized respiratory syncytial virus monoclonal antibodies: MEDI-493 and RSHZl9. J Infect Dis. 1999;180:35–40. doi: 10.1086/314846. [DOI] [PubMed] [Google Scholar]
- 4.Maynard JA, Maassen CBM, Leppla SH, Brasky K, Patterson JL, Iverson BL, Georgiou G. Protection against anthrax toxin by recombinant antibody fragments correlates with antigen affinity. Nat Biotechnol. 2002;20:597–601. doi: 10.1038/nbt0602-597. [DOI] [PubMed] [Google Scholar]
- 5.Putnam WS, Li J, Haggstrom J, Ng C, Kadkhodayan-Fischer S, Cheu M, Deniz Y, Lowman H, Fielder P, Visich J, et al. Use of quantitative pharmacology in the development of HAE1, a high-affinity anti-IgE monoclonal antibody. AAPS J. 2008;10:425–30. doi: 10.1208/s12248-008-9045-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu H, Pfarr DS, Johnson S, Brewah YA, Woods RM, Patel NK, White WI, Young JF, Kiener PA. Development of motavizumab, an ultra-potent antibody for the prevention of respiratory syncytial virus infection in the upper and lower respiratory tract. J Mol Biol. 2007;368:652–65. doi: 10.1016/j.jmb.2007.02.024. [DOI] [PubMed] [Google Scholar]
- 7.Verhoeyen M, Milstein C, Winter G. Reshaping human antibodies: grafting an antilysozyme activity. Science. 1988;239:1534–6. doi: 10.1126/science.2451287. [DOI] [PubMed] [Google Scholar]
- 8.Makabe K, Nakanishi T, Tsumoto K, Tanaka Y, Kondo H, Umetsu M, Sone Y, Asano R, Kumagai I. Thermodynamic consequences of mutations in vernier zone residues of a humanized anti-human epidermal growth factor receptor murine antibody, 528. J Biol Chem. 2008;283:1156–66. doi: 10.1074/jbc.M706190200. [DOI] [PubMed] [Google Scholar]
- 9.Schlapschy M, Gruber H, Gresch O, Schäfer C, Renner C, Pfreundschuh M, Skerra A. Functional humanization of an anti-CD30 Fab fragment for the immunotherapy of Hodgkin’s lymphoma using an in vitro evolution approach. Protein Eng Des Sel. 2004;17:847–60. doi: 10.1093/protein/gzh098. [DOI] [PubMed] [Google Scholar]
- 10.Schlapschy M, Fogarasi M, Gruber H, Gresch O, Schäfer C, Aguib Y, Skerra A. Functional humanization of an anti-CD16 Fab fragment: obstacles of switching from murine lambda to human lambda or kappa light chains. Protein Eng Des Sel. 2009;22:175–88. doi: 10.1093/protein/gzn066. [DOI] [PubMed] [Google Scholar]
- 11.McCafferty J, Griffiths AD, Winter G, Chiswell DJ. Phage antibodies: filamentous phage displaying antibody variable domains. Nature. 1990;348:552–4. doi: 10.1038/348552a0. [DOI] [PubMed] [Google Scholar]
- 12.Breitling F, Dübel S, Seehaus T, Klewinghaus I, Little M. A surface expression vector for antibody screening. Gene. 1991;104:147–53. doi: 10.1016/0378-1119(91)90244-6. [DOI] [PubMed] [Google Scholar]
- 13.Clackson T, Hoogenboom HR, Griffiths AD, Winter G. Making antibody fragments using phage display libraries. Nature. 1991;352:624–8. doi: 10.1038/352624a0. [DOI] [PubMed] [Google Scholar]
- 14.Hoogenboom HR, Griffiths AD, Johnson KS, Chiswell DJ, Hudson P, Winter G. Multi-subunit proteins on the surface of filamentous phage: methodologies for displaying antibody (Fab) heavy and light chains. Nucleic Acids Res. 1991;19:4133–7. doi: 10.1093/nar/19.15.4133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schofield DJ, Pope AR, Clementel V, Buckell J, Chapple SDj, Clarke KF, Conquer JS, Crofts AM, Crowther SRE, Dyson MR, et al. Application of phage display to high throughput antibody generation and characterization. Genome Biol. 2007;8:R254. doi: 10.1186/gb-2007-8-11-r254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dübel S, Stoevesandt O, Taussig MJ, Hust M. Generating recombinant antibodies to the complete human proteome. Trends Biotechnol. 2010;28:333–9. doi: 10.1016/j.tibtech.2010.05.001. [DOI] [PubMed] [Google Scholar]
