Abstract
Caenorhabditis elegans is a powerful in vivo model in which transgenesis is highly developed. However, while the analysis of biological phenomena often require the expression of more than one protein of interest, no reliable tool exists to ensure efficient concomitant and equivalent expression of more than two polypeptides from a single promoter. We report the use of viral 2A peptides, which trigger a “ribosomal-skip” or “STOP&GO” mechanism during translation, to express multiple proteins from a single vector in C. elegans. Although none of the viruses known to infect C. elegans contain 2A-like sequences, our results show that 2A peptides allow the production of separate functional proteins in all cell types and at all developmental stages tested in the worm. In addition, we constructed a toolkit including a 2A-based polycistronic plasmid and reagents to generate 2A-tagged fosmids. 2A peptides constitute an important tool to ensure the delivery of multiple polypeptides in specific cells, enabling several novel applications such as the reconstitution of multi-subunit complexes.
Keywords: 2A peptide, stoichiometric delivery of multiple polypeptides, Caenorhabditis elegans, SL2, internal ribosomal entry site (IRES)
COMPLEX biological mechanisms usually involve multiple factors, and engineering expression of more than one protein of interest is often required but, although transgenesis is routinely used in Caenorhabditis elegans (Rieckher et al. 2009), no reliable tool exists to ensure the efficient concomitant and stoichiometric co-expressions of proteins. Previous attempts to overcome this issue (e.g., multiple transformations, multiple promoters::ORFs in a unique vector, or, in mammals, the use of proteolytic cleavage sites) have had limited success or efficiency, and associated technical limitations such as transcriptional interference, promoter suppression, or the expression of exogenous proteases often are toxic for cells (de Felipe 2002). In addition, a C. elegans endogenous internal ribosomal entry site (IRES)-like sequence has been used for bicistronic expression (Li and Wang 2012). However, the IRES sequence commonly used in mammals to allow co-expression of two polypeptides results in nonstoichiometric expression and creates disproportionate protein levels (de Felipe 2002; Szymczak and Vignali 2005). Endogenous bicistronic expression cassettes (resulting from trans-splicing, hereafter called SL2) have also been used in C. elegans to express two products from a single promoter (Macosko et al. 2009; Kagias et al. 2012), but the expression levels may vary and the production of multiple proteins (e.g., more than two) remains uncertain. For these reasons we decided to develop the 2A viral peptide technology for C. elegans. 2As have been used to deliver multiple proteins from a single ORF for various vertebrate models and very recently in Drosophila (Gonzalez et al. 2011; Diao and White 2012). Several studies have shown that the 2A system allows robust production of several proteins in a near-stochiometric manner (Szymczak et al. 2004; Torres et al. 2010). 2A are viral peptides encompassing the conserved consensus GDVExNPGP (Luke et al. 2008). When the ribosome encounters a 2A sequence, a “ribosomal-skip” or “STOP&GO” occurs and a first polypeptide is released while translation of the messenger RNA (mRNA) continues (Atkins et al. 2007; Doronina et al. 2008). Using this unique property, different polypeptides of interest can be cloned in frame between sequences encoding 2A viral peptides. From different viruses, four 2A peptides were discovered: F2A from the FMD virus, T2A from the Thosea asigna virus, E2A from the equine rhinitis A virus, and P2A from porcine teschovirus-1 (Luke et al. 2008). Even if 2A viral peptides seem efficient in a wide range of eukaryotic models (El Amrani et al. 2004; Szymczak et al. 2004; Doronina et al. 2008; Diao and White 2012), there is no evidence proving that this technology could function in C. elegans. Indeed, many 2A sequences have been described in mammals or insect viruses (Luke et al. 2008) but no 2A sequences have been described in viruses infecting the worm (Felix et al. 2011; Franz et al. 2012), nor did we find the 2A consensus sequence in the C. elegans genome by blast analysis, raising the possibility that C. elegans ribosome could be insensitive to such a viral hijacking strategy.
Materials and Methods
Molecular cloning
All the primers used and all the molecular constructs generated for this study are summarized in supporting information, Table S2. Standard methods were used for strain handling, maintenance, and genetic analysis. The relevant transgenic lines used in this study are described in Table S3.
The DNA sequence [acc65i::F2A::age1::T2A::not1::E2A::xho1::P2A::xba1] was synthesized by the Genscript Company and was then used to clone different ORFs in frame inside.
GFP was amplified from pPD122.53 (with Andy Fire Kit) with primers 1 and 2 and then digested with Acc65i and inserted into the F-T-E-P2A-pUC57 vector. his-24::mCherry was amplified from pD4H1Cherry vector (kindly provided by S. Kim Lab, Stanford University) with primers 3 and 4 and cloned in XbaI/SpeI in GFP::F-T-E-P2A-pUC57 vector. col-34 promoter (gatacagctagcgac. . . aaaccaccactgcataca) was subcloned and inserted in front of GFP with EcoRI sites.
