Abstract
The Philadelphia Neurodevelopmental Cohort (PNC) is a large-scale, NIMH funded initiative to understand how brain maturation mediates cognitive development and vulnerability to psychiatric illness, and understand how genetics impacts this process. As part of this study, 1,445 adolescents ages 8–21 at enrollment underwent multimodal neuroimaging. Here, we highlight the conceptual basis for the effort, the study design, and measures available in the dataset. We focus on neuroimaging measures obtained, including T1-weighted structural neuroimaging, diffusion tensor imaging, perfusion neuroimaging using arterial spin labeling, functional imaging tasks of working memory and emotion identification, and resting state imaging of functional connectivity. Furthermore, we provide characteristics regarding the final sample acquired. Finally, we describe mechanisms in place for data sharing that will allow the PNC to become a freely available public resource to advance our understanding of normal and pathological brain development.
Keywords: neuroimaging, development, adolescence, connectome, MRI, fMRI, brain, resting-state
INTRODUCTION
Major mental illnesses are increasingly conceptualized as developmental disorders (Paus et al., 2008); 75% of all psychiatric disorders begin before age 24 (Kessler et al., 2005). Therefore, understanding the neurobiological origin of mental illness is predicated upon knowledge of how the brain develops normally, and how abnormal brain development mediates vulnerability to psychiatric symptoms (Insel, 2009). Accordingly, data regarding how both genetics and the environment “bend the curve” of brain development to confer both risk and resilience are of paramount importance. Such an endeavor requires large-scale samples with data that spans multiple levels of analysis, including genetics, neuroimaging, as well as psychiatric and cognitive assessment.
In response to this challenge, as part of the American Reinvestment and Reconstruction Act of 2009, the National Institutes of Mental Health funded an ambitious two-year collaborative study between the Center for Applied Genomics (CAG) at the Children’s Hospital of Philadelphia (CHOP; PI: Hakon Hakonarson) and the Brain Behavior Laboratory at the University of Pennsylvania (Penn, PI: Raquel E. Gur). The study design leveraged existing resources at CAG, including a subject pool of approximately 50,000 children, adolescents, and young adults who had previously been genotyped and had provided consent to be re-contacted for future research. As part of the Philadelphia Neurodevelopmental Cohort (PNC), 9,428 children ages 8–21 at enrollment were evaluated with a detailed cognitive and psychiatric assessment. A sub-sample of 1,445 participants received multi-modal neuroimaging at Penn.
Here we describe the study design and methods of the neuroimaging component of the PNC. We summarize other study components, which will be fully described elsewhere. We focus in this report on the neuroimaging recruitment process, neuroimaging methods, and informatics systems of the PNC. We conclude by discussing the PNC in relation to other large-scale neuroimaging initiatives, describe the data-sharing policies of the PNC, and introduce ongoing and planned follow-up studies. Taken together, the PNC will form a valuable, publically available resource for the study of both normal and pathological human brain development.
STUDY OVERVIEW
The PNC is a large-scale initiative that seeks to describe how genetics impact trajectories of brain development and cognitive functioning in adolescence, and understand how abnormal trajectories of development are associated with psychiatric symptomatology. Accordingly, psychiatric and cognitive phenotyping was performed on a sample of n=9,428 participants ages 8–21; a sub-sample (n=1,445) of these participants received multimodal neuroimaging as described here (Figure 1). All participants were drawn from a pool of approximately 50,000 subjects who had already been genotyped by the Center for Applied Genomics at the Children’s Hospital of Philadelphia. The participants were from the greater Philadelphia area and selected at random after stratification by sex, age, and ethnicity. Participants had been previously enrolled in a genomics study at CAG and they and/or their parents had provided informed consent (assent) to be re-contacted for participation in additional studies such as this one. The institutional review boards of both the University of Pennsylvania and the Children’s Hospital of Philadelphia approved all study procedures.
Figure 1.
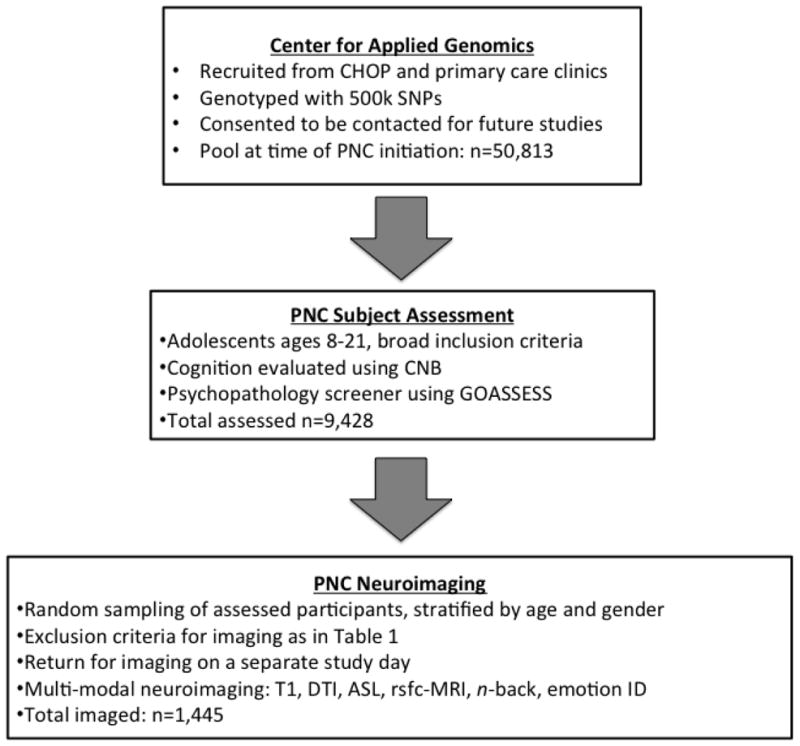
Overall study design.
Initial Participant Contact and Study Inclusion Criteria
Participants were first mailed a letter that described the study, followed by a telephone call. The purpose of the phone call, which followed a prescribed script, was to establish that the potential participant is still interested in participation and was able to participate by meeting the following minimal inclusion criteria: (a) able to provide signed informed consent (for participants under age 18 assent and parental consent were required); (b) English proficiency; and (c) physically and cognitively able to participate in an interview and computerized neurocognitive testing. The inclusion bar was set at a minimal level in order to ensure that the child can provide useful data, but children at this stage were not otherwise screened out for any specific medical or psychiatric disorder. Thus, the overall sample consists of children who came for pediatric care, gave blood for genomic studies, and consented to be contacted for future studies. Most subjects came for primary care in one of the many CHOP-affiliated clinics throughout the Delaware Valley, but the sample could be somewhat enriched by children with more complicated illnesses who received care at CHOP. Thus, the overall sample was not screened for neurological or other deficits except for such that would result in damage severe enough to cause failure to meet the inclusion criteria (e.g., pervasive developmental disorder, mental retardation, or intracranial lesions that impact the sensory, motor or mental ability to be tested). However, participants with medical problems that could impact brain function were excluded from neuroimaging (see below). Notably, the sample is not enriched by people with behavioral disorders or those who seek out participation in research by responding to advertisements. Cognitive and psychiatric assessment was conducted at home (68.8% of participants) or in the laboratory (31.2%), according to family and subject preference.
Inclusion criteria for neuroimaging
Genotyped participants who completed the initial cognitive and psychiatric phenotyping were potentially eligible for enrollment in the neuroimaging arm of the study. However, subjects were only enrolled in the neuroimaging portion of the study if they did not meet certain additional exclusion criteria. These included medical problems that could impact brain function or compromise the ability to complete the neuroimaging tasks, claustrophobia, or implanted ferrous metal (see Table 1 for details). Neuroimaging was performed in coordination with psychiatric and cognitive assessment on a separate study visit, so that on average subjects were imaged 3.3 months after assessment was completed.
Table 1.
Subject exclusion criteria
| Medical history | Severe general medical problems, including but not limited to: cancer, cerebral meningitis, cystic fibrosis, immunological conditions (e.g., lupus, common variable immunodeficiency), lead poisoning, severe liver or kidney problems, sickle cell anemia |
| Neurological/endocrine conditions | Epilepsy, stroke, loss of consciousness for more than 5 minutes, major neurodevelopmental disorders (e.g., autism), brain tumor or injury, reflex neurovascular dystrophy, Marfan syndrome, thyroid problems, Turner syndrome |
| Factors affecting ability to complete MRI tasks | History of difficulty completing cognitive battery on laptop, impaired vision or hearing. |
| Unverified metal exposure | Welding without safety goggles, injury of metallic object without proper treatment |
| General MRI contraindications | Biomedical implants, current pregnancy, dental work (e.g., braces), neurological tic disorders severe enough to prevent staying still in scanner, piercing that was not removable, known abnormal brain anatomy, significant number of amateur tattoos |
GENOMICS
All genotyping was performed at the Center for Applied Genomics as previously described. Of the 1,445 samples recruited for the imaging studies 657 were genotyped on the Illumina HumanHap 610 array; 399 on the Illumina HumanHap 550 array; 281 on the Illumina Human Omni Express array and 108 on the Affymetrix Axiom array. Samples were recruited randomly from the pool of genotyped samples, so it is unlikely that there is a genotype/phenotype platform bias. All genetic data will be imputed to the same 1KGP reference. However, data from different platforms (e.g., Affymetrix versus Illumina) will be analyzed separately and then combined using meta-analysis.