- 17.Hust M, Meyer T, Voedisch B, Rülker T, Thie H, El-Ghezal A, Kirsch MI, Schütte M, Helmsing S, Meier D, et al. A human scFv antibody generation pipeline for proteome research. J Biotechnol. 2011;152:159–70. doi: 10.1016/j.jbiotec.2010.09.945. [DOI] [PubMed] [Google Scholar]
- 18.Söderlind E, Simonsson AC, Borrebaeck CAK. Phage display technology in antibody engineering: design of phagemid vectors and in vitro maturation systems. Immunol Rev. 1992;130:109–24. doi: 10.1111/j.1600-065X.1992.tb01523.x. [DOI] [PubMed] [Google Scholar]
- 19.Thie H, Toleikis L, Li J, von Wasielewski R, Bastert G, Schirrmann T, Esteves IT, Behrens CK, Fournes B, Fournier N, et al. Rise and fall of an anti-MUC1 specific antibody. PLoS One. 2011;6:e15921. doi: 10.1371/journal.pone.0015921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hanes J, Jermutus L, Weber-Bornhauser S, Bosshard HR, Plückthun A. Ribosome display efficiently selects and evolves high-affinity antibodies in vitro from immune libraries. Proc Natl Acad Sci U S A. 1998;95:14130–5. doi: 10.1073/pnas.95.24.14130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhao X-L, Chen W-Q, Yang Z-H, Li J-M, Zhang S-J, Tian L-F. Selection and affinity maturation of human antibodies against rabies virus from a scFv gene library using ribosome display. J Biotechnol. 2009;144:253–8. doi: 10.1016/j.jbiotec.2009.09.022. [DOI] [PubMed] [Google Scholar]
- 22.Wang Z, Kim G-B, Woo J-H, Liu YY, Mathias A, Stavrou S, Neville DM., Jr. Improvement of a recombinant anti-monkey anti-CD3 diphtheria toxin based immunotoxin by yeast display affinity maturation of the scFv. Bioconjug Chem. 2007;18:947–55. doi: 10.1021/bc0603438. [DOI] [PubMed] [Google Scholar]
- 23.Siegel RW. Antibody affinity optimization using yeast cell surface display. Methods Mol Biol. 2009;504:351–83. doi: 10.1007/978-1-60327-569-9_20. [DOI] [PubMed] [Google Scholar]
- 24.Daugherty PS, Chen G, Olsen MJ, Iverson BL, Georgiou G. Antibody affinity maturation using bacterial surface display. Protein Eng. 1998;11:825–32. doi: 10.1093/protein/11.9.825. [DOI] [PubMed] [Google Scholar]
- 25.Fukuda I, Kojoh K, Tabata N, Doi N, Takashima H, Miyamoto-Sato E, Yanagawa H. In vitro evolution of single-chain antibodies using mRNA display. Nucleic Acids Res. 2006;34:e127. doi: 10.1093/nar/gkl618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martineau P. Error-prone polymerase chain reaction for modification of scFvs. Methods Mol Biol. 2002;178:287–94. doi: 10.1385/1-59259-240-6:287. [DOI] [PubMed] [Google Scholar]
- 27.Thie H, Voedisch B, Dübel S, Hust M, Schirrmann T. Affinity Maturation by Phage Display [Internet]. In: Dimitrov AS, herausgeber. Therapeutic Antibodies. Humana Press; 2009 [zitiert 2013 Aug 5]. Seite 309–22.Available from: http://link.springer.com/protocol/10.1007/978-1-59745-554-1_16
- 28.Irving RA, Kortt AA, Hudson PJ. Affinity maturation of recombinant antibodies using E. coli mutator cells. Immunotechnology. 1996;2:127–43. doi: 10.1016/1380-2933(96)00044-9. [DOI] [PubMed] [Google Scholar]
- 29.Low NM, Holliger PH, Winter G. Mimicking somatic hypermutation: affinity maturation of antibodies displayed on bacteriophage using a bacterial mutator strain. J Mol Biol. 1996;260:359–68. doi: 10.1006/jmbi.1996.0406. [DOI] [PubMed] [Google Scholar]
- 30.Lewis L, Lloyd C. Optimisation of antibody affinity by ribosome display using error-prone or site-directed mutagenesis. Methods Mol Biol. 2012;805:139–61. doi: 10.1007/978-1-61779-379-0_9. [DOI] [PubMed] [Google Scholar]
- 31.Yang WP, Green K, Pinz-Sweeney S, Briones AT, Burton DR, Barbas CF., 3rd CDR walking mutagenesis for the affinity maturation of a potent human anti-HIV-1 antibody into the picomolar range. J Mol Biol. 1995;254:392–403. doi: 10.1006/jmbi.1995.0626. [DOI] [PubMed] [Google Scholar]
