Abstract
Aims: FUsed in sarcoma (FUS) is a multifunctional DNA/RNA-binding protein that possesses diverse roles, such as RNA splicing, RNA transport, DNA repair, translation, and transcription. The network of enzymes and processes regulated by FUS is far from being fully described. In this study, we have focused on the mechanisms of FUS-regulated manganese superoxide dismutase (MnSOD) gene transcription. Results: Here we demonstrate that FUS is a component of the transcription complex that regulates the expression of MnSOD. Overexpression of FUS increased MnSOD expression in a dose-dependent manner and knockdown of FUS by siRNA led to the inhibition of MnSOD gene transcription. Reporter analyses, chromatin immunoprecipitation assay, electrophoretic mobility shift assay, affinity chromatography, and surface plasmon resonance analyses revealed the far upstream region of MnSOD promoter as an important target of FUS-mediated MnSOD transcription and confirmed that FUS binds to the MnSOD promoter and interacts with specificity protein 1 (Sp1). Importantly, overexpression of familial amyotropic lateral sclerosis (fALS)-linked R521G mutant FUS resulted in a significantly reduced level of MnSOD expression and activity, which is consistent with the decline in MnSOD activity observed in fibroblasts from fALS patients with the R521G mutation. R521G-mutant FUS abrogates MnSOD promoter-binding activity and interaction with Sp1. Innovation and Conclusion: This study identifies FUS as playing a critical role in MnSOD gene transcription and reveals a previously unrecognized relationship between MnSOD and mutant FUS in fALS. Antioxid. Redox Signal. 20, 1550–1566.
Introduction
FUsed in sarcoma/translocated in liposarcoma (FUS/TLS) is a DNA and RNA binding protein that is normally localized in the nucleus but is often redistributed to cytoplasm in familial amyotropic lateral sclerosis (fALS) cases with FUS mutations (31). It has been demonstrated that FUS plays important roles in the regulation of gene transcription (34, 48), DNA binding, mRNA splicing (60), DNA repair, and cell proliferation (5). Although FUS/TLS knockout mice exhibit perinatal lethality (23), male sterility, and radiation sensitivity (30), the molecular targets of FUS/TLS have not been identified.
In this study, we have identified FUS as an interacting partner of nucleophosmin (NPM), which we previously established as a coactivator with NF-κB for transactivation of manganese superoxide dismutase (MnSOD) (SOD2) (12). However, the role of FUS in regulating MnSOD is not well understood. Here we investigated the role of FUS in the regulation of MnSOD gene transcription. MnSOD is a primary antioxidant enzyme present in mitochondria. Human MnSOD is coded by a single-copy gene consisting of five exons interrupted by four introns with typical splice junctions (53). The basal promoter of MnSOD exhibits multiple transcription factor binding motifs containing specificity protein 1 (Sp1) and AP-2 binding sites. Functional studies in cell lines with different Sp1 protein levels suggest that Sp1 is essential for the constitutive expression of the MnSOD gene (58). In addition to the regulatory elements in the 5′-flanking region, the MnSOD gene contains enhancer elements in the second intron that contains NF-κB, C/EBP, and NF-1 transcription factor binding sites (26, 57). Activation of Sp1 and NF-κB is important for the induction of MnSOD under oxidative stress conditions in a variety of diseases, including cancer and neurodegeneration (13, 38).
Innovation.
Our findings demonstrate that FUsed in sarcoma (FUS) regulates the basal level of manganese superoxide dismutase (MnSOD), the principal enzyme responsible for the removal of superoxide in mitochondria. We have identified FUS as a component of the transcription complex that regulates the expression of MnSOD by binding to a FUS recognition element on the human MnSOD gene. The data suggest not only a role for FUS in ROS detoxifying cellular systems, but also suggest the possibility that dysregulation of the MnSOD gene is a mediator of mutant FUS toxicity in mitochondria. This study is the first to implicate a mitochondrial-mediated mechanism by which FUS, may participate in the progression of fALS.
Abundant evidence supports the critical role of MnSOD as a cytoprotective enzyme both in vivo and in vitro. MnSOD knockout mice develop cardiomyopathy and die within 10–15 days after birth (33), and treatment with SOD mimetics can protect MnSOD knockout mice from systemic toxicity and neonatal death (41). Conversely, transgenic mice overexpressing human MnSOD are protected from inflammation (54), cardiotoxic drugs (62), and pathological and physiological conditions leading to neuronal injury (28).
ALS is an age-associated human neurodegenerative disease affecting cortical and spinal motor neurons (44). ALS is typically manifested by progressive muscle atrophy and weakness leading to death due to respiratory paralysis within 3–5 years. Although the critical sequence of molecular events leading to ALS is not clear, recurring themes of ALS pathogenesis include mitochondrial dysfunction, protein misfolding and aggregation, axonal transport defects and glutamate excitotoxicity. Approximately 90% of ALS cases are sporadic and the other 10% of ALS cases are familial (fALS), usually with an autosomal dominant pattern of inheritance (24). Several genes have been implicated in familial ALS, including SOD1 (encoding Cu/Zn superoxide dismutase), ANG (angiogenin), VAPB (vesicle associated membrane protein B), TARDP (TAR DNA binding protein TDP-43), FUS/TLS (42) and more recently expansion of a noncoding hexanucleotide repeat in the C9orf72 gene (9). FUS was originally identified by its fusion to C/EBP homologous protein (CHOP), a member of the CCAAT/enhancer-binding protein, family of transcription factors in human myxoid liposarcoma with t(12;16) chromosomal translocation (11). FUS/TLS shares a high homology with Ewing's sarcoma and TATA-binding protein associated factor II (TAFII); which are collectively called FUS Ewing's sarcoma (FET) family proteins (1). The C-terminal of FUS is composed of multiple domains of peptide sequences that are often found in RNA-binding proteins. The RNA-binding sequences known as RNA recognition motif (RRM) are relatively conserved and are flanked by two regions rich in Arg-Gly-Gly repeats (RGG domains) separated by a zinc finger region. FUS can also bind with double stranded DNA via the C-terminal region (46). In the N-terminus, the FET family of proteins contains a glutamine-, serine-, and tyrosine-rich region that functions as a transcriptional activator when fused to a heterologous DNA binding domain (39). However, the cellular targets of these genes have not been identified.
In the present study, we investigated the roles of wild-type and mutant FUS in MnSOD gene activation using cell culture models and skin biopsy samples from fALS patients with the R521G FUS mutation. The key finding of this study is that FUS regulates MnSOD gene transcription and the FUS mutation in fALS causes impaired MnSOD expression and reduced enzymatic activity. We further elucidated the detailed molecular mechanism by which FUS regulates MnSOD gene expression.
Results
Identification of FUS in MnSOD transcription complex by proteomic analysis
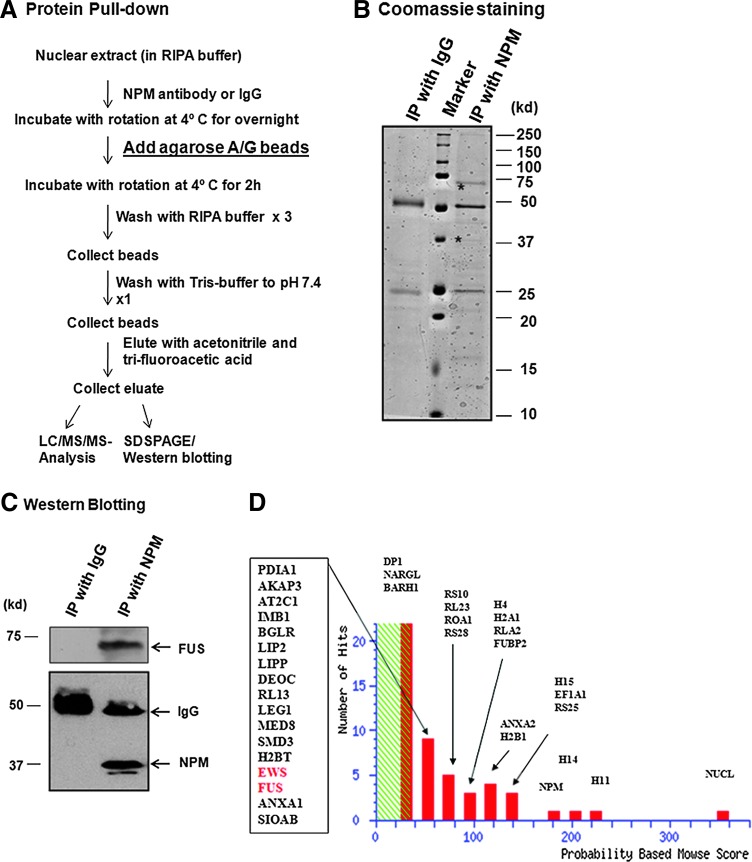
We previously showed that NPM, a RNA binding protein, physically interacts with the MnSOD promoter and enhancer regions, facilitates transcription factor bindings and coactivates gene transcription (12, 15). To identify proteins interacting with NPM in the transcription complex, nuclear extracts were immunoprecipitated with NPM antibody, or normal immunoglobulin (IgG) as a control coupled to agarose A/G beads, and the immune complexes were eluted as illustrated in Figure 1A. Eluted protein complexes were then subjected to sodium dodecyle sulphate-polyacrylamide gel electrophoresis (SDS-PAGE) electrophoresis and visualized by Coomassie brilliant blue staining (Fig. 1B). The specificity of immunoprecipitation was verified using normal IgG instead of the NPM antibody. To verify the presence of NPM and FUS protein in the immunoprecipitated complex, the proteins separated on the SDS-PAGE gels were transferred to nitrocellulose membranes and subjected to Western blotting analysis using the NPM or FUS antibody. As shown in Figure 1C, the NPM antibody immunoprecipitated the NPM protein along with FUS, but the nonimmune IgG did not. To identify proteins in the immunoprecipitate, the whole complex (eluted fraction) was digested with trypsin and analyzed by mass spectrometry. LC-MS/MS analysis identified several proteins abundantly present in the immunoprecipitate. The significant hits obtained by MS/MS analysis are shown in Figure 1D. The identified peptide fragments of NPM, nucleolin and FUS are presented in Table 1.
FIG. 1.
Identification of FUsed in sarcoma (FUS) in nucleophosmin (NPM) immunocomplex. Transcription factor as immune-complexes were pulled down from the nuclear extracts obtained from hepatocellular carcinoma (HepG2) cells by immunoprecipitation with the NPM antibody for LC/MS/MS analysis and sodium dodecyle sulphate-polyacrylamide gel electrophoresis (SDS-PAGE) analysis (A). The proteins were separated on SDS-polyacrylamide gel and stained with Coomassie brilliant blue. The immunoprecipitated bands with a molecular weight that corresponds to NPM or FUS are marked by an asterisk (B). The presence of NPM and FUS in the immunocomplex was detected by Western blotting using monoclonal antibody to NPM (1:1000) and polyclonal antibody for FUS (1:1000), respectively (C). Identities of proteins or group of proteins that are present in the immunocomplex are shown (D). To see this illustration in color, the reader is referred to the web version of this article at www.liebertpub.com/ars
Table 1.
