Abstract
Bacillus subtilis RecA is important for spore resistance to DNA damage, even though spores contain a single non-replicating genome. We report that inactivation of RecA or its accessory factors, RecF, RecO, RecR and RecX, drastically reduce survival of mature dormant spores to ultrahigh vacuum desiccation and ionizing radiation that induce single strand (ss) DNA nicks and double-strand breaks (DSBs). The presence of non-cleavable LexA renders spores less sensitive to DSBs, and spores impaired in DSB recognition or end-processing show sensitivities to X-rays similar to wild-type. In vitro RecA cannot compete with SsbA for nucleation onto ssDNA in the presence of ATP. RecO is sufficient, at least in vitro, to overcome SsbA inhibition and stimulate RecA polymerization on SsbA-coated ssDNA. In the presence of SsbA, RecA slightly affects DNA replication in vitro, but addition of RecO facilitates RecA-mediated inhibition of DNA synthesis. We propose that repairing of the DNA lesions generates a replication stress to germinating spores, and the RecA·ssDNA filament might act by preventing potentially dangerous forms of DNA repair occurring during replication. RecA might stabilize a stalled fork or prevent or promote dissolution of reversed forks rather than its cleavage that should require end-processing.
INTRODUCTION
A DNA double-strand break (DSB) represents a potentially lethal form of DNA damage in all living organisms (1,2). These lesions, which mainly occur as by-products of normal cell metabolism, can arise from external sources, such as exposure to ionizing radiation (IR), and chemical mutagens (1,3). Cells rely on two major DSB repair pathways, homologous recombination (HR) and non-homologous end joining (NHEJ). Many cells can also use a minor pathway known as single-strand DNA annealing to repair two-ended DSBs (2,4–8). DSB repair via error-free HR requires many steps including, but not limited to DSB recognition, long-range end-processing and the presence of an intact homologous template. In eukaryotes error-free HR is constrained to the S and G2 phases of the cell cycle and in bacteria occurs during vegetative growth, when replication is active and the homologous sister chromatid (chromosome) is present (4,6–12). In contrast, by NHEJ, two-ended DSBs can be joined, with minimal end-processing. NHEJ is the predominant form of DSB repair during G1 in eukaryotes, or during growth phases when bacteria have a single genomic copy (5,9,13–15). When classical NHEJ is impeded, single-strand DNA annealing, which relies on redundant 3′-or 5′-end-processing functions, and on terminal tracts with discrete homology, can anneal the DNA ends followed by ligation (6–8,11,14,15). Thus, depending on the organism and growth environment, cells have several strategies to repair DSBs that have occurred.
In certain bacteria including Bacillus subtilis, nutritional starvation triggers the formation of two types of cells: a larger mother cell, and smaller prespore or forespore, each of which contains a single genome equivalent (reviewed in 16). During spore development asymmetric cell division is followed by different morphological events and eventually liberation of a dormant spore that has only one non-replicating genome copy (17,18). Bacillus subtilis dormant spores protect their chromosome from DNA damage due to its specific structure (19,20) and process DNA damage under unfavourable metabolic conditions (21,22). The mechanisms used for dormant B. subtilis spores to remove DSBs generated by IR (e.g., X-rays) or single-stranded nicks generated by ultrahigh vacuum (UHV) desiccation upon spore germination are poorly understood (19,23–27). In the presence of two-ended DSBs, the spore-encoded NHEJ system reconnects the broken ends, and it depends on two proteins, YkoV (also termed Ku), which binds to dsDNA ends and YkoU (LigD) a protein that processes and ligates the dsDNA ends (21,27,28). Dormant spores lacking RecA are sensitive to DNA DSBs (27,29,30). This result is interesting because spores should have one genomic DNA copy precluding a role for RecA in HR. Furthermore, spores defective in NHEJ and lacking RecA are more sensitive to DNA DSBs than null recA (ΔrecA) spores (27,28,31,32). Therefore, the role of RecA in spore resistance to DSBs is unclear.
In bacteria RecA is the central player in HR (10,12,33). RecA, with the help of accessory proteins (e.g., RecF, RecO, RecR and RecX), nucleates and polymerizes on the single stranded (ss) DNA coated by a single-stranded binding protein (SSB/SsbA), forming a RecA·ssDNA nucleoprotein filament (NPF) (34–38). Up to now, five functions were reported for Escherichia coli RecA·ssDNA NPF. First, it catalyses DNA strand exchange reaction in the presence of an intact homologous template (10). Second, it promotes induction of the SOS response by promotion of the autocatalytic cleavage of the LexA repressor (39,40). Third, RecA (41–43) can displace the replicase (DNA polymerase III) when lesions or obstacles are encountered (44). Fourth, RecA mediates activation of UmuD′ by mediating autocatalytic cleavage of UmuD (45). Fifth, the RecA·ssDNA NPF participates in SOS mutagenesis by activating Pol V (45,46). In B. subtilis only two of these five RecA functions have been documented (9,47). Here, the RecA·ssDNA NPF formed with the help of mediators (e.g., RecF, RecO, RecR) and modulators (e.g., RecF, RecX, etc.) is involved in homology search and DNA strand exchange, in the presence of an intact homologous DNA molecule during recombinational repair, and it also plays an important role in the SOS response (48,49). Among the SOS-regulated genes, no homologue of E. coli UmuD (UmuDEco) has been detected in the B. subtilis genome, therefore the existence of a UmuD-like function in this bacterium is still an open question (50), and the role of RecA·ATP·Mg2+ in the generation of an active mutasomal complex remains to be documented (45).
We aimed to investigate how RecA contributes to spore survival after DSBs or nicks generated by X-rays or UHV treatment, respectively, in the presence of one genome copy. We report that in the absence of DSB recognition (recN spores) or end-processing (addAB, recQ, recS, recJ or addA recJ mutant spores) germinated spores are as sensitive to X-ray treatment as the wild-type (wt) strain. The absence of RecA or its accessory factors (RecF, RecO, RecR, RecX) render germinated spores extremely sensitive to X-rays and UHV treatment, suggesting that DNA repair in the spore requires a RecA·ssDNA NPF rather than DSB repair via DNA strand exchange. Spores impeded in SOS induction were sensitive to X-rays and UHV treatments, suggesting that increased levels of RecA or of an unknown modulator (e.g., PcrA) might be also involved in spore survival. In the presence of ATP, RecA cannot compete with SsbA for ssDNA binding, and slightly inhibits PolC-DnaE-promoted DNA synthesis. SsbA blocks RecA polymerization on ssDNA, and in vitro RecO is sufficient to overcome the SsbA barrier. RecA can nucleate and filament on the RecO·ssDNA·SsbA complexes and it can inhibit leading and lagging strand DNA synthesis in the presence of RecO. Our results establish an intimate connection between resistance of dormant spores, RecA·ssDNA NPF formation on SsbA-coated ssDNA tracts, and the inhibition of DNA synthesis during spore germination. We propose that a RecA·ssDNA NPF should work as a replicase auxiliary protein, and that the RecA·ssDNA NPF might act by preventing potentially dangerous forms of DNA repair occurring during a replication stress. RecA by inhibition of DNA replication might stabilize a stalled fork or prevent or promote dissolution of a reversed fork rather than its cleavage; that should require end-processing.
MATERIALS AND METHODS
Bacterial strains and spore preparation
All bacterial strains used in this study are derivatives of BG214 or 168 strains and are listed in Supplementary Table S1. Bacillus subtilis strains were grown overnight to stationary phase in the NB medium supplemented with appropriate antibiotic. For spore preparation, 200 µl of overnight cultures were plated evenly on Schaeffer’s sporulation medium (SSM) plates (51) and plates were stored at 37°C for 7 days. The fully formed spores were harvested and purified by adjusted protocol (52). The purified spores were resuspended in 5 ml of desterilized water and stored until final usage at 4°C.
Spore X-ray irradiation
The 200 µl of purified spores were transferred to PCR tubes and irradiated at room temperature (RT) with X-rays (200 kV/15 mA) generated by an X-ray tube (Gulmay Xstrahl RS225 A, Gulmay Medical Inc., GA, USA). The appropriate dilutions of treated and untreated spore samples were plated on NB agar plates in order to measure spore survival by counting colony-forming units (CFUs). The survival curves were obtained by plotting the logarithm of survival percentage versus applied X-ray radiation dose. Each X-ray irradiation experiment was repeated at least three times and the data presented are expressed as average values with standard deviations. Additionally, spore resistance after X-ray radiation was expressed as D10-value, which represents the X-ray dose that reduces spore survival to 10%. The D10-values of treated spores were compared statistically using Student’s t-test and differences with P-values of ≤0.05 were considered statistically significant (32,53).
Spores UHV desiccation
Spore samples consisted of air-dried spore monolayers immobilized on 7-mm quartz discs were exposed 7 days to UHV produced by an ion-getter pumping system (400 l/s; Varian SpA, Torino, Italy) reaching a final pressure of 3 × 10−6 Pa (32,54,55). The spores immobilized on quartz discs were recovered by 10% aqueous polyvinyl alcohol solution as described previously (32,53). The appropriate dilutions of treated and untreated spore samples were plated on NB agar plates in order to count CFUs as a measure of spore survival. The CFUs of untreated spore samples were represented as 100% survival. The UHV experiment was performed in triplicate. The CFUs of UHV-treated spores were divided with the average CFU-value of untreated spore samples in order to obtain the survival after UHV. The data presented are expressed as average values with standard deviations. The percentage of survivals of treated spores was compared statistically using Student’s t-test and differences with P-values of ≤0.05 were considered statistically significant (32).
RecA nucleation and filament formation
A continuous ATP hydrolysis assay was used to analyse the extent of RecA nucleation and filament formation on ssDNA (56). SsbA, RecO and RecA proteins and the pGEM3-Zf(+) ssDNA substrate were purified as described (57,58). RecA protein concentration is expressed as moles of protein as monomers, RecO as dimers and SsbA as tetramers (56). The rate of ssDNA-dependent RecA-mediated ATP hydrolysis and the nucleation lag time were measured in buffer A (50 mM Tris–HCl [pH 7.5], 1 mM DTT, 90 mM NaCl, 10 mM MgOAc [magnesium acetate], 50 µg/ml BSA, 5% glycerol) containing 5 mM ATP for 25 min at 37°C as described (56). The nucleation time was deduced from the time-intercept of a linear regression of the steady state portion of data in ATP hydrolysis assays as reported (56,59).