- 32.Balint RF, Larrick JW. Antibody engineering by parsimonious mutagenesis. Gene. 1993;137:109–18. doi: 10.1016/0378-1119(93)90258-5. [DOI] [PubMed] [Google Scholar]
- 33.Barbas CF, 3rd, Hu D, Dunlop N, Sawyer L, Cababa D, Hendry RM, Nara PL, Burton DR. In vitro evolution of a neutralizing human antibody to human immunodeficiency virus type 1 to enhance affinity and broaden strain cross-reactivity. Proc Natl Acad Sci U S A. 1994;91:3809–13. doi: 10.1073/pnas.91.9.3809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rajpal A, Beyaz N, Haber L, Cappuccilli G, Yee H, Bhatt RR, Takeuchi T, Lerner RA, Crea R. A general method for greatly improving the affinity of antibodies by using combinatorial libraries. Proc Natl Acad Sci U S A. 2005;102:8466–71. doi: 10.1073/pnas.0503543102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jermutus L, Honegger A, Schwesinger F, Hanes J, Plückthun A. Tailoring in vitro evolution for protein affinity or stability. Proc Natl Acad Sci U S A. 2001;98:75–80. doi: 10.1073/pnas.98.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marks JD. Antibody affinity maturation by chain shuffling. Methods Mol Biol. 2004;248:327–43. doi: 10.1385/1-59259-666-5:327. [DOI] [PubMed] [Google Scholar]
- 37.Schier R, Bye J, Apell G, McCall A, Adams GP, Malmqvist M, Weiner LM, Marks JD. Isolation of high-affinity monomeric human anti-c-erbB-2 single chain Fv using affinity-driven selection. J Mol Biol. 1996;255:28–43. doi: 10.1006/jmbi.1996.0004. [DOI] [PubMed] [Google Scholar]
- 38.Yoshinaga K, Matsumoto M, Torikai M, Sugyo K, Kuroki S, Nogami K, Matsumoto R, Hashiguchi S, Ito Y, Nakashima T, et al. Ig L-chain shuffling for affinity maturation of phage library-derived human anti-human MCP-1 antibody blocking its chemotactic activity. J Biochem. 2008;143:593–601. doi: 10.1093/jb/mvn009. [DOI] [PubMed] [Google Scholar]
- 39.Kobayashi N, Oyama H, Kato Y, Goto J, Söderlind E, Borrebaeck CAK. Two-step in vitro antibody affinity maturation enables estradiol-17beta assays with more than 10-fold higher sensitivity. Anal Chem. 2010;82:1027–38. doi: 10.1021/ac902283n. [DOI] [PubMed] [Google Scholar]
- 40.Marks JD, Griffiths AD, Malmqvist M, Clackson TP, Bye JM, Winter G. By-passing immunization: building high affinity human antibodies by chain shuffling. Biotechnology (N Y) 1992;10:779–83. doi: 10.1038/nbt0792-779. [DOI] [PubMed] [Google Scholar]
- 41.He M, Taussig MJ. Antibody-ribosome-mRNA (ARM) complexes as efficient selection particles for in vitro display and evolution of antibody combining sites. Nucleic Acids Res. 1997;25:5132–4. doi: 10.1093/nar/25.24.5132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bird RE, Hardman KD, Jacobson JW, Johnson S, Kaufman BM, Lee SM, Lee T, Pope SH, Riordan GS, Whitlow M. Single-chain antigen-binding proteins. Science. 1988;242:423–6. doi: 10.1126/science.3140379. [DOI] [PubMed] [Google Scholar]
- 43.Hust M, Jostock T, Menzel C, Voedisch B, Mohr A, Brenneis M, Kirsch MI, Meier D, Dübel S. Single chain Fab (scFab) fragment. BMC Biotechnol. 2007;7:14. doi: 10.1186/1472-6750-7-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schirrmann T, Menzel C, Hust M, Prilop J, Jostock T, Dübel S. Oligomeric forms of single chain immunoglobulin (scIgG) MAbs. 2010;2:73–6. doi: 10.4161/mabs.2.1.10784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jäger V, Büssow K, Wagner A, Weber S, Hust M, Frenzel A, Schirrmann T. High level transient production of recombinant antibodies and antibody fusion proteins in HEK293 cells. BMC Biotechnol. 2013;13:52. doi: 10.1186/1472-6750-13-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ames RS, Tornetta MA, Deen K, Jones CS, Swift AM, Ganguly S. Conversion of murine Fabs isolated from a combinatorial phage display library to full length immunoglobulins. J Immunol Methods. 1995;184:177–86. doi: 10.1016/0022-1759(95)00086-P. [DOI] [PubMed] [Google Scholar]