Sequences of Trypsin-Produced Peptide Fragments from Nucleophosmin Mediated Immunoprecipitates by MS/MS-Analysis
| Human Nucleophosmin |
| MQASIEKGGSLPK |
| MTDQEAIQDLWQWRK |
| MTDQEAIQDLWQWR |
| MSVQPTVSLGGFEITPPVVLR |
| Human Nucleolin |
| AIRLELQGPR |
| GFGFVDFNSEEDAK |
| GLSEDTTEETLKESFDGSVR |
| TLVLSNLSYSATEETLQEVFEK |
| QKVEGTEFTTAFNLFVGNLNFNK |
| RNA-binding protein FUS |
| AAIDWFDGK |
| ADFNRGGGNGRGGR |
| GGMGGSDRGGFNKFGGPR |
Impairment of MnSOD enzyme activity in fALS
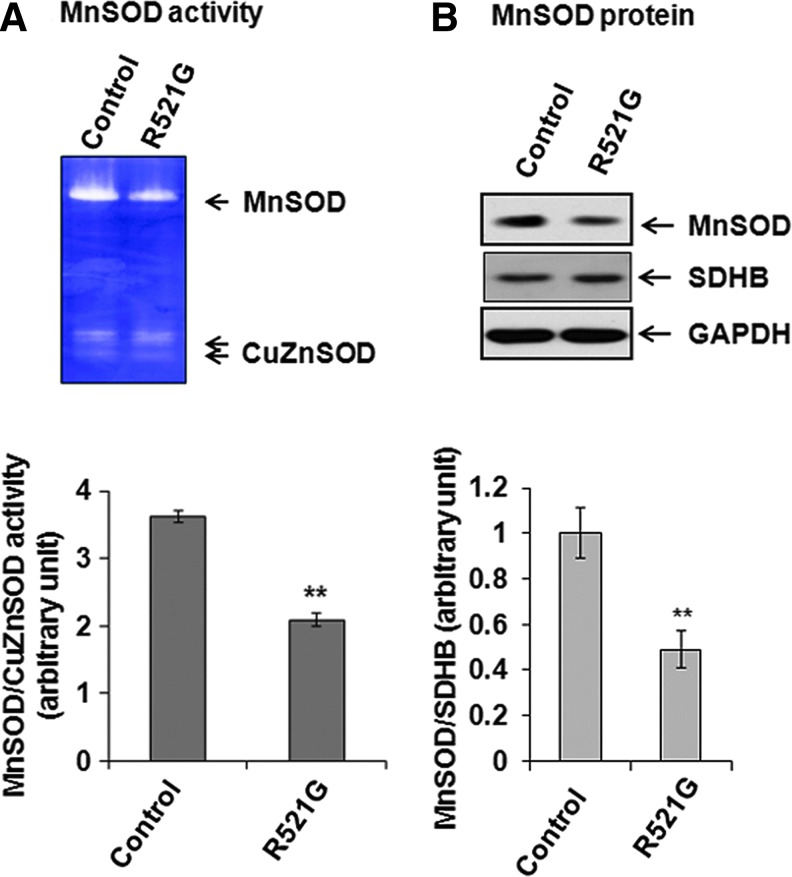
To probe the relationship between MnSOD and fALS, we collected skin biopsy samples from three individual patients carrying the R521G mutation in the FUS gene who were treated at the University of Kentucky, and three non-ALS control subjects. Using these biopsy samples, we established primary fibroblast cultures, and determined MnSOD protein levels and enzyme activity by Western blotting and activity gel electrophoresis, respectively. The samples were also examined for FUS mutation at position 1561 (R521G) in exon 15, where it has recently been demonstrated to be present in some families with fALS (31). The R521G mutation was verified in all three symptomatic patients and the levels of MnSOD activity (Fig. 2A) in cultured fibroblasts derived from these fALS patients were reduced as compared to the non-ALS controls. The reduction in MnSOD activity is related to the reduction of MnSOD protein level (Fig. 2B). That the reduced level of MnSOD protein is not due to the loss of mitochondria but is due to the reduction of protein expression in fALS patients was confirmed by referencing the levels of succinate dehydrogenase. These results indicate that MnSOD enzymatic activity is compromised in fALS patients with the R521G mutation in FUS. The reduced MnSOD activity and protein levels are consistent with the possibility that FUS may participate in the transcriptional control of MnSOD identified by proteomic analysis.
FIG. 2.
Levels of manganese superoxide dismutase (MnSOD) in amyotropic lateral sclerosis (ALS) patients. Human skin biopsy samples collected from the familial ALS (fALS) patients or from normal individuals were minced into small pieces in modified Eagle's medium (MEM) cell culture medium, and then transferred onto primary culture dish. After the tissues were attached to the flask, MEM was removed and supplemented with MEM containing 20% fetal bovine serum, 20 U/ml penicillin-streptomycin, 2 mM glutamine (Invitrogen). Cells at 80%–90% confluence were used for experiments. The cell lysates of the primary human skin fibroblasts obtained from ALS patients with R521G mutation in FUS gene or from normal individuals were subjected to 12.5% polyacrylamide native gel for MnSOD activity, which was then detected by staining the gel with nitro-blue-tetrazolium dye (A, top panel). The bands were densitometrically scanned and normalized with CuZnSOD (A, bottom panel). The cell lysates were also subjected to SDS-PAGE followed by Western blotting using MnSOD specific antibody to detect MnSOD protein levels (B, top panel). The protein levels were quantified by densitometric scanning of MnSOD bands followed by normalization with succinate dehydrogenase (SDHB) as an internal control (B, bottom panel). Each data point represents mean±SD of three independent samples and significant difference as compared to respective controls is indicated by **p<0.01. To see this illustration in color, the reader is referred to the web version of this article at www.liebertpub.com/ars
Mutation of FUS abrogates MnSOD gene transcription and enzymatic activity
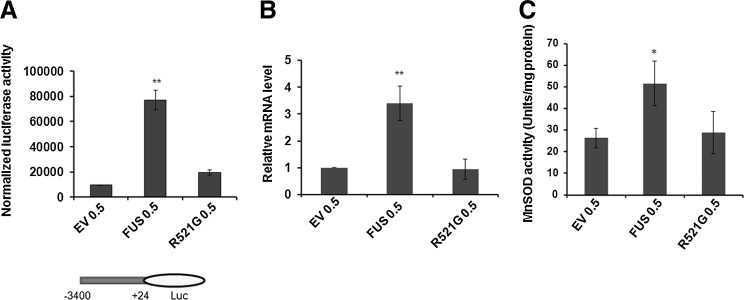
To directly determine how FUS plays a role in MnSOD transcription and whether the mutated FUS gene found in fALS can perturb MnSOD transcription, we overexpressed wild-type FUS cDNA or FUS cDNA carrying the mutation at position 1561 in exon 15 (C>G), which corresponds to the arginine to glycine mutation at amino acid 521 (R521G). The arginine residue in FUS at position 521 is highly conserved in a variety of species (Supplementary Fig. S1; Supplementary Data are available online at www.liebertpub.com/ars). After transient transfection of JB6 cells with a p3X-Flag-CMV10/FUS expression vector containing wild-type or mutant FUS along with MnSOD promoter construct, we examined MnSOD promoter activity. Overexpression of wild-type FUS increased the levels of MnSOD luciferase reporter gene activity (Fig. 3A) and mRNA (Fig. 3B), whereas overexpression of the mutant FUS did not. To examine whether the increase in MnSOD level led to the production of an active MnSOD, we tested the effects of FUS on MnSOD activity. As shown in Figure 3C, MnSOD enzyme activity increased significantly in JB6 cells overexpressing wild-type FUS but not in cells transfected with the mutant FUS. These results demonstrate that compared to wild-type FUS, the R521G mutant FUS is incapable of activating MnSOD gene transcription.
FIG. 3.
Mutation in FUS gene abrogates MnSOD transcription. Luciferase activity of the MnSOD promoter (−3401 to +24)-driven pGl3 reporter gene after transfection with wild-type or mutant FUS expression vector (A). Relative level of MnSOD mRNA was determined by real-time PCR after normalization with GAPDH mRNA levels as loading control (B). MnSOD enzyme activity (Units/mg protein) was measured in whole cell extract after transfection of wild-type FUS or mutant FUS expression vector in JB6 cells (C). Each data point represents mean±SD of three independent experiments and significant difference as compared to respective controls is indicated by *p<0.05 and **p<0.01.
Role of FUS in MnSOD gene expression
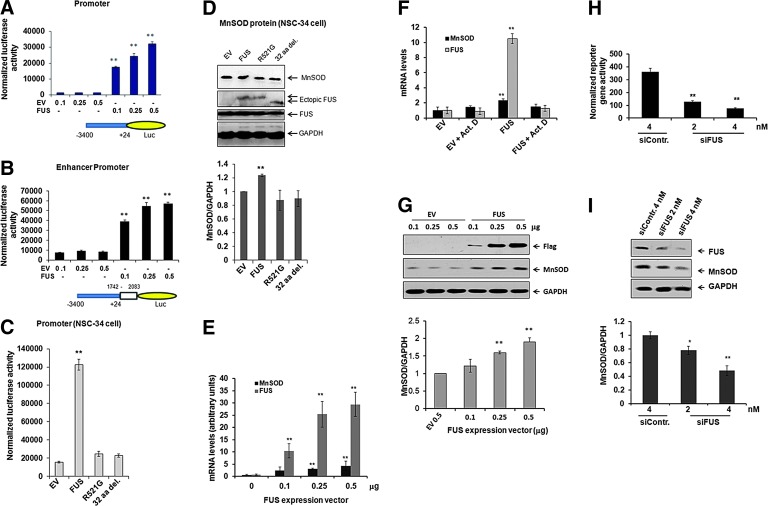
To investigate the role of FUS in MnSOD gene expression, JB6 cells were transiently transfected with different concentrations of FUS expression vector and MnSOD promoter with or without the I2E enhancer constructs (57). Overexpression of FUS with the promoter and enhancer constructs led to a dose-dependent increase of MnSOD gene transcription as measured by luciferase activity, which was not observed with transfection of empty vectors without the FUS gene sequence (Fig. 4A, B). We verified these findings in motor neuron-like NSC-34 cells after transfection with wild-type or mutant FUS expression vector together with MnSOD promoter-driven luciferase reporter vector. Consistent with the findings in JB6 cells, expression of wild-type FUS significantly increased the reporter gene activity and MnSOD protein levels in NSC-34 cells (Fig. 4C). In contrast, expression of mutant FUS (R521G) or deletion of 32 amino acids from the C-terminal was unable to induce MnSOD reporter gene activity or protein levels (Fig. 4C, D). To verify the role of FUS as a transcriptional regulator for MnSOD, we transfected the FUS expression vector into JB6 cells and measured the MnSOD mRNA levels. As shown in Figure 4E, FUS overexpression led to a significant increase in MnSOD mRNA level as detected by quantitative real-time PCR, indicating that FUS-mediated induction of MnSOD gene expression is a transcriptional event. This result was corroborated by measuring MnSOD mRNA levels in FUS-transfected cells with or without actinomycin D, an inhibitor of de novo mRNA synthesis. Overexpression of FUS significantly increased MnSOD mRNA levels as compared to the empty vector transfection control; however, the increase was blocked completely by pretreatment with actinomycin D (Fig. 4F). These results indicate that FUS-mediated induction of MnSOD gene expression is a result of increased mRNA synthesis and is not due to mRNA stabilization. Overexpression of FUS also resulted in dose-dependent increases of FUS and MnSOD proteins as detected by Western blotting using the Flag and MnSOD antibodies, respectively, which was not observed with the transfection of the empty vector (Fig. 4G). These results indicate that FUS induces MnSOD gene transcription resulting in increased protein expression.
