Abstract
An unusual ΔE693 mutation in the amyloid precursor protein (APP) producing a β-amyloid (Aβ) peptide lacking glutamic acid at position 22 (Glu22) was recently discovered, and dabbed the Osaka mutant (ΔE22). Previously, several point mutations in the Aβ peptide involving Glu22 substitutions were identified and implicated in the early onset of familial Alzheimer’s disease (FAD). Despite the absence of Glu22, the Osaka mutant is also associated with FAD, showing a recessive inheritance in families affected by the disease. To see whether this aggregation-prone Aβ mutant could directly relate to the Aβ ion channel-mediated mechanism as observed for the wild type (WT) Aβ peptide in AD pathology, we modeled Osaka mutant β-barrels in a lipid bilayer. Using molecular dynamics (MD) simulations, two conformer ΔE22 barrels with the U-shaped monomer conformation derived from NMR-based WT Aβ fibrils were simulated in explicit lipid environment. Here, we show that the ΔE22 barrels obtain the lipid-relaxed β-sheet channel topology, indistinguishable from the WT Aβ1–42 barrels, as do the outer and pore dimensions of octadecameric (18-mer) ΔE22 barrels. Although the ΔE22 barrels lose the cationic binding site in the pore which is normally provided by the negatively charged Glu22 side-chains, the mutant pores gain a new cationic binding site by Glu11 at the lower bilayer leaflet, and exhibit ion fluctuations similar to the WT barrels. Of particular interest, this deletion mutant suggests that toxic WT Aβ1–42 would preferentially adopt a less C-terminal turn similar to that observed for Aβ17–42, and explains why the solid state NMR data for Aβ1–40 point to a more C-terminal turn conformation. The observed ΔE22 barrels conformational preferences also suggest an explanation for the lower neurotoxicity in rat primary neurons as compared to WT Aβ1–42.
Keywords: Aβ mutant, point mutation, Alzheimer’s, toxic oligomer, β-sheet channel, molecular dynamics simulation
INTRODUCTION
A rare mutation in the amyloid precursor protein (APP) with a deletion of glutamic acid, ΔE693, was first identified in patients of Japanese pedigree.1 In clinical studies on propositus, it was discovered that the variant of APP is closely linked to pathogenesis of Alzheimer’s disease (AD) with symptoms similar to AD-type dementia. The ΔE693 mutation in APP produces a form of β-amyloid (Aβ) peptide that lacks a Glu22 residue, ΔE22, which is known as the Osaka mutant. Initial studies in vitro suggested that the mutant did not form fibrils, but presented subcellular oligomers in transfected cells.2 Subsequent studies in vivo showed that transgenic mice exhibited age-dependent intraneuronal Aβ oligomerization without extracellular amyloid deposits.3
In contrast to earlier reports that the Osaka mutant (ΔE22) did not form fibrils,1–3 recent studies, however, demonstrated that the mutant peptides derived from Aβ1–42 and Aβ1–40 have strong tendency to form fibrils faster than those of wild type (WT) Aβ peptides.4–6 The analysis of secondary structure dynamics showed that elimination of Glu22 from both Aβ1–42 and Aβ1–40 (below referred to ΔE22-Aβ1–42 and ΔE22-Aβ1–40, respectively) substantially increase β-sheet formation propensities.4 Subsequent fibril morphology study demonstrated that both ΔE22-Aβ1–42 and ΔE22-Aβ1–40 form short protofibrillar and fibrillar structures with high conformational stability. Mixture of ΔE22-Aβ1–40 and WT Aβ1–40 also produced fibrils with morphology similar to that of fibril made of pure ΔE22-Aβ1–40, indicating that ΔE22-Aβ1–40 fibrils served as a seed for WT Aβ1–40 fibril elongation.5 In rat primary neuron cultures, ΔE22-Aβ1–40 was neurotoxic, while WT Aβ1–40 was found to be nontoxic.6 In contrast, ΔE22-Aβ1–42 was less toxic than WT Aβ1–42.
The presence of Glu22 point substitutions in the Aβ peptide implicates an early onset of familial Alzheimer’s disease (FAD) and cerebral amyloid angiopathy (CAA).7 These point mutations include the E22Q, associated with Hereditary Cerebral Hemorrhage with Amyloidosis ‘Dutch type’ (HCHWA-D);8 the E22G, known as the Arctic mutation;9,10 and the E22K, known as the Italian mutation.11 The Arctic mutant increased bilayer disruption due to high hydrophobicity. The Italian mutant was observed to increase the rate of aggregation12 as did the Dutch mutant which aggregated faster than WT Aβ.13 These mutants formed fibrils in solution morphologically similar to those formed by WT Aβ peptides and presented polymorphic aggregates on a lipid membrane.12 Taken together, these results emphasized the importance of the point substitution at Aβ position 22 in FAD and CAA.
Unlike point substitutions, the Osaka mutant eliminated Glu22 in patients of FAD. This recently described unusual variant of Aβ mutation prompted us to interrogate the biological properties of the mutant for its structure and function in the cell membrane. Here, we modeled octadecameric (18-mer) Aβ barrels of ΔE22-Aβ1–42 (below we refer to “ΔE22” throughout the text) and WT Aβ1–42 peptides in a lipid bilayer containing 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC). In our Aβ barrels, both mutant and WT peptides adopted the U-shaped motif of β-strand-turn-β-strand predicted by simulations14 and found in NMR experiments.15,16 Overall, we observed that the ΔE22 barrels present similar morphologies and dimensions as observed for the WT Aβ1–42 barrels, suggesting that they share features of monomer folding and aggregation into a toxic oligomer state. Due to the deletion of a charged residue, the hydrophobicity of the mutant is increased, and the increased hydrophobicity can cause faster kinetics of nucleation and membrane insertion leading to toxic channel formation. We suggest that the conformational similarity of the mutant with the WT Aβ peptide reflects functional similarity in the disease stage. As the WT Aβ peptide in the aggregation state is highly polymorphic,17–19 the mutant would explore conformational space along the aggregation pathway of the highly polymorphic conformational states. Our simulations provide a possible conformational species of the barrels which is more highly populated by the Osaka mutant in atomic detail, to help drug discovery efforts targeting the mutant and WT Aβ peptides.
MATERIALS AND METHODS
Recruiting Aβ Monomer Conformations
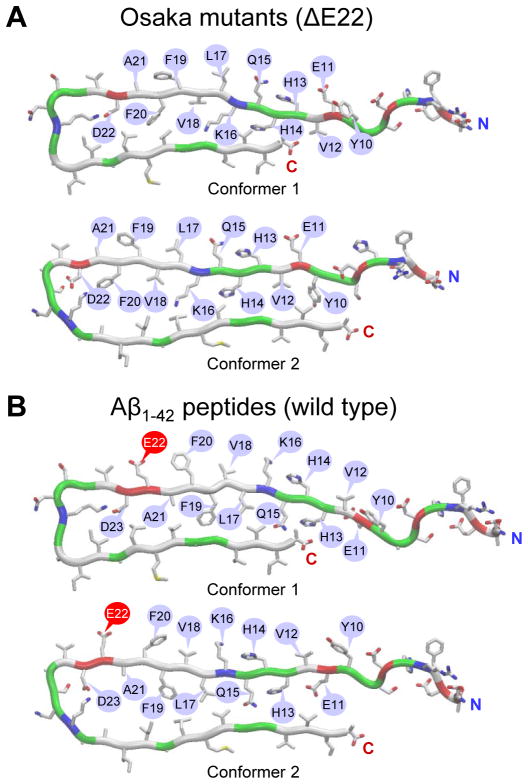
Two Aβ monomer conformations with the β-strand-turn-β-strand motif (known as U-shaped or β-arcade20 structures) were extracted from Aβ1–42 fibrils, where the structure was defined by hydrogen/deuterium-exchange NMR data, side-chain packing constraints from pair-wise mutagenesis, ssNMR and EM (PDB code: 2BEG),15 and small Aβ1–40 protofibrils (PDB codes: 2LMN and 2LMO),16 where the structure was based on the ssNMR model. In both structures, the N-terminal coordinates, residues 1–16 for the former and 1–8 for the latter structure, are missing due to disorder. We used the Aβ1–16 coordinates, in the absence of Zn2+ (PDB code: 1ZE7),21 for the missing portions of the peptides. For each combination of the N-terminal structure with the U-shaped motifs, two Aβ1–42 conformers were generated.22–25 Conformer 1 has a turn at Ser26-Ile31, and conformer 2 at Asp23-Gly29. In the latter conformer, two C-terminal residues, Ile41 and Ala42 were added to create Aβ1–42. For convenience, we divide both WT Aβ conformers into four domains: N-terminal chain (residues 1–16 and 1–8 for conformer 1 and 2, respectively), pore-lining strand (residues 17–25 and 9–22 for conformer 1 and 2, respectively), turn (residues 26–31 and 23–29 for conformer 1 and 2, respectively), and C-terminal strand (residues 32–42 and 30–42 for conformer 1 and 2, respectively).