Constructs to individually assess each 2A:
All the different col-34p::GFP::2A::his-24::mCherry constructs (pSJ6171-76) were generated by successive reverse PCRs using primers 5–12. P2A was mutated by site-directed mutagenesis using primers 13 and 14 to create P2A* (pSJ6175). The SL2 sequence was inserted by Megawhop (Miyazaki 2011) using primers 15 and 16 (pSJ6176). The myo-3 promoter was inserted by Megawhop (Miyazaki 2011) using primers 17 and 18 (pSJ6177-82). The ugt-22 promoter was amplified from pSJ317 and assembled by PCR fusion to GFP::T2A::mcherry::let-858 3′ UTR using primers 19+7 for PCR1, primers 20+21 for PCR2, and primers 22+24 for the final fusion PCR that was subcloned to give pSJ6187. F25B3.3 promoter was amplified from pOH273 (kindly provided by O. Hobert Lab, Columbia University) and assembled by PCR fusion to GFP::T2A::mcherry::let-858 3′ UTR using primers 19+7 for PCR1, primers 20+21 for PCR2, and primers 22+23 for the final fusion PCR that was subcloned to give pSJ6183. pSJ6188 was obtained by performing triple fusion PCR using primers 40–47 and M13R/T7 for nested PCRs, and the his-72 promoter and 3′ UTR were amplified from pDONRP4P1Rhis-72p and pDONRP2RP3his-723’UTR (kind gifts from P. Meister, Bern University).
Construct to express multiple markers and rescue Y-to-PDA TD event:
All the different marker modules were added to the previously described col-34p::GFP::F-T-E-P2A::His-24::mCherry::pUC57 vector to create pSJ6184. mCherry::PH (Pleckstrin Homology domain) was amplified from pSJ704 with primers 25 and 26 and then inserted in AgeI sites. The emr-1 gene was amplified from N2 genomic DNA with primers 30 and 31, fused to cerulean, and emr-1::cerulean was inserted by Megawhop (Miyazaki 2011). Finally, sem-4::HA complementary DNA (cDNA) (Kagias et al. 2012) was amplified with primers 32 and 33 (−HA tag included) and inserted into the NotI restriction site.
Fosmid tagging:
pBALU9, pBALU13, and pBALU20 (Tursun et al. 2009) were modified by PCR to remove the intergenic sequences and add T2A. pSJ6113 was generated by modifying pBALU9 with primers 34 and 35. pSJ6112 was generated by modifying pBALU13 with primers 34 and 36. pSJ6170 was generated by modifying pBALU20 with primers 34 and 37. pSJ6185 was generated according to Tursun et al. (2009), and the sequence NLS::mCherry::T2A was amplified from pSJ6170 using primers 38 and 39 and was used to tag fosmid WRM0622aC02 containing the ceh-6 gene (kindly provided by D. Moerman and J. Perkins, University of British Columbia).
Generic 2A containing plasmid:
pSJ6186 was made by inserting [Acc65i-F2A-Age1-T2A-Not1-E2A-Xho1-P2A-Xba1] in pSJ901 into SacI/StuI. pSJ901 contains the pPD95.75 backbone from the Andy Fire Kit as well as a multiple cloning site (MCS) and unc-54 3′ UTR, (see map and sequence in Figure S6).
Worm injections
Arrays and lines obtained after injections are summarized in Table S3. Plasmid pSJ6171-76 (col-34p) was injected at 10 ng⋅µl-1 with 50 ng⋅µl-1 of odr-1::RFP as co-injection marker (kind gift of C. I. Bargmann and P. Sengupta) and 200 ng⋅µl-1 of pBluescript. pSJ6177-82 (myo-3p) was injected at 5 ng⋅µl-1 with 200 ng⋅µl-1 pBluescript. ugt-22p and F25B33p::GFP::2A::His-24::mCherry constructs were injected at 5 ng⋅µl-1 with 100 ng⋅µl-1 of pRF-4 (rol-6) co-injection marker. pSJ6184 was injected at 10 ng⋅µl-1 with 50 ng⋅µl-1 of odr-1::RFP and 200 ng⋅µl-1 of pBluescript to obtain IS2070 and IS2008. pSJ6185 was injected at 20 ng⋅µl-1 with 5 ng⋅µl-1 of myo-3::GFP co-injection marker and 50 ng⋅µl-1 of pBluescript. pSJ6188 was injected at 1 ng⋅µl-1 along with 2 ng⋅µl-1 pRF4 and 120 ng⋅µl-1 sonicated genomic DNA.
Microscopy and imaging
DIC and epifluorescence observations were performed on in vivo worms using a Zeiss Z1 imager microscope. The presence and possible colocalization of the fluorescent markers at the plasma membrane, nuclear membrane, and in the cytoplasm and/or the nucleus were scored in individual cells expressing the 2A construct. Unexpected localization, possibly due to a “self-cleavage” defect, was scored irrespective of the intensity levels. To determine with more precision the localization of each marker, and the colocalization of GFP and HISTONE::mCherry in the nucleus, which could be indicative of the presence of a fusion protein between GFP and HISTONE::mCherry, a Leica SP5 and SP2 MP confocal microscopes were used. The presence of the motor neuron named PDA was scored as previously described (Zuryn et al. 2010).
Western blots
For each condition, one 100-mm enriched peptone plate [1.2 g sodium chloride, 20 g peptone, 25 g agar in 1 liter—autoclaved and cooled to 55°, after which 1 ml cholesterol (5 mg/ml in EtOH), 1 ml 1 M MgSO4, and 25 ml 1 M potassium phosphate (pH 6.0) were added] with 50 µg/ml streptomycin, and HB101 bacteria was used to grow worms. The entire plate was harvested before starvation with M9 buffer (3 g KH2PO4, 6 g Na2HPO4, 5 g NaCl, 1 ml 1 M MgSO4, H2O to 1 liter) and washed two times with M9 and one time with H2O. Pellets were resuspended in 1 ml of sample buffer 1×, snap-frozen in liquid nitrogen, and sonicated 3 × 4 min at 5–8% intensity (MSE Soniprep 150 Plus with exponential probe). Before loading on 10% electrophoresis gels, samples were boiled for 5 min at 100° and centrifuged 13,000 × g for 15 min. GFP was detected using the mouse monoclonal α-GFP (MAB3580-Chemicon) antibody (1/2000e dilution), and mCherry was detected by the mouse monoclonal α-mCherry (1C51, NovusBio) antibody (1/1000e dilution) and a chicken–α-mouse HRP (1/10,000e dilution to minimize background noise). For HA tag detection, an α-HA-HRP-conjugated antibody (6E2, Cell Signaling) was used. For normalization, an α-α-tubulin (DM1A, Sigma) antibody was used.