FIG. 4.
FUS increases MnSOD expression. Luciferase activity of pGl3 reporter vector driven by the MnSOD promoter (−3401 to +24) (A) or MnSOD promoter/enhancer (−3401 to +24/1742 to 2083) construct (B) was measured in JB6 cells. MnSOD promoter (−3401 to +24) driven luciferase activity (C) and MnSOD protein levels (D) were determined in NSC-34 cells after transfection with wild-type or mutant FUS expression vector. MnSOD mRNA levels after FUS transfection were determined by Real-time PCR [briefly, real-time PCR was performed with a Light Cycle System (Roche Diagnostics) according to the manufacturer's protocol]. Real time PCR in the presence or absence of actinomycin D (0.5 μg/ml) (E, F). The level of MnSOD proteins in JB6 cells with or without FUS overexpression was also detected by Western blotting (G, top panel), and the protein bands were densitometrically scanned and graphed (G, bottom panel). The effect of endogenous FUS on MnSOD gene expression was evaluated by measuring the MnSOD promoter (−3401 to +24)-driven luciferase reporter gene activity in JB6 cells after cotransfection with FUS siRNA (H). Endogenous levels of FUS and MnSOD proteins were measured by Western blot (I, top panel), and the MnSOD protein bands were quantified and normalized with the band intensity of GAPDH (I, bottom panel). Each data point represents mean±SD of three independent experiments and significant difference as compared with respective controls is indicated by *p<0.05 or **p<0.01. To see this illustration in color, the reader is referred to the web version of this article at www.liebertpub.com/ars
To further verify the role played by FUS in MnSOD gene expression, JB6 cells were transfected with control siRNA or FUS siRNA. Transfection of FUS siRNA at different concentrations suppressed MnSOD gene expression in a dose-dependent manner as indicated by MnSOD reporter gene activity (Fig. 4H) and protein level (Fig. 4I). The suppression is statistically significant compared to transfection with the control siRNA. Transfection of FUS siRNA but not the control siRNA suppressed the level of endogenous FUS and MnSOD proteins with no suppressing effect on the nontarget gene product GAPDH (Fig. 4I). These results suggest that the endogenous FUS gene is as critical to activating MnSOD gene expression as the ectopically expressed FUS.
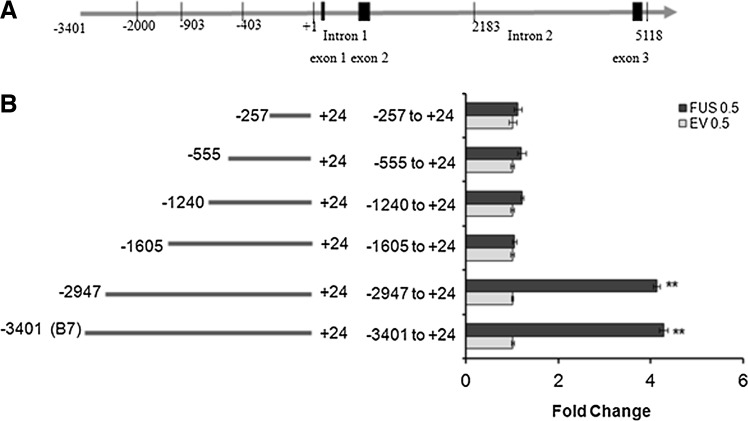
Identification of FUS responsive sites in the MnSOD promoter
To identify FUS binding site(s) in the MnSOD promoter, we prepared a series of deletion constructs of the MnSOD promoter, which we cotransfected with FUS expression vector or empty vector along with renilla luciferase as an internal control. Cotransfection of the long fragment of the MnSOD promoter and FUS expression vector resulted in increased luciferase activity as compared with the empty vector cotransfection control. The cotransfection of the 5′-region of MnSOD promoter constructs containing nucleotides (−3401 to +24) and (−2947 to +24) showed approximately a four-fold increase of luciferase activity as compared to that of the empty vector cotransfection control (Fig. 5B). Cotransfection of the 5′region of MnSOD promoter constructs containing the shorter nucleotides (−1605 to +24), (−1240 to +24), (−555 to +24), or (−257 to +24) with the FUS expression vector failed to induce luciferase activity (Fig. 5B). These results indicate that FUS-responsive site(s) are located in the far upstream region of the MnSOD gene promoter between nucleotides (−2947 to −1605).
FIG. 5.
Identification of FUS-mediated response in MnSOD promoter. Schematic description of MnSOD gene showing the promoter region (A) and various lengths of MnSOD promoter were subcloned in PGl3 luciferase vector (B). DNAs were cotransfected with FUS expression vector for 48 h. Cells were then collected for luciferase activity measurement, which was used as a surrogate measure for MnSOD gene transcription. Data were normalized and expressed in fold change relative to empty vector transfected control (B).
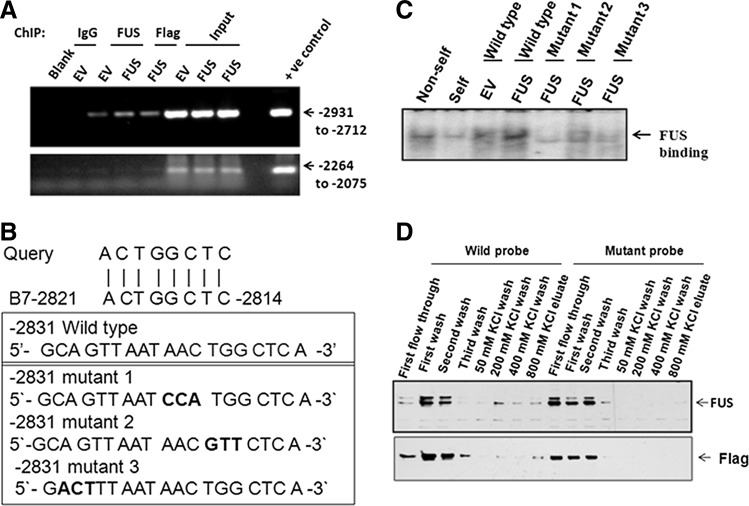
To identify the putative FUS binding site on the MnSOD promoter, we first performed chromatin immunoprecipitation (ChIP) assay using FUS or Flag antibody with the nuclear extracts prepared from cells with or without FUS overexpression. For this purpose, we cross-linked DNA protein complexes, which were subjected to ChIP using FUS or Flag antibody, as well as nonimmune control antibody. As shown in Figure 6A, ChIP with FUS or Flag antibody and subsequent PCR amplification detected a 220-bp MnSOD promoter fragment (−2931 to −2712), and FUS overexpression resulted in a higher level of PCR product compared to empty vector transfection control. The ChIP assay performed using the nonimmune antibody was unable to pull down the promoter fragment (Fig. 6A). Although reporter activity data suggest that FUS may bind anywhere between −2947 to −1605, PCR amplification of −2264 to −2075 fragment showing no bands after ChIP assay suggests that FUS binds between −2931 to −2712 in the MnSOD promoter (Fig. 6A).
FIG. 6.
FUS binds with MnSOD promoter. The binding of FUS to the promoter of MnSOD gene was evaluated by chromatin immunoprecipitation (ChIP) assay as described under “Experimental Procedures.” Briefly, chromatins were immunoprecipitated by FUS antibody or immunoglobulin (IgG) as control and immunoprecipitated DNA was amplified by PCR using primers targeted to the promoter region (−2931 to −2712 and −2264 to −2075) of MnSOD gene. Total genomic DNA obtained after transfection was parallel-amplified by using the same primer as input control (A). Predicted putative FUS binding element in MnSOD promoter and three mutated oligonucleotides were designed to use as probe (B). Oligonucleotides were radioactively labeled with [32P]ATP and T4 polynucleotide kinase as described previously (23). Electrophoretic mobility shift assay (EMSA) was carried out to separate DNA-protein complexes on 6% polyacrylamide native gel. The arrow points to protein-DNA complexes (C). The biotinylated oligonucleotides of wild-type and mutant type probes were used for DNA protein interaction by affinity chromatography followed by Western blotting using anti-FUS or anti-Flag antibody (D).
FUS is known to have a potential DNA binding site with a nucleotide sequence of ACTGGCTC (2). A homology search identified this specific nucleotide sequence at the far upstream region of the MnSOD promoter (Fig. 6B). To verify FUS binding to this nucleotide sequence, we made a wild-type probe with this nucleotide sequence and three mutant variants (Fig. 6B) for electrophoretic mobility shift assay (EMSA) using nuclear extracts prepared from cells with or without FUS transfection. The results demonstrate that FUS protein binds to the double stranded DNA of the wild-type probes even in the presence of a 100-fold excess of nonspecific DNA (non-self-lane in Fig. 6C), and binding is suppressed in the presence of self-competitor or cold oligonucleotide (self-lane in Fig. 6C). Transfection of FUS increased the DNA binding of the nuclear extract as compared to empty vector transfection control. All three mutations in this oligonucleotide sequence significantly suppressed FUS protein binding (Fig. 6C). These results confirm that FUS specifically binds with double stranded DNA in the MnSOD promoter with the nucleotide sequence of ACTGGCTC and the alterations in this nucleotide sequence significantly reduce FUS binding to the MnSOD promoter.
To confirm the identity of FUS protein bound to DNA, the wild-type or mutant-1 double stranded DNA probes as described in Figure 6B were labeled with biotin and immobilized on streptavidine columns. Nuclear extract prepared from cells overexpressing FUS was subjected to DNA affinity chromatography to purify the FUS protein. The FUS protein that bound to the biotinylated DNA probes was eluted and subjected to SDS-PAGE electrophoresis followed by Western blot analysis using FUS or Flag antibody. The results show that FUS protein was eluted from the column with biotinylated wild-type DNA probes but was not eluted from similarly prepared mutant DNA probes (Fig. 6D, Supplementary Fig. S2).
To verify the interaction of FUS with DNA, the interaction between FUS and the wild-type or mutant double stranded DNA probes was further studied by surface plasmon resonance (SPR) analysis. Affinity purified FUS protein bound to the wild-type probe with very high affinity, whereas binding to the modified probe was remarkably lower compared to the binding with the wild-type probe (Table 2). The detailed binding kinetics of the affinity-purified FUS to wild-type or mutant probe is shown in Supplementary Fig. S3.
Table 2.
Kinetics of Purified FUsed in Sarcoma Binding to MnSOD Promoter
| Type of ligand | Ka | Kd | KD | Chi2 |
|---|---|---|---|---|
| (1/ms) | (1/s) | (M) | RU | |
| Wild type probe | 7.11E+03 | 7.70E-04 | 1.08E-07 | 11.82 |
| Mutant probe | 2.41E+00 | 8.12E-04 | 3.37E-04 | 10.65 |
The biotinylated wild type or mutant type oligonucleotides were used for DNA protein interaction by Surface Plasmon Resonance analysis.