SsbA and RecO (1 SsbA/33 nt and 1 RecO/50 nt) proteins did not exhibit ATP hydrolysis activity when compared to the mock reaction in the absence of both proteins or when BSA was added instead of both proteins (data not shown). The orders of addition of 3199-nt pGEM3-Zf(+) ssDNA (10 µM in nt), the purified proteins (RecA, SsbA and RecO) and their concentrations are indicated in the text.
DNA synthesis assay
A B. subtilis replisome was reconstituted in vitro with purified SsbA, DnaG, DnaC, PriA, DnaD, DnaB, DnaI, PolC, DnaE, τ, δ, δ′ and β subunits of the replicase and the mini-circular DNA template as described (60). PolC, DnaE, DnaG, PriA, δ and δ′ are expressed as moles of protein monomers, β as dimers, τ, DnaB and DnaD as tetramers, and DnaC and DnaI as hexamers (60). Reaction conditions for DNA synthesis assays were: 15 nM DnaE, 20 nM PolC, 8 nM DnaG, 25 nM τ, 25 nM δ, 25 nM δ′, 24 nM β, 30 nM DnaC, 15 nM PriA, 50 nM DnaD, 100 nM DnaB, 40 nM DnaI, 5 nM mini-circular DNA template in molecules, 350 µM ATP, 100 µM CTP, GTP and UTP, 48 µM dNTPs (excepts 18 µM dCTP or dGTP for the leading- and lagging-strand DNA synthesis, respectively) and 15 µCi/reaction [α-32P]dCTP or [α-32P]dGTP, all in BsRC buffer (60). The DNA template was a 409-bp circle containing a 396-nt tail previously described (60) that it has a strong (50:1) GC strand bias, so labelled dCTP was incorporated, mostly, into the nascent leading strand, and labelled dGTP into the lagging strand (60).
An enzyme mix containing all protein components except SsbA, RecA, RecO was prepared in buffer BsRC at 4°C. The substrate mix contained the DNA substrate and 90 nM SsbA, 1 µM RecA, 0.1 µM RecO as indicated, and prepared on ice. The enzyme mix was added to the substrate mix and reactions were then pre-incubated for 5 min at 37°C in presence of 5 µM ATPγS and rNTPs except ATP. The reactions were initiated by the addition of dNTPs and ATP and aliquots were withdrawn at the indicated times and incubated for 20 min with an equal volume of a stop mix consisting of 40 mM Tris–HCl (pH8.0), 0.2% w/v SDS, 100 mM EDTA and 50 µg/ml proteinase K. Samples were applied onto Sephadex G-50 columns to eliminate non-incorporated dNTPs. The extent of DNA synthesis in leading- and lagging-strands was then quantified by scintillation counting as described (61). For the analysis of the size of replication products, samples were brought to 50 mM NaOH, 5% v/v glycerol and 0.05% bromphenol blue and fractionated in alkaline 0.45% agarose gels for ∼5 h at 60 V as described (60). Alkaline agarose gel buffer consisted of 30 mM NaOH and 0.5 mM EDTA. Gels were fixed in 7% (w/v) trichloroacetic acid, dried, autoradiographed on storage phosphor screens and analysed with Quantity One (Bio-Rad) software.
RESULTS AND DISCUSSION
Experimental system
RecA was found to be important for processing the DNA damage of fully formed B. subtilis spores upon germination (27,32). Spores contain a single non-replicating genome (17,18) and recombinational repair requires an intact homologous template (6–9,47), suggesting a function for RecA other than recombinational repair. To investigate the importance of HR functions in spore survival we have used fully formed spores with inactivated recombination gene products involved in DSB recognition (ΔrecN), long-range end-processing (addA5, addA5 addB72, ΔrecJ, ΔrecQ, ΔrecS and addA5 ΔrecJ), RecA accessory factors that act before (ΔrecO, ΔrecR and recF15) or during homology search (ΔrecX, recF15 and ΔrecX recF15) and the recombinase itself (ΔrecA). In addition, we analysed the role of the SOS induction in spore survival (lexA (Ind-) (Supplementary Table S1). As a control of genuine two-ended DSB repair we have inactivated NHEJ (ΔykoV, also termed ku) (Supplementary Table S1). The spores were subjected to X-ray radiation, which induces two-ended DSBs, and UHV treatment, which induces single-strand nicks that were then converted into one-ended DSBs during spore germination.
Spore resistance after X-ray radiation and UHV treatment depends on RecA
X-ray radiation generates two-ended DSBs that should be bound by YkoV (Ku), and should be processed and ligated by the YkoU (LigD) enzyme (27,28). We have previously reported that the ΔykoV single or ykoV ykoU double mutation render spores sensitive to DSB induction by X-ray radiation (32,53,62,63). As revealed in Supplementary Figure S1A, the ΔykoV single mutation render spores sensitive to DSB induction by IR. The ΔykoV mutant spores, however, showed similar survival as wt spores after UHV exposure for 7 days (Supplementary Figure S1B, Table 1). It is likely that the majority of the DSBs formed upon UHV treatment for 7 days and detected upon germination were ssDNA nicks that cannot be repaired by NHEJ.
Table 1.
Survival characteristics of B. subtilis spores after X-ray radiation and UHV
| Genotype | Dose (Gy) to D10a | D10REM/D10wt | % Surv. UHVb | SREM/Swt |
|---|---|---|---|---|
| BG214 genetic background | ||||
| wt | 1062.2 ± 205.0 | 1 | 80.2 ± 3.4 | 1 |
| ΔrecA | 482.2 ± 48.0* | 0.5 | 28.9 ± 4.8* | 0.4 |
| ΔykoV | 571.7 ± 17.1* | 0.5 | 88.9 ± 17.6 | 1.1 |
| ΔykoV ΔrecA | 230.7 ± 102.9* | 0.2 | ND | ND |
| ΔrecN | 1006.9 ± 82.9 | 0.9 | 83.5 ± 18.8 | 1.0 |
| addA5 addB72 | 1077.5 ± 302.9 | 1.0 | 54.8 ± 16.8 | 0.7 |
| addA5 | 1303.8 ± 85.4 | 1.2 | 56.4 ± 28.5 | 0.7 |
| ΔrecJ | 1152.9 ± 123.3 | 1.1 | 100.4 ± 10.5 | 1.3 |
| addA5 ΔrecJ | 850.6 ± 171.7 | 0.8 | 39.6 ± 8.7* | 0.5 |
| ΔrecQ | 1188.9 ± 104.8 | 1.1 | 78.1 ± 12.0 | 1.0 |
| ΔrecS | 968.0 ± 132.2 | 0.9 | 83.2 ± 4.8 | 1.0 |
| recF15 | 479.0 ± 65.6* | 0.5 | 0.003 ± 0.005* | 0.00004 |
| ΔrecO | 384.7 ± 50.3* | 0.4 | 37.2 ± 4.9* | 0.5 |
| ΔrecR | 321.6 ± 77.6* | 0.3 | 20.9 ± 4.6* | 0.3 |
| ΔrecX | 1056.4 ± 239.1 | 1.0 | 19.1 ± 12.5* | 0.2 |
| ΔrecX recF15 | 523.4 ± 41.4* | 0.5 | 50.4 ± 22.4 | 0.6 |
| 168 genetic background | ||||
| wt | 1525.5 ± 671.0 | 1 | 96.0 ± 7.5 | 1 |
| recA | 391.4 ± 19.3* | 0.3 | 9.2 ± 3.4* | 0.1 |
| wtc | 1921.5 ± 541.1 | 1.3 | 100.3 ± 21.2 | 1.0 |
| lexA (Ind-)c | 508.4 ± 18.1* | 0.3 | 50.2 ± 9.4* | 0.5 |
Data are averages of at least three independent experiments with standard deviations. Asterisks indicate D10 or UHV survival values that were significantly different (P-values ≤ 0.05) from values for wt spores (BG214/168).
aThe dose that it reduced the survival of the given spore population to 10% is indicated (D10). bSurvival (%) after 7 days of UHV exposure with final pressure 3 × 10–6 Pa.
cBoth strains carry Δupp mutation (Supplementary Table S1).
REM, repair mutant spore; ND, not determined.
RecA is a non-essential product, but examination of recA mutants revealed extensive chromosomal fragmentation even in the absence of exogenous stress (64). Furthermore, exponentially growing recA cells accumulate RecN-YFP foci (a sensor of DSBs) in ∼30% of total cells, whereas RecN-YFP foci accumulation is rare in wt cells (<0.5%) (65) and recA colonies have ∼5-fold less cells than wt colonies (66,67). It is likely that RecA is required for genomic stability, and in its absence the replication fork might be more susceptible to breakage. Inactivation of RecA, which does not block spore formation, drastically sensitized dormant spores to X-ray radiation and UHV treatment (Supplementary Figure S1, Table 1). Supplementary Figure S1A shows that the ΔrecA mutant spores had considerable lower survival after X-ray radiation relative to wt spores. In comparison to the wt spores the ΔrecA mutant spores showed a significant decrease in the D10-value (∼2-fold) (Table 1). The experiments were performed in the BG190 genetic background that also lacks prophages, a conjugative transposon and it is impaired in the general stress response. Similar results were observed when the WN463 strain, which contains prophages, a conjugative transposon and it is proficient in the general stress response, was tested (Table 1). Supplementary Figure S1A shows that the ΔrecA mutant spores had considerable lower survival after X-ray radiation relative to wt spores. We have previously reported that YkoV and RecA contribute additively to spore resistance after X-ray radiation or high vacuum treatment (62,63). We corroborated the additive effect of these two mutations after X-ray radiation (Supplementary Figure S1A, Table 1).