Results and Discussion
Canonical 2A peptides efficiently allow the production of independent polypeptides in C. elegans
To assess whether the worm can be sensitive to 2A viral peptide technology, we tested the canonical 2A viral peptides used to deliver multiple products in various organisms (F2A, T2A, E2A, and P2A; see Figure S1). As a negative control, a single amino acid was mutated in P2A (NPGP to NPAP) to create the split-up inactive P2A* (Hahn and Palmenberg 1996). A SL2 trans-splicing sequence (Macosko et al. 2009; Kagias et al. 2012) was used as a control to follow the production of two independent products (Figure S1 and Figure S2). A GFP marker (addressed to the whole cell) was cloned in frame with each of the 2A peptides and HISTONE::mCherry (strictly addressed to the nucleus; see also Figure S1) under the col-34 promoter, which drives expression in rectal epithelial cells (Liu et al. 2009; Kagias et al. 2012). Constructs were injected, and several different lines were analyzed (Table S1; Table S2; Table S3).
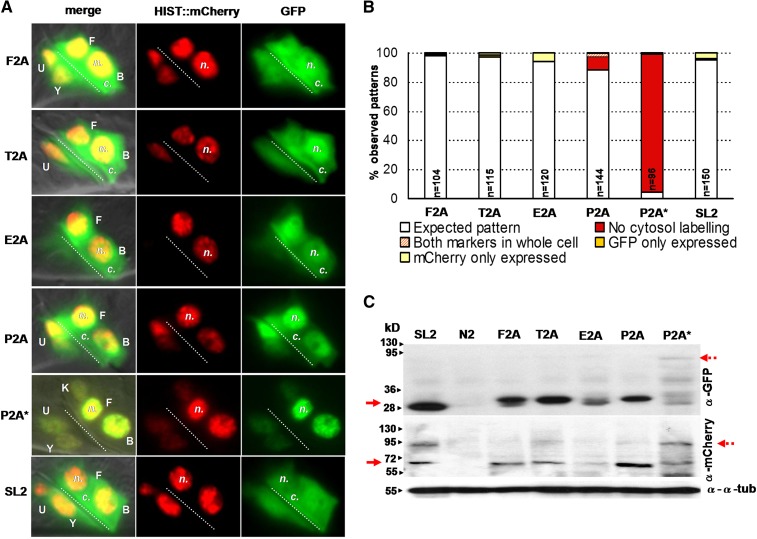
For all of the 2A viral peptides, a clearly different subcellular localization pattern was observed for GFP and HISTONE-mCherry. Indeed, GFP was found within the whole cell whereas mCherry exclusively localized to the nucleus. These observations suggest that F2A, T2A, E2A, and P2A peptides have the ability to trigger efficient self-cleavage in the worm (Figure 1A; Figure S1; Figure S2). A quantification of the localization patterns of these two proteins in rectal cells of early L1 worms is shown in Figure 1B and confirms the high level of self-cleavage efficiency for the four different 2As, as 88–98% of the cells, depending on the 2A sequence used, exhibited the expected localization pattern for two independent products (Figure S1). By contrast, the mutated version of P2A (P2A*) led to strictly nuclear GFP and mCherry, suggesting an abolition of the self-cleavage activity of P2A (Figure 1, A and B; Figure S1; Figure S2).
Figure 1.
Each 2A viral peptide allows the efficient production of two independent polypeptides in C. elegans. (A) Patterns of localization of GFP and HISTONE::mCherry in the rectal cells of L4 worms expressing col-34p::GFP::2A::histone::mCherry. GFP localizes within the whole cell whereas HISTONE-mCherry (HIST::mCherry) remains tightly restricted to the nucleus as expected if they are produced as two independent polypeptides, suggesting that all 2A peptides are correctly split up. For the mutant P2A* peptide, both markers are restricted to the nucleus as expected if the split-up activity is abolished. SL2, used as a positive control, triggers the production of two RNA species. In the merge column, the rectal cells B, F, U, K, and Y are labeled (capital letters); the nucleus (n.) and cytosol (c.) are labeled in all columns; the dotted line indicates the rectal slit; anterior is to the left and ventral to the bottom. (B) Quantification of the localization patterns observed for col-34p::GFP::2A::histone::mCherry in rectal cells of L1 worms; n, number of cells scored. (C) Western blot analysis showing the production of independent GFP and HISTONE-mCherry proteins. Red arrows on the left indicate GFP and HISTONE-mCherry bands detected at ∼30 and 60 kDa, respectively; dashed arrows on the right indicate noncleaved product. To obtain enough protein to perform the Western blot analyses, we used the myo-3 promoter, which drives expression in the 81 BWM cells of the worm; α-tubulin was used as a loading control; a SL2 trans-splicing line, resulting in the expression of two independent products, was used as a reference. Note that GFP migrates slightly slower in extracts from transgenic 2A lines as the majority of the 2A peptide remains fused to the C terminus of GFP. All transgenic lines obtained are listed in Table S3.