Generating the Osaka Mutant, ΔE22, from the Aβ Conformers by the Deletion of Glu22
To create the mutant, we removed the Glu22 residue from the pore-lining β-strand. As a result, pore-lining residues 10–21 for both conformers flipped their side-chains, while the other domains kept intact. Two ΔE22 conformers (Figure 1A), each derived from the WT Aβ1–42 conformers (Figure 1B) still retain the U-shaped structure with the β-strand-turn-β-strand motif. The 41-residue long mutant has the same turns as the corresponding WT peptides; Ser25-Ile30 for conformer 1 and Asp22-Gly28 for conformer 2. We similarly divide both mutant conformers into four domains: N-terminal chain (residues 1–15 and 1–7 for conformer 1 and 2, respectively), pore-lining strand (residues 16–24 and 8–21 for conformer 1 and 2, respectively), turn (residues 25–30 and 22–28 for conformer 1 and 2, respectively), and C-terminal strand (residues 31–41 and 29–41 for conformer 1 and 2, respectively).
Figure 1.
(A) Monomer conformations of the Osaka mutant (ΔE22) with different turns at Ser25-Ile30 (conformer 1) and Asp22-Gly28 (conformer 2), and (B) the wild type Aβ1–42 peptides with different turns at Ser26-Ile31 (conformer 1) and Asp23-Gly29 (conformer 2). Pore-lining residues are marked with blue callouts, and especially the Glu22 residues in the wild type Aβ1–42 peptides are marked with red callouts. In the peptide ribbon, hydrophobic, polar/Gly, positively charged, and negatively charged residues are colored white, green, blue, and red, respectively. Both termini of peptide are denoted as blue letter “N” for the N-terminus and red letter “C” for the C-terminus.
Construction of Aβ Barrels in the Lipid Bilayer
To construct the β-barrel structure, ΔE22 was inclined ~37° relative to the pore axis26 and then an 18-fold rotational symmetry operation was performed with respect to the pore axis creating an 18-mer ΔE22 barrel (Figure S1A and S1B of the Supporting Information). The WT Aβ1–42 barrel was also constructed in an analogous manner (see Figure S1C and S1D of the Supporting Information for comparison). We modeled the Aβ barrels with β-sheet structure by mimicking naturally-occurring β-barrels observed in transmembrane proteins that are found frequently in the outer membranes of bacteria, mitochondria, and chloroplasts. The β-barrel motif is a large β-sheet composed of an even number of β-strands. Some known structures of β-barrel membrane proteins have β-strands ranging in size from 8 to 22.27 The 18-mer Aβ barrel has 18 β-strands enclosing the solvated pore. This number is also in the range of the number of β-strands for natural β-barrels ranging from 8 to 22, which can form a β-barrel motif. Our previous simulations for Aβ channels indicate that different numbers of Aβ monomers could produce channels with different outer and pore dimensions.22–26,28–32 We found that Aβ channels obtained a preferred size range of 16–24 β-strands lining the pores.30,32 This range was also found to hold for other toxic β-sheet channels; K3 (a fragment of β2-microglobulin) channels with 24 β-strands33 and protegrin-1 (PG-1) channels with 16–20 β-strands.34,35 In this work, the outer/pore diameters of the WT 18-mer Aβ barrels are in good agreement with the experimental AFM ranges.23,25,36 The AFM experiments provide images of channels with a wide variety of sizes and shapes, but simulated Aβ barrels are limited to cover all ranges of channel sizes that are imaged by AFM.
To construct two layers of DOPC lipid in a unit cell, simple van der Waals (vdW) spheres representing lipid headgroups are placed in two parallel planes separated by the expected headgroup-to-headgroup DOPC lipid bilayer thickness.37,38 These planes can be regarded as membrane surfaces. Dynamics is performed on the spheres, constrained to their respective planes, with the embedded barrel structure held rigid. This planar harmonic constraint ensures that the vdW spheres are randomly distributed onto the planes and well packed around the Aβ barrel. The lipid bilayer is then constructed with headgroups at the positions of the vdW spheres. The DOPC lipid molecules were randomly selected from a library of preequilibrated liquid crystalline state lipids. After replacement of the vdW spheres with lipid molecules, a series of minimizations is performed to remove overlaps of the alkane chains and gradually relax the system. For DOPC, the cross-section area per lipid is 72.4 Å2 and headgroup distance across the bilayer is 36.7 Å at 30 °C.39 With a choice for the number of lipid molecules, the optimal value of lateral cell dimensions can be determined. The DOPC bilayer containing 420 lipids constitutes the unit cell with TIP3P water, added at both sides with lipid/water ratio of ~1/110. Updated CHARMM40 all-atom additive force field for lipids (C36)41 and the modified TIP3P water model42 were used to construct the set of starting points and to relax the systems to a production-ready stage. The system contains Mg2+, K+, Ca2+, and Zn2+ at the same concentration of 25 mM to satisfy a total cation concentration near 100 mM. The bilayer system containing an Aβ barrel, lipids, salts, and water has almost 210,000 atoms.
MD simulations employed the zwitterionic DOPC bilayer. All Aβ barrels were pre-assembled and simulated in the lipid bilayer. It has been known that anionic bilayers with negatively charged surfaces facilitate the interactions with Aβ peptides.43 However, once the peptides are inserted into the membrane core and subsequently assembled to form a channel, the hydrophobic interactions between lipid-facing residues in the channel and lipid tails should be an important factor in stabilizing the channel conformation. In our previous simulations, we have employed anionic lipid bilayer composed of DOPS/POPE in order to complement the electrophysiological recoding studies.22–25 The DOPS/POPE phospholipid combinations can form stable lipid bilayers, and allow conductance measurements in PLB experiments. However, we observed that there are no significant differences in the critical results of subunits formation in the channel conformation in zwitterionic and anionic bilayers.