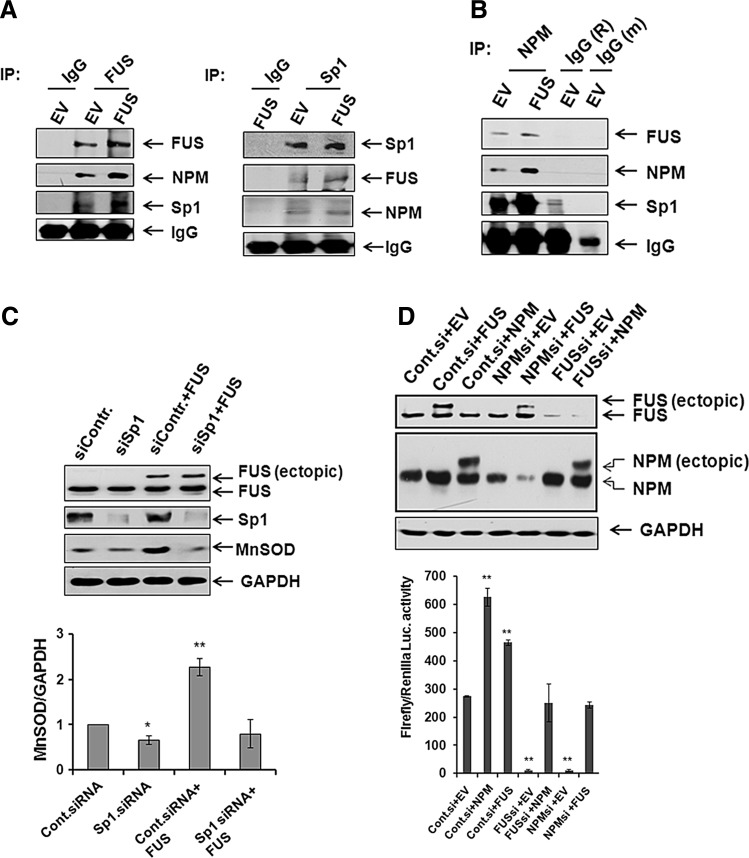
FUS mediates MnSOD expression via complex formation with Sp1 and NPM
We previously identified NPM as an interaction partner of Sp1 in the induction of MnSOD gene transcription (15). In this study, using a proteomic approach we identified FUS as a component of the immunocomplex with NPM and also identified the putative binding site of FUS on the MnSOD gene. Therefore, we tested the hypothesis that FUS is a component of the transcription complex regulating the expression of the MnSOD gene. Our immunoprecipitation data show that FUS antibody was able to immunoprecipitate Sp1 and NPM; reverse immunoprecipitation shows that Sp1 antibody was able to immunoprecipitate FUS and NPM (Fig. 7A), suggesting that FUS interacts with Sp1 and NPM. Reverse immunoprecipitation with the NPM antibody also shows that FUS, NPM and Sp1 were all present in the complex and further overexpression of FUS increased transcription complex formation (Fig. 7B). To determine whether FUS-mediated transcription of the MnSOD gene requires Sp1, we suppressed Sp1 expression with Sp1 siRNA and overexpressed FUS by transfecting the FUS expression vector. Transfection of Sp1 siRNA suppressed Sp1 protein and MnSOD protein expression (Fig. 7C). Cotransfection of FUS with control siRNA increased MnSOD protein level, whereas coexpression of FUS with Sp1 siRNA blocked MnSOD expression (Fig. 7C). These results demonstrate that FUS forms a complex with Sp1 and NPM and that Sp1 is required for the FUS-mediated induction of MnSOD expression. Similar experimental approaches further demonstrate that coexpression of FUS and NPM siRNA suppresses FUS mediated MnSOD gene transcription. Consistently, coexpression of NPM and FUS siRNA also attenuated NPM mediated MnSOD gene transcription (Fig. 7D). These results suggest that FUS, Sp1 and NPM form a functional complex for MnSOD gene transcription.
FIG. 7.
Association of FUS with NPM and specificity protein 1 (Sp1). In vivo association of FUS with NPM and Sp1 in isolated nuclear extract was detected by coimmunoprecipitation using FUS or Sp1 antibody. Coimmunoprecipitation by anti-rabbit IgG from nuclear extract was used as negative control (A). Reverse immunoprecipitation was also carried out using NPM antibody, and FUS and Sp1 were detected in the immunocomplex by Western blotting (B). Cells were transfected with Sp1 siRNA or control siRNA with or without FUS expression vector for 48 h. After the transfection, cell lysates were prepared and subjected to SDS-polyacrylamide gel electrophoresis (C). FUS, Sp1, and MnSOD proteins were detected by Western blotting using antibodies specific to FUS, Sp1, and MnSOD, respectively. GAPDH was used as loading control (top panel). MnSOD protein bands were densitometrically scanned and quantified using GAPDH as loading control (bottom panel). MnSOD promoter driven luciferase activity was measured after transfection of combined NPM siRNA and FUS expression vector or a combination of FUS siRNA and NPM expression vector in JB6 cells (D, bottom panel). The suppression of proteins after transfection of corresponding siRNA and overexpression of proteins after transfection of corresponding expression vector were confirmed by Western blotting (D, upper panel). Each data point represents the mean±SD of three independent experiments. Significant difference as compared with corresponding control is indicated by *p<0.05 and **p<0.01.
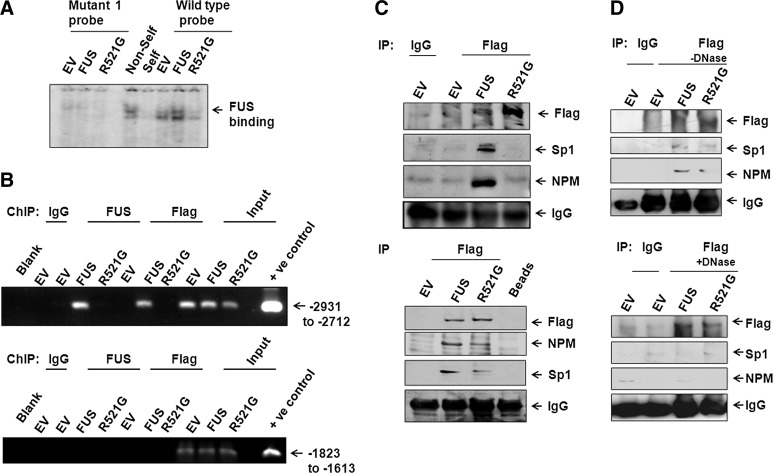
To determine whether the inability of mutated FUS to induce MnSOD transcription is due to loss in binding to the MnSOD promoter, we performed EMSA using nuclear extracts from cells transfected with wild-type FUS, mutant FUS, or empty vector. The nuclear extract from cells transfected with wild-type FUS displayed strong binding with the wild-type probe of the MnSOD promoter, whereas the binding activity of the nuclear extracts from cells transfected with mutated FUS was not different from that of the cells transfected with the empty vector. Mutations in the consensus nucleotide sequence in the MnSOD promoter for FUS binding abrogated the binding by the nuclear extracts prepared from cells transfected with either wild-type or mutated FUS (Fig. 8A). These data support the notion that FUS binds to a specific DNA sequence in the MnSOD promoter and that mutations in either the DNA sequence that is recognized by FUS, or the consensus coding sequence of the FUS gene, disrupt the binding of the FUS protein to its target DNA sequence in the MnSOD gene promoter. We also performed ChIP assay coupled with PCR amplification of the FUS binding region contained in the MnSOD promoter using nuclear extracts from cells overexpressing wild-type or mutant FUS. PCR amplification of a 220-bp promoter fragment (nucleotides −2931 to −2712) was achieved with transcription complexes that immunoprecipitated with FUS or Flag antibody from the FUS transfected cells but not with the transcription complex immunoprecipitated from cells transfected with mutant FUS (Fig. 8B, upper panel). PCR amplification of ChIP DNA products was not detected with primers encompassing nucleotides −1823 to −1613 within the MnSOD promoter (Fig. 8B, lower panel). Importantly, these results indicate that wild-type FUS specifically binds to the MnSOD promoter region and that mutation in the FUS gene abolishes its ability to interact with the MnSOD promoter region. To probe whether the inability of mutant FUS to bind to MnSOD promoter is due to the unavailability of the protein in the nucleus, we performed cell fractionation followed by Western blotting. Our results show that mutant FUS is located in the nucleus after overexpression (Supplementary Fig. S4). These results further demonstrate that the inability of mutant FUS to interact with the MnSOD promoter is not due to its absence in the nucleus.
FIG. 8.
Mutation in FUS abrogates its activity for MnSOD promoter binding and protein-protein interaction. EMSA was carried out with nuclear extracts from cells transfected with wild-type or mutant FUS using 32p labeled oligo-nucleotides as shown in Figure 6B as probe. The arrow points to protein-DNA complexes complexes (A). For ChIP assay, using FUS or Flag antibody or IgG as control and interacting DNAs were immunoprecipitated and amplified by PCR using primers targeted to the promoter region (−2931 to −2712) of MnSOD gene. Total genomic DNA obtained after transfection was parallel-amplified by using the same primer as input control (B). The specificity of FUS binding to the MnSOD promoter was verified by amplifying the same DNAs with the primer set selected from the untargeted region of MnSOD promoter (−1823 to −1613) (B). Coimmunoprecipitation experiment was performed using nuclear extract of cells transfected with wild-type or mutant FUS expression vector using Flag antibody (C). Flag, Sp1, and NPM was detected by Western blotting analysis using antibodies specific for these proteins and IgG was used as negative control. Equal sample loading was confirmed by probing the same membrane with anti-IgG (C, top panel). FUS interacting proteins were also affinity purified from the nuclear extracts of cells transfected with wild-type or mutant-type FUS expression vector or empty vector using anti-Flag affinity column. Flag binding proteins were eluted with 0.5 M glycine solution, analyzed by SDS-PAGE, and detected by Western blotting (C, bottom panel). Coimmunoprecipitation was performed with cell lysates incubated in the absence (D, top panel) or presence (D, bottom panel) of DNase for 1 h at 37°C.
Interaction of FUS with other transcription factors is DNA dependent
Although we established the interaction of FUS with NPM and Sp1 (Fig. 7A, B), the interaction of mutated FUS with these transcription factors remained to be determined. To address this question, we immunoprecipitated the transcription factors from both wild-type and mutant FUS overexpressing cells using the Flag antibody followed by Western blotting. The results show that the Flag antibody could immunoprecipitate Sp1 and NPM from cells overexpressing the wild-type FUS but not from cells overexpressing the mutant FUS. We confirmed that the Flag antibody immunoprecipitated Flag-tagged FUS protein from cells overexpressing either the wild-type or mutant FUS (Fig. 8C, upper panel). Nonimmune IgG, which was used as a control, did not immunoprecipitate any of the transcription factors. These results were further verified by an alternative method, in which the Flag antibody was impregnated in sepharose beads, packed in column, and used to pull down transcription factors from nuclear extracts obtained from cells overexpressing wild-type or mutant FUS. The Western blotting results acquired by using this method are shown in the lower panel of Figure 8C. Consistent with the earlier results, the Flag antibody was able to immunoprecipitate Flag-tagged FUS, SP1 and NPM from cells overexpressing wild-type FUS. In contrast, smaller amounts of Sp1 and NPM were immunoprecipitated with Flag-FUS from cells overexpressing the mutant FUS. The Flag antibody did not immunoprecipitate these transcription factors from the nuclear extracts of cells transfected with empty vector control (Fig. 8C, lower panel).
To determine whether FUS binding to DNA is required for FUS interaction with NPM and Sp1, we treated the cell lysates with DNase in vitro and performed immunoprecipitation. We confirmed that DNase treatment degraded DNA under the experimental conditions (Supplementary Fig. S5). Without DNase treatment, the Flag antibody was able to pull down Sp1 and NPM from cells overexpressing wild-type FUS (Fig. 8D, upper panel). After DNase treatment, the Flag antibody did not pull down either Sp1 or NPM, although Flag-tagged FUS was immunoprecipitated from cells overexpressing either the wild-type or the mutant FUS. Immunoprecipitation with nonimmune IgG did not show any band corresponding to Flag, Sp1, or NPM (Fig. 8D, lower panel). These results suggest that FUS-binding to DNA is required for FUS to interact with Sp1 and NPM and that the mutation in FUS significantly weakens such interactions. These findings support the conclusion that binding to MnSOD promoter DNA is required for FUS to interact with important transcription factors and enhance MnSOD gene transcription.