- 47.Huls GA, Heijnen IA, Cuomo ME, Koningsberger JC, Wiegman L, Boel E, van der Vuurst de Vries AR, Loyson SA, Helfrich W, van Berge Henegouwen GP, et al. A recombinant, fully human monoclonal antibody with antitumor activity constructed from phage-displayed antibody fragments. Nat Biotechnol. 1999;17:276–81. doi: 10.1038/7023. [DOI] [PubMed] [Google Scholar]
- 48.Liu B, Conrad F, Roth A, Drummond DC, Simko JP, Marks JD. Recombinant full-length human IgG1s targeting hormone-refractory prostate cancer. J Mol Med (Berl) 2007;85:1113–23. doi: 10.1007/s00109-007-0208-z. [DOI] [PubMed] [Google Scholar]
- 49.Pohl SC, Schwarz S, Frenzel A, Schirrmann T. A Cassette Vector System for the Rapid Cloning and Production of Bispecific Tetravalent Antibodies. Antibodies. 2012;1:19–38. doi: 10.3390/antib1010019. [DOI] [Google Scholar]
- 50.Menzel C, Schirrmann T, Konthur Z, Jostock T, Dübel S. Human antibody RNase fusion protein targeting CD30+ lymphomas. Blood. 2008;111:3830–7. doi: 10.1182/blood-2007-04-082768. [DOI] [PubMed] [Google Scholar]
- 51.Jostock T, Vanhove M, Brepoels E, Van Gool R, Daukandt M, Wehnert A, Van Hegelsom R, Dransfield D, Sexton D, Devlin M, et al. Rapid generation of functional human IgG antibodies derived from Fab-on-phage display libraries. J Immunol Methods. 2004;289:65–80. doi: 10.1016/j.jim.2004.03.014. [DOI] [PubMed] [Google Scholar]
- 52.Renaut L, Monnet C, Dubreuil O, Zaki O, Crozet F, Bouayadi K, Kharrat H, Mondon P. Affinity maturation of antibodies: optimized methods to generate high-quality ScFv libraries and isolate IgG candidates by high-throughput screening. Methods Mol Biol. 2012;907:451–61. doi: 10.1007/978-1-61779-974-7_26. [DOI] [PubMed] [Google Scholar]
- 53.Votsmeier C, Plittersdorf H, Hesse O, Scheidig A, Strerath M, Gritzan U, Pellengahr K, Scholz P, Eicker A, Myszka D, et al. Femtomolar Fab binding affinities to a protein target by alternative CDR residue co-optimization strategies without phage or cell surface display. MAbs. 2012;4:341–8. doi: 10.4161/mabs.19981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wezler X, Hust M, Helmsing S, Schirrmann T, Dübel S. Human antibodies targeting CD30(+) lymphomas. Hum Antibodies. 2012;21:13–28. doi: 10.3233/HAB-2012-0258. [DOI] [PubMed] [Google Scholar]
- 55.Falini B, Pileri S, Pizzolo G, Dürkop H, Flenghi L, Stirpe F, Martelli MF, Stein H. CD30 (Ki-1) molecule: a new cytokine receptor of the tumor necrosis factor receptor superfamily as a tool for diagnosis and immunotherapy. Blood. 1995;85:1–14. [PubMed] [Google Scholar]
- 56.Jordan E, Al-Halabi L, Schirrmann T, Hust M, Dübel S. Production of single chain Fab (scFab) fragments in Bacillus megaterium. Microb Cell Fact. 2007;6:38. doi: 10.1186/1475-2859-6-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Thie H, Binius S, Schirrmann T, Hust M, Dübel S. Multimerization domains for antibody phage display and antibody production. N Biotechnol. 2009;26:314–21. doi: 10.1016/j.nbt.2009.07.005. [DOI] [PubMed] [Google Scholar]