When spores were exposed to UHV we observed that the ΔrecA mutant spores showed significant decrease (3-fold) in survival when compared to wt spores (Table 1, Supplementary Figure S1B). From obtained results, we concluded that recA gene product is important for spore resistance to X-ray radiation and UHV treatment.
This result is in agreement with previous studies showing that NHEJ is an important DNA repair mechanism for spore resistance after X-ray (induction of two-ended DSBs). However, NHEJ plays a minor role in the repair of spore after exposure to UHV for 7 days treatment (induction of nicks) (Supplementary Figure S1B, Table 1) (21,32,53).
Spore resistance after X-ray radiation and UHV treatment is independent of RecN
In response to replication fork collapse, in exponentially growing cells, a complex molecular machinery involved in DNA repair is recruited, and cell proliferation is arrested (9,48). One of the first responders to the formed DSBs is the RecN protein. RecN, which assembles on one- or two-ended DSBs, choreographs the DSB response (65,68,69). Damage-inducible RecN, in concert with polynucleotide phosphorylase degrades the 3′-ends and specifically removes the 3′ dirty ends, and generates the substrate for long-range end-processing (70,71).
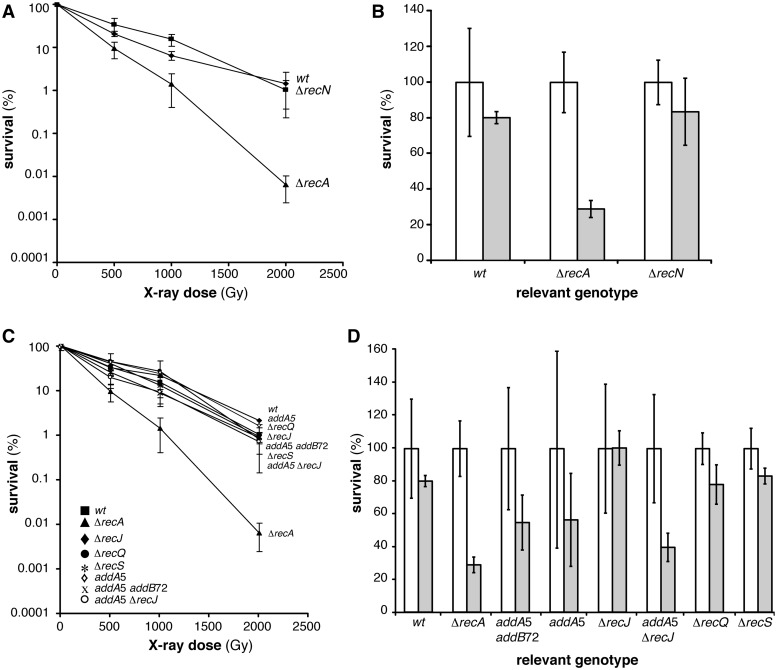
To determine whether RecN is necessary for spore resistance, we exposed dormant ΔrecN mutant spores to X-ray radiation and UHV (Table 1, Figure 1A and B). The survival pattern after X-ray radiation of ΔrecN mutant and wt spores was similar (Figure 1A, Table 1). Similar results were obtained after UHV treatment (Table 1, Figure 1B). These indicate that inactivation of RecN does not influence spore survival to X-ray radiation or UHV treatment.
Figure 1.
Survival of B. subtilis spores deficient in DSB recognition and end-processing after X-ray and UHV treatment. (A) X-ray dose-dependent survival of wt (filled square), ΔrecA (filled triangle) and ΔrecN mutant (filled diamond) spores. The wt and ΔrecA mutant spores were used as controls. (B) Viability (white column) and survival (grey column) after 7 days UHV exposure of wt, ΔrecA mutant and ΔrecN mutant spores. (C) X-ray dose-dependent survival of wt (filled square), ΔrecA (filled triangle), addA5 (open diamond), addA5 addB72 (cross), ΔrecJ (filled diamond), ΔrecQ (filled circle), ΔrecS (asterisk) or addA5 ΔrecJ (open circle) spores. (D) Untreated (white column) and treated (grey column) after 7 days UHV exposure of wt, ΔrecA, addA5, addA5 addB72, ΔrecJ, ΔrecQ, ΔrecS or addA5 ΔrecJ mutant spores.
Inactivation of end-resection does not influence spore resistance after DSBs
During exponential growth, after generation of one- or two-ended DSBs, basal and long-range end-processing steps are required for accurate DNA repair. (9,48). Long-range end-resection involves redundant pathways consisting of nuclease(s), DNA helicase(s) and a single-stranded binding protein (9,47,48). The AddAB helicase–nuclease complex (functional analogue of E. coli RecBCD, and of mycobacterial AdnAB), in concert with SsbA catalyses the formation of the 3′-tailed ssDNA (72,73). The RecA loading into the generated ssDNA via AddAB remains to be documented (72). The second long-range processing pathway includes the RecJ nuclease, a RecQ-like helicase (RecQ or RecS) and SsbA (47–49). Furthermore, the addA5 ΔrecJ double mutant strain is as sensitive to DNA damaging agents as exponentially growing recA cells (69).
To determine whether spore resistance after X-ray radiation and UHV treatment depends on the proteins involved in DSB end-processing, we exposed addA5, addA5 addB72, ΔrecQ, ΔrecS, ΔrecJ or addA5 ΔrecJ mutant spores to X-ray radiation and UHV (Table 1, Figure 1C and D). Figure 1C shows X-ray survival of spores with inactivated helicase functions (addA5, addA5 addB72, ΔrecQ and ΔrecS mutant spores) and nuclease functions (addA5, addA5 addB72, ΔrecJ and addA5 ΔrecJ mutant spores). Survival capability of addA5, addA5 addB72, ΔrecQ, ΔrecS, ΔrecJ or addA5 ΔrecJ mutant spores after X-ray radiation was comparable to wt spores, having similar survival patterns and D10-values (Table 1, Figure 1C). Survival levels of ΔrecQ, ΔrecS and ΔrecJ mutant spores obtained after UHV treatment were similar to wt survival levels (Table 1, Figure 1D). On the other hand, the introduction of addA5, addA5 addB72 and addA5 ΔrecJ mutations showed a moderate effect, implying a direct or indirect involvement of AddAB in spore resistance to UHV treatment (Table 1, Figure 1D).
The main conclusions from the presented results are that inactivation of functions involved in one (AddAB, RecQ, RecS or RecJ) or both (AddA and RecJ) end-resection pathways did not influence the DSB repair capability of spores exposed to X-ray radiation when compared to wt spores (Table 1, Figure 1C). Finally, single-strand DNA annealing, which relies on redundant end-processing functions and on terminal tracts with discrete homology (6,7,11,14,15), can be a very minor pathway, if at all, because absence of AddA and RecJ does not significantly impairs spore resistance to X-rays treatment (Figure 1C). The role of the AddAB complex in the survival of mutant spores upon UHV is poorly characterized. It is still unknown whether AddAB possesses a RecA loading function, as its E. coli counterpart (RecBCD) (72).
Spore survival depends on RecF, RecO, RecR and RecX
In exponentially growing cells concomitantly with long-range end-processing, RecN promotes tethering of DNA ends to form a repair centre (RC) (69), RecO, RecR and RecA are recruited to RecN-mediated RC, and the SOS response is induced (9,49). Then, RecA forms highly dynamic filamentous structures, termed threads, across the nucleoid and RecF forms a discrete focus in the nucleoid, which are coincident in time with RecX foci, in response to DNA damage. RecN co-localizes with the RecO, RecR, RecA and RecF focus (65,68,74). DNA damage-induced RecX foci co-localize with RecA threads that persisted for a longer time in the recX context (68,74), suggesting that RecO, RecR and RecF accessory proteins contribute to RecA nucleation onto SsbA-coated ssDNA, and that RecF and RecX contribute to RecA·ssDNA NPF formation (65,74).
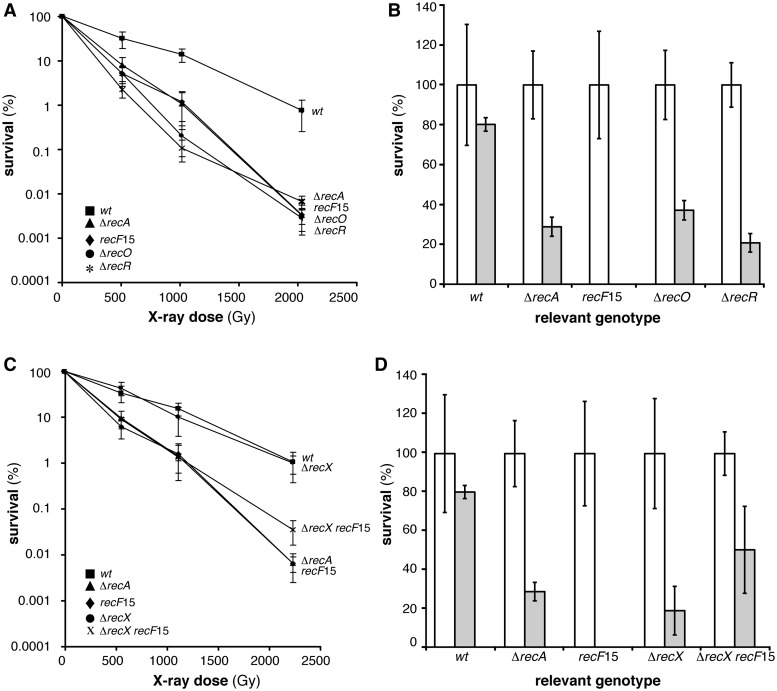
To investigate whether inactivation of RecA accessory proteins influence spore survival after DSBs, the recF15, ΔrecO, ΔrecR and ΔrecX mutant spores were exposed to X-ray radiation and UHV treatment (Table 1, Figure 2A–D). Inactivation of RecO, RecR or RecF rendered dormant spores equally extremely sensitive to X-ray radiation, with survival levels similar to ΔrecA mutant spores (Figure 2A). UHV treatment produced a drastic reduction of survival of recF15 mutant spores, whereas the ΔrecO or ΔrecR mutant spores showed survival level similar to the ΔrecA mutant spores (Table 1, Figure 2B). Since dormant spores have a single none replicating genome (17), and end-processing functions are not involved in spore resistance to DSBs (Figure 1), we have to assume that the major role of RecF, RecO and RecR is to promote RecA nucleation onto SsbA-coated ssDNA and subsequent filament growth during early steps of germination. Alternatively, RecF, RecO and RecR should protect the nascent DNA from degradation during spore germination, but absence of the degrading function (e.g., absence of RecJ and AddA) showed no phenotype (75). We favour the hypothesis that RecA loading on ssDNA (by the help of RecOR or RecFOR complexes) and filament growth (RecF) are essential for spore survival after X-ray radiation during spore germination and outgrowth.