Production Runs
We generated at least 10 different initial configurations for each Aβ barrel for the relaxation process in order to obtain the best initial configuration toward a starting point. In the pre-equilibrium stages, a series of minimizations was performed for the initial configurations to remove overlaps of the alkane chains in the lipids and to gradually relax the solvents around the harmonically restrained peptides. The initial configurations were gradually relaxed through dynamic cycles with electrostatic cutoffs (12 Å). The harmonic restraints were gradually diminished with the full Ewald electrostatics calculation and constant temperature (Nosé–Hoover) thermostat/barostat at 303 K. For t < 30 ns, our simulation employed the NPAT (constant number of atoms, pressure, surface area, and temperature) ensemble with a constant normal pressure applied in the direction perpendicular to the membrane. After t = 30 ns, the simulations employed the NPT ensemble. Production runs of 100 ns for the starting points with the NAMD code44 were performed on a Biowulf cluster at the NIH. Averages were taken after 30 ns, discarding initial transients. Analysis was performed with the CHARMM programming package.40
RESULTS
The Osaka Mutant (ΔE22) Adopts the β-Barrel Topology in the Lipid Bilayer
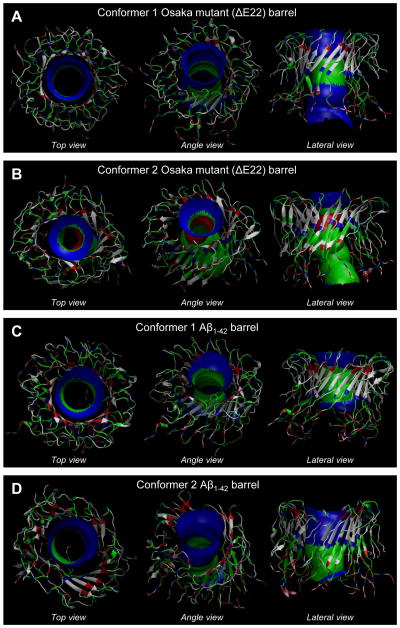
We performed 100 ns all-atoms molecular dynamics (MD) simulations on Aβ barrels, constructed using the Osaka mutant with complete elimination of Glu22 and WT Aβ1–42 peptides, embedded in a DOPC bilayer. Both ΔE22 and WT Aβ barrels comprising two different U-shaped conformers were initially pre-assembled as an annular shape. During the course of the simulations, no immediate peptide dissociation in the barrels was observed, but the peptides were gradually relaxed in the lipid bilayer (Figure S2A, B of the Supporting Information). Previous Aβ channel simulations showed that amyloid ion channels composed of truncated (Aβ17–42 (p3), Aβ9–42 (N9), and p3-F19P)26,28–32 and full-length (L-Aβ1–42, D-Aβ1–42, F19P, and F20C)22–25 Aβ peptides/mutants are heterogeneous compared to functional gated ion channels. In our current simulations, the mutant also presents heterogeneity in barrel conformations as observed in the WT barrels (Figure 2). Non-homogeneous peptide interactions with the surrounding environments, as evident by the independent peptide fluctuations, induce the heterogeneity in the mutant and WT barrel conformations (Figure S2C-F of the Supporting Information). In Table 1, the calculated outer dimensions and pore sizes are summarized for the 18-mer mutant and WT Aβ barrels. The outer/pore diameters are ~7.91/~2.1 and ~7.62/~1.52 nm for the conformer 1 and 2 ΔE22 barrels, and ~7.87/~1.98 and ~7.97/~2.18 nm for the conformer 1 and 2 Aβ1–42 barrels, respectively. Although the conformer 2 ΔE22 barrel presents slightly reduced outer diameter and significantly reduced pore size, those dimensions for the conformer 1 ΔE22 barrel are in the range of the WT barrel.
Figure 2.
Averaged pore structures calculated with HOLE50 embedded in the average barrel conformations during the simulations for the (A) conformer 1 and (B) conformer 2 Osaka mutant (ΔE22) barrels, and the (C) conformer 1 and (D) conformer 2 Aβ1–42 barrels. In the barrel structures with the ribbon representation, hydrophobic, polar/Gly, positively charged, and negatively charged residues are colored white, green, blue, and red, respectively. For the pore structures in the surface representation, red denotes pore diameter of d < 1.4 nm, green denotes pore diameter in the range, 1.4 nm = d = 2.0 nm, and blue denotes pore diameter of d > 2.0 nm.
Table 1.
The calculated outer and pore diameters, do and dp, with a description of the pore status, % of β-sheet secondary structure in the pore/C-terminal strands, and “straightness” of the strands by the β-strand order parameter using , where θα is the angle between the positional vectors connecting two Cα atoms and Nv is the total number of vector pairs for the residues in the pore/C-terminal strands for the Osaka mutant (ΔE22) and wild type Aβ1–42 barrels.
| Aβ barrels | do (nm) | dp (nm)* | Pore status | % of β-sheet pore/C-term | Sβ pore/C-term |
|---|---|---|---|---|---|
| Conformer 1 Osaka mutant (ΔE22) | ~7.91 | ~2.10 | Opened | 41.6/2.4 | 0.58/0.45 |
| Conformer 2 Osaka mutant (ΔE22) | ~7.62 | ~1.52 | Reduced | 46.6/5.1 | 0.57/0.43 |
| Conformer 1 Aβ1–42 (wild type) | ~7.87 | ~1.98 | Opened | 46.0/3.8 | 0.56/0.45 |
| Conformer 2 Aβ1–42 (wild type) | ~7.97 | ~2.18 | Opened | 50.4/5.8 | 0.57/0.45 |
Pore diameters are averaged along the pore axis within the cutoffs defined by the height of pore.
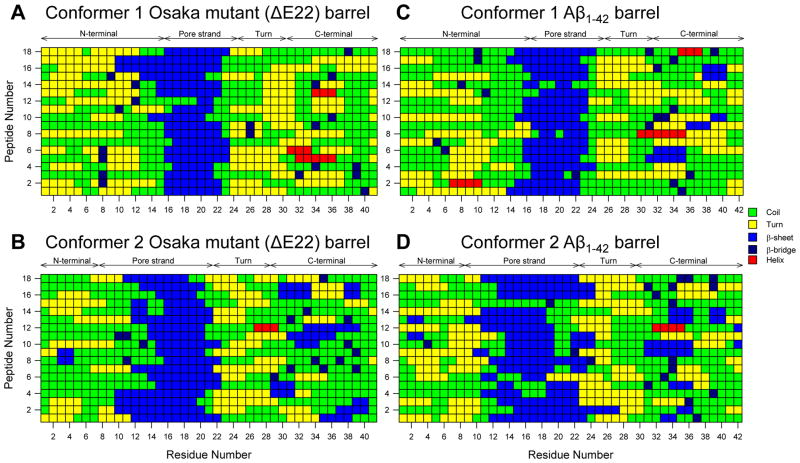
When assembled in a barrel, the U-shaped peptides form two layered annular β-sheets. The pore-lining strands form a β-sheet with intermolecular backbone hydrogen bonds (H-bonds), tightly encompassing a solvated pore, as evident by the secondary structure analysis (Figure 3). However, the outer β-sheet is not so preserved because of the longer H-bonding pair distance between the C-terminal strands due to the larger curvature. In contrast to the pore-lining strands with high β-sheet content, the C-terminal strands have low β-sheet content, suggesting that the evolution of secondary structure is rather dynamic. For the same conformers, the ΔE22 barrels exhibit slightly lower percentage of the β-sheet secondary structure for the membrane embedded pore and the C-terminal strands than the Aβ1–42 barrels (Table 1). However, these differences are very subtle. In addition, both mutant and WT barrels have similar β-strand order parameters for the membrane embedded portions, suggesting analogous barrel conformations in the lipid bilayer.
Figure 3.
The description of secondary structure by STRIDE51 averaged during the simulations for the (A) conformer 1 and (B) conformer 2 Osaka mutant (ΔE22) barrels, and the (C) conformer 1 and (D) conformer 2 Aβ1–42 barrels.