- 58.Kirsch MI, Hülseweh B, Nacke C, Rülker T, Schirrmann T, Marschall H-J, Hust M, Dübel S. Development of human antibody fragments using antibody phage display for the detection and diagnosis of Venezuelan equine encephalitis virus (VEEV) BMC Biotechnol. 2008;8:66. doi: 10.1186/1472-6750-8-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pelat T, Hust M, Hale M, Lefranc M-P, Dübel S, Thullier P. Isolation of a human-like antibody fragment (scFv) that neutralizes ricin biological activity. BMC Biotechnol. 2009;9:60. doi: 10.1186/1472-6750-9-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pelat T, Hust M, Laffly E, Condemine F, Bottex C, Vidal D, Lefranc M-P, Dübel S, Thullier P. High-affinity, human antibody-like antibody fragment (single-chain variable fragment) neutralizing the lethal factor (LF) of Bacillus anthracis by inhibiting protective antigen-LF complex formation. Antimicrob Agents Chemother. 2007;51:2758–64. doi: 10.1128/AAC.01528-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schütte M, Thullier P, Pelat T, Wezler X, Rosenstock P, Hinz D, Kirsch MI, Hasenberg M, Frank R, Schirrmann T, et al. Identification of a putative Crf splice variant and generation of recombinant antibodies for the specific detection of Aspergillus fumigatus. PLoS One. 2009;4:e6625. doi: 10.1371/journal.pone.0006625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schmiedl A, Breitling F, Winter CH, Queitsch I, Dübel S. Effects of unpaired cysteines on yield, solubility and activity of different recombinant antibody constructs expressed in E. coli. J Immunol Methods. 2000;242:101–14. doi: 10.1016/S0022-1759(00)00243-X. [DOI] [PubMed] [Google Scholar]
- 63.Nowakowski A, Wang C, Powers DB, Amersdorfer P, Smith TJ, Montgomery VA, Sheridan R, Blake R, Smith LA, Marks JD. Potent neutralization of botulinum neurotoxin by recombinant oligoclonal antibody. Proc Natl Acad Sci U S A. 2002;99:11346–50. doi: 10.1073/pnas.172229899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Phumyen A, Jumnainsong A, Leelayuwat C. Improved binding activity of antibodies against major histocompatibility complex class I chain-related gene A by phage display technology for cancer-targeted therapy. J Biomed Biotechnol. 2012;2012:597647. doi: 10.1155/2012/597647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Xie MH, Yuan J, Adams C, Gurney A. Direct demonstration of MuSK involvement in acetylcholine receptor clustering through identification of agonist ScFv. Nat Biotechnol. 1997;15:768–71. doi: 10.1038/nbt0897-768. [DOI] [PubMed] [Google Scholar]
- 66.Durocher Y, Perret S, Kamen A. High-level and high-throughput recombinant protein production by transient transfection of suspension-growing human 293-EBNA1 cells. Nucleic Acids Res. 2002;30:E9. doi: 10.1093/nar/30.2.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Schirrmann T, Büssow K. Transient Production of scFv-Fc Fusion Proteins in Mammalian Cells [Internet]. In: Kontermann R, Dübel S, herausgeber. Antibody Engineering. Berlin, Heidelberg: Springer Berlin Heidelberg; 2010 [zitiert 2013 Mai 22]. Seite 387–98.Available from: http://www.springerprotocols.com/Abstract/doi/10.1007/978-3-642-01147-4_30
- 68.Sambrook J, Russell DW. Molecular Cloning: A Laboratory Manual, 3 Vol. 0003 Aufl. Cold Spring Harbor Laboratory; 2000. [Google Scholar]
- 69.Rondot S, Koch J, Breitling F, Dübel S. A helper phage to improve single-chain antibody presentation in phage display. Nat Biotechnol. 2001;19:75–8. doi: 10.1038/83567. [DOI] [PubMed] [Google Scholar]
- 70.Hust M, Steinwand M, Al-Halabi L, Helmsing S, Schirrmann T, Dübel S. Improved microtitre plate production of single chain Fv fragments in Escherichia coli. N Biotechnol. 2009;25:424–8. doi: 10.1016/j.nbt.2009.03.004. [DOI] [PubMed] [Google Scholar]
- 71.Li J, Menzel C, Meier D, Zhang C, Dübel S, Jostock T. A comparative study of different vector designs for the mammalian expression of recombinant IgG antibodies. J Immunol Methods. 2007;318:113–24. doi: 10.1016/j.jim.2006.10.010. [DOI] [PubMed] [Google Scholar]