Figure 2.
Survival of B. subtilis spores deficient in RecA loading after X-ray and UHV treatment. (A) X-ray dose-dependent survival of wt (filled square), ΔrecA (filled triangle), recF15 (filled diamond), ΔrecO (filled circle) or ΔrecR (asterisk) spores. (B) Viability (white column) and survival (grey column) after 7 days UHV exposure of wt, ΔrecA, recF15, ΔrecO or ΔrecR spores. (C) X-ray dose-dependent survival of wt (filled square), ΔrecA (filled triangle), recF15 (filled diamond), ΔrecX (filled circle) or ΔrecX recF15 (cross) spores. (C) Untreated (white column) and treated (grey column) after 7 days UHV exposure of wt, ΔrecA, recF15, ΔrecX or ΔrecX recF15 spores.
Recent in vivo data showed that RecX facilitates, whereas RecF delays RecA-mediated LexA self-cleavage with subsequent induction of the SOS response, and in the absence of both proteins (i.e., in recF15 ΔrecX cells) SOS induction is wt-like, but the double mutant strain is blocked in DNA repair, suggesting that RecF and RecX have counteracting functions (74). Similarly, RecXEco disassembles the RecA·ssDNA NPF and RecF stabilizes its assembly or prevents its disassembly in vitro (76). The recF15 or ΔrecX mutant spores were more sensitive to UHV treatment than ΔrecA spores (Figure 2B and D, Table 1).
To test whether the dynamic assembly and subsequent disassembly of RecA filaments contributed to spore survival, the sensitivity of the ΔrecX recF15 mutant spores was measured (Figure 2C and D). The absence of RecX did not sensitized spores to X-ray radiation when compared to wt spores. The ΔrecX mutation partially suppressed the sensitivity to X-ray or UHV in the ΔrecX recF15 context (Figure 2C and D).
Spore resistance after X-ray radiation and UHV treatment depends on the SOS response
In exponentially growing cells, mutation of recF, recO or recR shows a drastically reduction in the DNA repair capacity (77–79), and results in a delay and reduction of SOS induction (80). However, these phenotypes are suppressed by specific mutations in the recA gene or by expressing heterologous SSB proteins in the background (81). Under similar conditions, the absence of RecX reduces DNA repair, but facilitates SOS induction (74). The absence of both RecF and RecX renders exponentially growing cells extremely sensitive to DNA damaging agents, but the SOS induction is similar to wt cells (74). The results obtained in the previous section suggested that the role of RecA in spore survival is not only the induction of SOS response, because the double recF15 ΔrecX mutant has normal levels of SOS induction, but is not spore repair proficient.
Constitutive levels of RecA are sufficient to facilitate the repair of one- or two-ended DSBs via HR during vegetative growth (82). However, the SOS regulon was expressed in germinating spores exposed to space conditions (29,83), which are known to cause DSBs in spores (26).
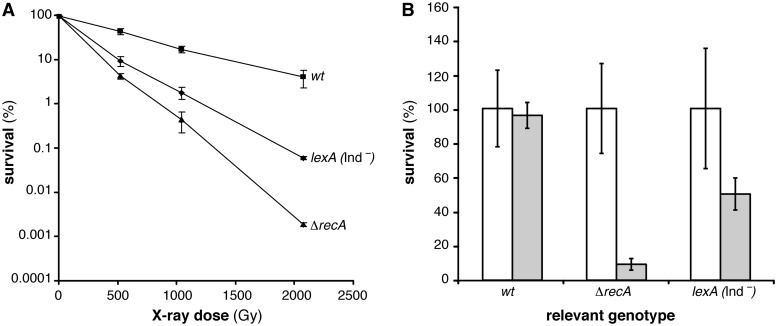
To test whether some SOS-regulated function is needed for spore resistance after DSB introduction we used a specific lexA mutant variant, lexA (Ind-), resulting in a non-cleavable LexA repressor and therefore unable to induce the SOS response (82). The lexA (Ind-) mutant spores and related wt spores carry Δupp mutation, but the Δupp mutation does not reduce spore survival after X-ray radiation (Table 1). As revealed in Figure 3A the absence of SOS significantly reduced X-ray survival of lexA (Ind-) mutant spores with the survival level similar to ΔrecA mutant spores when spores were treated with an X-ray dose of 500 Gy. With increasing dose, lexA (Ind-) mutant spores turned to be less sensitive than the ΔrecA mutant spores, but still very sensitive compared to wt spores (Figure 3A). Comparable results were obtained for D10-values, where lexA (Ind-) mutant spores showed slightly increased D10-value than the ΔrecA mutant spores, but they were still significantly decreased (3-fold) compared to the D10-values of the wt spores (Table 1). Similar results were obtained after UHV treatment where lexA (Ind-) mutant spores turned to be significantly sensitive than the wt spores (Figure 3B, Table 1). From obtained results we conclude that SOS response is important for spore survival after X-ray radiation and UHV. It is likely that increased RecA levels might contribute to spore survival.
Figure 3.
Survival of B. subtilis spores unable to induce the SOS response. (A) X-ray dose-dependent survival of wt (filled square), ΔrecA (filled triangle) and lexA (Ind-) (filled diamond) mutant spores. (B) Untreated (white column) and treated (grey column) after 7 days UHV exposure of wt, ΔrecA or lexA (Ind-) spores.
Among the SOS genes previously identified (84), only four of them correspond to genes whose roles in error-free recombinational repair, error-prone translesion synthesis or error-prone recombinational repair (recA, ruvAB, pcrA [uvrDEco]), polY2 [umuC]) have been identified (50,67,82,84,85). In addition, we cannot exclude that induction of other B. subtilis SOS genes with unknown function could be required for spore resistance to X-ray radiation or UHV treatment. Since spore survival is not affected in the addA5 ΔrecJ context and the involvement of RuvAB in spore resistance requires end-processing its requirement was tentatively ruled out. Pol Y2 shares homology with UmuCEco (50); and no homologue of UmuDEco has been detected in the B. subtilis genome, suggesting that a mutasome similar to the one described in E. coli does not exist in B. subtilis (see Introduction section). The contribution of PcrA cannot be analysed, because absence of PcrA renders cells synthetically lethal unless a mutation in recF, recO or recR accumulates in the background (86).
RecA·ATP·Mg2+ cannot nucleate and polymerize onto SsbA-coated ssDNA in vitro
Previously it has been shown that the essential SsbA (counterpart of SSBEco) is involved in different stages of DNA metabolism, including DNA repair, recombination and replication. The template for B. subtilis RecA assembly is SsbA-coated ssDNA in vivo, but RecA, in the ATP bound form, cannot nucleate on the SsbA·ssDNA complexes (87). However, RecA efficiently nucleates onto SsbA-coated ssDNA in the presence of dATP (56,58). The relationship between SsbA and RecA is complex; SsbA (SSB) helps RecA to self-assemble onto SsbA (SSB)-coated ssDNA by removing secondary structures, and competes with RecA for ssDNA binding (10,12,33). Genetic data revealed that a recA73 mutation partially overrides the recF, recO or recR defect (81), suggesting that this mutation conferred RecA the ability to remove SsbA from the ssDNA. To gain insight into the molecular basis of the inability of RecA to assemble onto SsbA-coated ssDNA, conditions where RecA efficiently assembles onto SsbA-coated ssDNA in the presence of ATP were investigated.
In the absence of SsbA, at a ratio of 1 RecA monomer/8 nt, RecA nucleation onto ssDNA is not apparently rate limiting and ATP hydrolysis increased with time approaching the levels previously observed, with a Kcat of 11.7 ± 0.3 min−1 (Supplementary Figure S2A) (87; B.C., T. Yadav and J.C.A., unpublished results). In the presence of limiting SsbA (1 SsbA/550 nt, 18 nM), RecA·ATP·Mg2+ nucleated onto ssDNA and polymerized on ssDNA with similar efficiency than in the absence of SsbA (10.6 ± 0.3 min−1 versus 11.7 ± 0.3 min−1) (Supplementary Figure S2A). At increased ratios (1 SsbA/275–133 nt, 37–75 nM), SsbA increased the lag time of the reactions, which correspond with the RecA binding onto ssDNA (nucleation step), and reduced the maximal rate of ATP hydrolysis (8.3 ± 0.2 min−1 and 7.2 ± 0.3 min−1, respectively), suggesting that limiting SsbA partially competed with RecA·ATP·Mg2+ for binding to ssDNA (Supplementary Figure S2A). In the presence of stoichiometric concentrations (1 SsbA /66 nt), SsbA-blocked RecA catalysed hydrolysis of ATP (1.4 ± 0.1 min−1) (Supplementary Figure S2A, 150 nM). Similar results were observed when saturating SsbA (1 SsbA/33 nt) concentrations were used (B.C., T. Yadav and J.C.A., unpublished results). It is likely that both RecA and SsbA bind competitively to ssDNA, and RecA cannot compete with SsbA bound to ssDNA as previously proposed (T. Yadav, B.C. and J.C.A., unpublished results).