PNC ASSESSMENTS
As every participant who underwent neuroimaging was recruited from the super-set of subjects for whom medical, cognitive, and psychiatric data was available, the phentoyping available for all imaged subjects is unusually deep for a study of this scale. While the focus of this paper is on the neuroimaging component of the study, the subject-level measures are briefly summarized here. (For further details on the cognitive and psychiatric assessment, see Gur et al. 2012 and Calkins et al; In Preparation).
Computerized Neurocognitive Battery
As previously described (Gur et al., 2012), the 1-hour Penn computerized neurocognitive battery (Penn CNB) was administered to all participants. The CNB consists of 14 tests that were adapted from tasks applied in functional neuroimaging studies to evaluate a broad range of cognitive domains. These domains include executive control (abstraction and mental flexibility, attention, working memory), episodic memory (verbal, facial, spatial), complex cognition (verbal reasoning, nonverbal reasoning, spatial processing), social cognition (emotion identification, emotion intensity differentiation, age differentiation) and sensori-motor and motor speed. Except for the latter two tests that only measure speed, each test provides measures of both accuracy and speed. As described in detail in Gur et al. (2012), the CNB is sensitive to both age and sex differences in this sample.
Psychiatric Assessment
Psychopathology was assessed using a computerized structured screener (GOASSESS) that was developed from a modified version of the Kiddie-Schedule for Affective Disorders and Schizophrenia The psychopathology screener allows symptom and criterion-related assessment of mood, anxiety, behavioral, eating disorders and psychosis spectrum symptoms and substance use history. Collateral informants were included for children <18. Quality control was maintained through rigorous training, certification and monitoring.
Finally, in contrast to the assessment described above that was administered on a separate study day from the imaging session, because of the known influence of anxiety on certain functional imaging phenotypes, anxiety was assessed using the State-Trait Anxiety Inventory (STAI; (Spielberger et al., 1970)). The STAI was administered both immediately before and after the scanning session using a web-based iPad © interface.
NEUROIMAGING RECRUITMENT
It is hard to overstate the logistical challenges involved in imaging nearly 1,500 adolescent participants in a 30-month period on a single scanner. From the outset, recruitment was among the biggest challenges of this study. The recruitment strategy was adapted based on initial experience, where 30% of subjects scheduled for imaging did not arrive for their appointment.
Prior research indicates that this no-show rate, while high, is not atypical. In clinical research, reported rates of no-show range from 10–30% (Goldman et al., 1982; Lehmann et al., 2007; Neal et al., 2001). The present study had many “risk-factors” for a high no-show rate, including an adolescent sample, a racially and socioeconomically diverse subject pool, and a wide geographic catchment area (Lehmann et al., 2007; Neal et al., 2001). In response to these obstacles, the recruitment process was comprehensively revised and a dedicated imaging recruitment team, whose sole responsibility was to recruit subjects and manage the imaging schedule was established. This restructuring significantly increased the number of subjects scheduled and also lowered the no-show rate. Key adjustments included evening and weekend scanning to accommodate adolescent schedules, as well as a system of over-booking imaging slots based on no-show rate data to ensure full utilization of available scanning slots.
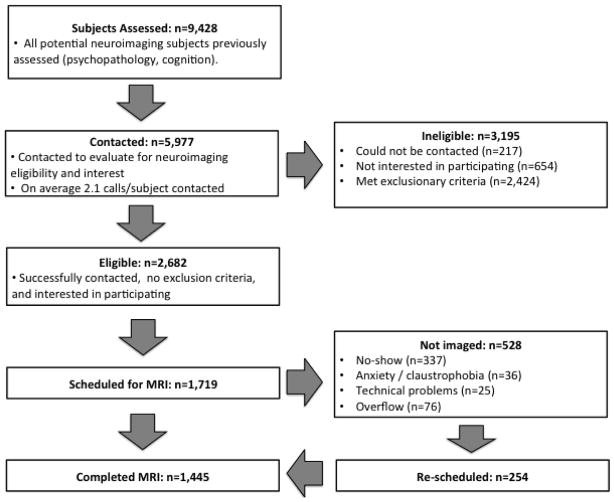
In total, PNC neuroimaging recruiters contacted nearly 6,000 of 8,500 eligible participants who completed psychiatric and cognitive assessment (Figure 2). Of the 1,409 subjects scheduled for MRI as part of the revised recruitment strategy, only 16% did not arrive for their scheduled appointment, representing a nearly 50% decline in the no-show rate. As displayed in Figure 3, the final sample imaged (n=1,445) included a broad range of subjects in the critical late childhood through adolescent period; the sample was well balanced at each age bin by sex (Figure 3A). Furthermore, the sample included relatively even proportions of Caucasians and African-Americans (Figure 3B); the diversity of the participants in the PNC is one of its major strengths.
Figure 2.
Schematic of recruitment process.
Figure 3.
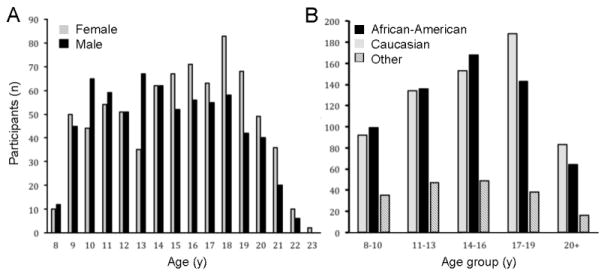
Final sample composition (n=1,445) by age and sex (A) or race (B).
NEUROIMAGING
In contrast to several other recent large-scale neuroimaging efforts (Biswal et al., 2010; Brown et al., 2012; Jack et al., 2008; Schumann et al., 2010), all imaging data from the PNC was acquired at a single site, on a single scanner, in a short period of time that did not span any software or hardware upgrades. Conversely, unlike other large-scale single-site studies (Nooner et al., 2012; Van Essen et al., 2012), due to the demanding recruitment goals and the short study timeline, there was not a dedicated development phase. Accordingly, product sequences were used, with the only exceptions being the perfusion and B0 mapping sequences, which were based on customer written routines (see below). The MRI protocol was comprised of scans designed to obtain information on brain structure, perfusion, structural connectivity, resting state functional connectivity, working memory function, and emotion identification. A measurement of static magnetic field inhomogeneity (B0 map) was also performed. The parameters of each sequence are described in Table 2. All scans were acquired with a straight magnet axial orientation (i.e. non-oblique). The total scanning time of the entire protocol was 50 minutes, 32 seconds. Scanner stability was monitored routinely over an 18-month period by calculating the mean temporal SNR using one of the BOLD sequences (fractal n-back sequence, see below) with a standard Siemens cylindrical phantom doped with nickel sulfate (see Figure 4).
Table 2.
Sequence parameters.
| Sequence | TR/TE/TI (ms) | FOV RL/AP (mm) | Matrix RL/AP/slices | Slice thick/gap (mm) | Flip angle (deg) | Reps | GRAP PA factor | BW/pixel (Hz) | PE direction | Acq time |
|---|---|---|---|---|---|---|---|---|---|---|
| MPRA GE | 1810/3.5/1100 | 180/240 | 192/256/160 | 1/0 | 9 | - | 2 | 130 | RL | 3:28 |
| PCASL | 4000/15/- | 220/220 | 96/96/20 | 5/1 | 90/180 | 80 | 2 | 2604 | AP | 5:32 |
| B0 Map | 1000/2.69+5.27/- | 240/240 | 64/64/44 | 4/0 | 60 | - | - | 500 | AP | 1:04 |
| N-Back | 3000/32/- | 192/192 | 64/64/46 | 3/0 | 90 | 231 | - | 2056 | AP | 11:39 |
| Emotion ID | 3000/32/- | 192/192 | 64/64/46 | 3/0 | 90 | 210 | - | 2056 | AP | 10:36 |
| DTI | 8100/82 | 240/240 | 128/128/70 | 2/0 | 90/180/180 | 35 | 3 | 2170 | AP | 5:24 |
| DTI | 8100/82 | 240/240 | 128/128/70 | 2/0 | 90/180/180 | 36 | 3 | 2170 | AP | 5:32 |
| Resting FC | 3000/32/- | 192/192 | 64/64/46 | 3/0 | 90 | 124 | - | 2056 | AP | 6:18 |
Figure 4.