To analyze more precisely the self-cleaving efficiency mediated by 2A peptides, Western blot analyses were performed (Figure 1C). F2A, T2A, E2A, and P2A lines showed a highly efficient self-cleavage activity: independent bands were detected for GFP and HISTONE-mCherry (Figure 1C), comparable to the bands seen using the SL2 line. For P2A*, a product was observed at ∼95 kDa with α-GFP and α-mCherry antibodies, which likely corresponds to a noncleaved product composed of both proteins. Consistent with previous studies, we observed a high amount of protein degradation of the P2A* product (Hahn and Palmenberg 1996), which correlated with the decrease in expression observed using confocal microscopy (Figure S2). A barely detectable band that could correspond to a potential uncleaved product was sometimes observed using the α-mCherry antibody in T2A, E2A, and P2A lines (Figure 1C), suggesting that, for these 2A viral peptides, a small residual fraction of noncleaved product may exist, as reported in some other models (Kim et al. 2011). In sum, we found that each of the canonical F2A, T2A, E2A, and P2A viral peptides work efficiently in C. elegans.
2A peptides are working at all developmental stages and tissues tested
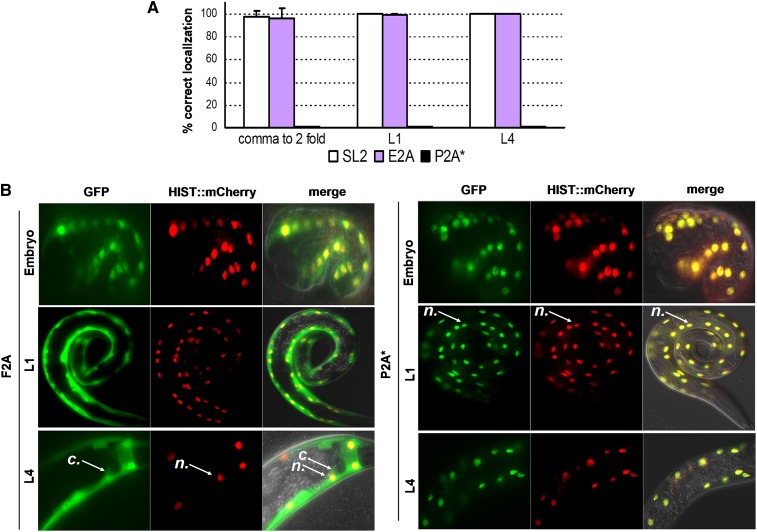
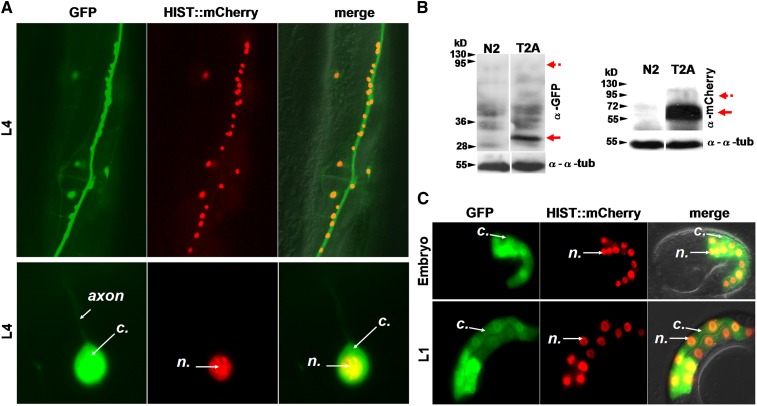
We next examined whether self-cleavage is observed at different developmental stages and in most tissues. To test whether 2As can work all along the development of the worm, we generated and scored a myo-3p::GFP::E2A::histone::mCherry line. GFP localized to the whole cell whereas HISTONE::mCherry remained exclusively in the nucleus from the embryonic comma to the L4 stage, suggesting an efficient self-cleavage of both markers at all stages (Figure 2A and Figure S3). In the negative control P2A* lines, no split between GFP and mCherry was observed, and both markers were found in the nucleus (Figure 2B). We confirmed these results using another 2A sequence (myo-3p::GFP::F2A::histone::mCherry lines, Figure 2B). To extend our results and assess their efficiency in the main tissues of the worm in addition to epithelial (Figure 1A and Figure S2) and muscle (Figure 1C and Figure 2, A and B) cells, the ugt-22p [intestinal cells (Liu et al. 2009)], F25B3.3p [pan-neuronal (Chen et al. 2011)], and his-72p [ubiquitous (Liu et al. 2009)] promoters were used. We found that the T2A peptide was efficiently active in neural cells in both epifluorescence microscopy and Western blot analyses (Figure 3, A and B). No residual noncleaved product was observed (Figure 3B). The T2A peptide was also validated for endodermal tissue (Figure 3C). Finally, using the ubiquitous his-72 promoter, we found that F2A allowed the production of two separate polypeptides in all cells examined (Figure S4). In sum, we showed that the canonical 2A viral peptides were efficiently used to deliver two products of interest throughout development in most, if not all, tissues.
Figure 2.
Each 2A viral peptide allows the efficient production of two independent polypeptides at different developmental stages. (A) Quantification at different developmental stages of the expected localization patterns (GFP in the whole cell, HISTONE::mCherry in the nucleus) observed in lines expressing myo-3p::GFP::E2A::histone::mCherry, myo-3p::GFP::P2A*::histone::mCherry, or myo-3p::GFP::SL2::histone::mCherry. (B) Patterns of localization of GFP and HISTONE::mCherry in muscle cells of myo-3p::GFP::F2A::histone::mCherry or myo-3p::GFP::P2A*::histone::mCherry transgenic worms at the indicated developmental stages. n, nucleus; c, cytosol. See Table S3 for information on the different transgenic lines obtained.