RecO was necessary to overcome the inhibition of RecA assembly onto SsbA-coated ssDNA. In the presence of stoichiometric SsbA concentrations (1 SsbA/66 nt) RecA cannot nucleate, but addition of RecO, as low as 1 RecO/100 nt, to the preformed SsbA·ssDNA complex, significantly stimulated RecA nucleation and RecA·ssDNA filament formation (Supplementary Figure S2B). Similar results were observed when RecO and saturating SsbA (1 SsbA/33 nt) concentrations were used (B.C., T. Yadav and J.C.A., unpublished results).
RecA inhibits DNA replication in the absence of SsbA
Previously, it has been shown that: (i) RecAEco binds and releases a halted replicase at a replication fork stalled by the inactivation of the replicative helicase (DnaB6) in vivo (43). Here, replicase clearance is not affected by the absence of RecFEco, RecOEco or RecREco function (43) suggesting that RecA can assemble onto the SSBEco-coated lagging-strand template forming short RecA filaments in the absence of accessory factors in vivo. (ii) RecAEco alone strongly inhibited the advance of a replication fork when added prior initiation of DNA synthesis (42). To test whether B. subtilis RecA inhibits the replisome during DNA synthesis we have used a reconstituted in vitro replication system (60,61).
The Firmicutes replicase is composed of two distinct polymerases (PolC and DnaE), the τ, δ and δ′ subunits of the clamp loader, and the sliding clamp processivity factor β (60,88). The B. subtilis core replicase [PolC-τ-δ-δ′-β] acts in concert with other nine proteins (DnaC helicase, and its loading system composed by DnaB, DnaD, DnaI and PriA, in addition to the DnaG primase and SsbA) (60). To test whether the replisome could directly load RecA as it proceeds to unwound DNA or if RecA could bind to the ssDNA tracts in a direct competition with SsbA during replisome progression we have measured DNA synthesis (60) in the presence or absence of SsbA and/or RecA proteins (Figure 4, Supplementary Figure S2).
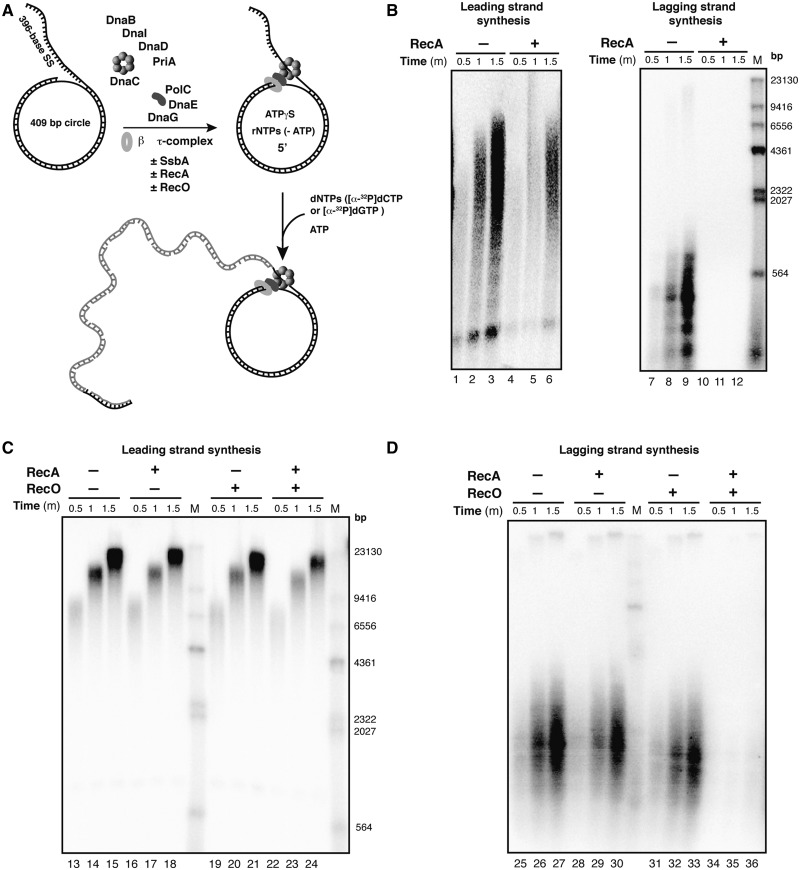
Figure 4.
RecA·ssDNA NPF inhibits DNA synthesis in the presence of RecO. (A) Diagram of the DNA template and the B. subtilis replisome assembly scheme in the presence/absence (±) of SsbA, RecA and/or RecO. The mini-circular DNA template has an asymmetric G:C ratio (50:1) between the two strands, permitting quantification of leading- and lagging-strand synthesis by measuring radioactive dCMP and dGMP incorporation. (B) Time course of the leading- and lagging-strand synthesis by the reconstituted replisome in the absence of SsbA and without (lanes 1–3 and 7–9) or with RecA (1 µM, lanes 4–6 and 10–12). (C) Time course of the leading-strand synthesis by a replisome containing SsbA (0.09 µM) (lanes 13–24), with RecA (1 µM, lanes 16–18), with RecO (0.1 µM, lanes 19–21) and in the presence of both RecO and RecA (lanes 22–24). (D) Time course of the lagging-strand synthesis by a replisome containing SsbA (lanes 25–36), in the presence of RecA (lanes 28–30), RecO (lanes 31–33) or both (34–36). M indicates the 3′-labelled HindIII-digested λ DNA used as a size marker.
In the absence of RecA, leading- and lagging-strand synthesis was 2-fold reduced in the absence of SsbA when compared with the DNA synthesis observed in the presence of SsbA (Figure 4A, lanes 1–3 and 7–9 versus Figure 4C, lanes 13–15 and 25–27, and Supplementary Figure S3A versus Supplementary Figure S3B) (61). This is consistent with the observation that in the DNA substrate used, the DNA helicase can self-assemble by threading over the exposed 5′-end of the flap of the forked substrates in the absence of SsbA (61,89). In the absence of SsbA, addition of RecA reduced leading-strand synthesis ∼1.5-fold, but blocked lagging-strand synthesis (Figure 4B, lanes 4–6 and 10–12, and Supplementary Figure S3A). It is likely that RecA bound to ssDNA can compete with the DNA helicase for substrate binding. These results are similar to the ones described in vivo and in vitro E. coli DNA replication in the presence of the SSB protein (42,43).
In the presence of SsbA, the addition of RecA did not significantly alter leading-strand synthesis, but reduced lagging-strand synthesis ∼1.5-fold (Figure 4C and D, lanes 16–18 and 28–30, and Supplementary Figure S3B). It is likely that RecA, bound to the dATP present in the reaction mix (60), can at least partially assemble on SsbA-coated ssDNA and it can reduce lagging-strand synthesis (56,58).
RecO-mediated RecA filament growth onto ssDNA inhibits DNA replication in vitro
In the previous section, it has been shown that RecA cannot be efficiently loaded and polymerize onto the SsbA-coated ssDNA substrate in the presence of ATP and in the absence of the RecO accessory factor.
To determine whether RecO could participate with RecA in the modulation of the activity of the replisome, we tested the effect of adding RecO (1 RecO/20 nt) and RecA (1 RecA/3 nt) to a replication reaction containing SsbA (Figure 4C and D). The result shows that RecO did not affect DNA synthesis in the presence of SsbA (Figure 4C and D, Supplementary Figure S3B). The weak insolubility of RecR and the strong one of RecF, and the buffers used for maintaining them in a soluble (or partially soluble) state (90,91) restrained us to include them in the complex in vitro replication reaction to analyse their contribution.
In the presence of sub-stoichiometric amounts of RecA and RecO a significant inhibition of leading-strand DNA synthesis was observed (Figure 4C, lanes 23–26 and Supplementary Figure S3B). Addition of RecO- and RecA-blocked lagging-strand synthesis (Figure 4D, lanes 34–36 and Supplementary Figure S3B). It is likely that RecO facilitates the efficient nucleation and RecA filament growth onto SsbA-coated ssDNA. Conversely, RecAEco filament formation was not a pre-requisition for replicase dislodging in vivo (43). Here, upon inactivation of the hexameric helicase, RecAEco nucleates probably onto the lagging strand, in the absence of the RecFOREco complex, and facilitates the dislodging of the replisome, perhaps by facilitating fork reversal (43). Alternatively, thermal DnaBEco inactivation leads to uncoupling of the moving polymerases with the DNA helicase, and any asynchrony might lead to the generation of large tracts of protein-free ssDNA to which RecAEco can be loaded with no mediator requirement.
CONCLUSIONS
We have shown that spore resistance after X-ray radiation or UHV treatment depends on RecA, its accessory proteins, namely RecF, RecO, RecR and RecX, that facilitate RecA nucleation and filament growth onto ssDNA, and the induction of the SOS response (Table 1). However, dormant spores have a single non-replicating genome (17,18), and recombinational repair is constrained by the requirement of an intact homologous template (10,33). How can we rationalize the need of RecA and its accessory factors for spore resistance to DSBs in the absence of a homologous template? From the results obtained previously and in this work we propose that: (i) an uncharacterized DNA damage checkpoint function(s) should inhibit DNA end-resection (which represents the primary regulatory step towards HR), increase end-protection and contribute to the choice of NHEJ during spore survival; (ii) the two-ended DSBs were sealed by NHEJ, and the ssDNA nicks should be repaired by specialized repair pathways (Table 1) (13,14,20,47); (iii) during germination the replisome might encounter ‘barriers’ or base damage in the DNA template (92), leading to replication fork stalling; and (iv) a RecA·ssDNA NPF formed on ssDNA, with the contribution of RecO, RecR, RecF and RecX might be crucial for spore survival by stabilizing a stalled replication fork. This activity appears to be separate from RecA-mediated DNA pairing and strand exchange.