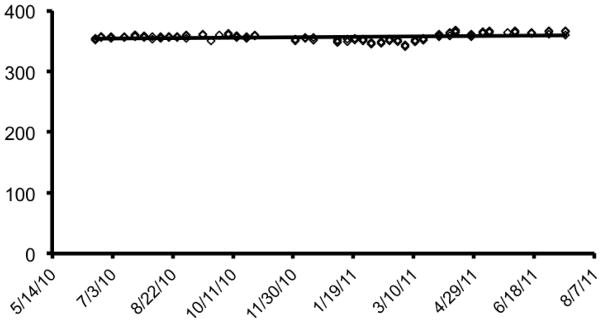
Scanner stability. Scanner stability was monitored by calculating the mean temporal SNR of the fractal n-back BOLD sequence with a standard Siemens cylindrical phantom doped with nickel sulfate.
Mock scanner
Prior to scanning, in order to acclimate subjects to the MRI environment and to help subjects learn to remain still during the actual scanning session, a mock scanning session was conducted using a decommissioned MRI scanner and head coil. Mock scanning was accompanied by acoustic recordings of the noise produced by gradient coils for each scanning pulse sequence. During these sessions, feedback regarding head movement was provided using the MoTrack motion tracking system (Psychology Software Tools, Inc, Sharpsburg, PA). Motion feedback was only given during the mock scanning session.
MRI Scanner
All MRI scans were acquired on a single 3T Siemens TIM Trio whole-body scanner located in the Hospital of the University of Pennsylvania. The system operated under the VB17 revision of the Siemens software. Signal excitation and reception was obtained using a quadrature body coil for transmit and a 32-channel head coil for receive. Gradient performance was 45mT/m, with a maximum slew rate of 200 T/m/s.
Magnetic Field Mapping
The main magnetic field (i.e. B0) was spatially mapped using a double-echo, gradient-recalled echo (GRE) sequence. Both magnitude and phase images were selected for image reconstruction since it is the phase signal which contains information about the magnetic field. Care was taken to ensure that the B0 shim settings were identical for this acquisition and subsequent BOLD scans. Furthermore, the Siemens advanced shim option was selected. This option performs multiple passes of the automated shim current optimization, resulting in improved magnetic field homogeneity across the brain. Since the Siemens product B0 mapping sequence did not support this option at the time this study was begun, a user-modified version of the multi-echo GRE sequence that enabled this feature was used instead. The field-of-view of this scan was chosen to be larger than that of the BOLD scans so that the obtained field map covered all of the volume of interest in all BOLD runs.
Structural MRI
Brain structural imaging was obtained using a magnetization prepared, rapid-acquisition gradient-echo (MPRAGE) sequence. Receive coil (i.e. B1) shading was reduced by selecting the Siemens prescan normalize option, which corrects for B1 inhomogeneity based on a body coil reference scan. Image quality assessment (QA) was performed using visual inspection, which primarily focused on identifying excessive subject motion (Table 3).
Table 3.
Images acquired and yield after QA.
| Sequence | Acquired (n) | Passed QA (n) |
|---|---|---|
| MPRAGE | 1445 | 13321 |
| pCASL | 1365 | 13301,2 |
| n-Back | 1316 | 12592 |
| Emotion ID | 1355 | 12952 |
| DTI | 1279 | 12251 |
| Resting FC | 1275 | 10281,* |
visual inspection
QA threshold of mean relative displacement >0.5mm
In ongoing analyses, due to the deleterious effects of motion on connectivity data, a more stringent exclusion criteria of MRD >0.2 mm or >20 displacements over 0.2 mm has been used, resulting in a sample of n=1018
Functional MRI
Both task-based and resting-state BOLD scans were acquired with a single-shot, interleaved multi-slice, gradient-echo, echo planar imaging (GE-EPI) sequence. In order to reach steady-state signal levels, the sequence performed two additional dummy scans at the start of the sequence. These scans were not saved to the image database. The imaging volume was sufficient to cover the entire cerebrum of all subjects, starting superiorly at the apex. In some subjects, the inferior portion of the cerebellum could not be completely included within the imaging volume. The selection of imaging parameters was driven by the goal of achieving whole brain coverage with acceptable image repetition time (i.e. TR = 3000ms). A voxel resolution of 3×3×3mm with 46 slices was the highest obtainable resolution that satisfied those constraints. Higher spatial resolution could have been obtained by adopting parallel imaging acceleration (e.g. GRAPPA), but pilot studies revealed undesirable decreases in BOLD activation with this option.
These acquisition parameters were used in three separate runs, including two task-related scans and one resting-state scan. Tasks were selected to probe working memory and affective functioning, which have been implicated in a wide range of psychiatric disorders. Tasks were administered in a counter-balanced order across the course of the study. As a probe of working-memory function, we used a fractal version of the standard n-back task (Ragland et al., 2002). The task was chosen because it has been shown to be a reliable probe of the executive system, and has the advantage of not being contaminated by lexical processing abilities that also evolve during development (Brown et al., 2005; Schlaggar et al., 2002). The task involved presentation of complex geometric figures (fractals) for 500ms, followed by a fixed interstimulus interval of 2500ms. This occurred under three conditions: 0-back, 1-back, and 2-back, producing different levels of WM load. In the 0-back condition, participants responded with a button press to a specified target fractal. For the 1-back condition, participants responded if the current fractal was identical to the previous one; in the 2-back condition, participants responded if the current fractal was identical to the item presented two trials previously. Each condition consisted of a 20-trial block (60 s); each level was repeated over three blocks. The target-foil ratio was 1:3 in all blocks with 45 targets and 135 foils overall. Visual instructions (9 s) preceded each block, informing the participant of the upcoming condition. The task included a total of 72s of rest while a fixation crosshair was displayed, which was distributed equally in three blocks of 24s at beginning, middle, and end of the task. Total working memory task duration was 11.6 minutes.
The emotion identification task is an extension of prior studies in our laboratory (Gur et al., 2007; Gur et al., 2002). It employs a fast event-related design with a jittered inter-stimulus interval (ISI). Subjects viewed 60 faces displaying neutral, happy, sad, angry, or fearful expressions, and were asked to label the emotion displayed. Briefly, the stimuli were color photographs of actors (50% female) who volunteered to participate in a study on emotion. Actors were coached by professional directors to express a range of facial expressions. For the present task, a subset of intense expressions was selected based on high degree of accurate identification (80%) by raters. Each face was displayed for 5.5 seconds followed by a variable ISI of 0.5 to 18.5 seconds, during which a complex crosshair (that matched the faces’ perceptual qualities) was displayed. Total emotion identification task duration was 10.5 minutes.
During the resting-state scan, a fixation cross was displayed as images were acquired. Subjects were instructed to stay awake, keep their eyes open, fixate on the displayed crosshair, and remain still. Total resting state scan duration was 6.2 minutes.
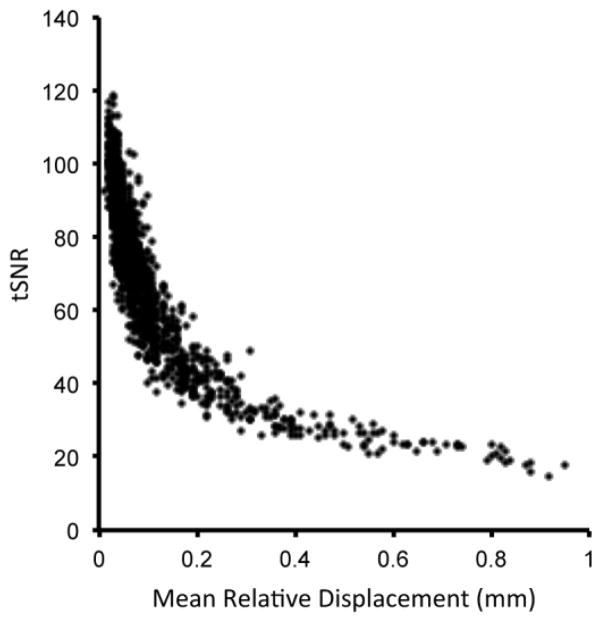
BOLD image quality was extensively assessed through custom written software that calculated the following QA metrics: temporal signal-to-noise ratio (tSNR), subject motion, global signal spike rate, and global signal drift. Voxel-wise tSNR was computed for all brain voxels by dividing the mean time course voxel amplitude by its standard deviation. Overall imaging session tSNR was computed as the average tSNR over all brain voxel. Subject motion was computed using the motion parameter estimations returned by the FSL mcflirt routine. The six motion parameters at each time point were converted to a time course measure of the relative RMS voxel displacement (Jenkinson et al., 2002). Finally, the temporal average of this time course displacement signal was used to represent overall subject motion for the session. This metric is termed the mean relative displacement (MRD), and is expressed in mm. As seen in Figure 5 strong relationship was present between tSNR and subject motion. This relationship persisted even when tSNR was computed from the motion correction image data. Earlier, we (Satterthwaite et al., 2012b) and others (Power et al., 2011; Van Dijk et al., 2011) have demonstrated that subject motion is of particular concern for rsfc-MRI data. Optimized processing techniques substantially mitigate the impact of motion (Satterthwaite et al., 2013); nonetheless a more stringent inclusion criteria for imaging data quality may be advisable (see Table 3).