Interactions of the Osaka Mutant (ΔE22) in the Lipid Environment
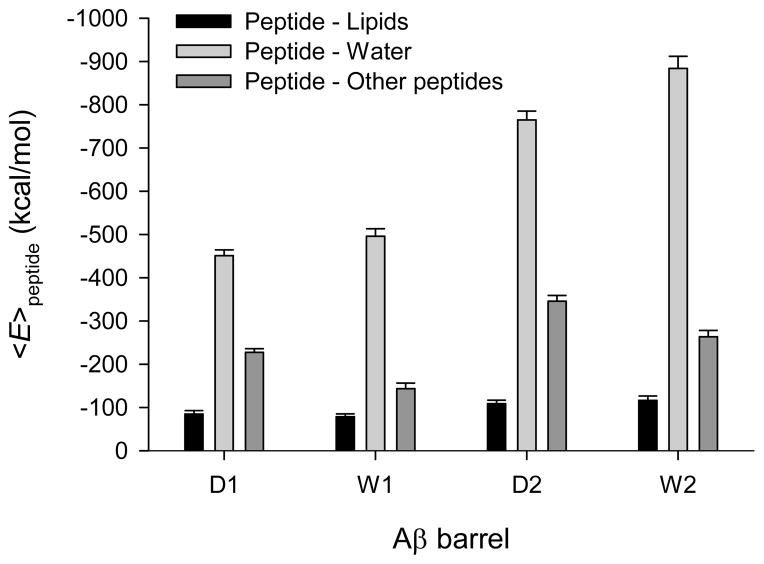
To observe how the deletion of Glu22 affects peptide interactions in such complex lipid environments, we calculated the interaction energy for each peptide with the lipids, water, and other peptides, and then averaged over the number of peptides in the barrel, yielding per peptide interaction energy (Figure S3 of the Supporting Information). For comparison, in the calculation only membrane embedded portions of the peptides, i.e. residues 17–41 and 9–41 for the conformer 1 and 2 ΔE22, respectively, and residues 17–42 and 9–42 for the conformer 1 and 2 Aβ1–42 peptides, respectively, were considered. These portions of the peptides represent the intrinsic U-shape obtained from the experimental coordinates.15,16 Figure 4 shows averaged peptide interaction energy obtained from the per peptide interaction energy averaged over time. In the comparison of the same conformers between mutant and WT barrels no apparent energy difference from the mutation in the peptide/lipid interaction can be observed, since the lipid interaction energy of the mutant is similar to that of the WT peptide. This suggests that the mutation site is located at the solvated pore, far from lipids, and each conformer mutant has the lipid-contacting C-terminal strand sequentially identical to the corresponding conformer WT peptide (Figure 1). However, energy differences from the mutation in the peptide/water and peptide/peptide interactions are immediately apparent. Comparison of corresponding conformers of mutant and WT Aβ1–42 barrels indicates that each conformer ΔE22 barrel exhibits relatively weaker peptide/water (mainly pore water) and relatively stronger peptide/peptide interactions. The energy contributions of the mutation in the peptide/water interaction are ~−44.7 and ~−119.1 kcal/mol per peptide for the conformer 1 and 2 ΔE22 barrels, respectively. In the peptide/peptide interaction, they are ~83.8 and ~82.5 kcal/mol per peptide for the conformer 1 and 2 ΔE22 barrels, respectively. The negative sign in the energy contribution denotes “loss” of attraction, while the positive sign corresponds to “gain” of attraction. The conformer 2 ΔE22 loses attraction more than the conformer 1 ΔE22 in the peptide/water interaction, suggesting that the conformer 2 ΔE22 barrel slightly reduces the size of the water pore (Table 1). However, both conformer mutants similarly gain attraction in the peptide/peptide interaction, since the repulsive force between the negatively charged Glu22 side-chains in the water pore disappears from the peptide/peptide interaction upon deletion of the residue.
Figure 4.
Averaged peptide interaction energy for the conformer 1 and 2 Osaka mutant (ΔE22) barrels (denoted as D1 and D2, respectively), and the conformer 1 and 2 wild type Aβ1–42 barrels (denoted as W1 and W2, respectively).
Ion Activities in the Osaka Mutant (ΔE22) Pore
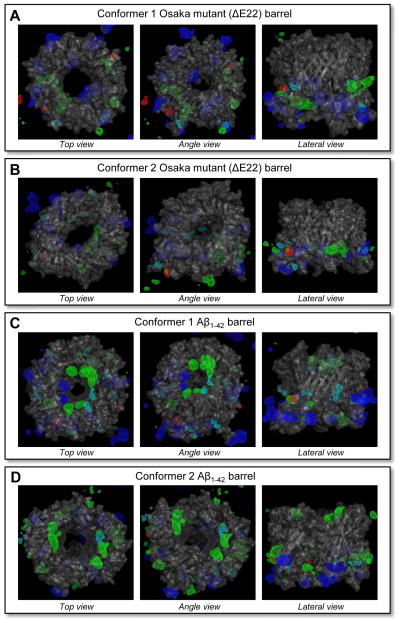
In the pores of WT Aβ channel/barrel, the negatively charged Glu22 side-chains can attract cations into the pore, forming a cationic ring.26,28,29 However, this is not the case for the ΔE22 barrel in the absence of the Glu22 residue. To locate the ionic binding sites in the mutant barrel, we calculated three-dimensional (3D) density maps of ions around Aβ barrels (Figure 5). Highly populated ionic binding sites are enclosed by 3D meshes with the colors for Mg2+ (green), K+ (red), Ca2+ (blue), Zn2+ (cyan), and Cl− (gray). As expected, in the ΔE22 barrel pores the cationic ring at the vestige of Glu22 binding site is invisible. In contrast to the mutant barrels, Aβ1–42 barrels present a cationic binding site at a cluster of Glu22 side-chains. In addition to the cationic binding site, the WT pores provide an anionic binding site at a cluster of Lys16 side-chains. This is not the case for the mutant barrels, since the Lys16 side-chains are located behind the solvated pore (Figure 1). However, in the pores of ΔE22 barrels, cations strongly interact with the Glu11 side-chains at the lower bilayer leaflet, suggesting that the side-chains serve as an emerging cationic binding site.
Figure 5.
Three-dimensional density map of Mg2+ (green mesh), K+ (red mesh), Ca2+ (blue mesh), Zn2+ (cyan mesh), and Cl− (gray mesh) for the (A) conformer 1 and (B) conformer 2 Osaka mutant (ΔE22) barrels, and the (C) conformer 1 and (D) conformer 2 Aβ1–42 barrels. Averaged channel structure is shown as the ribbon and transparent surface representations in gray. Density map indicates populated interaction sites for the ions, each with the same probability of 0.01 for cations and the probability of 0.03 for Cl−.
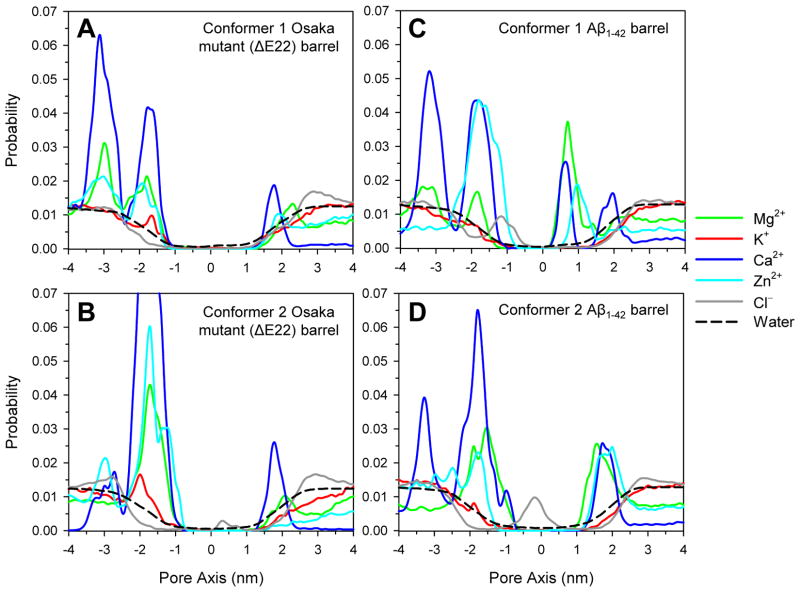
To obtain the probability distribution for ions across the bilayer, the 3D density maps of ions are projected onto a plane (Figure 6). The highly populated ion binding sites are reflected in peaks in the probability distribution curves. In the simulations, we observed that the ions mainly bind to the charged side-chains of the peptide with a strong electrostatic attraction. In particular, the cations also interact with a phosphate group in the lipid head and the C-terminus of peptide, reflecting that peaks at z = ~±2.0 nm represent those cations interacting with lipid head groups. For the Aβ1–42 barrels, peaks at z = ~1.0 and ~−1.2 nm for the conformer 1, and at z = ~1.8 and ~−0.2 nm for the conformer 2 correspond to the Glu22 cationic and Lys16 anionic binding sites in the pore, respectively. However, for both conformer ΔE22 barrels, those peaks are absent from the probability distributions for ions in the pore. Instead, peaks at z = ~−2.9 and ~−1.4 nm emerge at the lower bilayer leaflet for the conformer 1 and 2 ΔE22 barrels, respectively, representing a highly populated cation binding site at a cluster of Glu11 side-chains.