FUS-mediated induction of MnSOD protects cells from toxicity
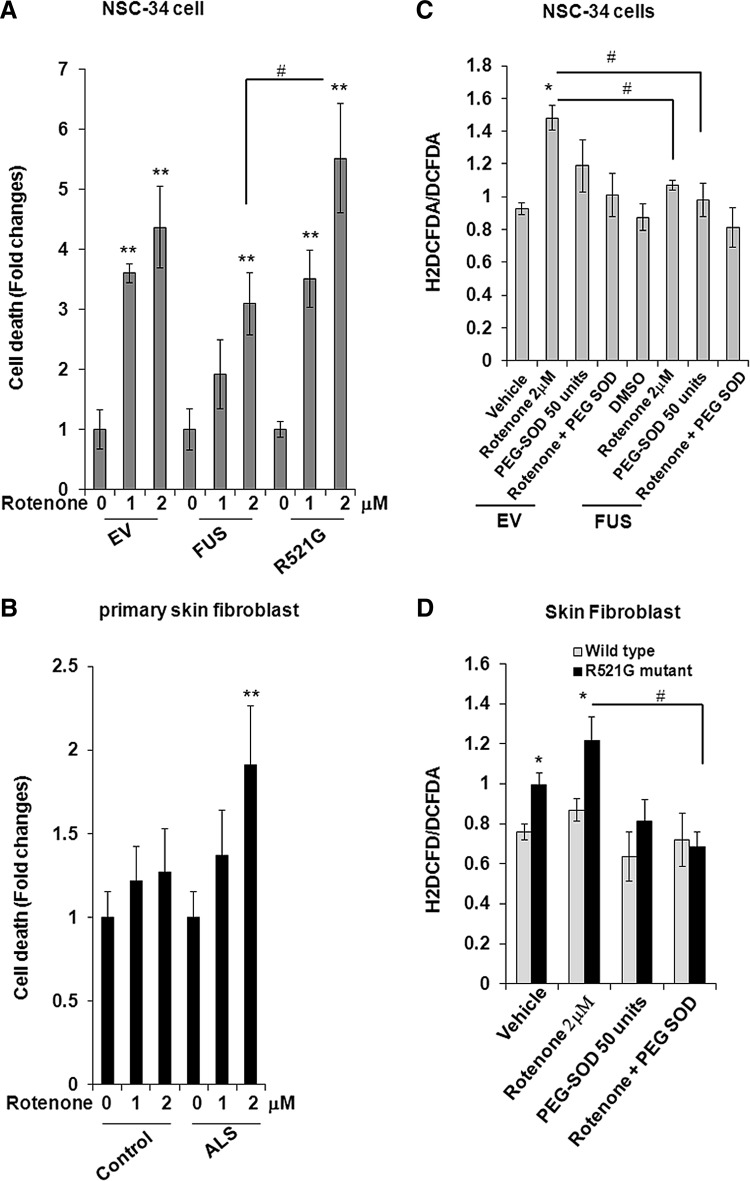
To evaluate the significance of FUS-mediated MnSOD induction, motor neuron-like NSC-34 cells were transfected with either wild-type FUS or mutant FUS expression vectors. The biological significance of FUS-mediated MnSOD expression was determined by performing cytotoxicity assay after treatment with the cytotoxic drug rotenone that targets mitochondria. We used Trypan blue exclusion assay to determine the rotenone-mediated toxicity in cells transfected with the wild-type or mutant FUS. Treatment with rotenone significantly increased cell death in a dose-dependent manner which is markedly protected by overexpression of FUS. Overexpression of mutant FUS in NSC-34 cells exhibited a higher level of cell death after rotenone treatment (Fig. 9A). These findings were further verified in primary skin fibroblasts after treatment with rotenone. Consistently, skin fibroblast from fALS patients harboring mutant FUS showed significant and dose-dependent increase in cell death after exposure to rotenone (Fig. 9B). These results suggest that wild-type but not mutant FUS provides significant protection against the cytotoxic effect of rotenone (Fig. 9A, B).
FIG. 9.
Overexpression of FUS protects against rotenone induced cytotoxicity. NSC-34 cells were transfected with wild-type or mutant FUS expression vector followed by treatment with rotenone for 24 h. Trypan blue exclusion assay was performed to detect the dead cells (A). Cultured primary skin fibroblast cells that were obtained from fALS patients or from normal subjects were treated with the indicated concentration of rotenone and the Trypan blue exclusion assay was performed 24 h after treatment (B). Both live and dead cells were counted, and the ratio of dead to total cells was calculated and expressed as fold changes. H2DCFDA and DCFDA fluorescence was measured in NEC-34 cells (C) and skin fibroblast (D) after treatment with rotenone. Cells were also treated with polyethylene glycol-SOD 1 h before rotenone treatment. Each data point represents mean±SD of three to four samples and significant difference compared to respective control is indicated by *p<0.05 and **p<0.01; different from treatment #p<0.05.
To further investigate whether FUS-mediated protection of cell death is related to mitochondrial phenomena, we measured the DCFDA and H2DCFDA in NSC-34 cells after overexpression of FUS combined with rotenone treatment. Treatment of rotenone significantly increased the H2DCFDA/DCFDA ratio which is attenuated by FUS overexpression and/or polyethylene glycol (PEG)-SOD pretreatment (Fig. 9C). Similarly, primary skin fibroblasts from fALS patients showed a higher level of H2DCFDA/DCFDA ratio compared to normal control fibroblasts. Pretreatment of PEG-SOD suppressed the rotenone induced oxidative stress in mutant FUS harboring skin fibroblasts (Fig. 9D). Additionally, we measured the reduced and oxidized glutathione (GSSG) in NSC-34 cells after overexpression of the wild-type or mutant FUS expression vector. Consistent with DCFDA data, overexpression of wild-type FUS led to the reduction of GSSG and an increase in GSH:GSSG ratio (Supplementary Fig. 6B). Consistently, GSSG levels were higher in primary fibroblasts from fALS patients compared to normal counterparts. GSH levels remained unchanged in fALS patients compared to controls resulting in decreased GSH:GSSG ratio (Supplementary Fig. 6C).
Discussion
MnSOD is the primary antioxidant enzyme that removes superoxide radicals in mitochondria. The expression of MnSOD is essential for the survival of all aerobic organisms; thus, elucidating the mechanisms of MnSOD regulation is very important for understanding the well-being of normal cells and tissues, especially such tissues as motor neurons that have a high energy demand. Reactive oxygen species (ROS) play important roles in maintaining the prooxidant and antioxidant states of cells. The cellular stress response is associated with the activation of prosurvival pathways that are under the control of protective genes called vitagenes (reviewed in 6). MnSOD may participate as a vitagene to protect the cells via the dismutation of superoxide radicals. In addition, dismutation of superoxide to hydrogen peroxide after enzymatic reaction of MnSOD can also lead to the expression of other vitagenes, such as thioredoxin further contributes to protection against cellular damage (7). In this context, the FUS mediated induction of MnSOD may be considered a component of the vitagenes network. The association of the FUS mutation with MnSOD expression that results in a compromised level of MnSOD activity in fALS disease may affect the vitagenes network, leading to enhanced cellular damage.
The current report identifies and characterizes the FUS-responsive element in the promoter of the MnSOD gene with an emphasis on the role of FUS in the transcriptional regulation of MnSOD gene expression. FUS mutations have recently been reported in Type 6 fALS (52). We have demonstrated that skin fibroblasts derived from fALS patients carrying the R521G mutation in FUS had significantly decreased levels of MnSOD, suggesting that MnSOD and mitochondria may play an important role in the pathogenesis of ALS in this subset of patients with the R521G FUS mutation.
Transcriptional activation of the MnSOD gene has been shown to be dependent on several transcription factors, including Sp1 and NF-κB (29, 47). Previously, we identified the minimal promoter in the MnSOD gene, which is located at 555-bp upstream from the start site that binds Sp1 and Ap2 and is required for its basal transcription function (58, 61). Mutagenesis study of this region demonstrated that Sp1 is essential for the transcription of the MnSOD gene (58). The MnSOD gene also contains an enhancer element located in the second intron responsible for NF-κB-mediated gene transcription (26, 57). Overexpression of FUS resulted in a several-fold increase in the luciferase activity driven by the MnSOD promoter; however, the presence of an enhancer in the reporter gene construct did not play an additional role in FUS-mediated MnSOD gene transcription, suggesting that FUS-mediated MnSOD transactivation is not dependent on NF-κB sites in the second intron. Using a variety of cell lines, Fuji and Taniguchi (19) reported that MnSOD gene transcription can be activated in a promoter-dependent manner in NF-κB unresponsive cells. This observation is consistent with our finding that the intronic element is not an essential factor for the induction of the MnSOD gene by FUS. Our observation that FUS-mediated MnSOD induction occurred only when we used a long MnSOD promoter (construct [−3401 to +24] or [−2947 to +24], Figure 5B) suggests that the FUS-responsive element is located in the far 5′-upstream region of the MnSOD gene.
It has been demonstrated that ACTGGCTC (reverse sequence) in the form of double stranded DNA in the macrophage specific cerebrospinal fluid receptor-1 gene can bind with FUS protein (25). Blast homology search analysis identified that the GGCTC sequence was present in MnSOD promoter at about 2.8-kb upstream from the start site. Our experimental results establish that the FUS-responsive element in the promoter region between (−2947 to 24) (Fig. 5B) is functional. Consistent with this finding, in vivo ChIP assay data confirm that FUS interacts with the MnSOD promoter at the upstream region (Fig. 6A). Although we observed that FUS DNA binding was increased by overexpressing FUS, as determined by EMSA, we could not supershift the binding complex with the FUS antibody. Therefore, we took an alternative approach: we used the affinity purified FUS with double stranded DNA probes containing wild-type or mutant oligonucleotide sequences of the FUS response element, and established the binding of FUS to double stranded DNA. It has been reported that the RRM, together with a glycine rich region and zinc finger motifs, are critical for RNA and DNA binding (4). Our findings that FUS binds with DNA and that a fALS mutation (R521G) in the C-terminal region disrupts DNA binding suggest that the C-terminal region of FUS is also crucial for its binding to DNA. The binding of FUS to the promoter region has been implicated in the induction of gene transcription (11, 48). FUS also functions as a gene transcription modulator by binding to a number of gene specific transcription factors. The glycine-rich region and zinc finger motifs of FUS are capable of binding with transcription factors, such as NF-κB and transcription factor II D complex as a component of RNA polymerase II preinitiation complex (32), thereby participating in transcription. However, that FUS functions as a specific modulator of MnSOD gene expression was previously unknown. Our coimmunoprecipitation results indicate that full-length FUS interacts with Sp1 and NPM (Figs. 7 and 8). In addition, our finding that FUS physically interacts with Sp1 and NPM is consistent with FUS-mediated regulation of MnSOD basal transcription.