Figure 5.
Relationship between tSNR and subject motion (mean relative displacement) in fractal n-back task (n=1,316).
Diffusion-weighted MRI
Diffusion weighted imaging (DWI) scans for the purpose of measuring apparent water diffusion were obtained using a twice-refocused spin-echo (TRSE) single-shot EPI sequence. The sequence employs a four-lobed diffusion encoding gradient scheme combined with a 90-180-180 spin-echo sequence designed to minimize eddy-current artifacts (Reese et al., 2003). The sequence consisted of 64 diffusion-weighted directions with b = 1000 s/mm2, and 7 scans with b = 0 s/mm2. The imaging volume was prescribed in straight magnet axial orientation with the top most slice just superior to the apex.
DWI is typically a poorly tolerated sequence, primarily due to the gradient induced table vibrations. In order to reduce the continuous duration for which the subject was required to tolerate the scan, the DWI sequence was broken into two separate imaging runs. Consequently, a 64-direction set (Jones et al., 2002) was divided into two independent sets, each with 32 diffusion-weighted directions (see Supplementary Material). Each sub-set was chosen to be maximally independent, such that they separately sampled the surface of a sphere. In addition, direction set 1 contained 3 b=0 acquisitions, and direction set 2 contained 4 b=0 acquisitions.
Image QA of the DWI data was primarily assessed by visual inspection (Table 3). Rarely, two artifacts were noted in the DTI data. Less common was image striping caused by sub-optimal gradient performance, which was the result of mechanical vibrations at the interface of the gradient cables and the magnet bore. On several occasions during the course of this study, these connections required either replacement or repair by Siemens service engineers. More commonly, images failed QA due to signal dropout caused by the interaction of subject motion and diffusion encoding.
Perfusion MRI
Brain perfusion was imaged using a custom written pseudo-continuous arterial spin labeling (pCASL) sequence (Wu et al., 2007). The sequence used a single-shot spin-echo EPI readout. Parallel acceleration (i.e. GRAPPA factor = 2) was used to reduce the minimum achievable echo time. The arterial spin labeling parameters were: label duration = 1500 ms, post label delay = 1200 ms, labeling plane = 90 mm inferior to the center slice. The sequence alternated between label and control acquisitions for a total of 80 acquired volumes (40 label and 40 control), with the first acquired volume being a label. The slices were acquired in ascending, non-interleaved order to avoid slice ordering confounds associated with interleaved schemes. In order to ensure that all slices had a similar post-label delay, slices were acquired in a compressed scheme immediately following the post-label delay, as opposed to distributing the slice acquisitions evenly throughout the TR period.
Perfusion image QA was assessed using the same tSNR and subject motion measures described for the BOLD scans (Table 3), with the addition of a visual QA of each image. While spin-echo pCASL has the advantage of a higher SNR than gradient-echo pCASL (Elliott et al., In Preparation), due to the large chemical shift of fat in the phase-encoding direction, it was observed that residual fat signal resulted in erroneous CBF quantitation, primarily in inferior occipital regions. We are developing methods to mitigate this effect, which will be described in detail in a separate report (Elliott et al., In Preparation).
INFORMATICS AND DATA MANEGMENT
Given the large quantity of data and the rapid timeline of the study, systematic procedures for data management and automated quality assurance were of critical importance. Here, we highlight several of the innovative solutions deployed as part of the PNC, including systems used for subject recruitment, image transfer, image QA and archiving, and data tracking (Figure 6).
Figure 6.
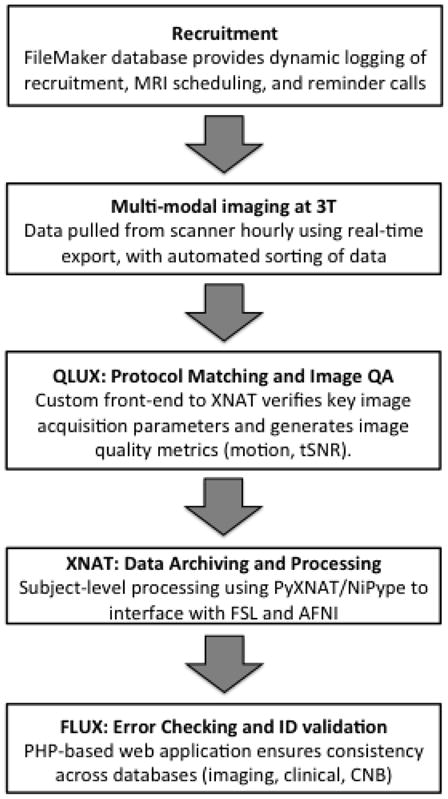
PNC informatics.
Recruitment
Given the ambitious recruitment goals, information systems to ensure an organized recruitment effort were necessary. As part of the revised recruitment strategy (see above), each communication with the participant and/or parent/guardian was logged digitally within a custom FileMaker database. This database contained all information necessary to determine eligibility and exclusion criteria, including fields for demographics, MRI compatibility, and medical exclusion criteria. All participant contacts were logged along with any relevant notes and outcomes (e.g. excluded, not interested, scheduled). Current status in the study was clearly indicated and dynamically updated (for example, “Trying to Schedule,” “Scheduled”, “Completed”, etc). Participants were scheduled into open imaging slot using a customized iCal © server that was updated every 5 minutes.
Real-Time Image Export
All dicom images generated by the MRI scanner were transferred automatically to an external hard drive using the Siemens real-time export feature. This feature sends dicom images from each sequence immediately upon completion of the scan, without the need of user interaction. Additionally the dicom files are sorted and named into a file structure allowing unambiguous identification of the subject, scan date and image series. The file structure was automatically backed up to a remote storage location via automated scripts executed nightly.
Custom XNAT Instance with Protocol Matching
The dicom image data was imported into a customized instance of the XNAT imaging informatics platform (Marcus et al., 2007). This included a customized front-end that checked the incoming dicom files for adherence to a template MRI protocol. This front-end (called QLUX) was written in the java programming language, and compared key imaging parameters (e.g. TR, TE, resolution, flip angle, etc.) of the incoming data to a pre-defined template in order to identify any errors or deviations from the scanning procedure. Scanning sessions that successfully matched the template were then automatically imported into the XNAT database. Data sets that contained errors in the protocol were flagged for manual review. Image quality metrics (motion, tSNR) were calculated automatically. The QLUX interface also associated subject responses in the fMRI tasks to the imaging data within the XNAT database. Log files from fMRI tasks were scored by a custom, hand-validated Java-based application that uses an XML description of the task stimuli, possible responses, and the classification of responses to calculate and store the scores within XNAT. Basic image processing utilized tools that are part of FSL (Jenkinson et al., 2012) and AFNI (Cox, 1996), and was completed within the XNAT framework using NiPype (Gorgolewski et al., 2011) and PyXNAT (Schwartz et al., 2012). Highly accurate registration of T1-weighted structural images to template space was achieved using DRAMMS (Ou et al., 2011). Additional processing will be tailored to the specific goals of a given analysis and be discussed in detail elsewhere.
Error checking and ID validation
A custom PHP-Based Web Application (called FLUX) was developed that alerts team members if an MRI has been completed but was not uploaded into the XNAT instance. FLUX also functions to remind the acquisition team members if assessment data was uploaded to the central FileMaker GOASSESS instance. In addition, the FLUX system mines the CNB, GOASSESS, XNAT, and enrollment databases for digit transposition, omission, or addition errors by combining the most commonly entered information in each (IDs, Age, Date of Birth, Gender) to ensure consistency across all data types. In cases where the data mismatches occurred, team members were alerted for manual review.
DISCUSSION
While the PNC is notable in many respects, it is neither the only nor the largest study of neurodevelopment. Below we consider the PNC in relation to other prior or ongoing efforts, describe PNC data-sharing policies, and introduce ongoing and planned follow-up studies.
Relationship to Other Large-Scale Neuroimaging Initiatives
At the outset, neuroimaging studies were typically of very small size, and used to localize within-subject perceptual (Kwong et al., 1992; Ogawa et al., 1992) or cognitive (Casey et al., 1995; Braver et al., 1997) manipulations. Studies of individual or group differences required larger sample sizes, but these were frequently still feasible for a single investigator. However, as neuroimaging research increasingly aims to parse cognitive function and psychiatric pathology on a dimensional basis (Insel et al., 2010) and relate brain-imaging phenotypes to genomics (Pine et al., 2010; Bigos and Weinberger, 2010), much larger sample sizes are required.