Figure 6.
Probability distribution functions for Mg2+ (green line), K+ (red line), Ca2+ (blue line), Zn2+ (cyan line), Cl− (gray line), and water (black dashed line) as a function of the distance along the pore center axis for the (A) conformer 1 and (B) conformer 2 Osaka mutant (ΔE22) barrels, and the (C) conformer 1 and (D) conformer 2 Aβ1–42 barrels.
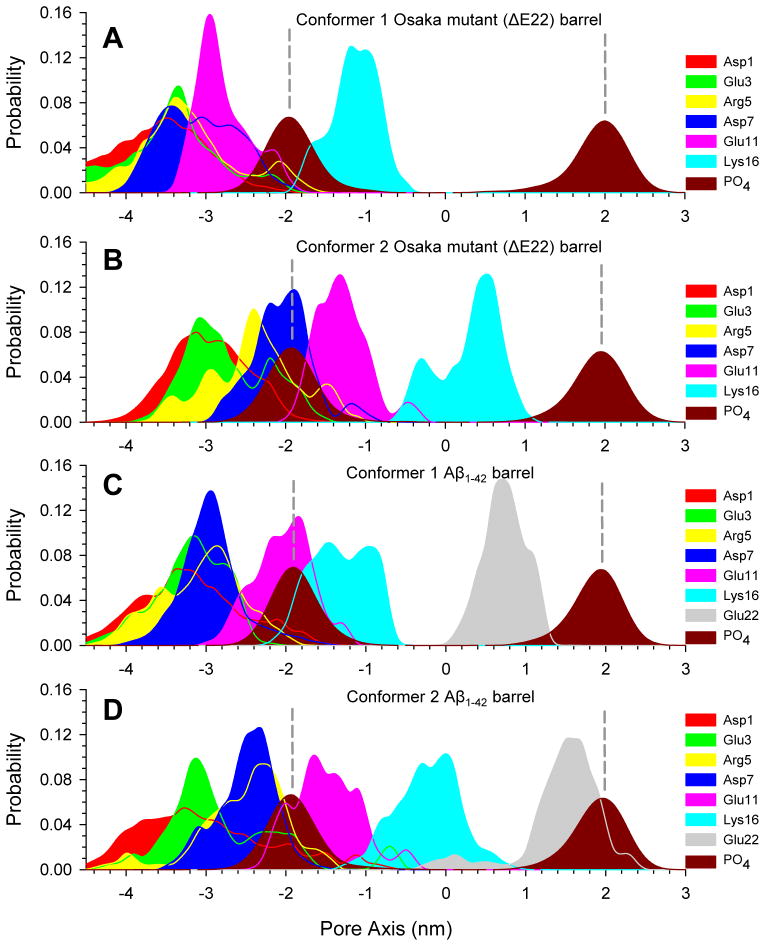
The locations of the peak in the 2D ion distribution curves are qualitatively consistent with the charged side-chain locations across the bilayer. To locate the charged groups in the Aβ barrels, the probability distributions for few selected charged groups in the peptides are calculated (Figure 7). The peaks in the distribution curves reflect the highly populated locations of the charged groups across the bilayer. During the simulations, we observed that four charged side-chains, Asp1, Glu3, Arg5, and Asp7 in the N-terminus of the peptides are mainly located in the bulky water area below the lower bilayer leaflet. Although these residues attract ions, they do not participate in the pore formation. In the ΔE22 barrels, the Glu11 side-chains are located at z = ~−2.9 and ~−1.4 nm for the conformer 1 and 2 barrels, respectively, attracting cations. In the Aβ1–42 barrels, they are located at z = ~−2.0 and ~−1.5 nm for the conformer 1 and 2 barrels, respectively. It can be seen that Glu11 side-chain locations are highly correlated with the peaks in Figure 6 for the highly populated cationic binding sites across the bilayer. In the mutant barrels, the Glu11 side-chain points toward the water pore, while in the WT barrels it is directed away from the pore (see the peptide topology in Figure 1). This indicates that in the mutant barrels the Glu11 side-chains circularly cluster to form a negatively charged ring with cations screening, serving as a cationic binding site. The circular clustering by the Glu11 side-chains at the lower bilayer leaflet further encompasses the solvated pore, causing an extension of the pore. Thus, the lengths of both conformer mutant pores are relatively longer than the corresponding conformer WT pores (Figure 2). We observed that several mutant monomers participate in the β-sheet formation (Figure 3). At the anionic biding site, the positively charged Lys16 side-chains can attract Cl− in the pore. In the mutant barrels, the Lys16 side-chains are located at z = ~−1.0 and ~0.5 nm for the conformer 1 and 2 barrels, respectively. In the WT barrels, they are located at z = ~−1.5 and ~−0.2 nm for the conformer 1 and 2 barrels, respectively. While WT pores provide the anionic binding site, it is absent from the mutant pores since the Lys16 side-chains are not present in the water pore. Thus, for the mutant barrels, the associated peak representing the anionic binding site in Figure 6 is missing, since the Lys16 side-chains are exposed in the hydrophobic core between the β-sheets. In contrast, in the WT barrel, the Lys16 side-chains point toward the water pore, forming the anionic binding site.
Figure 7.
Probability distribution functions for selected charged residues, Asp1 (red), Glu3 (green), Arg5 (yellow), Asp7 (blue), Glu11 (pink), Lys16 (cyan), and Glu22 (gray), and for the phosphate group of lipid head, PO4 (dark red), as a function of the distance along the pore center axis for the (A) conformer 1 and (B) conformer 2 Osaka mutant (ΔE22) barrels, and the (C) conformer 1 and (D) conformer 2 Aβ1–42 barrels.
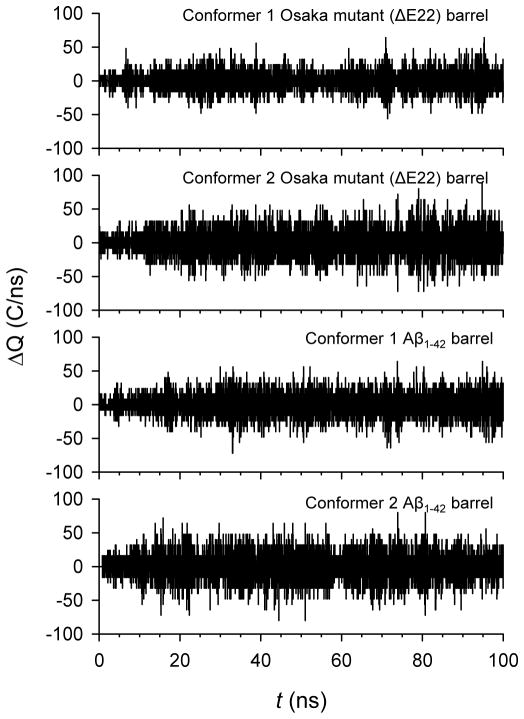
Both ΔE22 and Aβ1–42 barrels present the solvated pore, wide enough for the ion conductance. To observe ion fluctuation across the pore, we calculated the change in the total charge in the pore as a function of the simulation time. In the calculation, a pore height cutoff along the pore axis is |z| < 1.5 nm was used (Figure 8). A larger pore height cutoff, |z| < 1.8 nm, was also considered (Figure S4 of the Supporting Information). The pore height cutoff with |z| < 2.0 nm ensures the charge fluctuations including only a contribution from the ions in the middle of the pore. For both conformer mutant barrels, we observed that charge fluctuations in the total charges are similar to those for the corresponding conformer WT barrels. The charge fluctuations with the larger pore height cutoff also show a similar pattern, suggesting that the ΔE22 barrel is ion permeable in the membrane.
Figure 8.
Change in total charge in the pore as a function of the simulation time for the conformer 1 and 2 Osaka mutant (ΔE22) barrels (1st and 2nd rows), and the conformer 1 and 2 Aβ1–42 barrels (3rd and 4th rows). The pore height with cutoff along the pore axis, −1.5 < z < 1.5 nm was used.