Figure 3.
2A viral peptides allow the efficient production of two independent polypeptides in different tissues. (A) Localization pattern of GFP and HISTONE::mCherry in neuronal cells in transgenic lines expressing a F25B3.3p::GFP::T2A::histone::mCherry construct. GFP localizes within the whole cell whereas HISTONE-mCherry remains tightly restricted to the nucleus. n, nuclear; c, cytosol. The ventral midbody area of the worm is shown. The worm is oriented vertically with its tail to the bottom. (B) Western blot analysis showing the production of independent GFP and mCherry proteins from a T2A construct in neuronal cells. The GFP and HISTONE-mCherry products, detected at 30 and 60 kDa, respectively, are indicated by red arrows; dotted arrows indicate the expected size for an uncleaved product; α-tubulin was used as a loading control. (C) Localization pattern of GFP and HISTONE::mCherry in intestinal cells of embryos and L1 larvae in lines expressing a ugt-22p::GFP::T2A::histone::mCherry construct. GFP localizes within the whole cell whereas HISTONE-mCherry remains tightly restricted to the nucleus. n, nuclear; c, cytosol. The midbody area of L1 larvae oriented vertically with the tail to the bottom is observed. See Table S3 for information on the different transgenic lines obtained.
2A peptides can simultaneously deliver five distinct functional products
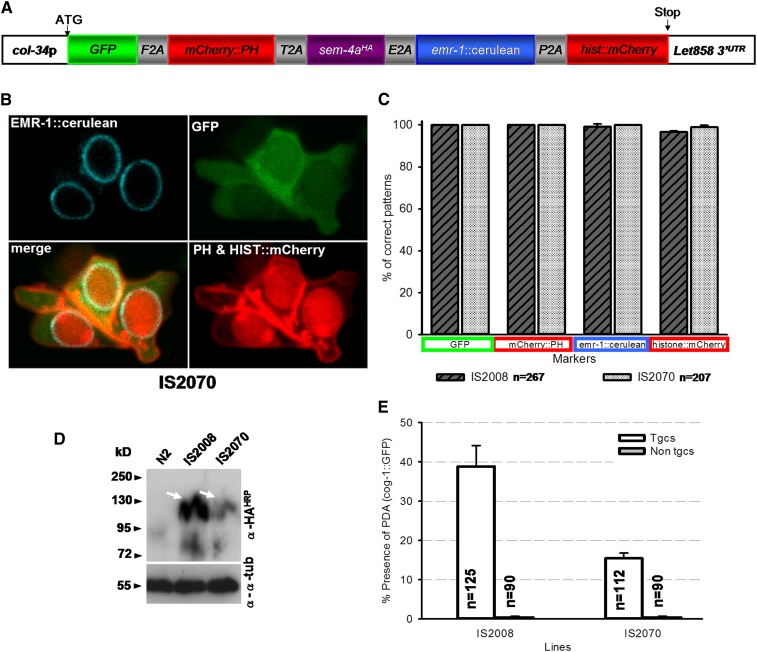
After validation of each of the 2A viral peptides for the worm model, we decided to test their potential to deliver several functional proteins. Four fluorescent proteins, addressed to different cellular compartments (Figure 4A), were cloned in frame with the F2A, T2A, E2A, and P2A peptides, and a rescuing cDNA was incorporated in the middle of the construct (pSJ6184, Figure 4A). For this proof-of-principle experiment, we decided to rescue a specific biological process: the Y-to-PDA transdifferentiation event, a model developed and characterized in the laboratory, where a single rectal epithelial cell called Y becomes a motorneuron called PDA during the L2 stage (Richard et al. 2011; Hajduskova et al. 2012; Zuryn et al. 2012) We have shown that SEM-4 (a homolog of SALL) is crucial for the initiation of this reprogramming event as a member of a NODE-like complex (Jarriault et al. 2008; Kagias et al. 2012). The sem-4(n1971) mutant, in which Y-to-PDA is 100% defective, can be rescued by expressing col-34p::sem-4a::HA::SL2::mCherry (Kagias et al. 2012). Confocal imaging confirmed the correct subcellular localization of each fluorescent protein in two independent transgenic lines transformed with pSJ6184, suggesting that each of the different products were separated and correctly addressed to the expected cellular compartment (Figure 4, B and C). In the two lines, a single band at ∼100 kDa was detected for SEM-4a::HA by Western blot (Figure 4D), which allowed the rescue of the Y-to-PDA mutant phenotype of the sem-4(n1971) mutant (Figure 4E). Thus, delivering multiple products coupled to different 2A peptides within the cell of interest does not impair the ability of sem-4 to rescue the Y-to-PDA process, and 2A viral peptides (F2A, T2A, E2A, and P2A) can be used to deliver simultaneously five different functional proteins in the worm.
Figure 4.