The replisome is inherently tolerant after collision with DNA lesions (44), and RecA·ssDNA NPF formation is required to inhibit replication fork progression upon stress, and this novel RecA activity is necessary for recovery during germination. RecF, RecO, RecR and RecX, which enable RecA assembly on SsbA-coated ssDNA in vivo, are required for spore resistance. Then, a RecA·ssDNA NPF promotes the LexA self-cleavage and induction of the SOS response, and works as a genuine auxiliary component of the replisome (Figure 4) by preventing recombinational repair to occur (Figure 1). The potential SOS-induced contributor(s) to spore resistance are RecA itself, PcrA and Pol Y2 (see above), although the potential contribution of SOS genes of unknown function cannot be ruled out. Pol Y2, however, does not contribute to spore survival upon induction of DSBs, at least after H2O2 treatment (93). The role of PcrA is to dismantle a RecA·ssDNA NPF (94,95), but the absence of PcrA renders cells synthetically lethal, and for viability mutations in recF, recO or recR gene accumulated in the background (86). The phenotype of the pcrA (recF) mutation precluded any further studies to address the potential mechanism of PcrA in spore resistance to DSBs.
Why RecA accessory proteins are required? RecA·ATP·Mg2+, which cannot compete with SsbA bound to ssDNA (Supplementary Figure S2A), has some effect on DNA synthesis even in the presence of SsbA, but stoichiometric concentrations of RecO and RecA inhibit DNA synthesis (Figure 4C and D). In vitro RecO is sufficient to overcome the kinetic barriers imposed by SsbA on RecA assembly onto ssDNA, and facilitates RecA nucleation and filament growth (Supplementary Figure S2B), although genetic data support that in vivo the RecFOR complex is required for RecA·ssDNA NPF assembly (9).
The RecA·ssDNA NPF by inhibition of DNA replication might prevent potentially dangerous forms of DNA repair from occurring during replication of germinating spores. Alternatively, RecA contributes to convert the stalled fork to a ‘chicken-foot’ structure, which is subject to cleavage, causing DSBs (96). If fork reversal takes place we have to assume that the RecA·ssDNA NPF might stabilize a stalled fork, preventing or promoting dissolution of a reversed forks rather than its cleavage, which should require end-processing functions for recovery, and end-processing mutants were only slightly impaired in spore survival. On the basis of an Occam’s razor-like rationale we assume that: (i) the replication speed thresholds dictate RecA assembly on the ssDNA; (ii) a RecA·ssDNA filament, which is crucial for spore survival upon generation of DSBs, inhibits replication fork progression in a manner that resembles replication through barriers (e.g., fragile sites, etc.); and (iii) RecA assembled on ssDNA triggers SOS induction, and inhibits DNA replication. It is likely that PcrA or any other helicase might dismantle the RecA·ssDNA NPF, followed by de novo re-started and high-speed replication.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online, including [97–99].
FUNDING
DLR-FuE-Projekt ISS Nutzung in der Biodiagnostik, Programm RF-FuW, Teilprogramm 475 (to G.R. and R.M.); Ministerio de Economía y Competitividad, Dirección General de Investigación [BFU2012-39879-C02-01 to J.C.A., BFU2012-39879-C02-02 to S.A.]. Funding for open access charge: Ministerio de Economia y Competividad, Dirección General de Investigación: DLR-FuE-Projekt ‘ISS Nutzung in der Biodiagnostik’, Programm RF-FuW, Teilprogramm 475.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Prof. C. McHenry (Univ. of Colorado, Boulder) for providing us with materials for in vitro replication, Prof. L. A. Simmons (Univ. of Michigan) for strains and helpful discussions, and we thank Andrea Schröder, Chiara Marchisone and Petra Schwendner for their excellent technical assistance. Ralf Moeller gratefully acknowledges DLR’s Forschungssemesterprogramm Ignacija Vlašić expresses gratitude to DLR’s ‘Gastwissenschaftlerprogramm’.
REFERENCES
- 1.Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993;362:709–715. doi: 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- 2.Ciccia A, Elledge SJ. The DNA damage response: making it safe to play with knives. Mol. Cell. 2010;40:179–204. doi: 10.1016/j.molcel.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Friedberg EC, Walker GC, Siede W, Wood RD, Schultz RA, Ellenberger T. DNA Repair and Mutagenesis. Washington, DC: ASM Press; 2006. [Google Scholar]
- 4.Branzei D, Foiani M. Maintaining genome stability at the replication fork. Nat. Rev. Mol. Cell Biol. 2010;11:208–219. doi: 10.1038/nrm2852. [DOI] [PubMed] [Google Scholar]
- 5.Lieber MR. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu. Rev. Biochem. 2010;79:181–211. doi: 10.1146/annurev.biochem.052308.093131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.San Filippo J, Sung P, Klein H. Mechanism of eukaryotic homologous recombination. Annu. Rev. Biochem. 2008;77:229–257. doi: 10.1146/annurev.biochem.77.061306.125255. [DOI] [PubMed] [Google Scholar]
- 7.West SC. Molecular views of recombination proteins and their control. Nat. Rev. Mol. Cell Biol. 2003;4:435–445. doi: 10.1038/nrm1127. [DOI] [PubMed] [Google Scholar]
- 8.Wyman C, Kanaar R. DNA double-strand break repair: all's well that ends well. Annu. Rev. Genet. 2006;40:363–383. doi: 10.1146/annurev.genet.40.110405.090451. [DOI] [PubMed] [Google Scholar]
- 9.Ayora S, Carrasco B, Cardenas PP, Cesar CE, Canas C, Yadav T, Marchisone C, Alonso JC. Double-strand break repair in bacteria: a view from Bacillus subtilis. FEMS Microbiol. Rev. 2011;35:1055–1081. doi: 10.1111/j.1574-6976.2011.00272.x. [DOI] [PubMed] [Google Scholar]
- 10.Kowalczykowski SC, Dixon DA, Eggleston AK, Lauder SD, Rehrauer WM. Biochemistry of homologous recombination in Escherichia coli. Microbiol. Rev. 1994;58:401–465. doi: 10.1128/mr.58.3.401-465.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pâques F, Haber JE. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 1999;63:349–404. doi: 10.1128/mmbr.63.2.349-404.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cox MM. Motoring along with the bacterial RecA protein. Nat. Rev. Mol. Cell Biol. 2007;8:127–138. doi: 10.1038/nrm2099. [DOI] [PubMed] [Google Scholar]
- 13.Pitcher RS, Brissett NC, Doherty AJ. Nonhomologous end-joining in bacteria: a microbial perspective. Annu. Rev. Microbiol. 2007;61:259–282. doi: 10.1146/annurev.micro.61.080706.093354. [DOI] [PubMed] [Google Scholar]
- 14.Shuman S, Glickman MS. Bacterial DNA repair by non-homologous end joining. Nat. Rev. Microbiol. 2007;5:852–861. doi: 10.1038/nrmicro1768. [DOI] [PubMed] [Google Scholar]
- 15.Symington LS, Gautier J. Double-strand break end resection and repair pathway choice. Annu. Rev. Genet. 2011;45:247–271. doi: 10.1146/annurev-genet-110410-132435. [DOI] [PubMed] [Google Scholar]
- 16.Piggot PJ, Hilbert DW. Sporulation of Bacillus subtilis. Curr. Opn. Microbiol. 2004;7:579–586. doi: 10.1016/j.mib.2004.10.001. [DOI] [PubMed] [Google Scholar]
- 17.Lindsay JA, Murrell WG. Changes in density of DNA after interaction with dipicolinic acid and its possible role in spore heat resistance. Curr. Microbiol. 1985;12:329–334. [Google Scholar]
- 18.Errington J. Septation and chromosome segregation during sporulation in Bacillus subtilis. Curr. Opn. Microbiol. 2001;4:660–666. doi: 10.1016/s1369-5274(01)00266-1. [DOI] [PubMed] [Google Scholar]
- 19.Setlow P. Spores of Bacillus subtilis: their resistance to and killing by radiation, heat and chemicals. J. Appl. Microbiol. 2006;101:514–525. doi: 10.1111/j.1365-2672.2005.02736.x. [DOI] [PubMed] [Google Scholar]
- 20.Setlow P. I will survive: DNA protection in bacterial spores. Trends Microbiol. 2007;15:172–180. doi: 10.1016/j.tim.2007.02.004. [DOI] [PubMed] [Google Scholar]
- 21.Wang ST, Setlow B, Conlon EM, Lyon JL, Imamura D, Sato T, Setlow P, Losick R, Eichenberger P. The forespore line of gene expression in Bacillus subtilis. J. Mol. Biol. 2006;358:16–37. doi: 10.1016/j.jmb.2006.01.059. [DOI] [PubMed] [Google Scholar]
- 22.Veening JW, Murray H, Errington J. A mechanism for cell cycle regulation of sporulation initiation in Bacillus subtilis. Genes Dev. 2009;23:1959–1970. doi: 10.1101/gad.528209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dose K, Bieger-Dose A, Kerz O, Gill M. DNA-strand breaks limit survival in extreme dryness. Origins Life Evol. B. 1991;21:177–187. doi: 10.1007/BF01809446. [DOI] [PubMed] [Google Scholar]
- 24.Dose K, Bieger-Dose A, Labusch M, Gill M. Survival in extreme dryness and DNA-single-strand breaks. Adv. Space Res. 1992;12:221–229. doi: 10.1016/0273-1177(92)90176-x. [DOI] [PubMed] [Google Scholar]
- 25.Potts M. Desiccation tolerance of prokaryotes. Microbiol. Rev. 1994;58:755–805. doi: 10.1128/mr.58.4.755-805.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nicholson WL, Munakata N, Horneck G, Melosh HJ, Setlow P. Resistance of Bacillus endospores to extreme terrestrial and extraterrestrial environments. Microbiol. Mol. Biol. Rev. 2000;64:548–572. doi: 10.1128/mmbr.64.3.548-572.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weller GR, Kysela B, Roy R, Tonkin LM, Scanlan E, Della M, Devine SK, Day JP, Wilkinson A, d'Adda di Fagagna F, et al. Identification of a DNA nonhomologous end-joining complex in bacteria. Science. 2002;297:1686–1689. doi: 10.1126/science.1074584. [DOI] [PubMed] [Google Scholar]