DISCUSSION
We performed explicit molecular dynamics (MD) simulations of octadecamer (18-mer) Osaka mutant (ΔE22) and wild type (WT) Aβ1–42 barrels in a DOPC bilayer. The monomer mutant conformation was derived from the Aβ1–42 peptide via a deletion of Glu22. Two U-shaped mutant conformers, with a turn at Ser25-Ile30 for conformer 1 and a turn at Asp22-Gly28 for conformer 2, inherited the same turn conformations from the Aβ1–42 conformers with slightly different turns, Ser26-Ile31 in conformer 1 and Asp23-Gly29 in conformer 2. However, the pore-lining residues invert their side-chain orientation due to the deletion of the residue. Here we show that the ΔE22 mutant forms a β-barrel-like channel in the lipid bilayer, with the membrane embedded conformation indistinguishable from the WT Aβ1–42 barrel. For the conformer 1 ΔE22 barrel, we obtained outer and pore diameters, ~7.91 and ~2.1 nm, respectively, indicating that the overall dimensions are consistent with the Aβ1–42 barrels in this study. Further, these dimensions are also in the range of the outer diameter, ~7.8 – 8.3 nm, and pore diameter, ~1.8 – 2.2 nm, for the same 18-mer Aβ1–42 barrels in the anionic bilayer composed of DOPS/POPE.22–25 However, for the conformer 2 ΔE22 barrel, we obtained outer and pore diameters, ~7.62 and ~1.52 nm, respectively, with the overall dimensions slightly reduced as compared to the Aβ1–42 barrels. The decrease in the pore diameter resulted from the inverted Lys16 side-chains that unfavorably reside in the central hydrophobic core between the β-sheets (z = ~0.5 nm). As a result, the unfavorable force exerted on the backbones of pore-lining residues slightly relocates them toward the pore axis. This causes reduction in the pore diameter and hence in the outer diameter of the barrel as well. In contrast, the conformer 1 ΔE22 barrel could preserve the overall dimensions consistent with the WT barrels, since the Lys16 side-chains are located very close to the amphipathic interface of the lipid bilayer at the lower bilayer leaflet (z = ~−1.0 nm), stretching to interact with the C-termini or phosphate groups of lipids. The heterogeneity in the dimension of ΔE22 barrels suggests that ΔE22 may be less toxic than WT Aβ1–42, since the mutant barrels with smaller pores would be populated. Recently, it was shown that ΔE22 exhibits less neurotoxicity than WT Aβ1–42 in rat primary neurons.6
It is interesting to note the implications for WT Aβ from the Osaka mutant barrels. The coordinates for conformer 1 Aβ1–42 were directly extracted from the NMR-based Aβ1–42 fibrils (PDB code: 2BEG).15 For the conformer 2 Aβ1–42, we obtained the coordinates from the Aβ1–40 protofibrils (PDB codes: 2LMN and 2LMO),16 and then added two C-terminal residues, Ile41 and Ala42, generating Aβ1–42. Although both conformers are the same U-shaped peptide with the β-strand-turn-β-strand motif, they can be distinguished by their different turns (Figure 1). This suggests that the turn at Asp23-Gly29 of the conformer 2 should belong to an intrinsic turn of Aβ1–40. A similar turn at Val24-Ala30 for Aβ1–40 was recently identified from a structural model of Aβ1–40 fibrils also using comprehensive ssNMR techniques.45 Thus, the conformer 2 Aβ1–42 adopting the Aβ1–40 turn topology is likely to be a relatively less populated conformation than the conformer 1 Aβ1–42, suggesting an explanation for the ssNMR observation for this turn for the Aβ1–40 peptide16 rather than the conformation of the Aβ1–42.15 If the WT Aβ1–40 were to adopt the turn conformation of Aβ1–42, then the C-terminal strand will be too short to create a stable U-shaped conformation. Thus, while the less C-terminal turn conformation of Aβ1–42 is more stable, the resulting shorter strand for the 1–40 sequence shifts the equilibrium toward the more C-terminal turn conformation. Recently, it was demonstrated that the C-terminal domain of Aβ1–42 showed a distinct conformational dynamics from that of Aβ1–40, which suggests that the Val36-Cly37 turn is the sine qua non of Aβ1–42.46 For the Osaka mutant the conformer with the less C-terminal turn is still more populated, since the shortened sequence with the Glu22 deletion affects the N-terminal strand, rather than the C-terminal, thus retaining an equilibrium favoring the Aβ1–42 turn.
The negatively charged Glu22 is implicated as a key site for point mutation, since amino acid substitutions at position 22 including Dutch (E22Q),8 Arctic (E22G),9,10 and Italian (E22K)11 mutants. While the Dutch mutation is closely associated with cerebral amyloid angiopathy (CAA),7 other mutants with a familial Alzheimer’s disease (FAD) linked point substitution at Glu22 are toxic species,13 suggesting that Glu22 plays a significant role in the pathogenesis of AD. However, in spite of the complete elimination of Glu22, rather than an amino acid substitution, Aβ peptide lacking Glu22 is still linked to FAD. Previously, we evaluated that in the solvated pore the charged side-chain of Glu22 plays an important role in conducting ions in the amyloid ion channel models. Our evaluation was supported by the observation that a circular cluster formed by the negatively charged Glu22 side-chains attracts cations into the pore and serves as a cationic binding site.26,28,29 The larger the ions population at the binding site, the higher the probability for ions to conduct through the water pore. However, in the absence of Glu22, the ΔE22 barrels still attract cations into the pore and show large charge fluctuations in the pore similar to the WT barrels. Although the ΔE22 barrels lose the negatively charged Glu22 side-chains at the upper bilayer leaflet, they gain the Glu11 side-chains at the lower bilayer leaflet. Thus, the emerging Glu11 side-chains attract cations into the pore and contribute to pore elongating with the β-sheet formation. To evaluate the biological role of a charged side-chain in the pore, we suggest that a circular cluster of charged side-chains can be formed with a help of ion screening. This minimizes the charge repulsions between the ions, with the circular assembly of side-chains strongly sustaining the backbone β-sheet formed by the pore-lining residues. With well-established pore, which is wide enough, ions can move freely through the water pore.
Even though the Osaka mutant that we have modeled also appears to adopt a toxic channel conformation consisting of a barrel organization of the U-shape motif, this does not necessarily imply that such conformational species are always the preferred conformational states. As we have already emphasized the amyloid landscape is highly heterogeneous17–19 and different conformations may be populated, including in membrane-permeated channels, suggesting that highly polymorphic conformations of Aβ channel could evolve from different seed formations.47–49 It is a challenge to infer all possible highly populated states for different mutants, under different conditions and membrane compositions.
To conclude, our MD simulations provide a membrane-bound conformation of the Osaka mutant barrel in atomic-level detail, illustrating that the multimeric β-barrel-like channel can be indistinguishable from the WT Aβ1–42 barrel. The U-shaped peptide with the β-strand-turn-β-strand motif supports the Osaka mutant barrel, suggesting the universality of the Aβ motif in aggregation. It has been known that due to the loss of charge, the mutant has higher hydrophobicity resulting in faster oligomerization and fibril formation.4–6 We speculate that high production of mutant oligomers can lead to toxic channel formation followed by oligomers insertion into the cell membrane in FAD. The mutant conformational dynamics along with the membrane insertion and channel formation have not been well elucidated. The conformational space of Aβ monomer, oligomer, and membrane-embedded channel states is highly polymorphic,17–19 with the mutant sharing these free energy landscapes, however with varied conformational preferences. Of particular interest, this deletion mutant suggests an explanation why the solid state NMR data for Aβ1–4016 presented a more C-terminal turn conformation versus Aβ1–42,15 and argues that the more toxic Aβ1–42 species would preferentially populate the less C-terminal turn. Those two extra residues at the C-terminus shift the free energy landscape toward the more stable Lührs et al.15 conformation. Aβ1–42 lacking Glu22 still retain these conformational preferences, since the deletion affects the conformation of the N-terminal, rather than the C-terminal strand.