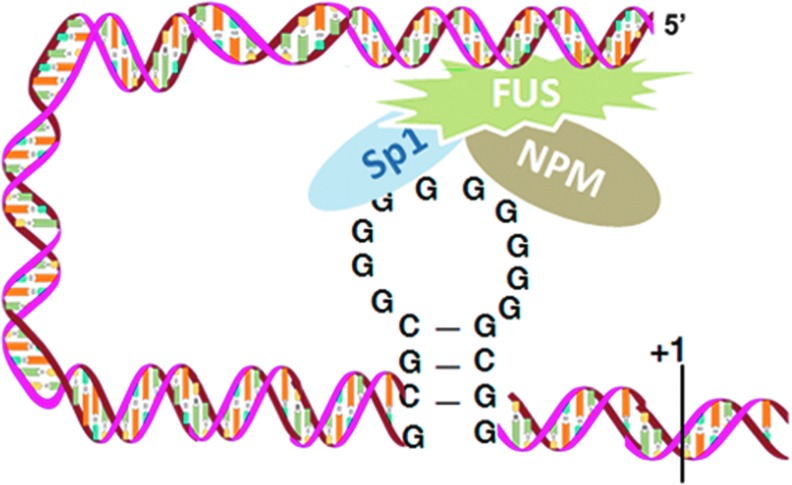
Interestingly, FUS interacts with both single and double stranded DNA (47). FUS promotes annealing of both single and double stranded DNA (possibly by binding with DNA) and facilitates D-loop formation (8). We and others have reported that the guanine cytosine (GC)-rich promoter of the MnSOD gene has a stem-loop structure containing Sp1 and NPM binding sites (18, 56). Since FUS is only responsive when it binds with the far upstream region of the MnSOD gene, FUS may link to the loop structure by interacting with Sp1 and NPM, which bind at the GC-rich region in the stem-loop structure. Together, our results support the notion that FUS induces MnSOD gene transcription by binding directly to DNA at (−2821 to −2814 bp) and indirectly in the stem-loop GC-rich structure via Sp1, which together are essential to maintain the basal level of MnSOD transcription (Fig. 10). Treatment with DNase indicates that the interaction of FUS with Sp1 or NPM is significantly diminished by the removal of DNA from the transcription complex, suggesting that DNA binding of FUS is required for the interaction of these transcription factors to increase MnSOD gene transcription. It has been demonstrated that many sequence-specific transcription factors bind to DNA and interact with each other. For instance, Sp1 and NF-κB bind with the HIV-1 promoter in a sequence-specific manner and interact with each other, leading to a cooperative induction of gene transcription (45). Recently, we found that both Sp1 and p53 bind to the MnSOD promoter, which negatively modulates MnSOD gene transcription (14, 17). Mutant FUS is unable to bind to DNA in the MnSOD promoter or interact with Sp1 and NPM, again suggesting that DNA binding and protein interaction are essential for FUS-mediated gene transcription.
FIG. 10.
The schematic depiction of FUS-mediated MnSOD gene transcription. FUS binds to a FUS binding element identified in the upstream MnSOD promoter and interacts with transcription factor Sp1 and nucleophosmin. This interaction leads to activation of MnSOD gene transcription. To see this illustration in color, the reader is referred to the web version of this article at www.liebertpub.com/ars
Increased oxidative stress has been shown to cause neuronal damage by gain-of-function mutations in Cu/Zn superoxide dismutase (SOD1) in Type 1 fALS (22). However, the role of MnSOD in fALS pathogenesis remains elusive. MnSOD is expressed in almost all mammalian cells, including neurons, and can be induced under various conditions (10, 16, 37, 50). We recently demonstrated that the molecular pathology of FUS mutation can be recapitulated in cell culture (20) and Drosophila (55) models. In the present study, we used JB6 mouse skin epithelial cells and NSC-34 motor neuron-like cells as cell culture systems to elucidate the underlying mechanisms for FUS-mediated MnSOD transcriptional regulation, which resulted in the first mechanistic insight into oxidative stress in mitochondria as a link to FUS mutation in fALS.
TDP-43 and FUS have been identified as being present in families with fALS. This information has led to increasing support for the idea that changes in alternative splicing are critical targets for ALS and other motor neuron diseases. Recent studies demonstrate that FUS binds to Survival Motor Neuron (SMN, which is absent in the infantile motor neuron disease, spinal muscular atropy) and to U1 in the small nuclear ribonuclear protein (RNP) (snRNP) pre-mRNA Spliceosome (21, 59). It appears that TDP-43 acts upstream and facilitates the assembly of the SMN complex. FUS protein binds to U1 and SMN in the snRNP complex which forms the pre-mRNA Spliceosome. FUS protein with ALS relevant mutations still bind to snRNP but divert the complex from the nucleus to the cytoplasm where it is trapped, resulting in functional changes in splice site recognition and exon skipping (21). Exon skipping of exon 7 in the SMN2 gene was observed after mitochondrial dysfunction, oxidative stress, and adenosine triphosphate depletion induced by paraquat exposure of human neuroblastoma cells (36). It is well-established that MnSOD is critical in modulating oxidative stress and maintaining proper mitochondrial function; thus, suppression of MnSOD is likely to augment the deleterious diversion of snRNP to the cytoplasm in disease-causing FUS mutations. Thus, FUS mutations may act negatively on three parallel, closely linked pathways involved in RNA metabolism by diverting FUS/snRNP complexes from the nucleus to the cytoplasm, by altering the binding of FUS to highly conserved introns in genes coding from RNA-binding proteins (43,49), and by reducing MnSOD gene transcription with downstream effects of increased cellular oxidative stress.
Because ALS is a disease associated with the impairment of motor neuron function and FUS mutations are associated with some fALS families, our experiments reveal a potential mechanism of mitochondrial dysfunction in FUS-mediated fALS involving chronically reduced basal levels of MnSOD. In contrast, however, other investigators have demonstrated that MnSOD levels are elevated in a sporadic ALS patient cell line that includes motor neurons, but those studies did not take into account the information about a specific FUS mutation (35, 40). It is possible that the elevated level of MnSOD observed in some sporadic ALS implies compensatory response due to high oxidative stress. We speculate that the chronically reduced basal levels of MnSOD would likely result in slow, cumulative oxidative damage to mitochondria eventuating in progressive dysfunction and eventual death of cortical and spinal motor neurons. This might be especially important in metabolically active neurons with extensive dendritic arbors and long axons. Dendrites and axons demand high metabolic rates and active long distance transport, which may render them particularly susceptible to oxidative stress. The insights from our work imply that mutant FUS-mediated Type 6 fALS might be amenable to such intervention measures as MnSOD mimetic drugs that reduce chronic oxidative stress in mitochondria.
Materials and Methods
Cell culture
The JB6 mouse skin epithelial cell line was originally provided by Dr. Nancy H. Colburn of the National Cancer Institute, Frederick, MD, and was maintained according to a previously described method (17). NSC-34 cells were provided by Dr. Haining Zhu of the University of Kentucky and were maintained according to published protocol (20).
Clinical material
Punch skin biopsies (3 mm) were obtained after informed consent from three males with symptomatic ALS who originated from an extended regional family with a documented R521G FUS mutation (Athena Diagnostics). The presence of R521G was confirmed by PCR amplification and direct sequencing of genomic DNA isolated from genomic DNA of skin fibroblast obtained from each individual. Two subjects are related as uncle/nephew and they are distant cousins of the third, all descendants of a common ancestor who died before 1869. Relationships were established by family history and were confirmed using public genealogy records (Ancestors.com). The mean age at onset was 49 years (36, 54, and 57 years). Two had limb-onset disease and one presented with respiratory failure requiring intubation. Clinically, subjects exhibited findings indicative of motor neuron degeneration in the cortex and spinal cord, which was confirmed on examination and electromyography (EMG) testing. Mean survival from onset of progressive weakness to death was 17 months (12, 14, and 25 months). Controls were four females and one male who were free of neurological disease (mean age 40.4 years; range 22–64 years). The study was approved by the Institutional Review Board of the University of Kentucky.
SOD activity assay using native gel electrophoresis
Human skin fibroblasts were collected, and then sonicated on ice for 30 s. Protein concentrations were estimated by the Bradford protein assay. One hundred micrograms of the total protein were loaded onto each lane of the gel, and the SOD activity was visualized by the nitroblue tetrazolium method (3).
Promoter constructs, transfection and luciferase assay
The original promoter and enhancer-reporter constructs were prepared in our laboratory as described previously (57) and transfected in cells after a lipofectamin® transfection protocol. Luciferase assay was performed by cotransfecting the cells with FUS expression vector and vector containing the enhancer (I2E) and promoter (−3400/+24) of the human MnSOD gene in pGL3 reporter vector. β-Galactosidase cDNA containing luciferase vector was used as an internal control. Forty-eight hours after transfection, cells were lysed and the samples were analyzed with the luciferase reporter assay system.
Mass spectrometry
All mass spectra reported in this study were acquired by the University of Kentucky Mass Spectrometry Facility as described previously (12). Briefly, the immunoprecipitated products were digested by trypsin (sequencing grade, Promega). The resulting peptides were extracted and LC-MS/MS spectra were acquired on a Finnigan LCQ “Classic” quadruple ion trap mass spectrometer (Finnigan C.). The data were submitted to an in-house MASCOT search engine for protein identification searching against the NCBI nonredundant protein database. All reported proteins showed multiple unique peptides with a MOWSE score >50.
RNA isolation, cDNA synthesis and reverse transcription PCR
Total RNA was isolated by using TRIzol reagent (Invitrogen) according to the manufacturer's protocol. The cDNA was generated using 0.4 μg of total RNA, oligo(dT) primer according to the manufacturer's instructions (Invitrogen), in a total volume of 20 μl. Two microliters of cDNA were amplified using the primer sets for MnSOD and GAPDH (primer sequence is available upon request). PCR products were separated on agarose gel and visualized by ethidium bromide.
Immunoprecipitation
Immunoprecipitation studies were carried out using nuclear extract and antibodies or normal IgG. Immunocomplexes were separated by protein A/G after centrifugation at 2500 rpm for 5 min. Immunocomplexes were eluted in the sample loading buffer, subjected to SDS-PAGE, and analyzed by Western blotting.
ChIP assays
ChIP assay was performed as described previously (17). Briefly, wild-type or mutant FUS overexpressed cells were cross-linked by formaldehyde and sonicated to obtain 200 to 1000 bp DNA fragments. The soluble chromatin fraction was precleaned with 20 μl of salmon sperm DNA/protein A-agarose beads and subjected to immunoprecipitation with the appropriate antibodies or normal IgG. DNA-immunocomplexes were eluted and reversal of cross-linking was performed to isolate DNA for PCR amplification.
Surface plasmon resonance
The interactions between biotinylated oligonucleotide (ligand) and purified FUS protein (analyte) were measured by SPR technique using ProteOn™ XPR36 instrument (Bio-Rad). The biotinylated double-stranded oligonucleotide was immobilized on sensor chips, which are precoated with streptavidine (Bio-Rad). Ligand capturing on the biosensor surface was performed according to manufacturer's instructions. Several different concentrations of analytes (pure FUS protein) were used to evaluate the dose- dependent binding. Data were acquired using ProteOn manager software, and the kinetic analysis was performed using the langumir 1:1 evaluation model.