- 28.de Vega M. The minimal Bacillus subtilis nonhomologous end joining repair machinery. PLoS One. 2013;8:e64232. doi: 10.1371/journal.pone.0064232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Setlow B, Setlow P. Role of DNA repair in Bacillus subtilis spore resistance. J. Bacteriol. 1996;178:3486–3495. doi: 10.1128/jb.178.12.3486-3495.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Carrasco B, Cozar MC, Lurz R, Alonso JC, Ayora S. Genetic recombination in Bacillus subtilis 168: contribution of Holliday junction processing functions in chromosome segregation. J. Bacteriol. 2004;186:5557–5566. doi: 10.1128/JB.186.17.5557-5566.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mascarenhas J, Sanchez H, Tadesse S, Kidane D, Krishnamurthy M, Alonso JC, Graumann PL. Bacillus subtilis SbcC protein plays an important role in DNA inter-strand cross-link repair. BMC Mol. Biol. 2006;7:20. doi: 10.1186/1471-2199-7-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moeller R, Stackebrandt E, Reitz G, Berger T, Rettberg P, Doherty AJ, Horneck G, Nicholson WL. Role of DNA repair by nonhomologous-end joining in Bacillus subtilis spore resistance to extreme dryness, mono- and polychromatic UV, and ionizing radiation. J. Bacteriol. 2007;189:3306–3311. doi: 10.1128/JB.00018-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Radding CM. Helical interactions in homologous pairing and strand exchange driven by RecA protein. J. Biol. Chem. 1991;266:5355–5358. [PubMed] [Google Scholar]
- 34.Sandler SJ, Clark AJ. RecOR suppression of recF mutant phenotypes in Escherichia coli K-12. J. Bacteriol. 1994;176:3661–3672. doi: 10.1128/jb.176.12.3661-3672.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Umezu K, Chi NW, Kolodner RD. Biochemical interaction of the Escherichia coli RecF, RecO, and RecR proteins with RecA protein and single-stranded DNA binding protein. Proc. Natl Acad. Sci. USA. 1993;90:3875–3879. doi: 10.1073/pnas.90.9.3875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Morimatsu K, Kowalczykowski SC. RecFOR proteins load RecA protein onto gapped DNA to accelerate DNA strand exchange: a universal step of recombinational repair. Mol. Cell. 2003;11:1337–1347. doi: 10.1016/s1097-2765(03)00188-6. [DOI] [PubMed] [Google Scholar]
- 37.Sakai A, Cox MM. RecFOR and RecOR as distinct RecA loading pathways. J. Biol. Chem. 2009;284:3264–3272. doi: 10.1074/jbc.M807220200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Morimatsu K, Wu Y, Kowalczykowski SC. RecFOR proteins target RecA protein to a DNA gap with either DNA or RNA at the 5' terminus: implication for repair of stalled replication forks. J. Biol. Chem. 2012;287:35621–35630. doi: 10.1074/jbc.M112.397034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Little JW. Mechanism of specific LexA cleavage: autodigestion and the role of RecA coprotease. Biochimie. 1991;73:411–421. doi: 10.1016/0300-9084(91)90108-d. [DOI] [PubMed] [Google Scholar]
- 40.Sassanfar M, Roberts JW. Nature of the SOS-inducing signal in Escherichia coli. The involvement of DNA replication. J. Mol. Biol. 1990;212:79–96. doi: 10.1016/0022-2836(90)90306-7. [DOI] [PubMed] [Google Scholar]
- 41.McInerney P, O'Donnell M. Replisome fate upon encountering a leading strand block and clearance from DNA by recombination proteins. J. Biol. Chem. 2007;282:25903–25916. doi: 10.1074/jbc.M703777200. [DOI] [PubMed] [Google Scholar]
- 42.Indiani C, Patel M, Goodman MF, O'Donnell ME. RecA acts as a switch to regulate polymerase occupancy in a moving replication fork. Proc. Natl Acad. Sci. USA. 2013;110:5410–5415. doi: 10.1073/pnas.1303301110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lia G, Rigato A, Long E, Chagneau C, Le Masson M, Allemand JF, Michel B. RecA-promoted, RecFOR-independent progressive disassembly of replisomes stalled by helicase inactivation. Mol. Cell. 2013;49:547–557. doi: 10.1016/j.molcel.2012.11.018. [DOI] [PubMed] [Google Scholar]
- 44.Yeeles JT, Marians KJ. The Escherichia coli replisome is inherently DNA damage tolerant. Science. 2011;334:235–238. doi: 10.1126/science.1209111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Patel M, Jiang Q, Woodgate R, Cox MM, Goodman MF. A new model for SOS-induced mutagenesis: how RecA protein activates DNA polymerase V. Crit. Rev. Biochem. Mol. Biol. 2010;45:171–184. doi: 10.3109/10409238.2010.480968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cox MM. Regulation of bacterial RecA protein function. Crit. Rev. Biochem. Mol. Biol. 2007;42:41–63. doi: 10.1080/10409230701260258. [DOI] [PubMed] [Google Scholar]
- 47.Lenhart JS, Schroeder JW, Walsh BW, Simmons LA. DNA repair and genome maintenance in Bacillus subtilis. Microbiol. Mol. Biol. Rev. 2012;76:530–564. doi: 10.1128/MMBR.05020-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Alonso JC, Cardenas PP, Sanchez H, Hejna J, Suzuki Y, Takeyasu K. Early steps of double-strand break repair in Bacillus subtilis. DNA Repair. 2013;12:162–176. doi: 10.1016/j.dnarep.2012.12.005. [DOI] [PubMed] [Google Scholar]
- 49.Carrasco B, Cardenas PP, Cañas C, Yadav T, César CE, Ayora S, Alonso JC. Dynamics of DNA Double-strand Break Repair in Bacillus subtilis. 2nd edn. Norfolk: Caister Academic Press; 2012. [Google Scholar]
- 50.Duigou S, Ehrlich SD, Noirot P, Noirot-Gros MF. Distinctive genetic features exhibited by the Y-family DNA polymerases in Bacillus subtilis. Mol. Microbiol. 2004;54:439–451. doi: 10.1111/j.1365-2958.2004.04259.x. [DOI] [PubMed] [Google Scholar]
- 51.Schaeffer P, Millet J, Aubert JP. Catabolic repression of bacterial sporulation. Proc. Natl Acad. Sci. USA. 1965;54:704–711. doi: 10.1073/pnas.54.3.704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nicholson WL, Setlow P. Sporulation, germination and outgrowth. In: Harwood CR, Cutting SM, editors. Molecular Biological Methods for Bacillus. Chichester, UK: J. Wiley & Sons; 1990. pp. 391–450. [Google Scholar]
- 53.Moeller R, Setlow P, Horneck G, Berger T, Reitz G, Rettberg P, Doherty AJ, Okayasu R, Nicholson WL. Roles of the major, small, acid-soluble spore proteins and spore-specific and universal DNA repair mechanisms in resistance of Bacillus subtilis spores to ionizing radiation from X rays and high-energy charged-particle bombardment. J. Bacteriol. 2008;190:1134–1140. doi: 10.1128/JB.01644-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Horneck G. Responses of Bacillus subtilis spores to space environment: results from experiments in space. Origins Life Evol. B. 1993;23:37–52. doi: 10.1007/BF01581989. [DOI] [PubMed] [Google Scholar]
- 55.Munakata N, Saitou M, Takahashi N, Hieda K, Morohoshi F. Induction of unique tandem-base change mutations in bacterial spores exposed to extreme dryness. Mutat. Res. 1997;390:189–195. doi: 10.1016/s0165-1218(97)00020-7. [DOI] [PubMed] [Google Scholar]
- 56.Yadav T, Carrasco B, Myers AR, George NP, Keck JL, Alonso JC. Genetic recombination in Bacillus subtilis: a division of labor between two single-strand DNA-binding proteins. Nucleic Acids Res. 2012;40:5546–5559. doi: 10.1093/nar/gks173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Carrasco B, Ayora S, Lurz R, Alonso JC. Bacillus subtilis RecU Holliday-junction resolvase modulates RecA activities. Nucleic Acids Res. 2005;33:3942–3952. doi: 10.1093/nar/gki713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Manfredi C, Carrasco B, Ayora S, Alonso JC. Bacillus subtilis RecO nucleates RecA onto SsbA-coated single-stranded DNA. J. Biol. Chem. 2008;283:24837–24847. doi: 10.1074/jbc.M802002200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hobbs MD, Sakai A, Cox MM. SSB protein limits RecOR binding onto single-stranded DNA. J. Biol. Chem. 2007;282:11058–11067. doi: 10.1074/jbc.M611007200. [DOI] [PubMed] [Google Scholar]
- 60.Sanders GM, Dallmann HG, McHenry CS. Reconstitution of the B. subtilis replisome with 13 proteins including two distinct replicases. Mol. Cell. 2010;37:273–281. doi: 10.1016/j.molcel.2009.12.025. [DOI] [PubMed] [Google Scholar]
- 61.Seco EM, Zinder JC, Manhart CM, Lo Piano A, McHenry CS, Ayora S. Bacteriophage SPP1 DNA replication strategies promote viral and disable host replication in vitro. Nucleic Acids Res. 2013;41:1711–1721. doi: 10.1093/nar/gks1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Moeller R, Reitz G, Li Z, Klein S, Nicholson WL. Multifactorial resistance of Bacillus subtilis spores to high-energy proton radiation: role of spore structural components and the homologous recombination and non-homologous end joining DNA repair pathways. Astrobiology. 2012;12:1069–1077. doi: 10.1089/ast.2012.0890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Moeller R, Schuerger AC, Reitz G, Nicholson WL. Impact of two DNA repair pathways, homologous recombination and non-homologous end joining, on bacterial spore inactivation under simulated martian environmental conditions. Icarus. 2011;215:204–210. [Google Scholar]