Supplementary Material
Acknowledgments
This project has been funded in whole or in part with Federal funds from the Frederick National Laboratory for Cancer Research, National Institutes of Health, under contract HHSN261200800001E. This research was supported [in part] by the Intramural Research Program of NIH, Frederick National Lab, Center for Cancer Research. This research was supported by the National Institutes of Health (National Institute on Aging AG028709 to RL). All simulations had been performed using the high-performance computational facilities of the Biowulf PC/Linux cluster at the National Institutes of Health, Bethesda, MD (http://biowulf.nih.gov).
Footnotes
Initial barrel structures of MD simulations, average root-mean-squared deviation (RMSD) and fluctuation (RMSF) for backbone atoms of each monomer, time series of peptide averaged interaction energy, and time series of change in total charge in the pore. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Tomiyama T, Nagata T, Shimada H, Teraoka R, Fukushima A, Kanemitsu H, Takuma H, Kuwano R, Imagawa M, Ataka S, et al. A new amyloid β variant favoring oligomerization in Alzheimer’s-type dementia. Ann Neurol. 2008;63:377–387. doi: 10.1002/ana.21321. [DOI] [PubMed] [Google Scholar]
- 2.Nishitsuji K, Tomiyama T, Ishibashi K, Ito K, Teraoka R, Lambert MP, Klein WL, Mori H. The E693Δ mutation in amyloid precursor protein increases intracellular accumulation of amyloid β oligomers and causes endoplasmic reticulum stress-induced apoptosis in cultured cells. Am J Pathol. 2009;174:957–969. doi: 10.2353/ajpath.2009.080480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tomiyama T, Matsuyama S, Iso H, Umeda T, Takuma H, Ohnishi K, Ishibashi K, Teraoka R, Sakama N, Yamashita T, et al. A mouse model of amyloid β oligomers: their contribution to synaptic alteration, abnormal tau phosphorylation, glial activation, and neuronal loss in vivo. J Neurosci. 2010;30:4845–4856. doi: 10.1523/JNEUROSCI.5825-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Inayathullah M, Teplow DB. Structural dynamics of the ΔE22 (Osaka) familial Alzheimer’s disease-linked amyloid β-protein. Amyloid. 2011;18:98–107. doi: 10.3109/13506129.2011.580399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cloe AL, Orgel JP, Sachleben JR, Tycko R, Meredith SC. The Japanese mutant Aβ (ΔE22-Aβ(1–39)) forms fibrils instantaneously, with low-thioflavin T fluorescence: seeding of wild-type Aβ(1–40) into atypical fibrils by ΔE22-Aβ(1–39) Biochemistry. 2011;50:2026–2039. doi: 10.1021/bi1016217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ovchinnikova OY, Finder VH, Vodopivec I, Nitsch RM, Glockshuber R. The Osaka FAD mutation E22Δ leads to the formation of a previously unknown type of amyloid β fibrils and modulates Aβ neurotoxicity. J Mol Biol. 2011;408:780–791. doi: 10.1016/j.jmb.2011.02.049. [DOI] [PubMed] [Google Scholar]
- 7.Selkoe DJ, Podlisny MB. Deciphering the genetic basis of Alzheimer’s disease. Annu Rev Genomics Hum Genet. 2002;3:67–99. doi: 10.1146/annurev.genom.3.022502.103022. [DOI] [PubMed] [Google Scholar]
- 8.Levy E, Carman MD, Fernandezmadrid IJ, Power MD, Lieberburg I, Vanduinen SG, Bots G, Luyendijk W, Frangione B. Mutation of the Alzheimer’s Disease Amyloid Gene in Hereditary Cerebral-Hemorrhage, Dutch type. Science. 1990;248:1124–1126. doi: 10.1126/science.2111584. [DOI] [PubMed] [Google Scholar]
- 9.Kamino K, Orr HT, Payami H, Wijsman EM, Alonso ME, Pulst SM, Anderson L, Odahl S, Nemens E, White JA, et al. Linkage and Mutational Analysis of Alzheimer-disease kindreds for APP Gene Region. Am J Hum Genet. 1992;51:998–1014. [PMC free article] [PubMed] [Google Scholar]
- 10.Nilsberth C, Westlind-Danielsson A, Eckman CB, Condron MM, Axelman K, Forsell C, Stenh C, Luthman J, Teplow DB, Younkin SG, et al. The ‘Arctic’ APP mutation (E693G) causes Alzheimer’s disease by enhanced Aβ protofibril formation. Nat Neurosci. 2001;4:887–893. doi: 10.1038/nn0901-887. [DOI] [PubMed] [Google Scholar]
- 11.Miravalle L, Tokuda T, Chiarle R, Giaccone G, Bugiani O, Tagliavini F, Frangione B, Ghiso J. Substitutions at codon 22 of Alzheimer’s Aβ peptide induce diverse conformational changes and apoptotic effects in human cerebral endothelial cells. J Biol Chem. 2000;275:27110–27116. doi: 10.1074/jbc.M003154200. [DOI] [PubMed] [Google Scholar]
- 12.Pifer PM, Yates EA, Legleiter J. Point Mutations in Aβ Result in the Formation of Distinct Polymorphic Aggregates in the Presence of Lipid Bilayers. Plos One. 2011;6:e16248. doi: 10.1371/journal.pone.0016248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Murakami K, Irie K, Morimoto A, Ohigashi H, Shindo M, Nagao M, Shimizu T, Shirasawa T. Neurotoxicity and physicochemical properties of Aβ mutant peptides from cerebral amyloid angiopathy - Implication for the pathogenesis of cerebral amyloid angiopathy and Alzheimer’s disease. J Biol Chem. 2003;278:46179–46187. doi: 10.1074/jbc.M301874200. [DOI] [PubMed] [Google Scholar]
- 14.Ma B, Nussinov R. Stabilities and conformations of Alzheimer’s β-amyloid peptide oligomers (Aβ16–22, Aβ16–35, and Aβ10–35): Sequence effects. Proc Natl Acad Sci U S A. 2002;99:14126–14131. doi: 10.1073/pnas.212206899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lührs T, Ritter C, Adrian M, Riek-Loher D, Bohrmann B, Doeli H, Schubert D, Riek R. 3D structure of Alzheimer’s amyloid-β(1–42) fibrils. Proc Natl Acad Sci U S A. 2005;102:17342–17347. doi: 10.1073/pnas.0506723102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Petkova AT, Yau WM, Tycko R. Experimental constraints on quaternary structure in Alzheimer’s β-amyloid fibrils. Biochemistry. 2006;45:498–512. doi: 10.1021/bi051952q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miller Y, Ma B, Nussinov R. Polymorphism in Alzheimer Aβ amyloid organization reflects conformational selection in a rugged energy landscape. Chem Rev. 2010;110:4820–4838. doi: 10.1021/cr900377t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ma B, Nussinov R. Selective molecular recognition in amyloid growth and transmission and cross-species barriers. J Mol Biol. 2012;421:172–184. doi: 10.1016/j.jmb.2011.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jang H, Connelly L, Arce FT, Ramachandran S, Lal R, Kagan BL, Nussinov R. Alzheimer’s disease: which type of amyloid-preventing drug agents to employ? Phys Chem Chem Phys. 2013;15:8868–8877. doi: 10.1039/c3cp00017f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kajava AV, Baxa U, Steven AC. β arcades: recurring motifs in naturally occurring and disease-related amyloid fibrils. Faseb J. 2010;24:1311–1319. doi: 10.1096/fj.09-145979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zirah S, Kozin SA, Mazur AK, Blond A, Cheminant M, Segalas-Milazzo I, Debey P, Rebuffat S. Structural changes of region 1–16 of the Alzheimer disease amyloid β-peptide upon zinc binding and in vitro aging. J Biol Chem. 2006;281:2151–2161. doi: 10.1074/jbc.M504454200. [DOI] [PubMed] [Google Scholar]