2A viral peptide technology allows simultaneous delivery of multiple independent and functional polypeptides in the worm. (A) Schematic view of the molecular construct encompassing five products fused in frame with F2A, T2A, E2A, and P2A. GFP is addressed to the whole cell, mCherry::PH (Pleckstrin Homology domain) is addressed to the cellular membrane, EMR-1::cerulean (Ce-emerin) is addressed to nuclear membrane (Lee et al. 2000), and HISTONE::mCherry is addressed to the nucleus. col-34p was used to drive expression in the rectal cells (Kagias et al. 2012). (B) Confocal imaging of rectal cells of early L1s expressing col-34p::GFP::F2A::mCherry-PH::T2A::sem-4A-HA::E2A::emr-1::cerulean::P2A::histone::mCherry. All the markers are addressed to the expected cellular compartments (see also Figure S5); the Y, B, and F rectal cells are shown; anterior is to the left and ventral to the bottom. (C) Quantification of the presence of all five markers at the correct compartment. We noted in one of the two lines (IS2008) that the fifth protein, HISTONE::mCherry, is not seen in 3.3% of the cells scored. n, total number of cells scored. (D) Western blot analysis showing the presence of SEM-4a::HA as one band at ∼100 kDa as expected (white arrow) in the two transgenic independent lines IS2008 and IS2070. (E) Rescuing efficiency of the sem-4(n1971) “no PDA” defect in the two independent lines IS2008 and IS2070 expressing col-34p::GFP::F2A::mCherry-PH::T2A::sem-4A-HA::E2A::emr-1::cerulean::P2A::histone::mCherry. Interestingly, the rescuing efficiency for each line correlated with the levels of expression of SEM-4a-HA detected by Western Blot. For each experiment, nontransgenic (Non tgcs) siblings were 100% PDA defective. n, number of animals scored.
A 2A-based toolkit for C. elegans
We built a toolkit to facilitate the use of the 2A viral technology in C. elegans. A plasmid pSJ6186 encompassing the in-frame F2A, T2A, E2A, and P2A sequences was generated (Figure S6) in which cloning of different products can easily be performed using unique restriction sites or using PCR-based cloning. In addition, as fosmids are useful reagents to follow gene or protein expression and new tagging methods have emerged (Tursun et al. 2009), we generated recombineering cassettes to insert T2A in frame with GFP (pSJ6113), YFP (pSJ6112), or mCherry (pSJ6170) in fosmids (Figure S6). As a proof of concept, using these reagents, we tagged the ceh-6 gene in frame with NLS::mCherry::T2A within a fosmid. The expression driven by the tagged fosmid recapitulates the pattern observed with antibody staining on the endogenous protein (Burglin and Ruvkun 2001), including expression in the rectal cells (Figure S7), providing a new useful reagent as smaller genomic fragments encompassing the ceh-6 locus did not recapitulate the CEH-6 antibody staining pattern.
C. elegans is a powerful model to perform genetic studies and microscopic observations that need to be coupled to innovative molecular biology techniques. However, there is a lack of tools to express multiple products simultaneously within the same cell. As we demonstrate, the 2A viral technology is a promising tool for multiple concomitant protein expression. We demonstrate that each of the four distinct 2A peptides used drives, with high efficiency, the production of separate polypeptides in worms and with no associated cytotoxic effect. In addition, they allowed the successful expression of five different functional proteins in the worm that were addressed to different subcellular localizations while at the same time rescuing a biological process, the Y-to-PDA transdifferentiation (Jarriault et al. 2008; Kagias et al. 2012).
Because 2A peptides are easy to use, reliable, and allow the simultaneous and near-stoichiometric production of multiple proteins, they represent a useful tool for nematode bi- or multi-cistronic transgenic or genome engineering approaches. The 2A polycistronic technology has several distinct advantages that distinguish it from other alternatives, such as endogenous trans-splicing-based bicistronic expression. First, 2A sequences are shorter than any other intergenic operon sequences facilitating their cloning. In addition, the same regulatory sequences, promoter and 3′ UTR regulate the expression of all of the ORFs in the construct, thus reducing the possibility of varying expression levels within the polycistron. Furthermore, these short 2A sequences have not been associated with the presence of potential cis-regulating elements. This is in contrast to the SL2 approach as operon sequences may be under the influence of cis-regulating elements (enhancers or silencers) that are difficult to predict (Pfleger et al. 2006). Moreover, when using an SL2-based approach, several additional parameters can impact on the expression levels of protein products, which could vary widely. For example, after trans-splicing, each subsequent mRNA molecule may have its own specific stability, and each transcript is read individually by different ribosomes, permitting variable translation. Nevertheless, SL2 sequences have successfully been used to express two distinct products in C. elegans although simultaneous expression of more than two genes remains to be demonstrated. As stated in the Introduction, an IRES-like sequence has also been tested in C. elegans, but further studies will be needed to assess the expression efficiency of the downstream product related to the upstream one (Li and Wang 2012). Similarly, IRES use is limited to bicistronic expression strategies and has been shown in other species to result in very different protein levels (de Felipe 2002).
However, there are two possible downsides associated with the use of 2A peptides. Indeed, an important aspect specific to the 2A strategy is that 2A peptides leave a proline on the downstream protein as well as a 17- to 22-amino acid “tag” on the upstream protein, a feature that could alter protein function. Studies in mammals have shown that, for proteins passing through the ER, inclusion of a furin proteinase cleavage site between the upstream protein and the 2A results in the efficient removal of the 2A peptide if the presence of a C-terminal peptide happens to be an issue (Fang et al. 2005). For proteins in which activity is impaired by the addition of a C-terminal peptide, one could envision converting this feature into an advantage and engineering a conditional change in protein activity by the addition of an endopeptidase site between the 2A peptide and the protein of interest and by spatially or temporally restricting expression of the endopeptidase. In addition, as specific antibodies recognizing these 2A tags have been developed, the 2A peptide left could be used to detect the upstream gene product or to distinguish a shift in protein size between the products of different alleles. Finally, we found that, in one of our transgenic lines carrying a polycistronic 2A transgene, the fifth protein, HISTONE::mCherry, is absent in cells in very rare instances (3.3%). However, in these cells, the other fluorescent markers are present and correctly localized with no apparent drop in their intensity, suggesting that translation of the downstream gene only is affected. Furthermore, the rare absence of a downstream product did not appear to negatively impact on the rescuing ability of one of the upstream gene products. It is not known why a protein corresponding to a downstream gene could be missing in these rare cells. It is possible that the structural properties of the N terminus of the downstream gene product, or the specific 2A sequence used last, may influence its translation.