High-performance liquid chromatography assay of GSH and GSSG
Cellular levels of reduced GSH and GSSG were measured from cell lysates by the quantitative High-performance liquid chromatography (HPLC) method as described previously (27). Briefly, cells were homogenized and sonicated, protein concentration was determined, and then an equal amount of protein (40 μg for GSH and 100 μg for GSSG) was used for GSH derivatization. Five percent trichloroacetic acid, 7.5 mmol/l of N-ethylmaleimide, and 100 mmol/l of dithiotheitol, in redox quenching buffer (5 mmol/l of diethylenetriaminepentaacetic acid, and 10 mmol/l of ascorbic acid, pH 6.8), were added to the sample. The GSH samples were incubated at 45°C for 15 min after addition of 50 mmol/l monobromobimane (MBB). For GSSG, samples were incubated at 45°C for 15 min before addition of MBB, and incubated at 45°C for 15 min after addition of MBB. The reaction was then stopped by adding 6 N HCl followed by centrifugation at 5000g for 5 min at 4°C. The supernatant was assayed for thiol–bimane fluorescence by reverse-phase HPLC using a linear gradient from 0 to 100% solvent B (50% methanol, 0.25% acetic acid in water) in solvent A (10% methanol, 0.25% acetic acid in water) over 28 min at a flow rate of 0.8 ml/min with fluorescence detection at Ex370/Em485 using the Waters 2475 Multi (lambda) fluorescence detector. Fluorescence intensities versus time of elution were quantified using Waters Breeze chromatography software v. 3.2 (Waters Corporation) and peak areas were integrated and converted to nmol GSH equivalents from the integrated areas under the GSH standard curve.
DCFDA fluorescence assay
DCFDA fluorescence assay was performed using oxidation sensitive carboxy-H2DCFDA (Invitrogen) and oxidation insensitive oxidized carboxy-DCFDA (Invitrogen) as described previously (51). To control the discrepancies among the number of cells seeded, dye uptakes between samples were taken into consideration by preloading oxidized-DCFDA to the samples. Briefly, equal numbers of cells were seeded in 48 well plates and grown for 24 h; after the necessary treatment the cells were then incubated with either H2DCFDA or DCFDA for 30 min, followed by washing twice with 1×PBS. Cellular fluorescence was then detected at a wavelength of Excitation 530 and Emission 580 using SpectraMax GEMNI fluorometer (Molecular Device). Fluorescence in cells preloaded with carboxy-H2DCFDA was normalized to that in cells preloaded with carboxy-DCFDA and data were presented as a H2DCFDA/DCFDA ratio.
Statistical analysis
Data were analyzed using student t-test for two group comparisons and one-way Analysis of Variance for multiple group comparison. Bonferroni's post-test multiple comparisons procedure was used to determine the statistical significance. Data shown represent mean±SD.
Supplementary Material
Abbreviations Used
- ALS
amytropic lateral sclerosis
- ATP
adenosine triphosphate
- ChIP
chromatin immunoprecipitation
- CHOP
C/EBP homologous protein
- CSF
cerebrospinal fluid
- EMG
electromyography
- EMSA
electrophoretic mobility shift assay
- EWS
Ewing's sarcoma
- fALS
familial amyotropic lateral sclerosis
- FET
FUS Ewing's sarcoma
- FUS
fused in sarcoma
- GC
guanine cytosine
- GSH
glutathione
- GSSG
oxidized glutathione
- IgG
immunoglobulin
- LCQ
quadrupole ion trap liquid chromatography
- MBB
monobromobimane
- MEM
modified Eagle's medium
- MnSOD
manganese super oxide dismutase
- NBT
nitro-blue-tetrazolium
- NF-κB
nuclear factor kappa B
- NPM
nucleophosmin
- PEG
polyethylene glycol
- RNP
ribonuclear protein
- ROS
reactive oxygen species
- RRM
RNA recognition motif
- RT-PCR
reverse transcriptase polymerase chain reaction
- SDS-PAGE
sodium dodecyle sulphate-polyacrylamide gel electrophoresis
- SMA
spinal muscular atropy
- SMN
Survival Motor Neuron
- Sp1
specificity protein 1
- SPR
surface plasmon resonance
- TAFII
TATA-binding protein associated factor II
- TLS
translocated in liposarcoma
Acknowledgments
We thank Mr. Fred Odago, Graduate Center of Toxicology, University of Kentucky, for assisting in SPR data acquisition by Proteon®. This work was supported, in part, by National Institutes of Health Grants CA 49797 and CA 73599 to Daret K. St. Clair; and RO1NS077284 to Haining Zhu. This work is also supported by an ALS Association grant (6SE340) to Haining Zhu and a pilot grant from the University of Kentucky Center for Clinical and Translational Science (CCTS) that is funded by Center for Research and Resources (NCRR) grant UL1RR033173. Studies also were supported by the Cynthia Shaw Crispen and Kevin Heidrich/Team 7 endowments, the Edward P. Evans Foundation.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Anderson MK, Stahlberg A, Arvidsson Y, Olofsson A, Semb H, Stenman G, Nilsson O, and Aman P. The multifunctional FUS, EWS and TAF15 proto-oncoproteins show cell type-specific expression pattern and involvement in cell spreading and stress response. BMC Cell Biol 11: 37, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baechtold H, Kuroda M, Sok J, Ron D, Lopez BS, and Akhmedov AT. Human 75-kDa DNA-pairing protein is identical to the pro-oncoprotein TLS/FUS and is able to promote D-loop formation. J Biol Chem 274: 34337–34342, 1999 [DOI] [PubMed] [Google Scholar]
- 3.Beauchamp C. and Fridovich I. Superoxide dismutase improved assays and an assay applicable to polyacrylamide gel. Anal Biochem 44: 276–287, 1971 [DOI] [PubMed] [Google Scholar]
- 4.Bertolotti A, Lutz Y, Heard DJ, Chambon P, and Tora L. hTAF(II)68, a novel RNA/ssDNA-binding protein with homology to the pro-oncoproteins TLS/FUS and EWS, is associated with both TFIID and RNA polymerase II. EMBO J 15: 5022–5031, 1996 [PMC free article] [PubMed] [Google Scholar]
- 5.Bertrand P, Akhmedov AT, Delacote F, Durrbach A, and Lopez BS. Human POMp75 is identified as the pro-oncoprotein TLS/FUS: both POMp75 and POMp100 DNA homologous pairing activities are associated to cell proliferation. Oncogene 18: 4515–4521, 1999 [DOI] [PubMed] [Google Scholar]
- 6.Calabrese V, Cornelius C, Dinkova-Kostova AT, Calabrese EJ, and Mattson MP. Cellular stress responses, the hormesis paradigm and vitagenes: novel targets for therapeutic intervention in neurodegenerative disorders. Antioxid Redox Signal 13: 1763–1811, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Calabrese V, Cornelius C, Dinkova-Kostova AT, Iavicoli I, Paola RD, Koverech A, Cuzzocrea S, Rizzarelli E, and Calabrese EJ. Cellular stress responses, hermetic phytochemicals and vitagenes in aging and longevity. Biochem Biophys Acta 1822: 753–783, 2012 [DOI] [PubMed] [Google Scholar]
- 8.Carrier F, Gatignol A, Hollander MC, Jeang KT, and Fornace AJ, Jr., Induction of RNA-binding proteins in mammalian cells by DNA-damaging agents. Proc Natl Acad Sci U S A 91: 1544–1558, 1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cozzolino M, Persaresi MG, Gerbino V, Grosskreutz J, and Carri MT. Amyotropic lateral sclerosis: new insights into underlying molecular mechanisms and opportunities for therapeutic intervention. Antioxid Redox Signal 17: 1277–1330, 2012 [DOI] [PubMed] [Google Scholar]
- 10.Crosti N, Bajer J, Serra A, Rigo A, Scarpa M, and Viglino P. Co-ordinated expression of MnSOD and CuZnSOD in human fibroblast. Exp Cell Res 160: 396–402, 1985 [DOI] [PubMed] [Google Scholar]
- 11.Crozat A, Aman P, Mandahl N, and Ron D. Fusion of CHOP to a novel RNA-binding protein in human myxoid liposarcoma. Nature 363: 640–644, 1993 [DOI] [PubMed] [Google Scholar]
- 12.Dhar SK, Lynn BC, Daosukho C, and St. Clair DK. Identification of nucleophosmin as an NF-κB co-activator for the induction of human SOD2 gene. J Biol Chem 279: 28209–28219, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dhar SK. and St. Clair DK. Manganese superoxide dismutase regulation and cancer. Free Rad Biol Med 52: 2209–2222, 2012 [DOI] [PubMed] [Google Scholar]
- 14.Dhar SK, Tangpong J, Chaiswing L, Oberley TD, and St. Clair DK. Manganese superoxide is a p53-regulated gene that switches cancers between early to advanced stages. Cancer Res 71: 1–12, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dhar SK, Xu Y, Chen Y, and St. Clair DK. Specificity protein 1-dependent p53-mediated suppression of human manganese superoxide dismutase gene expression. J Biol Chem 281: 21698–21709, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dhar SK, Xu Y, Noel T, and St. Clair DK. Chronic exposure to 12-O-tetradecanoylphorbol-13-acetate represses sod2 induction in vivo: the negative role of p50. Carcinogenesis 28: 2605–2613, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dhar SK, Xu Y, and St. Clair DK. Nuclear factor kappaB and specificity protein 1-dependent bi-directional regulation of the human manganese superoxide dismutase. J Biol Chem 284: 9835–9846, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Federici L, Arcovito A, Scaglione GL, Scaloni F, Sterzo CL, Matteo AD, Falini B, Giardina B, and Brunori M. Nucleophosmin C-terminal leukemia-associated domain interacts with G-rich quadruplex form in DNA. J Biol Chem 285: 3738–3749, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fuji J. and Taniguchi N. Phorbol ester induces manganese-superoxide dismutase in tumor necrosis factor-resistant cells. J Biol Chem 266: 23142–23146, 1991 [PubMed] [Google Scholar]
- 20.Gal J, Zhang J, Kwinter DM, Zhai J, Jia H, Jia J, and Xhu H. Nuclear localization sequence of FUS and induction of stress granules by ALS mutants. Neurobiol Aging 32: 27–40, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gerbino V, Carri MT, Cozzolino M, and Achsel T. Mislocalised FUS mutants stall spliceosomal snRNPs in the cytoplasm. Neurobiol Dis 55: 120–128, 2013 [DOI] [PubMed] [Google Scholar]
- 22.Gurney ME, Pu H, and Chiu Y. Motor neuron degeneration in mice that express a human Cu,Zn superoxide dismutase mutation. Science 264: 1772–1775, 1994 [DOI] [PubMed] [Google Scholar]
- 23.Hicks GG, Singh N, and Sing N. FUS deficiency in mice results in defective B-lymphocyte development and activation, high level of chromosomal instability and perinatal death. Nat Genet 24: 175–179, 2000 [DOI] [PubMed] [Google Scholar]
- 24.Horton WA, Eldridqe R, and Brody JA. Familial motor neuron disease. Evidence for at least three different types. Neurology 26: 460–465, 1976 [DOI] [PubMed] [Google Scholar]