Several large-scale imaging initiatives have been completed or are ongoing. Nonetheless, the diversity of imaging and subject data available and focus on neurodevelopment differentiates the PNC from existing resources. For example, the Alzheimer’s Disease Neuroimaging Initiative (ADNI; Jack et al., 2008) has become an incredible asset to the neuroimaging community, but primarily includes older adults. The Icelandic study of healthy aging provides a similarly ambitious resource (Harris et al., 2007) to study brain aging. Alternatively, the International Neuroimaging Data-sharing Initiative (INDI; Mennes et al., 2012) and the 1,000 Functional Connectomes Project (Biswal et al., 2010) have aggregated 1,000 freely-provided rsfc-MRI scans covering the lifespan from many contributing institutions. Despite heterogeneity in acquisition protocols and substantial site effects, recent work has demonstrated the power and utility of this approach (Biswal et al., 2010; Zuo et al., 2010). Additionally, the Genome Superstruct Project (Buckner et al., 2011; Choi et al., 2012; Yeo et al., 2011) has rapidly grown to be one of the largest imaging samples available through standardization of basic imaging sequences (structural, low-resolution DTI, rsfc-MRI) among multiple participating institutions and investigators, quickly amassing over 2000 scanning sessions from a mainly young adult sample. The ongoing Nathan Kline Institute Rockland Sample (NKI-RS; Nooner et al., 2012) will provide multi-modal neuroimaging and a very detailed phenotypic characterization in a sample of over 1,000 subjects covering the entire lifespan (ages 6–85). Perhaps most ambitiously, the Human Connectome Project (HCP; Van Essen et al., 2012) combines both cutting-edge methods development and a very large sample size (n=1,200 younger adult subjects from 300 sibships) to provide an unprecedented level of detail regarding the adult brain’s connectome. One notable feature that the PNC shares with both the NKI-RS and the HCP is that all data are collected on a single system, minimizing noise introduced by site-related variability.
While the studies described above primarily consider adult subjects, several other large-scale studies of brain development exist. These include collaborative efforts such as the Saungeny Youth Study (Pausova et al., 2007) and the NIH study of normal brain development (Evans and Brain Development Cooperative Group, 2006). Both of these are very large studies of neurodevelopment that primarily focused on structural neuroimaging measures. In contrast, ongoing studies such as Pediatric Imaging, Neurocognition, and Genetics Study (PING, Brown et al., 2012) and the IMAGEN study (Schumann et al., 2010) include multi-modal neuroimaging and a host of phenotypic measures. Both of these efforts are multi-site, but otherwise in aims and scope the PNC is closely related. Clearly, for the complex problems being studied, aggregation across multiple large-scale datasets will often be required.
Data Sharing
Establishing the PNC as a publicly available resource for the study of brain development was one of the principal aims of the initiative. As noted elsewhere (Biswal et al., 2010; Gorgolewski et al., 2013; Mennes et al., 2012; Milham, 2012; Nooner et al., 2012; Bis et al., 2012; Stein et al., 2012), data sharing is a prerequisite for the collaboration necessary to gain traction towards understanding complex phenomena such as the neurodevelopmental origins of psychiatric illness. Furthermore, the richness of the data that are part of the PNC are certain to outstrip the expertise of any single research group; appropriate utilization of the PNC as a resource will require the perspectives of many investigators with complementary expertise. Accordingly, all non-identifying data acquired as part of the PNC will be made public and freely available to qualified investigators through dbGaP (Mailman et al., 2007).
As for other dbGaP resources, access to detailed subject-level genotypic and phenotypic data will require that qualified investigators submit to a data usage agreement to guard subject confidentiality in accordance with the terms of the original informed consent document signed by subjects (Mailman et al., 2007). Data in dbGaP will include genomic data, summary measures from the CNB, item level data from the GOASSESS interview, anonymized Dicom images, and imaging task log files. The compressed size of a single subject’s data is approximately 250 MB. Potentially identifying data such as free text response fields from the clinical interview were removed prior to entry into dbGaP.
Ongoing Follow-up Studies and Future Directions
Notably, because imaged participants represented a random sub-sample of the super-set of subjects who were cognitively and clinically assessed, imaging data are not available for many individuals who described symptoms of interest, such as psychosis-spectrum symptoms or depression. Accordingly, two additional studies seek to acquire imaging data on participants who endorsed psychosis-spectrum symptoms (PIs: Gur and Hakonarson) or a history of depression (PIs: Gur and Merikangas) in their GOASSESS interview, thus providing a sample that is enriched for participants of particular interest. Through these efforts, approximately 200 additional participants with psychosis-spectrum symptoms and 150 participants with depression will be imaged using the protocol described here.
Despite the large scale and deep phenotyping of the PNC, one of the main limitations of this study is its cross-sectional design. As illustrated using both simulated data (Kraemer et al., 2000) and as seen in prior studies of neurodevelopment (Evans and Brain Development Cooperative Group, 2006; Gogtay et al., 2004; Raznahan et al., 2010; Giedd et al., 1999), longitudinal data are needed for understanding trajectories of normal and abnormal brain development. Accordingly, 200 typically developing adolescents are currently being followed with longitudinal imaging using the protocol described here; approximately 100 participants with psychosis-spectrum symptoms will likewise be re-imaged longitudinally.
While the PNC imaging protocol described above provides a great diversity of brain phenotypes in a brief, well-tolerated 1-hour scanning session, such time constraints inevitably led to other measures of interest not being collected. Accordingly, both typically developing (n=75) and psychosis-spectrum (n=75) participants will be recruited for a follow-up study that focuses on amygdala dysfunction and the circuitry of fear conditioning (P50MH096891; PI: RE Gur). Furthermore, in order to relate reward system dysfunction to dimensional symptoms of anhedonia across categorical boundaries of diagnosis (Insel et al., 2010), participants with mood and/or psychotic symptoms will be imaged using dedicated tasks to probe the reward system (n=100 total; K23MH098130; PI: Satterthwaite). Finally, olfactory dysfunction in a sample of participants who endorsed psychosis-spectrum symptoms will be evaluated using an innovative combination of behavioral, molecular (using tissue from nasal biopsy), and neuroimaging probes (e.g., dedicated imaging of the olfactory bulb; NIMH R01MH099156; PI: Turetsky). Together, the combination of longitudinal follow-up, targeted recruitment, and complementary measures obtained by additional protocols will substantially enhance the richness of data available.
Supplementary Material
Highlights.
The PNC is a large-scale study of neurodevelopment, with 1,445 subjects imaged
Measures span multi-modal MRI, genomics, and testing of cognition and psychopathology
The PNC will be a public resource to study normal and pathological brain development
Acknowledgments
FINANCIAL SUPPORT: Supported by RC2 grants from the National Institute of Mental Health MH089983 and MH089924, as well as T32 MH019112. Dr. Satterthwaite was supported by NIMH K23MH098130 and the Marc Rapport Family Investigator grant through the Brain & Behavior Research Foundation. Dr. Calkins was supported by NIMH K08MH79364. Genotyping was funded in part by an Institutional Development Award to the Center for Applied Genomics from The Children’s Hospital of Philadelphia and a donation from Adele and Daniel Kubert.
Many thanks to the acquisition team, including Jeff Valdez, Raphael Gerraty, Nicholas DeLeo, and Elliot Yodh. Thanks to Rosetta Chiavacci for study coordination, and to Larry Macy for systems support. Thanks to James Dickson for XNAT development.