The efficiency of producing two separate polypeptides in C. elegans, as judged by live fluorescence and Western blots, appears to be very high: for example, >90% of the cells expressing our constructs showed perfect subcellular localization of the individual fluorescent proteins. Because an uncleaved product can sometimes be detected, but not always, as already described in other species (Hasegawa et al. 2007; Provost et al. 2007; Gonzalez et al. 2011; Kim et al. 2011), we cannot exclude that, in rare cases, low amounts of an uncleaved product could be associated with a dominant negative activity. It should be noted that this has never been described, while the multi-subunit T-cell receptor, or cocktails of pluripotency factors, for example, have been delivered to cells with no adverse effects (Szymczak et al. 2004; Carey et al. 2009). Different efficiencies have been reported for a given 2A peptide in different species (Kim et al. 2011). This could be due to the phylogenetic distance between the species naturally infected by the 2A-containing virus and the species tested. Although some of the 2A peptides used in this study come originally from viruses infecting species that may be evolutionarily closer to C. elegans (for example, the T2A sequence is found in the T. asigna butterfly virus), we did not observe this to be correlated with their efficiency to produce separate polypeptides in C. elegans. It remains possible that 2A peptides found in viruses that would naturally infect C. elegans, if they exist, may ensure 100% self-cleavage efficiency in all cells. Because we found that 2A peptides are able to hijack the C. elegans ribosome that has probably never seen them, our results suggest additionally that the 2A peptide technology will likely be of use in other non-C. elegans nematode species where transgenesis and/or CRISPR/Cas9 genome engineering (Dickinson et al. 2013) are accessible.
Thus, 2A peptides constitute an invaluable tool for nematode researchers, and we can predict a number of interesting applications. For example, they will be the tool of choice to label a cell of interest while delivering one or more polypeptides without interfering with protein activity and to ensure that the products are expressed in the right cell(s); to label a different compartment in one cell and simultaneously assay multiple parameters; to underline the expression of different splicing products, for example, by tagging endogenous loci; to reconstitute a signaling cascade; or to express with a given stoichiometry different subunits of a complex or a receptor.
Supplementary Material
Acknowledgments
We thank B. Reina San Martin for helpful discussions; S. Zuryn, T. Daniele, M. C. Morin for the critical reading of this manuscript; P. Melenec for technical advice; and M. Koch and the Institut de Génétique et de Biologie Moléculaire et Cellulaire imaging facility for help with confocal microscopy. This work was supported by an award from the Fondation Générale de Santé/Académie des Sciences and by grants from the Fondation pour la Recherche Médicale (FRM–DRC20091217181), and the Association Française contre les Myopathies and the Fondation ARC pour la Recherche sur le Cancer (to S.J.). S.J. is an investigator of the Centre National de la Recherche Scientifique.
Footnotes
Communicating editor: O. Hobert
Literature Cited
- Atkins J. F., Wills N. M., Loughran G., Wu C. Y., Parsawar K., et al. , 2007. A case for “StopGo”: reprogramming translation to augment codon meaning of GGN by promoting unconventional termination (Stop) after addition of glycine and then allowing continued translation (Go). RNA 13: 803–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burglin T. R., Ruvkun G., 2001. Regulation of ectodermal and excretory function by the C. elegans POU homeobox gene ceh-6. Development 128: 779–790. [DOI] [PubMed] [Google Scholar]
- Carey B. W., Markoulaki S., Hanna J., Saha K., Gao Q., et al. , 2009. Reprogramming of murine and human somatic cells using a single polycistronic vector. Proc. Natl. Acad. Sci. USA 106: 157–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Fu Y., Ren M., Xiao B., Rubin C. S., 2011. A RasGRP, C. elegans RGEF-1b, couples external stimuli to behavior by activating LET-60 (Ras) in sensory neurons. Neuron 70: 51–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Felipe P., 2002. Polycistronic viral vectors. Curr. Gene Ther. 2: 355–378. [DOI] [PubMed] [Google Scholar]
- Diao F., White B. H., 2012. A novel approach for directing transgene expression in Drosophila: T2A-Gal4 in-frame fusion. Genetics 190: 1139–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson D. J., Ward J. D., Reiner D. J., Goldstein B., 2013. Engineering the Caenorhabditis elegans genome using Cas9-triggered homologous recombination. Nat. Methods 10: 1028–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doronina V. A., Wu C., de Felipe P., Sachs M. S., Ryan M. D., et al. , 2008. Site-specific release of nascent chains from ribosomes at a sense codon. Mol. Cell. Biol. 28: 4227–4239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Amrani A., Barakate A., Askari B. M., Li X., Roberts A. G., et al. , 2004. Coordinate expression and independent subcellular targeting of multiple proteins from a single transgene. Plant Physiol. 135: 16–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang J., Qian J. J., Yi S., Harding T. C., Tu G. H., et al. , 2005. Stable antibody expression at therapeutic levels using the 2A peptide. Nat. Biotechnol. 23: 584–590. [DOI] [PubMed] [Google Scholar]