- 25.Hume DA, Sasmono T, Himes SR, Sharma SM, Bronisz A, Constantin M, Ostrowski MC, and Ross IL. The Ewing sarcoma protein (EWS) binds directly to the proximal elements of the macrophase-specific promoter of the CSF-1 receptor (csf1r) gene. J Immunol 180: 6733–6742, 2008 [DOI] [PubMed] [Google Scholar]
- 26.Jones PL, Ping D, and Boss JM. Tumor necrosis factor alpha and interleukin-1beta regulate the murine manganese superoxide dismutase gene through a complex intronic enhancer involving C/EBP-beta and NF-kappaB. Mol Cell Biol 17: 6970–6990, 1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jungsuwadee P, Zhao T, Stolarczyk EI, Paumi CM, Butterfield DA, St. Clair DK, and Vore M. The G671V variant of MRP1/ABCC1 links doxorubicin-induced acute cardiac toxicity to disposition of the glutathione conjugate of 4-hydroxy-2-nonenal. Pharmacogenet Genomics 22: 273–284, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Keller JN, Kindy MS, Holtsberg FW, St. Clair DK, Yen H-C, Germeyer AM, Steiner SM, Bruce-Keller AJ, Hutchins JB, and Mattson MP. Mitochondrial manganese superoxide dismutase prevents neural apoptosis and reduces ischemic brain injury: suppression of peroxynitrite production, lipid peroxidation, and mitochondrial dysfunction. J Neurosci 18: 687–697, 1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kiningham KK, Xu Y, Daosukho C, Popova B, and St. Clair DK. Nuclear factor kappaB of superoxide dismutase 2. Biochem J 353: 147–156, 2001 [PMC free article] [PubMed] [Google Scholar]
- 30.Kuroda M, Sok J, Webb L, Baechtold H, Urano F, Yin Y, Chung P, De Rooij DG, Akhmedov A, Ashley T, and Ron D. Male sterility and enhances radiation sensitivity in TLS(-/-) mice. EMBO J 19: 453–462, 2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kwiatkowski TJ, Jr., Bosco DA, LeClerc AL, Tamrazian E, Vanderburg CR, Russ C, Davis A, Gilchrist J, Kasarakis EJ, Munsat T, Valdmains P, Rouleau GA, Hosler BA, Cortelli P, De Jong PJ, Yoshinaga T, Haines JL, Pericak-Vance MA, Yas J, Ticozzi N, Siddique T, McKenna-Yasek D, Sapp PC, Horvitz HR, Landers JE, and Brown RH, Jr., Mutations in the FUS/TLS genes on chromosome 16 causes familial amyotropic lateral sclerosis. Science 323: 1205–1208, 2009 [DOI] [PubMed] [Google Scholar]
- 32.Law WJ, Cann KL, and Hicks GG. TLS, EWS and TAF15: a model for transcriptional integration of gene expression. Brief Funct Genomic Proteomic 5: 8–14, 2006 [DOI] [PubMed] [Google Scholar]
- 33.Li Y, Huang TT, Carison EJ, Melov S, Ursell PC, Olson JL, Noble LJ, Yoshimura MP, Berger C, and Chan PH. Dilated cardiomyopathy and neonatal lethality in mutant mice lacking manganese superoxide dismutase. Nat Genet 11: 376–381, 1995 [DOI] [PubMed] [Google Scholar]
- 34.Liu X, Niu C, Ren J, Zhang J, Xie X, Zhu H, Feng W, and Gong W. The RRM domain of human fused in sarcoma protein reveals a non-canonical nucleic acid binding site. Biochim et Biophys Acta 1832: 375–385, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu Y, Brooks BR, Taniguchi N, and Hartmann HA. CuZnSOD and MnSOD immunoreactivity in brain stem motor neurons from amyotropic lateral sclerosis patients. Acta Neuropathol 95: 63–70, 1998 [DOI] [PubMed] [Google Scholar]
- 36.Maracchioni A, Totaro A, Angelini DF, Penta AD, Bernardi G, Carri MT, and Achsel T. Mitochondrial damage modulates alternative splicing in neuronal cells: implications for neurodeneration. J Neurochem 100: 142–153, 2007 [DOI] [PubMed] [Google Scholar]
- 37.Mariucci G, Ambrosini MV, Colarieti L, and Bruschelli G. Differential changes in Cu, Zn and Mn superoxide dismutase activity in developing rat brain and liver. Experimentia 46: 753–755, 1990 [DOI] [PubMed] [Google Scholar]
- 38.Martin LJ. Biology of mitochondria in neurodegenerative diseases. Prog Mol Biol Transl Sci 107: 355–415, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.May WA, Lessnick SL, Braun BS, Klemsz M, Lewis BC, Lunsford LB, Hromas R, and Denny CT. The Ewing's sarcoma EWS/FLI-1 fusion gene encodes a more potent transcriptional activator and is a more powerful transforming gene than FLI-1. Mol Cell Biol 13: 7393–7398, 1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McEachern G, Kassovska-Bratinova S, Raha S, Tarnopolsky MA, Turnbull J, Bourgeois J, and Robinson B. Manganese superoxide dismutase levels are elevated in a proportion of amyotropic lateral sclerotic patient cell lines. Biochem Biophys Res Commun 273: 359–363, 2000 [DOI] [PubMed] [Google Scholar]
- 41.Melov S, Schneider JA, Day BJ, Hinerfeld D, Coskun P, Mirra SS, Crapo JD, and Wallace DC. A novel neurological phenotype in mice lacking mitochondrial superoxide dismutase. Nat Genet 18: 159–163, 1998 [DOI] [PubMed] [Google Scholar]
- 42.Millecamps S, Salachas F, Cazeneuve C, Gordon P, Bricka B, Guillot-Noel L, Russaouen O, Bruneteau G, Pradat PE, Le Forestier N, Vandenberghe N, Danel-Brunaud V, Guy N, Thauvin-Robinet C, Lacomblez L, Couratier P, Hannequin D, Seilhean D, Le Ber I, Corcia P, Camu W, Brice A, Rouleau G, Leguem E, and Meininger V. SOD1, ANG, VAPB, TARDBP, and FUS mutations in familial amyotrophic lateral sclerosis: genotype-phenotype correlations. J Med Genet 47: 554–560, 2010 [DOI] [PubMed] [Google Scholar]
- 43.Nakaya T, Alexiou P, Maragkakis M, Chang A, and Mourelatos Z. FUS regulates genes coding for RNA-binding proteins in neurons by binding to their highly conserved introns. RNA 19: 498–509, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nelson LM. Epidemiology of ALS. Clin Neurosci 3: 327–331, 1995 [PubMed] [Google Scholar]
- 45.Perkins ND, Agranoff AB, Pascal E, and Nabel GL. An interaction between the DNA-binding domains of RelA(p65) and Sp1 mediates human immunodeficiency virus gene activation. Mol Cell Biol 14: 6570–6583, 1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Perrotti D, Bonatti S, Trotta R, Martinez R, Skorski T, Salomoni P, Grassilli E, Iozzo RV, Cooper DR, and Calabretta B. FUS/TLS, a pro-oncogene involved in multiple chromosomal translocations, is a novel regulator of BCR/ABL-mediated leukemogenesis. EMBO J 17: 4442–4455, 1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Porntadavity S, Xu Y, Kiningham K, Rangnekar VM, Prachayasitikul V, and St. Clair DK. TPA-activated transcription of the human MnSOD gene: role of transcription factor Sp1 and Egr-1. DNA Cell Biol 20: 473–481, 2001 [DOI] [PubMed] [Google Scholar]
- 48.Prasad DD, Ouchida M, Lee L, Rao VN, and Reddy ES. TLS/FUS fusion domain of TLS/FUS-erg chimeric protein resulting from the t(16:21) chromosomal translocation in human myeloid leukemia functions as a transcriptional activation domain. Oncogene 9: 3717–3729, 1994 [PubMed] [Google Scholar]
- 49.Schwartz JC, Ebmeier CC, Podell ER, Heimiller J, Taatjes DJ, and Cech TR. FUS binds the CTD of RNA polymerase II and regulates its phosphorylation at Ser2. Genes Dev 26: 2690–2695, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sompol P, Xu Y, Ittarat W, Daosukho C, and St. Clair DK. NF-kappaB-associated MnSOD induction protects against beta-amyloid-induced neuronal apoptosis. J Mol Neurosci 29: 279–288, 2006 [PubMed] [Google Scholar]
- 51.Sun Y, St. Clair DK, Xu Y, Crooks PA, and St. Clair WH. A NADPH oxidase dependent redox signaling pathway mediates the selective radiosensitization effect of parthenolide in prostate cancer cells. Cancer Res 70: 2880–2890, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vance C, Rogelj B, Hortobagyi T, De Vos KJ, Nishimura AL, Sreedharan J, Hu X, Smith B, Ruddy D, Wright P, Ganesalingam J, William KL, Tripati V, Al-Saraj S, Al-Chalabi A, Leigh PN, Blair IP, Nicholson G, Belleroche JD, Gallo JM, Miller CC, and Shaw CE. Mutations in FUS, an RNA processing protein, cause familial amyotropic lateral sclerosis type 6. Science 323: 1208–1211, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wan XS, Devalaraja MN, and St. Clair DK. Molecular structure and organization of the human manganese superoxide dismutase gene. DNA Cell Biol 13: 1127–1136, 1994 [DOI] [PubMed] [Google Scholar]
- 54.Wispe JR, Warner BB, Clark JC, Dey CR, Neuman J, Glasser SW, Crapo JD, Chang L, and Whitselt JA. Human Mn-superoxide dismutase in pulmonary epithelial cells of transgenic mice confers protection from oxygen injury. J Biol Chem 267: 23937–23941, 1992 [PubMed] [Google Scholar]
- 55.Xia R, Gal J, Zhu H, and Jia J. Motor neuron apoptosis and neuromuscular junction perturbation are prominent features in a Drosophila model of FUS-mediated ALS. Mol Neurodegener 24: 7–10, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xu Y, Fang F, Dhar SK, St. Clair WH, Kasarskis EJ, and St. Clair DK. The role of a single-stranded nucleotide loop in transcriptional regulation of the human sod2 gene. J Biol Chem 282: 15981–15994, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xu Y, Kiningham KK, Devalaraja MN, Yeh CC, Majima H, Kasarskis EJ, and St. Clair DK. An intronic NF-kappaB element is essential for induction of the human manganese superoxide dismutase gene by tumor necrosis factor-alpha and interleukin-1beta. DNA Cell Biol 18: 709–722, 1999 [DOI] [PubMed] [Google Scholar]
- 58.Xu Y, Porntadavity S, and St. Clair DK. Transcriptional regulation of the human manganese superoxide dismutase gene: the role of specificity protein 1 (Sp1) and activating protein-2. Biochem J 362: 401–412, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yamazaki T, Chen S, Yu Y, Yan B, Haertlein TC, Carrasco MA, Tapia JC, Zhai B, Das R, Lalancette-Hebert M, Sharma A, Chandran S, Sullivan G, Nishimura AL, Shaw CE, Gygi SP, Schneider NA, Maniatis T, and Reed R. FUS-SMN protein interactions link the motor neuron diseases ALS and SMA. Cell Rep 2: 799–806, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yang L, Embree LJ, Tsai S, and Hickstein DD. Oncoprotein TLS interacts with serin-arginine proteins involved in RNA splicing. J Biol Chem 273: 27761–27764, 1998 [DOI] [PubMed] [Google Scholar]
- 61.Yeh CC, Wan XS, and St. Clair DK. Transcriptional regulation of 5` proximal promoter of the human manganese superoxide dismutase gene. DNA Cell Biol 17: 921–930, 1998 [DOI] [PubMed] [Google Scholar]
- 62.Yen H-C, Oberley TD, Vichitbandha S, HO Y-S, and St. Clair DK. The protective role of manganese superoxide dismutase against adriamycin-induced acute cardiac toxicity in transgenic mice. J Clin Investig 98: 1253–1260, 1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.