Footnotes
DISCLOSURES: Authors report no disclosures.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bigos KL, Weinberger DR. Imaging genetics-days of future past. Neuroimage. 2010 doi: 10.1016/j.neuroimage.2010.01.035. [DOI] [PubMed] [Google Scholar]
- Bis JC, DeCarli C, Smith AV, van der Lijn F, Crivello F, Fornage M, Debette S, Shulman JM, Schmidt H, Srikanth V, Schuur M, Yu L, Choi SH, Sigurdsson S, Verhaaren BFJ, DeStefano AL, Lambert JC, Jack CR, Struchalin M, Stankovich J, Ibrahim-Verbaas CA, Fleischman D, Zijdenbos A, den Heijer T, Mazoyer B, Coker LH, Enzinger C, Danoy P, Amin N, Arfanakis K, van Buchem MA, de Bruijn RFAG, Beiser A, Dufouil C, Huang J, Cavalieri M, Thomson R, Niessen WJ, Chibnik LB, Gislason GK, Hofman A, Pikula A, Amouyel P, Freeman KB, Phan TG, Oostra BA, Stein JL, Medland SE, Vasquez AA, Hibar DP, Wright MJ, Franke B, Martin NG, Thompson PM, Nalls MA, Uitterlinden AG, Au R, Elbaz A, Beare RJ, van Swieten JC, Lopez OL, Harris TB, Chouraki V, Breteler MMB, De Jager PL, Becker JT, Vernooij MW, Knopman D, Fazekas F, Wolf PA, van der Lugt A, Gudnason V, Longstreth WT, Brown MA, Bennett DA, van Duijn CM, Mosley TH, Schmidt R, Tzourio C, Launer LJ, Ikram MA, Seshadri S Cohorts for Heart and Aging Research in Genomic Epidemiology Consortium. Common variants at 12q14 and 12q24 are associated with hippocampal volume. Nat Genet. 2012;44:545–551. doi: 10.1038/ng.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswal BB, Mennes M, Zuo XN, Gohel S, Kelly C, Smith SM, Beckmann CF, Adelstein JS, Buckner RL, Colcombe S, Dogonowski AM, Ernst M, Fair D, Hampson M, Hoptman MJ, Hyde JS, Kiviniemi VJ, Kötter R, Li SJ, Lin CP, Lowe MJ, Mackay C, Madden DJ, Madsen KH, Margulies DS, Mayberg HS, McMahon K, Monk CS, Mostofsky SH, Nagel BJ, Pekar JJ, Peltier SJ, Petersen SE, Riedl V, Rombouts SARB, Rypma B, Schlaggar BL, Schmidt S, Seidler RD, Siegle GJ, Sorg C, Teng GJ, Veijola J, Villringer A, Walter M, Wang L, Weng XC, Whitfield-Gabrieli S, Williamson P, Windischberger C, Zang YF, Zhang HY, Castellanos FX, Milham MP. Toward discovery science of human brain function. Proc Natl Acad Sci U S A. 2010;107:4734–4739. doi: 10.1073/pnas.0911855107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braver TS, Cohen JD, Nystrom LE, Jonides J, Smith EE, Noll DC. A parametric study of prefrontal cortex involvement in human working memory. Neuroimage. 1997;5:49–62. doi: 10.1006/nimg.1996.0247. [DOI] [PubMed] [Google Scholar]
- Brown TT, Kuperman JM, Chung Y, Erhart M, McCabe C, Hagler DJ, Venkatraman VK, Akshoomoff N, Amaral DG, Bloss CS, Casey BJ, Chang L, Ernst TM, Frazier JA, Gruen JR, Kaufmann WE, Kenet T, Kennedy DN, Murray SS, Sowell ER, Jernigan TL, Dale AM. Neuroanatomical assessment of biological maturity. Curr Biol. 2012;22:1693–1698. doi: 10.1016/j.cub.2012.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown TT, Lugar HM, Coalson RS, Miezin FM, Petersen SE, Schlaggar BL. Developmental changes in human cerebral functional organization for word generation. Cereb Cortex. 2005;15:275–290. doi: 10.1093/cercor/bhh129. [DOI] [PubMed] [Google Scholar]
- Buckner RL, Krienen FM, Castellanos A, Diaz JC, Yeo BTT. The organization of the human cerebellum estimated by intrinsic functional connectivity. J Neurophysiol. 2011;106:2322–2345. doi: 10.1152/jn.00339.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casey BJ, Cohen JD, Jezzard P, Turner R, Noll DC, Trainor RJ, Giedd J, Kaysen D, Hertz-Pannier L, Rapoport JL. Activation of prefrontal cortex in children during a nonspatial working memory task with functional MRI. Neuroimage. 1995;2:221–229. doi: 10.1006/nimg.1995.1029. [DOI] [PubMed] [Google Scholar]
- Choi EY, Yeo BTT, Buckner RL. The Organization of the Human Striatum Estimated By Intrinsic Functional Connectivity. J Neurophysiol. 2012 doi: 10.1152/jn.00270.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox RW. AFNI: software for analysis and visualization of functional magnetic resonance neuroimages. Comput Biomed Res. 1996;29:162–173. doi: 10.1006/cbmr.1996.0014. [DOI] [PubMed] [Google Scholar]
- Evans AC Brain Development Cooperative Group. The NIH MRI study of normal brain development. Neuroimage. 2006;30:184–202. doi: 10.1016/j.neuroimage.2005.09.068. [DOI] [PubMed] [Google Scholar]
- Giedd JN, Blumenthal J, Jeffries NO, Castellanos FX, Liu H, Zijdenbos A, Paus T, Evans AC, Rapoport JL. Brain development during childhood and adolescence: a longitudinal MRI study. Nat Neurosci. 1999;2:861–863. doi: 10.1038/13158. [DOI] [PubMed] [Google Scholar]
- Gogtay N, Giedd JN, Lusk L, Hayashi KM, Greenstein D, Vaituzis AC, Nugent TF, Herman DH, Clasen LS, Toga AW, Rapoport JL, Thompson PM. Dynamic mapping of human cortical development during childhood through early adulthood. Proc Natl Acad Sci U S A. 2004;101:8174–8179. doi: 10.1073/pnas.0402680101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman L, Freidin R, Cook EF, Eigner J, Grich P. A multivariate approach to the prediction of no-show behavior in a primary care center. Archives of Internal Medicine. 1982;142:563. [PubMed] [Google Scholar]
- Gorgolewski K, Burns CD, Madison C, Clark D, Halchenko YO, Waskom ML, Ghosh SS. Nipype: a flexible, lightweight and extensible neuroimaging data processing framework in python. Front Neuroinform. 2011;5:13. doi: 10.3389/fninf.2011.00013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorgolewski KJ, Margulies DS, Milham MP. Making data sharing count: a publication-based solution. Front Neurosci. 2013;7:9. doi: 10.3389/fnins.2013.00009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gur RC, Richard J, Calkins ME, Chiavacci R, Hansen JA, Bilker WB, Loughead J, Connolly JJ, Qiu H, Mentch FD, Abou-Sleiman PM, Hakonarson H, Gur RE. Age group and sex differences in performance on a computerized neurocognitive battery in children age 8–21. Neuropsychology. 2012;26:251–265. doi: 10.1037/a0026712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gur RC, Richard J, Hughett P, Calkins ME, Macy L, Bilker WB, Brensinger C, Gur RE. A cognitive neuroscience-based computerized battery for efficient measurement of individual differences: standardization and initial construct validation. J Neurosci Methods. 2010;187:254–262. doi: 10.1016/j.jneumeth.2009.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gur RC, Schroeder L, Turner T, McGrath C, Chan RM, Turetsky BI, Alsop D, Maldjian J, Gur RE. Brain activation during facial emotion processing. Neuroimage. 2002;16:651–662. doi: 10.1006/nimg.2002.1097. [DOI] [PubMed] [Google Scholar]
- Gur RE, Loughead J, Kohler CG, Elliott MA, Lesko K, Ruparel K, Wolf DH, Bilker WB, Gur RC. Limbic activation associated with misidentification of fearful faces and flat affect in schizophrenia. Arch Gen Psychiatry. 2007;64:1356–1366. doi: 10.1001/archpsyc.64.12.1356. [DOI] [PubMed] [Google Scholar]
- Hakonarson H, Grant SF, Bradfield JP, Marchand L, Kim CE, Glessner JT, Grabs R, Casalunovo T, Taback SP, Frackelton EC, Lawson ML, Robinson LJ, Skraban R, Lu Y, Chiavacci RM, Stanley CA, Kirsch SE, Rappaport EF, Orange JS, Monos DS, Devoto M, Qu HQ, Polychronakos C. A genome-wide association study identifies KIAA0350 as a type 1 diabetes gene. Nature. 2007;448:591–594. doi: 10.1038/nature06010. [DOI] [PubMed] [Google Scholar]
- Han C, McGue MK, Iacono WG. Lifetime tobacco, alcohol and other substance use in adolescent Minnesota twins: univariate and multivariate behavioral genetic analyses. Addiction. 1999;94:981–993. doi: 10.1046/j.1360-0443.1999.9479814.x. [DOI] [PubMed] [Google Scholar]