- Felix M. A., Ashe A., Piffaretti J., Wu G., Nuez I., et al. , 2011. Natural and experimental infection of Caenorhabditis nematodes by novel viruses related to nodaviruses. PLoS Biol. 9: e1000586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franz C. J., Zhao G., Felix M. A., Wang D., 2012. Complete genome sequence of Le Blanc virus, a third Caenorhabditis nematode-infecting virus. J. Virol. 86: 11940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez M., Martin-Ruiz I., Jimenez S., Pirone L., Barrio R., et al. , 2011. Generation of stable Drosophila cell lines using multicistronic vectors. Sci. Rep. 1: 75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn H., Palmenberg A. C., 1996. Mutational analysis of the encephalomyocarditis virus primary cleavage. J. Virol. 70: 6870–6875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajduskova M., Ahier A., Daniele T., Jarriault S., 2012. Cell plasticity in Caenorhabditis elegans: from induced to natural cell reprogramming. Genesis 50: 1–17. [DOI] [PubMed] [Google Scholar]
- Hasegawa K., Cowan A. B., Nakatsuji N., Suemori H., 2007. Efficient multicistronic expression of a transgene in human embryonic stem cells. Stem Cells 25: 1707–1712. [DOI] [PubMed] [Google Scholar]
- Jarriault S., Schwab Y., Greenwald I., 2008. A Caenorhabditis elegans model for epithelial-neuronal transdifferentiation. Proc. Natl. Acad. Sci. USA 105: 3790–3795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagias K., Ahier A., Fischer N., Jarriault S., 2012. Members of the NODE (Nanog and Oct4-associated deacetylase) complex and SOX-2 promote the initiation of a natural cellular reprogramming event in vivo. Proc. Natl. Acad. Sci. USA 109: 6596–6601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J. H., Lee S. R., Li L. H., Park H. J., Park J. H., et al. , 2011. High cleavage efficiency of a 2A peptide derived from porcine teschovirus-1 in human cell lines, zebrafish and mice. PLoS ONE 6: e18556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K. K., Gruenbaum Y., Spann P., Liu J., Wilson K. L., 2000. C. elegans nuclear envelope proteins emerin, MAN1, lamin, and nucleoporins reveal unique timing of nuclear envelope breakdown during mitosis. Mol. Biol. Cell 11: 3089–3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D., Wang M., 2012. Construction of a bicistronic vector for the co-expression of two genes in Caenorhabditis elegans using a newly identified IRES. Biotechniques 52: 173–176. [DOI] [PubMed] [Google Scholar]
- Liu X., Long F., Peng H., Aerni S. J., Jiang M., et al. , 2009. Analysis of cell fate from single-cell gene expression profiles in C. elegans. Cell 139: 623–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luke G. A., de Felipe P., Lukashev A., Kallioinen S. E., Bruno E. A., et al. , 2008. Occurrence, function and evolutionary origins of ‘2A-like’ sequences in virus genomes. J. Gen. Virol. 89: 1036–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macosko E. Z., Pokala N., Feinberg E. H., Chalasani S. H., Butcher R. A., et al. , 2009. A hub-and-spoke circuit drives pheromone attraction and social behaviour in C. elegans. Nature 458: 1171–1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazaki K., 2011. MEGAWHOP cloning: a method of creating random mutagenesis libraries via megaprimer PCR of whole plasmids. Methods Enzymol. 498: 399–406. [DOI] [PubMed] [Google Scholar]
- Pfleger B. F., Pitera D. J., Smolke C. D., Keasling J. D., 2006. Combinatorial engineering of intergenic regions in operons tunes expression of multiple genes. Nat. Biotechnol. 24: 1027–1032. [DOI] [PubMed] [Google Scholar]
- Provost E., Rhee J., Leach S. D., 2007. Viral 2A peptides allow expression of multiple proteins from a single ORF in transgenic zebrafish embryos. Genesis 45: 625–629. [DOI] [PubMed] [Google Scholar]
- Richard J. P., Zuryn S., Fischer N., Pavet V., Vaucamps N., et al. , 2011. Direct in vivo cellular reprogramming involves transition through discrete, non-pluripotent steps. Development 138: 1483–1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieckher M., Kourtis N., Pasparaki A., Tavernarakis N., 2009. Transgenesis in Caenorhabditis elegans. Methods Mol. Biol. 561: 21–39. [DOI] [PubMed] [Google Scholar]
- Szymczak A. L., Vignali D. A., 2005. Development of 2A peptide-based strategies in the design of multicistronic vectors. Expert Opin. Biol. Ther. 5: 627–638. [DOI] [PubMed] [Google Scholar]
- Szymczak A. L., Workman C. J., Wang Y., Vignali K. M., Dilioglou S., et al. , 2004. Correction of multi-gene deficiency in vivo using a single ‘self-cleaving’ 2A peptide-based retroviral vector. Nat. Biotechnol. 22: 589–594. [DOI] [PubMed] [Google Scholar]
- Torres V., Barra L., Garces F., Ordenes K., Leal-Ortiz S., et al. , 2010. A bicistronic lentiviral vector based on the 1D/2A sequence of foot-and-mouth disease virus expresses proteins stoichiometrically. J. Biotechnol. 146: 138–142. [DOI] [PubMed] [Google Scholar]
- Tursun B., Cochella L., Carrera I., Hobert O., 2009. A toolkit and robust pipeline for the generation of fosmid-based reporter genes in C. elegans. PLoS ONE 4: e4625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuryn S., Le Gras S., Jamet K., Jarriault S., 2010. A strategy for direct mapping and identification of mutations by whole-genome sequencing. Genetics 186: 427–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuryn S., Daniele T., Jarriault S., 2012. Direct cellular reprogramming in Caenorhabditis elegans: facts, models, and promises for regenerative medicine. Wiley Interdiscip. Rev. Dev. Biol. 1: 138–152. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.