- 64.Michel B, Boubakri H, Baharoglu Z, LeMasson M, Lestini R. Recombination proteins and rescue of arrested replication forks. DNA Repair. 2007;6:967–980. doi: 10.1016/j.dnarep.2007.02.016. [DOI] [PubMed] [Google Scholar]
- 65.Kidane D, Sanchez H, Alonso JC, Graumann PL. Visualization of DNA double-strand break repair in live bacteria reveals dynamic recruitment of Bacillus subtilis RecF, RecO and RecN proteins to distinct sites on the nucleoids. Mol. Microbiol. 2004;52:1627–1639. doi: 10.1111/j.1365-2958.2004.04102.x. [DOI] [PubMed] [Google Scholar]
- 66.Sanchez H, Carrasco B, Cozar MC, Alonso JC. Bacillus subtilis RecG branch migration translocase is required for DNA repair and chromosomal segregation. Mol. Microbiol. 2007;65:920–935. doi: 10.1111/j.1365-2958.2007.05835.x. [DOI] [PubMed] [Google Scholar]
- 67.Sanchez H, Kidane D, Reed P, Curtis FA, Cozar MC, Graumann PL, Sharples GJ, Alonso JC. The RuvAB branch migration translocase and RecU Holliday junction resolvase are required for double-stranded DNA break repair in Bacillus subtilis. Genetics. 2005;171:873–883. doi: 10.1534/genetics.105.045906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kidane D, Graumann PL. Dynamic formation of RecA filaments at DNA double strand break repair centers in live cells. J. Cell Biol. 2005;170:357–366. doi: 10.1083/jcb.200412090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sanchez H, Kidane D, Cozar MC, Graumann PL, Alonso JC. Recruitment of Bacillus subtilis RecN to DNA double-strand breaks in the absence of DNA end processing. J. Bacteriol. 2006;188:353–360. doi: 10.1128/JB.188.2.353-360.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cardenas PP, Carrasco B, Sanchez H, Deikus G, Bechhofer DH, Alonso JC. Bacillus subtilis polynucleotide phosphorylase 3'-to-5' DNase activity is involved in DNA repair. Nucleic Acids Res. 2009;37:4157–4169. doi: 10.1093/nar/gkp314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cardenas PP, Carzaniga T, Zangrossi S, Briani F, Garcia-Tirado E, Deho G, Alonso JC. Polynucleotide phosphorylase exonuclease and polymerase activities on single-stranded DNA ends are modulated by RecN, SsbA and RecA proteins. Nucleic Acids Res. 2011;39:9250–9261. doi: 10.1093/nar/gkr635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yeeles JT, Dillingham MS. The processing of double-stranded DNA breaks for recombinational repair by helicase-nuclease complexes. DNA Repair. 2010;9:276–285. doi: 10.1016/j.dnarep.2009.12.016. [DOI] [PubMed] [Google Scholar]
- 73.Niu H, Raynard S, Sung P. Multiplicity of DNA end resection machineries in chromosome break repair. Genes Dev. 2009;23:1481–1486. doi: 10.1101/gad.1824209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cardenas PP, Carrasco B, Defeu Soufo C, Cesar CE, Herr K, Kaufenstein M, Graumann PL, Alonso JC. RecX facilitates homologous recombination by modulating RecA activities. PLoS Genet. 2012;8:e1003126. doi: 10.1371/journal.pgen.1003126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chow KH, Courcelle J. RecO acts with RecF and RecR to protect and maintain replication forks blocked by UV-induced DNA damage in Escherichia coli. J. Biol. Chem. 2004;279:3492–3496. doi: 10.1074/jbc.M311012200. [DOI] [PubMed] [Google Scholar]
- 76.Lusetti SL, Hobbs MD, Stohl EA, Chitteni-Pattu S, Inman RB, Seifert HS, Cox MM. The RecF protein antagonizes RecX function via direct interaction. Mol. Cell. 2006;21:41–50. doi: 10.1016/j.molcel.2005.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Alonso JC, Tailor RH, Luder G. Characterization of recombination-deficient mutants of Bacillus subtilis. J. Bacteriol. 1988;170:3001–3007. doi: 10.1128/jb.170.7.3001-3007.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Alonso JC, Stiege AC, Luder G. Genetic recombination in Bacillus subtilis 168: effect of recN, recF, recH and addAB mutations on DNA repair and recombination. Mol. Gen. Genet. 1993;239:129–136. doi: 10.1007/BF00281611. [DOI] [PubMed] [Google Scholar]
- 79.Fernandez S, Kobayashi Y, Ogasawara N, Alonso JC. Analysis of the Bacillus subtilis recO gene: RecO forms part of the RecFLOR function. Mol. Gen. Genet. 1999;261:567–573. doi: 10.1007/s004380051002. [DOI] [PubMed] [Google Scholar]
- 80.Gassel M, Alonso JC. Expression of the recE gene during induction of the SOS response in Bacillus subtilis recombination-deficient strains. Mol. Microbiol. 1989;3:1269–1276. doi: 10.1111/j.1365-2958.1989.tb00277.x. [DOI] [PubMed] [Google Scholar]
- 81.Alonso JC, Luder G. Characterization of recF suppressors in Bacillus subtilis. Biochimie. 1991;73:277–280. doi: 10.1016/0300-9084(91)90213-k. [DOI] [PubMed] [Google Scholar]
- 82.Simmons LA, Goranov AI, Kobayashi H, Davies BW, Yuan DS, Grossman AD, Walker GC. Comparison of responses to double-strand breaks between Escherichia coli and Bacillus subtilis reveals different requirements for SOS induction. J. Bacteriol. 2009;191:1152–1161. doi: 10.1128/JB.01292-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Nicholson WL, Moeller R, Protect Team and Horneck G. Transcriptomic responses of germinating Bacillus subtilis spores exposed to 1.5 years of space and simulated martian conditions on the EXPOSE-E experiment PROTECT. Astrobiology. 2012;12:469–486. doi: 10.1089/ast.2011.0748. [DOI] [PubMed] [Google Scholar]
- 84.Au N, Kuester-Schoeck E, Mandava V, Bothwell LE, Canny SP, Chachu K, Colavito SA, Fuller SN, Groban ES, Hensley LA, et al. Genetic composition of the Bacillus subtilis SOS system. J. Bacteriol. 2005;187:7655–7666. doi: 10.1128/JB.187.22.7655-7666.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Fernandez De Henestrosa AR, Ogi T, Aoyagi S, Chafin D, Hayes JJ, Ohmori H, Woodgate R. Identification of additional genes belonging to the LexA regulon in Escherichia coli. Mol. Microbiol. 2000;35:1560–1572. doi: 10.1046/j.1365-2958.2000.01826.x. [DOI] [PubMed] [Google Scholar]
- 86.Petit MA, Ehrlich D. Essential bacterial helicases that counteract the toxicity of recombination proteins. EMBO J. 2002;21:3137–3147. doi: 10.1093/emboj/cdf317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Carrasco B, Manfredi C, Ayora S, Alonso JC. Bacillus subtilis SsbA and dATP regulate RecA nucleation onto single-stranded DNA. DNA Repair. 2008;7:990–996. doi: 10.1016/j.dnarep.2008.03.019. [DOI] [PubMed] [Google Scholar]
- 88.Bruck I, O'Donnell M. The DNA replication machine of a Gram-positive organism. J. Biol. Chem. 2000;275:28971–28983. doi: 10.1074/jbc.M003565200. [DOI] [PubMed] [Google Scholar]
- 89.Manhart CM, McHenry CS. The PriA replication restart protein blocks replicase access prior to helicase assembly and directs template specificity through its ATPase activity. J. Biol. Chem. 2013;288:3989–3999. doi: 10.1074/jbc.M112.435966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Alonso JC, Stiege AC, Dobrinski B, Lurz R. Purification and properties of the RecR protein from Bacillus subtilis 168. J. Biol. Chem. 1993;268:1424–1429. [PubMed] [Google Scholar]
- 91.Ayora S, Alonso JC. Purification and characterization of the RecF protein from Bacillus subtilis 168. Nucleic Acids Res. 1997;25:2766–2772. doi: 10.1093/nar/25.14.2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Durkin SG, Glover TW. Chromosome fragile sites. Annu. Rev. Genet. 2007;41:169–192. doi: 10.1146/annurev.genet.41.042007.165900. [DOI] [PubMed] [Google Scholar]
- 93.Rivas-Castillo AM, Yasbin RE, Robleto E, Nicholson WL, Pedraza-Reyes M. Role of the Y-family DNA polymerases YqjH and YqjW in protecting sporulating Bacillus subtilis cells from DNA damage. Curr. Microbiol. 2010;60:263–267. doi: 10.1007/s00284-009-9535-3. [DOI] [PubMed] [Google Scholar]
- 94.Fagerburg MV, Schauer GD, Thickman KR, Bianco PR, Khan SA, Leuba SH, Anand SP. PcrA-mediated disruption of RecA nucleoprotein filaments – essential role of the ATPase activity of RecA. Nucleic Acids Res. 2012;40:8416–8424. doi: 10.1093/nar/gks641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Park J, Myong S, Niedziela-Majka A, Lee KS, Yu J, Lohman TM, Ha T. PcrA helicase dismantles RecA filaments by reeling in DNA in uniform steps. Cell. 2010;142:544–555. doi: 10.1016/j.cell.2010.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Seigneur M, Bidnenko V, Ehrlich SD, Michel B. RuvAB acts at arrested replication forks. Cell. 1998;95:419–430. doi: 10.1016/s0092-8674(00)81772-9. [DOI] [PubMed] [Google Scholar]
- 97.Ceglowski P, Luder G, Alonso JC. Genetic analysis of recE activities in Bacillus subtilis. Mol Gen Genet. 1990;222:441–445. doi: 10.1007/BF00633853. [DOI] [PubMed] [Google Scholar]
- 98.Fernández S, Sorokin A, Alonso JC. Genetic recombination in Bacillus subtilis 168: effects of recU and recS mutations on DNA repair and homologous recombination. J Bacteriol. 1998;180:3405–3409. doi: 10.1128/jb.180.13.3405-3409.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Alonso JC, Shirahige K, Ogasawara N. Molecular cloning, genetic characterization and DNA sequence analysis of the recM region of Bacillus subtilis. Nucleic Acids Res. 1990;18:6771–6777. doi: 10.1093/nar/18.23.6771. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.