- Harris TB, Launer LJ, Eiriksdottir G, Kjartansson O, Jonsson PV, Sigurdsson G, Thorgeirsson G, Aspelund T, Garcia ME, Cotch MF, Hoffman HJ, Gudnason V. Age, Gene/Environment Susceptibility-Reykjavik Study: multidisciplinary applied phenomics. Am J Epidemiol. 2007;165:1076–1087. doi: 10.1093/aje/kwk115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Insel T, Cuthbert B, Garvey M, Heinssen R, Pine DS, Quinn K, Sanislow C, Wang P. Research domain criteria (RDoC): toward a new classification framework for research on mental disorders. Am J Psychiatry. 2010;167:748–751. doi: 10.1176/appi.ajp.2010.09091379. [DOI] [PubMed] [Google Scholar]
- Insel TR. Translating scientific opportunity into public health impact: a strategic plan for research on mental illness. Arch Gen Psychiatry. 2009;66:128–133. doi: 10.1001/archgenpsychiatry.2008.540. [DOI] [PubMed] [Google Scholar]
- Jack CR, Bernstein MA, Fox NC, Thompson P, Alexander G, Harvey D, Borowski B, Britson PJ, Whitwell LJ, Ward C, Dale AM, Felmlee JP, Gunter JL, Hill DLG, Killiany R, Schuff N, Fox-Bosetti S, Lin C, Studholme C, DeCarli CS, Krueger G, Ward HA, Metzger GJ, Scott KT, Mallozzi R, Blezek D, Levy J, Debbins JP, Fleisher AS, Albert M, Green R, Bartzokis G, Glover G, Mugler J, Weiner MW. The Alzheimer’s Disease Neuroimaging Initiative (ADNI): MRI methods. J Magn Reson Imaging. 2008;27:685–691. doi: 10.1002/jmri.21049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkinson M, Bannister P, Brady M, Smith S. Improved optimization for the robust and accurate linear registration and motion correction of brain images. Neuroimage. 2002;17:825–841. doi: 10.1016/s1053-8119(02)91132-8. [DOI] [PubMed] [Google Scholar]
- Jenkinson M, Beckmann CF, Behrens TEJ, Woolrich MW, Smith SM. FSL. Neuroimage. 2012;62:782–790. doi: 10.1016/j.neuroimage.2011.09.015. [DOI] [PubMed] [Google Scholar]
- Jones DK, Williams SCR, Gasston D, Horsfield MA, Simmons A, Howard R. Isotropic resolution diffusion tensor imaging with whole brain acquisition in a clinically acceptable time. Hum Brain Mapp. 2002;15:216–230. doi: 10.1002/hbm.10018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufman J, Birmaher B, Brent D, Rao U, Flynn C, Moreci P, Williamson D, Ryan N. Schedule for Affective Disorders and Schizophrenia for School-Age Children-Present and Lifetime Version (K-SADS-PL): initial reliability and validity data. J Am Acad Child Adolesc Psychiatry. 1997;36:980–988. doi: 10.1097/00004583-199707000-00021. [DOI] [PubMed] [Google Scholar]
- Kessler RC, Berglund P, Demler O, Jin R, Merikangas KR, Walters EE. Lifetime prevalence and age-of-onset distributions of DSM-IV disorders in the National Comorbidity Survey Replication. Arch Gen Psychiatry. 2005;62:593–602. doi: 10.1001/archpsyc.62.6.593. [DOI] [PubMed] [Google Scholar]
- Kobayashi H, Nemoto T, Koshikawa H, Osono Y, Yamazawa R, Murakami M, Kashima H, Mizuno M. A self-reported instrument for prodromal symptoms of psychosis: testing the clinical validity of the PRIME Screen-Revised (PS-R) in a Japanese population. Schizophr Res. 2008;106:356–362. doi: 10.1016/j.schres.2008.08.018. [DOI] [PubMed] [Google Scholar]
- Kraemer HC, Yesavage JA, Taylor JL, Kupfer D. How can we learn about developmental processes from cross-sectional studies, or can we? Am J Psychiatry. 2000;157:163–171. doi: 10.1176/appi.ajp.157.2.163. [DOI] [PubMed] [Google Scholar]
- Kwong KK, Belliveau JW, Chesler DA, Goldberg IE, Weisskoff RM, Poncelet BP, Kennedy DN, Hoppel BE, Cohen MS, Turner R. Dynamic magnetic resonance imaging of human brain activity during primary sensory stimulation. Proc Natl Acad Sci U S A. 1992;89:5675–5679. doi: 10.1073/pnas.89.12.5675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann TNO, Aebi A, Lehmann D, Balandraux Olivet M, Stalder H. Missed appointments at a Swiss university outpatient clinic. Public Health. 2007;121:790–799. doi: 10.1016/j.puhe.2007.01.007. [DOI] [PubMed] [Google Scholar]
- Mailman MD, Feolo M, Jin Y, Kimura M, Tryka K, Bagoutdinov R, Hao L, Kiang A, Paschall J, Phan L, Popova N, Pretel S, Ziyabari L, Lee M, Shao Y, Wang ZY, Sirotkin K, Ward M, Kholodov M, Zbicz K, Beck J, Kimelman M, Shevelev S, Preuss D, Yaschenko E, Graeff A, Ostell J, Sherry ST. The NCBI dbGaP database of genotypes and phenotypes. Nat Genet. 2007;39:1181–1186. doi: 10.1038/ng1007-1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcus DS, Olsen TR, Ramaratnam M, Buckner RL. The Extensible Neuroimaging Archive Toolkit: an informatics platform for managing, exploring, and sharing neuroimaging data. Neuroinformatics. 2007;5:11–34. doi: 10.1385/ni:5:1:11. [DOI] [PubMed] [Google Scholar]
- Mennes M, Biswal BB, Castellanos FX, Milham MP. Making data sharing work: The FCP/INDI experience. Neuroimage. 2012 doi: 10.1016/j.neuroimage.2012.10.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milham MP. Open neuroscience solutions for the connectome-wide association era. Neuron. 2012;73:214–218. doi: 10.1016/j.neuron.2011.11.004. [DOI] [PubMed] [Google Scholar]
- Miller TJ, Cicchetti D, Markovich PJ, McGlashan TH, Woods SW. The SIPS screen: a brief self-report screen to detect the schizophrenia prodrome. Schizophr Res. 2004;70:s78. [Google Scholar]
- Miller TJ, McGlashan TH, Rosen JL, Somjee L, Markovich PJ, Stein K, Woods SW. Prospective diagnosis of the initial prodrome for schizophrenia based on the Structured Interview for Prodromal Syndromes: preliminary evidence of interrater reliability and predictive validity. Am J Psychiatry. 2002;159:863–865. doi: 10.1176/appi.ajp.159.5.863. [DOI] [PubMed] [Google Scholar]
- Neal RD, Lawlor DA, Allgar V, Colledge M, Ali S, Hassey A, Portz C, Wilson A. Missed appointments in general practice: retrospective data analysis from four practices. Br J Gen Pract. 2001;51:830–832. [PMC free article] [PubMed] [Google Scholar]
- Nooner KB, Colcombe SJ, Tobe RH, Mennes M, Benedict MM, Moreno AL, Panek LJ, Brown S, Zavitz ST, Li Q, Sikka S, Gutman D, Bangaru S, Schlachter RT, Kamiel SM, Anwar AR, Hinz CM, Kaplan MS, Rachlin AB, Adelsberg S, Cheung B, Khanuja R, Yan C, Craddock CC, Calhoun V, Courtney W, King M, Wood D, Cox CL, Kelly AMC, Di Martino A, Petkova E, Reiss PT, Duan N, Thomsen D, Biswal B, Coffey B, Hoptman MJ, Javitt DC, Pomara N, Sidtis JJ, Koplewicz HS, Castellanos FX, Leventhal BL, Milham MP. The NKI-Rockland Sample: A Model for Accelerating the Pace of Discovery Science in Psychiatry. Front Neurosci. 2012;6:152. doi: 10.3389/fnins.2012.00152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa S, Tank DW, Menon R, Ellermann JM, Kim SG, Merkle H, Ugurbil K. Intrinsic signal changes accompanying sensory stimulation: functional brain mapping with magnetic resonance imaging. Proc Natl Acad Sci U S A. 1992;89:5951–5955. doi: 10.1073/pnas.89.13.5951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou Y, Sotiras A, Paragios N, Davatzikos C. DRAMMS: Deformable registration via attribute matching and mutual-saliency weighting. Med Image Anal. 2011;15:622–639. doi: 10.1016/j.media.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paus T, Keshavan M, Giedd JN. Why do many psychiatric disorders emerge during adolescence? Nat Rev Neurosci. 2008;9:947–957. doi: 10.1038/nrn2513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pausova Z, Paus T, Abrahamowicz M, Almerigi J, Arbour N, Bernard M, Gaudet D, Hanzalek P, Hamet P, Evans AC, Kramer M, Laberge L, Leal SM, Leonard G, Lerner J, Lerner RM, Mathieu J, Perron M, Pike B, Pitiot A, Richer L, Séguin JR, Syme C, Toro R, Tremblay RE, Veillette S, Watkins K. Genes, maternal smoking, and the offspring brain and body during adolescence: design of the Saguenay Youth Study. Hum Brain Mapp. 2007;28:502–518. doi: 10.1002/hbm.20402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pine DS, Ernst M, Leibenluft E. Imaging-genetics applications in child psychiatry. J Am Acad Child Adolesc Psychiatry. 2010;49:772–782. doi: 10.1016/j.jaac.2009.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Power JD, Barnes KA, Snyder AZ, Schlaggar BL, Petersen SE. Spurious but systematic correlations in functional connectivity MRI networks arise from subject motion. Neuroimage. 2011;59:2142–2154. doi: 10.1016/j.neuroimage.2011.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragland JD, Turetsky BI, Gur RC, Gunning-Dixon F, Turner T, Schroeder L, Chan R, Gur RE. Working memory for complex figures: An fMRI comparison of letter and fractal< xh: i> n</xh: i>-back tasks. Neuropsychology. 2002;16:370. [PMC free article] [PubMed] [Google Scholar]