- 22.Capone R, Jang H, Kotler SA, Connelly L, Teran Arce F, Ramachandran S, Kagan BL, Nussinov R, Lal R. All-D-Enantiomer of β-Amyloid Peptide Forms Ion Channels in Lipid Bilayers. J Chem Theory Comput. 2012;8:1143–1152. doi: 10.1021/ct200885r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Connelly L, Jang H, Arce FT, Capone R, Kotler SA, Ramachandran S, Kagan BL, Nussinov R, Lal R. Atomic force microscopy and MD simulations reveal pore-like structures of all-D-enantiomer of Alzheimer’s β-amyloid peptide: relevance to the ion channel mechanism of AD pathology. J Phys Chem B. 2012;116:1728–1735. doi: 10.1021/jp2108126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Capone R, Jang H, Kotler SA, Kagan BL, Nussinov R, Lal R. Probing structural features of Alzheimer’s amyloid-β pores in bilayers using site-specific amino acid substitutions. Biochemistry. 2012;51:776–785. doi: 10.1021/bi2017427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Connelly L, Jang H, Arce FT, Ramachandran S, Kagan BL, Nussinov R, Lal R. Effects of point substitutions on the structure of toxic Alzheimer’s β-amyloid channels: atomic force microscopy and molecular dynamics simulations. Biochemistry. 2012;51:3031–3038. doi: 10.1021/bi300257e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jang H, Arce FT, Ramachandran S, Capone R, Lal R, Nussinov R. β-Barrel topology of Alzheimer’s β-amyloid ion channels. J Mol Biol. 2010;404:917–934. doi: 10.1016/j.jmb.2010.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schulz GE. The structure of bacterial outer membrane proteins. Biochim Biophys Acta. 2002;1565:308–317. doi: 10.1016/s0005-2736(02)00577-1. [DOI] [PubMed] [Google Scholar]
- 28.Jang H, Zheng J, Nussinov R. Models of β-amyloid ion channels in the membrane suggest that channel formation in the bilayer is a dynamic process. Biophys J. 2007;93:1938–1949. doi: 10.1529/biophysj.107.110148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jang H, Zheng J, Lal R, Nussinov R. New structures help the modeling of toxic amyloidβ ion channels. Trends Biochem Sci. 2008;33:91–100. doi: 10.1016/j.tibs.2007.10.007. [DOI] [PubMed] [Google Scholar]
- 30.Jang H, Arce FT, Capone R, Ramachandran S, Lal R, Nussinov R. Misfolded amyloid ion channels present mobile β-sheet subunits in contrast to conventional ion channels. Biophys J. 2009;97:3029–3037. doi: 10.1016/j.bpj.2009.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jang H, Arce FT, Ramachandran S, Capone R, Azimova R, Kagan BL, Nussinov R, Lal R. Truncated β-amyloid peptide channels provide an alternative mechanism for Alzheimer’s disease and Down syndrome. Proc Natl Acad Sci U S A. 2010:6538–6543. doi: 10.1073/pnas.0914251107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jang H, Arce FT, Ramachandran S, Capone R, Lal R, Nussinov R. Structural convergence among diverse, toxic β-sheet ion channels. J Phys Chem B. 2010;114:9445–9451. doi: 10.1021/jp104073k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mustata M, Capone R, Jang H, Arce FT, Ramachandran S, Lal R, Nussinov R. K3 fragment of amyloidogenic β2-microglobulin forms ion channels: implication for dialysis related amyloidosis. J Am Chem Soc. 2009;131:14938–14945. doi: 10.1021/ja9049299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Capone R, Mustata M, Jang H, Arce FT, Nussinov R, Lal R. Antimicrobial protegrin-1 (PG-1) forms ion channels: MD simulation, AFM, and electrical conductance studies. Biophys J. 2010;98:2644–2652. doi: 10.1016/j.bpj.2010.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jang H, Ma B, Lal R, Nussinov R. Models of toxic β-sheet channels of protegrin-1 suggest a common subunit organization motif shared with toxic Alzheimer β-amyloid ion channels. Biophys J. 2008;95:4631–4642. doi: 10.1529/biophysj.108.134551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Quist A, Doudevski I, Lin H, Azimova R, Ng D, Frangione B, Kagan B, Ghiso J, Lal R. Amyloid ion channels: a common structural link for protein-misfolding disease. Proc Natl Acad Sci U S A. 2005;102:10427–10432. doi: 10.1073/pnas.0502066102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Woolf TB, Roux B. Molecular dynamics simulation of the gramicidin channel in a phospholipid bilayer. Proc Natl Acad Sci USA. 1994;91:11631–11635. doi: 10.1073/pnas.91.24.11631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Woolf TB, Roux B. Structure, energetics, and dynamics of lipid-protein interactions: A molecular dynamics study of the gramicidin A channel in a DMPC bilayer. Proteins. 1996;24:92–114. doi: 10.1002/(SICI)1097-0134(199601)24:1<92::AID-PROT7>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 39.Kucerka N, Tristram-Nagle S, Nagle JF. Structure of fully hydrated fluid phase lipid bilayers with monounsaturated chains. J Membr Biol. 2005;208:193–202. doi: 10.1007/s00232-005-7006-8. [DOI] [PubMed] [Google Scholar]
- 40.Brooks BR, Bruccoleri RE, Olafson BD, States DJ, Swaminathan S, Karplus M. Charmm - a program for macromolecular energy, minimization, and dynamics calculations. J Comp Chem. 1983;4:187–217. [Google Scholar]
- 41.Klauda JB, Venable RM, Freites JA, O’Connor JW, Tobias DJ, Mondragon-Ramirez C, Vorobyov I, MacKerell AD, Jr, Pastor RW. Update of the CHARMM all-atom additive force field for lipids: validation on six lipid types. J Phys Chem B. 2010;114:7830–7843. doi: 10.1021/jp101759q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Durell SR, Brooks BR, Bennaim A. Solvent-induced forces between two hydrophilic groups. J Phys Chem. 1994;98:2198–2202. [Google Scholar]
- 43.Wong PT, Schauerte JA, Wisser KC, Ding H, Lee EL, Steel DG, Gafni A. Amyloid-β membrane binding and permeabilization are distinct processes influenced separately by membrane charge and fluidity. J Mol Biol. 2009;386:81–96. doi: 10.1016/j.jmb.2008.11.060. [DOI] [PubMed] [Google Scholar]
- 44.Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kale L, Schulten K. Scalable molecular dynamics with NAMD. J Comp Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bertini I, Gonnelli L, Luchinat C, Mao J, Nesi A. A new structural model of Aβ40 fibrils. J Am Chem Soc. 2011;133:16013–16022. doi: 10.1021/ja2035859. [DOI] [PubMed] [Google Scholar]
- 46.Roychaudhuri R, Yang M, Deshpande A, Cole GM, Frautschy S, Lomakin A, Benedek GB, Teplow DB. C-terminal turn stability determines assembly differences between Aβ40 and Aβ42. J Mol Biol. 2013;425:292–308. doi: 10.1016/j.jmb.2012.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tofoleanu F, Buchete NV. Molecular interactions of Alzheimer’s Aβ protofilaments with lipid membranes. J Mol Biol. 2012;421:572–586. doi: 10.1016/j.jmb.2011.12.063. [DOI] [PubMed] [Google Scholar]
- 48.Tofoleanu F, Buchete NV. Alzheimer Aβ peptide interactions with lipid membranes: fibrils, oligomers and polymorphic amyloid channels. Prion. 2012;6:339–345. doi: 10.4161/pri.21022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jang H, Connelly L, Arce FT, Ramachandran S, Kagan BL, Lal R, Nussinov R. Mechanisms for the Insertion of Toxic, Fibril-like β-Amyloid Oligomers into the Membrane. J Chem Theory Comput. 2013;9:822–833. doi: 10.1021/ct300916f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Smart OS, Goodfellow JM, Wallace BA. THE PORE DIMENSIONS OF GRAMICIDIN-A. Biophys J. 1993;65:2455–2460. doi: 10.1016/S0006-3495(93)81293-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Frishman D, Argos P. Knowledge-based protein secondary structure assignment. Proteins. 1995;23:566–579. doi: 10.1002/prot.340230412. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.