- Raznahan A, Lee Y, Stidd R, Long R, Greenstein D, Clasen L, Addington A, Gogtay N, Rapoport JL, Giedd JN. Longitudinally mapping the influence of sex and androgen signaling on the dynamics of human cortical maturation in adolescence. Proc Natl Acad Sci U S A. 2010;107:16988–16993. doi: 10.1073/pnas.1006025107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reese TG, Heid O, Weisskoff RM, Wedeen VJ. Reduction of eddy-current-induced distortion in diffusion MRI using a twice-refocused spin echo. Magn Reson Med. 2003;49:177–182. doi: 10.1002/mrm.10308. [DOI] [PubMed] [Google Scholar]
- Satterthwaite TD, Elliott MA, Gerraty RT, Ruparel K, Loughead J, Calkins ME, Eickhoff SB, Hakonarson H, Gur RC, Gur RE, Wolf DH. An improved framework for confound regression and filtering for control of motion artifact in the preprocessing of resting-state functional connectivity data. Neuroimage. 2013;64:240–256. doi: 10.1016/j.neuroimage.2012.08.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satterthwaite TD, Ruparel K, Loughead J, Elliott MA, Gerraty RT, Calkins ME, Hakonarson H, Gur RC, Gur RE, Wolf DH. Being right is its own reward: load and performance related ventral striatum activation to correct responses during a working memory task in youth. Neuroimage. 2012a;61:723–729. doi: 10.1016/j.neuroimage.2012.03.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satterthwaite TD, Wolf DH, Loughead J, Ruparel K, Elliott MA, Hakonarson H, Gur RC, Gur RE. Impact of in-scanner head motion on multiple measures of functional connectivity: Relevance for studies of neurodevelopment in youth. Neuroimage. 2012b;60:623–632. doi: 10.1016/j.neuroimage.2011.12.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlaggar BL, Brown TT, Lugar HM, Visscher KM, Miezin FM, Petersen SE. Functional neuroanatomical differences between adults and school-age children in the processing of single words. Science. 2002;296:1476–1479. doi: 10.1126/science.1069464. [DOI] [PubMed] [Google Scholar]
- Schumann G, Loth E, Banaschewski T, Barbot A, Barker G, Büchel C, Conrod PJ, Dalley JW, Flor H, Gallinat J, Garavan H, Heinz A, Itterman B, Lathrop M, Mallik C, Mann K, Martinot JL, Paus T, Poline JB, Robbins TW, Rietschel M, Reed L, Smolka M, Spanagel R, Speiser C, Stephens DN, Ströhle A, Struve M IMAGEN consortium. The IMAGEN study: reinforcement-related behaviour in normal brain function and psychopathology. Mol Psychiatry. 2010;15:1128–1139. doi: 10.1038/mp.2010.4. [DOI] [PubMed] [Google Scholar]
- Schwartz Y, Barbot A, Thyreau B, Frouin V, Varoquaux G, Siram A, Marcus DS, Poline JB. PyXNAT: XNAT in Python. Front Neuroinform. 2012;6:12. doi: 10.3389/fninf.2012.00012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spielberger CD, Gorsuch RL, Lushene RE. Manual for the state-trait anxiety inventory. 1970. Manual for the state-trait anxiety inventory. [Google Scholar]
- Stein JL, Medland SE, Vasquez AA, Hibar DP, Senstad RE, Winkler AM, Toro R, Appel K, Bartecek R, Bergmann, Bernard M, Brown AA, Cannon DM, Chakravarty MM, Christoforou A, Domin M, Grimm O, Hollinshead M, Holmes AJ, Homuth G, Hottenga J-J, Langan C, Lopez LM, Hansell NK, Hwang KS, Kim S, Laje G, Lee PH, Liu X, Loth E, Lourdusamy A, Mattingsdal M, Mohnke S, Maniega SM, Nho K, Nugent AC, O’Brien C, Papmeyer M, Pütz B, Ramasamy A, Rasmussen J, Rijpkema M, Risacher SL, Roddey JC, Rose EJ, Ryten M, Shen L, Sprooten E, Strengman E, Teumer A, Trabzuni D, Turner J, van Eijk K, van Erp TGM, van Tol M-J, Wittfeld K, Wolf C, Woudstra S, Aleman A, Alhusaini S, Almasy L, Binder EB, Brohawn DG, Cantor RM, Carless MA, Corvin A, Czisch M, Curran JE, Davies G, de Almeida MAA, Delanty N, Depondt C, Duggirala R, Dyer TD, Erk S, Fagerness J, Fox PT, Freimer NB, Gill M, Göring HHH, Hagler DJ, Hoehn D, Holsboer F, Hoogman M, Hosten N, Jahanshad N, Johnson MP, Kasperaviciute D, Kent JW, Kochunov P, Lancaster JL, Lawrie SM, Liewald DC, Mandl R, Matarin M, Mattheisen M, Meisenzahl E, Melle I, Moses EK, Mühleisen TW, Nauck M, Nöthen MM, Olvera RL, Pandolfo M, Pike GB, Puls R, Reinvang I, Rentería ME, Rietschel M, Roffman JL, Royle NA, Rujescu D, Savitz J, Schnack HG, Schnell K, Seiferth N, Smith C, Steen VM, Valdés Hernández MC, Van den Heuvel M, van der Wee NJ, Van Haren NEM, Veltman JA, Völzke H, Walker R, Westlye LT, Whelan CD, Agartz I, Boomsma DI, Cavalleri GL, Dale AM, Djurovic S, Drevets WC, Hagoort P, Hall J, Heinz A, Jack CR, Foroud TM, Le Hellard S, Macciardi F, Montgomery GW, Poline JB, Porteous DJ, Sisodiya SM, Starr JM, Sussmann J, Toga AW, Veltman DJ, Walter H, Weiner MW, Bis JC, Ikram MA, Smith AV, Gudnason V, Tzourio C, Vernooij MW, Launer LJ, DeCarli C, Seshadri S, Andreassen OA, Apostolova LG, Bastin ME, Blangero J, Brunner HG, Buckner RL, Cichon S, Coppola G, de Zubicaray GI, Deary IJ, Donohoe G, de Geus EJC, Espeseth T, Fernández G, Glahn DC, Grabe HJ, Hardy J, Hulshoff Pol HE, Jenkinson M, Kahn RS, McDonald C, McIntosh AM, McMahon FJ, McMahon KL, Meyer-Lindenberg A, Morris DW, Müller-Myhsok B, Nichols TE, Ophoff RA, Paus T, Pausova Z, Penninx BW, Potkin SG, Sämann PG, Saykin AJ, Schumann G, Smoller JW, Wardlaw JM, Weale ME, Martin NG, Franke B, Wright MJ, Thompson PM Enhancing Neuro Imaging Genetics through Meta-Analysis Consortium. Identification of common variants associated with human hippocampal and intracranial volumes. Nat Genet. 2012;44:552–561. doi: 10.1038/ng.2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dijk KRA, Sabuncu MR, Buckner RL. The influence of head motion on intrinsic functional connectivity MRI. Neuroimage. 2011;59:431–438. doi: 10.1016/j.neuroimage.2011.07.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Essen DC, Ugurbil K, Auerbach E, Barch D, Behrens TEJ, Bucholz R, Chang A, Chen L, Corbetta M, Curtiss SW, Della Penna S, Feinberg D, Glasser MF, Harel N, Heath AC, Larson-Prior L, Marcus D, Michalareas G, Moeller S, Oostenveld R, Petersen SE, Prior F, Schlaggar BL, Smith SM, Snyder AZ, Xu J, Yacoub E WU-Minn HCP Consortium. The Human Connectome Project: A data acquisition perspective. Neuroimage. 2012;62:2222–2231. doi: 10.1016/j.neuroimage.2012.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu WC, Fernández-Seara M, Detre JA, Wehrli FW, Wang J. A theoretical and experimental investigation of the tagging efficiency of pseudocontinuous arterial spin labeling. Magn Reson Med. 2007;58:1020–1027. doi: 10.1002/mrm.21403. [DOI] [PubMed] [Google Scholar]
- Yeo BTT, Krienen FM, Sepulcre J, Sabuncu MR, Lashkari D, Hollinshead M, Roffman JL, Smoller JW, Zöllei L, Polimeni JR, Fischl B, Liu H, Buckner RL. The organization of the human cerebral cortex estimated by intrinsic functional connectivity. J Neurophysiol. 2011;106:1125–1165. doi: 10.1152/jn.00338.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo XN, Kelly C, Di Martino A, Mennes M, Margulies DS, Bangaru S, Grzadzinski R, Evans AC, Zang YF, Castellanos FX, Milham MP. Growing together and growing apart: regional and sex differences in the lifespan developmental trajectories of functional homotopy. J Neurosci. 2010;30:15034–15043. doi: 10.1523/JNEUROSCI.2612-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.